LETTER
In their recent paper (1), Podnecky et al. identified novel clinically relevant co-trimoxazole resistance mutations in Burkholderia pseudomallei, the causative agent of melioidosis. Co-trimoxazole, a combination of sulfamethoxazole (SMX) and trimethoprim (TMP), is the best-studied and most widely used synergistic antimicrobial drug combination and is an essential component of melioidosis treatment. Podnecky et al. identified mutations in bpeT and bpeS from laboratory and clinical co-trimoxazole-resistant isolates of B. pseudomallei. Their elegant work demonstrated that mutations in bpeT or bpeS result in constitutive expression of the BpeEF-OprC efflux pump that confers co-trimoxazole resistance. The authors also provide the first report of folM mutations that confer SMX monoresistance, and yet, the biochemical basis for this novel molecular resistance mechanism was not fully explained.
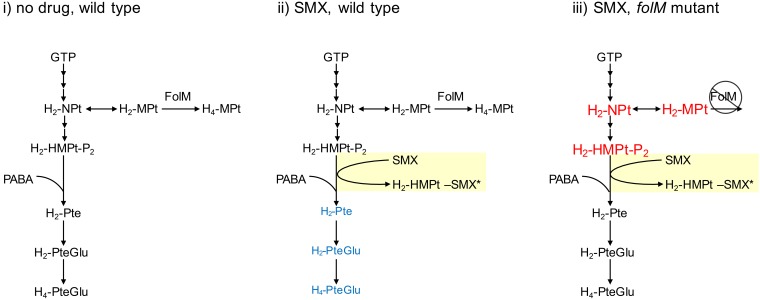
folM encodes dihydromonapterin reductase that catalyzes the final step in synthesis of tetrahydromonapterin (H4-MPt), a nonessential branched pathway from the folate biosynthesis pathway (Fig. 1). H4-MPt is a major pterin produced by Escherichia coli and likely many other bacterial species (2). Loss-of-function mutations in folM are expected to result in increased metabolic flux toward synthesis of the folate precursor dihydropterin pyrophosphate (H2-HMPt-P2).
FIG 1 .
Branched pathway for tetrahydrofolate and tetrahydromonapterin synthesis in B. pseudomallei (modified from Fig. 1 in the work of Podnecky et al. [1]). Blue text in scheme ii represents native metabolites that are expected to decrease in abundance relative to scheme i following treatment with SMX. Red text in scheme iii represents native metabolites that are expected to increase in abundance relative to scheme i due to folM mutation. The asterisk indicates an unmetabolizable product of SMX metabolism. Abbreviations: H2-NPt, 7,8-dihydroneopterin; H2-MPt, 7,8-dihydromonapterin; H4-MPt, tetrahydromonapterin; H2-HMPt-P2, 6-hydroxymethyl-7,8-dihydropterin pyrophosphate; PABA, para-aminobenzoic acid; H2-Pte, dihydropteroate; H2-PteGlu, dihydrofolate; H4-PteGlu, tetrahydrofolate; SMX, sulfamethoxazole.
SMX is typically regarded as an inhibitor of dihydropteroate synthase (FolP). However, it was recently demonstrated that SMX acts instead by competing with para-aminobenzoic acid (PABA) for ligation with H2-HMPt-P2 (Fig. 1) (3, 4). As a result, SMX forms dead-end complexes with H2-HMPt-P2 (H2-HMPt-SMX) and depletes the H2-HMPt-P2 pool and thereby inhibits dihydropteroate production through metabolic wasting (3–5). Consequently, SMX susceptibility is not impacted by the amount of “target” enzyme but is primarily influenced by the intracellular abundance of its cosubstrates PABA and H2-HMPt-P2. In contrast, the activity of TMP, a competitive inhibitor of dihydrofolate reductase, can be affected by both the intracellular abundance of substrate (dihydrofolate) and the amount of target enzyme.
We propose that the loss-of-function mutations in folM confer SMX resistance by increasing H2-HMPt-P2 production that mitigates SMX-driven metabolic wasting (Fig. 1). H2-HMPt-P2 overproduction is not sufficient to confer resistance to TMP because an equivalent amount of PABA would be required to increase dihydrofolate production. Based on this understanding of factors that govern susceptibility and resistance to SMX and TMP, we think that it is important to determine whether H2-HMPt-P2 is overproduced in the folM mutant. Further, it would seem worthwhile to determine whether folM mutations can confer resistance to TMP in PABA-overproducing strains. Resolving these standing questions is likely to reveal the biochemical basis for this novel antifolate drug resistance mechanism.
Footnotes
Citation Minato Y, Baughn AD. 2017. Subversion of metabolic wasting as the mechanism for folM-linked sulfamethoxazole resistance. mBio 8:e01769-17. https://doi.org/10.1128/mBio.01769-17.
REFERENCES
- 1.Podnecky NL, Rhodes KA, Mima T, Drew HR, Chirakul S, Wuthiekanun V, Schupp JM, Sarovich DS, Currie BJ, Keim P, Schweizer HP. 2017. Mechanisms of resistance to folate pathway inhibitors in Burkholderia pseudomallei: deviation from the norm. mBio 8:e01357-17. doi: 10.1128/mBio.01357-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pribat A, Blaby IK, Lara-Núñez A, Gregory JF, de Crécy-Lagard V, Hanson AD. 2010. FolX and FolM are essential for tetrahydromonapterin synthesis in Escherichia coli and Pseudomonas aeruginosa. J Bacteriol 192:475–482. doi: 10.1128/JB.01198-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palmer AC, Kishony R. 2014. Opposing effects of target overexpression reveal drug mechanisms. Nat Commun 5:4296. doi: 10.1038/ncomms5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yun MK, Wu Y, Li Z, Zhao Y, Waddell MB, Ferreira AM, Lee RE, Bashford D, White SW. 2012. Catalysis and sulfa drug resistance in dihydropteroate synthase. Science 335:1110–1114. doi: 10.1126/science.1214641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock L, Miller GH, Schaper KJ, Seydel JK. 1974. Sulfonamide structure-activity relationships in a cell-free system. 2. Proof for the formation of a sulfonamide-containing folate analog. J Med Chem 17:23–28. doi: 10.1021/jm00247a006. [DOI] [PubMed] [Google Scholar]