Abstract
Long non-coding RNAs (lncRNAs), which have emerged as important regulatory RNA molecules that have been implicated in carcinogenesis and cancer progression, may also serve as novel potential biomarkers for cancer diagnosis and prognosis. Our previous analysis has identified the lncRNA GHRLOS, the ghrelin antisense strand non-coding RNA gene, as one of the hub genes in the co-expression network of differentially expressed lncRNAs/mRNAs in colorectal cancer (CRC). Here, we further evaluate the expression of GHRLOS in CRC and explore its clinical significance. The expression of GHRLOS in 366 pairs of CRC and adjacent non-cancerous tissues was detected by quantitative RT-PCR assays. The results showed that the expression level of GHRLOS was significantly lower in CRC tissues than in matched non-cancerous tissues (P < 0.001). Decreased GHRLOS expression was observed in 54.4% (199/366) of cases, and was significantly correlated with the occurrence of lymph node metastasis (P = 0.033) and distant metastasis (P = 0.005). A Kaplan-Meier analysis demonstrated that decreased GHRLOS expression contributed to poor disease-free survival (log-rank test, P < 0.001) and overall survival (log-rank test, P < 0.001). Moreover, a multivariate Cox regression analysis revealed the decreased expression of GHRLOS as an independent prognostic marker of poor outcomes [disease-free survival: hazard ratio (HR) = 2.02, 95% confidence interval (CI) = 1.42-3.88; overall survival: HR = 1.96, 95% CI = 1.34-2.86] in CRC patients. In conclusion, our data suggest that the lncRNA GHRLOS might serve as a candidate biomarker of tumor metastasis and a prognostic indicator in CRC.
Keywords: long non-coding RNA, GHRLOS, colorectal cancer, clinical relevance, prognosis
Introduction
Cancers of the colon and rectum (colorectal) are some of the most commonly diagnosed malignant tumors in both males and females worldwide 1. Although improved colorectal cancer (CRC) screening and treatment modalities have provided substantial benefits with respect to patient outcome, CRC still constitutes a leading cause of cancer-related death 1. Up to half of CRC patients experience cancer relapse 2 and more than half of those at an advanced disease stage die from cancer recurrence after curative surgery for CRC 3. Therefore, it is urgent that novel molecular biomarkers that predict the recurrence and survival of CRC be identified.
About 80% of the human genome is dynamically and pervasively transcribed, mostly as non-protein-coding RNAs 4, 5. Long non-coding RNAs (lncRNAs) comprise a newly identified type of non-protein-coding RNAs that is greater than 200 nucleotides in length. All known lncRNAs can be classified into the following five subtypes according to their proximity to nearby protein-coding genes: sense, antisense, bidirectional, intronic, and intergenic 6, 7. The role of lncRNAs in biological processes has attracted increased attention, but this role has only partially been elucidated. As a novel regulatory RNA molecule, lncRNA has been suggested to regulate gene expression at the transcriptional, epigenetic, and post-transcriptional levels 6, and therefore, lncRNAs participate in various physiological and pathological processes, including carcinogenesis and cancer progression 8, 9. The aberrant expressions of certain lncRNAs, such as HOTAIR, CCAT1, and MALAT1, have been observed in different types of solid tumors 10-12. Furthermore, growing evidence has demonstrated the clinical significance of lncRNAs and has linked the dysregulation of lncRNA expression to cancer prognosis 13, 14. For example, HOTAIR, one of the well-studied lncRNAs, has been reported to be significantly associated with survival outcomes in patients with diverse cancers 15, including CRC 16. These findings indicate lncRNAs as an emerging source of novel potential biomarkers for cancer prognosis.
In a recent study, we identified a number of differentially expressed lncRNAs and mRNAs in CRC by screening their expression profiles, and after a co-expression gene network analysis of differentially expressed lncRNAs and mRNAs, we highlighted the importance of the lncRNA GHRLOS 17. GHRLOS was first identified from the opposite strand of the ghrelin gene GHRL by Seim and colleagues and was found to transcribe natural antisense transcripts of GHRL 18. GHRL encodes ghrelin, which is a multifunctional peptide hormone with roles not only in various physiological processes including appetite regulation, insulin release, and gut motility but also in cancer progression 19; GHRL also encodes other potentially active peptides such as obestatin 20, 21. GHRLOS has been shown to function as a non-coding RNA that is involved in regulation of the ghrelin axis due to its overlapping genomic arrangement with GHRL 22. The expression of GHRLOS has been demonstrated in a range of normal tissues and in the cells of various cancer types including CRC 22. However, the clinical value of GHRLOS in cancers has not yet been investigated.
Therefore, in the current study, we aimed to determine the expression pattern of GHRLOS in CRC tissues and in adjacent non-cancerous tissues and to elucidate the clinicopathological and prognostic significance of GHRLOS in CRC.
Materials and Methods
Patients and collection of tissue samples
The participants of this study were derived from the patients who underwent radical resection for CRC in the Department of General Surgery in Huashan Hospital (Shanghai, China) between 2007 and 2012. The inclusion criteria for participants were as follows: 1) the patients were diagnosed with pathologically confirmed CRC; 2) the patients were primarily treated with surgery; 3) the patients did not receive preoperative chemotherapy or radiotherapy; 4) the patients had complete medical records. Patients were excluded from this study if they met any of the following exclusion criteria: 1) advanced CRC patients with unresectable metastases who underwent palliative surgery; 2) insufficient tissues harvested for RNA isolation; 3) refused consent. Finally, 366 CRC patients were enrolled in this study. All clinicopathological data were obtained from the patients' medical records and the pathological staging was determined according to the American Joint Committee on Cancer TNM staging system for CRC. Cancer tissues were collected immediately after surgical removal. Corresponding adjacent non-cancerous colorectal tissues were obtained at a distance of ≥5 cm from the tumor margin and further confirmed by haematoxylin and eosin staining (Figure 1A). Fresh tissues, including both CRC tissues and adjacent non-cancerous tissues, were snap-frozen in liquid nitrogen and stored at -80 ℃ until RNA extraction. The present study was approved by the Medical Ethics Committee of Huashan Hospital, Fudan University and conducted according to the principles of Declaration of Helsinki. A written informed consent document was signed by each participant.
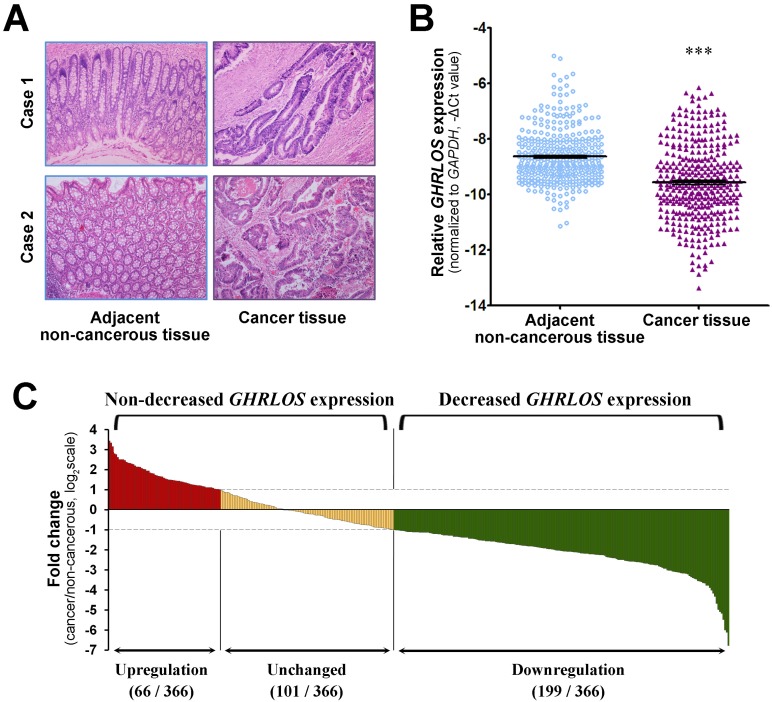
Figure 1.
Expression of the lncRNA GHRLOS in colorectal cancer. (A) The representative images of haematoxylin and eosin staining of cancer tissue and matched adjacent non-cancerous tissue from 2 CRC patients (original magnification, ×100). (B) The expression levels of GHRLOS were assessed by quantitative RT-PCR in cancer tissues and adjacent normal tissues from 366 patients. The expression levels of GHRLOS were normalized to the level of GAPDH. Note that the levels of GHRLOS in CRC tissues were significantly lower than those in adjacent non-cancerous tissues. ***, P < 0.001. (C) Relative GHRLOS expression (expressed as log2 fold change) in cancer tissues compared with matched non-cancerous tissues (n = 366). Patients with colorectal cancer were classified into either the decreased GHRLOS expression group (log2 fold change ≤ -1; n = 199) or the non-decreased GHRLOS expression group (log2 fold change > -1; n = 167) according to the changes in GHRLOS expression.
RNA extraction and quantitative RT-PCR
The total RNA was extracted from cancer and non-cancerous tissues using TRIzol reagent (Thermo Fisher Scientific) in accordance with the manufacturer's instructions. After quantification using a NanoDrop spectrophotometer, the isolated RNA (0.5 μg for each sample) was subjected to reverse transcription using the PrimeScript RT Reagent Kit (TaKaRa). The synthesized cDNA was used as a template for the quantitative PCR analysis of the expression of the lncRNA GHRLOS using a FastStart Universal SYBR Green Master kit (Roche Diagnostics). The thermal cycling program used was as follows: an initial step at 95 °C for 10 minutes, followed by 40 cycles of denaturation at 95 °C for 15 seconds and annealing and extension at 60 °C for 45 seconds. The melting curve of each PCR product was obtained by continuous fluorescence monitoring at a temperature gradient ramp from 60 to 95 °C. The quantitative PCR reactions were performed in triplicate to remove any outliers. The expression of GHRLOS was normalized to that of the internal reference gene GAPDH. The primers used were as follows: GHRLOS-F: 5'-TGGAAACTCCCCTAGCCACA; GHRLOS-R: 5'-GCATCTCTCCTCTGTTCCGT; Gapdh-F: 5'-ATCCTGGGCTACACTGAGCACC; Gapdh-R: 5'-AAGTGGTCGTTGAGGGCAATGC. The GHRLOS-specific primers, which were designed to encompass all validated transcript variants, were generated using the free online NCBI tool Primer-BLAST (available at https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
Measurement of endpoints
Patients were followed-up at intervals of approximately 3 months until death or until October 2016. Follow-up studies included a physical examination, laboratory tests, imaging analyses (including endoscopy, ultrasonography, computed tomography, magnetic resonance imaging, and positron emission tomography), and biopsy if necessary. Disease-free survival (DFS) is defined as the time from surgery until the detection of local recurrence or distant metastasis of CRC. Patients without recurrence were censored at the date of death or at the last follow-up. Overall survival (OS) is defined as the time from surgery until death or the last follow-up examination. Patients who were alive at their last follow-up were censored.
Statistical analysis
Paired Student's t test was used for the comparison of GHRLOS expression between cancer and non-cancerous tissues. A chi-square test was applied to evaluate the correlation of GHRLOS expression with the clinicopathological factors. Survival curves were plotted according to the Kaplan-Meier method, and the difference between survival curves was analyzed using the log-rank test. The Cox proportional hazards model was used in the univariate and multivariate survival analyses. Variables with a value of P < 0.05 by univariate analysis were included in the subsequent multivariate analysis. Statistical analyses were performed using the Statistical Program for Social Science (SPSS) software (version 12.0; SPSS); the level of statistical significance was set at 5%.
Results
The lncRNA GHRLOS is downregulated at a high frequency in CRC tissues
The expression level of the lncRNA GHRLOS in a total of 366 matched CRC tissues and adjacent non-cancerous tissues (Figure 1A) was determined by quantitative RT-PCR. As shown in Figure 1B, the expression levels of GHRLOS were significantly downregulated in CRC tissues compared with adjacent non-cancerous tissues (paired Student's t test, P < 0.001). Out of the 366 patients, 199 patients (54.4%) exhibited lower GHRLOS expression levels in cancer tissues compared with the corresponding non-cancerous tissues (fold change ≤ 0.5), while 101 patients (27.6%) exhibited similar expression levels in cancer tissues and non-cancerous tissues (0.5 < fold change < 2.0). Only 66 patients (18.0%) exhibited higher expression levels in cancer tissues compared with non-cancerous tissues (fold change ≥ 2.0; Figure 1C). Therefore, the results showed that GHRLOS was downregulated at a high frequency in CRC tissues.
Association between GHRLOS expression and the clinicopathological features of CRC patients
To explore the clinical value of decreased GHRLOS expression in CRC, the medical records of all 366 CRC patients were reviewed. The clinicopathological features are shown in Table 1. Based on the change in expression of GHRLOS in CRC tissues, all patients were divided into two groups, as follows (Figure 1C): the decreased GHRLOS expression group (fold change ≤ 0.5; n = 199) and the non-decreased GHRLOS expression group (fold change > 0.5; n = 167). The clinicopathological factors in these two groups were then analyzed. As shown in Table 1, decreased tumoral expression of GHRLOS was more frequently detected in CRC patients with lymph node metastasis (P = 0.033) and distant metastasis (P = 0.005). Decreased GHRLOS expression had a nearly significant association with poor histological tumor grade (P = 0.079) and deeper tumor invasion depth (P = 0.074). However, GHRLOS expression was not associated with the other clinicopathological factors included in our analyses, such as age, gender, tumor size and location, histological subtype of tumors, and serum CEA level (all P > 0.05). These results suggested a significant association between the decreased tumoral expression of GHRLOS and CRC metastasis.
Table 1.
GHRLOS expression and clinicopathological characteristics of patients with colorectal cancer
| Characteristics | Number of case | GHRLOS expression | P value | |
|---|---|---|---|---|
| Decreased (n=199) Number (%) |
Non-decreased (n=167) Number (%) |
|||
| Age | 0.465 | |||
| > 65 years | 159 | 83 | 76 | |
| ≤ 65 years | 207 | 116 | 91 | |
| Gender | 0.253 | |||
| Male | 196 | 112 | 84 | |
| Female | 170 | 87 | 83 | |
| Tumor site | 0.930 | |||
| Colon | 135 | 73 | 62 | |
| Rectum | 231 | 126 | 105 | |
| Tumor size | 0.475 | |||
| < 5 cm | 214 | 113 | 101 | |
| ≥ 5 cm | 152 | 86 | 66 | |
| Histological subtype | 0.281 | |||
| Adenocarcinoma | 341 | 188 | 153 | |
| Others | 25 | 11 | 14 | |
| Histological grade | 0.079 | |||
| Well/Moderate | 271 | 140 | 131 | |
| Poor | 95 | 59 | 36 | |
| Depth of tumor invasion | 0.074 | |||
| T1+T2 | 110 | 52 | 58 | |
| T3+T4 | 256 | 147 | 109 | |
| Lymph node metastasis | 0.033 | |||
| Absence | 217 | 108 | 109 | |
| Presence | 149 | 91 | 58 | |
| Distant metastasis | 0.005 | |||
| Absence | 315 | 162 | 153 | |
| Presence | 51 | 37 | 14 | |
| Serum CEA level | 0.136 | |||
| ≥ 10 ng/mL | 268 | 152 | 116 | |
| < 10 ng/mL | 98 | 47 | 51 | |
CEA, carcinoembryonic antigen.
Prognostic implications of GHRLOS expression in CRC patients
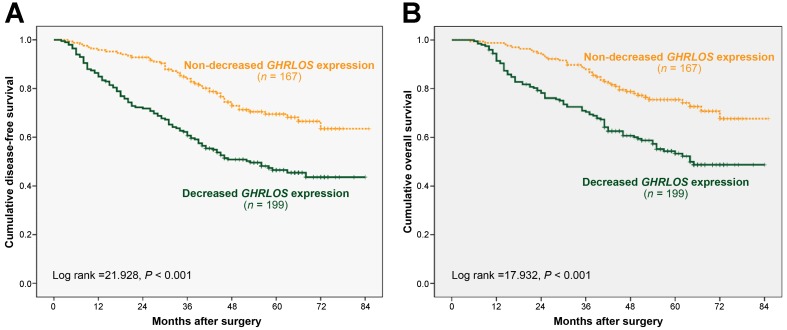
To determine the influence of GHRLOS expression on the prognosis and survival of patients with CRC, we attempted to evaluate the correlation of decreased GHRLOS expression with DFS and OS. Among the 366 CRC patients included in this study, 132 died during the follow-up period. After surgery for CRC, the follow-up time ranged from 5 to 85 months, and the median follow-up time was 56 months. The cumulative 5-year DFS and OS rates were 57% and 63%, respectively, for all included patients with CRC. Kaplan-Meier analyses showed that the CRC patients with decreased tumoral GHRLOS expression had a significantly worse DFS (log-rank test: P < 0.001) and OS (log-rank test: P < 0.001) compared with those with non-decreased GHRLOS expression (Figure 2), which suggested an adverse prognostic impact of decreased GHRLOS expression in CRC patients.
Figure 2.
Kaplan-Meier survival curves of colorectal cancer patients according to relative GHRLOS expression. (A) The disease-free survival curve and (B) the overall survival curve were analyzed based on the relative GHRLOS expression in 366 patients with colorectal cancer. Note that decreased GHRLOS expression (n = 199) was significantly associated with worse disease-free survival (log-rank test, P < 0.001) and overall survival (log-rank test, P < 0.001) compared with non-decreased GHRLOS expression (n = 167).
In addition, univariate and multivariate analyses using Cox proportional hazards models were performed for both DFS and OS. In the univariate analysis, decreased GHRLOS expression [hazard ratio (HR) = 2.20, 95% confidence interval (CI) 1.57-3.09, P < 0.001], larger tumor size, poor histological tumor grade, deeper tumor invasion depth, the presence of lymph node metastasis and distant metastasis, and higher CEA levels, were found to be significantly associated with worse DFS in CRC patients (Table 2). All of the above variables, including decreased GHRLOS expression (HR = 2.16, 95% CI 1.50-3.11, P < 0.001), were consistently associated with worse OS in the univariate analysis (Table 3). The significant prognostic variables were considered potential prognostic predictors and were then entered into the multivariate analyses. As shown in Tables 2 and 3, decreased expression of GHRLOS in cancer tissues maintained its significance as an independent prognostic factor for both DFS (HR = 2.02, 95% CI 1.42-3.88, P < 0.001) and OS (HR = 1.96, 95% CI 1.34-2.86, P = 0.001) in CRC patients. In addition, histological grade, depth of tumor invasion, lymph node metastasis, and distant metastasis were also shown to be significantly associated with shorter DFS or OS after controlling for other clinicopathological factors.
Table 2.
Univariate and multivariate analyses of disease-free survival in patients with colorectal cancer (Cox proportional hazards regression model)
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | |||||||
| > 65 vs ≤ 65 years | 0.81 | 0.59-1.12 | 0.198 | ||||
| Gender | |||||||
| Male vs Female | 0.91 | 0.66-1.25 | 0.568 | ||||
| Tumor site | |||||||
| Rectum vs Colon | 0.95 | 0.68-1.32 | 0.738 | ||||
| Tumor size | |||||||
| ≥ 5 vs < 5 cm | 1.54 | 1.11-2.12 | 0.009 | 1.33 | 0.95-1.86 | 0.096 | |
| Histological subtype | |||||||
| Adenocarcinoma vs others | 0.88 | 0.47-1.61 | 0.670 | ||||
| Histological grage | |||||||
| Poor vs Well/Moderate | 1.73 | 1.24-2.41 | 0.001 | 1.53 | 1.08-2.17 | 0.016 | |
| Depth of tumor invasion | |||||||
| T3+T4 vs T1+T2 | 2.56 | 1.70-3.87 | <0.001 | 1.65 | 1.07-2.56 | 0.024 | |
| Lymph node metastasis | |||||||
| Presence vs Adsence | 2.90 | 2.10-4.00 | <0.001 | 1.78 | 1.24-2.57 | 0.002 | |
| Distant metastasis | |||||||
| Presence vs Adsence | 6.79 | 4.77-9.67 | <0.001 | 4.31 | 2.91-6.38 | <0.001 | |
| Serum CEA level | |||||||
| ≥ 10 vs < 10 ng/mL | 1.50 | 1.07-2.10 | 0.019 | 1.30 | 0.91-1.86 | 0.144 | |
| GHRLOS expression | |||||||
| Decreased vs Non-decreased | 2.20 | 1.57-3.09 | <0.001 | 2.02 | 1.42-2.88 | <0.001 | |
HR, hazard ratio; CI, confidence interval; CEA, carcinoembryonic antigen.
Table 3.
Univariate and multivariate analyses for overall survival in patients with colorectal cancer (Cox proportional hazards regression model)
| Univariate analysis | Multivariate analysis | ||||||
|---|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P value | HR | 95% CI | P value | |
| Age | |||||||
| > 65 vs ≤ 65 years | 1.07 | 0.75-1.53 | 0.719 | ||||
| Gender | |||||||
| Male vs Female | 0.98 | 0.70-1.38 | 0.904 | ||||
| Tumor site | |||||||
| Rectum vs Colon | 1.21 | 0.83-1.77 | 0.312 | ||||
| Tumor size | |||||||
| ≥ 5 vs < 5 cm | 1.48 | 1.04-2.11 | 0.029 | 1.33 | 0.92-1.93 | 0.126 | |
| Histological subtype | |||||||
| Adenocarcinoma vs others | 0.66 | 0.36-1.18 | 0.160 | ||||
| Histological grage | |||||||
| Poor vs Well/Moderate | 1.64 | 1.15-2.35 | 0.006 | 1.56 | 1.07-2.27 | 0.020 | |
| Depth of tumor invasion | |||||||
| T3+T4 vs T1+T2 | 3.03 | 1.90-4.85 | <0.001 | 1.77 | 1.08-2.91 | 0.023 | |
| Lymph node metastasis | |||||||
| Presence vs Adsence | 3.63 | 2.55-5.19 | <0.001 | 2.41 | 1.62-3.58 | <0.001 | |
| Distant metastasis | |||||||
| Presence vs Adsence | 5.81 | 3.96-8.54 | <0.001 | 3.32 | 2.18-5.05 | <0.001 | |
| Serum CEA level | |||||||
| ≥ 10 vs < 10 ng/mL | 1.51 | 1.05-2.17 | 0.025 | 1.32 | 0.91-1.93 | 0.147 | |
| GHRLOS expression | |||||||
| Decreased vs Non-decreased | 2.16 | 1.50-3.11 | <0.001 | 1.96 | 1.34-2.86 | 0.001 | |
HR, hazard ratio; CI, confidence interval; CEA, carcinoembryonic antigen.
Discussion
Increased recognition and understanding of lncRNAs have defined these non-protein-coding RNAs as important components of tumor biology 23. Aberrant changes in lncRNA expression are seen across the spectrum of cancer progression and have been considered to be emerging potential biomarker candidates for patient outcomes 23. With the application of whole-transcriptome high-throughput screening, increasing numbers of lncRNAs have been found to be aberrantly expressed in CRC. Furthermore, some of the dysregulated lncRNAs, such as HOTAIR 16, NEAT1 24, and H19 25, have been demonstrated to be significantly associated with CRC prognosis, which indicates the prognostic importance of lncRNAs in CRC. In our previous study, we compared transcript expression profiles between CRC tissues and matched non-cancerous tissues and revealed a number of differentially expressed lncRNAs and mRNAs related to CRC 17. Further co-expression analysis of these aberrantly expressed lncRNAs and mRNAs identified several genes, including the lncRNA GHRLOS, as hub genes in the lncRNA/mRNA co-expression network in CRC 17. Nonetheless, currently, little is known about the clinical value of GHRLOS in CRC or in other types of cancer.
The current study analyzed the expression of GHRLOS in 366 pairs of CRC tissues and in matched adjacent non-cancerous tissues and demonstrated the high frequency of GHRLOS downregulation in cancer tissues, which suggests an important role of GHRLOS in CRC carcinogenesis. We further assessed the relationship between changes in GHRLOS expression and the clinicopathological features of CRC patients. It was found that decreased expression of GHRLOS was significantly associated with the presence of both lymph node metastasis and distant metastasis in CRC patients, which suggests a functional role of GHRLOS in the promotion of CRC metastasis. LncRNAs have been shown to be important regulators of the metastasis of various cancers including CRC 26-29. Further functional studies using GHRLOS-overexpressing or GHRLOS-knockdown cells are necessary to determine its influence on the migratory and invasive behavior of CRC cells. Furthermore, we revealed a significant association of decreased GHRLOS expression with poor survival outcomes (both DFS and OS) in CRC patients. Taken together, the above results indicate that GHRLOS may function as a tumor suppressor during CRC development and decreased GHRLOS expression promotes CRC progression via enhancing tumor metastasis. Therefore, GHRLOS may represent a potential therapeutic target for controlling cancer progression in patients with CRC, but this requires further functional studies. Importantly, the association between decreased GHRLOS expression and worse DFS or OS remained significant after adjustment for other clinicopathological factors, which highlights the clinical significance of GHRLOS as an independent prognostic predictor in CRC. These results suggest that measurements of GHRLOS expression may help identify CRC patients who are in high risk of early recurrence and death following curative surgery, and are candidates for receiving more aggressive treatment. Our findings, together with those of previous studies 16, 24, 30, 31, demonstrated the prognostic importance of various lncRNAs in CRC and supported lncRNAs as an emerging “gold mine” of prognostic biomarkers.
Despite the promising findings from the current analysis of clinical data, the biological function and mechanisms of GHRLOS in CRC were not investigated in this study. Since this lncRNA transcribes natural antisense RNA transcripts of the ghrelin gene, GHRLOS has been proposed to exert an important regulatory function in the ghrelin axis 22. Ghrelin, the most-studied product encoded by the ghrelin gene, is an important gastrointestinal peptide hormone characterized as the ligand of the growth hormone secretagogue receptor 21. Apart from its important physiological roles in the regulation energy homeostasis, ghrelin is also involved in the carcinogenesis and progression of various cancers, including both gastrointestinal and non-gastrointestinal cancers 19, 21, 32. In terms of CRC, the expression of ghrelin has been observed in both cancer cell lines and malignant tissue samples 33. Serum ghrelin levels were also found to be elevated in patients with colon cancers, which supports the idea that ghrelin is produced by tumors 34. Some studies have shown that ghrelin could promote the proliferation, as well as the migration and invasiveness of colorectal cancer cells 33, 35. On the contrary, the knockdown of the ghrelin receptor in colorectal cancer cells was shown to impair their growth in vitro and in vivo 36. These findings indicate ghrelin as an autocrine/paracrine factor that functions in the promotion of CRC progression. Given that GHRLOS was identified as an overlapping gene on the antisense strand of ghrelin and may act as a silencer of its overlapping gene, we speculate that decreased GHRLOS expression may promote CRC metastasis and progression via stimulating the ghrelin signaling. Further functional studies, as well as a correlation analysis between the expression of GHRLOS and the peptide hormone ghrelin in CRC tissues, are required to demonstrate this hypothesis.
To date, little is known about the mechanisms for the regulation of GHRLOS expression. Although GHRLOS transcripts have been suggested to be genuine products of RNA polymerase II-mediated transcription 22, the transcription factors that contribute to the regulation of GHRLOS transcription have not yet been identified. Interestingly, recent studies have revealed the involvement of several cancer-related transcription factors, such as HIF-1α and C/EBPα, in the regulation of the expression of various lncRNAs in cancer cells 37-39. These transcription factors are frequently dysregulated in cancers 40, 41. Thus, this provides a rational explanation for the aberrant expression of lncRNAs in cancers. Additional study is underway to identify the transcription factor that directly regulates the expression of GHRLOS, which may extend our understanding of the mechanism by which GHRLOS is downregulated in CRC tissues.
In conclusion, the present study demonstrated that the lncRNA GHRLOS is downregulated at a high frequency in human CRC tissues. To the best of our knowledge, for the first time, we revealed that GHRLOS may have clinical potential not only as a prospective biomarker for metastatic phenotype of CRC but also as a promising prognostic predictor in CRC. Our findings encourage further functional studies, which will explore its potential as a therapeutic target that may be used for the control of CRC metastasis and progression.
Acknowledgments
This study was funded by the Shanghai Municipal Commission of Health and Family Planning (201440271), and supported by grants from the Shanghai Natural Science Foundation (13ZR1405400) and the Startup Foundation of Huashan Hospital North (2015105).
Abbreviations
- LncRNA
long non-coding RNA
- CRC
colorectal cancer
- HR
hazard ratio
- CI
confidence interval
- DFS
disease-free survival
- OS
overall survival
- CEA
carcinoembryonic antigen.
References
- 1.Torre LA, Bray F, Siegel RL. et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Mirnezami A, Mirnezami R, Chandrakumaran K. et al. Increased local recurrence and reduced survival from colorectal cancer following anastomotic leak: systematic review and meta-analysis. Ann Surg. 2011;253:890–899. doi: 10.1097/SLA.0b013e3182128929. [DOI] [PubMed] [Google Scholar]
- 3.Edwards BK, Noone AM, Mariotto AB. et al. Annual Report to the Nation on the status of cancer, 1975-2010, featuring prevalence of comorbidity and impact on survival among persons with lung, colorectal, breast, or prostate cancer. Cancer. 2014;120:1290–1314. doi: 10.1002/cncr.28509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 5.Spizzo R, Almeida MI, Colombatti A. et al. Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene. 2012;31:4577–4587. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 7.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi X, Sun M, Liu H. et al. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339:159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 10.Gupta RA, Shah N, Wang KC. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mizrahi I, Mazeh H, Grinbaum R. et al. Colon Cancer Associated Transcript-1 (CCAT1) Expression in Adenocarcinoma of the Stomach. J Cancer. 2015;6:105–110. doi: 10.7150/jca.10568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ren D, Li H, Li R. et al. Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett. 2016;11:1621–1630. doi: 10.3892/ol.2016.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu M, Zou C, Pan L. et al. Long noncoding RNAs in digestive system cancers: Functional roles, molecular mechanisms, and clinical implications (Review) Oncol Rep. 2016;36:1207–1218. doi: 10.3892/or.2016.4929. [DOI] [PubMed] [Google Scholar]
- 14.Chen J, Miao Z, Xue B. et al. Long Non-coding RNAs in Urologic Malignancies: Functional Roles and Clinical Translation. J Cancer. 2016;7:1842–1855. doi: 10.7150/jca.15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serghiou S, Kyriakopoulou A, Ioannidis JP. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol Cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kogo R, Shimamura T, Mimori K. et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res. 2011;71:6320–6326. doi: 10.1158/0008-5472.CAN-11-1021. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Wu S, Li M. et al. LncRNA expression profiles reveal the co-expression network in human colorectal carcinoma. Int J Clin Exp Pathol. 2016;9:1885–1892. [Google Scholar]
- 18.Seim I, Collet C, Herington AC. et al. Revised genomic structure of the human ghrelin gene and identification of novel exons, alternative splice variants and natural antisense transcripts. BMC Genomics. 2007;8:298. doi: 10.1186/1471-2164-8-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nikolopoulos D, Theocharis S, Kouraklis G. Ghrelin's role on gastrointestinal tract cancer. Surg Oncol. 2010;19:e2–e10. doi: 10.1016/j.suronc.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 20.Seim I, Amorim L, Walpole C. et al. Ghrelin gene-related peptides: multifunctional endocrine / autocrine modulators in health and disease. Clin Exp Pharmacol Physiol. 2010;37:125–131. doi: 10.1111/j.1440-1681.2009.05241.x. [DOI] [PubMed] [Google Scholar]
- 21.Lin TC, Hsiao M. Ghrelin and cancer progression. Biochim Biophys Acta. 2017;1868:51–57. doi: 10.1016/j.bbcan.2017.02.002. [DOI] [PubMed] [Google Scholar]
- 22.Seim I, Carter SL, Herington AC. et al. Complex organisation and structure of the ghrelin antisense strand gene GHRLOS, a candidate non-coding RNA gene. BMC Mol Biol. 2008;9:95. doi: 10.1186/1471-2199-9-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Li Y, Chen W. et al. NEAT expression is associated with tumor recurrence and unfavorable prognosis in colorectal cancer. Oncotarget. 2015;6:27641–27650. doi: 10.18632/oncotarget.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han D, Gao X, Wang M. et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7:22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Z, Chen Z, Fan R. et al. Over-expressed long noncoding RNA HOXA11-AS promotes cell cycle progression and metastasis in gastric cancer. Mol Cancer. 2017;16:82. doi: 10.1186/s12943-017-0651-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma C, Wu G, Zhu Q. et al. Long intergenic noncoding RNA 00673 promotes non-small-cell lung cancer metastasis by binding with EZH2 and causing epigenetic silencing of HOXA5. Oncotarget. 2017;8:32696–32705. doi: 10.18632/oncotarget.16158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao X, Wen J, Gao P. et al. Overexpression of the long non-coding RNA, linc-UBC1, is associated with poor prognosis and facilitates cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2017;10:1017–1026. doi: 10.2147/OTT.S129343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Q, Yang L, Hu X. et al. Upregulated NNT-AS1, a long noncoding RNA, contributes to proliferation and migration of colorectal cancer cells in vitro and in vivo. Oncotarget. 2017;8:3441–3453. doi: 10.18632/oncotarget.13840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi D, Zheng H, Zhuo C. et al. Low expression of novel lncRNA RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal cancer. Med Oncol. 2014;31:31. doi: 10.1007/s12032-014-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi P, Xu MD, Ni SJ. et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nikolopoulos D, Theocharis S, Kouraklis G. Ghrelin: a potential therapeutic target for cancer. Regul Pept. 2010;163:7–17. doi: 10.1016/j.regpep.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 33.Waseem T, Javaid Ur R, Ahmad F. et al. Role of ghrelin axis in colorectal cancer: a novel association. Peptides. 2008;29:1369–1376. doi: 10.1016/j.peptides.2008.03.020. [DOI] [PubMed] [Google Scholar]
- 34.Nikolopoulos D, Theocharis S, Moutsios-Rentzos A. et al. The role of serum total ghrelin level elevation in colon cancer patients. J buon. 2014;19:388–393. [PubMed] [Google Scholar]
- 35.Lien GS, Lin CH, Yang YL. et al. Ghrelin induces colon cancer cell proliferation through the GHS-R, Ras, PI3K, Akt, and mTOR signaling pathways. Eur J Pharmacol. 2016;776:124–131. doi: 10.1016/j.ejphar.2016.02.044. [DOI] [PubMed] [Google Scholar]
- 36.Liu A, Huang C, Xu J. et al. Lentivirus-mediated shRNA interference of ghrelin receptor blocks proliferation in the colorectal cancer cells. Cancer Med. 2016;5:2417–2426. doi: 10.1002/cam4.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salle-Lefort S, Miard S, Nolin MA. et al. Hypoxia upregulates Malat1 expression through a CaMKK/AMPK/HIF-1alpha axis. Int J Oncol. 2016;49:1731–1736. doi: 10.3892/ijo.2016.3630. [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Gong Y, Jin B. et al. Long non-coding RNA urothelial carcinoma associated 1 induces cell replication by inhibiting BRG1 in 5637 cells. Oncol Rep. 2014;32:1281–1290. doi: 10.3892/or.2014.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou C, Ye L, Jiang C. et al. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015;36:9179–9188. doi: 10.1007/s13277-015-3453-8. [DOI] [PubMed] [Google Scholar]
- 40.Shih JW, Kung HJ. Long non-coding RNA and tumor hypoxia: new players ushered toward an old arena. J Biomed Sci. 2017;24:53. doi: 10.1186/s12929-017-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lourenco AR, Coffer PJ. A tumor suppressor role for C/EBPalpha in solid tumors: more than fat and blood. Oncogene. 2017;36:5221–5230. doi: 10.1038/onc.2017.151. [DOI] [PubMed] [Google Scholar]