Abstract
OBJECTIVES
Ovarian cancer leads to abdominal carcinomatosis and late stage (III/IV) diagnosis in 75% of patients. Three randomized phase III trials have demonstrated that intraperitoneal (IP) chemotherapy improves outcomes in epithelial ovarian cancer. While IP treatment is validated by clinical trials, there is a poor understanding of the mechanism(s) leading to the survival advantage other than the increased concentration of cytotoxic drugs within the tumor microenvironment. A better understanding of this process through analysis of dynamic biomarkers should promote novel approaches that may enhance tumor clearance. We propose this pilot study to confirm the feasibility of collecting serial peritoneal samples from implanted catheters in women receiving IP chemotherapy. We believe these specimens may be used for multiplex analysis to reveal unique biomarker fluctuations when compared to peripheral blood.
METHODS
From 13 women participating on GOG 252, 30 whole blood, 12 peritoneal fluid (PF), and 20 peritoneal wash (PW) with 30 mL saline were obtained. Samples were requested prior to the first three chemotherapy cycles. Samples were assessed for volume, cell populations, protein, RNA, and miRNA content changes.
RESULTS
Median volume for PF was 1.6 mL and 3.1 mL for PW. PW is a dilution of PF capable of capturing measurable biomarkers. Peritoneal aspirates contain a unique profile of biomarkers distinct from blood. miRNA undergo earlier alteration with chemotherapy than genes. Flow cytometry does not adequately capture biomarker fluctuations.
CONCLUSIONS
As a proof of principle study, this trial provides evidence that sampling the peritoneal cavity can be adapted for biomarker analysis.
Keywords: Ovarian cancer, IP chemotherapy, immunity, miRNA, IP wash, IP Fluid
INTRODUCTION
Ovarian cancer is the most lethal gynecologic cancer with over 22,000 new cases and 14,000 deaths in the United States per year ranking it fifth in cancer-related deaths for women.1 While the prognosis of ovarian cancer is recognized as poor, intraperitoneal (IP) chemotherapy has demonstrated a survival advantage in several Phase III clinical trials.2–4 Most frequently cited is the Gynecologic Oncology Group (GOG) protocol 172 (GOG-172) which noted an increased median survival of 65.6 months in the IP arm compared to 49.7 months in the intravenous (IV) group (p=0.03; RR 0.75 (0.58–0.97).3 Although proposed as the standard of care for women with optimally-debulked advanced disease, the mechanism(s) by which IP chemotherapy improves outcomes is poorly understood. We hypothesize that local-regional chemotherapy induces favorable modulation of the peritoneal tumor immune microenvironment during tumor involution. Testing this hypothesis requires adequate sampling of the peritoneal cavity. Previous studies from our group and others in women with recurrent ovarian cancer have demonstrated that peritoneal samples may be retrieved from the IP ports and analyzed for cytokines, chemokines, and cell populations.5–8 Despite this evidence, peritoneal sampling has not been actively pursued, particularly in women receiving primary treatment, leading to a disparity that could otherwise provide pertinent information on tumor biology during chemotherapy.
With the growing interest in immunotherapy and combination chemo-immune therapy, there is an urgent need to truly elucidate the molecular-level changes that occur with tumor involution during IP chemotherapy before moving forward with immunomodulating approaches.Given the toxicity to rapidly dividing immune cells, chemotherapeutics are generally thought of as immunosuppressive; however, the severe lymphopenia incurred with other cytotoxic therapies typically is not seen with treatment of ovarian cancer. Thus, as proposed by Lake and Robinson, chemotherapy might have a synergistic role with anti-tumor immunity.9 Nevertheless, these effects are postulated in the context of systemic drug administration and less is known about the local-regional changes post IP chemotherapy.
Though historically delivered via a Tenckhoff catheter, modern day IP chemotherapy is administered through an implanted peritoneal access device, which is sometimes a technically challenging approach depending on the type of device.10 A review of patients enrolled in GOG 172 suggested that ports with fenestrations may be more likely associated with complications such as obstruction, leading to 34% of patients receiving IP chemotherapy discontinuing treatment secondary to catheter-related complications.11 Subsequent patient series analyses have ranged widely in reported complications related to fenestrated catheters with some numbers as low as 3% from institutions with a high volume of IP catheters.12 Single-lumen vascular access devices are an alternative to fenestrated systems.10 A review of patients receiving IP chemotherapy at our institution noted a 6-cycle completion rate of 83% with no difference in discontinuation between single-lumen or fenestrated catheters.13 Unfortunately, insertion and management of the catheter and catheter-associated complications, along with concerns for IP drug toxicity, have limited widespread adoption of IP chemotherapy outside of academic institutions.4,14,15 In addition, it remains unclear which device would better facilitate obtaining IP samples as there have been no side-by-side quantitative and qualitative comparisons of the biological content of peritoneal samples (like peritoneal wash or fluid) to support their rationale and guide their applicability for translational research.
GOG 252 is the most recent phase III trial to evaluate the role of IP as compared to IV chemotherapy. Patients enrolled to GOG 252 and randomized to the IP arm were given the opportunity to participate in this translational protocol pilot study, GOG 271. This pilot study was designed to assess (i) if and how peritoneal samples can be obtained across different medical centers; (ii) to evaluate the utility of such specimens for multi-analyte bioassays commonly used to profile chemo-induced changes; and (iii) to contrast local-regional (PF/PW) with systemic (peripheral blood) molecular profiles.
MATERIALS AND METHODS
Sample Collection and Initial Analyses
This was a multi-institution IRB-approved trial in association with the GOG. Centers included the University of Oklahoma Health Sciences Center (Oklahoma City, OK), Women and Infants Hospital of Rhode Island (Providence, RI), Roswell Park Cancer Institute (Buffalo, NY), and Memorial Health System (Springfield, IL). Patients underwent primary surgery for ovarian, fallopian tube, or primary peritoneal carcinoma (EOC) at their local institution. Suitable candidates were those enrolled on GOG-252 and randomized to IP chemotherapy. This protocol enrolled women to IV paclitaxel 80mg/m2 on day 1, 8 and 15 along with IV carboplatin AUC 6 on day 1 and IV bevacizumab on day 1 or IV paclitaxel 80mg/m2 day 1, 8 and 15 along with IP carboplatin AUC 6 on day 1 and IV bevacizumab on day 1 or IV paclitaxel 135mg/m2 on day 1, IP cisplatin 75mg/m2 on day 2, IP paclitaxel 60mg/m2 on day 8 and IV bevacizumab on day 1. All arms received IV bevacizumab starting cycle 7 through 22. Eligible patients were those with stage II–IV EOC, either optimal or suboptimal residual disease following primary cytoreduction, and hematologic and metabolic parameters within institutional normal limits (NCT00951496). The adjoining translational study, GOG-271, was designed to collect biological specimens from women in the parent GOG-252 trial randomized to IP chemotherapy and enrolled 13 patients. The protocol requested collection of peritoneal fluid (PF) and/or peritoneal wash (PW) samples along with whole blood samples obtained at baseline (T0) and following the first (T1) and second (T2) cycles of chemotherapy. The blood and peritoneal samples were collected on the same day and were received within 24 hours of starting cycle 1, 2, or 3 of the patient’s chemotherapy, respectively. Peritoneal fluid is defined as fluid retrieved from the peritoneal cavity spontaneously during the initial port access. The tubing was cleared with 5 mL of saline prior to aspirating peritoneal fluid to avoid collecting only the fluid settled in the catheter tubing. Peritoneal wash is defined as the fluid obtained by flushing of IP catheter with 30 mL of saline and subsequently withdrawing the fluid. Samples were obtained by a clinician, de-identified, and shipped overnight to Magee-Womens Research Institute of Pittsburgh for processing and subsequent analyses. Upon arrival, the blood samples were centrifuged. The serum and plasma were aliquoted and frozen at −80°C. PBMCs were isolated by Ficoll separations (GE Healthcare Bio-Sciences), counted, and cryopreserved in 1:10 DMSO/FBS freezing medium overnight at −80°C before being transferred to vapor phase liquid nitrogen. IP fluid and wash samples with volumes of 1.5mL or less were used to create cytospins which were stored at −20°C. For samples with volumes greater than 1.5mL, 1.5mL was set aside for cytospins and the remainder of the sample was centrifuged. The supernatants were aliquoted and frozen at −80°C. The IP cells were counted and frozen in 1:10 DMSO/FBS freezing medium overnight at −80°C before being transferred to vapor phase liquid nitrogen. Patients were assigned a sequential study number based on order of sample receipt. Formalin-fixed, paraffin-embedded primary tumor was prepared at the collection sites and then sent to the GOG Tissue Bank.
Schematic of all sample processing and ensuing assays is presented in Supp. Figure 1. Volumes of the blood, PF, and PW samples were recorded. Cell viability assessed via the Trypan Blue exclusion method (Gibco, Grand Island, NY). Protein concentration in PF and PW supernatant samples (volume permitting) was obtained via Coomassie Plus (Bradford) Assay following the manufacturer’s protocol (Thermo Scientific Pierce, Rockford, IL)
Wright Stains
Cytospins were performed on the PF and PW samples using approximately 25,000 cells in 200 μL. The slides were fixed in acetone and dried overnight at room temperature. Staining solution was made of 0.3g Wright stain powder (Acros, New Jersey) dissolved in 100 mL methanol room temperature overnight then filtered through Whatman paper. Sorensen’s buffer (anhydrous KH2PO4 1.326g, anhydrous Na2PO4 0.516g, distilled water to 200mL, pH 6) was used as the buffer solution. Wright stain solution (750 μL) was applied to the cytospin slides for two minutes at room temperature followed by Sorensen’s buffer (1500 μL) for three minutes without decanting. The slides were rinsed in distilled water for 30 seconds then air-dried at room temperature. A coverslip was mounted with Cytoseal 60 (ThermoFisher, Kalamazoo, MI). Slides were read by two authors and in the event of discrepancy by a third author. Immune cell populations were classified as 0 (no cells present), 1+ (<10 cells), 2+ (10–30 cells), or 3+ (>100 cells) per high-power field.
Nanostring
Nanostring expression analyses were performed on miRNA isolated from plasma and PF/PW samples and on RNA samples from peripheral blood mononucleated cells (PBMC).16
miRNA profiling
As per our previously utilized protocols17, plasma or PF/PW samples were thawed and centrifuged to pellet cells out and the protocol was continued on the supernatant using the mirVana PARIS kit (Life Technologies, Carlsbad, CA). An equal volume of denaturing solution was added to the supernatant followed by acid-phenol:chloroform equal to the previous total volume for extraction. After centrifugation, the aqueous phase was transferred to a new microcentrifuge tube. For enrichment of small RNA, 100% room temperature ethanol was added at one-third the volume recovered of the aqueous phase. At this point, the mixture was processed through a series of filters and washed to remove contaminating large RNA. miRNA concentration was measured with a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific, Rockford, IL). The samples were prepared and analyzed with the NanoString nCounter miRNA Expression Assay (NanoString Technologies, Seattle, WA) comprising probes for 800 miRNAs run as per manufacturer’s protocols. Differential expression was identified with the edgeR Bioconductor package.18 The Venny 2.0 software was utilized to create the Venn diagrams.19
Immune gene profiling
Cryopreserved PBMCs were lysed with TRIzol (Life Technologies, Carlsbad, CA). Chloroform was added for phase separation and the aqueous phase containing RNA was transferred to a new tube. RNA was precipitated with isopropyl alcohol then washed with 75% ethanol in diethylpyrocarbonate (DEPC) water. Finally, the RNA was dissolved in RNAse free water and heated at 55 °C for 10 minutes. RNA concentration was measured with the NanoDrop Spectrophotometer and samples were prepared as per NanoString guidelines. The NanoString nCounter GX Human Immunology Kit for analysis of 511 immune genes was utilized. edgeR was used to identify the differentially expressed (DE) immune genes.18 The study has a small sample size and is relatively exploratory, so raw p-values without multiple comparison correction will be used to identify DE genes. Bioinformatics analyses (performed by TM and GT) revealed the top immune pathways most affected by chemotherapy through the use of QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN, Redwood City, CA). In the pathway analysis by IPA, the 511 immune genes were used as the background genome to avoid biased gene selection in the NanoString experiment. Venny 2.0 was used to create the Venn diagram.19
ELISA
Peritoneal fluid was diluted 1:1000 and 1:5000 in Millipore filtered water (EMD Millipore, Darmstadt, Germany). Peritoneal wash aliquots were diluted 1:250 in Millipore filtered water and also run at stock concentration. The samples were run in duplicate on the Human IL-6 Quantikine HS ELISA Kit (R&D Systems, Minneapolis, MN) per the manufacturer’s instructions and read at 490nm and 655nm on the BioRad iMark Microplate Reader using BioRad Microplate Manager Software Version 6.1 (Hercules, CA). After correcting for background, IL-6 concentrations were calculated in Microsoft Excel.
Flow cytometry
PBMCs were thawed, pelleted by centrifugation, and re-suspended in complete RPMI with L-glutamine (Corning, Manassas, VA plus 10% FBS, 1% nonessential amino acids, 1% penicillin/streptomycin, and 2-mercaptoethanol). After blocking for nonspecific staining with Fc block (BD Biosciences, San Jose, CA), cells were stained with fluorescent-labeled antibodies for CD3 (5 μL/sample, Alexa Fluor 700), CD4 (5 μL/sample, BV510), CD11b (5 μL/sample, V421), CD227 (20 μL/sample, FITC), CD274 (20 μL/sample, PE), CD279 (5 μL/sample, PE-CY7) all from BD Biosciences, and CD124 (5 μL/sample, APC) from BioLegend (San Diego, CA) for thirty minutes on ice in the dark. After washing with FACS buffer (BD Biosciences), stained cells were analyzed on a LSR II flow cytometer using the FACSDiva data analysis software (BD Biosciences).
For the intracellular staining protocol, cells first underwent T-cell activation with anti-CD3e/anti-CD28 incubation (BD Biosciences). After one hour incubation at 37°C in a 5% CO2 incubator, BD GolgiStop (Monensin) and BD GolgiPlug (Brefeldin A) were added per company recommendations along with PE-CD107a (LAMP1) (BD Biosciences) for a five hour incubation 37 °C in a 5% CO2 incubator. Cells were washed with FACS buffer then blocked for nonspecific staining with Fc block. Cells were surface stained with 5 μL/sample of each antibody for CD8 (Alexa Fluor 700), CD4 (BV510), and CD127 (BV421) from BD Biosciences and TCR α/β chain (PerCP/5.5), and CD336 (APC) from BioLegend for 30 minutes in the dark on ice then washed with FACS buffer. The FOXP3 Buffer Set from BD Biosciences was used per manufacturer’s recommendations for fixation and permeabilization of cells. This was followed by intracellular staining for IFN-γ (PE-CY7) and FOXP3 (Alexa Fluor 488) (BD Biosciences), 5 μL/sample per antibody. Stained cells were analyzed on a LSR II flow cytometer using the FACSDiva data analysis software.
Immunohistochemistry
Immunohistochemistry (IHC) assays and automated digital pathology analysis for CD3, CD4, and CD8 were performed at the Pathology Resource Network at Roswell Park Cancer Institute (RPCI). Whole slide sections were cut at 4–5µm, placed on charged slides, and dried at 60°C for one hour. Slides were cooled to room temperature, deparaffinized in three changes of xylene, and rehydrated using graded alcohols. For antigen retrieval, slides were heated in a steamer for 40 minutes in Target Retrieval Solution pH=9 (Dako) for the CD3 and CD8 assays, and 60 minutes for the CD4 assay. Slides were allowed to cool for 20 minutes and then endogenous peroxidase was quenched with aqueous 3% H2O2 for 10 minutes and washed with PBS-Tween. Slides were loaded on a DAKO autostainer (Dako, Glostrup, Denmark) and serum free protein block was applied for 5 minutes, blown off, and then the primary antibody was applied (CD3 1:100 for 30 minutes, CD4 1:50 for 60 minutes, CD8 1:75 for 30 minutes, all from Dako). A matched isotype was also applied on a replicate slide instead of primary antibody as a negative control. The EnVision+ horseradish peroxidase system (Dako) and DAB chromogen were used for visualization. Lastly, slides were counterstained with hematoxylin, dehydrated, cleared and cover slipped.
Aperio Slide Scanning and Image Analysis
Slides were digitally scanned using Aperio Scanscope (Aperio Technologies, Inc., Vista, CA) with 20× bright-field microscopy. These images are then accessible using Spectrum (Aperio Technologies, Inc., Vista, CA), a web-based digital pathology information management system. Slide images are automatically associated to a digital slide created in the Digital Slide table in Aperio eSlide Manager. Once slides are scanned, Aperio ImageScope version 11.2.0.780 (Aperio Technologies, Inc., Vista, CA) was used to view images for image analysis. Images were examined for quality and were amended as necessary. An annotation layer was created for each slide. Tumor regions were identified and annotated to appropriately represent the heterogeneity of staining for image analysis.
The Aperio platform was used to develop quantitative image analysis algorithm macros for the quantification of IHC slides. Briefly these algorithms used color de‐convolution to separate diaminobenzidine (DAB) from the haematoxylin counterstain thereby providing stain separation. A cytoplasmic algorithm was tailored to fine tune the lymphocytes detection using cellular, nuclear, and stain parameters, creating an algorithm macro based on the cell compartment location of the target protein. Separate macros were adjusted for each antibody target (CD3, CD4 and CD8) to optimize results. In this case the cytoplasmic algorithm was modified to detect and quantify the positive DAB staining cells (targeted lymphocytes). The results included the total number of positive cells and the area of analysis. Lastly, the number of lymphocytes positive for each antibody was reported per square millimeter using a simple conversion formula in Microsoft Excel.
RESULTS
Peritoneal wash and peritoneal fluid demonstrate high correlation of biological content
This multi-institutional study enrolled a total of n=13 patients at four medical centers, as described in Table 1. The majority of patients presented with stage III disease (75%). All patients had a complete response, as interpreted by a return to normal CA125 levels, other than one patient (8%) whose CA125 plateaued at 70 then increased. Blood samples were obtained more consistently (30 samples) than either PF (12 samples) or PW (20 samples) (Supp. Table 1). However, only 7 of 13 (54%) patients had blood samples from all three time points (T0-prior to chemotherapy, T1-after first cycle, and T2-after second cycle). Of those 7, 4 patients had paired peritoneal samples (PF or PW) from all three time points. At least one PF or PW sample, at one of the three time points, could be obtained in 11 of the 13 patients (84%). Of the 12 times PF was obtained (from 6 different patients), a paired PW from the same time point was obtained in 11 cases. The volume received from the peritoneal samples (PF or PW) varied widely from less than 100 μL to 34.5 mL (Supp. Table 1). Average volume received for PF was 6.9 mL (median 1.6 mL) and 7.5 mL (median 3.1 mL) for PW samples (p=0.85). Average volume received from patients with single-lumen IP catheters was 6.1 mL (median 1.1 mL) versus 12.4 mL (median 11.8 mL) from fenestrated catheters (p=0.16). Cells could be counted in 7 of 12 (58%) of PF samples with a median cell count of 0.9 million/mL (range 0.2–12.8 million/mL) and 11 of 20 (55%) of PW samples with a median cell count of 0.6 million/mL (range 0.2–62.4 million/mL). The remaining samples had inadequately low cell densities.
Table 1.
Patient Demographics
| Patients (N=13) | |
|---|---|
|
| |
| Institution (%) | |
| University of Oklahoma | 8 (62%) |
| Memorial Health System | 1 (8%) |
| Roswell Park | 2 (15 %) |
| Womens and Infants | 2 (15%) |
|
| |
| Median Age (range) | 62 (53–71) |
|
| |
| Stage (%)* | |
| I | 0 (0%) |
| II | 3 (25%) |
| III | 9 (75%) |
| IV | 0 (0 %) |
|
| |
| Pathology (%) | |
| Serous | 9 (69%) |
| Grade 1 | 3 (23%) |
| Grade 2 | 0 |
| Grade 3 | 6 (46%) |
| Clear Cell | 1 (8%) |
| Endometrioid | 3 (23%) |
|
| |
| Ascites at Primary Surgery (%) | |
| Yes | 5 (38%) |
| No | 8 (62%) |
|
| |
| Bowel Resection | |
| Yes | 2 (15%) |
| No | 11 (85%) |
|
| |
| Adhesive Disease | |
| Yes | 7 (54%) |
| No | 6 (46%) |
|
| |
| Extensive Retroperitoneal Dissection | |
| Yes | 5 (38%) |
| No | 8 (62%) |
|
| |
| Chemosensitive Tumor** | |
| Yes | 12 (92%) |
| No | 1 (8%) |
One patient was not fully staged.
CA125 was used as a surrogate to assess tumor response to chemotherapy. All tumors were deemed chemosensitive with a return to normal levels of CA125 other than one patient whose marker plateaued in the 70s then rose to 92.
Unlike the PW, which is retrieved upon exogenously adding saline solution, the PF represents spontaneous accumulation of secreted biological materials. Presence of PF is noted in many but not all patients and may be potentially due to enhanced local inflammatory reaction due to the catheter itself. To explore differences in biological content between PF (capturing reactions to both tumor and catheter) and PW (capturing mostly the tumor environment) in each patient, we compared cell densities and overall incidence of immune cells in paired PF/PW samples. Of the 7 PF samples with cells detected, 6 had paired PW samples. A higher cell density was found in the PF in 5 out of 6 pairs with a median of 0.9 vs 0.4 million cells/mL in PF vs PW, respectively (p=0.28). Due to low cell counts, inadequate for flow cytometry phenotyping, Wright stains were performed on the PF and PW samples to assess immune cell populations. Eight paired PF/PW samples (from 6 different patients, with samples from two time points assessed in patients 7 and 10, Supp. Figure 1) were analyzed. While the initial collected volumes and cell densities varied among patients, our results demonstrate that the incidence of white blood cells (lymphocytes, neutrophils, eosinophils etc) is similar in PF and PW, suggesting that, despite differences in collection method, a comparable cellular composition was found in these two samples types (Supp. Figure 2). Even with low volumes of fluid received and a low cell density, Wright stains can be adequately performed.
To further explore molecular content, we compared acellular supernatant protein concentration in five PF/PW pairs. All PF samples were slightly more concentrated (median 972 μg/mL) than PW samples (median 899 μg/mL) (p=0.09). One PW sample (Patient 1, T0) contained no measurable protein suggesting a collection failure or study error and was thus excluded from the analysis (Figure 1A). To assess whether pro-inflammatory markers are present in the PF or PW samples, we measured IL-6, a cytokine usually found in ovarian tumor microenvironment and ascites fluid.20 ELISA measurements demonstrate a higher concentration of IL-6 in PF versus the PW (Figure 1B) with an average of 5391.5 pg/mL in PF versus 1787.3 pg/mL in PW (p=0.19). Overall, these results from pairwise comparisons of PF and PW suggest that PF has a higher protein concentration and higher cellular density than PW, as would be expected from a dilution effect from the saline.
Figure 1.
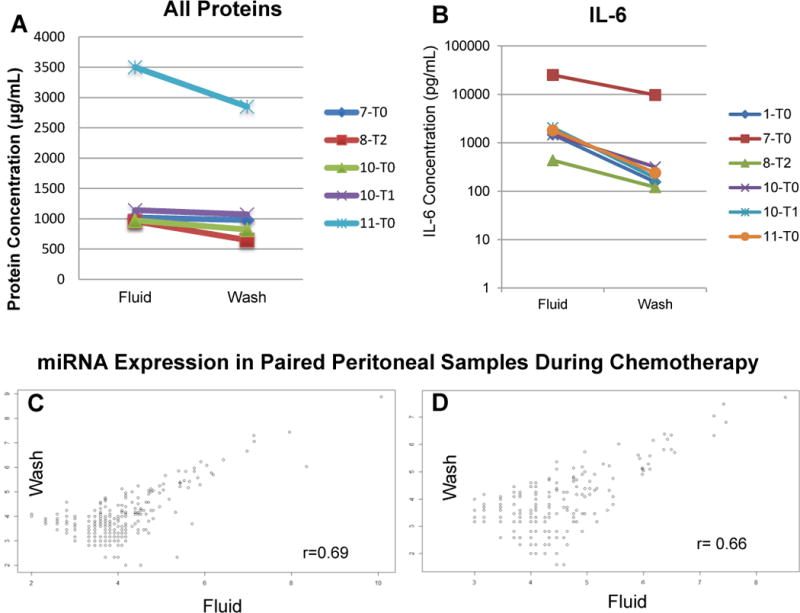
A. Protein concentration was evaluated by Coomassie Plus (Bradford) Assay following the manufacturer’s protocol for paired PF and PW samples. One wash specimen (patient 1, time point 0) had no measurable protein suggesting a collection failure or experiment error. The PF contained a higher protein concentration in each of the paired samples. B. An ELISA for the proinflammatory cytokine IL-6 similarly demonstrated higher levels in the PF versus PW when paired for patient and time point. C and D. Raw counts of miRNA were evaluated for Patient 10 at time point 0 and 1 respectively amongst paired wash and fluid samples. The high correlation between the miRNA suggests that a peritoneal wash captures the same biological information as the spontaneously aspirated fluid.
To test whether the PF and PW capture the same biological data and offer similar molecular profiles of the tumor microenvironment, we measured miRNA in paired PF/PW sets and performed correlation curves of miRNA counts, where a result of 1 would be a perfect correlation. In line with our hypothesis, we observed strong correlation of miRNA counts (n=800) in two paired PF/PW samples, collected from patient 10 at T0 and T1 (Figure 1C and 1D), r=0.69 and 0.66 respectively. These results suggest that PF represents a biological fluid rich in cells and secreted proteins and that PW may represent a more diluted version of PF. Given that spontaneous accumulation of PF may or may not occur, these results indicate that in the absence of PF, the collection of PW, which can be systematically performed, would allow for an assessment of the peritoneal cavity using biomarkers typically used to monitor tumor biology like immune cells, cytokines, and miRNA, among others.
Peritoneal samples and peripheral blood show distinct miRNA expression profiles
To assess if and how peritoneal samples, especially PW, could inform us on molecular changes in local-regional tumor microenvironment during chemotherapy and whether these changes are also captured systemically in patients’ peripheral samples, we profiled miRNAs in plasma (n=9 patients) and PF (n=1 patient) and PW (n=3 patients) at three time points using the NanoString nCounter miRNA Expression Assay (n=800 probes).
In plasma, a total of n=51 miRNAs were DE after the first round of chemo (T0 vs T1) and n=33 miRNAs were DE after the second chemotherapy (T1 vs T2), eight of which were common to both group comparisons (Venn diagram, intersection T0 vs T1 and T1 vs T2, Fig. 2A). Furthermore, once altered, the miRNA tended to remain expressed in the same direction (up or downregulated from baseline) throughout monitoring (Supp. Figure 3).
Figure 2.
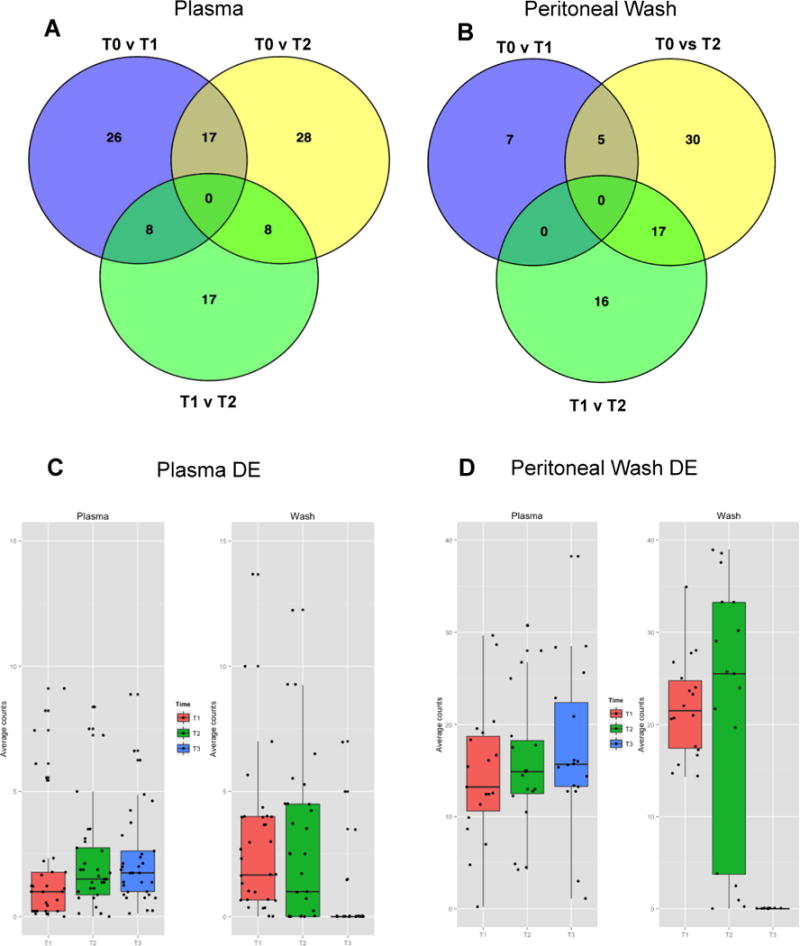
Plasma from n=9 (A) and PW samples from n=3 (B) were used for miRNA extraction at three time points (T0, T1, and T2 as described in Fig. 1). Significantly differentially expressed (DE) miRNA (p<0.05) were identified using the edgeR Bioconductor statistics package. The Venn diagrams show numbers of DE miRNA for all three group comparisons (T0 vs T1, T1 vs T2, and T0 vs T2 respectively). Complete lists of the DE mRNA from plasma and PW amongst two group comparisons are included as Supplemental Figures 2 and 3. More miRNA were found to be DE in plasma samples were an earlier alteration than PW. The 22 differentially expressed miRNA from PW samples and the 33 DE miRNA from plasma (Supplemental Figures 3 and 4) are not only unique to their environment but also display different patterns of expression in the alternate environment. C. The DE PW miRNA undergo significant fluctuation in the wash environment but are relatively stable when they are viewed in the plasma. D. The DE plasma miRNA appear to be altered in the opposite direction when viewed in the wash environment. Individual box plots for the PW and plasma DE miRNA viewed in each environment are included as Supplemental Figures 5–8.
In contrast, the majority of the miRNA that were differentially expressed in PW were altered after the second cycle of chemotherapy (n=12 in T0 v T1 and n = 33 in T1 v T2, Fig. 2B). This suggests that a larger number of plasma miRNAs may undergo changes early during treatment due to systemic chemo-induced effects, whereas changes in PF/PW may occur later and be reflective of more discrete local-regional changes in tumor biology, involving fewer miRNAs.
Strikingly, we observed no overlap between the local (PW) DE miRNAs and circulating (plasma) DE miRNA from the intersections of any of the two group comparisons (T0 vs T1, T1 vs T2 and T0 vs T2, respectively) (Supp. Figure 3 and 4). Furthermore, when assessing the individual miRNA from either the local or plasma samples, we observed little concordance in expression (Figure 2C and 2D). For example most (73%) of the 22 DE miRNA found in at least two group comparisons in the PW samples (i.e. intersection of at least of the two circles in Venn diagram, Figure 2B) seemed to remain unchanged when measured in plasma (Supp. Figure 5 and 6). A similar analysis of the n=33 plasma DE miRNA common to at least two group comparisons revealed that only five (15%) of all miRNAs demonstrated the same pattern of expression in the PW samples (Supp. Figure 7 and 8). Overall, these results demonstrate that changes in miRNAs resident in the peritoneal cavity may not be captured in the systemic circulation and that PF and/or PW are more suitable to accurately capture molecular changes unique to the tumor microenvironment.
Systemic profiling of PBMCs during chemotherapy shows discrete changes in immune genes
Previously considered immune suppressive, chemotherapeutic drugs (including cisplatin) are now increasingly recognized as being immune modulatory by increasing tumor immunogenicity. To explore whether chemo-induced immune modulation, expected to be most prevalent at the tumor site, can be also detected systemically during the course of chemotherapy, we profiled gene expression changes in PBMCs by Nanostring, using probes for n=511 immune genes RNA was extracted from PBMCs (n=7 patients) at baseline (T0) and after two rounds of chemotherapy (T1 and T2, respectively). Comparisons of DE genes between T0/T1, T1/T2 and T0/T2 retrieved 8, 35 and 15 differentially expressed genes respectively (Figure 3A). Twelve differentially expressed immune genes (p<0.05) common to at least two group comparisons were identified although there were no DE genes in a comparison of all three groups (Figure 3A and 3B). The first round of chemotherapy triggered fewer DE genes compared to the second cycle of chemotherapy. Importantly, several of the DE genes are killer cell immunoglobulin-like receptor (KIR) molecules involved in the NK signaling pathway, which was also found to be significantly dysregulated by Ingenuity Pathway Analysis (p=1.23E-03) (Figure 3C).
Figure 3.

NanoString analysis of PBMCs identified 12 differentially expressed (p<0.05) immune genes amongst two time point comparisons. A. After the first cycle of chemotherapy 8 immune genes were differentially expressed versus 35 after the second cycle of chemotherapy. At the circle intersections are the 12 genes shared within two-group comparisons. B. The majority of genes were downregulated during chemotherapy as denoted by the arrow direction. C. Combined IPA analysis of the genes (panel B) identified the top canonical pathways in which these genes are involved. Only one, a natural killer pathway, was significant and included two genes involved in inhibition and one in activation.
Overall, none of the DE immune genes revealed changes in known markers of lymphoid populations (such as CD3, CD4 and CD8) or their activation/suppression capacity (like FOXP3, PD-1, PD-L1). To test whether these markers showed any changes at protein level we performed multicolor flow cytometry on PBMCs (Supp. Figure 9). None of the phenotypic markers (for CD3,CD4,CD8, NK or T cells) or functional markers used (IFNɣ, LAMP-1, CD127) showed significant changes during treatment reinforcing the fact that flow cytometry of PBMCs, featuring commonly used immune markers, does not capture chemo-induced changes in the major immune cell subsets, at least at early time points during treatment.
Immunoscoring by Immunohistochemistry Remains a Useful Tool in Biomarker Analysis
CD3, CD4, and CD8 cell populations were analyzed within primary tumor sections by immunohistochemistry. While correlation analysis showed weaker correlations between CD3:CD4 (R = 0.607) and CD4:CD8 (R = 0.477) the CD3 and CD8 infiltration patterns were highly correlated (R = 0.987) as seen in Figure 4.
Figure 4.
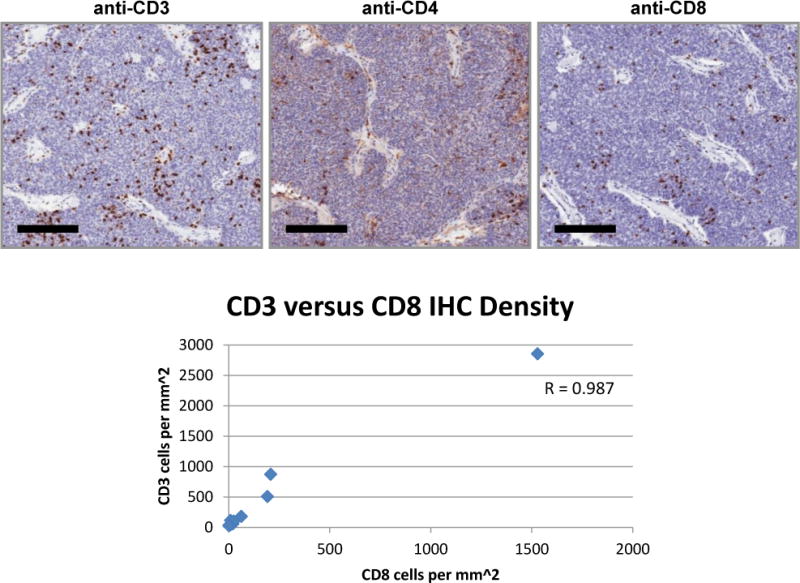
Infiltration pattern of T lymphocytes within tumor. Immunohistochemistry using antibodies specific for CD3, CD4, and CD8 was performed on tumor specimens from all patients enrolled on the trial. Representative IHC staining pattern from one patient. Bars represent 200 μm. CD3 and CD8 cell density was highly correlated with R = 0.987.
DISCUSSION
Ovarian cancer has a particularly insidious course with the majority of patients being diagnosed in late stages making the disease course difficult to study.21 In most cases, the tumors are chemo-sensitive and respond to platinum/taxane combination. Notably however, there is little knowledge of changes in the tumor microenvironment particularly in the process of tumor involution during serial chemotherapy treatment.22,23 This is partly due to difficulty in sampling the local-regional environment, given that the tumors develop in the peritoneal cavity. However, with intraperitoneal chemotherapy recommended as the standard of care for optimally-debulked patients, IP catheters provide an avenue to obtain potentially useful biomarkers.5–8 Furthermore, with the advent of personalized medicine, we postulate that IP ports can be used to retrieve clinical specimens that could be used to identify therapeutic targets, assess response to therapy, or make prognostic predictions.
This is the first study that reports results from standardized collection of peritoneal samples and clearly identifies utility of PF and PW samples. We demonstrate that although peritoneal samples may be of small volume, they typically contain cells and biomarkers sufficient for analysis. Additionally, as peritoneal samples may be difficult to obtain as tumors involute and ascites recedes with increasing cycles of chemotherapy, we have demonstrated that a peritoneal wash is an alternative collection sample. Findings from protein concentration analysis, cell counts, and miRNA assays demonstrate that a peritoneal wash can be considered a diluted sample of peritoneal fluid. Given that PF (a spontaneous fluid accumulation) may only be collected from a subset of patients, implementation of a standardized method of obtaining a PW can provide serial samples from patients in whom peritoneal fluid cannot be aspirated. Fenestrated IP catheters returned an average of double the volume versus single lumen catheters (12.4 mL versus 6.1 mL). While previous reports expressed concern for a higher incidence of complications related to fenestrated catheters11, more recent reviews contradict those findings.12,13 Although any implanted catheter poses potential risks for infection, adhesions, failure, or other complications, strictly with sample collection in mind our findings suggest it may be more advantageous to consider a fenestrated catheter in terms of higher volume returned. Only three patients had fenestrated catheters in this series. Two of those patients had no PF and the PW was unable to retrieve measurable cells, thus no conclusions can be drawn about the biological material retrieved via fenestrated catheters versus single-lumen.
NanoString results demonstrated that miRNA expression may be more reflective of early tumor involution with later PBMC gene expression occurring more slowly during the course of chemotherapy. While interpretation of the gene expression and pathways is outside the scope of this article and limited by the sample size, alterations in the natural killer pathway are an interesting finding and may provide information on the interplay between chemotherapy and the immune system in future studies. Further analysis of these changes in follow-up studies could identify biomarkers associated with tumor response or lack thereof. These differences in expression could be important targets as the concept of personalized medicine continues to evolve. Additionally, NanoString offers a wider array of biomarker testing as opposed to flow cytometry, where only a small set can be tested at one time, and is able to identify more discreet alterations as opposed to only phenotypic changes. Most striking is that we were able to demonstrate the difference in miRNA expression between the local tumor microenvironment and peripheral samples emphasizing the need for further exploration and evaluation of peritoneal samples.
The large focus of this manuscript is to provide evidence and encourage widespread adoption of IP sampling as a noninvasive tool to collect fluid and cellular content for biomarker analysis. That being said, the best conclusions from such an analysis would be made from expanding on knowledge and literature currently available. Thus, we would continue to encourage tissue collection at debulking and any subsequent procedures as done in this series. Of course, as demonstrated in this series, even under a strict protocol, samples are frequently not obtained. We would encourage institutions to consider a system-wide movement towards tissue and sample banking with dedicated staff to help ameliorate some of the barriers.
This study is mainly limited by its small sample size. While identification of conclusive changes in the tumor microenvironment during chemotherapy could be potentially identified through similar approaches, larger studies using a combination of systemic and local-regional samples are needed. Most significantly, this pilot study demonstrates that it is feasible to collect serial paired blood and intraperitoneal samples for evaluation via multiple platforms to explore tumor involution during chemotherapy and confirms that the local environment is distinctly unique and should be evaluated. Understanding tumor involution provides opportunities for new combination therapies during or immediately following chemotherapy that could increase progression-free and overall survival.
Supplementary Material
Supplemental Figure 1: Tumor tissue was preserved in formalin-fixed paraffin-embedded blocks from initial surgery for immunohistochemistry. Peripheral blood and peritoneal samples were requested at baseline and following the first and second cycles of chemotherapy. If peritoneal fluid (PF) could not be obtained spontaneously, a peritoneal wash (PW) with infusion of 30 mL normal saline was performed. Multiplex processing via NanoString (miRNA and immune gene expression profiles), ELISA (IL-6), Bradford assays, and multi-color flow cytometry were performed as described in Materials and Methods.
Supplemental Figure 2: Wright stains were performed on cytospins of peritoneal fluids and washes. Scoring was performed by two independent reviewers with the following scale: 0 for no stained cells, 1+ for 10 stained cells or less, 2+ for 10 to 30 stained cells, 3+ for 100 or more stained cells per high power field. Panel A demonstrates representative staining from Patient 10, time point 2 (scored as 3+), Peritoneal Fluid while Panel B is the corresponding Peritoneal Wash.
Supplemental Figure 3: 33 DE miRNA were identified between two group comparisons during analysis of miRNA extracted from plasma (Figure 2A). The majority (n=25) were altered after the first cycle of chemotherapy and remained expressed in the same direction throughout monitoring as denoted by the arrow direction.
Supplemental Figure 4. Analysis of miRNA extracted from PW samples (Figure 2B) noted 22 DE miRNA. In this environment, alterations typically occurred later than in plasma with 17 miRNA undergoing expression after the second cycle of chemotherapy.
Supplemental Figure 5. When observing the 22 DE PW miRNA in the plasma environment, the majority of the miRNA remain relatively stable during observation when compared to the fluctuations seen in Supplemental Figure 6.
Supplemental Figure 6. Individual boxplots of the 22 DE PW miRNA demonstrate the late alterations after the second cycle of chemotherapy. Conversely, these miRNA have relatively stable expression in the systemic environment (Supplemental Figure 5).
Supplemental Figure 7. Of the 33 DE miRNA from plasma, 25 underwent alteration after the first cycle of chemotherapy as demonstrated here in individual boxplots. These miRNA often had a pattern of expression in the opposite direction when viewed in the local environment (Supplemental Figure 8). Only five (see *) share the same pattern of expression.
Supplemental Figure 8. In addition to being uniquely expressed in the systemic environment, the 33 DE miRNA identified in plasma appear to be expressed in an opposite direction here in the wash samples when compared to the plasma samples (Supplemental Figure 7). Only five (see *) share the same pattern of expression.
Supplemental Figure 9: While NanoString analysis (Figure 3) was able to demonstrate alterations in genes during the course of chemotherapy, phenotypically the cells were not significantly different. Included here are two representative analyses by flow cytometry for CD4 (left) and CD8 (right) as well as a t Test analysis for various immune markers noting no significant fluctuations between time points 1 and 2.
Supplemental Table 1: Patients were assigned a study number 1–13 at enrollment. Histology included serous (S), endometrioid (E), and clear cell (C) disease. One patient did not have full staging (ND) but otherwise the majority of patients were Stage III. The catheters placed (SL= single-lumen or F=fenestrated) were based on institution and physician preference. 20 PW and 12 PF samples were obtained with the volume measured upon receipt recorded. Cell density (cells/mL) was evaluated by Trypan Blue exclusion counting. 30 blood samples were received as denoted by the X.
Acknowledgments
Dr. Shannon Grabosch received grants from the National Institutes of Health to the Gynecologic Oncology Group/NRG Oncology grants U10CA180822 and U1-CA180868, as well as National Institutes of Health Scaife Foundation, University of Pittsburgh School of Medicine Grant #P50 CA159981.
Dr. Robert Edwards receives money to his institution for P50 SPORE Grant which is shared with RPCI.
Dr. Kathleen Moore receives money for grant 1P50 CA159981 from the National Institute of Health and the Scaife Foundation at the University of Pittsburgh who funded the research – not paid to her institution. She also receives money to her institution for consultancy from Astra Zeneca (advisory board), Clovis (advisory board, steering committee), Immunogen (advisory board), Tesaro (steering committee), VBL Therapeutics (advisory boards) and Genentech Roche (advisory boards). She also received payment for development of educational presentations to her institution from Clovis and travel/accommodations/meeting expenses unrelated to activities listed paid to her institution from Onco Med.
Dr. Kunle Odunsi received money from the NIH for grant P50 CA159981 and a grant The Scaife Foundation at the University of Pittsburgh School of Medicine.
Dr. Mary Strange received grant funding to her institution for grant 1 P50 CA159981 from the NIH and also the Scaife Foundation at the University of Pittsburgh School of Medicine.
Dr. Amit Lugade received funding to his institute from NIH Grant P50 CA159981 and the Scaife Foundation at the University of Pittsburgh School of Medicine.
This study was supported by National Cancer Institute grants to the Gynecologic Oncology Group Administrative Office (CA 27469), the Gynecologic Oncology Group Statistical and Data Center (CA 37517), NRG Oncology 1 U10CA180822, NRG grant U1-CA180868, National Institutes of Health 1 P50 CA159981, and the Scaife Foundation at the University of Pittsburgh School of Medicine. The following Gynecologic Oncology institutions participated in this study: University of Oklahoma Health Sciences Center, Oklahoma City, OK, Roswell Park Cancer Institute, Buffalo, NY, Women and Infants Hospital, Providence, RI, and Memorial Medical Center, Springfield, IL.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Alberts DS, et al. Intraperitoneal cisplatin plus intravenous cyclophosphamide versus intravenous cisplatin plus intravenous cyclophosphamide for stage III ovarian cancer. N Engl J Med. 1996;335(26):1950–5. doi: 10.1056/NEJM199612263352603. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong DK, et al. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354(1):34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 4.Markman M, et al. Phase III trial of standard-dose intravenous cisplatin plus paclitaxel versus moderately high-dose carboplatin followed by intravenous paclitaxel and intraperitoneal cisplatin in small-volume stage III ovarian carcinoma: an intergroup study of the Gynecologic Oncology Group, Southwestern Oncology Group, and Eastern Cooperative Oncology Group. J Clin Oncol. 2001;19(4):1001–7. doi: 10.1200/JCO.2001.19.4.1001. [DOI] [PubMed] [Google Scholar]
- 5.Allavena P, et al. Intraperitoneal recombinant gamma-interferon in patients with recurrent ascitic ovarian carcinoma: modulation of cytotoxicity and cytokine production in tumor-associated effectors and of major histocompatibility antigen expression on tumor cells. Cancer Res. 1990;50(22):7318–23. [PubMed] [Google Scholar]
- 6.Francis P, et al. Phase I feasibility and pharmacologic study of weekly intraperitoneal paclitaxel: a Gynecologic Oncology Group pilot Study. J Clin Oncol. 1995;13(12):2961–7. doi: 10.1200/JCO.1995.13.12.2961. [DOI] [PubMed] [Google Scholar]
- 7.Lenzi R, et al. Phase II study of intraperitoneal recombinant interleukin-12 (rhIL-12) in patients with peritoneal carcinomatosis (residual disease < 1 cm) associated with ovarian cancer or primary peritoneal carcinoma. J Transl Med. 2007;5:66. doi: 10.1186/1479-5876-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vlad AM, et al. A phase II trial of intraperitoneal interleukin-2 in patients with platinum-resistant or platinum-refractory ovarian cancer. Cancer Immunol Immunother. 59(2):293–301. doi: 10.1007/s00262-009-0750-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lake RA, Robinson BW. Immunotherapy and chemotherapy–a practical partnership. Nat Rev Cancer. 2005;5(5):397–405. doi: 10.1038/nrc1613. [DOI] [PubMed] [Google Scholar]
- 10.Helm CW. Ports and complications for intraperitoneal chemotherapy delivery. BJOG. 119(2):150–9. doi: 10.1111/j.1471-0528.2011.03179.x. [DOI] [PubMed] [Google Scholar]
- 11.Walker JL, et al. Intraperitoneal catheter outcomes in a phase III trial of intravenous versus intraperitoneal chemotherapy in optimal stage III ovarian and primary peritoneal cancer: a Gynecologic Oncology Group Study. Gynecol Oncol. 2006;100(1):27–32. doi: 10.1016/j.ygyno.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 12.Black D, et al. Low risk of complications associated with the fenestrated peritoneal catheter used for intraperitoneal chemotherapy in ovarian cancer. Gynecol Oncol. 2008;109(1):39–42. doi: 10.1016/j.ygyno.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 13.Lesnock JL, et al. Completion of intraperitoneal chemotherapy in advanced ovarian cancer and catheter-related complications. Gynecol Oncol. 116(3):345–50. doi: 10.1016/j.ygyno.2009.11.009. [DOI] [PubMed] [Google Scholar]
- 14.Della Pepa C, et al. Ovarian cancer standard of care: are there real alternatives? Chin J Cancer. 34(1):17–27. doi: 10.5732/cjc.014.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tewari D, et al. Long-Term Survival Advantage and Prognostic Factors Associated With Intraperitoneal Chemotherapy Treatment in Advanced Ovarian Cancer: A Gynecologic Oncology Group Study. J Clin Oncol. doi: 10.1200/JCO.2014.55.9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.nCounter Applications and Solutions. 2017 [cited 2017 April 3]; Available from: http://www.nanostring.com/applications/
- 17.Suryawanshi S, et al. Plasma microRNAs as novel biomarkers for endometriosis and endometriosis-associated ovarian cancer. Clin Cancer Res. 19(5):1213–24. doi: 10.1158/1078-0432.CCR-12-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26(1):139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oliveros JC. Venny. An interactive tool for comparing lists with Venn’s diagrams. 2007–2015 [Google Scholar]
- 20.Lane D, et al. Prognostic significance of IL-6 and IL-8 ascites levels in ovarian cancer patients. BMC Cancer. 11:210. doi: 10.1186/1471-2407-11-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351(24):2519–29. doi: 10.1056/NEJMra041842. [DOI] [PubMed] [Google Scholar]
- 22.McCluggage WG, et al. Morphological effects of chemotherapy on ovarian carcinoma. J Clin Pathol. 2002;55(1):27–31. doi: 10.1136/jcp.55.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Musrap N, Diamandis EP. Revisiting the complexity of the ovarian cancer microenvironment–clinical implications for treatment strategies. Mol Cancer Res. 10(10):1254–64. doi: 10.1158/1541-7786.MCR-12-0353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Tumor tissue was preserved in formalin-fixed paraffin-embedded blocks from initial surgery for immunohistochemistry. Peripheral blood and peritoneal samples were requested at baseline and following the first and second cycles of chemotherapy. If peritoneal fluid (PF) could not be obtained spontaneously, a peritoneal wash (PW) with infusion of 30 mL normal saline was performed. Multiplex processing via NanoString (miRNA and immune gene expression profiles), ELISA (IL-6), Bradford assays, and multi-color flow cytometry were performed as described in Materials and Methods.
Supplemental Figure 2: Wright stains were performed on cytospins of peritoneal fluids and washes. Scoring was performed by two independent reviewers with the following scale: 0 for no stained cells, 1+ for 10 stained cells or less, 2+ for 10 to 30 stained cells, 3+ for 100 or more stained cells per high power field. Panel A demonstrates representative staining from Patient 10, time point 2 (scored as 3+), Peritoneal Fluid while Panel B is the corresponding Peritoneal Wash.
Supplemental Figure 3: 33 DE miRNA were identified between two group comparisons during analysis of miRNA extracted from plasma (Figure 2A). The majority (n=25) were altered after the first cycle of chemotherapy and remained expressed in the same direction throughout monitoring as denoted by the arrow direction.
Supplemental Figure 4. Analysis of miRNA extracted from PW samples (Figure 2B) noted 22 DE miRNA. In this environment, alterations typically occurred later than in plasma with 17 miRNA undergoing expression after the second cycle of chemotherapy.
Supplemental Figure 5. When observing the 22 DE PW miRNA in the plasma environment, the majority of the miRNA remain relatively stable during observation when compared to the fluctuations seen in Supplemental Figure 6.
Supplemental Figure 6. Individual boxplots of the 22 DE PW miRNA demonstrate the late alterations after the second cycle of chemotherapy. Conversely, these miRNA have relatively stable expression in the systemic environment (Supplemental Figure 5).
Supplemental Figure 7. Of the 33 DE miRNA from plasma, 25 underwent alteration after the first cycle of chemotherapy as demonstrated here in individual boxplots. These miRNA often had a pattern of expression in the opposite direction when viewed in the local environment (Supplemental Figure 8). Only five (see *) share the same pattern of expression.
Supplemental Figure 8. In addition to being uniquely expressed in the systemic environment, the 33 DE miRNA identified in plasma appear to be expressed in an opposite direction here in the wash samples when compared to the plasma samples (Supplemental Figure 7). Only five (see *) share the same pattern of expression.
Supplemental Figure 9: While NanoString analysis (Figure 3) was able to demonstrate alterations in genes during the course of chemotherapy, phenotypically the cells were not significantly different. Included here are two representative analyses by flow cytometry for CD4 (left) and CD8 (right) as well as a t Test analysis for various immune markers noting no significant fluctuations between time points 1 and 2.
Supplemental Table 1: Patients were assigned a study number 1–13 at enrollment. Histology included serous (S), endometrioid (E), and clear cell (C) disease. One patient did not have full staging (ND) but otherwise the majority of patients were Stage III. The catheters placed (SL= single-lumen or F=fenestrated) were based on institution and physician preference. 20 PW and 12 PF samples were obtained with the volume measured upon receipt recorded. Cell density (cells/mL) was evaluated by Trypan Blue exclusion counting. 30 blood samples were received as denoted by the X.


