Future enhancements, including more metadata about drugs and increasingly detailed efficacy and toxicity information, will continue to improve the value of this free source of chemotherapy drug and regimen information.
Abstract
Purpose:
Cancer care involves extensive knowledge about numerous chemotherapy drugs and chemotherapy regimens. This information is constantly evolving, and there has been no freely available, comprehensive, centralized repository of chemotherapy information to date.
Methods:
We created an online, freely accessible, ad-free, collaborative wiki of chemotherapy information entitled HemOnc.org to address the unmet need for a central repository of this information. This Web site was developed with wiki development software and is hosted on a cloud platform. Chemotherapy drug and regimen information (including regimen variants), as well as other information of interest to hematology/oncology professionals, is housed on the site in a fully referenced and standardized format. Accredited users are allowed to freely contribute information to the site.
Results:
From its inception in November 2011, HemOnc.org has grown rapidly and most recently has detailed information on 383 drugs and 1,298 distinct chemotherapy regimens (not counting variants) in 92 disease subtypes. There are regularly more than 2,000 visitors per week from the United States and international locations. A user evaluation demonstrated that users find the site useful, usable, and recommendable.
Conclusion:
HemOnc.org is now the largest free source of chemotherapy drug and regimen information and is widely used. Future enhancements, including more metadata about drugs and increasingly detailed efficacy and toxicity information, will continue to improve the value of the resource.
Introduction
Cancer medicine is a large and complex arena, with 120+ disease subtypes that are often treated in radically different ways.1 The body of knowledge surrounding the treatment of cancer has grown rapidly as conventional chemotherapeutics have been augmented and sometimes supplanted by novel therapies, such as immunotherapy and targeted agents. Because of this complexity, the field has become fractured, with clinical trials and resultant treatment paradigms becoming increasingly narrow in scope. This has resulted in narrow approvals by the US Food and Drug Administration, widespread off-label use,2 and a dauntingly large knowledge space. Chemotherapy treatments are complex, costly, and potentially highly toxic; thus, there are multiple stakeholders with an interest in accurate and timely information about dosages, sequencing of therapies, supportive medications, and durations of treatment. These include, but are not limited to, physicians and physician extenders, nurses, pharmacists, cost and resource decision makers, payers, vendors of computerized provider order entry software, and patients.
In November 2011, we began an experiment that was based on the fundamental question: How can we improve the sharing and access of information in cancer medicine? This question was inspired in part by the Web site Chemoregimen.com,3 which was a commonly used free source of chemotherapy regimen information and primary references. By 2011, it was evident that this Web site was no longer being updated, and it had become a partner with a commercial entity, the Monthly Prescribing Reference. Additionally, there were no means to correct or augment the information content of the Chemoregimen site. To our knowledge, there were no other noncommercial electronic media that offered a comparable information source for chemotherapy regimens and chemotherapeutics. Thus, we created HemOnc.org to provide an online, collaborative, wiki-based knowledge base for chemotherapy regimens, chemotherapeutics, and other related information of interest to hematology/oncology professionals.
Wikis are Web sites that allow people to collaborate on the Internet to create documents and knowledge bases.4 In traditional publishing, articles are written by a limited number of authors/editors, and once published, the content is fixed, except for infrequent erratum and retractions. Information flows in one direction, from the content creators to the content consumers. Conversely, wikis crowdsource information from a potentially much larger pool of contributors, who may add or edit information in an article in real-time to incrementally improve articles through continuous revision. This facilitates a more rapid and comprehensive means of knowledge dissemination. Information flows back and forth between the original content creators and content consumers.
Wikis retain past versions of pages, which allows for straightforward auditing, reversal of errors, and attribution of contributions. Wikis are accessible from any Internet-connected device, searchable, and have internally cross-linked information to connect pages to one another, which facilitates ease of information discovery for users. However, depending on the qualifications and expertise of contributors, wikis have been criticized for being vulnerable to the addition of inaccurate or poorly edited information. Furthermore, depending on how openly granted editing rights are, wikis can be prone to vandalism. Despite these potential disadvantages, this model has proven to be quite successful; the free online encyclopedia Wikipedia5 is perhaps the most well-known example of a wiki. The legitimacy of this approach has recently been bolstered by the finding that pharmacology information on Wikipedia is 99.7% accurate compared with textbooks of pharmacology.6
We were specifically interested in whether an open, collaborative, chemotherapy regimen knowledge base that would be more comprehensive and accurate than existing resources could be created using a wiki platform. This article reports on the progress to date and the initial successes and challenges, and includes the results of a user evaluation.
Methods
Needs Analysis
Before creation of the site, we informally polled oncology trainees and attending physicians to ascertain focus areas for development. One of the most frequently cited frustrations was the lack of a comprehensive, up-to-date chemotherapy regimen reference. Commonly used references were often years old and contained a limited set of references that sometimes did not match the listed regimen. One group's chemotherapy ordering system did not consistently list references for the regimens being ordered. As a result, trainees in particular believed that they were spending too much time trying to find rather than reading the primary literature. Clinicians who were questioned wanted to have a reference that could be used from anywhere and a way to easily record useful information or Web sites to be accessed at a later time. They expressed frustration about the limits of information sharing via only direct communication and conferences and wished to have a means to share information with a broader audience outside of their institution and invite collaboration. Approximately 18 months after the Web site's creation, we conducted a formal survey of its users through the Web site to elicit usability feedback and to learn about users' experiences with errors and inconsistencies found in other resources.
Knowledge base generation.
To appropriately frame the scope of HemOnc.org, we created an organizational matrix based on Zack's knowledge strategy.7,8 This strategy describes knowledge as a function of two axes: Levels and Categories. Categories include: declarative knowledge (termed know-what knowledge), procedural knowledge (know-how), and analytic knowledge (know-why). Levels include core knowledge, advanced knowledge, and innovative knowledge. The initial focus of development was to flesh out the core knowledge components as much as possible. To inform these elements, a wide range of source materials was used to identify data for inclusion in the site. A list of commonly used sources is available (http://hemonc.org/wiki/Sources). Content was manually created by clinicians with access rights to the site.
Drug and chemotherapy regimen information.
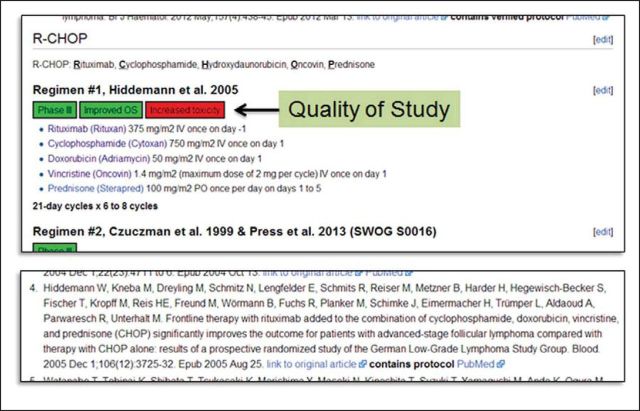
For chemotherapeutics and supportive medications included on the wiki, a page was created for each individual drug with general information about mechanism of action and specific information about safety, FDA approval history, common usage, and synonyms. Appendix Table A1 (online only) lists the current categories being applied to individual drugs. Information on antiemetic potential, which is a key concern for many chemotherapeutics, is accessible on the left sidebar of every page (http://hemonc.org/wiki/Antiemesis). For chemotherapy regimens included on the site, regimens are provided on a disease subtype–specific page and further classified by the context in which they were evaluated (eg, first-line metastatic, relapsed/refractory, adjuvant, and so on). An example is shown in Figure 1. Additional details about the information included on drug and chemotherapy regimen pages are available in the Appendix (online only).
Figure 1.
A portion of the rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP) regimen description for untreated follicular lymphoma. There are currently seven variants of this regimen for this context on HemOnc.org. Study design, efficacy, and toxicity information are shown directly above the regimen. All drug names are active links to the respective drug pages. The original reference is listed at the end of the regimen (references are listed in chronologic order), with links to the original article as well as the PubMed abstract page.
Other information.
In addition to a primary focus on chemotherapy drugs and regimens, HemOnc.org has several other content areas. These include sample order sets, lists of diagnosis and billing codes that are commonly used in hematology/oncology practice, reference tables for vesicant and irritant chemotherapeutics, performance status, corticosteroid conversions, and a large number of external links. There is also an extensive style guide (http://hemonc.org/wiki/Style_guide) to guide content contributors.
Platform and security.
HemOnc.org is hosted on a server within a data center run by Linode (Galloway, NJ), a computer hosting provider. The server runs on the Ubuntu Linux operating system (Canonical, London, United Kingdom) with Apache (Apache Software Foundation, Forest Hill, MD) as the Web server and MySQL (Oracle, Redwood City, CA) as the database. The Web site is powered by MediaWiki software (Wikimedia Foundation, San Francisco, CA). There are multiple security measures taken to ensure the integrity and performance of the system. The server is regularly patched with security updates to limit susceptibility to unauthorized access (hacking), and multiple offsite backups are regularly created to protect against data loss. Traffic for the site is routed through the CloudFlare content delivery network (CloudFlare, San Francisco, CA), which helps to improve speed and block traffic from spammers and bots (which include malicious software and/or humans that harvest user data to post or e-mail unsolicited, commercial, and/or malicious content). Asirra9 (Animal Species Image Recognition for Restricting Access; Microsoft, Redmond, WA) is used during the account creation process to ensure that creators are human rather than automated computer programs that are attempting to create spam accounts. Most importantly, the administrators verify the credentials of all users who wish to obtain access rights to add or modify content, and every account is manually approved.
Contributorship.
The Web site does not require visitors to login or have an account to use the site, but people who sign up for a free account have additional benefits, such as custom views. After creating an account, individuals must manually contact the administrators if they wish to have editing privileges. Editing privileges are generally restricted to hematology/oncology professionals (eg, physicians, nurse practitioners, physician assistants, and pharmacists). Thus far, 23 users have been granted editing privileges on the site. Of those 23 users, 10 have contributed content to HemOnc.org.
Results
Content Growth
The first version of the HemOnc.org Web site went live in November 2011. By December 2011, there was content on 66 distinct chemotherapy regimens in eight disease subtypes, and 132 drugs (all FDA approved). By June 2014, there were 1,298 distinct chemotherapy regimens in 58 disease subtypes that are further divided into 92 distinct entities (Appendix Table A2, online only), and 383 drugs (100 of which are in clinical trials). Figure 2 shows the growth of chemotherapy regimen and drug content over time. Annotation of chemotherapy regimens has also increased over time. In June 2013, less than 5% of regimens had information about study design; by February 2014, more than 99% of regimens had such information. The content of drug pages has also expanded over time, with information about FDA approval dates and indications, as well as synonyms, including RxNorm concept unique identifiers, for some drugs. For example, dexamethasone has three RxNorm concept unique identifiers (dexamethasone acetate [22690]; dexamethasone phosphate [235486]; dexamethasone sodium phosphate [48933]) and 529 synonyms. The latest enhancement to the site, which is still underway, is the addition of categorization metatags to drugs (Appendix Table A1).
Figure 2.
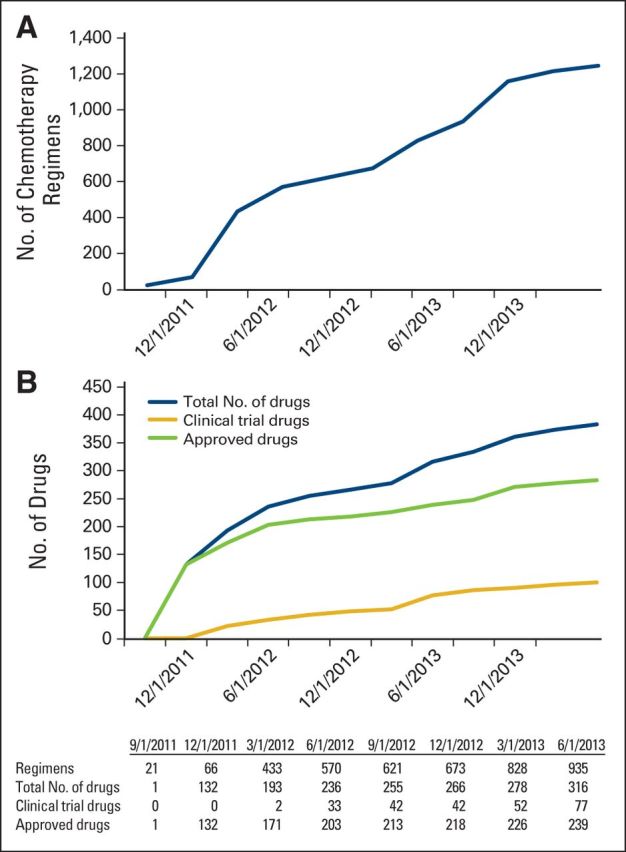
(A) The number of chemotherapy regimens for which information is available on HemOnc.org (count does not include variants of the same regimen). Counts were sampled at 3-month intervals. (B) The number of drugs (antineoplastic and supportive) for which information is available on HemOnc.org. Counts were sampled at 3-month intervals.
Visitors.
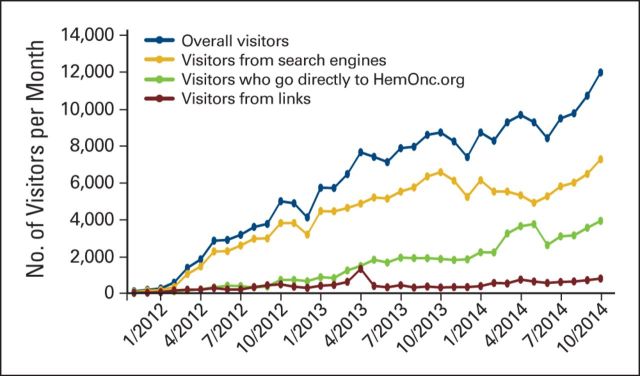
Figure 3 illustrates how the user traffic to HemOnc.org has changed during its existence. The site had 34,327 visits in 2012, and traffic increased by 159% in 2013, when there were 88,974 visits. From January 2014 through July 2014, there were 63,252 visits. Overall, 69.5% of users entered the site from search engines; 23.7% of users directly entered the address in their Web browsers or had it bookmarked; and 6.8% of users arrived via referral links from other sites. Over time, presumably as people became more familiar with the site, a greater percentage of them accessed it directly. For example, in July 2014, 32.5% of users directly accessed the site, 61.1% used a search engine, and 6.3% entered via a referral link.
Figure 3.
The number of visitors per month to HemOnc.org. The blue line represents the total number of visitors. The yellow, green, and brown lines represent the number of visitors per month who entered the site via search engines, directly entering the address in web browsers, and referral links from other sites, respectively.
User evaluation.
We conducted a survey to evaluate the usability of the site. The survey was open from May 1 through May 31, 2013. Survey data were collected and managed using REDCap (Research Electronic Data Capture) software hosted at Vanderbilt University.10 REDCap is a secure, Web-based application that is designed to support data capture for research studies, providing an intuitive interface for validated data entry, audit trails for tracking data manipulation and export procedures, automated export procedures for seamless data downloads to common statistical packages, and procedures for importing data from external sources. Because this survey was anonymous and HemOnc.org does not fall under the auspices of an institutional review board, institutional review board approval was not obtained. Participants were invited by site announcements, social media, and e-mails to hematology/oncology fellowship programs. Usability was assessed by 100-point Likert scales (0 being worst and 100 being best). Other data, including information about which other resources the respondents often used, were collected via multiple choice questions with optional free text entry.
There were 139 respondents, the majority of which were physicians (61%; 26% were nurses, 10% were pharmacists, and 3% were other). The respondents expressed that the site was useful (median score, 90; interquartile range [IQR], 76-99.5), usable (median score, 85; IQR, 69-98), and recommendable to colleagues (median score, 87; IQR, 72-99). All respondents (100%) reported using other references (eg, textbooks), but 70.5% reported accuracy issues with these references. The issues most commonly reported by respondents were: lack of precision needed to properly give the regimen (55%); schedule of administration incorrect (25%); dosage, including body-surface area–based dosing, missing or incorrect (23%); spelling/typographic errors (9%); and route of administration incorrect (5%). Interestingly, only 38 respondents (27%) reported using information provided through their electronic medical record (EMR).
Many respondents indicated a willingness to contribute to the site, including through indirect means such as e-mailing editors with corrections—although only four of 139 (3%) reported being active contributors. For those unwilling to contribute (23 respondents; 17%), the leading reason was a lack of time (15 of 23, 65%). Additional details are available in Appendix Table A3 (online only).
Discussion
HemOnc.org has been a success, as measured by the increase in content, number of visitors, and generally positive user satisfaction. Because of its wiki platform, new content can be added and adapted in real-time to provide whatever information is most interesting to its users. In addition to providing content to a wide variety of stakeholders who are participating in the care of patients with cancer, the site has enabled secondary investigations into the manner in which evidence for cancer treatment evolves over time. For example, we have used HemOnc.org as a content source to inform a treatment regimen network analysis of first-line treatments of chronic myelogenous leukemia,11,12 and have also conducted preliminary investigations into the reclassification of the cancer ontology as a function of treatment.13 Future work will expand on these preliminary studies; one goal is the automated synthesis of treatment guidelines.14 This is especially relevant for disease contexts for which there are large numbers of published regimens, such as the first-line treatment of chronic lymphocytic leukemia (30 regimens on HemOnc.org, 19 of which have been evaluated in randomized controlled trials) or the treatment of metastatic erb-b2 receptor tyrosine kinase 2 (ERBB2/HER2) –negative breast cancer (40 regimens on HemOnc.org, 29 of which have been evaluated in randomized controlled trials).
Several challenges have emerged with this process, most notably, the lack of broad user participation with regard to updating content, and the challenges of dissemination. With respect to the former, it has been well documented that content generation and editing is nonuniform in social media, with a small number of users contributing disproportionately to projects such as Wikipedia.15,16 Medical professionals are typically well compensated for their time and have many conflicting priorities vying for their time, such as patient care, academic pursuits, and teaching. It may be appealing to some individuals to be able to instantly share information that is attributed to them on a well-trafficked, specialized site that their peers visit, but because there is no monetary reward for contributions and limited prestige is associated with an emerging Web site, user participation may remain a challenge. People who requested editing privileges and who did not subsequently contribute or stopped contributing often cited lack of time as a main factor.
Dissemination presents another set of challenges, primarily related to the fact that HemOnc.org is noncommercial and does not have a traditional marketing budget. Thus, awareness of the site is spread by traditional word-of-mouth, media exposure,17 reciprocal exposure on sites such as MyCancerGenome.org,18 and social media outlets such as Facebook and Twitter. It is notable that a majority of traffic arrives through search engines. The importance of search engine optimization cannot be overemphasized, and we have found some success here. For example, a search for the phrase vesicant chemotherapy on Google or Bing will return HemOnc.org as the first result (as of the writing of this article).
A future wish list for enhanced functionality includes improving usability on mobile platforms; enabling HL7 Infobutton technology19 so that drug and regimen information can be obtained by external EMRs; completion of efficacy and toxicity data for all regimens; verification of all unverified regimens; inclusion of more gray literature and abstract sources; determination of so-called outdated status for older regimens that are no longer commonly used; and increased outreach and engagement of patients. Additional information could be added on topics including the cost of treatments, drug shortages, and adverse events.
Many oncology practices use proprietary EMRs and oncology suites with built-in chemotherapy order sets, but they are limited by the release cycles of their vendors or expediency of their in-house order set writers. In a landscape in which the work of discovering new information is often internally duplicated many times over because of lack of information sharing across institutions, we believe that a free, public resource such as HemOnc.org can complement existing tools and be a valuable supplement for our field so that information can be rapidly disseminated and accessed.
In conclusion, HemOnc.org has become, over the course of approximately 3 years, the largest, freely available chemotherapy drug and regimen resource. We anticipate continued growth in content and invite all readers of this article to consider participation in this collaborative project.
Acknowledgment
We thank the contributors of HemOnc.org.
A preliminary version of the survey results was presented in poster format at the American Society of Clinical Oncology Quality Care Symposium, San Diego, CA, November 1 and 2, 2013.
Appendix
Drug information.
For chemotherapeutics and supportive medications included on the wiki, a page was created for each individual drug with, ideally, the following information: (1) general information, including a description of the mechanism of action, route of administration, extravasation information (when available for intravenous medications), and links to package inserts and Risk Evaluation and Mitigation Strategy information (when present); (2) patient drug information, including links to widely used patient drug information Web sites (Chemocare.com and UpToDate); (3) diseases for which the drug is commonly used; (4) date(s) of US Food and Drug Administration (FDA) approval, including the specific indication and any changes in indication; (5) synonyms, including brand name(s) and precise names in the RxNorm terminology (a systematic nomenclature for drugs developed by the National Library of Medicine; Nelson SJ, et al: J Am Med Inform Assoc 18:441-448, 2011); (6) structured categories based on mechanism of action, indication, and so on. For example, the drug carfilzomib [http://hemonc.org/wiki/Carfilzomib_(Kyprolis)] is categorized as a proteasome inhibitor, a multiple myeloma medication (on the basis of FDA indication; Herndon TM, et al: Clin Cancer Res 19:4559-4563, 2013), a Waldenström macroglobulinemia medication (on the basis of positive phase II trial results; Treon SP, et al: Blood 124:503-510, 2014), and a drug FDA approved in 2012. For drugs to be listed on the wiki, they should have FDA or European Medicines Agency approval or have promising published results (generally defined as phase II results that are sufficiently positive to warrant a phase III trial). Investigational drugs are denoted as such and additionally categorized into an investigational agents category. Discontinued drugs (eg, gemtuzumab ozogamicin) are categorized as such; clinical trial drugs failing in the evaluation stage are removed from the wiki when the failure is clearly disclosed by the manufacturer.
Regimen information.
For chemotherapy regimens included on the site, regimens are listed on a disease subtype–specific page and further classified by the context in which they were evaluated, for example, first-line metastatic, relapsed/refractory, adjuvant, and so on. Under each major contextual subheading, regimens are listed in alphabetical order by their commonly known abbreviation (eg, R-CHOP [rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone]) or by individual drug names in the absence of a commonly known abbreviation. Synonyms are listed as well as the Unified Medical Language System structured concept (Bodenreider O: Nucleic Acids Res 32:D267-D270, 2004) when available (eg, Unified Medical Language System concept unique identifier C0393023 represents R-CHOP). Each variant of a regimen is listed separately, with the primary author and study consortium name, if there is one, listed (eg, there are seven variants of R-CHOP listed for untreated diffuse large B-cell lymphoma; the first variant listed was used by the LNH-98.5 [Coiffier B, et al: N Engl J Med 346:235-242, 2002] and LNH03-6B [Delarue R, et al: Lancet Oncol 14:525-533, 2013] study consortiums). Directly under each individual variant, a colored box containing information about the level of evidence for the regimen is displayed, using a green-yellow-red traffic light metaphor. Generally, green represents regimens that have been evaluated in a randomized fashion (eg, phase III studies), yellow represents regimens that have been evaluated in at least 20 patients in a nonrandomized fashion (eg, phase II studies, large pilots), and red represents regimens that have been evaluated in fewer than 20 patients or have been evaluated retrospectively. The chemotherapy regimen is then listed by individual drug (with a link to the individual drug page) with dosage, route, infusion time, and day(s) of administration listed. Missing or ambiguous information in the primary article is denoted, except for infusion time, which is usually missing and therefore simply omitted when not available. Supportive medications reported in the article are also listed (these are often omitted or ambiguous). Finally, the length of cycles and the total number of cycles are listed. Primary references are then listed accompanying each regimen, with the following special tags as needed: Abstract (no manuscript has yet been published to our knowledge); Update (an update to a previously published regimen); Retrospective; Meta-Analysis. Once a regimen has been published in a peer-reviewed publication, abstracts are hidden from view. Links to original articles as well as the PubMed citation are provided for every reference, and references for a full publication, rather than an abstract or other early publication, that was reviewed and confirmed as accurate by an authorized contributor are denoted as “contains verified protocol.” References that contain regimen information that could not be thoroughly reviewed —such as publications to which the contributors did not have subscription access—are denoted as “contains protocol.”
Table A1.
The Main Drug Categories and Subcategories in HemOnc.org
| Categories |
|---|
| Cytotoxic chemotherapy |
| Alkylating agents |
| Anthracyclines |
| DNA synthesis inhibitors |
| Nitrogen mustards |
| Nitrosureas |
| Nucleic acid analogs |
| Platinum agents |
| Proteasome inhibitors |
| Taxanes |
| Microtubule inhibitors |
| Topoisomerase inhibitors |
| Vinca alkaloids |
| Antimetabolites |
| Antifolates |
| Purine analogues |
| Pyrimidine analogues |
| Kinase inhibitors |
| AAK inhibitors |
| ALK inhibitors |
| Bcr-Abl inhibitors |
| BRAF inhibitors |
| BTK inhibitors |
| CDK inhibitors |
| EGFR inhibitors |
| FGFR inhibitors |
| FLT3 inhibitors |
| HDAC inhibitors |
| JAK inhibitors |
| KIT inhibitors |
| LYN inhibitors |
| MEK inhibitors |
| MET inhibitors |
| mTOR inhibitors |
| PDGFR inhibitors |
| PLK1 inhibitors |
| RET inhibitors |
| ROS1 inhibitors |
| SRC inhibitors |
| SYK inhibitors |
| TEK inhibitors |
| VEGF inhibitors |
| Corticosteroids and corticosteroid mimetics |
| Antiandrogens |
| Corticosteroid synthesis inhibitors |
| Androgen receptor inhibitors |
| 5 alpha-reductase inhibitors |
| GnRH agonists |
| GnRH antagonists |
| Aromatase inhibitors |
| Selective estrogen receptor modulators |
| Corticosteroids |
| Somatostatin analogs |
| Biologics |
| Antibody medications |
| Antibody-drug conjugates |
| Anti-HER2 medications |
| IL-6 inhibitors |
| Enzymes |
| Immunotherapy |
| Immunomodulatory drugs |
| Investigational and discontinued |
| Investigational |
| Discontinued |
| Supportive medications |
| Corticosteroids |
| Bisphosphonates |
| RANK ligand inhibitors |
| Antimicrobial |
| Antivirals |
| PCP prophylaxis |
| Chemotherapy protective agents |
| Radioactive agents |
| Alpha emitters |
| Radioimmunotherapy |
| Benign hematology medications |
| Hemostasis medications |
| Coagulation factors |
| Fibrinolysis inhibitors |
| Direct thrombin inhibitors |
| Factor Xa inhibitors |
| Heparins |
| Low-molecular-weight heparins |
| Phosphodiesterase inhibitors |
| Cyclooxygenase inhibitors |
| P2Y12 ADP inhibitors |
| Chelators |
| Hematopoietic growth factors |
| Erythrocyte growth factors |
| Granulocyte growth factors |
| Megakaryocyte growth factors |
| Miscellaneous |
| Retinoids |
| Vitamins |
| Immunosuppresants |
| Vasopressin analogs |
| Medications by cancer subtype |
| Acute lymphocytic leukemia medications |
| Acute myeloid leukemia medications |
| Acute promyelocytic leukemia medications |
| Aggressive non-Hodgkin lymphoma medications |
| Anal cancer medications |
| Basal cell and squamous cell skin cancer medications |
| Bladder cancer medications |
| Bone cancer medications |
| Breast cancer medications |
| Cancer of unknown primary medications |
| Castleman's disease medications |
| CNS cancer medications |
| CNS lymphoma medications |
| Cervical cancer medications |
| Chronic lymphocytic leukemia and small lymphocytic lymphoma medications |
| Chronic myelogenous leukemia medications |
| Chronic myelomonocytic leukemia medications |
| Colon cancer medications |
| Esophageal cancer medications |
| Essential thrombocythemia medications |
| Follicular lymphoma medications |
| Gastric cancer medications |
| Hairy cell leukemia medications |
| Head and neck cancer medications |
| Hepatobiliary cancer medications |
| HIV-associated lymphoma medications |
| Hodgkin lymphoma medications |
| Hodgkin lymphoma, nodular lymphocyte–predominant medications |
| Immune thrombocytopenic purpura medications |
| Light-chain amyloidosis medications |
| Mantle cell lymphoma medications |
| Marginal zone lymphoma medications |
| Melanoma medications |
| Mesothelioma medications |
| Multiple myeloma medications |
| Myelodysplastic syndrome medications |
| Myelofibrosis medications |
| Non-Hodgkin lymphoma medications |
| Non–small-cell lung cancer medications |
| Neuroendocrine tumor medications |
| Ovarian cancer medications |
| Pancreatic cancer medications |
| Paroxysmal nocturnal hemoglobinuria medications |
| Penile cancer medications |
| Polycythemia vera medications |
| Prostate cancer medications |
| Rectal cancer medications |
| Renal cancer medications |
| Sarcoma medications |
| Small-cell lung cancer medications |
| T-cell lymphoma medications |
| Testicular cancer medications |
| Thymoma medications |
| Thyroid cancer medications |
| Transplantation medications |
| Uterine cancer medications |
| Waldenström macroglobulinemia medications |
| Medications by year of approval |
| Specific year of initial approval (eg, 2010) |
NOTE. There are 12 main drug categories. One medication can belong to as many subcategories as are relevant.
Abbreviations: AAK, Aurora A kinase; ADP, adenosine diphosphate; ALK, anaplastic lymphoma kinase; BRAF, serine/threonine-protein kinase B-Raf; BTK, Bruton's tyrosine kinase; CDK, cyclin-dependent kinase; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; FLT3, fms-related tyrosine kinase 3; GnRH, gonadotropin releasing hormone; HDAC, histone deacetylase; HER2, human epidermal growth factor receptor 2; IL-6, interleukin 6; JAK, Janus kinase; KIT, tyrosine-protein kinase c-Kit; LYN, tyrosine-protein kinase Lyn; MEK, mitogen-activated protein kinase kinase; MET, hepatocyte growth factor receptor; mTOR, mammalian target of rapamycin; PCP, pneumocystis pneumonia; PDGFR, platelet-derived growth factor receptor; PLK1, polo-like kinase 1; RET, tyrosine-protein kinase ret; ROS1, proto-oncogene tyrosine-protein kinase ROS; SRC, proto-oncogene tyrosine-protein kinase Src; SYK, spleen tyrosine kinase; TEK, angiopoietin-1 receptor; VEGF, vascular endothelial growth factor.
Table A2.
Disease Subtypes Currently Represented on HemOnc.org, With No. of Regimens for Each Subtype (excludes regimen variants)
| Subtype | No. of Regimens |
|---|---|
| Benign hematology | |
| Aplastic anemia | 1 |
| Autoimmune thrombocytopenic purpura | 6 |
| Castleman's disease | 1 |
| Paroxysmal nocturnal hemoglobinuria | 2 |
| Malignant hematology | |
| Acute lymphocytic leukemia | 23 |
| Ph positive | |
| Ph negative | |
| Acute myeloid leukemia | 41 |
| Acute promyelocytic leukemia | 22 |
| Aggressive non-Hodgkin lymphoma | 68 |
| DLBCL | |
| Burkitt's lymphoma | |
| Primary mediastinal B-cell lymphoma | |
| Gastric DLBCL | |
| Chronic lymphocytic leukemia and small lymphocytic lymphoma | 55 |
| Chronic myelogenous leukemia | 25 |
| Chronic phase | |
| Accelerated phase | |
| Blast crisis | |
| Chronic myelomonocytic leukemia | 1 |
| CNS lymphoma | 7 |
| Essential thrombocythemia | 2 |
| Follicular lymphoma | 51 |
| Hairy cell leukemia | 5 |
| HIV-associated lymphoma | 9 |
| Hodgkin lymphoma | 26 |
| Hodgkin lymphoma, nodular lymphocyte-predominant | 8 |
| Large granular lymphocytic leukemia | 2 |
| Light-chain amyloidosis | 14 |
| Mantle cell lymphoma | 29 |
| Marginal zone lymphoma | 20 |
| Multiple myeloma | 52 |
| Myelodysplastic syndrome | 10 |
| Myelofibrosis | 6 |
| Polycythemia vera | 1 |
| T-cell lymphoma | 28 |
| Anaplastic large-cell lymphoma | |
| Cutaneous T-cell lymphoma | |
| Extranodal NK/T-cell lymphoma, nasal type | |
| NK/T-cell lymphoma | |
| Peripheral T-cell lymphoma NOS | |
| Waldenström macroglobulinemia | 11 |
| Other | |
| Transplantation conditioning regimens | 18 |
| Autologous stem-cell transplantation | |
| Allogeneic stem-cell transplantation | |
| Solid oncology | |
| Anal cancer | 4 |
| Basal cell and squamous cell skin cancer | 8 |
| Basal cell carcinoma | |
| Squamous cell carcinoma | |
| Bladder cancer | 27 |
| Bone cancer | 28 |
| Chondrosarcoma | |
| Ewing's sarcoma | |
| Osteosarcoma | |
| Malignant fibrous histiocytoma of bone | 94 |
| Breast cancer | |
| HER2 negative | |
| HER2 positive | |
| CNS cancer | 35 |
| Anaplastic glioma | |
| Glioblastoma multiforme | |
| Oligodendroglioma | |
| Supratentorial astrocytoma | |
| Cervical cancer | 27 |
| Colon cancer | 34 |
| Esophageal cancer | 79 |
| Gastric cancer | 6 |
| Head and neck cancer | 31 |
| Hepatobiliary cancer | 24 |
| Hepatocellular carcinoma | |
| Biliary tract cancer | |
| Melanoma | 27 |
| Mesothelioma | 11 |
| Neuroendocrine tumors | 29 |
| Adrenal gland tumors | |
| Carcinoid tumors | |
| Pancreatic neuroendocrine islet cell tumors | |
| Pheochromocytoma | |
| Non–small-cell lung cancer | 39 |
| Ovarian cancer | 31 |
| Pancreatic cancer | 21 |
| Penile cancer | 11 |
| Prostate cancer | 36 |
| Rectal cancer | 9 |
| Renal cancer | 18 |
| Sarcoma | 27 |
| Angiosarcoma | |
| GI stromal tumor | |
| Giant-cell tumor of bone | |
| Kaposi sarcoma | |
| Various other histologies | |
| Small-cell lung cancer | 27 |
| Testicular cancer | 17 |
| Pure seminoma | |
| Nonseminoma | |
| Thymoma | 9 |
| Thyroid cancer | 8 |
| Medullary | |
| Various other histologies | |
| Unknown primary | 14 |
| Adenocarcinoma or carcinoma NOS | |
| Squamous cell carcinoma | |
| Neuroendocrine | |
| Uterine cancer | 23 |
| Endometrioid | |
| Various other histologies | |
| Total | 1,298 |
Abbreviations: DLBCL, diffuse large B-cell lymphoma; HER2, human epidermal growth factor receptor 2; NK, natural killer; NOS, not otherwise specified.
Table A3.
Results of Questions From Usability Survey
| Questions and Answers | Respondents |
|
|---|---|---|
| No. | % | |
| Q1: Have you contributed to HemOnc.org? | ||
| A1: Yes | 4 | 3 |
| A2: No | 135 | 97 |
| Q2 (Multiple choices are allowed): HemOnc.org is a collaborative wiki. This type of site allows all of its users to work together and edit the pages you see. In what ways would you consider contributing to this site? | ||
| A1: Add example order sets | 18 | 13 |
| A2: Add new regimens | 23 | 17 |
| A3: Add new references | 31 | 22 |
| A4: Correct mistakes | 33 | 24 |
| A5: Create new pages that will be helpful to me in clinical practice, such as checklists for certain diseases | 27 | 19 |
| A6: Would not create an account and personally modify any information, but would e-mail an editor about errors, references/papers to be added, or other information to be included | 44 | 32 |
| A7: Would not be interested in contributing at all | 23 | 17 |
| Q3 (Only visible if A7 is checked for Q2, above; multiple choices are allowed): Which of the following best describes the reason for your unwillingness to contribute? | ||
| A1: Fear of introducing errors | 6 | 26 |
| A2: HemOnc.org not likely to be around for long | 1 | 4 |
| A3: I'm not tech-savvy enough | 6 | 26 |
| A4: Information is complete already | 1 | 4 |
| A5: Lack of knowledge (real or perceived) | 3 | 13 |
| A6: Not enough time | 15 | 65 |
| A7: Not worth my time | 2 | 9 |
| A8: Too steep of a learning curve | 0 | 0 |
| Q4 (Multiple choices are allowed): Please choose which of the below, if any, would help you start contributing (or contribute more, if you're already contributing) | ||
| A1: Continuing medical education (CME) or similar professional development recognition | 38 | 27 |
| A2: A different user interface | 9 | 6 |
| A3: Inclusion in future publications based on number of contributions | 15 | 11 |
| A4: Recognition as a content expert (eg, section editor) | 21 | 15 |
| A5: Reminder emails | 22 | 16 |
| A6: A small monetary compensation for each contribution | 21 | 15 |
| A7: None of the above | 47 | 34 |
| A8: Other | 5 | 4 |
NOTE. These questions specifically focused on the degree to which users have contributed, reasons for noncontribution, and barriers to contribution. Except for Q1, answers may not sum up to 100% because the questions were multiple choice, and leaving a question unanswered was permitted.
Abbreviations: A, answer; Q, question.
Authors' Disclosures of Potential Conflicts of Interest
Disclosures provided by the authors are available with this article at jop.ascopubs.org.
Author Contributions
Conception and design: Jeremy L. Warner, Peter C. Yang
Financial support: Jeremy L. Warner
Administrative support: Jeremy L. Warner
Collection and assembly of data: All authors
Data analysis and interpretation: Jeremy L. Warner, Peter C. Yang
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
HemOnc.org: A Collaborative Online Knowledge Platform for Oncology Professionals
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jop.ascopubs.org/site/misc/ifc.xhtml.
Jeremy L. Warner
No relationship to disclose
Andrew J. Cowan
Stock or Other Ownership: Doximity
Consulting or Advisory Role: Doximity
Aric C. Hall
No relationship to disclose
Peter C. Yang
Stock or Other Ownership: Merck, Pfizer, Cyclacel
References
- 1.Kleihues P, Sobin LH: World Health Organization classification of tumors Cancer 88:2887,2000 [DOI] [PubMed] [Google Scholar]
- 2.Levêque D: Off-label use of anticancer drugs Lancet Oncol 9:1102–1107,2008 [DOI] [PubMed] [Google Scholar]
- 3.Chemoregimen.com. Home of all chemotherapy regimens. http://chemoregimen.com/
- 4.Wagner C: Wiki: A technology for conversational knowledge management and group collaboration Commun Assoc Inf Syst 13:58,2004 [Google Scholar]
- 5.Wikipedia. Wikipedia. http://en.wikipedia.org/wiki/Wikipedia.
- 6.Kräenbring J Monzon Penza T Gutmann J, etal: Accuracy and completeness of drug information in Wikipedia: A comparison with standard textbooks of pharmacology PLoS One 9:e106930,2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zack MH: Developing a knowledge strategy Calif Manage Rev 41:125–145,1999 [Google Scholar]
- 8.Zack M, McKeen J, Singh S: Knowledge management and organizational performance: An exploratory analysis J Knowl Manage 13:392–409,2009 [Google Scholar]
- 9.Elson J Douceur JR Howell J, etal: Asirra: A Captcha that exploits interest-aligned manual image categorization. Proceedings of the Association for Computing Machinery Conference on Computer and Communications Security Citeseer 366–374,2007 [Google Scholar]
- 10.Harris PA Taylor R Thielke R, etal: Research electronic data capture (REDCap): A metadata-driven methodology and workflow process for providing translational research informatics support J Biomed Inform 42:377–381,2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warner J, Yang P, Alterovitz G: Automated synthesis and visualization of a chemotherapy treatment regimen network Stud Health Technol Inform 192:62–66,2013 [PMC free article] [PubMed] [Google Scholar]
- 12.Warner JL, Yang P, Alterovitz G: Reversal of medical practices Mayo Clin Proc 88:1182–1183,2013 [DOI] [PubMed] [Google Scholar]
- 13.Gao M Warner J Yang P, etal: On the Bayesian derivation of a treatment-based cancer ontology 210–218,2014Proceedings of the American Medical Informatics Association Summit on Clinical Research Informatics [PMC free article] [PubMed] [Google Scholar]
- 14.Yu P, Artz D, Warner J: Electronic health records (EHRs): Supporting ASCO's vision of cancer care Am Soc Clin Oncol Educ Book 34:225–231,2014 [DOI] [PubMed] [Google Scholar]
- 15.Lerman K: User participation in social media: Digg study 255–258,20072007 IEEE/WIC/ACM International Conferences on Web Intelligence and Intelligent Agent Technology Workshops [Google Scholar]
- 16.Kittur A Chi E Pendleton BA, etal: Power of the few vs. wisdom of the crowd: Wikipedia and the rise of the bourgeoisie World Wide Web 1:19,2007 [Google Scholar]
- 17.Butcher L: How health IT is changing the practice of oncology: HemOnc.org—Sharing oncology information easily Oncology Times 35:16,2013 [Google Scholar]
- 18.Swanton C: My Cancer Genome: A unified genomics and clinical trial portal Lancet Oncol 13:668–669,2012. 22748256 [Google Scholar]
- 19.Del Fiol G Huser V Strasberg HR, etal: Implementations of the HL7 Context-Aware Knowledge Retrieval (“Infobutton”) standard: Challenges, strengths, limitations, and uptake J Biomed Inform 45:726–735,2012 [DOI] [PMC free article] [PubMed] [Google Scholar]