Abstract
Chl1 DNA helicase promotes sister chromatid cohesion and associates with both the cohesion establishment acetyltransferase Eco1/Ctf7 and the DNA polymerase processivity factor PCNA that supports Eco1/Ctf7 function. Mutation in CHL1 results in precocious sister chromatid separation and cell aneuploidy, defects that arise through reduced levels of chromatin-bound cohesins which normally tether together sister chromatids (trans tethering). Mutation of Chl1 family members (BACH1/BRIP/FANCJ and DDX11/ChlR1) also exhibit genotoxic sensitivities, consistent with a role for Chl1 in trans tethering which is required for efficient DNA repair. Chl1 promotes the recruitment of Scc2 to DNA which is required for cohesin deposition onto DNA. There is limited evidence, however, that Scc2 also directs the deposition onto DNA of condensins which promote tethering in cis (intramolecular DNA links). Here, we test the ability of Chl1 to promote cis tethering and the role of both Chl1 and Scc2 to promote condensin recruitment to DNA. The results reveal that chl1 mutant cells exhibit significant condensation defects both within the rDNA locus and genome-wide. Importantly, chl1 mutant cell condensation defects do not result from reduced chromatin binding of condensin, but instead through reduced chromatin binding of cohesin. We tested scc2-4 mutant cells and similarly found no evidence of reduced condensin recruitment to chromatin. Consistent with a role for Scc2 specifically in cohesin deposition, scc2-4 mutant cell condensation defects are irreversible. We thus term Chl1 a novel regulator of both chromatin condensation and sister chromatid cohesion through cohesin-based mechanisms. These results reveal an exciting interface between DNA structure and the highly conserved cohesin complex.
Introduction
Structural changes to the genome that occur over the cell cycle are fundamental yet mysterious features that underlie many cellular events. During G1 phase of the cell cycle, chromatin compaction and higher order DNA assemblies termed TADS (topological associated domains) are largely regional [1], [2]. These cis-based (intramolecular) and trans-based (intermolecular) tetherings of DNA segments must remain dynamic to allow for plasticity and appropriate transcriptional responses to external cues such as changes in temperature, nutrient levels and signaling factors [1], [3–5]. During S phase, trans tethers are established specifically between the products of chromosome replication, termed sister chromatids. These trans tethers remain stable and thus identify chromatids as sisters until anaphase onset. Cis tethers established during prophase also are stable—maintaining fully condensed and disentangled chromosomes through mitosis. These cis tethers are required for high fidelity chromosome segregation and the positioning of chromosomes away from the cytokinetic furrow. In an impressive coopting of function through evolution, each of these tethering activities in combination are mediated by SMC (stability of minichromosomes or structural maintenance of chromosomes) complexes that include cohesins (Smc1, Smc3, Mcd1/Scc1/RAD21, Pds5, Scc3/Irr1/SA1,2 and Sororin in vertebrate cells) and condensins (Smc2/Cut14, Smc4/Cut3, Ycs4/Cnd1/DPY-28, Ycg1/Cdn3/CAP-G1, Brn1/Cdn2/DPY-26) [1], [2], [6], [7].
Divisions between SMC complex functions are not always distinct. For instance, it is well established that cohesins form both cis and trans tethers that function in DNA replication, repair, chromosome segregation, chromatin condensation and transcription regulation [1], [2]. Thus, mutations of cohesin pathways produce aneuploidy, are tightly correlated with numerous cancers and directly result in severe developmental maladies that include Robert Syndrome, Cornelia de Lange Syndrome and Warsaw Breakage Syndrome [2], [8], [9]. Condensins on the other hand, which primarily tether DNA segments in cis conformations, provide for longitudinal chromatin compaction, removal of DNA catenations, chromosomal disentanglement, and dosage compensation [6], [7]. Mutations of condensation pathways result in T cell lymphomas, colon cancer, microcephaly, and are predictors of cancer survivorship [10–14]. Mechanistically, convincing evidence suggests that both cohesins and condensins entrap individual DNA segments within a topologically closed structure. In turn, DNA segment tethering requires oligomerization of the appropriate SMC complexes, although little is known regarding how these oligomerization steps are directed toward either cis or trans conformations [1], [15–17].
The targeting and deposition of cohesins and condensins onto DNA represents a critical regulatory mechanism that spans a wide range of cellular activities, but remains largely undefined. What is clear is that cohesin deposition onto DNA requires the loader complex comprising Scc2/NIPBL and Scc4/MAU-2 [18–22]. One particular study, however, implicated Scc2,4 in the recruitment of condensin to DNA, a finding largely based on fluorescent intensity levels performed on chromosome spreads [23]. In yeast, Scc2,4 recruitment to DNA is regulated at the level of DNA structure and requires the conserved Chl1 DNA helicase [24–26]. At least during S phase, Scc2 deposition appears coordinated with DNA replication fork progression given that Chl1 physically interacts with numerous DNA replication fork factors (PCNA, Rad27/FEN1, MCMs) and the S phase acetyltransferase Eco1/Ctf7 [24], [25], [27–30]. Thus, Chl1 DNA helicase appears as the earliest regulator identified to date of Scc2 and cohesin recruitment to DNA.
Despite the wealth of evidence that Chl1 is critical for sister chromatid trans-tethering [27], [31–34], a role for Chl1 in cis-tethering remains untested. The issue is a critical one given that mutations in the Chl1 human homologs ChlR1/DDX11 and BACH1/BRIP1/FANCJ collectively result in Warsaw Breakage Syndrome, Fanconi anemia, cell aneuploidy and breast and ovarian cancers [27], [31], [32], [34–40]. Moreover, the extent to which Chl1 DNA helicase regulation of Scc2 translates to both cohesin and condensin recruitment to chromatin is unknown, revealing a significant deficit in our understanding of these clinically relevant processes. Here, we report that Chl1 and Scc2 are indeed regulators of genome-wide condensation, but that these roles occur independent of condensin binding to DNA and instead rely primarily on cohesin function.
Materials and methods
Yeast strains and strain construction
Saccharomyces cerevisiae strains used in this study are listed in Table 1. GFP-tagging and deletion of genes were performed following a published protocol [41].
Table 1. Yeast strains used in this study.
| Strain name | Genotype | Reference |
|---|---|---|
| YBS1019 | MATa; S288C | [25] |
| YBS1041 | MATa; chl1::KAN; S288C | [27] |
| YBS2020 | MATa; NET1:GFP:HIS3; w303 | For this study |
| YBS2078 | MATa; lacOs::YLR003c-1; lacOs::MMP1; LacI-GFP; w303 | Y2869 [23] |
| YBS2079 | MATa; chl1::TRP; lacOs::YLR003c-1; lacOs::MMP1; LacI-GFP; w303 | For this study |
| YBS2080 | MATa; NET1:GFP:HIS3; chl1::TRP; w303 | For this study |
| YDS101 | MATa; SMC2:3HA:KanMX6; w303 | For this study |
| YDS104 | MATa; SMC2:3HA:KanMX6; chl1::TRP; w303 | For this study |
| YDS108 | MATa; SMC2:3HA:KanMX6; scc2-4; w303 | For this study |
| YMM551 | MATa; scc2-4; can1-100; w303 | [91] |
rDNA condensation assays
rDNA condensation assays were performed using Net1-GFP as previously described with the following modifications [23], [42]. Briefly, cells were cultured to log phase (OD600 between 0.2 to 0.4), then incubated for 2.5 hours at 23°C in rich YPD medium supplemented with alpha-factor. The resulting synchronized G1 cells were collected, washed, resuspended in fresh YPD supplemented with nocodazole, and incubated for 3 hours at 23°C. The resulting preanaphase cells were fixed by incubation in 3.7% paraformaldehyde for 10 min at 30°C. GFP signals were then assayed microscopically. Cell cycle progression was confirmed by detection of DNA content using flow cytometry as described [42].
rDNA condensation was independently assessed using a streamlined condensation assay adapted from a published FISH protocol [43–45]. Briefly, log phase cells (OD600 between 0.2 to 0.4) were incubated for 2.5 hours at 23°C in rich YPD medium supplemented with alpha-factor. The resulting synchronized G1 cells were collected, washed, resuspended in fresh YPD supplemented with nocodazole, and incubated for 3 hours at 23°C (where appropriate, additional temperature shifts are described within each experimental design). The resulting preanaphase cells were fixed by incubation in 37% formaldehyde for 2 hours at 23°C. Cells were washed with distilled water and resuspended in buffer (1 M sorbitol, 20 mM KPO4, pH 7.4), then spheroplasted by the addition of beta-mercaptoethanol and Zymolyase 100T and incubation for 1 hour at 23°C. The resulting spheroplasted cells were placed onto poly-L-lysine coated slides prior to addition of 0.5% Triton X-100 and 0.5% SDS solution. The slides were then incubated in 3:1 methanol:acetic acid solution and stored at 4°C until completely dry. Slides were then treated with RNase in 2X SSC buffer (0.3 M NaCl, 30 mM Sodium Citrate, pH 7.0) followed by washes in 2X SSC and then by a series of cold ethanol washes. Slides were then incubated at 72°C in 70% formamide with 2X SSC followed by a series of cold ethanol washes. DNA masses were detected by DAPI staining and assayed microscopically. Cell cycle progression was confirmed by detection of DNA content using flow cytometry as described [42].
Chromosome arm condensation assay
Chromosome arm condensation assays were performed in yeast cells that contained two lacO repeats, one integrated telomere-proximal on the left arm and another integrated centromere-proximal on the right arm of chromosome XII. Each LacO cassette was monitored microscopically through detection of lacI-GFP [23]. Condensation assays and quantification were performed as previously described with the following modifications [23]. Briefly, log phase cells (OD600 between 0.2 to 0.4) were incubated for 2.5 hours at 23°C in rich YPD medium supplemented with alpha-factor. The resulting synchronized G1 cells were collected, washed, resuspended in fresh YPD supplemented with nocodazole and incubated for 3 hours at 23°C. The resulting preanaphase cells were fixed by incubation in 3.7% paraformaldehyde for 10 min at 30°C. Wildtype and chl1 mutant cells that contained a single GFP dot reflected sister chromatids positioned vertical to the z-axial focal plane and were thus excluded from analysis. We also excluded cells that contained three or four GFP loci, since the cohesion defect at one or both loci made it difficult to determine which GFP dot represented an intra- or inter-sister chromatid locus. Distances between two GFP dots were quantified microscopically with images captured using iVision. Cell cycle progression was confirmed by detection of DNA content using flow cytometry as described [42].
Chromatin binding assay
Nocodazole arrested cells were harvested and processed for chromatin binding assay [25]. Briefly, the densities of 50 ml cultures were normalized to an OD600 between 0.4–0.6. Cells were spun down and washed with 25 ml cold sterile water, followed by a wash in 1.2 M sorbitol. Pellets were resuspended in 1 ml CB1 buffer (50 mM Sodium citrate, 40 mM EDTA, 1.2 M sorbitol, pH 7.4) prior to the addition of 125 μl of spheroplast solution (125 μl CB1, 50 μl zymolase, 5 μl BME) and incubation with gentle shaking for 1 hour at 23°C. The spheroplast suspensions were supplemented with protease inhibitor cocktail, washed 2X with 1.2 M cold sorbitol, resuspended in 425 μl of 1.2 M cold sorbitol and snap frozen in liquid Nitrogen. Frozen samples were thawed on ice prior to the addition of 50 μl lysis buffer (500 mM Lithium acetate, 20 mM MgSO4, 200 mM HEPES, pH 7.9) and 20 μl of 25% Triton-X-100. Whole cell extract (WCE) fractions were collected and denatured by the addition of an equal volume of 2X Laemmli buffer, boiled for 5 minutes and then snap frozen. The remaining lysates were centrifuged at 12,000 g for 15 minutes. Supernatants consisting of soluble fractions were collected and denatured by the addition of an equal volume of 2X Laemmli buffer. Pellets were resuspended in Lysis buffer with 150 mM NaCl and centrifuged at 15,000 g for 15 minutes. Chromatin bound fractions were obtained by suspending the resulting pellets 1.2 M sorbitol and then denatured by the addition of an equal volume of 2X Laemmli buffer. Whole cell extract, soluble and chromatin bound fractions were resolved by SDS-PAGE electrophoresis and analyzed by Western blot using anti-HA (1:2000) (Santa Cruz), anti-PGK (1:20000) (Invitrogen) with goat anti mouse HRP (1:50000) (Bio-Rad) or by anti-H2B (1:2000) (Santa Cruz) or 1:60000 (Abcam), anti-Mcd1 [46] in combination with goat anti rabbit HRP (1:50000) (Bio-Rad) and ECL prime (GE Healthcare) for visualization.
Results
Chl1 DNA helicase promotes rDNA condensation
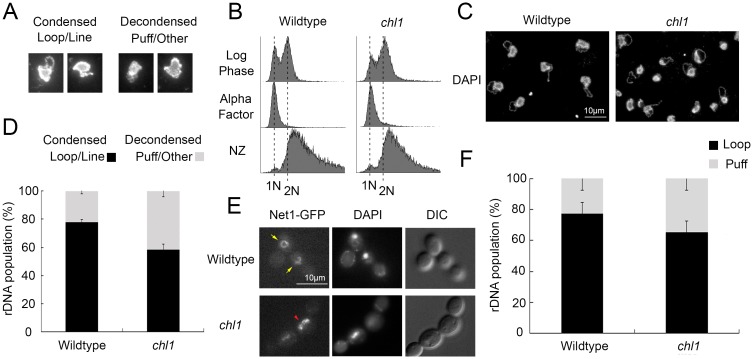
Chl1 DNA helicase is critical for Scc2 recruitment to DNA [25], but the extent through which SMC-dependent DNA compaction is regulated through Chl1 remains untested. Here, we exploit the structural changes that rDNA undergoes across the cell cycle. In yeast, rDNA comprises up to 150 copies of linearly arrayed 9 kb sequence that form a diffuse and amorphous puff-like structure during G1 and condense into a discrete line and loop-like structure during mitosis [43], [44]. [47], [48]. Fluorescence in situ Hybridization (FISH) is a well-established methodology for detecting structural changes of rDNA loci, but is both time intensive and involves sequential application of three different antibodies and labeled probe [43], [44]. Previously, we developed and validated a streamlined condensation assay based on FISH but one that produces exquisite imaging of rDNA in the absence of antibodies and hybridization of labeled probe (Fig 1A) [45]. To assess the impact of Chl1 helicase on rDNA structure, wildtype and chl1 deletion cells were synchronized in G1 using medium supplemented with alpha factor, washed and released into fresh medium supplemented with nocodazole for 3 hours. The resulting synchronized pre-anaphase cells were then processed to quantify the status of rDNA condensation (Fig 1B). The results confirm that wildtype cells exhibit high levels (78%) of tightly condensed (loop/line-like) rDNA loci while chl1 mutant cells exhibit significantly lower levels (58%) of condensed rDNA loci. In fact, chl1 mutant cells exhibited nearly twice the frequency of decondensed rDNA than wildtype cells (Fig 1C and 1D).
Fig 1. Chl1 helicase promotes rDNA condensation.
A) Representative examples of micrographs that highlight condensed (loop and line) and decondensed (amorphous puff-like and other non-discrete configuration) rDNA structures. B) Flow cytometer of DNA content at times indicated throughout the experimental procedure. Cells were maintained in nocodazole for 3 hours at 23°C post-alpha factor arrest. C) Chromosome mass and rDNA detected using DAPI in wildtype (YBS1019) and chl1 mutant (YBS1041) strains. D) Quantification of condensed (loop/line) and decondensed (puff/other) rDNA populations in wildtype and chl1 mutant cells. Data quantified from 3 biological replicates, 100 cells for each strain analyzed per replicate and statistical analysis performed using Student's T-test (p = 0.005). E) rDNA structures visualized using Net1-GFP, genome DNA detected using DAPI, and cell morphology images obtained using Differential Interference Contrast (DIC) microscopy. Yellow arrows indicate condensed rDNA loop/line and red arrowhead indicates decondensed rDNA puff. F) Quantification of condensed (loop) and decondensed (puff) rDNA populations in wildtype (YBS2020) and chl1 mutant (YBS2080) cells. Data quantified from 3 biological replicates, 100 cells for each strain analyzed per replicate and statistical analysis performed using Student's T-test (p = 0.006). Statistical significant differences (*) are based on p < 0.05.
Chl1 DNA helicase is not essential for DNA replication, but we were nonetheless concerned that loss of Chl1 might produce a minor cell cycle delay that could be misinterpreted as a condensation defect. We assessed this possibility in three ways. First, we assessed cells after only 2.5 hours of preanaphase synchronization in medium supplemented with nocodazole. The results obtained by flow cytometry clearly reveal that both wildtype and chl1 mutant cells are synchronized at this early step in the arrest protocol (S1 Fig). Thus, any imperceptible cell cycle delays will be fully compensated for by the additional 30 minutes of synchronization in the procedure described above. Second, we exploited the well-established changes in yeast cell morphology in which G1 cells are unbudded, S phase entry typically correlates with bud emergence, and subsequent G2 and M phases denoted by increased bud growth [49]. Our results reveal nearly identical large budded populations of wildtype and chl1 mutant cells after 3 hours arrest in nocodazole (S1 Fig). Third, we quantified the minor 1N DNA peak in the preanaphase arrested cultures. The results reveal that 9.6% of wildtype cells exhibit a 1N DNA content while only 6.2% chl1 mutant cells exhibit a 1N DNA content. Thus, chl1 mutant cells arrest in preanaphase as efficiently as wildtype cells, negating the model that the two-fold increase in decondensed rDNA is due to cell cycle progression defects (S1 Fig).
Net1 is an rDNA binding protein such that Net1-GFP is another established method from which to monitor for architectural changes within the rDNA locus [23], [26], [42], [45], [50]. To independently assess for rDNA condensation defects in chl1 mutant cells, Net1-GFP transformants of wildtype and chl1 deletion cells were synchronized in pre-anaphase (S1 Fig), and the status of rDNA condensation (loops/lines versus puffs) quantified as previously described [23], [42–44]. As expected, wildtype cells exhibited high levels (77%) of condensed (loop/line-like) rDNA loci. In contrast, chl1 mutant cells exhibited significantly lower levels (65%) of condensed rDNA loci and over a 50% increase in the number of rDNA puff structures, compared to wildtype cells (Fig 1E and 1F). Importantly, the impact of chl1 mutation on rDNA structure occurs in the absence of shifting to elevated temperatures (37°C), a procedure that significantly impacts rDNA structure even in wildtype cells [45]. These combined results therefore reveal that Chl1 promotes condensation of the rDNA locus.
Chl1 helicase promotes arm condensation
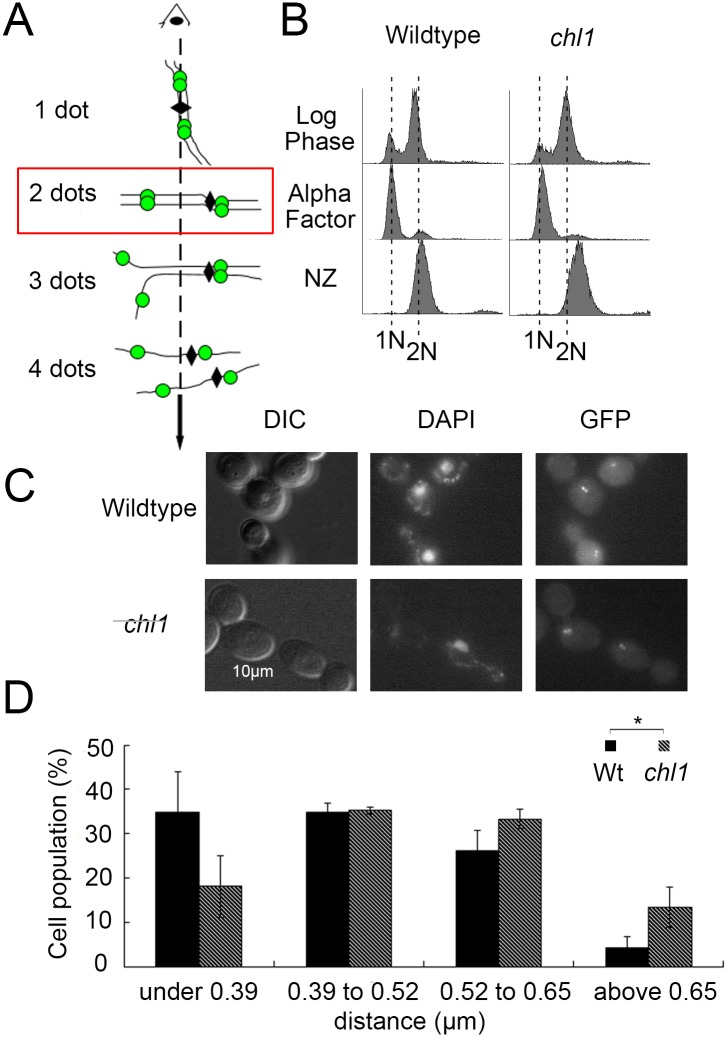
rDNA is unique in architecture, associated factors, transcription regulation, and recombination frequency compared to the remainder of the genome [51]. Thus, it became important to assess if Chl1 is an rDNA-specific regulator of condensation or instead impacts condensation genome-wide. We obtained from the lab of Dr. Frank Uhlmann a chromosome arm condensation assay strain that contains two lacO repeats, one integrated telomere-proximal on the left arm and another integrated centromere-proximal on the right arm of chromosome XII [23]. Each LacO cassette is detectable through lacI-GFP binding such that the inter-GFP distance allows for quantification of chromosome arm condensation (Fig 2A). Isogenic chromosome arm condensation assay strains, except for deletion of CHL1, were synchronized in G1 using alpha factor, washed and released into fresh medium supplemented with nocodazole (Fig 2B). The resulting pre-anaphase synchronized cells were then fixed in paraformaldehyde and the disposition of arm condensation quantified by measuring the distance between GFP loci. As expected, chl1 mutant cells exhibit cohesion defects [27], [32] and we therefore encountered a range of detectable GFP loci. We thus limited our analysis to wildtype and chl1 mutant cells that contained only 2 GFP dots and in which both were resolvable within a single focal plane (Fig 2A). We found a wide range of inter-GFP distances both in wildtype and chl1 mutant cells. Regardless, the results reveal that 70% of wildtype cells exhibited inter-GFP distances under 0.52 μm. In contrast, only 53% of chl1 mutant cells exhibited inter-GFP distances under 0.52 μm. In fact, chl1 mutant cells exhibited inter-GFP distances above 0.65 μm at roughly 3 times the frequencies of wildtype cells (Fig 2C and 2D). These results reveal for the first time that Chl1 plays a genome-wide role in chromosome condensation.
Fig 2. Chl1 helicase promotes chromosome arm condensation.
A) Schematic of chromosome conformations and GFP-labeled loci (dashed line indicates Z-axial microscope orientation, solid line indicates sister chromatid, diamond indicates centromere, green dot indicate GFP locus). Red box indicates cells analyzed. B) Flow cytometer data of DNA content throughout the experiment. Cells were maintained in nocodazole for 3 hours at 23°C post-alpha factor arrest. C) Micrographs of representative fields of view that include GFP loci, genomic mass (DAPI) and cell morphology (DIC). D) Distribution of distances measured between GFP dots in wildtype (YBS2078) and chl1 mutant (YBS2079) cells. Data obtained from 3 biological replicates, 100 cells for each strain analyzed per replicate and statistical analysis performed using Student's T-test. p = 3.862E-6 indicates the significant differences between the average distance (0.44 μm) in wildtype cells versus the average distance (0.50 μm) in chl1 mutant cells. Statistical significant differences (*) are based on p < 0.05.
Chl1 and Scc2 function in condensation independent of condensin deposition
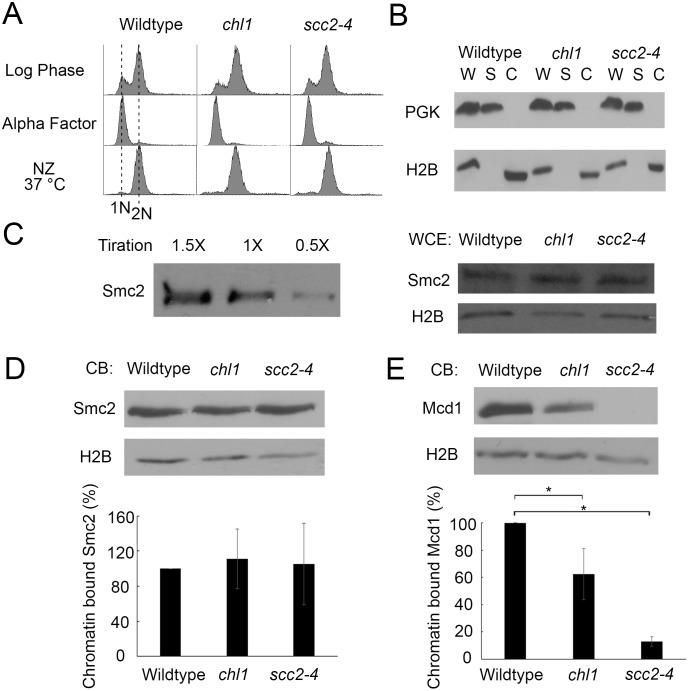
What is the mechanism through which Chl1 and Scc2 function in condensation? Chl1 is well-documented as a cohesion regulator that is critical for Scc2 recruitment to chromatin [25]. In turn, Scc2 is essential for cohesin deposition onto chromatin, but a role for Scc2 in condensin deposition remains controversial [19], [23]. Thus, it became critical to differentiate between models that Chl1 promotes chromatin compaction through either Scc2-dependent regulation of condensins, cohesins, or both. We first tested whether the condensation defect produced in chl1 mutant cells occurs through the reduction of condensin deposition onto DNA. Condensin subunit Smc2 was epitope-tagged as the sole source of Smc2 function in both wildtype and chl1 mutant cells. We included scc2-4 cells in our analyses so that we could directly compare the roles of Chl1 and Scc2 on condensin deposition. We then exploited Triton X-100 cell fractionation assays previously used to demonstrate chromatin-association of a spectrum of factors that include Ctf7/Eco1, cohesin subunits, DNA replication initiators and fork stabilization proteins [25], [52–57]. Wildtype, chl1 and scc2-4 single mutant strains each expressing Smc2-HA were synchronized in G1 (alpha factor), washed and released at 37°C (non-permissive for scc2-4) into fresh medium supplemented with nocodazole (Fig 3A). We validated the cell fractionation procedure using Phosphoglycerokinase (PGK) and Histone 2B (H2B) as cytosolic and chromatin fiduciary markers, respectively. The results show efficient enrichment of H2B, and undetectable levels of PGK, in Triton-X-100 insoluble chromatin bound fractions (Fig 3B). Smc2 titration demonstrates that protein loading is within the linear range of detection (Fig 3B). We first compared the total levels of Smc2-HA, normalized to H2B levels, in whole cell extractions obtained from wildtype, chl1 and scc2-4 single mutant cells. The results from whole cell extracts document that Smc2-HA levels are unaffected in either chl1 or scc2-4 mutations, compared to wildtype cells (Fig 3C). We then compared the levels of chromatin-bound Smc2-HA. The result revealed that chromatin-bound Smc2-HA levels in both chl1 and scc2-4 mutant cells are not reduced, compared to wildtype cells (Fig 3D). Thus, chl1 and scc2-4 single mutant cells exhibit significant condensation defects despite full retention of chromatin-bound condensin. These results document that the condensation defects exhibited by chl1 and scc2-4 single mutant cells occur largely independent of changes in condensin deposition onto DNA.
Fig 3. Chl1 helicase promotes chromosome condensation through cohesin, but not condensin, regulation.
A) Flow cytometer data of DNA content throughout the experiment. Cells were maintained in nocodazole for 3 hours at 37°C post-alpha factor arrest. B) Fractionation of preanaphase-arrested wildtype (YDS101), chl1 (YDS104) and scc2-4 (YDS108) cells. Phosphoglycerate kinase (PGK) and Histone 2B (H2B) indicate levels of cytoplasmic and chromatin-bound proteins, respectively, in whole cell extracts (W), cytoplasmic soluble fractions (S) and chromatin bound fractions (C). C) Left: Titration of Smc2-HA indicates 1X sample concentration is in the linear range of detection. Right: Whole cell extracts of Smc2-HA in wildtype, chl1 and scc2-4 cells. H2B is shown as internal loading control. All samples reflect 1X concentration levels. D) Top: Chromatin-bound (CB) fraction of Smc2-HA in wildtype, chl1 and scc2-4 cells. Chromatin-bound H2B levels are shown as internal loading control. Bottom: Quantification of Smc2-HA binding to chromatin in chl1 and scc2-4 mutant cells, based on the ratio of Smc2-HA to H2B levels and normalized to wildtype levels of Smc2-HA obtained from 3 biological replicates. E) Top: Chromatin-bound fraction of Mcd1 in wildtype, chl1 and scc2-4 cells. Chromatin-bound H2B levels are shown as loading controls. Bottom: Quantification of Mcd1 binding to chromatin in chl1 and scc2-4 mutant cells, based on the ratio of Mcd1 to H2B levels and normalized to wildtype levels obtained from 3 biological replicates. Statistical analysis was performed using one-way ANOVA followed by post-hoc Tukey HSD Test. (p = 0.024 for chromatin bound Mcd1 in wildtype versus chl1 mutant cells. p = 0.001 for chromatin bound Mcd1 in wildtype cells versus scc2-4 mutant cells). Statistical significant differences (*) are based on p < 0.05.
Chl1 and Scc2 function in condensation through cohesins
Having eliminated reduced condensin deposition as a central mechanism through which chl1 and scc2-4 mutants produce condensation defects, we turned to cohesin deposition. Wildtype, chl1 and scc2-4 single mutant strains were released from G1 into 37°C medium supplemented with nocodazole and the resulting preanaphase cells assayed for cohesin deposition. As before, we confirmed the Triton X-100 cell fractionation assay using PGK and H2B as cytosolic and chromatin fiduciary markers, respectively (Fig 3B). We and others previously ascertained that Mcd1 levels in whole cell lysates are unaffected in either chl1 or scc2-4 mutant cells [19], [25]. Thus, we compared the levels of chromatin bound Mcd1, normalized to H2B levels, in wildtype and chl1 and scc2-4 single mutant cells using a previously validated Mcd1/Scc1-directed antibody generously provided by Dr. Vincent Guacci of the Koshland Lab [44]. As expected from prior studies, Mcd1 binding to DNA to significantly reduced in chl1 (38%) and scc2-4 (82%) single mutant cells (Fig 3E). Notably, the reductions in Mcd1 binding mirrored the severity of the condensation defect, strongly indicative of a dose-dependent cohesin mechanism (Fig 3E). Independently, we repeated the assessment of Smc2-HA in these chromatin fractions now validated for scc2-4 inactivation through reduced cohesin levels. Our results confirm that Scc2 inactivation has negligible effects on condensin deposition.
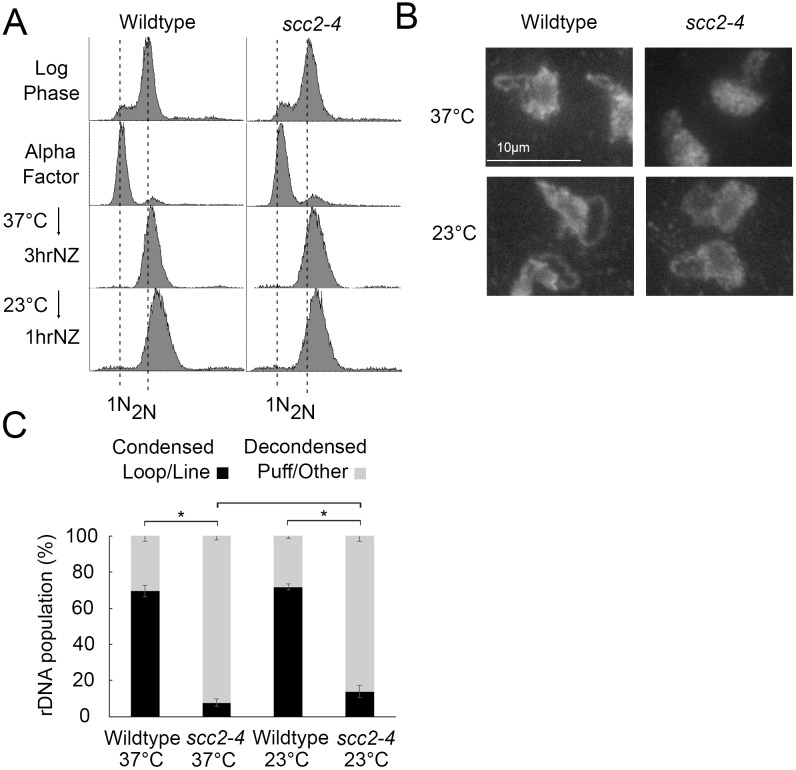
While rDNA condensation defects are completely reversible in condensin mutant cells, condensation defects are irreversible in cohesin mutants [58]. If Scc2 promotes condensation only through cohesin deposition, then condensation should be irreversible in scc2-4 mutant cells. To test this prediction, wildtype and scc2-4 mutant strains were released from G1 into 37°C medium supplemented with nocodazole for 3 hours and the resulting preanaphase cultures then shifted back to 23°C for 1 hour (Fig 4A). Cell samples harvested both during the preanaphase arrest at 37°C and after the shift down to 23°C were then assessed for the disposition of rDNA condensation as previously described [45]. As expected, preanaphase wildtype cells arrested at 37°C exhibited high levels (69%) of condensed (loop-like) rDNA loci while scc2-4 mutant cells instead exhibited significantly low levels (8%) of condensed rDNA loci (Fig 4B). Upon shifting down to 23°C, preanaphase wildtype cells continued to exhibit high levels (71%) of condensed (loop-like) rDNA loci. Importantly, scc2-4 mutant cells also exhibited significantly low levels (14%) of condensed rDNA loci (Fig 4C). The predominantly irreversible condensation defect in scc2-4 mutant cells mirrors that of cohesin mutants and is distinct from the complete rescue of condensation defects exhibited in condensin mutant cells following the same regimen [58]. The combination of these results strongly suggest that the condensation defects exhibited by chl1 and scc2-4 mutant cells occur predominantly through a reduction in cohesin, but not condensin, chromatin association.
Fig 4. Condensation is irreversible in scc2-4 mutants.
A) Flow cytometer data reveals DNA content throughout the experimental analyses. Cells were maintained in nocodazole, post-alpha factor release, for 3 hours at 37°C followed by an additional 1 hour at 23°C. B) Chromosome mass and rDNA structures detected using DAPI in wildtype and scc2-4 mutant strains. C) Quantification of condensed (loop/line) and decondensed (puff/other) rDNA populations in wildtype (YBS1019) and scc2-4 mutant (YMM551) cells. Quantifications and statistical analyses of rDNA condensation were obtained from 3 biological replicates for each strain (wildtype and scc2-4 mutant cells) in which each replicate included 100 cells for each strain. Statistical analyses of condensed populations were performed using one-way ANOVA followed by post-hoc Tukey HSD Test (p = 0.001 for wildtype versus scc2-4 mutant cell rDNA condensation at 37°C; p = 0.001 for wildtype versus scc2-4 mutant cell rDNA condensation at 23°C; p = 0.890 for wildtype cell rDNA condensation at 37°C versus 23°C; p = 0.301 for scc2-4 mutant cell rDNA condensation at 37°C versus 23°C). Statistical significant differences (*) are based on p < 0.05.
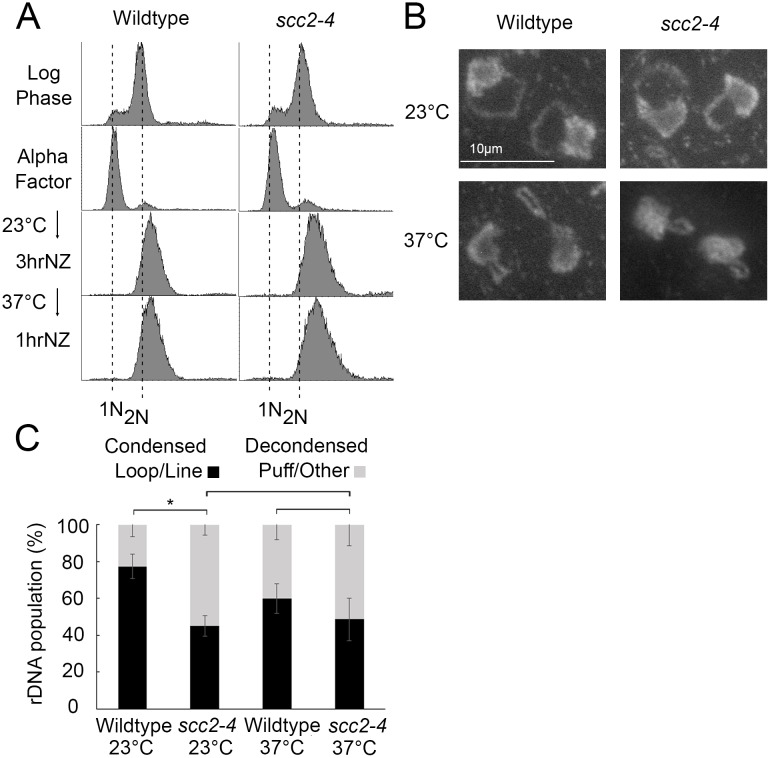
Scc2 plays a mitotic role in arm condensation but not rDNA condensation
Scc2 inactivation during S phase causes severe chromosome condensation defects and cell lethality [19]. Intriguingly, there is limited evidence that Scc2 inactivation specifically during M phase also produces chromosome arm condensation defects, even while cells retain high viability [23]. We were intrigued by the possibility that scc2-4 cell viability, during M phase inactivation, might be explained if rDNA remains condensed even while chromosome arms decondense. To test this possibility, wildtype and scc2-4 mutant strains were synchronized in G1 in medium supplemented with alpha factor, released into 23°C medium supplemented with nocodazole for 3 hours, and the resulting preanaphase cultures then shifted to 37°C for 1 hour to inactivate scc2-4 specific during M phase (Fig 5A). Cell samples were processed to determine the status of rDNA condensation as previously described [45]. As expected, preanaphase wildtype cells maintained at 23°C exhibited high levels (77%) of condensed (loop-like) rDNA loci. Even at this temperature permissive for cell viability, scc2-4 mutant cells exhibited surprisingly modest levels (45%) of condensed rDNA loci (Fig 5B and 5C). Preanaphase wildtype cells shifted to 37°C retained high levels (60%) of condensed rDNA, albeit with shorter rDNA loops as previously described [45]. Importantly, preanaphase scc2-4 mutant cells shifted to 37°C similarly retained its modest level of condensed rDNA (48%) loci (Fig 5B and 5C). These results reveal that Scc2 is not required for condensation maintenance of rDNA during M phase, in contrast to the role played by Scc2 in condensation along chromosome arms [23]. Thus, Scc2-dependent cohesin roles in condensation differentially effects rDNA and chromosome arm loci during mitosis.
Fig 5. Scc2 is dispensable for condensation maintenance during M phase.
A) Flow cytometer data reveals DNA content throughout the experimental analyses. Cells were maintained in nocodazole, post-alpha factor release, for 3 hours at 23°C followed by an additional 1 hour at 37°C. B) Chromosome mass and rDNA structures detected using DAPI in wildtype and scc2-4 mutant strains. C) Quantification of condensed (loop/line) and decondensed (puff/other) rDNA populations in wildtype (YBS1019) and scc2-4 mutant (YMM551) cells. Quantifications and statistical analyses of rDNA condensation were obtained from 3 biological replicates for each strain (wildtype and scc2-4 mutant cells) in which each replicate included 100 cells for each strain. Statistical analyses of condensed populations were performed using one-way ANOVA followed by post-hoc Tukey HSD Test (p = 0.014 for wildtype cell versus scc2-4 mutant cell rDNA condensation at 23°C; p = 0.804 for wildtype cell versus scc2-4 mutant cell rDNA condensation at 37°C; p = 0.133 for wildtype cell rDNA condensation at 23°C versus 37°C; p = 0.878 for scc2-4 mutant cell rDNA condensation at 23°C versus 37°C). Statistical significant differences (*) are based on p < 0.05.
Discussion
Analyses of Chl1 helicase family members are of immediate clinical relevance. Mutations in CHL1 human homologs BACH1/BRIP/FANCJ and ChlR1/DDX11 helicases collectively result in Warsaw Breakage Syndrome, Fanconi anemia, breast and ovarian cancers [27], [35–37], [40], [59–61]. A link between the Chl1 helicase family and global changes in chromatin structure, however, remained untested. The first major revelation of the current study is that Chl1 is an important factor in promoting genome-wide chromosome condensation. Intriguingly, we found that chl1 mutants exhibit both chromosome arm and rDNA condensation defects, but at relatively moderate levels. This suggests that additional factors may support Chl1 in condensation reactions (including Scc2 and cohesin deposition onto DNA) and that cohesin-dependent condensation is taken place in S phase. Regardless, our findings extend the role of Chl1 beyond trans tethering (required for sister chromatid cohesion and DNA repair) to now include cis tethering [27], [31–34]. This distinction is critical in that cis tethering during G1 is thought to stabilize intramolecular DNA loops through which regulatory elements (enhancers, promoters, insulators) are brought into registration and thus deploy developmental transcription programs [1], [2]. Moreover, cis tethering also mediates both regional and genome-wide compaction reactions throughout the cell cycle—the latter of which is required for chromosome segregation. While the current study is unique in identifying a role for Chl1 DNA helicase in chromatin condensation, we note that mutation of other helicases (MCM7, MCM10, FBH1) in C. elegans or Drosophila models produced chromosome condensation defects, but effects attributed to incomplete replication and often with minimal effects on chromosome segregation [62–66]. Our findings suggest a reevaluation of current models may be warranted.
How does Chl1 DNA helicase promote DNA condensation? A second revelation of the current study is that Chl1, and its downstream target Scc2, function predominantly through cohesin-based condensation. In the current study, we simultaneously monitored both cohesin and condensin chromatin binding levels and found that only cohesins are adversely affected in chl1 and scc2-4 mutant strains. Intriguingly, a prior study found that scc2-4 inactivation during a mitotic-arrest produced a modest condensin binding defect, but an effect predicated on perceived changes in fluorescent intensity levels obtained from chromosome spreads. Notably, that study provided inconsistent data regarding reductions in condensin recruitment to loci assessed by chromatin immunoprecipitations [23]. While our current findings diminish the role of Scc2 (and Chl1) in condensin recruitment, we note that mutation of the RNA helicase Vasa, which produce condensation defects in mitotic germ-line Drosophila cells, exhibit reduced recruitment of the condensin SMC capping factor Barren to DNA [67]. Thus, it will be critical to elucidate the extent through which different model cell systems prioritize the use of SMC complexes to drive chromatin compaction. Assessing these possibilities is complicated, however, due to evidence that condensin recruitment may be mediated indirectly through reduced cohesin recruitment [58].
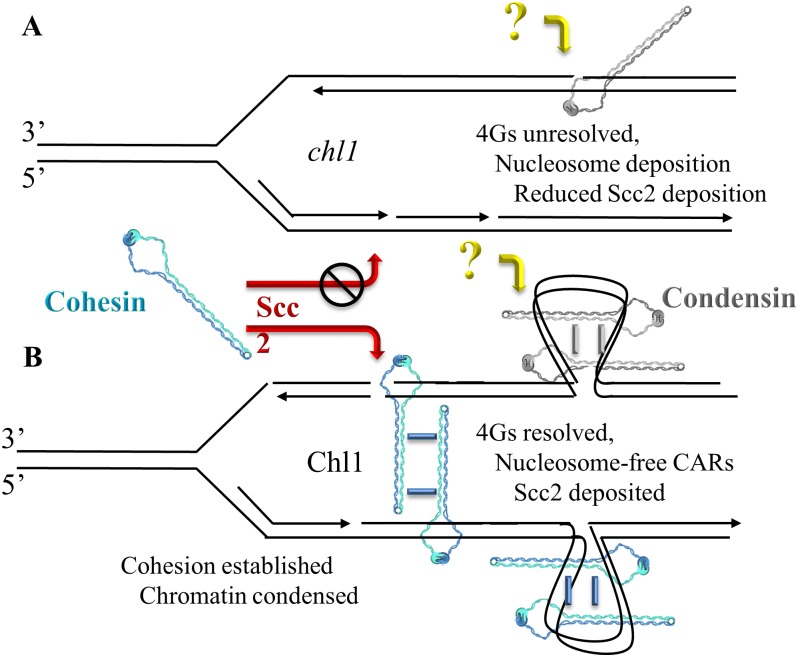
Elucidating the mechanism through which Chl1 promotes both Scc2 and cohesin recruitment to DNA to mediate cohesion (trans tethering) and condensation (cis tethering) remains an important issue in cell biology. Elegant biochemical findings reveal that Chl1 family members resolve secondary DNA structures such as G4s, triple helices, and 5’ forked/flapped duplexes thought to arise either immediately behind the DNA replication fork or occur within specific loci throughout the genome [28], [34]. [68–74]. That these secondary DNA structures can be resolved in a post-fork context is strongly supported by findings that both Chl1 expression and chromatin binding peak during S phase and that Chl1 binds to numerous replication factors (such as Ctf4, Eco1, Fen1, Ctf18 and PCNA) that act in in conjunction with or immediately behind DNA polymerase [24], [27], [28], [30], [52], [54]. More recent findings posit that Scc2,4 binds DNA to maintain nucleosome-free domains onto which cohesins are later deposited [75]. Based on evidence that Chl1 family members disrupt streptavidin binding to biotinylated single-strand DNA oligonucleotides in vitro and resolve DNA secondary structures such as G quadruplexes (G4s) and triple DNA helices [70], [73], [76], we posit that Chl1 helicase actively promotes nucleosome-free domains that promote Scc2 and subsequent cohesin deposition (Fig 6). This helicase-based model in which DNA structure modulates Scc2 recruitment may equally apply to protein-based adaptors of Scc2,4 that include the elongation factor Paf1, Mediator transcription scaffold complex, the pre-Replication Complex (pre-RC), Ctf19/COMA kinetochore complex [77–89].
Fig 6. Chl1 DNA helicase functions in condensation.
A) In the absence of Chl1, condensation defects occur despite normal recruitment of condensin (‘?’ reflects that condensin deposition, but not cohesin deposition, occurs despite Scc2 inactivation). We hypothesize that secondary DNA structures (such as G4s and nucleosomes) reduce both Scc2 and cohesin recruitment, resulting in cohesion and condensation defects. B) Chl1 activities (resolution of DNA secondary structures, histone displacement, etc) provides for both Scc2 and cohesin recruitment, resulting in sister chromatid cohesion (trans tethering) and chromatin condensation (cis tethering). Cohesin and condensin oligomerization are shown as one of several possible mechanisms of cohesion and condensation [1], [90].
Supporting information
A) Morphology quantification for both nocodazole-arrested wildtype and chl1 mutant cells (N = 100 cells for each strain). B) 1N peak quantification for both wildtype and chl1 mutant cells arrested in nocodazole for 3 hours at 23°C. C) Flow cytometer data reveals DNA contents in wildtype and chl1 mutant cells after nocodazole arrest at 23°C for 2.5 hours and 3 hours. D) Flow cytometer data reveals DNA content in wildtype and chl1 mutant cells analyzed in Fig 1E and 1F.
(TIF)
Acknowledgments
We thank Skibbens laboratory members (Caitlin Zuilkoski, Michael Mfarej, Divya Sirdeshpandi, Chris Geigler), Cassimeris laboratory and Cassbens members for helpful discussion throughout this process, and Maria Ocampo-Hafalla and Dr. Frank Uhlmann and Vincent Guacci and Doug Koshland for kindly sharing yeast strains and reagents.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Institute of General Medical Sciences, 1R15GM110631-01 to Robert Skibbens, PhD. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Skibbens RV. Of Rings and Rods: Regulating Cohesin Entrapment of DNA to Generate Intra- and Intermolecular Tethers. PLoS Genet. 2016;12(10): e1006337 doi: 10.1371/journal.pgen.1006337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dorsett D, Merkenschlager M. Cohesin at active genes: a unifying theme for cohesin and gene expression from model organisms to humans. Curr Opin Cell Biol. 2013;25(3): 327–333. doi: 10.1016/j.ceb.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martin RM, Cardoso MC. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J. 2010;24(4): 1066–1072. doi: 10.1096/fj.08-128959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piskadlo E, Oliveira RA. Novel insights into mitotic chromosome condensation. F1000Res. 2016;5: 1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsang CK, Li H, Zheng XS. Nutrient starvation promotes condensin loading to maintain rDNA stability. EMBO J. 2007;26(2): 448–458. doi: 10.1038/sj.emboj.7601488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26(15): 1659–1678. doi: 10.1101/gad.194746.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeppsson K, Kanno T, Shirahige K, Sjögren C. The maintenance of chromosome structure: positioning and functioning of SMC complexes. Nat Rev Mol Cell Biol. 2014;15(9): 601–614. doi: 10.1038/nrm3857 [DOI] [PubMed] [Google Scholar]
- 8.Mannini L, Musio A. The dark side of cohesin: the carcinogenic point of view. Mutat Res. 2011;728(3): 81–87. [DOI] [PubMed] [Google Scholar]
- 9.Mehta GD, Kumar R, Srivastava S, Ghosh SK. Cohesin: functions beyond sister chromatid cohesion. FEBS Lett. 2013;587(15): 2299–2312. doi: 10.1016/j.febslet.2013.06.035 [DOI] [PubMed] [Google Scholar]
- 10.Feng XD, Song Q, Li CW, Chen J, Tang HM, Peng ZH, et al. Structural maintenance of chromosomes 4 is a predictor of survival and a novel therapeutic target in colorectal cancer. Asian Pac J Cancer Prev. 2014;15(21): 9459–9465. [DOI] [PubMed] [Google Scholar]
- 11.Yin L, Jiang LP, Shen QS, Xiong QX, Zhuo X; Zhang LL, et al. NCAPH plays important roles in human colon cancer. Cell Death Dis. 2017;8(3): e2680 doi: 10.1038/cddis.2017.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woodward J, Taylor GC, Soares DC, Boyle S, Sie D, Read D, et al. Condensin II mutation causes T-cell lymphoma through tissue-specific genome instability. Genes Dev. 2016;30(19): 2173–2186. doi: 10.1101/gad.284562.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perche O, Menuet A, Marcos M, Liu L, Pâris A, Utami KH, et al. Combined deletion of two Condensin II system genes (NCAPG2 and MCPH1) in a case of severe microcephaly and mental deficiency. Eur J Med Genet. 2013;56(11): 635–641. doi: 10.1016/j.ejmg.2013.07.007 [DOI] [PubMed] [Google Scholar]
- 14.Martin CA, Murray JE, Carroll P, Leitch A, Mackenzie KJ, Halachev M, et al. Deciphering Developmental Disorders Study, Wood AJ, Vagnarelli P, Jackson AP. Mutations in genes encoding condensin complex proteins cause microcephaly through decatenation failure at mitosis. Genes Dev. 2016;30(19): 2158–2172. doi: 10.1101/gad.286351.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber S, Haering CH, Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112: 765–777. [DOI] [PubMed] [Google Scholar]
- 16.Cuylen S, Metz J, Haering CH. Condensin structures chromosomal DNA through topological links. Nat Struct Mol Biol. 2011;18(8): 894–901. doi: 10.1038/nsmb.2087 [DOI] [PubMed] [Google Scholar]
- 17.Kalitsis P, Zhang T, Marshall KM, Nielsen CF, Hudson DF. Condensin, master organizer of the genome. Chromosome Res. 2017;25(1): 61–76. doi: 10.1007/s10577-017-9553-0 [DOI] [PubMed] [Google Scholar]
- 18.Rollins RA, Morcillo P, Dorsett D. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics. 1999;152(2): 577–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, et al. Cohesin's binding to chromosomes depends on a separate complex consisting of Scc2 and Scc4 proteins. Mol Cell. 2000;5(2): 243–254. [DOI] [PubMed] [Google Scholar]
- 20.Tonkin ET, Wang TJ, Lisgo S, Bamshad MJ, Strachan T. NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet. 2004. June;36(6): 636–641. doi: 10.1038/ng1363 [DOI] [PubMed] [Google Scholar]
- 21.Krantz ID, McCallum J, DeScipio C, Kaur M, Gillis LA, Yaeger D, et al. Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet. 2004;36(6): 631–635. doi: 10.1038/ng1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seitan VC, Banks P, Laval S, Majid NA, Dorsett D, Rana A, et al. Metazoan Scc4 homologs link sister chromatid cohesion to cell and axon migration guidance. PLoS Biol. 2006;4(8): e242 doi: 10.1371/journal.pbio.0040242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, et al. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22(16): 2215–2227. doi: 10.1101/gad.1675708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudra S, Skibbens RV. Sister chromatid cohesion establishment occurs in concert with lagging strand synthesis. Cell Cycle. 2012;11(11): 2114–2121. doi: 10.4161/cc.20547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudra S, Skibbens RV. Chl1 DNA helicase regulates Scc2 deposition specifically during DNA-replication in Saccharomyces cerevisiae. PLoS One. 2013;8(9): e75435 doi: 10.1371/journal.pone.0075435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lopez-Serra L, Lengronne A, Borges V, Kelly G, Uhlmann F. Budding yeast Wapl controls sister chromatid cohesion maintenance and chromosome condensation. Curr Biol. 2013;23(1): 64–69. doi: 10.1016/j.cub.2012.11.030 [DOI] [PubMed] [Google Scholar]
- 27.Skibbens RV. Chl1p, a DNA Helicase-Like Protein in Budding Yeast, Functions in Sister-Chromatid Cohesion. Genetics. 2004;166(1): 33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farina A, Shin JH, Kim DH, Bermudez VP, Kelman Z, Seo YS, et al. Studies with the Human Cohesin Establishment Factor, ChlR1 association of ChlR1 with CTF18-RFC and Fen1. J Biol Chem. 2008;283(30): 20925–20936. doi: 10.1074/jbc.M802696200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borges V, Smith DJ, Whitehouse I, Uhlmann F. An Eco1-independent sister chromatid cohesion establishment pathway in S. cerevisiae. Chromosoma (2013; 122(1–2): 121–134. doi: 10.1007/s00412-013-0396-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samora CP, Saksouk J, Goswami P, Wade BO, Singleton MR, Bates PA, et al. Ctf4 Links DNA Replication with Sister Chromatid Cohesion Establishment by Recruiting the Chl1 Helicase to the Replisome. Mol Cell. 2016;63(3): 371–384. doi: 10.1016/j.molcel.2016.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petronczki M, Chwalla B, Siomos MF, Yokobayashi S, Helmhart W, Deutschbauer AM, et al. Sister-chromatid cohesion mediated by the alternative RF-CCtf18/Dcc1/Ctf8, the helicase Chl1 and the polymerase-alpha-associated protein Ctf4 is essential for chromatid disjunction during meiosis II. J Cell Sci. 2004;117(16): 3547–3559. [DOI] [PubMed] [Google Scholar]
- 32.Mayer ML, Pot I, Chang M, Xu H, Aneliunas V, Kwok T, et al. Identification of protein complexes required for efficient sister chromatid cohesion. Mol Biol Cell. 2004;15(4): 1736–1745. doi: 10.1091/mbc.E03-08-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci. 2006;119(23): 4857–4865. [DOI] [PubMed] [Google Scholar]
- 34.Inoue A, Li T, Roby SK, Valentine MB, Inoue M, Boyd K, et al. Loss of ChlR1 helicase in mouse causes lethality due to the accumulation of aneuploid cells generated by cohesion defects and placental malformation. Cell Cycle. 2007;6(13): 1646–1654. [DOI] [PubMed] [Google Scholar]
- 35.Van der Lelij P, Chrzanowska KH, Godthelp BC, Rooimans MA, Oostra AB, Stumm M, et al. Warsaw breakage syndrome, a cohesinopathy associated with mutations in the XPD helicase family member DDX11/ChlR1. Am J Hum Genet. 2010;86(2): 262–266. doi: 10.1016/j.ajhg.2010.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005;8(3): 255–265. [DOI] [PubMed] [Google Scholar]
- 37.Rafnar T, Gudbjartsson DF, Sulem P, Jonasdottir A, Sigurdsson A, Jonasdottir A, et al. Mutations in BRIP1 confer high risk of ovarian cancer. Nat Genet. 2011;43(11): 1104–1107. doi: 10.1038/ng.955 [DOI] [PubMed] [Google Scholar]
- 38.Peng M, Litman R, Jin Z, Fong G, Cantor SB. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene. 2006;25(15): 2245–53. [DOI] [PubMed] [Google Scholar]
- 39.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105(1): 149–160. [DOI] [PubMed] [Google Scholar]
- 40.Cantor S, Drapkin R, Zhang F, Lin Y, Han J, Pamidi S, et al. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc Natl Acad Sci U S A. 2004;101(8): 2357–2362. doi: 10.1073/pnas.0308717101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1988;14(10): 953–961. [DOI] [PubMed] [Google Scholar]
- 42.Tong K, Skibbens RV. Pds5 regulators segregate cohesion and condensation pathways in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2015;112(22): 7021–7026. doi: 10.1073/pnas.1501369112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guacci V, Hogan E, Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994;125(3): 517–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guacci V, Koshland D, Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91(1): 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shen D, Skibbens RV. Temperature-dependent regulation of rDNA condensation in Saccharomyces cerevisiae. Cell Cycle. 2017; 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Noble D, Kenna MA, Dix M, Skibbens RV, Unal E, Guacci V. Intersection between the regulators of sister chromatid cohesion establishment and maintenance in budding yeast indicates a multi-step mechanism. Cell Cycle. 2006;5(21): 2528–2536. [DOI] [PubMed] [Google Scholar]
- 47.Lavoie BD, Hogan E and Koshland D. In vivo dissection of the chromosome condensation machinery reversibility of condensation distinguishes contributions of condensin and cohesin. J Cell Biol. 2002;156(5): 805–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi T. How does genome instability affect lifespan? Genes Cells. 2011. June; 16(6): 617–624. doi: 10.1111/j.1365-2443.2011.01519.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52(4): 536–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, et al. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97(2): 245–256. [DOI] [PubMed] [Google Scholar]
- 51.Nomura M. Ribosomal RNA genes, RNA polymerases, nucleolar structures, and synthesis of rRNA in the yeast Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 2001;66: 555–565. [DOI] [PubMed] [Google Scholar]
- 52.Skibbens RV, Corson BL, Koshland D, Hieter P. Ctf7p is essential for sister chromatid cohesion and links mitotic chromosome structure to the DNA replication machinery. Genes Dev. 1999;13(3): 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tóth A, Ciosk R, Uhlmann F, Galova M, Schleiffer A, Nasmyth K. Yeast Cohesin complex requires a conserved protein, Eco1p(Ctf7), to establish cohesion between sister chromatids during DNA replication. Genes Dev. 1999;13(3): 320–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moldovan GL, Pfander B, Jentsch S. PCNA controls establishment of sister chromatid cohesion during S phase. Mol Cell 2006;23: 723–732. doi: 10.1016/j.molcel.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 55.Méndez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20(22): 8602–8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Leman AR, Noguchi C, Lee CY, Noguchi E. Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci. 2010;123(5): 660–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tong K, Skibbens RV. Cohesin without cohesion: a novel role for Pds5 in Saccharomyces cerevisiae. PLoS One. 2014;9(6): e100470 doi: 10.1371/journal.pone.0100470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lavoie BD, Hogan E, Koshland D. In vivo requirements for rDNA chromosome condensation reveal two cell-cycle-regulated pathways for mitotic chromosome folding. Genes Dev. 2004;18(1): 76–87. doi: 10.1101/gad.1150404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amann J, Kidd VJ, Lahti JM. Characterization of putative human homologues of the yeast chromosome transmission fidelity gene, CHL1. J Biol Chem. 1997;272(6): 3823–3832. [DOI] [PubMed] [Google Scholar]
- 60.Levitus M, Waisfisz Q, Godthelp BC, de Vries Y, Hussain S, Wiegant WW, et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat Genet. 2005;37(9): 934–935. doi: 10.1038/ng1625 [DOI] [PubMed] [Google Scholar]
- 61.Gupta R, Sharma S, Sommers JA, Kenny MK, Cantor SB, Brosh RM Jr. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110(7): 2390–2398. doi: 10.1182/blood-2006-11-057273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sonneville R, Craig G, Labib K, Gartner A, Blow JJ. Both Chromosome Decondensation and Condensation Are Dependent on DNA Replication in C. elegans Embryos. Cell Rep. 2015;12(3): 405–417. doi: 10.1016/j.celrep.2015.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Christensen TW, Tye BK. Drosophila Mcm10 Interacts with Members of the Prereplication Complex and Is Required for Proper Chromosome Condensation. Mol Biol Cell. 2003;14(6): 2206–2215. doi: 10.1091/mbc.E02-11-0706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chmielewski JP, Henderson L, Smith CM, Christensen TW. Drosophila Psf2 has a role in chromosome condensation. Chromosoma. 2012;121(6): 585–596. doi: 10.1007/s00412-012-0383-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petrova B, Dehler S, Kruitwagen T, Hériché JK, Miura K, Haering CH. Quantitative Analysis of Chromosome Condensation in Fission Yeast. Mol Cell Biol. 2013;335: 984–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsutsui Y, Kurokawa Y, Ito K, Siddique MS, Kawano Y, Yamao F, et al. Multiple Regulation of Rad51-Mediated Homologous Recombination by Fission Yeast Fbh1. PLoS Genet. 2014;10(8): e1004542 doi: 10.1371/journal.pgen.1004542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pek JW, Kai T. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol. 2011;21(1): 39–44. doi: 10.1016/j.cub.2010.11.051 [DOI] [PubMed] [Google Scholar]
- 68.Hirota Y, Lahti JM. Characterization of the enzymatic activity of hChlR1, a novel human DNA helicase. Nucleic Acids Res. 2000;28(4): 917–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu Y, Shin-ya K, Brosh RM Jr. FANCJ Helicase Defective in Fanconia Anemia and Breast Cancer Unwinds G-Quadruplex DNA To Defend Genomic Stability. Mol Cell Biol. 2008;28(12): 4116–4128. doi: 10.1128/MCB.02210-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu Y, Sommers JA, Khan I, de Winter JP, Brosh RM Jr. Biochemical characterization of Warsaw breakage syndrome helicase. J Biol Chem. 2012;287(2): 1007–1021. doi: 10.1074/jbc.M111.276022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kuryavyi V, Patel DJ. Solution structure of a unique G-quadruplex scaffold adopted by a guanosine-rich human intronic sequence. Structure. 2010;18(1): 73–82. doi: 10.1016/j.str.2009.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bharti SK, Sommers JA, George F, Kuper J, Hamon F, Shin-Ya K, et al. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J Biol Chem. 2013;288(39): 28217–2829. doi: 10.1074/jbc.M113.496463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bharti SK, Khan I, Banerjee T, Sommers JA, Wu Y, Brosh RM Jr. Molecular functions and cellular roles of the ChlR1 (DDX11) helicase defective in the rare cohesinopathy Warsaw breakage syndrome. Cell Mol Life Sci. 2014;71(14): 2625–2639. doi: 10.1007/s00018-014-1569-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guo M, Hundseth K, Ding H, Vidhyasagar V, Inoue A, Nguyen C-H, et al. A distinct triplex DNA unwinding activity of ChlR1 helicase. J Bio Chem. 2015;290: 5174–5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lopez-Serra L, Kelly G, Patel H, Stewart A, Uhlmann F. The Scc2-Scc4 complex acts in sister chromatid cohesion and transcriptional regulation by maintaining nucleosome-free regions. Nat Genet. 2014;46(10): 1147–1151. doi: 10.1038/ng.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bochman ML, Paeschke K, Zakian VA. DNA secondary structures: stability and function of G-quadruplex structures. Nat Rev Genet. 2012;13(11): 770–780. doi: 10.1038/nrg3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gillespie PJ, Hirano T. Scc2 couples replication licensing to sister chromatid cohesion in Xenopus egg extracts. Curr Biol. 2004;14: 1598–1603. doi: 10.1016/j.cub.2004.07.053 [DOI] [PubMed] [Google Scholar]
- 78.Takahashi TS, Yiu P, Chou MF, Gygi S, Walter JC. Recruitment of Xenopus Scc2 and cohesin to chromatin requires the pre-replication complex. Nat Cell Biol. 2004;6(10): 991–996. doi: 10.1038/ncb1177 [DOI] [PubMed] [Google Scholar]
- 79.Bermudez VP, Farina A, Higashi TL, Du F, Tappin I, Takahashi TS, et al. In vitro loading of human cohesin on DNA by the human Scc2-Scc4 loader complex. Proc Natl Acad Sci U S A. 2012;109(24): 9366–9371. doi: 10.1073/pnas.1206840109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Misulovin Z, Schwartz YB, Li XY, Kahn TG, Gause M, MacArthur S, et al. Association of cohesin and Nipped-B with transcriptionally active regions of the Drosophila melanogaster genome. Chromosoma.2008;117(1): 89–102. doi: 10.1007/s00412-007-0129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Muto A, Ikeda S, Lopez-Burks ME, Kikuchi Y, Calof AL, Lander AD, et al. Nipbl and mediator cooperatively regulate gene expression to control limb development. PLoS Genet. 2014;10(9): e1004671 doi: 10.1371/journal.pgen.1004671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314): 430–435. doi: 10.1038/nature09380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zakari M, Trimble Ross R, Peak A, Blanchette M, Seidel C, Gerton JL. The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity. PLoS Genet. 2015;11(7): e1005308 doi: 10.1371/journal.pgen.1005308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fernius J, Nerusheva OO, Galander S, Alves Fde L, Rappsilber J, Marston AL. Cohesin-Dependent Association of Scc2/4 with the Centromere Initiates Pericentromeric Cohesion Establishment. Curr Biol. 2013;23(7): 599–606. doi: 10.1016/j.cub.2013.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Natsume T, Müller CA, Katou Y, Retkute R, Gierliński M, Araki H, et al. Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment. Mol Cell. 2013;50(5): 661–674. doi: 10.1016/j.molcel.2013.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Unal E, Arbel-Eden A, Sattler U, Shroff R, Lichten M, Haber JE, et al. DNA damage response pathway uses histone modification to assemble a double-strand break-specific cohesin domain. Mol Cell. 2004;16(6): 991–1002. doi: 10.1016/j.molcel.2004.11.027 [DOI] [PubMed] [Google Scholar]
- 87.Unal E, Heidinger-Pauli JM, Kim W, Guacci V, Onn I, Gygi SP, et al. A molecular determinant for the establishment of sister chromatid cohesion. Science. 2008;321(5888): 566–569. doi: 10.1126/science.1157880 [DOI] [PubMed] [Google Scholar]
- 88.Ström L, Lindroos HB, Shirahige K, Sjögren C. Postreplicative recruitment of cohesin to double-strand breaks is required for DNA repair. Mol Cell. 2004;16(6): 1003–1015. [DOI] [PubMed] [Google Scholar]
- 89.Ström L, Sjögren C. Chromosome segregation and double-strand break repair—a complex connection. Curr Opin Cell Biol. 2007;19(3): 344–349. [DOI] [PubMed] [Google Scholar]
- 90.Rankin S, Dawson DS. Recent advances in cohesin biology. F1000Res. 2016;5: 1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maradeo ME, Garg A, and Skibbens RV. Rfc5p regulates alternate RFC complex functions in sister chromatid pairing reactions in budding yeast. Cell Cycle. 2010; 9(21): 4370–4378. doi: 10.4161/cc.9.21.13634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Morphology quantification for both nocodazole-arrested wildtype and chl1 mutant cells (N = 100 cells for each strain). B) 1N peak quantification for both wildtype and chl1 mutant cells arrested in nocodazole for 3 hours at 23°C. C) Flow cytometer data reveals DNA contents in wildtype and chl1 mutant cells after nocodazole arrest at 23°C for 2.5 hours and 3 hours. D) Flow cytometer data reveals DNA content in wildtype and chl1 mutant cells analyzed in Fig 1E and 1F.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.