ABSTRACT
TP53 is one of the most commonly mutated genes in human cancers. Unlike other tumor suppressors that are frequently deleted or acquire loss-of-function mutations, the majority of TP53 mutations in tumors are missense substitutions, which lead to the expression of full-length mutant proteins that accumulate in cancer cells and may confer unique gain-of-function (GOF) activities to promote tumorigenic events. Recently, mutant p53 proteins have been shown to mediate metabolic changes as a novel GOF to promote tumor development. There is a strong rationale that the GOF activities, including alterations in cellular metabolism, might vary between the different p53 mutants. Accordingly, the effect of different mutant p53 proteins on cancer cell metabolism is largely unknown. In this study, we have metabolically profiled several individual frequently occurring p53 mutants in cancers, focusing on glycolytic and mitochondrial oxidative phosphorylation pathways. Our investigation highlights the diversity of different p53 mutants in terms of their effect on metabolism, which might provide a foundation for the development of more effective targeted pharmacological approaches toward variants of mutant p53.
KEYWORDS: cancer, EMT, OxPhos, glycolysis, metabolism, mutant p53
INTRODUCTION
TP53, encoding the tumor suppressor protein p53, is one of the most frequently mutated genes in human tumors. While a key consequence of TP53 mutations is inactivation of the wild-type (wt) function, many TP53 mutations in tumors are missense substitutions which may lead to the expression of full-length proteins that are often accumulated at high levels in cancer cells (1) and carry dominant phenotypes distinct from those caused by the loss of wt p53 function. Such phenotypes, described as mutant p53 oncogenic gain of function (GOF), include increased cell proliferation, chemoresistance, antiapoptotic functions, increased cell migration, and invasion (2–6).
Altered metabolism is one of the key biomarkers for cancer (7–9). In particular, enhanced glucose metabolism under aerobic conditions is a common feature of many tumors to meet the high biosynthetic demand of rapidly dividing cells (10). Recent studies have reported compelling evidence for a novel GOF of mutant p53 proteins in promoting metabolic changes (11–13). These studies suggest that metabolic pathways normally regulated by wt p53 are altered by mutant p53 to facilitate the supply of metabolites required for proliferation. Although six missense mutations (hot-spot mutations) account for about 30% of all TP53 mutations in human cancers, there are more than 1,500 distinct p53 mutants that have been reported in various cancer types (http://p53.iarc.fr/). However, the vast majority of the current mutant p53 GOF properties, including altered metabolism, have been described using hot-spot missense mutations, including p53R175H, p53R248Q, and p53R273H. For example, while the p53R175H mutant has been shown to metabolically utilize exogenous pyruvate in cancer cells to promote survival during glucose starvation (14), the p53R273H mutant has been shown to inhibit the expression of phase 2 detoxifying enzymes to promote survival under oxidative stress (15). Further, p53R175H, p53R248Q, and p53R273H mutants have been demonstrated to stimulate enhanced glucose uptake (12). Genome-wide expression analysis has identified the mevalonate pathway to be highly upregulated in mutant p53 expressing MDA-MB-231 (p53R280K) and MDA-MB-468 (p53R273H) breast cancer cell lines, and mutations in p53 have been correlated with high expression of sterol biosynthesis genes and lipid metabolism in these human breast tumors (16). Additionally, endogenous mutant p53 was shown to regulate the nucleotide pools transcriptionally by inducing the expression of multiple genes involved in the de novo nucleotide synthesis and salvage pathways in breast cancer cells (11).
While the role of wt p53 in regulating many aspects of metabolism, such as glycolysis, mitochondrial oxidative phosphorylation, glutaminolysis, and lipid metabolism, is established, currently the potential metabolic effects of distinct mutant p53 proteins is largely unknown. Considering that in vivo studies demonstrate mutant p53 GOF activities (2, 17–20), along with in vitro observations that even a seemingly subtle difference of one amino acid, such as R248W versus R248Q, can have a large impact on mutant p53 function, further provides a rationale for the need to resolve the contribution of individual mutant p53 proteins on metabolism.
In the present study, we investigated the effect of different p53 mutant proteins on metabolic pathways in experimental models of human cancer and normal cells. We present data showing significant differences among these mutants on metabolic pathways, including glycolysis and oxidative phosphorylation (OxPhos). In addition, we demonstrate that even the same amino acid substitutions at the same position in the p53 protein can have dramatically different phenotypic effects in terms of metabolism depending on the origin of the cell line in which it is expressed. Taken together, results of our study reveal distinct metabolic profiles of different mutant p53 proteins.
RESULTS
The effect of various mutant p53 proteins on glycolysis.
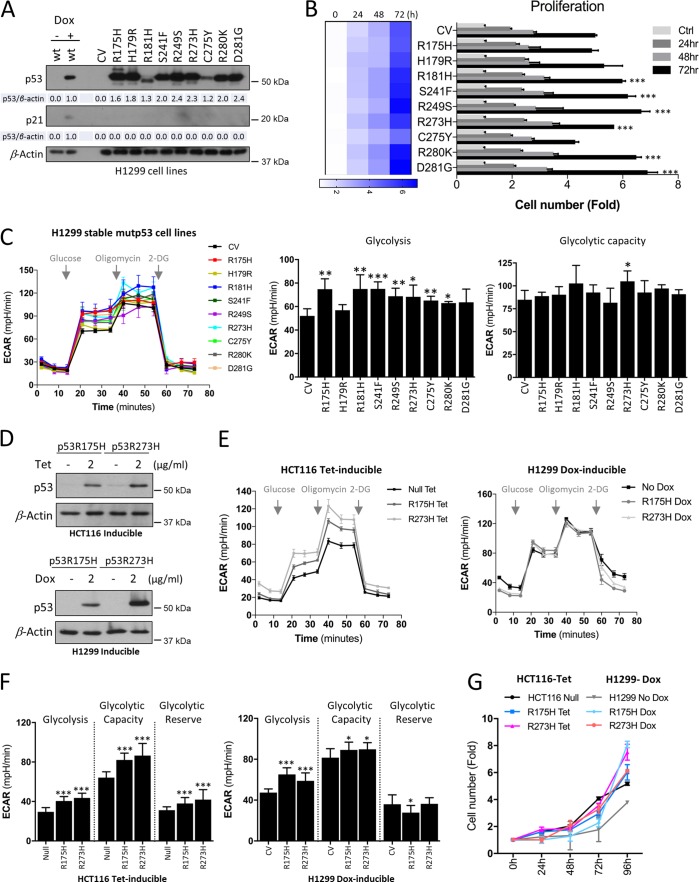
To study the potential metabolic alterations imposed by distinct mutant p53 variants, we generated a set of isogenic clones of human non-small-cell lung cancer H1299 cell lines that express nine different mutational p53 proteins. H1299 cells that stably express p53R175H, p53H179R, p53R181H, p53S241F, p53R249S, p53R273H, p53C275Y, p53R280K, and p53D281G mutants were compared to parental cells carrying a control vector (CV) (5) (Fig. 1A). The parental H1299 cells display a homozygous partial deletion of the TP53 gene and are null in their p53 expression, thus excluding the possibility that mutant p53 would act through dominant-negative (DN) effects on wild-type (wt) p53. Instead, the inducible expression system was employed in H1299 cells to express wt p53 induced by doxycycline (Dox). The different p53 mutant proteins displayed variable expression levels in H1299 cells (Fig. 1A). Compared to the Dox-induced wt p53 level, most mutants showed higher expression levels. However, regardless of their expression status, as expected, all of the mutants showed significant loss of ability to upregulate the cyclin-dependent kinase inhibitor p21WAF1/CIP1, a transcriptional target of wt p53, compared to the control. Subsequently, we studied the effect of mutant p53 expression on H1299 cell proliferation. While most p53 mutants showed no or only a slight increase in cell proliferation, the p53C275Y mutant displayed marginally decreased growth rates compared to the CV cells at 72 h (Fig. 1B), indicating that there is no significant difference in proliferation between most of the mutants. Further, to assess the potential effect on glycolytic function, the extracellular acidification rate (ECAR) was measured in real time, comparing the nine mutant p53 forms to the CV. ECAR is a surrogate measure to monitor lactate export, a terminal product of glycolysis. Consistent with previous findings, our analysis confirmed that the tumor hot-spot mutants p53R175H and p53R273H stimulated glycolysis (Fig. 1C) (12). Glycolysis was similarly observed to be higher in most of the other mutants, but not all mutants showed a significant increase in ECAR in H1299 cells (Fig. 1C), as the p53H179R and p53D281G mutants displayed no significant changes in glycolysis compared to the CV cells. However, there were no significant differences in the maximal glycolytic capacity (Fig. 1C) in most of the mutant p53-expressing H1299 cells compared to the CV cells. Only the p53R273H mutant displayed significant values for both glycolytic rates and maximal glycolytic capacity (Fig. 1C).
FIG 1.
Effect of mutant p53 proteins on glycolysis. (A) Western blots showing the expression levels of p53 and p21 (a transcriptional target of wt p53) in H1299 control (CV) and stable expression of indicated mutant p53 proteins. β-Actin was used as a loading control (n = 4). Densitometry analyses of the bands are indicated over β-actin. (B) Cell expansion assay was performed over 3 days to analyze proliferation in stable H1299 cells. Statistical significance is shown for 72 h. (C) Extracellular flux analyzer was used to measure glycolysis and the maximal glycolytic capacity with extracellular acidification rates (ECAR). (D) Western blot analysis of tetracycline (Tet)- or doxycycline (Dox)-induced expression of p53R175H and p53R273H mutants in HCT116 or H1299 cells, respectively. β-Actin was used as a loading control. (E) Mutant expression was induced as described for panel D, and ECAR were analyzed in HCT116 and H1299 inducible cell lines. (F) ECAR of glycolysis, maximal glycolytic capacity, and glycolytic reserve in inducible HCT116 or H1299 cells. (G) Cell expansion assay was performed over 4 days to analyze proliferation in inducible HCT116 and H1299 cells. Data are expressed as means ± SD (n = 3 to 7). Statistical significance is shown above the level for the control. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; two-tailed Student's t test.
To understand if the metabolic changes associated with the distinct mutant p53 proteins are due to adaptation of cancer cells to their stable expression or whether similar effects could be observed during direct induction of the mutant proteins, we utilized two independent cell systems (H1299 and HCT116) engineered with inducible expression systems of the two hot-spot p53 mutants, p53R175H and p53R273H (Fig. 1D). Compared to the parental H1299 cells that are null, the colon cancer HCT116 cells normally express low levels of wt p53; thus, the inducible expression system was employed in a p53-null HCT116 background. The expression of the mutants in H1299 and HCT116 cell lines was induced by Dox and tetracycline (Tet), respectively (Fig. 1D). For both cancer cell lines, analysis of glycolytic function showed enhanced glycolysis and maximal glycolytic capacity subsequent to induction of p53R175H and p53R273H mutants for 48 h, similar to effects of stable expression of these mutants in H1299 cells (Fig. 1E and F). However, compared to their stable counterparts, the inducible p53R175H and p53R273H mutants displayed more accelerated cell proliferation rates than the control cells up to 96 h (Fig. 1G), suggesting that they are more metabolically active.
Increased metabolic activity of cancer cells upon induced mutant p53 expression.
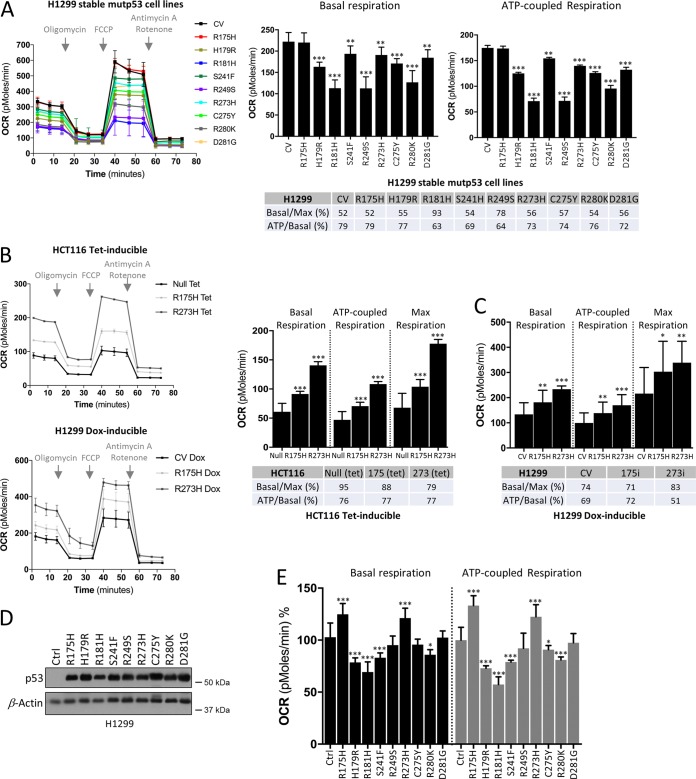
Beyond elevated glycolytic rates in tumors, several studies have demonstrated that the majority of tumor cells similarly have the capacity to sustain high fuel oxidation and ATP production in mitochondria (21). To investigate the role of distinct mutant p53 proteins on mitochondrial metabolism, oxygen consumption rate (OCR), which is a key parameter of mitochondrial function (22), was monitored in real time in the stable H1299 cell line panel. In contrast to the glycolytic functions, our analysis showed suppressed basal mitochondrial and ATP-coupled respiration of the majority of the mutants compared to the control cells, except for the p53R175H mutant, which displayed no significant difference (Fig. 2A, table). The p53R181H and p53R249S mutants displayed the most suppressive effect on OxPhos. In addition, while both the CV and most mutants respired at about half of their maximal capacity in the basal state (approximately 52%), the p53R181H mutant cells respired at their maximal capacity (93%) and p53R249S showed an increased (78%) ratio of basal respiration compared to its maximal capacity (Fig. 2A). Nonetheless, there were no significant differences between the mutants in terms of the fraction of O2 consumed to generate ATP (Fig. 2A, table). These observations suggest that various p53 mutants influence respiration through distinct mechanisms.
FIG 2.
Effect of mutant p53 proteins on mitochondrial metabolism. (A to C) Measurement of mitochondrial oxygen consumption rates (OCRs) in stable H1299 cell lines (A) and in tetracycline (Tet)- or doxycycline (Dox)-induced expression of p53R175H and p53R273H mutants in HCT116 or H1299 cells, respectively (B and C). OCR values of maximal respiration and ATP generated per consumed oxygen level were calculated. (D) Western blot of p53 in H1299 control (CV) cells with transient expression of the indicated mutant p53 proteins. β-Actin was used as a loading control. (E) Mutant p53 expression was induced as described for panel D, and mitochondrial OCRs were analyzed. Data are expressed as means ± SD (n = 3). Statistical significance is shown above the level for the control. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; two-tailed Student's t test.
We next analyzed the OCR/ECAR ratio of the cell line panel (data not shown). All mutant-expressing cells showed low OCR/ECAR ratios compared to the control cells, indicating their relatively higher reliance on glycolysis. Further, since mutant p53 proteins were not equally expressed in H1299 cells (Fig. 1A), we compared their expression level to those of either ECAR or OCR readouts. However, the observed distinct metabolic profiles of mutant p53 proteins did not correlate with their expression status. For instance, the low-expression mutants p53R181H, p53C275Y, and p53R280K still showed increased glycolysis, similar to mutants with high expression levels (Fig. 1A and C and 2A).
Compared to the stable H1299 cell lines, the functional measurement of mitochondrial parameters (basal respiration, ATP production, and maximal respiration) in the inducible p53R175H and p53R273H mutants revealed contrasting results. While only a minor effect of stable expression of p53R175H and p53R273H mutants could be observed on OxPhos (Fig. 2A, table), the inducible expression of these mutants in both H1299 and HCT116 cell lines caused significant increases in basal and ATP-coupled respiration (Fig. 2B and C), which is in line with the observed increased proliferation rates for these cells. Since control cells, HCT116 null and H1299 CV, were also treated with Tet and Dox, respectively, the possibility that experimental variations like these reagents could influence the cell line and metabolism was excluded. These data suggest that while glycolytic changes are kept consistent between stable and inducible mutant p53 expression in cancer cells, the mitochondrial energy metabolism pathway might be affected by the type of experimental system used for mutant p53 expression, which may affect the timing of transcriptional profiles regulated by mutant p53; hence, in the cell panel of the stable H1299 cells, no significant changes were observed in terms of cell growth. Our observations in the inducible mutant cancer cells are in agreement with the rationale that the increased cell proliferation correlates with enhanced glycolytic and mitochondrial activity. To gain insight into the differences between stable and inducible expression of the mutants, we studied the effect of the nine mutant p53 proteins that were used to create the stable cell lines and measured the OCR when they were transiently expressed in the H1299 cells (Fig. 2D). This analysis revealed the same trend of increased mitochondrial respiration for p53R175H and p53R273H mutants as that observed in the inducible systems. However, p53H179R, p53R181H, and p53S241F mutants still displayed suppressed basal and ATP-coupled respiration, while the remaining mutants did not show significant changes compared to the CV (Fig. 2E). Since the transient expression of p53R175H and p53R273H mutants responded in a fashion similar to that of the inducible H1299 cell lines, we concluded that stable H1299 cell lines have adapted to ectopic mutant p53 expression. Constitutive activation of oncogenes may change the adaptive capabilities; thus, it is likely that in stable H1299 cancer cells the actual effect of mutant p53 expression is masked by potential compensatory mechanisms, which may have suppressed its effect in terms of mitochondrial metabolism.
Metabolic characteristics of cancer cells that express endogenous mutant p53.
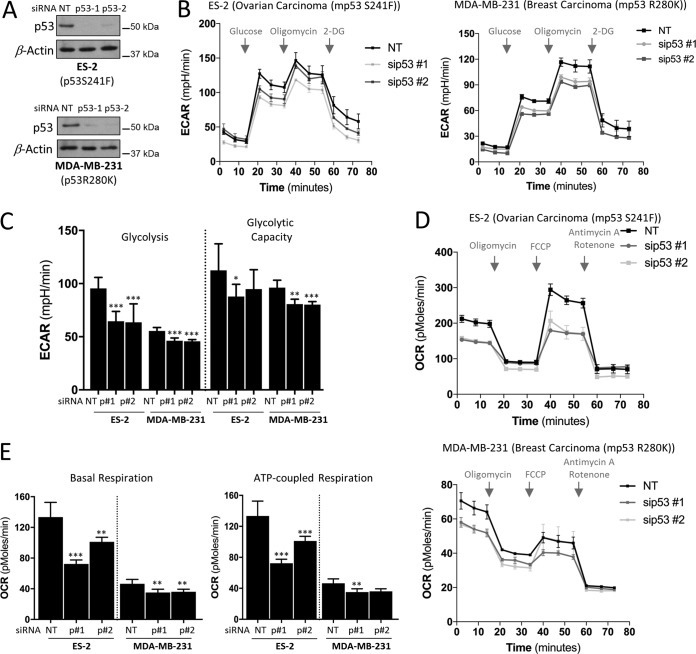
We next turned our attention to cells derived from tumors with endogenous mutant p53 expression. To assess the contribution of endogenous mutant p53, we silenced its expression using two independent short interfering RNAs (siRNAs) in ovarian ES-2 (p53S241F) and breast MDA-MB-231 (p53R280K) cancer cells (Fig. 3A), which normally express these mutants at high levels. Suppression of mutant p53 in both cells lines significantly reduced glycolysis, whereas the maximal glycolytic capacity was either not or marginally affected (Fig. 3B and C). A more pronounced effect of mutant p53 knockdown was observed on both basal and ATP-coupled respiration (Fig. 3D and E). In addition, knockdown of mutant p53 led to decreased maximal respiration capacity (Fig. 3D). Consistent with our observations from the inducible mutant p53 experiments presented in Fig. 1 and 2, these results demonstrate that mutant p53 proteins not only positively stimulate glycolytic functions but also significantly enhance the OxPhos in cancer cells. However, these data also suggest that since direct induction of mutant p53 or suppression of endogenous mutant p53 expression displays more pronounced effects on metabolic pathways than stable expression of the same mutants, cancer cells may have adapted differently to endogenous mutant p53 compared to stable ectopic mutant expression. It is also likely that cancer cells with endogenous mutant p53 show dependency on mutant p53, and suppression of its levels affects cells drastically.
FIG 3.
Effects of endogenous mutant p53 proteins on metabolism. (A) Western blots showing the NT (nontargeting) and p53 siRNA knockdown efficiency by two independent siRNAs targeting p53 in ovarian cancer ES-2 (p53S241F) and breast cancer MDA-MB-231 (p53R280K) cell lines. β-Actin was used as a loading control. (B and C) Cells were depleted of mutant p53 proteins as described for panel A and prior to analysis of the ECAR (B) and ECAR values of glycolysis and maximal glycolytic capacity (C). 2-DG, 2-deoxyglucose. (D) OCR analysis upon siRNA depletion of endogenous p53 levels as described for panel A. (E) OCR values of basal respiration and of ATP generated per consumed oxygen. Data are expressed as means ± SD (n = 3). Statistical significance is shown above the level of the control. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; two-tailed Student's t test.
Mutant p53 proteins exert different effects on metabolism in normal cells.
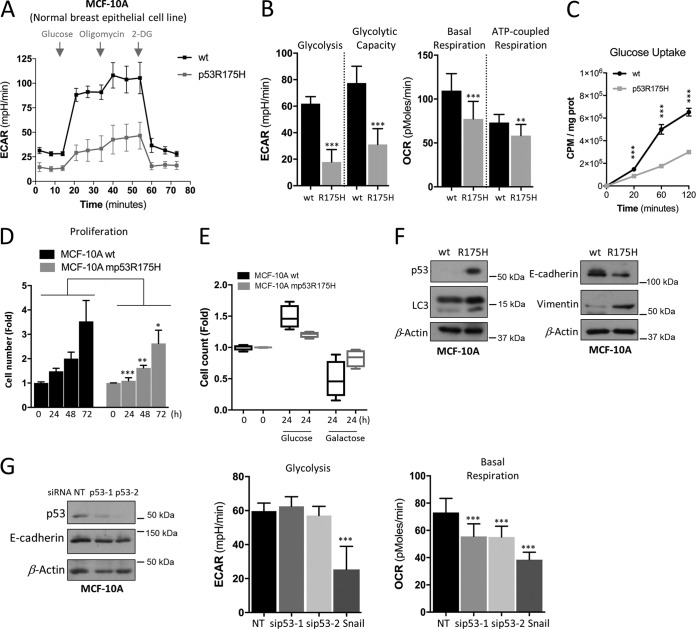
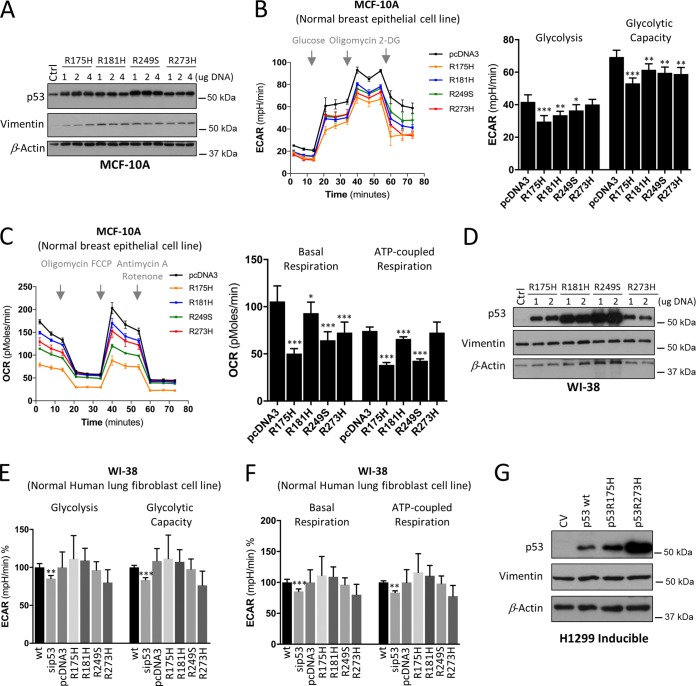
To gain further insights into the role of distinct mutant p53 proteins on metabolism, we studied their effect on nontumorigenic human cell lines. To this end, we first utilized MCF-10A, which is a spontaneously immortalized but nontransformed mammary epithelial cell line that contains wt p53. Metabolic pathways of parental MCF-10A and MCF-10A cells expressing the hot-spot mutant p53R175H cells were analyzed. MCF-10A stably expressing a tumor-derived hot-spot mutant (p53R175H) was created as described in reference 5, which excludes the dominant-negative activity of mutants on wt p53. Unexpectedly, unlike this mutant's effect in cancer cells and compared to the parental cells, our analysis showed that the effect of p53R175H expression was markedly altered, as it significantly suppressed glycolysis and glycolytic capacity in MCF-10A cells (Fig. 4A). Furthermore, beyond repressed glycolytic pathways, mutant p53 expression reduced mitochondrial basal and ATP-coupled respiration in MCF-10A cells (Fig. 4B). Given that both metabolic pathways are linked and might be affected by fuel limitation, we examined the cellular uptake of glucose. In contrast to the parental cells, mutant p53 cells showed a significant reduction in the ability of glucose uptake in a time-dependent manner (Fig. 4C), indicating that in MCF-10A the expression of mutant p53R175H limits glucose utilization. Combined, these data predict that the mutant p53R175H MCF-10A cells should display a lower growth rate and be less sensitive when exposed to galactose media. Indeed, mutant p53R175H cells displayed less proliferative capacity and were less affected by galactose than the wt MCF-10A cells (Fig. 4D and E).
FIG 4.
p53R175H mutant in nontumorigenic MCF-10A cells suppresses both glycolysis and mitochondrial metabolism. (A and B) Seahorse measurements of ECAR and OCR in parental (wt) and stable mutant p53 (p53R175H) expressing normal breast epithelial cell line MCF-10A cells. (C) Glucose uptake in parental (wt p53) and mutant p53R175H MCF-10A cells. (D) Cell expansion assay performed over 3 days to analyze proliferation in wt and stably expressing mutant p53R175H MCF-10A cells. (E) The effect of galactose on cell expansion in wt and stably expressing mutant p53R175H MCF-10A cells compared to glucose. (F) Western blots showing p53 expression, E-cadherin and vimentin (EMT markers), and LC-3 (autophagy marker). β-Actin was used as a loading control. (G) Western blots showing the NT (nontargeting) and p53 siRNA knockdown efficiency of p53 and E-cadherin expression in wt MCF-10A cells and measurements of ECAR and OCR following siRNA-mediated knockdown compared to that of NT or stably expressing Snail MCF-10A cells. ECAR values of glycolysis and maximal glycolytic capacity, as well as of basal versus maximal respiration and of ATP generated per consumed oxygen, are presented. Data are expressed as means ± SD (n = 3). Statistical significance is shown above the level for the control. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; two-tailed Student's t test.
It is known that p53 plays an essential role in epithelial-mesenchymal transition (EMT) and stemness (23–25), conditions during which cells are metabolically dormant. We first confirmed by Western blotting that mutant p53 stable cell lines represent an EMT model by analyzing the expression of EMT markers with decreased E-cadherin and increased vimentin (Fig. 4F). Furthermore, we observed increased LC3-II lipidation in mutant p53-expressing MCF-10A cells, indicative of autophagy activation, which is known to be triggered under nutritionally deprived and dormant conditions (Fig. 4F). We therefore measured the metabolic profile of MCF-10A cells that stably express Snail (snail1), a transcription factor that promotes EMT. These cells showed very similar effects on glycolysis and OxPhos (Fig. 4G) compared to cells stably expressing mutant p53R175H (Fig. 4A). Further, to understand if these effects were caused by loss of wt activity in MCF-10A cells, we used two different siRNAs against p53 (Fig. 4G). However, neither E-cadherin expression nor glycolysis was affected following p53 knockdown, whereas basal respiration was decreased (Fig. 4G), indicating that the robust glycolytic alterations observed in MCF-10A cells are due to GOF of mutant p53 and not the loss of its wt activity.
To further substantiate these observations, we transiently expressed the p53R175H, p53R181H, p53R249S, and p53R273H mutants in WI-38, a normal nontransformed human lung fibroblast cell line, in addition to MCF-10A cells (Fig. 5A and D). These mutants represent different classes of mutant proteins that displayed differential effects on metabolism, as shown in Fig. 1 and 2. We were able to observe the same marked effect of decreased metabolic activity in transient expression of p53R175H as that of the stable expression of this mutant in MCF-10A cells (Fig. 5B and C). Similarly, the expression of p53R181H, p53R249S, and p53R273H mutants displayed suppressed behaviors of glycolytic and mitochondrial functions (Fig. 5B and C).
FIG 5.
Effect of mutant p53 proteins on glycolysis and mitochondrial metabolism in normal cells. (A) Western blots showing the expression of p53 and vimentin following transient expression of p53 mutants (p53R175H, p53R181H, p53R249S, and p53R273H) using 1, 2, or 4 μg DNA, as indicated, in normal breast epithelial MCF-10A cells. β-Actin was used as a loading control. (B and C) MCF-10A cells as described for panel A were analyzed for ECAR (B) and OCR (C). (D) Western blots showing the expression of p53 and vimentin following transient expression of p53 mutants (p53R175H, p53R181H, p53R249S, and p53R273H) using 1 or 2 μg DNA, as indicated, in the normal nontransformed lung fibroblast WI-38 line. β-Actin was used as a loading control. (E and F) Measurements of ECAR (E) and OCR (F) following siRNA-mediated knockdown and p53 mutant overexpression compared to that of wt or pcDNA3 in WI-38 cells. (G) Western blots showing the expression of p53 and vimentin in Dox-induced expression of wt and p53R175H and p53R273H mutants in H1299 cells. Data are expressed as means ± SD (n = 3). Statistical significance is shown above the level for the control (wt or pcDNA). *, P < 0.05; **, P < 0.001; ***, P < 0.0001; two-tailed Student's t test.
In WI-38 cells, the p53 mutants did not influence glycolysis while the expression of R249H stimulated OxPhos (Fig. 5E and F), whereas suppression of wt p53 decreased mitochondrial respiration, as observed in MCF-10A cells (Fig. 4G). Since loss of wt p53 caused a similar effect on basal respiration in both cell lines, this effect likely is caused by the loss of wt activity. In addition, glycolysis in MCF-10A and WI-38 cells was not similarly affected by the expression of the mutant p53 proteins (Fig. 5B and E). This is likely due to the fact that none of the p53 mutants promoted EMT in WI-38 cells, as revealed by a lack of increase in vimentin levels (Fig. 5D) compared to that of the MCF-10A cells (Fig. 5A). Vimentin levels were also unchanged in inducible H1299 cell lines (Fig. 5G). Thus, our study suggests that the GOF activities, such as promoting OxPhos or EMT, differs even for the same mutant, including p53R175H and p53R273H mutants, depending on the cell line in which it is expressed. Moreover, upregulation of basal respiration by p53R249S is more likely to be a GOF than a DN capacity compared to the p53R175H, p53R181H, and p53R249S mutants, which may possess their metabolic alterations in a cancer cell-type-dependent manner.
Taken together, these findings reveal unexpected contrasting metabolic alterations imposed by the mutant p53 proteins in the H1299 and HCT116 cancer cell lines compared to normal MCF-10A and WI-38 cells.
Effect of mutant p53 proteins on mitochondrial functions.
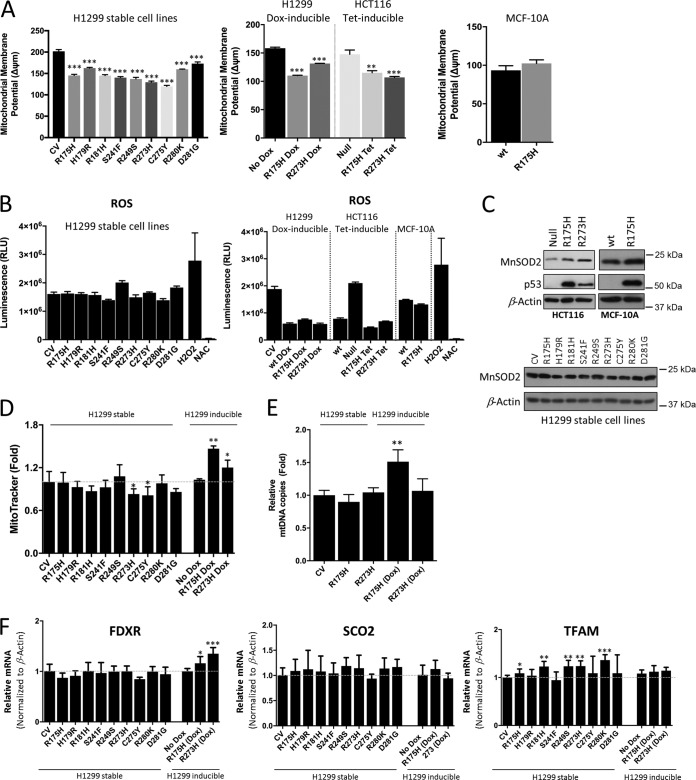
To understand the underlying mechanisms that might explain the observed changes in the mitochondrial metabolism between the cell lines tested, we analyzed parameters that may affect the efficiency of mitochondrial respiration, including mitochondrial membrane potential, reactive oxygen species (ROS) production, mitochondrial content, and biogenesis. Analysis of mitochondrial membrane potential revealed that mutant p53-expressing stable H1299 cells displayed lower membrane potential than the CV cells (Fig. 6A). However, since the inducible mutant p53 H1299 cell lines also displayed the same response of decreased mitochondrial membrane potential (Fig. 6A), we concluded that changes in the mitochondrial membrane potential do not correlate with the metabolic differences observed between the stable and inducible mutant expression in cancer cells.
FIG 6.
Impact of mutant p53 on mitochondrial functions. (A) Mitochondrial membrane potential measured by TMRE in the indicated cell lines. (B) Measurements of intracellular ROS production in the indicated cell lines. H2O2 and NAC were used as positive and negative controls, respectively. (C) Western blots showing the expression of MnSOD2 and p53 in the inducible p53R175H and p53R273H HCT116 and wt and stable p53R175H MCF-10A cells, as well as in the stable mutant p53-expressing H1299 cells. β-Actin was used as a loading control. (D) Mitochondrial content measurement using MitoTracker. (E) Mitochondrial DNA copy number analysis in the inducible and stable mutant p53-expressing H1299 cell lines. (F) qPCR data showing the expression of TFAM (mitochondrial biogenesis), SCO2 (complex I-IV assembly), and FDXR (initiates electron transfer within the electron transport chain) in the indicated mutant p53-expressing H1299 cells. Data are expressed as means ± SD (n = 3). Statistical significance is shown above the level for the control. *, P < 0.05; **, P < 0.001; ***, P < 0.0001; two-tailed Student's t test.
Mitochondria are the main sites for intracellular ROS formation (26). Excessive ROS production can damage mitochondrial DNA and oxidize proteins and lipids, limiting the efficiency of the electron transport chain (22). In line with this, when induced, the p53R175H and p53R273H mutants, which displayed enhanced mitochondrial respiration, exhibited significantly lower cellular ROS (Fig. 6B). However, this was not observed in the stable mutant H1299 cells. Furthermore, given that cells displaying high mitochondrial activity often counterbalance ROS by an enhanced antioxidant capacity (27, 28), we could show an increase in the expression of the p53-inducible central mitochondrial antioxidant MnSOD2 in the p53R175H and p53R273H mutants compared to null p53 cells (Fig. 6C). The MnSOD2 level was unchanged in the stable mutant H1299 cells as well as in mutant p53R175H MCF-10A cells (Fig. 6C). This suggests a more efficient electron transport machinery in the inducible mutants than in stable cells. Hence, the inducible p53R175H and p53R273H mutants may have a more efficient electron transport machinery, protected from ROS by increased levels of antioxidants like MnSOD2, and therefore display increased mitochondrial respiration (Fig. 2C, OCR).
A further explanation for enhanced oxidative phosphorylation in the inducible mutant p53 cells could be that they contain more mitochondria or have an increase in their biogenesis. To interrogate this, we measured mitochondrial content (Fig. 6D), mitochondrial DNA copy number (Fig. 6E), and the expression of key factors that affect mitochondrial respiration at different layers (Fig. 6F), including TFAM (mitochondrial biogenesis), SCO2 (complex I-IV assembly), and FDXR (initiates electron transfer within the electron transport chain). Combined, these analyses reveal that while both inducible p53R175H and p53R273H mutants displayed a slight increase in mitochondrial content, as shown by MitoTracker staining (Fig. 6D), only the inducible p53R175H displayed higher mitochondrial DNA copy number, whereas there were no major differences in mitochondrial biogenesis (TFAM) or complex assembly (SCO2) between the stable and inducible H1299 cells. In addition, FDXR (initiates electron transfer within the electron transport chain), which contributes to electron transfer within the respiratory complexes, seems to be enhanced in the inducible p53R175H and p53R273H mutants (Fig. 6F). Thus, a more efficient electron transfer ultimately can lead to a more efficient electron transport chain.
DISCUSSION
Altered metabolism is a characteristic of cancer and a key contributor to tumor development (29). Thus, metabolic characterization is essential for better understanding of the phenotypic changes induced by tumorigenic mutations. Studies have already reported the effect of mutant p53 regulation in cancer cell metabolism compared to that of wt p53 (30). However, descriptions of potential metabolic differences of mutant p53 protein variants have been relatively limited. In this study, we have used assays to metabolically characterize individual mutant p53 proteins by measuring glycolytic and mitochondrial metabolic functions in various cell types that express different p53 mutants. We found that mutant p53 proteins have differential phenotypic impacts on these central metabolic pathways, and p53 mutants differentially regulate metabolism in different cellular contexts.
Metabolic regulation by mutant p53 proteins has been linked to the promotion of glycolysis through distinct mechanisms as a novel gain of function (12). While our data support this notion in general, not all mutants tested in this study displayed increased glycolysis in cultured cells. Furthermore, our data demonstrate that even the same amino acid substitutions at the same position in the p53 protein can have dramatically different phenotypic effects in terms of glycolysis. For example, in line with previous findings, while the hot-spot p53R175H and p53R273H mutants (both of which are common tumor-derived mutants) are able to confer enhanced glycolysis when induced, overexpressed (in p53-null cells), or endogenously expressed in cancer cells, when expressed in nontumorigenic human cell lines these mutants displayed dramatically different properties, which is highlighted by the lessened glycolytic phenotypes. Thus, our findings demonstrate that the metabolic activity of mutant p53-harboring cancer cells, either tumor-derived p53 mutants or by induced expression in cancer cells, differ in normal cells with ectopic expression of the same mutant p53 variant. This suggests that p53 mutants do not acquire and possess the same metabolic GOF and do not display biological effects equally in all cultured cells. Correspondingly, while our study supports the generality of the mutant p53 metabolic GOF in cancer cells, our data further indicate that different aspects of metabolism are regulated in different cell or tissue types (cancer versus normal), and some metabolic GOFs of mutant p53 might be cancer cell type specific and not a general response.
Another major metabolic difference we observed between the mutant p53 proteins was their effect on mitochondrial energy metabolism. When stably expressed, most p53 mutant cells showed suppressed basal mitochondrial respiration. To some extent, these mutants have lost wt p53 tumor-suppressive activity, and since wt p53 enhances mitochondrial oxidative phosphorylation and mutant p53 proteins often regulate the same cellular biological processes with opposite effects, it is easy to think that these metabolic changes are not truly neomorphic or GOF activities. We should also keep in mind that constitutive activation of oncogenes may change the adaptive capabilities; thus, it is likely that in stable mutant cancer cells the GOF effect of some mutant p53 variants is masked by compensatory mechanisms, which may have drastically suppressed p53's effect on metabolism. Accordingly, similar to our observations in cancer cells with endogenous mutants, when the same p53 mutant protein was transiently expressed, OxPhos was significantly stimulated by the p53R175H and p53R273H mutants, indicating that the OxPhos effects were not caused by loss of wt activity. This shows that some mutant p53 proteins not only positively stimulate glycolytic functions but also significantly enhance the OxPhos in cancer cells, which is in line with experimental studies of tissue samples from patients with the Li-Fraumeni syndrome who carry germline mutations in TP53 and in a mouse model of the syndrome that support this finding of increased mitochondrial function in vivo (31, 32). This is in line with the idea that beyond elevated glycolytic rates, most cancer cell lines similarly have the capacity to sustain high fuel oxidation and ATP production in mitochondria (21). However, not all mutants stimulated OxPhos. This indicated that while glycolytic changes opposed by mutant p53 can be kept consistent in cancer cells, the mitochondrial energy metabolism pathways may be affected by the type of mutant p53. Thus, based on the mutant's mitochondrial effects, it is tempting to speculate that there is a metabolic heterogeneity in terms of mutant p53 metabolic GOF with mutants with increased (p53R175H and p53R273H) or decreased (p53H179R, p53R181H, and p53S241F) OxPhos in cancer cells.
Furthermore, our findings demonstrate that the hot-spot mutants display increased metabolic activity in cancer cells, but when they are induced in normal cells they can display contrasting effects. This may be caused by another GOF activity of mutant p53, such as promoting EMT. Although cancer cells may display other oncoproteins that can cooperate with mutant p53 proteins, our data show that even the exact same p53 mutants can have opposing impacts on metabolism in different cell models. Mutant p53 proteins are likely to drive changes through distinct metabolic pathways, and some of their GOF effects might be cancer cell specific and not a general response seen in nontransformed cells.
Altogether, our findings highlight the importance of recognizing that not all p53 mutants are alike. Keeping in mind that analysis of human breast cancer patients to evaluate the prognostic impact of TP53 mutations in breast cancer showed that specific missense mutations actually correlated with a significantly poorer prognosis compared with those of other missense mutations (39), it is of great importance to recognize that not all p53 mutations are equivalent. This distinction is important to avoid making generalizations about mutant p53 proteins, not only as a conceptual distinction but also for clinical implications, as multiple studies involving cancer patients revealed that different p53 mutations were associated with diverse prognostic values. Targeting cancer bioenergetics is a promising and rapidly rising direction for anticancer therapy. The differential effect on metabolism of mutant p53 proteins might provide prognostic information and a foundation for development of more effective targeted clinical implications.
MATERIALS AND METHODS
Cell lines and culture conditions.
The human non-small-cell lung carcinoma H1299 cell lines, colon cancer HCT116 cell lines, and breast cancer MDA-MB-231 cell lines were cultured as previously described (33) in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS) (Gibco), 100 U/ml penicillin and 100 U/ml streptomycin (Sigma-Aldrich), and 2% (wt/vol) glutamine (Sigma-Aldrich). The normal lung fibroblast WI-38 cell line was cultured in the same medium as that mentioned above but supplemented with 1% MEM nonessential amino acid solution (Sigma-Aldrich). The normal epithelial breast MCF-10A cells were cultured in DMEM–F-12 (Gibco) supplemented with 5% (vol/vol) horse serum (Gibco), 20 ng/ml epidermal growth factor (EGF) (Peprotech), 0.5 mg/ml hydrocortisone (Sigma-Aldrich), 100 ng/ml cholera toxin (Sigma-Aldrich), 10 μg/ml insulin (Sigma-Aldrich), and 100 U/ml penicillin and 100 U/ml streptomycin (Sigma-Aldrich). The ovarian cancer ES-2 cell line was cultured in RPMI 1640 medium (Sigma-Aldrich) supplemented with 10% (vol/vol) heat-inactivated FBS (Gibco), 100 U/ml penicillin and 100 U/ml streptomycin (Sigma-Aldrich), and 2% (wt/vol) glutamine (Sigma-Aldrich). Cells were grown in a humidified 5% CO2 atmosphere at 37°C and maintained in logarithmic growth phase.
siRNAs and plasmids.
All siRNAs (scramble control, or nontargeting [NT], and p53) were purchased from Shanghai GenePharma Co., Ltd. Forty nanomolar siRNA was used, and the transfection was performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. The expression level of p53 protein was monitored 48 to 72 h posttransfection by Western blotting. The following oligonucleotides (5′ to 3′) were used for siRNA experiments: NT siRNA sense, UUCUCCGAACGUGUCACGUTT; antisense, ACGUGACACGUUCGGAGAATT; siRNA p53 #1 sense, CCCGGACGAUAUUGAACAATT; siRNA p53 #1 antisense, UUGUUCAAUAUCGUCCGGGTT; siRNA p53 #2 sense, CCACCAUCCACUACAACUATT; siRNA p53 #2 antisense, UAGUUGUAGUGGAUGGUGGTT.
Transient transfections were performed with Lipofectamine 2000 reagent (Invitrogen) or ViaFect reagent (Promega), as recommended by the manufacturer, and protein expression was allowed for 24 to 48 h before experimental procedures. An empty vector, pcDNA3.1 (Invitrogen), was used as a negative control. p53 mutant vectors exhibited the following single-amino-acid substitutions: R175H, R181H, R249S, and R273H. For the generation of stable H1299, cells were transfected with an empty vector (PCB6+) or constructs expressing the single-amino-acid substitutions R175H, H179R, R181H, S241F, R249S, R273H, C275Y, R280K, and D281G, with Effectene reagent (Qiagen). Cells were selected by medium containing 600 μg/ml G418. p53 mutants were created using site-directed mutagenesis, as previously described for p53175H or p53273H (5) or using the following oligonucleotides: 179H-R Fw, GTGAGGCGCTGCCCCCACCgTGAGCGCTGCTCAGATAGC; 179H-R Rev, GCTATCTGAGCAGCGCTCACGGTGGGGGCAGCGCCTCAC; 181R-H Fw, GCGCTGCCCCCACCATGAGCaCTGCTCAGATAGCGATGG; 181R-H Rev, CCATCGCTATCTGAGCAGTGCTCATGGTGGGGGCAGCGC; 249R-S Fw, GCATGGGCGGCATGAACCGGAGtCCCATCCTCACCATCATC; 249R-S Rev, GATGATGGTGAGGATGGGACTCCGGTTCATGCCGCCCATGC; 241S-F Fw, TACATGTGTAACAGTTtCTGCATGGGCGGCATGAACCGG; 241S-F Rev, CCGGTTCATGCCGCCCATGCAGAAACTGTTACACATGTA; 248 R-Q Fw, TGCATGGGCGGCATGAACCaGAGGCCCATCCTCACCATC; 248R-Q Rev, GATGGTGAGGATGGGCCTCTGGTTCATGCCGCCCATGCA; 280 R-T Fw, GTTTGTGCCTGTCCTGGGAcAGACCGGCGCACAGAGGAAG; 280 R-T Rev, CTTCCTCTGTGCGCCGGTCTGTCCCAGGACAGGCACAAAC; 275 C-Y Fw, GAGGTGCGTGTTTGTGCCTaTCCTGGGAGAGACCGGCGC; 275 C-Y Rev, GCGCCGGTCTCTCCCAGGATAGGCACAAACACGCACCTC; 281 D-E Fw, GTGCCTGTCCTGGGAGAGAGCGGCGCACAGAGGAA; 281 D-E Rev, TTCCTCTGTGCGCCGCTCTCTCCCAGGACAGGCAC.
SNAI1 (NM_005985) human cDNA was used (OriGene). MCF-10A cells stably expressing an empty vector (PCB6+) or constructs expressing p53175H (in PCB6+; 72R polymorphism) with Effectene reagent (Qiagen) were created as described in reference 5. HCT116 cell lines express the inducible pcDNA4-Flag-HA-p53(R175H) or pcDNA4-Flag-HA-p53(R273H) vector.
Cell expansion and cell death assay.
Cell proliferation was assessed by monitoring the number of cells over time through cell counting using a hemocytometer. A total of 10,000 cells/well were seeded in 6-well plates in triplicates on day one, and cell numbers were monitored every 24 h for up to 3 days. Cell number/survival was measured using the ToxiLight bioassay (Thermo Fisher Scientific) or the CellTiter-Glo luminescent cell viability assay according to the manufacturer's instructions.
Measurements of mitochondrial parameters.
ROS measurements were performed using the ROS-Glo H2O2 assay (Promega) according to the manufacturer's instructions. Luminescence was determined with a GloMax multiluminometer. The average relative light units (RLU) and standard deviations from triplicate samples (SD) were calculated. Hydrogen peroxide (400 μM) and 5 mM N-acetylcysteine (NAC) were used as positive and negative controls, respectively. For mitochondrial content of the cells, cells were stained with MitoTracker green (Molecular Probes) according to the manufacturer's instructions for 30 min using 200 nM, washed, and analyzed using a plate reader (excitation wavelength, 490 nm; emission wavelength, 516 nm). Mitochondrial membrane potential was determined using tetramethylrhodamine ethyl ester (TMRE), and cells were stained with 25 nM TMRE, subsequently washed with phosphate-buffered saline (PBS), and analyzed using a plate reader (excitation wavelength, 549 nm; emission wavelength, 575 nm).
Determination of glycolytic function.
The extracellular acidification rate (ECAR) was determined as previously described (34–36) and measured in real time using the XFp extracellular flux analyzer (Seahorse Bioscience), which allows the measurement of all relevant glycolysis parameters (glycolysis, glycolytic capacity, and glycolytic reserve). Cells were seeded in an XFp cell culture miniplate at a concentration of 8,000 to 20,000 cells per well in their specified growth media. The next day, cells were washed twice with 180 μl of XF base medium (Seahorse Bioscience), followed by a 15-min incubation in the same medium. Cells were analyzed using the Seahorse XFp glycolysis stress test kit (Seahorse Bioscience) by following the manufacturer's instructions. Briefly, after baseline measurement, the following injections were made: 10 mM glucose, 1 μM oligomycin, and 50 mM 2-deoxyglucose.
Mitochondrial respiration measurements.
The oxygen consumption rate (OCR) was determined, as previously described (34, 35), in real time using the XFp extracellular flux analyzer (Seahorse Bioscience), which allows the measurement of many relevant parameters (total respiration, basal mitochondrial respiration, ATP-linked respiration, proton leak, maximal respiration, and spare respiratory capacity). Cells were seeded in an XFp cell culture miniplate at a concentration of 8,000 to 20,000 cells per well in their specified growth media. The next day, cells were washed twice with 180 μl of XF base medium (Seahorse Bioscience), followed by a 15-min incubation in the same medium. Cells were analyzed using the Seahorse XFp Cell Mito stress test kit according to the manufacturer's instructions. Briefly, after baseline measurement, the following injections were made: 1 μM oligomycin, 0.5 μM carbonyl cyanide-4 (trifluoromethoxy) phenylhydrazone (FCCP), and 0.5 μM rotenone/antimycin A.
Glucose uptake assay.
Glucose uptake was performed as previously described (37), with some modifications. Cells were seeded at a density of 1 × 105 cells/well overnight and then starved for 5 h in DMEM without glucose, glutamine, pyruvate, and FBS for 5 h. The medium then was replaced with 1 ml of medium per well containing 10 μM 2-deoxyglycose (Sigma-Aldrich) and 1 μCi (20 nM) [3H]2-deoxyglucose (American Radiolabeled Chemicals, Inc.). Cells were incubated with labeled 2-deoxyglucose for 20, 60, and 120 min, and then they were rinsed three times with chilled PBS and lysed in ice-cold 0.4N NaOH. The lysates were mixed with Emulsifier-Safe scintillation liquid (Perkin-Elmer), and 2-deoxyglucose uptake was quantified using a scintillation counter. Data were determined as counts per minute (cpm) and normalized by total amount of protein galactose.
RNA extraction and cDNA synthesis.
Total RNA was extracted using the RNAqueous phenol-free total RNA isolation kit (Ambion, Life Technologies) according to the manufacturer's instructions. One microgram of RNA was used to synthesize cDNA with the IScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Approximately 200 ng of the cDNA sample was analyzed by quantitative PCR (qPCR) using Maxima qPCR SYBR green master mix (Thermo Fisher Scientific, Waltham, MA) and amplified using the 7500 real-time PCR system (Applied Biosystems, Foster City, CA). The ΔΔCT method (where CT is threshold cycle) was used to determine the relative mRNAs expression after normalization with β-actin as a reference gene. The following oligonucleotides were used: total PGC-1a Fw, 5′-CAGCCTCTTTGCCCAGATCTT-3′; total PGC-1a Rv 5′-TCACTGCACCACTTGAGTCCAC-3′; TFAM Fw, 5′-ATTCCAAGAAGCTAAGGGTG-3′; TFAM Rv, 5′-CAGAGTCAGACAGATTTTTCC-3′; SCO2 Fw, 5′-TGGGTGCTGATGTACTTTGGC-3′; SCO2 Rv, 5′-ACAGTCTTGGGTGGAAGTCCTG-3′; FDXR Fw, 5′-GTCCGTCTGAGTGGGACTTT-3′; FDXR Rv, 5′-GAGGAGAGACGCTGGAAGAG-3′; β-actin Fw, 5′-GCAAGCAGGAGTATGACGAG-3′; β-actin Rv, 5′-CAAATAAAGCCATGCCAATC-3′.
Mitochondrial/genomic DNA assay.
DNA was extracted with the QIAamp DNA minikit (Qiagen) according to the manufacturer's instructions. Two nanograms of total DNA input was loaded into a plate with prealiquoted PCR primer pairs from NovaQUANT human mitochondrial and nuclear DNA ratio kit (Merck) to perform reverse transcription-PCR (RT-PCR) using Maxima qPCR SYBR green master mix (Thermo Fisher Scientific, Waltham, MA). The relative copy number of each gene was estimated based on differences in the CT (ΔCT) values of four targets (ND1, BECN1, ND6, and NEB).
Antibodies.
The following primary antibodies were used: anti-p53 (DO-1) (sc-126; Santa Cruz Biotechnology), anti-E-cadherin (24E10) (3195; Cell Signaling Technology), anti-vimentin (D21H3) (5741; Cell Signaling Technology), anti-β-actin (ACTBD11B7) (sc-81178; Santa Cruz Biotechnology), anti-LC3 (L7543; Sigma-Aldrich), and anti-p21 (F-5) (sc-6246; Santa Cruz Biotechnology). Secondary antibodies used for Western blotting were Pierce goat anti-mouse IgG (H+L), peroxidase conjugated, and Pierce goat anti-rabbit IgG (H+L), peroxidase conjugated (both from Thermo Scientific). All antibodies were diluted according to the manufacturer's recommendations.
Gel electrophoresis and Western blotting.
Cells were harvested, washed in PBS (Sigma-Aldrich), and lysed for 30 min on ice in radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with complete mini protease inhibitor cocktail (Roche Diagnostics). The lysates were centrifuged at 10,000 rpm for 10 min to separate the insoluble material, followed by a measurement of protein concentration using the bicinchoninic acid (BCA) assay (Pierce). Equal amounts of protein from each sample were mixed with Laemmli loading buffer, boiled for 5 min, and subjected to SDS-PAGE. Ten to 15% acryl amide gels were used to separate proteins of different molecular masses. Membranes were blocked for 1 h with 5% nonfat milk in PBS at room temperature and subsequently probed with the primary antibody of interest overnight at 4°C. Blots were revealed by enhanced chemiluminescence (ECL) (Bio-Rad). The level of mutant p53 proteins stably expressed in H1299 cells was quantified by Image J (version 1.42q).
Statistical analyses.
All graphs were designed using GraphPad Prism 6. Quantitative data, represented by bar graphs from at least three independent experiments in triplicates, are shown and expressed as means ± SD followed by analysis of variance (38). Some results are presented as fold change over control levels (dimethyl sulfoxide). In all experiments, treatment groups (mutants) were compared with a control group (CV) unless otherwise shown.
ACKNOWLEDGMENTS
We are grateful for the kind gifts from Xinbin Chen (inducible mutant p53 HCT116 cell lines). We are grateful to Per Hydbring for valuable comments on the manuscript.
This work was supported by grants from the Swedish Research Council (VR), The Swedish Society for Medical Research (SSMF), The Malin and Lennart Philipson Foundation, The Ragnar Söderberg Stiftelse, Jeanssons Stiftelse, O. E. och Edla Johanssons Vetenskapliga Stiftelse, Åke Wiberg Stiftelse, Magnus Bergwalls Stiftelse, Stiftelsen Lars Hiertas Minne, and Karolinska Institute. A.G.-C. was supported by a Ramón y Cajal fellowship (RYC-2014-15792) from the Spanish Ministerio de Economía y Competitividad.
M.E., G.A., A.T.O., A.L.Q., D.S., and A.G.-C. performed the experiments. All authors analyzed and interpreted the data. P.A.M. and M.P.I. created stable H1299 mutant p53 and MCF-10A cells and contributed intellectually to the project. H.V.-N. wrote the manuscript, with input from all authors. H.V.-N. and E.N. designed and financed the study. H.V.-N. supervised the research.
We have no conflicts of interest to declare.
REFERENCES
- 1.Kupryjanczyk J, Thor AD, Beauchamp R, Merritt V, Edgerton SM, Bell DA, Yandell DW. 1993. p53 gene mutations and protein accumulation in human ovarian cancer. Proc Natl Acad Sci U S A 90:4961–4965. doi: 10.1073/pnas.90.11.4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119:847–860. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. 1994. Tumor spectrum analysis in p53-mutant mice. Curr Biol 4:1–7. [DOI] [PubMed] [Google Scholar]
- 4.Oren M, Rotter V. 2010. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller PA, Caswell PT, Doyle B, Iwanicki MP, Tan EH, Karim S, Lukashchuk N, Gillespie DA, Ludwig RL, Gosselin P, Cromer A, Brugge JS, Sansom OJ, Norman JC, Vousden KH. 2009. Mutant p53 drives invasion by promoting integrin recycling. Cell 139:1327–1341. doi: 10.1016/j.cell.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 6.Muller PA, Vousden KH. 2014. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koppenol WH, Bounds PL, Dang CV. 2011. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 8.DeBerardinis RJ, Chandel NS. 2016. Fundamentals of cancer metabolism. Sci Adv 2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Lunt SY, Vander Heiden MG. 2011. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol 27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 11.Kollareddy M, Dimitrova E, Vallabhaneni KC, Chan A, Le T, Chauhan KM, Carrero ZI, Ramakrishnan G, Watabe K, Haupt Y, Haupt S, Pochampally R, Boss GR, Romero DG, Radu CG, Martinez LA. 2015. Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat Commun 6:7389. doi: 10.1038/ncomms8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C, Liu J, Liang Y, Wu R, Zhao Y, Hong X, Lin M, Yu H, Liu L, Levine AJ, Hu W, Feng Z. 2013. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun 4:2935. doi: 10.1038/ncomms3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou G, Wang J, Zhao M, Xie TX, Tanaka N, Sano D, Patel AA, Ward AM, Sandulache VC, Jasser SA, Skinner HD, Fitzgerald AL, Osman AA, Wei Y, Xia X, Songyang Z, Mills GB, Hung MC, Caulin C, Liang J, Myers JN. 2014. Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol Cell 54:960–974. doi: 10.1016/j.molcel.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavez-Perez VA, Strasberg-Rieber M, Rieber M. 2011. Metabolic utilization of exogenous pyruvate by mutant p53 (R175H) human melanoma cells promotes survival under glucose depletion. Cancer Biol Ther 12:647–656. doi: 10.4161/cbt.12.7.16566. [DOI] [PubMed] [Google Scholar]
- 15.Kalo E, Kogan-Sakin I, Solomon H, Bar-Nathan E, Shay M, Shetzer Y, Dekel E, Goldfinger N, Buganim Y, Stambolsky P, Goldstein I, Madar S, Rotter V. 2012. Mutant p53R273H attenuates the expression of phase 2 detoxifying enzymes and promotes the survival of cells with high levels of reactive oxygen species. J Cell Sci 125:5578–5586. doi: 10.1242/jcs.106815. [DOI] [PubMed] [Google Scholar]
- 16.Freed-Pastor WA, Mizuno H, Zhao X, Langerod A, Moon SH, Rodriguez-Barrueco R, Barsotti A, Chicas A, Li W, Polotskaia A, Bissell MJ, Osborne TF, Tian B, Lowe SW, Silva JM, Borresen-Dale AL, Levine AJ, Bargonetti J, Prives C. 2012. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hanel W, Marchenko N, Xu S, Yu SX, Weng W, Moll U. 2013. Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ 20:898–909. doi: 10.1038/cdd.2013.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, El-Naggar AK, Lozano G. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 19.Liu DP, Song H, Xu Y. 2010. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene 29:949–956. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song H, Hollstein M, Xu Y. 2007. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol 9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 21.Ahn CS, Metallo CM. 2015. Mitochondria as biosynthetic factories for cancer proliferation. Cancer Metab 3:1. doi: 10.1186/s40170-015-0128-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vakifahmetoglu-Norberg H, Ouchida AT, Norberg E. 2017. The role of mitochondria in metabolism and cell death. Biochem Biophys Res Commun 482:426–431. doi: 10.1016/j.bbrc.2016.11.088. [DOI] [PubMed] [Google Scholar]
- 23.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, Liu M, Chen CT, Yu D, Hung MC. 2011. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol 13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim NH, Kim HS, Li XY, Lee I, Choi HS, Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, Rowe RG, Lee S, Maher CA, Weiss SJ, Yook JI. 2011. A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol 195:417–433. doi: 10.1083/jcb.201103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. 2011. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med 208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norberg E, Orrenius S, Zhivotovsky B. 2010. Mitochondrial regulation of cell death: processing of apoptosis-inducing factor (AIF). Biochem Biophys Res Commun 396:95–100. doi: 10.1016/j.bbrc.2010.02.163. [DOI] [PubMed] [Google Scholar]
- 27.Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, Zhou F, Green MR, Chen L, Monti S, Marto JA, Shipp MA, Danial NN. 2012. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. 2004. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer Res 64:2350–2356. doi: 10.1158/0008-5472.CAN-2287-2. [DOI] [PubMed] [Google Scholar]
- 29.Pavlova NN, Thompson CB. 2016. The emerging hallmarks of cancer metabolism. Cell Metab 23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Zhang C, Hu W, Feng Z. 2015. Tumor suppressor p53 and its mutants in cancer metabolism. Cancer Lett 356:197–203. doi: 10.1016/j.canlet.2013.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang PY, Ma W, Park JY, Celi FS, Arena R, Choi JW, Ali QA, Tripodi DJ, Zhuang J, Lago CU, Strong LC, Talagala SL, Balaban RS, Kang JG, Hwang PM. 2013. Increased oxidative metabolism in the Li-Fraumeni syndrome. N Engl J Med 368:1027–1032. doi: 10.1056/NEJMoa1214091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang PY, Li J, Walcott FL, Kang JG, Starost MF, Talagala SL, Zhuang J, Park JH, Huffstutler RD, Bryla CM, Mai PL, Pollak M, Annunziata CM, Savage SA, Fojo AT, Hwang PM. 2017. Inhibiting mitochondrial respiration prevents cancer in a mouse model of Li-Fraumeni syndrome. J Clin Investig 127:132–136. doi: 10.1172/JCI88668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vakifahmetoglu-Norberg H, Kim M, Xia HG, Iwanicki MP, Ofengeim D, Coloff JL, Pan L, Ince TA, Kroemer G, Brugge JS, Yuan J. 2013. Chaperone-mediated autophagy degrades mutant p53. Genes Dev 27:1718–1730. doi: 10.1101/gad.220897.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia HG, Najafov A, Geng J, Galan-Acosta L, Han X, Guo Y, Shan B, Zhang Y, Norberg E, Zhang T, Pan L, Liu J, Coloff JL, Ofengeim D, Zhu H, Wu K, Cai Y, Yates JR, Zhu Z, Yuan J, Vakifahmetoglu-Norberg H. 2015. Degradation of HK2 by chaperone-mediated autophagy promotes metabolic catastrophe and cell death. J Cell Biol 210:705–716. doi: 10.1083/jcb.201503044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu R, Galan-Acosta L, Norberg E. 2015. Glucose metabolism provide distinct prosurvival benefits to nonsmall cell lung carcinomas. Biochem Biophys Res Commun 460:572–577. doi: 10.1016/j.bbrc.2015.03.071. [DOI] [PubMed] [Google Scholar]
- 36.Zhang B, Zheng A, Hydbring P, Ambroise G, Ouchida AT, Goiny M, Vakifahmetoglu-Norberg H, Norberg E. 2017. PHGDH defines a metabolic subtype in lung adenocarcinomas with poor prognosis. Cell Rep 19:2289–2303. doi: 10.1016/j.celrep.2017.05.067. [DOI] [PubMed] [Google Scholar]
- 37.Peng X, Gimenez-Cassina A, Petrus P, Conrad M, Ryden M, Arner ES. 2016. Thioredoxin reductase 1 suppresses adipocyte differentiation and insulin responsiveness. Sci Rep 6:28080. doi: 10.1038/srep28080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vaux DL. 2012. Research methods: know when your numbers are significant. Nature 492:180–181. [DOI] [PubMed] [Google Scholar]
- 39.Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J, Theillet C, Rodriguez C, Lidereau R, Bieche I, Varley J, Bignon Y, Uhrhammer N, Winqvist R, Jukkola-Vuorinen A, Niederacher D, Kato S, Ishioka C, Hainaut P, Borresen-Dale AL. 2006. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12:1157–1167. [DOI] [PubMed] [Google Scholar]