Abstract
Huanglongbing (HLB) in citrus infected by Candidatus Liberibacter asiaticus (CLas) has caused tremendous losses to the citrus industry. No resistant genotypes have been identified in citrus species or close relatives. Among citrus varieties, rough lemon (Citrus jambhiri) has been considered tolerant due to its ability to produce a healthy flush of new growth after infection. The difference between tolerance and susceptibility is often defined by the speed and intensity of a plant’s response to a pathogen, especially early defense responses. RNA-seq data were collected from three biological replicates of CLas- and mock-inoculated rough lemon and sweet orange at week 0 and 7 following infection. Functional analysis of the differentially expressed genes (DEGs) indicated that genes involved in the mitogen activated protein kinase (MAPK) signaling pathway were highly upregulated in rough lemon. MAPK induces the transcription of WRKY and other transcription factors which potentially turn on multiple defense-related genes. A Subnetwork Enrichment Analysis further revealed different patterns of regulation of several functional categories, suggesting DEGs with different functions were subjected to reprogramming. In general, the amplitude of the expression of defense-related genes is much greater in rough lemon than in sweet orange. A quantitative disease resistance response may contribute to the durable tolerance level to HLB observed in rough lemon.
Introduction
Huanglongbing (HLB) or citrus greening is one of the most destructive plant diseases in the world.1 The disease is caused by the bacterium, Candidatus Liberibacter asiaticus (CLas), a phloem-inhabiting, α-proteobacteria, and is transmitted by the insect vector, Asian citrus psyllid (Diaphorina citri).2 It is generally recognized that after infection or inoculation, CLas bacteria migrate through the phloem and accumulate there, resulting in the formation of sieve plugs, which contribute to HLB symptoms.3 Since no toxins, cell wall degrading enzymes or specialized secretion systems have been identified in the CLas genome, it is believed that the disease symptoms are the result of host metabolic imbalances brought about by nutrient depletion or interference with nutrient transport.4 The relationship between plant hosts and their associated microbes is essentially guided by a plant innate immunity system. Plants have evolved two layers of immune systems, pathogen-associated (or microbe-associated) molecular patterns (PAMP)-triggered immunity (PTI) and effector-triggered immunity. PTI is mediated by pattern-recognition receptors (PRRs) that recognize PAMPs, whereas effector-triggered immunity is mediated by resistance (R) proteins that recognize pathogen effectors.5 The role of these immunity systems in citrus greening disease and the transcriptomic response of citrus is poorly understood, especially in regards to the early defense response of rough lemon to HLB infection.
No source of HLB resistance has been identified in citrus species or closely-related relatives. Folimonova et al.6 classified the response of 30 citrus genotypes to HLB from sensitive to tolerant. They classified Eureka lemon and Persian lime as tolerant, with little or no HLB symptoms and no strong correlation between bacterial titer and disease severity. Zhang et al.7 also reported that HLB-affected scions of lemon had a higher titer of CLas, survival rate, and pathogen transmission rate than pummelo (Citrus maxima) scions. Citrus limon Burm. F., represented by ‘true’ lemons such as ‘Eureka’, contains genome contributions from three species, (C. maxima, C. reticulata, and C. medica) and is distinct from rough lemon, though related.8 Our previous study also showed that rough lemon is tolerant of HLB.9 Once rough lemon trees are infected and symptomatic, they can be rejuvenated by the continued growth of new shoots with few or no foliar symptoms of the disease, and they repeat this cycle for many growing seasons. In contrast, sweet orange exhibits continuous growth inhibition and eventual dieback.9
Understanding the differences in response of the tolerant and susceptible citrus genotypes to HLB is essential for developing genetic strategies that can produce tolerant, or perhaps even resistant, varieties. Most of transcriptomic studies of citrus-CLas interactions have been conducted on different tissues (leaves, fruit and root) of sweet orange,10–13 and roots of tangerine;14 while only two studies have focused on the leaves of tolerant genotypes of rough lemon9 and US-897.10 US-897, however, is an intergeneric hybrid of Citrus spp. and Poncirus spp, and may not provide a complete understanding of HLB response in citrus varieties.9 Using a comparative approach for understanding plant-microbe interactions can be complicated because the response and effect of candidate genes in tolerant genotypes may differ in the genetic background of susceptible trees due to epistasis. Previous studies have indicated that resistance and susceptibility can often depend on the speed and magnitude of the defense mechanism employed and the ability of the pathogen to suppress host response.4,15,16 It has been reported that infection of sweet orange with CLas does not lead to a significant induction of defense-related genes at the early stages of the infection process.10,11 Therefore, it is important to learn if HLB-tolerant rough lemon recognizes the presence of CLas at an early stage of infection and whether or not a defense response is elicited. Leaves of citrus trees in the field can remain asymptomatic and the titer of CLas by reverse transcription-quantitative PCR (RT-qPCR) can remain undetectable as long as two years after the initial psyllid transmission of CLas due to the need for the presence of a high bacterial titer for detection.17 False negatives and false positives are common due to low titers of bacteria inside the phloem, the presence of PCR amplification inhibitors in the phloem sap, and non-specific amplification.18 CLas in greenhouse-inoculated plants can remain undetectable for 5–9 weeks 10 or 20 weeks.9 In a time course study of CLas infection, Fan et al.9 reported that a greater number of stress response genes were distinctively modulated in rough lemon than in sweet orange. The study, however, did not identify any genes at a significant false-discovery rate (FDR) rate in the early stages (5 weeks) of the infection. In another study, no significant induction of defense-related genes was observed in sweet orange inoculated to CLas over a 5- to 9-week period of time,10. Both of the mentioned studies utilized an Affymetrix citrus microarray for their transcriptome analysis. However, it is not known how many unique citrus genes are actually represented in the chip. Misleading interpretations of microarray results can also occur due to non-specific hybridization.14 Next-generation RNA sequencing technology, however, can reveal rare and unknown transcripts, thus offering a more precise and accurate picture of the transcriptome.13,19
In the present study, a whole-genome transcriptional analysis of rough lemon and sweet orange leaves was conducted at 7 weeks post-inoculation in order to identify genes that were induced as part of an early response to CLas. Leaf samples collected immediately after inoculation (week 0) were used as a baseline, which was not done in previous transcriptome studies of CLas in citrus.4,10,11 By using week 0 as a baseline, the natural variation among biological replications could be minimized and the number of genes whose expression was significantly affected by CLas could be maximized. The response of mock-inoculated and CLas-inoculated rough lemon and sweet orange were separately analyzed and the two datasets were subsequently compared, in order to identify biological mechanisms associated with each genotype and infection versus mock-inoculation. The transcriptome analysis identified statistically significant differentially expressed genes (DEGs) between rough lemon and sweet orange in response to CLas. A distinct difference in the defense response between rough lemon and sweet orange in CLas-inoculated leaves was observed. Not only were two distinct sets of DEGs identified but greater amplitude in the profile of defense response genes in rough lemon, relative to sweet orange, was also observed. The present study provides a comprehensive overview of the early transcriptional reprogramming that occurs in rough lemon in response to CLas.
Materials and methods
Plant material and experimental design
Young trees were inoculated as described by Fan et al.9 Briefly, two-year-old seedlings of rough lemon (C. jambhiri Lush.) and ‘Madam Vinous’ sweet orange (C. sinensis L Osb.) were graft-inoculated with 3–4 cm bud wood from CLas-infected Carrizo citrange (C. sinensis×P. trifoliata L. Raf.) trees maintained under greenhouse conditions in order to provide an inoculum source for various HLB-related experiments; Carrizo was used as the inoculum source because it is immune to citrus tristeza virus (CTV), a common contaminant in field source trees of CLas. Control seedlings were grafted with 3–4 cm budwood from healthy Carrizo trees. All these plants were kept in a United States Department of Agriculture Animal and Plant Health Inspection Service and Center for Disease Control-approved and secured greenhouse at the University of Florida, Citrus Research and Education Center, Lake Alfred. Leaf samples were collected from the middle of the plant crown and approximately 60–80 cm from the inoculation site. Three biological replicates were produced for each species in each treatment (Figure 1). Reverse transcription-quantitative PCR (RT-qPCR) was performed to confirm the presence of CLas in the inoculum source and in inoculated plants as previously described.8 Four fully expanded leaves were sampled separately from CLas-inoculated plants and mock-inoculated plants (used as controls) of each species at 0 and 7 weeks after inoculation. Leaves were immediately frozen in liquid nitrogen and stored at −80 °C until further use. Three biological replicates were analyzed for each condition (Figure 1). In total, 12 plants and 24 leaf samples were collected (2 species×2 treatments×3 replications×2 time points). Selection of week 7 as the time point representing early response to CLas was based on results of a preliminary pilot experiment conducted to determine the time point at which the maximum number of DEGs changing in expression was first initiated in response to CLas.
Figure 1.
Phenotype of sweet orange and rough lemon trees 18 months after CLas-inoculation.
PCR detection of CLas
DNA from leaf midribs and petioles was extracted using the Plant DNeasyMini Kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. RT-qPCR assays were performed as previously described.20 Amplifications were performed in an Agilent Mx3005P (Agilent Technology) real-time PCR system using the Brilliant III Ultra-Fast QPCR Master Mix (Agilent Technology Inc, Waldbronn, Germany). Plants were considered PCR-positive when CT (cycle threshold) values were below 30.
RNA extraction and sequencing library construction
Total RNA from the 24 samples described above was extracted using TRIzol® Reagent following the manufacturer’s protocol. RNA was further purified using the TURBO DNA-free™ kit to eliminate genomic DNA. A NanoDrop Spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA) and gel electrophoresis were used to assess RNA quality and quantity. RNA quality was further assessed using an Agilent 2100 Bioanalyzer (Aglient Technologies Inc, Waldbronn, Germany). Ribosomal RNA was removed from the total RNA using a Ribo-Zero rRNA removal kit for using plant seed/plant leaf following the manufacturer’s protocol. Subsequently, 50 ng of the purified RNA from each sample were used for library construction utilizing an Epicentre ScriptSeq v2 RNA-Seq library preparation kit according to the user’s guide.
Illumina RNA library construction was performed at the Interdisciplinary Center for Biotechnology Research (ICBR) Gene Expression Core, University of Florida (UF). Briefly, rRNA-depleted RNA was fragmented by incubation at 85 °C, and then reverse transcribed using random primers containing a 5ʹ-tagging sequence. A 3ʹ-tag was added using a terminal-tagging reaction resulting in Di-tagged, single-stranded cDNA. Following purification, the di-tagged cDNA was amplified by limited-cycle PCR, in order to add the Illumina adaptor sequences. The amplified libraries were purified using Agencourt AMPure beads (Beckman Coulter, catalog # A63881). The library size and mass was assessed by analysis in the Bioanalyzer. Typically, a 200–2000 broad library peak was observed with the highest peak at ~500 bp. Quantitative PCR was used to validate the library’s functionality, using the KAPA library quantification kit (Kapa Biosystems, catalog number: KK4824). Finally, the libraries were pooled in equimolar concentration and sequenced on an Illumina 2×100 HiSeq 2000 (Illumina Inc., San Diego, CA, USA)
RNA-seq data analysis
Raw sequencing reads were initially cleaned as follows. Ambiguous residues were trimmed off from both sides of the sequence. Bases with a Phred quality below 20 from the 3ʹ end of the sequence were removed. Reads shorter than 40 bases or those containing >10 bases with a quality rating below 20 were also discarded. In addition, reads consisting of repetitive single bases that accounted for >60% of the length at the 5ʹ or 3ʹ end were also discarded. Approximately 50–65% of reads were retained after the initial processing, providing 45–75 million clean reads for each of the 24 samples (Table 1). The Citrus clementina genome v1.0 (JGI) genomic sequence (http://www.citrusgenomedb.org/species/clementina/genome1.0) was used as reference genome for mapping the reads. Sequencing reads from each sample were mapped independently to the reference sequences using gmap v3 (http://research-pub.gene.com/gmap/src/gmap-gsnap-2012-07-20.v3.tar.gz).21 This step successfully mapped all of the cleaned reads to the genome; resulting in a total of ~56.3% of the reads that were uniquely mapped to the genome.
Table 1. Differentially expressed genes (⩾4 log2 fold change) in rough lemon and sweet orange at week 7 compared to week 0.
| Transcript name | Best-hit-arabi-name | Gene name | Fold | FDR | Description |
|---|---|---|---|---|---|
|
Rough lemon inoculated | |||||
| Ciclev10025910m | AT5G67080 | MAPKKK19 | 5.71 | 0.000 | Mitogen-activated protein kinase 19 |
| Ciclev10010340m | AT2G46330 | AGP16 | 5.41 | 0.000 | Arabinogalactan protein 16 |
| Ciclev10026376m | AT1G19180 | JAZ1 | 5.26 | 0.001 | Jasmonate-zim-domain protein 1 |
| Ciclev10028254m | AT2G27690 | CYP94C1 | 5.21 | 0.017 | Cytochrome P450, family 94, subfamily C, polypeptide 1 |
| Ciclev10017198m | AT1G30135 | JAZ8 | 5.20 | 0.001 | Jasmonate -zim-domain protein 8 |
| Ciclev10011844m | AT3G52450 | PUB22 | 5.09 | 0.018 | Plant U-box 22 |
| Ciclev10022337m | AT2G45760 | BAP2 | 5.08 | 0.012 | BON association protein 2 |
| Ciclev10001822m | AT4G24570 | DIC2 | 5.00 | 0.001 | Dicarboxylate carrier 2 |
| Ciclev10008930m | AT1G80840 | WRKY40 | 4.94 | 0.000 | WRKY D-binding protein 40 |
| Ciclev10026527m | AT1G19180 | JAZ1 | 4.93 | 0.009 | Jasmonate-zim-domain protein 1 |
| Ciclev10021038m | AT4G11070 | WRKY41 | 4.76 | 0.001 | WRKY family transcription factor |
| Ciclev10021738m | AT4G11070 | WRKY41 | 4.75 | 0.001 | WRKY family transcription factor |
| Ciclev10007843m | AT5G48150 | PAT1 | 4.71 | 0.010 | GRAS family transcription factor |
| Ciclev10017141m | Unknown | 4.63 | 0.008 | ||
| Ciclev10022199m | AT4G11070 | WRKY41 | 4.62 | 0.001 | WRKY family transcription factor |
| Ciclev10028930m | AT4G34410 | RRTF1 | 4.53 | 0.027 | Redox responsive transcription factor 1 |
| Ciclev10002273m | AT1G01720 | ATAF1 | 4.49 | 0.007 | NAC (No Apical Meristem) domain transcriptional regulator |
| Ciclev10002277m | AT1G01720 | ATAF1 | 4.49 | 0.007 | NAC (No Apical Meristem) domain transcriptional regulator |
| Ciclev10022487m | AT1G21010 | F9H16.19 | 4.38 | 0.000 | |
| Ciclev10028253m | AT2G27690 | CYP94C1 | 4.38 | 0.025 | Cytochrome P450, family 94, subfamily C, polypeptide 1 |
| Ciclev10001956m | AT1G01720 | ATAF1 | 4.37 | 0.008 | NAC (No Apical Meristem) domain transcriptional regulator |
| Ciclev10019990m | AT1G73805 | AT1G73805 | 4.35 | 0.000 | Calmodulin binding protein-like |
| Ciclev10029464m | AT5G59820 | RHL41 | 4.35 | 0.025 | C2H2-type zinc finger family protein |
| Ciclev10020744m | AT2G46400 | WRKY46 | 4.28 | 0.000 | WRKY D-binding protein 46 |
| Ciclev10019920m | AT4G11280 | ACS6 | 4.24 | 0.000 | 1-Aminocyclopropane-1-carboxylic acid (acc) synthase 6 |
| Ciclev10033110m | AT2G31945 | AT2G31945 | 4.17 | 0.011 | |
| Ciclev10015210m | AT1G61800 | GPT2 | 4.15 | 0.011 | Glucose-6-phosphate/phosphate translocator 2 |
| Ciclev10015938m | AT1G61800 | GPT2 | 4.13 | 0.012 | Glucose-6-phosphate/phosphate translocator 2 |
| Ciclev10015941m | AT1G61800 | GPT2 | 4.13 | 0.012 | Glucose-6-phosphate/phosphate translocator 2 |
| Ciclev10005570m | AT5G51190 | MWD22.13 | 4.11 | 0.000 | Integrase-type D-binding superfamily protein |
| Ciclev10015511m | AT1G61800 | GPT2 | 4.09 | 0.013 | Glucose-6-phosphate/phosphate translocator 2 |
|
Rough lemon mock-inoculated | |||||
| Ciclev10017183m | AT4G15248 | AT4G15248 | 4.79 | 0.043 | B-box type zinc finger family protein |
|
Sweet orange inoculated | |||||
| Ciclev10003623m | AT1G09155 | PP2-B15 | 15.00 | 0.019 | Phloem protein 2-B15 |
| Ciclev10010533m | AT5G11330 | F2I11.220 | 15.00 | 0.041 | FAD/D(P)-binding oxidoreductase family protein |
| Ciclev10013424m | AT1G04360 | F19P19.21 | 15.00 | 0.004 | RING/U-box superfamily protein |
| Ciclev10013832m | AT5G38830 | K15E6.3 | 15.00 | 0.039 | Cysteinyl-tRNA synthetase, class Ia family protein |
| Ciclev10023904m | AT4G08850 | AT4G08850 | 15.00 | 0.014 | Leucine-rich repeat receptor-like protein kise family |
| Ciclev10024342m | AT1G73805 | AT1G73805 | 15.00 | 0.009 | Calmodulin binding protein-like |
| Ciclev10029883m | Unknown | 15.00 | 0.044 | ||
| Ciclev10013766m | AT5G51990 | CBF4 | 7.63 | 0.002 | C-repeat-binding factor 4 |
| Ciclev10015568m | AT3G02040 | SRG3 | 7.41 | 0.000 | senescence-related gene 3 |
| Ciclev10010503m | AT1G53270 | F12M16.17 | 7.32 | 0.003 | ABC-2 type transporter family protein |
| Ciclev10014954m | AT3G27640 | AT3G27640 | 7.31 | 0.017 | Transducin/WD40 repeat-like superfamily protein |
| Ciclev10007406m | AT2G25600 | SPIK | 6.16 | 0.040 | Shaker pollen inward K+ channel |
| Ciclev10028254m | AT2G27690 | CYP94C1 | 6.07 | 0.003 | Cytochrome P450, family 94, subfamily C, polypeptide 1 |
| Ciclev10028253m | AT2G27690 | CYP94C1 | 5.89 | 0.001 | Cytochrome P450, family 94, subfamily C, polypeptide 1 |
| Ciclev10013853m | AT3G47570 | AT3G47570 | 5.44 | 0.006 | Leucine-rich repeat protein kise family protein |
| Ciclev10011844m | AT3G52450 | PUB22 | 5.42 | 0.007 | Plant U-box 22 |
| Ciclev10024282m | AT5G17680 | MVA3.30 | 5.37 | 0.046 | disease resistance protein (TIR-NBS-LRR class) family |
| Ciclev10026096m | AT1G17710 | F11A6.5 | 5.30 | 0.008 | Pyridoxal phosphate phosphatase-related protein |
| Ciclev10017198m | AT1G30135 | JAZ8 | 5.21 | 0.001 | Jasmonate-zim-domain protein 8 |
| Ciclev10013223m | Unknown | 5.18 | 0.047 | ||
| Ciclev10011408m | AT1G26810 | GALT1 | 5.14 | 0.011 | Galactosyltransferase1 |
| Ciclev10032298m | AT4G00430 | PIP1;4 | 5.09 | 0.000 | Plasma membrane intrinsic protein 1;4 |
| Ciclev10019961m | AT3G07250 | AT3G07250 | 5.07 | 0.001 | Nuclear transport factor 2 (NTF2) family protein |
| Ciclev10004868m | AT5G54010 | K19P17.18 | 5.02 | 0.002 | UDP-Glycosyltransferase superfamily protein |
| Ciclev10007591m | AT5G04860 | MUK11.18 | 4.94 | 0.033 | |
| Ciclev10013749m | AT1G68450 | T2E12.4 | 4.94 | 0.023 | VQ motif-containing protein |
| Ciclev10028305m | AT4G34131 | UGT73B3 | 4.94 | 0.000 | UDP-glucosyl transferase 73B3 |
| Ciclev10032278m | AT5G05600 | MOP10.14 | 4.92 | 0.013 | 2-Oxoglutarate and Fe(II)-dependent oxygese superf |
| Ciclev10031811m | AT5G05600 | MOP10.14 | 4.81 | 0.007 | 2-Oxoglutarate and Fe(II)-dependent oxygese superf |
| Ciclev10031910m | AT5G05600 | MOP10.14 | 4.79 | 0.009 | 2-Oxoglutarate and Fe(II)-dependent oxygese superf |
| Ciclev10017141m | Unknown | 4.76 | 0.002 | ||
| Ciclev10021923m | AT4G25470 | CBF2 | 4.76 | 0.016 | C-Repeat/DRE binding factor 2 |
| Ciclev10022337m | AT2G45760 | BAP2 | 4.75 | 0.012 | BON association protein 2 |
| Ciclev10007652m | AT5G04860 | MUK11.18 | 4.71 | 0.041 | |
| Ciclev10008003m | AT5G04860 | MUK11.18 | 4.67 | 0.038 | |
| Ciclev10002675m | AT1G35210 | T32G9.25 | 4.60 | 0.011 | |
| Ciclev10031320m | AT2G44500 | F4I1.31 | 4.59 | 0.001 | O-fucosyltransferase family protein |
| Ciclev10013649m | AT4G26850 | VTC2 | 4.57 | 0.000 | mannose-1-phosphate guanylyltransferase |
| Ciclev10005385m | AT1G47990 | GA2OX4 | 4.50 | 0.008 | gibberellin 2-oxidase 4 |
| Ciclev10023861m | AT4G28460 | F20O9.140 | 4.48 | 0.013 | |
| Ciclev10013959m | AT4G12010 | F16J13.80 | 4.46 | 0.003 | Disease resistance protein (TIR-NBS-LRR class) family |
| Ciclev10006910m | AT3G16520 | UGT88A1 | 4.44 | 0.019 | UDP-glucosyl transferase 88A1 |
| Ciclev10005376m | AT5G49330 | MYB111 | 4.44 | 0.019 | myb domain protein 111 |
| Ciclev10029690m | AT2G21650 | MEE3 | 4.40 | 0.000 | Homeodomain-like superfamily protein |
| Ciclev10023483m | AT4G01950 | GPAT3 | 4.40 | 0.004 | Glycerol-3-phosphate acyltransferase 3 |
| Ciclev10022989m | AT1G03020 | F10O3.16 | 4.36 | 0.044 | Thioredoxin superfamily protein |
| Ciclev10010348m | AT1G19210 | T29M8.8 | 4.33 | 0.016 | Integrase-type D-binding superfamily protein |
| Ciclev10029663m | AT2G21650 | MEE3 | 4.33 | 0.000 | Homeodomain-like superfamily protein |
| Ciclev10000083m | AT1G75640 | F10A5.16 | 4.32 | 0.000 | Leucine-rich receptor-like protein kise family protein |
| Ciclev10020246m | AT4G00770 | T18A10.2 | 4.26 | 0.019 | |
| Ciclev10005777m | AT5G43650 | BHLH92 | 4.23 | 0.018 | Basic helix-loop-helix (bHLH) D-binding superfamily |
| Ciclev10032519m | AT5G13790 | AGL15 | 4.23 | 0.002 | AGAMOUS-like 15 |
| Ciclev10029774m | Unknown | 4.22 | 0.001 | ||
| Ciclev10030876m | AT1G10560 | PUB18 | 4.18 | 0.000 | Plant U-box 18 |
| Ciclev10013961m | AT5G17680 | MVA3.30 | 4.11 | 0.008 | Disease resistance protein (TIR-NBS-LRR class) family |
| Ciclev10029578m | AT1G58420 | F9K23.5 | 4.09 | 0.014 | Uncharacterized conserved protein UCP031279 |
| Ciclev10033986m | AT4G39670 | T19P19.60 | 4.08 | 0.020 | Glycolipid transfer protein (GLTP) family protein |
| Ciclev10022303m | AT5G52600 | MYB82 | 4.03 | 0.000 | myb domain protein 82 |
| Ciclev10033777m | AT1G24430 | F21J9.9 | 4.03 | 0.033 | HXXXD-type acyl-transferase family protein |
|
Sweet orange mock-inoculated | |||||
| Ciclev10029484m | UnKnown | 4.74 | 0.033 | ||
| Ciclev10029585m | unknown | 4.65 | 0.036 | ||
Abbreviation: FDR, false discovery rate.
Gene expression values were determined as follows. The number of mapped reads for each individual gene was counted using an in house perl script. The gene annotation file in GFF format was downloaded from Phytozome v1.0 http://www.phytozome.net (Phytozome: Cclementina 182 v1.0.gene.gff3.gz). The DESeq package in R was used for gene expression analysis.22 DESeq uses the negative binomial distribution, with variance and mean linked by local regression, to model the null distribution of the count data. Significant up- and downregulated genes were selected using two cutoffs: an adjusted P value of 0.05 and a minimum fold-change of 2.0. Gene sets from each combined sample and treatment were annotated using Blast2GO[67] to assign Gene Ontology (GO) terms to each gene. Lists of transcripts that were differentially expressed using a false-discovery rate (FDR) <0.05 in the pairwise comparisons were used in the Fisher’s Exact Test in Blast2GO in order to identify GO terms that were significantly over-represented. The biological interpretation of the DEGs was further assessed by assigning the genes to metabolic pathways using the Kyoto Encyclopedia of Genes and Genomes (KEGG).23 Subnetwork enrichment analyses were run on the data set using Pathway Studio 10.0.24
Reverse transcription-quantitative PCR
Nine DEGs that were identified by RNA-seq as either up- or downregulated in response of CLas were selected for validation by RT-qPCR. The RT-qPCR analyses were performed in two steps. First strand cDNA was synthesized from 0.3 μg of total RNA using an Affinityscript QPCR cDNA Synthesis Kit (Agilent Technologies), according to the manufacturer's recommendations. Primers for the twelve selected genes were designed using Primer-BLAST software at NCBI (http://www.ncbi.nlm.nih.gov/tools/primer-blast/), and are listed in Supplementary Table S1 of Supplementary File 6. RT-qPCR was performed using a Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies), following the manufacturer's recommended procedures. GAPDH was used as a reference gene to normalize the expression of the other analyzed genes. PCR reactions were performed using 2 μl of cDNA, 0.25 nM of each primer, and 10 μl of 2× SYBRGreen PCR master mix (Aglient Technologies Inc, Santa Clara, CA, USA) in a 20-μl volume. A negative control was included using water as a template for each primer pair. Specificity of the amplification reactions was checked by post-amplification dissociation curves and by sequencing the reaction products. The fluorescent intensities were used to determine relative mRNAlevels with MxPro software.
Results
CLas detection and HLB symptom development
Leaf samples collected at week 7 did not exhibit the characteristic blotchy mottle appearance and were RT-qPCR negative for CLas. Continued RT-qPCR analysis for CLas was conducted every two weeks after inoculation. Positive confirmation of CLas in inoculated plants was not obtained in rough lemon and sweet orange until 23 weeks post inoculation. Even after 8 months, however, rough lemon did not exhibit any signs of growth inhibition, and continued growth of new shoots with few or no symptoms was observed; however, the typical blotchy mottled appearance was commonly found on mature, older leaves (Figure 1). In contrast, severe levels of blotchiness were observed on mature and older leaves of CLas- inoculated sweet orange and growth was significantly inhibited, with the rare production of new shoots toward the end of the experiment (Figure 1). CLas was not detected by RT-qPCR in samples from any of the mock-inoculated rough lemon or sweet orange throughout the entire experiment.
Comparative RNA-seq analysis
A comparison of the RNA-seq data of the tolerant rough lemon and susceptible sweet orange was conducted using samples collected in week 7. RNA-seq data obtained at week 0 were used as a baseline. A total of 24 cDNA libraries (2 treatments×2 varieties×3 replications×2 time points) were sequenced, generating 1274.9 million reads and 740.4 million unique reads were assigned (Supplementary File 1, Supplementary Table S1). The reads were trimmed and aligned to the Citrus clementina genome available on the Phytozome website (http://www.phytozome.net). Expressed genes and transcript isoforms were identified and annotated using the C. clementina genome v.1 assembly consisting of 301.4 Mb spread over 1398 scaffolds. RNA-seq analysis was independently performed using a pair-wise comparison between week 7 and the baseline week 0 for inoculated and mock-inoculated rough lemon and sweet orange using DEseq for each treatment.22
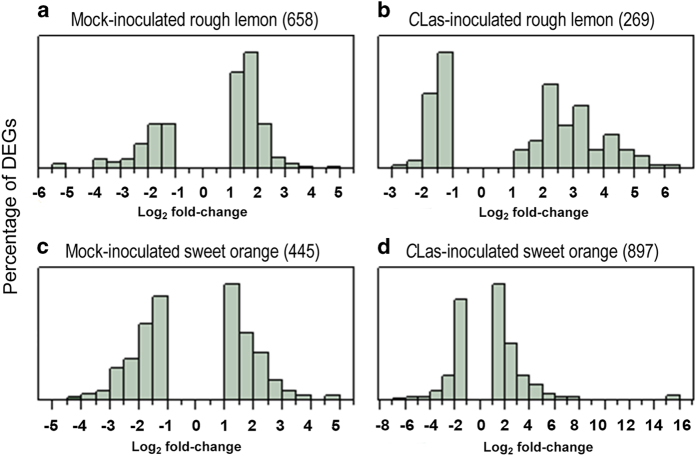
A total of 33 930 unique transcripts were identified and quantified but 1884 genes did not have any corresponding Arabidopsis orthologs (Supplementary File 2, Supplementary Table S1). Due to duplications of genes matching a single Arabidopsis identifier, only 14,393 Arabidopsis identifiers were obtained. A total of 3266 DEGs were identified in the combined inoculated and mock-inoculated rough lemon and sweet orange by a pair-wise comparison between week 7 and week 0 (Supplementary File 2, Supplementary Table S2). More specifically, 1518 and 1129 DEGs were identified in the mock-inoculated and CLas-inoculated samples of rough lemon, respectively. For sweet orange, 1944 and 2396 DEGs were identified in the mock-inoculated and CLas-inoculated samples, respectively. Further analysis indicated that 860 DEGs overlapped between the mock- and CLas-inoculated samples of rough lemon (Figure 2a), whereas 1499 DEGs overlapped between mock- and CLas-inoculated samples of sweet orange (Figure 2b). A total of 2024 DEGs were selected for further functional analysis after the removal of mock- and CLas-inoculated overlapping genes (Supplementary File 2, Supplementary Table S3). The removal of the overlapping genes resulted in a total of 658 DEGs and 269 DEGs for mock- and CLas-inoculated samples of rough lemon, respectively, and 445 DEGs and 897 DEGs for mock- and CLas-inoculated samples of sweet orange (Supplementary File 2, Supplementary Table S3). Among the 269 DEGs in inoculated rough lemon samples, 76 were upregulated⩾3-fold (log2), abbreviated as log-fold change (LFC), while no DEGs with this magnitude of downregulation were observed (Figure 3). CLas-inoculated samples of sweet orange contained 897 DEGs, of which 119 DEGs were upregulated ⩾3 LFC, while 11 DEGs were found to be upregulated ⩾3 LFC among the 445 DEGs identified in mock-inoculated samples. Genes related to the defense response, such as those in the MAPK signaling pathway, WRKY transcription factors, and other pathogenesis-related genes were identified among the highly expressed (⩾4 LFC) genes in the CLas-inoculated samples of rough lemon (Table 1). Upregulated genes greatly outnumbered downregulated genes in both CLas-inoculated samples of sweet orange and rough lemon (Figure 3). The number of downregulated DEGs was slightly higher than upregulated DEGs in mock-inoculated samples of sweet orange, while the number of upregulated DEGs was much greater than the number of downregulated DEGs in CLas-inoculated samples of sweet orange. A total of 29 and 25% of the DEGs in samples of CLas-inoculated rough lemon and sweet orange had a LFC⩾3, respectively, whereas only 1% of the DEGs in mock-inoculated samples of rough lemon and sweet orange were observed to have an LFC⩾3. These results indicate that overall gene transcription levels were higher in CLas-inoculated samples than in mock-inoculated samples of rough lemon and sweet orange.
Figure 2.
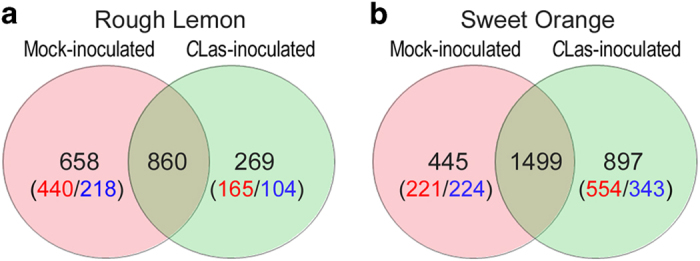
Venn diagrams of differentially expressed genes in mock- and CLas-inoculated rough lemon (a) and sweet orange (b). The number of significantly up-regulated genes is shown (first) in red, and significantly downregulated genes (second) in blue.
Figure 3.
Distribution of significant differentially expressed genes (DEGs): (a) CLas-inoculated rough lemon; (b) Mock-inoculated rough lemon; (c) CLas-inoculated sweet orange; (d) Mock-inoculated sweet orange.
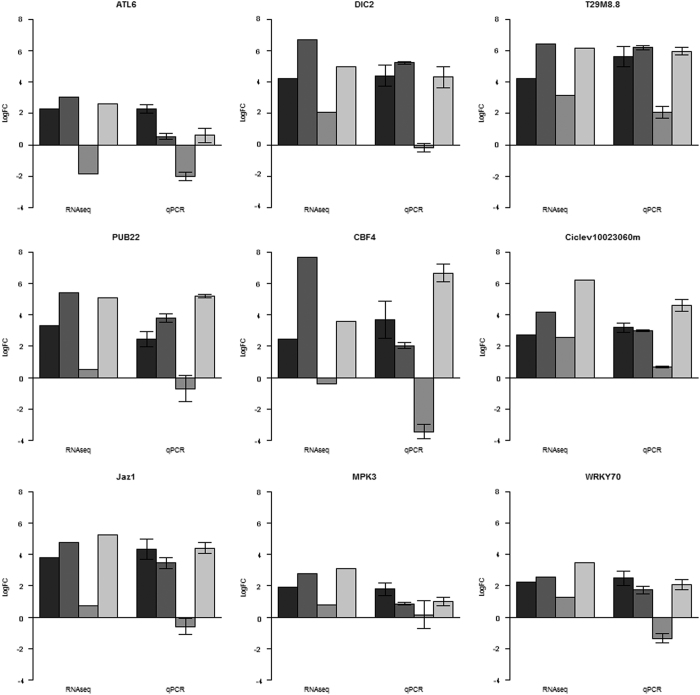
Expression levels of 9 defense response-related genes were analyzed by RT-qPCR in order to confirm the results obtained by RNA-seq analysis. As shown in Figure 4, the relative levels of expression of the investigated genes were consistent between the RT-qPCR and RNA-seq data, with a few exceptions but within error range, indicating that the RNA-seq data were reliable.
Figure 4.
Relative expression of nine differentially expressed genes (DEGs) associated with disease resistance response as determined by RT-qPCR. Standard error bars are provided. ATL6: ubiquitin-protein ligase; DIC2: dicarboxylate carrier 2; T29M8.8: ethylene-responsive transcription factor; PUB22: ubiquitin-protein ligase PUB22; CBF4: DEHYDRATION-RESPO; Ciclev10023060m: hypothetical protein; JAZ1: JASMONATE-ZIM-DOMAIN PROTEIN; MPK3: MITOGEN-ACTIVATED PROTEIN KINASE 3; WRKY70: WRKY transcription factor 70.
Functional categorization and subnetwork enrichment analysis
The Fisher Extraction Test and subnetwork enrichment analysis (SNEA), as provided in Blast2GO and Pathway Studio (Elsevier/Ariadne Genomics), were used to determine the specific Gene Ontology (GO) terms affected by CLas infection. GO terms associated with hormones, defense response, as well as stomatal and photosynthetic acclimation, were significantly over-represented based on the Fisher’s pairwise comparisons of CLas-inoculated and mock-inoculated genes in both rough lemon and sweet orange samples (Supplementary File 3, Supplementary Tables S1 and S2). Only 1 GO term (FDR⩽0.05), response to chitin, was identified in sweet orange, while 19 GO terms were identified in rough lemon samples (Table 2). Three of the top GO terms in rough lemon were response to chitin, ethylene biosynthetic process, and respiratory burst involved in defense response and response to molecules of bacterial origin (Table 2). Although a lower number of DEGs were identified in rough lemon than in sweet orange, more defense response GOs were associated with the transcriptomic response of rough lemon to CLas infection than in sweet orange, and may reflect the ‘tolerance’ response observed in rough lemon. The results of the Fisher’s Test indicated that ‘ATP binding’ was the most significantly enriched GO molecular function term in rough lemon, perhaps reflecting the significant number of genes under transcriptional regulation during the early response of rough lemon to CLas infection (Supplementary File 3, Supplementary Table S1). No statistically significant GO terms in the molecular function category were found in the sweet orange RNA-seq data (Supplementary File 3, Supplementary Table S2). SNEA uses a global expression regulatory network extracted from the entire PubMed database and full-text journals to extract regulatory networks. Using the non-parametric Mann-Whitney test, SNEA identified significant (P⩽0.05), over-represented (P⩽0.05) ontologies among the up- and downregulated DEGS that indicate an enrichment of each sub-network. A total of 32 and 44 ontologies associated with upregulated DEGs in CLas-inoculated rough lemon and sweet orange samples were identified, while 26 and 22 ontologies associated with downregulated DEGs were found in rough lemon and sweet orange, respectively (Supplementary File 4, Supplementary Tables S1 and S3). The top 3 ontologies for upregulated genes in CLas-inoculated samples of rough lemon were associated with defense response, biotic stress, and plant defense (Table 3a). In comparison, plant immunity, jasmonate response (JA), and shade avoidance were the top ontology terms identified in CLas-inoculated samples of sweet orange (Table 3b). Among downregulated DEGs, root phototropism, seed width, and cell expansion were the top 3 ontologies identified in CLas-inoculated samples of rough lemon; whereas root phototropism, response to ethylene stimulus and greening were the top ontologies identified in sweet orange (Tables 3a and b). In regards to mock-inoculated samples of rough lemon, photosynthesis, reductive pentose-phosphate cycle, and seed length were the top three ontologies identified for upregulated DEGs. On the other hand, plant response, nitrogen metabolism, and biotic stress were the top ontologies identified for downregulated DEGs. With respect to mock-inoculation of sweet orange, cuticle development, response to auxin stimulus, and ion homeostasis were the top ontologies identified for upregulated genes, whereas meristem development, meristem initiation, and leaf size were the top ontologies for downregulated genes (Supplementary File 4, Supplementary Tables S2 and S4).
Table 2. Fisher's exact test with FDR (<0.05) between CLas- or inoculation and mock-inoculated rough lemon and sweet orange.
| GO-ID | Term | FDR |
|---|---|---|
|
CLas inoculated versus mock inoculated rough lemon | ||
| GO:0010200 | Response to chitin | 0.000 |
| GO:0009693 | Ethylene biosynthetic process | 0.000 |
| GO:0010288 | Response to lead ion | 0.000 |
| GO:0002679 | Respiratory burst involved in defense response | 0.000 |
| GO:0002237 | Response to molecule of bacterial origin | 0.010 |
| GO:0009684 | Indoleacetic acid biosynthetic process | 0.013 |
| GO:0010105 | Negative regulation of ethylene-activated signaling pathway | 0.016 |
| GO:0009736 | Cytokinin-activated signaling pathway | 0.017 |
| GO:0006569 | Tryptophan catabolic process | 0.024 |
| GO:0050832 | Defense response to fungus | 0.029 |
| GO:2000038 | Regulation of stomatal complex development | 0.040 |
| GO:2000037 | Regulation of stomatal complex patterning | 0.040 |
| GO:0080001 | Mucilage extrusion from seed coat | 0.040 |
| GO:0048358 | Mucilage pectin biosynthetic process | 0.040 |
| GO:0009690 | Cytokinin metabolic process | 0.040 |
| GO:0009643 | Photosynthetic acclimation | 0.040 |
| GO:0007276 | Gamete generation | 0.040 |
| GO:0009401 | Phosphoenolpyruvate-dependent sugar phosphotransferase system | 0.040 |
|
CLas inoculated versus mock inoculated sweet orange | ||
| GO:0010200 | Response to chitin | 0.039 |
Abbreviation: FDR, false discovery rate.
Table 3. Top 10 biological processes, as determined by sub-network enrichment analysis, associated with genes in rough lemon that were significantly (P<0.05) upregulated or downregulated in response to CLas infection.
| Ontology | Gene name | P-value |
|---|---|---|
|
Upregulated biological processes | ||
| Defense response | RBOHD, MPK3, SOBIR1, WRKY70, RHL41, ATAF1, ERF-1, WRKY33, JAZ1, BAP2, WRKY40, STZ,BT1 | 1.77E−06 |
| Biotic stress trait | ERF-1, WRKY33, MKK9, WRKY40, STZ | 1.30E−05 |
| Plant defense | RBOHD, MPK3, SOBIR1, WRKY70, ERF-1, NAC062, WRKY33, MKK9, WRKY40, STZ | 3.15E−05 |
| Plant immunity | MPK3, SOBIR1, ATAF1, ERF-1, NAC062, WRKY40 | 5.92E−05 |
| Leaf senescence | SAUL1, WRKY70, ATAF1, JAZ8, MKK9, RAP2.4 | 4.41E−04 |
| Jasmonate response | WRKY70, ERF-1, JAZ1, CML37 | 5.24E−04 |
| Stomatal movement | RBOHD, MPK3, NCED3, CYP707A1, WRKY70, ATAF1, ERF-1, WRKY46 | 5.30−04 |
| Jasmonate metabolism | WRKY70, ERF-1 | 6.75E−04 |
| Response to ethylene stimulus | MPK3, ACS6, ERF-1, MKK9, RAP2.4 | 7.33E−04 |
| Abiotic stress | MPK3, RHL41, ATAF1, NAC062, WRKY33, STZ | 8.21E−04 |
|
Downregulated biological processes | ||
| Root phototropism | NPH4, PKS1 | 7.59E−05 |
| Seed width | NPH4, ARF8 | 1.16E−04 |
| Cell expansion | ERS1, CKI1, NPH4, ARF8, TUB1 | 1.33E−04 |
| Hypocotyl growth | CYP83B1, NPH4, PIL1, ARF8, PKS1 | 1.88E−04 |
| Hypocotyl shape | NPH4, PKS1 | 4.81E−04 |
| Flower development | ERS1, SEX1, DME, PIL1, ARF8, LUH | 1.81E−03 |
| Fruit set | NPH4, ARF8 | 2.59E−03 |
| Shade avoidance | CYP83B1, PIL1 | 2.69E−03 |
| Response to ethylene stimulus | ERS1, NPH4, EBF1 | 3.17E−03 |
| De-etiolation | CYP83B1, PIL1 | 3.92E−03 |
Table 4. Top 10 biological processes, as determined by sub-network enrichment analysis, associated with genes in sweet orange that were significantly (P<0.05) upregulated or downregulated in response to CLas infection.
| Ontology | Gene name | P-value |
|---|---|---|
|
Upregulated biological processes | ||
| Plant immunity | GLIP1, MPK3, SOBIR1, EFR, ERF-1, NAC062, WRKY22, SNC4 | 3.43E−04 |
| Jasmonate response | EFR, ERF-1, JAZ1, MYC2, ASK2, CML37 | 3.60E−04 |
| Shade avoidance | COP1, PIF7, MYC2, BEE1 | 1.20E−03 |
| K+ Import/homeostasis | RBOHD, SOS1, CHX20, ZIFL1 | 1.59E−03 |
| Cold acclimation | MPK3, PPDK, COP1, RHL41, PHO1, CBF2, HSFA2,MYC2, CBF4 | 1.69E−03 |
| Disease resistance | GLIP1,RBOHD,MPK3,SOBIR1,CCR1,VTC2,WIN2,PTF1,HSC70-1,HSP90.1,RPM1 | 1.92E−03 |
| Plant defense | GLIP1,RBOHD,MPK3,SOBIR1,CCR1,FAB1,ERF-1,NAC062,MYC2,WRKY22,RPM1,YSL3,AAT1 | 2.26E−03 |
| Stomatal movement | RBOHD,MPK3,PPDK,PUB18,COP1,NRT1.1,ERF-1,NHX2,HSC70-1,ALMT9,CHX20,ZIFL1 | 3.01E−03 |
| Floral organ abscission | MPK3,HSL2,AGL15 | 3.45E−03 |
| Cryptochrome response | COP1,SUB1 | 4.18E−03 |
|
Downregulated biological processes | ||
| Root phototropism | GN,PKS1 | 0.0006 |
| Response to ethylene stimulus | EFE,ERS1,EBF1,EIN3,ETR2 | 0.0019 |
| Greening | PORA,EIN3,RR12,PKS1 | 0.0030 |
| Hypocotyl shape | GN,PKS1 | 0.0041 |
| Vascularization | ZLL,GN,RR12 | 0.00427 |
| Leaf size | AVP1,TCTP,ETR2 | 0.0054 |
| Hypocotyl growth | MUR4,CYP83B1,EIN3,FHY1,ETR2,PKS1 | 0.00631 |
| Gene silencing | EFE,DME,ZLL,EMB25 | 0.0071 |
| Cation transport | AVP1,CNGC1 | 0.0076 |
| Translation elongation | MTO1,TCTP | 0.0076 |
Transcription factors
A total of 68 transcription factors were identified (Table 4). Eighteen transcription factors (TFs) were identified in CLas-inoculated and mock-inoculated samples of rough lemon, all of which were related to plant immunity, and 12 of which had a transcription level ⩾3 LFC. Only one TF⩾3 LFC was observed in mock-inoculated rough lemon. ERF-1 was the only TF found in both CLas- and mock-inoculated samples of rough lemon, and was upregulated in CLas-inoculated samples and downregulated in mock-inoculated samples of rough lemon. A total of 34 TFs were identified in CLas-inoculated samples of sweet orange and 7 in mock-inoculated samples. CLas-inoculated samples of rough lemon had 15 upregulated versus 3 downregulated TFs, whereas 26 upregulated versus 10 down regulated TFs were identified in CLas-inoculated sweet orange. Only EIN3 was found to be downregulated in both CLas- and mock-inoculated sweet orange. Several TFs families, such as ATAF1, bHLH, ERF, MYC2, MYB, and WRKY, are specifically associated with regulating plant defense and immunity.25 A greater number of defense-related TFs were found in CLas-inoculated samples than in mock-inoculated samples. ERF-1, MYC2, RAP2.4, TFIID, and WRKY70 were associated with upregulation of jasmonate and ethylene metabolism. WRKY TFs exhibited stronger expression in CLas-inoculated rough lemon than in sweet orange (Table 4).
Table 5. Fold-change of identified transcription factors.
| Name | Mock inoculated rough lemon | CLas inoculated rough lemon | Mock inoculated sweet orange lemon | CLas inoculated sweet orange |
|---|---|---|---|---|
| CBF4 | 7.6274 | |||
| T29M8.8 | 4.3308 | |||
| BHLH92 | 4.2331 | |||
| MYB82 | 4.0349 | |||
| NF-YA1 | 3.4703 | |||
| EMB1444 | 3.2101 | |||
| BEE1 | 3.1625 | |||
| ERF-1 | 3.2017 | 3.0451 | ||
| WRKY41 | 4.7115 | 3.0081 | ||
| AT3G20640 | 2.9462 | |||
| HSFA2 | 2.7241 | |||
| WRKY27 | 2.4071 | |||
| HSF4 | 2.3725 | |||
| PIF7 | 1.9969 | |||
| WRKY22 | 1.963 | |||
| MYC2 | 1.8374 | |||
| NF-YA7 | 1.3202 | 1.8007 | ||
| F15H21.12 | 2.0221 | 1.7584 | ||
| F6F3.7 | 1.7225 | |||
| WLIM1 | 1.5062 | |||
| T13M11.1 | 1.4698 | |||
| TFIID | 1.4263 | |||
| TCP20 | 1.4007 | |||
| ARF3 | 1.376 | |||
| PTF1 | 1.3533 | |||
| LRL1 | 1.1847 | |||
| T4I9.13 | −1.2194 | |||
| EIN3 | −1.4775 | −1.2078 | −1.3138 | |
| AT3G57795 | −1.3328 | −1.3829 | ||
| AT1G05805 | −1.5484 | |||
| MUD21.1 | −1.5583 | |||
| MRH10.19 | −2.1213 | −1.6206 | ||
| HMGB6 | −1.738 | |||
| MBK20.1 | −2.6197 | |||
| WRKY40 | 4.9433 | |||
| PAT1 | 4.7133 | |||
| RRTF1 | 4.5266 | |||
| ATAF1 | 4.4503 | |||
| WRKY46 | 4.2845 | |||
| MWD22.13 | 4.1084 | |||
| WRKY33 | 3.7237 | |||
| WRKY70 | 3.4712 | |||
| ERF9 | 3.3143 | |||
| ERF1 | −1.9025 | 3.1845 | ||
| RAP2.4 | 2.282 | |||
| MUG13.28 | 1.2747 | |||
| GT2 | −1.1792 | |||
| PIL1 | −2.1031 | |||
| MYB66 | 3.2872 | |||
| F20P5.26 | 2.6354 | |||
| CGA1 | 2.3686 | |||
| HSFA6B | 2.0545 | |||
| ILR3 | 1.877 | |||
| T6K21.80 | 1.7872 | |||
| NF-YC2 | 1.6739 | |||
| TTG2 | 1.5884 | |||
| UNE10 | 1.4733 | |||
| GBF3 | 1.4358 | |||
| YAB2 | 1.2666 | |||
| BZIP60 | 1.2192 | |||
| WRKY42 | −1.6272 | |||
| AT3G10330 | −2.3198 | |||
| AGL8 | −3.5222 | |||
| AP2 | 1.2999 | |||
| AS1 | 1.6832 | |||
| HSFC1 | −2.0526 | |||
| IAA8 | 1.6307 | |||
| T24P15.19 | −1.8958 |
Highly-expressed DEGs
DEGs involved in signal transduction were overwhelmingly observed and highly expressed in CLas-inoculated leaves of both rough lemon and sweet orange (Table 1). The AT3G47570 gene, which encodes a protein that functions in protein phosphorylation in the transmembrane receptor protein tyrosine kinase signaling pathway, was the gene most hit (117 times) by citrus sequences obtained in the RNA-seq results. The R-gene, AT2G20142 (TIR domain family protein), and RBOHD (respiration burst) were both more highly upregulated in rough lemon than in sweet orange. Two MAPK genes (MPK3 and MKK9) were upregulated (LFC 3.1 and 1.51, respectively), in rough lemon, whereas only MPK3 (LFC=2.79) was upregulated in sweet orange. MAPKKK19, JAZ8, JAZ1, PUB22 (associated with respiratory burst involved in defense response), DIC2 (ethylene biosynthetic process), AT1G73805 (salicylic acid (SA) mediated signaling pathway), CBF4 (abscisic acid signaling), CYP94C1 (signal transduction), MPK3, WRKYs (WRKY 40, WRKY 41, WRKY 46, WRKY 33, WRKY 70), ACS6, ERF-1 and ERF9, all of which are involved in signaling transduction pathway, were in the top 50 upregulated (LFC⩾3) DEGs in CLas-inoculated samples of rough lemon. The most highly upregulated (LFC⩾3) genes in CLas-inoculated sweet orange included a few DEGs involved in signaling pathways, such as AT1G73805 (SAR-deficient 1), CBF4, CYP94C1 and JAZ8, as well as other genes, such as SRG3 (glycerol metabolic process), F12M16.17 (transporter), AT3G27640 (DNA dependent transporter), and PP2-B15 (phloem protein) (Table 1, Supplementary File 2, Supplementary Table S3). The WRKY transcription factor, WRKY 41 (LFC=3.00), was ranked 89 among the most highly expressed DEGs CLas-inoculated sweet orange, while WRKY40 (LFC=4.94) was ranked 10 in the list of most highly upregulated genes in CLas-inoculated rough lemon. CKI1 (cytokinin mediated signaling), EBF1 (negative mediated ethylene signaling), MJB24.15 (brassinosteroid stimulus), PIL1 (red light signaling), CYP71B37, and F4H6.15 (hypothetical protein) were among the most highly downregulated DEGs in CLas-inoculated rough lemon. MMI9.1 (plant invertase/pectin methylesterase inhibitor), UGE1 (UGE1 (UDP-D-glucose/UDP-D-galactose 4-epimerase 1), AHP4 (cytokinin mediated signaling), PRP2 (cell wall organization), and AT3G62950 (cell redox homeostasis) were among the most highly downregulated DEGs in CLas-inoculated sweet orange.
Discussion
The current study examined transcriptomic changes associated with HLB during the early stage of CLas infection in HLB-tolerant rough lemon and HLB-susceptible sweet orange. Detection of CLas in citrus is delayed after initial inoculation; trees remain visibly asymptomatic until the bacterial titer reaches levels sufficient for reliable detection by RT-qPCR.17 Infection of CLas-inoculated plants of rough lemon and sweet orange could not be confirmed by RT-qPCR in our study until 23 weeks after the plants had been inoculated. A previous study reported that HLB could be detected in sweet orange at 5–9 weeks after inoculation under greenhouse conditions.10 The discrepancy in the time needed for confirmation in the two studies is likely associated with the use of different inoculum sources with different titers of bacteria, and possibly differences in plant age and growing conditions. Stover et al.26 found that inoculation with Kuharske citrange results in lower CLas titers than other inoculum sources such as lemon and sweet oranges. Since both Kuharske and Carrizo are citranges, the inoculum used in our study could take a longer time to be detected by qPCR than ‘Lisbon’ lemon in the earlier study. Identifying the proper time point to collect samples to characterize an early response to infection can also be problematic. In our study, bulk samples were collected at 0, 5, 7, 9, 12 and 17 weeks in order to determine the appropriate time point to represent an early response. Preliminary RNA-seq data indicated that the number of DEGs changing in expression was maximized at week 7; as a result, this time point was selected for detailed sequencing and analysis (data not shown).
An appropriate statistical test is needed to determine whether or not observed differences in the number of obtained sequences (read counts) of a specific gene is significant, that is, whether the difference is greater than what would be expected due to natural random variation.22 In addition, it is also critical to design the experiment so that natural variation (not due to a treatment effect) in gene expression is minimized and the number of identified DEGs is maximized. Previous studies directly comparing CLas- and mock-inoculated plants, or tolerant and susceptible genotypes, may have not sufficiently reduced biological variation after inoculation at week 0. As a result, a low number of early-stage DEGs were identified.4,10,11,27 In the present study, gene expression in week 0 was used as a baseline in order to reduce the level of random variation between individual plants. To identify the DEGs induced by CLas, only DEGs that were uniquely expressed in CLas-inoculated and not in mock-inoculated samples at week 7 were selected for comparative analysis. Using this approach, we identified a greater number of DEGs in response to CLas inoculation than by direct pairwise comparisons of CLas-inoculated rough lemon and sweet orange at week 7. This approach has not been used in previous transcriptome studies of CLas infected citrus. However, we would not identify as DEGs those genes which had high absolute levels of expression in both week 0 and week 7 after inoculation. We speculate that some of these genes might play important roles against CLas, although these genes were not necessarily induced by CLas.
Gene expression associated with tolerance and susceptibility to CLas
Plant disease resistance is generally divided into two categories, qualitative resistance, determined by major R-genes, and quantitative resistance, determined by multiple genes with minor effects.28 The first tier of defense, PTI, is triggered by the perception of PAMP/DAMPs by membrane-anchored PRRs, which is then followed by the activation of a MAPK cascade and downstream transcription factors, leading to immune responses. The second tier of defense is elicited by pathogen effectors via an interaction with an R protein (effector-triggered immunity), where the interaction between R protein and a pathogen effector oscillates between compatible and incompatible reactions over time. Plant pathogens are broadly divided into biotrophs and necrotrophs. Plant defense against biotrophic pathogens is largely due to major gene resistance.29 CLas is an obligate biotrophic pathogen and currently no resistant citrus varieties has been identified. R-gene-mediated resistance usually induces a hypersensitive response (HR), which is thought to combat biotrophic pathogens by restricting their access to water and nutrients.29 R gene-mediated resistance also activates SA-dependent signaling, leading to an activation of a string of presumed defense effector genes. The activation of SA signaling occurs throughout the plant to establish systemic acquired resistance (SAR) against subsequent pathogen infections.29 During SAR, deposition of callose and lignin occurs in plant cell walls, and plants acquire the ability to mount a rapid HR. Fan et al.9 reported that callose-plugged phloem sieve elements and inhibition of phloem loading were observed in CLas-inoculated leaves of both rough lemon and sweet orange, but that phloem transport was less affected in rough lemon than in sweet orange. In our study, phloem protein 2 (PP2–15) was identified as one of the most highly upregulated genes in CLas-infected sweet orange, but not in CLas-infected rough lemon. Several other studies also reported that phloem protein 2 (PP2) genes are upregulated in HLB-infected leaves.4,9–11,14 In addition to being involved in the differentiation of vascular tissue, a lectin-like protein (PP2) is associated with the plugging of sieve plates in response to wounding and as a defense against pathogens and insects.30 Accumulation of PP2 in sieve plates, in conjunction with phloem necrosis and blockage of the translocation stream, appears to be a major factor in disease symptom development of citrus greening.11 PP2-B15 was the most highly upregulated gene in CLas-inoculated roots of ‘Sanhu’ red tangerine at 50 days post inoculation. Zhong et al.14 speculated that PP2-like genes haves an active role in defense against the invading bacteria after a plant has been infected with CLas.
In our study, SNEA specifically identified ‘defense response’ (P=1.77E−06) and ‘plant immunity’ (P=0.0003) as over-represented ontologies among the upregulated genes (Table 3a and 3b) in CLas-inoculated leaves of rough lemon and sweet orange, respectively. Ontologies for ‘jasmonate response’ and ‘stomatal movement’ were also significantly over-represented in CLas-inoculated rough lemon. The accumulation of callose, which is synthesized between the cell wall and the plasma membrane, as well as stomatal closure, are classic markers of PTI.31 In addition, SA, JA, and ethylene hormones are induced during PTI. DEGs related to R gene-mediated resistance, which activates an SA-dependent signaling pathway, were not found in our study. EDS1and PAD4, which play important roles in SA signaling, and NPR1, a master regulator of SA,32 were also not significantly induced in any of the samples analyzed in the current study. Wang 33,41 indicated that CLas contains CLIBASIA_00255, which encodes a salicylate hydroxylase that can convert salicylic acid (SA) into catechol, a metabolite that does not induce a resistance response. ‘AT1G73805’, Systemic Acquired Resistance Deficient 1 (SARD1,), however, was highly expressed in CLas-inoculated samples of rough lemon (LFC=4.35) and sweet orange (LFC=9.00). SARD1, a plant-specific DNA-binding protein, is a key positive regulator of SA synthesis, and was induced after exposure to SA and JA.34 Knocking out SARD1 compromises both basal resistance and SAR.31 Further study is needed to understand the mechanism of SARD1 in citrus defense system. The SNEA analysis of the DEGs identified in the present study, specifically identified ‘hypersensitive response’ and ‘cell death’ as over-represented ontologies (P⩽0.05) among upregulated DEGs in CLas-inoculated leaves of both rough lemon and sweet orange, but these ontologies were not identified in mock-inoculated samples of rough lemon and sweet orange (Supplementary File 4). A total of 4 and 7 DEGs associated with hypersensitive response and cell death were identified in CLas-inoculated leaves of rough lemon, respectively, whereas 8 and 16 DEGs associated with hypersensitive response and cell death were identified in CLas-inoculated leaves of sweet orange, respectively. Among the identified DEGs, BAP2, GLIP1 and ACL5, associated with inhibition of cell death, were found in CLas-inoculated samples of sweet orange, whereas only BAP2 was found in CLas-inoculated samples of rough lemon. Arabidopsis C2 domain proteins, BAP1, and its homologue BAP2, negatively regulate biotic and abiotic cell death.35 ACL5 controls Arabidopsis xylem morphogenesis through the prevention of premature cell death.36 Treatment of Arabidopsis plants with GLIP1 protein systemically inhibited cell death in distant leaves inoculated with A. brassicicola, where cell death would be otherwise strongly induced.37 MPK3, PLA2A, RBOHD, RPM1, SAUL1, SOBIR1, VTC2, and WRKY22, all of which are associated with hypersensitive response and cell death, were upregulated in CLas-inoculated leaves of sweet orange, whereas SOBIR1, MKK9, RBOHD, and MPK3, also associated with hypersensitive response and cell death, were upregulated in CLas-inoculated leaves of rough lemon.
Starch, which accumulates extensively in photosynthetic cells, as well as in phloem elements and vascular parenchyma cells of leaf blades and petioles, has also been observed to accumulate in the xylem parenchyma and phelloderm of HLB-affected ‘Valencia’ orange trees but not in control (non-infected) samples.38 Albrecht and Bowman4 found that glucose-6-phosphate/phosphate transporter (GPT2), which mediates the import of glucose-6-phosphate, an essential substrate for starch biosynthesis, was more highly upregulated in infected ‘Cleopatra’ mandarin than in sweet orange. In the present study, GPT2 expression was significantly induced to a high degree in infected rough lemon but not in sweet orange. A total of 19 enzymes belonging to the starch and sugar metabolism pathway were identified. Fourteen were identified in CLas-inoculated sweet orange and 5 in mock-inoculated sweet orange, whereas 6 sugar and starch metabolism enzymes were identified in mock-inoculated rough lemon and 2 in CLas-inoculated rough lemon. Synthase (ec:2.4.1.12), diphosphatase (ec:3.6.1.9), and glycogenase (ec:3.2.1.1) were also found to be upregulated in CLas-inoculated leaves of sweet orange, while saccharogen amylase (ec:3.2.1.2) was upregulated in mock-inoculated sweet orange. Synthases (ec:2.4.1.12, and ec:2.4.1.14) were upregulated in mock-inoculated rough lemon, while hexokinase type IV glucokinase (ec:2.7.1.1) and fructokinase (phosphorylating) (ec:2.7.1.4) were found in Clas-inoculated samples of rough lemon (Supplementary File 5, Supplementary Tables S1 and S2). Although no symptoms of HLB were observed in inoculated leaves of rough lemon and sweet orange, a greater number of DEGs related to sugar and starch metabolism were found in inoculated leaves of sweet orange than in inoculated rough lemon leaves (Supplementary File 5, Supplementary Tables S1 and S2). These data indicate that transcriptomic reprogramming in carbohydrate metabolism and defense is occurring in response to CLas even before symptoms are visibly observed or positive RT-qPCR results for HLB were found. Previous studies have observed that many genes involved in photosynthesis are repressed in response to HLB, most likely due to increased accumulation of sucrose/glucose levels in leaves.10,11,39 In our study, SNEA analysis indicated that photosynthesis (P=0.001), as an ontology term, was the most significant in DEGs obtained from mock-inoculated rough lemon, whereas defense response (P=1.77 E−06), as an ontology term, was the most significant term among the DEGs obtained for CLas-inoculated rough lemon (Supplementary File 4, Supplementary Table S2). These data suggest that a dramatic shift from photosynthesis- to defense-related gene expression occurs in rough lemon as an early response to CLas. In previous proteome studies, it was found that accumulation of starch synthase increased, but the production of photosynthesis-related proteins decreased in infected rough lemon,40 and most significantly, upregulated proteins of infected sweet orange were involved in stress/defense response.39
Regulation of WRKY expression by MAPKs
The peptide flg22, a bacterial flagella protein recognized by most plants, activates a MAPK cascade which then leads the activation of WRKY-type transcription factors, key regulators of plant defense.41 Flagella in CLas, however, have not been observed in any of the numerous electron micrographs of these bacteria infecting plants and psyllids.2 The reduced genome of CLas and their transmission by psyllids may allow it to avoid PTI. CLas, however, still possesses 57 genes coding for products that function in cell envelope biogenesis and the outer membrane, including lipopolysaccharides, and most flagellar genes, which might function as PAMPs.33 Elongation factor Tu (EF-Tu) is one of the most abundant bacterial proteins and is recognized as a PAMP by Arabidopsis.42 The plant PRR for EF-Tu is the LRR-RLK EF-Tu receptor (EFR), which belongs to the same subfamily (LRRXII) as FLS2.42 In this study, upregulation (LFC=2.84) of EFR was only found in inoculated sweet orange. No expression of FLS2 was observed in CLas-inoculated leaves of either rough lemon or sweet orange.
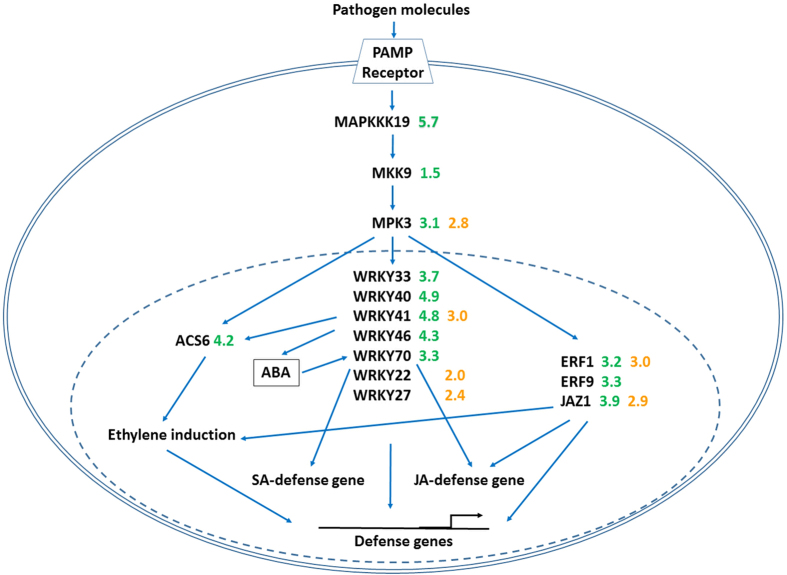
During PTI, activation of the MAPK cascade leads to the activation of WRKY-type transcription factors and other key regulators of plant immunity.43 It is thought that the MAPK cascade regulates plant immunity through the activation of defense-related genes via direct phosphorylation of downstream transcription factors, such as WRKYs and ERFs.44,45 MKK9 is upstream of MPK3, and MAPKKK19 is upstream of MKK9 (Figure 5). In our study rough lemon exhibited a stronger MAPK response than sweet orange (Table 1). After the perception of flg22, MPK6 activates ethylene biosynthesis through the phosphorylation of ACS6.46 ACS6 was highly upregulated in CLas-inoculated rough lemon but not sweet orange. We also observed the strong upregulation of ERF1 (LFC=3.18) and ERF9 (LFC=3.31) in CLas-inoculated leaves of rough lemon (Tables 3a and b, Figure 5). In response to pathogen attack, ET and JA cooperate through transcriptional induction of ET response factor 1 (ERF1).47 Activation of the MAPK cascade induces members of the WRKY family of transcription factors and defense-related genes 48 in tobacco.49 For example, WRKY22 is activated via a MAPKcascade induced by flg22.50 In our study, WRKY22 expression was only found in sweet orange, however, the upregulation was only moderate (LFC=1.96). These data were much lower than the upregulation of group 1 WRKY (WRKY33, WRKY70 and WRKY40) TFs in rough lemon. Emerging evidence has indicated that group I WRKY transcription factors, which contain a conserved motif in the N-terminal region, are also activated by MAPK-dependent phosphorylation, underlining their importance in plant immunity.51
Figure 5.
MAPK cascades as central signaling components in citrus pathogen defense. Upregulated genes in rough lemon are shown in green, and upregulated genes in sweet orange are shown in orange.
Our study indicates that WRKY transcription factor genes may play an important role in the tolerance to HLB exhibited by rough lemon. A total of eight WRKYs were identified in our RNA-seq analysis; with five in CLas-inoculated rough lemon and three in sweet orange. The magnitude of upregulation of all the WRKY genes in CLas-inoculated rough lemon was highly significant (LFC⩾3), while only WRKY41 approached an increase of LFC=3.00 in CLas-inoculated sweet orange. Higashi et al.52 indicated that WRKY41 may be a key regulator in the crosstalk between the salicylic acid and jasmonic acid pathways.52 In our study, WRKY41 was upregulated in inoculated samples of both rough lemon and sweet orange. In response to mock-inoculation, however, only rough lemon exhibited downregulation (LFC=−1.62) of WRKY42 and no significant difference was observed in sweet orange. The magnitude of the changes in the expression level of all of the WRKYs was higher in CLas-inoculated rough lemon than in CLas-inoculated sweet orange (Tables 3a and b). In Arabidopsis, WRKY46 was specifically induced by salicylic acid (SA) and the biotrophic pathogen Pseudomonas syringae.30,53 The WRKY33 transcription factor in Arabidopsis is required for resistance to necrotrophic fungal pathogens.30 Previous studies have presented evidence indicating that WRKY70 regulates plant disease resistance,53 and identified WRKY70 as a node of convergence for integrating salicylic acid (SA)- and jasmonic acid (JA)-mediated signaling events during plant response to bacterial pathogens.30 Over-expression of WRKY70 in Arabidopsis resulted in the suppression of several JA responses, including expression of a subset of JA-responsive and Alternaria brassicicola responsive genes. SA-induced expression of WRKY46, WRKY70, and WRKY53 is mainly dependent on NPR1.30 In our study, expression of NPR1 did not respond significantly to either mock- or CLas-inoculation, however, WRKY46 and WRKY70 were strongly upregulated in CLas-inoculated rough lemon. Martinelli et al.54 reported that several WRKY members were more highly upregulated in fully ripe fruits than in young leaves of Clas infected sweet orange.54
ABA-induced activation of MAPKs in rough lemon
MAPK cascades (Figure 5) have also been shown to be involved in ABA signaling.55 In the present study, MAK 3, MAPKKK19 and MKK9 were all significantly upregulated in CLas-inoculated rough lemon (Supplementary Files, Supplementary Figures S1 and S2), and may play a role in the ABA signaling pathway as suggested in a previous Arabidopsis study. Blast2GO also indicated strong over representation of an ABA signaling GO term in CLas-inoculated rough lemon (Supplementary File 6). MPK3 is activated by both H2O2 and ABA in Arabidopsis seedlings, and overexpression of MPK3 increased ABA sensitivity to ABA-induced post-germination arrest of growth.56 MPK9, which is preferentially expressed in guard cells, is also activated by ABA and has been shown to mediate ABA signaling in guard cells.57 MPK3/MPK6 negatively regulates ABA signaling in plants.58 The WRKY superfamily of TFs is the major regulator of plant defense and SA-mediated signaling, but also participates in ABA-mediated signaling.59 AtWRKY40 has been reported to be a negative regulator of ABA signaling during seed germination and also interacts with AtWRKY18 and AtWRKY60 to inhibit the expression of stress-responsive genes.53 In our study, WRKY40 was highly upregulated (LFC=4.9) in CLas-inoculated rough lemon. In addition, it was the most highly expressed among all of the upregulated WRKYs in CLas-inoculated rough lemon, but was not expressed in inoculated sweet orange. An and Mou32 stated that although ABA is known to play a crucial role in adaptation to abiotic stress, its role in biotic stress responses is less understood. In general, however, ABA is considered to be a negative regulator of disease resistance.55
Rapid production of reactive oxygen species (ROS) is one of the early detectable events following ABA perception.60 Plant recognition of PAMPs also induces the rapid and transient production of ROS in an oxidative burst.55 Interestingly, components of an ABA-activated MAPK cascade are also activated by ROS, suggesting that ABA and ROS may converge at the MAPK level in regulating stomatal closure.55 Previous studies of the hypersensitive response (HR) in pepper revealed that the activation of HR-induced cell death and defense responses is linked to signal transduction pathways that are coordinated by the activity of defense signaling molecules, such as ROS and plant defense hormones.61 In our current study, PUB22 (respiration burst in defense) was highly upregulated in both CLas-inoculated rough lemon (LFC=5.08) and sweet orange (LFC=5.42). RBOHD (respiratory burst oxidase-D) was also upregulated in CLas-inoculated rough lemon (LFC=3.15) and sweet orange (LFC=2.88). ROS mediate ABA signaling in guard cells.62 Although most of the links between ABA and MAPKs are poorly understood, it is evident that these pathways are part of the complex cellular signaling network that exists in plants to integrate various environmental cues, such as pathogen challenges, nutrient status or developmental programs.55 AtRbohD and AtRbohF were initially described in Arabidopsis as key components of plant defense. RBOH-dependent ROS generation is associated with pathogen recognition during the oxidative burst linked to the perception of MAMPs/PAMPs and during the HR, which is coupled to the recognition of pathogen avirulence factors through resistance proteins.37
Quantitative defense signaling
In a review, Eulgem16 stated that R-mediated resistance, basal defense, and SAR are related defense systems that share similar regulatory components and are effective against biotrophic pathogens. Differences between transcript profiles associated with R-mediated resistance and basal defense are quantitative rather than qualitative.16 Our results indicate that defense-response-related DEGs may be mediated by both basal defense and R-mediated resistance mechanisms. Distinct sets of DEGs were induced in CLas-inoculated rough lemon and sweet orange. We observed that the overall global mRNA expression (upregulated and downregulated) profiles in CLas-inoculated and mock-inoculated rough lemon and sweet orange were similar (Supplementary File 2, Supplementary Table S1). However, the amplitude of expression of the defense response in susceptible sweet orange was lower than in the tolerant rough lemon (Figure 3, Table 5). Resistance is regulated by changes in the levels of response of a variety of defense mechanisms and by quantitative enhancements that make the defense response more effective.15,16,63 The signaling mechanism that controls the activation of defense mechanisms consists of a highly interconnected network. Differences in the level of resistance often lie in the speed and intensity of specific defense responses. Resistant plants often respond more rapidly and vigorously to pathogens than susceptible plants. Hence, it is important to understand how plants sense the presence of a pathogen and initiate a subsequent response. Although the expression profile of up and downregulated genes in our study was similar in tolerant rough lemon and susceptible sweet orange, the proportion of downregulated DEGs with an LFC⩽−3 was 0.0% and 5.8% in rough lemon and sweet orange, respectively, while the proportion of upregulated DEGs with an LFC⩾3 was 29.0% and 25.1% in CLas-inoculated rough lemon and sweet orange, respectively. In mock-inoculated rough lemon and sweet orange, however, the proportion of DEGs that were down regulated was 3.8% and 1.2% in rough lemon and sweet orange, while the proportion of upregulated genes was 1.0% and 1.1%, respectively. RT-qPCR demonstrated that the transcription amplitude of 9 selected genes was higher in CLas-inoculated than in mock-inoculated treatments. In general, these data were consistent with the expression levels obtained from the RNA-seq data. The LFC values of T29M8.8, AGP16, PUB22, CBF, AT1G32928, JAZ1, MPK3 and WRKY70 were also higher in CLas-inoculated rough lemon than in the CLas-inoculated sweet orange. It should be noted, however, that there was a slight discrepancy between the expression levels of some genes in the RNA-seq data versus the RT-qPCR data, including DIC2, EXL2, and HSP90.1. Our results demonstrated that a higher number of DEGs with an LFC⩾3 were present in CLas-inoculated rough lemon sampled at week 7 post-inoculation. Activation of resistance and defense responses is stronger and faster in CLas-inoculated rough lemon than in CLas-inoculated sweet orange. Mock-inoculated plants of both rough lemon and sweet orange exhibited fewer WRKYs and defense-related genes than CLas-inoculated plants. These findings suggest that sweet orange exhibits an inability to suppress effectively the pathogen, resulting in the compatibility of the interaction between the host and pathogen. Maleck et al.64 found that defined sets of up-regulated genes are more highly expressed in an accelerated manner during incompatible interactions. Tao et al.15 proposed a mechanism that is commonly used in both R-gene and basal defense responses, where signal input to gene expression output occurs in a quantitatively determined manner. This mechanism is likely to involve regulators commonly used by both defense systems, such as WRKYs and defense response genes (Tables 4 and 5).
Table 6. Fold-change of identified defense response-related genes.
| Gene | Mock inoculated rough lemon | CLas inoculated rough lemon | Mock inoculated sweet orange lemon | CLas inoculated sweet orange |
|---|---|---|---|---|
| ATAF1 | 4.5 | |||
| ATL6 | −1.8 | 0.3 | ||
| BAP2 | 4.2 | 4.8 | ||
| BT1 | 2.2 | |||
| CCR1 | 2.8 | |||
| EFR | ||||
| ERF-1 | 2.8 | |||
| GLIP1 | 3.2 | 3.0 | ||
| JAZ1 | 2.1 | |||
| MPK3 | 3.9 | 2.9 | ||
| MYC2 | 3.1 | 2.8 | ||
| RBOHD | 1.8 | |||
| RHL41 | 3.2 | 2.9 | ||
| RPM1 | 4.3 | 3.9 | ||
| SOBIR1 | 2.1 | 1.9 | ||
| STZ | 2.9 | 2.6 | ||
| WRKY22 | 3.3 | |||
| WRKY33 | 2.0 | |||
| WRKY40 | 3.7 | |||
| WRKY70 | 4.9 |
Conclusions
The current study presents a comparative transcriptional analysis of the response of rough lemon and sweet orange to mock-inoculation and CLas-inoculation at 7 weeks post-inoculation in order to identify the early response to CLas infection in tolerant (rough lemon) and susceptible (sweet orange) trees. Rough lemon and sweet orange may recognize CLas via a pathogen recognition receptor, and activate plant immune systems even before positive RT-qPCR results can be obtained for CLas. Our collective results indicate that quantitative disease resistance may contribute to the durable tolerance to HLB exhibited by rough lemon, characterized by distinct transcriptional regulation of genes in various functional categories. Signaling pathways used by different defense systems appear to converge and target overlapping gene sets. The defense response DEGs mediated by basal defense and R-mediated resistance mechanisms were accelerated and amplified by CLas inoculation. Some transcription factors have been proven to play a pivotal role in disease resistance. Regulatory circuits linking signaling and gene regulation are emerging which suggest that a complex interplay between transcriptional activators and repressors fine-tunes the expression of the defense-related transcriptome. The current study provides new insights into the complex network of transcriptional regulation that occurs during the early response of rough lemon and sweet orange to CLas.
Acknowledgments
This work was supported by grants from the Citrus Research and Development Foundation Inc. (CRDF), on behalf of the Florida citrus industry.
Footnotes
Supplementary Information for this article can be found on the Horticulture Research website (http://www.nature.com/hortres)
The authors declare no conflict of interest.
References
- Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald P et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol 2012; 13: 614–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bove JM. Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol 2006; 88: 7–37. [Google Scholar]
- Zheng Z-L, Zhao Y. Transcriptome comparison and gene coexpression network analysis provide a systems view of citrus response to 'Candidatus Liberibacter asiaticus' infection. BMC Genomics 2013; 14: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht U, Bowman KD. Transcriptional response of susceptible and tolerant citrus to infection with Candidatus Liberibacter asiaticus. Plant Sci 2012; 185: 118–130. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature 2006; 444: 323–329. [DOI] [PubMed] [Google Scholar]
- Folimonova SY, Robertson CJ, Garnsey SM, Gowda S, Dawson WO. Examination of the responses of different genotypes of citrus to Huanglongbing (Citrus Greening) under different conditions. Phytopathology 2009; 99: 1346–1354. [DOI] [PubMed] [Google Scholar]
- Zhang MQ, Powell CA, Guo Y, Doud MS, Duan YP. A graft-based chemotherapy method for screening effective molecules and rescuing Huanglongbing-affected citrus plants. Phytopathology 2012; 102: 567–574. [DOI] [PubMed] [Google Scholar]
- Curk F, Ollitrault F, Garcia-Lor A, Luro F, Navarro L, Ollitrault P. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann Bot 2016; 117: 565–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Chen C, Yu Q, Khalaf A, Achor DS, Brlansky RH et al. Comparative transcriptional and anatomical analyses of tolerant rough lemon and susceptible sweet orange in response to 'Candidatus Liberibacter asiaticus' infection. Mol Plant Microbe Interact 2012; 25: 1396–1407. [DOI] [PubMed] [Google Scholar]
- Albrecht U, Bowman KD. Gene expression in Citrus sinensis (L.) Osbeck following infection with the bacterial pathogen Candidatus Liberibacter asiaticus causing Huanglongbing in Florida. Plant Sci 2008; 175: 291–306. [Google Scholar]
- Kim JS, Sagaram US, Burns JK, Li JL, Wang N. Response of sweet orange (Citrus sinensis) to 'Candidatus Liberibacter asiaticus' infection: microscopy and microarray analyses. Phytopathology 2009; 99: 50–57. [DOI] [PubMed] [Google Scholar]
- Liao HL, Burns JK. Gene expression in Citrus sinensis fruit tissues harvested from huanglongbing-infected trees: comparison with girdled fruit. J Exp Bot 2012; 63: 3307–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinelli F, Uratsu SL, Albrecht U, Reagan RL, Phu ML, Britton M et al. Transcriptome profiling of citrus fruit response to Huanglongbing disease. PLoS ONE 2012; 7: e38039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Cheng C-Z, Jiang N-H, Jiang B, Zhang Y-Y, BM-L Hu et al. Comparative transcriptome and iTRAQ proteome analyses of citrus root responses to Candidatus Liberibacter asiaticus infection. PLoS ONE 2015; 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Xie ZY, Chen WQ, Glazebrook J, Chang HS, Han B et al. Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 2003; 15: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci 2005; 10: 71–78. [DOI] [PubMed] [Google Scholar]
- Slisz AM, Breksa AP, Mishchuk DO, McCollum G, Slupsky CM. Metabolomic analysis of citrus infection by 'Candidatus Liberibacter' reveals insight into pathogenicity. J Proteome Res 2012; 11: 4223–4230. [DOI] [PubMed] [Google Scholar]
- Tatineni S, Sagaram US, Gowda S, Robertson CJ, Dawson WO, Iwanami T et al. In planta distribution of 'Candidatus Liberibacter asiaticus' as revealed by polymerase chain reaction (PCR) and real-time PCR. Phytopathology 2008; 98: 592–599. [DOI] [PubMed] [Google Scholar]
- Van Verk MC, Hickman R, Pieterse CM, Van Wees SC. RNA-seq: revelation of the messengers. Trends Plant Sci 2013; 18: 175–179. [DOI] [PubMed] [Google Scholar]
- Li WB, Hartung JS, Levy L. Quantitative real-time PCR for detection and identification of Candidatus Liberibacter species associated with citrus Huanglongbing. J Microbiol Methods 2006; 66: 104–115. [DOI] [PubMed] [Google Scholar]
- Wu TD, Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics 2010; 26: 873–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010; 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000; 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics 2003; 19: 2155–2157. [DOI] [PubMed] [Google Scholar]
- Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol 2015; 206: 932–947. [DOI] [PubMed] [Google Scholar]
- Stover E, Hall DG, Shatters RG, Moore GA. Influence of citrus source and test genotypes on inoculations with Candidatus Liberibacter asiaticus. HortScience 2016; 51: 805–809. [Google Scholar]
- Mafra V, Martins PK, Francisco CS, Ribeiro-Alves M, Freitas-Astua J, Machado MA. Candidatus Liberibacter americanus induces significant reprogramming of the transcriptome of the susceptible citrus genotype. BMC Genomics 2013; 14: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: the world of quantitative disease resistance. Trends Plant Sci 2009; 14: 21–29. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu Rev Phytopathol 2005; 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Hu Y, Dong Q, Yu D. Arabidopsis WRKY46 coordinates with WRKY70 and WRKY53 in basal resistance against pathogen Pseudomonas syringae. Plant Sci 2012; 185-186: 288–297. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lubberstedt T, Xu M. The genetic and molecular basis of plant resistance to pathogens. J Genet Genomics 2013; 40: 23–35. [DOI] [PubMed] [Google Scholar]
- An C, Mou Z. Salicylic acid and its function in plant immunity. J Integr Plant Biol 2011; 53: 412–428. [DOI] [PubMed] [Google Scholar]
- Wang N, Trivedi P. Citrus Huanglongbing: a newly relevant disease presents unprecedented challenges. Phytopathology 2013; 103: 652–665. [DOI] [PubMed] [Google Scholar]
- Jung IJ, Hwang JE, Han SM, Kim DS, Ahn JW, Choi HI et al. Molecular dissection of the response of the rice Systemic Acquired Resistance Deficient 1 (SARD1) gene to different types of ionizing radiation. Int J Radiat Biol 2017, 1–9. [DOI] [PubMed]
- Sakamoto M, Tomita R, Kobayashi K. A protein containing an XYPPX repeat and a C2 domain is associated with virally induced hypersensitive cell death in plants. FEBS Lett 2009; 583: 2552–2556. [DOI] [PubMed] [Google Scholar]
- Muniz L, Minguet EG, Singh SK, Pesquet E, Vera-Sirera F, Moreau-Courtois CL et al. ACAULIS5 controls Arabidopsis xylem specification through the prevention of premature cell death. Development 2008; 135: 2573–2582. [DOI] [PubMed] [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE et al. Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 2005; 17: 2832–2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxeberria E, Gonzalez P, Achor D, Albrigo G. Anatomical distribution of abnormally high levels of starch in HLB-affected Valencia orange trees. Physiol Mol Plant Pathol 2009; 74: 76–83. [Google Scholar]
- Fan J, Chen C, Yu Q, Brlansky RH, Li Z-G, Gmitter FG Jr. Comparative iTRAQ proteome and transcriptome analyses of sweet orange infected by ‘Candidatus Liberibacter asiaticus’. Physiol Plant 2011; 143: 235–245. [DOI] [PubMed] [Google Scholar]
- Nwugo CC, Duan YP, Lin H. Study on citrus response to Huanglongbing highlights a down-regulation of defense-related proteins in lemon plants upon 'Ca. Liberibacter asiaticus' infection. PLoS ONE 2013; 8: e67442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicaise V, Roux M, Zipfel C. Recent advances in PAMP-triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiol 2009; 150: 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JDG, Boller T et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 2006; 125: 749–760. [DOI] [PubMed] [Google Scholar]
- Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol 2009; 150: 1648–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Shan L, He P. Microbial signature-triggered plant defense responses and early signaling mechanisms. Plant Sci 2014; 228: 118–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol 2013; 51: 245–266. [DOI] [PubMed] [Google Scholar]
- Liu YD, Zhang SQ. Phosphorylation of 1-aminocyclopropane-1-carboxylic acid synthase by MPK6, a stress-responsive mitogen-activated protein kinase, induces ethylene biosynthesis in Arabidopsis. Plant Cell 2004; 16: 3386–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asselbergh B, De Vleesschauwer D, Hofte M. Global switches and fine-tuning—ABA modulates plant pathogen defense. Mol Plant Microbe Interact 2008; 21: 709–719. [DOI] [PubMed] [Google Scholar]
- Rawat N, Kiran SP, Du D, Gmitter FG Jr., Deng Z. Comprehensive meta-analysis, co-expression, and miRNA nested network analysis identifies gene candidates in citrus against Huanglongbing disease. BMC. Plant Biol 2015; 15: 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Zhang S. Activation of a mitogen-activated protein kinase cascade induces WRKY family of transcription factors and defense genes in tobacco. Plant J 2004; 38: 142–151. [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002; 415: 977–983. [DOI] [PubMed] [Google Scholar]
- Banerjee A, Roychoudhury A. WRKY proteins: signaling and regulation of expression during abiotic stress responses. Sci World J 2015; 2015: 807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashi K, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana. Mol Genet Genomics 2008; 279: 303–312. [DOI] [PubMed] [Google Scholar]
- Jiang JJ, Ma SH, Ye NH, Jiang M, Cao JS, Zhang JH. WRKY transcription factors in plant responses to stresses. J Integr Plant Biol. 2017; 59: 86–101. [DOI] [PubMed] [Google Scholar]
- Martinelli F, Reagan RL, Uratsu SL, Phu ML, Albrecht U, Zhao W et al. Gene regulatory networks elucidating Huanglongbing disease mechanisms. PLoS ONE 2013; 8: e74256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danquah A, de Zelicourt A, Colcombet J, Hirt H. The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 2014; 32: 40–52. [DOI] [PubMed] [Google Scholar]
- Lu C, Han MH, Guevara-Garcia A, Fedoroff NV. Mitogen-activated protein kinase signaling in postgermination arrest of development by abscisic acid. Proc Natl Acad Sci USA 2002; 99: 15812–15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Wang R-G, Mao G, Koczan JM. Identification of drought tolerance determinants by genetic analysis of root response to drought stress and abscisic acid. Plant Physiol 2006; 142: 1065–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock AK, Willmann R, Kolb D, Grefen L, Lajunen HM, Bethke G et al. The Arabidopsis mitogen-activated protein kinase phosphatase PP2C5 affects seed germination, stomatal aperture, and abscisic acid-inducible gene expression. Plant Physiol 2010; 153: 1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Abu Qamar S, Chen Z, Mengiste T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J 2006; 48: 592–605. [DOI] [PubMed] [Google Scholar]
- Kwak JM, Mori IC, Pei ZM, Leonhardt N, Torres MA, Dangl JL et al. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 2003; 22: 2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HW, Hwang BK. Molecular and cellular control of cell death and defense signaling in pepper. Planta 2015; 241: 1–27. [DOI] [PubMed] [Google Scholar]
- Jammes F, Song C, Shin D, Munemasa S, Takeda K, Gu D et al. MAP kinases MPK9 and MPK12 are preferentially expressed in guard cells and positively regulate ROS-mediated ABA signaling. Proc Natl Acad Sci USA 2009; 106: 20520–20525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri F. A global view of defense gene expression regulation—a highly interconnected signaling network. Curr Opin Plant Biol 2004; 7: 506–511. [DOI] [PubMed] [Google Scholar]
- Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA et al. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat Genet 2000; 26: 403–410. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.