Abstract
Prunus mira Koehne, an important economic fruit crop with high breeding and medicinal values, and an ancestral species of many cultivated peach species, has recently been declared an endangered species. However, basic information about genetic diversity, population structure, and morphological variation is still limited for this species. In this study, we sampled 420 P. mira individuals from 21 wild populations in the Tibet plateau to conduct a comprehensive analysis of genetic and morphological characteristics. The results of molecular analyses based on simple sequence repeat (SSR) markers indicated moderate genetic diversity and inbreeding (A = 3.8, Ae = 2.5, He = 0.52, Ho = 0.44, I = 0.95, FIS = 0.17) within P. mira populations. STRUCTURE, GENELAND, and phylogenetic analyses assigned the 21 populations to three genetic clusters that were moderately correlated with geographic altitudes, and this may have resulted from significantly different climatic and environmental factors at different altitudinal ranges. Significant isolation-by-distance was detected across the entire distribution of P. mira populations, but geographic altitude might have more significant effects on genetic structure than geographic distance in partial small-scale areas. Furthermore, clear genetic structure, high genetic differentiation, and restricted gene flow were detected between pairwise populations from different geographic groups, indicating that geographic barriers and genetic drift have significant effects on P. mira populations. Analyses of molecular variance based on the SSR markers indicated high variation (83.7% and 81.7%), whereas morphological analyses revealed low variation (1.30%–36.17%) within the populations. Large and heavy fruits were better adapted than light fruits and nutlets to poor climate and environmental conditions at high altitudes. Based on the results of molecular and morphological analyses, we classified the area into three conservation units and proposed several conservation strategies for wild P. mira populations in the Tibet plateau.
Introduction
Prunus mira Koehne (2n = 2x = 16) is a perennial woody plant that belongs to the genus Prunus of the family Rosaceae [1]. It is native to China, and it is an ancestral species of many cultivated peach species, including P. davidiana, P. kansuensis, and P. ferganensis [2–4]. P. mira is also considered a valuable “living fossil” of peach species because of its long life cycle (over 1000 years). Its fruits are rich in nutrients (vitamin C, calcium, and ferrum), fatty acids (oleic acid, linoleic acid, cetylic acid, and octadecanoic acid), and active medicinal ingredients (arbutin). Therefore, in addition to supplying fruits, P. mira is used in Chinese traditional medicine to treat irregular menses, fractures, and congestion [5]. Considering that P. mira has high tolerance to environmental stress (drought, cold, and barren soil) and high yields, it has great potential with regard to peach breeding [6,7].
P. mira is widely distributed along the Yarlung Zangbo Grand Canyon and its tributary basins (Parlung Zangbo Basin and Nyang River Basin) in the Tibet plateau [1,8]. Shannan and Linzhi, located in the middle and lower reaches of the Yarlung Zangbo Grand Canyon, are the two areas where P. mira densely inhabits [3,7]. Shanan is characterized by a plateau temperate arid climate, with a mean altitude of 3700 m. Linzhi has a tropical humid and semi-humid climate, with a mean altitude of 3000 m. Owing to different climates and landscapes, the P. mira wild resources from different areas develop different phenological phases [9,10]. For instance, the flowering time of P. mira is March 15–25 in Shannan and March 9–20 in Linzhi. Recent field investigations revealed that morphological characteristics such as fruit size, nutlet size, and nutlet surfaces, were considerably different between P. mira wild resources in Shannan and those in Linzhi. Previous studies [7,8,11,12] revealed high genetic and morphological variations within wild P. mira resources, as well as many excellent breeding resources in the Shannan and Linzhi areas. Unfortunately, because of deforestation, over-harvest, and road building, the natural distribution and population size of P. mira have been remarkably reduced [5,6]. Although several studies have shown genetic diversity and morphological variations in P. mira [6–8], they were conducted using a small number of individuals from a small-scale district, and the results provided very limited information on genetic and morphological variations in this species. A comprehensive study of P. mira genetic and morphological variations is still lacking. The knowledge of genetic and morphological variations can reveal the evolutionary history and existing situations associated with a plant species [13–15]. Therefor, studying the genetic diversity, population structure, and morphological variation of P. mira is crucial for the protection of this endangered species. For decades, numerous molecular markers such as single nucleotide polymorphisms (SNP), random amplified polymorphisms DNA (RAPDs), simple sequence repeats (SSRs), and inter-simple sequence repeats (ISSRs), have been developed to analyze the genetic diversity and population structure of plant species [16–19]. Among these markers, SSRs have been the first choice for studies of genetic diversity and population structure, because of their excellent characteristics, including high polymorphism, co-dominant inheritance, and low inquiry of DNA [20]. Furthermore, SSRs were found to be highly transferable among Prunus species [21,22]. Therefore, numerous SSRs that were developed in related species can be effective tools for conducting genetic analysis of P. mira. Morphological classification is a traditional and intuitive means of characterizing plant species, and it has been widely used to evaluate morphological variation and differentiation within and among populations [23]. The same data can be used to conduct various types of morphological analyses to provide valuable information on morphological variations for plant conservation.
Therefore, in this study, we used 25 SSR markers and 11 morphological characteristics to assess the genetic diversity, population structure, and morphological variation of 21 wild P. mira populations that were collected from the Tibetan plateau. This study aimed to (1) comprehensively reveal the genetic diversity and population structure of P. mira; (2) estimate morphological variation among and within populations; and (3) propose effective conservation strategies for the studied populations.
Materials and methods
Materials
The leaf samples, fruits, and nutlets of 420 individuals were collected from 21 wild P. mira populations along the Yarlung Zangbo Grand Canyon and its tributary basin in the Tibet plateau (S1 Fig, Table 1). The longitude, latitude, and altitude of each individual were detected using a global position system. For molecular analyses, 20 individuals were sampled from each population, with a minimum distance of 50 m between any two individuals. Healthy and young leaves were collected and immediately preserved with silica gel for DNA extraction. For morphological analyses, 20 fruits and 20 nutlets were randomly selected from each individual. Finally, 8400 fruits and 8400 nutlets were collected to analyze the morphological variation within and among P. mira populations.
Table 1. Location and sample size of the 21 populations of Prunus mira Koehne used in this study.
| Population | Population ID | Sample size | Latitude (N) | Longitude (E) | Altitude (m) | Habitats |
|---|---|---|---|---|---|---|
| Yusong, Tibet | P1 | 20 | 29°33′–29°37′ | 96°19′–96°22′ | 2921–2958 | Valley |
| Bengga, Tibet | P2 | 20 | 29°34′–29°51′ | 96°50′–96°54′ | 2931–2962 | Valley |
| Zhongsha, Tibet | P3 | 20 | 29°35′–29°54′ | 95°44′–95°56′ | 2939–2972 | Valley |
| Danniang, Tibet | P4 | 20 | 29°43′–29°58′ | 95°23′–95°36′ | 2944–2963 | Valley |
| Langga, Tibet | P5 | 20 | 30°01′–30°36′ | 95°35′–95°49′ | 2937–2966 | Hillside terraces |
| Luxia, Tibet | P6 | 20 | 30°06′–30°11′ | 95°09′–95°13′ | 3232–3244 | Roadside |
| Jieguo, Tibet | P7 | 20 | 30°01′–30°59′ | 94°47′–94°56′ | 3198–3228 | Around rural house |
| Qiangna, Tibet | P8 | 20 | 29°45′–29°53′ | 94°34′–94°59′ | 3295–3311 | High slope |
| Caiba, Tibet | P9 | 20 | 29°57′–30°05′ | 94°24′–94°57′ | 3286–3302 | Around rural house |
| Bujiu, Tibet | P10 | 20 | 29°16′–29°50′ | 94°04′–94°22′ | 3129–3169 | Roadside |
| Duodang, Tibet | P11 | 20 | 29°43′–29°49′ | 94°18′–94°26′ | 3238–3256 | Hillside terraces |
| Gongzhong, Tibet | P12 | 20 | 29°30′–29°44′ | 94°32′–94°39′ | 3111–3152 | Around rural house |
| Baga, Tibet | P13 | 20 | 29°49′–29°55′ | 93°56′–94°04′ | 3294–3338 | Roadside |
| Gangga, Tibet | P14 | 20 | 29°20′–29°29′ | 94°16′–94°24′ | 2953–2979 | Hillside terraces |
| Zhuomu, Tibet | P15 | 20 | 29°16′–29°24′ | 93°13′–93°53′ | 3359–3394 | Roadside |
| Taohua, Tibet | P16 | 20 | 29°54′–30°01′ | 93°30′–93°41′ | 3297–3318 | High slope |
| Xiake, Tibet | P17 | 20 | 29°31′–29°39′ | 92°43′–92°54′ | 3661–3703 | Hillside terraces |
| Semai3, Tibet | P18 | 20 | 29°15′–29°23′ | 92°30′–92°39′ | 3778–3793 | Valley |
| Semai6, Tibet | P19 | 20 | 29°22′–29°30′ | 92°00′–92°59′ | 3836–3844 | Valley |
| Taohua2, Tibet | P20 | 20 | 29°25′–29°31′ | 91°18′–91°32′ | 3802–3839 | High slope |
| Tunmi, Tibet | P21 | 20 | 29°08′–29°22′ | 92°05′–92°17′ | 3667–3708 | Hillside terraces |
In this study, the 21 sampled populations covered the entire distribution of P. mira in the Tibet plateau. Among these populations, P1, P2, P3, P4, and P5 were located in Yarlung Zangbo Grand Canyon National Nature Reserve, and P14 was distributed in the Qomolangma National Nature Reserve. Our study was permitted and approved by these authorities. Furthermore, P8, P11, P16, P17, P18, P19, P20, and P21 were distributed in the wild, and P6, P10, P13, and P15 were distributed around roadsides, which were taken for the wild species. No specific permits were required for collecting these samples from the wild, and no specific permissions were required for these locations or activities. Furthermore, P7, P9, and P12 were distributed around rural houses, and were collected with the permission of private land owners. No endangered or protected species were sampled. All populations could be divided into two groups according to their geographic distribution. The Linzhi group (G1) contained 16 populations (P1–P16). Among these populations, P1, P2, P3, P4, P5, and P14 were distributed in low-altitude areas (2921–2979 m), and P6, P7, P8, P9, P10, P11, P12, P13, P15, and P16 were located in medium-altitude areas (3111–3394 m; Table 1). The Shannan group (G2) included the five remaining populations (P17–P21), which were distributed in the high-altitude areas (3661–3844 m).
DNA concentration, SSR analysis, and polymerase chain reaction amplification
The total genomic DNA of all dried leaves was extracted using the CTAB method [24], and DNA concentration and quality were tested using a 1% agarose gel. In all, 25 microsatellite markers, developed from three species related to P. mira, were selected to determine the genetic diversity and population structure of P. mira, including the 22 loci from P. persica [25–30], one locus from P. cerasus [31], and two loci from P. avium [32] (S1 Table). The forward primers of all SSR markers were assembled using M13 (5′-TGTAAAACGACGGCCAGT-3′), which contained one of four fluorescent dyes—FAM, NED, VIC, and PET. Polymerase chain reaction (PCR) amplification was conducted in a 20 μL reaction volume that contained 20 ng DNA, 0.2 μL 2× Tab Master Mix, 2 μL 10× buffer, 0.5 μL dNTP (0.25 mol·L-1), 0.5 μL 10% DMSO, 14.8 μL ddH2O, and 0.5 μL of each primer (0.8 μmol·L-1). The PCR protocol was as follows: 95°C for 5 min; followed by 30 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s; and a final extension at 72°C for 5 min. PCR products were analyzed using an ABI 3500XL (Applied Biosystems, USA) DNA Analyzer. The amplicon fragments were sized using Gene-Marker (Soft Genetics LLC, USA), and the details are shown in S2 Table.
Genetic analysis
GenAlEx version 6.501 [33] was used to convert size data into various formats for genetic analysis. Based on the Bayesian method, we used STRUCTURE version 2.3.4 [34] to determine the population structure of the 21 P. mira populations. Twenty independent runs were performed for each set, with a K value between 1 and 21, length of burn-in period of 1 × 105 iterations, and 1 × 105 MCMC iterations. No prior information about populations was used. The method described by Evanno et al. [35] was used to calculate the distribution of delta K (ΔK) using the online website STRUCTURE HARVESTER [36] (http://taylor0.biology.ucla.edu/struct_harvest/). Permutations of the most likely results among all runs for each K were performed in CLUMPP [37], and the final figure was visualized using Distruct software [38].
GENELAND [39,40] is a powerful way to detect genetic boundaries in R (2011). This method was used to estimate the number of clusters and their spatial patterns. The analysis was based on an uncorrelated frequency model, and was conducted over 10 replicates for each K value (1–10). The null allele models were selected to infer the number of clusters using 1,000,000 MCMC iterations, of which every 1000th was retained. The replicates with the highest mean logarithm of posterior probability were used to compute the posterior probabilities of population membership for each pixel of the spatial domain with a burn-in of 500.
P. mira genetic variation was estimated by conducting an analysis of molecular variance (AMOVA) in Arlequin version 3.5 [41]. This analysis subdivided all individuals into 21 populations and K genetic clusters. Three hierarchical divisions were identified based on the genetic variance: within populations, among populations within genetic clusters, and among genetic clusters using a nonparametric permutation procedure that incorporated 10,000 iterations. An unrooted unweighted pair-group method with arithmetic means (UPGMA) tree was constructed using Nei’s genetic distance [42] in NTSYS PC version 2.10 [43]. Mantel tests were conducted between the genetic distance (FST/(1—FST)) and geographic distance (km) in GenAlEx version 6.501.
GenAlEx version 6.501 was used to calculate the following genetic indices for loci, populations, and genetic clusters identified in the STRUCTURE analysis: allele number (A), effective allele number (Ae), private allele number (Np), Shannon’s information index (I), expected heterozygosity (He), observed heterozygosity (Ho), gene flow (Nm), and F-statistics indices (FIS, FIT, and FST). The Hardy-Weinberg equilibrium (HWE) was tested for each locus and population using Arlequin version 3.1 with 100,000,000 steps in the Markov chain and 100,000 dememorization steps. Polymorphism information content (PIC) was calculated using the following formula [44]:
where Pi and Pj are the frequency of the amplified alleles, and n is the number of alleles at each SSR locus.
Morphologic analysis
In all, 20 fruits and 20 nutlets were randomly selected per individual to measure and calculate the following 11 morphological characteristics: fruit weight, fruit length, fruit width, fruit diameter, fruit shape index (fruit length/fruit width), nutlet weight, nutlet length, nutlet width, nutlet diameter, nutlet shape index (nutlet length/nutlet width), and nutlet surface. The weights of fruits and nutlets were tested using an electronic balance (accuracy of 0.001 g). The lengths, widths, and diameters of fruits and nutlets were measured using a digital Vernier caliper (accuracy of 0.001 mm). The mean values of all morphological characteristics were used to construct an unrooted UPGMA tree in NTSYS PC version 2.10 based on Nei’s genetic distance (1978). IBM SPSS Statistics 19 software was used to conduct Duncan’s multiple comparison analysis, calculate coefficients of variation (CV, %), and draw the boxplots of the 11 morphological characteristics. For clustering and statistical analyses, the three types of nutlet surfaces were classified as follows: 1 (smooth), 2 (shallow groove), and 3 (deep groove).
Results
Characteristics of SSR markers
Twenty-five SSR markers were selected to identify genotype 420 P. mira individuals. A total of 214 alleles were detected across all loci, ranging from four (UDP96-019) to 15 (BPPCT-025 and PMS67), with an average of 8.6 alleles per locus (S3 Table). The effective number of alleles (Ae) was between 1.5 (UDP96-019) and 7.5 (BPPCT-025), with an average of 3.6. The mean expected heterozygosity (He) was 0.52, and it exceeded the observed heterozygosity (Ho = 0.46). F-statistics showed moderate FST (0.15) and FIS (0.13) values across the 25 loci, indicating moderate genetic differentiation across all sites. In addition, Shannon’s information index (I) ranged between 0.18 (Pchgms 4) and 2.07 (BPPCT-025), with an average of 1.19. The polymorphism information content (PIC) was between 0.82 (BPPCT-025) and 0.32 (Pchgms4), with an average of 0.62. Of the 25 SSR markers, six (UDP98-412, Pchgms4, BPPCT-004, UDP98-405, UDP98-408, and UDP96-019) were moderately informative (0.25 < PIC < 0.50), and 19 were highly informative (PIC > 0.50), indicating their high potential regarding the genetic diversity analysis of P. mira populations.
Genetic structure and differentiation
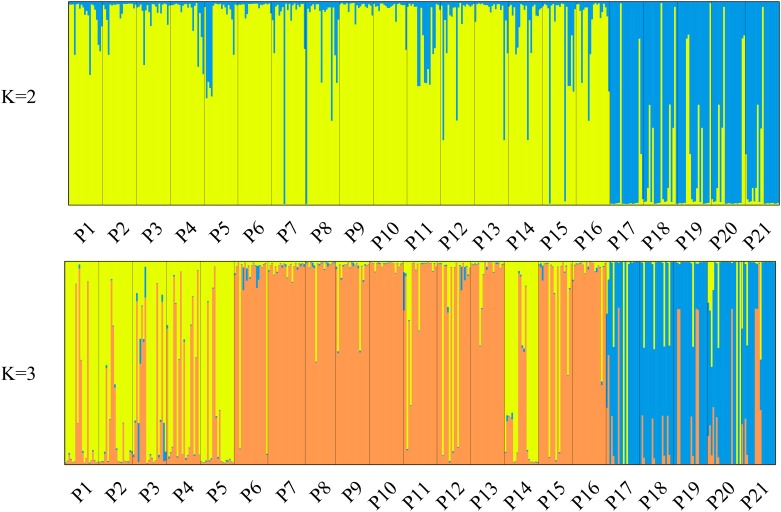
The genetic structure of 21 P. mira populations was tested using Bayesian clustering methods in STRUCTURE version 2.3.4. The classification of populations was highly correlated with their geographic altitudes. When K = 2, 16 low- and medium-altitude populations (P1–P16) from Linzhi group and five high-altitude populations (P17–P21) from Shannan group were assigned to two different clusters with high ancestry values (Q ≥ 0.80; Fig 1, S4 Table). When K = 3 and Q ≥ 0.60, 21 populations could be clearly assigned to three genetic clusters (Fig 1, S4 and S5 Tables). Cluster I consisted of six low-altitude populations (P1, P2, P3, P4, P5, and P14) from the Linzhi group. Of the 120 individuals from these populations, 88 individuals (73.33%) were assigned to Cluster I; 25 individuals (20.84%), to Cluster II; and seven individuals (5.83%) were mixed (Q < 0.60). Cluster II included 10 medium-altitude populations in the Linzhi group (P6, P7, P8, P9, P10, P11, P12, P13, P15, and P16). Among these populations, only 11 individuals (5.50%) were assigned to Cluster I, and the Q values of six individuals (3.00%) were below 0.60. Cluster III included five high-altitude populations (P17, P18, P19, P20, and P21) from the Shannan group. Among these populations, only 13 individuals (13.00%) were assigned to two other clusters, and 15 (15.00%) were mixed. The maximum value of ΔK was observed at K = 3 (S2 Fig), indicating that the 21 populations were potentially assigned to three clusters.
Fig 1. Clustering of 21 P. mira populations based on STRUCTURE analyses.
Population structure of 21 P. mira populations at K = 2 and K = 3. Each individual is shown as a vertical line divided into segments representing the estimated membership proportion; different genetic clusters were inferred using STRUCTURE. At K = 2, high-altitude populations (P17–21) were separated from the medium- and low altitude populations. Furthermore, medium-altitude (P6–13, P15, P16) and low-altitude (P1–5, P14) populations were assigned to two distinct clusters at K = 3.
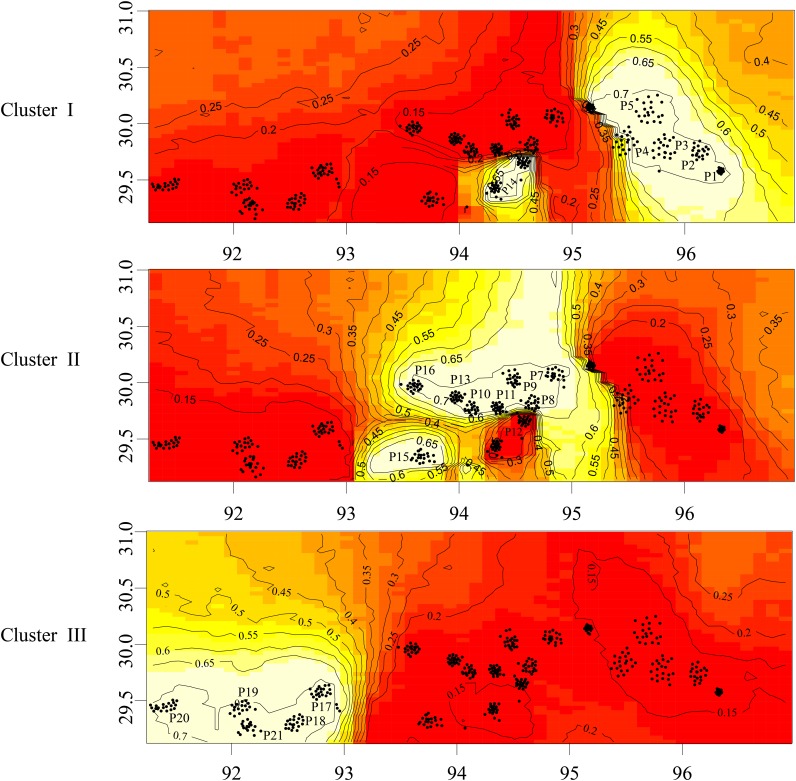
We further investigated the spatial patterns among the 21 P. mira populations based on an uncorrelated frequency model in GENELAND. This analysis inferred three distinct clusters that were the same as those identified in the STRUCTURE analysis (Fig 2). Spatial patterning indicated substantial genetic boundaries among low-, medium- and high-altitude populations.
Fig 2. Clustering of 21 P. mira populations based on GENELAND analyses.
Maps of posterior probabilities of population membership at K = 3 were inferred using GENELAND. The three maps show three clusters that were consistent with the three genetic clusters identified by the STRUCTURE analyses.
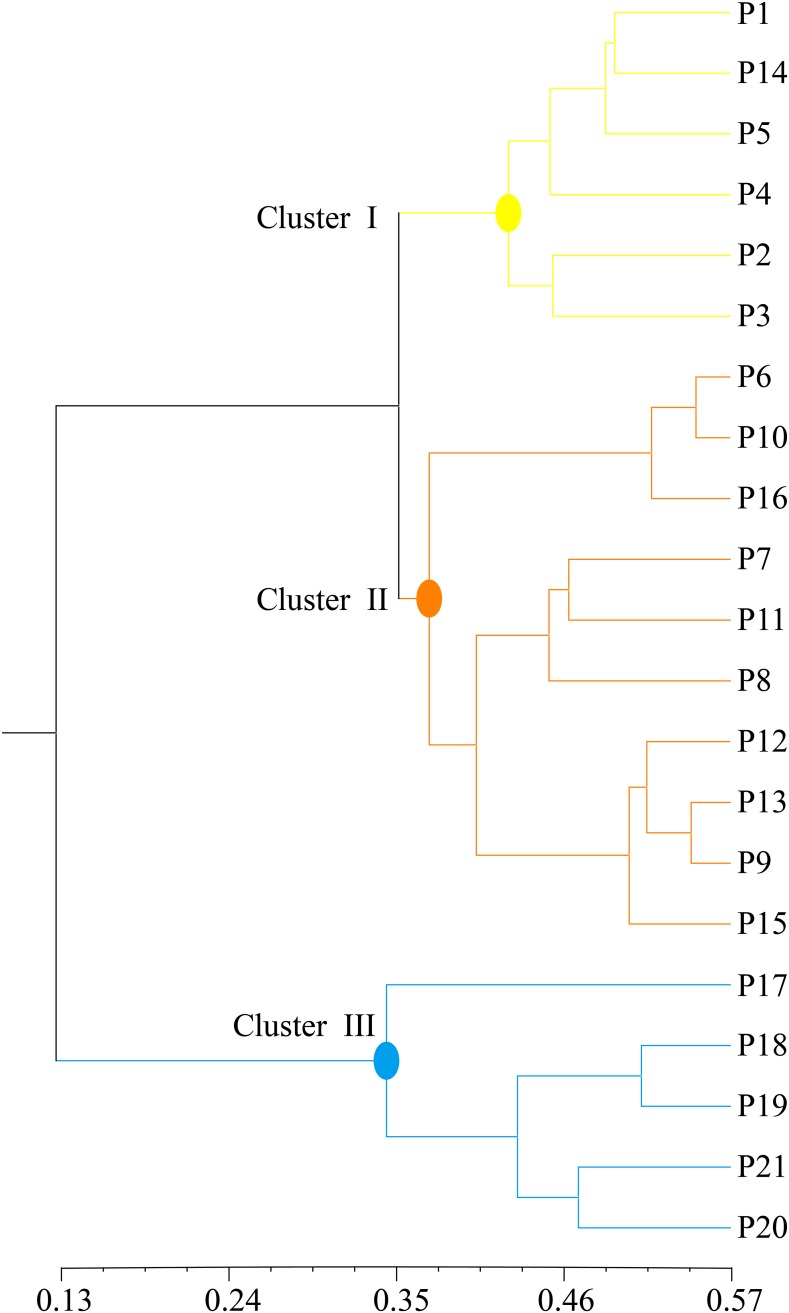
The unrooted UPGMA tree also denoted three clusters that were consistent with those of STRUCTURE and GENELAND analyses (Fig 3). Therefore, we divided the 21 P. mira populations into three genetic clusters for subsequent analyses.
Fig 3. Clustering of 21 P. mira populations based on UPGMA analyses.
Dendrogram of 21 populations of P. mira resulting from the UPGMA cluster analysis based on Nei’s genetic distance, which was obtained from SSR markers.
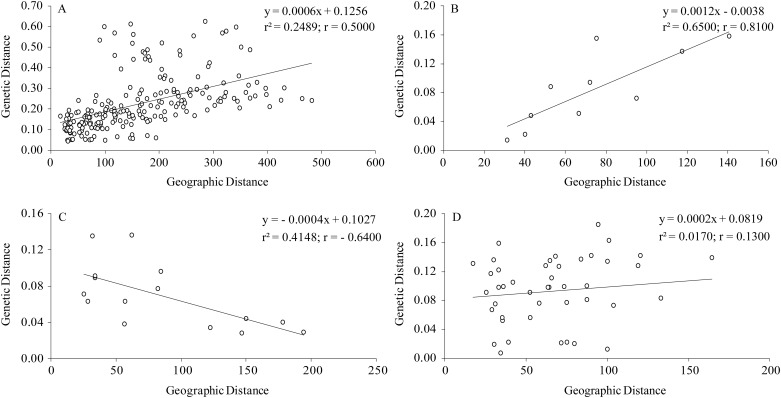
The Mantel test revealed a moderately positive correlation between genetic and geographic distance (r = 0.50, P = 0.01) across the entire distribution of P. mira populations (Fig 4A). This result indicated significant isolation-by-distance (IBD) among all populations. We also tested the pattern of IBD within each genetic cluster. Three separate Mantel tests indicated that a highly positive correlation between geographic and genetic distance still existed within Cluster III (r = 0.81, P = 0.02; Fig 4B), but it was absent within Cluster I (r = -0.64, P = 0.12) and Cluster II (r = 0.13, P = 0.19; Fig 4C and 4D).
Fig 4. Correlation between genetic distance and geographical distance.
(A) Among all populations, (B) among five populations within Cluster III, (C) among six populations within Cluster I, and (D) among 10 populations within Cluster II.
To investigate genetic differentiation in P. mira, we conducted an AMOVA and calculated pairwise differentiation values (FST) and pairwise gene flow values (Nm) among all populations and the three genetic clusters. Genetic differentiation between the 21 populations was significant and high as evidenced by a global FST value of 0.16 (P = 0.01; Table 2). The pairwise FST and pairwise Nm values ranged from 0.01 to 0.36 and from 0.44 to 20.33, respectively (S6 Table). Low genetic differentiation and frequent gene flow were detected between pairwise populations from the same genetic cluster. Populations in Cluster III were significantly and greatly differentiated from the populations in the other two clusters. Among the three genetic clusters, genetic differentiation was also significant and high (FST = 0.18, P = 0.01; Table 2). The differentiation between Clusters I and II was significant, but very slight (FST = 0.03, P = 0.01), whereas the genetic differentiation between Cluster III and the other clusters was significant and slightly high (FST = 0.14, P = 0.01 and FST = 0.13, P = 0.01; S7 Table). Furthermore, an AMOVA based on the 21 populations revealed 83.7% of variation within populations, and that based on the three genetic clusters detected 81.7% of variation within populations, 13.6% among the three clusters, and 4.7% among the populations within each cluster (Table 2). These results supported high genetic variation within the P. mira populations.
Table 2. Analysis of molecular variance (AMOVA) based on twenty-five SSR markers.
| Source of variation | d.f. | Sum of squares | Variance components | Percentage of variation | Fixation index |
|---|---|---|---|---|---|
| Among 21 populations | |||||
| Among populations | 20 | 1172.81 | 1.30 Va | 16.3 | FST = 0.16 |
| Within populations | 819 | 5480.55 | 6.69 Vb | 83.7 | |
| Total | 839 | 6653.36 | 7.99 | 100.0 | |
| Among three genetic clusters | |||||
| Among clusters | 2 | 610.07 | 1.91 Va | 13.6 | FST = 0.18 |
| Among the populations within each cluster | 18 | 562.74 | 0.57 Vb | 4.7 | FSC = 0.05 |
| Within populations | 819 | 5480.55 | 6.69 | 81.7 | FCT = 0.14 |
| Total | 839 | 6653.36 | 9.17 | 100.0 |
FST: variance among the coefficients of individuals relative to the total variance. FSC: variance among subpopulations within groups. FCT: variance among groups relative to the total variance.
Genetic diversity
The mean A, Ae, He, Ho, and I values of the 21 populations were 3.8, 2.5, 0.52, 0.44, and 0.95, respectively, indicating moderate genetic diversity in P. mira populations (Table 3). The highest genetic diversity was detected in P17, and the lowest diversity was observed in P18. In addition to P4, P17, and P20, the remaining 18 populations had private alleles (Np), which ranged from one to four alleles per population. Interestingly, P18 had the highest number of private alleles (Np = 4), although it contained the lowest genetic diversity. Genetic diversity was also estimated within each genetic cluster (Table 3). Cluster II contained the highest genetic diversity, followed by Cluster I, and Cluster III had the lowest genetic diversity.
Table 3. Genetic diversity estimations in all populations and the three genetic clusters.
| Population ID | Sample size | A | Ae | I | He | Ho | FIS | Np | HWE |
|---|---|---|---|---|---|---|---|---|---|
| P1 | 20 | 3.3 ± 0.6 | 2.4 ± 0.5 | 0.89 ± 0.09 | 0.52 ± 0.05 | 0.43 ± 0.05 | 0.18 ± 0.09 | 1 | * |
| P2 | 20 | 3.8 ± 0.5 | 2.3 ± 0.4 | 0.92 ± 0.11 | 0.52 ± 0.08 | 0.44 ± 0.05 | 0.16 ± 0.04 | 1 | -- |
| P3 | 20 | 3.8 ± 0.4 | 2.6 ± 0.3 | 1.00 ± 0.13 | 0.57 ± 0.09 | 0.49 ± 0.08 | 0.13 ± 0.06 | 1 | -- |
| P4 | 20 | 3.6 ± 0.5 | 2.2 ± 0.3 | 0.89 ± 0.08 | 0.50 ± 0.07 | 0.45 ± 0.03 | 0.11 ± 0.04 | 0 | -- |
| P5 | 20 | 3.5 ± 0.6 | 2.2 ± 0.5 | 0.91 ± 0.04 | 0.53 ± 0.05 | 0.44 ± 0.03 | 0.17 ± 0.08 | 2 | -- |
| P6 | 20 | 3.8 ± 0.4 | 2.5 ± 0.3 | 0.95 ± 0.09 | 0.53 ± 0.04 | 0.47 ± 0.04 | 0.12 ± 0.07 | 2 | * |
| P7 | 20 | 3.5 ± 0.5 | 2.6 ± 0.4 | 1.00 ± 0.07 | 0.58 ± 0.05 | 0.47 ± 0.03 | 0.19 ± 0.07 | 2 | * |
| P8 | 20 | 3.5 ± 0.6 | 2.3 ± 0.4 | 0.90 ± 0.07 | 0.52 ± 0.05 | 0.44 ± 0.04 | 0.15 ± 0.07 | 1 | -- |
| P9 | 20 | 3.6 ± 0.6 | 2.5 ± 0.4 | 0.99 ± 0.08 | 0.57 ± 0.05 | 0.43 ± 0.03 | 0.24 ± 0.08 | 2 | * |
| P10 | 20 | 3.6 ± 0.5 | 2.4 ± 0.5 | 0.94 ± 0.07 | 0.54 ± 0.04 | 0.44 ± 0.03 | 0.19 ± 0.07 | 2 | * |
| P11 | 20 | 4.2 ± 0.6 | 2.7 ± 0.4 | 1.05 ± 0.09 | 0.58 ± 0.04 | 0.49 ± 0.04 | 0.15 ± 0.05 | 2 | -- |
| P12 | 20 | 4.1 ± 0.7 | 2.6 ± 0.5 | 1.03 ± 0.08 | 0.57 ± 0.04 | 0.42 ± 0.04 | 0.26 ± 0.05 | 2 | * |
| P13 | 20 | 4.2 ± 0.6 | 2.5 ± 0.4 | 0.99 ± 0.08 | 0.54 ± 0.04 | 0.45 ± 0.04 | 0.16 ± 0.06 | 2 | * |
| P14 | 20 | 4.3 ± 0.7 | 2.6 ± 0.5 | 1.03 ± 0.08 | 0.57 ± 0.04 | 0.46 ± 0.04 | 0.19 ± 0.05 | 1 | -- |
| P15 | 20 | 4.1 ± 0.7 | 2.5 ± 0.5 | 1.00 ± 0.08 | 0.56 ± 0.03 | 0.42 ± 0.05 | 0.25 ± 0.04 | 1 | * |
| P16 | 20 | 3.9 ± 0.7 | 2.4 ± 0.4 | 0.94 ± 0.09 | 0.53 ± 0.04 | 0.42 ± 0.04 | 0.20 ± 0.06 | 1 | -- |
| P17 | 20 | 6.7 ± 0.5 | 4.7 ± 0.6 | 1.57 ± 0.05 | 0.63 ± 0.02 | 0.56 ± 0.01 | 0.11 ± 0.03 | 0 | -- |
| P18 | 20 | 2.1 ± 0.4 | 1.7 ± 0.5 | 0.49 ± 0.09 | 0.30 ± 0.05 | 0.22 ± 0.05 | 0.26 ± 0.10 | 4 | * |
| P19 | 20 | 2.5 ± 0.4 | 1.8 ± 0.4 | 0.58 ± 0.09 | 0.34 ± 0.05 | 0.32 ± 0.05 | 0.06 ± 0.10 | 3 | * |
| P20 | 20 | 3.6 ± 0.4 | 2.3 ± 0.5 | 0.99 ± 0.07 | 0.52 ± 0.04 | 0.43 ± 0.04 | 0.17 ± 0.03 | 0 | -- |
| P21 | 20 | 4.9 ± 0.4 | 2.3 ± 0.4 | 1.01 ± 0.07 | 0.51 ± 0.04 | 0.47 ± 0.03 | 0.08 ± 0.03 | 2 | -- |
| Mean | 20 | 3.8 ± 0.5 | 2.5 ± 0.4 | 0.95 ± 0.08 | 0.52 ± 0.05 | 0.44 ± 0.04 | 0.17 ± 0.06 | 1.50 | -- |
| Cluster I | 120 | 5.7 ± 0.6 | 2.6 ± 0.4 | 1.06 ± 0.09 | 0.56 ± 0.05 | 0.48 ± 0.05 | 0.15 ± 0.06 | 5 | -- |
| Cluster II | 200 | 6.0 ± 0.5 | 2.8 ± 0.6 | 1.13 ± 0.07 | 0.59 ± 0.04 | 0.49 ± 0.03 | 0.16 ± 0.10 | 6 | -- |
| Cluster III | 100 | 8.3 ± 0.6 | 2.5 ± 0.5 | 1.03 ± 0.06 | 0.53 ± 0.04 | 0.47 ± 0.04 | 0.12 ± 0.09 | 7 | -- |
A: Number of alleles; Ae: Number of effective alleles; I: Shannon’s information index; He: Expected heterozygosity; Ho: Observed heterozygosity; FIS: Inbreeding coefficient; Np: Number of private alleles. HWE: Hardy-Weinberg Equilibrium;
*P < 0.05 significant; -- non-significant.
The inbreeding coefficient (FIS) ranged from 0.06 in P19 to 0.26 in P12 and P18, with an average of 0.17 (Table 3). The expected heterozygosity was higher than the observed heterozygosity within each population. These results indicated moderate inbreeding within P. mira populations. In addition, 10 of the 21 populations (P1, P6, P7, P9, P10, P12, P13, P15, P18, and P19) significantly deviated from the HWE.
Morphological clustering and variation
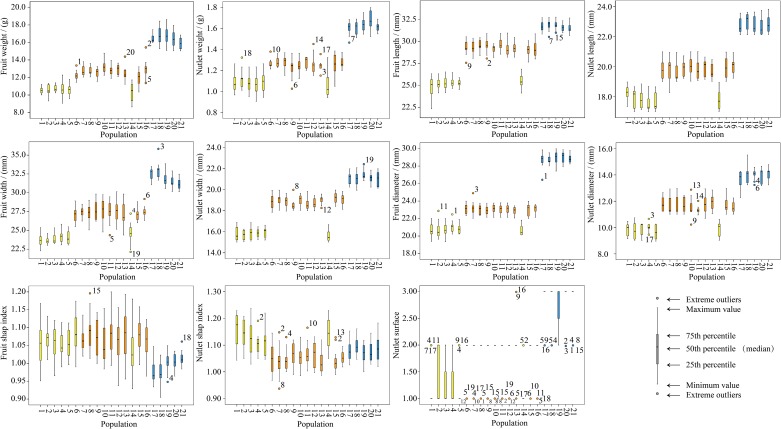
We measured and calculated 11 morphological characteristics of 8400 fruits and 8400 nutlets from the 21 populations, and we used these morphological data to analyze morphological variation among and within P. mira populations. An unrooted UPGMA tree was constructed using the dataset based on the 11 morphological characteristics. Similar to the results based on SSR markers, the 21 populations were assigned to three distinct clusters according to their altitudes (S3 Fig).
When the 21 populations were divided into three clusters based on the morphological data, with the exception of the fruit shape index, the remaining 10 morphological characteristics exhibited a wide range of variation and significant differences among the different clusters (S8 Table). The nutlet surface of most individuals was smooth in Cluster I, shallow groove in Cluster II, and deep groove in Cluster III (S4 Fig, S9 Table). Most of the morphological characteristics increased as the geographic altitudes of populations increased. For instance, low-altitude populations in Cluster I had the lightest and smallest fruits and nutlets, and high-altitude populations from Cluster III contained the heaviest and largest fruits and nutlets. Furthermore, all characteristics exhibited no or low differences among the populations within each cluster (S8 Table, Fig 5). Among the six populations in Cluster I, five characteristics (fruit weight, fruit diameter, nutlet width, nutlet diameter, and nutlet surface) exhibited no significant differences, and the remaining showed low morphological variations. Among the 10 populations from Cluster II, no significant variation was detected for four characteristics (fruit width, fruit diameter, nutlet weight, and nutlet surface), but a low degree of differences was observed for seven characteristics. In Cluster III, only four characteristics (fruit width, fruit shape index, nutlet shape index, and nutlet diameter) indicated low differences among the five populations.
Fig 5. Boxplots based on 11 morphological characteristics.
The coefficients of variation (VC) within populations ranged from 1.30% to 36.17% (S8 Table). The F values among populations were higher than those within populations for all morphological characteristics. These results indicated that most of the variation occurred among populations, which was not consistent with our molecular analysis results.
Discussion
Knowledge of genetic diversity and morphological variation can provide basic information that can aid our understanding of plant evolutionary history and the protection of extant wild resources. In this study, we sampled 420 P. mira individuals from 21 wild populations to study their genetic diversity, population structure, and morphological variation by conducting molecular analyses and morphological evaluations.
Population structure and differentiation
The STRUCTURE and GENELAND analyses based on SSR markers revealed three genetic clusters, which were highly correlated with geographic altitudes. The most likely reason for this result is that altitude differences led to significant climatic and environmental differences in water, soil, light, and temperature among the populations distributed at different altitude ranges. Climatic and environmental factors can significantly influence plant geographic distributions, evolutionary patterns, and phenological phases [45]. According to the results of our field investigation, P. mira populations at high altitudes usually flower five to 14 days later than populations at low altitudes. This phenomenon can significantly reduce the gene flow through pollen, and it can shape clear structures among populations that were distributed at different altitudinal ranges.
Mantel et al. [46] suggested that large geographic distances would limit pollen and seed dispersal among populations. In this study, moderately positive IBD was detected among all P. mira populations. However, although we conducted separate Mantel tests within clusters, different results were revealed. All of these results indicated that geographic distance is not a crucial factor for the current genetic composition in partial small-scale areas. Regarding populations from Clusters I and II, we inferred that altitude gradients might have more significant effects on population structure than geographic distance. For instance, P14 was not assigned to Cluster II based on pairwise geographic distances, but was instead grouped with Cluster I because of its geographic altitude.
Our findings indicated that Cluster III was significantly and highly differentiated from the other two clusters. Five populations from Cluster III and 16 populations from Cluster I and Cluster II are distributed in Shannan and Linzhi, respectively. In the central region of the Tibet plateau, numerous mountains, rivers, and valleys shape geographic barriers between the two geographic groups, thereby leading to the observed high genetic differentiation. However, low gene flow values (Nm < 1) were observed between pairwise populations from different geographic regions, thus indicating that the gene flow cannot combat genetic differentiation caused by genetic drift [16, 18]. According to Slaktin et al. [47], the founder effect is an important form of genetic drift in isolated populations, so the genetic drift caused by “founder effects” might have affected the genetic structures of P. mira populations.
Genetic diversity of populations
Molecular analyses revealed moderate genetic diversity (A = 3.8, Ae = 2.5, Ho = 0.44, He = 0.52, I = 0.95) in P. mira populations. The genetic diversity of P. mira in this study was higher than that previously reported for P. mira (A = 1.7, Ae = 1.4, I = 0.35) [48] and other related peach species (A = 2.7, Ae = 2.1, Ho = 0.32–0.34, He = 0.23–0.46) [49,50]. Theoretically, the genetic diversity of endangered species should be low, but this was not the case for P. mira. Moderate or even high levels of genetic diversity have been reported in some endangered species [51,52]. Regarding P. mira, high adaption to various environments and wide distributions might have helped shape and preserve the genetic diversity of populations under poor living conditions [53,54]. For example, more than 60% of Tibetan areas are mountains, and the impact of topography and elevation led to significant climatic and environmental differences in water, soil, light, and temperature, thus resulting in a number of distinct and natural geographical areas. Overall, the natural ecological conditions of the Tibet region are poor because the plateau climate is characterized by thin air, low air pressure, reduced oxygen, thin soil, low rainfall, stronger solar radiation, perennial ice and snow, and frost weathering [55]. The effective population size of P. mira is relatively large, but more recent periods of excessive logging and fragmentation due to habitat destruction may have led to the initial loss of genetic diversity, despite the short period of fragmentation. Therefore, this factor might be also responsible for the moderate genetic diversity in P. mira.
Positive inbreeding coefficients were detected within each population and at each locus, indicating the significant excess of homozygotes in P. mira. Deforestation and geographic isolation have led to the heavy fragmentation of P. mira habitats [6,48,55]. Habitat fragmentation can limit genetic exchange among populations, and it can increase inbreeding within a population [56–58]. Thus, moderate inbreeding in P. mira could be attributed to habitat fragmentation. Previous studies [55] revealed that human activities such as over-harvesting and deforestation had significant effects on wild P. mira resources near rural areas in the Tibetan plateau. In this study, we found 7 populations near rural areas that deviated from HWE, thus indicating that recent human activities might have significant effects on these P. mira populations.
Genetic and morphological variation
Perennial woody plant species are expected to preserve high variation within populations because of their long life cycles [59–61]. An AMOVA based on SSR markers revealed that most of the total genetic variation (83.7% and 81.7%) occurred within P. mira populations, which is higher than that reported previously based on SSR (81.0%), ISSR (71.0%), and AFLP (74.48%) data [7,48]. This discrepancy can be attributed to the fact that the number of individuals analyzed in this study was considerably greater than that in previous studies. For P. mira, an outcrossing system and long life cycle (over 1000 years) can improve gene flow and preserve shared ancestral genetic information among populations, thus significantly improving genetic variation within populations.
Both molecular and morphological data revealed three clusters that were correlated with geographic altitudes. Interestingly, our morphological analyses showed fewer differences within populations, and this was contrary to our molecular analysis results. The morphological variation among the three clusters was higher than that within clusters. P. mira individuals are distributed over a narrow altitude range within each population, although they are widely distributed in the Tibet plateau. Combined with our molecular results, we concluded that the morphological differences may be caused by the relatively isolated distributions and various climates and environments associated with P. mira populations from different geographic altitudes. For instance, high-altitude populations contained larger and heavier fruits and nutlets than the medium- and low- altitude populations. These results were consistent with the findings of Pluess et al. [62] in that large seeds could be retained at high-altitude areas because of their high seedling rates under poor environmental conditions.
Conservation strategy
Studying genetic diversity, population structure, and morphological variations could provide basic information about plant conservation [63]. Although the levels of genetic diversity of P. mira populations were moderate, some populations still exhibited high risk of genetic variation loss, and human activities and climates had significant effects on most P. mira populations. Therefore, there is also an urgent need to conduct P. mira resource conservation.
The analyses based on SSR markers and morphological characteristics revealed three completely consistent clusters among the 21 wild P. mira populations. Each cluster contained unique genetic and morphological variations and had different evolutionary histories. Therefore, the conservation efforts should be divided into three units that correspond to the three genetic and morphological clusters. Because the six populations in Cluster I were found in national nature reserves, more attention needs to be paid to the other clusters. Furthermore, in situ conservation should prioritize populations with high genetic diversity in Clusters II and III such as P11 and P17. Because human activities have had significant effects on many P. mira populations (P6, P7, P9, P10, P12, P13, and P15), we propose the strengthening of conservation awareness in local farmers and the hanging of placards to reduce deforestation and prevent continuing habitat deterioration. We also suggest ex situ conservation for individuals with unique traits and populations with high inbreeding to prevent the potential loss of genetic variation. Furthermore, improving the genetic diversity within populations requires the monitoring of crossing among populations.
Conclusions
This study comprehensively analyzed the genetic diversity, population structure, and morphological variation of 21 wild P. mira populations in the Tibet plateau. Moderate levels of genetic diversity were detected in P. mira populations. Significant homozygote excess was observed within each population, and this might be the result of sibling mating caused by habitat fragmentation. Furthermore, human activities might be responsible for the deviation of numerous P. mira populations from the HWE. Both molecular and morphological analyses assigned the 21 populations to three clusters, which were significantly correlated with the geographic altitudes. Because of the presence of geographic barriers and genetic drift, populations in Shannan were highly differentiated from populations in Linzhi. Lastly, we formed three conservation units and proposed several conservation strategies for these studied populations.
Supporting information
(TIF)
(TIF)
(TIF)
The nutlet surfaces of 420 P. mira individuals can be divided into three clusters based on visual inspection. The first cluster was smooth (P1–P5, P14), the second cluster exhibited nutlet surfaces with shallow grooves (P6, P7, P8, P9, P10, P11, P12, P13, P15, and P16), and the third cluster displayed nutlet surfaces with deep grooves (P17, P18, P19, P20, and P21).
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to Tiezhu Li, Huimin Liu, Zhongmao Jiang, Xuchun Zhu, and Xiaolin Du for contributing to the collection of plant materials and to Jing Zhang for technical assistance, English correction, and useful suggestions.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the National Science and Technology Pillar Program during the 12th Five-year Plan Period (2013BAD14B02) to Tana Wuyun.
References
- 1.Yü DJ. Taxonomy of fruit trees in China. Agricultural Press, Beijing; 1979. [Google Scholar]
- 2.Peng M, Guan FC, Tao L, Li RQ, Wang C, Meng FJ. Analysis of genetic relationship on Amygdalus mira (koehne) Ricker with other peach species using simple sequence repeat (SSR). Biochemical Systematics and Ecology. 2015; 62: 98–105. [Google Scholar]
- 3.Guo GN, Wang LR, Yan ZL, Zhu GR, Fang WC. Study on the variation and evolution of Peach and its related wild species by Pollen morphology. Journal of fruit science. 2006; 23: 664–669. [Google Scholar]
- 4.Guo JY. RAPD analysis of germplasm resources on peach. Northwest A&F University; Yangling, Shaanxi, China, 2002; 1–3. [Google Scholar]
- 5.Dong GZ. The investigation of Prunus mira Koehne in Tibet. Quarterly Forest by-product and Speciality in China. 1991; 3: 44–45. [Google Scholar]
- 6.Tian Y, Xing C, Cao Y, Wang C, Guan FC, Li RQ, et al. Evaluation of genetic diversity on Prunus mira Koehne by using ISSR and RAPD markers. Biotechnol Biotec Eq. 2015; 29: 1053–1061. doi: 10.1080/13102818.2015.1064780 [Google Scholar]
- 7.Li TF, Liu JR, Xie YN, Wang QY, Meng FJ. Analysis of genetic diversity in Prunus mira Koehne ex Sargent populations using AFLP markers. Plant Syst Evol. 2014; 300: 475–482. doi: 10.1007/s00606-013-0896-5 [Google Scholar]
- 8.Guan FC, Wang SP, Li RQ, Peng M, Meng FJ. Genetic diversity of wild peach (Prunus mira Koehne kov et. Kpst) from different altitudes in the Tibetan plateau by pollen morphous and RAPD markers. HortScience. 2014; 49: 1017–1022. [Google Scholar]
- 9.Lu J, Lan XZ. The characteristics of the rare and endangered Tibetan medicinal plant resources in shannan region. Journal of Natural Resource. 2013; 11: 1977–1987. doi: 10.11849/zrzyxb.2013.11.014 [Google Scholar]
- 10.Ge LW, Pan G, Ren DZ, Du YJ, Zheng XL. Forest carbon storage, carbon density, and their distribution characteristics in Linzhi area of Tibet, China. Chinese Journal of Applied Ecology, February 2013; 24: 319–325. [PubMed] [Google Scholar]
- 11.Liu L, Meng FJ. Fruit quality analysis of Amygalus mira in Tibet. Journal of Anhui Agri. Sci. 2013, 41: 13740–137413. [Google Scholar]
- 12.Zeng XL. Study on the diversity of the wild Amygalus mira in Tibet. Anhui Agri. Sci. Bull. 2016; 22: 12–15. [Google Scholar]
- 13.Kramer AT, Havens K. Plant conservation genetics in a changing world. Trends Plant Sci. 2009; 14: 599–607. doi: 10.1016/j.tplants.2009.08.005 [DOI] [PubMed] [Google Scholar]
- 14.Allendorf FW, Luikart G. Conservation and the genetics of populations. Wiley, New York: 2009. [Google Scholar]
- 15.Abebe TD, Bauer AM, Léon J. Morphological diversity of Ethiopian barleys (Hordeum vulgare L.) in relation to geographic regions and altitudes. Hereditas. 2010; 147: 154–164. doi: 10.1111/j.1601-5223.2010.02173.x [DOI] [PubMed] [Google Scholar]
- 16.Singh N, Choudhury DR, Singh AK, Kumar S, Srinivasan K, Tyagi RK, et al. Comparison of SSR and SNP markers in estimation of genetic diversity and population structure of Indian Rice varieties. PLoS ONE. 2013; 8: 1524–1528. doi: 10.1371/journal.pone.0084136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramakrishnan M, Ceasar SA, Duraipandiyan V, A-dhabi NA, Ignacimuthu S. Using molecular markers to assess the genetic diversity and population structure of finger millet (Eleusine coracana (L.) Gaertn.) from various geographical regions. Genet Resour Crop Evol. 2016; 63: 361–376. doi: 10.1007/s10722-015-0255-1 [Google Scholar]
- 18.Berlin S, Trybush SO, Fogelqvist J, Gyllenstrand N, Hallingbäck HR, Ähman I, et al. Genetic diversity, population structure and phenotypic variation in European Salix viminalis L. (Salicaceae). Tree Genet Genomes. 2014; 10:1595–1610. doi: 10.1007/s11295-014-0782-5 [Google Scholar]
- 19.Pinheiro LR, Rabbani ARC, Siliva AVCD, Lédo ADS, Pereira KLG, Diniz LEC. Genetic diversity and population structure in the Brazilian Cattleya labiate (Orchidaceae) using RAPD and ISSR markers. Plant Syst Evol. 2012; 298: 1815–1825. doi: 10.1007/s00606-012-0682-9 [Google Scholar]
- 20.Du QZ, Wang BW, Wei ZZ, Zhang DQ, Li BL. Genetic diversity and population structure of Chinese White poplar (Populus tomentosa) revealed by SSR markers. J Hered. 2012; 103: 853–862. doi: 10.1093/jhered/ess061 . [DOI] [PubMed] [Google Scholar]
- 21.Mourad M, Jordi GM, Audergon JM, Arús P. Prunus microsatellite marker transferability across rosaceous crops. Tree Genet Genomes. 2010; 6: 689–700. doi: 10.1007/s11295-010-0284-z [Google Scholar]
- 22.Dettori MT, Micali S, Giovinazzi J, Scalabrin S, Verde I, Cipriani G. Mining microsatellites in the peach genome: development of new long-core SSR markers for genetic analyses in five Prunus species. Springerplus. 2015; 4: 337 doi: 10.1186/s40064-015-1098-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raggi L, Bitocchi E, Russi L, Marconi G, Sharbel TF, Veronesi F, et al. Understanding genetic diversity and population structure of a Poa pratensis worldwide collection through morphological, nuclear and chloroplast diversity analysis. PLoS ONE. 2015: 10: e0124709 doi: 10.1371/journal.pone.0124709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987; 19: 11–15. [Google Scholar]
- 25.Sosinski B, Gannavarapu M, Hager LD, Beck LE, King GJ, Ryder CD, et al. Characterization of microsatellite markers in peach [Prunus persica (L.) Batsch]. Theor Appl Genet. 2000; 101:421–428. doi: 10.1007/s001220051499 [Google Scholar]
- 26.Wang Y, Georgi LL, Zhebentyayeva TN, Reighard GL, Scorza R, Abbott AG. High-throughput targeted SSR marker development in peach (Prunus persica). Genome. 2002; 45: 319–328. doi: 10.1139/G01-153 . [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T, Mochida K, Imai T, Shi YZ, Ogiwara I, Hayashi T. Microsatellite markers in peach [Prunus persica (L.) Batsch] derived from an enriched genomic and cDNA libraries. Mol Ecol. 2002; 2(6): 298–301. doi: 10.1046/j.1471-8286.2002.00242.x [Google Scholar]
- 28.Dirlewanger E, Cosson P, Tavaud M, Aranzana J, Poizat C, Zanetto A, et al. Development of microsatellite markers in peach [Prunus persica (L.) Batsch] and their use in genetic diversity analysis in peach and sweet cherry (Prunus avium L.). Theor Appl Genet, 2002; 105(6): 127–138. doi: 10.1007/s00122-002-0867-7 . [DOI] [PubMed] [Google Scholar]
- 29.Cipriani G, Lot G, Huang WG, Marrazzo MT, Peterlunger E, Testolin R. AC/GT and AG/CT microsatellite repeats in peach [Prunus persica (L) Batsch]: isolation, characterisation and cross-species amplification in Prunus. Theor Appl Genet. 1999; 99: 65–72. doi: 10.1007/s001220051209 [Google Scholar]
- 30.Testolin R, Marrazzo T, Cipriani G, Quarta R, Verde I, Dettori MT, et al. Microsatellite DNA in peach (Prunus persica L. Batsch) and its use in fingerprinting and testing the genetic origin of cultivars. Genome, 2000; 43(3): 512–520. doi: 10.1139/g00-010 . [PubMed] [Google Scholar]
- 31.Xie H, Sui Y, Chang FQ, Xu Y, Ma RC. SSR allelic variation in almond (Prunus dulcis Mill.). Theor Appl Genet. 2006; 112: 366–372. doi: 10.1007/s00122-005-0138-5 . [DOI] [PubMed] [Google Scholar]
- 32.Cantini C, Iezzoni AF, Lamboy WF, Boritzki M, Struss D. DNA Fingerprinting of Tetraploid Cherry Germplasm Using Simple Sequence Repeats. JASHS March. 2001; 126: 205–209. [Google Scholar]
- 33.Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics. 2012; 28: 2537–2539. doi: 10.1093/bioinformatics/bts460 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Resour. 2007; 7: 574–578. doi: 10.1111/j.1471-8286.2007.01758.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005; 14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x . [DOI] [PubMed] [Google Scholar]
- 36.Earl D, VonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012; 4: 359–361. doi: 10.1007/s12686-011-9548-7 [Google Scholar]
- 37.Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007; 23: 1801–1806. doi: 10.1093/bioinformatics/btm233 . [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg NA. DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes. 2004; 4: 137–138. doi: 10.1046/j.1471-8286.2003.00566.x [Google Scholar]
- 39.Safner T, Miller MP, McRae BH, Fortin MJ, Manel S. Comparison of Bayesian clustering and edge detection methods for inferring boundaries in landscape genetics. International Journal of Molecular Sciences. 2011;12: 865–889. doi: 10.3390/ijms12020865 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao LJ, Wei SJ, Hoffmann AA, Wen JB, Chen M. Rapid genetic structuring of populations of the invasive fall webworm in relation to spatial expansion and control campaigns. Diversity and Distributions. 2016; 9:1–12. doi: 10.1111/ddi.12486 [Google Scholar]
- 41.Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010; 10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x [DOI] [PubMed] [Google Scholar]
- 42.Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978; 89: 583–590. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohlf FJ. NTSYS-PC: Numerical Taxonomy and Multivariate Analysis System Version 1. 80 Setauket New York: Distribution by Exeter Soft Ware; 1994. [Google Scholar]
- 44.Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980; 32: 314–331. . [PMC free article] [PubMed] [Google Scholar]
- 45.Bemmels JB, Title PO, Ortego J, Knowles LL. Tests of species-specific models reveal the importance of drought and ABC model selection. Mol Ecol. 2016; 19: 4889–4906. doi: 10.1111/mec.13804 [DOI] [PubMed] [Google Scholar]
- 46.Mantel S. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 2003; 18: 189–197. doi: 10.1016/S0169-5347(03)00008-9 [Google Scholar]
- 47.Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993; 47: 264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x [DOI] [PubMed] [Google Scholar]
- 48.Xing C, Tian Y, Guan F, Meng FJ. Evaluation of genetic diversity in Amygdalus mira (Koehne) ricker using SSR and ISSR markers. Plant Syst Evol. 2015; 301: 1055–1064. [Google Scholar]
- 49.Aranzana MJ, Abbassi EK, Howad W, Arús P. Genetic variation, population structure and linkage disequilibrium in peach commercial varieties. BMC Genet. 2010; 69: 1–13. doi: 10.1186/1471-2156-11-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tahan O, Geng Y, Zeng L, Dong S, Chen F, Chen J, et al. Assessment of genetic diversity and population structure of Chinese wild almond, Amygdalus nana, using EST- and genomic SSRs. Biochem Syst Ecol. 2009; 37: 146–153. doi: 10.1016/j.bse.2009.02.006 [Google Scholar]
- 51.Jia J, Zeng LQ, Gong X. High genetic diversity and population differentiation in the critically endangered plant species Trailliaedoxa gracilis (Rubiaceae). Plant Mol Biol Rep. 2016; 1: 327–338. doi: 10.1007/s11105-015-0924-4 [Google Scholar]
- 52.Davidnm M, Marka MP. Endangered species in small habitat patches can possess high genetic diversity: the case of the Tana River red colobus and mangabey. Conserv Genet. 2010; 11: 1725–1735. doi: 10.1007/s10592-010-0065-0 [Google Scholar]
- 53.Wang Z, Kang M, Liu HB, Gao J, Zhang ZD, Li YY, et al. High-Level Genetic Diversity and Complex Population Structure of Siberian Apricot (Prunus sibirica L.) in China as Revealed by Nuclear SSR Markers. PLoS ONE. 2014; 9: 1–13. doi: 10.1371/journal.pone.0087381 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hamrick JK. Isozymes and the Analysis of Genetic Structure in Plant Populations Isozymes in Plant Biology. London: Chapman and Hall; 1990; 87–105. [Google Scholar]
- 55.Fang JP, Zhong C, Zhong GH. The age structure of Tibetan Purnus mira Koehne Kovet. Kpsl population in Tibet Linzhi region. china forestry science and technology. 2008; 22: 52–53. [Google Scholar]
- 56.Aguilar R, Quesada M, Ashworth L, Herreriasdiego Y, Lobo J. Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol Ecol. 2008; 17: 5177–5188. doi: 10.1111/j.1365-294X.2008.03971.x [DOI] [PubMed] [Google Scholar]
- 57.Leimu R, Vergeer P, Angeloni F, Ouborg J. Habitat fragmentation, climate change, and inbreeding in plants. Ann Ny Acad Sci. 2010; 1195: 84–98. doi: 10.1111/j.1749-6632.2010.05450.x [DOI] [PubMed] [Google Scholar]
- 58.Breed MF, Gardner MG, Ottewell KM, Navarro CM, Lowe AJ. Shifts in reproductive assurance strategies and inbreeding costs associated with habitat fragmentation in Central American mahogany. Ecol Lett. 2012; 15: 444–452. doi: 10.1111/j.1461-0248.2012.01752.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hamrick J, Godt M. Effects of life history traits on genetic diversity in plant species. Phil. Trans R Soc Lond B. 1996; 351: 1291–1298. doi: 10.1098/rstb.1996.0112 [Google Scholar]
- 60.Gillies ACM, Navarro C, Lowe AJ, Newton AC, Hernandez M, Wilson J, et al. Genetic diversity in Mesoamerican populations of mahogany (Swietenia macrophylla), assessed using RAPDs. Heredity. 1999; 83: 722–732. . [DOI] [PubMed] [Google Scholar]
- 61.Yeh FC, Chong DKX, Yang RC. RAPD variation within and among natural populations of trembling aspen (Populus tremuloides Michx.) from Alberta. J Hered. 1995; 86: 454–460. . [DOI] [PubMed] [Google Scholar]
- 62.Pluess AR, Schutz W, Stucklin J. Seed weight increase with altitude in Swiss Alps between related species but not among populations of individual species. Oecologia. 2005; 144: 55–61. doi: 10.1007/s00442-005-0047-y [DOI] [PubMed] [Google Scholar]
- 63.Newton AC, Allunutt TR, Gillies ACM, Lowe AJ, Ennos RA. Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol. 1999; 14: 140–145. doi: 10.1016/S0169-5347(98)01555-9 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
The nutlet surfaces of 420 P. mira individuals can be divided into three clusters based on visual inspection. The first cluster was smooth (P1–P5, P14), the second cluster exhibited nutlet surfaces with shallow grooves (P6, P7, P8, P9, P10, P11, P12, P13, P15, and P16), and the third cluster displayed nutlet surfaces with deep grooves (P17, P18, P19, P20, and P21).
(TIF)
(XLSX)
(XLSX)
(XLSX)
(XLS)
(XLS)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.