Abstract
The heterogeneous course, severity, and treatment responses among patients with atopic dermatitis (AD; eczema) highlight the complexity of this multifactorial disease. Prior studies have used traditional typing methods on cultivated isolates or sequenced a bacterial marker gene to study the skin microbial communities of AD patients. Shotgun metagenomic sequence analysis provides much greater resolution, elucidating multiple levels of microbial community assembly ranging from kingdom to species and strain-level diversification. Here, we analyze microbial temporal dynamics from a cohort of pediatric AD patients sampled throughout the disease course. Species-level investigation of AD flares showed greater Staphylococcus aureus-predominance in patients with more severe disease and S. epidermidis-predominance in patients with less severe disease. At the strain-level, metagenomic sequencing analyses demonstrated clonal S. aureus strains in more severe patients and heterogeneous S. epidermidis strain communities in all patients. To investigate strain-level biological effects of S. aureus, we topically colonized mice with strains isolated from AD patients and controls. This cutaneous colonization model demonstrated S. aureus strain-specific differences in eliciting skin inflammation and immune signatures characteristic of AD patients. Specifically, S. aureus isolates from AD patients with more severe flares induced epidermal thickening and expansion of cutaneous Th2 and Th17 cells. Integrating high-resolution sequencing, culturing, and animal models demonstrated how functional differences of staphylococcal strains may contribute to the complexity of AD disease.
Atopic dermatitis (AD, eczema) is a common inflammatory skin disorder in industrialized countries, affecting 10–30% of children (1). Patients with AD suffer from chronic, relapsing, intensely itchy, inflamed skin lesions, and have an increased likelihood of developing asthma and/or hay fever (2). AD is a complex multifactorial disease in which epidermal barrier impairment, type 2 immunity, and skin microbes are each thought to play a causative role (1). Over 30 susceptibility loci have been associated with AD, including mutations in the gene encoding the skin barrier protein filaggrin (FLG) (3) and genes linked to the immune system (4).
In addition to host genetic susceptibility, the relationship between AD and skin bacteria is well-recognized clinically. Patients with AD are often treated with varying combinations of antimicrobial approaches (e.g. antibiotics and dilute bleach baths) and anti-inflammatory or immunosuppressive medications (5). The efficacy of these antimicrobial treatments is associated with decreases in staphylococcal relative abundances (6, 7). Staphylococcus aureus commonly colonizes AD skin and has been studied using colony-counting, sequence typing methods (e.g. pulsed field gel electrophoresis, SpA typing) of selected cultivated isolates (8–11), and more recently, amplicon-based (marker gene) sequencing of the 16S rRNA gene (6, 7, 12, 13). However, sequence typing and amplicon sequencing methods are unable to distinguish between genetically distinct strains as determined by whole genome sequencing (14, 15). By contrast, shotgun metagenomic sequencing of skin samples from healthy individuals provided deeper resolution and demonstrated the multiphyletic composition of commensal Staphylococcus (16).
With an increasing appreciation of functional differences between strains within a single species, we performed shotgun metagenomic sequencing of AD patient skin samples to capture the full genetic potential and strain-level differences of the skin microbiome throughout the course of disease. We confirmed an increase of staphylococcal species during disease flares in our cohort and more deeply explored the S. aureus and S. epidermidis strain diversity of each patient. To test the functional consequence of strain-level differences between patients, we isolated staphylococcal strains from patients and healthy controls and investigated the cutaneous and immunologic effects when applied topically in a mouse model.
Bacterial communities shift during AD disease progression
To examine the relationship between the skin microbiota and AD, eleven children with moderate-to-severe AD and seven healthy children were recruited to the NIH Clinical Center between June 2012 and March 2015 (Table S1,S2). As AD has a chronic relapsing course, patients were sampled at stable/typical disease state (baseline/B), worsening of disease (flare/F) and 10–14 days after initiation of treatment using a combination of skin-directed therapies (post-flare/PF). Since the use of topical medications on AD skin alters skin bacterial communities (6, 7), baseline samples were defined as those collected from subjects in their routine disease state who refrained from using skin-directed antimicrobial and anti-inflammatory treatments for seven days, a duration of time determined based on prior findings (6). Flares were defined as timepoints when patients experienced worsening in the clinical severity of their typical AD, had not used skin-directed antimicrobial and anti-inflammatory treatments for seven days, and did not have clinical skin infection (i.e. yellow crusts or pustules). At each timepoint, disease severity was determined with objective SCORAD (SCORing Atopic Dermatitis), a validated clinical severity assessment tool (17–19). Subjects were sampled bilaterally at sites of disease predilection, the inner elbow (antecubital crease/Ac) and behind the knees (popliteal crease/Pc), along with five additional sites to investigate defined areas with different skin physiologies (fig. S1). Due to the clinical severity of their AD, six of the eleven patients experienced exacerbations of their skin disease with the seven-day skin preparation regimen and could not provide baseline timepoint samples, reflecting the spectrum of the natural course of AD. Because the skin microbial dysbiosis during AD flares was of greatest interest, the majority of the analyses focused on comparisons between flare and post-flare timepoints. In total, we performed shotgun metagenomic sequencing of 422 samples, generating 191 Gb of microbial sequence data from 27 AD patient visits and 7 healthy control visits (Table S3). During patient flares, AD disease severity was significantly elevated as indicated by higher mean objective SCORAD (38 ± 2.9) as compared to baseline (9.4 ± 1.6, p < 4.5×10−4) and post-flare (11 ± 1.6, p < 2.8×10−6) (Fig. 1A).
Fig. 1. Bacterial communities shift during AD disease progression.
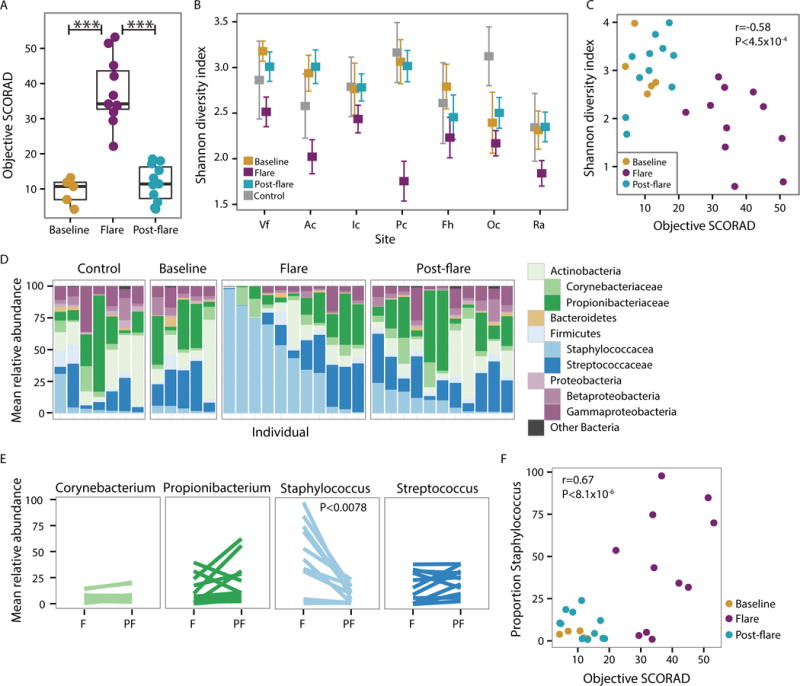
(A) Objective SCORAD for each patient at baseline, flare, and post-flare. Higher SCORAD corresponds to more severe disease. *** P<0.001 (B) Mean Shannon diversity +/− SEM in controls and AD disease states. Colors correspond to disease state. Volar forearm (Vf), antecubital crease (Ac), inguinal crease (Ic), popliteal crease (Pc), forehead (Fh), occiput (Oc), and retroauricular crease (Ra). (C) Shannon diversity versus objective SCORAD for combined antecubital (Ac) and popliteal creases (Pc) (AcPc) of AD patients. Partial correlation (adjusting for disease state). (D) Mean relative abundance of bacterial genera in AcPc for controls and AD disease states. (E) Mean relative abundance of predominant genera in AcPc for disease states, Flare (F) and Post-flare (PF). (F) Proportion of Staphylococcus versus objective SCORAD for AcPc of AD patients, partial correlation (adjusting for disease state).
To compare the microbial community composition across timepoints, we mapped microbial reads to a multi-kingdom reference database. As seen in healthy adults (16, 20, 21), bacteria was the most predominant kingdom across timepoints and body sites (fig. S2, Table S4), Malassezia species, particularly M. restricta and M. globosa, predominated the fungal communities (fig. S3, Table S5), and eukaryotic DNA viral communities were mostly polyomaviruses or papillomaviruses depending on the individual (fig. S4). No significant differences in the fungal or viral components over time were identified; therefore, we focused on bacterial communities that demonstrated the greatest shifts in this cohort (fig. S5, Table S6). We first determined the Shannon diversity index, an ecological measure of richness (total number of bacterial species) and evenness (relative proportion of the bacterial species), to evaluate the overall community structure/composition across body sites and timepoints. During flares, sites of AD predilection (Ac and Pc) exhibited a marked reduction in Shannon diversity compared to baseline, post-flare, and healthy controls, a trend observed to a lesser extent across other sites (Fig. 1B). Since changes in bacterial diversity were most pronounced at the sites of disease predilection and Ac/Pc have similar microbial communities (21), we averaged these sites per subject and used the composite “AcPc” for subsequent analyses. Similar to our previous analysis of microbial diversity in an AD patient cohort (6), the partial correlation between objective SCORAD and Shannon diversity, adjusting for disease state, was significantly inversely correlated (r = −0.58, p =4.5×10−4)(Fig. 1C), indicating that reduced skin bacterial diversity corresponds to worse disease severity, primarily at sites of disease predilection (fig. S5A).
To determine which taxa were contributing to the loss of diversity, we compared the relative abundances of the most prominent taxa (Fig. 1D and fig. S5B). Of the four most prominent genera in the AcPc, only Staphylococcus was significantly increased in flares (45 ± 10.2%) as compared to post-flares (9.2 ± 2.4%, p < 0.0078) and healthy controls (6.6 ± 4.1%, p < 0.033) (Fig. 1E). This increase in Staphylococcus relative abundances was positively correlated with objective SCORAD (r=0.67, p < 8.1×10−6)(Fig. 1F), indicating that severe AD was associated with higher staphylococcal relative abundances at sites of disease predilection. In addition, there was a positive correlation for the forehead, retroauricular crease, and volar forearm (fig. S5C), sites that can be affected in more severe disease. However, differences in Corynebacterium, Propionibacterium, and Streptococcus relative abundances between flares and post-flares were not statistically significant (Fig. 1E).
AD flare severity associated with specific staphylococcal species
To further examine the positive correlation between Staphylococcus and AD disease course (22), we identified the relative abundances of staphylococcal species including S. aureus, S. epidermidis, S. hominis, and S. capitis (Fig. 2A and fig. S6). Only relative abundances of S. aureus were significantly increased from flares (28 ± 8.8%) to post-flares (2.3 ± 0.8%, p < 0.014)(Fig. 2B). While S. epidermidis relative abundances were also higher during flares (13 ± 5.4%) as compared to post-flares (3.7 ± 1.4%), results did not reach statistical significance. For all patients, relative abundances of S. aureus were positively correlated with objective SCORAD (r = 0.73, p < 1.×10−7), while S. epidermidis was not correlated (Fig. 2C and fig. S7). This association between S.aureus and AD severity (23) has been observed in prior studies. Neither S. hominis nor S. capitis demonstrated significant shifts in relative abundances between timepoints (Fig. 2B) or were correlated with disease severity (fig. S7).
Fig. 2. Staphylococcal species increase during AD disease flare.
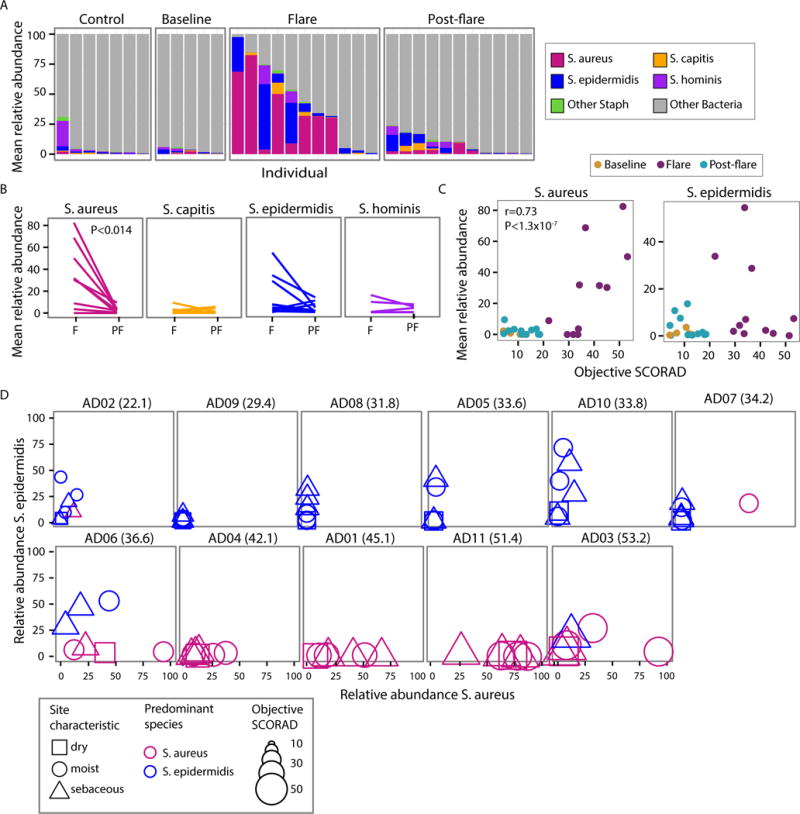
(A) Mean relative abundance of staphylococcal species within the total bacterial population in combined antecubital (Ac) and popliteal creases (Pc) (AcPc) of AD patients and controls. (B) Mean relative abundance of most abundant Staphylococcus species in AcPc for disease states, Flare (F) and Post-flare (Pf). (C) Correlation of S. aureus (left) and S. epidermidis (right) mean relative abundance and objective SCORAD for AcPc of patients, partial correlation (adjusting for disease state). (D) Comparison of S. aureus to S. epidermidis relative abundance by patient for all sites. Patient’s objective SCORAD indicated in parenthesis. Shape corresponds to physiological characteristic of the body site, color to the predominant species, and size to the magnitude of disease severity (objective SCORAD). Patients in the top row have a higher predominance of S. epidermidis, while bottom row patients are S. aureus-predominant.
To explore further the relationship between disease severity and staphylococcal species, we sorted the patients by their objective SCORAD and plotted the relative abundances of S. aureus and S. epidermidis at flare (Fig. 2D). We observed a trend whereby patients with more severe AD flares (objective SCORAD 45 ± 3.0) had higher relative abundances of S. aureus (Fig 2D bottom row, fig. S8, and Table S7). In contrast, patients with less severe AD flares, and lower objective SCORAD (31 ± 1.9, p < 0.004 in comparison to the more severe flares), had higher relative abundances of S. epidermidis (Fig. 2D top row) across sampled sites. Specifically, more severe AD flare patients had relative abundances of 34 ± 8.7% S. aureus with 7.4 ± 4.2% S. epidermidis and less severe AD flare patients had relative abundances of 3.8 ± 1.7% S. aureus with 13 ± 3.9% S. epidermidis averaged across all sites during flare. The range of S. aureus relative abundances based upon sequencing was variable: 3 of 11 patients had no S. aureus on their skin and 3 of 11 had relative abundances of S.aureus on their skin exceeding 50%, similar to prior studies of S. aureus relative abundances on AD skin.
To compare these metagenomic results with more traditional studies, we cultured bacteria from skin and nares swabs collected concurrently with genomic samples. Cultures of S. aureus from skin clinical samples correlated with the microorganism detection by sequencing. Of note, two less severe AD flare patients were culture-positive for S. aureus only in their nares, a common site of carriage. The S. aureus culture-positive rates in this cohort was consistent with other studies (6–8, 11–13). The genomic analyses were internally consistent with cultivation results and both supported the strong association between AD disease severity and S. aureus.
Monoclonal S. aureus strains observed in AD
While the differential association of S. aureus and S. epidermidis with AD severity defined an intriguing feature of disease heterogeneity, the underlying strain communities of these species during the disease course remained unknown. Two alternative scenarios could underlie microbial shifts in a disease flare: a) all strains equally increase in relative abundance or b) particular strain(s) bloom and drive the increase. This distinction is important as individual strains may exhibit functional differences. In previous studies, this question could not be addressed, as traditional typing and amplicon-based sequencing methods may differentiate clonal complexes but miss gene content and single nucleotide variant (SNV) variants (14, 15). In contrast, shotgun metagenomics provides resolution of microbial communities at a strain and SNV level (24). We used our previously validated strain-tracking approach to identify strains of S. aureus and S. epidermidis present on our AD patients (16, 21). For S. aureus strain-tracking, microbial reads were mapped against a database composed of 215 S. aureus genomes, of which 61 representatives are shown in Figure 3A.
Fig. 3. S. aureus-predominant individuals are often colonized with a single S. aureus strain.
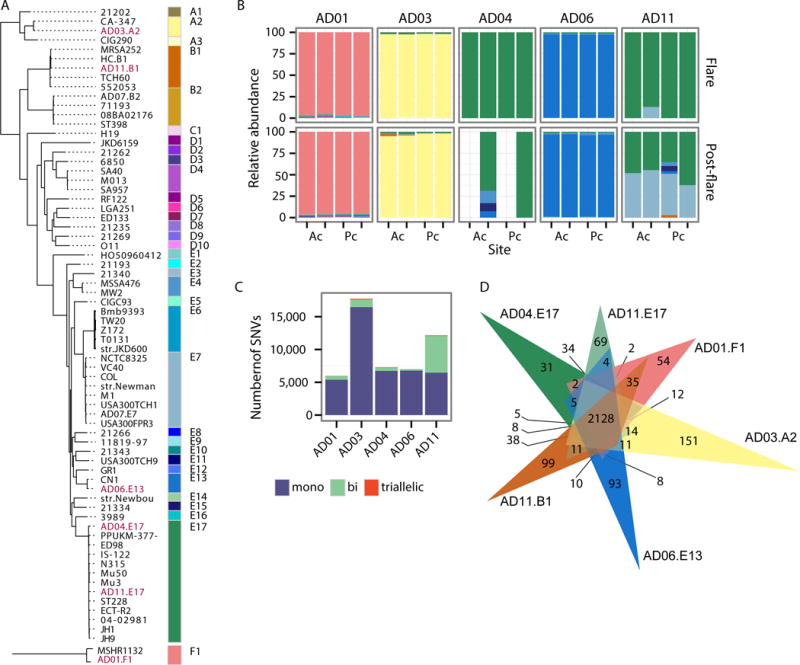
(A) Dendogram of 61 representative S. aureus strains based on SNVs in the core genome. Strains labeled in red were isolated from patients in (B). Colored blocks correspond to genomes of the same clade. Phylogenetically distant clade F1 is shown as an outgroup as it was recently reclassified as S. argenteus (32). (B) For S. aureus-predominant individuals, S. aureus clade relative abundances in bilateral antecubital (Ac) and popliteal creases (Pc) for AD disease states, flare and post-flare. Colors correspond to those in (A). (C) For combined samples of all sites/timepoints of individuals in (B), barcharts show the number of SNVs per individual that are mono, bi, and triallelic. (D) Venn diagram showing the number of genes shared between isolates from patients in (B), indicated in red in (A).
In contrast to the heterogenous communities of P. acnes and S. epidermidis strains observed in healthy adult skin (16, 21), the more severe AD patients were strikingly colonized with a single clade of S. aureus during disease flares (patients exhibited no clinical signs of infection) (Fig. 3B, fig. S9, and Table S8). In 4 of the 5 severe AD flare patients, this colonization with a single strain persisted in the post-flare but at notably lower mean relative abundances. AD patient AD11 was the exception, colonized by 3 different clades of S. aureus (E17, E7, and B1) with only clade E17 predominating during a flare. Notably, these more severe AD patients were colonized with distinct S. aureus clades. This supports previous studies demonstrating AD patients do not share a single dominant S. aureus clone (11, 25–27). The variation in the clonal S. aureus-clades colonizing AD patients raises the possibility that this heterogeneity may contribute to the differential course and/or therapeutic responses of AD patients.
To confirm our strain-tracking results, we used a complementary approach in which SNVs were identified in the S. aureus core genome (1.9 Mbps shared between all sequenced S. aureus). To power this analysis, we combined all sites and timepoints for each patient. In total, we identified 38,867 variant positions in the S. aureus core or ~10,000 SNPs per patient. We then used the degree of polyallelism in each individual to infer genetic heterogeneity or the presence of multiple S. aureus strains. We calculated the number of mono, bi, and triallelic SNVs for each patient (Fig. 3C). Consistent with strain-tracking results, SNVs in clonal S. aureus-colonized AD patients were monoallelic at 93% of sites, while heterogeneous patient AD11’s SNVs were monoallelic at only 53% of sites.
S. aureus isolates cultured from each of the more severe AD flare patients underwent whole genome sequencing to confirm that the cultured patient isolates grouped into the respective clades predicted by strain-tracking of the metagenomic data (Fig. 3A). Colony-picking from cultured swabs for patient AD11 isolated a representative from the dominant E17 and the non-dominant B1 clade. Based on standard sensitivity-testing methods and whole genome sequencing analysis, five of the six S. aureus isolates from the more severe AD patients were methicillin-sensitive S. aureus (MSSA), consistent with higher incidences of MSSA than methicillin-resistant S. aureus (MRSA) cultivated from AD patient skin (28–30).
Comparative genomic analysis of these six S. aureus strains revealed extensive heterogeneity in the gene content as predicted based on the initial mapping of the shotgun metagenomics sequences to disparate phylogenetic clades. The genome of a single S. aureus isolate encodeŝ2,500 genes of which ~85% (2128 genes) are present in every strain’s genome and constitute the functional core (Fig. 3D); the remaining ~300 genes derive from the flexible pangenome comprised of 1,020 genes. We looked for functional enrichment in noncore versus core genes to identify pathways that were the most variable between our isolates (Table S9). In doing so, we identified the KEGG pathways ko05150 Staphylococcus aureus infection, ko00906 Carotenoid biosynthesis, and ko01501 beta-Lactam resistance as functionally enriched in the variable component of the pangenome. With a targeted search, enterotoxin genes, previously shown to exacerbate AD (31), were differentially present in the 6 AD patient strains of S. aureus. The 5 genes in the carotenoid biosynthesis pathway were present in all genomes but AD01.F1; this isolate is most closely related to strain MSHR1132 that was recently reclassified as Staphylococcus argenteus and can be visually distinguished by its white versus yellow pigment (32). Finally, variability of genes in the beta-Lactam resistance family, including the mec cassette, was consistent with our previous result that only isolate AD11.E17 was an MRSA. Overall, this strain-level gene variation generates additional questions regarding the potential role of specific strains on disease pathogenesis and host factors on clonal strain selection.
Heterogeneous S. epidermidis strain communities
To further address the microbial community structure, we explored whether AD patients harbored heterogenous communities of S. epidermidis on skin. For S. epidermidis strain-tracking, microbial reads were mapped against a database composed of 61 sequenced, phylogenetically diverse S. epidermidis genomes (Fig. 4A). As seen with healthy adults (21) and children, AD patients’ S. epidermidis communities at both flares and post-flares were composed of multiple different strains from diverse clades of the phylogenetic tree (Fig. 4B. fig. S10, and Table S10). This directly contrasts with the identification of clonal S. aureus communities. This heterogenous S. epidermidis strain diversity was observed for both the more severe and less severe AD flare patients (fig. S10). However, analysis of the S. epidermidis strain composition in this cohort and our previous cohort of healthy adults (21) revealed a clustering of the less severe AD flare patients (Fig. 4C). Specifically, unsupervised clustering and principal coordinate analyses both identified S. epidermidis clades A29 and A30 as contributing to the clustering of the less severe AD patients and clade A20 as contributing to the clustering of the healthy adults (Fig. 4D). In contrast, the S. epidermidis strain diversity in healthy control children and more severe flare patients were intermixed.
Fig. 4. S. epidermidis-predominant individuals are colonized by a heterogenous community of S. epidermidis strains.
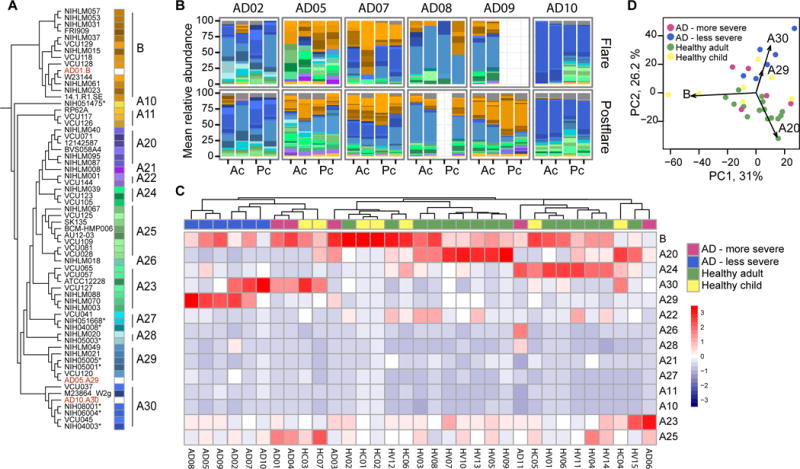
(A) Dendogram of S. epidermidis strains based on SNVs in the core genome. Strains isolated from patients in our study are labeled in red. Similar colors represent closely related strains that were grouped into 14 clades. Starred (*) isolates are nosocomial in origin (B) For S. epidermidis-predominant individuals, S. epidermidis strain relative abundances in combined antecubital (Ac) and popliteal creases (Pc) for AD disease states, flare and post-flare. Colors correspond to those in (A). (C) Heatmap shows mean relative abundance of each clade across all sites in S. aureus and S. epidermidis-predominant AD patients, healthy adults (HA), and healthy children (HC). (D) In principal component analysis, clades A20, A29, and A30 drive separation between S. epidermidis-predominant AD patients and healthy adults.
S. epidermidis clades A29 and A30 were enriched in strains originally collected from nosocomial infections rather than as skin commensals (33)(indicated with *s in Fig. 4A). Comparative genomic analysis of nosocomial isolates and the other strains revealed higher relative abundances of the SCCmec cassette (33), which encodes genes necessary for methicillin-resistance, in the nosocomial isolates. To further evaluate the S. epidermidis strains in this cohort, isolates were cultured from swabs collected from less severe patients AD05 and AD10. Whole genome sequencing confirmed the patient isolates as members of the A29 and A30 clade, respectively (Fig. 4A in red). Consistent with the trend of increased antibiotic resistance genes observed through genomic analysis, these patient isolates were methicillin-resistant S. epidermidis. A potential explanation for the overrepresentation of isolates genomically similarto nosocomial strains in less severe AD flare patients may be that these S. epidermidis strains outcompete commensals and/or S. aureus in inflammatory or non-steady-state conditions or that antibiotic usage in these patients may have selected for antibiotic resistance genes.
Strain-specific differences in cutaneous immune response in a murine model
While S. aureus has been tightly linked with AD, it is still debated whether S. aureus is a cause or effect, i.e. whether S. aureus can elicit and/or worsen AD skin disease or is a bystander that flourishes with increased access to extracellular matrix or other products of inflammation in eczematous skin (34, 35). Observing that individual strains of S. aureus predominated during AD flares in our more severe flare patients, we sought to investigate if these clonal strains elicited a biological response distinct from other strains of staphylococci. Harnessing the combined power of shotgun metagenomic sequencing of clinical samples and whole genome sequencing of bacteria cultivated from concurrently collected skin swabs, we next analyzed a) if strains associated with AD flares would be sufficient to elicit skin inflammation in the absence of any known genetic predisposition or prior barrier disruption and b) if there were strain-specific differences. To test this, we topically applied staphylococcal strains cultivated from AD patients and healthy controls onto intact skin of C57BL/6 wild-type mice with a method previously developed to test the immune response to skin commensals (Fig. 5A) We individually tested 10 phylogenetically distinct S. aureus isolates: six cultivated directly from the flared skin of patients with more severe flares, two from the skin of less severe patient AD07’s flare timepoint, one S. aureus from a healthy control, and a common pathogenic S. aureus USA300 FPR3757 isolate (highlighted in red in fig. S9A). In addition, we tested three S. epidermidis isolates from AD patients: a representative from the clades A29 and A30 which predominated in the skin of less severe AD patients, and a representative from the ubiquitous B clade (highlighted in red in fig. S10A). In contrast to the non-inflammatory responses observed following association with either skin commensals (36, 37) or AD patient S. epidermidis isolates, topical application of the S. aureus isolates, particularly those associated with more severe AD flare patients, was sufficient to induce epidermal thickening and inflammatory responses (Fig. 5B,C, fig. S11A) as well as immune cell infiltrate composed of neutrophils and eosinophils (Fig. 5D, fig. S11B,C). Interestingly, the USA300 isolate, commonly used as a representative S. aureus in functional experiments, induced only a modest immune response as compared to many of the isolates cultivated from severe AD patients, underscoring the importance of utilizing matched clinical isolates.
Fig. 5. Topical application of AD isolates induce AD-like immune responses in murine models.
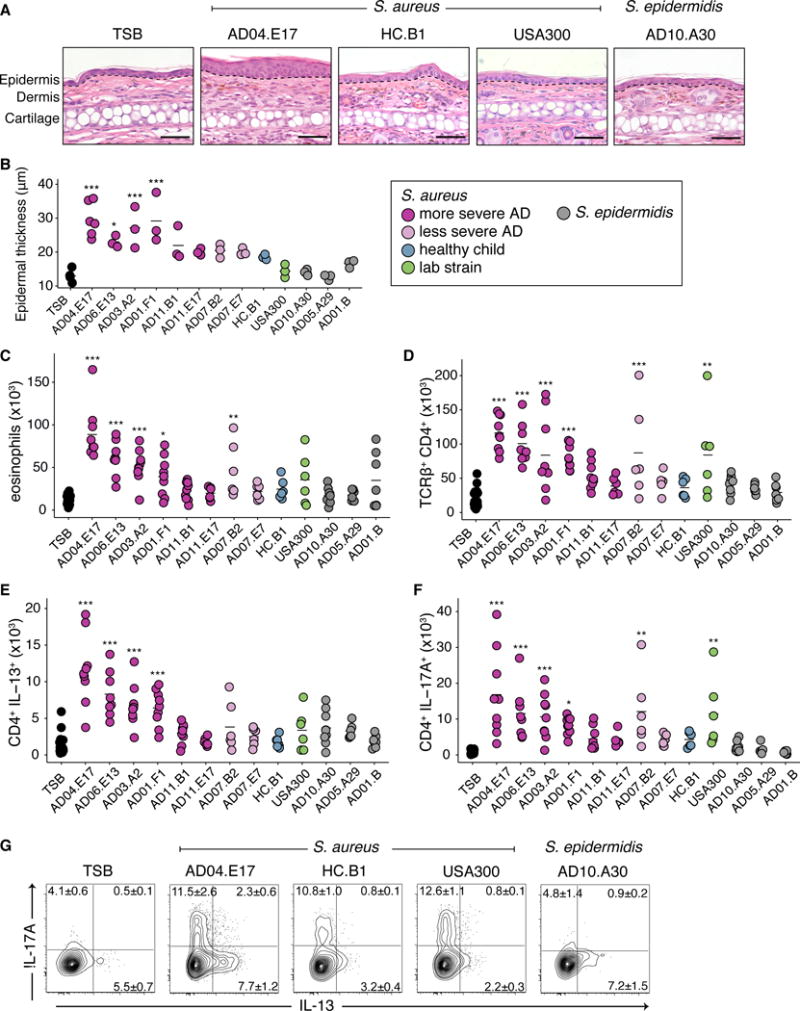
(A) Mice were topically associated with staphylococcal monocultures every other day 4 times before sacrifice on the 8th day. (B) Representative histological images of the ear pinnae of mice associated with tryptic soy broth TSB, S. aureus AD04.E17, HC.B1, USA300, or S. epidermidis A10.A30. Dotted line indicates separation between the epidermidis and dermis. Scale bar 50 μm. (C) Epidermal thickness of ears post topical association of patient AD isolates. Color indicates origin and species of the isolate. (D) Absolute numbers of skin eosinophil (E) Absolute numbers of skin TCRβ+ CD4+ cells. (F) Absolute numbers of skin IL-13+ CD4+ cells. (G) Absolute numbers of skin IL-17A+ CD4+ cells. (H) Frequencies of IL-13+ and IL-17A+ CD4+ cells from mice in (B). Results are cumulative data from 2 or 3 independent experiments, 3 mice per group. *p<0.05, **p<0.01, ***p<0.001 as calculated by ANOVA with multiple comparison correction.
In addition to innate immune cells, infiltration of T cell receptor (TCR) αβ+ and γδlow cells were also observed (fig. S12A) in mice colonized only with specific S. aureus strains. The majority of TCRβ+ cells were CD4+ (Fig. 5E) with variable effector potential, depending on the associated isolate. Notably, four S. aureus isolates from more severe AD flare patients induced production of the cytokine interleukin-13 (IL-13) (Fig. 5F), which is commonly associated with allergic inflammation. Cutaneous Th17 cells were also identified when mice were colonized with the four IL-13 inducing strains, in addition to AD07.B2 and USA300 (Fig. 5G). Recent reports have identified the presence of Th17 cells in AD lesions (38, 39), particularly in the Asian patient populations (40). Similar to CD4+ T cells, the γδ T cells of mice associated with specific strains of S. aureus isolates also had the potential to make higher levels of interleukin-17A (IL-17A) (fig. S12B). Notably, four of the S. aureus isolates [2 from more severe flare patient AD11 (AD11.B1, AD11.E17), 1 from less severe flare patient AD07 (AD07.E7), and the isolate from a healthy child (HC.B1)] induced minimal immune responses in all categories. Overall, association of S. aureus strains isolated from more severe AD flare patients to wild-type mice without prior barrier disruption induced immune responses in the skin that were significantly greater than those induced with S. epidermidis or S. aureus isolates from less severe AD flare patients or controls. Thus, these findings suggest specific strains of S. aureus may be sufficient to elicit and/or exacerbate skin inflammation as part of AD disease pathogenesis.
Discussion
AD is a complex disease with many contributing factors including skin barrier integrity, innate and adaptive immunity, and the microbiome. The heterogeneity of the course, severity, and clinical response in AD patients underscores the diversity of phenotypic presentations, as well as the probable differences in disease pathogenesis, within this one diagnosis. In addition to the various genetic susceptibility loci for AD, deeper investigation into the skin microbiome could provide better understanding of the microbial heterogeneity of AD and its potential contributions to disease.
While there have been many efforts to identify bacteria in AD skin, studies have generally relied on methods which do not distinguish microbes beyond the species-level or can misclassify genomically distinct clones (14, 15). Here, we combined shotgun metagenomic sequencing of clinical samples with whole genome sequencing of patient-derived isolates to investigate the microbial communities of AD skin down to the strain and SNV-level resolution. Since topical anti-inflammatory and anti-microbial treatments alter the skin microbiota (6, 7), the baseline and flare timepoints in this cohort were strictly defined by skin preparatory regimens to capture the natural history of the skin disease and to avoid potential confounders. As compared to healthy controls, the AD patients exhibited striking skin bacterial dysbiosis during flares. This dysbiosis was related to the increased relative abundance of staphylococci, consistent with prior cohorts. Based on the disease severity (defined by objective SCORAD) during flares, we observed a strong correlation between severe AD flares and S. aureus relative abundances. These findings demonstrated that despite the relatively small numbers of subjects in this study, our cohort of patients is representative of other published patient cohorts as defined by validated diagnostic criteria.
Shotgun metagenomic sequencing enables strain-level examination of microbes within the broad microbial community of bacteria, fungi, and viruses. Strain-tracking identified striking outgrowth of clonal S. aureus strains in the skin of flaring AD patients with more severe disease; these same strains persisted post-flare at lower relative abundances. Other methods have examined whether S. aureus expansion in the skin of AD flares was related to either proportional increases in the entire community of S. aureus strains or the increase of a single or a few dominant clones; however, these studies were limited by the inability to examine these possibilities in the context of the whole skin microbial community. While the fungal and viral communities were not significantly different in this study, expansion of reference databases/genomes and studies into the microbial ‘dark matter’ in metagenomic data may provide further insights into AD microbiota. Our findings demonstrate that AD skin flares in patients with more severe disease are tightly linked with clonal S. aureus isolates.
In addition to characterizing strain communities during the course of AD, we found that less severe AD patients were colonized with more methicillin-resistant strains while the more severe AD patients were primarily colonized with methicillin-sensitive strains. While methicillin resistance is not as common in AD as would be predicted based on the high rates of S. aureus colonization in this disease, the finding of MSSA and MRSE predominance may contribute to differential responses to therapies in AD patients (41). The contrasts between S. aureus and S. epidermidis observed in this study likely also relate to the differences in microbial genetics and population dynamics at both the species and strain levels. Additional investigations of these microbiome phenotypic differences may improve the understanding of AD pathogenesis and lead to more targeted therapeutics. Birth cohort studies may address whether these patients acquired bacterial strains from family members and/or environmental sources as part of microbial inheritance (42). Testing of S. aureus strains in gnotobiotic mice, similar to Bacteroides gut commensal studies, may functionally address whether colonization by clonal S. aureus occurs through limited exposure or colonization resistance (43).
Using strains isolated from the skin of AD flares and a healthy control as well as a known laboratory strain, we examined the potential biological differences between staphylococcal strains. In a murine model without prior skin barrier disruption and with intact immunity, S. aureus strains from flare timepoints in more severe AD patients were sufficient to induce manifestations of skin inflammation, such as epidermal thickening and cutaneous infiltration of Th2 and Th17 cells. The magnitude of different immunologic effects varied depending on the isolated strain but was not strictly related to the disease severity of the source patient. Notably, murine colonization with either isolate AD11.B1 or AD11.E17 induced minimal immune responses even though patient AD11 has an objective SCORAD of 51.4. However, AD11 is also heterozygous for a null mutation in the FLG gene (S757X), suggesting that AD11’s strains of S. aureus may be immunogenic in the setting of an impaired skin barrier, which previous studies have shown allows S. aureus to breach the epidermis into the dermis where it can trigger expression of proinflammatory cytokines (44). Caveats of these findings in the murine model are the relatively small number of isolates from this cohort that were fully sequenced and studied in the murine model and the observation of varied host responses when testing isolates from the same clade (AD04 and AD11), highlighting the need to examine a larger number of isolates including strains from similar and different clades and from healthy individuals and AD patients. An important additional limitation is the recognition that this murine model as well as others do not recapitulate the multiple complexities of human AD.
In mouse models, S. aureus enterotoxins have been shown to act as superantigens that can initiate Th17 responses (45), while S. aureus δ-toxin can induce degranulation of mast cells (46). These genes were both present in the non-inflammatory S. aureus isolates indicating strain-variability exists not only in gene content but also gene expression. Since healthy control-associated S. aureus strains were limited in our cohort due to the small percentage of healthy individuals colonized with S. aureus, future studies with additional S. aureus isolates from healthy individuals are necessary to tease apart the mechanisms underlying functional differences between S. aureus strains. In the context of prior studies demonstrating cutaneous immunologic responses to skin commensals (36, 37) and exacerbation of eczematous skin in AD mouse models by S. aureus (36, 37, 46, 47), our findings demonstrate that staphylococcal strains may play an important role in AD disease progression in a strain-specific manner.
In this study, we used shotgun metagenomic sequencing to examine strain-level microbial compositions of AD skin coupled with whole genome sequencing of patient isolates. With increasing recognition of highly individualized skin microbiomes (16), the presence of patient-specific strains underscores the individuality of the disease course and therapeutic response and may represent an opportunity for precision medicine. Our functional studies with cutaneous colonization of AD patient-associated strains of S. aureus and S. epidermidis demonstrated strain-specific differences in the ability to elicit histologic and immunologic alterations. AD typically has an age of onset in the first year of life when the human immune system is developing and being tuned by the endogenous microbial community. Recent studies have shown that early exposures can modulate host immunity to subsequent exposure and induce tolerance (48, 49). Thus, in light of the known links between severe AD and subsequent development of asthma and hay fever (“the atopic march”), targeted modulation of an AD patient’s particular staphylococcal strains has the potential to ameliorate the broader development of atopic disorders.
Supplementary Material
One Sentence Summary.
Genomic and functional analyses of staphylococcal strain specificity reveal roles for microbes in human atopic dermatitis skin pathogenesis.
Acknowledgments
We thank Pamela Thomas, Morgan Park, Elizabeth A. Kennedy, Sheila Phang, Amynah Pradhan, Valentina S. Pillai, Irina Bozhenko (Harris Corporation employee working under contract with NCI) and Kimberly Beacht for underlying efforts; Segre lab, Mark C. Udey, and Keisuke Nagao for helpful discussions. This study utilized the high-performance computational capabilities of the NIH Biowulf Linux cluster. IGS Analysis Engine at University of Maryland School of Medicine provided structural and functional annotation of genomes.
Funding: Supported by NHGRI, NCI and NIAID Intramural Research Programs. Sequencing was funded by grants from National Institutes of Health (4UH3AR057504-02).
Footnotes
Author contributions: A.L.B., Y.B, J.A.S. and H.H.K. designed study and drafted manuscript. Sequencing was carried out by NISC. A.L.B. analyzed microbial sequence data. C.D., S.K.B.C., O.J.H., S.C., W.N. performed experiments.
Data and materials availability: The sequencing data for this study are linked to NCBI Bioproject 46333.
References
- 1.Eyerich K, Eyerich S, Biedermann T. The Multi-Modal Immune Pathogenesis of Atopic Eczema. Trends in immunology. 2015;36:788–801. doi: 10.1016/j.it.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 2.Bantz SK, Zhu Z, Zheng T. The Atopic March: Progression from Atopic Dermatitis to Allergic Rhinitis and Asthma. Journal of clinical & cellular immunology. 2014;5 doi: 10.4172/2155-9899.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.E. A. Genetics, C. Lifecourse Epidemiology Eczema, C. Australian Asthma Genetics, A. Australian Asthma Genetics Consortium. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nature genetics. 2015;47:1449–1456. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, Goudie DR, Sandilands A, Campbell LE, Smith FJ, O’Regan GM, Watson RM, Cecil JE, Bale SJ, Compton JG, DiGiovanna JJ, Fleckman P, Lewis-Jones S, Arseculeratne G, Sergeant A, Munro CS, El Houate B, McElreavey K, Halkjaer LB, Bisgaard H, Mukhopadhyay S, McLean WH. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nature genetics. 2006;38:441–446. doi: 10.1038/ng1767. [DOI] [PubMed] [Google Scholar]
- 5.Huang JT, Abrams M, Tlougan B, Rademaker A, Paller AS. Treatment of Staphylococcus aureus colonization in atopic dermatitis decreases disease severity. Pediatrics. 2009;123:e808–814. doi: 10.1542/peds.2008-2217. [DOI] [PubMed] [Google Scholar]
- 6.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD, Program NCS, Murray PR, Turner ML, Segre JA. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome research. 2012;22:850–859. doi: 10.1101/gr.131029.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez ME, Schaffer JV, Orlow SJ, Gao Z, Li H, Alekseyenko AV, Blaser MJ. Cutaneous microbiome effects of fluticasone propionate cream and adjunctive bleach baths in childhood atopic dermatitis. Journal of the American Academy of Dermatology. 2016;75:481–493 e488. doi: 10.1016/j.jaad.2016.04.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leyden JJ, Marples RR, Kligman AM. Staphylococcus aureus in the lesions of atopic dermatitis. The British journal of dermatology. 1974;90:525–530. doi: 10.1111/j.1365-2133.1974.tb06447.x. [DOI] [PubMed] [Google Scholar]
- 9.Chiu LS, Chow VC, Ling JM, Hon KL. Staphylococcus aureus carriage in the anterior nares of close contacts of patients with atopic dermatitis. Archives of dermatology. 2010;146:748–752. doi: 10.1001/archdermatol.2010.129. [DOI] [PubMed] [Google Scholar]
- 10.Bonness S, Szekat C, Novak N, Bierbaum G. Pulsed-field gel electrophoresis of Staphylococcus aureus isolates from atopic patients revealing presence of similar strains in isolates from children and their parents. Journal of clinical microbiology. 2008;46:456–461. doi: 10.1128/JCM.01734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DW, Park JY, Park KD, Kim TH, Lee WJ, Lee SJ, Kim J. Are there predominant strains and toxins of Staphylococcus aureus in atopic dermatitis patients? Genotypic characterization and toxin determination of S. aureus isolated in adolescent and adult patients with atopic dermatitis. The Journal of dermatology. 2009;36:75–81. doi: 10.1111/j.1346-8138.2009.00592.x. [DOI] [PubMed] [Google Scholar]
- 12.Seite S, Flores GE, Henley JB, Martin R, Zelenkova H, Aguilar L, Fierer N. Microbiome of affected and unaffected skin of patients with atopic dermatitis before and after emollient treatment. Journal of drugs in dermatology: JDD. 2014;13:1365–1372. [PubMed] [Google Scholar]
- 13.Drago L, De Grandi R, Altomare G, Pigatto P, Rossi O, Toscano M. Skin microbiota of first cousins affected by psoriasis and atopic dermatitis. Clinical and molecular allergy: CMA. 2016;14:2. doi: 10.1186/s12948-016-0038-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salipante SJ, SenGupta DJ, Cummings LA, Land TA, Hoogestraat DR, Cookson BT. Application of whole-genome sequencing for bacterial strain typing in molecular epidemiology. Journal of clinical microbiology. 2015;53:1072–1079. doi: 10.1128/JCM.03385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ugolotti E, Larghero P, Vanni I, Bandettini R, Tripodi G, Melioli G, Di Marco E, Raso A, Biassoni R. Whole-genome sequencing as standard practice for the analysis of clonality in outbreaks of meticillin-resistant Staphylococcus aureus in a paediatric setting. The Journal of hospital infection. 2016;93:375–381. doi: 10.1016/j.jhin.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 16.Oh J, Byrd AL, Park M, Program NCS, Kong HH, Segre JA. Temporal stability of the human skin microbiome. Cell. 2016;165:854–866. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams HC, Burney PG, Strachan D, Hay RJ. The U.K. Working Party’s Diagnostic Criteria for Atopic Dermatitis. II. Observer variation of clinical diagnosis and signs of atopic dermatitis. The British journal of dermatology. 1994;131:397–405. doi: 10.1111/j.1365-2133.1994.tb08531.x. [DOI] [PubMed] [Google Scholar]
- 18.Kunz B, Oranje AP, Labreze L, Stalder JF, Ring J, Taieb A. Clinical validation and guidelines for the SCORAD index: consensus report of the European Task Force on Atopic Dermatitis. Dermatology. 1997;195:10–19. doi: 10.1159/000245677. [DOI] [PubMed] [Google Scholar]
- 19.Oranje AP, Glazenburg EJ, Wolkerstorfer A, de Waard-van der Spek FB. Practical issues on interpretation of scoring atopic dermatitis: the SCORAD index, objective SCORAD and the three-item severity score. The British journal of dermatology. 2007;157:645–648. doi: 10.1111/j.1365-2133.2007.08112.x. [DOI] [PubMed] [Google Scholar]
- 20.Findley K, Oh J, Yang J, Conlan S, Deming C, Meyer JA, Schoenfeld D, Nomicos E, Park M, N. I. H. I. S. C. C. S. Program. Kong HH, Segre JA. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, Segre JA. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514:59–64. doi: 10.1038/nature13786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Current allergy and asthma reports. 2015;15:65. doi: 10.1007/s11882-015-0567-4. [DOI] [PubMed] [Google Scholar]
- 23.Tauber M, Balica S, Hsu CY, Jean-Decoster C, Lauze C, Redoules D, Viode C, Schmitt AM, Serre G, Simon M, Paul CF. Staphylococcus aureus density on lesional and nonlesional skin is strongly associated with disease severity in atopic dermatitis. The Journal of allergy and clinical immunology. 2016;137:1272–1274. e1271–1273. doi: 10.1016/j.jaci.2015.07.052. [DOI] [PubMed] [Google Scholar]
- 24.Chng KR, Tay AS, Li C, Ng AH, Wang J, Suri BK, Matta SA, McGovern N, Janela B, Wong XF, Sio YY, Au BV, Wilm A, De Sessions PF, Lim TC, Tang MB, Ginhoux F, Connolly JE, Lane EB, Chew FT, Common JE, Nagarajan N. Whole metagenome profiling reveals skin microbiome-dependent susceptibility to atopic dermatitis flare. Nature microbiology. 2016;1:16106. doi: 10.1038/nmicrobiol.2016.106. [DOI] [PubMed] [Google Scholar]
- 25.Yeung M, Balma-Mena A, Shear N, Simor A, Pope E, Walsh S, McGavin MJ. Identification of major clonal complexes and toxin producing strains among Staphylococcus aureus associated with atopic dermatitis. Microbes and infection/Institut Pasteur. 2011;13:189–197. doi: 10.1016/j.micinf.2010.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Lomholt H, Andersen KE, Kilian M. Staphylococcus aureus clonal dynamics and virulence factors in children with atopic dermatitis. The Journal of investigative dermatology. 2005;125:977–982. doi: 10.1111/j.0022-202X.2005.23916.x. [DOI] [PubMed] [Google Scholar]
- 27.Hoeger PH, Lenz W, Boutonnier A, Fournier JM. Staphylococcal skin colonization in children with atopic dermatitis: prevalence, persistence, and transmission of toxigenic and nontoxigenic strains. The Journal of infectious diseases. 1992;165:1064–1068. doi: 10.1093/infdis/165.6.1064. [DOI] [PubMed] [Google Scholar]
- 28.Suh L, Coffin S, Leckerman KH, Gelfand JM, Honig PJ, Yan AC. Methicillin-resistant Staphylococcus aureus colonization in children with atopic dermatitis. Pediatric dermatology. 2008;25:528–534. doi: 10.1111/j.1525-1470.2008.00768.x. [DOI] [PubMed] [Google Scholar]
- 29.Hsiang MS, Shiau R, Nadle J, Chan L, Lee B, Chambers HF, Pan E. Epidemiologic Similarities in Pediatric Community-Associated Methicillin-Resistant and Methicillin-Sensitive Staphylococcus aureus in the San Francisco Bay Area. Journal of the Pediatric Infectious Diseases Society. 2012;1:200–211. doi: 10.1093/jpids/pis061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaptini C, Quinn S, Marshman G. Methicillin-resistant Staphylococcus aureus in children with atopic dermatitis from 1999 to 2014: A longitudinal study. The Australasian journal of dermatology. 2015 doi: 10.1111/ajd.12371. [DOI] [PubMed] [Google Scholar]
- 31.Strange P, Skov L, Lisby S, Nielsen PL, Baadsgaard O. Staphylococcal enterotoxin B applied on intact normal and intact atopic skin induces dermatitis. Archives of dermatology. 1996;132:27–33. [PubMed] [Google Scholar]
- 32.Tong SY, Schaumburg F, Ellington MJ, Corander J, Pichon B, Leendertz F, Bentley SD, Parkhill J, Holt DC, Peters G, Giffard PM. Novel staphylococcal species that form part of a Staphylococcus aureus-related complex: the non-pigmented Staphylococcus argenteus sp. nov. and the nonhuman primate-associated Staphylococcus schweitzeri sp. nov. International journal of systematic and evolutionary microbiology. 2015;65:15–22. doi: 10.1099/ijs.0.062752-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conlan S, Mijares LA, Program NCS, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Otto M, Kong HH, Murray PR, Segre JA. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome biology. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho SH, Strickland I, Boguniewicz M, Leung DY. Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. The Journal of allergy and clinical immunology. 2001;108:269–274. doi: 10.1067/mai.2001.117455. [DOI] [PubMed] [Google Scholar]
- 35.Kuusela P. Fibronectin binds to Staphylococcus aureus. Nature. 1978;276:718–720. doi: 10.1038/276718a0. [DOI] [PubMed] [Google Scholar]
- 36.Naik S, Bouladoux N, Wilhelm C, Molloy MJ, Salcedo R, Kastenmuller W, Deming C, Quinones M, Koo L, Conlan S, Spencer S, Hall JA, Dzutsev A, Kong H, Campbell DJ, Trinchieri G, Segre JA, Belkaid Y. Compartmentalized control of skin immunity by resident commensals. Science. 2012;337:1115–1119. doi: 10.1126/science.1225152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naik S, Bouladoux N, Linehan JL, Han SJ, Harrison OJ, Wilhelm C, Conlan S, Himmelfarb S, Byrd AL, Deming C, Quinones M, Brenchley JM, Kong HH, Tussiwand R, Murphy KM, Merad M, Segre JA, Belkaid Y. Commensal-dendritic-cell interaction specifies a unique protective skin immune signature. Nature. 2015;520:104–108. doi: 10.1038/nature14052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suarez-Farinas M, Dhingra N, Gittler J, Shemer A, Cardinale I, de Guzman Strong C, Krueger JG, Guttman-Yassky E. Intrinsic atopic dermatitis shows similar TH2 and higher TH17 immune activation compared with extrinsic atopic dermatitis. The Journal of allergy and clinical immunology. 2013;132:361–370. doi: 10.1016/j.jaci.2013.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koga C, Kabashima K, Shiraishi N, Kobayashi M, Tokura Y. Possible pathogenic role of Th17 cells for atopic dermatitis. The Journal of investigative dermatology. 2008;128:2625–2630. doi: 10.1038/jid.2008.111. [DOI] [PubMed] [Google Scholar]
- 40.Noda S, Suarez-Farinas M, Ungar B, Kim SJ, de Guzman Strong C, Xu H, Peng X, Estrada YD, Nakajima S, Honda T, Shin JU, Lee H, Krueger JG, Lee KH, Kabashima K, Guttman-Yassky E. The Asian atopic dermatitis phenotype combines features of atopic dermatitis and psoriasis with increased TH17 polarization. The Journal of allergy and clinical immunology. 2015;136:1254–1264. doi: 10.1016/j.jaci.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 41.Bath-Hextall FJ, Birnie AJ, Ravenscroft JC, Williams HC. Interventions to reduce Staphylococcus aureus in the management of atopic eczema: an updated Cochrane review. The British journal of dermatology. 2010;163:12–26. doi: 10.1111/j.1365-2133.2010.09743.x. [DOI] [PubMed] [Google Scholar]
- 42.Faith JJ, Colombel JF, Gordon JI. Identifying strains that contribute to complex diseases through the study of microbial inheritance. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:633–640. doi: 10.1073/pnas.1418781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakatsuji T, Chen TH, Two AM, Chun KA, Narala S, Geha RS, Hata TR, Gallo RL. Staphylococcus aureus exploits epidermal barrier defects in atopic dermatitis to trigger cytokine expression. The Journal of investigative dermatology. 2016 doi: 10.1016/j.jid.2016.05.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Macias ES, Pereira FA, Rietkerk W, Safai B. Superantigens in dermatology. Journal of the American Academy of Dermatology. 2011;64:455–472. doi: 10.1016/j.jaad.2010.03.044. quiz 473–454. [DOI] [PubMed] [Google Scholar]
- 46.Nakamura Y, Oscherwitz J, Cease KB, Chan SM, Munoz-Planillo R, Hasegawa M, Villaruz AE, Cheung GY, McGavin MJ, Travers JB, Otto M, Inohara N, Nunez G. Staphylococcus delta-toxin induces allergic skin disease by activating mast cells. Nature. 2013;503:397–401. doi: 10.1038/nature12655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, Kong HH, Amagai M, Nagao K. Dysbiosis and Staphylococcus aureus Colonization Drives Inflammation in Atopic Dermatitis. Immunity. 2015;42:756–766. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scharschmidt TC, Vasquez KS, Truong HA, Gearty SV, Pauli ML, Nosbaum A, Gratz IK, Otto M, Moon JJ, Liese J, Abbas AK, Fischbach MA, Rosenblum MD. A Wave of Regulatory T Cells into Neonatal Skin Mediates Tolerance to Commensal Microbes. Immunity. 2015;43:1011–1021. doi: 10.1016/j.immuni.2015.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Du Toit G, Roberts G, Sayre PH, Bahnson HT, Radulovic S, Santos AF, Brough HA, Phippard D, Basting M, Feeney M, Turcanu V, Sever ML, Gomez Lorenzo M, Plaut M, Lack G, L. S. Team Randomized trial of peanut consumption in infants at risk for peanut allergy. The New England journal of medicine. 2015;372:803–813. doi: 10.1056/NEJMoa1414850. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


