Abstract
Venomous animals use venom, a complex biofluid composed of unique mixtures of proteins and peptides, to act on vital systems of the prey or predator. In bees, venom is solely used for defense against predators. However, the venom composition of bumble bees (Bombus sp.) is largely unknown. The Thoracobombus subgenus of Bombus sp. is a diverse subgenus represented by 14 members across Turkey. In this study, we sought out to proteomically characterize the venom of five Thoracobombus species by using bottom-up proteomic techniques. We have obtained two-dimensional polyacrylamide gel (2D-PAGE) images of each species’ venom sample. We have subsequently identified the protein spots by using matrix assisted laser desorption ionization/time-of-flight mass spectrometry (MALDI-TOF MS). We have identified 47 proteins for Bombus humilis, 32 for B. pascuorum, 60 for B. ruderarius, 39 for B. sylvarum, and 35 for B. zonatus. Moreover, we illustrated that intensities of 2DE protein spots corresponding to putative venom toxins vary in a species-specific manner. Our analyses provide the primary proteomic characterization of five bumble bee species’ venom composition.
Keywords: bumble bees, toxins, MALDI-TOF MS, proteomics, 2D-PAGE, venom
1. Introduction
In venomous animals, the use of venom or poison is one of the most essential mechanisms for either capturing prey or defending oneself against predators. In competing for resources, venom is an adaptive trait and an example of convergent evolution. It is a complex biofluid secreted by the venom gland and is composed of a unique mixture of proteins, enzymes, peptides, and biogenic amines that act on vital systems of the recipient [1]. In real bees (Apoidea), the largest family of bees with over 20,000 members [2], venom is solely used for defense against predators [3].
Among Apoidea, the majority of venom studies are focused on the honey bee Apis mellifera (A. mellifera). Bombus sp., which also belongs in the same family as A. mellifera, are pollinators for various native and cultivated plants [4]. The venom composition of Bombus sp. is poorly understood.
Previous studies on Bombus sp. venom have mainly focused on characterization and isolation of highly abundant single molecules using bio-assays and chemical sequencing via Edman degradation [5,6,7,8,9,10,11,12,13,14,15,16]. Among the described peptides, bombolitins are unique to Bombus sp. and enhance phospholipase A2 in liposome hydrolysis, in a similar fashion to melittin found in A. mellifera venom and crabrolin and mastoparan found in vespid Vespa crabro venom [17]. Phospholipase A2 [18,19,20], serine proteases [6,8,10,11,13,20], serine protease inhibitors [14,15], acid phosphatases [18,19,20], arginine kinase [21], hyaluronidase [19], putrescine [22], citrate [23], defensin [24], and acethylcoline [25] have also been isolated from Bombus sp. venom.
Recently, the whole genome sequencing of European large earth bumblebees B. (Bombus) terrestris and B. impatiens have become publicly accessible [26]. The accessibility to the genome sequence, combined with the use of mass spectrometry, paved the way for deeply analyzing the venom proteome composition of B. terrestris [20]. This study has identified 55 new molecules in the B. terrestris venom. Moreover, 72% of identified molecules had homologs in A. mellifera venom, indicating that while there are some species-specific differences, these two bee species share major defense mechanisms. B. terrestris is the only member of the Bombus sp. whose venom proteome composition has been characterized.
In bumble bees, color patterns show similarity among species or are highly variable within species. Therefore, classification based on coat color can be unreliable [27]. This is especially true for the Thoracobombus subgenus, which consists of 14 species in Turkey that are morphologically difficult to separate. To overcome this, we have previously used landmark-based geometric morphometrics on their wing structure [28]. Other approaches, such as the use of pheromone analysis [29,30,31,32] and DNA-based molecular techniques [33,34,35,36,37,38,39] have also been used in differentiating bumble bee species.
Turkey is one of the countries with the highest number and diversity of bumble bee species in the in the West-Palearctic region [28]. The main aim of this study is to determine and compare the venom protein profiles of different Thoracobombus species in Turkey using bottom-up proteomic strategies. With the adoption of these approaches, not only will we characterize the venom proteome of the Thoracobombus species for the first time, but we will also highlight the proteomic differences within the subgenus that may help in differentiating between the species.
2. Results
2.1. 2D-PAGE Analysis of Thoracobombus Venom
The 2DE gel images were obtained from each venom sample (Figure S1).
In the Thoracobombus match set, 87 protein spots were manually matched in 2DE images of B. humilis, B. pascuorum, B. sylvarum, B. ruderarius, and B. zonatus venom. Spot quantities were calculated and significant spots were detected statistically. Among these protein spots, 21 spots were present in all species. Common and differentially expressed protein spots are summarized (Table S2).
2.2. Mass-Spectrometry of Thoracobombus Venom
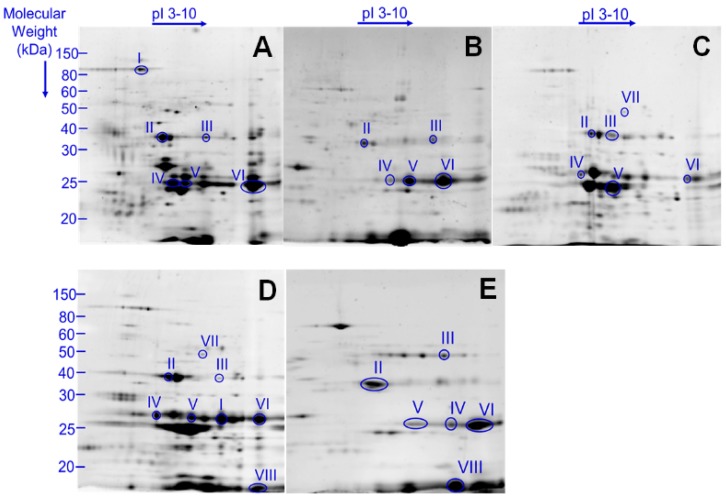
Using 2DE gel images, we have spectrometrically characterized 47 proteins for B. humilis, 32 for B. pascuorum, 60 for B. ruderarius, 39 for B. sylvarum, and 35 for B. zonatus (Figure S1 and Tables S3–S7). Putative toxins are categorized according to their function and subcellular location [20] (Figure 1). Toxins previously identified in B. terrestris and A. mellifera venoms are used as references (Table 1) [20,40]. Major putative toxins phospholipase A2, venom protease, venom acid phosphatase, and hyaluronidase were detected in venom of all five analyzed Thoracobombus species. Arginine kinase, a known component of spider and parasitoid wasp venom, was also identified in all studied species. In contrast, serine protease snake (B. ruderarius and B. sylvarum), peptidases (B. humilis and B. sylvarum), and kunitz-type serine protease inhibitor (B. humilis and B. sylvarum) were only detected in certain species.
Figure 1.
2DE gel images of Thoracobombus venom. Representative images of venom profile of (A) B. humilis; (B) B. pascuorum; (C) B. ruderarius; (D) B. sylvarum; (E) B. zonatus. Gel analyses were conducted in triplicates. Putative venom toxins marked with roman numerals: I: Peptidase I *, Aminopeptidase N **; II: Venom acid phosphatase Acph1-like; III: Hyaluronidase 1; IV: Venom protease; V: Arginine Kinase; VI: Phospholipase A2; VII: Serine protease snake; VIII: Kunitz-type serine protease inhibitor. (See Table 1).
Table 1.
Putative toxins identified in Thorocabombus venom. Toxin names and the corresponding 2D gel electrophoresis (2DE) gel spots are indicated. The presence of the identified toxin (and/or its homologs/ortholog) in B. terrestris and A. mellifera venom is presented.
| Putative Toxin Family | Spot | Toxin Name | B. terrestris | A. mellifera |
|---|---|---|---|---|
| Peptidase/protease | I | Peptidase I *, Aminopeptidase N ** | Dipeptidyl peptidase IV, Serine carboxypeptidase, Prolylcarboxypeptidase |
Dipeptidyl peptidase IV, Serine carboxypeptidase, Prolylcarboxypeptidase |
| IV | Venom protease | |||
| VII | Serine protease snake | Serine protease 1–6 CUB serine protease | Serine protease snake, CLIP serine protease, CUB serine protease | |
| Esterase | II | Venom acid phosphatase Acph1-like | Acid phosphatase | Acid phosphatase 1 |
| V | Arginine Kinase | |||
| VI | Phospholipase A2 | Phospholipase A2-1, Phospholipase A2-2 |
Phospholipase A2-1, Phospholipase A2-2 |
|
| Carbohdyrate metabolism | III | Hyaluronidase 1 | Hyaluronidase | Hyaluronidase |
| Serine protease inhibitor | VIII | Kunitz-type serine protease inhibitor | Serpin 1–3 | Serpin 1–2 |
* B. sylvarum, ** B. humilis.
2.3. Species Specific Expression of Putative Venom Toxins
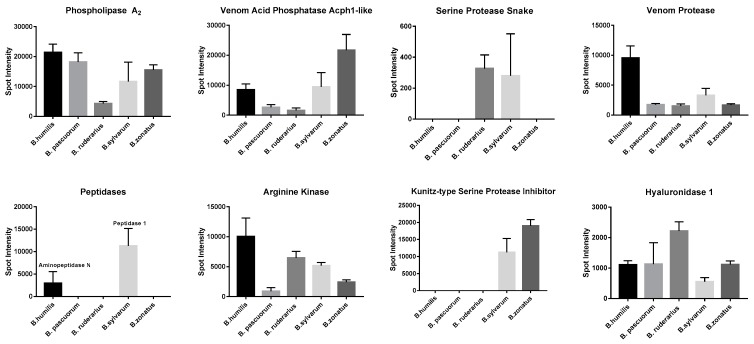
Next, we compared the intensities of 2DE gel spots corresponding to putative toxins in each sample. Several toxins displayed species-specific expression patterns (Figure 2). The expression of phospholipase A2, was the lowest in B. ruderarius venom among the studied species. B. zonatus venom displayed the highest venom acid phosphatase expression. B. pascuorum and B. ruderarius were characterized by a relatively low venom acid phosphatase expression. B. humilis and B. pascuorum displayed the most and the least abundant arginine kinase profile, respectively. B. humilis displayed the highest venom protease expression. Hyaluronidase 1 expression was more than two folds higher in B. ruderarius venom, compared to the other species.
Figure 2.
Relative 2DE gel spot intensities of putative toxins in Thoracobombus venom.
Intensities, as calculated by PDQuest Software, are presented as mean ± standard deviation (3 replicates).
3. Discussion
Identification of the Thoracobombus subgenus represents a challenge as its members are morphologically similar. In our previous study, we had applied landmark-based geometric morphometrics to distinguish its members from each other more accurately. In the current study, we provide the primary proteomic characterization of the venom profile of five bumble bee species belonging to this subgenus. Several putative venom toxins are detected in Thoracobombus which are in accordance with B. terrestris and A. mellifera venom. Furthermore, we outline the species-specific expression patterns of putative toxins.
The defensive behavior of social groups of Hymenoptera is similar to each other. Venom is used for protecting the colonies rather than for capturing prey. Therefore, the venom compositions of these groups are expected to have common overall pattern. It is illustrated that the venom profile of B. terrestris and A. mellifera largely overlap, while also retaining certain species-specific differences.
Accordingly, in Thoracobombus venom, we have identified putative venom toxins collectively used by several Hymenoptera species to debilitate predators such as phospholipase A2, arginine kinase, acid phosphatase (venom acid phosphatase Acph-1), serine protease (serine protease snake), peptidases (peptidase 1 and aminopeptidase N), hyaluronidase (hyaluronidase 1), and serine protease inhibitors (kunitz-type serine protease inhibitor).
While the Thoracobombus venom composition may be similar to other Hymenoptera species in terms of major toxins identified, it is important to note that the venom may be tailored for the individual species’ needs with regards to abundancies of protein families present. Accordingly, a change in the protein family profile of snake venom has been described upon nutritional shift [41].
Here, we have compared the 2DE gel images obtained from different Thoracobombus species and highlighted the species-specific expression differences of venom proteins. In particular, our results indicate that expression levels of putative toxins may be used in differentiating between species. Within the Thoracobombus subgenus, B. humilis is morphologically very similar to B. pascuorum, while B. ruderarius and B. sylvarum resemble each other in terms of morphology and coat color. On the other hand, B. zonatus with its characteristic yellow coat color is easily distinguished from others. We have collected B. ruderarius and B. sylvarum at the highest altitudes (>2000 m) and B. zonatus at the lowest (~1000 m). Interestingly, we have found that morphologically similar species differentiate in their expression of putative venom toxins, even in a similar habitat.
In future studies, different populations of same species can be used to assess the direct effect of altitude, vegetation, and prey population on the venom composition of bees. One of the main limitations of this study is the lack of genome sequences of the studied species, which hinders the database matching for analyzed samples. In this study we utilized 2DE gel images and MALDI-TOF mass spectrometry for assessing the expression levels of putative toxins, which may have its disadvantages. For example, we could not detect bombolitin, unique bumble bee peptides, in our samples. Bombolitins are very small peptides (10 kilo Daltons), and we may have failed to detect bombolitins and other very small peptides with our equipment. More sensitive quantitative mass-spectrometry approaches, such as liquid chromatography (LC-MS/MS), fragmentation- and label-free quantitation of the corresponding protein may be necessary to increase the sensitivity of the study and confirm the correlation between the microenvironment and the venom profile of bumble bees.
Profiling bumble bee venom can have pharmaceutical benefits. Venom peptides can be orally active and transported through the blood–brain barrier and through mammalian cell membranes. Commercial venom-derived drugs are available for the treatment of major diseases such as diabetes, stroke, and hypertension [42]. Bumble bee venom may have the potential to serve as an alternative source of these drugs, which are mostly snake venom-derived. Understanding the species-specific expression patterns of toxins can also benefit the specific-compound-oriented pharmacological analyses.
In conclusion, we provide the primary proteomic characterization of a diverse bumble bee subgenus and illustrate species-specific differences in toxin expression.
4. Methods
4.1. Venom Extraction
B. humilis (Illiger), B. pascuorum (Scopoli), B. ruderarius (Müller), B. sylvarum (Linnaeus), and B. zonatus (Smith) were collected from the Ankara, Bolu, Çankırı, and Kayseri provinces from the Middle Anatolian Region during June, July, August, and September in 2013 and 2014 (Table S1), with permission from the Republic of Turkey Ministry of Forestry and Water Affairs Directorate of Nature Conservation & National Parks and the Republic of Turkey Ministry of Food, Agriculture & Livestock. Live specimens were transported to the Hacettepe University Department of Biology for venom extraction. For venom extraction, the venom reservoir was removed from the stinger in a sterile environment with forceps and the membrane was disrupted afterwards. From each individual, this method yielded 2–5 µL of venom, which was collected in a cryotube and transported in liquid nitrogen. Venom samples belonging to the same species were pooled and stored at −80 °C until use.
4.2. Proteomic Analysis
4.2.1. Protein Quantification and Two-Dimensional Gel Electrophoresis
Venom of the aforementioned bumble bee species was collected from 20 individuals per species. Protein concentrations of the venom samples were obtained by using the Bradford assay [43]. Venom was dissolved in 20 µL rehydration buffer [7 M urea, 2 M thiourea, 4% CHAPS (w/v), 1% ampholytes (pH 3–10), 10 mM DTT, and trace amount of bromophenol blue]. Two-dimensional polyacrylamide gel (2D-PAGE) was performed in three technical replicates as previously described [44] and a total of 50 µg of protein was loaded per gel. For minimizing the –SH oxidation of proteins responsible for the vertical and horizontal tailing in 2D gels, alkylation and reduction were performed by equilibrating IPG strips in equilibration buffer I (6 M urea, 0.375 M Tris-HCl, 2% SDS, 20% glycerol, 2% DTT) and equilibration buffer II (6 M urea, 0.375 M Tris-HCl, 2% SDS, 20% glycerol, 2.5% Iodoacetamide, bromophenol blue).
4.2.2. 2D-PAGE Analysis
The 2D gel electrophoresis (2DE) gels from venom samples of 5 different Thoracobombus species were stained with OrioleTM Fluoroscent Gel Stain (Bio-Rad, Munich, Germany). The gels were analyzed using PDQuest Software Advanced 8.0.1 (Bio-Rad, San Diego, CA, USA). The PDQuest software examines each spot on every gel image and picks a master gel containing most spots. All spots in each gel image are then matched onto the master gel. Each post is annotated with an identification number (SSP). For quantifying spots, four different normalization options are performed, including total density in gel images, local regression, total quantity in valid spot, and mean of log ratios. In the present study, 2DE image analysis is performed to both detect common and/or unique spots between samples and quantify spot intensity, which accounts for the amount of protein in that spot. All spots were matched manually and ‘total density in gel images’ normalization was performed. If the same putative toxin was identified in different spots on the gel, then the spot with the highest score and proximity to theoretical pI/mW was chosen for analyses. Two-way ANOVA was performed to detect significant differences between spot intensities (IBM Corp. Released 2010. IBM SPSS Statistics for Windows, Version 19.0, Armonk, NY, USA).
4.2.3. In-Gel Enzymatic Digestion, Matrix-Assisted Laser Desorption Ionization/Time of Flight (MALDI-TOF) Mass Spectrometry and Peptide Mass Fingerprinting (PMF) Analysis
In-gel trypsinization of protein spots was performed as previously described [44]. Obtained peptides were loaded on MALDI-TOF plates using ZipTip® Pipette Tips (Merck KGaA, Millipore, Darmstadt, Germany). Tryptic peptides were dissolved in sample solvent containing 0.1% TFA and 50% acetonitrile and mixed with an equal volume of matrix solution containing alpha-cyano-4-hydroxycinnamic acid, 0.1% TFA, and 75% acetonitrile. An amount of 1.5 µL of these mixtures was spotted onto a target plate and analyzed with MALDI-TOF mass spectrometer (Micromass, Waters, Manchester, UK) operated in positive-ion reflectron mode. Obtained spectra were then examined in MASCOT search engine (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=PMF). Organisms taxonomically related to bumble bees were selected, as bumble bees are not present in the database. “Other Metazoa” was selected as the taxonomical group, as it is a diverse group including all venomous animals as well as Drosophila sp., the taxonomically closest species to bumble bees. This database includes venom toxins from venomous metazoans such as bees (including B. terrestris and B. impatiens), spiders, scorpions, and snakes. Both Swissprot and NCBInr databases were used to maximize the number of hits. Hits with MASCOT scores less than 15 were excluded.
Acknowledgments
This study was part of the Ph.D. thesis of NPB and was funded by Hacettepe University Scientific Research Projects Coordination Unit. The experiments were performed entirely at the Ankara University Biotechnology Institute. We are grateful to Seçil Karahisar Turan, Selen Peker, Naşit İğci, Beycan Ayhan, and Hatice Yıldızhan for their experimental assistance, and Gautam Kao for revising the manuscript.
Supplementary Materials
The following are available online at www.mdpi.com/2072-6651/9/11/362/s1, Figure S1: 2DE gel images of Thoracobombus venom. Representitive images of venom profile of (A) B. humilis, (B) B. pascuorum, (C) B. ruderarius, (D) B. sylvarum, and (E) B. zonatus. Gel analyses were conducted in triplicates. Marked spots are spectrometrically identified (See: Tables S3–S7), Table S1: Sample Collection. Locations, number of specimens, and plants species where the bumble bee species are collected from, Table S2: Common and differentially expressed protein spots in Thoracobombus venom. Spots intensities are compared in B. humilis, B. pascuorum, B. ruderarius, B. sylvarum, and B. zonatus venom, Table S3: List of identified proteins in B. humilis venom, Table S4: List of identified proteins in B. pascuorum venom, Table S5: List of identified proteins in B. ruderarius venom, Table S6: List of identified proteins in B. sylvarum venom, Table S7: List of identified proteins in B. zonatus venom.
Author Contributions
N.P.B. and D.O.D. conceived and designed the experiments; N.P.B. performed the experiments; N.P.B., D.O.D., and M.B.B. analyzed the data; N.P.B. and M.B.B. wrote the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Calvete J.J. Venomics: Digging into the evolution of venomous systems and learning to twist nature to fight pathology. J. Proteom. 2009;72:121–126. doi: 10.1016/j.jprot.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 2.Fitzgerald K.T. Chapter 49—Insects—Hymenoptera A2—Peterson. Michael E. In: Talcott P.A., editor. Small Animal Toxicology. 3rd ed. W.B. Saunders; Saint Louis, MO, USA: 2013. pp. 573–588. [Google Scholar]
- 3.Arbuckle K. Evolutionary Context of Venom in Animals. In: Malhotra A., editor. Evolution of Venomous Animals and Their Toxins. Springer; Dordrecht, The Netherlands: 2017. pp. 3–31. [Google Scholar]
- 4.Free J. Insect Pollination of Crops. Academic Press; London, UK: 1993. p. 684. [Google Scholar]
- 5.Choo Y.M., Lee K.S., Yoon H.J., Je Y.H., Lee S.W., Sohn H.D., Jin B.R. Molecular cloning and antimicrobial activity of bombolitin, a component of bumblebee Bombus ignitus venom. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2010;156:168–173. doi: 10.1016/j.cbpb.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Choo Y.M., Lee K.S., Yoon H.J., Kim B.Y., Sohn M.R., Roh J.Y., Je Y.H., Kim N.J., Kim I., Woo S.D., et al. Dual function of a bee venom serine protease: Prophenoloxidase-activating factor in arthropods and fibrin (ogen) olytic enzyme in mammals. PLoS ONE. 2010;5:e10393. doi: 10.1371/journal.pone.0010393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choo Y.M., Lee K.S., Yoon H.J., Qiu Y., Wan H., Sohn M.R., Sohn M.R., Sohn H.D., Jin B.R. Antifibrinolytic role of a bee venom serine protease inhibitor that acts as a plasmin inhibitor. PLoS ONE. 2012;7:e32269. doi: 10.1371/journal.pone.0032269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choo Y.M., Qiu Y., Yoon H.J., Lee K.Y., Sohn H.D., Jin B.R. Enzymatic properties of a bee venom serine protease from the bumblebee Bombus ignitus. J. Asia Pac. Entomol. 2011;14:249–251. doi: 10.1016/j.aspen.2011.03.009. [DOI] [Google Scholar]
- 9.Choo Y.M., Yoon H.J., Jin B.R. Effects of the bumblebee (Bombus ignitus) venom serine protease inhibitor on serine protease and phospholipase A 2 of B. ignitus venom. J. Asia Pac. Entomol. 2012;15:543–545. doi: 10.1016/j.aspen.2012.05.011. [DOI] [Google Scholar]
- 10.Kim J.S., Choi J.Y., Lee J.H., Park J.B., Fu Z., Liu Q., Tao X., Jin B.R., Skinner M., Parker B.L., et al. Bumblebee venom serine protease increases fungal insecticidal virulence by inducing insect melanization. PLoS ONE. 2013;8:e62555. doi: 10.1371/journal.pone.0062555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu Y., Choo Y.M., Yoon H.J., Jia J., Cui Z., Wang D., Kim D.H., Sohn H.D., Jin B.R. Fibrin (ogen) olytic activity of bumblebee venom serine protease. Toxicol. Appl. Pharmacol. 2011;255:207–213. doi: 10.1016/j.taap.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 12.Qiu Y., Choo Y.M., Yoon H.J., Jin B.R. Molecular cloning and antibacterial activity of bombolitin isolated from the venom of a bumblebee, Bombus terrestris. J. Asia Pac. Entomol. 2012;15:21–25. doi: 10.1016/j.aspen.2011.08.007. [DOI] [Google Scholar]
- 13.Qiu Y., Choo Y.M., Yoon H.J., Jin B.R. Molecular cloning and fibrin (ogen) olytic activity of a bumblebee (Bombus hypocrita sapporoensis) venom serine protease. J. Asia Pac. Entomol. 2012;15:79–82. doi: 10.1016/j.aspen.2011.09.002. [DOI] [Google Scholar]
- 14.Qiu Y., Lee K.S., Choo Y.M., Kong D., Yoon H.J., Jin B.R. Molecular cloning and antifibrinolytic activity of a serine protease inhibitor from bumblebee (Bombus terrestris) venom. Toxicon. 2013;63:1–6. doi: 10.1016/j.toxicon.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Wan H., Kim B.Y., Lee K.S., Yoon H.J., Lee K.Y., Jin B.R. A bumblebee (Bombus ignitus) venom serine protease inhibitor that acts as a microbial serine protease inhibitor. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2014;167:59–64. doi: 10.1016/j.cbpb.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Favreau P., Menin L., Michalet S., Perret F., Cheneval O., Stöcklin M., Bulet P., Stöcklin R. Mass spectrometry strategies for venom mapping and peptide sequencing from crude venoms: Case applications with single arthropod specimen. Toxicon. 2006;47:676–687. doi: 10.1016/j.toxicon.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 17.Argiolas A., Pisano J.J. Bombolitins, a new class of mast cell degranulating peptides from the venom of the bumblebee Megabombus pennsylvanicus. J. Biol. Chem. 1985;260:1437–1444. [PubMed] [Google Scholar]
- 18.Hoffman D.R., El-Choufani S.E., Smith M.M., De Groot H. Occupational allergy to bumblebees: Allergens of Bombus terrestris. J. Allergy Clin. Immunol. 2001;108:855–860. doi: 10.1067/mai.2001.119029. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman D.R., Jacobson R.S. Allergens in hymenoptera venom XXVII: Bumblebee venom allergy and allergens. J. Allergy Clin. Immunol. 1996;97:812–821. doi: 10.1016/S0091-6749(96)80159-X. [DOI] [PubMed] [Google Scholar]
- 20.Van Vaerenbergh M., Debyser G., Smagghe G., Devreese B., de Graaf D.C. Unraveling the venom proteome of the bumblebee (Bombus terrestris) by integrating a combinatorial peptide ligand library approach with FT-ICR MS. Toxicon. 2015;102:81–88. doi: 10.1016/j.toxicon.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Barkan N.P., Demiralp D.O., Aytekin A.M. Comparison of Venom Extraction Methods of Bombus (Bombus) terrestris (Hymenoptera: Apidae: Bombus) by Using Bottom-up Proteomic Strategies. Curr. Proteom. 2015;12:80–86. doi: 10.2174/157016461202150903111853. [DOI] [Google Scholar]
- 22.Piek T. Venoms of the Hymenoptera: Biochemical, Pharmacological and Behavioural Aspects. Elsevier; London, UK: 2013. [Google Scholar]
- 23.Fenton A.W., West P.R., Odell G.V., Hudiburg S.M., Ownby C.L., Mills J.N., Scroggins B.T., Shannon S.B. Arthropod venom citrate inhibits phospholipase A2. Toxicon. 1995;33:763–770. doi: 10.1016/0041-0101(95)00019-I. [DOI] [PubMed] [Google Scholar]
- 24.Rees J.A., Moniatte M., Bulet P. Novel antibacterial peptides isolated from a European bumblebee, Bombus pascuorum (Hymenoptera, Apoidea) Insect Biochem. Mol. Biol. 1997;27:413–422. doi: 10.1016/S0965-1748(97)00013-1. [DOI] [PubMed] [Google Scholar]
- 25.Piek T., Veldsema-Currie R., Spanjer W., Mantel P. Acetylcholine and an unidentified, muscle-contracting factor in the venom of the bumblebee, Bombus terrestris L. Comp. Biochem. Physiol. Part C Comp. Pharmacol. 1983;75:351–356. doi: 10.1016/0742-8413(83)90204-9. [DOI] [PubMed] [Google Scholar]
- 26.Sadd B.M., Barribeau S.M., Bloch G., de Graaf D.C., Dearden P., Elsik C.G., Gadau J., Grimmelikhuijzen C.J., Hasselmann M., Lozier J.D., et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams P. The distribution of bumblebee colour patterns worldwide: Possible significance for thermoregulation, crypsis, and warning mimicry. Biol. J. Linn. Soc. 2007;92:97–118. doi: 10.1111/j.1095-8312.2007.00878.x. [DOI] [Google Scholar]
- 28.Barkan N.P., Aytekin A.M. Systematical studies on the species of the subgenus Bombus (Thoracobombus) (Hymenoptera: Apidae: Bombus Latreille) in Turkey. Zootaxa. 2013;3737:167–183. doi: 10.11646/zootaxa.3737.2.5. [DOI] [PubMed] [Google Scholar]
- 29.Terzo M., Valterová I., Rasmont P. Atypical Secretions of the Male Cephalic Labial Glands in Bumblebees: The Case of Bombus (Rhodobombus) mesomelasGerstaecker (Hymenoptera, Apidae) Chem. Biodivers. 2007;4:1466–1471. doi: 10.1002/cbdv.200790124. [DOI] [PubMed] [Google Scholar]
- 30.Terzo M., Valterova I., Urbanova K., Rasmont P. De la nécessité de redécrire les phéromones sexuelles des mâles de bourdons [Hymenoptera: Apidae, Bombini] publiées avant 1996 pour leur utilisation en analyse phylogénétique. Phytoprotection. 2003;84:39–49. doi: 10.7202/007806ar. [DOI] [Google Scholar]
- 31.Urbanová K., Valterová I., Rasmont P., Terzo M. Znaèkovací feromony: Slovení sekretu labiální vlázy samcù Bombus magnus. [Marking pheromones of bumblebees: Composition of the labial gland secretion of males of Bombus magnus] Chemické Listy. 2002;96:918–919. [Google Scholar]
- 32.Hovorka O., Valterova I., Rasmont P., Terzo M. Male cephalic labial gland secretions of two bumblebee species of the subgenus Cullumanobombus (Hymenoptera: Apidae: Bombus latreille) and their distribution in central Europe. Chem. Biodivers. 2006;3:1015–1022. doi: 10.1002/cbdv.200690099. [DOI] [PubMed] [Google Scholar]
- 33.Cameron S., Hines H., Williams P. A comprehensive phylogeny of the bumble bees (Bombus) Biol. J. Linn. Soc. 2007;91:161–188. doi: 10.1111/j.1095-8312.2007.00784.x. [DOI] [Google Scholar]
- 34.Cameron S.A., Hines H.M., Williams P.H. Molecular phylogeny of the bumble bee subgenus Pyrobombus (Hymenoptera: Apidae: Bombus) with insights into gene utility for lower-level analysis. Invertebr. Syst. 2006;20:289–303. [Google Scholar]
- 35.Cameron S.A., Williams P.H. Phylogeny of bumble bees in the New World subgenus Fervidobombus (Hymenoptera: Apidae): Congruence of molecular and morphological data. Mol. Phylogenet. Evol. 2003;28:552–563. doi: 10.1016/S1055-7903(03)00056-3. [DOI] [PubMed] [Google Scholar]
- 36.Kawakita A., Sota T., Ascher J.S., Ito M., Tanaka H., Kato M. Evolution and phylogenetic utility of alignment gaps within intron sequences of three nuclear genes in bumble bees (Bombus) Mol. Biol. Evol. 2003;20:87–92. doi: 10.1093/molbev/msg007. [DOI] [PubMed] [Google Scholar]
- 37.Kawakita A., Sota T., Ito M., Ascher J.S., Tanaka H., Kato M., Roubik D.W. Phylogeny. historical biogeography, and character evolution in bumble bees (Bombus: Apidae) based on simultaneous analysis of three nuclear gene sequences. Mol. Phylogenet. Evol. 2004;31:799–804. doi: 10.1016/j.ympev.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 38.Du Q., Bi G., Zhao E., Yang J., Zhang Z., Liu G. Complete mitochondrial genome of Bombus terrestris (Hymenoptera: Apidae) Mitochondrial DNA. 2016;27:4455–4456. doi: 10.3109/19401736.2015.1089568. [DOI] [PubMed] [Google Scholar]
- 39.Williams P.H., An J., Brown M.J.F., Carolan J.C., Goulson D., Huang J., Ito M. Cryptic Bumblebee Species: Consequences for Conservation and the Trade in Greenhouse Pollinators. PLoS ONE. 2012;7:e32992. doi: 10.1371/journal.pone.0032992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Vaerenbergh M., Debyser G., Devreese B., de Graaf D.C. Exploring the hidden honeybee (Apis mellifera) venom proteome by integrating a combinatorial peptide ligand library approach with FTMS. J. Proteom. 2014;99:169–178. doi: 10.1016/j.jprot.2013.04.039. [DOI] [PubMed] [Google Scholar]
- 41.Gibbs H.L., Sanz L., Chiucchi J.E., Farrell T.M., Calvete J.J. Proteomic analysis of ontogenetic and diet-related changes in venom composition of juvenile and adult Dusky Pigmy rattlesnakes (Sistrurus miliarius barbouri) J. Proteom. 2011;74:2169–2179. doi: 10.1016/j.jprot.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 42.King G.F. Venoms as a platform for human drugs: Translating toxins into therapeutics. Expert Opin. Biol. Ther. 2011;11:1469–1484. doi: 10.1517/14712598.2011.621940. [DOI] [PubMed] [Google Scholar]
- 43.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 44.Igci N., Demiralp D.O. A preliminary investigation into the venom proteome of Macrovipera lebetina obtusa (Dwigubsky, 1832) from Southeastern Anatolia by MALDI-TOF mass spectrometry and comparison of venom protein profiles with Macrovipera lebetina lebetina (Linnaeus, 1758) from Cyprus by 2D-PAGE. Arch. Toxicol. 2012;86:441–451. doi: 10.1007/s00204-011-0763-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.