Abstract
Anandins A (1) and B (2), two rare steroidal alkaloids, were isolated from the fermentative broth of a marine actinobacteria Streptomyces anandii H41-59. The gross structures of the two alkaloids were elucidated by spectroscopic methods including HR-ESI-MS, and NMR. Their absolute configurations were confirmed by single-crystal X-ray diffraction analysis and comparison of their experimental and calculated electronic circular dichroism spectra, respectively. Anandin A exhibited a moderate inhibitory effect against three human cancer cell lines MCF-7, SF-268, and NCI-H460 with IC50 values of 7.5, 7.9, 7.8 μg/mL, respectively.
Keywords: Streptomyces anandii, anandins A and B, steroidal alkaloids, cytotoxicity
1. Introduction
Because of the increasing difficulty to discover new bioactive compounds from terrestrial sources and the structure diversity of marine metabolites, many researchers have great interest in investigating secondary metabolites from marine-derived organisms [1]. Marine organisms are regarded as a prolific resource of novel bioactive metabolites, including a vast array of macrolide, cyclic peptides, pigments, polyketides, terpenes, steroids and alkaloids, but only a few steroidal alkaloids [2]. Steroidal alkaloids are a class of alkaloids with the basic steroidal skeleton containing a nitrogen atom, either in a ring or in a side chain. Structurally, these alkaloids can be classified into three major groups according to their carbon skeleton, namely, pregnane alkaloids, cholestane alkaloids and C-nor-D-homosteroidal alkaloids. Accumulated evidence in previous studieshas demonstrated that steroidal alkaloids and their glycosides have a wide range of bioactivities, such as antimicrobial, cytotoxic, anti-HIV, anti-inflammation, and antinociceptive activities [3,4,5].
Steroidal alkaloids are well-known metabolites of certain terrestrial plants, mainly from Solanaceae, Liliaceae, Apocynaceae and Buxaceae families [6,7,8]. The first steroidal alkaloid was isolated from the berries of Solanum nigrum almost 200 years ago [9]. However, steroidal alkaloid was not found from marine organisms until 1984 when two antimicrobial plakinamines were isolated from sponge Plakina sp. [10]. Later, four novel cortistatins exhibiting highly selective anti-proliferative activity were isolated from Corticium sp. [11], and cytotoxic cephalostatins were discovered from Cephalodiscus gilchristi [12]. Compared to those from plants, the marine steroidal alkaloids were relatively few, but with diverse chemistry structures.
Streptomyces are known for their ability to produce novel structural and bioactive metabolites [13,14,15]. While searching for bioactive compounds from marine actinomycete, we encountered a strain of actinomycete Streptomyces anandii H41-59, the crude extract of which showed strong antifungal activity against Candida albicans. In aprevious report, we have isolated three new ergosterols and ten known ones from culture broth of the strain H41-59 [16]. Further investigation on the broth of the same marine actinomycete led to the isolation of two novel ergostane-type steroidal alkaloids, named anandins A (1) and B (2). In addition, the antibioticand cytotoxic activities of the two steroidal alkaloids were tested in the present study. Herein, details of the isolation, structure elucidation, configuration assignment, and bioactivities of the new metabolites are described.
2. Results and Discussion
2.1. Elucidation of New Compounds
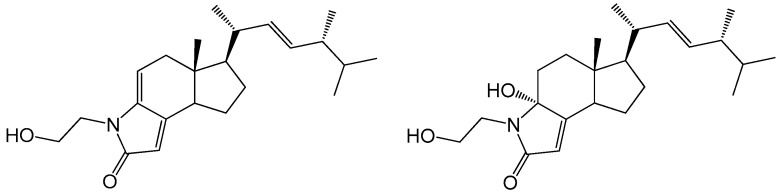
Actinomycete Streptomyces anandii H41-59 was isolated from a sea sediment sample from the mangrove zone in the South China Sea. An ethyl acetate partition of the ethanol extract was subjected to silica gel and LH-20 column chromatography, followed by RP-HPLC purification to yield two rare steroidal alkaloids (Figure 1).
Figure 1.
Chemical structure of anandins A (1) and B (2).
Compound 1 was obtained as a colorless plate crystal and given a molecular formula of C23H35NO2. Its seven degrees of unsaturation were determined on the basis of HR-ESI-MS [M + H]+ ion at m/z 358.2752 (C23H36NO2, calcd. 358.2741) (see Figure S1-8). The 1H NMR spectrum (see Figure S1) of 1 measured in acetone-d6 (Table 1) revealed the presence of five characteristic steroid methyl groups including a methyl singlet at δH 0.71 (Me-13) and four methyl doublets at δH 1.08 (Me-15), 0.84 (Me-20), 0.87 (Me-21) and 0.95 (Me-22). The 13C NMR and DEPT spectra indicated the presence of 23 carbon signals including a carbonyl carbon at δC 171.1 (C-2), six olefinic carbons at δC 115.3 (C-3), 152.0 (C-4), 141.5 (C-5), 107.4 (C-6), 136.4 (C-16), and 133.1 (C-17), a quaternary carbon at δC 47.0 (C-8), five methine carbons at δC 49.4 (C-9), 55.6 (C-12), 41.1 (C-14), 43.8 (C-18) and 33.9 (C-19), one oxygenated methylene at δC 61.6 (C-2′), four methylenes at δC 40.8 (C-7), 29.7 (C-10), 23.1 (C-11), 20.8 (C-1′), and five methyl carbons at δC 12.4 (C-13), 21.5 (C-15), 20.1 (C-20), 20.4 (C-21) and 18.2 (C-22). Further analyses of the 1D NMR and HSQC data suggested that compound 1 was more likely to be a highly degraded sterol.
Table 1.
1H and 13C NMR data of 1 and 2 (δ in ppm, J in Hz).
| Position | 1 a | 2 b | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 2 | - | 171.1 | - | 171.1 |
| 3 | 5.63 (br.s) | 115.3 | 5.52 (d, 1.9) | 116.5 |
| 4 | - | 152.0 | - | 164.9 |
| 5 | - | 141.5 | - | 89.3 |
| 6 | 5.60 (m) | 107.4 | 1.65 (m), 1.92 (m) | 36.0 |
| 7 | 2.63 (m), 2.34 (dd, 2.5, 17.5) | 40.8 | 2.23 (m), 1.60 (m) | 35.7 |
| 8 | - | 47.0 | - | 49.1 |
| 9 | 2.68 (m) | 49.4 | 2.65 (m) | 50.1 |
| 10 | 1.48 (m), 1.93 (m) | 29.7 | 1.46 (m), 1.91 (m) | 29.6 |
| 11 | 1.51 (m), 1.94 (m) | 23.1 | 1.58 (m), 1.65 (m) | 22.0 |
| 12 | 1.59 (m) | 55.6 | 1.53 (m) | 56.3 |
| 13 | 0.71 (s) | 12.4 | 0.60 (s) | 12.1 |
| 14 | 2.13 (m) | 41.1 | 2.11 (br. s) | 41.2 |
| 15 | 1.08 (d, 6.9) | 21.5 | 1.08 (d, 6.9) | 21.5 |
| 16 | 5.27 (dd, 7.7, 15.1) | 136.4 | 5.28 (dd, 7.6, 14.6) | 136.4 |
| 17 | 5.30 (dd, 7.2, 15.1) | 133.1 | 5.30 (dd, 7.2, 14.6) | 133.1 |
| 18 | 1.90 (m) | 43.8 | 1.90 (m) | 43.8 |
| 19 | 1.51 (m) | 33.9 | 1.50 (m) | 33.9 |
| 20 | 0.84 (d, 6.9) | 20.1 | 0.85 (d, 6.9) | 20.1 |
| 21 | 0.87 (d, 6.9) | 20.4 | 0.88 (d, 6.9) | 20.4 |
| 22 | 0.95 (d, 6.8) | 18.2 | 0.96 (d, 6.9) | 18.2 |
| 1′ | 3.64 (m) | 42.8 | 3.32 (m), 3.60 (m) | 42.5 |
| 2′ | 3.63 (m) | 61.1 | 3.66 (m) | 62.3 |
| 5-OH | 5.12 (br.s) | - | ||
| 2′-OH | 3.87 (br.s) | - | 4.36 (br.s) | - |
a Measured in CD3COCD3 at 300 MHz for 1H and 75 MHz for 13C NMR; b Measured in CD3COCD3 at 600 MHz for 1H and 125 MHz for 13C NMR.
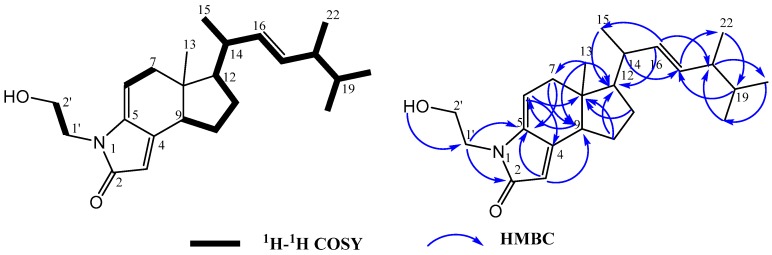
The H-H COSY spectrum of 1 gave several spin systems which belong to substructures as shown in Figure 2 with bold lines. The HMBC correlations from a sp2methine proton at δH 5.27 (1H, dd, H-16) to carbons at δC 21.5 (C-15, 3J) and 43.8 (C-18, 3J), and from a sp2methine proton at δH 5.30 (1H, dd, H-17) to carbon at δC 41.1 (C-14, 3J), confirmed the presence of a characteristic side chain of ergosterol [16]. A methyl proton at δH 0.71 (3H, s, Me-13) displayed four HMBC correlations to carbons at δC 40.8 (C-7, 3J), 47.7 (C-8, 2J), 49.4 (C-9, 3J) and 55.6 (C-12, 3J), interpreted as direct attachment to the quaternary C-8. The HMBC correlations from the proton at δH 1.93 (1H, m, H-10) to carbons at δC 47.7 (C-8, 3J) and 55.6 (C-12, 3J), from the proton at δH 5.60 (1H, m, H-6) to carbons at δC 47.7 (C-8, 3J) and 152.0 (C-4, 3J), from protons at δH 2.63 (2H, m, H-7)/2.34 (dd, 2.5, 17.5) to carbons at δC 141.5 (C-5, 3J), 49.4 (C-9, 3J) and 55.6 (C-12, 3J), confirmed the substructure of the B ring and C ring of ergosterol. The HMBC correlations from the proton at δH 5.63 (1H, br.s, H-3) to carbons at δC 171.1 (C-2, 2J), 141.5 (C-5, 3J) and 49.4 (C-9, 3J), from protons at δH 3.64 (2H, m, H-1′) to carbons at 171.1 (C-2, 3J) and 141.5 (C-5, 3J), and from protons at δH 3.63 (2H, m, H-2′) to carbon at δH 171.1 (C-2, 4J), confirmed the presence of an α,β-unsaturated γ-lactam and two methylene carbons linked to γ-lactam and the hydroxyl group.
Figure 2.
Key 1H-1H COSY and HMBC correlations of 1.
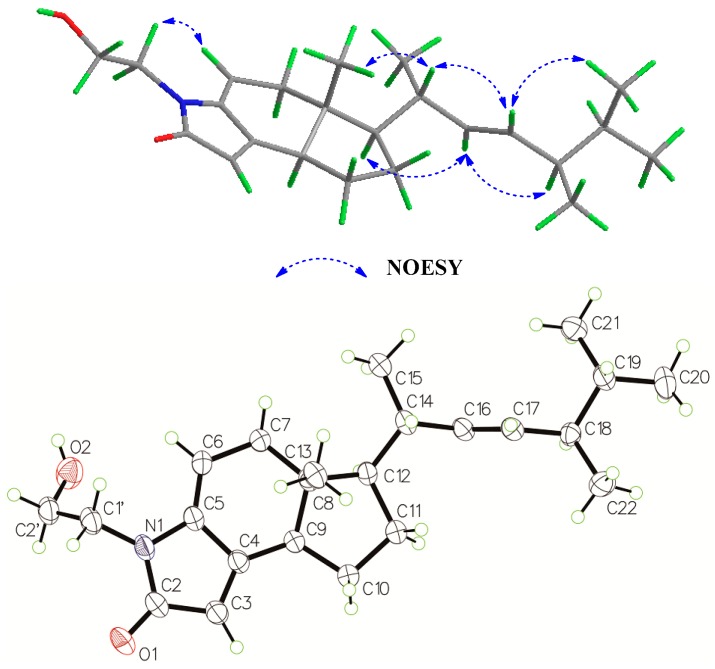
The relative configuration of 1 was determined on the basis of the analysis of 1H-1H coupling constants and the NOESY information (Figure 3). Large coupling constants of 15.1 Hz between H-16 and H-17, and the NOESY correlations between H-17 and H-14, between H-17 and H-20/H-21, between H-16 and H-12, and between H-16 and H-18 indicated the E configuration of the double bond in the side chain. A correlation was also observed between H-13β and H-14, confirming the β-orientation of H-14. The absolute configuration of compound 1 was unequivocally defined as (16E,8R,12R,14S,18R) by single crystal X-ray diffraction analysis (see Table S1) using Cu Ka radiation with Flack and Hooft parameters of 0.1 (2) and 0.15 (6), respectively. Compound 1 is a new steroidal alkaloid, named as anandin A.
Figure 3.
Key NOESY correlations and X-ray ORTEP of 1.
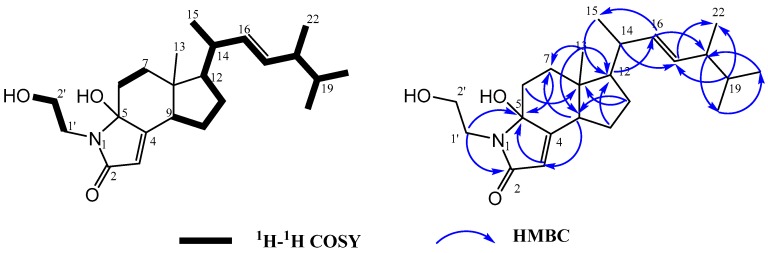
Compound 2 was isolated as white powder. Its molecular formula was determined as C23H37NO3 with six degrees of unsaturation, by the HR-ESI-MS [M + H]+ ion at m/z 376.2860 (C23H38NO3, calcd. 376.2853) (see Figure S2-8). The 13C NMR and DEPT (see Figure S2) spectroscopic data of 2 measured in acetone-d6 (Table 1) indicated the presence of 23 carbon signals for a carbonyl carbon at δC 171.1 (C-2), four olefinic carbons at δC 116.5 (C-3), 164.9 (C-4), 136.4 (C-16), 133.1 (C-17), one quaternary signal at δC 49.1 (C-8), one oxygenated methylene at δC 62.3 (C-2′), five methine carbons at δC 50.1 (C-9), 56.3 (C-12), 41.2 (C-14), 43.8 (C-18) and 33.9 (C-19), five methyl groups at δC 12.1 (C-13), 21.5 (C-15), 20.1 (C-20), 20.4 (C-21) and 18.2 (C-22). The 1H and 13C NMR spectroscopic data of 2 were comparable to those of 1, suggesting that 2 is an analogue of 1 with an additional oxygenated quaternary carbon and the absence of a double bond. The gross structure of 2 was further elucidated by analysis of COSY and HMBC spectrum data (Figure 4). The substructure of γ-hydroxy-α,β-unsaturated γ-lactam was deduced by the HMBC correlations from H-3 (δH 5.52, br.s, 1H) to C-2 (δC 171.1, 2J) and C-5 (δC 89.3, 3J), from H-1′ (δH 3.60, 3.32) to C-2 (δC 171.1, 3J) and C-5 (δC 89.3, 3J), from H-2′ (δH 3.66, m, 2H) to C-2 (δC 171.1, 4J), and from H-7 (δH 2.23/1.60, m, 2H) to C-5 (δC 89.3, 3J). The HMBC correlation from H-9 (δH 2.65, m, 1H) to C-5 (δC 89.3, 3J), C-3 (δC 116.5, 3J) and C-12 (δC 56.3, 3J), from H-11 (δH 1.65/1.58, m, 2H) to C-9 (δC 50.1, 3J), from H-9 (δH 2.65, m, 1H) to C-5 (δC 89.3, 3J) and C-3 (δC 116.5, 3J), confirmed the substructure of the B ring and the saturated C ring of ergosterol. Other HMBC correlations permitted further confirmation of the carbon skeleton of 2.
Figure 4.
Key 2D NMR correlations of 2.
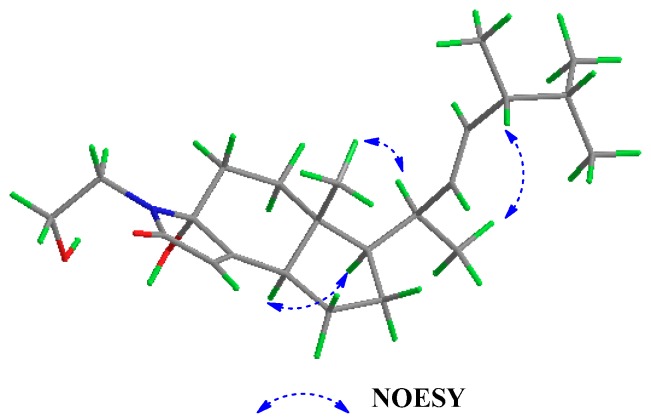
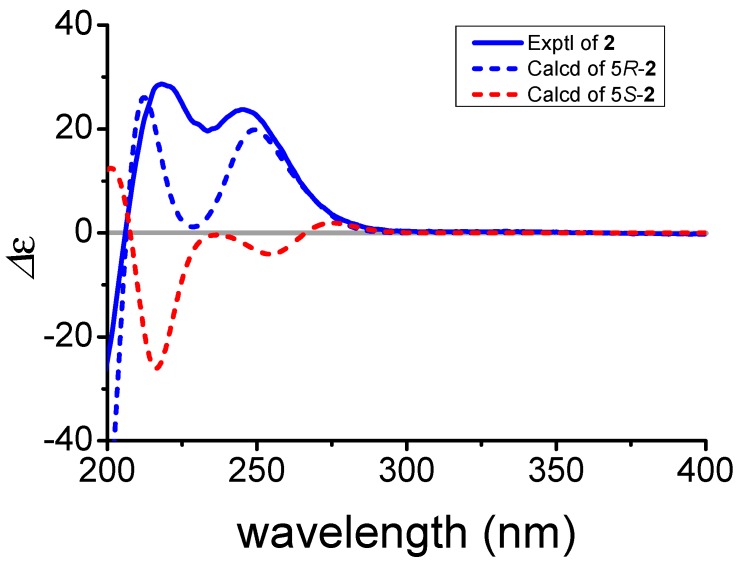
In the NOESY spectrum, NOE correlations (Figure 5) of H-13 with H-14, and H-9 with H-12, confirmed the β-orientation of the side chain and the α-orientation of H-12, respectively. A large coupling constant (14.7 Hz) between H-16 and H-17 indicated the E configuration of the double bond between C-16 and C-17. The relative configuration at C-14 and C-18 was tentatively assigned to be identical to those in 1 based on similar NMR spectrum data and biogenetic consideration. The orientation of 5-OH (δH 5.12) was tentatively defined to be α, because the comparison of the experimental and calculated CD spectra (Figure 6) facilitated assignment of the absolute configuration of 2 as 16E,5R,8R,12R,14S,18R. Compound 2 is named as anandin B.
Figure 5.
Key NOESY correlations of 2.
Figure 6.
Experimental and calculated ECD spectra of 2.
Anandins A (1) and B (2) are two rare steroidal alkaloids possessing a unique highly degraded ergosterol skeleton and a conjugated γ-lactam moiety. Why do two novel conjunction-type steroidal alkaloids exist in the ethanol extract of the mycelium material of Streptomyces anandii H41-59? With further investigation, we found that a cholesterol oxidase (Cho), usually observed among Gram-positive G+C-rich actinobacteria, is able to oxidize and degrade steroids [17]. In a previous study, we discovered a series of ergosterols including three new ones [16]. Thus, we considered that anandins A (1) and B (2) were probably derived from a highly degraded ergosterol [18,19].
2.2. Bioactivities of 1 and 2
Anandins A (1) and B (2) were evaluated in a wide panel of biological assays, including antimicrobial activity against Candida albicans, Escherichia coli, Staphylococcus aureus, Bacillus sp. and Dickey azeae, and anti-proliferative activity against human breast adenocarcinoma cell line MCF-7, human glioblastoma cell line SF-268 and human lung cancer cell line NCI-H460 by the methods described below. As shown in Table 2, both compounds 1 and 2 were shown to be active against three cancer cell lines. As a result, compound 1 exhibited moderate cytotoxicity against MCF-7, SF-268 and NCI-H460 with IC50 values of 7.5, 7.9, 7.8 μg/mL, respectively. However, both the compounds were inactive against the tested strains at the concentration of 20 μg/mL, even if the crude extract displayed moderate antimicrobial activity (inhibiting the zone of 16 mm against C. albicans with 6 mm paper discs at the concentration of 20 μg/mL).
Table 2.
Cytotoxicities of 1 and 2 (IC50: μg/mL).
| Cell Line | 1 | 2 | cis-Dichlorodiamine Platinum |
|---|---|---|---|
| MCF-7 | 7.5 | >50 | 4.0 |
| SF-268 | 7.9 | >50 | 41.0 |
| NCI-H460 | 7.8 | >50 | 25.1 |
Concentration range: 1.56–100 μg/mL; IC50: half maximal inhibitory concentration.
3. Experimental Section
3.1. General
Melting points were measured on an X-5 micro-MP apparatus (Huayan Corporation, Shanghai, China), uncorrected. Optical rotations were measured with a JASCO digital polarimeter (JASCO Corporation, Tokyo, Japan). UV spectra were measured on a JASCO V-550 UV/VIS spectrometer (JASCO Corporation, Tokyo, Japan). IR data were recorded with a Nicolet Impact 410-FTIR instrument (Thermo, San Jose, CA, USA) in KBr pellets. HR-ESI-MS were acquired on an Agilent 6210 LC/MSD TOF mass spectrometer (Agilent Technologies, Santa Clara, CA, USA). NMR spectra were measured on a Bruker AV-300 and AV-600 spectrometer (Bruker Instrument, Inc., Zurich, Switzerland). Chemical shifts were expressed in δ (ppm) and referenced to the NMR solvent used. X-ray crystallographic analysis was performed on an X calibur, sapphires, Gemini ultra diffractometer (Oxford Diffraction Ltd., Tokyo, Japan). The crystal was kept at 173.00(10) K during data collection. Using Olex2, the structure was solved with the ShelXS structure solution program using Direct Methods and refined with the ShelXL refinement package using Least Squares minimisation. HPLC was performed on an Agilent 1200 HPLC system (Agilent Technologies, Palo Alto, CA, USA) equipped with a diode array detector, using a column A (Ultimate XB-C18, 5 μm, 4.6 × 250 mm, Welch, Potamac, MA, USA) for analysis and a semi-preparative HPLC column B (Ultimate XB-C18, 5 μm, 10 × 250 mm, Welch, Potamac, MA, USA) for purification. Open column chromatography was performed on silica gel (300–400 mesh, Qingdao Haiyang Chemical Group Corporation, Qingdao, China). Sephadex LH-20 (25–100 mm) was purchased from Pharmacia (Uppsala, Sweden). HSGF254 silica gel TLC plates (0.2 mm thickness, 200 × 200 mm, Qingdao Marine Chemicals Co., Qingdao, China) were used for routine analysis of fractions. Strains Candida albicans, Escherichia coli, Staphylococcus aureus, Bacillus sp., and Dickey azeae were from the Institute of New Drug Research (Guangzhou, China) in our college.
3.2. Strain Isolation and Identification
The isolation and activation of actinomycete strain H41-59; the identification of morphological characteristics, physiological and chemical properties; the molecular genetic analysis and information on storing the strain are mentioned in our previous report [16].
3.3. Fermentation and Isolation
Fermentation of strain H41-59 including condition and media compositions were reported in our earlier paper [16]. The extract and partition procedure from fermentation material for the EtOAc extract were introduced in the same paper. The ethyl acetate-soluble extract (50 g) was dissolved in chloroform and loaded on the silica gel column (1.5 kg, 300–400 mesh, Qingdao, China) after filtration. A stepwise gradient elution of petroleum ether-EtOAc (10:0, 9:1, 8:2, 7:3, 6:4, 5:5, 4:6, 2:8 and 0:10 (v/v)) was used, and 20 fractions (Fr-1 to Fr-20) were obtained through TLC analysis and combination of fractions with same TLC pattern. Fr-6 was separated on Sephadex LH-20 (2 × 200 cm, CH2Cl2-MeOH, 1:1) and purified via semi-preparative HPLC (MeOH-H2O, 90:10) to yield two new natural products, namely, 1 (10 mg) and 2 (3 mg).
Compound 1 (anandin A): colorless plate crystal; M.P. = 133–135 °C; −13.5° (c 0.25, CHCl3); UV (MeOH) λmax (logε): 204.6 (3.88), 263.8 (3.73) nm; IR (KBr) νmax: 3382, 2958, 2930, 2871, 1666, 1650, 1372, 1300, 1191, 1111, 1015, 981, 620 cm−1; 1H NMR (CD3COCD3, 300 MHz) and 13C NMR (CD3COCD3, 75 MHz), see Table 1; HR-TOF-ESI-MS (positive) m/z 358.2752 [M + H]+ (calcd. for C23H36NO2, 358.2741).
Compound 2 (anandin B): white powder; M.P. = 129–131 °C; −15.1° (c 0.25, CHCl3); UV (MeOH) λmax (logε): 208.2 (3.78) nm; IR (KBr) νmax: 3423, 2929, 2853, 1650, 1542, 1461, 1381, 1190, 1110, 620 cm−1; 1H NMR (CD3COCD3, 600 MHz) and 13C NMR (CD3COCD3, 150 MHz), see Table 1; HR-ESI-MS: m/z 376.2860 [M + H]+ (calcd. for C23H38NO3, 376.2846).
3.4. Single-Crystal X-ray Data for Anandin A (1)
Crystal data (CCDC No. 1544792) for 1: C23H35NO2, M = 357.52, T = 100(2) K, monoclinic, space group C2, a = 21.7844 (3) Å, b = 7.2393 (11) Å, c = 13.1475 (20) Å, α = 90.00°, β = 93.2128 (13)°, γ = 90.00°, V = 2070.15 (5) Å3, Z = 4, μ (Cu Kα) = 0.555 mm−1, 16,181 reflections measured, 3274 independent reflections (Rint = 0.0282). The final R1 values were 0.0286 (I > 2σ (I)). The final wR (F2) values were 0.0747 (I > 2σ (I)). The final R1 values were 0.0290 (all data). The final wR (F2) values were 0.0750 (all data). The goodness of fit on F2 was 1.058. Flack parameter = 0.1 (2). Hooft parameter = 0.15 (6).
3.5. ECD Calculation
ECD calculations of Anandium B (2): The systematic random conformational analysis of two possible stereoisomers (5R and 5S) of 2 was performed in the SYBYL 8.1 program by using a MMFF94s molecular force field, which afforded 40 and 42 conformers for 5R and 5S respectively, with an energy cutoff of 10 kcal mol−1 to the global minima. All the obtained conformers were further optimized using DFT at the B3LYP/6-31+G (d) level in gas phase by using Gaussian09 software (Gaussian, Inc., Wallingford, CT, USA), and eight conformers of each stereoisomer were selected. All of the optimized stable conformers were used for TDDFT (cam-B3LYP/6-31+G (d)) computation of the excited states at the same levels, with consideration of the first 50 excitations. The overall ECD curves were weighted by the Boltzmann distribution of each conformer (with a half-band width of 0.25 eV), with a UV correction of 10 nm. The calculated ECD spectra of 5R and 5S of 2 were subsequently compared with the experimental spectra, respectively. The ECD spectra were produced by SpecDis 1.6 software (T. Bruhn, Y. Hemberger, A. Schaumlöffel, G. Bringmann, SpecDis version 1.6, University of Wuerzburg, Wuerzburg, Germany).
3.6. Biological Activities
3.6.1. Antimicrobial Activity
Antimicrobial evaluation of compounds 1 and 2 was performed following the same method and procedure as in a previous paper [16].
3.6.2. Cytotoxicity Assay
Three cancer cell lines MCF-7, SF-268, and NCI-H460 were used for measuring the cytotoxicities of compounds 1 and 2. For details on the procedure, we refer readers to our previous paper [16]. The 50% inhibition concentrations (IC50) of compounds 1 and 2 against the test cells were calculated using Origin 8 software (OriginLab, Northampton, MA, USA).
4. Conclusions
Anandins A (1) and B (2) were obtained from the fermented mycelia of Sterptomyces anandii H41-59. They have the same basic skeleton, and belong to a rare type of steroidal alkaloids. The structures of 1 and 2 were elucidated on the basis of extensive spectroscopic data including HR-ESI-MS, NMR, and X-ray crystallography for 1, and calculation chemistry for 2. To the best of our knowledge, 1 and 2 are the second reported compounds and second and third compounds with this type of structure. Anandin A (1) exhibited moderate in vitro inhibitory activity against MCF-7, SF-268, and NCI-H460 cell lines by the MTT method. Neither 1 nor 2 showed any antibiotic activities against the test pathogenic microorganisms.
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (No. 81373320), the Foundation of Guangdong Natural Sciences Committee (No. 2015A030312002), and Guangzhou Science and Technology Project (No. 201704030120), China. The 2D NMR experiments were measured with the assistance of the Xiangxue Pharmaceutical Factory Co., Ltd. X-ray crystallography was performed by Guo-qiang Li at Analytical and Testing Center Jinan University. Zhizhen Zhang at Zhejiang University read the manuscript.
Abbreviations
| COSY | Two dimensional 1H correlation |
| DEPT | Distortionless enhancement by polarization transfer |
| HPLC | High performance liquid chromatography |
| HMBC | 1H-detected heteronuclear multiple-bond correlation |
| HR-ESI-MS | High resolution electrospray ionization mass spectrometry |
| HSQC | 1H-detected heteronuclear single-quantum coherence |
| MTT | 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide |
| NMR | Nuclear magnetic resonance |
| NOESY | Nuclear overhauser effect spectroscopy |
| TLC | Thin layer chromatography |
Supplementary Materials
The following are available online at http://www.mdpi.com/1660-3397/15/11/355/s1, Figure S1: Spectra of compound 1; Figure S2: Spectra of compound 2; Table S1: X-ray diffraction data compound 1.
Author Contributions
Yang-Mei Zhang, Bai-Lian Liu and Guang-Xiong Zhou conceived of and designed the experiments. Yang-Mei Zhang, Bai-Lian Liu, Hong-Yu Li, Ying Zhang, Ting-Ting Zhang, Da-Yuan Sun, Bi-Run Lin performed the experiments. Yang-Mei Zhang, Xin-Heng Zheng and Bai-Lian Liu analyzed the spectroscopic data. Xiao-Jun Huang calculated the ECD data. Hong-Yu Li measured the IC50 on cytotoxicity. Yang-Mei Zhang and Guang-Xiong Zhou wrote the manuscript. Yang-Mei Zhang and Bai-Lian Liu equally contributed to this work, Bai-Lian Liu is co-first author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Podar M., Reysenbach A.L. New opportunities revealed by biotechnological explorations of extremophiles. Curr. Opin. Biotechnol. 2006;17:250–255. doi: 10.1016/j.copbio.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 2.Blunt J.W., Copp B.R., Keyzers R.A., Munro M.H.G., Prinsep M.R. Marine natural products. Nat. Prod. Rep. 2015;32:116–211. doi: 10.1039/C4NP00144C. [DOI] [PubMed] [Google Scholar]
- 3.Rahman A., Choudhary M.I. Chemistry and biology of steroidal alkaloids. In: Cordell G.A., editor. The Alkaloids: Chemistry and Biology. Volume 50. Elsevier; New York, NY, USA: 1998. pp. 61–108. [Google Scholar]
- 4.Jiang Q.W., Chen M.W., Cheng K.J., Yu P.Z., Wei X., Shi Z. Therapeutic potential of steroidal alkaloids in cancer and other diseases. Med. Res. Rev. 2016;36:119–143. doi: 10.1002/med.21346. [DOI] [PubMed] [Google Scholar]
- 5.Zhai H.Y., Zhao C., Zhang N., Jin M.N., Tang S.A., Qin N., Kong D.X., Duan H.Q. Alkaloids from Pachysandra terminalis inhibit breast cancer invasion and have potential for development as antimetastasis therapeutic agents. J. Nat. Prod. 2012;75:1305–1311. doi: 10.1021/np300207c. [DOI] [PubMed] [Google Scholar]
- 6.Ur-Rahman A., Choudhary M.I. Diterpenoid and steroidal alkaloids. Nat. Prod. Rep. 1997;14:191–203. doi: 10.1039/np9971400191. [DOI] [PubMed] [Google Scholar]
- 7.Li H.J., Jiang Y., Li P. Chemistry, Bioactivity and geographical diversity of steroidal alkaloids from the Liliaceae Family. Nat. Prod. Rep. 2006;23:735–752. doi: 10.1039/b609306j. [DOI] [PubMed] [Google Scholar]
- 8.Ata A., Andersh B.J. Buxus steroidal alkaloids: Chemistry and biology. Alkaloids Chem. Biol. 2008;66:191–213. doi: 10.1016/s1099-4831(08)00203-4. [DOI] [PubMed] [Google Scholar]
- 9.Cárdenas P.D., Sonawane P.D., Heinig U., Bocobza S.E., Burdman S., Aharoni A. The bitter side of the nightshades: Genomics drives discovery in Solanaceae steroidal alkaloid metabolism. Phytochemistry. 2015;113:24–32. doi: 10.1016/j.phytochem.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Rosser R.M., Faulkner D.J. Two steroidal alkaloids from a marine sponge, Plakina sp. J. Org. Chem. 1984;49:5157–5160. doi: 10.1021/jo00200a029. [DOI] [Google Scholar]
- 11.Aoki S., Watanabe Y., Sanagawa M., Setiawan A., Kotoku N., Kobayashi M. Cortistatins A, B, C, and D, anti-angiogenic steroidal alkaloids, from the marine sponge Corticium simplex. J. Am. Chem. Soc. 2006;128:3148–3149. doi: 10.1021/ja057404h. [DOI] [PubMed] [Google Scholar]
- 12.George R.P., Masuo I., Yoshiaki K., Claude D., Nigel C., Margaret L.N., Delbert L.H. Isolation and structure of the hemichordate cell growth inhibitors cephalostatins 2, 3, and 4. J. Chem. Soc. Chem. Commun. 1988;13:865–867. [Google Scholar]
- 13.Toumatia O., Yekkour A., Goudjal Y., Riba A., Coppel Y., Mathieu F., Sabaou N., Zitouni A. Antifungal properties of an actinomycin D-producing strain, Streptomyces sp. IA1, isolated from a Saharan soil. J. Basic Microbiol. 2015;55:221–228. doi: 10.1002/jobm.201400202. [DOI] [PubMed] [Google Scholar]
- 14.Triton T.R., Yee G. The anticancer agent adriamycin can be actively cytotoxic without entering cells. Science. 1982;217:248–250. doi: 10.1126/science.7089561. [DOI] [PubMed] [Google Scholar]
- 15.Kino T., Hatanaka H., Miyata S., Inamura N., Nishiyama M., Yajima T., Goto T., Okuhara M., Kohsaka M., Aoki H. FK-506, a novel immunosuppressant isolated from a Streptomyces. II. Immunosuppressive effect of FK-506 in vitro. J. Antibiot. 1987;40:1256–1265. doi: 10.7164/antibiotics.40.1256. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y.M., Li H.Y., Hu C., Sheng H.F., Zhang Y., Lin B.R., Zhou G.X. Ergosterols from the culture broth of marine Streptomyces anandii H41-59. Mar. Drugs. 2016;14:84. doi: 10.3390/md14050084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drzyzga O., Fernández de Las H.L., Morales V., Navarro L.J.M., Perera J. Cholesterol degradation by Gordonia cholesterolivorans. Appl. Environ. Microbiol. 2011;77:4802–4810. doi: 10.1128/AEM.05149-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dewick P.M. Medicinal Natural Products. John Wiley & Sons, Ltd.; London, UK: 2005. pp. 232–236. [Google Scholar]
- 19.Mansoor T.A., Hong J., Lee C.O., Bae S.J., Im K.S., Jung J.H. Cytotoxic sterol derivatives from a marine sponge Homaxinella sp. J. Nat. Prod. 2005;68:331–336. doi: 10.1021/np0496690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.