Abstract
Background
Sperm DNA is protected against fragmentation by a high degree of chromatin packaging. It has been demonstrated that proper chromatin packaging is important for boar fertility outcome. However, little is known about the molecular mechanisms underlying differences in sperm DNA fragmentation. Knowledge of sequence variation influencing this sperm parameter could be beneficial in selecting the best artificial insemination (AI) boars for commercial production. The aim of this study was to identify genes differentially expressed in testis tissue of Norwegian Landrace and Duroc boars, with high and low sperm DNA fragmentation index (DFI), using transcriptome sequencing.
Results
Altogether, 308 and 374 genes were found to display significant differences in expression level between high and low DFI in Landrace and Duroc boars, respectively. Of these genes, 71 were differentially expressed in both breeds. Gene ontology analysis revealed that significant terms in common for the two breeds included extracellular matrix, extracellular region and calcium ion binding. Moreover, different metabolic processes were enriched in Landrace and Duroc, whereas immune response terms were common in Landrace only. Variant detection identified putative polymorphisms in some of the differentially expressed genes. Validation showed that predicted high impact variants in RAMP2, GIMAP6 and three uncharacterized genes are particularly interesting for sperm DNA fragmentation in boars.
Conclusions
We identified differentially expressed genes between groups of boars with high and low sperm DFI, and functional annotation of these genes point towards important biochemical pathways. Moreover, variant detection identified putative polymorphisms in the differentially expressed genes. Our results provide valuable insights into the molecular network underlying DFI in pigs.
Electronic supplementary material
The online version of this article (10.1186/s12917-017-1279-x) contains supplementary material, which is available to authorized users.
Keywords: Transcriptome profiling, Sperm DNA integrity, Differential expression
Background
Analysis of sperm parameters is important for predicting boar fertility and the outcome of artificial insemination (AI) in pig production. The classical way of evaluating sperm parameters is subjective scoring of viability, motility, concentration and morphology, to identify ejaculates with poor fertilization potential [1, 2]. However, this is insufficient for accurate prediction of the boar’s reproductive capacity, since the sperm cells must have additional qualities to fertilize the oocytes and since it is a subjective score. Combining several assays is suggested to better predict the fertility of an ejaculate [3]. For example, combining sperm morphology parameters and evaluation of DNA chromatin integrity has been found to be related to field fertility, as measured by farrowing rate in pigs [4].
During the last phase of spermatogenesis, spermiogenesis, the DNA of sperm cells is tightly packed by protamine and results in a condensed chromatin structure [5]. This leaves the DNA protected against degradation during transport through the male and female reproductive tracts until fertilization. Altered sperm chromatin structure is associated with DNA fragmentation and the degree of sperm DNA fragmentation is shown to be correlated to fertility in different species [4, 6–13]. This parameter is a much more objective marker of sperm quality and function than standard subjective microscopic evaluations [14, 15]. The sperm chromatin structure assay (SCSA) is a flow cytometry-based method that measures abnormal chromatin structure, as an increased acid-induced degradation of sperm DNA in situ [11]. More specifically, the acid denatures DNA at the sites of DNA breaks, which again reflects chromatin integrity status. The SCSA thereafter measures the relationship between double-stranded (i.e. condensed chromatin) and single-stranded (i.e. denatured) DNA for each sperm cell. This relationship is quantified by the DNA Fragmentation Index (DFI) [12]. Previous studies in pigs showed that DFI was significantly associated with litter size [8]. Moreover, DFI is found to be an important parameter for predicting normal development of the embryo [11, 16] and is also associated with abortion in humans [17].
Although the amount of sperm DFI is shown to influence fertility outcome, little is known about the underlying molecular mechanisms. Differentially expressed proteins have been identified in human seminal plasma and spermatozoa [18, 19]. Studies in humans have also showed that a truncated form of KIT tyrosine kinase, expressed in testis, causes higher amounts of DNA damage in sperm cells [20]. Moreover, depletion of excision repair cross-complementing gene 1 (ERCC1) and tumor suppressor gene p53 in mouse testis resulted in increased DNA breaks in sperm cells [21]. Recent studies indicate that the main reason of DFI in sperm is apoptosis, likely triggered by an impairment of chromatin maturation in the testis and by oxidative stress during the transit in the male genital tract [22].
The goal of this study was to use transcriptome sequencing to examine differential gene expression in testis tissue of boars with high and low sperm DFI. Testis tissue was chosen because chromatin condensation and DNA packaging in sperm cells occurs during testicular spermatogenesis [5, 23]. The biological functions of the differentially expressed genes were also investigated and a search for putative polymorphisms in the differentially expressed genes was performed. The results obtained in this study highly contribute to the knowledge of the molecular mechanisms underlying DNA fragmentation.
Methods
Animals and phenotypes
The sperm DFI was determined in a total of 241 Landrace and 302 Duroc AI boars in this study. All the boars were housed individually in pens sized approximately two by three meters and fed the same commercial diet. Nine Landrace and eleven Duroc boars were selected for transcriptome profiling based on their extreme high/low DFI values (Table 1). The boars’ age at semen sample collection ranged from 221 to 1000 days (mean = 310 days, standard deviation (SD) = 84.5). The sperm-rich fraction of the ejaculates was collected with the “gloved hand technique” at the Norsvin AI center (Hamar, Norway), similar to other studies recently published [24, 25]. From each of the boars, samples from up to six different ejaculates were analyzed, and the mean of the measurements was used as the final score. The ejaculates were diluted to a concentration of 28 × 106 spermatozoa per ml, according to the normal routines of the AI center at each date. The ejaculates were shipped as regular semen doses to commercial swine producers for the use within the next four days. From each individually diluted ejaculate, a sample of approximately 12 mL was transferred to a plastic tube. The samples were stored at 18 °C for 48 to 96 h depending on day of the week, before they were frozen in −80 °C until used for the DFI analysis. Boars were culled according to normal culling procedures at the AI station. From these boars, the testicle tissue samples were collected at the slaughter line. A piece of sample was collected from the middle part of one of the testicles, approximately 3 × 1.5 cm in size, immediately frozen in liquid N2, and thereafter stored at −80 °C until used for RNA extraction.
Table 1.
DFI measurements for the different boars used in this study
| Group | Boar | n(ejaculates) | DFI mean | DFI SD |
|---|---|---|---|---|
| Landrace low | L1 | 2 | 0.62% | 0.10% |
| L2 | 1 | 0.73% | ||
| L3 | 1 | 0.82% | ||
| L4 | 3 | 1.50% | 0.33% | |
| L5 | 4 | 1.52% | 0.68% | |
| Landrace high | L6 | 1 | 6.41% | |
| L7 | 2 | 5.47% | 1.69% | |
| L8 | 6 | 7.26% | 5.11% | |
| L9 | 7 | 8.07% | 3.85% | |
| Duroc low | D1 | 1 | 1.05% | |
| D2 | 2 | 1.07% | 0.37% | |
| D3 | 2 | 1.08% | 0.34% | |
| D4 | 1 | 1.11% | ||
| D5 | 3 | 1.13% | 0.35% | |
| Duroc high | D6 | 1 | 4.13% | |
| D7 | 1 | 4.77% | ||
| D8 | 1 | 4.14% | ||
| D9 | 1 | 4.69% | ||
| D10 | 1 | 5.63% | ||
| D11 | 3 | 5.36% | 3.64% |
The number of ejaculates and mean DFI value with SD is presented for each boar
DFI measurements
The SCSA protocol was performed using Cell Lab Quanta™ SC MPL (Beckman Coulter, Fullerton, CA, USA), equipped with a 22 mW argon laser with excitation at 488 nm, according to the procedure described by Evenson and Jost [13] with modifications [26]. The method is based on DNA staining properties of acridine orange (AO) which fluoresces green and red when binding to native dsDNA and denatured ssDNA, respectively. Frozen samples were thawed at 37 °C and diluted to a concentration of 2 × 106 sperm cells/mL in TNE buffer (10 mM Tris-HCL, 0.15 M NaCl, 1 mM EDTA, pH 7.4) to a final volume of 200 μL. Immediately afterwards, 400 μL of acid detergent solution (0.38 M NaCl, 80 mM HCL, 0.1% (w/v) Triton X-100, pH 1.2) was added. After exactly 30 s, 1.2 mL of AO staining solution (0.6 μg/mL AO (A3568, Life Technologies, OR, USA) in a buffer containing 37 mM citric acid, 0.126 M Na2HPO4, 1.1 μM EDTA, 0.15 M NaCl, pH 6) was added, and the sample was further incubated at room temperature in the flow cytometer. The sample was run in setup mode until 3 min after addition of the acid detergent solution, and then the acquisition of data was started. For each sample, 5000 events were collected with a flow rate of ~200 events/s. Prior to the analysis, the flow cytometer was AO saturated by running an AO equilibration solution (1.2 mL AO staining solution and 400 μL acid detergent solution) through the system for 5 min. The green fluorescence was collected by a 525 nm band pass filter, while the red fluorescence was collected by a 670 nm long pass filter. Prior to analysis and after every 10th sample, a reference sample was thawed, prepared and analyzed in the same way as the experimental samples to ensure the stability of the instrument and the laser throughout the experiment. The X-mean channel value was set to 125 ± 5 and Y-mean channel value was set to 425 ± 5. To identify the spermatozoa, a combination of electronic volume (EV)- and side scatter (SS)- signals were used, as described by Standerholen et al. [27]. The percentage of red and green fluorescence was determined using the Cell Lab Quanta™ SC MPL Analysis software package (Beckman Coulter, Software Version 1.0 A). Based on the ratio of red/(red + green), the DFI-value was calculated.
RNA extraction and sequencing
Total RNA for RNA sequencing was extracted from testicle tissue using the RNeasy Midi Kit from Qiagen according to the manufacturer’s instruction (Qiagen, CA, USA). Concentrations were measured using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, DE, USA) and the RNA quality was examined by the 28S:18S rRNA ratio using the RNA 6000 Nano LabChip® Kit on 2100 Bioanalyzer (Agilent Technologies, CA, USA). All samples displayed a 260/280 ratio > 1.8 and RNA integrity numbers (RIN) >8.5. RNA sequencing was done using Illumina HiSeq 2000 by the Norwegian Sequencing Centre at Ullevål Hospital (http://www.sequencing.uio.no). and generated 50 basepair single end reads. TruSeq RNA v2 was used for non-stranded library preparation, V3 clustering and sequencing reagents were used according to manufacturer’s instructions. Sample amount of 2 μg RNA was used as input, and 4 min fragmentation at 94 °C was employed. Image analysis and base calling were performed using Illumina’s RTA software version 1.17.21.3. Reads were filtered to remove those with low base call quality using Illumina’s default chastity criteria. The FASTQC software was used for quality control of raw sequence data (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). All reads had a per base sequence quality Phred score above 27 for all positions and were considered high quality. The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) [28] and are accessible through GEO Series accession number GSE74934.
Differential expression
The high quality reads were mapped to the Sus scrofa genome build 10.2 using the software TopHat v.2.0.12 [29] and default parameters. The Picard AddOrReplaceReadGroups program (http://broadinstitute.github.io/picard/) was used to assign unique IDs to the files. Gene prediction coordinates (release 10.2.75) were obtained from the ENSEMBL web site (http://www.ensembl.org). Mapped reads were sorted and indexed using Samtools v.1.1 [30] and the software HTSeq [31] was used with the stranded = no option to calculate the number of reads mapped to each gene. The R software package “edgeR” v.3.2.4 from Bioconductor was used to analyze the data [32] [see Additional file 6 for code]. The breeds were analyzed separately and the boars were divided into “high” and “low” groups based on their DFI values. The package assumes that the data follow a negative binomial distribution and it uses raw counts without correcting for gene length as this bias is assumed to be the same in all samples. Filtering was done to keep genes that reached at least one count per million in at least half of the samples. A heatmap was made for the differentially expressed genes between the high (bad) and low (good) DFI groups using the heatmap function in R (default parameters).
Statistical analysis
Normalization was done using the trimmed mean of the M values method [33] as implemented in “edgeR”. Moreover, tagwise dispersion was applied to estimate separate mean-variance relationships for the individual genes, and the generalized linear model likelihood test ratio method was employed to test for differential expression. The resulting p-values were adjusted for multiple testing by the Benjamini and Hochberg algorithm [34] and the level of significance for differentially expressed genes was set to an false discovery rate (FDR) of 0.05.
Gene ontology
Gene enrichment analyses make it easier to get an overview of functions that are overrepresented in gene expression datasets. Gene ontology (GO) tools can conveniently assign genes to different terms in the three categories “Molecular Function”, “Cellular Component” and “Biological Process”. In order to map all differentially expressed genes to corresponding GO terms, the R package “goseq” was applied [35]. The Wallenius approximation method was used to account for gene length bias before each GO term was tested for over-representation and under-representation of significant genes. The Benjamini and Hochberg algorithm [34] was used to correct for multiple testing and GO terms were considered significantly enriched at a 0.05 FDR cutoff.
Variant calling
Variant calling was done within breed using Samtools v.1.1 mpileup and bcftools call [30], and the Integrative Genomics Viewer (IGV) was used to visually inspect putative polymorphisms [36]. Using Samtools v.1.1 bcftools filter, variants (single nucleotide polymorphisms (SNPs)/insertions and deletions (indels)) were filtered to include only those having an alternate allele count of at least two, minor allele frequency above 0.01 and a read depth above 10. Moreover, only polymorphisms in differentially expressed genes were considered. The detected variants were annotated using SnpEff v.4.1 to classify variants (such as missense, nonsense, synonymous, stop gain/loss) and their impact (high, moderate, low, modifier) [37, 38]. Variants causing frameshift mutations or affecting start or stop codons are considered to have high impact, whereas variants e.g. in 3’UTR get the lowest impact (modifier). SnpSift was used to extract relevant information from list of variants files [39]. SNP validation was performed in-silico by matching putative polymorphism positions to known pig dbSNP entries [40]. SNPs not present in the database were considered novel. The putative variants identified in differentially expressed genes of this study have been deposited to the European Nucleotide Archive (EVA) under accession number PRJEB22189. For validation purposes, 15 of the high impact variants were genotyped using the KASP SNP genotyping system platform (KBiosciences, Herts, UK) using the 20 animals from the RNA-seq as well as 18 other pigs from Norsvin’s boar testing station (nine from each breed), which are relatives to the RNA-seq boars. SNP validation was also performed in an independent next generation sequencing dataset of related boars [41]. The putative high impact variants were compared by sequence position, reference and alternate alleles to polymorphisms identified in this dataset. Corresponding variants were considered validated.
Results
Mapping
Gene expression in testis tissue from Landrace and Duroc boars, with high and low sperm DFI, was analyzed by transcriptome sequencing. The mean (± SD) of the DFI values for the low and high groups were 1.04% (± 0.44%; n = 5) and 6.80% (± 1.12%; n = 4) in Landrace and 1.09% (± 0.03%; n = 5) and 4.79% (± 0.62%; n = 6) in Duroc, respectively (Table 1). The sequence data was maximum 50 basepair reads and the total number of sequenced reads per animal ranged from 59.6 to 95.0 million of which on average 76.7% of the reads were uniquely mapped to the current porcine genome assembly (Sus scrofa build 10.2). Altogether, 22,059 genes in Landrace and 21,717 in Duroc had at least one count in at least one sample. After filtering, 14,609 (66.2%) and 14,713 (67.7%) genes were used for differential expression analysis in Landrace and Duroc, respectively.
Differential expression
A total of 308 genes in Landrace and 374 genes in Duroc were significantly differentially expressed in testis tissue from boars with high and low sperm DFI [see Additional file 1 and Additional file 2 for Landrace and Duroc, respectively]. Of these genes, 71 were common for the two breeds (Table 2). The most significant differentially expressed gene in Landrace and Duroc was actin ACTA1 (FDR = 2.89e-09 and logarithmic fold change (logFC) = −1.78) and serum amyloid precursor SAA4 (FDR = 1.90e-06 and logFC = −0.68), respectively. In Landrace, ACTA1 was also the most down-regulated gene in the high DFI group, whereas neurexophilin NXPH2 showed the highest up-regulation (FDR = 6.11e-04 and logFC = 3.44). In Duroc, L-dopachrome tautomerase DCT showed the most down-regulation (FDR = 2.65e-02 and logFC = −0.94), whereas metallopeptidase ADAMTS4 was most significantly up-regulated (FDR = 1.88e-02 and logFC = 2.60). The majority of differentially expressed genes (94% and 78% in Landrace and Duroc, respectively) showed increased expression in the high DFI group compared to the low DFI group [see Additional file 5]. In addition to the annotated genes described below, genes encoding functionally uncharacterized proteins were differentially expressed in both breeds and they are included in the results tables with their corresponding Ensembl ID.
Table 2.
Differentially expressed genes common for the two breeds Landrace and Duroc
| Gene symbol | Gene name | FDR L | FDR D |
|---|---|---|---|
| ACER2 | alkaline ceramidase 2 | 1.61E-02 | 3.74E-02 |
| ACTN4 | alpha-actinin | 1.52E-02 | 1.34E-02 |
| APP | amyloid beta A4 protein | 2.73E-02 | 1.34E-02 |
| ATG4A | autophagy related 4A, cysteine peptidase | 4.02E-03 | 2.13E-02 |
| BGN | Biglycan | 2.64E-02 | 1.88E-02 |
| BMP1 | bone morphogenetic protein 1 | 4.60E-02 | 1.09E-02 |
| SERPING1 | Serpin family G member 1 | 2.68E-02 | 1.89E-02 |
| GLMP | Glycosylated lysosomal membrane protein | 3.66E-03 | 3.59E-02 |
| C1R | complement component 1, r subcomponent | 4.29E-02 | 2.13E-02 |
| C4A | Sus scrofa complement C4 (C4), mRNA | 2.86E-02 | 1.84E-03 |
| CA4 | carbonic anhydrase IV | 4.06E-02 | 1.44E-02 |
| CAT | Catalase | 1.01E-02 | 1.24E-02 |
| CDC42EP1 | CDC42 effector protein (Rho GTPase binding) 1 | 4.93E-02 | 2.42E-02 |
| CITED1 | Sus scrofa Cbp/p300-interacting transactivator | 2.31E-02 | 1.11E-02 |
| ENSSSCG00000001711 | Uncharacterized protein | 3.89E-02 | 2.72E-02 |
| COL3A1 | collagen, type III, alpha 1 | 4.40E-02 | 3.59E-02 |
| COPZ2 | coatomer protein complex, subunit zeta 2 | 1.04E-02 | 4.75E-02 |
| CPED1 | cadherin-like and PC-esterase domain containing 1 | 4.51E-03 | 2.72E-02 |
| CSF1R | colony stimulating factor 1 receptor | 3.86E-02 | 2.92E-02 |
| CTDSPL | CTD small phosphatase-like | 1.52E-02 | 1.66E-02 |
| CTSA | cathepsin A | 2.43E-02 | 2.71E-02 |
| CTSB | Sus scrofa cathepsin B (CTSB) | 1.26E-02 | 9.65E-03 |
| CTSH | Sus scrofa cathepsin H (CTSH), mRNA | 2.07E-02 | 3.84E-02 |
| CYP11A1 | Sus scrofa cytochrome P450, family 11, subfamily A, polypeptide 1 | 1.58E-02 | 4.56E-02 |
| ENSSSCG00000028912 | Uncharacterized protein | 3.02E-02 | 2.72E-02 |
| CFD | Complement factor D | 4.02E-02 | 1.98E-02 |
| DNASE1L1 | deoxyribonuclease I-like 1 | 4.51E-03 | 3.59E-02 |
| ECHDC3 | enoyl CoA hydratase domain containing 3 | 1.21E-03 | 2.04E-02 |
| ENSSSCG00000024587 | Uncharacterized protein | 2.49E-02 | 2.64E-02 |
| ENSSSCG00000028244 | Uncharacterized protein | 2.41E-02 | 4.91E-02 |
| EDNRA | endothelin receptor type A | 4.56E-02 | 2.42E-02 |
| EFEMP2 | EGF containing fibulin-like extracellular matrix protein 2 | 4.86E-02 | 1.66E-02 |
| EHD2 | EH-domain containing 2 | 9.26E-03 | 1.20E-02 |
| ENSSSCG00000011239 | Uncharacterized protein | 3.85E-02 | 9.09E-03 |
| ENSSSCG00000021406 | Uncharacterized protein | 1.35E-02 | 4.85E-02 |
| ENSSSCG00000025934 | Uncharacterized protein | 2.75E-02 | 7.72E-03 |
| ENSSSCG00000029074 | Uncharacterized protein | 1.61E-02 | 3.58E-02 |
| EPHX1 | Sus scrofa epoxide hydrolase 1, microsomal (xenobiotic) (EPHX1) | 1.61E-02 | 1.11E-02 |
| FAH | fumarylacetoacetate hydrolase (fumarylacetoacetase) | 1.66E-02 | 8.16E-03 |
| FAM213A | family with sequence similarity 213, member A | 4.93E-02 | 1.01E-02 |
| FCGRT | Sus scrofa Fc fragment of IgG, receptor, transporter, alpha | 3.42E-02 | 4.56E-02 |
| FDXR | NADPH:adrenodoxin oxidoreductase, mitochondrial | 1.61E-02 | 1.29E-02 |
| FGFR1 | Fibroblast growth factor receptor 1 | 4.69E-02 | 1.21E-02 |
| ENSSSCG00000022236 | Uncharacterized protein | 4.02E-02 | 1.52E-02 |
| ENSSSCG00000002797 | Uncharacterized protein | 4.29E-02 | 2.22E-02 |
| GRK5 | G protein-coupled receptor kinase 5 | 4.60E-02 | 2.87E-03 |
| GSDMD | gasdermin D | 4.11E-02 | 1.36E-02 |
| ENSSSCG00000000620 | Uncharacterized protein | 2.01E-02 | 3.84E-03 |
| ITM2C | Sus scrofa integral membrane protein 2C | 4.71E-02 | 4.75E-02 |
| ENSSSCG00000004207 | Uncharacterized protein | 3.99E-02 | 2.36E-02 |
| LAMB2 | laminin, beta 2 (laminin S) | 1.23E-02 | 2.81E-02 |
| LAMC3 | laminin, gamma 3 | 1.30E-02 | 2.15E-02 |
| LIPA | lipase A, lysosomal acid, cholesterol esterase | 4.50E-02 | 3.84E-02 |
| ENSSSCG00000023235 | Uncharacterized protein | 2.63E-02 | 1.23E-02 |
| MAOB | monoamine oxidase B | 3.62E-02 | 2.50E-02 |
| NEU1 | sialidase 1 (lysosomal sialidase) | 4.11E-02 | 4.56E-02 |
| ENSSSCG00000022516 | Uncharacterized protein | 2.23E-02 | 4.97E-02 |
| PGAM1 | phosphoglycerate mutase 1 (brain) | 1.50E-02 | 1.29E-02 |
| PLCB1 | phospholipase C, beta 1 (phosphoinositide-specific) | 4.06E-02 | 4.75E-02 |
| PRDX2 | peroxiredoxin 2 | 4.02E-02 | 4.69E-02 |
| ENSSSCG00000016522 | Uncharacterized protein | 5.88E-03 | 1.20E-02 |
| SERTAD1 | SERTA domain containing 1 | 3.22E-02 | 4.35E-02 |
| ENSSSCG00000011357 | Uncharacterized protein | 3.87E-02 | 4.11E-02 |
| SLC1A5 | solute carrier family 1 (neutral amino acid transporter), member 5 | 4.99E-02 | 3.91E-02 |
| SLC41A1 | solute carrier family 41 (magnesium transporter), member 1 | 4.29E-02 | 7.39E-03 |
| SLC44A2 | Choline transporter-like protein 2 | 4.02E-02 | 1.12E-02 |
| SLC44A4 | Sus scrofa solute carrier family 44, member 4 (SLC44A4), mRNA | 4.98E-02 | 3.50E-02 |
| TMEM176B | transmembrane protein 176B | 4.40E-02 | 7.46E-03 |
| TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 2.64E-02 | 5.56E-03 |
| TPM4 | tropomysin alpha-4 chain | 5.77E-03 | 3.10E-02 |
| VIM | Vimentin | 4.02E-02 | 1.12E-02 |
Genes differentially expressed in both breeds are presented with gene symbol, gene name and significance level (FDR) for Landrace (L) and Duroc (D)
Gene ontology
Functional characterization of differentially expressed genes revealed an overrepresentation of genes with roles in the cellular components “extracellular matrix” and “extracellular region” for both Landrace and Duroc. Results of the GO classification of the differentially expressed genes are shown in Fig. 1. The molecular function “calcium ion binding” was also enriched in both breeds. In addition, “cholesterol metabolic process” and “oxidation-reduction process” were Duroc specific whereas “collagen catabolic process”, “hydrolase activity” and “proteolysis” were Landrace specific. Moreover, immune system ontologies were Landrace specific.
Fig. 1.
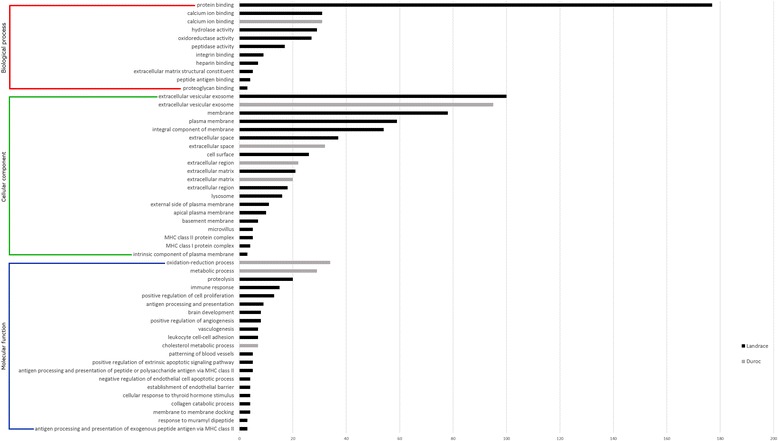
Gene ontology classification of the differentially expressed genes. The figure shows the GO enrichments of the differentially expressed genes in terms of the biological process, cellular component and molecular function classes
Variant calling
Variant detection identified 1501 and 1751 putative polymorphisms in differentially expressed genes in Landrace and Duroc, respectively, out of which 91 and 88% had an existing dbSNP entry [see Additional files 3 and 4]. In Landrace/Duroc, most of the polymorphisms (610/731) in differentially expressed genes were synonymous SNPs (Table 3). Of the polymorphisms in differentially expressed genes, 4/17 in Landrace/Duroc were high impact variants, predicted to cause frameshifts or a change in start or stop codon. 15 of the high impact variants were chosen for validation using the KASP SNP Genotyping system. Five of the SNPs were successfully validated, including four of the ones with previous dbSNP entries (see Additional file 7]. Ten of the detected high impact variants, including one with an existing dbSNP entry, failed validation. When comparing the variants to an independent next generation sequencing dataset, the same result was found. The differentially expressed genes with validated high impact variants were RAMP2, GIMAP6, ENSSSCG00000000712, ENSSSCG00000009348 and ENSSSCG00000028326.
Table 3.
Effects of putative SNPs
| SNP effect | After filtration | Joined with gene expression results | ||
|---|---|---|---|---|
| Landrace | Duroc | Landrace | Duroc | |
| 3’UTR | 17,741 | 17,715 | 573 | 627 |
| 5’UTR | 3622 | 3691 | 62 | 102 |
| Frameshift | 453 | 516 | 3 | 12 |
| Missense | 9946 | 10,424 | 267 | 246 |
| Splice acceptor | 16 | 17 | 0 | 0 |
| Splice donor | 12 | 12 | 0 | 0 |
| Splice site region | 338 | 362 | 3 | 7 |
| Start lost | 13 | 8 | 1 | 0 |
| Stop gained | 68 | 63 | 0 | 3 |
| Stop lost | 27 | 34 | 0 | 2 |
| Synonymous | 21,060 | 21,273 | 610 | 731 |
| SNP impact | ||||
| High | 600 | 663 | 4 | 17 |
| Moderate | 10,006 | 10,486 | 269 | 250 |
| Low | 21,974 | 22,200 | 623 | 752 |
| Modifier | 23,251 | 23,293 | 676 | 807 |
SNP effect according to SnpEff for putative polymorphisms detected in Landrace and Duroc. The results presented are after filtration and joined with differentially expressed genes. Some SNPs have more than one predicted effect
Discussion
Chromatin condensation and DNA packaging in sperm cells occur during testicular spermatogenesis, and altered chromatin structure is associated with sperm DFI. High levels of sperm DFI has been associated with decreased fertility, however, the molecular mechanisms contributing to alterations in sperm DFI is not clear. In the present study, we explored gene expression differences in testis between groups of boars with high and low sperm DFI and investigated the gene enrichments associated with the results. The experiment was performed in two different breeds, Landrace and Duroc, and 308 and 374 genes were found differentially expressed in Landrace and Duroc, respectively. Of these genes, 71 were found to be common for the two breeds, which means they are likely to be essential for alterations in sperm DFI. The Landrace specific and Duroc specific differentially expressed genes might reflect breed specific mechanisms in chromatin condensation and DFI level with regards to these two breeds. Breed differences in DFI have also previously been found in boars as well as bulls [42, 43]. The GO terms “extracellular matrix”, “extracellular region” and “calcium ion binding” were significant for both breeds and differentially expressed genes belonging to these pathways are discussed in more detail below. None of the differentially expressed pathways were found to overlap with pathways previously identified for spermatogenesis in Large White, Duroc and Meishan pigs [44, 45], indicating that we have identified pathways related to DFI and not general spermatogenesis.
Genes enriched in “extracellular matrix” and “extracellular region”
The seminiferous tubules in testis contain Sertoli and germ cells and direct progression of spermatogenesis. The “extracellular matrix”, an enriched GO term in both Landrace and Duroc, plays a significant role in regulating spermatogenesis because Sertoli and germ cells are structurally and hormonally supported by extracellular matrix during their development in the seminiferous tubules [46]. To complete spermatogenesis, germ cells must migrate across the seminiferous epithelium while still attached to the nourishing Sertoli cells, a process controlled by restructuring events at cell junctions known as ectoplasmic specialization [46, 47]. This is the stage where DNA compaction and chromatin condensation occur [48]. These junctions are located in the “extracellular region” [47], another enriched GO term in both breeds. The results suggest that genes involved in different stages of spermatogenesis affect DNA fragmentation in sperm cells.
Laminins and collagens are important building blocks of the extracellular matrix in testis and they act together with proteases, protease inhibitors, cytokines and focal adhesion components to regulate membrane proteins [46]. Two genes of the laminin family (LAMB2 and LAMC3) and one of the collagen family (COL3A1) were found up-regulated in the high DFI group in both breeds in this study. Both pre-collagens and laminins are processed by bone morphogenetic protein 1 (BMP1) [49], which was also up-regulated in the high DFI condition in both breeds. Furthermore, genes of the collagen family were exclusively up-regulated in the high DFI group in one of the breeds (COL1A1 in Duroc and COL1A2, COL4A1, COL4A2 and COL14A1 in Landrace). The differential expression of the laminin and collagen genes might suggest that the structure of the extracellular matrix, where the sperm cells are attached during development, could influence chromatin condensation and hence DFI level. This is also supported by the differential expression of genes encoding other components of the extracellular matrix such as the cytokines tumor necrosis factor (TNF) alpha and interleukins. TNFα regulates germ cell apoptosis, Leydig cell steroidogenesis and junction dynamics in the testes [46] and it has also been shown to induce sperm damage such as DNA fragmentation [50]. TNF member TNFAIP3 was up-regulated in the high DFI group in both breeds in this study. Additionally, breed specific up-regulation in the high DFI group was found for genes of this family (TNFSF10 and TNFRSF12A in Landrace and LITAF in Duroc). Interleukin IL1R1 was up-regulated in the high DFI group in Landrace. This is in agreement with previous findings, where IL1R1 protein was associated with DFI in human sperm and seminal plasma [18, 19].
Genes encoding fibulins, proteases, protease inhibitors and cathepsins, all interacting with components of the extracellular matrix, were also differentially expressed in this study. Fibulins are extracellular matrix glycoproteins that modulate cellular behavior and function and are involved in binding of laminin and calcium [51, 52]. In this study EGF containing fibulin-like EM protein 2 (EFEMP2, also known as FBLN4) was up-regulated in the high DFI group in both breeds whereas fibulins FBLN5 and EFEMP1 (also known as FBLN3) were up-regulated in Duroc. Furthermore, extracellular matrix protein 1 (ECM1), known to interact with fibulins and laminins [53], was up-regulated in Duroc. The ECM1 protein has previously been found associated with sperm DNA fragmentation in human seminal plasma [19], supporting the findings of this study. Matrix metallopeptidases (MMPs) and MMP inhibitors (TIMPs) are proteases and protease inhibitors, respectively. They are capable of degrading different components of the extracellular matrix, like laminins and collagen, and thereby regulate spermatogenesis [46, 54]. A disintegrin and metalloproteases (ADAMs) regulate spermatogenesis by cleaving growth factors and cytokines from the extracellular matrix [54]. In this study, MMP2, MMP19, TIMP1 and ADAMTS9 were up-regulated in the high DFI group in Landrace whereas TIMP3, ADAM33 and ADAMTS4 were up-regulated in Duroc. ADAMTS4 was the most up-regulated gene in Duroc in this study indicating an important role for proteases in DNA fragmentation of sperm cells, possibly by interrupting with the testicular extracellular matrix stability. Cathepsins contribute in protein degradation in the extracellular matrix by cleaving collagens and laminins [55]. The cathepsin members CTSA, CTSB and CTSH were found up-regulated in the high DFI group of both breeds. Additionally, CTSC, CTSL and CTSS were up-regulated in Landrace. Interestingly, CTSL has been linked to chromatin decondensation in sea urchin embryos [56] and CTSA has been shown to affect sperm motility in rats [57]. Moreover, CTSB, CTSC, CTSD, CTSL and CTSS are all involved in testis tissue restructuring during spermatogenesis in rats [58].
Peroxiredoxins are located in the ectoplasmic specialization and encode redox proteins, which protect sperm cells from oxidative stress that cause DNA damage such as DNA fragmentation [59]. In this study, peroxiredoxin PRDX2 was up-regulated in the high DFI group in both breeds whereas PRDX3 was up-regulated in the high DFI group in Duroc. Furthermore, glutathione peroxidase GPX3 was up-regulated in the high DFI group in Landrace. These results are supported by previous findings in human, where levels of peroxiredoxin members PRDX1 and PRDX6 have been associated with sperm DNA integrity [60]. The differentially expressed gene GPX3 is interesting since glutathione peroxidases can work both as redox proteins and to mediate disulfide bridging, which stabilizes sperm chromatin [61].
Actins are important components of the ectoplasmic specialization of the seminiferous tubules [46, 47] and are involved in the development of mature sperm through several processes, including chromatin remodeling [62, 63]. The ACTN4 was up-regulated in high DFI boars of both breeds. In Landrace, three additional actin and actin-binding proteins were found to be differentially expressed (ABLIM1, ACTA1 and ACTA2). ACTA1 was down-regulated in the high DFI group, whereas the other actin members were up-regulated, indicating different functions of these actin members when developing DFI in the testis. It was the most down-regulated of the differentially expressed genes in Landrace, suggesting an important role for this gene in DFI levels of this breed. In Duroc, coronin acting binding protein 1B (CORO1B) and demantin actin binding protein (DMTN) were up-regulated whereas capping protein (actin filament) muscle Z-line, alpha 3 (CAPZA3) was down-regulated. The significance of different actin genes between the two breeds could imply breed specialized mechanisms, however, this needs to be further investigated.
In this study, genes encoding extracellular matrix compounds such as collagens, laminins, fibulins and cytokines were differentially expressed. Moreover, peroxiredoxins and actins of the ectoplasmic specialization were up- and down-regulated. Genes involved in regulation of these compounds, like proteases, protease inhibitors and cathepsins, were also differentially expressed. The results confirm previous findings, as well as reporting a number of new genes, highlighting the importance of testicular steroidogenesis in the outcome of sperm DFI. In this study, a major part of the differentially expressed genes were up-regulated. A hypothesis explaining this could be that deficiencies of the extracellular matrix makes the cell compensate by up-regulating gene expression.
Genes enriched in “calcium ion binding”
The GO term calcium ion binding was significantly enriched in both breeds and calcium uptake in sperm is known to be important for the regulation of fertility by affecting sperm maturation, motility, capacitation and the acrosome reaction [64, 65]. A role for calcium in chromatin condensation and DFI is less described, however, the calcium permeable ion channels proteins VDAC2 and VDAC3 have previously shown significant association with DFI in human sperm [18] and fertility in boars [24]. Moreover, along with chromatin condensation in spermatogenesis, the sperm cell redundant nuclear envelope evolves, which has been proposed a role in calcium ion storage [64]. This could explain the significance of the “calcium ion binding” enrichment in both breeds of this study.
The up-regulation of voltage-dependent anion channel gene VDAC1 in the high DFI group in Duroc is interesting as the proteins VDAC2 and VDAC3 has been associated with DNA fragmentation in human sperm [18]. Moreover, abnormal regulation of different calcium channels has previously been shown to negatively affect sperm function [66]. A number of other genes involved in reproduction related processes where calcium influx plays a role, like hyperactivation, capacitation, the acrosome reaction and fertilization [63], were differentially expressed in this study (PLCB1 in both breeds, PLCZ1, DLD and PLD1 in Duroc, and PDGFRB, CAPN1, PLA2G4A, NPR1 and RAPGEF3 in Landrace). The up-regulation of all these genes in boars with high sperm DFI could imply an interrupted function of calcium mediated regulation, which would affect the fertilizing capability of these sperm cells after being ejaculated. Further studies are needed, however, to clarify the role of testicular calcium signaling in sperm DFI levels.
Variant detection
An advantage of calling genomic variants from transcriptome sequencing data is that it directly allows for detection of polymorphisms in transcribed regions and is an efficient way to discover putative causative SNPs. Variant detection requires sufficient coverage with high quality sequence reads in order to distinguish true polymorphisms from sequencing errors. Filtering on sequencing depth might have removed polymorphisms in low expression genes, however, visualization by IGV showed likely false positive variants if this filtering was not done. This is in agreement with another study showing that the majority of false positive SNPs occur at sites with less than 10X coverage [67]. Comparing our detected polymorphisms with variants in dbSNP showed that 91 and 88% of our putative polymorphisms in Landrace and Duroc had a corresponding dbSNP entry, respectively. However, only five of the predicted high impact variants had an existing dbSNP entry and a validation study was therefore conducted to test 15 of the putative high impact SNPs. The results showed that high impact variants in the differentially expressed genes RAMP2, GIMAP6, ENSSSCG00000000712, ENSSSCG00000009348 and ENSSSCG00000028326 are particularly interesting for sperm DNA fragmentation in boars. Failure to validate ten of the variants shows that SNP detection in short read sequencing data can produce false positives. It has been shown that a number of factors can contribute to false positive SNPs in sequence data, including quality of the reference sequence, read length, choice of mapper and variant caller, mapping stringency and filtering of SNPs [68]. The importance of a high quality reference genome was highlighted in Ribeiro et al. (2015) [68] and we know that the reference genome used in this study has its limitations [69]. Approximately 90% overlap of our identified SNPs and previously identified SNPs does however indicate that our pipeline works, but that caution should be taken especially for variants with no supporting evidence. The identical results of validation using a PCR-based method (KASP) and in silico in an independent dataset could suggest that the latter is equally good in those cases where datasets are available.
Although many of the putative polymorphisms identified are located outside open reading frames or cause synonymous changes, they may be in linkage disequilibrium to other causative mutations. Moreover, studies have also shown that synonymous SNPs may have functional effects by affecting mRNA stability or by translation suppression [70, 71].
Conclusions
The present study identified whole genome expression differences in testis tissue between boars with high and low levels of sperm DFI. Moreover, putative polymorphisms were detected in the differentially expressed genes. The results of this study show that differentially expressed genes of steroidogenic pathways, where the chromatin condensation and DNA packaging occurs, are important for the outcome of DFI levels in ejaculated spermatozoa. Transcriptome sequencing analysis showed that the major changes at transcription level in the testicle of pig concerning sperm DFI were related to the functional categories “extracellular matrix”, “extracellular region” and “calcium ion binding”. Variant detection showed that predicted high impact SNPs in RAMP2, GIMAP6 and three uncharacterized genes are particularly interesting for the trait. The candidate genes identified in this study provide a valuable resource to identify molecular markers for sperm DFI, for use in selection towards improved sperm quality.
Additional files
Differentially expressed genes for DFI in Landrace. The results are presented with Ensembl gene id, gene symbol, gene name, fold change and significance level (FDR). (XLSX 31 kb)
Differentially expressed genes for DFI in Duroc. The results are presented with Ensembl gene id, gene symbol, gene name, fold change and significance level (FDR). (XLSX 34 kb)
High quality SNPs occurring in differentially expressed genes in Landrace. The SNPs are presented with Ensembl gene id, gene name, FDR value of differentially expressed gene, chromosome (SSC), position, reference allele and alternate allele, as well as effect, impact according to SnpEff and dbSNP ID. (XLSX 100 kb)
High quality SNPs occurring in differentially expressed genes in Duroc. The SNPs are presented with Ensembl gene id, gene name, significance level (FDR) of differentially expressed gene, chromosome (SSC), position, reference allele and alternate allele, as well as effect, impact according to SnpEff and dbSNP ID. (XLSX 117 kb)
Heatmap of the differentially expressed genes for DFI. The differentially expressed genes in testis of A) Duroc and B) Landrace boars with high (bad) and low (good) sperm DFI ordered by hierarchical clustering show higher (red) and lower (yellow) expression of genes in the two DFI groups. (TIFF 59 kb)
The edgeR source code used for testing for differential expression. (TXT 2 kb)
Putative high impact SNPs in differentially expressed genes. Putative high impact SNPs in differentially expressed genes presented with breed, gene name, position, FDR and log fold change. Validation by KASP SNP Genotyping System (N.A. is for SNPs not tested). (DOCX 12 kb)
Acknowledgements
The authors wish to thank BioBank AS for collecting and storing the samples, the Centre for Integrative Genetics (CIGENE) at the Norwegian University of Life Sciences for providing lab and computer facilities and the Norwegian Sequencing Centre at Ullevål for performing the sequencing. We also want to thank the semen collectors at Topigs Norsvin in Hamar for taking samples for DFI analyses and Anne Guri Marøy for performing KASP genotyping.
Funding
This study is financed by the Norwegian Research Council (Grant number 207568) and Norsvin SA. The funding bodies had no role in the design of the study or collection, analysis, and interpretation of data or in writing the manuscript.
Availability of data and materials
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) [29] and are accessible through GEO Series accession number GSE74934.
Abbreviations
- ABLIM1
Actin binding LIM protein 1
- ACTA1
Actin, alpha 1, skeletal muscle
- ACTA2
Actin, alpha 2, smooth muscle, aorta
- ACTN4
Actinin alpha 4
- ADAM33
ADAM metallopeptidase domain 33
- ADAMs
A disintegrin and metalloproteases
- ADAMTS4
ADAM metallopeptidase with thrombospondin type 1 motif, 4
- ADAMTS9
ADAM metallopeptidase with thrombospondin type 1 motif, 9
- AI
Artificial insemination
- AO
Acridine orange
- BMP1
Bone morphogenetic protein 1
- CAPN1
Calpain 1
- CAPZA3
Capping actin protein of muscle Z-line alpha subunit 3
- COL14A1
Collagen type XIV alpha 1 chain
- COL1A1
Collagen type I alpha 1 chain
- COL1A2
Collagen type I alpha 2 chain
- COL3A1
Collagen type III alpha 1 chain
- COL4A1
Collagen type IV alpha 1 chain
- COL4A2
Collagen type IV alpha 2 chain
- CORO1B
Coronin 1B
- CTSA
Cathepsin A
- CTSB
Cathepsin B
- CTSC
Cathepsin C
- CTSH
Cathepsin H
- CTSL
Cathepsin L
- CTSS
Cathepsin S
- DCT
Dopachrome tautomerase
- DFI
DNA fragmentation index
- DLD
Dihydrolipoamide dehydrogenase
- DMTN
Dematin actin binding protein
- ECM1
Extracellular matrix protein 1
- EFEMP1
EGF containing fibulin-like extracellular matrix protein 1
- EFEMP2
EGF containing fibulin-like extracellular matrix protein 2
- ERCC1
Excision repair cross-complementing gene 1
- EV
Electronic volume
- FBLN4
Fibulin 4
- FBLN5
Fibulin 5
- FDR
False discovery rate
- GEO
Gene expression omnibus
- GIMAP6
GTPase, IMAP family member 6
- GO
Gene ontology
- GPX3
Glutathione peroxidase 3
- IGV
Integrative genomics viewer
- IL1R1
Interleukin 1 receptor type 1
- Indel
Insertion/deletion
- LAMB2
Laminin, beta 2
- LAMC3
Laminin, gamma 3
- LITAF
Lipopolysaccharide-induced TNF
- logFC
Logarithmic fold change
- MMP19
Matrix metallopeptidase 19
- MMP2
Matrix metallopeptidase 2
- MMPs
Matrix metallopeptidases
- NPR1
Natriuretic peptide receptor 1
- NXPH2
Neurexophilin 2
- PDGFRB
Platelet-derived growth factor receptor beta
- PLA2G4A
Phospholipase A2 group IVA
- PLCB1
Phospholipase C beta 1
- PLCZ1
Phospholipase C zeta 1
- PLD1
Phospholipase D1
- PRDX2
Peroxiredoxin 2
- PRDX6
Peroxiredoxin 6
- RAMP2
Receptor activity modifying protein 2
- RAPGEF3
Rap guanine nucleotide exchange factor 3
- RIN
RNA integrity number
- SAA4
Serum amyloid A4, constitutive
- SCSA
Sperm chromatin structure assay
- SD
Standard deviation
- SNP
Single nucleotide polymorphism
- SS
Side scatter
- TIMP1
TIMP metallopeptidase inhibitor 1
- TIMP3
TIMP metallopeptidase inhibitor 3
- TIMPs
MMP inhibitors
- TNF
Tumor necrosis factor
- TNFAIP3
Tumor necrosis factor, alpha-induced protein 3
- TNFRSF12A
TNF receptor superfamily member 12A
- UTR
Untranscribed region
- VDAC1
Voltage dependent anion channel 1
- VDAC2
Voltage dependent anion channel 2
- VDAC3
Voltage dependent anion channel 3
Authors’ contributions
MvS conducted RNA extraction, analyzed the data and drafted the paper. NHT was involved in the DFI analysis, the SNP detection analysis and contributed to writing the paper. AHG was involved in planning the project, organizing the DFI analysis and contributed to writing the paper, FDM was involved in planning the project, organizing the DFI analysis and contributed to writing the paper, EBS and TTZ conducted DFI analysis, DIV was involved in supervision and contributed to writing the paper, EG was involved in planning the project, supervision and contributed to writing the paper. All authors read and approved the final manuscript.
Ethics approval
All animals were cared for according to laws, internationally recognized guidelines and regulations controlling experiments with live animals in Norway according to the rules given by Norwegian Animal Research Authority (The Animal Protection Act of December 20th, 1974, and the Animal Protection Ordinance Concerning Experiments with Animals of January 15th, 1996). The animals used in this study were AI boars kept as a routine by Norsvin’s breeding program. The semen samples were standard procedure whereas tissue samples were taken after slaughter, and no ethics committee approval was needed. Norsvin’s trained technicians obtained all the semen samples and BioBank AS (Hamar, Norway) obtained tissue samples, following standard routine monitoring procedures and relevant guidelines. No animal experiment has been performed in the scope of this research.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12917-017-1279-x) contains supplementary material, which is available to authorized users.
Contributor Information
Maren van Son, Email: maren.van.son@norsvin.no.
Nina Hårdnes Tremoen, Email: nina.tremoen@inn.no.
Ann Helen Gaustad, Email: ann-helen.gaustad@norsvin.no.
Frøydis Deinboll Myromslien, Email: froydis.myromslien@inn.no.
Dag Inge Våge, Email: daginge.vage@nmbu.no.
Else-Berit Stenseth, Email: elseberit.stenseth@inn.no.
Teklu Tewoldebrhan Zeremichael, Email: teklu.zeremichael@inn.no.
Eli Grindflek, Email: eli.grindflek@norsvin.no.
References
- 1.Foxcroft GR, Dyck MK, Ruiz-Sanchez A, Novak S, Dixon WT. Identifying useable semen. Theriogenology. 2008;70:1324–1336. doi: 10.1016/j.theriogenology.2008.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Gadea J. Sperm factors related to in vitro and in vivo porcine fertility. Theriogenology. 2005;63:431–444. doi: 10.1016/j.theriogenology.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 3.Graham JK. Assessment of sperm quality: a flow cytometric approach. Anim Reprod Sci. 2001;68(3–4):239–247. doi: 10.1016/S0378-4320(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 4.Tsakmakidis IA, Lymberopoulos AG, Khalifa TAA. Relationship between sperm quality traits and field-fertility of porcine semen. J Vet Sci. 2010;11(2):151–154. doi: 10.4142/jvs.2010.11.2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rathke C, Baarends WM, Awe S, Renkawitz-Pohl R. Chromatin dynamics during spermiogenesis. Biochim Biophys Acta. 1839;2014:155–168. doi: 10.1016/j.bbagrm.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 6.Sun JG, Jurisicova A, Casper RF. Detection of deoxyribonucleic acid fragmentation in human sperm: correlation with fertilization in vitro. Biol Reprod. 1997;56:602–607. doi: 10.1095/biolreprod56.3.602. [DOI] [PubMed] [Google Scholar]
- 7.Didion BA, Kasperson KM, Wixon RL, Evenson DP. Boar fertility and sperm chromatin structure status: a retrospective report. J Androl. 2009;30(6):655–660. doi: 10.2164/jandrol.108.006254. [DOI] [PubMed] [Google Scholar]
- 8.Boe-Hansen GB, Christensen P, Vibjerg D, Nielsen MBF, Hedeboe AM. Sperm chromatin structure integrity in liquid stored boar semen and its relationships with field fertility. Theriogenology. 2008;69:728–736. doi: 10.1016/j.theriogenology.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Ballachey BE, Hohenboken WD, Evenson DP. Heterogeneity of sperm nuclear chromatin structure and its relationship to fertility of bulls. Biol Reprod. 1987;36:915–925. doi: 10.1095/biolreprod36.4.915. [DOI] [PubMed] [Google Scholar]
- 10.Broekhuijse MLWJ, Sostaric E, Feitsma H, Gadella BM. Relation of flow cytometric sperm integrity assessments with boar fertility performance under optimized field conditions. J Anim Sci. 2012;90(12):4327–4336. doi: 10.2527/jas.2012-5040. [DOI] [PubMed] [Google Scholar]
- 11.Evenson DP, Darzynkiewicz Z, Melamed MR. Relation of mammalian sperm chromatin heterogeneity to fertility. Science. 1980;210(4474):1131–1133. doi: 10.1126/science.7444440. [DOI] [PubMed] [Google Scholar]
- 12.Evenson D, Wixon R. Meta-analysis of sperm DNA fragmentation using the sperm chromatin structure assay. Reprod BioMed Online. 2006;12(4):466–472. doi: 10.1016/S1472-6483(10)62000-7. [DOI] [PubMed] [Google Scholar]
- 13.Evenson DP, Thompson L, Jost L. Flow cytometric evaluation of boar semen by the sperm chromatin structure assay as related to cryopreservation and fertility. Theriogenology. 1994;41:637–651. doi: 10.1016/0093-691X(94)90174-H. [DOI] [PubMed] [Google Scholar]
- 14.Zini A, Kamal K, Phang D, Willis J, Jarvi K. Biologic variability of sperm DNA denaturation in infertile men. Urology. 2001;58(2):258–261. doi: 10.1016/S0090-4295(01)01180-3. [DOI] [PubMed] [Google Scholar]
- 15.Evenson DP, Larson KL, Jost LK. Sperm chromatin structure assay: its clinical use for detecting sperm DNA fragmentation in male infertility and comparisons with other techniques. J Androl. 2002;23:25–43. doi: 10.1002/j.1939-4640.2002.tb02599.x. [DOI] [PubMed] [Google Scholar]
- 16.Ahmadi A, Ng S-C. Fertilizing ability of DNA-damaged spermatozoa. J Exp Zool. 1999;284(6):696–704. doi: 10.1002/(SICI)1097-010X(19991101)284:6<696::AID-JEZ11>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 17.Zhao J, Zhang Q, Wang Y, Li Y. Whether sperm deoxyribonucleic acid fragmentation has an effect on pregnancy and miscarriage after in vitro fertilization/intracytoplasmic sperm injection: a systematic review and meta-analysis. Fertil Steril. 2014;102(4):998–1005. doi: 10.1016/j.fertnstert.2014.06.033. [DOI] [PubMed] [Google Scholar]
- 18.Intasqui P, Camargo M, Del Giudice PT, Spaine DM, Carvalho VM, Cardozo KHM, Cedenho AP, Bertolla RP. Unraveling the sperm proteome and post-genomic pathways associated with sperm nuclear DNA fragmentation. J Assist Reprod Genet. 2013;30:1187–1202. doi: 10.1007/s10815-013-0054-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Intasqui P, Camargo M, Del Giudice PT, Spaine DM, Carvalho VM, Cardozo KHM, Zylbersztejn DS, Bertolla RP. Sperm nuclear DNA fragmentation rate is associated with differential protein expression and enriched functions in human seminal plasma. BJU Int. 2013;112(6):835–843. doi: 10.1111/bju.12233. [DOI] [PubMed] [Google Scholar]
- 20.Muciaccia B, Sette C, Paronetto MP, Barchi M, Pensini S, D'Agostino A, Gandini L, Geremia R, Stefanini M, Rossi P. Expression of a truncated form of KIT tyrosine kinase in human spermatozoa correlates with sperm DNA integrity. Hum Reprod. 2010;25(9):2188–2202. doi: 10.1093/humrep/deq168. [DOI] [PubMed] [Google Scholar]
- 21.Paul C, Povey JE, Lawrence NJ, Selfridge J, Melton DW, Saunders PT. Deletion of genes implicated in protecting the integrity of male germ cells has differential effects on the incidence of DNA breaks and germ cell loss. PLoS One. 2007;2(10):e989. doi: 10.1371/journal.pone.0000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muratori M, Tamburrino L, Marchiani S, Cambi M, Olivito B, Azzari C, Forti G, Baldi E. Investigation on the origin of sperm DNA fragmentation: role of apoptosis, immaturity and oxidative stress. Mol Med. 2015;21:109–122. doi: 10.2119/molmed.2014.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Vries M, Ramos L, Housein Z, de Boer P. Chromatin remodelling initiation during human spermiogenesis. Biology Open. 2012;1:446–457. doi: 10.1242/bio.2012844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwon WS, S-A O, Kim Y-J, Rahman MS, Park YJ, Pang MG. Proteomic approaches for profiling negative fertility markers in inferior boar spermatozoa. Sci Rep. 2015;5:13821. doi: 10.1038/srep13821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwon WS, Rahman MS, Lee J-S, Yoon S-J, Park YJ, Pang MG. Discovery of predictive biomarkers for litter size in boar spermatozoa. Mol Cell Proteomics. 2015;14(5):1230–1240. doi: 10.1074/mcp.M114.045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boe-Hansen GB, Ersbøll AK, Greve T, Christensen P. Increased storage time of extended boar semen reduces sperm DNA integrity. Theriogenology. 2005;63:2006–2019. doi: 10.1016/j.theriogenology.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 27.Standerholen FB, Myromslien FD, Kommisrud E, Ropstad E, Waterhouse KE. Comparison of electronic volume and forward scatter principles of cell selection using flow cytometry for the evaluation of acrosome and plasma membrane integrity of bull spermatozoa. Cytometry Part A. 2014;85A(8):719–728. doi: 10.1002/cyto.a.22474. [DOI] [PubMed] [Google Scholar]
- 28.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Subgroup GPDP. The sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anders S, Pyl PT, Huber W. HTSeq - a python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal Of The Royal Statistical Society Series B. 1995;57(1):289–300. [Google Scholar]
- 35.Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Ruden DM, Lu X. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants within the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26(16):2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, Lu X. Using drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012;3:35. doi: 10.3389/fgene.2012.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Son M, Enger EG, Grove H, Ros-Freixedes R, Kent MP, Lien S, Grindflek E. Genome-wide association study confirm major QTL for backfat fatty acid composition on SSC14 in Duroc pigs. BMC Genomics. 2017;18(1):369. doi: 10.1186/s12864-017-3752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bochenek M, Smorag Z. The level of sperm DNA fragmentation in bulls of different breeds. Ann Anim Sci. 2010;10(4):379–384. [Google Scholar]
- 43.Saravia F, Núñez-Martìnez I, Morán JM, Soler C, Muriel A, Rodríguez-Martínez H, Peña FJ. Differences in boar sperm head shape and dimensions recorded by computer-assisted sperm morphometry are not related to chromatin integrity. Theriogenology. 2007;68(2):196–203. doi: 10.1016/j.theriogenology.2007.04.052. [DOI] [PubMed] [Google Scholar]
- 44.Song H, Zhu L, Li Y, Ma C, Guan K, Xia X, Li F. Exploiting RNA-sequencing data from the porcine testes to identify the key genes involved in spermatogenesis in large white pigs. Gene. 2015;573(2):303–309. doi: 10.1016/j.gene.2015.07.057. [DOI] [PubMed] [Google Scholar]
- 45.Ding H, Luo Y, Liu M, Huang J, Xu D. Histological and transcriptome analyses of testes from Duroc and Meishan boars. Sci Rep. 2016;6:20758. doi: 10.1038/srep20758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siu MKY, Cheng CY. Extracellular matrix and its role in spermatogenesis. Adv Exp Med Biol. 2008;636:74–91. doi: 10.1007/978-0-387-09597-4_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee NPY, Cheng CY. Ectoplasmic specialization, a testis-specific cell-cell actin-based adherens junction type: is this a potential target for male contraceptive development. Hum Reprod Update. 2004;10(4):349–369. doi: 10.1093/humupd/dmh026. [DOI] [PubMed] [Google Scholar]
- 48.O'Donnell L. Mechanisms of spermiogenesis and spermiation and how they are disturbed. Spermatogenesis. 2014;4(2):e979623. doi: 10.4161/21565562.2014.979623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trackman PC. Diverse biological functions of extracellular collagen processing enzymes. J Cell Biochem. 2005;96(5):927–937. doi: 10.1002/jcb.20605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perdichizzi A, Nicoletti F, La Vignera S, Barone N, D'Agata R, Vicari E, Calogero AE. Effects of tumor necrosis factor-α on human sperm motility and apoptosis. J Clin Immunol. 2007;27(2):152–162. doi: 10.1007/s10875-007-9071-5. [DOI] [PubMed] [Google Scholar]
- 51.de Vega S, Iwamoto T, Yamada Y. Fibulins: Multiple roles in matix structures and tissue functions. Cell Mol Life Sci. 2009;66:1890–1902. doi: 10.1007/s00018-009-8632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Timp R, Sasaki T, Kostka G, Chu M-L. Fibulins: a versatile family of extracellular matrix proteins. Nat Rev Mol Cell Biol. 2003;4:479–489. doi: 10.1038/nrm1130. [DOI] [PubMed] [Google Scholar]
- 53.Sercu S, Lambeir AM, Steenackers E, El Ghalbzouri A, Geentjens K, Sasaki T, Oyama N, Merregaert J. ECM1 interacts with fibulin-3 and the beta 3 chain of laminin 332 through its serum albumin subdomain-like 2 domain. Matrix Biol. 2009;28(3):160–169. doi: 10.1016/j.matbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 54.Li MWM, Mruk DD, Lee WM, Cheng CY. Cytokines and junction restructuring events during spermatogenesis in the testis: an emerging concept of regulation. Cytokine Growth Factor Rev. 2009;20:329–338. doi: 10.1016/j.cytogfr.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fonovic M, Turk B. Cysteine cathepsins and extracellular matrix degradation. Biochim Biophys Acta Gen Subj. 2014;1840(8):2560–2570. doi: 10.1016/j.bbagen.2014.03.017. [DOI] [PubMed] [Google Scholar]
- 56.Morin V, Sanchez A, Quiñones K, Huidobro JG, Iribarren C, Bustos P, Puchi M, Genevière AM, Imschenetzky M, Cathepsin L. Inhibitor I blocks mitotic chromosomes decondensation during cleavage cell cycles of sea urchin embryos. J Cell Physiol. 2008;216(3):790–795. doi: 10.1002/jcp.21459. [DOI] [PubMed] [Google Scholar]
- 57.Hermo L, Korah N, Gregory M, Liu LY, Cyr DG, D'Azzo A, Smith CE. Structural alterations of epididymal epithelial cells in cathepsin A-deficient mice affect the blood-epididymal barrier and lead to altered sperm motility. J Androl. 2007;28(5):784–797. doi: 10.2164/jandrol.107.002980. [DOI] [PubMed] [Google Scholar]
- 58.Mathur PP, Grima J, Mo M-Y, Zhu L-J, Aravindan GR, Calcagno K, O'Bryan M, Chung S, Mruk D, Lee WM, et al. Differential expression of multiple cathepsin mRNAs in the rat testis during maturation and following lonidamine induced tissue restructuring. Biochem Mol Biol Int. 1997;42(2):217–233. doi: 10.1080/15216549700202611. [DOI] [PubMed] [Google Scholar]
- 59.O'Flaherty C. Peroxiredoxins: hidden players in the antioxidant defence of human spermatozoa. Basic and Clinical Andrology. 2014;24:4. doi: 10.1186/2051-4190-24-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gong S, San Gabriel MC, Zini A, Chan P, O'Flaherty C. Low amounts and high thiol oxidation of peroxiredoxins in spermatozoa from infertile men. J Androl. 2012;33(6):1342–1351. doi: 10.2164/jandrol.111.016162. [DOI] [PubMed] [Google Scholar]
- 61.Noblanc A, Kocer A, Chabory E, Vernet P, Saez F, Cadet R, Conrad M, Drevet JR. Glutathione peroxidases at work on Epididymal spermatozoa: an example of the dual effect of reactive oxygen species on mammalian male fertilizing ability. J Androl. 2011;32(6):641–650. doi: 10.2164/jandrol.110.012823. [DOI] [PubMed] [Google Scholar]
- 62.Sun X, Kovacs T, YJ H, Yang WX. The role of actin and myosin during spermatogenesis. Mol Biol Rep. 2011;38(6):3993–4001. doi: 10.1007/s11033-010-0517-0. [DOI] [PubMed] [Google Scholar]
- 63.Ickowicz D, Finkelstein M, Breitbart H. Mechanism of sperm capacitation and the acrosome reaction: role of protein kinases. Asian Journal of Andrology. 2012;14:816–821. doi: 10.1038/aja.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Darszon A, Nishigaki T, Beltran C, Treviño CL. Calcium channels in the development, maturation, and function of spermatozoa. Physiol Rev. 2011;91:1305–1355. doi: 10.1152/physrev.00028.2010. [DOI] [PubMed] [Google Scholar]
- 65.Rahman MS, Kwon WS, Pang MG. Calcium influx and male fertility in the context of the sperm proteome: an update. Biomed Res Int. 2014:841615. [DOI] [PMC free article] [PubMed]
- 66.Kwon WS, Park YJ, Mohamed ESA, Pang MG. Voltage-dependent anion channels are a key factor of male fertility. Fertil Steril. 2013;99(2):354–361. doi: 10.1016/j.fertnstert.2012.09.021. [DOI] [PubMed] [Google Scholar]
- 67.Quinn EM, Cormican P, Kenny EM, Hill M, Anney R, Gill M, Corvin AP, Morris DW. Development of strategies for SNP detection in RNA-seq data: application of lymphoblastoid cell lines and evaluation using 1000 genomes data. PLoS One. 2013;8(3):e58815. doi: 10.1371/journal.pone.0058815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ribeiro A, Golicz A, Hackett CA, Milne I, Stephen G, Marshall D, Flavell AJ, Bayer M. An investigation of causes of false positive single nucleotide polymorphisms using simulated reads from a small eukaryote genome. BMC Bioinformatics. 2015;16:382. doi: 10.1186/s12859-015-0801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Warr A, Robert C, Hume D, Archibald AL, Deeb N, Watson M. Identification of low-confidence regions in the pig reference genome (Sscrofa10.2) Front Genet. 2015;6:338. doi: 10.3389/fgene.2015.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12(3):205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 71.Clop A, Marcq F, Takeda H, Pirottin D, Tordoir X, Bibè B, Bouix J, Caiment F, Elsen J-M, Eychenne F, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet. 2006;38:813–818. doi: 10.1038/ng1810. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differentially expressed genes for DFI in Landrace. The results are presented with Ensembl gene id, gene symbol, gene name, fold change and significance level (FDR). (XLSX 31 kb)
Differentially expressed genes for DFI in Duroc. The results are presented with Ensembl gene id, gene symbol, gene name, fold change and significance level (FDR). (XLSX 34 kb)
High quality SNPs occurring in differentially expressed genes in Landrace. The SNPs are presented with Ensembl gene id, gene name, FDR value of differentially expressed gene, chromosome (SSC), position, reference allele and alternate allele, as well as effect, impact according to SnpEff and dbSNP ID. (XLSX 100 kb)
High quality SNPs occurring in differentially expressed genes in Duroc. The SNPs are presented with Ensembl gene id, gene name, significance level (FDR) of differentially expressed gene, chromosome (SSC), position, reference allele and alternate allele, as well as effect, impact according to SnpEff and dbSNP ID. (XLSX 117 kb)
Heatmap of the differentially expressed genes for DFI. The differentially expressed genes in testis of A) Duroc and B) Landrace boars with high (bad) and low (good) sperm DFI ordered by hierarchical clustering show higher (red) and lower (yellow) expression of genes in the two DFI groups. (TIFF 59 kb)
The edgeR source code used for testing for differential expression. (TXT 2 kb)
Putative high impact SNPs in differentially expressed genes. Putative high impact SNPs in differentially expressed genes presented with breed, gene name, position, FDR and log fold change. Validation by KASP SNP Genotyping System (N.A. is for SNPs not tested). (DOCX 12 kb)
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (GEO) [29] and are accessible through GEO Series accession number GSE74934.


