Abstract
Objective
Previous studies have suggested an association between gout susceptibility and common dysfunctional variants in ATP-binding cassette transporter subfamily G member 2/breast cancer resistance protein (ABCG2/BCRP), including rs72552713 (Q126X) and rs2231142 (Q141K). However, the association of rare ABCG2 variants with gout is unknown. Therefore, we investigated the effects of rare ABCG2 variants on gout susceptibility in this study.
Methods
We sequenced the exons of ABCG2 in 480 patients with gout and 480 healthy controls (Japanese males). We also performed functional analyses of non-synonymous variants of ABCG2 and analysed the correlation between urate transport function and scores from the protein prediction algorithms (Sorting Intolerant from Tolerant (SIFT) and Polymorphism Phenotyping v2 (PolyPhen-2)). Stratified association analyses and multivariate logistic regression analysis were performed to evaluate the effects of rare and common ABCG2 variants on gout susceptibility.
Results
We identified 3 common and 19 rare non-synonymous variants of ABCG2. SIFT scores were significantly correlated with the urate transport function, although some ABCG2 variants showed inconsistent scores. When the effects of common variants were removed by stratified association analysis, the rare variants of ABCG2 were associated with a significantly increased risk of gout (OR=3.2, p=6.4×10−3). Multivariate logistic regression analysis revealed that the size effect of these rare ABCG2 variants (OR=2.7, p=3.0×10−3) was similar to that of the common variants, Q126X (OR=3.4, p=3.2×10−6) and Q141K (OR=2.3, p=2.7×10−16).
Conclusions
This study revealed that multiple common and rare variants of ABCG2 are independently associated with gout. These results could support both the ‘Common Disease, Common Variant’ and ‘Common Disease, Multiple Rare Variant’ hypotheses for the association between ABCG2 and gout susceptibility.
Keywords: gout, gene polymorphism, epidemiology, arthritis
Key messages.
What is already known about this subject?
Common dysfunctional variants (Q126X and Q141K) of ABCG2 are risk factors for gout/hyperuricaemia, which supports the ‘Common Disease, Common Variant (CDCV)’ hypothesis.
What does this study add?
Multiple common and rare variants of ABCG2 are independently associated with gout.
This study supports both the ‘CDCV’ and ‘Common Disease, Multiple Rare Variant’ hypotheses for the association between ABCG2 and gout susceptibility.
How might this impact on clinical practice?
These findings indicate that genotyping the rare variants of ABCG2 along with its common variants (Q126X and Q141K) is essential for evaluating the individual risk for gout.
Introduction
Gout is the most common form of inflammatory arthritis and is caused by hyperuricaemia. Many previous studies have indicated that dysfunctional variants (rs72552713 (Q126X) and rs2231142 (Q141K)) of the gene encoding ATP-binding cassette transporter subfamily G member 2/breast cancer resistance protein (ABCG2/BCRP) increase the risk of gout1–3 and hyperuricaemia.1 4 Approximately 80% of Japanese patients with gout have been reported to possess either the Q126X or Q141K variant of ABCG2,1 and these variants increased the risk of gout conferring an OR of more than 3.1 3 Thus, the effects of common ABCG2 variants on gout susceptibility are very strong, whereas any effect of rare ABCG2 variants is still unknown.
In this study, we first identified common and rare non-synonymous variants of ABCG2 by target exon sequencing of genomes from Japanese male patients with gout and healthy controls. Second, we evaluated the non-synonymous variants using three different protein prediction algorithms. Third, we performed molecular analyses of the urate transport function and evaluated the correlation between the functional analyses and the scores from the protein prediction algorithms. Finally, we performed association analyses between the ABCG2 variants and gout susceptibility.
Methods
Patients and controls
This study was approved by the institutions’ Ethical Committees (National Defense Medical College, National Institute of Genetics and Nagoya University). All protocols were in accordance with the Declaration of Helsinki, and written informed consent was obtained from all participants. For the study, 480 Japanese males with gout were recruited from the Ryougoku East Gate Clinic (Tokyo, Japan). All patients were clinically diagnosed with primary gout according to the criteria established by the American College of Rheumatology.5 Patients with inherited metabolic disorders, including Lesch-Nyhan syndrome, were excluded from this study. As the control group, 480 healthy Japanese males without hyperuricaemia (serum uric acid (SUA) levels >7.0 mg/dL) or history of gout were recruited from the participants of the Shizuoka Study, which is a part of the Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study).6 The mean age (±SD) of the case and control groups was 46.2 years (±9.8) and 52.3 years (±7.9), respectively, and their mean body mass index (±SD) was 25.3 kg/m2 (±3.7) and 23.2 kg/m2 (±2.6), respectively.
Targeted sequencing
Genomic DNA was extracted from the whole peripheral blood cells of the participants.7 We performed targeted exon sequencing of ABCG2 with a pool and capture method described in a previous study.8 Briefly, the extracted DNA was quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) on FilterMax F5 Multi-Mode Microplate Readers (Molecular Devices, Sunnyvale, California, USA). Twenty nanogram of DNA was simultaneously fragmented and ligated with adapters using the SureSelect QXT Library Prep Kit (Agilent Technologies, Santa Clara, California, USA). The 96 fragmented libraries with distinct indexed adapters were pooled in equimolar amounts. Target enrichment was then performed using the SeqCap EZ choice system (Roche Diagnostics, Tokyo, Japan). A DNA probe set complementary to the target region was designed using NimbleDesign (https://design.nimblegen.com). The libraries were sequenced on an Illumina HiSeq 2500 platform in rapid run mode with 2×150 bp paired-end modules (Illumina).
Variant calling and annotation
The generated sequences were aligned to a human reference genome (hg19) using BWA9 and converted to the BAM format for subsequent analysis using SAMtools.10 The aligned reads were processed using Picard tools (broadinstitute.github.io/picard) for removing the PCR duplicates. Local realignment and base quality recalibration were implemented using GATK.11 12 Genetic variations, including single nucleotide variants (SNVs) and short insertions and deletions (indels), were determined using HaplotypeCaller walker of GATK.11 12 Functional annotation of the identified variants was implemented using ANNOVAR.13
Functional analysis
To estimate the functions of the ABCG2 variants, we used the protein prediction algorithms, Sorting Intolerant from Tolerant (SIFT)14 and Polymorphism Phenotyping v2 (PolyPhen-2).15 For PolyPhen-2, both PolyPhen-2 HumVar and PolyPhen-2 HumDiv15 were used. PolyPhen-2 HumVar is designed for distinguishing mutations with drastic effects from abundant mildly deleterious alleles, whereas PolyPhen-2 HumDiv is targeting rare alleles at loci potentially involved in complex phenotypes.16 To evaluate these protein algorithms, we performed functional analysis of ABCG2-mediated urate transport for previously identified variants (N208S, N299S, E311K, L447V, S486N and V516M) in our laboratories as well as reported variants whose vesicles were available (V12M, Q141K, V178I, G268R, P269S, S441N, G462R, V508I and A634V). We analysed the correlation between the urate transport function and the scores from these protein prediction algorithms. In addition, we performed functional analysis of one frameshift deletion (F506SfsX4) and four nonsense variants (Q126X, E334X, R575X and C608X).
Functional assays were performed to determine the urate transport activity of each ABCG2 variant, as described in previous studies.1 17 Briefly, using site-directed mutagenesis, vectors expressing the different ABCG2 variants were generated from a myc-ABCG2 wild-type (WT)/pcDNA3.1(+) plasmid that was prepared in our previous study.1 Plasma membrane vesicles were isolated from human embryonic kidney-derived cells that were transiently transfected with the vectors expressing the ABCG2 variants using a standard method18 or with empty vector as a control using general lipofection methods.1 19 The isolated membrane vesicles were stored at −80°C until use. Expression of ABCG2 protein in the membrane vesicles was examined by immunoblotting with an anti-myc antibody (see online supplementary figure S1) according to our previous studies.1 17 Then, using a rapid filtration technique,17 the [14C]-urate transport assay was performed for the ABCG2-expressing or control membrane vesicles. Based on the radioactivity detected in the membrane vesicles, the urate transport activity was calculated based on the formula; incorporated clearance (μL/mg protein/min)=incorporated urate level (DPM/mg protein/min)/urate level in the incubation mixture (DPM/μL). By subtracting the urate transport activity in the absence of ATP from that in the presence of ATP, ATP-dependent urate transport activity was also determined.
rmdopen-2017-000464supp001.docx (853.5KB, docx)
Statistical analysis
Statistical analyses were performed using SPSS v.22.0J (IBM Japan, Tokyo, Japan).
We selected missense and nonsense SNVs and indels in the exons of ABCG2 based on the DNA reference sequence NM_004827 for association analyses and excluded synonymous SNVs in ABCG2 exons as well as variants in introns or untranslated regions. Rare variants and common variants were defined when the minor allele frequencies (MAF) were <1% and ≥1%, respectively. MAF in the Japanese population was determined based on the Japanese in Tokyo (JPT) population from the 1000 Genomes Project data (http://www.internationalgenome.org/).20 We analysed multiple rare non-synonymous variants using the Collapsing method.21 If a rare allele was present at any of the variant sites, an individual was regarded as a rare variant carrier and as a non-carrier otherwise.21 Since the effects of common dysfunctional variants of ABCG2, Q126X and Q141K, on gout susceptibility were very strong,1 we performed stratified association analyses between rare non-synonymous variants of ABCG2 and gout susceptibility by its common variants. The χ2 test was used for association analyses. Furthermore, multivariate logistic regression analysis was performed between the rare non-synonymous variants and the common variants (Q126X and Q141K) of ABCG2. The dominant model of the Collapsing method21 was applied for rare variants of ABCG2, whereas the additive codominant model was applied for the common variants.
Results
Details of all the non-synonymous variants of ABCG2 found in the cases and the control samples by targeted exon sequencing are shown in table 1. We identified 3 common and 19 rare non-synonymous variants of ABCG2. Genotype counts of the common non-synonymous variants (V12M, Q126X and Q141K) are shown in online supplementary table S1. For the identified rare variants, we observed 33 carriers among patients with gout and 18 carriers among the controls (table 1); no participants were found to be homozygous for rare variants.
Table 1.
Non-synonymous variants of ABCG2 found in this study
| Type of variant | rs number | Position* | Change in DNA sequence† |
AA change | Case (n)‡ | MAF in case (%) | Control (n)‡ | MAF in control (%) |
| Common variant | rs2231137 | 89061114 | G34A | V12M | 118 | 12.3 | 180 | 18.8 |
| rs72552713 | 89052957 | C376T | Q126X | 55 | 5.73 | 23 | 2.40 | |
| rs2231142 | 89052323 | C421A | Q141K | 325 | 33.9 | 218 | 22.7 | |
| Rare variant | 89052998 | C335T | P112L | 6 | 0.625 | 4 | 0.417 | |
| rs149106245 | 89052361 | A383T | D128V | 2 | 0.208 | 1 | 0.104 | |
| rs201006821 | 89052299 | G445C | A149P | 0 | 0 | 1 | 0.104 | |
| rs199753603 | 89052289 | T455C | M152T | 1 | 0.104 | 0 | 0 | |
| 89052255 | G489C | R163S | 1 | 0.104 | 0 | 0 | ||
| rs746311704 | 89042944 | G532A | V178I | 1 | 0.104 | 1 | 0.104 | |
| rs200190472 | 89039366 | C736T | R246X | 1 | 0.104 | 2 | 0.208 | |
| rs34678167 | 89039297 | C805T | P269S | 3 | 0.313 | 2 | 0.208 | |
| 89039275 | A827G | Y276C | 0 | 0 | 1 | 0.104 | ||
| rs750972998 | 89034567 | 1079_1081delAGA | K360del | 2 | 0.208 | 1 | 0.104 | |
| 89022427 | G1322A | S441N | 3 | 0.313 | 0 | 0 | ||
| rs752408502 | 89020584 | G1384A | G462R | 1 | 0.104 | 0 | 0 | |
| rs192169063 | 89020503 | T1465C | F489L | 8 | 0.833 | 3 | 0.312 | |
| rs868217328 | 89018737 | 1515delC | F506SfsX4 | 3 | 0.313 | 0 | 0 | |
| 89018730 | G1522A | V508I | 2 | 0.208 | 0 | 0 | ||
| rs548254708 | 89016686 | C1723T | R575X | 0 | 0 | 2 | 0.208 | |
| rs200933122 | 89013532 | T1822C | C608R | 2 | 0.208 | 0 | 0 | |
| rs748531218 | 89013495 | A1859G | D620G | 1 | 0.104 | 0 | 0 | |
| 89013453 | C1901T | A634V | 1 | 0.104 | 0 | 0 | ||
| Total of rare variant carriers§ | 33 | 18 | ||||||
| Total of participants | 480 | 480 |
For all rare variants, there were only heterozygous and no homozygous participants.
*Positions refer to the GRCh37 assembly.
†Nucleotide numbering is based on the DNA reference sequence NM_004827.
‡Summary count of participants with homozygous or heterozygous variants.
§Count of participants with one or more rare variants.
AA, amino acid; ABCG2, ATP-binding cassette transporter subfamily G member 2; MAF, minor allele frequency.
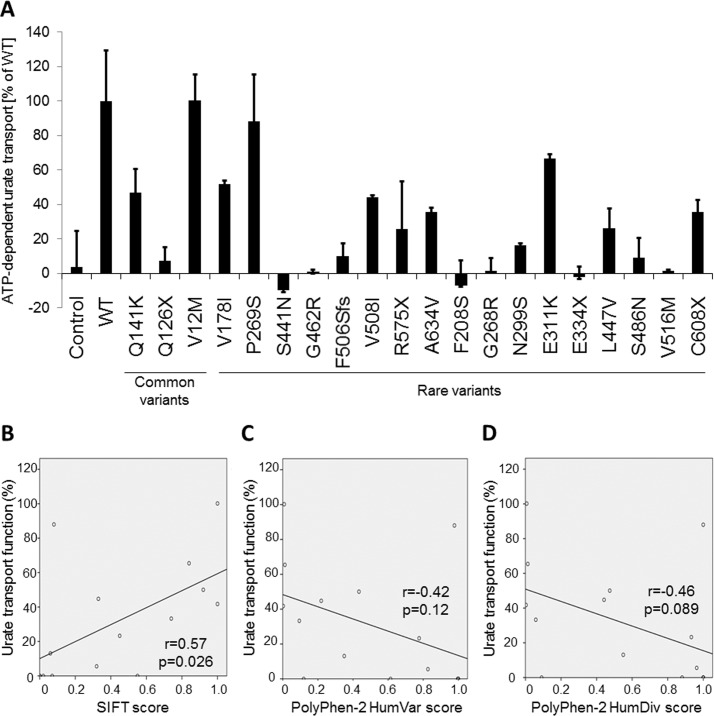
Results of the functional analysis of 19 non-synonymous variants of ABCG2 are shown in figure 1A and table 2. The ATP-dependent urate transport activity was almost completely eliminated in several missense variants (S441N, G462R, F208S, G268R, S486N and V516M), nonsense variants (Q126X, R575X and E334X) and a frameshift deletion (F506SfsX4) of ABCG2 (figure 1A). In contrast, V12M and P269S variants did not show altered urate transport activity. The other rare variants addressed in this study exhibited lower transport activity as compared with WT. Based on the relationship between the protein level and urate transport activity of each ABCG2 variant (see online supplementary figures S1 and S2), the diminished function of each ABCG2 variant could primarily depend on the quantitative changes in the ABCG2 protein on the plasma membrane. On the other hand, several variants (L447V, N299S, S486N, V516M and S441N) would affect the intrinsic ability of ABCG2 as a urate transporter. Of note, the quantitative changes must have been due to the presence of each variant because the membrane vesicles were prepared by using certain procedure established in previous reports.1 18 In addition, previous reports showed that three variants (Q126X, S441N and F506SfsX4) disrupt the transport activity of ABCG2, whereas other three variants (V12M, A149P and P269S) have little effect on the transport activity.1 18
Figure 1.
Functional analyses of ATP-binding cassette transporter subfamily G member 2 (ABCG2) variants. (A) Data from ATP-dependent urate transport analyses of ABCG2 variants are presented as the mean function (%) relative to the activity of the wild type (WT) ABCG2 transporter. Transport function was almost completely abolished in several missense variants (S441N, G462R, F208S, G268R, S486N and V516M) and two nonsense variants (Q126X and E334X) of ABCG2. Transport function was also diminished in F506SfsX4 (F506Sfs), a frameshift deletion of ABCG2. In contrast, no remarkable changes in urate transport activity were observed in the V12M and P269S variants. (B) Sorting Intolerant from Tolerant (SIFT) scores were significantly correlated (r=0.57, p=0.026) with the results of the urate transport analyses. (C and D) The PolyPhen-2 HumVar and HumDiv scores showed non-significant correlation, although these scores showed a tendency toward decrease urate transport function (r=−0.42, p=0.12 and r=−0.46, p=0.089, respectively), based on the results of the functional analyses.
Table 2.
ATP-dependent urate transport activity and scores of protein prediction algorithms for each ABCG2 variant
| rs number | AA change | Transport function (%) | SIFT | PolyPhen-2 HumVar |
PolyPhen-2 HumDiv |
| rs2231137 | V12M* | 100.2 | 1 | 0.008 | 0.007 |
| rs72552713 | Q126X* | 3.6 | N/A | N/A | N/A |
| rs2231142 | Q141K* | 44.8 | 0.33 | 0.219 | 0.442 |
| rs746311704 | V178I* | 50.0 | 0.92 | 0.434 | 0.474 |
| rs34678167 | P269S* | 88.0 | 0.08 | 0.978 | 1 |
| S441N* | 0 | 0.55 | 0.119 | 0.091 | |
| rs752408502 | G462R* | 0 | 0.07 | 0.997 | 1 |
| rs868217328 | F506SfsX4* | 6.6 | N/A | N/A | N/A |
| V508I* | 41.8 | 1 | 0.002 | 0.003 | |
| rs548254708 | R575X* | 22.8 | N/A | N/A | N/A |
| A634V* | 33.3 | 0.74 | 0.094 | 0.058 | |
| rs1061018 | F208S | 0 | 0 | 1 | 1 |
| G268R | 0 | 0 | 1 | 1 | |
| N299S | 13.1 | 0.06 | 0.350 | 0.550 | |
| E311K | 65.4 | 0.84 | 0.012 | 0.013 | |
| rs3201997 | E334X | 0 | N/A | N/A | N/A |
| L447V | 23.3 | 0.45 | 0.777 | 0.933 | |
| rs780310265 | S486N | 5.5 | 0.32 | 0.827 | 0.963 |
| V516M | 0 | 0.02 | 0.614 | 0.881 | |
| C608X | 33.2 | N/A | N/A | N/A |
*These variants were detected through exonic sequencing analysis in this study. For rare variants, there were no homozygous participants.
AA, amino acid; ABCG2, ATP-binding cassette transporter subfamily G member 2; N/A, not applicable; PolyPhen-2, Polymorphism Phenotyping v2; SIFT, Sorting Intolerant from Tolerant.
The scores from SIFT were significantly correlated with the urate transport function (figure 1B; r=0.57, p=0.026). On the other hand, the scores of PolyPhen-2 were not significantly correlated; these scores tended to decrease as the urate transport function increased (figure 1C,D; r=−0.42, p=0.12 in PolyPhen-2 HumVar; r=−0.46, p=0.089 in PolyPhen-2 HumDiv). These correlations suggested that the protein prediction algorithms were useful for predicting changes in the transport function of ABCG2; however, some variants of ABCG2 showed inconsistent scores (table 2; P269S and S441N). Therefore, we determined the effects of ABCG2 variants using the following protocol: (1) when functional data were available, we used the results of urate transport analysis and (2) when functional data were not available, we used the scores from SIFT and PolyPhen-2. V12M and P269S variants were excluded from these analyses because they did not significantly decrease urate transport activity. The V12M variant did not show a significant association with gout susceptibility in the multivariate logistic regression analysis with the other common variants, Q126X and Q141K (table 3). In addition, the A149P variant was also excluded because it was not likely to cause functional changes, according to the scores of SIFT (1), PolyPhen-2 HumVar (0) and PolyPhen-2 HumDiv (0) (see online supplementary table S2). It was shown in a previous study through functional analysis that the A149P variant did not significantly affect the drug transport activity of ABCG2.18
Table 3.
Multivariate logistic regression analysis for gout susceptibility with three common variants of ABCG2
| Variables | β | OR (95% CI) | p Value |
| V12M | −0.15 | 0.86 (0.66 to 1.1) | 0.27 |
| Q126X | 1.1 | 3.0 (1.8 to 5.1) | 2.3×10−5 |
| Q141K | 0.78 | 2.2 (1.8 to 2.7) | 5.7×10−13 |
Each variant was adjusted by the other two variants in this analysis.
ABCG2, ATP-binding cassette transporter subfamily G member 2.
To evaluate the effects of rare variants by removing the effects of the common variants, stratified association analyses were performed, as shown in table 4. When the gout susceptibility was analysed for samples without Q126X or Q141K, rare non-synonymous variants of ABCG2 were found to increase the gout risk conferring OR to 3.2 (p=6.4×10−3, table 4). In the multivariate logistic regression analysis as well, rare non-synonymous variants of ABCG2 were found to be associated with gout susceptibility independent of the Q126X and Q141K variants (table 5). In addition, the effect size of rare variants of ABCG2 (OR=2.7, 95% CI 1.4 to 5.2, p=3.0×10−3) was similar to that of common variants Q126X (OR=3.4, 95% CI 2.0 to 5.6, p=3.1×10−6) and Q141K (OR=2.3, 95% CI 1.9 to 2.9, p=2.7×10−16) (table 5). Thus, our findings indicated that multiple common and rare variants of ABCG2 are strongly and independently associated with gout susceptibility.
Table 4.
Stratified association between rare non-synonymous variants of ABCG2 and gout susceptibility by common variants of ABCG2, Q126X and Q141K
| Case | Control | |||||||
| Sample set | Number | Carrier* | Frequency (%)† | Number | Carrier* | Frequency (%)† | p Value | OR (95% CI) |
| All | 480 | 30 | 6.3 | 480 | 15 | 3.1 | 0.022 | 2.1 (1.1 to 3.9) |
| Without Q126X | 425 | 30 | 7.1 | 457 | 15 | 3.3 | 0.011 | 2.2 (1.2 to 4.2) |
| Without Q126X or Q141K | 131 | 14 | 10.7 | 247 | 9 | 3.6 | 6.4×10-3 | 3.2 (1.3 to 7.5) |
Only non-synonymous SNVs or indels with minor allele frequency less than 1% were considered rare non-synonymous variants in this analysis.
Since the P268S variant of ABCG2 did not decrease urate transport activity, it was excluded from this analysis.
The A149P variant of ABCG2 was also excluded from this analysis due to its scores of SIFT and PolyPhen-2 as well as a previous report of functional analysis.18
*The number of carriers with rare non-synonymous variants of ABCG2.
†The percentage of cases or controls carrying rare non-synonymous variants of ABCG2.
ABCG2, ATP-binding cassette transporter subfamily G member 2; indels, short insertions and deletions; PolyPhen-2, Polymorphism Phenotyping v2; SIFT, Sorting Intolerant from Tolerant; SNVs, single nucleotide variants.
Table 5.
Multivariate logistic regression analysis of gout susceptibility with rare and common variants of ABCG2
| Variables | β | OR (95% CI) | p Value |
| Rare variant | 0.99 | 2.7 (1.4 to 5.2) | 3.0×10−3 |
| Q126X | 1.21 | 3.4 (2.0 to 5.6) | 3.1×10−6 |
| Q141K | 0.85 | 2.3 (1.9 to 2.9) | 2.7×10−16 |
Each variant was adjusted by the other two variants in this analysis.
Since the P268S variant of ABCG2 did not decrease urate transport activity, it was excluded from this analysis.
The A149P variant of ABCG2 was also excluded from this analysis due to its scores of SIFT and PolyPhen-2 as well as a previous report of functional analysis.18
ABCG2, ATP-binding cassette transporter subfamily G member 2; PolyPhen-2, Polymorphism Phenotyping v2; SIFT, Sorting Intolerant from Tolerant.
Discussion
Common dysfunctional variants (Q126X and Q141K) of ABCG2, a urate transporter gene, have been shown to be strongly associated with gout susceptibility1 2 and age of onset of gout.3 Q126X, a common variant in the Japanese population, is a rare variant in Caucasian and African-American populations, whereas Q141K is a common variant in all these populations.22 The MAF of Q126X and Q141K are 0.024 and 0.322, respectively in the Japanese population (JPT from the 1000 Genomes Project).20
ABCG2 excretes urate from the intestine and kidney, and its dysfunction causes ‘extrarenal urate underexcretion type23’ and/or ‘renal urate underexcretion type24’ gout. ABCG2-mediated intestinal urate excretion was also discovered in humans recently after investigation of haemodialysis and acute gastroenteritis patients.25 It was also shown that increased SUA could be a useful marker of intestinal impairment, because increased SUA of gastroenteritis patients was explained both by dehydration and by impaired intestinal epithelium which excretes urate via ABCG2.25 In addition, ABCG2 variants have been shown to have stronger effects on the risk of hyperuricaemia than major environmental risk factors such as obesity and heavy drinking.4
In recent genome-wide association studies (GWASs) of clinically defined gout,26–28 the ABCG2 locus showed the most significant association with gout susceptibility. The ABCG2 locus was also the most significantly detected locus in GWASs of gout in Caucasian populations.29 30 These findings indicate that common variants of ABCG2 are extremely important in gout pathogenesis.
In addition to Q126X and Q141K, V12M is another common non-synonymous variant of ABCG2.1 A haplotype frequency analysis in a previous study showed that the minor alleles of V12M, Q126X and Q141K were not simultaneously present in one haplotype.1 23 24 In other words, these three variants are in complete linkage disequilibrium. Therefore, the minor allele of V12M seemingly has a protective effect on gout susceptibility, although it does not have an actual effect. In this study, we performed both univariate and multivariate logistic regression analyses of gout susceptibility in common variants of ABCG2 (V12M, Q126X and Q141K). Indeed, V12M showed a significant association with gout susceptibility only in the univariate logistic regression analysis (see online supplementary table S3), although it was no more statistically significant after adjustment for Q126X and Q141K genotypes in the multivariate logistic regression analysis (p=0.27, table 3). This finding was also consistent with the results of the molecular functional analysis,1 in which the V12M variant did not show decreased urate transport (figure 1A). These findings showed that V12M had no significant protective effects against gout susceptibility, as shown in table 3.
Molecular functional analysis of transporters is useful for in vitro quantitative assessment of functional changes caused by non-synonymous variants and for evaluating the scores from the protein prediction algorithms. While functional analysis of the ABC transporters by a vesicle system is powerful, it takes considerable effort to analyse all the rare variants identified by targeted exon sequencing. Therefore, in this study, we performed functional analyses for half of the non-synonymous variants whose vesicles were available detected through the targeted exon sequencing (n=11, table 2) and previously identified rare variants (n=9, table 2). We then compared the results of the functional analyses with the scores from SIFT and PolyPhen-2. Based on these results, we predicted the functional changes of other missense variants (n=9) (see online supplementary table S2) with the protein prediction algorithms. We considered that it was reasonable to use molecular functional analyses and protein prediction algorithms in combination. We propose this as a suitable model for analysing rare variants of transporter genes to evaluate individual genetic risks for common diseases including gout.
Previous studies have indicated that common variants of ABCG2 are risk factors for gout/hyperuricaemia.1–4 These studies supported the ‘Common Disease, Common Variant (CDCV)’ hypothesis, which states that common genetic variants are the major contributors to genetic susceptibility to common diseases.31 32 In contrast, recent genetic studies have suggested that multiple rare variants also play important roles in several complex genetic diseases such as early-onset myocardial infarction33 and Alzheimer’s disease,34 which support the ‘Common Disease, Multiple Rare Variant (CDMRV)’ or ‘Common Disease, Rare Variant (CDRV)’ hypothesis.32 In this study, gene-based stratified association analyses revealed an association between rare non-synonymous variants of ABCG2 and gout susceptibility. In addition, we used logistic regression analysis to evaluate the effect of both common and rare variants of ABCG2 on gout susceptibility. Results of this study support the CDMRV hypothesis for ABCG2 and gout, whereas many previous studies1–3 26 27 as well as this study have also supported the CDCV hypothesis. We found that multiple rare variants as well as common variants of ABCG2 are independently associated with gout risk. These results further indicate that ABCG2 is a key molecule in the pathogenesis of gout. Evaluating the risk associated with common and rare variants of individual genes could help in developing precision medicine or personalised genome medicine for common diseases such as gout. Our results showed that genotyping the rare variants of ABCG2 along with its common variants (Q126X and Q141K) is essential for evaluating the individual risk for gout. Although the rare variants of ABCG2 showed highly significant association with gout susceptibility in this study, the number of the participants is small compared with other association analyses of rare genetic variants.33 34 Further studies are required in order to find other important rare variants of ABCG2 and perform more detailed analyses.
In summary, our findings revealed that multiple common and rare variants of ABCG2 could cause gout. Thus, this study could support both ‘CDCV’ and ‘CDMRV’ hypotheses, and we proposed the novel ‘Common Disease, Multiple Common and Rare Variant’ model for the association between ABCG2 and gout.
Acknowledgments
We thank all the participants for their generous involvement in this study. We also thank members of the Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) Shizuoka Field for supporting the study. We are indebted to J. Kajiwara, J. Kitayama, R. Sugimoto, K. Gotanda, Y. Morimoto, M. Miyazawa, M. Takahashi and T. Chiba for genetic analysis. We are also indebted to A. Tokumasu, K. Ooyama, M. Naito, A. Hishida, N. Hamajima and H. Tanaka for sample collection. We also thank T. Shimizu and A. Toyoda for helpful discussions.
Footnotes
Contributor: THigashino, TT, HNakaoka and YToyoda contributed equally.
Contributors: THi, TT, HNakao, KH, NS and HMa conceived and designed this study. BS and KY assisted with research design. KW, RO and HO collected samples and analysed clinical data. THi, HNakao, SS, MS, MK, AN, AA, YTa, YK, KY, KH, II, NS and HMa performed genetic analysis. TT, YTo, HMi and YI performed functional analysis. THi, HNakao, HNakas, TN and HMa performed statistical analyses. BS, KY, KH, THo, KI, HS, TRM and NS provided intellectual input and assisted with the preparation of the manuscript. THi, TT, HNakao, YTo and HMa wrote the manuscript. All authors have read and approved the final version of the manuscript.
Funding: This study was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, including MEXT KAKENHI (Nos 25293145 and 15K15227), Grants-in-Aid for Scientific Research on Priority Areas (No 17015018) and Innovative Areas (Nos 221S0001 and 221S0002) and JSPS KAKENHI Grants (Nos 16H06277 and 16H06279), the Ministry of Health, Labour and Welfare of Japan, the Ministry of Defense of Japan, the Japan Society for the Promotion of Science, the Kawano Masanori Memorial Foundation for Promotion of Pediatrics, the Gout Research Foundation of Japan and the Health Research Council of NZ.
Competing interests: TT, TN, KI, HS, NS and HMa have a patent pending based on the work reported in this paper.
Patient consent: Obtained.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: No additional data are available.
References
- 1. Matsuo H, Takada T, Ichida K, et al. . Common defects of ABCG2, a high-capacity urate exporter, cause gout: a function-based genetic analysis in a Japanese population. Sci Transl Med 2009;1:5:ra11. [DOI] [PubMed] [Google Scholar]
- 2. Woodward OM, Köttgen A, Coresh J, et al. . Identification of a urate transporter, ABCG2, with a common functional polymorphism causing gout. Proc Natl Acad Sci U S A 2009;106:10338–42. 10.1073/pnas.0901249106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Matsuo H, Ichida K, Takada T, et al. . Common dysfunctional variants in ABCG2 are a Major cause of early-onset gout. Sci Rep 2013. 2014;3:3 10.1038/srep02014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakayama A, Matsuo H, Nakaoka H, et al. . Common dysfunctional variants of ABCG2 have stronger impact on hyperuricemia progression than typical environmental risk factors. Sci Rep 2014;4:5227 10.1038/srep05227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wallace SL, Robinson H, Masi AT, et al. . Preliminary criteria for the classification of the acute arthritis of primary gout. Arthritis Rheum 1977;20:895–900. 10.1002/art.1780200320 [DOI] [PubMed] [Google Scholar]
- 6. Hamajima N, Group JMS; J-MICC Study Group. The Japan Multi-Institutional Collaborative Cohort Study (J-MICC Study) to detect gene-environment interactions for Cancer. Asian Pac J Cancer Prev 2007;8:317–23. [PubMed] [Google Scholar]
- 7. Sakiyama M, Matsuo H, Shimizu S, et al. . Common variant of leucine-rich repeat-containing 16A (LRRC16A) gene is associated with gout susceptibility. Hum Cell 2014;27:1–4. 10.1007/s13577-013-0081-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ahmadloo S, Nakaoka H, Hayano T, et al. . Rapid and cost-effective high-throughput sequencing for identification of germline mutations of BRCA1 and BRCA2. J Hum Genet 2017;62:561–7. 10.1038/jhg.2017.5 [DOI] [PubMed] [Google Scholar]
- 9. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li H, Handsaker B, Wysoker A, et al. . The sequence alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McKenna A, Hanna M, Banks E, et al. . The Genome analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20:1297–303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. DePristo MA, Banks E, Poplin R, et al. . A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 2011;43:491–8. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ng PC, Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res 2003;31:3812–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sunyaev S, Ramensky V, Koch I, et al. . Prediction of deleterious human alleles. Hum Mol Genet 2001;10:591–7. 10.1093/hmg/10.6.591 [DOI] [PubMed] [Google Scholar]
- 16. Adzhubei I, Jordan DM, Sunyaev SR, et al. . Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet 2013;Chapter 7::Unit7.20 10.1002/0471142905.hg0720s76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miyata H, Takada T, Toyoda Y, et al. . Identification of febuxostat as a new strong ABCG2 inhibitor: potential applications and risks in clinical situations. Front Pharmacol 2016;7:518 10.3389/fphar.2016.00518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kondo C, Suzuki H, Itoda M, et al. . Functional analysis of SNPs variants of BCRP/ABCG2. Pharm Res 2004;21:1895–903. 10.1023/B:PHAM.0000045245.21637.d4 [DOI] [PubMed] [Google Scholar]
- 19. Stiburkova B, Miyata H, Závada J, et al. . Novel dysfunctional variant in ABCG2 as a cause of severe tophaceous gout: biochemical, molecular genetics and functional analysis. Rheumatology 2016;55:191–4. 10.1093/rheumatology/kev350 [DOI] [PubMed] [Google Scholar]
- 20. Auton A, Brooks LD, Durbin RM, et al. . A global reference for human genetic variation. Nature 2015;526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 2008;83:311–21. 10.1016/j.ajhg.2008.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sakiyama M, Matsuo H, Takada Y, et al. . Ethnic differences in ATP-binding cassette transporter, sub-family G, member 2 (ABCG2/BCRP): genotype combinations and estimated functions. Drug Metab Pharmacokinet 2014;29:490–2. 10.2133/dmpk.DMPK-14-SC-041 [DOI] [PubMed] [Google Scholar]
- 23. Ichida K, Matsuo H, Takada T, et al. . Decreased extra-renal urate excretion is a common cause of hyperuricemia. Nat Commun 2012;3:764 10.1038/ncomms1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Matsuo H, Nakayama A, Sakiyama M, et al. . ABCG2 dysfunction causes hyperuricemia due to both renal urate underexcretion and renal urate overload. Sci Rep 2014;4:3755 10.1038/srep03755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsuo H, Tsunoda T, Ooyama K, et al. . Hyperuricemia in acute gastroenteritis is caused by decreased urate excretion via ABCG2. Sci Rep 2016;6:31003 10.1038/srep31003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsuo H, Yamamoto K, Nakaoka H, et al. . Genome-wide association study of clinically defined gout identifies multiple risk loci and its association with clinical subtypes. Ann Rheum Dis 2016;75:652–9. 10.1136/annrheumdis-2014-206191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li C, Li Z, Liu S, et al. . Genome-wide association analysis identifies three new risk loci for gout arthritis in Han Chinese. Nat Commun 2015;6:7041 10.1038/ncomms8041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakayama A, Nakaoka H, Yamamoto K, et al. . GWAS of clinically defined gout and subtypes identifies multiple susceptibility loci that include urate transporter genes. Ann Rheum Dis 2017;76:869–77. 10.1136/annrheumdis-2016-209632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sulem P, Gudbjartsson DF, Walters GB, et al. . Identification of low-frequency variants associated with gout and serum uric acid levels. Nat Genet 2011;43:1127–30. 10.1038/ng.972 [DOI] [PubMed] [Google Scholar]
- 30. Köttgen A, Albrecht E, Teumer A, et al. . Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013;45:145–54. 10.1038/ng.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet 2001;17:502–10. 10.1016/S0168-9525(01)02410-6 [DOI] [PubMed] [Google Scholar]
- 32. Schork NJ, Murray SS, Frazer KA, et al. . Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev 2009;19:212–9. 10.1016/j.gde.2009.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Do R, Stitziel NO, Won HH, et al. . Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature 2015;518:102–6. 10.1038/nature13917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cruchaga C, Karch CM, Jin SC, et al. . Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer's disease. Nature 2014;505:550–4. 10.1038/nature12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
rmdopen-2017-000464supp001.docx (853.5KB, docx)