Abstract
Exosomes are extracellular vesicles of endocytic origin, which function in intercellular communication. Our previous studies indicate that exosomes released from M. tuberculosis infected macrophages contain soluble mycobacterial proteins. However, it was unclear how these secreted proteins were targeted to exosomes. In this study we determined that exosome production by the murine macrophage cell line RAW264.7 requires the endosomal sorting complexes required for transport (ESCRT) and that trafficking of mycobacterial proteins from phagocytosed bacilli to exosomes was dependent on protein ubiquitination. Moreover, soluble mycobacterial proteins when added exogenously to RAW264.7 or human HEK 293 cells were endocytosed, ubiquitinated and released via exosomes. This suggested that endocytosed proteins could be recycled from cells through exosomes. This hypothesis was supported using the tumor–associated protein He4 which when endocytosed by RAW264.7 or HEK 293 cells was transported to exosomes in an ubiquitin-dependent manner. Our data suggest that ubiquitination is a modification sufficient for trafficking soluble proteins within the phagocytic/endocytic network to exosomes.
Introduction
Tuberculosis (TB) is a leading causes of death globally, with an estimated 1.4 million deaths annually. Mycobacterium tuberculosis (M.tb), the causative agent of TB, is an intracellular pathogen that primarily infects macrophages. Despite its intracellular “lifestyle,” M. tuberculosis, and other members of the mycobacterial species, shed and release bacterial components which gain access to the immune system (1). Evidence suggests that one mechanism which facilitates the release of mycobacterial proteins into the extracellular milieu is through exosomes (2) (3). Exosomes are membrane-bound vesicles of endocytic origin that are 30–100nm in sizes which have been shown to function in intercellular communication, immune cell activation, and serve as a source of disease biomarkers. In the context of an M.tb infection, exosomes have been shown to carry mycobacterial components including over 40 bacterial proteins many of which are known immuno-dominate antigens (4). Moreover, these exosomes when used as a vaccine can protect mice against an aerosolized infection (5). Exosomes carrying various pathogen- or cancer-derived antigens were also found to elicit a protective immune response (6). While much of the published work has characterized the host T cell response to antigenic proteins present within exosomes, we know little about the mechanism by which these proteins are targeted to exosomes.
Exosome biogenesis begins with the invagination of endosomal membranes to form intraluminal vesicles (ILV). While most late endosomes, or multivesicular bodies (MVB), fuse with lysosomes for degradation, some traffic to the plasma membrane, which upon fusion with the membrane, release the intraluminal vesicles as exosomes. Despite their discovery nearly 30 years ago, the mechanism for biogenesis is still not well understood. Evidence suggest that biogenesis may vary within a cell and between cell types (7) (8) (9). To date, several models have been suggested in mediating the formation of MVBs and sorting of protein cargo into the ILVs including the endosomal sorting complex required for transport (ESCRT) machinery (10) (11) (12) (13). Two such protein complexes, ESCRT-0 and ESCRT-1 have been shown to be involved in MVB protein sorting and loading in dendritic and neuroglia cells as well as other cell types. Protein sorting by ESCRT-0 and ESCRT-1 is mediated by a ubiquitin-interacting motifs, that are found on the Hrs and Tsg101 subunits, which constitute part of ESCRT-0 and ESCRT-1 complexes respectively. Through its ubiquitin interacting domains, ESCRT-0 clusters ubiquitinated proteins for delivery into MVBs (14). ESCRT-0 subsequently recruits ESCRT-1 to the endosomal membrane, which in turn recruits the remaining members of the ESCRT machinery; ESCRT-II and ESCRT-III (15) (16). It is believed that filaments formed by ESCRT-III ultimately promote invagination of the membrane and IVL formation. While a general model for exosome biogenesis through the ESCRT proteins has been elucidated, identifying specific proteins trafficked to exosomes through this mechanism has been difficult, particularly for non-membrane proteins.
Previously we reported that of the mycobacterial proteins identified on exosomes, the vast majority were experimentally shown to be secreted, soluble proteins (4). Interestingly, macrophages treated with culture filtrate protein (CFP), which contains mycobacterial proteins secreted or shed in culture, release exosomes containing many of the same mycobacterial proteins that are on exosomes following a M. tuberculosis infection. This finding suggests that these soluble mycobacterial proteins have the necessary “signal” to be trafficked to the MVB during exosome biogenesis independent of their entry mechanism into macrophages. The data presented in this study suggest that ubiquitination of both mycobacterial- as well as host-derived soluble proteins within the phagocytic/endocytic network is an important mechanism for their trafficking to MVBs and onto exosomes. It also suggest that endocytosed proteins’ can be “recycled” back to the extracellular environment by incorporation into exosomes.
Material and Methods
Bacterial strains and media
The mouse macrophage cell line RAW264.7 and human HEK293 cells was maintained in DMEM supplemented with 10% FBS, 10 mM sodium pyruvate, and 25 mM HEPES. M. tuberculosis H37Rv was grown in Middlebrook 7H9 broth supplemented with oleic albumin dextrose catalase until mid-logarithmic growth phase and frozen down as stocks in growth media plus 15% glycerol. Prior to use, the bacterial stocks were thawed and the mycobacteria were de-clumped by a brief sonication and passed through a syringe fitted with a 27-gauge needle at least 10 times.
siRNA transfections in RAW mouse macrophages
5×105 RAW 264.7 cells were cultured overnight in 6-well plates in DMEM supplemented with 10% FCS. Prior to transfection, cells were washed three times with 1X PBS and replenished with DMEM containing exosome-depleted FCS, produced by spinning the FCS at 100,000×g for 1 hour to remove exosomes. The transfection mix consisted of 75nM of target siRNA in 100uL of DMEM containing 10% HiPerfect (Qiagen). siRNA for Tsg101, Hrs, Park2 and control used were siGENOME SMARTpools (Dharmacon). The oligonucleotides used for the knockdowns are as follows. Tsg101 primers: 5′-CCGCUUAGAUCAAGAAGUAUU-3′, 5′-CGUAAACAGUUCCAGCUAAUU-3′, 5′-UACAAUCCCAGUGCGUUAUU-3′, 5′-UGUCAUCGCUAUGUACAAAUU-3′. Hrs primers: 5′-AGAGACAAGUGGAGGUAA-3′, 5′-UUUACCUCCACUUGUCUC-3′, 5′-GCACGUCUUUCCAGAAUUCAA-3′, 5′-ACAAGAACCCACACGUC-3′. Parkin primers: 5′-GGACUACAUGAUUCGACGUCAACUG-3′, 5′-GGAAACAUCAGUAGCUUUGCACCUG-3′, 5′-UUGCUUAGACUGUUUCCACUUAUAC-3′, 5′-GAUGACUAAACCUGACAGAA-3′. Knockdown was confirmed by western blot using 10ug of whole cell lysate.
M. tuberculosis H37Rv infection or CFP treatment of RAW 264.7 cells and isolation of exosomes
Confluent layer of RAW264.7 cells were infected with M. tuberculosis H37Rv or left untreated. Prior to infection, the bacteria were complement opsonized using normal horse serum for 2 hours and infected at a MOI that achieved 80% infectivity as determined by uptake assay (17) The RAW264.7 macrophages were infected with bacteria for 4 hours before being washed three times with 1X PBS. The cells were cultured in DMEM supplemented with 10% exosomes-depleted FCS. Cell culture supernatants were harvested at 72 hours post infection for exosomes isolation. For CFP treatment, RAW264.7 cells were seeded in Ti-175 tissue culture flasks and treated with CFP (20 mg/mL) in 20 mL of DMEM supplemented with 10% exosome-depleted FCS. After 16h, culture supernatant was harvested for exosome purification. CFP was purchased from BEI Resources (Manassas, VA) and was original made by Karen Dobos, Colorado State University as described in the product sheet (catalog # NR-14825).
For exosomes isolation, culture supernatants were centrifuged at 300 × g for 10 min at 4°C to remove debris. Cleared culture supernatants were filtered through 0.22-μm polyethersulfone filters (Nalgene). Filtered supernatants were centrifuged at 10,000 × g for 30 min at 4°C and again at 100,000 × g for 1 h to pellet the exosome-enriched vesicle population.
Sucrose gradient prepared exosomes
Raw 267.4 cells were infected with Mycobacterium smegmatis expressing wild-type M.tb HspX or K85R M.tb HspX for 72 hours. Cell culture supernatants were centrifuged at 300 × g for 10 min at 4°C to remove debris. Cleared culture supernatants were filtered through 0.22-μm polyethersulfone filters (Nalgene). Filtered supernatants were centrifuged at 10,000 × g for 30 min at 4°C and again at 100,000 × g for 1 h to pellet the exosomes. Exosomes were purified on linear sucrose gradient. The exosomes formed a distinct ring based on their density (1.13 and 1.18 g/ml) and were carefully recovered from the gradient. In the absence of a distinct ring, gradient fractions 5, 6, and 7 were collected, pooled, and washed in 1X PBS.
Western Blotting and antibodies
Exosomes (10 μg) were resuspended in 1X PBS with protease inhibitors. The suspension was mixed with Laemmli buffer, heated at 95°C for 5 min, and chilled on ice for 5 min before loading onto SDS gel. Whole cell lysates were prepared in RIPA buffer. Suspension was mixed with Laemmli buffer, heated at 95°C for 5 min. before loading onto SDS gel. Immunoblots probed with antibodies for proteins: ubiquitin (P4D1, 1:1000, Santa Cruz), Tsg101 (C-2, 1:1000, Santa Cruz), Hrs (V-20, 1:500, Santa Cruz), Tubulin (T5293, Sigma), CFP (C192, 1:1000, ATCC), Kat-G (IT-42, 1:20, ATCC), His (1:500, Santa Cruz), GroES (SA-12, 1:20, ATCC), HspX (IT-20, 1:15, ATCC), Park2 (ARP43038, 1:500, Aviva). Primary antibody incubation was followed with HRP-conjugated secondary antibodies (1:25,000, Pierce) and detected using enhanced chemiluminecence kit (Pierce).
Immunoprecipitation of ubquitinated proteins
In brief, ubiquitin antibody (P4D1) was conjugated to Separose A/G beads. Conjugation mixture containing: 2μg antibody, 40μL of Sepharose A/G beads (Santa Cruze), and 450 μL of 1xPBS containing 1% MPI was incubated for 1 hour at 4°C. Beads were centrifuged at 1500 × G for 5 minute to remove supernatant. Beads were blocked at 4°C for 3 hours with cold 1% BSA in 1X PBS containing MPI. Beads were centrifuged at 1500 × g for 5 minutes to remove supernatant. Exosomes were lysed by incubating with blocking buffer containing 0.1% Triton-X100. Exosome lysate (50 μg) were added to beads and incubated overnight at 4°C. Supernatant was removed and beads washed three times in 1X PBS containing MPI. Ubiquitinated proteins were removed from beads by heating for 5 minutes at 95°C and analyzed by western blot.
Plasmids and Constructs
All primers for are listed below. Wild-Type HspX Forward Primer: 5′-CTCGAGTGGCCACCACCCTTCCC-3′. Wild-type HspX Reverse Primer: 5′-CATATGTCAGTTGGTGGACCGGATCTG-3′. Mutant Lys47 HspX Forward Primer: 5′-CTGGAAGACGAGATGAGAGAGGGGCGCTACGAGGTACGC-3′. Mutant Lys47 HspX Reverse Primer: 5′-AAGCTCCGCGCGTACCTCGTAGCGCCCCTCTCTCATCTC-3′. Mutant Lys58 HspX Forward Primer: 5′-GGGGTCGACCCCGACAGAGACGTCGACATTATGGTCCGC-3′. Mutant Lys58 HspX Reverse Primer: 5′-CTGACCATCGCGGACCATAATGTCGACGTCTCTGTCGGG-3′. Mutant Lys85 HspX Forward Primer: 5′ GAGCGCACCGAGCAGAGAGACTTCGACGGTCGCTCGGAA-3′. Mutant Lys85 HspX Forward Primer: 5′-GAGCGCACCGAGCAGAGAGACTTCGACGGTCGCTCGGAA-3′. Mutant Lys85 HspX Reverse Primer: 5′-GTACGCGAATTCCGAGCGACCGTCGAAGTCTCTCTGCTC. E. coli BL21 and DH5α were grown in LB agar and liquid culture. Plasmid containing mycobacteria proteins were grown in liquid media with 100mM Ampicillin (Sigma) for selection. The He4 expressing plasmid was purchased from ThermoScientific. Constructs for the expression of GroES and HspX point mutant proteins in E. coli were created the QuikChange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer’s instructions. Following mutagenesis, the resulting constructs were verified by DNA sequencing.
Protein Expression and Purification
In brief, pellets from 250 ml cultures were dissolved in 10 ml lysis buffer containing 50 mM Tris-HCl pH 8.0, 300 mM NaCl, 0.5 M urea with protease inhibitor cocktail (Sigma). Resuspended cells were sonicated two times, each for 5 min (with a 0.3 s pulse and 0.7 s rest) at 5 min intervals to prevent overheating, using a “Fisherbrand Sonicator” at 45% amplitude. The supernatant was collected after centrifugation (30,000×g) for 30 minutes at 4°C, and loaded onto a nickel affinity sepharose (NiAC) column. After washing with 10 column volumes of 50 mM Tris-HCl pH 8.0, 300 mM NaCl and 20 mM imidazole, proteins were eluted in 50 mM Tris-HCl, 100–150 mM NaCl and 250–500 mM imidazole.
His-tag Removal from recombinant GroES
His-tag was removed from recombinant GroES by thrombin digestion. Target protein was incubated with .5U/uL thrombin for 4 hours at 22°C. Following digestion, thrombin was removed from sample by a benzamidine sepharose column.
Treatment of cells with endocytosis inhibitor Dynasore
RAW264.7 cells were plated at 1×106 cells/mL in 6-well plates and allowed to attach overnight. Cells were treated for 30 minutes with 80uM Dynasore (Sigma-Aldrich) or equivalent volume of the DMSO vehicle control. Cells were washed with PBS and fresh culture media was added. Cells were treated with 40ug/mL of the His-HspX recombinant protein for 4 hours. Cells were again washed with PBS to remove any free recombinant protein and exosome-depleted culture media was added. After 16 hours, exosomes were harvested from cell culture supernatant as described above and whole cell lysates were generated by the addition of RIPA buffer. Samples were analyzed by western blot for His-tagged HspX and Lamp-1.
Transfection of M. smegmatis
Wildtype and mutant HspX constructs were subcloned into pMV261 transfection plasmid. M. Smegmatis culture was grown in 7H9 mediate until OD reached log phase growth. Pelleted cells were washed, and resuspended in 10% (v/v) glycerol in PBS. Concentrated bacteria were mixed with 100ng of plasmid DNA and electroporated at 2.5kV, 1000ohms, and 25uF in 0.2cm electroporation cuvettes. Cells were diluted with fresh 7H9 media and incubated for 5 hours at 37C. Following incubation, cells were plated and screened after 3 days.
Immunofluorescence Staining and Microscopy
Cultured Raw264.7 cells were seeded on glass coverslips in 24-well plates. Cells were treated with 40μg/mL of His-tagged HspX for 3 and 6 hours. Cells were fixed and stained and F-actin distribution was visualized by staining with FITC-phalloidin (Molecular Probes). The anti-Lamp1 was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD and maintained at The University of Iowa, Department of Biology. The antibody at 1:250 dilution was used to label endosomal compartments and the His-tag antibody (1:100, Biolegends) was used to label the HspX protein. Cells were visualized with a Nikon fluorescent microscope coupled to a Bio-Rad MRC 1024 scanning confocal three-channel system.
Results
ESCRT-machinery is necessary for exosomes biogenesis in macrophages
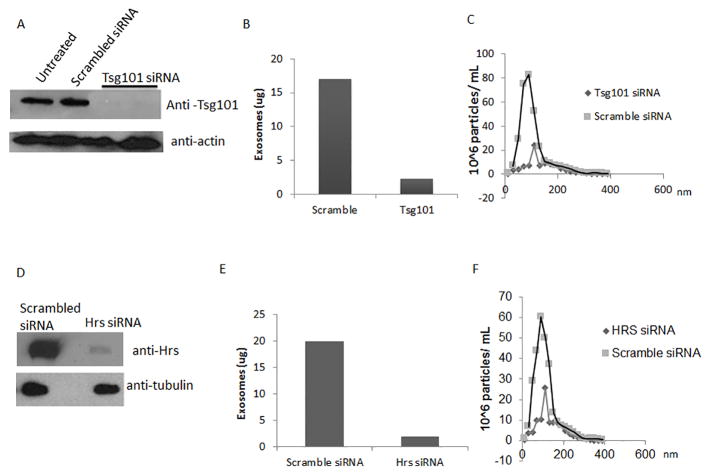
Various studies have identified proteins involved in exosome biogenesis in specific cell types; however, there is limited information on how proteins are trafficked to MVB/exosomes in macrophages. This is an important consideration since the published data suggest that different cells may use different mechanisms for MVB biogenesis (7) (8). Therefore we evaluated the requirement for ESCRT-0 and ESCRT-1 in macrophage exosome biogenesis. Using siRNA knockdown, we targeted Tsg101 and Hrs, the ubiquitin-binding domain of ESCRT-1 and ESCRT-0, respectively. RAW264.7 cells were treated for 48 hours with four independent siRNA oligos against either a specific target or with a scramble control. Knockdown efficiency of Tsg101 and Hrs were assayed by Western blot and was greater than 95% for both proteins (Fig. 1A and 1D). No significant difference in cell numbers was observed for the different treatment groups (data not shown). Exosomes were isolated from cell culture supernatants of the transfected macrophages and protein concentrations determined using a bicinchoninic acid assay (BCA). Exosome protein concentration was reduced greater than 85% as a result of Tsg101 and Hrs knockdown compared to siRNA scramble control (Fig. 1B and 1E). Since the protein assay cannot discriminate between a decrease in the number of exosomes and a decrease in the amount of protein per exosome we used particle tracking to quantify the total number of vesicles secreted by the Raw264.7 cells. As shown in Figures 1C and 1F we observed a >80% decrease in vesicle concentration from cells transfected with Tsg101 and Hrs specific silencing RNAs compared to cells treated with scramble oligonucleotides, which matched our BCA data.
Figure 1. Hrs and Tsg101 are required for exosome biogenesis in Raw264.7 cells.
Raw 264.7 cells were treated for 48 hours with 325ng of siRNA directed against either Hrs or Tsg101. Scrambled siRNAs were used as a control. (A and D) Raw 264.7 cell lysates were loaded onto a SDS-page gel and knockdown efficiencies determined by western blot using antibodies to Hrs and Tsg101. Exosomes were purified and the total protein content quantified by BCA (B and E) and particle number quantified by Nanosight analysis (C and F).
Exosomes from infected macrophages contain ubiquitinated proteins
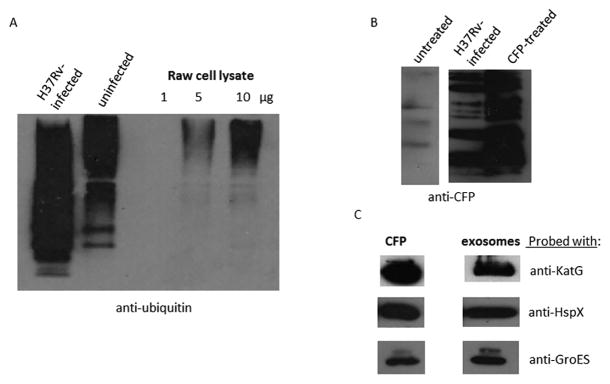
The results from the Tsg101 and Hrs knockdown experiments indicate that the ESCRT machinery is required for production of exosomes in macrophages suggesting that ubiquitination is an important mechanism for protein trafficking to MVBs and exosomes. Therefore, we evaluated exosomes from H37Rv-infected and uninfected cells for the presence of ubiquitinated cargo. As shown in figure 2A exosomes were enriched for ubiquitinated proteins relative to cell lysate and exosomes from infected macrophages appeared to contain a higher number of ubiquitinated proteins at lower molecular weight. To determine if any of the ubiquitinated proteins were mycobacterial in origin we first isolated then lysed exosomes from infected macrophages and performed an immuno-precipitation using an antibody that recognizes mono-ubiquitinated proteins. The pull-down was probed for mycobacterial proteins using a polyclonal antibody made against M.tb culture filtrate proteins (CFP), which has previously been shown to recognize many of the M.tb proteins present in exosomes (18). As shown in figure 2B a number of immuno-precipitated proteins were identified using the polyclonal antibody made against the CFP and this was specific to exosomes from infected cells. To gain some insight into which mycobacterial proteins are pull-downed with the ubiquitin antibody, we first analyzed M.tb proteins previously identified in exosomes for predicted ubiquitination sites. KatG, HspX and GroES were all identified as having a number of lysine that could be ubiquitinated. Therefore, we probed the proteins from the pull-down for these mycobacterial proteins and found all three to be present (Fig. 2C).
Figure 2. Mycobacterial proteins within exosomes can be immuno-precipitated using an ubiquitin antibody.
(A) Exosomes from uninfected and H37Rv-infected macrophages (10μg) and Raw 264.7 whole cell lysate at the concentrations shown were loaded on an SDS-page gel and probed for the presence of mono-ubiquitin by western blot. (B) Purified exosomes from untreated, H37Rv-infected and CFP pulsed Raw264.7 cells were lysed and the ubiquitinated proteins were immuno-precipitated. The protein lysates from the immuno-precipitation were probed for the presence of mycobacterial proteins using a polyclonal antibody made against the culture filtrate proteins (CFP) of M. tuberculosis. (C) Pull-down lysate was probed for the presence of specific mycobacterial proteins: KatG, HspX, and GroES. CFP was also probed with the various antibodies as a positive control. CFP; M. tuberculosis culture filtrate protein.
In our previous studies we determined that exosomes released from macrophages treated with M.tb CFP also contained mycobacterial proteins, sharing much of the same proteins present in exosomes isolated from directly infected cells (4). This suggest that mycobacteria proteins may contain the necessary “signal” to be trafficked to exosomes upon entry into the cell, whether through phagocytosis or endocytosis. In support of this hypothesis we found exosomes derived from CFP-treated macrophages also contained ubiquitinated proteins (Supplementary Figure 1A). Moreover, exosomes released from CFP-treated macrophages when lysed and immunoprecipitated for ubiquitinated proteins showed a number of specific bands when probed with the mycobacterial antibody made against CFP (Fig. 2B).
Inhibiting ubiquitination blocks trafficking of mycobacterial proteins to exosomes
Given ubiquitinated proteins could be detected in exosomes, we examined the dependency of ubiquitination for protein sorting in macrophages. Ubiquitination is mediated by the actions of ubiquitin activating enzyme (E1), a ubiquitin conjugating enzyme (E2), and a ubiquitin ligase (E3) (19). Previous studies have shown that the drug compound, PYR-41 effectively prevents ubiquitination in the cells through inhibition of ubiquitin thioester bond formation within E1 (20). Treating macrophages with 50mM of PYR-41 resulted in a 30 to 40% reduction in exosome production as defined by BCA and Nanosight (Fig. 3A and B). Furthermore, pre-treatment of CFP-treated Raw264.7 cells with PYR-41 resulted in exosomes which lacked mycobacterial proteins detectable by the CFP polyclonal antibody (Fig. 3C).
Figure 3. Ubiquitination is required for mycobacterial protein trafficking to exosomes.
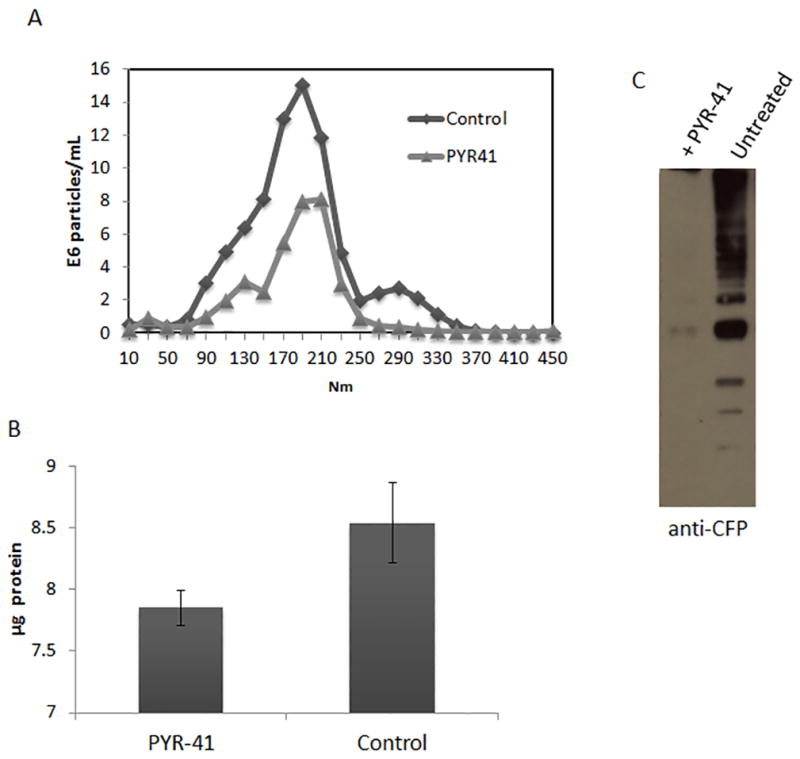
Raw264.7 cells were treated for 2 hours with 50 nM of the ubiquitination inhibitor PYR-41. Cells were washed and treated with M. tuberculosis H37Rv culture filtrate proteins (CFP) for 16 hours. (A) Nanosight analysis of vesicle concentration from untreated and PYR-41 treated Raw 264.7 cells. (B) BCA analysis of vesicle concentration from untreated and PYR-41 treated cells. (C) Raw cells were left untreated or treated with 50 ρmoles of PYR-41 for two hours and subsequently treated with 20μg/mL of CFP. Purified exosomes were assayed for the presence of mycobacterial proteins by western blot using a polyclonal antibody made against the M. tuberculosis CFP.
Trafficking of GroES and HspX to exosomes requires ubiquitination
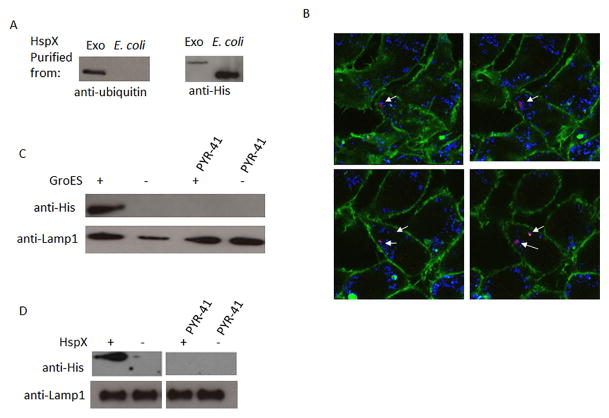
While the above results suggest ubiquitination is required for mycobacterial protein trafficking to exosomes, it is not clear whether this is due to direct ubiquitination of the mycobacterial proteins or their association with ubiquitinated host proteins. To address this question, HspX and GroES were expressed and purified as His-tag fusion proteins. The purified fusion proteins were added to Raw264.7 cells and exosomes were isolated 16 hours post-treatment. Based on the presence of HspX and GroES in exosomes following treatment of macrophages with CFP we anticipated that the His-tagged fusion proteins would be endocytosed and trafficked to MVBs and into exosomes. Indeed, we observed both proteins in exosomes following their addition to macrophages suggesting that each protein has the “signal” for trafficking to MVBs/exosomes (Fig. 4A–D). We also found that GroES purified from M.tb culture supernatant and His-tagged-GroES with the tag removed were also trafficked to exosomes when added to macrophages indicating that the His-tag was not responsible for the trafficking (Supplementary Fig. 1B). Immunofluorescence staining of macrophages post-treatment with His-HspX indicate its intracellular localization and partial co-localization with the late endosomal/MVB marker LAMP1 (Fig. 4B). Interestingly, when we purified the His-tagged proteins from exosomes using nickel-resin we were able to detect the protein using a His-tag antibody and an antibody that recognizes mono-ubiquitinated protein (Fig. 4A). Of note, the 8 KDa shift in protein size observed by western blot for the fusion protein isolated from exosomes compared to originally purified His-HspX from E. coli corresponds to the size expected from a mono-ubiquitination event. These results suggest that HspX and GroES are endocytosed, ubiquitinated and trafficked to exosomes. However, to determine if ubiquitination was necessary, macrophages were pretreated with E1 inhibitor PYR-41 and as shown in figures 4C and D the presences of the His-tagged GroES and HspX in exosomes was lost when cells were treated with ubiquitin inhibitor.
Figure 4. His-tagged HspX and GroES when added to Raw264.7 cells are ubiquitinated and packaged into exosomes.
(A) Raw 264.7 macrophages were pulsed with recombinant His-tagged HspX and exosomes were isolated from culture supernatant 16 hours post-treatment. The recombinant protein from lysed exosomes was purified using a Ni-based resin. Also shown is the original His-tagged HspX purified from E. coli. The Ni-column purified proteins were probed for the presence of ubiquitin and for the His-tagged protein. (B) Confocal microscopy analysis of Raw 264.7 macrophages that were pulsed with recombinant His-tagged HspX. (Cy5: Lamp-1, Texas-Red: poly His, FITC: actin). Arrows indicate staining of His-tagged protein. (C and D) Raw cells were treated with 50 pmoles of PYR-41 for 2 hours or left untreated, pulsed with recombinant His-tagged GroES or HspX and supernatants collected after 16 hours for exosome purification. Exosomes were probed for His-tagged proteins. Lamp-1 was used as a loading control.
Ubiquitination occurs at lysine residues within proteins; furthermore, it has been shown that mutation of lysine residues can inhibit proper ubiquitin-tagging and trafficking within a cell (21). To identify which lysine residue on HspX was ubiquitinated and required for exosomal trafficking, we first used an in silico analysis of the HspX protein sequence to identify potential ubiquitation sites. We identified three lysine residues that gave high probability as sites for ubiquitation. Using site-directed mutagenesis, lysine to arginine substitutions were generated in HspX at amino acid positions 47, 58 and 85. Following purification of native and mutated His-tagged HspX, the recombinant proteins were added to naïve macrophages and exosomes purified 16 hours post-treatment. Despite similar concentrations for all HspX mutants within the treated macrophages, the K85R HspX mutant was not present in exosomes (Fig. 5A). Together, these studies suggest that HspX is directly ubiquitinated and that this modification is required for sorting into exosomes.
Figure 5. Lysine 85 on HspX is required for its packaging into exosomes when added exogenously or expressed by M. smegmatis.
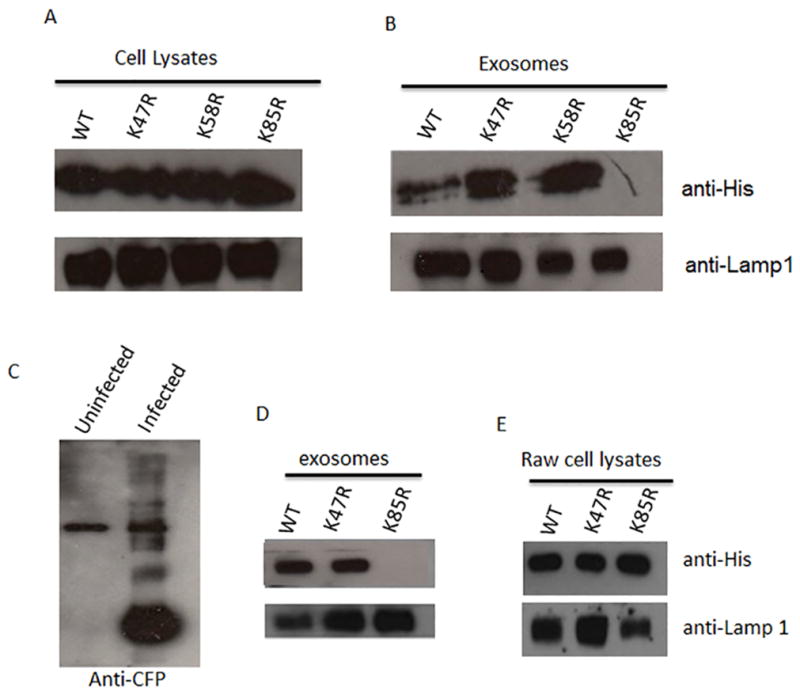
Raw 267.4 cells were seeded in a 6-well plate and treated with 40μg/mL of either purified wild-type His-HspX, His-HspX K47R, His-HspX K58R, or His-HspX K85R. After a 16 hour incubation culture media was removed and used as the starting material for exosome purification. Purified exosomes and the corresponding cells were lysed to obtain total protein. (A and B) Exosomes (5μg) and cell lysates (10μg) were assayed for presence of the wild-type and mutant His-HspX by western blot using a polyclonal His-tag antibody. (C) Exosomes from uninfected or M. smegmatis infected Raw264.7 cells were purified, lysed and the ubiquitinated proteins immuno-precipitated. The ubiquitinated proteins were probed for the presence of mycobacterial proteins using the polyclonal antibody made against the culture filtrate proteins (CFP) of M. tuberculosis. (D and E) Raw 267.4 cells were infected with M. smegmatis expressing wild-type HspX, K47R HspX, or K85R HspX for 72 hours. Exosomes were purified from the culture supernatant and the exosome lysate (10μg) and the cell lysate (20μg) were assayed for presence of the wild-type and mutant His-HspX proteins by western blot using a polyclonal His-tag antibody. Lamp-1 was used as a loading control.
Macrophages infected with M. smegmatis expressing WT or K47R HspX but not K85R HspX release exosomes containing the His-tagged HspX fusion protein
While our evidence suggest that HspX and GroES are ubiquitinated upon endocytosis by macrophages and that this ubiquination is required for trafficking to exosomes, it is unclear if this mechanism extends to trafficking of mycobacterial proteins during a natural infection. Therefore, due to the ease of expressing mycobacterial proteins in the non-pathogenic fast–growing M. smegmatis relative to M.tb, we generated M. semgmatis clones that express either WT, K47R or K85R HspX. However, prior to evaluating the trafficking of the recombinant protein, it was necessary to determine whether mycobacterial proteins are transported to exosomes during an M. smegmatis infection of Raw264.7 cells as has been observed for other mycobacteria (e.g. M. avium, M.tb and M. bovis BCG) (3). As shown in figure 5B exosomes derived from M. smegmatis infected macrophages also contained mycobacterial proteins that could be pulled-down with an anti-ubiquitin antibody. Based on these results, we infected Raw 267.4 cells with M. smegmatis expressing WT, K47R or K85R HspX and the exosomes from the cell culture supernatant were purified by ultracentrifugation 72 hours post-infection. Exosomes and cell lysates were assayed for the presence of the wild type and mutant His-tagged HspX proteins by western blot using the His-tag antibody. As was observed using purified protein, macrophages infected with M. smegmatis expressing the K85R mutant released exosomes that lacked this HspX while bacilli expressing either WT or K47R HspX contained exosomes with the recombinant protein (Fig. 5C). The results were not due to a loss of K85R expression as wild type, K47R, and K85R HspX were found to be present at similar expression levels within infected macrophage cell lysate (Figure 5C). To confirm that the HspX that we observed was associated with exosomes and not protein aggregates that co-purified, we further purified the exosomes released from M. smegmatis infected macrophages by sucrose gradient and analyzed the exosomes for HspX. Using these highly purified exosomes we again observed HspX in vesicles released from M. smegmatis expressing the WT but not the HspX K85R mutant (Supplementary Fig. 2).
Trafficking of HspX into exosomes requires clathrin-mediated endocytosis
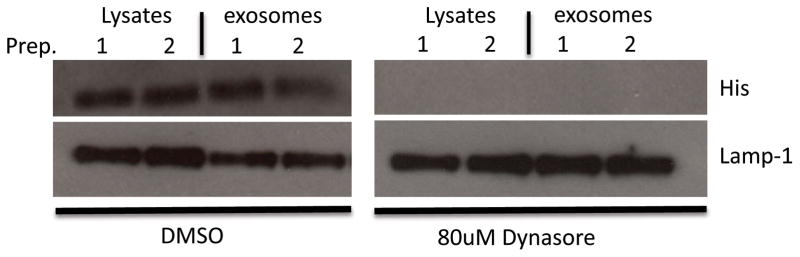
The data above indicates that ubiquitination of HspX is required for its targeting to exosomes suggesting that ubiquitination occurs within an endosomal compartment. However, these studies did not directly evaluate the mechanism of HspX uptake. To assess whether HspX is endocytosed and therefore present within an endosomal compartment, we treated cells with Dynasore, an inhibitor of clathrin-mediated endocytosis and showed that the inhibitor blocked uptake of HspX and consequently blocked transport of HspX to exosomes (Fig. 6).
Figure 6. Trafficking of HspX to exosomes was dependent on clathrin-mediated endocytosis of the recombinant protein.
Raw 264.7 cells were pre-treated with the endocytosis inhibitor Dynasore followed by the addition of recombinant His-tagged HspX. Cells were washed 4 hours post HspX treatment and incubated for an additional 16 hours. Cell lysates were analyzed by western blot for His-tagged HspX and Lamp-1. Exosomes were isolated from the culture media and probed for His-tagged HspX and Lamp-1 by western blot. Shown are the results of duplicate experiments.
Ubiquitin-mediated transport of endocytosed proteins to exosomes is not limited to mycobacterial proteins
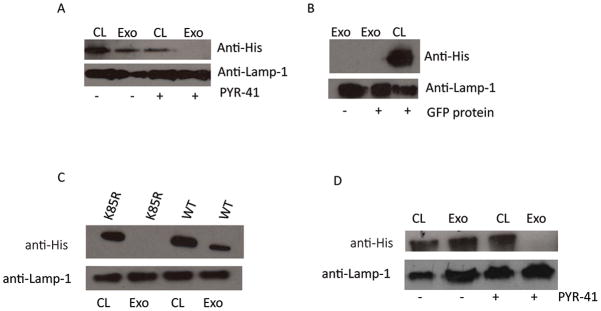
Our results suggest that soluble proteins upon endocytosis by macrophages can be trafficked to MVBs and released on exosomes if they are properly ubiquitinated. However, this hypothesis is based on the analysis of just mycobacterial proteins; specifically HspX and GroES. Therefore, to determine if this mechanism is more broadly applicable to endocytosed proteins, we tested a different soluble antigen, He4, a well-characterized cancer protein. Based on our prediction analysis, He4 contains lysine residues that are potential ubiquitinated. A His-tagged He4 fusion protein was generated and purified and added to macrophages using the same protocol as performed for the mycobacterial proteins. The exosomes isolated from the He4-treated macrophages contained the His-tagged protein and this localization was ubiquitin-dependent as its trafficking was inhibited by macrophage pre-treatment with PYR-41 (Fig. 7A). This sorting of endocytosed proteins into exosomes is relatively specific as His-tagged GFP when added to macrophages was endocytosed but remained intracellular and was not released via exosomes (Fig. 7B).
Figure 7. His-tagged He4 when added to Raw264.7 or HEK293 cells is packaged into exosomes in an ubiquitin-dependent manner.
(A) Raw 264.7 cells pre-treated with PYR-41 for 2 hours of left untreated were pulsed with recombinant He4 protein for 16 hours. Exosomes were purified from the culture supernatant and the exosomal proteins and cell lysates were probed for His-tagged He4. (B) Raw 264.7 cells were pulsed with recombinant GFP protein for 16 hours. Exosomes purified from the culture supernatant were lysed and along with the cell lysates probed for the presence of His-tagged GFP. (C) HEK293 cells were pulsed with recombinant wild-type HspX or the K85R HspX. (D) HEK293 cells pre-treated with PYR-41 for 2 hours or left untreated were pulsed with His-tagged He4. After 16 hours the HEK293 culture media was removed for exosomes purification and the exosomal proteins and the cell lysates were probed for the presence of His-tagged recombinant protein. Lamp-1 served as a loading control. CL; cell lysate, Exo; exosomes.
Trafficking of HspX and He4 into exosomes derived from HEK cells is dependent on ubiquitation
Our findings indicate that for at least a subset of soluble proteins, ubiquitination is required for their sorting to exosomes. However, these studies were limited to a macrophage cell line and whether this ubiquitin-dependent transport applied to other cell-types was unknown. Therefore, purified WT or K85R mutant HspX were incubated with HEK293 cells; a human embryonic kidney cell line. As described above, exosomes were purified from the cell supernatant and the cell lysates were collected as controls for cellular uptake. Similar to the macrophage results, the WT but not the K85R HspX was present in exosomes released from HEK293 cells (Figure 7C). Furthermore, exosomes derived from HEK293 cells treated with He4 contained the recombinant protein but He4 was absent when cells were pretreated with PYR-41 (Figure 6D). Therefore our data suggest that ubiquitin-dependent trafficking of soluble proteins within the endocytic/phagocytic pathway to exosomes is not restricted to macrophages.
The E3 ligase Parkin is not responsible for the ubiquitination of HspX
Previous studies identified parkin as an E3 ligase that can poly-ubiquitinate M.tb (22). Parkin, while primarily characterized for its poly-ubiquitination function, also has mono-ubiquitination activity. Therefore we hypothesized that parkin may be responsible for the ubiquitation of the mycobacterial proteins. We were successful in knocking down >90% of the parkin expression in Raw264.7 cells. However, this did not lead to a diminished incorporation of HspX into exosomes when added exogenous to the Raw264.7 cells (Supplementary Fig. 3) indicating that parkin is not the E3 ligase responsible for HspX ubiquitination.
Discussion
As an intracellular pathogen, M. tuberculosis has limited exposure to various immune components including antibodies and complement. Nonetheless the immune system reacts efficiently and appropriately in most cases, as evidenced by greater than 90% of those infected with M.tb never developing clinical disease. A key aspect to our understanding of the immune response to infection is to know the mechanisms of how and where antigens are processed and presented. Recent evidence suggest that exosomes serve as a sources of antigen during infections and may play an important role in T cell activation and antigen cross-priming (23). Exosomes are vesicles of 30–100nm in size that are secreted from cells of both hematopoietic and non-hematopoietic origin and in general function in intercellular communication. Previously, we have reported that exosomes modulate both the innate and acquired immune response during an M.tb infection (3) (24) and shown that exosomes containing TB antigens can be used as an effective vaccine against an aerosolized M.tb infection using a mouse model (5). These results suggest that exosomes may provide a unique approach to TB vaccine development. However, a number of challenges remain including developing exosomes with specific mycobacterial antigens. Therefore, to bioengineer exosomes as a TB vaccine or as a vaccine in general we need a better understanding how proteins are sorted into exosomes.
We have identified over 40 different mycobacterial proteins on exosomes derived from M.tb-infected macrophages and many of the same proteins from CFP-pulsed macrophages (4). Of the mycobacterial proteins identified on exosomes, 95% were experimentally shown to be secreted, soluble proteins (4). In the present study we showed that host ubiquitation is required for this sorting and demonstrated that HspX and GroES are directly ubiquitinated by macrophages and that this provides the signal necessary for trafficking and loading into exosomes. Surprisingly, this sorting seemed to be independent of entry mechanisms as proteins were trafficked to exosomes in an ubiquitin-dependent manner whether added as free protein and therefore taken in by an endocytic route or expressed by a mycobacteria which gains entry by phagocytosis. This suggest that in both cases the soluble mycobacterial protein gains access to the E1, E2 and E3 ligases. Whether the same ligases are involved is unclear but the data with the HspX K85R mutant which was absent from exosomes when expressed by M. smegmatis or added as exogenous protein, suggest a homologous ubiquitination process.
Ubiquitination by E3 ligases predominately occurs within the cytosol (19) and there are a number cytosolic proteins present within exosomes. Presumably, some of these proteins are targeted through an ubiquitin-dependent/ESCRT-mediated pathway. This hypothesis is supported by the presence of ubiquitinated luminal proteins within exosomes (25). There is even more evidence to support transport of plasma-membrane receptors to MVBs/exosomes through an ubiquitin-dependent process (26). Our data suggest that endocytosed or phagocytosed proteins can also be trafficked to exosomes. In the context of an infection we and others have shown that some microbial components including proteins are trafficked to exosomes (3) (27). However, none of these studies defined a mechanism of how this targeting was mediated. One possibility is that these antigens, particularly in the context of direct M.tb infection gain access to the cytosol, perhaps through pores formed within the phagosome in an ESX-1 dependent manner (28) and are ubiquitinated by the cytosolic E3. However, identifying mycobacterial proteins within the cytosol has been a challenge. Moreover, mycobacterial proteins can be found on exosomes derived from M. bovis BCG-infected macrophages. This strain lacks a genomic region called RD1 which contains a number of genes essential for ESX-1-mediated protein release from the phagosome (28). In addition, our data using an inhibitor of clathrin-mediated endocytosis indicates that the endocytic process is required for uptake of soluble HspX and there is no known mechanism that would transport HspX from an endosomal compartment to the cytosol. The retention of HspX in an endosomal compartment is supported by our fluorescent microscope data.
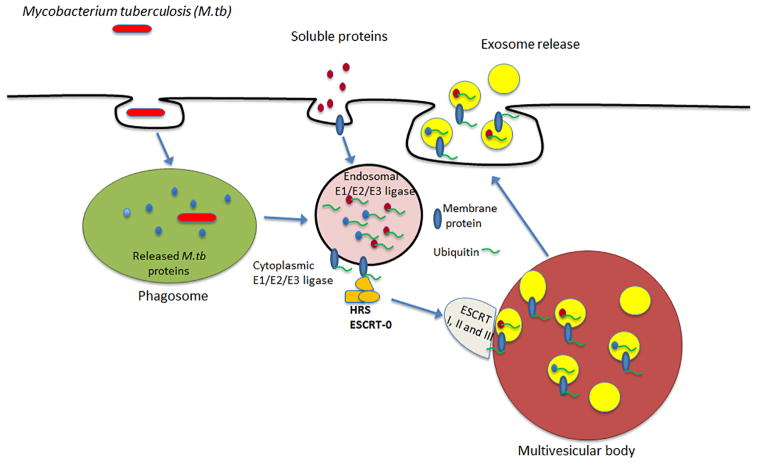
The second possibility is that soluble antigens are first ubiquitinated within the endocytic network. Interestingly, a number of E3 ligases have been identified within the endocytic network (29) (30) (see table S1 for a more complete list) and likely more still remain to be defined. Furthermore, proteomic analysis of exosomes have shown that they too contain all the necessary machinery required for ubiquitination to occur, including both E1 and E2, and different members of E3 ligase family; suggesting their propensity for being confined within endocytic compartments (31) (32). Therefore we hypothesize and our data supports that the endocytosed and phagocytosed proteins are sorted into endocytic vesicles which have the machinery for ubiquitination (Fig. 8); however, the E3 ligase responsible has yet to be identified. Due to their ability to target a myriad of proteins, there are many E3 ligases within a mammalian cell. Current estimates suggest as many as 1,000 E3 ligases within a single cell (33). Nevertheless, we hypothesized that the E3 ligase Park2 (parkin) may be involved since it has been shown to poly-ubiquitinate M.tb (22). However, knockdown of parkin in macrophages did not alter the transport of soluble HspX. Therefore, additional studies into the ligases contained within endocytic compartments is needed. Moreover, how ubiquitinated proteins within the luman of an endocytic vesicle are recognized by components of the ESCRT machinery, which are cytoplasmic, or by other host components for packaging into exosomes is presently unclear.
Figure 8.
An illustration highlighting ubiquitination as a mechanism for the transport of proteins from a phagosome or endosome to an MVB and into the intraluminal vesicles for release in exosomes.
It is generally accepted that monoubiquitin of a lysine residue can target a protein to the MVB through both an ESCRT dependent and independent mechanisms (15) (34). Through the use of inhibitors and lysine point mutants, we show that HspX when added to macrophages requires ubiquitination for trafficking to exosomes. Interestingly, this was true for the trafficking of HspX in both macrophages and HEK293 cells, a kidney cell line. This data suggests that endocytosed/soluble proteins which are mono-ubiquitinated might be trafficked to exosomes as a general mechanism. In light of this observation, we evaluated whether ubiquitination also served to traffic other soluble proteins to the MVB. Our analysis of He4, a soluble cancer antigen, indicated that it behaved similarly to the mycobacterial proteins both in macrophages and HEK293 cells and was trafficked to exosomes in an ubiquitin-dependent fashion. This suggest that endocytosed proteins can be “recycled” by cells through exosomes, a process not previously described and this has implications into how normally secreted proteins may be repackaged for extracellular release. However, additional studies are needed to determine how general this mechanism is for endocytosed proteins and what role it may have under physiological and pathological conditions.
In summary our data indicates that within the endocytic/phagocytic network ubiquitination functions as a tag for protein delivery to exosomes. What ligases are responsible for this ubiquitination remains unknown, but likely involves multiple E3 ligases within endosomal compartments. This previously undescribed mechanism of packaging and re-release of extracellular proteins through exosomes may have consequence for protein function as their presence in exosomes may lead to their different distribution and cellular response. To what extent secreted proteins are repackaged for release through exosomes and the functional consequences of this exosome localization remain to be defined.
Supplementary Material
Acknowledgments
We kindly acknowledge Dr. Yong Cheng for the His tagged GroES construct that was used for subsquent cloning and GroES protein expression.
Footnotes
Funds for this work were provided by the grant AI052439 from the National Institute of Allergy and Infectious Diseases
Authorship Contributions
V. Smith designed and executed most of the experiments described in this study and co-wrote the manuscript. L. Jackson performed some of the immuno-precipitation experiments shown. J. Schorey helped in the design of the experiments and co-wrote the manuscript.
References
- 1.Killick KE, Ni Cheallaigh C, O’Farrelly C, Hokamp K, MacHugh DE, Harris J. Receptor-mediated recognition of mycobacterial pathogens. Cellular microbiology. 2013;15:1484–1495. doi: 10.1111/cmi.12161. [DOI] [PubMed] [Google Scholar]
- 2.Beatty WL, Ullrich HJ, Russell DG. Mycobacterial surface moieties are released from infected macrophages by a constitutive exocytic event. Eur J Cell Biol. 2001;80:31–40. doi: 10.1078/0171-9335-00131. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar S, Shinagawa K, Castellino FJ, Schorey JS. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood. 2007;110:3234–3244. doi: 10.1182/blood-2007-03-079152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giri PK, Kruh NA, Dobos KM, Schorey JS. Proteomic analysis identifies highly antigenic proteins in exosomes from M. tuberculosis-infected and culture filtrate protein-treated macrophages. Proteomics. 2010;10:3190–3202. doi: 10.1002/pmic.200900840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng Y, Schorey JS. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur J Immunol. 2013 doi: 10.1002/eji.201343727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez-Vazquez C, Villarroya-Beltri C, Mittelbrunn M, Sanchez-Madrid F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunological reviews. 2013;251:125–142. doi: 10.1111/imr.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamai K, Tanaka N, Nakano T, Kakazu E, Kondo Y, Inoue J, Shiina M, Fukushima K, Hoshino T, Sano K, Ueno Y, Shimosegawa T, Sugamura K. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochemical and biophysical research communications. 2010;399:384–390. doi: 10.1016/j.bbrc.2010.07.083. [DOI] [PubMed] [Google Scholar]
- 8.Stuffers S, Sem Wegner C, Stenmark H, Brech A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic. 2009;10:925–937. doi: 10.1111/j.1600-0854.2009.00920.x. [DOI] [PubMed] [Google Scholar]
- 9.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 10.Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- 11.Davies BA, Lee JR, Oestreich AJ, Katzmann DJ. Membrane protein targeting to the MVB/lysosome. Chemical reviews. 2009;109:1575–1586. doi: 10.1021/cr800473s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Metcalf D, Isaacs AM. The role of ESCRT proteins in fusion events involving lysosomes, endosomes and autophagosomes. Biochemical Society transactions. 2010;38:1469–1473. doi: 10.1042/BST0381469. [DOI] [PubMed] [Google Scholar]
- 13.Hurley JH. The ESCRT complexes. Critical reviews in biochemistry and molecular biology. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raiborg C, Stenmark H. Hrs and endocytic sorting of ubiquitinated membrane proteins. Cell structure and function. 2002;27:403–408. doi: 10.1247/csf.27.403. [DOI] [PubMed] [Google Scholar]
- 15.Katzmann DJ, Babst M, Emr SD. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 2001;106:145–155. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 16.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Developmental cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 17.Yadav M, Schorey JS. The beta-glucan receptor dectin-1 functions together with TLR2 to mediate macrophage activation by mycobacteria. Blood. 2006;108:3168–3175. doi: 10.1182/blood-2006-05-024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonnenberg MG, Belisle JT. Definition of Mycobacterium tuberculosis culture filtrate proteins by two-dimensional polyacrylamide gel electrophoresis, N-terminal amino acid sequencing, and electrospray mass spectrometry. Infection and immunity. 1997;65:4515–4524. doi: 10.1128/iai.65.11.4515-4524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komander D, Rape M. The ubiquitin code. Annual review of biochemistry. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li CC, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Inhibitors of ubiquitin-activating enzyme (E1), a new class of potential cancer therapeutics. Cancer research. 2007;67:9472–9481. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura S, Roth JA, Mukhopadhyay T. Multiple lysine mutations in the C-terminal domain of p53 interfere with MDM2-dependent protein degradation and ubiquitination. Molecular and cellular biology. 2000;20:9391–9398. doi: 10.1128/mcb.20.24.9391-9398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150:803–815. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bobrie A, Colombo M, Raposo G, Thery C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 24.Giri PK, Schorey JS. Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PloS one. 2008;3:e2461. doi: 10.1371/journal.pone.0002461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buschow SI, Liefhebber JM, Wubbolts R, Stoorvogel W. Exosomes contain ubiquitinated proteins. Blood cells, molecules & diseases. 2005;35:398–403. doi: 10.1016/j.bcmd.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 26.Shields SB, Oestreich AJ, Winistorfer S, Nguyen D, Payne JA, Katzmann DJ, Piper R. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. The Journal of cell biology. 2009;185:213–224. doi: 10.1083/jcb.200811130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhatnagar S, Schorey JS. Exosomes released from infected macrophages contain Mycobacterium avium glycopeptidolipids and are proinflammatory. The Journal of biological chemistry. 2007;282:25779–25789. doi: 10.1074/jbc.M702277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Champion PA, Cox JS. Protein secretion systems in Mycobacteria. Cellular microbiology. 2007;9:1376–1384. doi: 10.1111/j.1462-5822.2007.00943.x. [DOI] [PubMed] [Google Scholar]
- 29.Hassink G, Slotman J, Oorschot V, Van Der Reijden BA, Monteferrario D, Noordermeer SM, Van Kerkhof P, Klumperman J, Strous GJ. Identification of the ubiquitin ligase Triad1 as a regulator of endosomal transport. Biology open. 2012;1:607–614. doi: 10.1242/bio.2012778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santonico E, Belleudi F, Panni S, Torrisi MR, Cesareni G, Castagnoli L. Multiple modification and protein interaction signals drive the Ring finger protein 11 (RNF11) E3 ligase to the endosomal compartment. Oncogene. 2010;29:5604–5618. doi: 10.1038/onc.2010.294. [DOI] [PubMed] [Google Scholar]
- 31.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 32.Carayon K, Chaoui K, Ronzier E, Lazar I, Bertrand-Michel J, Roques V, Balor S, Terce F, Lopez A, Salome L, Joly E. Proteolipidic composition of exosomes changes during reticulocyte maturation. The Journal of biological chemistry. 2011;286:34426–34439. doi: 10.1074/jbc.M111.257444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nature reviews Cancer. 2006;6:776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 34.Stringer DK, Piper RC. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. The Journal of cell biology. 2011;192:229–242. doi: 10.1083/jcb.201008121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.