ABSTRACT
Three-dimensional (3D) chromatin organization is important for proper gene regulation, yet how the genome is remodeled in response to stress is largely unknown. Here, we use a highly sensitive version of chromosome conformation capture in combination with fluorescence microscopy to investigate Heat Shock Protein (HSP) gene conformation and 3D nuclear organization in budding yeast. In response to acute thermal stress, HSP genes undergo intense intragenic folding interactions that go well beyond 5′-3′ gene looping previously described for RNA polymerase II genes. These interactions include looping between upstream activation sequence (UAS) and promoter elements, promoter and terminator regions, and regulatory and coding regions (gene “crumpling”). They are also dynamic, being prominent within 60 s, peaking within 2.5 min, and attenuating within 30 min, and correlate with HSP gene transcriptional activity. With similarly striking kinetics, activated HSP genes, both chromosomally linked and unlinked, coalesce into discrete intranuclear foci. Constitutively transcribed genes also loop and crumple yet fail to coalesce. Notably, a missense mutation in transcription factor TFIIB suppresses gene looping, yet neither crumpling nor HSP gene coalescence is affected. An inactivating promoter mutation, in contrast, obviates all three. Our results provide evidence for widespread, transcription-associated gene crumpling and demonstrate the de novo assembly and disassembly of HSP gene foci.
KEYWORDS: interchromosomal clustering, budding yeast, chromatin, chromosome conformation capture (3C), gene coalescence, gene looping, heat shock protein genes, Hsf1, live cell imaging, nuclear architecture, transcription factories
INTRODUCTION
It is becoming increasingly clear that gene expression can be modulated by the three-dimensional (3D) organization of chromatin (reviewed in references 1 and 2). When normal chromosome topology is disrupted, developmental malformations or cancer may be the consequence (3–5). As revealed by high-resolution chromatin contact mapping techniques (chromosome conformation capture [3C] and its derivatives), genomes of higher eukaryotes are organized into discrete structural and regulatory units of 100 kb to 1 Mb, termed topologically associating domains (TADs). These DNA loop structures serve to insulate active euchromatin from repressive heterochromatin (reviewed in reference 6). Shorter chromatin loops found within TADs have been shown to bring distant enhancer elements into physical contact with the gene promoters that they regulate, and these looped structures confine the activity of an enhancer to a single gene or set of genes within the TAD (reviewed in reference 2).
In addition to enhancer-promoter looping, other genome structural changes have been observed during gene activation, including colocalization of actively transcribed genes within a limited number of discrete sites of nascent RNA production, termed “transcription factories.” Such clustering has been observed chiefly at coregulated mammalian genes in erythroid cell- and lymphoid cell-specific lineages (7–10), as well as those induced by the cytokine tumor necrosis factor alpha (TNF-α) (11). Gene colocalization is thought to reflect a requirement for high local concentrations of RNA polymerase (Pol) II and other transcription and pre-mRNA processing factors to boost expression (12). Intergenic interactions revealed by 3C-based techniques that may reflect a more complex layer of Pol II gene regulation has not, to our knowledge, been seen in nonmammalian systems (for examples, see references 13 to 16).
In the budding yeast Saccharomyces cerevisiae, whose genomic organization of regulatory elements and genes is more compact than that of mammals, enhancer (upstream activation sequence [UAS])-promoter looping has been observed with transgenes (17). Gene loops that juxtapose promoter and terminator regions of various constitutive and inducibly expressed native genes have also been seen (18–20). These structures are thought to be a product of transcription that physically connects 5′-end and 3′-end machineries (19) and have been shown to suppress antisense transcription and confer transcriptional memory (21–23). Nonetheless, the existence of 5′-3′ gene loops remains controversial (24). Extensive regional and higher-order folding of individual yeast chromosomes has also been observed. Using a technique termed Micro-C, chromosomal interaction domains (CIDs) were identified that span ∼5 kb and encompass 1 to 3 genes (25). Recently, 200-kb TADs were identified in yeast that appear to be formally analogous to their counterparts in mammals (26).
Interchromosomal interactions, principally between centromeres, telomeres, origins of DNA replication, and sites of chromosomal breakpoints, also have been observed in yeast (13, 16). There is also evidence for tRNA gene clustering in this organism (27, 28), although this may stem from their predominant pericentric location and not from direct interactions (16). Interallelic clustering of actively transcribed genes, as well as that of coregulated transgenes, has been suggested based on fluorescence microscopy (29–31). However, whether such clustering represents bona fide interchromosomal interactions or simply subnuclear colocalization cannot be distinguished due to the limited resolution (∼500 nm) of such analyses. Moreover, as most previous studies have focused on the normal physiological state, it is unknown how transcriptional reprogramming that occurs in response to stress might affect yeast genome organization and structure.
The response to thermal stress in yeast is one of the most dynamic examples of transcriptional control known. Within 1 to 5 min of temperature upshift (30°C to 39°C), dramatic changes in protein-DNA interactions take place within HSP gene promoters, and these are accompanied by equally dramatic increases in transcription. Occupancy of the gene-specific activator heat shock factor 1 (Hsf1) is strongly increased within the UAS regions of most target genes (32–36). Accompanying this are striking increases in Mediator, SAGA, and Pol II occupancy of HSP gene promoters (36, 37) and gene-wide disassembly of nucleosomes (37–39). Certain of these phenomena, including SAGA and Mediator occupancy and gene-wide nucleosome disassembly, are transient, returning to pre-heat shock levels within 30 to 60 min (36–38, 40).
Here, we have exploited this dynamic transcriptional system to test the hypothesis that coordinated gene activation in response to thermal stress involves rearrangement of genome structure. To do so, we used a highly sensitive version of 3C in combination with fluorescence microscopy to evaluate the chromosomal conformation and genomic organization of yeast HSP genes. We found that not only do HSP genes form loops between their 5′ and 3′ ends upon transcriptional induction, their 5′/3′-regulatory and coding regions engage in intense intragenic interactions, a phenomenon that we term “crumpling.” Accompanying these dynamic looping and crumpling interactions is the evanescent physical clustering of both chromosomally linked and unlinked HSP genes. Our observations represent the first example of stress-mediated coalescence of transcriptionally activated genes revealed by 3C, and supported by live imaging, in any organism.
RESULTS
HSP genes undergo concerted intragenic interactions in response to heat shock.
To investigate conformational changes within HSP genes that occur during heat shock (HS), we used a modified version of the 3C technique (20, 41) that we term TaqI-3C. Briefly, control or heat shock-induced cells were formaldehyde cross-linked, and chromatin was isolated and digested with the 4-bp cutter TaqI, whose sites are favorably distributed throughout the bodies and regulatory regions of the genes we evaluated (Fig. 1A; see Fig. S1A and S2A and B in the supplemental material). Following proximity ligation of TaqI-cut DNA, the abundance of cross-link-dependent ligation products was determined using quantitative PCR (qPCR). A key to our approach is the fact that we normalized 3C signals for the efficiency of cleavage at each cut site, a critical though rarely used control. Indeed, we observed a wide range of variation in TaqI digestion efficiencies among tested genomic regions and time points of heat shock (e.g., see Fig. S3). Additional controls are described in Materials and Methods. As we describe below, the TaqI-3C approach afforded unprecedented insight into the dynamic restructuring and nuclear organization of heat shock-responsive genes.
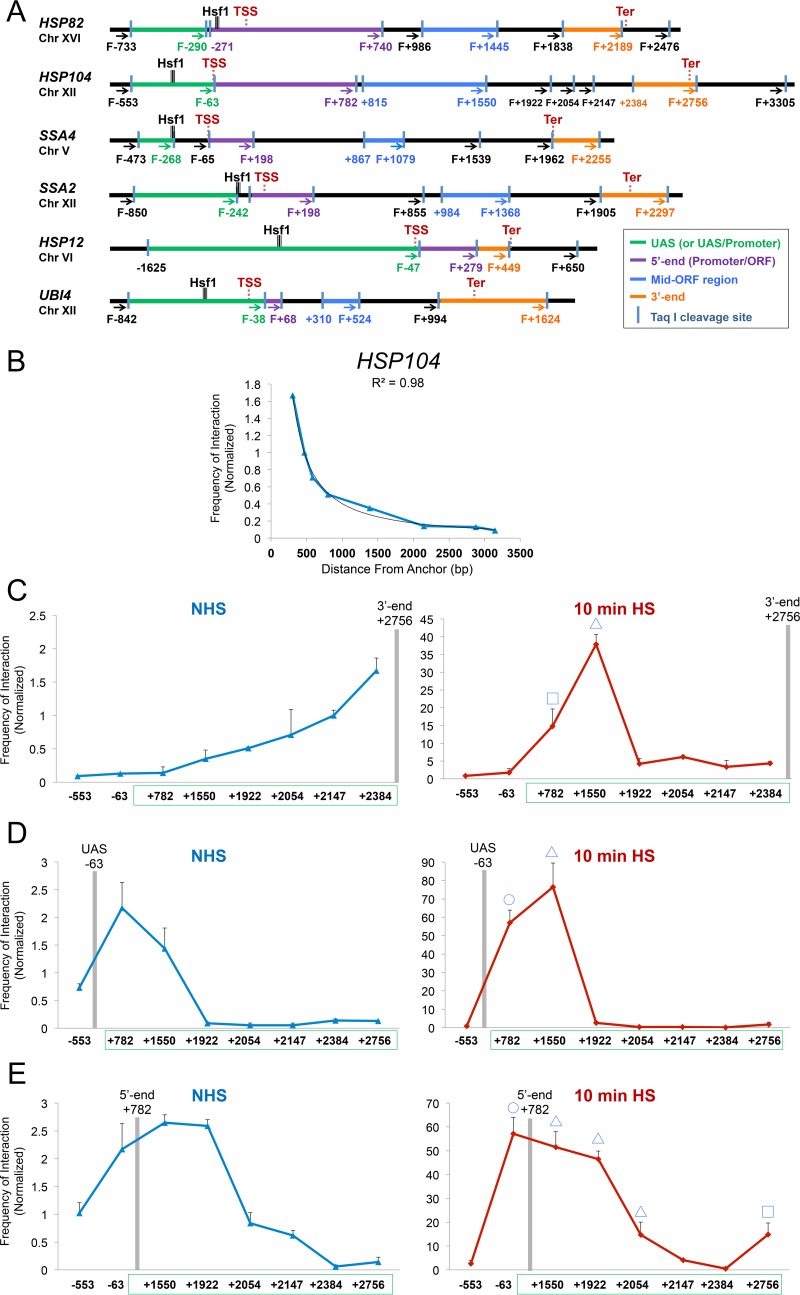
FIG 1.
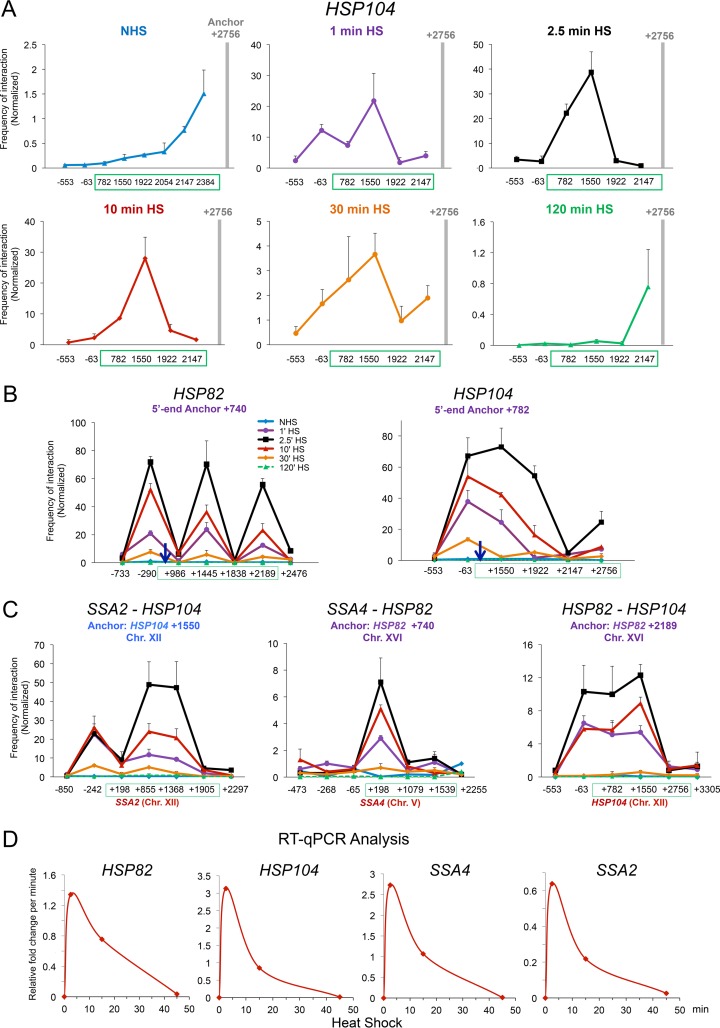
TaqI-3C reveals heat shock-dependent interactions between regulatory and coding regions. (A) Physical maps of HSP genes evaluated in this study. Coordinates correspond to TaqI sites; numbering is relative to the ATG codon (+1). Primers used for 3C analysis were sense strand identical (forward [F]) and positioned proximal to TaqI sites as indicated (arrows). UAS, 5′-end, mid-ORF, and 3′-end TaqI fragments are color-coded as indicated. Also indicated are locations of transcription start sites (TSS) and termination sites (Ter) (65, 66). Sites of Hsf1 occupancy were determined by ChIP sequencing (unpublished data). (B) Chromosomal contacts detected by TaqI-3C at a representative HSP gene exponentially decay with distance under noninducing conditions. The plot depicts frequency of contacts detected in cross-linked chromatin isolated from NHS cells sequentially digested with TaqI and ligated with T4 DNA ligase as described in Materials and Methods. The +2756 primer (3′ end) of HSP104 was used as an anchor. A regression curve (power function) to fit the data is also shown. Data are derived from two independent biological replicates (n = 2; qPCR = 4 for each primer combination). (C) Nonspecific contacts detected by TaqI-3C are strongly overridden by heat shock. Plots depict normalized interaction frequencies between the 3′-end anchor and the indicated loci within HSP104 in NHS and 10-min HS cells (cross-linking performed at 30°C and 39°C, respectively, as is the case throughout). Note the difference in scale. Green box, HSP104 transcribed region. A square signifies 5′-3′ gene looping, and a triangle signifies gene crumpling. Shown are means and standard deviations (SD); n = 2; qPCR = 4. (D) Analysis performed as described for panel C, except that the plots depict interaction frequencies between the UAS anchor and the indicated loci within HSP104. A circle signifies UAS-promoter looping; other symbols are as described for panel C. (E) Analysis as described for panel D, except the HSP104 5′ end served as an anchor. Here and for previous panels, a BY4742 background strain was used.
To determine whether HSP genes form loops between their 5′ and 3′ ends or engage in other intragenic contacts, we used TaqI-3C to query the relative frequencies with which the terminator (3′ end) of a representative HSP gene, HSP104, interacts with its coding and upstream regions in log-phase haploid cells. This strategy revealed that under non-heat shock (NHS; 30°C) conditions, only nonspecific contacts between the anchor region and neighboring regions were detectable, with their frequency decaying in accord with the approximate 1/L scaling of chromosomal contacts (Fig. 1B and C, left), as previously observed for other 3C-based methods (for examples, see references 25, 41, and 42). However, following a 10-min, 39°C heat shock, such nonspecific contacts were dwarfed by robust interactions between the terminator and other, more distant loci, particularly the 5′ end (+782) and midcoding region (+1550) (Fig. 1C, right). 3C analysis using the terminator regions of HSP82, SSA4, and HSP12 as anchors gave similar results (Fig. 2A).
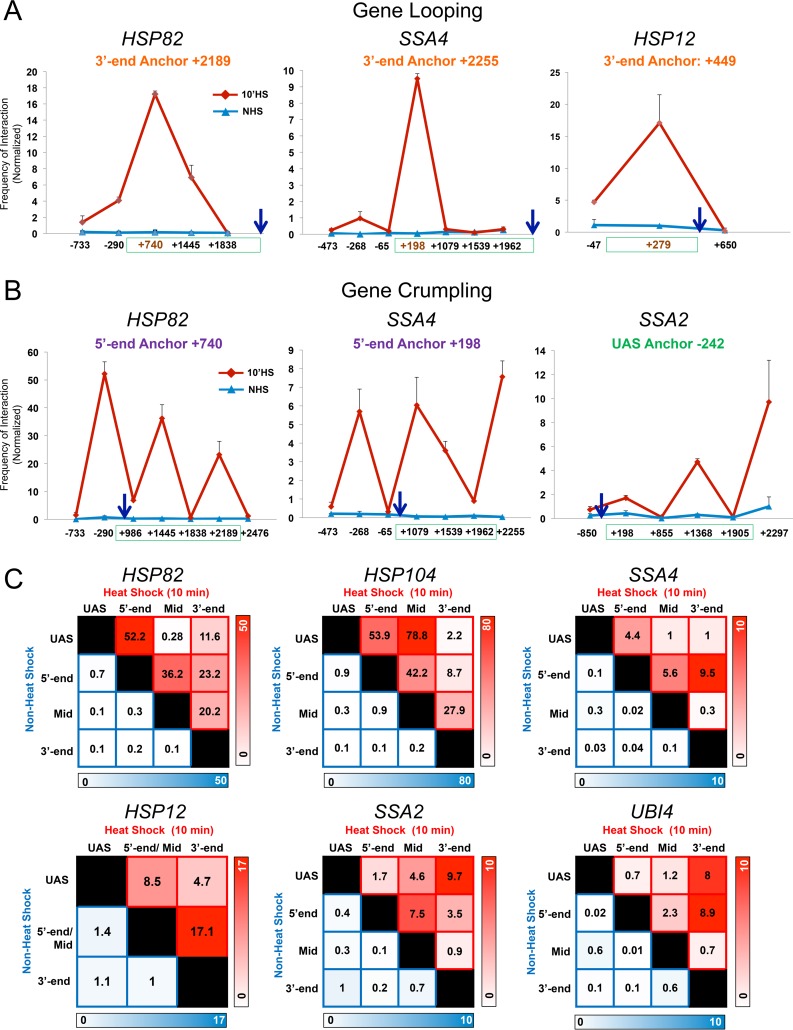
FIG 2.
Heat shock-dependent looping and crumpling is a general feature of HSP genes. (A) HSP genes form prominent 5′-3′ gene loops in response to heat shock. Depicted are normalized 3C interaction frequencies at representative HSP genes under NHS or 10-min HS conditions (strain BY4741). In each case the anchor primer (arrow), corresponding to the gene's 3′ end, was paired with primers abutting TaqI sites located along the coding and upstream regions (red coordinate on x axis corresponds to the TaqI fragment at the gene's 5′ end). In each graph, the green box spans the coding region. Shown are means and SD; n = 2; qPCR = 4. (B) HSP genes crumple in response to heat shock. Intragenic interactions within the indicated genes were detected under NHS and HS states using the indicated anchor primers. Symbols and analysis were the same as those described for panel A. (C) Matrix summaries of intragenic contact frequencies of HSP genes under control and inducing conditions. The upper right triangle corresponds to the frequency of interaction between indicated loci in 10-min heat-shocked cells (red shading); the lower left triangle corresponds to their frequency of interaction in NHS cells (blue shading). The intensity of color is proportional to the frequency of interaction. Regions within each gene are defined in the legend to Fig. 1A. For each pairwise test, n = 2 and qPCR = 4.
Use of UAS and promoter (5′-end) loci as anchors gave further evidence for strong regulatory region-coding region contacts. In the case of HSP104, infrequent contacts under the NHS state were overwhelmed by intense interactions between each anchor fragment and the midcoding region (+1550) following heat shock. Similar heat shock-dependent intragenic contacts were observed at other HSP genes (Fig. 2B; summarized in Fig. 2C). We propose the term gene “crumpling” to describe the DNA looping that takes place between the 5′-regulatory region (UAS/promoter)-coding region and 3′-regulatory region (terminator)-coding region of active genes. As we demonstrate below (see Fig. 8), gene crumpling can be functionally uncoupled from gene looping.
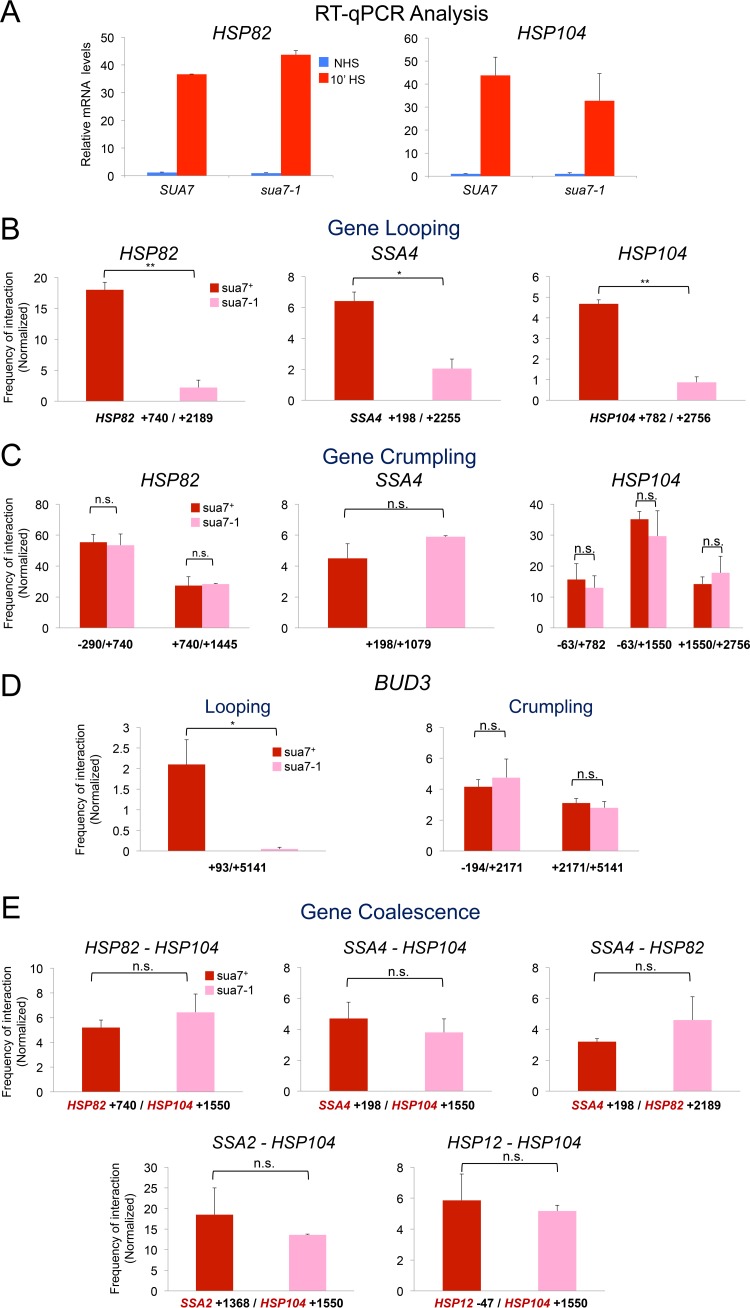
FIG 8.
sua7-1 functionally uncouples gene looping from crumpling and coalescence. (A) HSP82 and HSP104 mRNA levels in isogenic SUA7+ and sua7-1 strains under NHS and 10-min-HS conditions. For each transcript, expression levels were normalized to those in noninduced SUA7+ cells. Depicted are means and SD (n = 2; qPCR = 4). (B) 5′-3′ gene looping at representative HSP genes in SUA7+ and sua7-1 strains following 10 min of HS (analysis conducted as described for Fig. 2A). Depicted are means and SD; n = 2; qPCR = 4; *, P < 0.05; **, P < 0.01. (C) Gene crumpling at representative HSP genes in SUA7+ and sua7-1 strains following 10 min of HS as described for panel B. n.s., P > 0.1. (D) 5′-3′ looping of BUD3 is abolished in an sua7-1 mutant, yet the gene still crumples. (Left) 3C signal between 5′-end/+93 and 3′-end/+5141 regions of BUD3 in SUA7+ and sua7-1 cells (10-min HS state). (Right) Crumpling interactions between regulatory (UAS or 3′ end) and coding regions of BUD3 evaluated similarly. *, P < 0.05; n.s., P > 0.5. (E) Representative cis and trans intergenic interactions in SUA7+ and sua7-1 strains following 10 min of HS. Analysis was performed as described for panel D. n.s., P > 0.3.
In addition to gene looping and crumpling, we observed that the UAS and promoter regions (−63 and +782, respectively) of HSP104 frequently contacted one another (Fig. 1D and E). Detection of UAS-promoter contacts suggests that, as is the case in metazoans, proteins bound to enhancer sequences of natural genes engage in direct interaction with proteins located at promoters, consistent with earlier observations of yeast transgenes (17). Heat shock-dependent UAS-promoter loops were evident at genes in which the pertinent restriction sites were as close as ∼0.3 kb apart, attesting to the specificity of the TaqI-3C method. Our observations therefore are consistent with earlier demonstrations of 5′-3′ gene looping of actively transcribed yeast genes (18–20) yet reveal the existence of additional, typically more intense, intragenic folding interactions. Collectively, these observations suggest that multiple intralocus interactions take place upon transcriptional induction.
Intragenic folding interactions are cross-link dependent yet heat shock independent.
The above-described observations suggest the existence of concerted intragenic folding interactions at HSP genes distinct from the nonspecific short-range contacts between neighboring regions observed previously (13, 16, 25). These earlier studies found that cis interactions fall off exponentially with increasing distance, a phenomenon that we observe as well (Fig. 1B and C, left). To demonstrate that the contacts we detected were dependent on prior formaldehyde-induced cross-linking, we asked whether 3C interactions could be detected in non-cross-linked chromatin, yet we observed only background levels of interaction (S. Chowdhary and D. S. Gross, unpublished observations). Therefore, intragenic looping interactions can be detected only in chromatin that is cross-linked prior to its isolation.
To investigate how general crumpling is, we asked whether it could be detected at constitutively transcribed genes. For this purpose, we investigated the chromosomal conformation of BUD3, previously shown to loop between its 5′ and 3′ ends (19), as well as two highly expressed metabolic genes, PDC1 and FAS2 (see Fig. S2 for physical maps). Indeed, TaqI-3C detects prominent 5′-3′ gene loops within BUD3 (+93/+5141) as well as within FAS2 (+245/+5779) (Fig. 3A). We also detected prominent crumpling between the BUD3 coding region (+2171) and its terminator (+5141) (Fig. 3A, center and right), as well as between the 5′- and 3′-regulatory regions of PDC1 and FAS2 and their linked coding regions. Such interactions were maintained following a 10-min heat shock (and unaffected by cross-linking at the higher temperature) (Fig. 3B). These observations, combined with those discussed above, suggest that gene crumpling is a general property of actively transcribed Pol II genes.
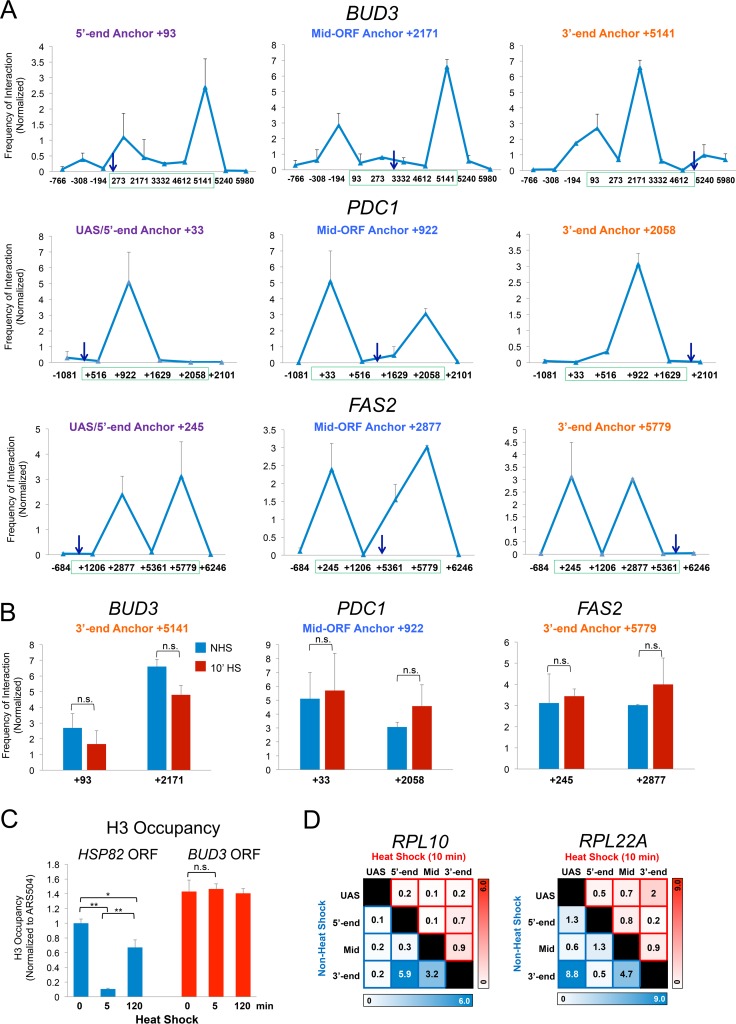
FIG 3.
TaqI-3C detects intragenic interactions within constitutively expressed genes. (A) Normalized interaction frequencies within BUD3, PDC1, and FAS2 in control cells detected using the indicated anchors (arrows). These were paired with primers abutting TaqI sites located along upstream, coding, and downstream regions. The green box spans the transcribed region of each gene. Gene regions are defined in Fig. S2A in the supplemental material. Shown are means and SD; n = 2; qPCR = 4. (B) Intragenic contact frequencies within constitutively expressed genes do not significantly change following shift to 39°C. The indicated 3C interactions within BUD3, PDC1, and FAS2 were assayed in NHS and 10-min HS cells. n.s., not significant; P = 0.1 to 0.4 (BUD3), P = 0.3 to 0.8 (PDC1), and P = 0.4 to 0.8 (FAS2). Two-tailed t test was used here and elsewhere unless specified otherwise. (C) H3 occupancy within HSP82 and BUD3 coding regions prior to or following 5 or 120 min of heat shock. H3 abundance was determined using ChIP-qPCR as described in Materials and Methods. **, P < 0.01; *, P < 0.05; n.s., P > 0.5 (one-way analysis of variance test). (D) Intragenic contact frequencies detected within ribosomal protein-coding genes RPL10 and RPL22A under NHS and 10-min-HS conditions. Representations are the same as those described in the legend to Fig. 2C.
Are crumpling interactions linked to nucleosome displacement? While HSP genes undergo significant displacement of histones during acute heat shock (37–39), a phenomenon that we confirm here (Fig. 3C, blue bars), H3 density within the BUD3 coding region remains high under both non-heat shock and heat shock conditions (Fig. 3C, red). Thus, both nucleosome-depleted and nucleosome-containing regions crumple in actively transcribed genes, arguing that nucleosome displacement is not a prerequisite for crumpling.
As an additional control, we asked whether TaqI-3C could detect looping/crumpling contacts within ribosomal protein genes whose constitutive transcription is strongly downregulated in response to acute heat shock (43) (D. Pincus, personal communication). Strong intragenic interactions were observed within both RPL22A and RPL10 under NHS conditions (Fig. 3D, blue squares), yet such contacts were substantially reduced following 10 min of heat shock (Fig. 3D, red squares). Thus, ribosomal protein genes behave oppositely of HSP genes: they crumple under NHS conditions yet decrumple following heat shock. This argues that crumpling is not a nonspecific response of chromatin to elevated temperature but instead reflects a biologically meaningful gene conformation that correlates with transcriptional activity. Moreover, it is seen at a variety of genes, including constitutively expressed, heat shock-induced, and heat shock-repressed genes.
HSP genes coalesce into transcriptionally active foci upon heat shock.
Having established the utility of TaqI-3C as a method to decipher 3D gene conformation, we asked if HSP genes engaged in intergenic interactions with each other. This might be expected if formation of Pol II transcription factories containing coregulated genes is conserved between mammals and yeast. The absence of previous 3C-based evidence for such intranuclear assemblies may stem from the fact that earlier studies examining global genome contacts in S. cerevisiae focused on the nonstressed expression state and/or were insufficiently sensitive (13, 16, 25) (see Discussion).
Initially, we tested for the presence of intrachromosomal looping interactions between HSP104 and SSA2, genes that lie within a 10-kb locus on chromosome (Chr.) XII. Consistent with their residing within distinct CIDs (25) (Fig. S1B), no significant interaction could be detected between the two genes in control cells (Fig. 4A, left, blue plot). However, following a 10-min HS, they engaged in extensive and robust interactions (red plot), and these were both cross-link and ligation dependent (Chowdhary and Gross, unpublished). These and other intergenic interactions (summarized in Fig. 4B) suggest that HSP104 and SSA2 are driven into a transcriptionally active chromosomal interaction domain in response to heat shock.
FIG 4.
HSP genes engage in striking, heat shock-dependent cis and trans intergenic interactions. (A) HSP genes engage in frequent intra- and interchromosomal interactions in response to acute heat shock. (Top left) Normalized 3C interaction frequencies between the mid-ORF (+1550) region of HSP104 and the indicated regions within SSA2 under NHS and 10-min-HS conditions. (Top right) As described above, except 3C interactions were quantified between the HSP82 5′ end (+740) and indicated regions within HSP104. Analysis and symbols are the same as those described in the legend to Fig. 2A. Shown are means and SD; n = 2; qPCR = 4 for each primer combination. (Bottom) DNA sequence confirmation of novel joints formed between the HSP104 +1550 and SSA2 +855 TaqI sites and HSP82 +740 and HSP104 +1550 TaqI sites (indicated by silver and blue globes, respectively). (B) Matrix summary of intergenic contact frequencies between chromosomally linked HSP genes in 10-min-heat-shocked cells. Normalized interaction frequencies, determined as described for panel A, are presented for the indicated loci. n = 2; qPCR = 4 for each primer combination. (C) Interchromosomal interaction frequencies of representative HSP genes presented in pairwise combinations as described for panel B. HSP104, Chr. XII; HSP82, Chr. XVI; SSA4, Chr. V; HSP12, Chr. VI; SSA2, Chr. XII. (D) Intergenic contacts between the constitutively expressed BUD3 gene and heat shock-inducible HSP genes under the indicated conditions. Anchor primers corresponding to the 5′ end of HSP82 or mid-ORF of HSP104 (as indicated) were paired with primers located along the BUD3 coding and upstream regions. Shown are means and SD; n = 2; qPCR = 4.
We next asked if HSP intergenic interactions could occur in trans and found compelling evidence for this in every pairwise test that we conducted. For example, the promoter-containing region of HSP82, located on Chr. XVI, contacted specific loci within HSP104 (Fig. 4A, right), while the midcoding region and 3′ end of HSP82 contacted multiple loci within SSA4, located on Chr. V (summarized in Fig. 4C). All such contacts were strictly heat shock dependent, and distinctive patterns of intergenic interaction were seen in all pairwise tests. Select intra- and intergenic interactions were confirmed by DNA sequencing (Fig. 4A) (Chowdhary and Gross, unpublished); all pairwise reactions additionally were shown to result in a single PCR product (Chowdhary and Gross, unpublished). Importantly, as very similar looping, crumpling, and coalescence interactions were observed in an unrelated genetic background (W303; A. S. Kainth and D. S. Gross, unpublished observations), they most likely represent fundamental features of these genes.
Constitutively transcribed BUD3 does not detectably coalesce.
Is gene coalescence a general feature of transcriptionally active genes? As a preliminary test of the idea, we asked whether constitutively transcribed BUD3, which looped and crumpled under both NHS and HS conditions (Fig. 3A and B), engaged in physical interactions with representative HSP genes. However, as shown in Fig. 4D, no interchromosomal interaction could be detected between BUD3 and representative HSP genes under either condition. This observation suggests that coalescence is not characteristic of all active Pol II genes (see Discussion).
HSP gene looping, crumpling, and coalescence interactions are dynamic and correlate with transcriptional activity.
In response to heat shock, HSP genes undergo dramatic changes in chromatin structure as well as in general transcription factor and coactivator occupancy. These alterations, including gene-wide displacement of nucleosomes and robust recruitment of Hsf1, Mediator, SAGA, and Pol II, are strikingly rapid, as such alterations generally are detectable within 60 s and peak within 2.5 min (36–39). They are also transient, as both histone and nonhistone protein occupancy typically returns to pre-heat shock levels within 90 min. To determine whether the intra- and intergenic interactions detected at HSP genes were similarly dynamic, we tested them at intervals over a heat shock time course, taking advantage of the instantaneous nature of our heat shock protocol as well as the fact that formaldehyde-mediated cross-linking permits snapshots of chromosome conformation at any instant in time.
We observed that all three phenomena, looping, crumpling, and coalescence, were remarkably dynamic and took place in parallel. Stimulus-dependent cis interactions overwhelmed distance-dependent stochastic interactions within 60 s of heat shock (Fig. 5A). Indeed, both cis and trans interactions were detectable within 60 s, the earliest time point evaluated, and all reached their maxima within 2.5 min (Fig. 5A to C, purple and black plots, respectively). Maximum intensities of gene looping and crumpling (including UAS-promoter interactions) were several hundredfold greater than those detectable under NHS conditions. Moreover, intrachromosomal interactions were >200-fold more frequent than those detected in the NHS state, and interchromosomal interactions were up to 100-fold more frequent (Fig. 5C). However, following 10 min at 39°C, the frequency of both intragenic and intergenic interactions had diminished (red plots), and as a general rule they were close to background by 30 min and indistinguishable from it by 120 min (orange and green plots, respectively).
FIG 5.
HSP genes dynamically loop, crumple, and coalesce during heat shock. (A) Heat shock rapidly overrides nonspecific contacts between the anchor and nearby loci, as detected by TaqI-3C. Illustrated is the frequency of intragenic interactions detected within HSP104 over a heat shock time course (note the difference in scale between time points). Analysis and presentation were as described for Fig. 1C. (B) Kinetics of HSP82 and HSP104 looping and crumpling in heat-shocked cells. (Left) 5′-End anchor of HSP82 was paired to the indicated loci upstream, within or downstream of the gene. Normalized 3C interaction frequencies were determined at the times indicated following instantaneous, 30°C to 39°C heat shock. (Right) As above, except the 5′ anchor of HSP104 was paired to the indicated loci. Shown are means and SD; n = 2; qPCR = 4 for each primer combination. (C) Kinetics of intergenic interactions between chromosomally linked and unlinked HSP genes. Analysis and presentation were as described for panel B. (D) Kinetics of HSP gene transcription during heat shock. Depicted is the relative fold change per minute of the indicated mRNAs following a 30°C to 39°C upshift, as deduced from steady-state (RT-qPCR) measurements. Data are consistent with genomic run-on analysis (67).
Importantly, kinetics of accumulation of HSP mRNA are consistent with the idea that these conformational changes correlate with transcriptional activity. At the majority of these genes, maximal fold increase in transcript levels occurs within the first 2.5 min of heat shock and diminishes thereafter (Fig. 5D). Consistent with this, we have previously observed that Pol II occupancy at HSP genes peaks within 2.5 min of heat shock, plateaus over the next 10 to 15 min, and returns to near-basal levels by 90 min (36, 37, 39).
Single-cell analysis reveals dynamic interactions between HSP genes in thermally stressed cells.
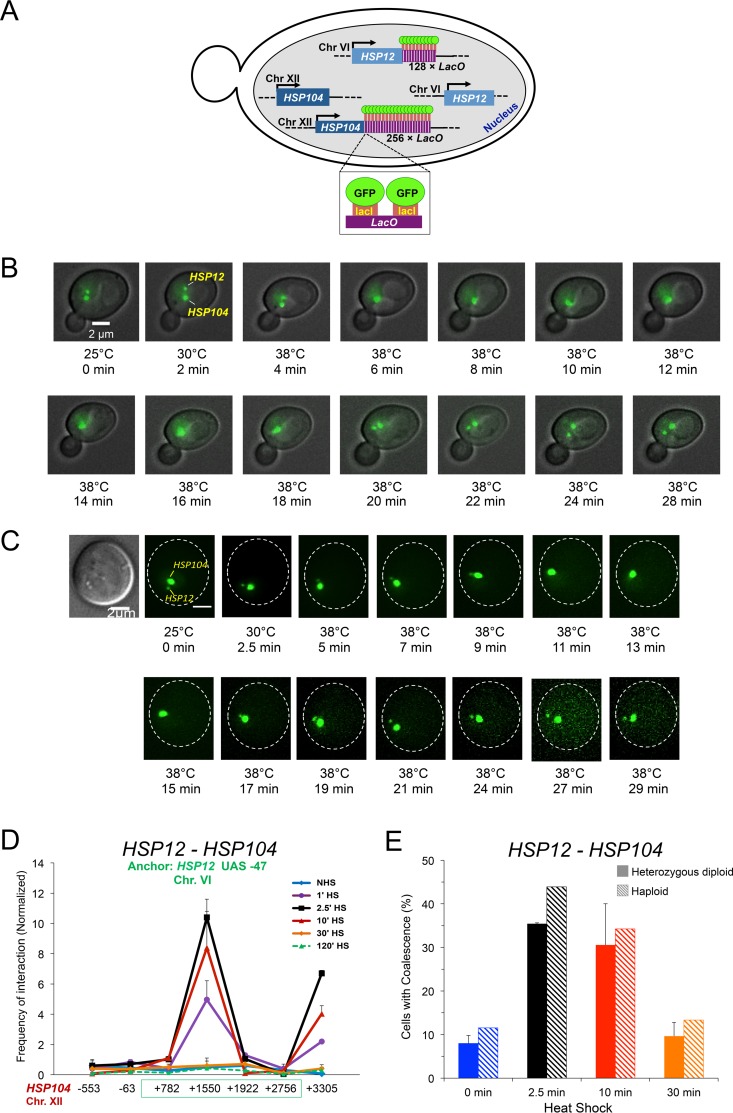
The 3C results described above suggest that concerted looping, crumpling, and coalescence interactions take place within and between HSP genes in cells exposed to heat shock. We sought to demonstrate one striking aspect of this phenomenon, interchromosomal HSP gene coalescence, using an independent approach. To do so, we created a diploid strain bearing fluorescently tagged alleles of HSP104 and HSP12. This was achieved through integration of 3′-flanking lacO arrays at each gene combined with ectopic expression of LacI-green fluorescent protein (GFP) (schematically illustrated in Fig. 6A). We then used fluorescence microscopy to assess the relative disposition of the two genes in live cells. As depicted in Fig. 6B, HSP104-lacO256 and HSP12-lacO128 were spatially separated within the interphase nucleus of a representative cell under noninducing conditions (25°C), consistent with the two genes lying on separate chromosomes. Following the application of heat, the two genes coalesced into a single focus within ∼2 min of reaching 38°C (6-min thermal exposure). Such coalescence dissipated 12 to 14 min later. A second example gave similar results, in which coalescence followed by dissociation occurred over a period of ∼12 min after reaching 38°C (Fig. 6C). Although in neither case was the response as rapid as that seen in the 3C assay, sequential clustering and dissociation of HSP104 and HSP12 are consistent with 3C analysis of unlinked HSP genes generally and for HSP12 and HSP104 in particular (Fig. 6D). The slower kinetics in the microscopy analysis likely arises from the fact that there is a gradual, rather than instantaneous, increase in temperature.
FIG 6.
Single-cell analysis demonstrates that HSP12 and HSP104 transiently coalesce in response to heat shock. (A) Schematic of ASK702, a diploid strain heterozygous for HSP12-lacO128 and HSP104-lacO256 and expressing LacI-GFP. (B) Live-cell fluorescence microscopy of an ASK702 diploid cell prior to and following application of heat for the times and temperatures indicated. The large and small green dots denote HSP104-lacO256 and HSP12-lacO128, respectively. Images were taken across nine planes in the z direction with an interplanar distance of 0.5 μm. Shown is a representative image for each time point. (C) Live-cell microscopy of an ASK702 cell responding to heat shock, as described for panel B. The first image was acquired using differential interference contrast (DIC). (D) TaqI-3C analysis of the kinetics of HSP12-HSP104 interaction, conducted as described for panel C. (E) Fraction of cells exhibiting HSP104-HSP12 coalescence. Mid-log-phase diploid and haploid cells (solid and hatched bars, respectively) were subjected to 30°C to 39°C heat shock for the indicated times and then fixed. For diploids (ASK706), 50 to 70 cells were evaluated per time point (means and SD are depicted; n = 2). For haploids (JTY001), 70 to 80 cells were evaluated per time point (n = 1).
To address how frequently HSP104-lacO256 and HSP12-lacO128 coalesce within the population, we performed microscopic analysis of cells that were subjected to instantaneous 30°C to 39°C heat shocks for 0, 2.5, 10, or 30 min and then fixed. This revealed the presence of HSP12-HSP104 coalescence in 30 to 35% of cells subjected to either a 2.5- or 10-min heat shock. Far fewer 0-min- or 30-min-heat shock-induced cells evinced similar clustering (Fig. 6E, solid bars). In light of 3C data suggesting negligible interaction between these two genes under the latter two conditions (Fig. 6D, blue and orange plots), it is possible that coincidental overlap of the two loci accounts for the 5 to 10% rate of coalescence observed under such conditions. Importantly, a parallel analysis of haploid cells subjected to similar treatment gave virtually identical results (Fig. 6E, hatched bars).
A promoter mutation obviates looping, crumpling, and coalescence.
To strengthen the link between transcription and looping, crumpling, and coalescence of HSP genes, we examined the effect of a 19-bp chromosomal substitution of the TATA box at HSP82, present in an allele termed hsp82-ΔTATA. This mutant retains its ability to transcriptionally activate in response to heat shock, albeit at 5% of the level of HSP82+ (38, 39). Pol II occupancy, revealed by chromatin immunoprecipitation (ChIP) analysis of its largest subunit, likewise is ∼20-fold reduced over the promoter, open reading frame (ORF), and 3′ untranslated region (UTR) in cells heat shocked for 10 min (Fig. 7A). Notably, occupancy of Hsf1 was not affected (Fig. 7B). On the other hand, hsp82 looping and crumpling interactions (signified by peaks at +2189 and +1445, respectively) were dramatically reduced in heat-shocked cells (Fig. 7C), while hsp82 coalescence with either HSP104 or SSA4 was obviated (Fig. 7D). In contrast, coalescence frequency of WT HSP genes in the same cells was unaffected (Fig. 7E). Taken together, these observations indicate that structural rearrangements of an HSP gene are linked to its ability to transcribe, and that neither binding of Hsf1 to the gene's upstream region nor the normal coalescence of other HSP genes is sufficient to elicit these changes in conformation or genome organization.
FIG 7.
A 19-bp core promoter mutation abolishes hsp82 looping, crumpling, and coalescence. (A) Pol II occupancy within the indicated regions of HSP82 and hsp82-ΔTATA prior to or following 10 min of heat shock. Abundance of Rpb1 was determined in isogenic HSP82+ and hsp82-ΔTATA strains (SLY101 and DSG118, respectively) using ChIP-qPCR. n = 2; qPCR = 4. (B) Hsf1 occupancy of the UAS regions of HSP82 and hsp82-ΔTATA prior to or following 10 min of heat shock. Hsf1 ChIP-qPCR was performed as described for panel A. n = 2; qPCR = 4; n.s., P > 0.05. (C) Looping and crumpling interactions within HSP82 and hsp82-ΔTATA in 10-min-heat-shocked SLY101 and DSG118 cells, respectively. Frequency of 3C interaction, as detected by TaqI-3C, between the +740 promoter anchor and the indicated loci is depicted. n = 2; qPCR = 4; ***, P < 0.001; *, P < 0.05; n.s., P > 0.2. (D) Coalescence tests between HSP82 or hsp82-ΔTATA and representative HSP genes in 10-min HS cells. n = 2; qPCR = 4; *, P < 0.05. (E) As described for panel D, except coalescence between WT HSP genes at the indicated loci in the same strains is depicted. n = 2; qPCR = 4; n.s., P > 0.3.
Gene looping can be functionally uncoupled from crumpling and coalescence.
Finally, we asked if either Pol II gene crumpling or HSP gene coalescence were functionally linked with 5′-3′ gene looping. This possibility seemed likely, given the tight temporal linkage between both novel phenomena and looping (Fig. 5A to C). The E62K missense mutation in transcription factor TFIIB, located in its B-finger domain and termed sua7-1, previously has been shown to affect transcription start site selection and obviate 5′- to 3′-end gene looping at a number of genes (44–46). Consistent with previous observations, we found that this mutation had little or no effect on Pol II transcription, although it strongly suppressed promoter-terminator looping of heat-shock-inducible HSP genes as well as of BUD3 (Fig. 8A, B, and D). This is in accord with previous observations of BLM10, GAL10, HEM3, SAC3, and SEN1 (46). Strikingly, crumpling of these same genes was not affected (Fig. 8C and D), and HSP gene coalescence, between either linked or nonlinked genes, was also not affected (Fig. 8E). This result indicates that crumpling and coalescence can be functionally uncoupled from 5′- to 3′-end gene looping and implicates a distinct biological role for these novel conformations and genome rearrangements.
DISCUSSION
Both heat shock-induced and constitutively transcribed genes form promoter-terminator loops and engage in extensive intragenic interactions.
Using TaqI-3C, we found that transcriptionally active, Pol II-transcribed genes in S. cerevisiae not only form 5′- to 3′-end gene loops similar to those previously described but also engage in extensive intragenic folding interactions, i.e., they crumple. In the case of HSP genes, the frequency of formation of both gene loops and intragenic folds strongly correlates with transcription. Likewise, ribosomal protein genes decrumple concomitantly with their transcriptional downregulation. Therefore, crumpling is not unique to HSP genes and is not unique to the heat shock state. These observations point to the widespread existence of a heretofore unappreciated phenomenon, intragenic crumpling, that accompanies the transcriptional activation of protein-coding genes. As the genes evaluated here are largely intronless, they are unlikely to undergo intragenic interactions linked to RNA splicing (47). On the other hand, UAS/promoter-coding region contacts might be related to interactions observed between enhancer/promoter elements and coding regions of model mammalian genes during their synchronous transcription (48). Upon release of paused Pol II, enhancer/promoter contacts were observed within the gene body that correlated with progression of elongating Pol II. While we did not see this in our kinetic analysis, a caveat is that the genes evaluated were comparatively small (1 to 3 kb), thus making capture of early elongating complexes difficult.
As 3C data represent population averages, it is possible that individual genes do not engage in multiple intragenic interactions. Instead, only one or a few intragenic folding interactions may take place in each gene at a specific moment, and the multiple interactions detected by 3C represent a composite average of the cell population. However, recent evidence derived from genome architecture mapping (a technique unrelated to 3C) suggests that multiple interactions can occur within a single intramolecular complex (49). Thus, it is formally possible that crumpling reflects the presence of multiple intragenic folds within individual, transcriptionally active genes.
We conducted formaldehyde-mediated cross-linking of heat shock-induced cells at 39°C, thereby ensuring an accurate snapshot of chromatin contacts occurring at that temperature and transcriptional state. While efficiency of formaldehyde cross-linking is increased at higher temperatures, this is unlikely to play a measurable role in the dramatic conformational changes detected at HSP and non-HSP genes. First, constitutively transcribed genes cross-linked at different temperatures exhibit similar 3C signals (Fig. 3B). Second, the frequency of HSP gene looping, crumpling, and coalescence contacts vary dramatically during a heat shock time course (Fig. 5A to C), despite the fact that formaldehyde cross-linking was conducted at 39°C in all cases. This reveals the dynamic aspect of these structural phenomena, whose intensity correlates not with temperature but rather with transcription. Third, perturbations such as a TFIIB point mutation or a TATA box deletion suppress HSP gene looping, crumpling, and/or coalescence; these effects were readily detectable at 39°C (Fig. 7C and D and 8B).
A related issue is our selection of TaqI to fragment chromatin. We chose TaqI due to the frequent and favorable disposition of its recognition sites across the genes we examined (Fig. 1A; see also Fig. S1 and S2 in the supplemental material). However, as TaqI requires elevated temperature for its activity (60°C was used in our experiments), a potential concern is that extended incubation at this temperature might cause reversal of cross-links. Empirical data mute this concern. In addition to controls revealing that cross-linking of chromatin is essential to obtaining above-background 3C signals, the fact that dramatically different frequencies of interaction are detected in heat-shocked versus non-heat-shocked cells, as well as during a heat shock time course (as argued above), suggest that if reversal of cross-linking takes place during the TaqI-3C procedure, it is relatively insignificant.
Rapid and robust trans interactions take place between HSP genes in response to heat shock.
In addition to widespread Pol II gene crumpling, a striking finding is that HSP genes engage in rapid and extensive trans interactions with each other concomitantly with their activation. Interchromosomal interactions include those between UAS and promoter regions suggestive of trans regulation; these have also been observed in mammalian systems where similarly regulated genes colocalize into transcription factories (for examples, see references 9, 10, and 50). Beyond transvection-like phenomena, the HSP genes examined here engage in intricate physical interactions involving both regulatory and coding regions that, to our knowledge, have no precedent. Such interactions are also highly dynamic, closely paralleling the dynamic transcriptional activation/attenuation cycle of these genes.
In support of the 3C data are complementary findings obtained from fluorescence microscopy of cells bearing lacO-tagged HSP12 and HSP104 genes. Such orthogonal analysis confirms the rapid yet evanescent clustering of unlinked Hsf1 target genes in response to heat stress. They extend our 3C analysis by demonstrating that HSP gene clustering is relatively common, detectable in 35 to 40% of heat shock-induced cells. We suggest that this colocalization rate is consistent with the notion that all activated HSP genes cluster within a single intranuclear focus. This conclusion is based on two further considerations: (i) the rate of HSP12-HSP104 colocalization in heat-shocked haploid cells is very similar to that observed in the heterozygous diploid (Fig. 6E), consistent with alleles colocalizing in the diploid analysis; and (ii) the probability that a given HSP gene transcriptionally activates in response to heat shock may be less than 100%. Pertinent to the latter point, we have evaluated the frequency of expression of a chromosomal HSP-EGFP fusion gene and found that it was transcriptionally induced in ∼60% of heat-shocked cells (Kainth and Gross, unpublished). This observation is consistent with a stochastic transcriptional response similar to that described for other inducible genes, whose alleles tend to coexpress in the same cells (51–53), and with the bursting pattern of enhancer function observed in Drosophila melanogaster (54). If HSP12 and HSP104 likewise activate stochastically, then a 0.6 × 0.6 coalescence rate (35 to 40%) would be the maximum frequency expected.
The intergenic interactions between HSP genes provide strong support for the de novo assembly model of transcription factories (55), especially in light of the fact that trans interactions are undetectable prior to heat shock. In this regard, our results resemble those of Cisse et al., who used superresolution imaging to evaluate the nuclear distribution of Pol II molecules in human U2OS cells under nonstimulating and serum-stimulating conditions (56). These investigators observed transient formation of Pol II clusters whose mean lifetime increased an order of magnitude (∼5 to 50 s) upon serum stimulation. These dynamic, Pol II-containing assemblies bear a strong similarity to our observations of rapid assembly and disassembly of transcriptionally active HSP gene foci in response to thermal stress and, taken together with the noncoalescing phenotype of hsp82-ΔTATA, suggest that coalescence is the consequence rather than the cause of induced levels of HSP gene transcription.
Our results provide a contrast to previous work in Drosophila that failed to detect any alteration in nuclear organization of HSP genes in response to heat shock (14, 15). While TAD structures rapidly reorganize in response to thermal stress, such reorganization involves Polycomb-mediated repression of Pol II-transcribed non-HSP genes (15). On the other hand, HSP genes have been observed to relocate upon heat shock to the nuclear pore in Caenorhabditis elegans or move to nuclear speckles in mammalian cells (57–59). How these observations may relate to our 3C and live imaging data of yeast HSP genes is unclear, but they raise the possibility that HSP gene coalescence is linked to relocalization of these genes to a specific nuclear compartment.
Crumpling and coalescence can be functionally uncoupled from promoter-terminator gene looping.
Consistent with earlier observations, we found that an E62K missense mutation in the general transcription factor TFIIB strongly diminishes 5′-3′ looping of active Pol II genes. Strikingly, however, this mutation has little or no effect on gene crumpling and does not impact HSP gene coalescence. It also has little effect on transcription. The effect of sua7-1 on gene looping has been proposed to arise from loss of TFIIB contacts with one or more subunits of the CPF 3′-end processing complex (46). The fact that crumpling of both heat shock-inducible and constitutively transcribed genes, as well as coalescence of HSP genes, are unaffected by this mutation argues that these conformational phenomena are mechanistically separable from gene looping.
Dynamic coalescence may be a distinctive property of HSP genes.
As mentioned above, Hsf1-activated HSP genes coalesce with each other through both intra- and interchromosomal contacts. However, as shown here, they fail to interact with the constitutively transcribed BUD3 gene. Likewise, HSP genes fail to engage in detectable interaction with heat shock-activated genes regulated by an alternative activator, Msn2/Msn4 (S. Chowdhary, A. S. Kainth, and D. S. Gross, unpublished data). Thus, coalescence appears not to be a general property of transcriptionally activated yeast genes. Hi-C analysis of the yeast interactome likewise failed to uncover evidence for trans interactions between Pol II-regulated genes under conditions corresponding to the NHS state in our experiments (13, 16, 25).
Finally, we note that transcription-induced relocalization of yeast genes from the nucleoplasm to the nuclear periphery, and specifically to nuclear pore complexes (NPC) (21, 60–62), may be related to our observations. Work of Brickner and colleagues has suggested that, upon activation, similarly regulated genes (typically transgenes or homologues) can relocate to the nuclear periphery, where they appear to cluster, based on fluorescence microscopy analyses (∼500-nm resolution) (29, 31). However, whether these observations are related to those reported here using the sensitive TaqI-3C technique, which (like other 3C-based methods) has a resolution of 1 to 5 nm (1) and has revealed the presence of intricate and extensive physical interactions between unlinked HSP genes dispersed on multiple chromosomes and at a variety of latitudes, is unclear. Future work will be necessary to address this and other intriguing questions.
MATERIALS AND METHODS
Yeast strains.
The diploid strain ASK702 (HSP104-lacO256 HSP12-lacO128 GFP-lacI) was made by crossing ICY33 and a MATα derivative of DBY255 (haploid strains generously provided by D. G. Brickner and J. H. Brickner, Northwestern University). For fluorescence microscopy of fixed cells, the strains ASK706 and JTY001 were used. ASK706 is a derivative of ASK702 bearing the mCherry-tagged NPC protein Pom34. JTY001 is a haploid spore derivative of ASK706. In DSG118, 19 bp of the TATA box region of HSP82 was chromosomally replaced with random sequence (namely, TAAAACATATAAATATGCA [boldface indicates the DNA sequence of the gene's TATA box within the larger DNA sequence that was altered by the 19-bp mutation] was replaced with GATCTCCTTTAGCTTCTCG), creating an allele termed hsp82-ΔTATA, as previously described (38). The sua7-1 strain, YMH124, harbors a G184A point mutation within SUA7 (verified by DNA sequencing; generous gift of M. Hampsey, Rutgers University).
BY4741 was used for most molecular assays presented in this paper; exceptions to this are noted in the figure legends. A complete list of strains is provided in Table 1.
TABLE 1.
Yeast strains
| Strain name | Genotype | Source or reference |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| BY4742-Hsf1-AA | BY4741 MATα tor1-1 fpr1Δ RPL13A-FKBP12::NAT MET15+ HSF1-FRB-yEGFP::KAN-MX | F. Holstege |
| SLY101 | MATα ade− can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 cyh2r | 68 |
| DSG118 | SLY101 hsp82-ΔTATA | 38 |
| ICY33 | MATa ade2-1 can1-100 leu2-3,112 trp1-1 ura3-1 his3-11,15::GFP-lacI::HIS3 HSP12-lacO128::URA3 SEC63-MYC×13::TRP1 | J. Brickner |
| DBY255 | MATa ade2-1 can1-100 leu2-3,112 trp1-1 ura3-1 his3-11,15::GFP-lacI::HIS3 HSP104-lacO256::TRP1 SEC63-MYC×13::KAN-MX | 29 |
| ASK701 | DBY255 MATα | This study |
| ASK702 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15::GFP-lacI::HIS3/his3-11,15::GFP-lacI::HIS3 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3-1/ura3-1 HSP12-lacO128::URA3/HSP12+ HSP104-lacO256::TRP1/HSP104+ SEC63-MYC×13::TRP1/SEC63-MYC×13::KAN-MX | This study |
| ASK706 | ASK702 POM34-mCherry::NAT/POM34+ | This study |
| JTY001 | MATa ade2-1 can1-100 his3-11,15::GFP-lacI::HIS3 leu2-3,112 trp1-1 ura3-1 HSP12-lacO128::URA3 HSP104-lacO256::TRP1 SEC63-MYC×13::KAN-MX POM34-mCherry::NAT | This study |
| YMH14 | MATα cyc1-5000 cyc7-67 ura3-52 leu2-3,112 | 69 |
| YMH124 | YMH14 sua7-1 | 69 |
Culture conditions.
For most assays, cells were grown at 30°C in YPDA (yeast extract-peptone-dextrose [YPD] supplemented with 0.002% adenine) medium to mid-log density (A600 = 0.65 to 0.8). At that point, a portion of the culture was maintained at 30°C (non-heat-shocked [NHS] sample), while the remainder (heat shocked [HS]) was subjected to an instantaneous 30°C to 39°C upshift by mixing with an equal volume of prewarmed medium (55°C) in a shaking 39°C water bath for durations indicated in the figures. For 3C and ChIP, heat shock was terminated at the elevated temperature via addition of formaldehyde to a final concentration of 1% for 15 min; excess formaldehyde was quenched through addition of glycine (final concentration [Cf] = 135 mM). The NHS sample was treated similarly at 30°C.
3C.
Chromosome conformation capture (3C) was conducted using a modification of published protocols (20, 41) that incorporated the following features: (i) use of a 4-bp cutter, TaqI (recognizes T^CGA, where ^ indicates a cleavage site); (ii) normalization to percent digestion under each experimental condition (see Fig. S3 in the supplemental material for representative examples); (iii) quantification of 3C signal using quantitative PCR (qPCR); (iv) use of a no-template control; (v) normalization to a noncut region of the genome; (vi) normalization to purified genomic DNA similarly cleaved and ligated; and (vii) normalization to a no-ligation control. In addition, we confirmed that chromatin-specific looping interactions detected in this study, intragenic as well as intergenic, were dependent on prior formaldehyde cross-linking of cells and that a single PCR product arose from each pairwise reaction. Especially notable is the normalization to percent digestion. Efficiency of digestion of individual TaqI sites varies across a given locus (Fig. S3A and B) and can vary according to the physiological state of the cell (e.g., TaqI sites within HSP104 in cells exposed to heat stress for 0, 1, 2.5, 10, 30, and 120 min [Fig. S3A]).
Cells were grown, heat shocked, and formaldehyde cross-linked as described above. Cross-linked cells from a 50-ml culture were harvested and subjected to glass bead lysis for two cycles (20 min each) of vigorous vortexing at 4°C. A fraction of the crude chromatin lysate (typically 10% of the total) was digested with 200 U of TaqI (New England BioLabs) at 60°C for 7 h. TaqI was subsequently heat inactivated (80°C for 20 min) in the presence of SDS at a final concentration of 1%. Excess SDS was quenched via addition of Triton X-100 (Cf = 1%); the digested chromatin fragments were centrifuged and the pellet was resuspended in 100 μl of Tris-HCl (pH 7.5). Ligation was performed with 7× diluted TaqI-digested chromatin using 10,000 cohesive end units of Quick T4 DNA ligase (M2200L; New England BioLabs) at 25°C for 2 h. The sample then was digested with RNase (final concentration of 30 ng/μl; Sigma) at 37°C for 20 min. Proteinase K (final concentration of 70 ng/μl; Sigma) digestion was performed at 65°C for 12 h in the presence of 0.1% SDS. The 3C DNA template then was extracted using phenol-chloroform and ethanol precipitated in the presence of glycogen.
3C signals were quantified by real-time qPCR (7900HT real-time PCR system [Applied Biosystems]; Power SYBR green PCR master mix obtained from Thermo Fisher Scientific Corporation) using tandem primer combinations listed in Table S1. Use of tandem primers to generate 3C signals minimizes the possibility that PCR products obtained result from cross-link-independent ligation (20). For each tested gene, four anchors were selected and paired with primers to “walk” across the length of the gene. For intergenic 3C tests, four anchors selected for each gene were paired with primers arrayed along the length of the partner gene. Representative interactions are summarized in 3C line graphs and interaction matrices provided throughout the manuscript. Select amplicons were analyzed by agarose gel electrophoresis and/or DNA sequencing (Eurofins Genomics). Examples are presented in Fig. 4A (sequencing primers are presented in Table S3).
Variability in absolute 3C interaction frequencies for a given primer combination/heat shock condition reported in different figures may stem from the fact that such data arose from independent experiments; in contrast, data used to generate a specific panel were obtained from two biologically independent samples processed in parallel. Thus, such samples shared in common chromatin preparation, enzymes (TaqI and T4 DNA ligase), and other components of the 3C reaction. Therefore, slight variations in TaqI digestion, extent of cross-linking, or ligation frequency may account for the 2- to 3-fold difference in 3C values sometimes seen. Relative differences between 3C signals were nonetheless largely maintained between independent analyses.
A complete description of the TaqI-3C technique will be published elsewhere.
ChIP.
ChIP was performed essentially as described previously (36). As described above, cells were grown in YPDA medium to an A600 of 0.65 to 0.8 at 30°C and then subjected to an instantaneous heat shock at 39°C for the duration indicated. Heat shock treatments were terminated as described above. Cross-linked cells from a 50-ml culture then were harvested and subjected to glass bead lysis at 4°C for 30 min using vigorous vortexing. The crude chromatin lysate thus obtained was sonicated to an average size of ∼0.25 kb using 40 cycles of sonication (with 30-s on/off pulses; Diagenode Biorupter Plus). Twenty percent of the sonicated chromatin was incubated with 1 μl of anti-H3 globular domain antibody (ab1791; Abcam), 1 μl of anti-Rpb1 antibody (36), or 1 μl of anti-Hsf1 antibody (34). Following immunoprecipitation, RNA and proteins were removed by the addition of DNase-free RNase and proteinase K as described above. The ChIP template was extracted using phenol-chloroform, ethanol precipitated, and quantified by qPCR. The quantity of ChIP DNA detected by each locus-specific primer pair combination (Table S4) was deduced from interpolation of a standard curve generated using genomic DNA.
To correct for variation in the yield of the extracted ChIP templates, the signal obtained for each locus-specific primer combination was normalized to the corresponding signal obtained from the input DNA using the same primer pair. The input DNA was prepared using 10% of the total sonicated cross-linked chromatin, which was subjected to proteinase K treatment for 12 h at 65°C followed by phenol-chloroform extraction and ethanol precipitation.
Reverse transcription-qPCR (RT-qPCR).
S. cerevisiae cells were grown in YPDA at 30°C to an A600 of 0.65 to 0.8 and subjected to heat shock as described above. To terminate transcription, cells were treated with 20 mM sodium azide. Total RNA then was isolated using an RNeasy kit (74204; Qiagen). cDNA was synthesized using the high-capacity cDNA reverse transcription kit from Applied Biosystems (4368814). The template was 0.5 to 2 μg of purified RNA, and random primers were used; the reaction mix was incubated at 37°C for 2 h. The reaction mix then was 20-fold diluted, and 5 μl of this diluted cDNA template was used in qPCR. Relative cDNA levels were determined using the ΔΔCT method (64). In order to correct for variation in the yield of cDNA templates, the amplified signal from SCR1 Pol III transcript was used as a normalization control. To determine fold change per minute in mRNA levels, mean mRNA levels (derived from two independent biological samples for each time point) were normalized to that of the previous time point and were then divided by the time elapsed in minutes. Primer combinations used in mRNA measurements are listed in Table S5.
Fluorescence microscopy.
For live cell imaging, cells were grown at 30°C to early log phase in synthetic dextrose complete (SDC) medium supplemented with 0.1 mg/ml adenine. One microliter of a cell pellet was placed onto a patch of 2% agarose (prepared in SDC supplemented with 0.1 mg/ml adenine) on a glass slide and covered with a coverslip. The objective, in contact with the glass slide, was heated from 25°C to 38°C, and images were taken at 2- to 3-min intervals. For fixed-cell imaging, cells were grown at 30°C to early log phase in YPDA, subjected to instantaneous heat shock at 39°C for the indicated times (Fig. 6E), and then fixed in 1% formaldehyde for 10 min. Following washing with phosphate-buffered saline (PBS) (pH 7.4), a small quantity was transferred to a patch of 2% agarose (prepared in PBS) on a glass slide.
Images were taken through an Olympus Ach 100×/1.25-numeric-aperture (NA) or Olympus UPlanFl 100×/1.3-NA objective using a CoolSNAP HQ charge-coupled-device camera. Images (binned 2 by 2) were acquired across 9 planes on the z axis with an interplanar distance of 0.5 μm. An 89021 filter set (Chroma Technology) was used for imaging GFP. Slidebook, version 4 (Intelligent Imaging Innovations), was used to control camera acquisition and the z-axis stepping motor (Ludl Electronic products). Images were analyzed with ImageJ (1.48v). For coalescence analysis, cells with large buds typically had one or both genes replicated, as indicated by more than two green fluorescent spots. Such cells were not used in our analysis. Nine planes in the z direction, covering the entire depth of nuclei, were inspected for location of tagged genomic loci. A cell was scored positive for coalescence if the two green spots were not resolvable in all nine planes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Jayamani Anandhakumar for experimental assistance; David Pincus for helpful suggestions, stimulating discussions, and sharing unpublished data; Denes Hnisz for critical reading of an earlier version of the manuscript; Kelly Tatchell for assistance with fluorescence microscopy; Mike Hampsey and B. N. Singh for advice on the 3C procedure; and Jason and Donna Brickner, Mike Hampsey, and Frank Holstege for generous gifts of yeast strains.
S.C. and D.S.G. conceived the project. S.C., A.S.K., and D.S.G. designed the experiments. S.C. and A.S.K. performed the experiments. S.C., A.S.K., and D.S.G analyzed the data. S.C. and A.S.K. made the figures. D.S.G. wrote the paper, with assistance from S.C. and A.S.K.
This work was supported by grants from the National Science Foundation, Division of Molecular and Cellular Biosciences, awarded to D.S.G. (MCB-1025025 and MCB-1518345), and by Ike Muslow predoctoral fellowships awarded to S.C. and A.S.K.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00292-17.
REFERENCES
- 1.Dekker J, Mirny L. 2016. The 3D genome as moderator of chromosomal communication. Cell 164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hnisz D, Day DS, Young RA. 2016. Insulated neighborhoods: structural and functional units of mammalian gene control. Cell 167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupianez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, Santos-Simarro F, Gilbert-Dussardier B, Wittler L, Borschiwer M, Haas SA, Osterwalder M, Franke M, Timmermann B, Hecht J, Spielmann M, Visel A, Mundlos S. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161:1012–1025. doi: 10.1016/j.cell.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hnisz D, Weintraub AS, Day DS, Valton AL, Bak RO, Li CH, Goldmann J, Lajoie BR, Fan ZP, Sigova AA, Reddy J, Borges-Rivera D, Lee TI, Jaenisch R, Porteus MH, Dekker J, Young RA. 2016. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science 351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suva ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wendt KS, Grosveld FG. 2014. Transcription in the context of the 3D nucleus. Curr Opin Genet Dev 25:62–67. doi: 10.1016/j.gde.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 7.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. 2004. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 8.Brown JM, Leach J, Reittie JE, Atzberger A, Lee-Prudhoe J, Wood WG, Higgs DR, Iborra FJ, Buckle VJ. 2006. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol 172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet 42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park SK, Xiang Y, Feng X, Garrard WT. 2014. Pronounced cohabitation of active immunoglobulin genes from three different chromosomes in transcription factories during maximal antibody synthesis. Genes Dev 28:1159–1164. doi: 10.1101/gad.237479.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Papantonis A, Kohro T, Baboo S, Larkin JD, Deng B, Short P, Tsutsumi S, Taylor S, Kanki Y, Kobayashi M, Li G, Poh HM, Ruan X, Aburatani H, Ruan Y, Kodama T, Wada Y, Cook PR. 2012. TNFalpha signals through specialized factories where responsive coding and miRNA genes are transcribed. EMBO J 31:4404–4414. doi: 10.1038/emboj.2012.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feuerborn A, Cook PR. 2015. Why the activity of a gene depends on its neighbors. Trends Genet 31:483–490. doi: 10.1016/j.tig.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. 2010. A three-dimensional model of the yeast genome. Nature 465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. 2007. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol Cell 28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Lyu X, Hou C, Takenaka N, Nguyen HQ, Ong CT, Cubenas-Potts C, Hu M, Lei EP, Bosco G, Qin ZS, Corces VG. 2015. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell 58:216–231. doi: 10.1016/j.molcel.2015.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rutledge MT, Russo M, Belton JM, Dekker J, Broach JR. 2015. The yeast genome undergoes significant topological reorganization in quiescence. Nucleic Acids Res 43:8299–8313. doi: 10.1093/nar/gkv723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dobi KC, Winston F. 2007. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol Cell Biol 27:5575–5586. doi: 10.1128/MCB.00459-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Sullivan JM, Tan-Wong SM, Morillon A, Lee B, Coles J, Mellor J, Proudfoot NJ. 2004. Gene loops juxtapose promoters and terminators in yeast. Nat Genet 36:1014–1018. doi: 10.1038/ng1411. [DOI] [PubMed] [Google Scholar]
- 19.Ansari A, Hampsey M. 2005. A role for the CPF 3′-end processing machinery in RNAP II-dependent gene looping. Genes Dev 19:2969–2978. doi: 10.1101/gad.1362305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh BN, Ansari A, Hampsey M. 2009. Detection of gene loops by 3C in yeast. Methods 48:361–367. doi: 10.1016/j.ymeth.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan-Wong SM, Wijayatilake HD, Proudfoot NJ. 2009. Gene loops function to maintain transcriptional memory through interaction with the nuclear pore complex. Genes Dev 23:2610–2624. doi: 10.1101/gad.1823209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laine JP, Singh BN, Krishnamurthy S, Hampsey M. 2009. A physiological role for gene loops in yeast. Genes Dev 23:2604–2609. doi: 10.1101/gad.1823609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan-Wong SM, Zaugg JB, Camblong J, Xu Z, Zhang DW, Mischo HE, Ansari AZ, Luscombe NM, Steinmetz LM, Proudfoot NJ. 2012. Gene loops enhance transcriptional directionality. Science 338:671–675. doi: 10.1126/science.1224350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh TS, Fudenberg G, Goloborodko A, Rando OJ. 2016. Micro-C XL: assaying chromosome conformation from the nucleosome to the entire genome. Nat Methods 12:1009–1011. doi: 10.1038/nmeth.4025. [DOI] [PubMed] [Google Scholar]
- 25.Hsieh TH, Weiner A, Lajoie B, Dekker J, Friedman N, Rando OJ. 2015. Mapping nucleosome resolution chromosome folding in yeast by Micro-C. Cell 162:108–119. doi: 10.1016/j.cell.2015.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eser U, Chandler-Brown D, Ay F, Straight AF, Duan Z, Noble WS, Skotheim JM. 2017. Form and function of topologically associating genomic domains in budding yeast. Proc Natl Acad Sci U S A 114:E3061–E3070. doi: 10.1073/pnas.1612256114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson M, Haeusler RA, Good PD, Engelke DR. 2003. Nucleolar clustering of dispersed tRNA genes. Science 302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. 2008. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev 22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brickner DG, Ahmed S, Meldi L, Thompson A, Light W, Young M, Hickman TL, Chu F, Fabre E, Brickner JH. 2012. Transcription factor binding to a DNA zip code controls interchromosomal clustering at the nuclear periphery. Dev Cell 22:1234–1246. doi: 10.1016/j.devcel.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Randise-Hinchliff C, Coukos R, Sood V, Sumner MC, Zdraljevic S, Meldi Sholl L, Brickner DG, Ahmed S, Watchmaker L, Brickner JH. 2016. Strategies to regulate transcription factor-mediated gene positioning and interchromosomal clustering at the nuclear periphery. J Cell Biol 212:633–646. doi: 10.1083/jcb.201508068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brickner DG, Sood V, Tutucci E, Coukos R, Viets K, Singer RH, Brickner JH. 2016. Subnuclear positioning and interchromosomal clustering of the GAL1-10 locus are controlled by separable, interdependent mechanisms. Mol Biol Cell 27:2980–2993. doi: 10.1091/mbc.E16-03-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giardina C, Lis JT. 1995. Dynamic protein-DNA architecture of a yeast heat shock promoter. Mol Cell Biol 15:2737–2744. doi: 10.1128/MCB.15.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Erkine AM, Magrogan SF, Sekinger EA, Gross DS. 1999. Cooperative binding of heat shock factor to the yeast HSP82 promoter in vivo and in vitro. Mol Cell Biol 19:1627–1639. doi: 10.1128/MCB.19.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekinger EA, Gross DS. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105:403–414. doi: 10.1016/S0092-8674(01)00329-4. [DOI] [PubMed] [Google Scholar]
- 35.Hahn JS, Hu Z, Thiele DJ, Iyer VR. 2004. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24:5249–5256. doi: 10.1128/MCB.24.12.5249-5256.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, Gross DS. 2013. Mediator recruitment to heat shock genes requires dual Hsf1 activation domains and Mediator Tail subunits Med15 and Med16. J Biol Chem 288:12197–12213. doi: 10.1074/jbc.M112.449553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kremer SB, Gross DS. 2009. SAGA and Rpd3 chromatin modification complexes dynamically regulate heat shock gene structure and expression. J Biol Chem 284:32914–32931. doi: 10.1074/jbc.M109.058610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao J, Herrera-Diaz J, Gross DS. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol Cell Biol 25:8985–8999. doi: 10.1128/MCB.25.20.8985-8999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang H, Gao L, Anandhakumar J, Gross DS. 2014. Uncoupling transcription from covalent histone modification. PLoS Genet 10:e1004202. doi: 10.1371/journal.pgen.1004202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erkina TY, Erkine AM. 2006. Displacement of histones at promoters of Saccharomyces cerevisiae heat shock genes is differentially associated with histone H3 acetylation. Mol Cell Biol 26:7587–7600. doi: 10.1128/MCB.00666-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 42.Liu C, Wang C, Wang G, Becker C, Zaidem M, Weigel D. 2016. Genome-wide analysis of chromatin packing in Arabidopsis thaliana at single-gene resolution. Genome Res 26:1057–1068. doi: 10.1101/gr.204032.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade JT, Hall DB, Struhl K. 2004. The transcription factor Ifh1 is a key regulator of yeast ribosomal protein genes. Nature 432:1054–1058. doi: 10.1038/nature03175. [DOI] [PubMed] [Google Scholar]
- 44.Pinto I, Ware DE, Hampsey M. 1992. The yeast SUA7 gene encodes a homolog of human transcription factor TFIIB and is required for normal start site selection in vivo. Cell 68:977–988. doi: 10.1016/0092-8674(92)90040-J. [DOI] [PubMed] [Google Scholar]
- 45.Pinto I, Wu WH, Na JG, Hampsey M. 1994. Characterization of sua7 mutations defines a domain of TFIIB involved in transcription start site selection in yeast. J Biol Chem 269:30569–30573. [PubMed] [Google Scholar]
- 46.Singh BN, Hampsey M. 2007. A transcription-independent role for TFIIB in gene looping. Mol Cell 27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 47.Moabbi AM, Agarwal N, El Kaderi B, Ansari A. 2012. Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci U S A 109:8505–8510. doi: 10.1073/pnas.1112400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee K, Hsiung CC, Huang P, Raj A, Blobel GA. 2015. Dynamic enhancer-gene body contacts during transcription elongation. Genes Dev 29:1992–1997. doi: 10.1101/gad.255265.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beagrie RA, Scialdone A, Schueler M, Kraemer DC, Chotalia M, Xie SQ, Barbieri M, de Santiago I, Lavitas LM, Branco MR, Fraser J, Dostie J, Game L, Dillon N, Edwards PA, Nicodemi M, Pombo A. 2017. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543:519–524. doi: 10.1038/nature21411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papantonis A, Larkin JD, Wada Y, Ohta Y, Ihara S, Kodama T, Cook PR. 2010. Active RNA polymerases: mobile or immobile molecular machines? PLoS Biol 8:e1000419. doi: 10.1371/journal.pbio.1000419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zenklusen D, Larson DR, Singer RH. 2008. Single-RNA counting reveals alternative modes of gene expression in yeast. Nat Struct Mol Biol 15:1263–1271. doi: 10.1038/nsmb.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. 2006. Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raser JM, O'Shea EK. 2004. Control of stochasticity in eukaryotic gene expression. Science 304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fukaya T, Lim B, Levine M. 2016. Enhancer control of transcriptional bursting. Cell 166:358–368. doi: 10.1016/j.cell.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Buckley MS, Lis JT. 2014. Imaging RNA polymerase II transcription sites in living cells. Curr Opin Genet Dev 25:126–130. doi: 10.1016/j.gde.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cisse II, Izeddin I, Causse SZ, Boudarene L, Senecal A, Muresan L, Dugast-Darzacq C, Hajj B, Dahan M, Darzacq X. 2013. Real-time dynamics of RNA polymerase II clustering in live human cells. Science 341:664–667. doi: 10.1126/science.1239053. [DOI] [PubMed] [Google Scholar]
- 57.Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. 2013. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol 200:589–604. doi: 10.1083/jcb.201207024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khanna N, Hu Y, Belmont AS. 2014. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol 24:1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jolly C, Vourc'h C, Robert-Nicoud M, Morimoto RI. 1999. Intron-independent association of splicing factors with active genes. J Cell Biol 145:1133–1143. doi: 10.1083/jcb.145.6.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brickner JH, Walter P. 2004. Gene recruitment of the activated INO1 locus to the nuclear membrane. PLoS Biol 2:e342. doi: 10.1371/journal.pbio.0020342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Casolari JM, Brown CR, Drubin DA, Rando OJ, Silver PA. 2005. Developmentally induced changes in transcriptional program alter spatial organization across chromosomes. Genes Dev 19:1188–1198. doi: 10.1101/gad.1307205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM. 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441:774–778. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 63.Reference deleted. [Google Scholar]
- 64.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang C, Pugh BF. 2009. A compiled and systematic reference map of nucleosome positions across the Saccharomyces cerevisiae genome. Genome Biol 10:R109. doi: 10.1186/gb-2009-10-10-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castells-Roca L, Garcia-Martinez J, Moreno J, Herrero E, Belli G, Perez-Ortin JE. 2011. Heat shock response in yeast involves changes in both transcription rates and mRNA stabilities. PLoS One 6:e17272. doi: 10.1371/journal.pone.0017272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee S, Gross DS. 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol Cell Biol 13:727–738. doi: 10.1128/MCB.13.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Goel S, Krishnamurthy S, Hampsey M. 2012. Mechanism of start site selection by RNA polymerase II: interplay between TFIIB and Ssl2/XPB helicase subunit of TFIIH. J Biol Chem 287:557–567. doi: 10.1074/jbc.M111.281576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.