Abstract
Our study sought to clarify the effects of microRNA‐139‐5p (miR‐139‐5p) in the tumorigenesis and progression of oral squamous cell carcinoma (OSCC) by regulating HOXA9. MiR‐139‐5p and HOXA9 expression in OSCC tissues, tumour adjacent tissues, OSCC cells and normal cells were tested by qRT‐PCR. SAS and CAL‐27 cell lines were selected in among four OSCC cell lines and then transfected with miR‐139‐5p mimics, pEGFP‐HOXA9 and cotransfected with miR‐139‐5p mimics + pEGFP‐HOXA9. We used MTT, colony formation, transwell and wound healing assays to analyse cell viability, proliferation, invasion and migration. The target relationship between miR‐139‐5p and HOXA9 was verified by luciferase reporter assay and Western blot, respectively. MiR‐139‐5p was down‐regulated, whereas HOXA9 was up‐regulated in OSCC tissues and cells. The proliferation, invasion and migration ability of SAS and CAL‐27 cells in miR‐139‐5p mimics group were significantly weaker than those in the control group and the miR‐NC group (P < 0.01). MiR‐139‐5p can negatively regulate HOXA9. The proliferation, invasion and migration of SAS and CAL‐27 cells in the miR‐139‐5p mimics + pEGFP‐HOXA9 group were not significantly different from those in the blank control and negative control groups (P > 0.05). Our results indicated that miR‐139‐5p could directly inhibit HOXA9, which might be a potential mechanism in inhibiting the proliferation, invasiveness and migration of OSCC cells.
Keywords: oral squamous cell carcinoma, miR‐139‐5p, HOXA9
Introduction
Oral cancer originates from the epithelial cells of the oral cavity, which is widely known as one of the most frequently developed malignant tumours among head and neck carcinomas 1. Pathologically, over 90% of all patients suffering from oral cancer are diagnosed as OSCC with well or moderately differentiation 2. When it comes to the treatment of OSCC, we often give priority to surgery with adjuvant radiotherapy or systemic chemotherapy based on the pathological type, the disease stage, patients’ physical state and other correlated factors 3. In spite of advanced therapy, the 5‐year survival rate of OSCC remains unsatisfactory and even lower than 50% when diagnosed during the middle or late stage of the disease 4. These following cancers are malignant, and have a tendency to attack locally, they are relative with a high risk of recurrence 5 .The poor prognosis of OSCC can be attributed to the comprehensive factors including the late diagnosis, radiation resistance, recurrence at the primary site and frequent distant metastasis after treatment 6, 7. The mechanism that is responsible for tumour cells migration and invasion is complicated, which remains unknown. Therefore, more efforts need to make in accounting the potential molecular mechanism for OSCC tumorigenesis.
MicroRNAs (miRNAs) are a large family of non‐coding endogenous RNA molecules, consisting of 18‐22 nucleotides 8. MiRNAs execute functions in RNA silencing by binding to the 3′‐untranslated region (UTR) of the target messenger RNAs (mRNAs) 9. In the past decades, emerging evidence has demonstrated that miRNAs play important roles in diverse biological processes including cell proliferation, metastasis, survival, autophagy and differentiation 10, 11. A large sum of miRNAs has been described as cancer genes and their deregulations have been measured in a series of tumours, including OSCC 12. Furthermore, recent studies have revealed that miRNAs serve as either onco‐miRNAs or tumour suppressors during cancer progression and tumorigenesis 13, 14. Previous research has demonstrated that a variety of miRNAs that are abnormally expressed in OSCC, such as miR‐301b, miR‐96, miR‐194, miR‐221 and miR‐139‐5p 15, 16, 17, 18, 19. MiR‐139‐5p has been reported to be down‐regulated in various carcinomas, such as bladder cancer, colorectal cancer, prostate cancer and breast cancer 20, 21, 22, 23. Besides, it was documented to participate in regulating cell proliferation and invasion of human non‐small cell lung cancer 24, oesophageal squamous cell carcinoma 25, hepatocellular carcinoma 26, glioblastoma multiforme 19 and so on. However, studies about the regulatory role of miR‐135‐5p for OSCC cells remained limited; therefore, the clinical value and prognostic potential of miR‐139‐5p for OSCC still needed to be further investigated. The targeted relationship of miR‐139‐5p and HOXA9 has been confirmed in other disorders, such as acute myeloid leukaemia 27, and HOXA9 has been indicated to be involved in development of OSCC 28. Moreover, HOXA9 also participated in the development of other carcinomas. For instance, Li et al. detected that HOXA9 had essential oncogenic effect in leukaemia 29. Besides, Liu et al. found that the expression of HOXA9 could be enhanced by miR‐196b in chronic myeloid leukemogenesis 30. Mammalian HOX genes are organized in four clusters (HOXA, HOXB, HOXC and HOXD), each containing nine to 11 genes 31. HOXA9, 180‐nucleotide in length, is a member of mammalian HOX family 32. HOXA9 is required to maintain the development of proper limb, skeletal, mammary gland, urogenital tract and kidney 33. HOXA9 also plays a role in normal myeloid, erythroid and lymphoid haematopoiesis 34. The overexpression of HOXA9 remarkably expands haematopoietic stem cells, suggesting its function in early haematopoiesis and thus, it might affect the angiogenesis during the tumorigenesis process 32.
Here, we quantified the expression of miR‐139‐5p and HOXA9 mRNA in 40 OSCC tissue samples by qRT‐PCR. MiR‐139‐5p was significantly down‐regulated in both tissues and cells, whereas HOXA9 was completely opposite. Dual luciferase reporter assay verified that miR‐139‐5p directly targeted HOXA9. Furthermore, cellular assays were, respectively, performed in vitro to detect the role that miR‐139‐5p plays in a series of cell activities. In all, we found that miR‐139‐5p severed as an anti‐oncogenic role in OSCC and suppressed cell proliferation, invasion and migration through inhibiting HOXA9.
Materials and methods
Patients and samples
Forty cases of OSCC tissues and corresponding adjacent tissues were obtained from OSCC patients who underwent OSCC resection without receiving preoperative chemotherapy or radiation at Henan Provincial People's Hospital from January 2014 to January 2016. Among the patients, there were 24 men and 16 women, and the average age of the patients was around 49.8 ± 5.3 years old. The OSCC tissues were all pathologically identified as OSCC, and the numbers of high‐differentiated, medium‐differentiated and poor‐differentiated cases were, respectively, 13, 8 and 19. According to the tumour node metastasis (TNM) staging, the included OSCC tissues were classified as stage I (n = 5), stage II (n = 10), stage III (n = 15) and stage IV (n = 10). Tissue samples were immediately frozen in liquid nitrogen and stored at −80°C until analysis. The study was approved by the ethics committee of Henan Provincial People's Hospital. All patients included in this study have signed consent forms.
Cell culture
The normal oral keratinocyte NOK cell line and human OSCC cell lines SAS, TCA8113, KON were supplied by Henan Provincial People's Hospital cell centre and CAL‐27 cell line was purchased from ATCC. All cells were all cultured in 10% foetal bovine serum (FBS) dulbecco's modified eagle medium (DMEM) at 37°C in a humidified atmosphere with 5% CO2.
Quantitative real‐time PCR (qRT‐PCR)
Total RNA was extracted from the tumour tissues and adjacent tissues or cells using Trizol RNA extraction kit. Then, we reverse‐transcribed the RNAs to cDNAs using RT Kit (Fermentas, Waltham, MA, USA), and cDNAs were amplified using PCR Kit (Invitrogen, Carlsbad, CA, USA). We listed the used primers in Table 1. The mRNA expressions were the relative expression compared with U6 and GAPDH expressions in the same samples. U6 served as the internal control for miR‐139‐5p, and GAPDH served as the internal control for HOXA9. And the method was used to calculate their relative expressions.
Table 1.
The sequences of primers used in qRT‐PCR
| Primer | Sequence (5′–3′) |
|---|---|
| miR‐139‐5p (F) | 5′‐ GCCTCTACAGTGCACGTGTCTC ‐3′ |
| miR‐139‐5p (R) | 5′‐ CGCTGTTCTCATCTGTCTCGC ‐3′ |
| U6 (F) | 5′ ‐ CTCGCTTCGGCAGCACA ‐3′ |
| U6 (R) | 5′‐ AACGCTTCACGAATTTGCGT‐3′ |
| HOXA9 (F) | 5′‐ CCACGCTTGACACTCACACT ‐3′ |
| HOXA9 (R) | 5′‐ CAGTTCCAGGGTCTGGTGTT ‐3′ |
| GAPDH (F) | 5′‐ ACAACTTTGGTATCGTGGAAGG ‐3′ |
| GAPDH (R) | 5′‐ GCCATCACGCCACAGTTTC ‐3′ |
Cell transfection
Cells were seeded into six‐well plates at 30% concentration in 2 ml of supplemented DMEM and cultured for 24 hrs. Then 50 nM of miR‐139‐5p mimics (miR‐mimics), mimics control (miR‐NC), pEGFP‐N1‐3FLAG‐HOXA9‐GFP plasmids (pEGFP‐HOXA9), empty pEGFP plasmids (pEGFP‐NC) and miR‐139‐5p mimics + pEGFP‐HOXA9 (50 nM; Shanghai GenePharma Co. Ltd., Shanghai, China) were, respectively, transfected to SAS and CAL‐27 cells using Lipofectamine 2000 according to the instructions of manufacturer (Invitrogen).Cells in the control group were without transfection. After post‐transfection for 48 hrs, cells were harvested.
MTT assay
Cells of each group were plated into 96‐well plates at 5 × 103 cells/well. Then after incubation of the cells for 24, 48 and 72 hrs, respectively, we added 20 μl MTT solutions (5 mg/ml) to each well. After incubated for 4 hrs, we added 150 μl DMSO to each well in order to promote the dissolution of crystals. Cell viability was detected at 0, 24, 48 and 72 hrs, and the OD was measured on a microplate reader at 490 nm.
Colony formation assay
Cells of each group were plated into six‐well plates at a concentration of 500 cells/well after transfection for 24 hrs. After the cells adhered to the wall, 0.1% DMSO was applied to act on the cells for 10 days. After being washed with PBS, the cells were fixed by 4% paraformaldehyde, and stained with 5% crystal violet. Then the numbers of colonies were recorded.
Transwell assay
Cells of each group were plated into the upper Transwell chamber at a concentration of 1 × 105 cells/ml, and 600 μl 10% FBS DMEM was added to the lower chamber. After incubation for 24 hrs, the membrane was fixed by 70% ethanol and stained with 0.1% crystal violet. Finally, the number of cells that passed across the membrane was counted in six randomly selected fields under the microscope.
Wound healing assay
Cells of each group were plated in 35 mm2 culture dish at 8 × 105 cells/dish. When the cells grew to a degree that 90% cells were merged, lines were drawn at the bottom of dish with a mark as markers, and scratched at the cell layers of each group with a 200 μl sterile pipette. After cells were incubated for 0, 24 or 48 hrs later, photographs were taken, respectively. The intersection was considered as the observation point, and the open wound rate was measured using a microscope.
Luciferase reporter assay
The wild‐type and mutated sequences of HOXA9 3 ‘UTR were cloned into pGL3‐M (Promega, Madison, WI, USA) using Xba I and Pst I restriction sites. PGL3‐HOXA9‐3′UTR‐wt and pGL3‐HOXA9‐3 ‘UTR‐mut recombinant vectors were generated and verified by sequencing. MiR‐139‐5p mimics or mimics control was cotransfected into cells with pGL3‐HOXA9‐3 ‘UTR‐wt or pGL3‐HOXA9‐3 ‘UTR‐mut. After the transfection of 48 hrs, luciferase activity of cells was detected using dual luciferase reporter gene system.
Western blotting
After culturing the transfected cells for 48 hrs, the total protein was extracted. The electrophoretic separation with 10% dodecyl sulphate, sodium salt‐polyacrylamide gel electrophoresis (SDS‐PAGE) was conducted, and each lane was loaded with 50 μg of total proteins. The total proteins were then transferred to polyvinylidene fluoride (PVDF) membrane which was subsequently blocked for 1.5 hrs using 5% skim milk. Primary antibodies of HOXA9 and GAPDH (Abcam, Cambridge, MA, USA) diluted at 1:1000 were added onto the membrane and stained overnight at 4°C. After the membrane was washed three times using TBST, goat anti‐rabbit HRP horseradish peroxidase‐labelled antibodies (1:1000) were added and incubated. electro‐chemi‐luminescence (ECL) was used to develop the film.
Data analysis and statistical methods
Receiver operating characteristic (ROC) curve analysis was performed using SPSS 15.0 (IBM, Armonk, New York, USA) to calculated for evaluated the predictive ability of miR‐139‐5p to distinguish OSCC patients from normal patients. Statistical analyses were conducted using SPSS 21.0 and GraphPad Prism 6.0 (GraphPad Software Inc., San Diego, CA). Measurement data were recorded as mean ± S.D. The comparison between two groups was analysed using t‐test, whereas one‐way anova test was used for the analyses among more than two groups, followed by post hoc Bonferroni's test. P < 0.05 was statistically significant.
Results
MiR‐139‐5p was under‐expressed in OSCC tissues and diverse OSCC cells
The miR‐139‐5p level in OSCC tissues and adjacent tissues was measured by qRT‐PCR, which indicated that miR‐139‐5p in OSCC tissues was remarkably decreased compared to adjacent tissues (P < 0.01, Fig. 1A). MiR‐139‐5p expression was also remarkably down‐regulated in four OSCC cell lines SAS, TCA8113, KON and CAL‐27 compared with the normal cell line (P < 0.01, Fig. 1B). The succeeding experiments were conducted in SAS cell line in which miR‐139‐5p expression was the lowest. The expression of HOXA9 mRNA in OSCC tissue samples and adjacent tissues was compared by qRT‐PCR and results demonstrated a remarkable increasing of HOXA9 mRNA in OSCC tissues compared to adjacent tissues (P < 0.01, Fig. 1C). HOXA9 mRNA expression level was remarkably up‐regulated in four OSCC cells SAS, TCA8113, KON and CAL‐27 compared with the normal cell line (P < 0.01, Fig. 1D). Further analysis in Table 2 showed that miR‐139‐5p expression was strongly associated with tumour size and clinical stage, whereas HOXA9 level was correlated to pathological differentiation according to Fisher's exact test (n = 40). Probably limited by small sample size, no significant association among miR‐139‐5p, HOXA9, age, gender and LN metastasis was obtained.
Figure 1.
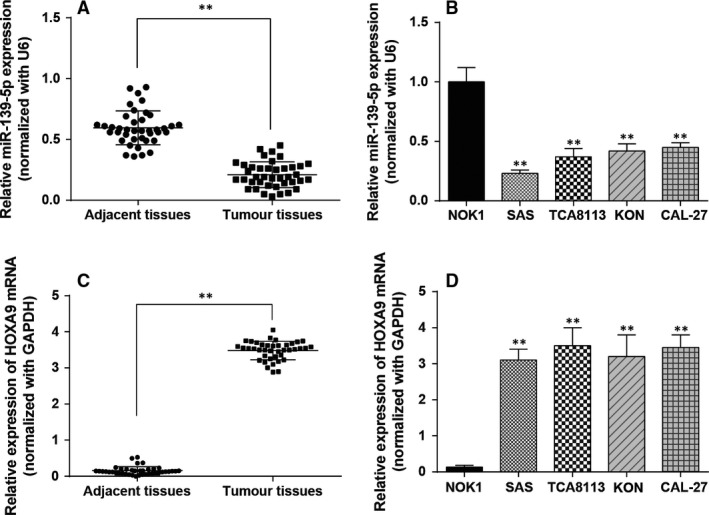
Expression levels of miR‐139‐5p and HOXA9. (A) MiR‐139‐5p was detected with qRT‐PCR in OSCC tissues and adjacent non‐tumorous tissues. (B) MiR‐139‐5p was measured by qRT‐PCR in OSCC cells versus normal cells. (C) The HOXA9 mRNA expression was measured by qRT‐PCR in OSCC tissues and adjacent non‐tumorous tissues. (D) The HOXA9 mRNA expression was detected with RT‐PCR in OSCC cells versus normal cells. Data are expressed as mean ± S.D. **P < 0.01, compared with the adjacent tissues or the normal cell line NOK1. #P < 0.05, compared with the SAS cell line. OSCC: oral squamous cell carcinoma.
Table 2.
The baseline characteristics of OSCC patients included (n = 40)
| Characteristic | Case. | miR‐139‐5p expression | P value | HOXA9 expression | P value | ||
|---|---|---|---|---|---|---|---|
| Low | High | Low | High | ||||
| Age | |||||||
| ≤50 | 17 | 7 | 10 | 0.5231 | 11 | 6 | 0.2003 |
| >50 | 23 | 13 | 10 | 9 | 14 | ||
| Gender | |||||||
| Male | 24 | 14 | 10 | 0.3332 | 13 | 11 | 0.7475 |
| Female | 16 | 6 | 10 | 7 | 9 | ||
| Tumour size | |||||||
| ≤4 cm | 26 | 9 | 17 | 0.0187 | 15 | 11 | 0.3203 |
| >4 cm | 14 | 11 | 3 | 5 | 9 | ||
| Differentiation | |||||||
| High/Moderate | 21 | 8 | 13 | 0.2049 | 15 | 6 | 0.0104 |
| Poor | 19 | 12 | 7 | 5 | 14 | ||
| Clinical stage | |||||||
| I–II | 15 | 4 | 11 | 0.0484 | 7 | 8 | 1 |
| III–IV | 25 | 16 | 9 | 13 | 12 | ||
| LN metastasis | |||||||
| No | 23 | 13 | 10 | 0.5231 | 10 | 13 | 0.5231 |
| Yes | 17 | 7 | 10 | 10 | 7 | ||
| 20 | 20 | 20 | 20 | ||||
LN: Lymph node; HOXA9: homeobox A9.
P value was obtained with Fisher's exact test.
P < 0.05 represents significant differences.
Bold type indicates statistically significant difference.
MiR‐139‐5p inhibited the viability, proliferation, invasion and migration of SAS and CAL‐27 cell lines
Firstly, we generated ROC curves and found that miR‐139‐5p exhibited AUC of 0.985, HOXA9 exhibited AUC of 1 (Fig. 2A), indicating sufficient power to distinguish OSCC tissues from adjacent tissues. Then, results of qRT‐PCR suggested that the miR‐139‐5p expression in miR‐139‐5p mimics group is significantly higher than that in control and miR‐NC groups in both cell lines(**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 2B). Next, results of MTT assay showed that cells of miR‐139‐5p mimics group had significantly lower viability than control and miR‐NC groups (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 2C), suggesting that miR‐139‐5p inhibited the viability of SAS and CAL‐27 cell lines Colony formation assay was used to detect the colony forming ability. Similar to the results of MTT assay, as shown in Figure 2D, both cell lines in miR‐139‐5p mimics group had fewer colony formations than control groups (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line), indicating that miR‐139‐5p could inhibit SAS and CAL‐27 cells proliferation.
Figure 2.

MiR‐139‐5p suppresses proliferation of OSCC cells. (A) Receiver operating curves(ROC) comparing miR‐139‐5p and HOXA9 level in OSCC tissues and adjacent tissues. (B) MiR‐139‐5p was detected with qRT‐PCR in each group including control, miR‐negative control (NC) and miR‐mimics of SAS and CAL‐27 cell lines. (C) Cell viability was analysed by MTT assay. (D) Cell proliferation was analysed by colony formation assay. Data are expressed as mean ± S.D. **P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line. Control: control group, cells were without transfection. MiR‐NC: miR‐NC group, cells transfected with mimics control. MiR‐mimics: miR‐mimics group, cells transfected with miR‐139‐5p mimics.
Wound healing assay was used to assess the migration of SAS and CAL‐27 cells. The results suggested that open wound rate of cells in the control and miR‐NC groups surpassed that in the miR‐139‐5p mimics group (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 3A). The results revealed that miR‐139‐5p inhibited the migration of SAS and CAL‐27 cells. And the invasion of SAS and CAL‐27 cells was measured by Transwell assay. The results of transwell assay indicated that cells in the miR‐139‐5p mimics group had weaker invasiveness than cells in the control groups (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 3B), showing that miR‐139‐5p inhibited the invasion of SAS and CAL‐27 cells.
Figure 3.
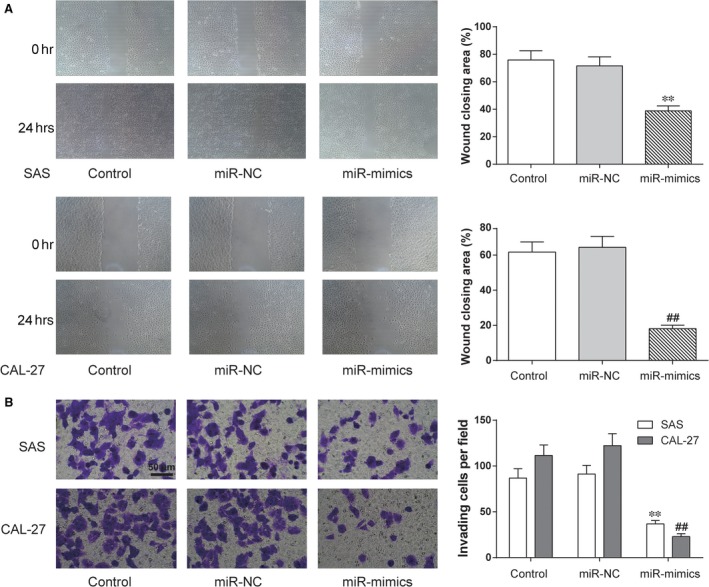
MiR‐139‐5p suppresses migration and invasion of OSCC cells. (A) After replenishing miR‐139‐5p expression in SAS and CAL‐27 cells, cell migration was assessed by wound healing assay. (B) Cell invasion ability was analysed by Transwell assay. **P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line. Control: control group, cells were without transfection. MiR‐NC: miR‐NC group, cells transfected with mimics control. MiR‐mimics: miR‐mimics group, cells transfected with miR‐139‐5p mimics.
HOXA9 is the target of miR‐139‐5p
HOXA9 was predicted to be a target of miR‐139‐5p with the possible binding sequence ACUGUAGA in HOXA9 3′UTR by the online database TargetScan Human 7.0 (Fig. 4A). The luciferase activity of HOXA9‐wt 3′UTR + miR‐139‐5p mimics decreased significantly (P < 0.01, Fig. 4B), while that of HOXA9‐mut 3′UTR + miR‐139‐5p mimics was not significantly different from the control groups. It suggested that miR‐139‐5p can directly inhibit HOXA9.
Figure 4.
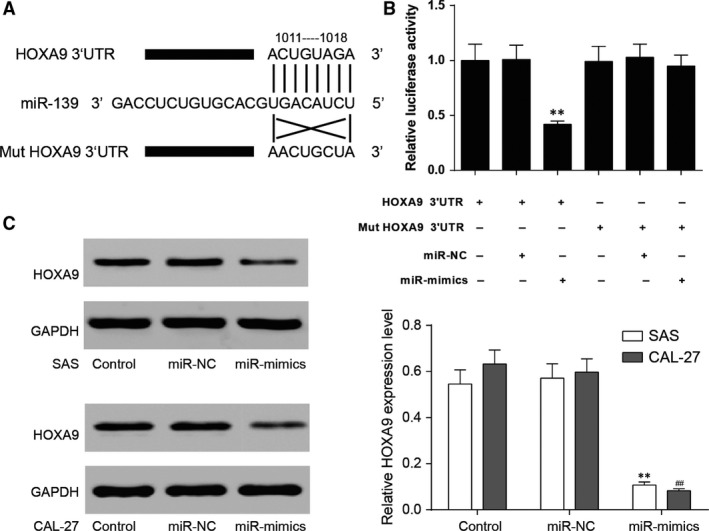
MiR‐139‐5p targeted HOXA9. (A) The predicted binding site of miR‐139‐5p was in the 3 ‘UTR of HOXA9. (B) HEK293 cells were transfected with either miR‐139‐5p mimics or miR‐ negative control (NC) and HOXA9‐wt or HOXA9‐mut. The luciferase activity was analysed after 48 hrs. (C) The protein expression of HOXA9 in SAS and CAL‐27 cell lines was detected by Western blot. GAPDH was served as an internal control. Data are expressed as mean ± S.D. **P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line. Control: control group, cells were without transfection. MiR‐NC: miR‐NC group, cells transfected with mimics control. MiR‐mimics: miR‐mimics group, cells transfected with miR‐139‐5p mimics.
The suppression effect of miR‐139‐5p on HOXA9 was evaluated at protein levels by Western blot. HOXA9 protein level was down‐regulated in SAS and CAL‐27 cells transfected with miR‐139‐5p mimics in comparison with the control groups, further verifying the inhibition of miR‐139‐5p on HOXA9 (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 4C).
MiR‐139‐5p inhibited the viability of SAS and CAL‐27 cells by suppressing HOXA9
Firstly, the results of qRT‐PCR demonstrated that the miR‐139‐5p expression in miR‐139‐5p mimics + pEGFP‐HOXA9 group was remarkable higher than that of other groups (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 5A). It turned out that the expression of miR‐139‐5p was not affected by the overexpression of HOXA9. Then, in Figure 5B and C, HOXA9 mRNA expression and protein level were significantly up‐regulated in pEGFP‐HOXA9 group compared with the pEGFP‐NC1 group (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line), while HOXA9 protein level was almost identical in miR‐139‐5p mimics + pEGFP‐HOXA9 and pEGFP‐NC2 groups. The results showed that miR‐139‐5p inhibited the expression of HOXA9.
Figure 5.
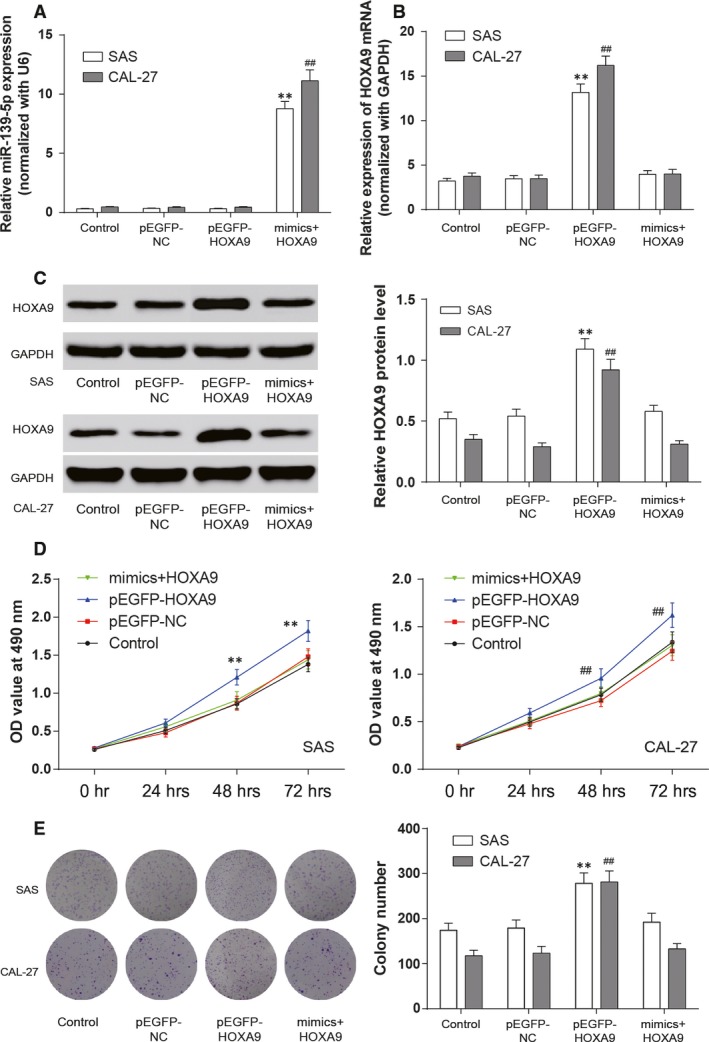
MiR‐139‐5p suppressed the proliferation of OSCC cells by targeting HOXA9. (A and B) Expression of miR‐139‐5p and HOXA9 mRNA was detected with qRT‐PCR in each group. (C) The protein level of HOXA9 in SAS and CAL‐27 cell lines was assessed by Western blot. GAPDH was used as an internal control. (D) Cell viability was analysed by MTT assay. (E) Cell proliferation was detected through colony formation assay. Data are expressed as mean ± S.D. **P < 0.01 compared with control in SAS cell line, ##P < 0.01 compared with control in CAL‐27 cell line. Control: control group, cells were without transfection. pEGFP‐NC: pEGFP‐NC group, cells transfected with empty pEGFP plasmids. pEGFP‐HOXA9: pEGFP‐HOXA9 group, cells transfected with pEGFP‐N1‐3FLAG‐HOXA9‐GFP plasmids. MiR‐139‐5p mimics + pEGFP‐HOXA9: miR‐139‐5p mimics + pEGFP‐HOXA9 group, cells cotransfected with miR‐139‐5p mimics and pEGFP‐HOXA9.
In MTT assay, the overexpression of HOXA9 (pEGFP‐HOXA9 group) significantly increased cell viability, whereas the viability of SAS and CAL‐27 cells in miR‐139‐5p mimics + pEGFP‐HOXA9 group were not significantly different from that in corresponding controls (P > 0.05, Fig. 5D). These findings indicated that miR‐139‐5p could eliminate the increased cell viability of OSCC cells induced by the overexpression of HOXA9.
MiR‐139‐5p inhibited the proliferation, invasion and migration of SAS and CAL‐27 cells by suppressing HOXA9
The cell proliferation of miR‐139‐5p on HOXA9 was assessed by colony formation assay. Compared with the control and pEGFP‐NC groups, the pEGFP‐HOXA9 group formed more colonies while the colony formation of SAS and CAL‐27 cells in miR‐139‐5p mimics + pEGFP‐HOXA9 group were not different from that in corresponding control groups (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 5E).
The cell migration of SAS and CAL‐27 cells was detected by wound healing assay. As shown in Figure 6A, the open wound rate of cells in the pEGFP‐HOXA9 group was higher than that in the control groups. No significant difference in the open wound rate was observed among the miR‐139‐5p mimics + pEGFP‐HOXA9 group, the the control group and the pEGFP‐NC group (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line).
Figure 6.
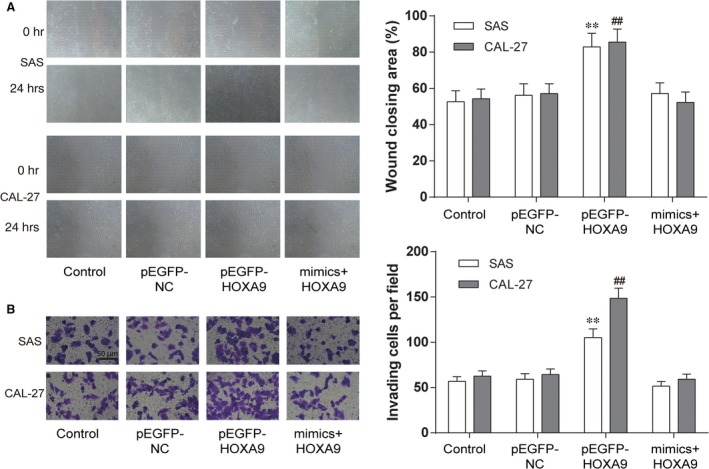
MiR‐139‐5p attenuated migration and invasion of OSCC cells by targeting HOXA9. (A) At 48 hrs after transfection, cell migration was assessed by wound healing assay. (B) Cell invasion was analysed by Transwell assay. Data are expressed as mean ± S.D. **P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line. Control: control group, cells were without transfection. pEGFP‐NC: pEGFP‐NC group, cells transfected with empty pEGFP plasmids. pEGFP‐HOXA9: pEGFP‐HOXA9 group, cells transfected with pEGFP‐N1‐3FLAG‐HOXA9‐GFP plasmids. MiR‐139‐5p mimics + pEGFP‐HOXA9: miR‐139‐5p mimics + pEGFP‐HOXA9 group, cells cotransfected with miR‐139‐5p mimics and pEGFP‐HOXA9.
Then Transwell assay was conducted to confirm the suppression effect of miR‐139‐5p on HOXA9 in cell invasion. The pEGFP‐HOXA9 group had stronger invasiveness in comparison with the control group and the pEGFP‐NC group. There was no remarkable difference in the invasiveness of SAS and CAL‐27 cells among the miR‐139‐5p mimics + pEGFP‐HOXA9 group, the control group and the pEGFP‐NC group (**P < 0.01 compared with control in SAS cell line, ## P < 0.01 compared with control in CAL‐27 cell line, Fig. 6B).
Discussion
More attention is paying to the explosion of potential molecular mechanism responsible for OSCC due to its increased incidence in both developed and developing countries 35, 36, 37, 38. Among previous studies, most abnormally expressed miRNAs were discovered in the OSCC. For instance, Stuart Hunt et al. found that miR‐124 suppressed OSCC motility by targeting ITGB1 39. Shiah et al. demonstrated that down‐regulated miR‐329 and miR‐410 promoted the proliferation and invasion of OSCC cells by targeting wnt‐7b 40. Wang et al. detected that miR‐433 inhibits OSCC cell growth and metastasis by targeting HDAC6 41. Of note, a number of target genes can be regulated by a single miRNA instantly. For example, it has been reported that miR‐139‐5p suppresses the non‐small cell lung cancer cell proliferation and induces apoptosis via inhibiting c‐Met 42. Besides, miR‐139‐5p is able to repress the activation of AMFR and NOTCH1 in colorectal cancer 43. Also, miR‐139‐5p functions as a master regulator of glioblastoma metastasis through targeting ZEB1 and ZEB2 19. Furthermore, the increased expression level of miR‐139‐5p can lead to suppressed cell viability and proliferation in various cancers through cell cycle arrest, which induces more cells arrested at G0/G1 or S phase 44, 45. Besides, cell cyclin involving E1, D1, MMP2 and MMP9 is significantly changed in cell transfected with miR‐139‐5p mimics 25, 46. Furthermore, a decrease in the apoptotic protein such as Bax 47, 48 and an increase in the anti‐apoptotic protein including Bcl‐2 as well as Mcl‐1 47, 49, regulated by miR‐139‐5p, also participates in the tumour process. In the present study, miR‐139‐5p was found down‐regulated in the OSCC sample tissues, and its up‐regulation suppressed the viability, proliferation, migration and invasiveness of OSCC cells. Notably, emerging evidence has highlighted that miR‐139‐5p served as a tumour suppressor in various carcinomas, including bladder cancer, colorectal cancer, prostate cancer and breast cancer 20, 21, 22, 23. Besides, miR‐139‐5p has also been confirmed as a prognostic element for progression of breast cancer 50. Therefore, the suppressor role of miR‐139‐5p in OSCC proposed in our study was convincing, and we should explore it further.
To better understand the precise molecular mechanism that is responsible for the antitumour effects of miR‐139‐5p in OSCC, we searched potential target genes of miR‐139‐5p using TargetScan (http://www.targetscan.org). HOXA9 was predicted to harbour one highly conservative miR‐139‐5p binding sites in the sequence of 3′UTR. With the use of dual luciferase reporter assays, HOXA9 was eventually identified as a direct target for miR‐139‐5p. The increase in miR‐139‐5p expression was accompanied by the down‐regulated HOXA9 expression. QRT‐PCR results revealed that HOXA9 was up‐regulated in SAS cells, which was inconsistent with some of the previous studies. Ruiz et al. found that HOXA9 was under‐expressed in cervical cancer cells and its restoration decreased the proliferation, migration and expression of epithelial‐to‐mesenchymal transition (EMT) genes 51. Hwang JA et al. detected that HOXA9 inhibits lung cancer cells migration and its hypermethylation is closely linked with the recurrence in non‐small cell lung cancer 52. Miso et al. found that the HMGA2‐TET1‐HOXA9 signalling was coordinately regulated in breast cancer. Both TET1 and HOXA9 suppressed breast cancer development in nude mice 53. On the contrary, HOXA9 was believed to simulate ovarian cancer progression by inducing peritoneal macrophages to acquire an M2 tumour‐promoting phenotype 54. Similarly, Ko et al. asserted that the HOXA9 expression in epithelial ovarian cancer cells promised a comfortable environment for tumour growth 55. The work performed by Pojo et al. discovered that HOXA9 promoted human glioblastoma initiation and aggressiveness 56. In terms of the promoter role of HOXA9 during OSCC development and progression, our results verified that miR‐139‐5p suppressed OSCC cell viability, proliferation, invasion and migration by directly inhibiting HOXA9. We also found that the effects of HOXA9 could be deteriorated by exogenous miR‐139‐5p. But whether HOXA9 contributed to the alteration of tumour microenvironments need further proved.
Still, there are several limitations in our study. Due to the resource constraint of clinical samples, only 40 pairs of human OSCC tissues and matched adjacent tissues were involved in the present study. Such a small sample size might result in minor errors in the relative expression of miR‐139‐5p. We intended to future research to verify the above conclusions. Besides, the correlated signalling pathway might help explain the molecular mechanism of anti‐oncogenic effect of miR‐139‐5p on OSCC cells. Moreover, we verified the anti‐oncogenic effect of miR‐139‐5p in SAS OSCC cell line in vitro, while the other OSCC cell lines as well as the experiments in vivo (i.e. in established animal models) were lacking. In addition, the role of miR‐139‐5p and HOXA9 in other mechanisms, such as EMT, also needed to be further investigated, which could provide evidence for discovering novel targets for OSCC. As a result, further researches should pay attention to resolving the above limitations.
Conclusion
In summary, the present study demonstrated that miR‐139‐5p, functioning as a suppressor in the tumorigenesis process, suppressing OSCC cell mobility by targeting HOXA9 directly. The relative expression of miR‐139‐5p and HOXA9 was quantified by qRT‐PCR. Dual luciferase reporter gene assay verified that HOXA9 was a target for miR‐139‐5p. In vitro assays performed with OSCC cells further detected the role of miR‐139‐5p played during tumorigenesis. Combined with previous studies on miR‐139‐5p, we believe that miR‐139‐5p might be a crucial molecular that participates in the OSCC tumorigenesis. Anyway, we hope that the results might provide a therapeutic strategy for the treatment of OSCC.
Funding
None.
Author Contributions
Kai Wang and Jun Jin performed the research; Tengxiao Ma and Hongfeng Zhai analysed and interpreted the data; Kai Wang designed the research study and wrote themanuscript.
Conflict of interest
None.
Acknowledgements
Reference
- 1. Torre LA, Bray F, Siegel RL, et al Global cancer statistics, 2012. CA Cancer J Clin. 2015; 65: 87–108. [DOI] [PubMed] [Google Scholar]
- 2. Zini A, Czerninski R, Sgan‐Cohen HD. Oral cancer over four decades: epidemiology, trends, histology, and survival by anatomical sites. J Oral Pathol Med. 2010; 39: 299–305. [DOI] [PubMed] [Google Scholar]
- 3. Huang SH, O'Sullivan B. Oral cancer: current role of radiotherapy and chemotherapy. Med Oral Patol Oral Cir Bucal. 2013; 18: e233–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ganly I, Patel S, Shah J. Early stage squamous cell cancer of the oral tongue–clinicopathologic features affecting outcome. Cancer. 2012; 118: 101–11. [DOI] [PubMed] [Google Scholar]
- 5. Duz MB, Karatas OF, Guzel E, et al Identification of miR‐139‐5p as a saliva biomarker for tongue squamous cell carcinoma: a pilot study. Cell Oncol (Dordr). 2016; 39: 187–93. [DOI] [PubMed] [Google Scholar]
- 6. Walden MJ, Aygun N. Head and neck cancer. Semin Roentgenol. 2013; 48: 75–86. [DOI] [PubMed] [Google Scholar]
- 7. Sinha N, Mukhopadhyay S, Das DN, et al Relevance of cancer initiating/stem cells in carcinogenesis and therapy resistance in oral cancer. Oral Oncol. 2013; 49: 854–62. [DOI] [PubMed] [Google Scholar]
- 8. Turchinovich A, Weiz L, Langheinz A, et al Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011; 39: 7223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010; 11: 597–610. [DOI] [PubMed] [Google Scholar]
- 10. Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012; 149: 515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012; 33: 2018–25. [DOI] [PubMed] [Google Scholar]
- 12. Chen Z, Yu T, Cabay RJ, et al miR‐486‐3p, miR‐139‐5p, and miR‐21 as biomarkers for the detection of oral tongue squamous cell carcinoma. Biomark Cancer. 2017; 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bertoli G, Cava C, Castiglioni I. MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics. 2015; 5: 1122–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Plummer PN, Freeman R, Taft RJ, et al MicroRNAs regulate tumor angiogenesis modulated by endothelial progenitor cells. Can Res. 2013; 73: 341–52. [DOI] [PubMed] [Google Scholar]
- 15. Funamizu N, Lacy CR, Parpart ST, et al MicroRNA‐301b promotes cell invasiveness through targeting TP63 in pancreatic carcinoma cells. Int J Oncol. 2014; 44: 725–34. [DOI] [PubMed] [Google Scholar]
- 16. Guo Y, Ren MS, Shang C, et al MTSS1 gene regulated by miR‐96 inhibits cell proliferation and metastasis in tongue squamous cellular carcinoma Tca8113 cell line. Int J Clin Exp Med. 2015; 8: 15441–9. [PMC free article] [PubMed] [Google Scholar]
- 17. Chi H. miR–194 regulated AGK and inhibited cell proliferation of oral squamous cell carcinoma by reducing PI3K‐Akt‐FoxO3a signaling. Biomed Pharmacother. 2015; 71: 53–7. [DOI] [PubMed] [Google Scholar]
- 18. He S, Lai R, Chen D, et al Downregulation of miR‐221 inhibits cell migration and invasion through targeting Methyl‐CpG binding domain protein 2 in human oral squamous cell carcinoma cells. Biomed Res Int. 2015; 2015: 751672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yue S, Wang L, Zhang H, et al miR‐139‐5p suppresses cancer cell migration and invasion through targeting ZEB1 and ZEB2 in GBM. Tumour Biol. 2015; 36: 6741–9. [DOI] [PubMed] [Google Scholar]
- 20. Yonemori M, Seki N, Yoshino H, et al Dual tumor‐suppressors miR‐139‐5p and miR‐139‐3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016; 107: 1233–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu H, Yin Y, Hu Y, et al miR‐139‐5p sensitizes colorectal cancer cells to 5‐fluorouracil by targeting NOTCH‐1. Pathol Res Pract. 2016; 212: 643–9. [DOI] [PubMed] [Google Scholar]
- 22. Pang C, Liu M, Fang W, et al MiR‐139‐5p is increased in the peripheral blood of patients with prostate cancer. Cell Physiol Biochem. 2016; 39: 1111–7. [DOI] [PubMed] [Google Scholar]
- 23. Zhang HD, Sun DW, Mao L, et al MiR‐139‐5p inhibits the biological function of breast cancer cells by targeting Notch1 and mediates chemosensitivity to docetaxel. Biochem Biophys Res Comm. 2015; 465: 702–13. [DOI] [PubMed] [Google Scholar]
- 24. Xu W, Hang M, Yuan CY, et al MicroRNA‐139‐5p inhibits cell proliferation and invasion by targeting insulin‐like growth factor 1 receptor in human non‐small cell lung cancer. Int J Clin Exp Pathol. 2015; 8: 3864–70. [PMC free article] [PubMed] [Google Scholar]
- 25. Liu R, Yang M, Meng Y, et al Tumor‐suppressive function of miR‐139‐5p in esophageal squamous cell carcinoma. PLoS One. 2013; 8: e77068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qiu G, Lin Y, Zhang H, et al miR‐139‐5p inhibits epithelial‐mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem Biophys Res Comm. 2015; 463: 315–21. [DOI] [PubMed] [Google Scholar]
- 27. Krowiorz K, Ruschmann J, Lai C, et al MiR‐139‐5p is a potent tumor suppressor in adult acute myeloid leukemia. Blood Cancer J. 2016; 6: e508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guerrero‐Preston R, Soudry E, Acero J, et al NID2 and HOXA9 promoter hypermethylation as biomarkers for prevention and early detection in oral cavity squamous cell carcinoma tissues and saliva. Cancer Prev Res. 2011; 4: 1061–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Z, Huang H, Chen P, et al miR‐196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL‐rearranged leukaemia. Nat Commun. 2012; 3: 688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu Y, Zheng W, Song Y, et al Low expression of miR‐196b enhances the expression of BCR‐ABL1 and HOXA9 oncogenes in chronic myeloid leukemogenesis. PLoS One. 2013; 8: e68442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Boncinelli E, Acampora D, Pannese M, et al Organization of human class I homeobox genes. Genome. 1989; 31: 745–56. [DOI] [PubMed] [Google Scholar]
- 32. Lawrence HJ, Christensen J, Fong S, et al Loss of expression of the Hoxa‐9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005; 106: 3988–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kmita M, Tarchini B, Zakany J, et al Early developmental arrest of mammalian limbs lacking HoxA/HoxD gene function. Nature. 2005; 435: 1113–6. [DOI] [PubMed] [Google Scholar]
- 34. Lawrence HJ, Helgason CD, Sauvageau G, et al Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997; 89: 1922–30. [PubMed] [Google Scholar]
- 35. Liu C, Wang Z, Wang Y, et al MiR‐338 suppresses the growth and metastasis of OSCC cells by targeting NRP1. Mol Cell Biochem. 2015; 398: 115–22. [DOI] [PubMed] [Google Scholar]
- 36. Shao Y, Qu Y, Dang S, et al MiR‐145 inhibits oral squamous cell carcinoma (OSCC) cell growth by targeting c‐Myc and Cdk6. Cancer Cell Int. 2013; 13: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xu P, Li Y, Yang S, et al Micro‐ribonucleic acid 143 (MiR‐143) inhibits oral squamous cell carcinoma (OSCC) cell migration and invasion by downregulation of phospho‐c‐Met through targeting CD44 v3. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015; 120: 43–51. [DOI] [PubMed] [Google Scholar]
- 38. Zeng Q, Tao X, Huang F, et al Overexpression of miR‐155 promotes the proliferation and invasion of oral squamous carcinoma cells by regulating BCL6/cyclin D2. Int J Mol Med. 2016; 37: 1274–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hunt S, Jones AV, Hinsley EE, et al MicroRNA‐124 suppresses oral squamous cell carcinoma motility by targeting ITGB1. FEBS Lett. 2011; 585: 187–92. [DOI] [PubMed] [Google Scholar]
- 40. Shiah SG, Hsiao JR, Chang WM, et al Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt‐7b. Can Res. 2014; 74: 7560–72. [DOI] [PubMed] [Google Scholar]
- 41. Wang XC, Ma Y, Meng PS, et al miR‐433 inhibits oral squamous cell carcinoma (OSCC) cell growth and metastasis by targeting HDAC6. Oral Oncol. 2015; 51: 674–82. [DOI] [PubMed] [Google Scholar]
- 42. Sun C, Sang M, Li S, et al Hsa‐miR‐139‐5p inhibits proliferation and causes apoptosis associated with down‐regulation of c‐Met. Oncotarget. 2015; 6: 39756–92. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43. Song M, Yin Y, Zhang J, et al MiR‐139‐5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell. 2014; 5: 851–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dai S, Wang X, Li X, et al MicroRNA‐139‐5p acts as a tumor suppressor by targeting ELTD1 and regulating cell cycle in glioblastoma multiforme. Biochem Biophys Res Comm. 2015; 467: 204–10. [DOI] [PubMed] [Google Scholar]
- 45. Zhang L, Dong Y, Zhu N, et al microRNA‐139‐5p exerts tumor suppressor function by targeting NOTCH1 in colorectal cancer. Mol Cancer. 2014; 13: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gu W, Li X, Wang J. miR‐139 regulates the proliferation and invasion of hepatocellular carcinoma through the WNT/TCF‐4 pathway. Oncol Rep. 2014; 31: 397–404. [DOI] [PubMed] [Google Scholar]
- 47. Li RY, Chen LC, Zhang HY, et al MiR‐139 inhibits Mcl‐1 expression and potentiates TMZ‐induced apoptosis in glioma. CNS Neurosci Ther. 2013; 19: 477–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bae S, Lee EM, Cha HJ, et al Resveratrol alters microRNA expression profiles in A549 human non‐small cell lung cancer cells. Mol Cells. 2011; 32: 243–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng K, Chen DS, Wu YQ, et al MicroRNA expression profile in RAW264.7 cells in response to Brucella melitensis infection. Int J Biol Sci. 2012; 8: 1013–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cava C, Bertoli G, Ripamonti M, et al Integration of mRNA expression profile, copy number alterations, and microRNA expression levels in breast cancer to improve grade definition. PLoS One. 2014; 9: e97681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Alvarado‐Ruiz L, Martinez‐Silva MG, Torres‐Reyes LA, et al HOXA9 is underexpressed in cervical cancer cells and its restoration decreases proliferation, migration and expression of epithelial‐to‐mesenchymal transition genes. Asian Pac J Cancer Prev. 2016; 17: 1037–47. [DOI] [PubMed] [Google Scholar]
- 52. Hwang JA, Lee BB, Kim Y, et al HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non‐small cell lung cancer. Mol Carcinog. 2015; 54(Suppl 1): E72–80. [DOI] [PubMed] [Google Scholar]
- 53. Sun M, Song C‐X, Huang H, et al HMGA2/TET1/HOXA9 signaling pathway regulates breast cancer growth and metastasis. Proc Natl Acad Sci. 2013; 110: 9920–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ko SY, Ladanyi A, Lengyel E, et al Expression of the homeobox gene HOXA9 in ovarian cancer induces peritoneal macrophages to Acquire an M2 tumor‐promoting phenotype. Am J Pathol. 2014; 184: 271–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ko SY, Barengo N, Ladanyi A, et al HOXA9 promotes ovarian cancer growth by stimulating cancer‐associated fibroblasts. J Clin Invest. 2012; 122: 3603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pojo M, Goncalves CS, Xavier‐Magalhaes A, et al A transcriptomic signature mediated by HOXA9 promotes human glioblastoma initiation, aggressiveness and resistance to temozolomide. Oncotarget. 2015; 6: 7657–74. [DOI] [PMC free article] [PubMed] [Google Scholar]


