ABSTRACT
We present an overview of how members of the oral microbiota respond to their environment by regulating gene expression through two-component signal transduction systems (TCSs) to support conditions compatible with homeostasis in oral biofilms or drive the equilibrium toward dysbiosis in response to environmental changes. Using studies on the sub-gingival Gram-negative anaerobe Porphyromonas gingivalis and Gram-positive streptococci as examples, we focus on the molecular mechanisms involved in activation of TCS and species specificities of TCS regulons.
KEYWORDS: Oral bacteria, two-component transduction systems, gene regulation, virulence genes, biofilm formation, Porphyromonas gingivalis, streptococci
Introduction
How bacteria respond to environmental changes is regulated, in part, by two-component signal transduction systems (TCSs). These systems have been studied extensively in pathogens and environmental organisms with the goal of deciphering the activating signals and the genes induced in the response, i.e. the regulon of each system. Such studies include both classical and high-throughput technologies ranging from bacterial genetics to next-generation sequencing. In this review, we use studies on two well-known oral bacteria, Porphyromonas gingivalis and Streptococcus mutans, to illustrate the approaches used to understand how their TCSs work and the genes they regulate. Furthermore, strain- and species-specific variations have been discovered as well as alternate strategies that have evolved to compensate for gene loss.
Oral microbiota
The generic term ‘oral microbiota’ refers to a large collection of different microbial communities whose development, maturation, and control are modulated by a wide range of environmental conditions. Shedding oral mucosa and hard tooth surfaces formed by enamel and/or dentin present different receptors for microbial adhesion, particular biophysical conditions (e.g. pH, temperature, moist, oxygen tension), as well as exposure to diverse host defence factors [1,2]. Dental biofilms contribute to robust microbial communities organized in a complex extracellular matrix that not only provides a structural scaffold but also protects the biofilm organisms from host and exogenous antimicrobial functions [3,4]. As in most biofilms [5], the composition and structure of the extracellular matrix of dental biofilms are dynamically modulated by the colonizing bacteria in response to local biophysical conditions and the availability of substrates that support its synthesis and other metabolic functions [6]. The biophysical and immunological challenges that affect biofilm formation on oral surfaces are site-specific. Micro-organisms in both supra- and sub-gingival plaque are exposed to differing temperatures, pH, redox potential, and availability of nutrients from diet, blood, or tissue fluids, as well as host cells and molecules present in saliva and serum [2,3]. Supra-gingival biofilms must adapt to fluctuations in nutrient availability in saliva, higher redox potentials, and oxidative stress, as well as host defence functions present in whole saliva and, to a lesser extent, in the serum-like gingival exudate (gingival crevicular fluid: GCF). On the other hand, sub-gingival biofilms depend less on saliva for nutrients than on the flow and composition of the GCF, and are adapted to low redox potentials and anaerobiosis [2,3]. Both supra- and sub-gingival biofilms may become dysbiotic under certain environmental and host conditions, which can promote dental caries or periodontal disease, respectively. The shift to a dysbiotic community involves changes in physiologic functions of the community members in response to the environmental conditions. To survive in such continuously changing oral environments, micro-organisms use regulatory systems to sense and rapidly respond to stimuli derived from other members of the microbial community, host functions, and exogenous factors, as addressed in the sections below. A major challenge is to identify the signals and functions that account for shifts in microbial communities from homeostasis to dysbiosis.
This review focuses on two members of the dental biofilms, S. mutans and P. gingivalis, because these species are consistently involved in dysbiotic processes associated with dental caries and periodontal diseases [7–9]. In part, because these species are relatively easy to grow and manipulate genetically, studies on these two oral pathogens led to the discovery of sophisticated mechanisms of virulence, several of which seem to be expressed by a limited group of species. Identification of these virulence functions is compatible with the notion that pathogens can disturb microbial-host homeostasis and cause infections depending on the host and environmental conditions [10–12]. For example, P. gingivalis expresses gingipain cysteine proteinases that subvert host immune functions and promote dysbiosis, giving rise to claims that it is a ‘keystone’ pathogen [13]. On the other hand, S. mutans expresses specific types of glucosyltransferases for the production of a stable glucan biofilm scaffold from sucrose, which favours the assembly of three-dimensional acidic microenvironments within the biofilm matrix, and the emergence of aciduric and acidogenic species, which characterizes cariogenic biofilms [14]. However, the expression of genes for virulence and metabolic adaptation to stresses promoted by dysbiotic processes are not constitutive, but modulated by the environmental stimuli. As we will address through this review, most of the genes involved in virulence of S. mutans and P. gingivalis are controlled through regulatory signal transduction systems, which allow microbial cells to sense local conditions and to respond readily to specific signals by altering their gene expression profile. Some of these responses may promote dysbiosis and increase the pathogenic potential of the microbial community. Therefore, defining the environmental signals that activate bacterial TCSs is an important step in order to understand the molecular mechanisms involved in the pathogenesis of biofilm-associated oral diseases.
Bacterial signal transduction systems
In nature, bacteria are subjected to changes in local pH, osmotic pressure, temperature, redox potential, nutrient availability, and exposure to toxic chemicals. In order to survive these challenges, bacteria must be able not only to communicate with each other but also to perceive and respond to environmental signals. To cope successfully with all these selective pressures, bacteria have evolved simple but highly efficient signal transduction systems to regulate gene expression to match the specific challenge. Bacteria utilize three major classes of signal transduction systems: (1) one-component systems; (2) TCSs; and (3) phosphorelay systems [15]. One-component systems respond to intracellular signals whereby a cytosolic protein binds to a cytosolic ligand and undergoes a conformational change that allows binding to the promoter region of a target gene; such binding results in either activation or repression of gene expression. TCSs are primarily (but not exclusively) involved in adaptation to external stimuli. Each system is formed by a pair of proteins, a transmembrane sensor protein, and a cytoplasmic response regulator (RR). The sensor protein is phospho-activated in response to a specific environmental trigger, and then in a subsequent phosphorelay the activation signal is transferred to a cognate intracellular RR. The regulons of each TCS are variable and may include one operon to hundreds of genes [16]. The third signal transduction system could be defined as a variant of the TCS in the sense that the activation of the RR by the sensor protein involves multiple steps of transference of the phosphate group to intermediary proteins [17]. Additional phosphate signalling activities with similarities to eukaryotic pathways have been detected in prokaryotes including P. gingivalis [18,19].
TCS and phosphorelay systems are mostly abundant in prokaryotic organisms, but are also present in Archaea, and in some eukaryotic organisms [20]. However, the TCSs of eukaryotes are involved in more complex signalling cascades, likely evolved in function of the large size and complex intracellular structure [15,17]. Bacterial species can express several different types of TCS, which differ in structure depending on the types of signals sensed (normally chemical ligands) and in the set of target genes affected. The numbers of TCSs expressed by a species appears to correlate with the size of its genomes and more interestingly with the extent of diversity of the natural niches [21,22]. Table 1 shows the numbers of TCSs expressed for some to the most common bacterial species present in supra-gingival and sub-gingival biofilms. These TCSs could be predicted by sequence homology based on their domain architecture and on its genetic organization. Most of the genes encoding a TCS sensor protein (a histidine kinase, HK) form an operon with genes encoding its cognate RR and sometimes other accessory component. There are, though, exceptions in that an HK-encoding gene is not associated with a corresponding RR gene, or vice versa. These solitary HKs or RRs are also known as ‘orphan’ components. As observed with obligate intracellular species, e.g. Mycoplasma spp. [22], oral species that are mostly restricted to intracellular compartments of the host and/or to anoxic conditions, e.g. Actinobacillus actinomycetencomitans and P. gingivalis, have a limited number (two to seven) of complete TCSs (Table 1). On the other hand, bifidobacteria and streptococci, which are adapted to diverse oral sites, including different mucosal sites and supra-gingival or sub-gingival biofilms, harbour 11–15 different and complete TCSs (Table 1). Interestingly, Tannerella forsythia and Treponema denticola harbour a higher number of orphan HKs and RR than complete TCS, and evolutionary analysis of these genes might help to explain their biological significance. Although Table 1 includes representative strains of selected species, the number of TCSs may vary between strains of a species. As for example, the total number of TCSs identified in the genomes of S. mutans strains varies from 12 to 14 [23], and certain TCSs are less prevalent, while others are highly conserved. For example, the TCS ScnKR, likely involved in the regulation of bacteriocin production, is absent is a sub-set of S. mutans strains [23]. Molecular studies of several of these TCSs in particular S. mutans strains were previously reviewed [24]. Evolutionary analyses of TCSs indicate that bacterial lineages might acquire novel TCSs by gene duplication and lateral gene transfer under specific selective pressures. These ‘novel’ TCSs evolve to assume specific functions, likely to avoid unsuitable cross-talk with other pathways [22].
Table 1.
TCSs in the genomes of oral bacteria.
| No. of complete TCS |
No. of orphan HK, HHK, RR, HRR |
||
|---|---|---|---|
| Species and strain | Genome size (bp) | p2csa;MiST2b | p2cs; MiST2 |
| Aggregatibacter actinomycetemcomitans, D7S-1 | 2,309,073 | 2;2 | 13 (8HK, 5RR); |
| 6 (2HK, 3,1HHK) | |||
| Bifidobacterium longum ATCC 15,697 (JCM 1222) | 2,828,958 | 14;NA | 16 (8HK, 8RR); |
| NA | |||
| Fusobacterium nucleatum subsp. Nucleatum ATCC25586 | 2,268,272 | 6;6 | 3 (1 HK, 2 RR); |
| 4 (2HK, 2RR) | |||
| Lactobacillus acidophilus NCFM | 1,993,560 | 8;6 | 0; |
| 2 (2RR) | |||
| Porphyromonas gingivalis W83 | 2,343,476 | 4;4 | 4 (2HK, 2RR); |
| 4 (1HK,2RR,1HHK) | |||
| Porphyromonas gingivalis ATCC 33277 | 2,354,886 | 3; 3 | 5 (1HHK, 1HK, 3RR); |
| 5 (1HK,1HHK, 3RR) | |||
| Prevotella intermedia 17 | 2,119,790 | 2;4 | 9 (5 HK, 4 RR); 7 (3HK, 2HHK, 2RR) |
| Rothia dentocariosa, ATCC 17931 | 2,506,025 | 6;8 | 6 (3 HK, 3 RR); |
| 3 (2HK, 1RR) | |||
| Streptococcus gordonii Challis | 2,196,662 | 14;13 | 3 (1 HK, 2 RR); |
| 4 (1HK, 3RR) | |||
| Streptococcus mitis B6 | 2,146,611 | 15;13 | 2 (1 HK, 1 RR); |
| 4 (1HK, 3RR) | |||
| Streptococcus mutans UA159 (serotype c) | 2,032,925 | 14;12 | 1 (1 RR); |
| 3 (1HK, 2RR) | |||
| Streptococcus mutans LJ23 (serotype k) | 2,015,626 | 13;12 | 1 (1 RR); |
| 2 (2RR) | |||
| Streptococcus oralis Uo5 | 1,958,690 | 11;9 | 1 (1 RR); |
| 4 (1HK, 3RR) | |||
| Streptococcus salivarius CCHSS3 | 2,217,184 | 13;11 | 6 (2 HK, 4 RR); |
| 6 (6RR) | |||
| Streptococcus sanguinis SK36 | 2,388,435 | 14;13 | 1 (1 RR); |
| 2 (2RR) | |||
| Tannerella forsythia ATCC 43,037 | 3,405,521 | 7;8 | 13 (8 HK, 5 RR); |
| 8 (1HK, 5HHK, 2RR) | |||
| Treponema denticola ATCC 35,405 | 2,843,201 | 5;3 | 10 (6 HK, 4 RR); |
| 11 (4HK, 1HHK, 5RR, 1HRR) |
Data obtained from ap2cs database URL:http://www.p2cs.org/ (Barakat M, Ortet P, Whitworth DE 2011 P2CS: a database of prokaryotic two-component systems. Nucleic Acids Res.39 (Database issue):D771-6. doi: 10.1093/nar/gkq1023) and b Mist2 database (URL:http://mistdb.com) (Ulrich LE and Zhulin IB. The MiST2 database: a comprehensive genomics resource on microbial signal transduction. 2010 Nucleic Acids Res. 38 (Database issue):D401-7. doi: 10.1093/nar/gkp940.).
How TCSs work
The basic structure of a typical TCS consists of a transmembrane sensor protein, normally an HK with an extracellular (or membrane embedded) sensor domain and a conserved intracellular kinase core [16,17]. An external signal recognized by the sensor domain promotes ATP-dependent autophosphorylation at the histidine (H) residue of the kinase core. The phosphate group is then transferred to the cognate intracellular RR at its regulatory domain, normally to an aspartate residue (D), promoting conformational changes. This activated RR then binds to the regulatory regions upstream to the promoters of its target genes, inducing or repressing their transcription. HKs may also have phosphatase activities, to modulate the phosphorylation level through their cognate RR. Figure 1 illustrates two types of TCS, with basic architectural domains of the HKs and cognate RR. Most of the HKs are homodimeric proteins containing multiple functional domains linked by flexible hinges. The N-terminal sensor domain (input domain) is the most variable, since it defines the specificity for the environmental ligand and includes one to several transmembrane domains (TMs). The highly conserved dimerization and histidine phosphotransferase domain (DHp, also known as HisKA) is contained in the intracellular compartment and is responsible for autophosphorylation at the conserved His residue and for the phosphotransfer reactions (the transmitter domain). Next to the DHp is the conserved C-terminal catalytic and ATP-binding domain (HATPase-c; histidine kinase-like ATPase C-terminal domain), typical of several ATP-binding proteins. In addition to this basic domain architecture, HKs vary in the number and types of accessory domains (normally located between the input and the DHp domains), which assist in the transmission of the input signal and sometimes detect intracellular stimuli. Some of the common accessory domains are PAS (a domain common in Per-Arnt-Sim proteins), GAF (a domain common in cGMP-specific phosphodiesterases, adenyl cyclases and the bacterial transcriptional regulator FhlA), and HAMP (a domain common in HKs, adenyl cyclases, methyl-accepting proteins and phosphatases) [25]. The conserved domain architecture of RR includes the N-terminal receiver domain (REC) containing a conserved D residue that receives the phosphoryl group required for activation of the regulatory domain (output domain). In addition to HKs containing the sensor and transmitter domains (Figure 1(a)), there are hybrid HKs (HHKs). As exemplified in Figure 1(b), the HHK has a receiver domain for intermediary phosphotransfer (histidine phosphotranferase-mediated) before final relay to the receiver domain of the cognate RR. Some HHKs (called unorthodox HKs) also contain a histidine phosphotransferase domain (HPT). The P2CS database classified HKs into three major families, classical HKs, HHKs, and unorthodox HKs, as well as the CheA-like HKs [22,26]. There are also hybrid RRs (HRRs) containing HK domains [27,28]. There are 35 different families of RRs based on different types of output domains [22,26,29]. Approximately 50% of the activated RRs undergo homodimerization involving conserved residues at their receiver domains, and dimer formation is required for interaction with the regulatory regions of the target genes, which may contain tandem and inverted repeats [16,22].
Figure 1.
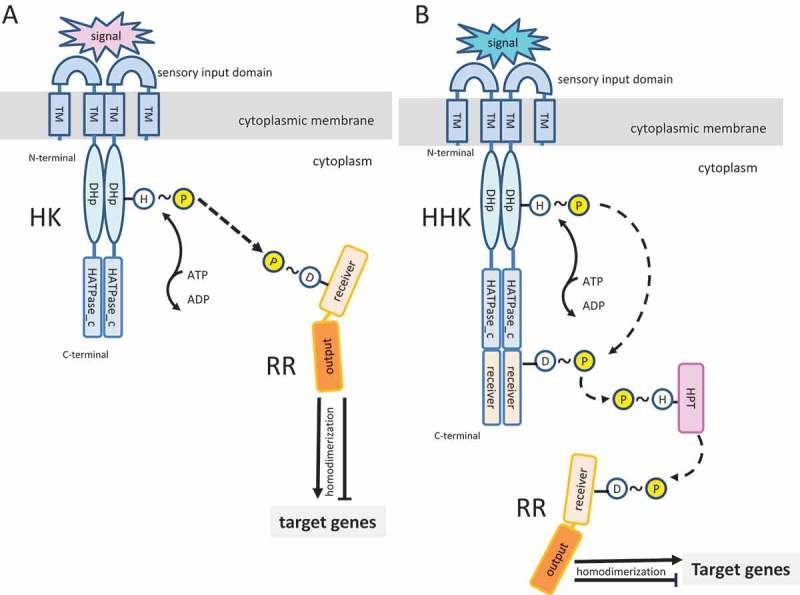
Schematic representation of histidine kinases (HK) and cognate response regulators (RR) of two-component systems (TCS). (A) Basic TCS consisting of a homodimeric HK with N-terminal extracytoplasmic sensory input and transmembrane (TM) domains. The cytoplasmic C-terminus contains the conserved DHp (dimerization and histidine phosphotransferase) and catalytic and ATP-binding (HATPase_c) domains. Although not shown in this simplified diagram, a variable number of cytoplasmic accessory domains of different classes may also be found in HK. Upon recognition of a specific environmental signal by the sensory domain, the HK undergoes autophosphorylation through HATPase_c-mediated transference of a phosphate group from ATP to the conserved histidine (H) in the DHp domain. This phosphate group is then transferred to a conserved amino acid (frequently an aspartate; D) in the receiver domain of the RR. This phosphorylation promotes conformational changes, frequently homodimerization, to permit interactions of the RR output domain with the regulatory regions of target genes. These interactions may activate or inhibit gene transcription. (B) Example of a hybrid HK (HHK), involved in more complex phosphorelay systems. In the schematic diagram, the HK contains a C-terminal receiver domain that receives the phosphate group and is involved in a phosphotransfer to the RR via a histidine phosphotransferase (HPT) intermediate.
Although sequence analyses of bacterial genomes reveals a diverse array of TCSs based on their conserved domain architecture, there is still limited information about their biological functions, and few studies have identified the stimuli responsible for HK activation [29]. Experimental studies and comparative genome analysis indicate that arrangement and topology of the N-terminal sensor and TM domains of HKs correlate with the type of stimuli sensed [30]. Based on the analyses of close to 4500 HKs, these sensor proteins were classified into three major groups. The first group includes HKs with an extracellular input domain, which is flanked by at least two TM domains. Basic HKs (Figure 1) fall into this large group and are involved in sensing extracellular (or periplasmic) stimuli, including nutrients and other soluble chemicals. The second group is characterized by N-terminal transmembrane regions formed by two to 20 membrane-spanning domains linked by short sequences, and thus with no clear extracellular sensor domain. This group seems to be involved in sensing stimuli directed to the membrane sites, e.g. membrane stresses, membrane transport and electrochemical gradients, or stimuli derived from other membrane components. HKs of TCSs involved in quorum-sensing in Gram-positive bacteria are within this second group. The third group of HKs is characterized by input domains at the cytoplasmic compartment. Examples of stimuli sensed by this group of HKs include cytoplasmic solutes or signalling proteins, stimuli transmitted through transmembrane proteins, or membrane-diffusible compounds [29]. Domain architectures of N-terminal and C-terminal regions of HKs of these three groups have been used to classify HKs of the three major groups into a relatively small number of sub-groups [29]. Few representatives of these sub-groups were functionally characterized, highlighting the need of functional studies on TCS. Some of these sub-groups analysed in oral bacteria are addressed in the next sections.
Because several bacterial species express a large number of different TCSs, it is important to understand how a particular RR is only activated by its cognate HK and no other HK. Several molecular and genetic studies have addressed this question [22,30,31]). There is evidence that each HK has a preferred affinity to its cognate RR, because the specificity of the HK–RR interaction involves the hypervariable regions flanking the conserved aspartate of the RRs. Mutations on these hypervariable regions are shown to affect the efficiency of interaction between a specific HK and the cognate RR [30,31]. A second factor affecting the specificity of the HK–RR interaction is the ratio of HK and RR production in the cell in response to a certain stimulus. Most of the operons encoding TCSs are self-regulated by the encoding TCS, implying that activation of an HK by its specific stimuli will also enhance the expression of this HK and RR [30]. In addition, because the genes encoding HK and RR of a specific TCS are expressed from the same operon, appropriate ratios of HK and RR production result in efficient interaction between these proteins. Specificities of HK localization at the cell membranes appear to avoid contact with HKs of other TCS. For example, some HKs are preferentially localized at cell poles, while others are restricted to other cell-surface regions, potentially avoiding physical interactions between non-cognate partners. Finally, because HKs commonly have phosphatase activity to dephosphorylate the cognate RR, they can control non-specific phosphorylation reactions by other TCSs or by low-molecular-weight cytoplasmic phosphoryl donors (e.g. acetyl phosphate; AcP) in the absence of the specific stimulus. Thus, there are several mechanisms by which cross-talk between different TCSs is avoided, ensuring that each TCS is activated only by its specific stimulus. On the other hand, because bacteria may need to respond to multiple signals simultaneously, the activities of multiple TCSs have to be integrated, and mechanisms of cross-talk between different TCSs and other transcriptional regulators are also required [30,31]. As addressed below, there is increasing evidence of TCS interactions with different classes of kinases and phosphatases, such as eukaryotic-like serine/threonine and low-molecular-weight kinases and phosphatases.
According to the predicted model of oral biofilm formation, streptococci are substrate-adhering pioneer bacteria, fusobacteria are bridging organisms, and P. gingivalis is a late colonizer that was shown by direct imaging to bind to both Streptococcus and Fusobacterium [32]. A series of studies examined proteomic and gene expression of P. gingivalis, Streptococcus gordonii, and F. nucleatum in a simulated biofilm in which resting cocultures were incubated for 18 h under anaerobic conditions [33–35]. While the proteome of P. gingivalis generally reflected its non-growing state, the expression of haemin uptake protein HmuR was upregulated during coculture, and an hmuR mutant showed decreased survival in the community [33]. Furthermore, P. gingivalis appeared to have a strong species-specific ‘dominant’ effect on the proteome of S. gordonii. It was suggested that the increased oxidative stress response of S. gordonii may protect P. gingivalis from secreted peroxide [34]. In addition, asaccharolytic P. gingivalis appeared to reduce glycolysis in S. gordonii and F. nucleatum [35] possibly to avoid production of acids that could inhibit P. gingivalis metabolism and or growth. A more recent RNA-seq analysis of P. gingivalis–S. gordonii coculture showed increased expression of genes in the P. gingivalis OxyR and RprY regulons, consistent with protection against peroxide produced by S. gordonii [36].
P. gingivalis FimSR TCS
P. gingivalis fimbriae are associated with biofilm formation [37] and binding to other bacteria [38]. Production of fimbriae is regulated by the FimS-FimR TCS encoded by PGN_0904-PGN_0903 in strain ATCC 33277, and PG 1432-PG1431 in strain W83. In ATCC 33277, expression of fimA, encoding the fimbrilin protein subunit of fimbriae, is positively regulated by the FimR RR, i.e. it is an activator of fimA expression. It was demonstrated that FimR controlled the expression of several genes including five clustered around the fimA locus. Gene-expression analyses of mutant strains revealed a transcriptional cascade with FimR activating expression of the first gene of the cluster that encodes a key regulatory protein [39]. In addition, comparative analyses of fimbriate type strain ATCC 33277 and fimbria-deficient strain W83 revealed differences in their fimS loci encoding the FimS HK. FimS from W83 is non-functional because of a defective kinase domain that has a truncated conserved G3 box motif and so is unable to bind ATP for phosphorylation. Thus, in spite of possessing a fimA gene, strain W83 does not produce a fimA transcript or a FimA protein. Introduction of the functional fimS gene from 33,277 restored production, but not polymerization, of endogenous FimA subunits in W83 [40]. Recent work from Nishikawa focused on mutant analysis of the FimS periplasmic sensor region that contains eight tetratricopeptide (TPR) repeat regions. The TPR motif consists of up to 16 tandem-repeats of 34 amino acid consensus residues and mediates the assembly of multiprotein complexes and protein–protein interactions, implying that the environmental signal for the FimRS TCS might be a protein [41].
RR RprY
In P. gingivalis, RprY (PGN_1186 in ATCC 33277, PG1089 in W83) is an ‘orphan’ RR because the usual close genetic linkage to a candidate cognate HK gene is absent. Using several experimental approaches (direct DNA-RprY binding screens, ChIP-on-Chip, and electromobility shift assays) it was established that the RprY regulon included genes associated with oxidative stress, iron transport, and sodium translocation [42]. RprY bound to the promoter of the first gene (nqrA) in an operon encoding Na+-translocating NADH: ubiquinone oxidoreductase, a sodium pump possibly involved in energy production by P. gingivalis from anaerobic respiration [43]. Indeed, RprY also bound to the nqrA promoters from Bacteroides fragilis and Vibrio cholera [42]. In the latter, NQR enhances cholera toxin production and is involved in generating intracellular superoxide [44,45].
The role of sodium (Na+) depletion as a potential environmental signal to activate RprY function was further investigated [46]. In Escherichia coli, an RprY–LacZ fusion protein was induced specifically by Na+ depletion; however, P. gingivalis rprY and oxyR mutants were unable to grow in a similar medium. Microarray-based comparative transcription profiling of P. gingivalis parent and rprY mutant strains grown under Na+ replete and depleted conditions confirmed the involvement of RprY in the oxidative stress response. These results were supported by EMSA assays that showed RprY binding not only to the promoter of alkyl hydroperoxidase, which detoxifies peroxide, but also to the promoters of several protein chaperones that protect against oxidative stress, i.e. groES, clpB, and dnaK. In addition, these experiments showed RprY bound to its own promoter, and so it was autoregulated.
As noted above, in P. gingivalis regulator RprY is an orphan. Indeed, the cognate HK, RprX, is present in all other oral and enteric Bacteroidetes except P. gingivalis, which does not contain proteins with any similarity to RprX. However, the gene adjacent to rprY encodes a protein acetyltransferase (pat: PGN_1185 in ATCC 33277); the pat and rprY genes have the same transcription start site and are co-transcribed [47]. Based on protein homology, Pat is a GCN5-related acetyltransferase (GNAT), proteins that were originally identified for their role in modification of eukaryotic histone proteins; however, recently, similarly acetylated proteins were identified in prokaryotes [48,49]. N ε-Lysine acetylation of proteins is effected by acetyl CoA synthase and a GNAT that uses acetyl CoA as the acetyl donor with the concomitant release of CoA [50]. Acetyl CoA can be derived from pyruvate or from acetate via phosphorylation by acetate kinase (AckA); acetate phosphate is then converted to acetyl CoA by phosphotransacetylase (Pta). An interesting and potentially evolutionary adaptation is that in P. gingivalis, the genes for AckA and Pta are just upstream from those for Pat and RprY. This is not the case in oral Bacteroidetes that contain the rprX gene.
The acetylation modification is reversed by a deacetylase, and the bacterial CobB sirtuin was identified as responsible for this function [51]. Based on protein homology, we identified CobB (PGN_004) with 58% exact and 73% positive identity to the CobB protein from Bacteroides thetaiotaomicron (Accession number NP_811887). The findings that pat and rprY had the same transcriptional start prompted a study to determine whether acetylation, a post-translational modification, altered the regulatory functions of RprY in strain ATCC 33277 [47]. Western blots of cell extracts probed with anti-acetyl-lysine or anti-RprY antibodies showed that the parent strain contained acetylated RprY, which was absent in rprY mutant extracts. In addition, RprY could be chemically acetylated in vitro in a reaction that was dependent on acetyl CoA as the acetyl donor and recombinant Pat; the latter was not self-acetylated. Based on these observations, it was hypothesized that Pat functioned as the modifier of RprY protein in vitro. The results also suggested that acetylated RprY does not transfer acetyl groups to other proteins, indicating that it does not possess acetyltranferase activity. CobB, PGN_0004 in ATCC 33277, is a Sir2 family nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase, and the acetylation level of RprY was significantly reduced in the presence of CobB and NAD+, while nicotinamide, a CobB inhibitor, reduced CobB-dependent deacetylation of RprY. Not surprisingly, the data obtained from electromobility shift assays (EMSA) indicated that acetylated RprY showed reduced binding to the promoter of nqrA, and addition of the CobB deacetylase restored binding. On the other hand, the presence of increasing concentrations of AcP (phosphate donor) resulted in increased incorporation of the phosphorylated protein into RprY–DNA complexes. Consistent with these data, in vitro chromatin immunoprecipitation (ChIP) assays to quantify binding of non-acetylated and acetylated RprY to sonicated genomic DNA fragments of P. gingivalis showed that binding of DNA to acetylated RprY was reduced approximately twofold compared with the unacetylated RprY control, while phosphorylation of RprY increased DNA binding at least threefold compared with the control. Thus, the DNA-binding ability of RprY was regulated by both acetylation and phosphorylation with the potential to alter the expression of virulence genes.
The relationship between acetylation of RprY and gene expression under Na+-depleted conditions was investigated [47]. Compared with extracts from cells from normal medium, those from Na+-depleted medium contained 60% less RprY, which was highly acetylated. To determine whether the repressor function of RprY was impaired in ATCC 33277 parent cells harvested from Na+-depleted medium, the relative expression of two promoter targets of RprY, i.e. nqrA and rprY itself, was measured. By QRT-PCR, expression of the rprY transcript was reduced in cells from Na+-depleted medium, consistent with the Western data, while expression of nqrA increased almost sevenfold, consistent with expression in the rprY mutant. Taken together, these results suggest that RprY may be a constitutively expressed repressor and that, in the absence of a cognate HK, its function is modulated by the acetylation activity of Pat. The exact role of Na+ depletion on expression of RprY is still unknown, although the rprY mutant has the same growth phenotype as an oxyR mutant during depletion, suggesting a role in the oxidative stress response [46]. Interestingly, it was recently demonstrated that the NQR pump of V. cholerae is a generator of ROS, specifically associated with reduced a flavin adenine dinucleotide cofactor of subunit F (nqrF) of the complex [45].
haeSR system
Iron is an essential nutrient for survival of P. gingivalis, and the majority of iron in the human body is stored in haemoglobin as haem (iron complexed with protoporphyrin IX). P. gingivalis has a number of mechanisms for acquiring haem from haemoglobin and other host proteins such as degradation and haem binding by gingipains, followed by transport into cells by outer membrane receptors with a high affinity for haem. Preliminary in vivo ChIP-on-chip experiments established that the TCS encoded by PGN_0752-0753 (strain ATCC 33277) and PG0719-0720 (strain W83) was involved in haemin/iron acquisition because immunoprecipitates with anti-regulator (PG0720) antibody enriched for promoters associated with haemin transport. Therefore, the TCS was named HaeRS (for haemin). A mutant in the PGN_0752 HK (haeS) from ATCC 33277 could not be generated using DNA primers designed from the W83 genome sequence, and in a previous study it was noted that the genomic region surrounding this TCS was highly divergent between the strains [52]. Following publication of the genome sequence of ATCC 33277 [53], a comparison of these regions revealed a 2.529 kbp deletion in ATCC 33277 at the locus homologous to haeSR (PG0719-720) in W83. Thus, ATCC 33277 is a naturally occurring mutant in the TCS. Correspondingly, expression of the HKs and RRs was dramatically reduced, resulting in slower growth of the strain, but transfer of the functional haeS (PG0720) from strain W83 restored expression of the TCS and growth in the ATCC 33277 transconjugant [54].
Based on ChIP-seq data, HaeR binds at least two classes of promoters, depending on the specific concentrations of haemin used in growth media. In the first class, the number of bound HaeR sequences increased with the haemin concentration, implying that expression of these genes was induced by haemin. The gene cluster hmuYRSTUV is the best-characterized haemin transport system in P. gingivalis. Among the targets of HaeR identified by ChIP-seq were 5ʹ untranslated sequences upstream of PGN_0704 (ihtA), PGN_0687 (htrA) and PGN_0556 (hmuS). RR binding to these sequences was confirmed by EMSA, so we conclude that the three loci, consistent with their previously established roles in haemin transport, are directly regulated by PGN_0753 [55–57].
In the second class of HaeR promoter targets, the number of bound promoter sequences decreased in cells grown with increased haemin concentration, suggesting haemin repressed expression of the corresponding genes. Included in this class are several TonB-dependent receptors that transport haem across the outer membrane, and ABC transporters carry haem across the periplasm and inner membrane [58]. Gingipain cysteine proteinases comprise an important class of proteins that play a role in haem acquisition by releasing and binding haem from haemoglobin and other host proteins. The ChIP-seq data show that the promoter regions of RgpA and Kgp were enriched under haem-deficient and -limiting conditions, and HaeR bound to the promoter regions of both genes, indicating direct regulation. The first indication of Kgp involvement in haem accumulation came from a key genetic study showing that kgp mutant colonies did not present the normal black-pigmentation phenotype owing to haem adsorption at the cell surface [59]. Subsequent work showed that both proteinases were responsible for the capture of haemoglobin, its degradation and release, and conversion of haem to m-oxo bishaem aggregates [60,61]. Most recently, gingipain and HmuY activities have been linked together in the release of haem from proteins degraded by gingipains and its capture by HmuY [62].
In summary, the HaeR regulon includes a number of iron uptake/acquisition genes that encode transporters and metabolic functions, and HaeR acts as an activator or repressor depending on the target gene and the haemin concentration in growth media. Collectively, the ChIP-seq data suggest that the TCS is induced by low concentrations of haemin as indicated by increased expression and binding of HaeR to promoters in haemin-depleted or -limited conditions. Finally, it was established that the HaeSR regulon includes, and HaeR directly regulates expression of Kgp and RgpA, multifunctional virulence factors of P. gingivalis.
Genes associated with haemin acquisition predominated in the HaeR regulon, raising the question of whether haemin was the environmental signal that HaeS senses in order to activate the system. Because P. gingivalis ATCC 33277 is a naturally occurring haeS deletion strain, the environmental signal for HaeS was defined in an ATCC 33277 chimeric strain, TR719, which expresses the complete haeS gene (PG0719) from W83 and restores function to the HaeSR system of ATCC 33277 [54]. HaeS is a 427-amino-acid sensor kinase protein containing a conserved DHp (dimerization and histidine phosphotransfer) domain with H226 as the predicted phosphorylation site, an HATPase-c domain, and the periplasmic sensor region is 104 amino acids in length. Two in vitro approaches established that haemin bound to both the periplasmic and cytoplasmic domains of HaeS, but not to HaeR, the negative control [63]. Haemin bound to the HaeS dimer (101 kDa) in vivo after purification with haemin-conjugated agarose. The periplasmic domain of HaeS contains seven tyrosine residues, five of which are conserved in HKs of species most closely related to P. gingivalis, and two non-conserved histidine residues. Histidine and tyrosine residues bind iron and haemin with high affinity, and a point mutation Y88A showed a significant reduction in haemin binding.
HaeS is a bifunctional HK predicted to phosphorylate and dephosphorylate its cognate RR [12] and belongs to a subfamily that contains a specific and highly conserved amino acid motif, E/DxxN/T, adjacent to the phospho-accepting histidine H226. The E/D residue is required for kinase and N/T for phosphatase activities, respectively [64,65]. Experimentally, it was demonstrated that the recombinant cytoplasmic fragment of HaeS uses ATP as the phosphodonor to autophosphorylate the conserved histidine 226 residue, while a recombinant cytoplasmic fragment of HaeS carrying the H226A point mutation was not phosphorylated. The cytoplasmic domain of HaeS also contains the conserved ExxT motif associated with phosphatase activity as demonstrated in vitro by the ability of this domain to reduce phosphorylation of HaeR-P [63]. Thus, the cytoplasmic fragment of HaeS behaves as a phosphatase and dephosphorylates HaeR, and thus has the potential to modulate the function of the regulator. It is proposed that extracellular haemin binds with high affinity to the periplasmic domain of HaeS under haemin-limited conditions activating production of enzymes and transporters to acquire more haemin from host proteins. Haemin binds with low affinity to the cytoplasmic domain of HaeS under haemin-replete conditions allowing tighter regulatory control over the phosphorylation site and therefore activity of HaeR.
PorXY
P. gingivalis is the most-studied anaerobe in the oral cavity, in part because they produce gingipains, a class of cysteine proteinases that play important roles in the nutrition, physiology, and virulence of the organism (reviewed in [66]). The study of these enzymes led to advances in the development of growth conditions, genetic systems, and tissue and animal models of infection for the bacterium. Arg-gingipains cleave proteins after arginine residues (RgpA and RgpB), and lys-gingipain (Kgp) cleaves after lysine residues. While RgpB (81.2 kDa) contains only catalytic activity, RgpA and Kgp are larger proteins (108.8 and 186.8 kDa, respectively) that possess N-terminal catalytic activities as well as extensive C-terminal adhesin domains that contain significant regions of sequence homology within individual peptides of the RgpA and Kgp adhesin domains. Gingipains are also associated with the black colony phenotype of P. gingivalis grown on blood plates, and after screening a P. gingivalis transposon library, a non-pigmented mutant was obtained in the porT gene that produced but was unable to secrete gingipains [67,68,69]. In addition to understanding the biochemistry and numerous functions associated with gingipain components, a key question has been: how are these large proteins secreted? Genomic analyses revealed that proteins related to PorT were associated with gliding motility in other species of Bacteroidetes, and mutant studies identified up to 10 orthologous and unique P. gingivalis genes associated with gingipain secretion by the Por Secretion System [70], later renamed the type nine (IX) secretion system because of conserved amino acid sequences characteristic of those secreted by the type IX system, present in gingipains and other P. gingivalis proteins [69]. The development of this research is comprehensively described in a recent review [66].
The PorXY genes have been identified as the TCS that regulates the type IX secretion system in P. gingivalis, whereby PorX (PGN_1019 in ATCC 33277 and PG0928 in W83) is the RR, and PorY (PGN_2001 in ATCC 33277 and PG0052 in W83) is the HK [69]. These authors reported that ECF SigP (PGN_0274 or PG0162), previously identified as associated with the production of gingipains [71,72], was also part of the PorXY TCS and directly interacted with the PorX RR. In another study, electromobility shift and DNA-protein co-purification assays showed that PorX does not bind directly to promoters of gene products that use the type XI secretion pathway, but rather binds to the cytoplasmic domain of PorL, part of the transmembrane complex. The authors suggest that PorX plays a role in the molecular machinery of secretion rather than direct regulation [73].
GppX
P. gingivalis contains a naturally occurring HK–RR hybrid protein, called GppX, PGN_1768 or PG1797, because it was associated with gingipain production and localization, and consequently pigment production [74]. A gppX mutant appeared to produce less arg- and lys-gingipain than parent cells, and most of that was released into the culture medium rather than retained inside or at the cell surface. Another study suggested that GppX acted as a repressor, since production of LuxS was upregulated in a gppX mutant [75]. A more comprehensive study of the GppX regulon comparing the tanscriptomes of gppX parent and mutant strains was carried out using RNA-seq [76]. An interesting finding was that the gppX mutation impacted expression of nearly half the genes that encoded hypothetical proteins. For example, transcription of hypothetical PGN_ 0151 was downregulated in the gppX mutant, and the regulator bound directly to the promoter of upstream gene PGN_0152, which is co-transcribed with PGN_0151 and activated transcription. Clearly, studies with GppX and P. gingivalis are at an early stage, but recent reports defined candidate functions and a gene target for the orthologue of GppX (TF0022) in the oral anaerobe T. forsythia [77]. Disruption of the gene led to bacterial aggregation in broth culture, and changes in the 2-D proteome profile of the mutant compared with the parent strain were observed; specfically, a glycosyltransferase (TF1061) was affected. This gene is part of a cluster that also contains an AmpG permease involved in transport of degraded murein peptides into cells. Well known for its fastidious growth requirement for N-acetylmuramic acid, T. forsythia scavenges this muropeptide precursor produced by other bacteria during cell-wall recycling. A recent report demonstrated that expression of the operon containing the ampG gene was downregulated in the gppX mutant, confirming a nutritional function for the regulator in T. forsythia physiology, and perhaps a larger role in cell-wall biogenesis [78].
Degeneracy of TCS in P. gingivalis
As suggested earlier, it is claimed that bacteria residing in secluded niches have fewer TCSs than those that grow in more diverse environments or have a complicated developmental cycle. This is an interesting issue for debate, and the obvious argument is that bacteria in every environment have evolved to optimize their fitness for survival. In P. gingivalis, each TCS studied in depth is somewhat atypical. As reported above, strain-specific mutations occurring in both FimRS and HaeSR render them non-functional or impaired. Therefore, studies comparing multiple strains such as that recently reported by [79] provide a valuable resource to discover more strain-specific differences that could impact infection.
TCSs of oral streptococci
The complex and heterogeneous group of oral streptococci is currently classified into five groups: (1) Mutans, (2) Salivarius, (3) Anginosus, (4) Sanguinus, and (5) Mitis [80]. These groups diverge in abundance, preferred sites of colonization, and expression of functions associated with oral and systemic infections, including dental caries, bacteraemia, and infectious endocarditis. Species that predominantly initiate colonization of the supra-gingival dental surfaces under healthy-associated conditions include the Mitis (Streptococcus mitis and Streptococcus oralis) and the Sanguinus groups (Streptococcus sanguinis and Streptococcus gordonii) [81,82]. Although S. mitis and S. sanguinis are pioneer colonizers of teeth, S. sanguinis is mostly restricted to dental surfaces, while S. mitis is abundant in a large range of mucosal and tooth sites, and may persist in significant numbers in biofilms associated with caries [83,84], a consequence of the high level of genetic heterogeneity within the species [85,86]. On the other hand, Mutans species S. mutans and S. sobrinus show a low abundance in health-associated biofilms and are poor initiators of tooth colonization [82], but predominate in biofilm communities in the presence of sucrose and acidic environments promoting the growth of the acidogenic and aciduric microbiota associated with dental caries [14,87,88]. Sequence homology analyses reveal that different streptococcal species including S. gordonii, S. sanguinis, and S. mutans share several TCSs controlling functions for bacterial persistence in the oral cavity [87,89]. However, molecular studies indicate that even highly conserved TCS, e.g. TCS VicRK, have species-specific functions in bacterial colonization and virulence as addressed below [90,91].
Clinical, animal, biochemical, genetic, and molecular studies on S. mutans, the major species affecting host-biofilm homeostasis during caries development, revealed a panel of virulence genes for the synthesis of highly stable water-insoluble glucan from sucrose (gtfB, gtfC) and other exopolysaccharides (ftf, gtfD, epsC), for bacterial binding to these polymers (gbpA, gbpB, gbpC), and for production and tolerance to acids from fermentable sugars and other stresses (including multiple metabolic and stress response genes), as addressed in several reviews [8,9,92–94]. Some of these gene functions are also associated with the ability of S. mutans to avoid host immune functions in systemic infections [95]. However, expression of these genes is not constitutive, and expression under different conditions varies between strains [95–97]. Increased expression of virulence functions was observed in strains associated with cariogenicity and systemic diseases (e.g. bacteraemia and infectious endocarditis) compared with strains isolated under healthy conditions [95,97]. Therefore, it is necessary to define the TCS regulatory circuits in S. mutans, as well as in other streptococcal species in oral biofilms in order to understand the environmental signals that trigger expression of virulence phenotypes. The large number of TCSs expressed by most oral streptococci (Table 1) poses a challenge to understand these complex regulatory functions, in part because multiple TCSs are coordinately activated in response to stimuli of specific growth conditions. However, interesting findings have been obtained by analysing TCSs that are highly conserved across streptococcal species, including the VicRK and CovRS TCS, which play important functions for coordinating bacterial growth with virulence functions for persistence in host sites.
TCS VicRK of streptococci
The TCS VicRK (also known as YycFG, and WalRK) is ubiquitous in the Firmicutes phylum, and one of the few TCSs essential for viability in several low GC Gram-positive bacteria. The system was first discovered in Bacillus subtilis as essential for cell division and growth because it directly regulates the ftsAZ operon [98,99]. Orthologues of vicR and vicK form operons with one to four accessory cistrons [100]. In B. subtilis, two of these cistrons (yycH and yycI) encode transmembrane proteins that negatively regulate YycFG (VicK) kinase activity, but the function of a third gene (yycJ) is still unclear [101]. A similar operon structure is found in most other Gram-positive genera, e.g. Staphylococcus, Enterococcus, Listeria, and Lactobacillus, except for Streptococcus spp. and Lactococcus lactis, in which the operons include only the yycJ orthologue, also known as vicX or walJ [100,102]. Proteins encoded by vicX are metal-dependent beta-lactamases of as-yet unknown function, although there is evidence suggesting that VicX modulates VicK activity in S. mutans, because vicX-defective strains showed altered phenotypes in VicRK-regulated functions [103]. VicX of S. pneumoniae is localized in the membrane, implying an interaction with the VicK sensor protein [102]. Figure 2 illustrates differences between B. subtilis and streptococcal species in the domain architecture of YycG (VicK) and ancillary proteins of the VicRK TCS (YycFG). In B. subtilis and other non-streptococcal species, the N-terminus of the HK includes two TM domains flanking an extracellular sensor loop, while the streptococcal VicK proteins have a single TM linked to a short extracellular peptide [102]. In a model for function of YycFG and associated ancillary proteins, an extracellular signal activates YycG by affecting inhibitory interactions of YycI and YycH. The lack of the extracellular loop of streptococcal VicK plus the absence of YycI and YycH orthologues suggests that VicK is activated by stimuli at the cytoplasmic membrane in a process involving VicX. As with YycG, VicK has two intracellular accessory domains, HAMP and PAS, next to the TM region. HAMP and PAS domain functions are not completely understood [101,104], although PAS domains respond to input signals including redox potential, oxygen, and small ligands, which could cross the cell membrane or affect cell metabolism [101,105].
Figure 2.
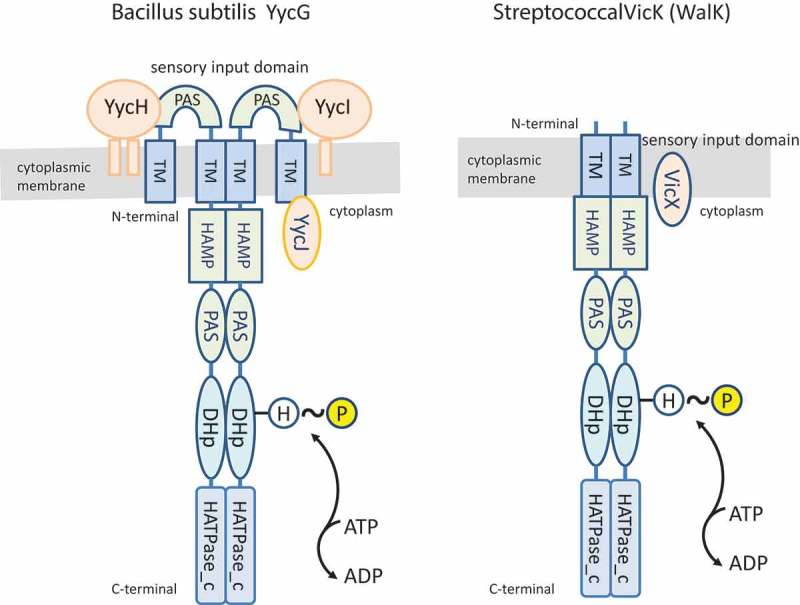
Comparison of the domain architecture of histidine kinases of the VicRK TCS. The B. subtilis VicK orthologue (known as YycG) has an extracellular loop in the sensory input domain that includes an accessory PAS domain. YycG is also associated with ancillary proteins YycH, YycI, and YYcJ, encoded by the five-cistron operon yycFGHIJ. A similar operon structure is found in most genera of Firmicutes, including Staphylococcus spp., Listeria spp., Enterococcus spp., and Lactobacillus spp. The streptococcal VicK is expressed from a three-cistronic vicRKX operon, which includes a single accessory cistron encoding VicX (an orthologue of YycJ), the VicR RR (vicR), and the histidine kinase VicK (vicK), also known as WalK. The streptococcal VicK lacks the extracellular loop in the sensory input domain. The cytoplasmic transmitter domains of the VicK and YycG histidine kinases share a similar domain architecture, which includes the accessory HAMP, PAS, DHp, and HATPase_c domains. The cognate RRs YycF of B. subtilis and VicR of streptococci are not shown in these schaems.
Figure 3 shows the structure of vicRKX operons and protein similarities in pathogenic and commensal species of oral streptococci. Analyses of VicK sequences using the SMART program detected a HAMP domain in S. gordonii and S. salivarius, but not in S. mutans, S. sanguinis, or S. mitis, reflecting low conservation of this domain. In addition, the N-terminus of VicK and part of the PAS domain are less conserved among species (Figure 3(c)). Whether diversity in these variable regions accounts for functional differences between streptococcal species remains to be investigated. VicR is highly conserved among streptococcal species with polymorphic sequences restricted to linker regions between receiver and output domains. VicR is in the OmpR/PhoB family of regulators in which the output domain forms a DNA-binding winged-helix domain. In B. subtilis and Staphylococcus aureus, both VicR and VicK are essential for bacterial viability [100]; on the other hand, only vicR is essential in S. pneumoniae [105], S. mutans [106,107], and S. sanguinis [90], and both vicR and vicK can be deleted in S. gordonii [91]. In S. pyogenes, insertional inactivation of vicR was possible, but the mutant was not viable in a mice model of infection [108]. The exact reasons for VicR and/or VicK essentially in different species are unknown, but a possibility is their role in the regulation of peptidoglycan (murein) biosynthesis, cell division and cell wall homeostasis [90,98,103,107,109–111]. Because vicK can be deleted in S. pneumoniae, S. mutans, and S. sanguinis, one possibility is that the VicR of these species could be phosphorylated by alternative pathways.
Figure 3.
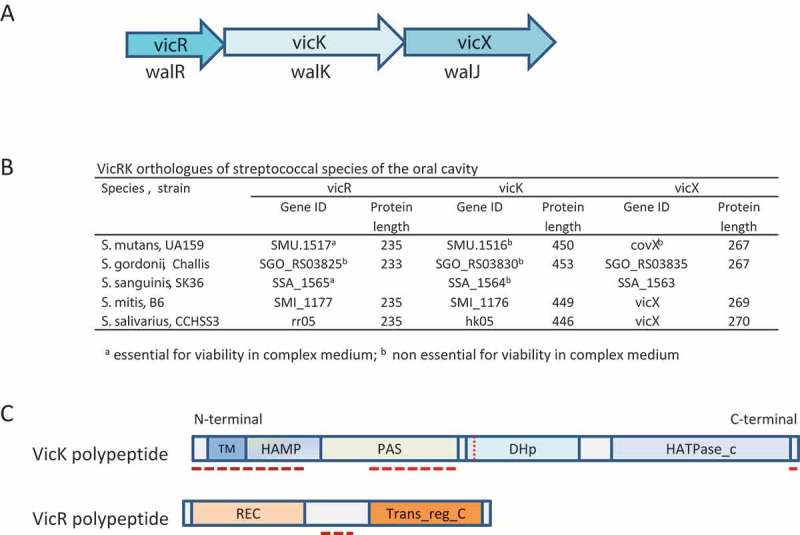
VicRK orthologues of oral streptococcal species. (a) Coding regions of the genes of the vicRKX operon represented by arrrows indicating the direction of transcription; orthologous genes found in streptocccal species are also called walRKJ. (b) Genes and protein length of VicRKX orthologues found in different species of oral streptococci. Gene ID numbers were obtained from GenBank (https://www.ncbi.nlm.nih.gov/gene). (c) Schematic representation of domain architectures of VicK and VicR polypeptides (coloured bars), which were determined using the SMART database (http://smart.embl-heidelberg.de/). Transmembrane (TM), HAMP, PAS, DHp, and catalitic HATPase_c domains are indicated within each coloured module. ClustalW multiple sequence alignment (http://www.genome.jp/tools-bin/clustalw) of amino acid sequences of streptococcal VicK and VicR proteins listed in Table B revealed regions with sequence diversity between species, as indicated by dashed red lines.
VicRK regulons of TCSs are diverse and species-specific
VicRK plays an important role in modulating cell division, cell-wall biosynthesis and homeostasis, and membrane integrity, as evidenced by the presence of several genes encoding autolysins and other murein hydrolases in the VicR regulons of B. subtilis [112], S. aureus [109], S. pneumoniae [105], S. mutans [107,113], and S. sanguinis [90]. Consistently, atypical long-chain phenotypes are found in vicK isogenic mutants obtained in S. pnemoniae, S. mutans, and S. sanguinis [90,105,106,111]. In VicR regulons, the most conserved target gene encodes PcsB (protein required for cell separation of group B Streptococcus) that was first identified in group B streptococci [114] and orthologue GbpB (glucan-binding protein B in S. mutans) [115,116]. Table 2 shows comparisons of VicR gene targets among streptococcal species of the oral cavity and oropharynx. PcsB/GbpB are cell-surface murein hydrolases with CHAP amidase domains and are part of the divisome, at least in S. pneumoniae [119–122]. PcsB/GbpB was implicated in the essentiality of VicRK in S. pneumoniae, where vicR deletion is possible only when pcsB is induced [119]. In S. mutans, GbpB/PcsB is required for surface binding to glucan exopolysaccharides synthesized from sucrose during biofilm formation [111].
Table 2.
Direct VicR gene targets that are positively (+) or negatively (–) regulated in different species of streptococci of the oropharynx.
| Streptococcal species |
||||||
|---|---|---|---|---|---|---|
| Gene name | Encoded protein or function | S. mutans | S. sanguinis | S. gordonii | S. pneumoniae | References |
| Cell-wall biogenesis, division, homeostasis | ||||||
| gbpB/pcsB | Glucan-binding protein B (GbpB)/protein for cell separation of group B streptococcus (PcsB) | (+) | (+) | ND | (+) | [90,105,111,117] |
| smu.2147c | LysM; murein hydrolase | (+) | WO | WO | WO | [107] |
| smu.2146c | Putative murein hydrolase | (+) | WO | WO | WO | [107] |
| smaA | Murein hydrolase | (–) | WO | WO | WO | [107] |
| smu.367 | Putative murein hydrolase | (+) | WO | WO | WO | [107] |
| cwdP | Putative murein hydrolase | WO | (+) | ND | WO | [90] |
| SSA_0094 | Putative murein hydrolase | WO | (+) | WO | WO | [90] |
| Smu_689 (atlA) | Autolysin | (+) | WO | WO | WO | [117] |
| Synthesis of exopolysaccharides from sucrose | ||||||
| gtfB | Glucosyltransferase B | (+) | WO | WO | WO | [106] |
| gtfC | Glucosyltransferase C | (+) | WO | WO | WO | [106] |
| ftf | Fructosyltransferase | (+) | WO | WO | WO | [106] |
| Competence | ||||||
| comCDE | (–) | (–)b | ND | (–)c | [90,110,113] | |
| Copper homeostasis | ||||||
| copY | Copper-responsive repressor | (–) | ND | ND | ND | [117] |
| Production of bacteriocin | ||||||
| nlmC | Mutacin V | (+) | WO | WO | WO | [113] |
| Transcriptional regulation | ||||||
| covR (gcrC) | Response regulator CovR | DBa | ND | ND | ND | [117] |
| Production of H2O2 | ||||||
| spxB | Pyruvate oxidase | WO | (+) | ND | ND | [90] |
| Evasion to complement immunity | ||||||
| pspA | Pneumococal surface protein A | WO | WO | WO | (+) | [105] |
| pepO | Endopeptidase O | (–) | ND | ND | ND | [118] |
| smu.399 | Putative C3-degrading proteinase | (+) | WO | WO | WO | [118] |
Orthologous proteins were considered as those showing ≥50% identity in the amino acid sequences over at least 80% of the length of the shortest protein.
WO: without orthologous gene/protein. ND: not determined. DB: evidence of VicR binding to the promoter region, without transcriptional analysis.
a No direct binding to covR promoter region observed in EMSA assays [107].
b Significant transcriptional changes of comE at late(–)log phase of growth; no evidence of direct VicR binding [90].
c Significant transcriptional changes in comE promoted by vicRKX downregulation in S. pneumoniae; no evidence of direct VicR binding [110].
In addition to GbpB/PcsB, several other non-essential murein hydrolases are directly regulated by VicR in S. mutans, including SMU_2146c, SMU.2157c (LysM), SmaA, SMU.367 [107], and the autolysin A (AtlA; smu_689) [117,123]. However, these proteins are species-specific. Similarly in S. sanguinis, the VicRKSs regulon includes genes for murein hydrolases SSA_0094 and SSA_0304 that are not expressed by S. mutans [90]. Most genes directly regulated by VicR in S. mutans are not present in the genomes of other streptococci and vice versa (Table 2). The S. mutans VicRK also induces expression of GtfB and GtfC, which are required for the synthesis of extracellular insoluble glucan [106,107] the major extracellular matrix component of S. mutans biofilms [124,125]. GtfB and GtfC are not expressed by commensal S. sanguinis or S. gordonii, although these species express Gtfs that synthesize different types of glucan [125]. The expression of glucan-binding proteins allows S. mutans to bind to glucan in biofilms, a mode of growth that affords evasion of complement [95], an important host-defence system present in saliva, GCF, and blood [126,127]. In S. mutans, VicRSm negatively regulates pepO and SMU_399, which encode proteases implicated in evasion of complement system [118]. In addition, VicRKSpn regulates pspA [105], which encodes a species-specific surface protein (PspA) involved in complement evasion [128]. In S. mutans, VicRSm also directly represses genes involved in competence and competence-induced bacteriocin (comCDE and nlmC) [113], and copY [117], part of the copYAZ operon required for copper homeostasis and regulation of membrane potential, competence, and biofilm formation [129]. In S. sanguinis, VicRKSs does not regulate gtfP, a unique gene encoding a glucosyltransferase for the synthesis of soluble glucan [90] but does regulate spxB encoding pyruvate oxidase, required for conversion of pyruvate, inorganic phosphate, and O2 to H2O2, CO2, and AcP [130]. Production of H2O2 by S. sanguinis inhibits the growth of the competitor species S. mutans [131]. By mechanisms not completely understood, H2O2 is also required for release of DNA (eDNA) to the extracellular milieu [132], where it is a major component of the extracellular matrix of S. sanguinis biofilms [90]. Thus, the VicRK regulons include species-specific genes involved in bacterial persistence in host niches, including immune evasion, inter-species competition, and biofilm formation.
What are the signals sensed by VicRK TCS?
The general role of VicRK in controlling the biosynthesis of peptidoglycan and cell division might imply that this TCS senses signals associated with peptidoglycan biosynthesis [100]. In B. subtilis, YycG (VicK) co-localized with FtsZ of the divisome, and it was proposed that VicKBs senses cell division directly [133]. S. pneumoniae, VicKSpn does not co-localize with the FtsZ ring at division septa, but is peripherally distributed during exponential growth [102]. However, VicRKSpn target protein, PcsB, localizes at the cell-division septa by interacting with FtsX, a component of the divisome [102,121]. It is possible that signals sensed by human streptococci are different from those that activate YycG of B. subtilis, present in soil and water habitats, microbial communities of plants, and gastrointestinal tracts of animals [134]. The domain architecture of streptococcal VicRK associated with molecular studies is compatible with the hypothesis that VicRK could be activated by stresses to the cell envelope that would promote changes in redox potentials sensed by the cytosolic PAS domain. In S. mutans, transcripts of vicRKX are found during the mid- and late-exponential phases of growth, but reduced during early-exponential growth [96,135]. Furthermore, vicRSm transcription is induced by vancomycin and other antibiotics that target peptidoglycan biosynthesis (ampicillin, penicillin G) or the cell membrane (polymyxin B) [135]. Increased transcript levels of vicRSm are observed during initial phases of biofilm growth in the presence of sucrose compared with planktonic cells (at mid-exponential growth) or to cells of biofilms formed in the absence of sucrose [107]. These findings suggest that VicRKSm is activated not only by intrinsic peptidoglycan growth and cell division, but also by processes associated with sucrose-dependent biofilm initiation. Consistent with these data, VicRSm directly induces expression of enzymes required for the synthesis of different sucrose-derived exopolysaccharides (GtfB, GtfC, Ftf) and glucan-binding (GbpB) [106,111,117].
To explore signals activating VicK in S. pneumoniae, strains expressing truncated versions of vicK were constructed to identify domains involved in VicK kinase activities. These studies revealed that VicK is a bifunctional sensor protein that alternates kinase and phosphatase activities, and that the phosphatase activity is dependent on the PAS domain [102,136]. There is also evidence that both forms of VicR, phosphorylated (P-VicR) and non-phosphorylated (NP-VicR), are capable of directly binding to a different set of target genes. P-VicR induces expression of PcsB and other murein hydrolases, but NP-VicR directly represses fabT, encoding a repressor of genes for fatty acid biosynthesis and, in turn, upregulates membrane synthesis [137]. Phosphatase activity has also been shown in the S. aureus VicK, also known as WalK [138]. Therefore, VicRK TCS apparently switches functions affecting the homeostasis of the cell envelope both by its kinase and phosphatase activities, and, in turn, by balancing the levels of P-VicR and NP-VicR. These findings would imply that streptococcal VicRK is constantly active, and perhaps its kinase and phosphatase functions are modulated by stimuli affecting cell growth and cell-envelope homeostasis.
Interactions of VicRK TCS with other regulatory systems
Bacteria inhabiting complex environments, such as the oral cavity, must integrate responses to a range of environmental stimuli [31]. Such integrated responses imply a certain degree of cross-talk. There is evidence that VicRK interacts with other regulatory circuits involved in cell-envelope homeostasis during challenges with a range of environmental stresses [107,138,139]. Some of the regulators found to interact or to cooperate with regulation of VicRK-gene targets include the orphan regulator CovR of S. mutans [107,117] and the TCS SaeSR of S. aureus [138]. In S. mutans, vicRK expression seems to be also indirectly induced by the TCS LiaSR [135], a TCS that regulates responses to cell-envelope stresses [140].
In S. mutans, there is a clear overlap between the regulons of VicRKSm and of CovRSm, the latter being an orphan RR in oral streptococci. The overlap includes genes for cell-wall division and biofilm formation (gbpB, lysM, gtfB, gtfC), with VicR positively regulating these genes and CovR acting as a repressor [107,141,142]. In EMSA assays, it was observed that VicRSm VicR and CovR co-bind the promoter regions of lysM, gbpB, and gtfC [107]. CovR is also a regulator of functions required for evasion to host immunity [95], and VicRSm/CovRSm co-regulatd genes include those required for evasion of complement and opsonophagocytosis, i.e. pepO and smu.399 [118,143]. These findings indicate that VicR/CovR cooperate to more precisely coordinate functions of cell division with biofilm formation and evasion of host immunity, two essential functions for S. mutans persistence in oral niches. In S. aureus, VicRKSau interacts with the SaeRS TCS to upregulate expression genes with similar functions, and so it is possible that VicRK systems may cross-talk with other TCSs involved in bacterial evasion of host immunity [138]. Direct approaches to establish the molecular mechanisms underlying these potential cross-talks remain to be performed.
In addition to potential cross-talk between TCS, studies on streptococci and other Gram-positive bacteria indicate that VicKR can interact with a transmembrane eukaryotic-like serine/threonine kinase (STK) [144,145] that functions in concert with cognate cytoplasmic eukaryotic-like serine/threonine phosphatases (STP) present in Gram-positive bacteria as STK–STP operons [146,147]. STK are transmembrane enzymes with extracellular domains containing one to five PASTA (penicillin-binding protein and serine/threonine kinase associated domain) repeats and a cytoplasmic kinase domain [146,148]. PASTA domains sense unlinked muropeptides and localize the enzyme complexes for peptidoglycan biosynthesis [148]. In S. pneumoniae, STKSpn co-localizes with FtsZ at the division septum [149]. STK also binds elongation factor Tu (EF-Tu) [146], which could potentially explain the reported interaction of GbpB/PcsB with EF-Tu in S. mutans [122]. In S. pyogenes, STKSpy phosphorylates VicRSpy [145], and in S. mutans, deletion of the gene encoding STK (pknB) affected transcription of smu.2146c [144], a VicR target, which encodes a protein harbouring a lysozyme-like domain [107]. STK is conserved among Gram-positive bacteria, including oropharyngeal streptococci, e.g. S. pneumoniae, S. gordonii, S. sanguinis, and S. mutans, and analysis of these orthologues might help decipher the VicRK network [147]. Analysis of the STK-STP system in S. mutans was investigated using an isogenic mutant in pknB, the STK-encoding gene [144,150,151]. The mutant showed reduced cariogenicity in a rat model of dental caries, and defects in biofilm formation, competence, production of bacteriocins, and sensitivity to acid stress [144,150] and to H2O2 produced by S. sanguinis [151]. Although the molecular mechanisms underlying this phenotype remain to be investigated, it is compatible with the evidence that STK PknB activates VicRSm in S. mutans [144]. The role of PknB in VicR phosphorylation might also explain the essentiality of vicR, but not of vicK in some oral streptococci, i.e. the sensor protein VicK might not be essential because VicR can be activated by PknB. Consistent with this hypothesis, it was not possible to obtain double mutants of vicK and pknB in S. mutans [144]. In addition to the roles of VicRK and STK-STP in bacterial responses to environmental stresses and host immune challenges, these systems also affect bacterial susceptibility to different classes of antibiotics. Accumulated mutations in vicRK and STK-encoding genes were detected among Staphylococcus strains resistant to vancomycin and daptomycin that target the cell wall and membrane, respectively [152–154]. Also, STK protein PknB (also called Stk1) appears to activate a transcriptional regulator of the antibiotic efflux pump NorA [146]. Further understanding of the molecular mechanisms involved in the interactions between these regulatory systems will increase interest in their potential as therapeutic targets to control infections.
TCS CovRS, and CovR as an orphan regulator
The TCS CovRS was identified in beta-hemolytic S. pyogenes by screening transposon mutants for defects in genes in the has operon that are required for the synthesis of hyaluronic acid capsule. The TCS was originally designated CsrRS (Csr: capsule synthesis regulator) [155,156] and then renamed CovRSSpy (cov; control of virulence) based on the demonstration that the system repressed multiple virulence genes, including streptokinase (ska), streptolysin S (sagA), and mitogenic factor (speMF) [157]. Later studies showed that CovRSSpy directly or indirectly regulates approximately 10–15% of the S. pyogenes genome [158,159]. The TCS CovRSSpy is atypical because of its repressor functions, although it also positively regulates some genes in its regulon. Deletion of covRSpy increases transcription of a panel of CovRSpy-repressed genes and also survival of S. pyogenes to opsonophagocytic killing by human PMNs in vitro [155,158].
Only the orphan RR CovR is conserved in oral streptococci and in S. pneumoniae in which it is known as RitR (repressor of iron transport) [160]. In S. mutans, the system was originally designated GcrR (for glucan-binding protein C regulator) following identification in a screen for genes that regulate aggregation between GbpC and dextran [161]. Later studies revealed that CovRSm negatively or positively regulates about 6.5% of the S. mutans genome [142]. More importantly, CovRSm directly represses a panel of genes required for the synthesis of water-soluble and water-insoluble glucan (gtfB, gtfC) and other exopolysaccharides derived or not from sucrose (epsC and ftf), as well as surface proteins GbpC and GbpB that bind to glucan [107,141,142]. Deletion of covR Sm reduced S. mutans cariogenicity in rat models by mechanisms not completely understood, but likely by affecting biofilm structure [162]. On the other hand, we recently showed that covR Sm deletion increases S. mutans’s capacity to stably bind sucrose-derived exopolysaccharides mediated by GbpC and EpsC, and that this property provides a capsule-like protection against opsonophagocytosis and killing by human PMNs, as well as bacterial survival in blood [95]. S. mutans strains isolated from systemic infections show low levels of covR Sm expression associated with derepressed transcription of CovRSm target genes (gbpB, gbpC and epsC), and increased resistance to opsonophagocytic killing in vitro, supporting the important role of CovRSm in the virulence of S. mutans [95]. CovRSm was also shown to repress complement system proteases directly or indirectly, suggesting that, as for S. pyogenes, CovRSm plays a role in host immune evasion and host invasiveness [143].
Signals that activate CovRS or orphan regulator CovR
CovRSpy is an RR of the OmpR family, and CovSSpy is part of the EnvZ family of HKs. CovSSpy contains two N-terminal transmembrane domains flanking a predicted external input domain of 150 amino acids, as well as the intracellular HAMP, DHp and C-terminal HATPase_c domains. There is evidence that CovSSpy is a bifunctional sensor protein, switching kinase or phosphatase activities depending on the environmental stimuli. Under growth conditions, CovSSpy acts as a kinase that phosphorylates CovR at conserved apartate (D-53) of the receiver domain, thus increasing its affinity for target gene promoters leading to repression of transcription [163]. Under environmental stress conditions including high temperature (40°C), low pH (≈6.0), and high salt concentration, CovSSpy acts as a phosphatase that dephosphorylates CovRSpy, which derepresses transcription of target genes [164]. Interestingly, although dephosphorylation of P-CovRSpy depends on CovSSpy, CovRSpy is still phosphorylated in covSSpy-defective strains, suggesting that CovRSpy is also phosphorylated by other TCS HK or additional kinases [164]. Because multiple stress conditions promote CovSSpy phosphatase activity, it was proposed that CovSSpy is responsive to general stresses affecting membrane structure [164]. On the other hand, there is also evidence that CovSSpy activities are modulated by specific external ligands, including Mg2+, but not other divalent cations, and the human antimicrobial peptide LL-37 at subinhibitory concentrations, but not other antimicrobial peptides [165–168]. CovRSpy is also phosphorylated at the threonine residue T-65 by an STK [145,169], similar to CovRSag of S. agalactiae [170]. Additional tyrosine/serine/threonine sites for CovRSpy phosphorylation were also suggested [171] indicating complexity of mechanisms for activation of this RR.
Studies on molecular mechanisms of CovR phosphorylation and activities in oral streptococci are few. Comparisons of the amino acid sequences of CovR orthologues between the beta-haemolytic species (S. pyogenes and S. agalactiae) with oral streptococci (Figure 4) revealed that S. salivarius CovR has both D-53 and T-65 phosphorylation sites. On the other hand, only the D-53 site is conserved in S. mutans, while neither the D-53 nor T-65 phosphorylation sites are present in CovR orthologues of streptococci of the Mitis and Sanguinus groups (Figure 4), implying different mechanisms of CovR activation between streptococcal species. In S. mutans, it was reported that in vitro phosphorylation of CovRSm does not increase the binding affinity to target promoters [141]. It was proposed that CovRSm-mediated regulation is dependent on the amounts of CovRSm present in cells regardless of the phosphorylation state; i.e. CovRSm would be always active and/or likely phosphorylated by small phosphodonors such as AcP [141,172]. The absence of CovS protein in S. mutans and amino acid differences between CovRSpy and CovRSmu [142] (Figure 4) account for this proposal. This hypothesis is compatible with our findings that S. mutans strains isolated from systemic infections that have increased resistance to opsonophagocytic killing by PMN express low levels of covRSm and upregulated CovRSm target genes compared with oral strains [95]. Mechanisms regulating covR Sm expression and the CovR regulon remain to be investigated in S. mutans strains. We have preliminary data indicating that BHI supplementation with human serum (20%) significantly reduces covR transcription in S. mutans strain UA159 [Mattos-Graner et al., unpublished data] suggesting that host components could induce covRSm downregulation and derepress transcription of genes for immune evasion such as pepO and smu.399, which are also regulated by the TCS VicRK [118]. Unpublished results reveal that pepO and smu.399 are also upregulated in the S. mutans covR-defective strain, connecting the TCS VicRK and CovR in modulating S. mutans evasion to host components [143]. Further studies are required to investigate serum-mediated signalling pathways by which CovR and VicRK regulate S. mutans responses to host stimuli, and cause oral and systemic infections.
Figure 4.

ClustalW multiple sequence alignment of the amino acid sequences of CovR orthologues of streptococcal species. Except for Streptococcus salivarius, CovR orthologues in oral streptococci and S. pneumoniae do not have the aspartate (D-35) and/or threonine (T-65) residues (marked in red) known to be phosphorylated by CovS histidine kinase and serine/threonine kinases, respectively, in the beta-hemolytic S. pyogenes and S. agalactiae species. The D-35 aspartate is conserved in S. mutans CovR. Sequence alignments were performed using the bioinformatics tool available at http://www.genome.jp/tools-bin/clustalw.
Concluding remarks and future work
Evolutionary analyses of bacterial signal transduction systems suggest that the number of TCSs in each oral species correlates with genome size and content, and the complexity of the environmental sites to which they have become adapted.
Although oral Bacteroidetes species have far fewer TCSs than their enteric relatives, they play vital roles such as protection against environmental stress and acquisition of essential nutrients. Oral streptococci are equipped with a considerably larger panel of TCSs, of which many are highly conserved across species. However, TCS orthologues have species-specific functions, as revealed by significant differences in their regulons as well as polymorphisms of regulatory proteins. VicRK and CovR are prototypic TCSs involved in coordination of cell growth and cell-wall homeostasis with fitness and persistence in host environments during streptococcal infections. Thus, TCSs have a central role in oral ecology.
As in all scientific fields, more knowledge about TCS regulation leads to more questions. For example, what are the support mechanisms provided by additional regulatory proteins such as one-component systems and extracytoplasmic-function σ factors (ECFs), as well as regulatory RNA species? Why do some systems degenerate and compensatory functions evolve? On a more practical level, identifying the regulons of each TCS will identify functional specificities in different species that could lead to the development of therapies to control local or systemic infections. Studies are necessary to understand the role of streptococcal TCS in modulating commensal to pathogenic interactions with the host, and in inter-species interactions. Increasing evidence that streptococcal TCSs are activated in response to host immune factors emphasizes the need to investigate TCS functions under host-like conditions, for example in the presence of serum, saliva, blood, and other host components.
Biographies
Renata O. Mattos-Graner is an Associate Professor of the Area of Microbiology and Immunology - Dept. of Oral Diagnosis; State University of Campinas, who investigates two-component systems in oral streptococci.
Margaret J. Duncan is a Senior Member of the Forsyth Institute, Cambridge, U.S.A., who investigates how environmental conditions affect the behavior of species often associated with periodontal diseases, Porphyromonas gingivalis, and dental caries, Streptococcus mutans.
Funding Statement
ROMG was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) (grants 15/12940-3, 12/51832-2, 12/50966-6, and 09/54182-7). MJD was supported by National Institutes of Health grants R01 DE24308 and R01 DE024468 (NIDCR).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1]. Faust K, Sathirapongsasuti JF, Izard J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8(7):e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2]. Devine DA, Marsh PD, Meade J.. Modulation of host responses by oral commensal bacteria. J Oral Microbiol. 2015;7(1):1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3]. Kolenbrander PE, Palmer RJ Jr, Periasamy S, et al. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–480. [DOI] [PubMed] [Google Scholar]
- [4]. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. [DOI] [PubMed] [Google Scholar]
- [5]. Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. [DOI] [PubMed] [Google Scholar]
- [6]. Xiao J, Klein MI, Falsetta ML, et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLoS Pathog. 2012;8(4):e1002623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7]. Teles R, Teles F, Frias-Lopez J, et al. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000. 2013;62(1):95–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Krzysciak W, Jurczak A, Koscielniak D, et al. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014;33(4):499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9]. Bowen WH. Dental caries - not just holes in teeth! A perspective. Mol Oral Microbiol. 2016;31(3):228–233. [DOI] [PubMed] [Google Scholar]
- [10]. Casadevall A, Pirofski LA. Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun. 1999;67(8):3703–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Casadevall A, Pirofski LA. Host-pathogen interactions: basic concepts of microbial commensalism, colonization, infection, and disease. Infect Immun. 2000;68(12):6511–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Hornef M. Pathogens, commensal symbionts, and pathobionts: discovery and functional effects on the host. Ilar J. 2015;56(2):159–162. [DOI] [PubMed] [Google Scholar]
- [13]. Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10(10):717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Klein MI, Hwang G, Santos PH, et al. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect Microbiol. 2015;5:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]. Cashin P, Goldsack L, Hall D, et al. Contrasting signal transduction mechanisms in bacterial and eukaryotic gene transcription. FEMS Microbiol Lett. 2006;261(2):155–164. [DOI] [PubMed] [Google Scholar]
- [16]. Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu RevMicrobiol. 2009;63:133–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]. Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. [DOI] [PubMed] [Google Scholar]
- [18]. Simionato MR, Tucker CM, Kuboniwa M, et al. Porphyromonas Gingivalis genes involved in community development with Streptococcus gordonii . Infect Immun. 2006;74:6419–6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19]. Maeda K, Tribble GD, Tucker CM, et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol Microbiol. 2008;69:1153–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20]. Schaller GE, Shiu SH, Armitage JP. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol. 2011;21(9):R320–R330. [DOI] [PubMed] [Google Scholar]
- [21]. Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188(12):4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22]. Capra EJ, Laub MT. Evolution of two-component signal transduction systems. Annu RevMicrobiol. 2012;66:325–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Song L, Sudhakar P, Wang W, et al. A genome-wide study of two-component signal transduction systems in eight newly sequenced mutans streptococci strains. BMC Genomics. 2012;13:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24]. Smith EG, Spatafora GA. Gene regulation in Streptococcus mutans: complex control in a complex environment. J Dent Res. 2012;91(2):133–141. [DOI] [PubMed] [Google Scholar]
- [25]. Wang S. Bacterial two-component systems: structures and signaling mechanisms In: Huang C, editor. Protein phosphorylation in human health. Rijeka: InTech; 2012. p. Ch. 15. [Google Scholar]
- [26]. Barakat M, Ortet P, Whitworth DE. P2CS: a database of prokaryotic two-component systems. Nucleic Acids Res. 2011;39(Database issue):D771–D776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Raghavan V, Groisman EA. Orphan and hybrid two-component system proteins in health and disease. Curr Opin Microbiol. 2010;13(2):226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Wuichet K, Cantwell BJ, Zhulin IB. Evolution and phyletic distribution of two-component signal transduction systems. Curr Opin Microbiol. 2010;13(2):219–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. Mascher T, Helmann JD, Unden G. Stimulus perception in bacterial signal-transducing histidine kinases. Microbiol Mol Biol Rev. 2006;70(4):910–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Bijlsma JJ, Groisman EA. Making informed decisions: regulatory interactions between two-component systems. Trends Microbiol. 2003;11(8):359–366. [DOI] [PubMed] [Google Scholar]
- [31]. Agrawal R, Sahoo BK, Saini DK. Cross-talk and specificity in two-component signal transduction pathways. Future Microbiol. 2016;11:685–697. [DOI] [PubMed] [Google Scholar]
- [32]. Mark Welch JL, Rossetti BJ, Rieken CW, et al. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113(6):E791–E800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Kuboniwa M, Hendrickson EL, Xia Q, et al. Proteomics of Porphyromonas gingivalis within a model oral microbial community. BMC Microbiol. 2009;9:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Hendrickson EL, Wang T, Dickinson BC, et al. Proteomics of Streptococcus gordonii within a model developing oral microbial community. BMC Microbiol. 2012;12:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Hendrickson EL, Wang T, Beck DAC, et al. Proteomics of Fusobacterium nucleatum within a model developing oral microbial community. Microbiologyopen. 2014;3(5):729–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]. Hendrickson EL, Beck DA, Miller DP, et al. Insights into dynamic polymicrobial synergy revealed by time-coursed RNA-seq. Front Microbiol. 2017;8:261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Kuboniwa M, Amano A, Hashino E, et al. Distinct roles of long/short fimbriae and gingipains in homotypic biofilm development by Porphyromonas gingivalis . BMC Microbiol. 2009;9:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38]. Maeda K, Nagata H, Kuboniwa M, et al. Identification and characterization of Porphyromonas gingivalis client proteins that bind to Streptococcus oralis glyceraldehyde-3-phosphate dehydrogenase. Infect Immun. 2013;81(3):753–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Nishikawa K, Yoshimura F, Duncan MJ. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol Microbiol. 2004;54(2):546–560. [DOI] [PubMed] [Google Scholar]
- [40]. Nishikawa K, Duncan MJ. Histidine kinase-mediated production and autoassembly of Porphyromonas gingivalis fimbriae. J Bacteriol. 2010;192(7):1975–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Nishikawa K. Mutational analysis of the periplasmic sensor domain of the Porphyromonas gingivalis FimS histidine kinase. Abstract 757. Boston (MA): American Society for Microbiology; 2016. [Google Scholar]
- [42]. Duran-Pinedo AE, Nishikawa K, Duncan MJ. The RprY response regulator of Porphyromonas gingivalis . Mol Microbiol. 2007;64(4):1061–1074. [DOI] [PubMed] [Google Scholar]
- [43]. Meuric V, Rouillon A, Chandad F, et al. Putative respiratory chain of Porphyromonas gingivalis . Future Microbiol. 2010;5(5):717–734. [DOI] [PubMed] [Google Scholar]
- [44]. Minato Y, Fassio SR, Reddekopp RL, et al. Inhibition of the sodium-translocating NADH-ubiquinone oxidoreductase [Na+-NQR] decreases cholera toxin production in Vibrio cholerae O1 at the late exponential growth phase. Microb Pathog. 2014;66:36–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45]. Muras V, Dogaru-Kinn P, Minato Y, et al. The Na+-translocating NADH: quinoneoxidoreductase enhances oxidative stress in the cytoplasm of Vibrio cholerae . J Bacteriol. 2016;198(17):2307–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46]. Krishnan K, Duncan MJ. Role of sodium in the RprY-dependent stress response in Porphyromonas gingivalis . PLoS One. 2013;8(5):e63180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47]. Li Y, Krishnan K, Duncan MJ. Acetylation of the RprY response regulator of Porphyromonas gingivalis. Under review. [Google Scholar]
- [48]. Soppa J. Protein acetylation in archaea, bacteria, and eukaryotes. Archaea. 2010;16:2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. Jones JD, O’Connor CD. Protein acetylation in prokaryotes. PubMed PMID: 21674803 Proteomics. 2011;11(15):3012–3022. DOI: 10.1002/pmic.201000812 [DOI] [PubMed] [Google Scholar]
- [50]. Hu LI, Lima BP, Wolfe AJ. Bacterial protein acetylation: the dawning of a new age. Mol Microbiol. 2010;77(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51]. Wang Q, Zhang Y, Yang C, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327(5968):1004–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52]. Chen T, Hosogi Y, Nishikawa K, et al. Comparative whole-genome analysis of virulent and avirulent strains of Porphyromonas gingivalis . J Bacteriol. 2004;186(16):5473–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53]. Naito M, Hirakawa H, Yamashita A, et al. Determination of the genome sequence of Porphyromonas gingivalis strain ATCC 33277 and genomic comparison with strain W83 revealed extensive genome rearrangements in P. gingivalis . DNA Res. 2008;15(4):215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54]. Scott JC, Klein BA, Duran-Pinedo A, et al. A two-component system regulates hemin acquisition in Porphyromonas gingivalis . PLoS One. 2013;8(9):e73351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55]. Lewis JP, Plata K, Yu F, et al. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology. 2006;152(Pt 11):3367–3382. [DOI] [PubMed] [Google Scholar]
- [56]. Dashper SG, Hendtlass A, Slakeski N, et al. Characterization of a novel outer membrane hemin-binding protein of Porphyromonas gingivalis . J Bacteriol. 2000;182(22):6456–6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57]. Slakeski N, Dashper SG, Cook P, et al. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol Immunol. 2000;15(6):388–392. [DOI] [PubMed] [Google Scholar]
- [58]. Nikaido H, Hall JA. Overview of bacterial ABC transporters. Methods Enzymol. 1998;292:3–20. [DOI] [PubMed] [Google Scholar]
- [59]. Okamoto K, Nakayama K, Kadowaki T, et al. Involvement of a lysine-specific cysteine proteinase in hemoglobin adsorption and heme accumulation by Porphyromonas gingivalis . J Biol Chem. 1998;273(33):21225–21231. [DOI] [PubMed] [Google Scholar]
- [60]. Smalley JW, Thomas MF, Birss AJ, et al. A combination of both arginine- and lysine-specific gingipain activity of Porphyromonas gingivalis is necessary for the generation of the micro-oxo bishaem-containing pigment from haemoglobin. Biochem J. 2004;379(Pt 3):833–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61]. Smalley JW, Birss AJ, Szmigielski B, et al. The HA2 haemagglutinin domain of the lysine-specific gingipain (Kgp) of Porphyromonas gingivalis promotes micro-oxo bishaem formation from monomeric iron(III) protoporphyrin IX. Microbiology. 2006;152(Pt 6):1839–1845. [DOI] [PubMed] [Google Scholar]
- [62]. Smalley JW, Byrne DP, Birss AJ, et al. HmuY haemophore and gingipain proteases constitute a unique syntrophic system of haem acquisition by Porphyromonas gingivalis . PLoS One. 2011;6(2):e17182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63]. Scott JC, Balasubramanian S, Duncan MJ. Characterization of HaeS, a hemin-binding histidine kinase of Porphyromonas gingivalis. Under revision. [Google Scholar]
- [64]. Huynh TN, Noriega CE, Stewart V. Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX. Proc Natl Acad Sci U S A. 2010;107(49):21140–21145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65]. Willett JW, Kirby JR. Genetic and biochemical dissection of a HisKA domain identifies residues required exclusively for kinase and phosphatase activities. PLoS Genetics. 2012;8(11):e1003084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66]. Veith PD, Glew MD, Gorasia DG, et al. Type IX Secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers. Mol Microbiol. 2017;106(1):35–53. [DOI] [PubMed] [Google Scholar]
- [67]. Sato K, Naito M, Yukitake H, et al. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A. 2010;107(1):276–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68]. Sato K, Sakai E, Veith PD, et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis . J Biol Chem. 2005;280(10):8668–8677. [DOI] [PubMed] [Google Scholar]
- [69]. Kadowaki T, Yukitake H, Naito M, et al. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci Rep. 2016;6:23288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70]. Veith PD, Nor Muhammad NA, Dashper SG, et al. Protein substrates of a novel secretion system are numerous in the Bacteroidetes phylum and have in common a cleavable C-terminal secretion signal, extensive post-translational modification, and cell-surface attachment. J Proteome Res. 2013;12(10):4449–4461. [DOI] [PubMed] [Google Scholar]
- [71]. Dou Y, Osbourne D, McKenzie R, et al. Involvement of extracytoplasmic function sigma factors in virulence regulation in Porphyromonas gingivalis W83. FEMS Microbiol Lett. 2010;312(1):24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72]. Dou Y, Aruni W, Muthiah A, et al. Studies of the extracytoplasmic function sigma factor PG0162 in Porphyromonas gingivalis . Mol Oral Microbiol. 2016;31(3):270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73]. Vincent MS, Durand E, Cascales E. The PorX response regulator of the Porphyromonas gingivalis PorXY two-component system does not directly regulate the Type IX Secretion genes but binds the PorL Subunit. Front Cell Infect Microbiol. 2016;6:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]. Hasegawa Y, Nishiyama S, Nishikawa K, et al. A novel type of two-component regulatory system affecting gingipains in Porphyromonas gingivalis . Microbiol Immunol. 2003;47(11):849–858. [DOI] [PubMed] [Google Scholar]
- [75]. James CE, Hasegawa Y, Park Y, et al. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis . Infect Immun. 2006;74(7):3834–3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76]. Hirano T, Beck DA, Wright CJ, et al. Regulon controlled by the GppX hybrid two component system in Porphyromonas gingivalis . Mol Oral Microbiol. 2013;28(1):70–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77]. Niwa D, Nishikawa K, Nakamura H. A hybrid two-component system of Tannerella forsythia affects autoaggregation and post-translational modification of surface proteins. FEMS Microbiol Lett. 2011;318(2):189–196. [DOI] [PubMed] [Google Scholar]
- [78]. Ruscitto A, Honma K, Veeramachineni VM, et al. Regulation and molecular basis of environmental muropeptide uptake and utilization in fastidious oral anaerobe Tannerella forsythia . Front Microbiol. 2017;8:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]. Chen T, Siddiqui H, Olsen I. In silico comparison of 19 Porphyromonas gingivalis strains in genomics, phylogenetics, phylogenomics and functional genomics. Front Cell Infect Microbiol. 2017;7:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80]. Facklam R. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev. 2002;15(4):613–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81]. Aas JA, Paster BJ, Stokes LN, et al. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. 2005;43(11):5721–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82]. Diaz PI, Chalmers NI, Rickard AH, et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl Environ Microbiol. 2006;72(4):2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83]. Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46(4):1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84]. Nyvad B, Kilian M. Comparison of the initial streptococcal microflora on dental enamel in caries-active and in caries-inactive individuals. Caries Res. 1990;24(4):267–272. [DOI] [PubMed] [Google Scholar]
- [85]. Kilian M, Poulsen K, Blomqvist T, et al. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One. 2008;3(7):e2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86]. Jensen A, Scholz CF, Kilian M. Re-evaluation of the taxonomy of the Mitis group of the genus Streptococcus based on whole genome phylogenetic analyses, and proposed reclassification of Streptococcus dentisani as Streptococcus oralis subsp. dentisani comb. nov., Streptococcus tigurinus as Streptococcus oralis subsp. tigurinus comb. nov., and Streptococcus oligofermentans as a later synonym of Streptococcus cristatus . Int J Syst Evol Microbiol. 2016;66(11):4803–4820. [DOI] [PubMed] [Google Scholar]
- [87]. Nobbs AH, Lamont RJ, Jenkinson HF. Streptococcus adherence and colonization. Microbiol Mol Biol Rev. 2009;73(3):407–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88]. Banas JA. Virulence properties of Streptococcus mutans . Front Biosci. 2004;9:1267–1277. [DOI] [PubMed] [Google Scholar]
- [89]. Patenge N, Fiedler T, Kreikemeyer B. Common regulators of virulence in streptococci. Curr Top Microbiol Immunol. 2013;368:111–153. [DOI] [PubMed] [Google Scholar]
- [90]. Moraes JJ, Stipp RN, Harth-Chu EN, et al. Two-component system VicRK regulates functions associated with establishment of Streptococcus sanguinis in biofilms. Infect Immun. 2014;82(12):4941–4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91]. Liu Y, Burne RA. Multiple two-component systems modulate alkali generation in Streptococcus gordonii in response to environmental stresses. J Bacteriol. 2009;191(23):7353–7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92]. Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. 1986;50(4):353–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93]. Burne RA. Oral streptococci… products of their environment. J Dent Res. 1998;77(3):445–452. [DOI] [PubMed] [Google Scholar]
- [94]. Lemos JA, Burne RA. A model of efficiency: stress tolerance by Streptococcus mutans . Microbiology. 2008;154(Pt 11):3247–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95]. Alves LA, Nomura R, Mariano FS, et al. CovR Regulates Streptococcus mutans susceptibility to complement immunity and survival in blood. Infect Immun. 2016;84(11):3206–3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96]. Stipp RN, Goncalves RB, Hofling JF, et al. Transcriptional analysis of gtfB, gtfC, and gbpB and their putative response regulators in several isolates of Streptococcus mutans . Oral Microbiol Immunol. 2008;23(6):466–473. [DOI] [PubMed] [Google Scholar]
- [97]. Mattos-Graner RO, Smith DJ, King WF, et al. Water-insoluble glucan synthesis by mutans streptococcal strains correlates with caries incidence in 12- to 30-month-old children. J Dent Res. 2000;79(6):1371–1377. [DOI] [PubMed] [Google Scholar]
- [98]. Fabret C, Hoch JA. A two-component signal transduction system essential for growth of Bacillus subtilis: implications for anti-infective therapy. J Bacteriol. 1998;180(23):6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99]. Fukuchi K, Kasahara Y, Asai K, et al. The essential two-component regulatory system encoded by yycF and yycG modulates expression of the ftsAZ operon in Bacillus subtilis . Microbiology. 2000;146(Pt7):1573–1583. [DOI] [PubMed] [Google Scholar]
- [100]. Dubrac S, Bisicchia P, Devine KM, et al. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol Microbiol. 2008;70(6):1307–1322. PubMed PMID: 19019149 DOI: 10.1111/j.1365-2958.2008.06483.x [DOI] [PubMed] [Google Scholar]
- [101]. Szurmant H, White RA, Hoch JA. Sensor complexes regulating two-component signal transduction. Curr Opin Struct Biol. 2007;17(6):706–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102]. Wayne KJ, Sham LT, Tsui HC, et al. Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae . J Bacteriol. 2010;192(17):4388–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103]. Senadheera MD, Lee AW, Hung DC, et al. The Streptococcus mutans vicX gene product modulates gtfB/C expression, biofilm formation, genetic competence, and oxidative stress tolerance. J Bacteriol. 2007;189(4):1451–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104]. Taylor BL, Zhulin IB. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63(2):479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105]. Ng WL, Tsui HC, Winkler ME. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae . J Bacteriol. 2005;187(21):7444–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106]. Senadheera MD, Guggenheim B, Spatafora GA, et al. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J Bacteriol. 2005;187(12):4064–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107]. Stipp RN, Boisvert H, Smith DJ, et al. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans . PLoS One. 2013;8(3):e58271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108]. Liu M, Hanks TS, Zhang J, et al. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR . Microbiology. 2006;152:967–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109]. Dubrac S, Boneca IG, Poupel O, et al. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus . J Bacteriol. 2007;189(22):8257–8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110]. Ng WL, Kazmierczak KM, Winkler ME. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol Microbiol. 2004;53(4):1161–1175. [DOI] [PubMed] [Google Scholar]
- [111]. Duque C, Stipp RN, Wang B, et al. Downregulation of GbpB, a component of the VicRK regulon, affects biofilm formation and cell surface characteristics of Streptococcus mutans . Infect Immun. 2011;79(2):786–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112]. Bisicchia P, Noone D, Lioliou E, et al. The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis . Mol Microbiol. 2007;65(1):180–200. [DOI] [PubMed] [Google Scholar]
- [113]. Senadheera DB, Cordova M, Ayala EA, et al. Regulation of bacteriocin production and cell death by the VicRK signaling system in Streptococcus mutans . J Bacteriol. 2012;194(6):1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114]. Reinscheid DJ, Gottschalk B, Schubert A, et al. Identification and molecular analysis of PcsB, a protein required for cell wall separation of group B streptococcus. J Bacteriol. 2001;183(4):1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115]. Mattos-Graner RO, Jin S, King WF, et al. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect Immun. 2001;69(11):6931–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116]. Smith DJ, Akita H, King WF, et al. Purification and antigenicity of a novel glucan-binding protein of Streptococcus mutans . Infect Immun. 1994;62(6):2545–2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117]. Ayala E, Downey JS, Mashburn-Warren L, et al. A biochemical characterization of the DNA binding activity of the response regulator VicR from Streptococcus mutans . PLoS One. 2014;9(9):e108027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118]. Alves LA, Harth-Chu EN, Palma TH, et al. The two-component system VicRK regulates functions associated with Streptococcus mutans resistance to complement immunity. Mol Oral Microbiol. 2017;32(5):419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119]. Ng WL, Robertson GT, Kazmierczak KM, et al. Constitutive expression of PcsB suppresses the requirement for the essential VicR (YycF) response regulator in Streptococcus pneumoniae R6. Mol Microbiol. 2003;50(5):1647–1663. [DOI] [PubMed] [Google Scholar]
- [120]. Sham LT, Barendt SM, Kopecky KE, et al. Essential PcsB putative peptidoglycan hydrolase interacts with the essential FtsXSpn cell division protein in Streptococcus pneumoniae D39. Proc Natl Acad Sci U S A. 2011;108(45):E1061–E1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121]. Bartual SG, Straume D, Stamsas GA, et al. Structural basis of PcsB-mediated cell separation in Streptococcus pneumoniae . Nat Commun. 3842;201(5):1–12. [DOI] [PubMed] [Google Scholar]
- [122]. Mattos-Graner RO, Porter KA, Smith DJ, et al. Functional analysis of glucan binding protein B from Streptococcus mutans . J Bacteriol. 2006;188(11):3813–3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123]. Ahn SJ, Burne RA. Effects of oxygen on biofilm formation and the AtlA autolysin of Streptococcus mutans . J Bacteriol. 2007;189(17):6293–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124]. Bowen WH, Koo H. Biology of Streptococcus mutans-derived glucosyltransferases: role in extracellular matrix formation of cariogenic biofilms. Caries Res. 2011;45(1):69–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125]. Yoshida Y, Konno H, Nagano K, et al. The influence of a glucosyltransferase, encoded by gtfP, on biofilm formation by Streptococcus sanguinis in a dual-species model. Apmis. 2014;122(10):951–960. [DOI] [PubMed] [Google Scholar]
- [126]. Andoh A, Fujiyama Y, Kimura T, et al. Molecular characterization of complement components (C3, C4, and factor B) in human saliva. J Clin Immunol. 1997;17(5):404–407. [DOI] [PubMed] [Google Scholar]
- [127]. Shelburne SA 3rd, Granville C, Tokuyama M, et al. Growth characteristics of and virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 2005;73(8):4723–4731128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128]. Ren B, Szalai AJ, Hollingshead SK, et al. Effects of PspA and antibodies to PspA on activation and deposition of complement on the pneumococcal surface. Infect Immun. 2004;72(1):114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129]. Singh K, Senadheera DB, Levesque CM, et al. The copYAZ operon functions in copper efflux, biofilm formation, genetic transformation, and stress tolerance in Streptococcus mutans . J Bacteriol. 2015;197(15):2545–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130]. Ramos-Montanez S, Tsui HC, Wayne KJ, et al. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae . Mol Microbiol. 2008;67(4):729–746. [DOI] [PubMed] [Google Scholar]
- [131]. Kreth J, Zhang Y, Herzberg MC. Streptococcal antagonism in oral biofilms: streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans . J Bacteriol. 2008;190(13):4632–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132]. Kreth J, Vu H, Zhang Y, et al. Characterization of hydrogen peroxide-induced DNA release by Streptococcus sanguinis and Streptococcus gordonii . J Bacteriol. 2009;191(20):6281–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133]. Fukushima T, Szurmant H, Kim EJ, et al. A sensor histidine kinase co-ordinates cell wall architecture with cell division in Bacillus subtilis . Mol Microbiol. 2008;69:621–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134]. Earl AM, Losick R, Kolter R. Ecology and genomics of Bacillus subtilis . Trends Microbiol. 2008;16(6):269–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135]. Tremblay YD, Lo H, Li YH, et al. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology. 2009;155(Pt9):2856–2865. [DOI] [PubMed] [Google Scholar]
- [136]. Gutu AD, Wayne KJ, Sham LT, et al. Kinetic characterization of the WalRKSpn (VicRK) two-component system of Streptococcus pneumoniae: dependence of WalKSpn (VicK) phosphatase activity on its PAS domain. J Bacteriol. 2010;192(9):2346–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137]. Mohedano ML, Amblar M, de la Fuente A, et al. The response regulator YycF inhibits expression of the fatty acid biosynthesis repressor FabT in Streptococcus pneumoniae . Front Microbiol. 2016;7:1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138]. Delaune A, Dubrac S, Blanchet C, et al. The WalKR system controls major staphylococcal virulence genes and is involved in triggering the host inflammatory response. Infect Immun. 2012;80(10):3438–3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139]. Downey JS, Mashburn-Warren L, Ayala EA, et al. In vitro manganese-dependent cross-talk between Streptococcus mutans VicK and GcrR: implications for overlapping stress response pathways. PLoS One. 2014;9(12):e115975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140]. Suntharalingam P, Senadheera MD, Mair RW, et al. The LiaFSR system regulates the cell envelope stress response in Streptococcus mutans . J Bacteriol. 2009;191(9):2973–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141]. Biswas S, Biswas I. Regulation of the glucosyltransferase (gtfBC) operon by CovR in Streptococcus mutans . J Bacteriol. 2006;188(3):988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142]. Dmitriev A, Mohapatra SS, Chong P, et al. CovR-controlled global regulation of gene expression in Streptococcus mutans . PLoS One. 2011;6(5):e20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143]. Alves LA, Harth-Chu EN, Mariano FS, et al. Identification of a novel protein PepO involved in susceptibility to complement immunity in Streptococcus mutans. San Francisco (CA): IADR; 2017. [Google Scholar]
- [144]. Banu LD, Conrads G, Rehrauer H, et al. The Streptococcus mutans serine/threonine kinase, PknB, regulates competence development, bacteriocin production, and cell wall metabolism. Infect Immun. 2010;78(5):2209–2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145]. Agarwal S, Agarwal S, Pancholi P, et al. Role of serine/threonine phosphatase (SP-STP) in Streptococcus pyogenes physiology and virulence. J Biol Chem. 2011;286(48):41368–41380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146]. Burnside K, Rajagopal L. Regulation of prokaryotic gene expression by eukaryotic-like enzymes. Curr Opin Microbiol. 2012;15(2):125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147]. Libby EA, Goss LA, Dworkin J. The eukaryotic-like Ser/Thr Kinase PrkC regulates the essential WalRK two-component system in Bacillus subtilis . PLoS Genet. 2015;11(6):e1005275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148]. Yeats C, Finn RD, Bateman A. The PASTA domain: a beta-lactam-binding domain. Trends Biochem Sci. 2002;27(9):438–440. [DOI] [PubMed] [Google Scholar]
- [149]. Giefing C, Jelencsics KE, Gelbmann D, et al. The pneumococcal eukaryotic-type serine/threonine protein kinase StkP co-localizes with the cell division apparatus and interacts with FtsZ in vitro . Microbiology. 2010;156(Pt 6):1697–1707. [DOI] [PubMed] [Google Scholar]
- [150]. Hussain H, Branny P, Allan E. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans . J Bacteriol. 2006;188(4):1628–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151]. Zhu L, Kreth J. Role of Streptococcus mutans eukaryotic-type serine/threonine protein kinase in interspecies interactions with Streptococcus sanguinis . Arch Oral Biol. 2010;55(5):385–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152]. Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus . Antimicrob Agents Chemother. 2006;50(6):2137–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153]. Mwangi MM, Wu SW, Zhou Y, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104(22):9451–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154]. Jansen A, Turck M, Szekat C, et al. Role of insertion elements and yycFG in the development of decreased susceptibility to vancomycin in Staphylococcus aureus . Int J Med Microbiol. 2007;297(4):205–215. [DOI] [PubMed] [Google Scholar]
- [155]. Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus . Mol Microbiol. 1998;30(1):209–219. [DOI] [PubMed] [Google Scholar]
- [156]. Bernish B. van de Rijn I. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J Biol Chem. 1999;274(8):4786–4793. [DOI] [PubMed] [Google Scholar]
- [157]. Federle MJ, McIver KS, Scott JR. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J Bacteriol. 1999;181(12):3649–3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158]. Graham MR, Smoot LM, Migliaccio CA, et al. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci U S A. 2002;99(21):13855–13860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159]. Graham MR, Virtaneva K, Porcella SF, et al. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166(2):455–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160]. Ulijasz AT, Falk SP, Weisblum B. Phosphorylation of the RitR DNA-binding domain by a Ser-Thr phosphokinase: implications for global gene regulation in the streptococci. Mol Microbiol. 2009;71(2):382–390. [DOI] [PubMed] [Google Scholar]
- [161]. Sato Y, Yamamoto Y, Kizaki H. Construction of region-specific partial duplication mutants (merodiploid mutants) to identify the regulatory gene for the glucan-binding protein C gene in vivo in Streptococcus mutans . FEMS Microbiol Lett. 2000;186(2):187–191. [DOI] [PubMed] [Google Scholar]
- [162]. Idone V, Brendtro S, Gillespie R, et al. Effect of an orphan response regulator on Streptococcus mutans sucrose-dependent adherence and cariogenesis. Infect Immun. 2003;71(8):4351–4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163]. Gusa AA, Gao J, Stringer V, et al. Phosphorylation of the group A Streptococcal CovR response regulator causes dimerization and promoter-specific recruitment by RNA polymerase. J Bacteriol. 2006;188(13):4620–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164]. Dalton TL, Scott JR. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes . J Bacteriol. 2004;186(12):3928–3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165]. Tran-Winkler HJ, Love JF, Gryllos I, et al. Signal transduction through CsrRS confers an invasive phenotype in group A Streptococcus . PLoS Pathog. 2011;7:e1002361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166]. Gryllos I, Levin JC, Wessels MR. The CsrR/CsrS two-component system of group A Streptococcus responds to environmental Mg2+ . Proc Natl Acad Sci USA. 2003;100:4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167]. Gryllos I, Grifantini R, Colaprico A, et al. Mg(2+) signalling defines the group A streptococcal CsrRS (CovRS) regulon. Mol.Microbiol. 2007;65:671–683. [DOI] [PubMed] [Google Scholar]
- [168]. Velarde J, Ashbaugh J, Wessels MR. The human antimicrobial peptide LL-37 binds directly to CsrS, a sensor histidine kinase of group A Streptococcus, to activate expression of virulence factors. J Biol Chem. 2014;289;36315–36324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169]. Horstmann N, Saldana M, Sahasrabhojane P, et al. Dual-site phosphorylation of the control of virulence regulator impacts group a streptococcal global gene expression and pathogenesis. PLoS Pathog. 2014;10:e1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170]. Rajagopal L, Vo A, Silvestroni A, et al. Regulation of cytotoxin expression by converging eukaryotic-type and two-component signalling mechanisms in Streptococcus agalactiae . Mol Microbiol. 2006;62(4):941–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171]. Kant S, Agarwal S, Pancholi P, et al. The Streptococcus pyogenes orphan protein tyrosine phosphatase, SP-PTP, possesses dual specificity and essential virulence regulatory functions. Mol Microbiol. 2015;97(3):515–540. [DOI] [PubMed] [Google Scholar]
- [172]. Biswas I, Drake L, Biswas S. Regulation of gbpC expression in Streptococcus mutans . J Bacteriol. 2007;189(18):6521–6531. [DOI] [PMC free article] [PubMed] [Google Scholar]


