ABSTRACT
It is well known that many bacteria can survive in a growth-arrested state for long periods of time, on the order of months or even years, without forming dormant structures like spores or cysts. How is such longevity possible? What is the molecular basis of such longevity? Here we used the Gram-negative phototrophic alphaproteobacterium Rhodopseudomonas palustris to identify molecular determinants of bacterial longevity. R. palustris maintained viability for over a month after growth arrest due to nutrient depletion when it was provided with light as a source of energy. In transposon sequencing (Tn-seq) experiments, we identified 117 genes that were required for long-term viability of nongrowing R. palustris cells. Genes in this longevity gene set are annotated to play roles in a number of cellular processes, including DNA repair, tRNA modification, and the fidelity of protein synthesis. These genes are critically important only when cells are not growing. Three genes annotated to affect translation or posttranslational modifications were validated as bona fide longevity genes by mutagenesis and complementation experiments. These genes and others in the longevity gene set are broadly conserved in bacteria. This raises the possibility that it will be possible to define a core set of longevity genes common to many bacterial species.
KEYWORDS: Rhodopseudomonas palustris, Tn-seq, growth arrest, starvation, stationary phase
IMPORTANCE
Bacteria in nature and during infections often exist in a nongrowing quiescent state. However, it has been difficult to define experimentally the molecular characteristics of this crucial element of the bacterial life cycle because bacteria that are not growing tend to die under laboratory conditions. Here we present and validate the phototrophic bacterium Rhodopseudomonas palustris as a model system for identification of genes required for the longevity of nongrowing bacteria. Growth-arrested R. palustris maintained almost full viability for weeks using light as an energy source. Such cells were subjected to large-scale mutagenesis to identify genes required for this striking longevity trait. The results define conserved determinants of survival under nongrowing conditions and create a foundation for more extensive studies to elucidate general molecular mechanisms of bacterial longevity.
INTRODUCTION
Microbiological research has focused primarily on understanding the physiology and dynamics of bacterial cells and populations during rapid growth. However, this rapid growth state is likely highly unusual in nature. It is known that many bacteria, including many pathogens, enter a growth-arrested state in which they remain viable for considerable periods of time (1–3). Good examples of this are Mycobacterium tuberculosis (4–6) and biofilm-forming organisms such as Pseudomonas aeruginosa (7–9). Growth arrest can be caused by nutrient or energy limitation or by other factors. While the physiology of fast-growing bacteria is well characterized, relatively little is understood about how cells maintain life under nongrowing conditions.
The two main established models of nongrowing bacteria are the dormant spore state, exemplified by Gram-positive Bacillus spp. and Clostridium spp. (10–12), and starvation-induced growth arrest in Gram-negative bacteria (6, 13). Nongrowing cultures of Gram-negative bacteria are typically established by supplying a resource such as carbon, nitrogen, or oxygen in a growth-limiting amount (6, 13). Studies of such cultures have informed us about general strategies that bacteria use to persist in a growth-arrested state, including scavenging nutrients, using endogenous storage compounds like glycogen, or degrading cellular components such as lipids to maintain viability (13, 14). In addition, some species have been shown to survive growth arrest by rerouting metabolic pathways to make energy generation more efficient (15–17). Many of these strategies are deployed to meet the imperative of maintaining an electrochemical gradient across the cell membrane, a requirement for viability (18). Growth arrest due to any sort of nutrient limitation will eventually impact the ability of a bacterium to generate the energy needed for this critical function. Because of the importance of having an energy supply to maintain viability, many species of bacteria exhibit a large decrease in viability following an initial period of growth arrest. This has been well documented for Escherichia coli, for which a small fraction of a growth-arrested population will survive after an initial die-off in a cycle of growth and death by feeding on the nutrients released from dead cells (1, 13, 14, 19). This failure of most cells to survive long-term starvation as a homogenous population may explain why E. coli has not served as a good model for discovery of bacterial longevity genes.
Several features of the physiology of the phototrophic alphaproteobacterium Rhodopseudomonas palustris suggest that it represents an ideal model for characterizing the molecular basis of bacterial survival in a nongrowing state. This bacterium can be put into a state of growth arrest due to nutrient limitation but not energy limitation (15, 20, 21). This would help remove from consideration genes that are important for cells to marshal resources to maintain an electrochemical gradient or to generate ATP to maintain viability. R. palustris generates ATP by cyclic photophosphorylation, and no portion of the carbon source that it is provided for biomass generation is used for energy (Fig. 1). Thus, R. palustris can enter into a growth-arrested state by carbon or nitrogen restriction, and cells remain viable for months when incubated in light (15, 20, 21). Here, we used transposon sequencing (Tn-seq) to identify genes required for growth-arrested R. palustris cells to remain viable. In complementary experiments, we analyzed the transcriptome of growth-arrested R. palustris. Over 100 genes were identified as essential for nongrowing R. palustris cells to maintain viability. Many members of this longevity gene set are conserved among bacteria, suggesting that common molecular mechanisms may support the longevity of diverse bacterial species.
FIG 1 .
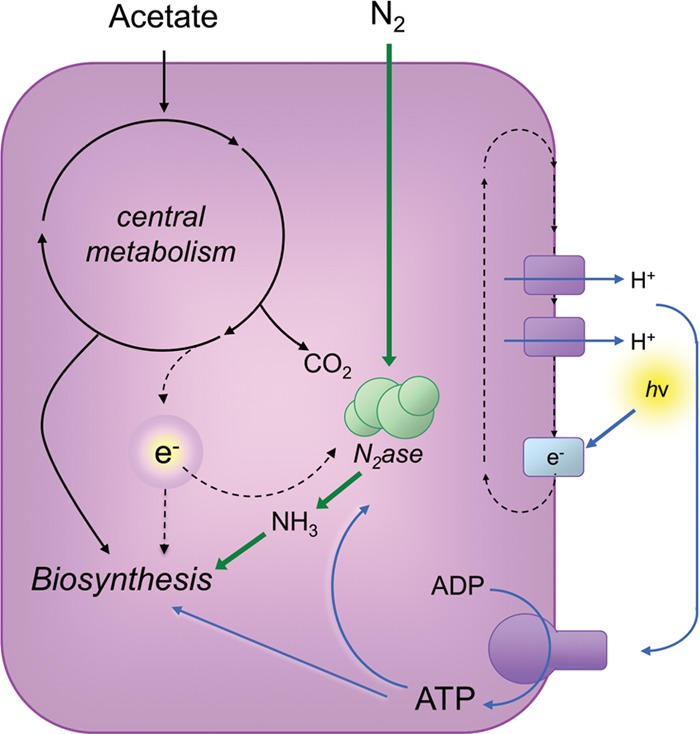
R. palustris metabolism. Cells for the experiments described in this article were grown anaerobically with light as the sole energy source and with acetate as the sole carbon source. Nitrogen gas (N2) or ammonia (NH3) was supplied as the nitrogen source. Under these conditions, a proton gradient generated by cyclic photophosphorylation provides energy for ATP synthesis. Acetate is used to support the production of cell biomass, and some electrons are diverted from acetate metabolism to nitrogenase (N2ase) for use in fixing N2 to NH3. In addition, some CO2 is released during acetate metabolism. In this work, cells were supplied with a growth-limiting amount of acetate, such that cells entered a growth-arrested state when all available acetate was exhausted. When in this state, R. palustris cells continue to generate ATP from light.
RESULTS
Establishment of growth-arrested R. palustris cell populations.
We grew cultures of R. palustris strain CGA009 anaerobically under conditions in which carbon but not energy was limited (Fig. 1). Under these conditions, nitrogen (N2 gas) and other resources, including energy from light, were provided in excess, while the carbon source, acetate, was provided in a growth-limiting amount. When acetate exhaustion resulted in growth arrest, we removed a subset of the cultures from light and incubated them in the dark for the duration of the experiment. The cultures that remained exposed to light remained fully viable over a period of 35 days, whereas the viability of dark cultures dropped by 4 orders of magnitude by approximately 10 days after the cells entered a growth-arrested state (Fig. 2). The results suggest that ATP generation through cyclic photophosphorylation is needed for long-term viability. However, these are population-level observations, and it is possible that the growth-arrested cultures that were incubated in light were continuously dying and growing in a cycle of balanced growth rather than maintaining continuous viability. To address this possibility, we examined growth-arrested cells by phase microscopy and after live/dead staining with propidium iodide. Propidium iodide permeates the membranes of dead, but not living, cells to stain DNA. Growth-arrested cells incubated in light were phase dense and had the same morphology as live cells, and we saw very few dead cells as assessed by propidium iodide staining. Cells incubated in the dark also tended to maintain their morphology, but many of the cells were less phase dense. Also, qualitative observations revealed that most dark-incubated cells stained red with propidium iodide after 10 days of growth arrest.
FIG 2 .
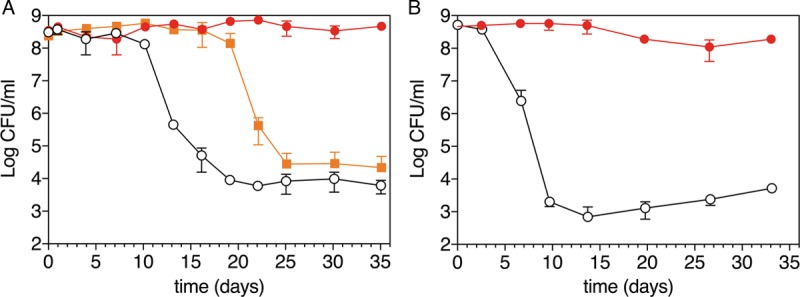
Viability of R. palustris after growth arrest. Growth arrest was initiated by two methods: (A) carbon limitation with N2 gas provided in excess as a nitrogen source or (B) carbon limitation with ammonium provided in excess as the nitrogen source. Growth-arrested cells were incubated anaerobically under constant light (red closed circles) or dark (black open circles) conditions. In panel A, cells were also exposed to 24 h of light following growth arrest and then placed in the dark for the remainder of the experiments (orange closed squares). Viability was measured by plating cells and counting CFU. The averages from two independent experiments are shown for each panel.
To investigate how the source of nitrogen impacts growth-arrested R. palustris survival, we carried out the same set of experiments with R. palustris cultures with 10 mM ammonia added. This concentration of ammonia represses the use of N2 gas as a nitrogen source by R. palustris (22). Growth-arrested cells provided with light as an energy source and ammonia as the nitrogen source (Fig. 2B) lost viability compared to cells that were given N2 gas as a nitrogen source (Fig. 2A); however, there was at most a 1-order-of-magnitude difference in the drop in viability between the two conditions over the 35-day experiments.
The transcriptional response of R. palustris to growth arrest.
We observed that if we incubated growth-arrested cultures in light for 24 h beyond the initiation of growth arrest and then covered the tubes with foil and incubated them in the dark, the cultures maintained stable viable counts for approximately 10 days longer than cultures that were immediately placed in the dark (Fig. 2A). We wondered if the ability of R. palustris to survive in the dark for an extended period after a relatively brief light treatment might depend on the initiation of a light-dependent protective transcriptional response that occurs when growth ceases.
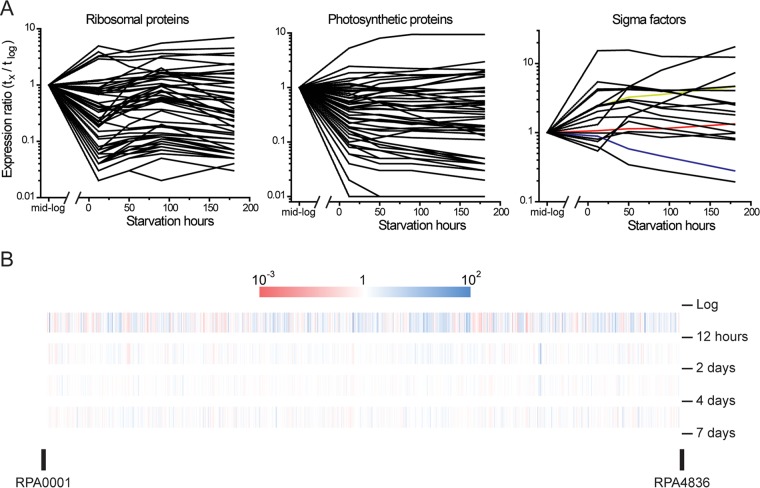
To address this, we extracted RNA from actively growing cells and from cells that were incubated in light for periods from 12 h to 180 h (0.5 to 7.5 days) after growth arrest and carried out transcriptome sequencing (RNA-seq). We found that cells responded rapidly to carbon starvation with most of the transcriptional responses occurring during the first 12 h after growth arrest. As seen in a previous study in which we looked at nitrogen starvation (15), genes encoding major functions that are normally highly expressed and needed for growth were turned down in expression (see Table S1 in the supplemental material). Included in this category are photosynthesis genes (RPA1505 to -1554), genes for ribosomal proteins, and genes encoding ATP synthase (Fig. 3A). There are 4,836 genes in the R. palustris genome (23). By the seventh day after growth arrest, the expression levels of 257 genes were more than 10-fold lower than actively growing cells. Another 120 genes increased more than 10-fold in expression during this interval (Fig. 3B; Table S1). These include genes for many of the 19 RNA polymerase sigma factor genes carried by R. palustris (Fig. 3A) and genes involved in nutrient acquisition, including many of the genes required for anaerobic benzoate and 4-hydroxybenzoate degradation (RPA0653 to -0673) and the phn gene cluster for phosphonate uptake and utilization (RPA0689 to -0702), for example (Table S1).
FIG 3 .
Transcriptional response of R. palustris to growth arrest. Cells were subjected to growth arrest by carbon limitation as described in the legend to Fig. 1A. (A) Expression levels of genes encoding ribosomal proteins, photosynthesis proteins, and sigma factors over time following growth arrest are shown. All numbers are normalized to each gene’s expression level during logarithmic growth. R. palustris encodes 19 RNA polymerase sigma factors. The expression levels of rpoH (RPA0367), rpoD (RPA1288), and rpoN (RPA0050) are shown in green, red, and blue, respectively. (B) Heat map of genome-wide expression level changes between the intervals. The numbers on the right are the intervals at which the samples were taken.
Transcriptome of R. palustris CGA009 at intervals following growth arrest due to carbon starvation. Download TABLE S1, XLSX file, 1.5 MB (1.5MB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tn-seq results. Fitness of R. palustris CGA009 genes at intervals after growth arrest due to carbon starvation. Download TABLE S2, CSV file, 1.8 MB (1.9MB, csv) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tn-seq screen to identify genes required for longevity.
Although many genes were elevated in expression following growth arrest, the approach of RNA-seq does not tell us if any of these genes are required for longevity. To identify longevity genes, we used Tn-seq, a high-throughput approach that takes advantage of next-generation sequencing to assess fitness defects and benefits conferred by transposon (Tn) insertion mutants within a saturating mutant pool. For this, we used a pool of R. palustris strain CGA009 transposon mutants described previously (24, 25) that consists of over 175,000 unique transposon insertion mutants. We inoculated the mutant pool into mineral medium containing a growth-limiting amount of acetate under continuous light illumination (as in Fig. 2A) and analyzed the effect of growth cessation on the makeup of the mutant pool. We sampled the cultures at the initiation of growth arrest and at 3, 7, 14, and 30 days post-growth arrest. We conducted two independent biological replicates of the experiment and processed the samples separately as described in Materials and Methods and previously (24). For each time point, genes with statistically significant (P < 0.05) read values below the mean in both replicates were noted. Cells with insertions in these genes, called reduced insertion fitness genes, were those present at a low frequency in the overall Tn pool. Genes with statistically significant values above the mean in both replicates were also identified. Mutants with Tn insertions in these genes were deemed to have increased insertion fitness. Genes with reduced and increased insertion fitness were recorded at the earliest experimental time point at which they met these criteria (see Tables S3 and S4 in the supplemental material). Genes that were previously classified as essential (P < 0.001) or that had reduced insertion fitness (P < 0.05) during aerobic growth in rich medium, or during anaerobic phototrophic growth in minimal medium, were excluded from our analysis (24, 25). Using this approach, we identified 117 genes that had reduced insertion fitness after 3 to 14 days of growth arrest (Table S3), comprising a longevity gene set.
R. palustris longevity genes: genes with reduced fitness 3 to 14 days after growth arrest. Download TABLE S3, XLSX file, 0.1 MB (70.2KB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes with increased insertion fitness 3, 7, 14, and 30 days after growth arrest. Download TABLE S4, XLSX file, 0.1 MB (70.8KB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Categories of longevity genes.
More than one-third of the longevity genes (43 out of 117) were hypothetical genes or genes that did not fit into a known functional category (Table 1; see Table S5 in the supplemental material) (23, 26, 27). We also identified 34 genes belonging to the general category of biosynthesis and metabolism in the longevity gene set. These genes are involved in diverse functions, including nitrogen fixation, cofactor synthesis, and lipid synthesis. We expect that some of the genes in this category would differ depending on the conditions used prior to growth arrest and on the growth-limiting nutrient used to establish growth arrest. For example, we would not expect nitrogen fixation genes in the longevity gene set were we to carry out the Tn-seq experiment with cells grown with ammonia rather than nitrogen gas as the nitrogen source. A functional group that was overrepresented in the longevity gene set was transporters and transport-related proteins. Fifteen longevity genes were found in this category, 11 of which were ATP-binding cassette (ABC) transporters, a class of transporters that has high affinity for its substrates. This was more than 12% of the longevity gene set, twice overrepresented compared to 293 transporters or transporter-related genes among all R. palustris loci (6%). As would be expected from extensive studies with E. coli, Bacillus subtilis, and other bacteria, the RelA-SpoT homologue (RSH) RPA2693 (annotated as relA) was found in our longevity gene set. RSH proteins are bifunctional enzymes that can synthesize and degrade the stringent response messenger (p)ppGpp (28, 29). (p)ppGpp regulates bacterial transcription globally in a number of ways in response to nutrient downshift (30–32). In addition, 10 genes classified as being involved in translation or posttranslational modification were identified as essential for R. palustris longevity in our Tn-seq screen. We verified the longevity functions of three of these genes in follow-up experiments described below.
TABLE 1 .
Functional categories of genes essential for R. palustris longevity
| Functional group | No. of genes (n = 117) | Functions of selected longevity genes |
|---|---|---|
| Biosynthesis and metabolism | 34 | Nitrogenase cofactor biosynthesis protein (NifB), alkaline phosphatase, hydrogenase/expression formation proteins, acetyl-CoA hydrolase |
| Hypothetical or unknown functional category | 43 | |
| DNA replication and repair | 2 | Smr protein/MutS2; Holliday junction resolvase-like protein |
| Cell wall/membrane biogenesis | 2 | Alanine racemase; UDP-N-acetylmuramoyl-l-alanyl-d-glutamate synthetase (MurD) |
| Signal transduction | 3 | Two-component sensor kinase (FixL), sensor signal transduction kinase of unknown function |
| Transcription | 8 | GTP-pyrophosphokinase (RelA), cold shock DNA-binding domain-containing protein (CspA) |
| Translation and posttranslational modification | 10 | tRNA uridine 5-carboxymethylaminomethyl modification protein (GidA), GTP-dependent nucleic acid-binding protein (EngD) |
| Transport | 15 | Twin-arginine translocation protein (TatB), high-affinity leucine-isoleucine-valine transport system periplasmic binding protein |
Functional categories of R. palustris longevity genes. Download TABLE S5, XLSX file, 0.04 MB (40.3KB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tn mutants that restore hydrogen gas uptake in R. palustris strain CGA009 were positively selected after growth arrest.
In addition to identifying genes with reduced insertion fitness during growth arrest, we also identified 46 genes with increased insertion fitness following growth arrest (Table S4). Among the most readily explicable of these was a regulatory gene that indirectly represses expression of the NiFe uptake hydrogenase encoded by R. palustris. We previously determined that a deletion mutation of RPA0980 (hoxJ) can override a regulatory mutation in our parent strain’s hydrogen uptake system such that cells acquire the ability to use H2 gas as an electron donor for carbon dioxide fixation (33). In our Tn-seq experiments, RPA0980::T24 insertion mutants outperformed all other mutants in the pool, and at 30 days post-growth arrest, they comprised 89 and 94% of the normalized reads in the two replicates, respectively. It is likely that these mutants used CO2 that accumulated in cultures when they oxidized acetate prior to growth arrest and H2 produced during nitrogen fixation to form biomass (see Fig. S1 in the supplemental material). At some point after day 14 of growth arrest, RPA0980::T24 insertion mutants outcompeted other Tn mutants sufficiently to start taking over the cultures.
Probable metabolic route to growth for RPA0980::T24 insertion mutants that were positively selected when cells were starved for carbon (acetate) under nitrogen-fixing conditions. These mutants allow a functional uptake hydrogenase enzyme to be expressed. CO2 produced from acetate prior to growth arrest and H2 produced along with ammonia as a product of nitrogenase can be used by the Calvin cycle to generate cell biomass. Download FIG S1, TIF file, 2.1 MB (2.2MB, tif) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Validation of selected longevity genes.
In any Tn-seq experiment, it is important to validate mutants that appear to be important for fitness under the condition being interrogated. Toward this end, we made in-frame deletions in the five candidate genes listed in Table 2. The deletion mutants were subjected to growth arrest due to (i) acetate depletion in a medium that included ammonium as the nitrogen source and (ii) nitrogen depletion imposed by replacing the N2 gas in the headspace of culture tubes with argon. In both situations, growth was arrested when cells reached an OD660 of between 0.45 and 0.55 and longevity was assessed by recovery of colony-forming units (CFU). Three out of the five strains that we tested, the ΔRPA0446, ΔRPA2580, and ΔRPA2698 deletion mutants, showed compromised longevities after growth arrest due to either carbon starvation or nitrogen starvation (Fig. 4A; see Fig. S2 in the supplemental material). These genes, which are homologous to genes that have been studied in E. coli, are annotated as encoding a 16S rRNA endonuclease, a protein l-isoaspartate O-methyltransferase, and a RAS-like GTPase involved in ribosome assembly (34–38). The ΔRPA2115 mutant, defective in cyanate hydratase (39), decreased slightly in viability after growth arrest due to carbon source limitation but maintained wild-type levels of viability under nitrogen starvation conditions. The fifth strain, the ΔRPA2183 mutant, which lacks a gene annotated to encode a DsbA thiolsulfide oxidoreductase (40), did not have a longevity phenotype (Fig. S2). The ΔRPA0446 and ΔRPA2580 mutants had wild-type growth rates on acetate when either ammonium or nitrogen gas was supplied as a nitrogen source. The ΔRPA2698 strain had a slightly lower growth rate than the wild type when ammonia was the nitrogen source (Table 3).
TABLE 2 .
Validation of candidate longevity genes
| Gene mutated | Annotation | Longevity phenotype |
|---|---|---|
| RPA0446 | Possible rRNA maturation endonuclease | Defective in survival after growth arrest |
| RPA2115 | Cyanate hydratase (CynS) | Slight defect in survival after carbon depletion-induced growth arrest |
| RPA2183 | DsbA thiol disulfide oxidoreductases | Wild-type longevity |
| RPA2580 | Protein l-isoaspartate O-methyltransferase | Defective in survival after growth arrest |
| RPA2698 | Possible GTP-binding protein involved in rRNA maturation | Defective in survival after growth arrest |
FIG 4 .
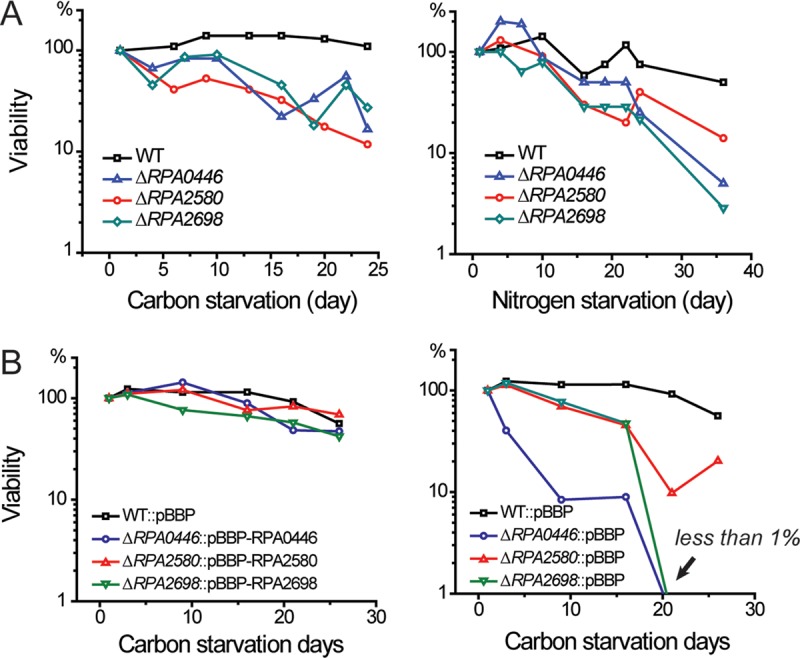
Longevity phenotypes of R. palustris wild-type and ΔRPA0446, ΔRPA0446, and ΔRPA2698 deletion mutant strains. Cells were subjected to growth arrest due to either carbon starvation or nitrogen starvation according to conditions described in the text, and cell viabilities were determined by measuring CFU. In both cases, argon was present as the headspace gas in culture tubes. (A) We show one representative set of survival curves for mutants. These experiments were repeated three times, and the full sets of survival curves are shown in Fig. S2. (B) Each mutant longevity phenotype was complemented by expressing the corresponding gene in trans. The empty vector control is also shown.
TABLE 3 .
Generation times of longevity mutants
| Strain | Generation time (h) for: |
|
|---|---|---|
| Nitrogen-fixing growtha | Non-nitrogen-fixing growthb | |
| WT | 10.7 ± 1.0 | 6.4 ± 0.2 |
| ΔRPA0446 mutant | 9.8 ± 0.7 | 7.2 ± 0.9 |
| ΔRPA2580 mutant | 12.1 ± 1.5 | 6.3 ± 0.1 |
| ΔRPA2698 mutant | 10.0 ± 1.0 | 8.1 ± 0.3 |
Nitrogen-fixing growth in NFM plus 20 mM sodium acetate.
Non-nitrogen-fixing growth in PM plus 10 mM sodium acetate.
Longevity phenotypes of R. palustris mutants compared to wild type. The results of all longevity trials are shown. The same set of survival curves for wild-type cells is shown in each panel. Download FIG S2, DOCX file, 0.4 MB (411.3KB, docx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The longevity defects of the ΔRPA0446, ΔRPA2580, and ΔRPA2698 strains were complemented by expressing the corresponding genes in trans (Fig. 4B). Gentamicin was included to maintain the complementing vector. Interestingly, ΔRPA0446 and ΔRPA2698 mutants carrying empty vector had a more severe longevity defect than mutant cells that did not carry the vector. Whereas 10 to 50% of mutant cells were viable after more than 20 days of carbon starvation in light (Fig. 4A; Fig. S2), less than 1% of either strain survived the same period of growth arrest when carrying an empty vector (Fig. 4B). One possibility is the mutant strains were hypersensitive to residual gentamicin present in the medium that was not inactivated by the gentamicin resistance gene carried on the plasmid.
DISCUSSION
Basta et al. have described genes essential for viability of P. aeruginosa during energy-limited growth arrest (41). As might be expected, a subset of these genes are important for energy generation. This article also summarizes work suggesting that energy limitation is a common cause of growth arrest for heterotrophic bacteria in natural environments. Some bacteria can circumvent this problem of energy-limited growth by generating energy from light. It has been suggested that marine bacteria use proteorhodopsin to capture light energy, providing a survival advantage in nutrient-scarce oceans (42–44). Indeed, expression of proteorhodopsin in the heterologous host Shewanella oneidensis helped it to survive significantly longer during stationary phase (45). We have demonstrated here and previously that R. palustris remains viable for weeks when not growing as long it is incubated in light, a condition that drives ATP synthesis (Fig. 1). This allowed us to apply the approach of Tn-seq in R. palustris to identify longevity genes. The annotations of these genes suggest that cells are metabolically active when in a growth-arrested state. Under these conditions, R. palustris is likely required to maintain protein synthesis and to repair and maintain chromosome integrity. The inventory of longevity genes that we compiled also suggests that it is important for cells to maintain transport functions—possibly to recapture nutrients that are leaked from growth-arrested cells that have degraded ribosomes and proteins that are no longer needed to support rapid growth.
In carrying out Tn-seq experiments to identify longevity genes, we made the unexpected finding that hoxJ mutants that were in our Tn pool dominated the growth-arrested cultures by day 30 of carbon starvation, presumably because such mutants could grow on CO2 and H2 that accumulated in the growth-arrested cultures due to acetate oxidation and nitrogen fixation (Fig. S1). Because of this, we analyzed our data for genes that had reduced insertion fitness in cultures at 3, 7, and 14 days following growth arrest, before the hoxJ mutants started taking over. To increase the stringency of our analysis, we validated selected longevity genes by testing their longevity phenotypes in cultures subjected to growth arrest by two methods: (i) acetate depletion in a medium that included ammonia rather than N2 gas as a nitrogen source and (ii) nitrogen depletion in a medium that had acetate in excess.
Global changes in transcription occurred in R. palustris in the first 12 h after growth arrest. We hypothesize that these changes helped cells adapt to growth arrest based on our findings that 24 h of exposure to light following growth arrest delayed the bulk cell death that occurred when cells were subsequently deprived of energy by incubation in the dark. In view of this, we were surprised to find that there was very little overlap between genes that were increased in expression following growth arrest and the set of genes that we identified as longevity genes by Tn-seq. There are several possible interpretations for this result. One is that genes that are more highly expressed following growth arrest may provide cells with an advantage, but the advantage is not sufficiently large for us to pick up the genes as essential for longevity using the rather stringent criteria of the Tn-seq screen. Also, the Tn-seq approach cannot identify genes that are functionally redundant with another gene. R. palustris encodes, for example, 16 extracytoplasmic function (ECF) RNA polymerase sigma factors, many of which were increased in expression following growth arrest, but none of these showed up in our longevity gene set (Fig. 3). This could be due to overlapping functions of these auxiliary sigma factors. The stationary-phase sigma factor rpoS, identified as a longevity gene in P. aeruginosa (41) and in E. coli (13, 46), is not present in R. palustris or other alphaproteobacteria.
We expect that our longevity gene set will vary depending on the growth-limiting nutrient used to establish growth arrest. Some transporter genes likely fall into this category. At the same time, some R. palustris genes—particularly those involved in core cellular functions—are likely to be important for longevity regardless of the specific condition used. In our analysis, longevity genes involved in DNA repair, tRNA modification, and the fidelity of protein synthesis were critically important after, but not before, growth arrest. This suggests that there is less margin for error for some mutants to remain viable when not growing compared to when they are actively growing. Longevity genes in this category tend to be conserved among bacteria.
The verified longevity gene RPA2580 is annotated as coding for a protein l-isoaspartyl O-methyltransferase (PIMT). PIMTs are found in most bacteria, but they have mainly been studied in animals and plants, where they function to repair aged proteins that can become nonfunctional due to the spontaneous deamidation and isomerization of aspartates and asparagines (36, 47–49). These modifications, which are one of the most common types of protein damage, result in the accumulation of isoapartate residues, which cause protein damage by generating kinks in polypeptide chains. One study in E. coli showed that mutants lacking the pcm gene encoding PIMT had decreased survival when challenged with an environmental stress, such as heat, salt, or hydrogen peroxide (50). Although it is tempting to conclude that RPA2580 functions in the repair of damaged proteins, this R. palustris protein is missing critical residues shown to be important for interaction with PIMT substrates. Thus, it will be important to verify the activity of this gene product in in vitro assays. Of the other two longevity genes that we verified, RPA0446 is a homologue of E. coli ybeY, coding for an RNase that influences the maturation of rRNAs and plays key roles in ribosome quality control (37, 38). The other gene, RPA2698, is annotated as a Ras-like small GTPase gene. Its product’s homologue, Era, is essential for E. coli growth and provides a checkpoint for cell cycle regulation (51) and ribosome assembly (34, 35). Depletion of Era from E. coli leads to morphological abnormalities (52, 53). It is interesting that the R. palustris ΔRPA0446 and ΔRPA2698 mutants had no obvious growth defects, when their homologues are essential for or strongly affect E. coli growth (38, 53). This could be due to differences in the host species, or it may be that the R. palustris proteins have different functions.
The results of these studies encourage us to believe that R. palustris is a good model microbe for use in identifying genes that are important for the longevity of diverse bacterial species. The most logical approach is to focus now on verifying and characterizing genes that are conserved in other bacteria and on testing the effects of mutations in homologous genes on the viabilities of other bacteria that are in a state of growth arrest. More than one-third of the R. palustris longevity gene set is comprised of genes of unknown functions. Many of these have homologues in other bacteria, providing interesting territory to explore in the future.
MATERIALS AND METHODS
Bacterial strains, growth, and incubation conditions.
R. palustris CGA009, a hupV mutant lacking uptake hydrogenase activity, was used as the wild-type strain for this study (33). Cells were grown under anaerobic conditions in light as described previously (12). Two previously described defined media were used: PM, which contains ammonium sulfate (10 mM) as a nitrogen source, and a nitrogen-free medium, NFM, which is identical to PM but lacks ammonium sulfate (54). Both media had N2 gas as a headspace in sealed cultures, except where noted. For the Tn-seq screen, 50-ml volumes of medium were placed into stoppered 170-ml serum bottles and the cultures were stirred slowly. In-frame deletion mutants of strain CGA009 were constructed as described previously (54). For genetic complementation experiments, R. palustris genes were cloned into plasmid pBBPgdh, with their native ribosome binding site maintained (55). The plasmids were mobilized into R. palustris strain CGA009 by conjugation with to E. coli S17-1 as described previously (54). Media were supplemented with kanamycin (100 μg/ml), gentamicin (100 μg/ml), and/or chloramphenicol (10 μg/ml) as indicated.
For carbon starvation experiments, R. palustris was grown anaerobically in light with constant stirring in either NFM or PM, as indicated, supplemented with 10 mM sodium acetate. The exhaustion of acetate (OD660 of ~0.55) induced carbon starvation; this point was designated 0 h, or t0. For nitrogen starvation experiments, R. palustris was grown anaerobically in light in NFM supplemented with 20 mM sodium acetate. At mid-log phase (OD660 of ~0.45), the headspace was exchanged with argon gas to induce nitrogen starvation; this point was designated 0 h poststarvation, or t0. For Tn-seq experiments, it was necessary to remove dead cells through an outgrowth step, as DNA sequencing techniques cannot differentiate between DNA from living versus dead cells. Samples of growth-arrested cells were plated on CA agar (PM supplemented with 02% Casamino Acids and 0.5% yeast extract) containing sodium acetate (20 mM) medium, and plates were incubated for 5 to 7 generations under aerobic conditions in ambient light. Nutrient agar (Difco) or CA agar plates were used to determine viable CFU, as indicated.
RNA-seq analysis of carbon-starved cells.
R. palustris was grown under nitrogen-fixing conditions with acetate as a limiting carbon source as described above. Control samples were removed during log phase (OD660 of 0.25). Samples were taken 12, 50, 90, and 180 h post-growth arrest. RNA was extracted from 5-ml samples using the RNeasy minikit (Qiagen), and samples were prepared for RNA-seq using previously described methods and the R. palustris-specific primer set v2 (15). RNA samples were sequenced, and the standard Illumina preprocessing pipeline was used for initial default filtering (Illumina, Inc.). Filtered reads were analyzed using Xpression, an RNA-seq bioinformatics pipeline developed by our laboratory (56).
Genetic analysis of R. palustris carbon starvation by Tn-seq.
The CGA009 T24 mutant library described previously (24) was grown under nitrogen-fixing carbon-limited conditions as described above; an aliquot (2 × 109 CFU) of the library was used as a culture inoculum, and the medium was supplemented with kanamycin. Cultures were sampled at t0, and at 3, 7, 14, and 30 days post-growth arrest to monitor OD660 and viable CFU and to prepare Tn-seq samples. For Tn-seq samples, culture aliquots (0.5 ml) were removed and immediately plated on large (15-cm) petri plates containing CA agar supplemented with 100 µg/ml kanamycin and 10 µg/ml chloramphenicol. Plates were incubated for 3 days at 30°C under aerobic conditions with low ambient light. Cells were scraped up and suspended in 1 ml sterile PM. An aliquot (10 µl) of the cell suspension was used to determine viable CFU. The remaining cell suspension was pelleted by centrifugation and stored at −80°C. Genomic DNA was extracted and Tn-seq samples were prepared for Illumina sequencing and sequenced as described previously (24), using the Tn circle method (57). Sequencing data were analyzed using a Tn-seq bioinformatics pipeline previously described by our laboratory (24).
Reduced- and increased-fitness knockouts were determined for each starvation time point as described previously (24). Briefly, for each biological replicate, the first 5% and last 10% read counts for each gene were discarded, and the central 5 to 90% read counts per gene were totaled and normalized to gene length (where 1 kb = 1 gene), resulting in reads per kilobase (RpK) values for each gene. Log2 RpK values were analyzed via a histogram and fit to a normal distribution using the nonlinear least-squares fit (Prism, GraphPad Software, Inc.), and the mean and standard deviation of the distribution were calculated. For each time point, genes with log2 RpK values falling outside a 95% confidence interval (P < 0.05 by two-tailed test) and below the mean in both biological replicates were considered genes with reduced insertion fitness, while those genes satisfying the same criteria above the mean were considered genes with increased insertion fitness. Genes with values not statistically significant from the mean were considered genes with neutral insertion fitness.
ACKNOWLEDGMENTS
We thank Mallory Beightol for technical assistance and the UW Genome Sciences Department for the use of the Covaris instrument, Samuel Miller for the use of the Qubit instrument, and Claudine Baraquet, Jose Mendiola, Amy Schaefer, Kathryn Fixen, Yanning Zheng, and Harley Pyles for helpful discussions.
The RNA-seq and Tn-seq data were obtained with funding from National Science Foundation grant MCB-11457304 to C.S.H. and C.M. Data analysis and all other experiments reported were supported by grant W911NF-15-1-0150 to C.S.H. from the US Army Research Office.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/mBio.01170-17.
Citation Pechter KB, Yin L, Oda Y, Gallagher L, Yang J, Manoil C, Harwood CS. 2017. Molecular basis of bacterial longevity. mBio 8:e01726-17. https://doi.org/10.1128/mBio.01726-17.
REFERENCES
- 1.Finkel SE. 2006. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol 4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 2.Bergkessel M, Basta DW, Newman DK. 2016. The physiology of growth arrest: uniting molecular and environmental microbiology. Nat Rev Microbiol 14:549–562. doi: 10.1038/nrmicro.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matin A, Auger EA, Blum PH, Schultz JE. 1989. Genetic basis of starvation survival in nondifferentiating bacteria. Annu Rev Microbiol 43:293–316. doi: 10.1146/annurev.mi.43.100189.001453. [DOI] [PubMed] [Google Scholar]
- 4.Watanabe S, Zimmermann M, Goodwin MB, Sauer U, Barry CE III, Boshoff HI. 2011. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog 7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao SP, Alonso S, Rand L, Dick T, Pethe K. 2008. The protonmotive force is required for maintaining ATP homeostasis and viability of hypoxic, nonreplicating Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 105:11945–11950. doi: 10.1073/pnas.0711697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parish T. 2003. Starvation survival response of Mycobacterium tuberculosis. J Bacteriol 185:6702–6706. doi: 10.1128/JB.185.22.6702-6706.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akiyama T, Williamson KS, Schaefer R, Pratt S, Chang CB, Franklin MJ. 2017. Resuscitation of Pseudomonas aeruginosa from dormancy requires hibernation promoting factor (PA4463) for ribosome preservation. Proc Natl Acad Sci U S A 114:3204–3209. doi: 10.1073/pnas.1700695114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Babin BM, Bergkessel M, Sweredoski MJ, Moradian A, Hess S, Newman DK, Tirrell DA. 2016. SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 113:E597–E605. doi: 10.1073/pnas.1514412113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kopf SH, Sessions AL, Cowley ES, Reyes C, Van Sambeek L, Hu Y, Orphan VJ, Kato R, Newman DK. 2016. Trace incorporation of heavy water reveals slow and heterogeneous pathogen growth rates in cystic fibrosis sputum. Proc Natl Acad Sci U S A 113:E110–E116. doi: 10.1073/pnas.1512057112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sunde EP, Setlow P, Hederstedt L, Halle B. 2009. The physical state of water in bacterial spores. Proc Natl Acad Sci U S A 106:19334–19339. doi: 10.1073/pnas.0908712106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cowan AE, Olivastro EM, Koppel DE, Loshon CA, Setlow B, Setlow P. 2004. Lipids in the inner membrane of dormant spores of Bacillus species are largely immobile. Proc Natl Acad Sci U S A 101:7733–7738. doi: 10.1073/pnas.0306859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higgins D, Dworkin J. 2012. Recent progress in Bacillus subtilis sporulation. FEMS Microbiol Rev 36:131–148. doi: 10.1111/j.1574-6976.2011.00310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zambrano MM, Siegele DA, Almirón M, Tormo A, Kolter R. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 14.Zinser ER, Kolter R. 2004. Escherichia coli evolution during stationary phase. Res Microbiol 155:328–336. doi: 10.1016/j.resmic.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 15.McKinlay JB, Oda Y, Rühl M, Posto AL, Sauer U, Harwood CS. 2014. Non-growing Rhodopseudomonas palustris increases the hydrogen gas yield from acetate by shifting from the glyoxylate shunt to the tricarboxylic acid cycle. J Biol Chem 289:1960–1970. doi: 10.1074/jbc.M113.527515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glasser NR, Kern SE, Newman DK. 2014. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton motive force. Mol Microbiol 92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gengenbacher M, Rao SP, Pethe K, Dick T. 2010. Nutrient-starved, non-replicating Mycobacterium tuberculosis requires respiration, ATP synthase and isocitrate lyase for maintenance of ATP homeostasis and viability. Microbiology 156:81–87. doi: 10.1099/mic.0.033084-0. [DOI] [PubMed] [Google Scholar]
- 19.Takano S, Pawlowska BJ, Gudelj I, Yomo T, Tsuru S. 2017. Density-dependent recycling promotes the long-term survival of bacterial populations during periods of starvation. mBio 8:e02336-16. doi: 10.1128/mBio.02336-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Odaa Y, Slagmana S, Meijerb WG, Forneya LJ, Gottschala JC. 2000. Influence of growth rate and starvation on fluorescent in situ hybridization of Rhodopseudomonas palustris. FEMS Microbiol Ecol 32:205–213. doi: 10.1111/j.1574-6941.2000.tb00713.x. [DOI] [PubMed] [Google Scholar]
- 21.Gosse JL, Engel BJ, Hui JC, Harwood CS, Flickinger MC. 2010. Progress toward a biomimetic leaf: 4,000 h of hydrogen production by coating-stabilized nongrowing photosynthetic Rhodopseudomonas palustris. Biotechnol Prog 26:907–918. doi: 10.1002/btpr.406. [DOI] [PubMed] [Google Scholar]
- 22.Oda Y, Samanta SK, Rey FE, Wu L, Liu X, Yan T, Zhou J, Harwood CS. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J Bacteriol 187:7784–7794. doi: 10.1128/JB.187.22.7784-7794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larimer FW, Chain P, Hauser L, Lamerdin J, Malfatti S, Do L, Land ML, Pelletier DA, Beatty JT, Lang AS, Tabita FR, Gibson JL, Hanson TE, Bobst C, Torres JL, Peres C, Harrison FH, Gibson J, Harwood CS. 2004. Complete genome sequence of the metabolically versatile photosynthetic bacterium Rhodopseudomonas palustris. Nat Biotechnol 22:55–61. doi: 10.1038/nbt923. [DOI] [PubMed] [Google Scholar]
- 24.Pechter KB, Gallagher L, Pyles H, Manoil CS, Harwood CS. 2015. Essential genome of the metabolically versatile alphaproteobacterium Rhodopseudomonas palustris. J Bacteriol 198:867–876. doi: 10.1128/JB.00771-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang J, Yin L, Lessner FH, Nakayasu ES, Payne SH, Fixen KR, Gallagher L, Harwood CS. 2017. Genes essential for phototrophic growth by a purple alphaproteobacterium. Environ Microbiol 19:3567–3578. doi: 10.1111/1462-2920.13852. [DOI] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martínez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, Latendresse M, Muñiz-Rascado L, Ong Q, Paley S, Schröder I, Shearer AG, Subhraveti P, Travers M, Weerasinghe D, Weiss V, Collado-Vides J, Gunsalus RP, Paulsen I, Karp PD. 2013. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 41:D605–D612. doi: 10.1093/nar/gks1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 29.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paul BJ, Berkmen MB, Gourse RL. 2005. DksA potentiates direct activation of amino acid promoters by ppGpp. Proc Natl Acad Sci U S A 102:7823–7828. doi: 10.1073/pnas.0501170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul BJ, Barker MM, Ross W, Schneider DA, Webb C, Foster JW, Gourse RL. 2004. DksA: a critical component of the transcription initiation machinery that potentiates the regulation of rRNA promoters by ppGpp and the initiating NTP. Cell 118:311–322. doi: 10.1016/j.cell.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Kriel A, Bittner AN, Kim SH, Liu K, Tehranchi AK, Zou WY, Rendon S, Chen R, Tu BP, Wang JD. 2012. Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol Cell 48:231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rey FE, Oda Y, Harwood CS. 2006. Regulation of uptake hydrogenase and effects of hydrogen utilization on gene expression in Rhodopseudomonas palustris. J Bacteriol 188:6143–6152. doi: 10.1128/JB.00381-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, Ji X. 2009. Structure of ERA in complex with the 3′ end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci U S A 106:14843–14848. doi: 10.1073/pnas.0904032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Britton RA. 2009. Role of GTPases in bacterial ribosome assembly. Annu Rev Microbiol 63:155–176. doi: 10.1146/annurev.micro.091208.073225. [DOI] [PubMed] [Google Scholar]
- 36.Fu JC, Ding L, Clarke S. 1991. Purification, gene cloning, and sequence analysis of an l-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J Biol Chem 266:14562–14572. [PubMed] [Google Scholar]
- 37.Jacob AI, Köhrer C, Davies BW, RajBhandary UL, Walker GC. 2013. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell 49:427–438. doi: 10.1016/j.molcel.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. 2010. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol 78:506–518. doi: 10.1111/j.1365-2958.2010.07351.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamblin AF, Fuchs JA. 1994. Functional analysis of the Escherichia coli K-12 cyn operon transcriptional regulation. J Bacteriol 176:6613–6622. doi: 10.1128/jb.176.21.6613-6622.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guilhot C, Jander G, Martin NL, Beckwith J. 1995. Evidence that the pathway of disulfide bond formation in Escherichia coli involves interactions between the cysteines of DsbB and DsbA. Proc Natl Acad Sci U S A 92:9895–9899. doi: 10.1073/pnas.92.21.9895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Basta DW, Bergkessel M, Newman DK. 2017. Identification of fitness determinants during energy-limited growth arrest in Pseudomonas aeruginosa. mBio 8:e01170-17. doi: 10.1128/mBio.01170-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gómez-Consarnau L, Akram N, Lindell K, Pedersen A, Neutze R, Milton DL, González JM, Pinhassi J. 2010. Proteorhodopsin phototrophy promotes survival of marine bacteria during starvation. PLoS Biol 8:e1000358. doi: 10.1371/journal.pbio.1000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- 44.Gómez-Consarnau L, González JM, Coll-Lladó M, Gourdon P, Pascher T, Neutze R, Pedrós-Alió C, Pinhassi J. 2007. Light stimulates growth of proteorhodopsin-containing marine Flavobacteria. Nature 445:210–213. doi: 10.1038/nature05381. [DOI] [PubMed] [Google Scholar]
- 45.Johnson ET, Baron DB, Naranjo B, Bond DR, Schmidt-Dannert C, Gralnick JA. 2010. Enhancement of survival and electricity production in an engineered bacterium by light-driven proton pumping. Appl Environ Microbiol 76:4123–4129. doi: 10.1128/AEM.02425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. doi: 10.1146/annurev-micro-090110-102946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lanthier J, Desrosiers RR. 2004. Protein l-isoaspartyl methyltransferase repairs abnormal aspartyl residues accumulated in vivo in type-I collagen and restores cell migration. Exp Cell Res 293:96–105. doi: 10.1016/j.yexcr.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 48.DeVry CG, Clarke S. 1999. Polymorphic forms of the protein l-isoaspartate (d-aspartate) O-methyltransferase involved in the repair of age-damaged proteins. J Hum Genet 44:275–288. doi: 10.1007/s100380050161. [DOI] [PubMed] [Google Scholar]
- 49.Ichikawa JK, Clarke S. 1998. A highly active protein repair enzyme from an extreme thermophile: the l-isoaspartyl methyltransferase from Thermotoga maritima. Arch Biochem Biophys 358:222–231. doi: 10.1006/abbi.1998.0830. [DOI] [PubMed] [Google Scholar]
- 50.Visick JE, Cai H, Clarke S. 1998. The l-isoaspartyl protein repair methyltransferase enhances survival of aging Escherichia coli subjected to secondary environmental stresses. J Bacteriol 180:2623–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Britton RA, Powell BS, Dasgupta S, Sun Q, Margolin W, Lupski JR, Court DL. 1998. Cell cycle arrest in Era GTPase mutants: a potential growth rate-regulated checkpoint in Escherichia coli. Mol Microbiol 27:739–750. doi: 10.1046/j.1365-2958.1998.00719.x. [DOI] [PubMed] [Google Scholar]
- 52.Gollop N, March PE. 1991. A GTP-binding protein (Era) has an essential role in growth rate and cell cycle control in Escherichia coli. J Bacteriol 173:2265–2270. doi: 10.1128/jb.173.7.2265-2270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Verstraeten N, Fauvart M, Versées W, Michiels J. 2011. The universally conserved prokaryotic GTPases. Microbiol Mol Biol Rev 75:507–542. doi: 10.1128/MMBR.00009-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fixen KR, Baker AW, Stojkovic EA, Beatty JT, Harwood CS. 2014. Apo-bacteriophytochromes modulate bacterial photosynthesis in response to low light. Proc Natl Acad Sci U S A 111:E237–E244. doi: 10.1073/pnas.1322410111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKinlay JB, Harwood CS. 2010. Carbon dioxide fixation as a central redox cofactor recycling mechanism in bacteria. Proc Natl Acad Sci U S A 107:11669–11675. doi: 10.1073/pnas.1006175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phattarasukol S, Radey MC, Lappala CR, Oda Y, Hirakawa H, Brittnacher MJ, Harwood CS. 2012. Identification of a p-coumarate degradation regulon in Rhodopseudomonas palustris by Xpression, an integrated tool for prokaryotic RNA-seq data processing. Appl Environ Microbiol 78:6812–6818. doi: 10.1128/AEM.01418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gallagher LA, Shendure J, Manoil C. 2011. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2:e00315-10. doi: 10.1128/mBio.00315-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transcriptome of R. palustris CGA009 at intervals following growth arrest due to carbon starvation. Download TABLE S1, XLSX file, 1.5 MB (1.5MB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Tn-seq results. Fitness of R. palustris CGA009 genes at intervals after growth arrest due to carbon starvation. Download TABLE S2, CSV file, 1.8 MB (1.9MB, csv) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
R. palustris longevity genes: genes with reduced fitness 3 to 14 days after growth arrest. Download TABLE S3, XLSX file, 0.1 MB (70.2KB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes with increased insertion fitness 3, 7, 14, and 30 days after growth arrest. Download TABLE S4, XLSX file, 0.1 MB (70.8KB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Functional categories of R. palustris longevity genes. Download TABLE S5, XLSX file, 0.04 MB (40.3KB, xlsx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Probable metabolic route to growth for RPA0980::T24 insertion mutants that were positively selected when cells were starved for carbon (acetate) under nitrogen-fixing conditions. These mutants allow a functional uptake hydrogenase enzyme to be expressed. CO2 produced from acetate prior to growth arrest and H2 produced along with ammonia as a product of nitrogenase can be used by the Calvin cycle to generate cell biomass. Download FIG S1, TIF file, 2.1 MB (2.2MB, tif) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Longevity phenotypes of R. palustris mutants compared to wild type. The results of all longevity trials are shown. The same set of survival curves for wild-type cells is shown in each panel. Download FIG S2, DOCX file, 0.4 MB (411.3KB, docx) .
Copyright © 2017 Pechter et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.