ABSTRACT
Human gut Bacteroides species produce different types of toxins that antagonize closely related members of the gut microbiota. Some are toxic effectors delivered by type VI secretion systems, and others are non-contact-dependent secreted antimicrobial proteins. Many strains of Bacteroides fragilis secrete antimicrobial molecules, but only one of these toxins has been described to date (Bacteroidales secreted antimicrobial protein 1 [BSAP-1]). In this study, we describe a novel secreted protein produced by B. fragilis strain 638R that mediated intraspecies antagonism. Using transposon mutagenesis and deletion mutation, we identified a gene encoding a eukaryotic-like ubiquitin protein (BfUbb) necessary for toxin activity against a subset of B. fragilis strains. The addition of ubb into a heterologous background strain conferred toxic activity on that strain. We found this gene to be one of the most highly expressed in the B. fragilis genome. The mature protein is 84% similar to human ubiquitin but has an N-terminal signal peptidase I (SpI) signal sequence and is secreted extracellularly. We found that the mature 76-amino-acid synthetic protein has very potent activity, confirming that BfUbb mediates the activity. Analyses of human gut metagenomic data sets revealed that ubb is present in 12% of the metagenomes that have evidence of B. fragilis. As 638R produces both BSAP-1 and BfUbb, we performed a comprehensive analysis of the toxin activity of BSAP-1 and BfUbb against a set of 40 B. fragilis strains, revealing that 75% of B. fragilis strains are targeted by one or the other of these two secreted proteins of strain 638R.
KEYWORDS: Bacteroides, antagonism, microbiota, ubiquitin
IMPORTANCE
We are just beginning to understand some of the important interactions that occur between microbes of the human gut microbiota that dictate the composition and abundance of its constituent members. The ability of one member to produce molecules that directly kill a coresident member has been shown among minor gut species and is just starting to be studied in the abundant Bacteroides species. Here, we show that some strains of Bacteroides fragilis have acquired a gene encoding a secreted eukaryotic-like ubiquitin protein with potent inhibitory activity against other B. fragilis stains. This is the first bacterially encoded ubiquitin-like molecule shown to function like a bacterial toxin. This molecule is an example of a gut symbiont acquiring and adapting a eukaryotic molecule likely to increase its competitiveness in the mammalian gut. Understanding antagonistic factors produced by abundant gut symbionts is an important prerequisite to properly engineer strains to colonize the gut for health benefits.
INTRODUCTION
The gut microbiota of healthy humans is comprised of many different microbes, with members of the order Bacteroidales being the most abundant Gram-negative bacteria. Numerous Bacteroidales species colonize the human gut simultaneously at high density, and colonization with more than one strain of the same Bacteroidales species is common (1, 2). Some factors that may account for the ability of so many closely related species and strains to colonize the same ecosystem include their ability to utilize different nutrients (3) or to prioritize the utilization of different nutrients (4, 5), their occupation of different spatial niches (6, 7), and their ability to cooperate in the utilization of dietary polysaccharides (8, 9). Bacteroidales species also physically contact each other in the human gut and have been shown to exchange more than 100 kb of DNA between strains with the transfer of a single conjugative element (10, 11).
Despite characteristics of the Bacteroidales that permit or promote cocolonization, these bacteria have also evolved mechanisms to antagonize each other. Antagonism or interference competition is likely an important factor dictating the composition of diverse microbial communities. Bacteroidales have been shown to elicit two different types of antagonistic systems: contact-dependent type VI secretion systems (T6SSs) (12–15) and secreted antimicrobial protein toxins (16, 17). Most Bacteroides fragilis strains have genetic loci encoding T6SSs (12), and some of these systems have been shown to antagonize nearly all gut Bacteroidales species tested (13). As T6SSs are contact dependent, this antagonism may largely occur when nutritional niches overlap and/or when dietary nutrients are limiting and Bacteroidales species are forced to utilize host mucins, one of the preferred carbon sources of B. fragilis (5).
In contrast to the B. fragilis T6SSs, the two identified Bacteroidales secreted (non-contact-dependent) antimicrobial proteins (BSAPs) each targets a subset of strains of the same species (16, 17). Both described BSAPs contain membrane attack/perforin (MACPF) domains found in immune molecules, such as complement components and perforin, that lyse bacteria or virally infected cells by pore formation. BSAP-1 and BSAP-2 are the only bacterially produced MACPF proteins shown to kill other bacteria. BSAP-1 is produced by a subset of B. fragilis strains and mediates its toxicity through pore formation following recognition of a specific outer membrane β-barrel protein on sensitive (non-BSAP-1-producing) B. fragilis strains. BSAP-2 is produced by a subset of Bacteroides uniformis strains and recognizes the lipopolysaccharide (LPS; core polysaccharide or short O antigen [O-ag]) of sensitive (non-BSAP-2-producing) B. uniformis strains. The genes encoding both BSAP-1 and BSAP-2 were acquired with adjacent genes encoding orthologs of their receptors, replacing the receptor and rendering the strain resistant to the newly acquired toxin (17).
In studying secreted antimicrobial molecules produced by B. fragilis, we found that several strains, such as 638R, inhibited the growth of many B. fragilis strains, whereas other strains had no secreted antimicrobial activity against the panel of strains analyzed (16). We also showed that a mutant in which the BSAP-1-encoding gene of B. fragilis 638R is deleted retains the ability to inhibit the growth of a subset of B. fragilis strains (16). The present study was designed to identify and characterize the additional secreted antimicrobial molecule of strain 638R. Here, we describe a novel eukaryotic-like ubiquitin molecule that mediates potent antimicrobial activity against B. fragilis strains.
RESULTS
Spectrum of intraspecies antagonism by secreted molecules of B. fragilis 638R.
We performed a comprehensive analysis using 40 B. fragilis strains from our collection to determine their sensitivity to BSAP-1 or other antimicrobial molecule(s) secreted by B. fragilis 638R. Partial or complete genome sequences were available for 11 of these B. fragilis strains, and we sequenced, assembled, and annotated the genomes of an additional six strains (see Table S1 in the supplemental material). Using wild-type 638R, the 638RΔ1646 (BSAP-1 gene deletion) mutant, and active purified His–BSAP-1, we determined the sensitivity profiles of these 40 strains (Table S1). As shown by the results in Fig. 1, there were four different patterns of sensitivity/resistance. Only seven of these strains were not susceptible to secreted molecules of 638R (Fig. 1A). Ten strains were sensitive to BSAP-1 only, as His–BSAP-1 produced a zone of inhibition and no inhibitory activity remained in the 638RΔ1646 mutant (Fig. 1B). Four strains were not antagonized by BSAP-1 but were inhibited by a different molecule(s) secreted by 638R (Fig. 1C), and 19 strains were antagonized by both BSAP-1 and an additional molecule(s) secreted by this strain (Fig. 1D).
FIG 1 .
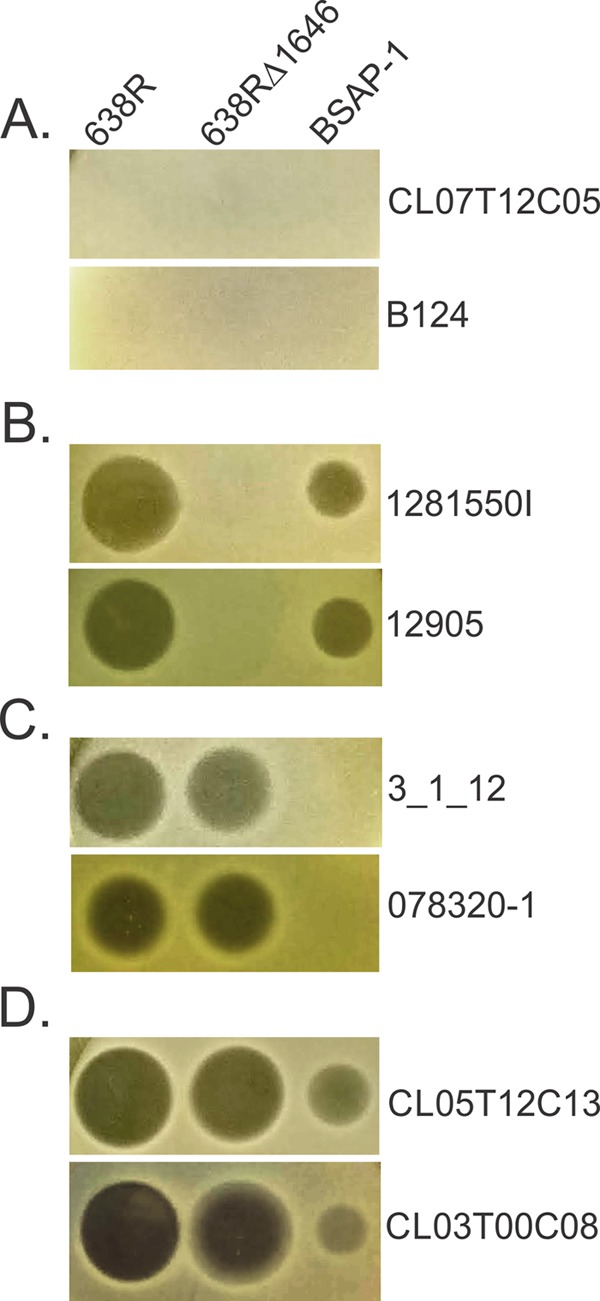
Agar spot assays of eight B. fragilis strains in overlays, showing zones of growth inhibition (dark spots) by secreted molecule(s) from wild-type strain 638R and 638RΔ1646 (BSAP-1 deletion mutant) and by purified, His-tagged BSAP-1. Results for two strains each, named to the right of the panels, are shown as examples of the four different patterns of sensitivity/resistance.
Ability of B. fragilis 638R, 638RΔ1646 mutant, and proteins to inhibit the growth of B. fragilis strains. Download TABLE S1, DOCX file, 0.01 MB (15.6KB, docx) .
Copyright © 2017 Chatzidaki-Livanis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
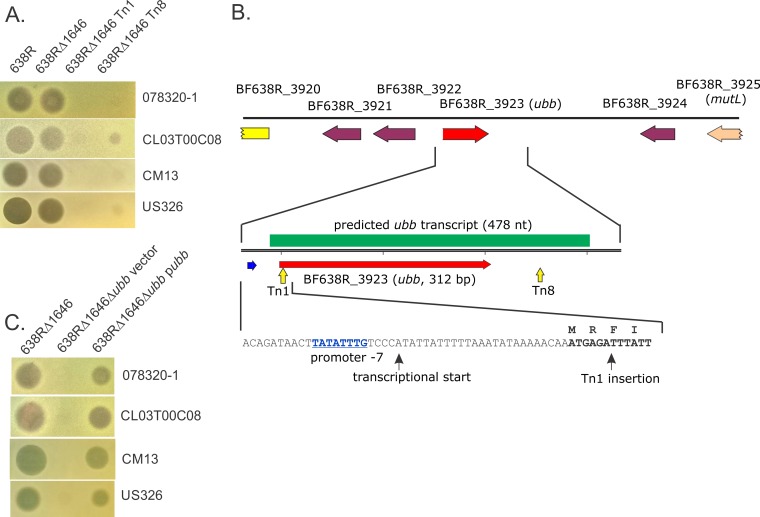
Identification of a gene necessary for inhibitory activity in 638RΔ1646.
To identify the molecule or molecules responsible for the second antimicrobial activity, we performed transposon mutagenesis of the 638RΔ1646 strain and screened for loss of inhibition of the sensitive B. fragilis strain CL03T00C08. Two transposon mutants of 638RΔ1646 were identified that were severely attenuated in their ability to inhibit the growth of this strain (Fig. 2A). In both of these mutants, the transposon inserted within the transcribed region of an unusual bacterial gene previously recognized as encoding a very close ortholog of eukaryotic ubiquitin, termed B. fragilis Ubb (BfUbb) (18, 19). One transposon (Tn1) inserted 6 bp into the predicted coding region of BF638R_3923 (ubb) (Fig. 2B) and abrogated its ability to antagonize strain CL03T00C08. The second mutant (Tn8) had the transposon inserted 76 bp downstream from ubb, resulting in severely attenuated activity. We analyzed our previously generated high-throughput RNA sequencing (RNA-Seq) data on in vitro-grown mid-log-phase B. fragilis NCTC 9343, which contains a ubb genetic region identical to that of strain 638R (18, 19). These analyses predicted that the ubb transcript begins just following the −7 region of the Bacteroides sigma 70 binding site (20) and terminates 140 bp downstream from the ubb stop codon (Fig. 2B). Based on this prediction, Tn8 has the transposon inserted within the ubb transcript. We tested whether these transposon mutants retained the ability to antagonize three other strains sensitive to 638RΔ1646 and found that growth inhibition of these strains was also abrogated or severely attenuated (Fig. 2A). The RNA-Seq data revealed that ubb is the 47th most highly expressed gene of the NCTC 9343 genome, with a fragments per kilobase of transcript per million mapped reads (FPKM) value of 7,453.
FIG 2 .
Identification of a gene necessary for antimicrobial activity of 638RΔ1646. (A) Agar spot assays showing results for two transposon mutants of 638RΔ1646 that lost inhibitory activity against four B. fragilis strains. (B) (Top) ORF map of the genetic region where the transposons insertions into the 638R genome resulted in loss of activity. (Bottom) Extent of the BF638R_3923 (ubb) transcript as predicted from analyses of RNA-Seq data. A perfect match with the −7 site of the promoter sequence recognized by the Bacteroides sigma 70 factor is shown (blue letters). (C) Agar spot assays showing the loss of secreted inhibitory activity in a ubb deletion mutant and the resulting phenotypes when the gene is added to the mutant in trans (pubb), as well as results for the vector control.
To confirm that the transposon insertions in the ubb transcript accounted for the loss of toxic activity, an internal deletion mutant with the deletion of ubb was created in the 638RΔ1646 background. This strain lost the ability to antagonize the four sensitive strains tested, and the toxic activity was restored when the gene was cloned into a Bacteroides expression vector and added to the deletion mutant in trans (Fig. 2C). Therefore, ubb is required for the antimicrobial activity against these strains.
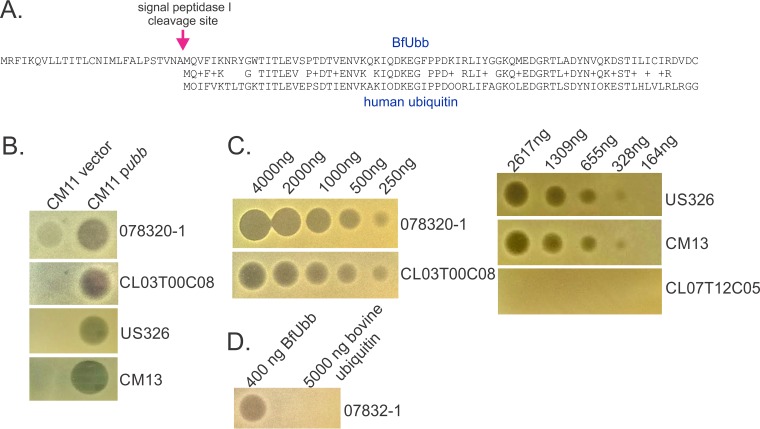
B. fragilis ubiquitin is the inhibitory factor.
BfUbb was previously shown to have a 27-amino-acid (aa) signal sequence and to be secreted extracellularly (18), even though the gene is annotated as a smaller open reading frame (ORF) without this sequence. Alignment of the mature 76-aa BfUbb with the 76-aa human ubiquitin shows that the similarity begins immediately after the signal sequence and the proteins are 84% similar along their lengths, with the exception of the last 4 amino acids (Fig. 3A). The fact that BfUbb is secreted suggests that it may mediate the toxic activity itself, either directly or indirectly. To eliminate the possibility that BfUbb may be modifying or acting on another gene or gene product of strain 638R that mediates the activity, we placed ubb in trans in B. fragilis strain CM11, which does not have BfUbb activity and is also not sensitive to it (Table S1). We found that the acquisition of ubb by strain CM11 conferred upon it the ability to antagonize BfUbb-sensitive strains (Fig. 3B). These data also suggest that BfUbb-producing strains do not require an immunity protein for protection, as CM11 was not noticeably affected by the addition of ubb.
FIG 3 .
BfUbb inhibits growth. (A) Alignment of BfUbb with human ubiquitin, showing the extended N-terminal SpI signal sequence of BfUbb. (B) Agar spot assays displaying the sensitivity/resistance profiles of four B. fragilis strains exposed to B. fragilis CM11 with an empty vector or ubb in trans. (C) Agar spot assays showing inhibition activities of dilutions of synthesized 76-aa BfUbb against four sensitive strains and one resistant strain. (D) Agar spot assay showing sensitivity of B. fragilis strain 0878320-1 to BfUbb versus purified bovine ubiquitin.
To further establish that BfUbb is the inhibitory factor of strain 638R, we had the 76-aa BfUbb peptide (lacking the signal sequence) synthesized and tested it in the assay. We found that the synthesized protein has very potent activity against sensitive strains, where as little as 250 ng of the protein resulted in a detectable zone of inhibition (Fig. 3C). Strain B. fragilis CL07T12C05, which is not affected by wild-type 638R, is not inhibited by the BfUbb peptide (Fig. 3C). In addition, purified 76-aa bovine ubiquitin, which is identical to human ubiquitin, has no activity against these strains, even when a much greater dose is used (Fig. 3D). We also tested the remaining 36 B. fragilis strains used in this study for growth inhibition by the BfUbb peptide. Of the 40 strains analyzed, 13 are inhibited by BfUbb (Table S1). All of these strains are antagonized by the 638RΔ1646 mutant, as expected (Table S1). Unlike our findings with BSAP-1, where the B. fragilis strains analyzed either produce the toxin or are sensitive to it, there are many B. fragilis strains that do not synthesize BfUbb and are also not sensitive to it (Table S1).
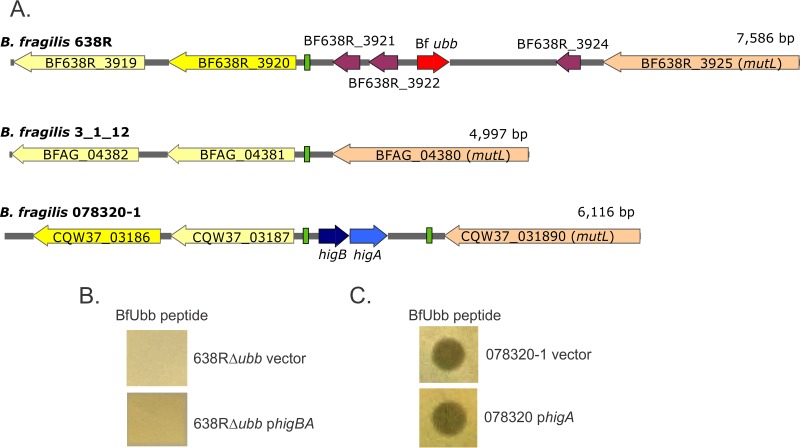
Species-wide analysis of the genetic architecture of the ubb region.
To investigate the diversity of the ubb genetic region among B. fragilis strains, we analyzed the ubb region or the corresponding region in strains lacking ubb. We analyzed 97 B. fragilis genomes contained in our curated genome database and retrieved and analyzed DNA from mutL to the gene encoding the first protein with a β-propeller motif (Fig. 4A). This analysis revealed that this area of the B. fragilis genome is heterogeneous, with three major genetic types identified (Fig. 4A). Each genome has a similar mutL; however, the DNA between strains begins to diverge significantly 39 bp downstream from this gene. Of the 97 genomes analyzed, 13 contain a ubb and surrounding DNA nearly identical to that of strain 638R (Fig. 4A, top). Seventy-three of the strains completely lack ubb and in its place have two genes encoding a toxin-antitoxin pair of the HigBA family (Fig. 4A, bottom). Eleven strains do not have any genes present in this region (Fig. 4A, middle). Each of the genomes encodes proteins with β-propeller motifs outside the divergent region (Fig. 4A, genes colored yellow), but these sequences are not conserved, even within a genetic type. A 52-bp direct-repeat element (Fig. 4A, green boxes) was identified flanking the higBA genes, one located 61 bp upstream from a β-propeller-encoding gene and the other 257 bp downstream from mutL. A single copy of this 52-bp element is present in the other two genetic types, in both cases 61 to 67 bp upstream from their respective β-propeller-encoding genes (Fig. 4A). The 52-bp element in ubb-containing genomes is an exact match with the element in the higBA genetic group. However, the element differs somewhat in the genomes with no genes in the region; for example, strain 3_1_12 has 8 mismatches. The other small genes unique to the ubb genetic region do not encode proteins that appear to be of eukaryotic origin: BF638R_3921 is a peptidase of the S41 superfamily, BF638R_3922 is the N terminus of a truncated β-propeller protein, and BF638R_3934 is a hypothetical protein with no predicted function. Comparisons of genomes within a genetic group revealed that the genomes without any genes between mutL and the downstream β-propeller-encoding gene (represented by strain 3_1_12 in Fig. 4A) are the most divergent within a group. To determine whether the ubb-containing genomes may be more phylogenetically related to each other than to the genomes of the other two genetic variants, we analyzed by BLAST all 97 genomes for their phylotype of five conserved genes that are commonly used for phylogenetic analyses: dnaJ, groEL, gyrB, recA, and rpoB. It was shown many years ago that the species B. fragilis is comprised of two genetic groups, with each group having distinct β-lactamase-encoding genes, one with cepA and the other containing cfiA (ccrA) (21–23). In these analyses, we also found that B. fragilis genomes segregate into two distinct branches and correlate perfectly with the presence of cepA or cfiA. In analyzing our three genetic types in the ubb region, we found that the ubb-containing genomes are indistinguishable from genomes containing the higBA toxin-antitoxin pair using this phylotyping method. Both genetic types contain cepA, and the five conserved genes are >99.6% identical to each other. However, the 11 B. fragilis genomes that do not have any genes in this region are all of the other subspecies, those which contain cfiA. The five conserved genes are only 90 to 92% identical to the orthologous gene in B. fragilis genomes containing ubb or higBA.
FIG 4 .
Heterogeneity of the B. fragilis genome in the ubb genetic region. (A) Gene maps of three representative B. fragilis strains showing the three predominant genetic types in the ubb or corresponding regions. B. fragilis strains each have a similar mutL (orange), and the DNA between each of the three genetic types begins to diverge 39 bp downstream from this gene. The genes colored yellow encode proteins with β-propeller motifs that are divergent even within a genetic type. The green boxes indicate a 52-bp element that is present in each of the three genetic types and is identical between strain 638R and 078320-1, with a few mismatches in strain 3_1_12. The B. fragilis 638R gene map (top) shows that ubb is contained in a genetic region likely acquired with three other small genes. The B. fragilis 3_1_12 gene map (middle) shows that a few B. fragilis genomes have no genes inserted in this region. The B. fragilis 078320-1 gene map (bottom) shows the most predominant B. fragilis genetic type, where higBA genes are present in the divergent region along with an additional copy of the 52-bp element downstream from mutL. (B) Agar spot assay of BfUbb peptide overlaid with B. fragilis CM11 containing an empty vector or the vector expressing higBA. (C) Agar spot assay of the BfUbb peptide overlaid with sensitive strain B. fragilis 078320-1 containing an empty vector or the vector overexpressing the antitoxin-encoding gene higA.
Both BSAP-1 and BSAP-2 target surface molecules in sensitive cells encoded by genes that were replaced by DNA containing the incoming BSAP-encoding gene. Therefore, the BSAP receptor genes are in the same genetic region of sensitive strains as the BSAP gene in producing strains. We therefore considered that BfUbb may be affecting the HigBA toxin-antitoxin (TA) system, which is not present in Bfubb-containing strains. Among possible mechanisms for such an activity is the binding of BfUbb to the HigA antitoxin so that it can no longer interact with the HigB toxin to prevent self-intoxication. Three lines of evidence suggest that the toxicity of BfUbb may not involve the HigBA TA system. We first attempted to delete either higBA or just the higB toxin gene from several sensitive B. fragilis strains. Despite repeated attempts, we were unable to construct these deletions, possibly due to the difficulties of creating deletion mutants in some B. fragilis strains or an intolerance to the deletion of these genes. In lieu of a deletion mutant, we cloned higBA into a Bacteroides expression vector with a constitutive promoter, thereby eliminating the normal regulation of these genes, and this construct was conjugated into strain 638RΔubb. The presence of constitutively expressed higBA did not render 638RΔubb sensitive to BfUbb (Fig. 4B). We also cloned higA into a vector for high gene expression in Bacteroides to determine whether excess quantities of the antitoxin could overcome the effects of BfUbb in a sensitive strain. We observed no difference in the ability of various quantities of BfUbb to antagonize B. fragilis 078320-1 in the agar spot assay when this sensitive strain contained either the empty vector or the highly expressed antitoxin gene (Fig. 4C, showing results for one concentration). Another finding indicating that the HigBA system may not be the target of BfUbb is that we identified four B. fragilis strains with higAB genetic regions nearly identical to those of other sensitive strains, and yet, these four strains are not sensitive to BfUbb (Table S1).
Analysis of human gut metagenomic data sets for ubb.
Among the 97 sequenced B. fragilis genomes in our curated genome collection, 13 genomes contain ubb. These analyses estimated the frequency of ubb in B. fragilis genomes at approximately 13%. To determine the frequency of ubb in the human gut microbiota, we analyzed the 3CGC human gut metagenomic set, a subset of the recently compiled integrated gene catalog (IGC) that contains 1,267 metagenomes, for the presence of ubb. We detected evidence of the species B. fragilis in 370 of these metagenomes, of which ubb was detected in 44 (Table S2). ubb was only detected in metagenomes identified as containing B. fragilis (Table S2). Based on these data, approximately 12% of metagenomes with evidence of B. fragilis contain ubb, a percentage relatively consistent with that of our sequenced genome collection. In addition, we found numerous metagenomes that contain both the BSAP-1 encoding gene and ubb and some metagenomes that contain one or the other. In total, 130 metagenomes contain at least one of the two secreted antimicrobial-protein-encoding genes, and 20 contain genes encoding both BSAP-1 and BfUbb.
Identification of B. fragilis and ubb, higA, higB, and BF638R_1646 (bsap1) in human gut metagenomes comprising subset 3CGC of the integrated gene catalog (IGC). Download TABLE S2, XLSX file, 0.1 MB (95.8KB, xlsx) .
Copyright © 2017 Chatzidaki-Livanis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
Ubiquitin is found in eukaryotic organisms from fungi to humans. Ubiquitin is attached to eukaryotic proteins by a series of three enzymes that create isopeptide bonds. Ubiquitination of these substrate proteins regulates their cellular fate in numerous and distinct ways. Ubiquitination or polyubiquitination of eukaryotic proteins can affect protein degradation (reviewed in reference 24), localization (25), activity, and interactions with other molecules (26). Ubiquitination of proteins has not been demonstrated in bacteria, largely due to the fact that most bacteria do not produce a eukaryotic-like ubiquitin. However, some pathogenic bacteria produce proteins that alter eukaryotic cell function by interfering with the ubiquitin signaling pathways in host cells (reviewed in reference 27). Therefore, coevolution of bacteria with eukaryotic hosts has resulted in ubiquitin-related processes involved in toxicity, where either the ubiquitin molecule itself is involved in antagonistic interactions or, as described above, bacterial effectors alter host ubiquitin processes.
B. fragilis is one of a few bacteria that encode a eukaryotic-like ubiquitin. BfUbb is an interesting ubiquitin ortholog with many similarities to human ubiquitin but with some important distinctions. One of the major differences is in the C termini of these molecules. The last 4 amino acids are distinct, and BfUbb lacks the critical terminal glycine residue involved in the isopeptide linkage to substrate proteins in the eukaryotic system. Therefore, BfUbb would not serve as a substrate for eukaryotic E3 ubiquitin ligase. Another distinction is that BfUbb is glycosylated; Patrick et al. (18) previously noted that BfUbb has two Bacteroidetes glycosylation sites (28). Our prior study showed that all analyzed secreted proteins of B. fragilis that have glycosylation motifs are in fact glycosylated (29), strongly suggesting that BfUbb is glycosylated. As synthetic BfUbb has potent activity, glycosylation of this protein is not necessary for its ability to antagonize target strains. We previously showed that glycosylation of some Bacteroides secreted proteins increases their stability (28); therefore, glycosylation of BfUbb may be important in the context of the bacterial cell.
One of the most interesting distinctions of BfUbb is the N-terminal signal sequence dictating its processing by signal peptidase I and subsequent secretion. The cleavage site of this signal peptide is located such that mature BfUbb aligns exactly with the first amino acid of its human counterpart (Fig. 2B). Although mammalian ubiquitin does not contain such a signal sequence, there are ubiquitin orthologs described in several nematode species with this feature. The best described is produced by the nematode Globodera rostochiensis, a parasite of plants that creates a syncytium for successful parasitism. The secreted ubiquitin of this organism is produced exclusively in the gland of the nematode and serves as an effector for host syncytium formation (30). In addition to the N-terminal signal sequence, the nematode ubiquitin has a 12-aa C-terminal extension that is cleaved from the ubiquitin molecule in the plant. The 12-aa peptide suppresses effector-triggered immunity. The remaining core ubiquitin molecule also plays a role in parasitism, possibly by perturbing ubiquitin levels, thereby affecting the host 26S proteasome (30). Therefore, the unique ubiquitin molecules produced by some nematodes are similar to BfUbb in that they are secreted and result in effector/toxic activity in recipient cells.
The source from which ubb was acquired is not clear from existing genomic sequences. There are several amoeba-infecting giant viruses that encode ubiquitin molecules (31, 32) that are the closest orthologs of mature BfUbb in the databases. One such ubiquitin-encoding giant virus was identified in human stool (33). Giant virus-infected cells are on occasion coinfected with small virophage that are eukaryotic viruses but have properties of prokaryotic phage and are predicted to transfer genes between giant viruses during coinfection (34). It is possible that such a virus or phage may have introduced an ortholog of ubb into the B. fragilis genome. We did not detect any obvious signs in the B. fragilis genomes to hint at how this region was acquired. However, we did identify a 52-bp element in the divergent regions of all three genetic variants (Fig. 4A). This 52-bp element is duplicated in higBA-containing genomes and flanks these genes. Therefore, this region may represent an integration site for chromosomal insertions.
How BfUbb antagonizes specific B. fragilis strains is not readily obvious from the protein sequence or genomic analyses. A potential target is the antitoxin protein of the HigBA toxin-antitoxin system that is present in the majority of non-ubb-containing B. fragilis strains. Our experimental and genomic data did not confirm a role for the HigBA TA system in the activity of BfUbb, but its involvement has also not been excluded. BSAP-1 and BSAP-2 each contain an MACPF domain and bind surface receptors, leading to pore formation. BfUbb may function by a completely different mechanism. For BSAP-1, B. fragilis strains typically produce the toxin or are sensitive to it. This occurs because BSAP-1 binds a surface protein necessary for gut colonization, and in BSAP-1-producing strains, the gene encoding the protein conferring sensitivity is replaced by an ortholog that serves its function in gut colonization but also renders the strain resistant to the BSAP-1. Therefore, most non-BSAP-1-producing strains have the BSAP-1 surface target by default. For BfUbb, no such pattern emerged, as we found many strains without ubb that were not sensitive to it. Our data also suggest that, similar to BSAP-1 and BSAP-2, BfUbb does not require an immunity protein to protect the producing cell. Therefore, it is likely that sensitive cells contain a specific molecule that BfUbb targets rather than certain B. fragilis strains producing an immunity protein for resistance. It is possible that BfUbb is transported into cells by a protein-specific nutrient uptake system, where it would act on an intracellular target rather than at the bacterial surface. Continued analysis of BfUbb will likely reveal a novel mechanism of action.
This is the first bacterially produced eukaryotic-like ubiquitin molecule shown to intoxicate bacterial cells, whether directly or indirectly. To date, the three described secreted antimicrobial proteins produced by Bacteroides all have eukaryotic-like features. We previously showed that Bacteroides species synthesize another mammalian-like enzyme, termed Fkp, that charges fucose with GDP for its addition to surface glycans of these bacteria. Fkp is necessary for the bacteria to colonize the mammalian gut (35). These secreted antimicrobial molecules are additional examples of these host-associated bacteria likely acquiring and adapting eukaryotic molecules to increase their fitness in the human gut.
The importance of antagonism to bacterial colonization of the mammalian gut is evident by the fact that a single B. fragilis strain produces not only a T6SS that targets nearly all Bacteroidales species (13) but also at least two secreted antimicrobial proteins to further antagonize strains of the same species. Identifying and characterizing molecules of gut bacteria that mediate competitive interactions will allow the rational design of engineered probiotic-type bacteria to successfully colonize the host to deliver health-promoting functions.
MATERIALS AND METHODS
Primers.
All primers used in this study are listed in Table S3 in the supplemental material.
Primers used in this study. Download TABLE S3, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2017 Chatzidaki-Livanis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial strains and growth conditions.
The Bacteroides strains used in this study were previously described (1, 36). All Bacteroides strains were grown in supplemented basal medium (37) or on supplemented brain heart infusion (BHIS) plates. Antibiotics (5 µg/ml erythromycin or 3 µg/ml tetracycline) were added where indicated below. Escherichia coli strains were grown in L broth or L plates with antibiotics added where appropriate (100 µg/ml ampicillin, 100 µg/ml trimethoprim, and 50 µg/ml kanamycin).
Agar spot test for growth inhibition analysis.
The ability of Bacteroides strains to inhibit the growth of other strains was assayed using the agar spot test (38). In brief, Bacteroides strains were resuspended from plates into phosphate-buffered saline (PBS) at a density of approximately 1010/ml, and 5-µl volumes were spotted onto BHIS plates and grown anaerobically at 37°C overnight. The bacteria were removed with swabs, and the residual bacteria remaining on the plates were killed by exposing them to chloroform vapor for 15 min. Strains to be tested for growth inhibition were grown to an optical density at 600 nm (OD600) of 0.6, and then 100-µl amounts were mixed with 4 ml top agar and overlaid onto the chloroform-treated plates. The zones of inhibition were analyzed after anaerobic overnight incubation at 37°C.
Growth inhibition assays using synthetic BfUbb, mammalian ubiquitin, and His-tagged BSAP-1.
The 76-aa BfUbb peptide corresponding to the mature molecule without the signal sequence was synthesized by LifeTein (Hillsborough, NJ). Agar overlay assays using synthetic BfUbb were performed the same as the regular agar spot assays except that the purified protein in PBS was added to the BHIS plates (2.6 µg in a 5-µl volume unless otherwise indicated), allowed to dry, and then overlaid with strains as described above. Purified bovine ubiquitin (76 aa, >98% pure) from erythrocytes was purchased from Sigma (U6253) and resuspended in PBS. Five micrograms of protein in 5 µl was spotted onto the BHIS plates for the overlay assay. The N-terminally His-tagged BSAP-1 protein was purified and processed as previously described (16). For the agar overlay assay, 2.5 µg of His-BSAP in a 5-µl volume of PBS was added to the plates and allowed to dry before performing the overlays.
Transposon mutagenesis and deletion of BF638R_3923 (ubb).
Random mutagenesis of B. fragilis 638RΔ1646 was performed using the transposon-containing plasmid pYT646b as described previously (39), using tetracycline selection. The insertion sites of transposon mutants were identified by cloning the junctional DNA as described previously (16, 39).
A deletion mutant with the deletion of ubb was constructed such that 255 bp of the 312-bp gene was removed. DNA segments upstream and downstream from the region to be deleted were PCR amplified, and the PCR products were digested with BamHI and EcoRI and cloned by three-way ligation into the BamHI site of pNJR6 (40). The resulting plasmid was conjugally transferred into wild-type B. fragilis 638R or 638RΔ1646 using helper plasmid R751, and cointegrates were selected by erythromycin resistance. Following growth under nonselective conditions, erythromycin-sensitive colonies were screened by PCR for the mutant genotype.
Cloning ubb into a conjugal expression vector.
ubb was PCR amplified using primers with BamHI ends (Table S3). The PCR product was digested with BamHI, cloned into the BamHI site of Bacteroides expression vector pFD340 (41), and screened for correct orientation in relation to the plasmid-borne promoter. The resulting plasmid was conjugated into B. fragilis 638RΔ1646 and B. fragilis CM11 by conjugal mating using an E. coli strain containing helper plasmid RK231 and selected by acquisition of erythromycin resistance.
Cloning of the higBA toxin-antitoxin genes.
The putative toxin-antitoxin genes HMPREF1067_00095 and -96 were PCR amplified from sensitive strain CL03T12C07, cloned into the BamHI site of pFD340, and screened for correct orientation in relation to the plasmid-borne promoter. The antitoxin gene (HMPREF1067_00096) was cloned into pMCL140 (42), a Bacteroides expression vector for high expression of cloned genes. Both plasmids were conjugated into Bacteroides strains as described above.
Determination of ubb transcript size and expression level.
We reanalyzed the two biological replicates of wild-type B. fragilis NCTC 9343 from our previously generated RNA-Seq data (13) by first adapter and quality trimming the two sets of paired-end reads using BBDuk (see “Genome sequencing of additional B. fragilis strains” below). Read alignment, transcript prediction, and statistical calculations, including normalization and measures of relative expression (fragments per kilobase of transcript per million mapped reads [FPKM]), were achieved using HISAT2 (version 2.1.0) (43), samtools (version 1.6) (44), and StringTie (version 1.1.3) (45). We used the sequence information for B. fragilis NCTC 9343 available from NCBI (GenBank accession number NC_003228) as a scaffold, except that we corrected the start coordinate of the ubb gene (BF9343_3779) to reflect the true beginning of the open reading frame.
Detection of B. fragilis and B. fragilis genes in human gut metagenomes.
Metagenomic analyses were performed using a subset (3CGC) of the recently compiled integrated gene catalog (IGC) (46). This subset comprises 1,267 human gut metagenomes. To detect B. fragilis in these metagenomes, we used DNA sequences of single-copy genes known previously to differentiate B. fragilis from other species, namely, dnaJ, groL, gyrB, recA, and rpoB. The DNA sequences of these five genes were collected from each of the three B. fragilis strains with unique genetic types in the ubb or corresponding region. The genes from B. fragilis 638R are BF638R_1741 (dnaJ), BF638R_3250 (groL), BF638R_0298 (gyrB), BF638R_1245 (recA), and BF638R_4052 (rpoB); the genes from B. fragilis 078320-1 are CQW37_03448 (dnaJ), CQW37_03630 (groL), CQW37_02683 (gyrB), CQW37_01563 (recA), and CQW37_01330 (rpoB); and the genes from B. fragilis 3_1_12 are BFAG_01078 (dnaJ), BFAG_03510 (groL), BFAG_02716 (gyrB), BFAG_00519 (recA), and BFAG_04295 (rpoB). These fifteen gene sequences were used as queries against a blastn database created using the makeblastdb program. The output from the blastn command (executed with switches -task megablast -evalue 1e−5 -dust no -best_hit_score_edge 0.05 -best_hit_overhang 0.25) was parsed, and the best hit (by highest bit score) returned from each metagenome was retained if that metagenome had a hit reaching the threshold levels indicated. Hits that survived the filter cutoff values were defined as evidence of the presence of B. fragilis in the subject metagenome (Table S2). The search for ubb, higBA, and the BSAP-1-encoding gene in the metagenomes was performed in the same way, using the DNA sequences of CQW37_03188 and CQW37_03189 (the higB and higA genes, respectively, from B. fragilis 078320-1), BF638R_3923 (the ubb gene from B. fragilis 638R, including the region encoding the heretofore unannotated signal sequence), and BF638R_1646 (the BSAP-1-encoding gene from B. fragilis 638R) as queries (Table S2).
Genome sequencing of additional B. fragilis strains.
Chromosomal DNA from B. fragilis strains 12905, CL04T03C20, US326, CM13, 1284, and 078320-1 was fragmented using the Covaris S2 instrument and analyzed for fragment distribution with a high-sensitivity D1K TapeStation machine and for sufficient quantity by a SYBR quantitative PCR (qPCR) assay. The DNA was sequenced using an Illumina MiSeq sequencer, producing paired-end reads of 150 bp. Genomic sequencing was performed by the Biopolymers Facility, Harvard Medical School. The raw paired Illumina reads were processed to remove adapter sequences and quality trimmed using BBDuk, part of the BBTools (version 37.50) suite of programs distributed by the Department of Energy’s Joint Genome Institute (https://jgi.doe.gov/data-and-tools/bbtools/). NCBI’s UniVec_Core database (build 10.0) was downloaded (ftp://ftp.ncbi.nlm.nih.gov/pub/UniVec), entries originating in GenBank were removed, and the Illumina reads were further screened against this data set using blastn, removing any read that returned a significant hit. The reads passing these screens (including orphans) were used to assemble the genomes. Velvet Optimizer (version 2.2.5, http://www.vicbioinformatics.com/software.velvetoptimiser.shtml) was utilized to determine the optimal k value (among other settings), and the genomes were assembled de novo using Velvet 1.2.10 (47). The draft genomes were annotated using an in-house-customized version of Prokka version 1.12 (48).
Accession numbers.
Genomes were deposited in GenBank under BioProject identification number (ID) PRJNA413027 and the following BioSample IDs: B. fragilis 1284, SAMN07735158; B. fragilis 12905, SAMN07735159; B. fragilis 078320-1, SAMN07735199; B. fragilis CL04T03C20, SAMN07735200; B. fragilis CM13, SAMN07735201; and B. fragilis US326, SAMN07735202.
ACKNOWLEDGMENTS
We declare that we have no conflicts of interest. We thank the BEI for providing some of the strains used in this study.
M.C.-L. performed experiments, analyzed data, and wrote the paper. M.J.C. performed all bioinformatics analyses and wrote the paper, K.G.R. performed experiments and analyzed data, R.R.G. performed experiments, J.M.C. performed experiments, and L.E.C. performed experiments, analyzed data, and wrote the paper.
This work was supported by Public Health Service grant R01AI093771 from the NIH/National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Chatzidaki-Livanis M, Coyne MJ, Roelofs KG, Gentyala RR, Caldwell JM, Comstock LE. 2017. Gut symbiont Bacteroides fragilis secretes a eukaryotic-like ubiquitin protein that mediates intraspecies antagonism. mBio 8:e01902-17. https://doi.org/10.1128/mBio.01902-17.
REFERENCES
- 1.Zitomersky NL, Coyne MJ, Comstock LE. 2011. Longitudinal analysis of the prevalence, maintenance, and IgA response to species of the order Bacteroidales in the human gut. Infect Immun 79:2012–2020. doi: 10.1128/IAI.01348-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjerke GA, Wilson R, Storrø O, Øyen T, Johnsen R, Rudi K. 2011. Mother-to-child transmission of and multiple-strain colonization by Bacteroides fragilis in a cohort of mothers and their children. Appl Environ Microbiol 77:8318–8324. doi: 10.1128/AEM.05293-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martens EC, Kelly AG, Tauzin AS, Brumer H. 2014. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. 2015. Symbiotic human gut bacteria with variable metabolic priorities for host mucosal glycans. mBio 6:e01282-15. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers TE, Pudlo NA, Koropatkin NM, Bell JS, Moya Balasch M, Jasker K, Martens EC. 2013. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol Microbiol 88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tropini C, Earle KA, Huang KC, Sonnenburg JL. 2017. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21:433–442. doi: 10.1016/j.chom.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakoff-Nahoum S, Coyne MJ, Comstock LE. 2014. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol 24:40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tuncil YE, Xiao Y, Porter NT, Reuhs BL, Martens EC, Hamaker BR. 2017. Reciprocal prioritization to dietary glycans by gut bacteria in a competitive environment promotes stable coexistence. mBio 8:e01068-17. doi: 10.1128/mBio.01068-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salyers AA, Shoemaker NB, Li LY. 1995. In the driver’s seat: the Bacteroides conjugative transposons and the elements they mobilize. J Bacteriol 177:5727–5731. doi: 10.1128/jb.177.20.5727-5731.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coyne MJ, Zitomersky NL, McGuire AM, Earl AM, Comstock LE. 2014. Evidence of extensive DNA transfer between Bacteroidales species within the human gut. mBio 5:e01305-14. doi: 10.1128/mBio.01305-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne MJ, Roelofs KG, Comstock LE. 2016. Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17:58. doi: 10.1186/s12864-016-2377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatzidaki-Livanis M, Geva-Zatorsky N, Comstock LE. 2016. Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc Natl Acad Sci U S A 113:3627–3632. doi: 10.1073/pnas.1522510113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wexler AG, Bao Y, Whitney JC, Bobay LM, Xavier JB, Schofield WB, Barry NA, Russell AB, Tran BQ, Goo YA, Goodlett DR, Ochman H, Mougous JD, Goodman AL. 2016. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc Natl Acad Sci U S A 113:3639–3644. doi: 10.1073/pnas.1525637113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hecht AL, Casterline BW, Earley ZM, Goo YA, Goodlett DR, Bubeck Wardenburg J. 2016. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 17:1281–1291. doi: 10.15252/embr.201642282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chatzidaki-Livanis M, Coyne MJ, Comstock LE. 2014. An antimicrobial protein of the gut symbiont Bacteroides fragilis with a MACPF domain of host immune proteins. Mol Microbiol 94:1361–1374. doi: 10.1111/mmi.12839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M, Comstock LE. 2016. Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio 7:e01055-16. doi: 10.1128/mBio.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patrick S, Jobling KL, O’Connor D, Thacker Z, Dryden DT, Blakely GW. 2011. A unique homologue of the eukaryotic protein-modifier ubiquitin present in the bacterium Bacteroides fragilis, a predominant resident of the human gastrointestinal tract. Microbiology 157:3071–3078. doi: 10.1099/mic.0.049940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patrick S, Blakely GW. 2012. Crossing the eukaryote-prokaryote divide: a ubiquitin homolog in the human commensal bacterium Bacteroides fragilis. Mob Genet Elem 2:149–151. doi: 10.4161/mge.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayley DP, Rocha ER, Smith CJ. 2000. Analysis of cepA and other Bacteroides fragilis genes reveals a unique promoter structure. FEMS Microbiol Lett 193:149–154. doi: 10.1111/j.1574-6968.2000.tb09417.x. [DOI] [PubMed] [Google Scholar]
- 21.Podglajen I, Breuil J, Casin I, Collatz E. 1995. Genotypic identification of two groups within the species Bacteroides fragilis by ribotyping and by analysis of PCR-generated fragment patterns and insertion sequence content. J Bacteriol 177:5270–5275. doi: 10.1128/jb.177.18.5270-5275.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutacker M, Valsangiacomo C, Piffaretti JC. 2000. Identification of two genetic groups in Bacteroides fragilis by multilocus enzyme electrophoresis: distribution of antibiotic resistance (cfiA, cepA) and enterotoxin (bft) encoding genes. Microbiology 146:1241–1254. doi: 10.1099/00221287-146-5-1241. [DOI] [PubMed] [Google Scholar]
- 23.Gutacker M, Valsangiacomo C, Bernasconi MV, Piffaretti JC. 2002. recA and glnA sequences separate the Bacteroides fragilis population into two genetic divisions associated with the antibiotic resistance genotypes cepA and cfiA. J Med Microbiol 51:123–130. doi: 10.1099/0022-1317-51-2-123. [DOI] [PubMed] [Google Scholar]
- 24.Kwon YT, Ciechanover A. 2017. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem Sci. doi: 10.1016/j.tibs.2017.09.002. [DOI] [PubMed] [Google Scholar]
- 25.MacGurn JA, Hsu PC, Emr SD. 2012. Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem 81:231–259. doi: 10.1146/annurev-biochem-060210-093619. [DOI] [PubMed] [Google Scholar]
- 26.Gilberto S, Peter M. 2017. Dynamic ubiquitin signaling in cell cycle regulation. J Cell Biol 216:2259–2271. doi: 10.1083/jcb.201703170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YH, Machner MP. 2017. Exploitation of the host cell ubiquitin machinery by microbial effector proteins. J Cell Sci 130:1985–1996. doi: 10.1242/jcs.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fletcher CM, Coyne MJ, Comstock LE. 2011. Theoretical and experimental characterization of the scope of protein O-glycosylation in Bacteroides fragilis. J Biol Chem 286:3219–3226. doi: 10.1074/jbc.M110.194506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chronis D, Chen S, Lu S, Hewezi T, Carpenter SCD, Loria R, Baum TJ, Wang X. 2013. A ubiquitin carboxyl extension protein secreted from a plant-parasitic nematode Globodera rostochiensis is cleaved in planta to promote plant parasitism. Plant J 74:185–196. doi: 10.1111/tpj.12125. [DOI] [PubMed] [Google Scholar]
- 31.Dornas FP, Assis FL, Aherfi S, Arantes T, Abrahão JS, Colson P, La Scola B. 2016. A Brazilian Marseillevirus is the founding member of a lineage in family Marseilleviridae. Viruses 8:76. doi: 10.3390/v8030076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas V, Bertelli C, Collyn F, Casson N, Telenti A, Goesmann A, Croxatto A, Greub G. 2011. Lausannevirus, a giant amoebal virus encoding histone doublets. Environ Microbiol 13:1454–1466. doi: 10.1111/j.1462-2920.2011.02446.x. [DOI] [PubMed] [Google Scholar]
- 33.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 34.La Scola B, Desnues C, Pagnier I, Robert C, Barrassi L, Fournous G, Merchat M, Suzan-Monti M, Forterre P, Koonin E, Raoult D. 2008. The virophage as a unique parasite of the giant Mimivirus. Nature 455:100–104. doi: 10.1038/nature07218. [DOI] [PubMed] [Google Scholar]
- 35.Coyne MJ, Reinap B, Lee MM, Comstock LE. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 307:1778–1781. doi: 10.1126/science.1106469. [DOI] [PubMed] [Google Scholar]
- 36.Comstock LE, Pantosti A, Kasper DL. 2000. Genetic diversity of the capsular polysaccharide C biosynthesis region of Bacteroides fragilis. Infect Immun 68:6182–6188. doi: 10.1128/IAI.68.11.6182-6188.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. 1991. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun 59:2075–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Avelar KE, Pinto LJ, Antunes LC, Lobo LA, Bastos MC, Domingues RM, Ferreira MC. 1999. Production of bacteriocin by Bacteriodes fragilis and partial characterization. Lett Appl Microbiol 29:264–268. doi: 10.1046/j.1365-2672.1999.00603.x. [DOI] [PubMed] [Google Scholar]
- 39.Tang YP, Malamy MH. 2000. Isolation of Bacteroides fragilis mutants with in vivo growth defects by using Tn4400', a modified Tn4400 transposition system, and a new screening method. Infect Immun 68:415–419. doi: 10.1128/IAI.68.1.415-419.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stevens AM, Shoemaker NB, Salyers AA. 1990. The region of a Bacteroides conjugal chromosomal tetracycline resistance element which is responsible for production of plasmidlike forms from unlinked chromosomal DNA might also be involved in transfer of the element. J Bacteriol 172:4271–4279. doi: 10.1128/jb.172.8.4271-4279.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith CJ, Rogers MB, McKee ML. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141–154. doi: 10.1016/0147-619X(92)90014-2. [DOI] [PubMed] [Google Scholar]
- 42.Chatzidaki-Livanis M, Weinacht KG, Comstock LE. 2010. Trans locus inhibitors limit concomitant polysaccharide synthesis in the human gut symbiont Bacteroides fragilis. Proc Natl Acad Sci U S A 107:11976–11980. doi: 10.1073/pnas.1005039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, Langmead B, Salzberg SL. 2015. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T, Juncker AS, Manichanh C, Chen B, Zhang W, Levenez F, Wang J, Xu X, Xiao L, Liang S, Zhang D, Zhang Z, Chen W, Zhao H, Al-Aama JY, Edris S, Yang H, Wang J, Hansen T, Nielsen HB, Brunak S, Kristiansen K, Guarner F, Pedersen O, Doré J, Ehrlich SD, MetaHIT Consortium, Bork P, Wang J. 2014. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 47.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Ability of B. fragilis 638R, 638RΔ1646 mutant, and proteins to inhibit the growth of B. fragilis strains. Download TABLE S1, DOCX file, 0.01 MB (15.6KB, docx) .
Copyright © 2017 Chatzidaki-Livanis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Identification of B. fragilis and ubb, higA, higB, and BF638R_1646 (bsap1) in human gut metagenomes comprising subset 3CGC of the integrated gene catalog (IGC). Download TABLE S2, XLSX file, 0.1 MB (95.8KB, xlsx) .
Copyright © 2017 Chatzidaki-Livanis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Primers used in this study. Download TABLE S3, DOCX file, 0.01 MB (14KB, docx) .
Copyright © 2017 Chatzidaki-Livanis et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.