Abstract
The addition of novel side chains at the 5-position of uracil is an effective means to increase chemical diversity of aptamers and hence the success rate for discovery of high-affinity ligands to protein targets. Such modifications also increase nuclease resistance, which is useful in a range of applications, especially for therapeutics. In this study, we assess the impact of these side chains on plasma pharmacokinetics of modified aptamers conjugated to a 40 kDa polyethylene glycol. We show that clearance from plasma depends on relative hydrophobicity: side chains with a negative cLogP (more hydrophilic) result in slower plasma clearance compared with side chains with a positive cLogP (more hydrophobic). We show that clearance increases with the number of side chains in sequences of ≥28 synthons, but this effect is dramatically diminished in shorter sequences. These results serve as a guide for the design of new therapeutic aptamers with diversity-enhancing side chains.
Keywords: : aptamer pharmacokinetics, modified nucleotides, side chains, plasma clearance
Introduction
The total plasma clearance of single-stranded oligonucleotides (SSOs), including aptamers, is the sum of the individual clearances due to metabolism, renal filtration, and irreversible tissue uptake [1–3]. Metabolic clearance of SSOs is largely driven by nuclease digestion occurring either in circulation or within tissues. To avoid this clearance pathway, nuclease resistance has been introduced by the addition of phosphorothioate backbones or by using sugar modifications such as locked nucleic acids or 2′-position modifications such as 2′-methoxy (2′-OMe), 2′-amino, 2′-fluoro, and 2′-methoxyethoxy [4–10]. Glomerular filtration can be reduced by attaching large moieties to the SSO such as a 40 kDa polyethylene glycol (PEG), cholesterol, diacyl lipids, or by increasing nonspecific binding to plasma proteins inherent to phosphorothioate backbones [4–10]. Tissue uptake of SSOs into mammalian cells such as liver cells has been shown to be a vesicular uptake process that is saturable [5,11]. SSOs do not cross the blood–brain barrier to an appreciable degree, but otherwise their tissue distribution is broad with major organs of distribution being the kidney, liver, spleen, bone marrow, lymph nodes, and adipocytes [5,12,13].
Despite such efforts, controlling clearance from plasma remains an important consideration for therapeutic aptamers since this feature influences the scope of indications for which aptamers represent sensible drug candidates. Rapid plasma clearance is especially problematic for aptamers whose targets are extracellular, which represents most therapeutic targets of aptamers to date. Even if tissue uptake is in equilibrium with the central compartment (tissue distribution), the exit rate from tissues is so slow as to keep aptamer plasma concentrations below therapeutic levels. The net effect has been to limit the use of therapeutic aptamers to local administration (e.g., compartments such as the eye [14]), or to parenteral delivery for indications where aptamers might have advantages over antibodies and other classes of drug candidates despite or because of their pharmacokinetic (PK) properties [10,15].
Recently, a new class of aptamers that contain modified uridines, designated Slow Off-Rate Modified Aptamer, or SOMAmer reagents, has been described [16]. Like side chains of proteins, modifications introduced at the 5-position of uracil are an effective means to increase the diversity of chemical groups for aptamer interaction with targets, and this has directly led to a greater success rate in SELEX (Systematic Evolution of Ligands by Exponential Enrichment) [16–21]. Among several types of moieties tested, aromatic functional groups such as those that resemble the side chains of phenylalanine and tryptophan, or bicyclic aromatic groups such as naphthalene have proven to be the most effective. Crystal structures of modified aptamers bound to three targets (platelet-derived growth factor, interleukin 6, and nerve growth factor) show that these side chains are involved in forming unique intramolecular structures as well as creating a surface for target interactions [17–19].
The goal of this study was to determine the PK properties of 5-uracil modified aptamers and to understand the effect of the side chains on plasma clearance. To accomplish this, we designed several series of aptamer test sequences with varying side chain hydrophobicity, number of side chains, and overall oligonucleotide length, for assessment of plasma clearance in rats following an intravenous (IV) bolus dose. To better isolate the effect of the side chains on clearance, test sequences contained 2′-OMe ribose at all nucleotide positions, except at three-carbon (C3) spacer positions, and included a 3′-terminal inverted deoxythymidine “cap” (3′-idT) and a 5′-terminal 40 kDa PEG (5′-PEG). Because all of these sequences were inactive due to the extensive 2′ modifications, potential on-target effects on PK properties were eliminated.
Our results clearly show that the side chains play a critical role in PEGylated aptamer clearance. Side chains with negative cLogP values (more hydrophilic) led to slower plasma clearance of aptamers compared with those with positive cLogP values (more hydrophobic). Furthermore, we demonstrate that plasma clearance of modified aptamers is directly proportional to the sequence length and number of side chains. Remarkably, the influence of these side chains on clearance is mitigated in shorter sequences. Based on these guidelines, we successfully identified examples of PEGylated modified aptamers with plasma clearance values similar to those of PEGylated aptamers without modified side chains.
Materials and Methods
Oligonucleotide synthesis
Oligonucleotides were synthesized by solid-phase methods using standard phosphoramidite chemistry [21–23]. The 5-position carboxamide-modified uridine nucleoside analogs were synthesized by reaction of the appropriate, commercially available, side-chain primary amine with the activated esters: 5′-O-dimethoxytrityl-5-trifluoroethoxycarbonyl-2′-deoxyuridine (2′-deoxy series [dU]) [24] or 5′-O-dimethoxytrityl-5-trifluoroethoxycarbonyl-2′-O-methyluridine (nuclease-resistant 2′-O-methyl series [mU]) [25] in acetonitrile (ACN; 1.2 equivalents of primary amine, 3 equivalents of triethylamine, 60°C, 4 h). The resulting modified nucleosides were converted into 5′-O-triphosphate reagents for enzymatic synthesis [26] and into 3′-O-(2-cyanoethyl-N,N-diisopropyl)phosphoramidite reagents for solid-phase chemical synthesis [27].
All sequences in every series contained a 3′-idT cap, were fully modified with 2′-OMe ribose, except for positions containing a C3 spacer in place of a nucleotide, and were conjugated without purification to a 40 kDa PEG (branched 2 × 20 kDa N-hydroxysuccinimide ester; JenKem Technology) through a 5′-photocleavable amino modifier (Glen Research). The length of the oligonucleotides denoted in each series does not include the idT cap. Analytical characterization of the oligonucleotides included verification of the preconjugated mass (required to be within 0.02%). Conjugated products were characterized for purity by ultra high-performance liquid chromatography (UPLC) and average purity was 84.4% (range 72.0%–90.3%). Details for measuring mass of preconjugated sequence and purity of PEGylated sequences are described in the Supplementary Materials and Methods, and Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/nat).
PK studies
Animal studies were conducted at Bolder BioPATH (Boulder, CO) in accordance with the Guiding Principles in the Use of Animals in Toxicology (8th edition). Male Sprague Dawley rats weighing between 200 and 300 g were obtained from Envigo (Indianapolis, IN) and were allowed to acclimate for at least 5 days after arrival. Animals were allowed food and water ad libitum. Aptamers were formulated in phosphate buffered saline, subjected to a heat/cool treatment as described elsewhere [23] and dosed based on the oligonucleotide weight only (not including mass of the PEG). Aptamers were administered at 1.2–2 mg/kg (exact dose for each sequence is listed in the Supplementary Table S2) by IV bolus injection through the tail vein (two animals per group). Dosing occurred near the beginning of the 12-h light cycle and the animal room was kept at 20°C–21°C. Venous blood samples were collected at various time points (predose to 24 h) into tubes containing ethylenediaminetetraacetic acid (EDTA) as anticoagulant. Samples were centrifuged at 3,600 g for 10 min at 4°C and frozen at −80°C within 30 min of collection.
Determination of aptamer concentrations in plasma
Aptamer concentrations were determined as described elsewhere [28]. Briefly, two volumes of 2-ethoxyethanol were added to a 50 μL aliquot of plasma. Samples were mixed and spun at 5,000 g for 5 min at room temperature to pellet proteins. Supernatants were recovered and centrifuged two additional times as before. A 10 μL aliquot of supernatant was injected onto a Waters ACQUITY® UPLC system (Waters, Milford, MA) consisting of an auto sampler (model UPA), a binary pump module (model UPB), a temperature-controlled column compartment (model UPM), and a photodiode array detector (model UPL). The system was equipped with an ACQUITY OST column (2.1 × 100 mm, 1.7 μm particle size; Waters) maintained at 80°C, with a flow rate of 0.2 mL/min. Detection was by absorbance at 260 nm. Mobile phase A consisted of a 95/5 ratio of 0.1 M triethylammonium acetate/ACN and mobile phase B consisted of 100% ACN. The column was maintained at 50% mobile phase B for the first minute followed by a gradient of 50% to 70% mobile phase B over 5 min. Quantification was performed by comparing the peak area of the sample to calibration standards. Standards and quality control samples were prepared in rat EDTA plasma and processed alongside PK study samples. The lower limit of quantification was 1.5 μg/mL.
cLogP
Values for the logarithm base 10 of the octanol/water partition coefficients (cLogP) of side chains were generated using a subroutine within the ChemBioDraw Ultra 13.0 software package (PerkinElmer, Utah). This package utilizes MOSES.logP, a computational calculator module based on Molecular Networks' cheminformatics platform MOSES (Molecular Networks GmbH, Erlangen, Germany). Additional details can be found at www.molecular-networks.com/moses
Chromatographic hydrophobicity index
Chromatographic hydrophobicity index (CHI) values of modified aptamers without 5′-PEG were determined using the UPLC method and were reported in units of UPLC retention time. PEGylated modified aptamers containing a photocleavable linker were cleaved (10 min at 385 nm using a Sylvania 350 Black Light at a distance of ∼6 inches) before analysis. 10 μL of each sample was injected onto a Waters ACQUITY OST column at 45°C, with a flow rate of 0.2 mL/min, and detection at 260 nm. Mobile phase A consisted of 50 mM ammonium acetate, pH ∼6.5, and mobile phase B was methanol. The column was maintained at 5% mobile phase B for the first minute and then at a gradient of 5% to 100% over 21 min.
PK analysis
PK parameters were determined using Summit PK Solutions 2.06 software (Summit Research Services, Montrose, CO). One-compartment models were used for determining half-lives. Plasma clearance was determined by noncompartmental analysis.
Statistical analysis
Mann–Whitney U tests (two sided) and linear regression analyses (1/y2 weighting) were performed using GraphPad Prism version 7.00 for Windows (GraphPad Software, La Jolla, CA).
Results
Base modifications enhance clearance in two representative aptamers
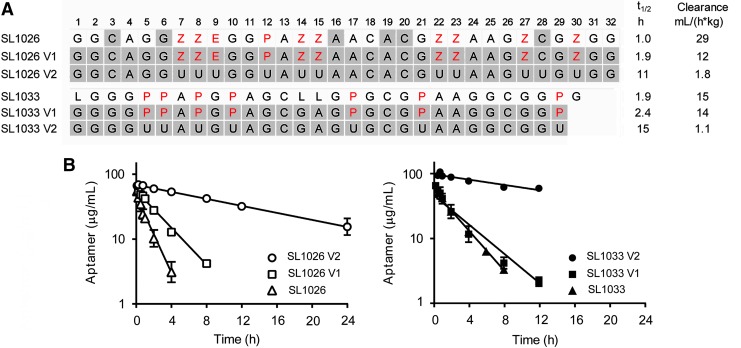
Our investigations focused on two previously reported modified DNA aptamers, SL1026 and SL1033, optimized for therapeutic use [23,29]. SL1026 is a 32-mer with three different side chains, N-benzylcarboxamide (Bn), N-(1-naphthylmethyl)carboxamide (Nap), and N-(phenyl-2-ethyl)carboxamide (Pe), whereas SL1033 is a 30-mer with Nap side chains (Fig. 1A). Furthermore, SL1026 has 2′-OMe substitutions at positions 3, 6, 16, 19, 20, and 28, whereas SL0133 has C3 spacers (L) at positions 1, 14, and 15. Both of these modified aptamers contain a 3′-idT cap to protect from 3′-exonucleases and a 5′-PEG (40 kDa) to prevent renal clearance. The PK properties of these aptamers were determined in male Sprague Dawley rats after an IV bolus dose. Plasma concentration–time curves showed a decrease in aptamer concentration with a half-life of 1.0 and 1.9 h for SL1026 and SL1033, respectively, with a corresponding clearance of 29 and 15 mL/(h*kg) (Fig. 1 and Supplementary Table S2). Although we have provided the full set of PK parameters for each of the sequences tested in Supplementary Table S2, we focus our comparative analysis on clearance (dose divided by the area under the concentration–time curve), since this parameter is computed by noncompartmental analysis of the data and is, therefore, independent of the assumptions inherent in any PK model.
FIG. 1.
Effect of side chains. (A) Sequences (5′→3′) of SL1026, SL1033, and variants thereof, utilized for determination of the effect of side chains on plasma PK. Standard single-letter designations (A, C, G, and U) are used for the unmodified dehydroxy nucleobases, whereas positions designated Z, E, and P show the locations of 5-(N-benzylcarboxamide), 5-[N-(2-phenylethyl)carboxamide], and 5-[N-(1-naphthylmethyl)carboxamide]-modified uridine (highlighted in red text). The positions containing 2′-OMe ribose are shaded gray. All sequences contain a 3′-idT cap and have a 40 kDa PEG attached to the 5′-terminus (not shown). SL1026 contains 2′-OMe ribose at positions 3, 6, 16, 19, 20, and 28, whereas SL1033 contains C3 spacer (L) at positions 1, 14, and 15. The variants (V1 and V2) are fully modified with 2′-OMe ribose. Plasma half-life (t1/2) and plasma clearance values are shown alongside each sequence. (B) Plasma concentration versus time curves. 2′-OMe, 2′-methoxy; PEG, polyethylene glycol; PK, pharmacokinetic.
To determine the effect of the side chains on plasma clearance, sequences were synthesized with (SL1026V1 and SL1033V1) and without (SL1026V2 and SL1033V2) side chains, based on the core sequences of SL1026 and SL1033. SL1033V1 and V2 are 29-mer sequences with no C3 spacers. These sequences (and all other test sequences reported herein) were further modified with 2′-OMe ribose at all positions except at positions with C3 spacers, a 3′-idT cap, and a 5′-PEG (40 kDa). The variants with side chains showed clearance of 12 and 14 mL/(h*kg) for SL1026V1 and SL1033V1, respectively. Removal of the side chains showed a substantial improvement in clearance for both sequences [1.8 and 1.1 mL/(h*kg) for SL1026V2 and SL1033V2, respectively] (Fig. 1). These results clearly highlight the impact of the side chains on aptamer clearance.
Hydrophobicity of side chains contributes to enhanced clearance
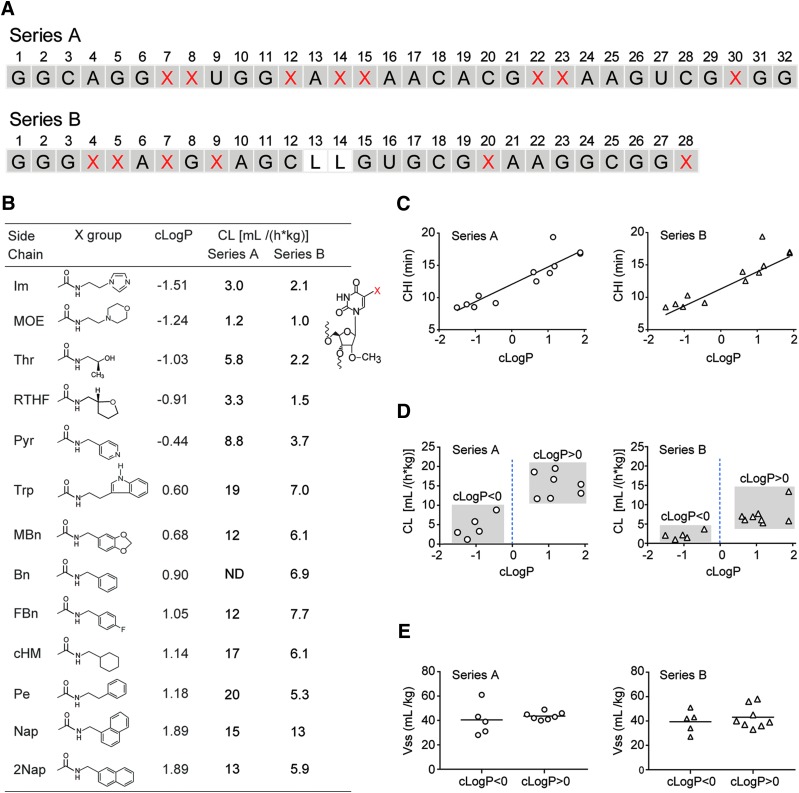
To determine the effect of various 5-uracil modifications on plasma PK, 12 sequences were designed based on SL1026. These sequences were 32 nucleotides in length (32-mers) and contained 8 modified nucleotides (Series A, Fig. 2A). Sequences differed from one another only by the type of side chain. For example, the MOE variant has a N-(morpholino-2-ethyl)carboxamide side chain at each of these eight positions, whereas the Nap variant has a (1-naphthylmethyl)carboxamide side chain at each of the eight positions. The cLogP of the side chains spanned a range predicted to partition primarily in the aqueous phase (−1.51) to those predicted to partition primarily in the octanol phase (1.89) (Fig. 2B). The cLogP values for the side chains were highly correlated with the independently measured CHI, which reflects the overall hydrophobicity of oligonucleotides that contain the modified side chains (Fig. 2C).
FIG. 2.
Effect of hydrophobicity of side chains. (A) Sequences (5′→3′) in Series A and B utilized for determination of the effect of different side chains (see below) on plasma PK. Standard single-letter designations (A, C, G, and U) are used for the unmodified dehydroxy nucleobases, whereas positions designated X highlighted in red text show the location of side chains. The positions containing 2′-OMe ribose are shaded gray. All sequences contain a 3′-idT cap, are fully modified with 2′-OMe ribose, except for positions containing a C3 spacer (L) in place of a nucleotide (positions 13 and 14 of Series B) and have a 40 kDa PEG attached to the 5′-terminus (not shown). (B) Structures of 13 different side chains are shown along with their cLogP and plasma clearance values obtained for sequences from Series A and B with each side chain. The abbreviations for the side chains (X group) are Im [(2-imidazolylethyl)carboxamide], MOE [(morpholino-2-ethyl)carboxamide], Thr [(R-2-hydroxypropyl)carboxamide], RTHF (R-tetrahydrofuranylmethylcarboxamide), Pyr [(4-pyridylmethyl)carboxamide], Trp [(3-indole-2-ethyl)carboxamide], MBn [(3,4-methylenedioxybenzyl)carboxamide], Bn (benzylcarboxamide), FBn [(4-fluorobenzyl)carboxamide], cHM [(1-cyclohexylmethyl)carboxamide], Pe [(2-phenylethyl)carboxamide], Nap [(1-naphthylmethyl)carboxamide], and 2Nap [(2-naphthylmethyl)carboxamide]. (C) Correlation between cLogP and CHI for Series A and B. (D) Rat plasma clearance (CL) values are plotted versus the cLogP values of the side chains. Data points are grouped by negative or positive cLogP values using gray boxes. (E) Volumes of distribution at steady state (Vss) are plotted for sequences with a negative cLogP or positive cLogP for Series A and B. Median values for each group are indicated with a horizontal bar. CHI, chromatographic hydrophobicity index.
Plasma PK results obtained following a single 1.2–2.0 mg/kg IV bolus dose in male Sprague Dawley rats showed that variants with side chains with a negative cLogP are cleared more slowly than those with a positive cLogP (P = 0.005) (Fig. 2B and 2D). Median (range) clearance was 3.3 mL/(h*kg) (1.2, 8.8) for the five variants with side chains having a negative cLogP and 15 mL/(h*kg) (12, 19) for the seven variants with side chains having a positive cLogP. We resisted the temptation to fit a model to this data set as there is no significant correlation between cLogP and clearance within the two subsets of data points; those with a negative cLogP or those with a positive cLogP (Fig. 2D). For example, a linear model produced slopes [95% confidence interval (CI)] of 4.1 (−4.9, 13) and −0.78 (−7.6, 6.1) for the negative and positive cLogP subsets, respectively, with corresponding coefficient of determination (r2) values of 0.41 and 0.017. A correlation may become clear when further data are available.
To verify these results, a second series of 13 sequences based on the SL1033 core sequence (Series B) was designed and tested. This series consisted of 28-mers each containing six modified nucleotides. The inclusion of C3 spacers in place of 2′-OMe nucleotides at positions 13 and 14 served to further differentiate this series from Series A (Fig. 2A). Plasma PK results, obtained as before, confirmed that modified aptamers containing side chains with a negative cLogP are cleared more slowly than those containing side chains with a positive cLogP (P = 0.01). Median (range) clearance was 2.1 mL/(h*kg) (1.0, 3.7) for the hydrophilic side chain variants and 6.5 mL/(h*kg) (5.3, 13) for the hydrophobic side chain variants (Fig. 2B and 2D). Again, no significant correlation was observed between cLogP and clearance within the subsets of sequences that contain side chains with either a negative or positive cLogP. The linear model produced slopes (95% CI) of 1.1 (−2.6, 4.8) and 2.1 (−2.1, 6.4) for the negative and positive cLogP subsets, respectively, with corresponding coefficient of determination (r2) values of 0.23 and 0.20.
As expected, the hydrophobicity of the side chain did not affect the volume of distribution at steady state (Vss), since these are large PEGylated molecules. Median (range) Vss was 39 (28, 61) and 43 mL/kg (39, 49) for Series A, and 42 (27, 51) and 40 mL/kg (33, 58) for Series B, with negative and positive cLogP values, respectively (Fig. 2E).
Regardless of the type of side chain, sequences within Series B were generally cleared more slowly than sequences within Series A. Because Series B sequences contained fewer modified bases (6 vs. 8) and fewer nucleotides (26 nucleotides and two C3 spacers vs. 32 nucleotides), we investigated the effect of both the number of side chains with a positive cLogP and aptamer length on clearance.
The number of hydrophobic side chains directly correlates with clearance
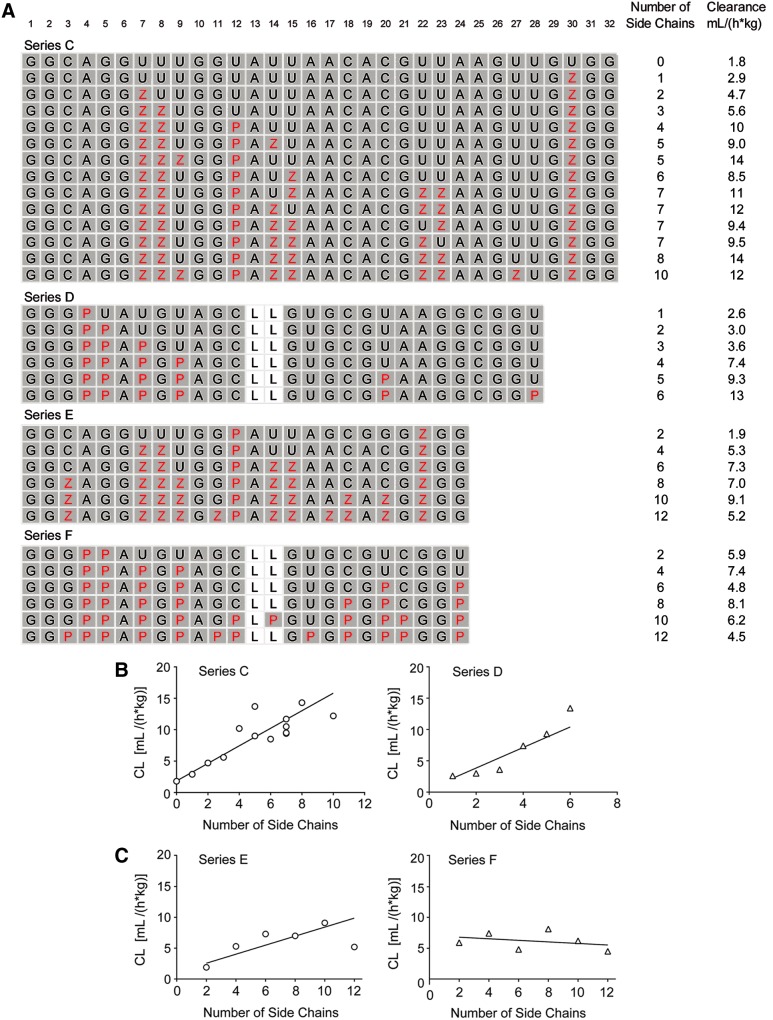
To determine the effect of the number of hydrophobic side chains on plasma clearance, we focused again on core sequences of SL1026 and SL1033. Two series were designed in which aptamer length was held constant, whereas the number of side chains was varied. Series C was created based on Series A in which the total number of Bn and Nap side chains was varied between 0 and 10. Similarly, Series D was created based on Series B in which the number of Nap side chains was varied between 1 and 6 (Fig. 3A). Plasma clearance values for both series were plotted versus the number of side chains (Fig. 3B). Results showed a distinct trend toward faster plasma clearance as the number of side chains increased. For Series C, consisting of 32-mers, a linear least-squares analysis of the data resulted in a reasonable fit with an r2 of 0.90. For this series, clearance increased by 1.4 mL/(h*kg) for every additional side chain [95% CI; 1.1, 1.7 mL/(h*kg)]. Similarly for Series D, consisting of 28-mers, a linear fit of the data yielded a coefficient of determination (r2) of 0.84. For this series, clearance increased by 1.6 mL/(h*kg) for every additional side chain [95% CI; 0.66, 2.6 mL/(h*kg)].
FIG. 3.
Effect of number of side chains. (A) Individual sequences (5′→3′) that make up Series C, D, E, and F designed for the determination of the effect of the number of side chains on rat plasma PK. Standard single-letter designations (A, C, G, and U) are used for the unmodified dehydroxy nucleobases, whereas positions designated Z and P (highlighted in red text) are as described in Fig. 1. The positions containing 2′-OMe ribose are shaded gray. All sequences contain a 3′-idT cap, are fully modified with 2′-OMe ribose except for positions containing a C3 spacer (L) in place of a nucleotide (positions 13 and 14 of Series D and F) and have a 40 kDa PEG attached to the 5′-terminus (not shown). The number of side chains and plasma clearance is shown next to each sequence. (B) Linear regression analyses of plasma clearance (CL) versus the number of side chains in Series C and D. (C) Linear regression analyses of plasma clearance (CL) versus the number of side chains in Series E and F.
Because of the correlation observed between clearance and number of side chains in 32-mer and 28-mer sequences, we also determined the effect in shorter sequences, which are often reported for aptamers. In this study, the number of side chains was varied between 2 and 12 in two series of 24-mers (Series E and F based on Series A and B, respectively) (Fig. 3A). Each of these series consisted of six sequences with Bn and Nap (Series E) or Nap (Series F) side chains.
The observed correlation between clearance and the number of side chains in these two series was considerably weaker than observed for longer sequences. For Series E, a linear fit of the data returned a coefficient of determination (r2) of 0.70, but clearance only increased by 0.73 mL/(h*kg) for every additional side chain [95% CI; 0.07, 1.4 mL/(h*kg)], whereas no statistically significant correlation was observed for Series F [slope (95% CI) −0.13 (−0.63, 0.38)] (Fig. 3C).
The length of aptamers with hydrophobic side chains also directly correlates with clearance
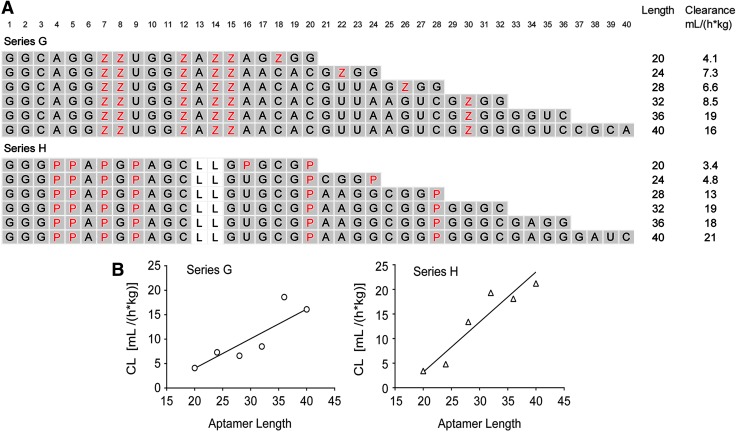
To further investigate the surprising effect of length on plasma clearance, we designed two new series of sequences in which the number of modified nucleotides with a positive cLogP was held constant while the length was varied. As described in Fig. 4A, these two series, designated Series G and H, were based on Series A and B, respectively. Both series contained six modified nucleotides (Bn or Nap), whereas the length was varied between 20 and 40 synthons. Results indicated a clear trend toward slower clearance as aptamer length decreased (Fig. 4B). Linear fits produced slopes (95% CI) of 0.61 (0.22, 1.0) and 1.0 (0.57, 1.5) for Series G and H, respectively, with corresponding r2 values of 0.82 and 0.91.
FIG. 4.
Effect of sequence length. (A) Individual sequences (5′→3′) that make up modified aptamer Series G and H designed for the determination of the effect of aptamer length on plasma PK. Standard single-letter designations (A, C, G, and U) are used for the unmodified dehydroxy nucleobases, whereas positions designated Z or P (highlighted in red text) are as described in Fig. 1. The positions containing 2′-OMe ribose are shaded gray. All sequences contain a 3′-idT cap, are fully modified with 2′-OMe ribose, except for positions containing a C3 spacer (L) in place of a nucleotide (positions 13 and 14 of Series H) and have a 40 kDa PEG attached to the 5′-terminus (not shown). Aptamer length and plasma clearance (CL) are shown alongside each sequence. (B) Linear regression analyses of plasma clearance versus aptamer length in Series G and H.
These results suggest that plasma clearance of modified aptamers can be slowed by using shorter sequences with fewer side chains. Based on these guidelines, we determined the plasma clearance of two active PEGylated modified aptamers that met these criteria: SL1028, a 17-mer with five side chains (Bn, Nap, and Pe) and two 2′-OMe substitutions [18], and SL1068, a 21-mer with five side chains (Nap) and eleven 2′-OMe substitutions. The clearance rate of SL1028 was 6.9 mL/(h*kg), whereas SL1068 was 4.4 mL/(h*kg) (Table 1). Both aptamers had clearance rates at the lower end of the spectrum, despite not being fully protected from endonuclease activity.
Table 1.
Pharmacokinetic Properties of Shorter Active Aptamers with Side Chains
| Modified aptamer | Aptamer length | Number of side chains | Number of 2′-OMe | t1/2 (h) | Clearance [mL/(h*kg)] |
|---|---|---|---|---|---|
| SL1028 | 17 | 5 | 2 | 3.9 | 6.9 |
| SL1068 | 21 | 5 | 11 | 9.6 | 4.4 |
2′-OMe, 2′-methoxy.
Discussion
The addition of 5-uracil modifications to aptamers has led to improvements in discovery success rate, target binding affinity, inhibition potency, and metabolic stability [19,23]. While each of these properties is beneficial for the development of therapeutic aptamers, the clinical potential of an aptamer drug will also depend on its PK properties, especially for indications requiring systemic administration and long-term treatment. Unlike antibodies, which have half-lives measured in weeks, aptamers typically have half-lives on the order of hours, and only after extensive modification of the backbone and termini to prevent renal filtration and degradation by nucleases. With features of both aptamers (backbone and nucleobases) and antibodies (side chains), we expected these 5-uracil modified aptamers to have PK properties at least comparable to those exhibited by aptamers with similar metabolic stability (or nuclease resistance), and were surprised to observe the rapid clearance of SL1026 and SL1033.
It is clear from data presented here that the side chains and the overall hydrophobic character they impart to the aptamer have a profound impact on the PK properties. Both the number and hydrophobicity of the side chains contribute to the clearance rate, as aptamers possessing side chains with positive cLogP values were cleared more quickly than those with negative cLogP values. Aptamer length is another important factor influencing plasma clearance. A positive correlation between sequence length and clearance rate was observed for Series C and D variants, with an apparent PK advantage for sequences less than about 24 nucleotides in length. Furthermore, the effect of the number of side chains with a positive cLogP on clearance was considerably smaller in the shorter sequences (≤24-mers) than in longer sequences (≥28-mers).
The effect of hydrophobicity on clearance is reminiscent of antibody–drug conjugates (ADC), where hydrophobic drugs and linkers have been shown to negatively impact the PK properties of the ADC. The potency of ADC is improved by increasing the drug-to-antibody ratio in vitro, but this leads to faster plasma clearance in vivo due to the increase in hydrophobicity of the molecule [30]. By manipulating linkers to reduce hydrophobicity, the required PK properties of these conjugates have been obtained. One proposed mechanism for this clearance effect is that hydrophobic ADC are cleared more rapidly by the reticuloendothelial system than unconjugated antibodies [31,32]. It is tempting to speculate that a similar mechanism may also be responsible for the observed effect for modified aptamers with more hydrophobic side chains, but assessing the source of this effect will require additional experiments. If such a cellular uptake clearance pathway is operative for modified aptamers, our data suggest that it may be a receptor-mediated interaction with the oligonucleotide portion of the molecule, where binding affinity is dependent on both aptamer length and the number of hydrophobic side chains. Cellular uptake may be dependent on binding to a specific receptor or through nonspecific binding to any number of targets on the cell surface. Saturable, size-dependent uptake of SSOs has been previously demonstrated, consistent with receptor-mediated endocytosis [11,33,34].
The studies presented here were performed with test sequences containing 2′-OMe modifications at all nucleotide positions and the aptamers were therefore inactive and possibly unstructured. To determine if the same set of guidelines apply to active, structured aptamers, we confirmed slower clearance rates for two 5-uracil modified aptamers from our collection with short lengths (17 and 21 nucleotides) and only five side chains. These examples demonstrate the ability to realize the benefits of the hydrophobic modifications without compromising plasma residence time.
These results have important implications for the design of new therapeutic aptamers with diversity-enhancing side chains. Plasma residence times may be increased through the development of shorter aptamers (≤24-mers) or those with more hydrophilic side chains, or both. Although aromatic side chains with a net positive cLogP have to date been more successful in SELEX than those with a net negative cLogP, conditions that better favor the selection of the latter have been relatively unexplored. Ironically, selection of shorter modified aptamers may be accomplished by further increasing the chemical diversity through SELEX libraries that contain 5-position modifications at both pyrimidines (uridines and cytidines), effectively doubling down on the number of side chains [35]. Such extensively modified aptamers will also allow the exploration of the effect of a mixture of hydrophilic and hydrophobic side chains on binding, metabolic stability, and PK, with the aim of identifying molecules with optimized drug-like properties.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the SomaLogic Process Chemistry and Analytical Sciences groups for synthesis and quality analysis of all oligonucleotides described in the article. SOMAmer® reagent is a registered trademark of SomaLogic, Inc.
Author Disclosure Statement
S.G., D.W.D., S.K.W., S.M.W., J.C.R., J.D.C., W.S.M., M.R.O., C.R.F., D.J.S., and N.J. are employees and stakeholders of SomaLogic, Inc. T.S., M.H., and Y.I. are employees and stakeholders of Otsuka Pharmaceutical Co., Ltd.
References
- 1.Dougan H, Lyster DM, Vo CV, Stafford A, Weitz JI. and Hobbs JB. (2000). Extending the lifetime of anticoagulant oligodeoxynucleotide aptamers in blood. Nucl Med Biol 27:289–297 [DOI] [PubMed] [Google Scholar]
- 2.Bouchard PR, Hutabarat RM. and Thompson KM. (2010). Discovery and development of therapeutic aptamers. Annu Rev Pharmacol Toxicol 50:237–257 [DOI] [PubMed] [Google Scholar]
- 3.Keefe AD, Pai S. and Ellington A. (2010). Aptamers as therapeutics. Nat Rev Drug Discov 9:537–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watson SR, Chang YF, O'Connell D, Weigand L, Ringquist S. and Parma DH. (2000). Anti-L-selectin aptamers: binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo. Antisense Nucleic Acid Drug Dev 10:63–75 [DOI] [PubMed] [Google Scholar]
- 5.Geary RS, Norris D, Yu R. and Bennett CF. (2015). Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87:46–51 [DOI] [PubMed] [Google Scholar]
- 6.Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C. and McCauley TG. (2004). Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res 21:2234–2246 [DOI] [PubMed] [Google Scholar]
- 7.Rusconi CP, Roberts JD, Pitoc GA, Nimjee SM, White RR, Quick G, Jr., Scardino E, Fay WP. and Sullenger BA. (2004). Antidote-mediated control of an anticoagulant aptamer in vivo. Nat Biotechnol 22:1423–1428 [DOI] [PubMed] [Google Scholar]
- 8.Lee CH, Lee SH, Kim JH, Noh YH, Noh GJ. and Lee. SW. (2015). Pharmacokinetics of a cholesterol-conjugated aptamer against the hepatitis C virus (HCV) NS5B protein. Mol Ther Nucleic Acids 4:e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haruta K, Otaki N, Nagamine M, Kayo T, Sasaki A, Hiramoto S, Takahashi M, Hota K, Sato H. and Yamazaki H. (2017). A novel PEGylation method for improving the pharmacokinetic properties of anti-interleukin-17A RNA aptamers. Nucleic Acid Ther 27:36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimjee SM, White RR, Becker RC. and Sullenger BA. (2017). Aptamers as Therapeutics. Annu Rev Pharmacol Toxicol 57:61–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koller E, Vincent TM, Chappell A, De S, Manoharan M. and Bennett CF. (2011). Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res 39:4795–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Souza EB, Cload ST, Pendergrast PS. and Sah DW. (2009). Novel therapeutic modalities to address nondrugable protein interaction targets. Neuropsychopharmacology 34:142–158 [DOI] [PubMed] [Google Scholar]
- 13.Kryza D, Debordeaux F, Azema L, Hassan A, Paurelle O, Schulz J, Savona-Baron C, Charignon E, Bonazza P, et al. (2016). Ex vivo and in vivo imaging and biodistribution of aptamers targeting the human matrix metalloprotease-9 in melanomas. PLoS One 11:e0149387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drolet DW, Green LS, Gold L. and Janjic N. (2016). Fit for the eye: aptamers in ocular disorders. Nucleic Acid Ther 26:127–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Que-Gewirth NS. and Sullenger BA. (2007). Gene therapy progress and prospects: RNA aptamers. Gene Ther 14:283–291 [DOI] [PubMed] [Google Scholar]
- 16.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, et al. (2010). Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One 5:e15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies DR, Gelinas AD, Zhang C, Rohloff JC, Carter JD, O'Connell D, Waugh SM, Wolk SK, Mayfield WS, et al. (2012). Unique motifs and hydrophobic interactions shape the binding of modified DNA ligands to protein targets. Proc Natl Acad Sci U S A 109:19971–19976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelinas AD, Davies DR, Edwards TE, Rohloff JC, Carter JD, Zhang C, Gupta S, Ishikawa Y, Hirota M, et al. (2014). Crystal structure of interleukin-6 in complex with a modified nucleic acid ligand. J Biol Chem 289:8720–8734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarvis TC, Davies DR, Hisaminato A, Resnicow DI, Gupta S, Waugh SM, Nagabukuro A, Wadatsu T, Hishigaki H, et al. (2015). Non-helical DNA triplex forms a unique aptamer scaffold for high affinity recognition of nerve growth factor. Structure 23:1293–1304 [DOI] [PubMed] [Google Scholar]
- 20.Rohloff JC, Gelinas AD, Jarvis TC, Ochsner UA, Schneider DJ, Gold L. and Janjic. N. (2014). Nucleic acid ligands with protein-like side chains: modified aptamers and their use as diagnostic and therapeutic agents. Mol Ther Nucleic Acids 3:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaught JD, Bock C, Carter J, Fitzwater T, Otis M, Schneider D, Rolando J, Waugh S, Wilcox SK. and Eaton BE. (2010). Expanding the chemistry of DNA for in vitro selection. J Am Chem Soc 132:4141–4151 [DOI] [PubMed] [Google Scholar]
- 22.Caruthers MH, Barone AD, Beaucage SL, Dodds DR, Fisher EF, McBride LJ, Matteucci M, Stabinsky Z. and Tang JY. (1987). Chemical synthesis of deoxyoligonucleotides by the phosphoramidite method. Methods Enzymol 154:287–313 [DOI] [PubMed] [Google Scholar]
- 23.Gupta S, Hirota M, Waugh SM, Murakami I, Suzuki T, Muraguchi M, Shibamori M, Ishikawa Y, Jarvis TC, et al. (2014). Chemically modified DNA aptamers bind interleukin-6 with high affinity and inhibit signaling by blocking its interaction with interleukin-6 receptor. J Biol Chem 289:8706–8719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nomura Y, Ueno Y. and Matsuda A. (1997). Site-specific introduction of functional groups into phosphodiester oligodeoxynucleotides and their thermal stability and nuclease-resistance properties. Nucleic Acids Res 25:2784–2791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito T, Ueno Y, Komatsu Y. and Matsuda A. (2003). Synthesis, thermal stability and resistance to enzymatic hydrolysis of the oligonucleotides containing 5-(N-aminohexyl)carbamoyl-2′-O-methyluridines. Nucleic Acids Res 31:2514–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig J, Eckstein F. (1989). Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J Org Chem 54:631–635 [Google Scholar]
- 27.Sanghvi YS, Guo Z, Pfundheller HM. and Converso A. (2000). Improved process for the preparation of nucleosidic phosphoramidites using a safer and cheaper activator. Org Process Res Dev 4:175–181 [Google Scholar]
- 28.Tucker CE, Chen LS, Judkins MB, Farmer JA, Gill SC. and Drolet DW. (1999). Detection and plasma pharmacokinetics of an anti-vascular endothelial growth factor oligonucleotide-aptamer (NX1838) in rhesus monkeys. J Chromatogr B Biomed Sci Appl 732:203–212 [DOI] [PubMed] [Google Scholar]
- 29.Hirota M, Murakami I, Ishikawa Y, Suzuki T, Sumida S, Ibaragi S, Kasai H, Horai N, Drolet DW, et al. (2016). Chemically modified interleukin-6 aptamer inhibits development of collagen-induced arthritis in cynomolgus monkeys. Nucleic Acid Ther 26:10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyon RP, Bovee TD, Doronina SO, Burke PJ, Hunter JH, Neff-LaFord HD, Jonas M, Anderson ME, Setter JR. and Senter PD. (2015). Reducing hydrophobicity of homogeneous antibody-drug conjugates improves pharmacokinetics and therapeutic index. Nat Biotechnol 33:733–735 [DOI] [PubMed] [Google Scholar]
- 31.Boswell CA, Mundo EE, Zhang C, Bumbaca D, Valle NR, Kozak KR, Fourie A, Chuh J, Koppada N, et al. (2011). Impact of drug conjugation on pharmacokinetics and tissue distribution of anti-STEAP1 antibody-drug conjugates in rats. Bioconjug Chem 22:1994–2004 [DOI] [PubMed] [Google Scholar]
- 32.Flygare JA, Pillow TH. and Aristoff P. (2013). Antibody-drug conjugates for the treatment of cancer. Chem Biol Drug Des 81:113–121 [DOI] [PubMed] [Google Scholar]
- 33.Loke SL, Stein CA, Zhang XH, Mori K, Nakanishi M, Subasinghe C, Cohen JS. and Neckers LM. (1989). Characterization of oligonucleotide transport into living cells. Proc Natl Acad Sci U S A 86:3474–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Juliano RL, Ming X. and Nakagawa O. (2012). Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug Chem 23:147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gawande BN, Rohloff JC, Carter JD, von Carlowitz I, Zhang C, Schneider DJ. and Janjic N. (2017). Selection of DNA aptamers with two modified bases. Proc Natl Acad Sci U S A 114:2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.