Abstract
High-throughput screening and activity-guided purification identified nicoyamycin A, a natural product comprised of an uncommon 3-methyl-1,4-dioxane ring incorporated into a desferrioxamine-like backbone via a spiroaminal linkage. Nicoyamycin A potently inhibits uropathogenic Escherichia coli growth in low iron medium, a promising step toward developing novel antibiotics to treat recalcitrant bacterial infections.
The intrinsic and acquired antibiotic resistance mechanisms among Gram-negative bacteria have stimulated intense efforts to discover novel therapeutics effective against these bacteria. The double layered membranes and multi-drug resistant efflux pumps impede the accumulation of drugs in bacterial cells.1 These innate attributes of Gram-negative bacteria, along with genetic mechanisms, have led to a rapid rise in the rates of antibiotic resistance and mortality associated with Gram-negative infections, causing alarm at the prospect of pan-antibiotic resistant bacteria.2 Uropathogenic E. coli (UPEC) causes 70–80% of all uncomplicated urinary tract infections (UTIs). The urgent challenge of UPEC is that one in forty women suffer recurrent UTIs, which requires frequent administration of antibiotics, sometimes even prophylactically for extended periods.3, 4 Thus, it is not surprising that multidrug resistance is steadily increasing among UPEC isolates. These trends limit treatment choices and foreshadow a costly shift from oral to injectable antibiotics for a common bacterial infections.2, 5–8
During infection, bacteria must scavenge iron from host stores to survive. As a protective mechanism the host sequesters iron, but in response to the nutrient limited environment, microbial pathogens express an array of iron acquisition systems that capture this critical metal co-factor from host stores.9 We previously demonstrated that UPEC iron-acquisition systems are up-regulated during infection and are required for full virulence in a murine model of UTI.10–21
High-throughput screens (HTS) for new antibiotic scaffolds to treat Gram-negative infections have typically utilized in vitro assays or tolC mutants to circumvent drug efflux.22 This increases the likelihood of discovering a molecule that binds to the desired target, but does not address the issue of overcoming the barrier of the Gram-negative cell envelope. Moreover, most chemical libraries utilized for HTS are comprised of synthetic small molecules with limited chemical diversity and ability to cross bacterial cell envelopes.23 Natural product libraries, however, offer a wider variety of novel drug scaffolds with diverse chemical features that may facilitate effective permeation of bacterial cell envelopes.23 Motivated by these advantages, we probed a unique natural product extract (NPE) library to identify new antibiotic scaffolds that naturally penetrate and accumulate in an unmodified UPEC isolate as a model Gram-negative pathogen.24 We specifically screened for molecules that inhibit the growth of UPEC in low-iron medium, analogous to the conditions encountered during infection.
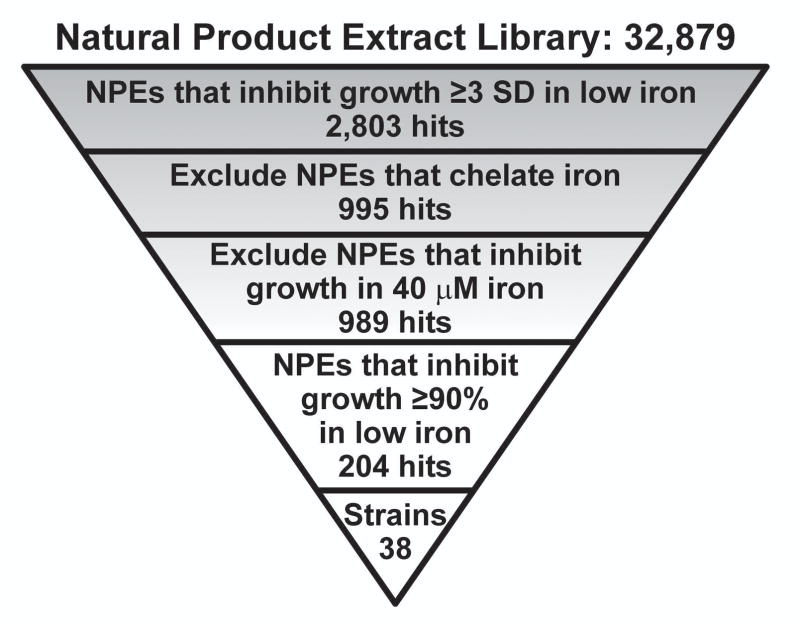
Wildtype E. coli CFT073, a model UPEC strain, was used to screen 32,879 NPEs in the UM Center for Chemical Genomics library. UPEC was prepared in MOPS minimal medium lacking iron (MOPS-Fe) and inoculated into wells containing the NPEs. Growth inhibition was monitored by measuring the optical density at 600 nm (OD600). 2,803 NPEs inhibited UPEC growth and were classified as active extracts (hits) (Fig. 1 and ESI).
Fig. 1.
A schematic overview of the HTS performed to identify NPEs that inhibit wildtype UPEC growth in low iron medium is presented. SD = standard deviation.
To identify hits targeting cellular processes, not simply depleting iron from the medium, NPEs were assayed for iron chelation properties using the chromazural S (CAS) reagent in a counter screen.25 NPEs containing an iron chelator were eliminated, resulting in 995 remaining hits (Fig. 1). These 995 hits were confirmed in triplicate growth assays. A secondary screen was simultaneously executed to ensure hits specifically inhibited growth under low iron conditions. Six NPEs that inhibited the growth of UPEC in the presence of iron were eliminated (Fig. 1). From the 989 hits, a prioritized list of 38 Streptomyces strains that produced active NPEs was compiled. These strains were revived and their NPEs regenerated from small-scale fermentations. The NPE from Streptomyces nicoyae (strain #34401-A3) inhibited UPEC growth in low iron conditions and represented the highest priority hit based on potency.
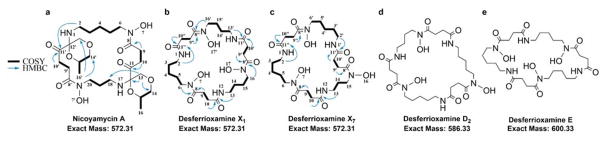
The active component(s) in the S. nicoyae extract were isolated using bioactivity-guided fractionation (ESI). We obtained an active fraction that contained a single pure molecule with an m/z of 573.3287 (ESI), which we named nicoyamycin A (NicA). NicA was purified as a light-yellow amorphous solid and possesses a molecular formula of C26H44N4O10. The planar structure of NicA was determined by an extensive multi-dimensional NMR analysis (Fig. 2a and ESI).
Fig. 2.
Structures of NicA and four desferrioxamines isolated from S. nicoyae. The structures of (a) NicA, (b) DesX1, and (c) DesX7 were solved using NMR and tandem MS. The structures of (d) DesD2 and (e) DesE were previously solved using NMR and the molecules were confirmed by tandem MS and 1H NMR.
1D NMR and gHSQCAD data, recorded in CD3OD, indicated the presence of at least six oxygenated carbons; also, four downfield methylene groups were observed, indicating linkage to amides (Fig. 2a). 1H NMR revealed at least one methyl group at δH 1.16 (d) and tell-tale methylene signals in the δH 1.80–2.78 region. Inspection of 13C NMR and gHMBC spectra identified six quaternary carbons, four of which were carbonyl carbons. gCOSY and gHMBC cross peaks revealed aliphatic chains extending from C-2 to C-6; C-9 to C-10; C-14 to C-16; and C-18 to C-20. gHMBC correlations between H-9/9′ to C-8/8′ and H-10/10′ to C-11/11′ confirmed the presence of succinyl groups. The uncommon 3-methyl-1,4-dioxane ring was deduced based on the downfield methine at δH-15/15′ 3.72 (δC-15/15′ 75.5) that showed gCOSY correlations with methyl and methylene groups (δH-16/16′ 1.16 and δH-14/14′ 3.48, respectively). And, H-14/14′ showed a gHMBC correlation with C-13/13′, which in turn had a gHMBC cross peak with C-12/12′ (δC-12/12′ 142.5), closing the dioxane ring. The methyl-dioxane ring is inserted between asymmetric diaminoalkyl-succinyl moieties via a spiroaminal motif as justified by the gHMBC correlation from H-13 to C-18, establishing the complete planar structure of NicA (Fig. 2a). 1H NMR data collected with NicA dissolved in DMSO-d6 identified two protons on secondary amines and two N-hydroxyl groups. The gCOSY collected with NicA dissolved in DMSO-d6 revealed coupling between H-1 and H-2, completing the ornithine moiety (Fig. 2a). The planar structure of NicA was confirmed by tandem MS (ESI). The compound consists of four stereocenters; to ascertain the absolute conformation, a chemical synthetic approach is required, owing to the extremely poor yield of NicA.
The diaminoalkyl-succinyl backbone of NicA is also reported in the desferrioxamine family of siderophores. Subsequent efforts to purify more NicA, yielded several desferrioxamines from S. nicoyae. Desferrioxamine X1 (DesX1), DesX7, DesD2, and DesE were all purified in quantities sufficient for structure confirmation and structure-activity relationship (SAR) studies. DesX1 and DesX7 were previously predicted based only on tandem MS profiles of NPEs from Erwinia amylovora cultured in low iron medium.26 DesX1 production from Streptomyces olivaceus Tű2718 cultures treated with exogenous 1,4-diaminobutane was also observed by MS.27‡ Here, both DesX1 and DesX7 were isolated from S. nicoyae cultured in rich medium without supplementation of pre-cursors. We confirmed that the previously predicted structures were correct using multi-dimensional NMR and tandem MS (Fig. 2b, 2c and ESI).26 The DesX1 and DesX7 NMR spectra were similar to NicA, except that the gHSQCAD spectrum showed only methylene groups, in agreement with a cyclic desferrioxamine structure. For DesX1, gCOSY and gHMBC cross peaks revealed aliphatic chains from C2 to C6; C9 to C10; and C13 to C16; integrating the aliphatic δH peaks in the DesX1 1H NMR spectrum indicated at least two diaminobutyl chains from C-13/13′ to C-16/C-16′ (Fig. 2b). The only observed difference between DesX1 and DesX7 was the integration of the aliphatic δH peaks suggesting that the diaminopentyl chain (C-2/2′ to C-6/6′) was present in a 2:1 ratio with the diaminopropyl chain (C-13 to C15) in DesX7 (Fig. 2c). The structures of DesD2 and DesE were confirmed by tandem MS and 1H NMR spectra (Fig. 2d, 2e and ESI).26, 28, 29 DesX1, DesX7, DesD2 and DesE are all tri-cyclic desferrioxamines, differing only in the length of the diaminoalkyl chains (Fig. 2).
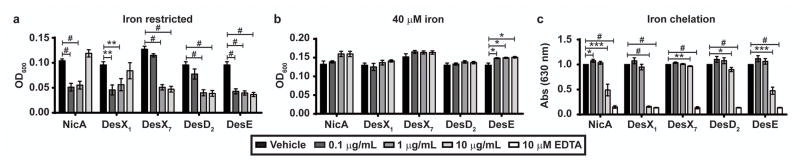
The isolated desferrioxamines were evaluated for activity against UPEC in MOPS-Fe and iron chelating properties, compared to NicA. No molecules inhibited UPEC growth in MOPS-Fe supplemented with 40 μM FeSO4 (Fig. 3B). The concentration of NicA, DesX1, and DesE that restricts UPEC growth to 50% of vehicle alone (IC50) is 0.1 μg/mL (~170 nM) (Fig. 3A). The IC50 of DesX7 and DesD2 are 1 μg/mL (~1.7 μM). At 10 μg/mL (17.5 μM), DesX1 chelates iron approximately at a 50% level compared to vehicle, DesD2 chelates iron at a 10% level, and DesX7 chelates iron at 3% compared to the control (Fig. 3C).
Fig. 3.
Antibacterial activity of NicA and other isolated molecules. UPEC was cultured for 18 h in (a) MOPS-Fe or (b) MOPS-Fe supplemented with 40 μM FeSO4 and the indicated pure molecule (x-axis). Growth was quantified by measuring OD600. (c) Iron chelation was quantified using the chrome azural S (CAS) assay and a decrease in absorbance at 630 nm represents iron chelation. The shading of the gray bars indicates the final concentration of the pure molecules. Shown is an average of 3 replicates. Error bars represent one standard deviation from the mean. Significance was calculated using an unpaired t test in Prism and determined using the Holm-Sidak method with alpha = 0.05. Each row was analyzed individually without assuming a consistent standard deviation where *, p < 0.05; **, p < 0.01; ***, p < 0.001; #, p < 0.0001.
In summary, we report the discovery of a novel molecule, nicoyamycin A, from S. nicoyae that restricts UPEC growth in low iron medium. DesX1, DesX7, DesD2, and DesE were also isolated from this strain and the predicted structures of DesX1 and DesX7 were validated by multi-dimensional NMR. The five compounds were used to conduct initial SAR studies. NicA, DesX1, and DesE are the most potent inhibitors of UPEC growth in low iron medium and the most potent iron chelators, suggesting that the primary mechanism could be due to iron chelation. However, DesX7 and DesD2 also inhibit UPEC growth in low iron, but are not strong iron chelators. Thus, as with other therapeutics with iron chelating properties (e.g. tetracycline, doxorubicin, 8-hydroxy-quinolines30–32), it is likely that the molecules described here have pleiotropic effects on UPEC growth.
Throughout the screening process, we intentionally eliminated hits with iron chelating activity (Fig. 1). The high frequency of iron chelating NPEs is likely due to the mixture of molecules present in NPEs and the inherent iron chelating properties of some natural products, which does not necessarily represent their primary biological functions.30, 31, 33–37 Despite our best efforts to eliminate metabolites that chelate iron, once single molecule purity was achieved, NicA was found to chelate iron in the CAS assay. It is possible that this property was initially masked due to the low concentration of NicA in the fractions, and the CAS assay was not sufficiently sensitive. Alternatively, there may have been appreciable levels of iron in the NPE samples and, therefore, NicA was metal-bound prior to being tested in the CAS assay. If the latter was the case, then the possibility that metal-bound NicA was present in appreciable quantities, yet still inhibited UPEC growth, suggests that the molecule exerts its effects by mechanisms other than iron chelation. This could also explain why at higher concentrations these molecules sometimes increase UPEC growth in low iron, by providing exogenous iron to the bacteria (Fig. 3A). Separating the iron chelation versus growth inhibition properties represents a primary challenge in future studies of NicA and its analogues.
The poorly understood balance between iron chelation and other mechanisms of action apply both to NicA, and to the cyclic desferrioxamines. It is not unprecedented that siderophores have multiple effects on the cell. Evidence supports that siderophore functions can also include the transport of other transition metals, heavy metals, and non-metals; protection from oxidative stress; intra- and inter-cellular signaling; and delivery of antibiotic warheads.36, 38, 39 A concerted effort is required to decipher the relative contributions of iron chelation and other cellular effects to the mechanism of action exerted by NicA and the desferrioxamines. Such studies, which will require total synthesis of the molecules and a medicinal chemistry approach, could also help determine the stereochemistry of NicA and potentially provide access to more potent antibiotics.
Complementary to understanding the activity of NicA and the desferrioxamines towards UPEC is the fascinating opportunity S. nicoyae offers for studying the biosynthesis of these metabolites. NicA and the desferrioxamines were all isolated from a Streptomyces isolate cultured in rich medium and as such, S. nicoyae is the first known Streptomyces strain to naturally produce the 1,3-diaminopropane containing NicA and DesX7. S. nicoyae production of NicA and DesX7 suggests that the desferrioxamine biosynthetic gene cluster found in S. nicoyae has unusual substrate flexibility and may act in concert with other enzymes to form the methyl-dioxane ring. Identifying and probing the S. nicoyae biosynthetic gene cluster responsible for NicA and desferrioxamine synthesis will provide an exciting resource for answering these intriguing biochemical questions.
The identification of nicoyamycin A with nanomolar potency against wildtype UPEC is a promising step towards developing novel antibiotics active against recalcitrant Gram-negative pathogens. Furthermore, this discovery has identified gaps in our understanding of the biological activity of siderophores and the mechanisms by which they are assembled. Not only has a new antibiotic scaffold been identified, but also a new Streptomyces species that may provide key insights to desferrioxamine biosynthesis, modification, and function. These unexpected benefits emphasize the importance of probing NPE libraries for new bioactive metabolites bearing unique structural characteristics.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH) R01 DK097362, R35 GM118101, and R01 AI116791; Fogarty International Center U01 TW007404; and University of Michigan’s Fast Forward Medical Innovation’s MTRAC for Life Sciences Innovation Hub. We are grateful to the Technical Office, CONAGEBIO, Ministry of the Environment and Telecommunications, Costa Rica for sample collection permits. LAM was supported by NIH T32 AI007528 and an ASM Career Development Grant for Postdoctoral Women; and is a Research Scholars Fellow for the American Urological Association administered by the Urology Care Foundation. DHS thanks the Hans W. Vahlteich Professorship for partial support of this research. The contents of this manuscript are that of the authors and do not necessarily represent the views of the NIH.
Footnotes
S. olivaceus Tű 2718 did not produce DesX7 when cultured with 1,3-diaminopropane, suggesting that the desferrioxamine biosynthetic cluster in S. olivaceus is not sufficiently flexible to accept the 1,3-diaminopropane subunit.27
There are no conflicts to declare.
Electronic Supplementary Information (ESI) includes supporting figures, NMR spectra, characterization data, and experimental procedures. See DOI: 10.1039/x0xx00000x
Notes and references
- 1.Tommasi R, Brown DG, Walkup GK, Manchester JI, Miller AA. Nat Rev Drug Discov. 2015;14:529–542. doi: 10.1038/nrd4572. [DOI] [PubMed] [Google Scholar]
- 2.WHO. Antimicrobial resistance: Global report on surveillance. Apr, 2014. [Google Scholar]
- 3.Beerepoot MJ, ter Riet G, Nys S, et al. Arch Intern Med. 2011;171:1270–1278. doi: 10.1001/archinternmed.2011.306. [DOI] [PubMed] [Google Scholar]
- 4.Barber AE, Norton JP, Spivak AM, Mulvey MA. Clin Infect Dis. 2013;57:719–724. doi: 10.1093/cid/cit284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gupta K, Hooton TM, Naber KG, Wullt B, Colgan R, Miller LG, Moran GJ, Nicolle LE, Raz R, Schaeffer AJ, Soper DE. Clinical infectious diseases: An official publication of the Infectious Diseases Society of America. 2011;52:e103–120. doi: 10.1093/cid/ciq257. [DOI] [PubMed] [Google Scholar]
- 6.Moura A, Nicolau A, Hooton T, Azeredo J. J Appl Microbiol. 2009;106:1779–1791. doi: 10.1111/j.1365-2672.2008.04115.x. [DOI] [PubMed] [Google Scholar]
- 7.Foxman B, Ki M, Brown P. Clinical infectious diseases: An official publication of the Infectious Diseases Society of America. 2007;45:281–283. doi: 10.1086/519267. [DOI] [PubMed] [Google Scholar]
- 8.Karlowsky JA, Hoban DJ, Decorby MR, Laing NM, Zhanel GG. Antimicrob Agents Ch. 2006;50:2251–2254. doi: 10.1128/AAC.00123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Subashchandrabose S, Mobley HL. Metallomics. 2015;7:935–942. doi: 10.1039/c4mt00329b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia EC, Brumbaugh AR, Mobley HL. Infect Immun. 2011;79:1225–1235. doi: 10.1128/IAI.01222-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torres AG, Redford P, Welch RA, Payne SM. Infec Immun. 2001;69:6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hagan EC, Lloyd AL, Rasko DA, Faerber GJ, Mobley HL. PLoS Pathog. 2010;6:e1001187. doi: 10.1371/journal.ppat.1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagan EC, Mobley HL. Mol Microbiol. 2009;71:79–91. doi: 10.1111/j.1365-2958.2008.06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russo TA, McFadden CD, Carlino-MacDonald UB, Beanan JM, Barnard TJ, Johnson JR. Infect Immun. 2002;70:7156–7160. doi: 10.1128/IAI.70.12.7156-7160.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. Infect Immun. 2014;82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Subashchandrabose S, Hazen TH, Brumbaugh AR, Himpsl SD, Smith SN, Ernst RD, Rasko DA, Mobley HL. P Natl Acad Sci USA. 2014;111:18327–18332. doi: 10.1073/pnas.1415959112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walters MS, Mobley HL. J Microbiol Meth. 2009;78:131–135. doi: 10.1016/j.mimet.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Snyder JA, Haugen BJ, Buckles EL, Lockatell CV, Johnson DE, Donnenberg MS, Welch RA, Mobley HL. Infect Immun. 2004;72:6373–6381. doi: 10.1128/IAI.72.11.6373-6381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alteri CJ, Hagan EC, Sivick KE, Smith SN, Mobley HLT. PLoS Pathog. 2009;5:e1000586. doi: 10.1371/journal.ppat.1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alteri CJ, Mobley HL. Infect Immun. 2007;75:2679–2688. doi: 10.1128/IAI.00076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson JR, Jelacic S, Schoening LM, Clabots C, Shaikh N, Mobley HL, Tarr PI. Infect Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yep A, McQuade T, Kirchhoff P, Larsen M, Mobley HLT. mBio. 2014;5:e01089–01013. doi: 10.1128/mBio.01089-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown DG, Lister T, May-Dracka TL. Bioorg Med Chem Lett. 2014;24:413–418. doi: 10.1016/j.bmcl.2013.12.059. [DOI] [PubMed] [Google Scholar]
- 24.Magarvey NA, Keller JM, Bernan V, Dworkin M, Sherman DH. Appl Environ Microb. 2004;70:7520–7529. doi: 10.1128/AEM.70.12.7520-7529.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwyn B, Neilands JB. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 26.Feistner GJ, Stahl DC, Gabrik AH. Org Mass Spectrom. 1993;28:163–175. [Google Scholar]
- 27.Meiwes J, Fiedler HP, Zähner H, Konetschny-Rapp S, Jung G. Appl Microbiol Biot. 1990;32:505–510. doi: 10.1007/BF00173718. [DOI] [PubMed] [Google Scholar]
- 28.Maehr H, Benz W, Smallheer J, Williams Thomas H. Z Naturforsch B. 1977;32:937. [Google Scholar]
- 29.Lee HS, Shin HJ, Jang KH, Kim TS, Oh KB, Shin J. J Nat Prod. 2005;68:623–625. doi: 10.1021/np040220g. [DOI] [PubMed] [Google Scholar]
- 30.Grenier D, Huot MP, Mayrand D. Antimicrob Agents Ch. 2000;44:763–766. doi: 10.1128/aac.44.3.763-766.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mjos KD, Cawthray JF, Jamieson G, Fox JA, Orvig C. Dalton T. 2015;44:2348–2358. doi: 10.1039/c4dt02934h. [DOI] [PubMed] [Google Scholar]
- 32.Prachayasittikul V, Prachayasittikul S, Ruchirawat S, Prachayasittikul V. Drug Des Dev Ther. 2013;7:1157–1178. doi: 10.2147/DDDT.S49763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park SR, Tripathi A, Wu J, Schultz PJ, Yim I, McQuade TJ, Yu F, Arevang CJ, Mensah AY, Tamayo-Castillo G, Xi C, Sherman DH. Nat Commun. 2016;7:10710. doi: 10.1038/ncomms10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Déjugnat C, Diat O, Zemb T. ChemPhysChem. 2011;12:2138–2144. doi: 10.1002/cphc.201100094. [DOI] [PubMed] [Google Scholar]
- 35.Hatcher HC, Singh RN, Torti FM, Torti SV. Future Med Chem. 2009;1 doi: 10.4155/fmc.4109.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Messa E, Carturan S, Maffè C, Pautasso M, Bracco E, Roetto A, Messa F, Arruga F, Defilippi I, Rosso V, Zanone C, Rotolo A, Greco E, Pellegrino RM, Alberti D, Saglio G, Cilloni D. Haematologica. 2010;95:1308–1316. doi: 10.3324/haematol.2009.016824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang KH, King ONF, Tumber A, Woon ECY, Heightman TD, McDonough MA, Schofield CJ, Rose NR. ChemMedChem. 2011;6:759–764. doi: 10.1002/cmdc.201100026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnstone TC, Nolan EM. Dalton T. 2015;44:6320–6339. doi: 10.1039/c4dt03559c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sidebottom AM, Johnson AR, Karty JA, Trader DJ, Carlson EE. ACS Chem Biol. 2013;8:2009–2016. doi: 10.1021/cb4002798. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.