SUMMARY
The human gut microbiota is engaged in multiple interactions affecting host health during the host's entire life span. Microbes colonize the neonatal gut immediately following birth. The establishment and interactive development of this early gut microbiota are believed to be (at least partially) driven and modulated by specific compounds present in human milk. It has been shown that certain genomes of infant gut commensals, in particular those of bifidobacterial species, are genetically adapted to utilize specific glycans of this human secretory fluid, thus representing a very intriguing example of host-microbe coevolution, where both partners are believed to benefit. In recent years, various metagenomic studies have tried to dissect the composition and functionality of the infant gut microbiome and to explore the distribution across the different ecological niches of the infant gut biogeography of the corresponding microbial consortia, including those corresponding to bacteria and viruses, in healthy and ill subjects. Such analyses have linked certain features of the microbiota/microbiome, such as reduced diversity or aberrant composition, to intestinal illnesses in infants or disease states that are manifested at later stages of life, including asthma, inflammatory bowel disease, and metabolic disorders. Thus, a growing number of studies have reported on how the early human gut microbiota composition/development may affect risk factors related to adult health conditions. This concept has fueled the development of strategies to shape the infant microbiota composition based on various functional food products. In this review, we describe the infant microbiota, the mechanisms that drive its establishment and composition, and how microbial consortia may be molded by natural or artificial interventions. Finally, we discuss the relevance of key microbial players of the infant gut microbiota, in particular bifidobacteria, with respect to their role in health and disease.
KEYWORDS: microbiome, microbiota, infants, metagenomics, virome, bifidobacteria, gut commensals, probiotics, gut microbiota
INTRODUCTION
General Features of the Infant Gut Microbiota
The human body harbors trillions of microbial cells whose coordinated actions are believed to be important for human life. Such microbial cell populations reach their highest density in the intestinal compartment, where they collectively form a complex microbial community known as the gut microbiota (1) which develops over the course of host infancy to eventually reach its adult form (2–4). Gut microbiota members may belong to any of the three domains of life, i.e., Archaea, Bacteria, and Eukarya, and also include viruses, and they are known to establish complex trophic relationships with each other and their human host, ranging from symbiosis to parasitism (5). The human gut microbiota is composed of autochthonous, also known as indigenous, microorganisms and allochthonous or transient microorganisms (6). In this context, only a relatively small number of (opportunistic) pathogens are considered to be members of the gut microbiota, residing unperturbed within the enteric host microbiota and becoming a health threat to the host only when the gut ecosystem is disturbed and the gut microbiota homeostasis becomes disrupted (see below).
The gastrointestinal microbiota composition may be affected by a number of environmental parameters, such as pH, oxygen levels/redox state, availability of nutrients, water activity, and temperature, enabling various populations to thrive and exert different activities while interacting with their environment, including that of the human host (7).
The abundant and diverse members of the human gut microbiota exert critical roles in the maintenance of human health by assisting in the breakdown of food substances so as to liberate nutrients that would otherwise be inaccessible to the host, by promoting host cell differentiation, by protecting the host from colonization of pathogens, and by stimulating/modulating the immune system. Various epidemiological studies have established a clear correlation between factors that disrupt the gut microbiota during childhood on the one hand and immune and metabolic disorders later in life on the other (8–10). Thus, there are increasing experimental data that support long-term health benefits elicited by the infant gut microbiota and that also implicate the early human gut microbiota in modulating risk factors related to particular adult health conditions (11). This realization has in turn fueled the development of strategies to influence the development, composition, and activities of the infant microbiota by use of nutraceutical products (e.g., probiotics and/or prebiotics).
An intriguing feature of the adult gut microbiota is that the development of such a microbial assemblage reaches a climax status represented by the establishment of a homeostasis among all its members (12). A wide range of factors can cause shifts in this microbiota balance, thereby disrupting the gut microbiota homeostasis and causing a so-called state of dysbiosis. There is controversy on the exact meaning of dysbiosis, simply because of the lack of an accurate description of a “normal” or healthy microbiota. Dysbiosis is usually associated with harmful effects and may have long-term consequences leading to disorders or diseases, including obesity, diabetes, and inflammatory bowel disease (IBD) (13–17). In addition, fluctuations occur in the gut microbiota composition throughout host development from infancy to early childhood, from young to aging adulthood, and during pregnancy.
Gut Microbiota Development and Dynamics
Each individual can be viewed as an island that consists of various habitats which are colonized by microbial communities and which follow rules that create and shape diversity in local assemblages, including dispersal, in situ diversification, environmental selection, and ecological drift (18, 19) (Fig. 1).
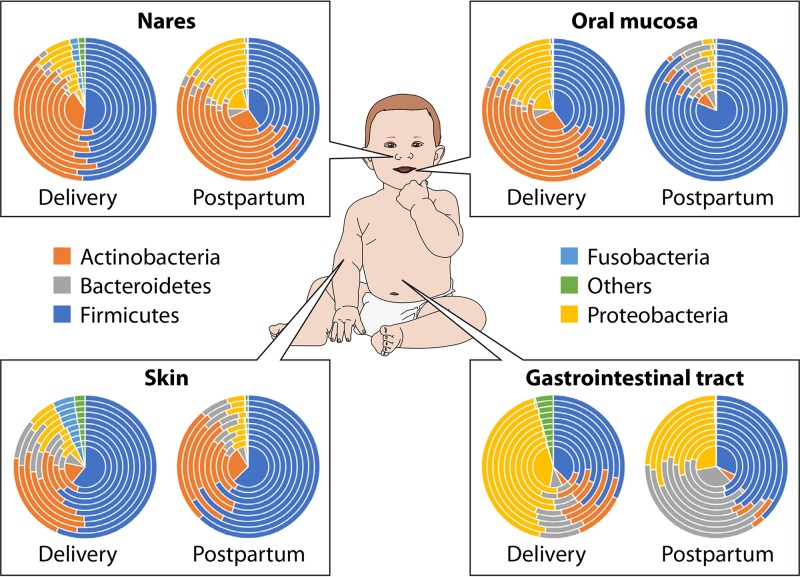
FIG 1.
Microbiota composition across the different infant body sites. A global overview of the relative abundances of key phyla of the infant microbiota composition in different body sites and at different stages of early life is shown. Concentric cake diagrams schematically represent interindividual variability.
Dispersal is a natural process causing an increase in diversity in local microbial communities, consistent with the view of the human body as an ecological “island,” an area of habitat, which is continually sampling the pool of available colonists (19). Another ecological process that impacts microbial communities is local diversification, which is based on rapid microbial adaptation via mutation or recombination (19). In this scenario, horizontal gene transfer events, often driven by phages, may represent one of the main forces responsible for microbial diversification, especially for those microbes that share the same ecological niche (20).
Environmental selection represents another key ecological process which shapes the human microbiota and which may be viewed as a “habitat filter” consisting of an assemblage of resources and conditions that allow and/or support growth of certain microorganisms but not others, underscoring the selection of microbial features that permit survival and growth in the host. Such a scenario assumes that the host determines the microbiota rather than the other way around. Notably, the overall profiles that can originate from environmental selection (niche-based interactions) may vary as a function of the spatial scale across which these processes occur (21).
In addition, the abundances of microbial taxa can change due to another ecological process, identified as ecological drift or demographic stochasticity. Such an ecological process is responsible for the disappearance of low-abundance species (e.g., antibiotic-sensitive strains) unless they gain a competitive advantage across a different ecological niche or become replenished by dispersal from outside the community (19).
Finally, the microbiota composition and relative abundance of each bacterial member are also influenced by phage predation, which acts as a very powerful force that impacts community structure and dynamics (see below).
The Holobiont Concept Applied to Infants
The human body is considered a host symbiont or holobiont, which acts as an ecosystem under selection to minimize conflict between individual members (22). The holobiont concept does not consider macrobes as autonomous individuals but as highly organized biological units, which not only consist of the entire eukaryotic cell arsenal that makes up the host's body but also include the microbiota contained on or within the host (23, 24) (Fig. 1). The holobiont concept emphasizes the important role of coevolution in the assembly and dynamics of the human ecosystem and highlights that long- and short-term selective pressures on the human microbiota are not inevitably aligned. In this context, genetic variation among the hologenomes, i.e., the combined genetic content of the host, its organelles, and its associated microbiota, may happen due to changes in the host genome or modifications of the genomes of the constituent symbiotic microorganisms (25, 26). Overall, the microbiomes, and consequently their encoded phenotypes, may change through variations in the relative abundances of specific microorganisms, through modification of the genomes of existing resident microorganisms, or through loss or gain of microbial symbionts into holobionts. Interestingly, the genetic variation that may occur within a microbiome greatly exceeds that of the host genome, while it also develops much more rapidly than that of the host genome. Therefore, microbial sources of hologenomic variation are potential targets for evolution, and the microbiome should consequently be considered in the overall study of human evolution.
An emerging perception from recent studies is that the microbial communities which belong to the holobiont are particularly important for host health during the establishment of the infant gut microbiota (27). This developmental trajectory involves crucial steps such as choreographed gut colonization by bacterial populations, dynamic alteration in the virome structure, and transkingdom interactions between host and microbial cells.
Projects Directed To Assess the Composition/Functionality of the Infant Gut Microbiota
As mentioned above, the microbial gut community plays an important role in human health (28). Alterations and aberrations in the gut microbiota composition during neonatal life, which represents the first month of life from the moment of birth, as well as during infancy, which spans 1 month until 2 years of age, have been associated with pediatric disorders and the onset of disease in later life (29). We may assume that the early gut microbiota contributes to disease progression later in life and that the foundation for a stable adult gut microbiota is already established in infancy. This notion explains the need for an in-depth comprehension of the infant microbiota composition and development, the interactions of microbiota members with each other and their infant host, and the mechanisms by which such host-microbiota interactions maintain gut homeostasis. Indeed, a number of research efforts, including the JPI-HDHL projects EarlyMicroHealth, EarlyVir, and GI-MDH and the French Epiflore, the Irish Infantmet, and the NIH-funded MOMS-PI projects, in addition to other publicly and privately funded initiatives, are aimed at understanding the factors that determine the infant intestinal microbiota composition, establishment, and development and their associated long-term health consequences.
In the following sections, we assess the current knowledge about the infant gut microbiota by analyzing the technical approaches employed to catalogue infant gut microbial consortia and reconstruct the functionalities exerted by these communities. In addition, we discuss the mechanisms responsible for development, transmission, establishment, and persistence of microorganisms in the infant gut and their implications for health in regard to early and long-lasting outcomes. We also analyze how the microbial consortia can be modulated by natural and/or artificial interventions. Furthermore, we discuss the relevance of some of the most dominant microbial members of the infant gut in terms of current knowledge regarding their biological role(s).
TECHNICAL APPROACHES FOR MICROBIOTA DETERMINATION
General Features
Although microorganisms are abundant and ubiquitous, we currently lack a fundamental mechanistic understanding of many of the key roles played by microorganisms in nature, including those that reside in the human body (30). Until the recent development of novel culturomics approaches (31), only a very small fraction of the human gut microbiota had been isolated and studied in pure culture (30). The presumption that a large proportion of the human gut microbiota was uncultured (32) prompted the development of culture-independent approaches, i.e., metagenomics, metatranscriptomics, and metaproteomics, to discover the identities, activities, and functional roles of the so-far-uncultivated members of the gut microbiota (Fig. 2). High-throughput sequencing of (a portion of) the 16S rRNA gene (i.e., 16S rRNA gene-based microbial profiling analysis) as a conserved phylogenetic marker represents the current standard methodology for profiling complex microbial communities, although shotgun metagenomics is progressively replacing 16S rRNA gene-based microbial profiling analysis (see below). The 16S rRNA gene-based microbial profiling approach relies on universal primers for amplification of single or multiple hypervariable regions of the 16S rRNA gene (33). Reads of the obtained amplicons, having been retrieved from a next-generation sequencing (NGS) platform, are processed using bioinformatic pipelines, such as the popular Qiime software suite (34) or Mothur (35), thus allowing the reconstruction of the microbial composition of the analyzed environmental sample. This methodology also facilitates identity assignment for unknown members of microbial communities through discrimination based on the sequences of their unique hypervariable regions (36). Furthermore, sequencing of a microbiome, an approach called metagenomics, has been developed to confirm both the phylogenetic and the functional gene repertoire of the gut microbiota (37). However, one of the limitations of metagenomic approaches is that the microbiome data do not provide information on whether or not genes are expressed at any given time. Other omics approaches have been developed to counteract these limitations, including the sequencing of the whole microbial RNA pool of a given sample, i.e., metatranscriptomics, or analysis of the overall protein content or proteome, i.e., metaproteomics. Notably, similar to the case for the metagenomics approach, the usefulness of the latter two technologies is limited by the fact that many genes or their homologs (and thus their products) are not functionally characterized. Finally, assessment of the (microbially) produced metabolites, i.e., metabolomics, will generate an overall signature representing microbial activities.
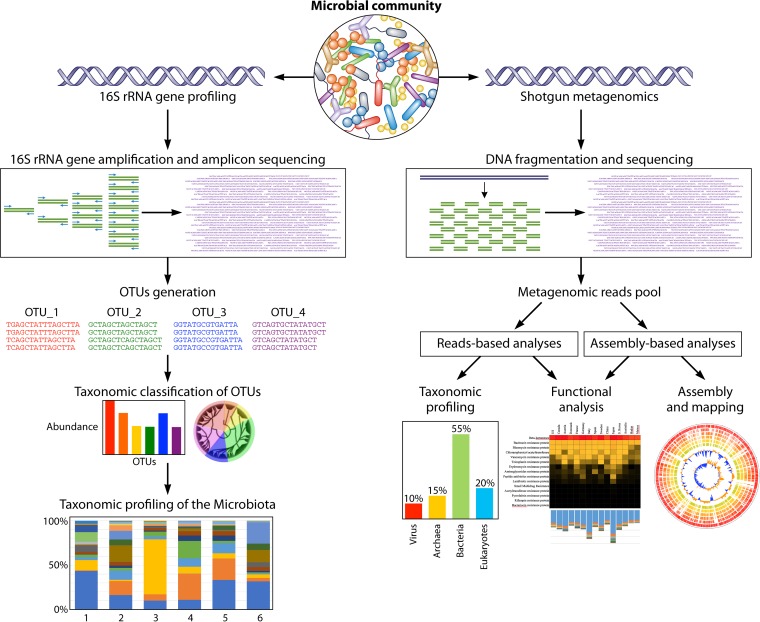
FIG 2.
General overview of the bioinformatic pipelines for the 16S rRNA gene microbial profiling and shotgun metagenomics. Starting from DNA extraction of a microbial community and subsequent sequencing, the pipeline generates taxonomic profiling of the microbiota and the reconstruction of microbial genomes with corresponding functional analyses of the genes.
The Gold Standard Methodology for Microbiota Determination
Many human gut microbiota studies have relied on 16S rRNA gene-based microbial profiling analyses. The 16S rRNA gene encompasses nine different variable regions, i.e., V1 to V9, each flanked by highly conserved DNA sequences that are suitable for PCR primer binding (38). However, no standard approach exists to select the most appropriate PCR primer pair that is equally efficient in amplifying part of the 16S rRNA-encoding gene for all taxa and phylotypes present in biological samples, and very often the decision to employ a particular primer pair is based on historic use, anecdotal evidence, or/and current literature (39–42). In addition, none of the currently available DNA sequencing technologies offers full-length gene sequencing at sufficient depth for cost-effective multiplexing of multiple samples in a single run.
As mentioned above, an alternative to human gut microbiota cataloguing through 16S rRNA gene microbial profiling is shotgun metagenomic sequencing. This approach bypasses gene-specific amplification and potentially sequences all (fragmented) DNA extracted from the analyzed environmental sample, including that from unclassified bacteria and viruses. Shotgun metagenomics provides substantially more information, including insights into functional aspects of the microbial community, than 16S rRNA gene-based microbial profiling. In this regard, it does not suffer from the potential bias of the amplification reaction required for 16S rRNA gene-based profiling. More specifically, shotgun data can be employed to explore the repertoire of genes participating in a wide range of metabolic processes, such as those involved in biosynthesis of compounds, e.g., short-chain fatty acids, or in the catabolism of nutrients, e.g., carbon sources. Functional classification of the shotgun metagenomic reads through the use of customized databases may also provide insights into a plethora of functional aspects of the gut microbiome, such as antibiotic resistance, degradation of conjugated bile salts, presence of (pro)phages, extracellular structures responsible for adhesion, and immunomodulation. Moreover, an assembly-based approach can be exploited to reconstruct complete or partial genomes of so-far-uncultivated taxa, enabling the exploration of what until recently was referred to as microbial dark matter (43).
However, interpretation of the enormous amount of data obtained from DNA sequencing of complex bacterial communities, such as those residing in the gastrointestinal tract (GIT), requires substantial processing power and bioinformatic pipelines for sequence information management, interrogation, and administration (30). Moreover, it should be mentioned that underpopulated reference databases and poor functional characterization of many genes considerably limit the usefulness of the metagenomic approaches employed to investigate the gut microbiota.
Novel NGS-Based, Cutting-Edge Approaches To Achieve a High-Definition Image of the Gut Microbiota Composition
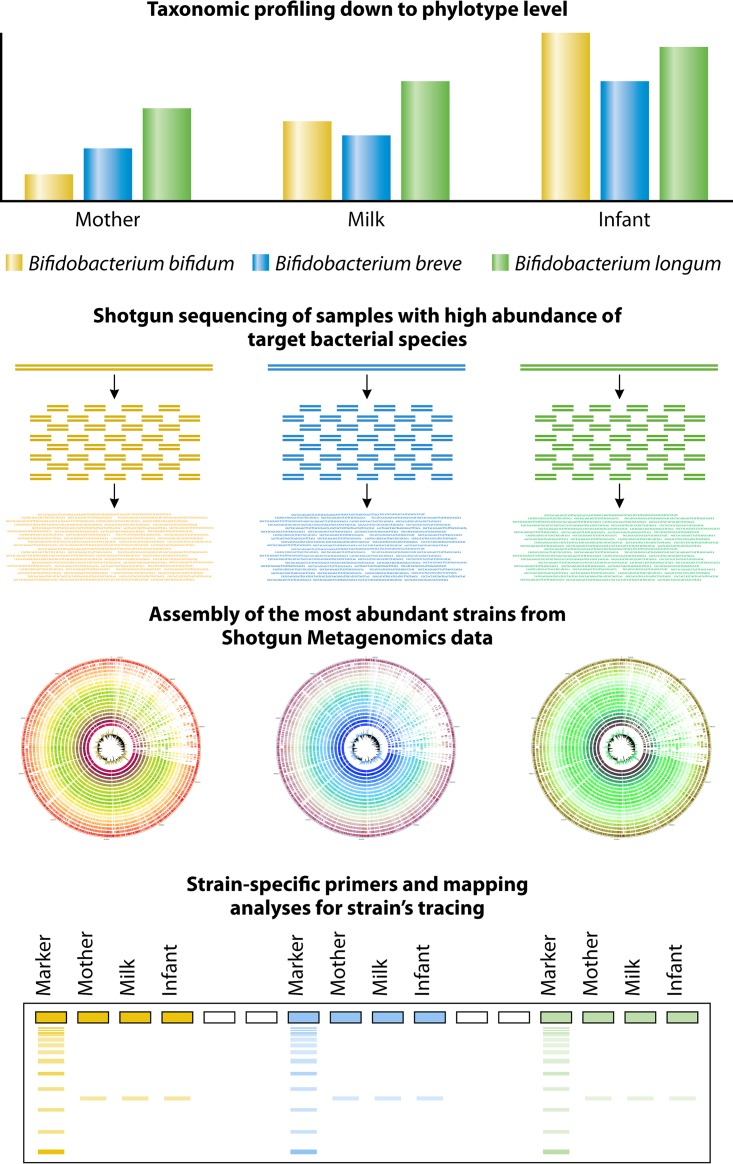
16S rRNA gene-based microbial profiling analyses provide insights into the composition of the human gut microbiota at a taxonomic level that is mostly higher than species level (44). Thus, in order to overcome this limitation and to obtain a more detailed image of the composition of the human gut microbiota, i.e., at the species or even subspecies level, it is necessary to target a molecular marker that is much more variable at the interspecies level than the 16S rRNA gene. The internally transcribed spacer (ITS) sequence, which represents a spacer region between the 16S rRNA and the 23S rRNA genes within the rRNA locus, represents a valuable genetic marker for such a purpose. An ITS-based protocol known as ITS bifidobacterial profiling analysis was applied to achieve a detailed image of bifidobacterial communities (45). This ITS-based approach can differentiate between closely related bifidobacterial taxa at the subspecies level and thus can resolve the bifidobacterial community composition in complex ecosystems, including the human gut (45, 46). In this context, the ITS bifidobacterial profiling approach was shown to identify bifidobacterial strains from the infant microbiota that apparently had been acquired by vertical transmission (from the corresponding mother) (47).
Complete genome analysis of the human gut microbiome implies decoding the complete genome sequence of each constituent strain. The possibility of achieving this goal is very challenging because of the complexity of the gut microbiota, which may include hundreds of operational taxonomic units (OTUs). In addition, the inability to simulate, under in vitro testing, the essential conditions of the ecological niches renders the cultivation of most members of the gut microbiota even more difficult. Single-cell genomics can productively contribute to the genomic characterization of the microbiome. Standard approaches to single-cell analyses involve the physical isolation of the microbial cell, followed by extraction of chromosomal DNA from each cell and amplification of its genomic content (48). Notably, single-cell genome sequences can be obtained directly from crude samples, thus generating reference genome sequences for those gut microorganisms that are recalcitrant to cultivation (49, 50) or that represent rare community members (51). However, the currently available single-cell approaches are still not particularly efficient, while the quality of the attained data and the possibility of contamination may skew output data compared to that obtained by standard genomic methods. Furthermore, single-cell data sets enable the recovery of only about 35% of the genomic data. Single-cell genomics, in particular if this technique can be further improved, is expected to fill important gaps in our understanding of the contents and structure of the human gut microbiome. Nonetheless, despite promising developments of microfluidic technologies for microbial single-cell analysis, actual implementation of this approach remains very challenging.
A recently applied approach to infer the gut microbiota composition at high resolution down to the strain level without performing any isolation and cultivation of bacterial strains involves the reconstruction of a genome sequence of an individual microbiota member from shotgun metagenomic data (52). Such an NGS approach not only provides taxonomic information about strain identity but also provides very useful data related to the genetic makeup of the organism, thereby providing metabolic and evolutionary insights (52).
An interesting tool aimed at determining the composition of the human gut microbiota at high resolution (down to the strain level) is named MetaPhlAn (53). This software relies on read mapping to a precomputed database of strain-specific marker genes generated through comparative analysis of all publicly available bacterial genome sequences. The main criticism of this approach is that only previously sequenced species can be profiled, thus ignoring the presence of as-yet-unknown/uncultured members in the population.
Culturomics Approaches
During the last decade, the above-mentioned culture-independent approaches have been applied mostly in order to dissect the human gut microbiota composition, whereas microbial cultivation techniques have, to a degree, been neglected (54). This has caused a substantial knowledge gap between bacterial species that reside in the human gut but have not yet been cultivated and those that have been isolated and cultivated (54). It has been reported that approximately 56% of gut bacteria detected by NGS approaches have cultured representatives (55, 56). With the advent of so-called culturomic approaches, this gap is being closed. Culturomics employs high-throughput cultivation conditions to investigate the human gut microbiota. Recently, various culturomics studies of human stool samples involved the formulation of complex growth media, which allowed the isolation and cultivation of a considerable number of novel gut microorganisms (57–59).
ESTABLISHMENT AND DEVELOPMENT OF THE INFANT INTESTINAL MICROBIOTA
Microbial Colonization of the Infant Intestine
Colonization of the infant gut represents the de novo assembly of a complex microbial community (19), a process that is influenced by several environmental and host factors (60) (Fig. 3). Until recently this colonization process was thought to begin at birth. However, this dogma of a sterile in utero environment has been challenged. A growing body of scientific evidence has provided indications of bacterial presence in the placenta, umbilical cord, and amniotic fluid in healthy full-term pregnancies (61–63). While these observations suggest that microbial exposure may start before delivery, thereby allowing colonization of the fetus with early pioneers derived from the maternal microbiota, several other studies have put forward arguments against such a possibility of in utero gut colonization (for further details, see elsewhere in this review) (64–66).
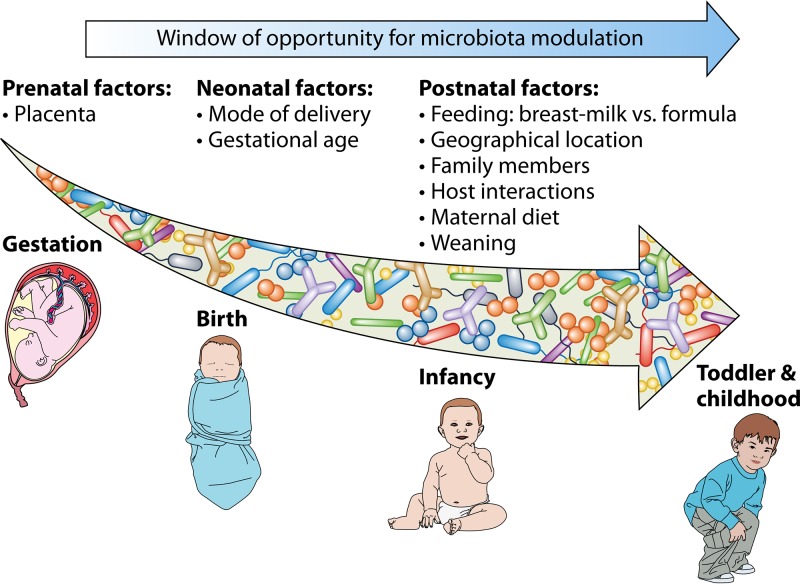
FIG 3.
Window of opportunity for microbiota modulation from gestation to childhood. The schematic representation shows a list of prenatal, neonatal, and postnatal factors that contribute to the bacterial gut composition in infants.
The development and maturation of gut microbiota constitute a dynamic and nonrandom process, in which positive and negative interactions between key microbial taxa take place (67, 68). This process is influenced by various perinatal conditions, such as mode of delivery, type of feeding, and antibiotic usage. Diet, the mother's age and metabolic status, and family genetics and lifestyle have also been reported to impact the infant microbiota, although these are more difficult to determine and quantify in humans. In the following section, we describe how these factors are thought to influence the development of the infant gut microbiota.
Main Drivers of the Microbial Colonization of the Infant Intestine
Mode of delivery.
As indicated above, in full-term infants, the delivery mode is recognized as an important driver of the early gut microbiota composition (69). Vaginally delivered infants come into contact with the maternal vaginal and fecal microbiota, which results in neonatal gut colonization by vagina-associated microbes such as Lactobacillus and Prevotella (70, 71). In contrast, caesarean section (C-section)-delivered infants are not directly exposed to maternal microbes and are thus more likely to become colonized by environmental microorganisms from maternal skin, the hospital staff, or the hospital environment (2, 60, 71–73). Several studies using different culture- and molecular-based methodologies, including the recently employed high-throughput sequencing technologies and metagenomics approaches, have described a deviating gut microbiota in these infants (2, 70, 73). Proteobacteria and Firmicutes were reported to be the main phyla represented during the first days of life, with Actinobacteria appearing in the feces of C-section-delivered babies at day 7 to 15 following birth (74). Infants born by C-section also show a reduced complexity of the gut microbiota and are less often colonized by microorganisms such as Bifidobacterium and Bacteroides, while being more frequently colonized by members of Clostridium sensu stricto (cluster I) and Clostridium difficile (70, 71, 74–79).
These differences between vaginally and C-section-delivered babies gradually decrease, but C-section-delivered infants remain more heterogeneous than vaginally born infants up to 12 months of life (73, 80). Notably, persistent differences in the gut microbiota between C-section- and vaginally delivered children have been detected in children as old as 7 years (77, 78, 81). In contrast, a very recent publication reported no discernible effect of C-section on the early microbiota beyond the immediate neonatal period (82, 83). The observed microbiota differences between vaginally delivered and C-section-delivered babies have been associated with the protective effect of natural birth, particularly since it has been suggested that C-section has long-term health implications. In fact, the levels of various cytokines have been shown to be remarkably reduced in infants born by C-section (76, 84), while C-section delivery has also been associated with an increased risk of immune disorders such as asthma (85), allergy (86), and type 1 diabetes (T1D) (87) and with a higher incidence of obesity (88). Notably, the finding that the mode of delivery impacts the health status throughout adulthood, while the effects on gut microbiota composition decrease after the first years of life, underlines the relevance of early gut microbiota in the maturation and development of the host's immune system.
Gestational age at birth.
Gestational age is another important factor in the establishment of the infant gut microbiota. Neonates are termed preterm when they are born prior to 37 full weeks of gestation (89). Preterm infants may initially, depending on the degree of prematurity, have to overcome serious health challenges. They often present with an immature gut and with immune, respiratory, and neurological issues, while they suffer from exposure to extensive antibiotic and other drug treatments. These neonates usually endure long stays in hospitals, frequently being put on artificial respiration and fed artificially or parenterally. All these factors are likely to interfere with the natural pattern of microbiota acquisition and development, thus resulting in an aberrant establishment or deviating composition of the intestinal microbiota. Several studies have reported differences in the fecal microbiotas of premature and full-term neonates. Preterm neonates exhibit delayed gut colonization with commensal anaerobic microbes, such as Bifidobacterium or Bacteroides, where instead their stools contain significantly higher levels of Enterobacteriaceae, Enterococcus, and other (opportunistic) pathogenic microorganisms than fecal material from full-term neonates (79, 90–94). Gram-positive bacteria, such as Staphylococcus, Enterococcus, and clostridia, dominate the gut microbiota of very premature infants during the first month of life, while Gram-negative microorganisms such as Enterobacteriaceae and Veillonella may be variably present in such cases (95). A pattern of colonization and succession of bacterial classes from Bacilli to Gammaproteobacteria to Clostridia was observed in a very-low-birth-weight (VLBW) premature population (96). In the latter study, the microbiota seemed to evolve with periods of abrupt population changes and with a common endpoint where the premature gut was shown to be colonized by anaerobes, particularly clostridia (96).
Although gestational age has been proposed to be the most important driver of the premature gut microbiota establishment, a huge interindividual variability is observed, likely related to the cooccurrence of a variety of factors cited above. It is important to underline that the aberrancies observed render the preterm infant microbiota more unstable than that of full-term equivalents, and a premature infant microbiota is believed to be associated with a delay in the transition to and establishment of an adult-type signature microbiota (97). These alterations may dramatically affect short- and long-term health. Indeed, the interaction between the altered premature neonatal microbiota and their immature immune system may cause inflammatory responses and facilitate infectious disease (98, 99). In fact, the composition of the intestinal microbiota of the preterm infant has been correlated to an increased risk of necrotizing enterocolitis (NEC) or sepsis (100–102), as discussed below. Moreover, the premature gut microbiota is different not only in composition but also in functionality. The main short-chain fatty acids (SCFAs) produced by the intestinal microbiota were found at lower levels in fecal samples from premature and VLBW infants than in the feces of full-term babies (92, 103). Metabolic pathways potentially affected by prematurity have also been identified by the use of functional inference analyses, with a higher frequency of genes related to xenobiotic biodegradation and metabolism and lipid metabolism and a lower frequency of genes related to energy metabolism and biosynthesis of cofactors and vitamins being present in fecal samples of premature infants than in those of full-term counterparts (103). Premature neonates were found to display an enrichment of bile acid derivatives, showing an altered lipid metabolism (79). Moreover, the metabolomes of urine samples from premature infants were shown to be higher in vitamins D and E (79).
Infant feeding mode.
Feeding type is another major factor determining early microbial colonization and, therefore, influencing the neonatal gut microbiota composition and gastrointestinal function. The differences in the gut microbial composition between breastfed and formula-fed infants are well documented (4, 104), with increased levels of bifidobacteria being present in the former group of infants. Breastfeeding provides a mix of nutrients and promicrobial and antimicrobial agents, which favors the development of a so-called “milk-oriented microbiota.” IgAs obtained from breast milk promote a regulatory and more “tolerogenic” immune system (105). Breast milk also contains human milk oligosaccharides (HMOs), which can selectively shape the growth and function of beneficial microbes (see below).
The gut microbiota of breastfed infants exhibits lower diversity than that of bottle-fed counterparts (106). Transcriptomic analyses of intestinal epithelial cells has shown that the infant feeding type also affects host gene expression, with breastfeeding enhancing transcription of genes that are associated with immunological and metabolic activities (73, 107). Formula-fed infants are exposed to different carbohydrates, bacteria, and (micro)nutrients, causing different microbial colonization patterns of the gut. In this context, several publications have reported that stools of breastfed infants contain higher levels of bifidobacteria and lactobacilli and lower levels of potential pathogens than those of their formula-fed counterparts, with the latter being associated with a more diverse gut microbiota that is dominated by staphylococci, Bacteroides, clostridia, enterococci, enterobacteria, and the genus Atopobium (78, 80, 106, 108–110). As a consequence of these microbiota differences, the levels of SCFAs are also different in the stools of breastfed versus formula-fed infants, with propionate and butyrate being present at higher levels in the latter group (111). Furthermore, it seems that infants fed with formula milk achieve an early divergence toward an adult-like microbiota composition (73).
During the exclusive milk-feeding period, the infant microbiota seems to fluctuate and the bacterial succession phenomenon continues, progressively diversifying until weaning, when it changes toward the adult-like microbiota in becoming more stable and complex (112–114). The impact of the weaning stage on microbiota development has been considerably less investigated than that of the early (exclusively milk) feeding stage (113, 114). During weaning, due to the complementary introduction of a variety of novel food substances and nutrients, the alpha diversity increases, resulting in the replacement of Proteobacteria and Actinobacteria by Firmicutes and Bacteroidetes phyla as the dominant members of the infant microbiota (112, 113). A survey of the gut microbiota development during the complementary feeding period, between the 9th and 18th months following birth, revealed an increase in the relative abundances of some major bacterial families, including Lachnospiraceae, Ruminococcaceae, Eubacteriaceae, Rikenellaceae, or Sutterellaceae (115). In contrast, the relative abundances of Bifidobacteriaceae, Actinomycetaceae, Veillonellaceae, Enterobacteriaceae, Lactobacillaceae, Enterococcaceae, Clostridiales incertae sedis XI, Carnobacteriaceae, and Fusobacteriaceae decreased during the transition from the infant to the toddler stage (115), which is in agreement with previous reports (3, 73, 80, 116, 117). Increased protein intake was shown to be correlated with an increase in Lachnospiraceae and a decrease in saccharolytic bacteria such as members of the Bifidobacteriaceae family, which are generally associated with breast milk and early infant feeding, while ingestion of fiber was demonstrated to be associated with higher levels of Prevotellaceae (115). Interestingly, two species, Faecalibacterium prausnitzii and Akkermansia muciniphila, which are either absent or present at very low levels during early infancy, increase in abundance to adult levels at 12 months and 24 months, respectively (3). In the latter case, the increase may reflect the gradual increase of the production of mucin, which is the main carbohydrate fermented by A. muciniphila and which is present at very low level during early infancy (see below).
The cessation of breastfeeding and the transition to more varied, solid foods is considered to cause an increase in alpha diversity of the infant gut microbiota (73, 115, 118). Moreover, the changeover from human milk to formula (i.e., bovine) milk also strongly influences the development of gut microbiota. Just 5 days after breast milk cessation, an increase in the relative abundances of the Bacteroides, Blautia, and Ruminococcus genera, among others, and a decrease in the relative abundances of Bifidobacterium, Lactobacillus, and enterobacteria have been observed, with an increase in alpha diversity and fecal pH (119). In addition, the observed bacterial diversity increase contributes to functional changes. An increase in total levels of SCFAs, in particular butyrate, has been reported (80, 113). The dietary shift from exclusively milk-based to solid foods induces the development of a mature microbiota with genes responsible for complex carbohydrate, starch, and xenobiotic degradation as well as vitamin production (113). The adult-like microbiota is functionally more complex and is structured to metabolize plant-derived polysaccharides from the adult diet, providing mutual benefits to host and microbe (116).
Maternal diet.
There is growing interest in understanding the effects of maternal body mass index (BMI) on the infant gut microbiota (72). Recently, it has been observed that the infant's fecal microbial composition is influenced by the BMI and weight gain of the mother during pregnancy (120). Overall, fecal Bacteroides and Staphylococcus concentrations were shown to be significantly higher in infants of overweight mothers during the first 6 months of life; on the other hand, bifidobacterial counts were determined to be higher in infants from nonobese mothers. These observations have not, however, been confirmed by other authors (121). Moreover, information regarding the impact of breastfeeding and its correlation with the mother's weight and number of children is currently not available. These parameters may act as confounding factors, thus indicating the need for further investigations.
Environment (family lifestyle and geographical location).
Family members and close relatives (siblings) have also been described as a relevant environmental factor that may influence the pattern of infant gut microbiota colonization (60), but as yet, definitive evidence of the effects of family size, structure, and birth order has yet to be established (72). Infants 1 month of age, who were recruited from the KOALA Birth Cohort Study in The Netherlands, with older siblings were shown to have a higher number of bifidobacteria in their gut microbiota than infants without siblings (78). It was also reported, in this case as part of the ALLERGYFLORA study, that infants without older siblings had increased proportions of non-Escherichia coli enterobacteria as well as clostridia in the gut but also a lower anaerobe-to-facultative anaerobe ratio (75). In a recent study performed with a Danish cohort, the presence of older siblings was shown to be associated with increased gut microbial diversity and richness during early childhood, while the presence of household pets had less-pronounced effects on the gut microbiota (122). The concept of the “sibling effect,” which may contribute to the substantiation of the hygiene hypothesis, remains controversial, and more studies in this area are needed.
Geographical location may also have an impact on the microbiota, as microbiota differences appear to be related to dietary patterns and lifestyle in a specific area (60). Additionally, different ethno-geographic populations have distinct regional diets and cultural practices (123). For example, children living in a rural village in Africa harbor a microbiota different from that of children living in an urban region in Italy (124), while several other studies have investigated the geographical effect, as linked to ethnicity and/or diet, on microbial diversity and composition (4, 112, 125–128). Fecal sample analyses of children living in an urban slum in Bangladesh point to a gut microbiota significantly different from that of children of the same age range in an upper-middle-class suburban community in the United States. In particular, children from these two different geographical locations had a distinct fecal bacterial community composition and structure, with the microbiota of Bangladeshi children being enriched in Prevotella and depleted in Bacteroides compared to that of U.S. children (126). Another analysis comparing southeastern African and northern European infants reported distinctions in bacterial group composition, with the genus Bifidobacterium and the group Bacteroides-Prevotella being present at a higher abundance in African children (125). Overall, it seems that home structure and family settings (rural versus urban) affect colonization of the gut microbiota after birth, although more studies are needed to establish the exact contributing factors.
Host genetics.
There is growing scientific evidence indicating that host genetics influences the acquisition and development of the infant gut microbiota (129–131). In this context, the contribution of host genotype in shaping the microbiota composition and structure has been assessed in human twins and family relatives. In this regard, a study with children younger than 10 years old reported higher levels of microbial similarity in genetically identical twins than in fraternal twins and unrelated controls (132). However, subsequent analyses performed by other authors did not identify significant differences in bacterial diversity between monozygotic and dizygotic twins (133, 134). Remarkably, a recent analysis of a large cohort (1,539 individuals; age range, 18 to 84 years) established a clear association between host genotype and the relative abundances of different bacterial taxonomies in adulthood. In that work, the authors (135) found that single nucleotide polymorphisms (SNPs) located in the LCT locus (responsible for human lactase production) are related to varying abundance of Bifidobacterium and found an association between host genetics and intake of dairy products. This highlights the need for further research on the interaction between human genotype, diet, and microbiota development.
Altogether, the almost infinite combinations of these environmental, family-associated, and genetic factors are responsible for the unique bacterial population harbored by the gut of each individual.
POTENTIAL MATERNAL-FETAL TRANSFER OF MICROBIOTA
Is There Actually a Maternal-Fetal Transfer of Microbiota?
The notion that the human fetal environment is sterile under physiological conditions (the “sterile womb paradigm”) has been an accepted dogma for decades. According to this concept, microbial colonization of the healthy newborn intestinal tract starts during and after birth, by both vertical (from the mother's microbiota) and horizontal transmission. Most studies that established the sterile womb paradigm employed traditional culture-based methods and microscopy, which, despite their limitations (such as their failing to detect viable but noncultivable microbes), are still considered valid today.
In contrast, many recent studies (most of them employing state-of-the-art, cultivation-independent techniques) have challenged this traditional view and have proposed that acquisition of the human microbiota begins in utero (see subsections below). If certain, this notion would change our understanding of gut microbiota acquisition and its role in human development. However, while it is possible that not all healthy babies are born sterile as was previously assumed, it is also true that the data that support the “in utero colonization hypothesis” must be taken with extreme caution, since most of them were obtained with particular methodological limitations (136).
For example, the detected bacterial DNA may belong to dead organisms, rather than viable microorganisms, which have been found in only very few studies in samples that originated from the fetal environment (66). Furthermore, it has been pointed out that the highly sensitive molecular techniques employed to study the low-abundance, fetus-related microbiome tend to detect contaminating microbes, thereby generating false-positive results. Avoiding contamination is nearly impossible when collecting samples related to the in utero environment within a clinical setting. Furthermore, the presence of contaminating DNA in PCR reagents, DNA extraction kits, and molecular biology-grade water (137, 138) is a particularly relevant challenge when working with samples that contain an extremely low (or no) microbial biomass, such as those obtained from the placenta, amniotic fluid, or meconium of healthy subjects. Therefore, if contaminating DNA is present and amplified during the PCR step, this will cause incorrect results and conclusions (139). A low level of (or no) bacterial target DNA in a sample has been shown to correlate with a higher proportion of bacterial sequences being attributable to contamination (64, 65). In this context, it has been reported that only an estimated 0.002 mg of bacterial DNA can be extracted from each one-gram placental tissue (63).
Contaminating DNA sequences typically correspond to water- and soil-associated bacterial genera, including Acinetobacter, Alcaligenes, Bacillus, Bradyrhizobium, Herbaspirillum, Legionella, Leifsonia, Mesorhizobium, Methylobacterium, Microbacterium, Novosphingobium, Pseudomonas, Ralstonia, Sphingomonas, Stenotrophomonas, and Xanthomonas. Interestingly, a high proportion of the taxa considered “the placenta microbiome” (including core members) in a highly cited publication overlaps with the microbial groups indicated above (63). In a recent study, placental samples from healthy deliveries and a matched set of controls (to check for the impact of contaminations) were subjected to 16S rRNA gene sequencing and microbiota analysis, which indicated that the microbiota data obtained from placental samples and controls could not be distinguished (64, 65, 140).
Although the presence of contaminating DNA has been acknowledged in the literature, its possible impact on 16S rRNA gene-based profiling and shotgun metagenomic analyses of samples that typically contain low biomass has not properly been taken into consideration in the currently available infant gut microbiomes (64, 65).
The presence of microorganisms in meconium and amniotic fluid is frequently considered evidence supporting the in utero colonization hypothesis. However, it has been argued that only a relatively small subset of such samples contains detectable microbes, which could be, at least partly, the result of postnatal colonization in the case of meconium samples or of prelabor rupture of membranes in that of amniotic fluid (66, 140, 141).
Finally, gnotobiology has been claimed as the strongest evidence against the existence of microbiomes in the fetal environment because of the ability to derive germfree animals via C-sections and subsequently raise the offspring in a sterile environment (66). This fact has to be taken in account, although on the other hand, it may be challenging to transfer results obtained in a germfree system to those derived from a conventional host (142).
In conclusion, concerns have been raised by the scientific community regarding the “in utero colonization hypothesis” because most of the studies employed molecular approaches that are unsuitable to study “low-biomass” microbial populations, lacked appropriate controls to account for contamination, and/or did not show bacterial viability (64, 66). However, the unambiguous presence of bacteria has occasionally been found in fetus-related samples (143). Although “in utero colonization” skeptics argue that this finding is due to subclinical conditions, it also indicates that fetal colonization may, at least occasionally, occur and that this subject (and its relation to maternal, fetal, and infant health) deserves further research. In addition, it is widely accepted that exposure of the fetal environment to microbial metabolites and compounds (including DNA) from the maternal microbiota may have a major impact on the pregnancy outcome and infant development (144–146). However, research regarding the role of viable bacterial cells or their DNA must be strictly controlled for DNA contamination during sample collection and processing in order to determine which observations are scientifically accurate. Recommendations to reduce the impact of contaminants in sequence-based, low-biomass microbiota studies have already been made (64, 65). The sections below should therefore be read with caution, keeping in mind that this is still a highly controversial area.
The Reproductive Microbiota before Pregnancy
As we learn more about the human microbiota, it appears that its complex interactions with the host occur on most epithelial and mucosal surfaces, even those belonging to organs that in the past were considered sterile under physiological conditions (Fig. 4). Until recently, the concept of a reproductive tract microbiota, playing active roles in health and disease, was limited to the vaginal cavity (147, 148). As an example, bacterial vaginosis, a condition that is characterized by a deviating or so-called dysbiosis state, is the most prevalent vaginal disorder and is associated not only with an increased incidence of intra-amniotic infection, a higher incidence of preterm delivery, and spontaneous abortion (149–152) but also with a reduced ability to conceive (153, 154).
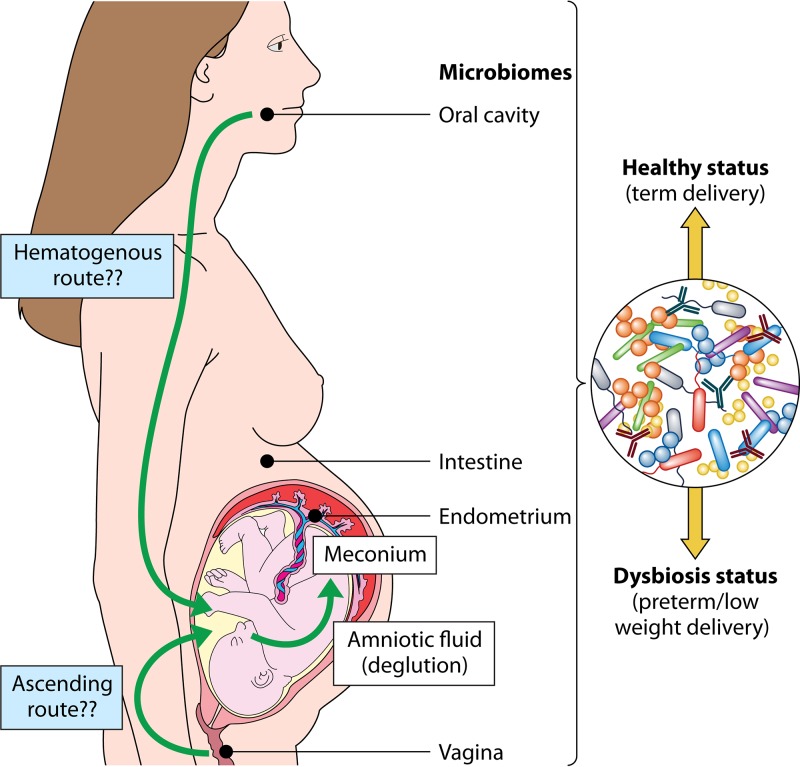
FIG 4.
Colonization routes of maternal microbiomes to the infant. The mother portrayal exhibits the maternal microbiome locations and the related routes that result in the vertical transmission of the microbiota to the infant.
In contrast to the case for the vagina and cervix, the anatomical site of conception and overall fetal environment, which encompass the fallopian tubes, endometrium, placenta, and amniotic fluid, were believed to be sterile (155). In fact, the notion that fetuses are sterile in utero and that microbial colonization of the newborn starts during and after birth had been widely accepted since the beginning of the 20th century. Thus, the search for microorganisms in samples from such environments was pertinent only when there were signs of infections related to adverse obstetric outcomes such as preterm rupture of membranes, chorioamnionitis, miscarriage, and preterm deliveries (62, 148, 156–163).
However, various publications have reported on the presence of a physiological microbiota at every stage and in every location related to human reproduction body compartments, including the reproductive tracts of both females (e.g., ovary, follicle, oocyte, fallopian tube, uterus, cervix, and vagina) and males (e.g., testes, semen/spermatozoa, prostate, and seminal glands), as well as fetal structures such as the placenta and umbilical cord (155, 164, 165). The understanding that these bacteria form their own biofilms in the human reproductive tracts, allowing complex interactions with the gametes, embryo, or fetus and with the maternal tissue interface, may provide new insights in the field of fertility and lead to advances in assisted reproductive technology (165).
The Endometrial Microbiome
As stated above, the uterus has traditionally been considered sterile in the absence of infection (166). In 1989, Hemsell isolated up to 231 bacterial species from 49 out of 55 endometrial samples collected from asymptomatic women without a history of previous uterine infection, providing the first deviating view on this (until then) uncontroversial subject (533). However, the number of subsequent studies dealing with the uterine microbiota in healthy women has remained very small, even after the availability of culture-independent techniques.
Recently, the microbiota composition of endometrial tissue and mucus samples from 19 nonpregnant women scheduled for hysteroscopy, yet without uterine anomalies, has been analyzed by 16S rRNA gene microbial profiling (167). Notably, such analyses highlighted the occurrence of bacteria in all samples, thus supporting the notion of a natural microbiota in these body compartments (167).
Furthermore, the endometrial microbiota in women undergoing single-embryo transfer was characterized (168) by 16S rRNA gene microbial profiling. Remarkably, microbial taxonomy assignments were carried out in samples from 33 patients, of whom 18 became pregnant and 15 did not. Several dominant bacterial genera, such as Flavobacterium and Lactobacillus, were present in both patient groups (women with or without ongoing pregnancy), while others appeared to vary by outcome. However, the differences in the relative abundances of these taxa between the patient groups did not reach statistical significance.
Recently, data from another study have reinforced the hypothesis that the composition of the endometrial microbiota influences the rate of success of implantation (169). Notably, the results of this work, involving patients undergoing in vitro fertilization, showed that the bacterial communities identified in the collected endometrial fluid and vaginal aspirate samples are distinct. Interestingly, the microbiota composition in the endometrial fluid could roughly be classified as “Lactobacillus dominated” or “non-Lactobacillus dominated,” where the presence of a non-Lactobacillus-dominated endometrial microbiota was associated with significant decreases in implantation, pregnancy, ongoing pregnancy, and live birth rates (169).
It is probable that the uterine microbiota influences the immune environment during conception, since it has been reported that the cytokines involved in endometrial receptivity and embryo development are affected by infection (170). An altered microbial consortia in endometrial fluid may trigger an inflammatory response in the endometrium compromising the success of embryo implantation, since a tight regulation of inflammatory mediators is required during the adhesion of the blastocyst to the endometrial wall (171).
Other recently published studies using culture-dependent and -independent techniques not only have reported the presence of live bacteria and bacterial DNA, respectively, in endometrial samples (172) but also have indicated that dysbiosis in the uterine microbiota may be related to a variety of adverse gynecological or obstetric outcomes, including endometritis, endometriosis (173), endometrial polyps (174), and endometrial cancer (175).
The process of conception and implantation is very complex, and recent findings suggest that reproductive success is defined not exclusively by endometrial histology and eukaryotic gene expression but also by the contribution of the microbiota residing in the reproductive tract (176).
Microbiotas of the Placenta and Meconium
Relatively few analyses have examined the uterine microbiota associated with healthy, full-term pregnancies, partly because of the enduring influence of the sterile womb paradigm but also because of technical and ethical issues that make it hard to obtain representative samples from healthy pregnancies before birth (89). However, the presence of bacteria in amniotic fluid was first reported in 1927 in samples collected during C-sections (161). Later, a culture-based study was able to isolate bacteria from 21% of placentas after noninfected, full-term deliveries (177). During the last decade, a more overt challenging of the in utero sterility dogma has led to an increasing number of reports describing the presence of bacteria or bacterial DNA in a healthy placental environment (161, 178, 179).
With the isolation of commensal bacteria in meconium samples from healthy neonates born by either vaginal delivery or caesarean section, the presumption of sterility of in utero fetuses has been challenged (180). Such findings suggest that full-term fetuses are not completely sterile and that a mother-to-fetus efflux of commensal bacteria mediated by the placenta may occur. Bacteria belonging to Enterococcus faecium, Propionibacterium acnes, Staphylococcus epidermidis and Streptococcus sanguinis were also isolated from umbilical cord blood of healthy neonates born by caesarean section (61). Bacterial counts ranged from 30 to 300 CFU/ml after an enrichment step, suggesting that the initial bacterial numbers in such samples must be extremely low. The identified species associated with the umbilical cord are naturally present in infants immediately after birth (181, 182), being generally regarded as commensals in healthy infant hosts.
In situ hybridization using a fluorescent probe targeting a highly conserved region of the 16S rRNA gene allowed the detection of bacteria in most fetal membrane samples (>73%) after term delivery (183).
A careful microbiological investigation indicated the occurrence of intracellular bacteria of diverse morphologies in the maternal basal plate in 27% of 195 sampled placentas, although no results beyond Gram classification and cell morphology were obtained in that work (143). No differences were observed between placental basal plates from preterm or term gestations, and intracellular bacteria were detected in placentas without clinical or pathological chorioamnionitis. These results were corroborated by a culture-based study, which identified bacteria in 16.4% of placentas from noninfected pregnancies (184).
Recently, application of whole-genome shotgun metagenomics to placental specimens collected under sterile conditions from 320 subjects suggested that the placenta harbors a low-abundance yet metabolically rich site-specific microbiome, composed largely of nonpathogenic commensals that belong to the Firmicutes, Tenericutes, Proteobacteria, Bacteroidetes, and Fusobacteria phyla (63). At the species level, Escherichia coli appears to dominate placental bacterial communities. Notably, the microbial profiles that have been found in the placenta and their associated genomic composition revealed intriguing similarities with those identified in the oral environment (185). Furthermore, the dominance of E. coli sequences suggests a direct or indirect connection between the placental microbiota and that of the maternal gut, where this species is a common and abundant microbial resident. Altogether, these data indicate that the entire maternal digestive tract, i.e., from the oral cavity to distal colon, plays a key role in placental colonization.
It has been found that the placental microbiota varies depending on the birth weight of full-term neonates, where the relative abundance of Lactobacillus sequences appears to be negatively associated with birth weight (186). Interestingly, the taxonomic profiles that seem to be associated with either term or preterm pregnancies are accompanied by changes in bacterium-encoded metabolic pathways, which are independent of delivery mode (185). It is well known that metabolic syndrome and obesity are linked with a state of inflammation and dysbiosis. In fact, obesity during pregnancy has been associated with macrophage accumulation and inflammation in the placenta (187–189) and also with preterm delivery (190–192). Recently, it has been shown that the placental microbiota varied among 320 women with spontaneous preterm birth depending on their excess gestational weight gain but not on obesity (193). Excess gestational weight gain was shown to be associated not only with significant changes in the placental microbiota (including decreased species richness) but also with alterations in the associated microbiome-encoded metabolic pathways (193).
Studies investigating nonbacterial components of the human reproductive microbiota are urgently required since they may play relevant, yet still unknown, roles in supporting or preventing successful and healthy reproduction upon interaction with bacteria and host cells. For example, it has been suggested that commensal bacteria that are present in or on the mucus layer covering the uterine epithelium promote induction of regulatory factors by trophoblast and decidual macrophages. In turn, macrophages would secrete antimicrobial products to control commensal overgrowth and prevent invasion by pathogenic bacteria. Recognition of bacterial products by trophoblasts enhances expression of anti-inflammatory factors, expands T regulatory cells, and promotes tolerance (194). However, viral infection at the implantation site inhibits the capacity of macrophages to control bacterial growth, thereby disturbing the symbiosis among microbiota, trophoblast, and immune cells at the implantation site and leading to an inflammatory condition responsible for preterm birth (195). In a murine model, infection of the placenta with murine herpesvirus 68 elicited a fetal inflammatory response and sensitized the mother to bacterial endotoxin, which in turn induced preterm labor (196). Consequently, it has recently been postulated that when trophoblast cells interact normally with commensal bacteria (194), their durable homeostatic relationship contributes to a fine regulatory tuning of the maternal-fetal interface. However, disturbances in this relationship may be the basis for an inflammatory state, which generally characterizes preterm birth and other adverse pregnancy outcomes.
Microbiotas of Amniotic Fluid and Meconium
Amniotic fluid surrounds and is continuously swallowed by fetuses. There are several findings supporting the lack of microbiological sterility of umbilical cord blood, amniotic fluid, or fetal membranes in human beings without any clinical or histological evidence of infection or inflammation (61, 197). These data are further corroborated by the isolation of viable bacteria in the first meconium belonging to the same or similar species previously isolated from umbilical cord blood (198). A recent 16S rRNA gene microbial profiling study reported that within the same mother-infant pair, the bacterial communities in meconium samples are very similar to those in the mother's placenta, regardless of the method of delivery, and that both differ from those found in the maternal vagina (199). From these pioneering studies, it is now generally accepted that the meconium harbors a complex microbial community and, similar to the case for the placenta microbiota, various studies have investigated the microbial diversity of meconium (141, 200–204).
Assessment of the microbiota composition of the first-secreted meconium from 15 healthy term infants following vaginal delivery indicated that about 66% of the infants carried viable bacteria in their meconium (141). Each of the neonates enrolled in this analysis contained between one and five microbial groups, with Bifidobacterium, Enterobacteriaceae, Enterococcaceae, and Bacteroides-Prevotella being the most prevalent. This analysis thus indicates that low numbers of bacteria are present in first-pass meconium samples from healthy, vaginally delivered, breastfed, term infants. Several studies have also detected DNAs from different bacteria in the meconium of healthy neonates (74, 200, 202, 204–206), further supporting the notion that gut microbial colonization begins before birth.
So far, there is only fragmentary information available regarding the influence of other maternal or infant health conditions on the bacterial communities in meconium. In this context, there is experimental evidence indicating that the overall bacterial content in meconium significantly differs depending on maternal health status (202). Specifically, the phylum Bacteroidetes and the genus Parabacteroides were enriched in the meconium of infants from mothers affected by diabetes, while there was a higher Proteobacteria abundance in the meconium of infants from nondiabetic mothers. It has also been suggested that meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants (200).
Overall, data from various independent laboratories indicate that the microbial taxa found in meconium samples are uniquely distinct from those found in subsequent fecal samples, irrespective of gestational age (201, 203, 207).
Origin of the Pregnancy-Related Microbiome
Several routes have been proposed to explain how bacteria are able to colonize the uterine cavity during pregnancy, including a retrograde pathway through the abdominal cavity or invasive procedures such as amniocentesis. Several possible routes have been put forward in the context of intrauterine infections and adverse pregnancy outcomes (159). In relation to healthy pregnancies, there are two main pathways that are currently being considered (179): (i) vertical ascension from the vagina and/or urinary tract and (ii) a hematogenous route through the placenta after translocation from the digestive tract (oral cavity and gut). However, the fact that some bacterial species (e.g., Lactobacillus salivarius, Streptococcus agalactiae, Streptococcus mitis, Enterococcus faecalis, or E. coli) can be found in more than one ecological niche within the same female has made it difficult to elucidate the origin of the bacteria that colonize the uterine environment.
Early investigations suggested that the vagina is the origin of pathogenic bacteria that ultimately reach the placenta and fetus through translocation across the choriodecidual plate (159, 160). This process is believed to start during the second trimester of pregnancy, although the actual timing is still unknown. The low frequency of detection of lactobacillus DNA in samples of meconium from infants born by C-section (204) suggests that the primary source of lactobacilli in the infant gut is mainly from the maternal vaginal and rectal microbiota during vaginal delivery, and this may explain, at least partly, the differences observed in the infant fecal microbiota depending on the delivery mode (70). In contrast to the case for placental samples, the finding of (high levels of) lactobacillus DNA in endometrial samples from nonpregnant women may reflect the technical difficulty, or even impossibility, in retrieving samples free of vaginal contamination when sampling devices are introduced through the vagina.
Although long-lasting paradigms in the context of preterm birth suggests that most intrauterine bacteria originate in the lower genital tract and ascend into an otherwise sterile intrauterine environment, many bacteria isolated or detected in the placenta with culture-dependent or independent techniques are not found in the urogenital tract but rather represent commensal species common to the digestive tract (179, 208). The alternative mechanism that may explain early colonization of the fetus represents hematogenously derived sources such as the maternal mouth (161, 209) and the maternal intestinal tract due to higher intercellular junctional permeability and/or dendritic cell transport (179).
The mechanisms by which digestive bacteria may translocate and reach this human niche are poorly understood. While the digestive epithelial barrier generally prevents microbial entry into the circulatory system, dendritic cells can actively penetrate the digestive tract epithelium, take up bacteria from the lumen, and transport live bacteria throughout the body as they migrate to lymphoid organs (210). To test whether maternal gut bacteria can be provisioned to fetuses in utero, two pioneering studies investigated whether oral administration of genetically labeled Enterococcus faecium to pregnant mice resulted in its presence in the amniotic fluid and meconium of term offspring after sterile C-section (61, 198). Remarkably, E. faecium with the genetic label was cultured from the amniotic fluid and meconium of pups from inoculated mothers but not from pups of control mice. In addition, other murine studies have reported not only significant similarities between the oral and placental microbial communities but also transmission of diverse oral bacteria to murine placenta, which further suggests that the placental microbiome may be established, at least partly, by hematogenous spread (211–215). In this context, a previous study carried out in pregnant women and focusing on the impact of oral microbiota composition on pregnancy outcome showed that certain bacteria, such as Actinomyces naeslundii, are associated with lower birth weight and earlier delivery, while others, such as lactobacilli, are positively correlated with a higher birth weight and later delivery date (216).
Many transient anatomical and physiological changes occur during pregnancy, thereby providing a suitable framework for the development of the fetus first and the neonate later. These changes affect virtually all systems, including the cardiovascular, respiratory, genitourinary, and digestive tracts. Interestingly, such adaptations may favor an increased bacterial translocation during late pregnancy and lactation (210, 217). Globally, this offers the possibility that modulation of the oral and gut microbiome during (pre)pregnancy may impact the pregnancy outcome and fetal and infant health.
HUMAN MILK OLIGOSACCHARIDES CONTRIBUTE TO SHAPING MICROBIAL COMMUNITIES
General Features
Human milk is a rich source of components that contribute to shaping the infant gut microbiota through a variety of mechanisms. After lactose and lipids, oligosaccharides are the third most abundant component of human milk. One liter of mature human milk contains 5 to 20 g of these complex sugars, which often exceeds the concentration of all human milk proteins combined. Oligosaccharide concentrations in colostrum, an early form of milk that is secreted in late pregnancy and shortly after delivery, are even higher.
Human Milk Oligosaccharides Are a Diverse Group of Complex Glycans
Human milk oligosaccharides (HMOs) consist of the five monosaccharide building blocks glucose (Glc), Galactose (Gal), N-acetylglucosamine (GlcNAc), fucose (Fuc), and the sialic acid N-acetyl-neuraminic acid (Neu5Ac). The combination of these building blocks in defined glycosidic linkages yields several dozen (up to more than 100) structurally distinct HMOs. All HMOs carry lactose (Galβ1-4Glc) at the reducing end. Lactose can be further elongated by the addition of β1-3- or β1-6-linked lacto-N-biose (Galβ1-3GlcNAc-, type 1 chain) or N-acetyllactosamine (Galβ1-4GlcNAc-, type 2 chain). Elongation with lacto-N-biose appears to terminate the chain, whereas N-acetyllactosamine can be further extended by the addition of one of the two disaccharides. A β1-6 linkage between two disaccharide units introduces chain branching. Lactose or the elongated and branched oligosaccharide chain can be fucosylated (addition of Fuc) in α1-2, α1-3, or α1-4 linkages and/or sialylated (addition of sialic acid Neu5Ac) in α2-3 or α2-6 linkages. For example, fucosylation of the terminal Gal in lactose in an α1-2 linkage yields 2′-fucosyllactose (2′FL). Sialylation of the terminal Gal in lactose in an α2-6 linkage yields 6′-sialyllactose (6′SL). Addition of two sialic acids to the tetrasaccharide lacto-N-tetraose (Galβ1-3GlcNAcβ1-3Galβ1-4Glc), one on the terminal Gal in an α2-3 linkage and one on the subterminal GlcNAc in an α2-6 linkage, yields an HMO called disialyllacto-N-tetraose (DSLNT). HMOs can carry fucose, sialic acid, both, or neither. Thus, structural diversity derives from both the complexity of the underlying backbone (chain elongation and branching) as well as modification (fucosylation and sialylation).
HMO Composition Varies between Women
The molecular structures of more than 100 different HMOs have been characterized, but it is important to note that total amount and composition are highly variable between different women. In other words, not every infant who receives human milk is exposed to the same set of HMOs with respect to total amount and structural composition. In fact, a recent cross-sectional, observational study revealed that the HMO composition produced by healthy women varies geographically (218). However, maternal genetic and environmental factors that determine HMO composition are not well understood. HMO fucosylation corresponds to the mother's secretor (Se) and Lewis (Le) blood group characteristics, which are determined by two genetic loci encoding the α1-2-fucosyltransferase FUT2 (encoded by the Se gene) and the α1-3/4-fucosyltransferase FUT3 (encoded by the Le gene) (219–225). Individuals with an active Se locus are classified as secretors. Milk of secretor women is abundant in 2′FL, lacto-N-fucopentaose 1 (LNFP I), and other α1-2-fucosylated HMOs. In contrast, nonsecretors lack a functional FUT2 enzyme, and their milk contains very low concentrations of α1-2-fucosylated HMOs. Individuals with an active Le locus are classified as Le positive. They express FUT3, which transfers Fuc in α1-4 linkage to subterminal GlcNAc on type 1 chains (226). In contrast, the milk of Le-negative women contains very low concentrations of these specific α1-4-fucosylated HMOs, e.g., LNFP II. Based on the combination of active or inactive FUT2 and FUT3 enzymes, HMO profiles can be roughly separated into four different groups: (i) Lewis-positive secretors (with active FUT2 and active FUT3), (ii) Lewis-negative secretors (with active FUT2 and inactive FUT3), (iii) Lewis-positive nonsecretors (with inactive FUT2 and active FUT3), and (iv) Lewis-negative nonsecretors (with inactive FUT2 and inactive FUT3). While FUT2- and FUT3-dependent fucosylation is almost an all-or-nothing phenomenon (the respective HMOs are either present or absent), differential expression of genes that encode other components of the cellular glycosylation machinery likely contributes to some of the more subtle variations in HMO composition between women as well as slight changes over the course of lactation. However, the influence of environmental exposures, such as maternal diet, exercise, and medical or recreational drugs, on HMO composition is currently unknown.
Once ingested, HMOs resist the low pH in the infant's stomach as well as digestion by pancreatic and brush border enzymes. HMOs are not degraded by the infant and thus reach the distal small intestine and colon in an intact form, where they are available to help shape microbial communities and host-microbe interactions.
HMOs Are Human Milk Prebiotics
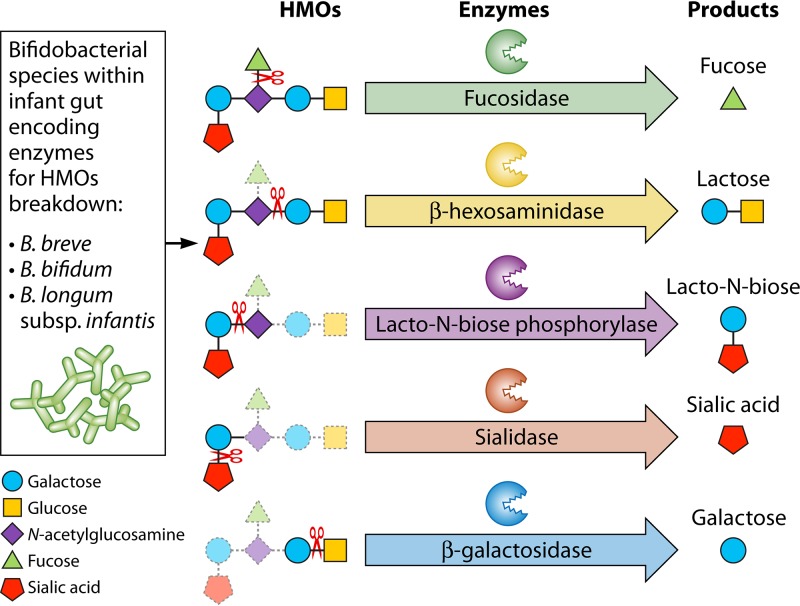
HMOs are considered natural prebiotic compounds because they actively stimulate the growth of specific members of the infant gut microbiota. In these terms, HMOs are often considered “bifidogenic,” since they specifically enhance growth of bifidobacteria, although it should be noted that only certain bifidobacterial taxa efficiently use HMOs as a sole carbon source (227–230). HMO utilization is conserved within the Bifidobacterium longum subsp. infantis lineage (231). Bifidobacteria that are associated with an adult microbiota, such as Bifidobacterium adolescentis, are unable to use the HMO core structures (lacto-N-tetraose [LNT] or lacto-N-neo-tetraose [LNnT]). Thus, it is important to note that the “bifidogenic” effect of HMOs is rather specific and favors B. longum subsp infantis, and in part a few other infant-associated bifidobacteria, but not all bifidobacteria alike (see below).
Other bacteria may also be able to utilize HMOs, in least in part, and thus HMO may have not only specific “bifidogenic” effects but prebiotic effects in general. It is important to note that the prebiotic effects of HMOs are likely structure specific, and HMOs may not always be fully interchangeable. For example, bacterium A may have a fucosidase to cleave fucose from the underlying HMO backbone, while bacterium B may not. Instead, bacterium B may produce sialidases that remove sialic acid. A diet rich in fucosylated HMOs would favor growth of bacterium A, while sialylated HMOs favor bacterium B. These fucosidases and sialidases are often structure specific in a way that 3′SL can be cleaved but 6′SL cannot or vice versa. The same is true for the underlying HMO backbone. Some bacteria can metabolize type 1 structures (terminal Galβ1-3GlcNAc); other bacteria prefer type 2 structures (Galβ1-4GlcNAc). Some bacteria may be able to metabolize branched HMOs, while other bacteria cannot metabolically access such structures.
Microbial Communities May Act in Concert To Fully Utilize HMOs
Only very few bacteria express the entire HMO-degrading machinery (e.g., B. longum subsp. infantis ATCC 15697) (232), while other bacteria can only cleave and metabolize specific elements of a complex HMO molecule (Fig. 5). However, microbial communities might be able to act in concert, sequentially degrade, and metabolize complex HMO structures in a team effort through cross-feeding activities (see below). A first set of bacteria may be able to cleave α1-2 linked fucose and expose an underlying HMO structure to the breakdown activities of other bacteria. In turn, a different set of bacteria that was unable to remove the terminal fucose now would be able to utilize the remaining HMO backbone. It is important to emphasize the structure specificity of prebiotics for several reasons. First, oligosaccharides that are structurally different from HMOs, mostly galacto-oligosaccharides (GOS) and fructo-oligosaccharides (FOS), were added to infant formula years ago in an attempt to introduce complex sugars that in part mimic the prebiotic effects of HMOs. GOS represent galactose oligomers, and FOS are fructose oligomers, not lacto-N-biose or N-acetyllactosamine oligomers like HMOs, and neither GOS nor FOS are fucosylated or sialylated. In fact, fructose itself is not part of human milk. It is easy to imagine that entirely different structures drive the enrichment of different microbial communities. Second, feeding one or two different specific HMOs such as 2′FL or LNnT instead of a mixture of dozens of structurally distinct HMOs is also likely to enrich different microbial communities, namely, those that can utilize 2′FL and LNnT. Other bacteria that specialize in sialic acid degradation would not find useful substrates in these specific HMOs. As a result, a balanced and diverse microbial community may suffer from overgrowth of a limited number of bacteria that thrive on these specific HMOs, while other bacteria are left at a disadvantage. Third and finally, since HMO composition varies between women, milk from different women has different effects on infant microbial communities.
FIG 5.
Chemical structures of human milk oligosaccharides and related enzymatic degradation. On the left are listed the bifidobacterial species that encode enzymes for HMO breakdown retrieved into the infant gut, while on the right are reported the products of the enzymatic reactions.
HMOs and Antimicrobial Effects
As described above, HMOs may exploit prebiotic features toward certain bacterial groups, while they may have opposite effects on others. For example, group B Streptococcus (GBS) stops growing in the presence of HMOs (233). This bacteriostatic effect seems to be linked to specific HMO structures that disrupt proper bacterial membrane glycosylation (233). In contrast, HMOs do not directly impact growth of Candida albicans yet alter hyphal morphology and length, which impacts the yeast's attachment to epithelial cells (234). Disseminated candidiasis is a frequent life-threatening infection in premature infants, with rates as high as 23% in those born at extremely low birth weights (<1,000 g) (235).
HMOs and Antiadhesive Properties
In addition to their prebiotic and antimicrobial effects, HMOs contribute to shaping the infant gut microbiota through various other mechanisms as well. Pathogens often need to attach to epithelial surfaces to be able to colonize and in some cases invade the host to cause disease. Pathogen attachment is often facilitated by protein-glycan interactions either when pathogens are covered by glycans that bind to proteins on epithelial cells or when pathogens express specific proteins that bind to glycans on the host cell surface (glycocalyx). HMOs resemble some of these glycans and serve as soluble analogs that can block pathogen attachment. For example, in vitro studies in tissue culture models revealed that Campylobacter jejuni binds to α1-2-fucosylated glycans and that 2′-fucosyllactose (2′FL) might be able to compete with cell surface glycans to block C. jejuni attachment. Accordingly, feeding α1-2-fucosylated HMOs reduces C. jejuni colonization in mice (236). Infants that receive human milk rich in α1-2-fucosylated HMOs are less likely to develop C. jejuni-associated diarrhea (237). Similar antiadhesive effects have been shown for enteropathogenic E. coli (EPEC) both in tissue culture and in mice (238). These effects are not limited to bacteria, as HMOs also block adhesion of Entamoeba histolytica, a protozoan parasite that infects 50 million people worldwide and causes amebiasis, which is responsible for over 100,000 deaths annually (239).
HMOs May Affect Microbial Communities in Niches Other than the Infant Gut
As mentioned above, there are multiple different mechanisms by which HMOs help to shape microbial communities in the infant gut: HMOs are prebiotics as well as antimicrobials that directly affect growth of specific bacteria, while they also act as antiadhesives that block the attachment of certain bacteria to epithelial cell surfaces, with potential consequences for colonization and invasion. In addition to influencing microbial communities and host-microbe interactions in the infant gut, HMOs may already have an effect on microorganisms in the oral cavity and upper respiratory tract, potentially even the skin, at least around the mouth. Furthermore, HMOs are absorbed in the infant gut, reach the systemic circulation, and are eventually excreted in an intact form with the infant's urine. Although the concentrations of HMOs in the urinary tract are much lower than those in the infant gut, it is possible that HMOs also impact microbial communities in the infant's urinary and genital tracts, with potential effects on health outcomes, e.g., reducing urinary tract infections (240).
The microbiota-shaping effects of HMOs might occur even earlier, before they reach the infant. Human milk itself is not sterile and contains bacterial communities that live within the milk matrix while still in the mammary gland, waiting to be expressed (241). Recent studies revealed that Firmicutes and Proteobacteria are the dominant bacterial phyla found in human milk samples, with Lactobacillus, Streptococcus, Pseudomonas, and Staphylococcus being the most abundant genera (242–245). In addition, the occurrence of bifidobacteria has very recently been demonstrated in human milk samples as well as in the milk of other mammalian species (246). Moreover, no statistically significant difference was observed between bacterial communities harbored by the milk of women who delivered at term or preterm, delivered vaginally or by caesarean section (C-section), or gave birth to boys or girls (242). Nevertheless, it must be underlined that the low bacterial abundance that is typical of human milk samples may lead to biases caused by bacterial contaminants (65). Milk microbiota and HMOs colocalize in the mammary gland between feeding or pumping episodes, potentially influencing each other through mechanisms similar to those that have been described for the infant gut. However, HMOs may act on microbial communities even earlier than that, potentially even before the baby is born.
The Use of Individual HMOs Alone May Bear Risks
Individual HMOs such as 2′FL or LNnT are now available on large scales to be used in a wide range of products with the goal to treat or even prevent diseases that are associated with dysbiosis. Future research needs to investigate whether or not the use of individual HMOs is harmful when they are used alone and not in concert with dozens and potentially hundreds of other oligosaccharides as they naturally occur in human milk.
It is important to emphasize that HMO composition varies between women. Thus, infants receive a distinct set of HMOs with their mother's milk, and the composition changes over the course of lactation. In that sense, human milk can be regarded as “personalized nutrition” and HMOs as “personalized prebiotics” that help shape a distinct infant gut microbiome dependent on what HMOs are present in the mother's milk. Every mother provides a distinct HMO mix to her infant(s), shaping distinct infant microbial communities with potential short- and long-term consequences.
Oligosaccharides in the Milk of Other Mammals
Oligosaccharides in the milk of many other mammals have been studied, but no other animal matches the large amount and high structural diversity of HMOs (reviewed in reference 247). Oligosaccharide concentrations in milk of most farm animals, including cows, goats, sheep, and pigs, are 100- to 1,000-fold lower than that in human milk, with a lower number of different oligosaccharides and a higher abundance of sialylated and a lower abundance of fucosylated oligosaccharides (248–252). Comparative analyses have shown that oligosaccharides in the milk of primates, including humans, are more complex and exhibit greater diversity than those in nonprimate milk (253, 254). In humans, 50 to 80% of the oligosaccharides are fucosylated, depending on the Se/Le group, which is followed by those in chimpanzees, at around 50%, and gorillas, with only 15%. Most other species, including cows, show very low levels of fucosylation (<1%). In humans, 10 to 30% of the oligosaccharides are sialylated, and similar values are found in chimpanzees, rhesus macaques, and gorillas. Interestingly, primate milk oligosaccharide cluster analysis does not follow primate phylogeny, suggesting an independent emergence of milk oligosaccharides, potentially driven by distinct pathogen exposures (254).
TRANSFER OF BACTERIA FROM MOTHER TO CHILD
Maternal Inheritance of Bacteria
According to the holobiont concept when applied to reproduction, not only is the eukaryotic body reproduced, but so is its symbiotic microorganisms, or at least part of them. Thus, birth may be viewed as the passage from one set of symbiotic relationships to another (255). According to this view, the two main partners are the mother and the conceptus (fetus and then child), with a third player in this symbiotic arrangement being represented by the mother's microbiota. The mother is a holobiont, whose associated microorganisms are actively metabolizing nutrients, while the maternal blood which the fetus receives may also be substantially modified by metabolic activities of the mother's microbiota (256, 257). Indeed, it has been shown, for example, that in mice almost all blood-derived serotonin is produced by symbiotic bacteria (256). Consequently, we can argue that the fetus is not free of the mother's symbiotic associations, even assuming the concept of sterility of the fetus.
Upon birth, the infant is moving from one set of symbiotic assemblies to another, transitioning from the mother's microbiota to its own (255). According to the commonly accepted scientific literature on this topic (although that is being challenged [see above]), the fetus is developing under sterile conditions in the amnion, and the microbial colonization occurs only when the amnion breaks and the conceptus moves through the birth canal (258). Then, according to this assumption, the fetus passes through the cervix and the vagina, and the microbiotas of these two body sites may colonize the newborn. Interestingly, based on the hologenome concept, the microbiome can be viewed as an additional set of inherited genes, (some of) which, together with the parental genes, are passed on from one generation to the following (259).
Such inheritance can be vertical in the case of a direct transmission of microbial genes from mother to offspring or horizontal in the case of acquisition of microbial genes from the environment. The mammalian newborn does not just leave the uterus and passively acquire a new microbiota, but the mother seems to actively transfer certain members of the microbiota to her offspring and may thus directly influence (e.g., by the selective nourishment with prebiotic milk compounds certain microorganisms such as bifidobacteria [see above]) the development of the new microbiota of her child.
From a bacterial perspective, a newborn baby represents an essentially uninhabited island, where the first colonizers are provided with a choice of settling options, thereby creating opportunities or restrictions for the next set of microbial colonizers (19). As discussed in other sections of this review, elements of the gut and vaginal microbiotas of the mother initiate new host-symbiont relationships, which are considered to be pivotal for the health of the new holobiont (260).
Vertically Transmitted Microorganisms
Recently, investigators have tried to answer how the infant gut microbiota is assembled soon after birth using metagenomic approaches. In this context, it has been reported that Lactobacillus rhamnosus LGG, which had previously been supplemented to their mothers, was detected by quantitative PCR (qPCR) in stool samples from infants (261). Notably, half of the children with detected LGG shared the same strain with their corresponding mothers and appeared to be heavily colonized by LGG at the very early stages of life (before 3 months of age) (261). However, the observed colonization with strain LGG seems to be transient, since no noticeable microbiota differences between probiotic and placebo groups were identified at 1 year of age.
Recently, a comparative analysis of fecal microbiota data obtained by 16S rRNA gene microbial profiling and involving 415 mothers and their children highlighted a highly shared microbial population for each mother-newborn pair (67). Notably, the number of phylotypes or OTUs that were shared between mothers and their children increased with age. In this context, the OTUs that are shared between mother-baby pairs were enriched in Bacteroidia and depleted of Clostridia, Gammaproteobacteria, and Erysipelotrichia (67). This indicates that the acquisition of the latter groups of bacteria, which represent dominant members of the adult gut microbiota (262), occurs during the very early phase of life (263). Furthermore, bifidobacteria appear to be subject to mother-baby transfer (47, 67). Specifically, shared OTUs that correspond to Bifidobacterium breve and Bifidobacterium bifidum were observed between mothers and their corresponding children, which were shown to persist until 3 months and 1 year of life, respectively (67). Similar findings were obtained in another recent study focusing on the identification of vertically acquired bifidobacterial strains (47) (Fig. 6). Such analyses involved a combination of shotgun metagenomics, ITS bifidobacterial profiling analyses (45), and culturomics and resulted in the isolation of two bifidobacterial strains, i.e., B. breve BBRI4 and Bifidobacterium longum subsp. longum BLOI2, which were identified in two mother-infant pairs and were also shown to persist in the infant gut until 6 months of age (47) (Fig. 6). Recently, a detailed profiling of bifidobacterial communities in 25 mother-offspring pairs through ITS bifidobacterial profiling analyses followed by an culturing approach revealed a large number of bifidobacterial strains that are commonly identified in mother and infant guts, as well as the corresponding human milk sample (264). These data are further reinforced by the identification of an identical scenario in other primate and nonprimate mammals, where bifidobacteria were shown to be commonly transmitted from a mother to her offspring and where the mother's milk represents an important means to drive such events (265).
FIG 6.
Strain-specific tracing of bifidobacteria from mother to infant. Each panel displays the protocol for shotgun sequencing of the high-abundance bacterial target and their corresponding tracing in mother and infant gut microbiota samples.
One may argue that bifidobacteria, despite their decline following weaning, persist following their initial transfer to the infant gut and are then maintained at (very) low levels in the adult gut, to be ultimately transferred to the next generation (47). Such a transfer process may be the consequence of millennia of strict coevolution between these bacteria and their mammalian host.
DYNAMICS OF THE MICROBIOTA COMPOSITION OF THE INFANT GUT
Dynamics of Colonization of the Infant Intestine
Immediately following birth, the neonatal intestine becomes rapidly colonized. As stated above, during this early postnatal period, facultative and aerotolerant microorganisms dominate the intestinal ecosystem. These microorganisms will reduce oxygen levels in the intestine, thereby facilitating the subsequent proliferation of a complex community dominated by anaerobic bacteria (74). In spite of this common pattern, the neonatal gut microbiota shows large interindividual differences, being more variable, over time and between individuals, than that of adults (4, 113). However, the high initial beta diversity is already reduced by 12 months of age (in contrast to alpha diversity, which is increased as a function of time), indicating that the neonatal community becomes more complex over time, while interindividual diversity progressively decreases to that typical of adults (73, 113). The exact age at which a stable adult-like gut microbiota structure is formed is still unclear, but generally this happens at an age of around 2.5 to 3 years (4, 113, 116). At this age, most bacterial groups have already reached a state of adult microbiota stability, whereas other microbial groups may still need more time to reach such a steady state (266). In fact, some differences seem to persist up to preadolescent age (267). Bäckhed et al. (73) observed that 1-year-old infants are more similar to their mothers in terms of microbiota composition and function than when they are younger yet have differences awaiting further maturation.
The dynamics of the colonization process shows differences depending on the perinatal factors present. While in full-term infants, delivery and feeding modes are reported to represent the major drivers of microbiota development, in preterm infants, the gestational age seems to have the biggest impact on the gut microbiota assembly process (73, 96). The existence of a microbiota “core” of OTUs in full-term babies, independently associated with delivery mode and lactation stage, has been reported, providing support for the Savage theory, which predicts the creation of a conserved stable microbiota, predicted to consist of approximately 30 OTUs, followed by a variable microbiota (74).
Due to the high instability of early-life gut microbiota, the high interindividual variability, and the multitude of factors (pre-, peri-, and postnatal) affecting the establishment of the microbiota, the definition of a “standard,” “normal,” or “healthy” infant gut microbiota is still difficult. Nevertheless, some general trends can be inferred from the different studies available. While the adult gut microbiota is dominated by members of the Firmicutes and Bacteroidetes phyla, the neonatal intestinal microbiota is initially represented by microorganisms from Proteobacteria and Actinobacteria, becoming more diverse later on with the rise of Firmicutes and Bacteroidetes (13, 268). Bifidobacterium has been considered to represent the dominant bacterial genus in the breastfed infant gut microbiota (41, 269), but recent studies have also shown a high occurrence of enterobacteria in such an infant population. It has been observed that Proteobacteria (mainly Enterobacteriaceae) dominate the infant intestinal microbiota during the first weeks, with bifidobacteria being the second microbial population, which then increases over time with a concomitant decrease of enterobacteria (93). These results were in concordance with observations in other cohorts, in which fecal samples from the first days of life were also characterized by high levels of Enterobacteriaceae, with samples from 6-month-old infants being dominated by Bifidobacterium, Collinsella, or Bacteroides (2, 270). Similarly, Yassour et al. (3) reported that Enterobacteriaceae were present at higher levels than Bifidobacterium in fecal samples of infants of up to 2 months of age, followed by replacement of the latter microbial group by members of the Lachnospiraceae and Ruminococcaceae families by 12 months of life. Members of the phylum Firmicutes were also detected in different studies, always showing a low abundance during the first weeks after birth (93, 270). Interestingly, it has been shown that members of the Firmicutes phylum, such as the families Staphylococcaceae, Clostridiaceae, Lachnospiraceae, and Veillonellaceae, are more numerous in breastfed infants than in formula-fed, full-term infants, whose microbiota was found to be dominated by enterobacteria up to 6 months after birth (271, 272). In contrast, a recent study showed a high abundance of Bifidobacterium from the first week following birth and lasting until the age of 6 months (79). According to other recent studies, another common intestinal bacterial phylum, Bacteroidetes, seems to be present, although at low levels, from the early stages after birth (3, 79, 93). In contrast, Koenig et al. (113) reported the absence of Bacteroides from the infant microbiota until the introduction of solid food in a single baby. Other differences between these studies may be due to the fact that the subjects originated from different countries and represented distinct populations, yet they may also be due to confounder factors, often present in the neonatal population, which may not have been considered, or to technical/methodological issues that significantly influence the results (see also above) (6). This variability observed for different studies calls for serious caution when interpreting and comparing published data, while it also illustrates the need for further investigations in order to determine how to best characterize the infant gut microbiota.
The Infant Intestinal Colonization Process: the Foundation of Health
As indicated above, during the initial colonization steps, the microbiota remains unstable and may suffer from sudden microbial succession phenomena that will continue until the infant is 2 to 3 years old, at which point the microbiota reaches a composition that resembles that of an adult microbiota (4). In spite of the extended duration of this stepwise microbiota evolution, we now know that microbiota-host cross talk is especially important during this period. These early moments of microbiota-host communication constitute key events that underpin appropriate maturation of the human host, with subsequent establishment and maintenance of the homeostasis of the host microbiota during early life, events that may have immediate and long-term health consequences (Fig. 7).
FIG 7.
Infant health status and microbiota establishment. The schematic representation shows several diseases related to the colonization of bacteria that are claimed to be pathogens.
The establishment of the early microbiota provides a massive antigenic stimulus necessary for the adequate maturation of the gut and associated immune system (273–275). This stimulus also affects the maturation of distal organs, affecting the host at systemic level (276–278). Thus, it is likely that the basis for a healthy microbiota throughout life is set up during its initial development, with the establishment and development of the microbiota being critical for the maturation of the host and long-term well-being (123, 260, 279). As an example of this, it was recently shown that fecal levels of IgA, an important characteristic potentially related to the risk of disease, may be determined by the presence of specific microbiota members (280). In this context, it seems logical to assume that the neonatal microbial colonization process is linked to the health of the infant and may represent a risk factor for disease later in life. Indeed, evidence is accumulating in this respect, with different studies reporting early microbiota alterations preceding disease development (281–284).
The key role of these early life microbiota events is demonstrated by in vivo trials involving murine models in which conventional animals are compared with germfree counterparts. These investigations have clearly demonstrated the serious health consequences caused by the absence of any microbiota-host interaction (285). Moreover, several authors have studied the effect of recolonizing germfree animals, at different ages, on restoring the parameters altered by the lack of microbial exposure. Interestingly, recolonizing animals in early life, as opposed to during adulthood, is needed to restore the altered phenotypes found in germfree models. For example, animals lacking a microbiota (germfree) were shown to exhibit increased levels of certain immune cells in the mucosae, a phenomenon that is reverted (to normal levels) when these animals are recolonized during early life, yet this reversion does not occur when recolonization is facilitated at adulthood (286).
Therefore, an altered early colonization pattern may represent a risk with immediate consequences for infant health and development yet may also present a risk for long-term effects (as discussed below). Another corollary is that the use of germfree mice colonized at some moment in time with human microbiota may be compromised by an altered sequence of immune priming events.
Infant Intestinal Microbiota and the Risk of Neonatal Pathologies
During the neonatal period, due to immune immaturity, the risk of early- or late-onset nosocomial infection is high (Fig. 7). Nosocomial sepsis in preterm infants is often related to the use of catheters, with Gram-positive microorganisms from the genus Staphylococcus being the main causative agent, followed by Gram-negative bacteria, mostly enterobacteria such as E. coli or Klebsiella (92, 93, 287). Moreover, especially in the case of premature neonates, this sepsis risk is exacerbated, as well as the risk of developing necrotizing enterocolitis (NEC), which is a very serious and often fatal condition (288). In a significant number of cases, infants who develop NEC will also develop sepsis, commonly caused by a dominant member of the gut microbiota of preterm infants such as enterobacteria (92, 93).
Although particular microorganisms have been proposed as causative agents (289), the etiology of NEC remains unclear. However, specific characteristics have been observed in the microbiota of infants developing NEC. These infants typically exhibit a reduced bacterial diversity in combination with increased levels of potentially pathogenic microorganisms (290, 291). However, the trials that have been performed so far have not identified a clear pattern of dysbiosis, though they have repeatedly identified an increased abundance of Proteobacteria as well as Clostridium perfringens preceding NEC development (291–295). Moreover, a potential protective role of high levels of bifidobacteria has been suggested (296). Specific metabolic pathways associated with NEC have been reported (288), and metabolome alterations have been observed in the sera of these infants (296).
It has been postulated that an intensified immune response caused by high levels of intestinal Proteobacteria may increase the risk of bacterial translocation and sepsis (98). Various metagenome-based studies have shown lower bacterial diversity and reduced levels of Bifidobacterium and Bacteroides, with a predominance of enterobacteria, in infants who develop late-onset sepsis compared with healthy counterparts (100–102). However, available evidence is still limited, and additional investigations are needed before drawing conclusions on the potential role that early microbiota alterations play in determining the risk of infection and sepsis.
Infant Intestinal Microbiota and Growth
The early microbiota may also impact infant malnutrition/growth impairment. During the last few years an increasing number of studies have reported associations between infant microbiota and neonatal growth (92, 297). Moreover, the microbiota of malnourished children was found to be different from that of their healthy counterparts, and experiments with microbiota transplantation into germfree animals have shown that receiving the microbiota from undernourished donors hampers weight gain (298). The same researchers have also employed different animal models to demonstrate a microbiota-dependent enhancement of growth after supplementation of the diet with certain (bovine) milk oligosaccharides (299). Several potential mechanisms may explain this microbiota-growth association, among them the production of growth hormones (300). Although more research is needed to fully understand the mechanistics of this phenomenon, the potential relationship between infant malnutrition and gut microbiota may constitute the basis for the development of microbiota modulation strategies aimed at restoring normal infant growth and development (301).
ROLE OF EARLY-LIFE MICROBIOTA IN PROGRAMMING FUTURE HEALTH
Impact of Early-Life Microbiota on Long-Lasting Physiological Effects
Microbial colonization of the human gut is believed to be responsible for the concurrent programming of our immune system and the simultaneous development of the intestinal tract and associated metabolism. A continuous dialogue between the microbiota and the host must occur in order to orchestrate these physiological processes. Therefore, intestinal dysbiosis may disrupt or modify this dialogue, which may in turn result in long-lasting physiological effects and health disorders (302). Among these, extensive research has been carried out regarding the potential effects of early-life microbiota on immune disorders. Some of the most convincing results come from animal experiments and point to a very close relationship between early exposure to microorganisms and development of immune pathologies. Although this relationship has been known for decades, some recent findings are decisively contributing to understand the mechanisms behind the long-term effects of our microbiota on (shaping) the immune response. In this regard, it has been shown that microbial factors regulate the activity of chemokine ligand CXCL16, which modulates the accumulation of invariant natural killer T cells in the colon and lungs, and that neonatal colonization of germfree mice with a conventional microbiota protects them from this accumulation (286). In this context, it has been suggested that the early-life microbiota triggers long-lasting effects, and the absence of such a microbial stimulus may induce later-life inflammatory responses related to IBD and asthma (286). Recently, the importance of a critical period in which disruption of the intestinal microbial balance can have long-lasting consequences in immune pathologies has been suggested, and the ordered establishment of an adequate dialogue between commensals and the mucosal surfaces of our body and its pivotal importance for the development of our immune defenses have been highlighted. This cross talk is facilitated by host-microbial interactions during the earliest days of life or even through microbial colonization during pregnancy, suggesting that disease risk is programmed during early life, including the prenatal period (273).
Allergy (Atopic Eczema and Asthma)
Among the immune pathologies related to a particular microbiota establishment, allergy, mainly in the form of atopic eczema and later asthma, has been linked to specific microbial features. Numerous epidemiological studies suggest that early development of the infant gut microbiota influences the risk of allergic diseases later in life (303). This has been attributed to an inappropriate development of gut microbiota and associated disruption of immune homeostasis during the first year of life (304).
Chronic recurring atopic eczema is the main symptom of atopic disease during the first years of life (305). Various research investigations involving infant cohorts have provided valuable information on the potential role of gut microbiota in the development of atopic eczema. More than a decade ago, pioneering analyses already reported microbiota alterations at an early age in infants who subsequently developed atopic diseases (283, 306). These investigations revealed differences, such as increased levels of clostridia and reduced levels of bifidobacteria, in the early-life gut microbiota composition of infants who did or did not develop atopic disease by the age of 2 years. Similarly, in a recent study, a reduced abundance of bifidobacteria and other intestinal anaerobes, such as Faecalibacterium, and an altered fecal metabolome were observed in 3-month-old infants who later developed atopy (determined at 2 years of age) or asthma (evaluated at 4 years) from a U.S. cohort (281). Another analysis using culture-independent PCR-based molecular techniques (quantitative real-time PCR) in a cohort of 957 infants showed that differences in the gut microbiota composition at the age of 1 month precede the manifestation of atopic symptoms within the first 2 years of life. In particular, the presence of E. coli was associated with a higher risk of developing eczema, and the presence of C. difficile was associated with several atopic outcomes, including eczema, wheeze, and allergic sensitization (307). Similarly, a nested case-control study based on quantitative real-time PCR and PCR-denaturing gradient gel electrophoresis (DGGE) analyses directed to explore the composition of the gut microbiota of a cohort of 646 infants revealed an association between the colonization by E. coli in infants of 1 month and the development of IgE-associated eczema within the first year of life (308). Subsequent studies have indicated that a reduced microbial diversity of early-life microbiota directly correlates with later development of atopic eczema. In two of these analyses, a correlation between a low microbiota diversity at 1 week and infants having atopic eczema at 12 and 18 months of age (309, 310). Next generation sequencing methodologies and metagenomic approaches have shown that infants with IgE-associated atopic eczema have a low microbiota diversity and a lower diversity of the bacterial phylum Bacteroidetes during the first month of life, compared with a control group of infants without allergic manifestations until 2 years of age was identified (311). Furthermore, a phylogenetic human intestinal tract chip (HITChip) was used to analyze the intestinal microbiota signatures associated with eczema symptoms in 6-month-old infants and the microbial changes associated with the physiology of this disease during the following 3 months. Such analyses highlighted that a decrease in the severity of eczema during the 3-month follow-up period was directly related to an increase of the butyrate-producing bacterium Coprococcus eutactus (312). Although these results do not correlate an early microbiota profile with a decrease of atopic eczema ailment, they indirectly point to the potential protective role of a butyrate-producing bacterium in the development of atopic eczema. In fact, it has been shown that a low relative abundance of butyrate producers precedes the development of atopic eczema (284).
A direct association of specific microbial patterns early in life with the development of asthma years later has not yet been unequivocally established, since genetic, epigenetic, and other environmental factors also affect the development of the disease. Nevertheless, it is becoming increasingly clear that the intestinal microbiota plays a crucial role in the perinatal programming of asthma (313). Animal trials have repeatedly reported the importance of the intestinal microbiota in determining the levels of immune cells and their recruitment to various tissues (286, 314, 315) or in the development of immune tolerance (316). As discussed below for metabolic diseases, antibiotic-induced modification of the early-life microbiota has been shown to increase the risk of allergic asthma in laboratory animals (317). In addition, epidemiological data sets have provided support for the notion that a link exists between perinatal antibiotic exposure and the risk of subsequent allergic disease development (318–320). In this regard, recent evidence suggests that the risk of suffering from asthma is higher in infants who exhibited gut microbiota dysbiosis during the first 100 days of life and that this risk is associated with particular bacterial groups. Analysis of the gut microbiota compositions of 319 subjects of a Canadian cohort showed that infants at risk of asthma display significantly decreased relative abundances of the genera Lachnospira, Veillonella, Faecalibacterium, and Rothia. Furthermore, these differences in abundance of bacterial taxa were linked to different levels of fecal bacterial metabolites. Inoculation of these bacteria in germfree mice reduced airway inflammation in their progeny, suggesting that some microbes play a causal role in the development of asthma (321). Furthermore, a reduced gut microbiota diversity during the first month of life is associated with a higher prevalence of asthma in 7-year-old children (322), and lower levels of Lachnospira and higher levels of Clostridium spp. at 3 months are positively associated with asthma risk at 4 years of age. These findings suggest that the ratio between these two genera can be used as a microbial biomarker to predict the risk of asthma development (323). Notably, colonization by Clostridium difficile detected at 1 month was positively associated with asthma at age 6 to 7 years (324). In addition, early IgA responses targeting the fecal microbiota during the first year of life differed between healthy children and children having asthma at up to 7 years of age (325).
Metabolic Disorders
The gut microbiota composition and function have been associated with obesity and obesity-related disorders. By increasing energy harvest, the so-called obesogenic microbiota regulates obesity behavior and peripheral metabolism. It has been suggested that different factors that impact gut microbiota establishment during infancy may contribute to the risk of obesity later in life (326). Microbiota-related obesity studies in animal models, particularly in rodents, have broadened our understanding of the role played by the gut microbiota in metabolic disorders. However, in recent years it has become clear that one should be cautious in extrapolating the results of animal studies to humans, thus highlighting the need for human clinical trials (327). In human beings, it has been suggested that early microbial patterns may predict overweight in children. In this context, it has been observed that the abundance of the bifidobacterial population at 6 and 12 months inversely correlates with overweight in 7-year-old children (282). Furthermore, in a large cohort study, quantitative PCR was used to determine the levels of several bacterial groups in 909 1-month-old infants, and body mass index (BMI) was reported from 1 to 10 years of age. That analysis showed that Bacteroides fragilis levels at 1 month of age are significantly associated with a higher BMI in children (328).
The maintenance of a properly functioning intestinal barrier seems to be critical for metabolic health, but different factors that disturb the microbial balance during early life play a pivotal role in overweight, obesity development, and child adiposity in later life. Among such factors, nutrition, maternal obesity, delivery mode, intestinal permeability, pathogenic infections, and antibiotic use have been highlighted (270, 329–331). In addition, recent literature also implicates microbiota-related epigenetic changes during early development (332). Furthermore, the impact of gut microbiota on brain developmental programming of obesity has also been suggested (333). Moreover, considerable attention has been drawn to investigating the role of early-life antibiotic therapy in being an important driver for subsequent development of metabolic diseases. Several analyses involving murine models have shown that altering the gut microbiota with antibiotics during early life will have long-lasting metabolic consequences, including adiposity, weight gain, insulin resistance, type 2 diabetes, and liver disease (334–336). In one of these studies, the authors observed that the effect on host metabolism was sustained over time, even when the microbiota alterations had disappeared following the discontinuation of the antibiotic treatment, which underlines the importance of microbiota-host interactions during early life. Nevertheless, a recent study encompassing a large cohort of more than 260,000 individuals proposed that childhood obesity is positively correlated only with untreated infections and not with antibiotic use during infancy (337). These above-mentioned analyses also demonstrated a causal role of the microbiota as opposed to an antibiotic effect, since such an antibiotic-modified microbiota, when transferred to germfree mice, was able to promote growth (335). Other murine trials have observed differences in the microbiota and host physiology depending on the antibiotic used (96). Notably, these analyses suggest that an association exists between early-life antibiotic-induced dysbiosis and alterations of host metabolism in later life. Epidemiological studies in humans have also demonstrated that antibiotic exposure is associated with long-term metabolic effects, including weight gain and obesity in children and adults, and this antibiotic-microbiota-obesity trinomial has attracted a substantial level of public interest (338). Investigations involving a large infant cohort showed an association between early-life antibiotic exposure and childhood obesity (339). In fact, it has been suggested that disturbances of the intestinal microbiota caused by antibiotics, during either prenatal or postnatal periods, increase the risk of becoming obese (340). However, more evidence from human epidemiological studies is needed to definitively establish a causative link between early-life antibiotic-induced dysbiosis and subsequent metabolic consequences in later life.
Other Long-Lasting Effects of the Infant Gut Microbiota
Early infant dysbiosis has also been associated with other chronic disorders that may manifest later on in life; in particular, IBD, irritable bowel syndrome (IBS), and type 1 diabetes (T1D) have received a lot of interest from the scientific community in recent years in this context.
T1D is an autoimmune disease characterized by the destruction of insulin-producing beta cells. Several environmental factors may affect T1D, and accumulating evidence supports the proposed role of intestinal microbiota in the development of this disease. Longitudinal studies in patients with T1D have shown a lower diversity and significant differences in the ratios of the most abundant intestinal phyla, including Firmicutes and Bacteroidetes, as well as a decreased abundance of the butyrate producer F. prausnitzii in diabetic children (341). Indeed, butyrate-producing species are more abundant in nondiabetic children, and it has been suggested that these bacteria play a key role in reducing the risk of developing T1D (342). Furthermore, how early intestinal colonization can influence the subsequent progression of T1D has also been investigated. Experiments in rodents have shown that early-life microbiota modifications, notably by low-dose or pulsed therapeutic antibiotic treatments, alter intestinal microbiota and T cell populations, increase the risk of T1D, and accelerate T1D development in a nonobese diabetic (NOD) murine model (343). Regarding human trials, recent findings suggest a role of early dysbiosis in future development of T1D (344). It has been shown that microbiota perturbations during early infancy may generate a proinflammatory environment that facilitates the development of autoimmune disease. In this regard, pioneering investigations by Kostic and coworkers reported different microbial patterns in the fecal microbiota of infants who later developed T1D compared with those who did not progress to diabetes (345). However, only a limited number of analyses have tried to correlate infant microbiota and T1D development. Therefore, the monitoring of human cohort studies, together with mechanistic studies, is needed to establish a causal relationship between microbiota and T1D.
Diseases that involve intestinal inflammatory symptoms have also been related to early-life microbiota colonization events. The notion that the human intestinal immune system is “trained” during early microbial colonization presumes a link between pioneering microbial inhabitants of the gut and subsequent intestinal inflammatory disease, notably IBD and IBS. A small number of trials have suggested that early-life shifts in the populations of specific bacterial groups precede onset of intestinal inflammatory diseases at later stages of life (346–349). However, convincing and consistent data about the long-term effects of early-life dysbiosis in the development of IBD and IBS in children and adults are scarce and do not allow the establishment of causality. Thus, future work with animal models and human epidemiological studies may shed light on microbiota-mediated immune and physiological responses that may result in the onset of these diseases.
MICROBIAL BIOMARKERS ASSOCIATED WITH THE CORE INFANT GUT MICROBIOTA
The Core Infant Gut Microbiota
Relative to the gut microbiota of adults or older children (age of >1 year), the infant gut microbiota exhibits low diversity, and the microbiota structure is generally unstable and highly dynamic (see above) (123). In contrast, the gut microbiota of adults is specific to an individual and is relatively stable (350). Nevertheless, bifidobacteria are typically found in large amounts in infants, particularly breastfed infants, and thus are considered a key member of the infant gut microbiota (41, 116, 123, 351).
Notably, despite the large variability at the intraindividual level during the period from initial assemblage of the infant gut microbiota to the establishment of an adult gut microbiota, the infant gut microbiota can be classified into six main types (Fig. 8) (114). Such infant gut microbiota types are determined according to the composition of the gut microbiota and the occurrence of dominant bacterial groups. In detail, these dominant groups encompass the following: group 1, consisting of Enterobacteriales; group 2, formed by Bacteroidales and Verrucomicrobiales; group 3, encompassing members of Selenomonadales as well as the Clostridiales genera Pseudoflavonifractor and Subdoligranum and Deltaproteobacteria Desulfovibrio; group 4, including all Pasteurellales; group 5, comprising most of the Clostridiales; and group 6, involving the Clostridiales genera Anaerostipes and Faecalibacterium and the Lactobacillales and Bifidobacteriales (114) (Fig. 8). Nevertheless, additional investigations that involve large data sets for infants from multiple geographic regions are needed to confirm these observations. Interestingly, the genera that dominate the infant gut microbiota in different individuals are represented by Bifidobacterium, Veillonella, Streptococcus, Citrobacter, Escherichia, Bacteroides, and Clostridium, which are also abundant in the gut microbiota of adults (114). Notably, further detailed analyses allowed the identification of an additional 10 genera that are shared between adults and infants, though at very different abundancies: Bacillus, Bacteroides, Bifidobacterium, Clostridium, Enterococcus, Escherichia, Eubacterium, Lactobacillus, Prevotella, Ruminococcus, and Streptococcus. These core bacteria of the gut microbiota include members of four of the phylogenetic groups described above (group 1, Escherichia; group 2, Bacteroides and Prevotella; group 5, Clostridium and Eubacterium; and group 6, Bifidobacterium, Enterococcus, Lactobacillus, Ruminococcus, and Streptococcus) (114).
FIG 8.
The infant gut core microbiota. A 16S rRNA gene-based tree involving the infant bacterial core microbiota is displayed. The colors of the branches indicate the six main phylogenetic groups of the infant gut microbiota. An electron microscopic image of the key infant gut bacterial taxon is displayed for each branch of the tree.
Bifidobacteria and the Gut Microbiota of Infants
As described above, bifidobacteria represent one of the dominant members of the core infant gut microbiota, and several ecological studies based on culture-dependent as well as culture-independent approaches have confirmed such findings (3, 41, 42, 67, 80, 117, 351–354). Interestingly, bifidobacteria are presumed to be particularly abundant in the colon, while they are identified at lower densities in the oral cavity (355). Bifidobacteria were first isolated from feces of a breastfed infant by Tissier in 1899 and are classified as a distinct genus, i.e., Bifidobacterium, belonging to the Actinobacteria phylum (356–358). The genus Bifidobacterium currently includes 59 different taxa, five of which have been isolated from fecal samples from the human gut (265, 359, 360). An ecological clustering of the currently described members of the Bifidobacterium genus distinguishes seven different ecological niches encompassing the GITs of humans, nonhuman mammals, birds, and social insects, wastewater, and human blood and oral cavity (352). Notably, these apparently unrelated ecological origins may represent an ecological niche that is common to all these habitats, represented by the fact that bifidobacterial hosts are social animals whose offspring enjoy parental care. Therefore, perhaps their ecological distribution is facilitated by direct transmission of bifidobacterial cells from mother/carer to newborn. Such a hypothesis has recently been experimentally confirmed by the identification of bifidobacterial strains that are common between mothers and their corresponding children (see above) (67, 354). Further detailed classification of bifidobacteria based on the ecological origin suggests the occurrence of (sub)species, such as Bifidobacterium animalis subsp. lactis/animalis, Bifidobacterium adolescentis, Bifidobacterium dentium, and Bifidobacterium catenulatum, that are found in different animals and which are therefore also known as cosmopolitan bifidobacterial species (361). In contrast, other species, such as Bifidobacterium bifidum, Bifidobacterium breve, and Bifidobacterium longum, appear to be less widely distributed than the above-mentioned species, and thus it was not possible to identify a strict host-specific adaptation of bifidobacterial species (265). Among those species identified in primates (including humans) it is possible to distinguish bifidobacterial taxa that are typically found in adults, e.g., B. adolescentis and B. catenulatum, while others are much more commonly found in the guts of breastfed infants, such as B. bifidum, B. breve, and B. longum subsp. infantis (41). Nevertheless, there does not seem to be a strict infant-versus-adult subdivision of bifidobacterial taxa. This makes sense in the context of vertical transfer of bifidobacterial species from mother to offspring, which also includes adult-type members such as B. adolescentis (362, 363).
Contribution of bifidobacteria to the infant host.
Due to their well-described saccharolytic features, bifidobacteria make a major metabolic contribution to their host through the degradation of diet-derived glycans and host-provided carbohydrates (known as host glycans and including mucins and HMOs) (364). The glycan-based metabolic activities exerted by bifidobacteria are pivotal in their establishment and persistence in the gut at the early stages of life (365). In this context, genome analyses of Bifidobacterium longum subsp. infantis ATCC 15697 and Bifidobacterium bifidum PRL2010 have revealed how these two microorganisms are able to utilize host-derived glycans (HMOs and mucin). In particular, the genome of B. longum subsp. infantis ATCC 15697, a strain that was originally isolated from a breastfed infant, contains genes that encode enzymes predicted to be involved in the degradation (such as fucosidase, sialidase, β-hexosaminidase, and β-galactosidase) and internalization (such as extracellular solute binding proteins and permeases of ABC transporter systems) of HMOs (232). Furthermore, the genome of this strain encompasses an operon involved in the metabolism of urea, an important source of nitrogen in human milk (232). Notably, maternal genotypes that determine fucosylation patterns of HMOs are implicated in the assemblage of infants' microbiota (366, 367). As mentioned above, specific infant-associated bifidobacteria such as B. longum subsp. infantis efficiently metabolize HMO components, e.g., lacto-N-tetraose (232, 368), although metabolic differences have been observed with respect to HMO utilization profiles between bifidobacterial species.
Mucin is another host-produced glycan that constitutes one of the main barriers covering the GIT mucosa. The main glycan components of these glycoproteins are N-acetylglucosamine, N-acetylgalactosamine, fucose, and galactose, and mucins are frequently covered by sialic acid and/or sulfate groups (369). In order to metabolize these glycoproteins, a number of specific glycosyl hydrolases are required, such as (i) endo-α-N-acetylgalactosaminidase, catalyzing the hydrolysis of O-glycosidic α-linkages between galactosyl β1-3-N-acetylgalactosamine and serine or threonine residues (370), and (ii) fucosidases, liberating the l-fucose from the oligosaccharide core of the mucin structure (371). Additional enzymes responsible for the complete breakdown of mucin encompass N-acetyl-β-hexosaminidases, β-galactosidases, and sialidases (372). Moreover, the core oligosaccharide structure of mucin is formed by galacto-N-tetraose, which is known to be metabolized by certain members of the human gut microbiota, including specific bifidobacterial species, into galacto-N-biose (368) and then transported in the cytoplasm through a specific ABC-type transporter, phosphorylated by a galacto-N-biose phosphorylase, and finally utilized in the glycolytic and glycoprotein metabolic pathways (372).
Only a few members of the human gut microbiota can directly access mucin as a C source, including Bifidobacterium bifidum and Akkermansia muciniphila (373–376). Notably, in silico genome analyses of B. bifidum PRL2010, a strain isolated from infant stool, revealed a gene set responsible for mucin metabolism, encoding extracellular enzymes that include sialidases, fucosidases, a putative cell wall-anchored endo-α-N-acetylgalactosaminidase, N-acetyl-β-hexosaminidases, and β-galactosidases (375). This genetic repertoire is part of the unique core genome of members of the B. bifidum species (377), providing an intriguing case of coevolution of a human gut commensal to the (human) intestine, where glycans produced by the host act as a carbon and energy source for the establishment and survival of certain bifidobacterial species within the human gut (375).
Interestingly, among the bifidobacterial communities that reside in the gut of human infants and adults, certain species are present, such as Bifidobacterium breve, which possess carbohydrate breakdown capabilities toward both dietary and host-derived glycans (378–382). Notably, while B. breve is not able to grow to any appreciable density on mucin or HMOs, host-derived mono-/oligosaccharides may become available through hydrolytic activities of other (bifido)bacteria present in the human gut microbiota through cross-feeding activities (364, 383–386).
Modulation of the infant gut microbiota by cross-feeding activities operated by bifidobacteria.
Within the human gut microbiota, bifidobacterial communities are believed to establish several trophic interactions with each other and with other members of the gut microbiota, leading to competition for or cooperative sharing of nutrients. Commonly, microbe-microbe interactions can either positively or negatively influence the fitness of the affected organisms (387) by the release of molecules in the environment (388, 389). Due to cross-feeding strategies, enteric microorganisms promote an expansion of their carbohydrate acquisition abilities, thereby positively influencing the ecological fitness of a specific proportion or even the overall gut microbiota (364). Cross-feeding actions in the gut are commonly exploited by primary microbial degraders such as bifidobacteria, which, due to partial extracellular hydrolysis of specific complex carbohydrates (e.g., host-produced glycans), provide monosaccharides and oligosaccharides to other microbial gut inhabitants (390). In addition, their fermentative metabolism of these carbohydrates generates end metabolites, such as acetate, which in turn may act as a substrate for secondary microbial degraders such as the butyrate-producing enteric bacteria (391–394).
Examples of cross-feeding activities in bifidobacterial communities have been experimentally demonstrated between two infant-type bifidobacteria (B. bifidum PRL2010 and B. breve UCC2003) when these bacteria were cultivated on sialyl lactose as the unique carbon source (384, 385). Recently, the cross-feeding activities of various bifidobacterial strains such as B. bifidum PRL2010, B. breve 12L, B. adolescentis 22L, and B. thermophilum JCM7017, when grown on plant-derived glycans such as starch and xylan, have been evaluated (383). Notably, cocultivation trials coupled with transcriptomic and metabolomic analyses revealed that the concurrent presence of the above-mentioned bifidobacterial strains results in enhanced metabolic activity of B. bifidum PRL2010, suggesting that PRL2010 cells benefit from the presence of other bifidobacterial strains.
The existence of cross-feeding activities between various bifidobacterial strains resembling the infant bifidobacterial communities, i.e., B. bifidum PRL2010, B. longum subsp. infantis ATCC 15697, B. adolescentis 22L, and B. breve 12L, has been further assessed under in vivo conditions in a conventional murine model (395). Remarkably, in this study, transcriptomic experiments coupled with metagenomic analyses of single, dual, or multiple associations of bifidobacterial strains uncovered cross-feeding behavior that caused an apparent expansion of the murine gut glycobiome toward its enzymatic potential related to the breakdown of plant-derived carbohydrates such as xylo-oligosaccharides, arabinoxylan, starch, and host glycan substrates. Furthermore, these analyses revealed distinct strategic responses by the different bifidobacterial strains toward glycans; for example, a “selfish” behavior was exhibited by B. longum subsp. infantis ATCC 15697 as it internalizes HMOs prior to degradation, thereby limiting opportunities for resource sharing by other enteric microorganisms. In contrast, B. bifidum PRL2010 actively participates in extracellular breakdown of host glycans and thus in the release of simple sugars that can in turn be utilized by other members of the (bifido)bacterial community (395).
How bifidobacteria interact with the human gut.
The molecular mechanisms by which bifidobacteria interact with the human host are still largely unexplored. However, bifidobacterial colonization at the early stages of life seems to play an important role in the host (see above). It has recently been demonstrated that many bifidobacterial species encode cell surface-associated exopolysaccharides (EPSs), cell surface-protruding proteinaceous appendages called fimbriae or pili, and/or a secreted serine protease inhibitor, all of which seem to be involved in host interactions (352, 396–401) (Fig. 9).
FIG 9.
Extracellular structures identified among members of the Bifidobacterium genus. The bifidobacterial phylogenetic tree is based on the core gene sequences conserved in each bifidobacterial (sub)species. Colored dots show the presence of the different extracellular structures distributed among the Bifidobacterium genus. The bifidobacterial colors are related to the ecological origin of each bifidobacterial (sub)species.
EPSs are carbohydrate polymers present as an extracellular layer covering the surfaces of various Gram-positive microorganisms (402). EPS can have two seemingly opposing functions: bacterial EPS can act as a virulence factor in particular diseases but may also elicit positive effects on human health by exploiting an immune modulatory effect on the host (403, 404). The EPS synthetized by certain lactic acid bacteria (LAB) can be classified in two groups: homopolysaccharides, consisting of just a single repeated monosaccharide type, and heteropolysaccharides, which are composed of a repeated oligosaccharide (405). Specifically, EPSs synthesized by bifidobacteria are composed of various monosaccharides, such as d-glucose, d-galactose, and l-rhamnose, and are thus classified as heteropolysaccharides (396). However, EPS gene clusters are not conserved in different Bifidobacterium species or even between strains of the same species. Several studies demonstrated that EPS production is linked with their ability to modulate the host immune response (398, 406) and/or their ability to shape the composition and activity of the gut microbiota (396). In this context, it has recently been shown that the EPS produced by B. breve UCC2003 cells positively modulates the small intestinal cell shedding response in IBD by reducing apoptotic signaling in the epithelial compartment. Notably, such novel findings imply that capsular structures of bifidobacteria play an important role in their host against pathological cell shedding (407). Furthermore, it has been shown that EPS produced by B. longum 35624 exerts an immune modulatory role by dampening proinflammatory host responses, while loss of EPS production results in the induction of local TH17 responses (408, 409).
Another key cell surface structure considered to be crucial in bifidobacterial interactions with its host, possibly with other members of the gut microbiota, is represented by pili or fimbriae (352). So far, in the various members of the genus Bifidobacterium two different types of pili have been described, the tight-adherence (Tad) pili and the sortase-dependent pili (353). Tad pili in bifidobacteria are encoded by a genetic locus which was originally found in the genome of B. breve UCC2003 (399). The main tad locus of UCC2003 is similar to that of the type IVb pilus-encoding gene cluster of Actinobacillus actinomycetemcomitans, though bifidobacteria harbor one tad gene which is separated from the main tad locus (399). The main tad locus of B. breve UCC2003 encodes proteins involved in pilus assembly and localization (TadB, TadA, TadC, and TadZ), as well as the Flp prepilin and two pseudopilins (TadE and TadF), while the second locus consists of a single gene, tadV, which is responsible for the processing of pilin precursors to mature pilin proteins (399). The main tad locus was identified through transcriptome analysis of B. breve UCC2003, which showed that upon murine colonization, transcription of this gene cluster is induced, while it is not transcribed under laboratory conditions. Thus, such extracellular structures may be produced only when B. breve UCC2003 is found in its natural environment (399). Notably, a mutational analysis of the tadA gene highlighted the crucial importance of the tad locus in the colonization and persistence of UCC2003 cells in the mammalian gut. The tad locus is highly conserved among sequenced bifidobacterial strains, which supports the hypothesis that such extracellular structures mediate host colonization by bifidobacteria (353, 399). Further extracellular appendages that are believed to be pivotal for colonization and/or the interaction of bifidobacterial cells with the mammalian host are represented by sortase-dependent pili (401). Such structures were shown to promote adhesion of bifidobacterial cells to human enterocytes through extracellular matrix (ECM) proteins as well as the interaction with the immune system of the host by eliciting a local proinflammatory response, which may be crucial in the first stages of life of the human host (401). In fact, the neonatal immune system is immature, and the presence of proinflammatory stimuli such as those exerted by bifidobacterial pili may be essential in developmental immunological programming. In this context, it is well known that decreased antigenic exposure has adverse effects on the budding immune system and increases the likelihood of developing atopic disorders (410).
Another protein that seems to be involved in bifidobacterium-host interaction is the serine protease inhibitor (serpin), which is a member of the serpin superfamily (411). Bifidobacterial serpins act as inhibitors of the human neutrophil and pancreatic elastases (411). Serpin-encoding genes in bifidobacteria are present in the genomes of only B. breve, B. longum, and B. dentium species (412). Functional analyses involving transcriptomics and targeted gene inactivation revealed that transcription of the serpin-encoding gene is enhanced when B. breve cultures are exposed to various host-derived proteases, such as pancreatic elastase, human neutrophil elastase, thrombin, papain, kallikrein, trypsin, α-antitrypsin, chymotrypsin, and plasmin (412, 413). This finding is highly relevant since many of these proteases are typically found within the human gut, and thus the presence of a protease inhibitor may provide an ecological advantage to bifidobacteria since serpin activity may protect them against these host proteases.
Bifidobacterial cells have also been characterized for the synthesis of an interspecies signaling molecule known as the quorum sensing molecule AI-2 (414, 415). Synthesis of AI-2 is modulated by an enzyme that forms an essential part of the activated methyl cycle, i.e., LuxS, which is involved in recycling S-adenosylhomocysteine (416). The LuxS-encoding gene belongs to the core genome of the genus Bifidobacterium (417) and has been molecularly characterized in B. breve UCC2003 (415). Insertional inactivation and complementation experiments indeed demonstrated that a functional luxS gene is necessary for bifidobacterial AI-2 synthesis. Furthermore, these analyses showed that the UCC2003 luxS mutant, compared to the UCC2003 wild-type strain, is less effective in iron sequestering and unable to colonize the murine gastrointestinal tract, while this mutant was also shown to confer less protection against Salmonella infection in a nematode model (415).
Other Members of the Core Infant Gut Microbiota
The Clostridia class.
Members of the genus Clostridium have recently been reclassified into several genera that all fall within the Clostridia class (418). These species are commonly found among microbial taxa present in the infant gut microbiota. So far, 72 different species of the Clostridia class have been isolated from the human gut (32). Among the infant gut microbiota-associated members of the Clostridium sensu stricto (cluster I) group, we should mention C. perfringens, while also worth mentioning is a member of the Peptostreptococcaceae family, Clostridiodes difficile, the recently reclassified Clostridium difficile (418, 419). Both of these species are known as pathogenic microorganisms that may cause bacteremia and pseudomembranous colitis, and their presence at high densities is interpreted as an indicator of an unhealthy microbiota (420). Historically, cultivation-based analyses have revealed that C. perfringens and other Clostridium sensu stricto members can be present in infants at densities of up to 107 CFU g−1 fecal content (421).
The Clostridia class also includes certain common human gut commensals such as species belonging to the Ruminococcaceae and Lachnospiraceae families, formerly known as Clostridium clusters IV and XIVa (422). Notably, the Ruminococcaceae and Lachnospiraceae families encompass highly diverse species, many of which produce SCFAs (423–425). Thus, a depletion of Ruminococcaceae and Lachnospiraceae is correlated with reduced production of SCFAs, e.g., butyrate, and the onset of IBD.
The genus Bacteroides.
Members of the Bacteroides genus are dominant components of the adult gut microbiota (262), although they may also be present in the infant gut microbiota, where their presence seems to be modulated by HMOs in a fashion similar to that for bifidobacteria (365). Notably, in mouse experiments, it has been shown that gut colonization by Bacteroides spp. is a result of the recognition and selection by the immune system of the host (426), mediated through Toll-like receptors (TLRs) (404) and other specific microbe-host interactions (427). Members of this genus are classified as saccharoclastic bacteria that are able to metabolize host-produced glycans, such as HMOs and mucins, but also complex plant polysaccharides such as starch, cellulose, xylans, and pectins (428, 429). Bacteroides species commonly possess proteolytic activity due to the action of extracellular proteases (430). Other key metabolic functions exploited by members of the Bacteroides genus encompass deconjugation of bile acids (431).
(i) How Bacteroides cells interact with the host.
Among the Bacteroides genus, B. fragilis has been described as a member that can produce multiple capsular polysaccharides (432), known as polysaccharide A (PSA), which act as important mediators of gut microbiota colonization, host-microbe cross talk, and/or immune modulation (433). In various Bacteroides species, capsular polysaccharides are predicted to alter the physical properties of cell surfaces and to play a key role in host-bacterial commensalism (434). Cells of B. fragilis are able to produce multiple capsular polysaccharides, and any experimental attempts to eliminate capsule-mediated protection against the host immune system lead to competitive defects of noncapsular mutants with subsequent spontaneous phenotypic reversion (432, 434–436). This reversion is explained by the finding that B. fragilis acapsular mutants are capable of reestablishing capsule production. The establishment of the production of other capsular polysaccharides restores the reduced fitness of acapsular mutants in the gut.
The genera Veillonella and Streptococcus.
A minor component of the core infant gut microbiota is represented by bacterial taxa belonging to the Veillonella genus. These bacteria are saccharolytic and utilize end products of carbohydrate fermentation (e.g., lactate) of other infant gut bacteria, such as Streptococcus spp. and Bifidobacterium spp., to produce propionate, forming an important trophic chain. This short-chain fatty acid is considered a beneficial product of the gut microbiota as it displays anti-inflammatory features, and influences glucose and energy homeostasis and increases insulin sensitivity (437).
Specific members of the genus Streptococcus also form part of the core infant gut microbiota and are among the first established bacteria in the infant gut, where they can be identified within the first 24 h following birth (200, 438).
The genus Collinsella.
Members of the genus Collinsella have recently been shown to reach high numbers when they are associated with an infant gut microbiota dominated by bifidobacteria (270). The Collinsella genus was identified from the reclassification of Eubacterium aerofaciens based on a 16S rRNA gene sequence divergence and the presence of a unique peptidoglycan type compared to other members of the genus Eubacterium (439). So far, the Collinsella genus includes five species, Collinsella aerofaciens, Collinsella intestinalis, Collinsella stercoris, Collinsella ihuae, and Collinsella tanakaei, which were all isolated from the human gastrointestinal tract (439–441). However, the biology of these bacteria is still largely ignored, and only recently a genome sequence of a member of this genus has been published (442).
The genus Lactobacillus.
Lactobacilli are known to be present in the infant gut microbiota, although they are present at lower numbers in the large intestine than the above-mentioned bacterial genera yet are present soon after delivery (438). In this context, lactobacilli belonging to the Lactobacillus gasseri, Lactobacillus ruminis, Lactobacillus casei, Lactobacillus reuteri, Lactobacillus sakei, Lactobacillus plantarum, and Lactobacillus brevis species were detected in the meconium, where their abundance is higher in vaginally delivered (VG) neonates than in caesarian section-delivered (CS) newborns (204). Follow-up studies of lactobacilli revealed that compared to that in VG infants, the rate of detection of the Lactobacillus genus remained significantly lower in CS infants at different time points during the first 6 months of life (204). It may be argued that vertical transmission of Lactobacillus species present in the maternal vaginal tract represents the origin of this early infant Lactobacillus microbiota component. In this context, preliminary genetic insights supporting the occurrence of the enzymatic arsenal for HMO metabolism have recently been discovered in the genomes of L. casei (443).
The genus Akkermansia.
A. muciniphila is the sole intestinal representative of the Verrucomicrobia that is present, though at very low levels, in the human gut since early life (444, 445). The presence of A. muciniphila has been correlated with intestinal integrity, and its relative abundance and absolute numbers are known to increase rapidly with age and specifically after weaning (446). Mouse experiments confirmed the effect of A. muciniphila on intestinal barrier function and demonstrated that its administration protects against diet-induced obesity (447). Recently, a mechanistic explanation involving TLR signaling via a specific pilus-associated protein has been provided (448, 449). These findings and the indications that A. muciniphila may ferment HMOs (450) make this species a relevant inhabitant in the early-life gut, even at low abundance, and a potential target for therapeutic developments aimed at increasing barrier function (451, 452).
MICROBIAL BIOMARKERS ASSOCIATED WITH INFANT HEALTH
General Features of Microbial Biomarkers
Alterations in the infant gut microbiota composition have been linked to multiple diseases and disorders (see above). Although the precise mechanisms and causalities of these associations still have to be uncovered, microbiota changes across various stages of disease/disorder might be utilized for novel diagnostic and prognostic tests, also known as microbial biomarkers. We categorize microbial markers in the following groups, which will be discussed further below: (i) microbiota maturation, (ii) microbial diversity, (iii) presence and abundance of bifidobacteria, (iv) presence and abundance of butyrogenic bacteria belonging to the Lachnospiraceae and Ruminococcaceae families, (v) concentration of SCFAs and pH, and (vi) microbial cell wall products.
Microbiota Maturation as a Marker for Health
Different aspects of the microbiota can be indicative of differences between healthy and disease states (453). Single microbial taxa or microbial products are preferable as health markers, since these make strategies for nutritional interventions or therapy most feasible. However, in many cases, if differences are found between a healthy microbiota and that of an ill person, the microbial biomarkers are not particularly specific. They are described as differences in community structure or differences in transition of the complete microbiota. Microbiota maturation has been defined as the rate at which the infant gut microbiota develops, as measured by age-dependent succession stages (2). A “mature” microbiota contains certain taxa that are biomarkers for the particular age group of the child/infant, while an “immature” or delayed microbiota resembles that of a younger child/infant (2). Delayed microbiota maturation is associated with physiological disturbances in the host. Typical factors that can hamper the process of microbiota maturation are premature birth as well as formula feeding, undernutrition, and antibiotic use (2, 73, 83, 454).
Microbial Diversity as a Biomarker for Health
In adults, microbial diversity is considered to represent a biomarker that can be associated with either health or disease. A decrease in microbial diversity, calculated as either Chao1, Simpson, Shannon, or phylogenetic diversity, is reported in states of disease. Notably, a decrease in metagenomic richness of the microbiota is described as a biomarker for metabolic syndrome, despite the fact that the sole evaluation of changes in biodiversity is not considered to be sufficient to reliably identify and confirm the presence of an ongoing pathological condition (455). In contrast, a healthy intestinal microbiota in early life is associated with a low microbial diversity (4, 269). Infants born preterm or by caesarean section show an even further reduced diversity compared to infants born vaginally at term. Milk typically makes up the complete nutrition of infants in the first months following birth. Depending on the milk source, either breast milk or formula, the microbiota is less or more diverse, respectively (456). HMOs in human breast milk render the microbiota low in diversity and richness and high in bifidobacteria (456). In this context, compared to that of a breastfed infant (of the same age), the microbiota of a formula-fed infant is usually higher in diversity and richness yet with lower abundance of bifidobacteria (73). During and after weaning, the changes that occur in microbial diversity are more pronounced in breastfed than in formula-fed infants (2). This is due to a rapid decrease of bifidobacteria, being replaced in relative abundance by Bacteroides spp. and members of the Lachnospiraceae and Ruminococcaceae families.
When the infant microbiota is exposed to antibiotics the diversity can drop to an even lower value compared to that of a reference non-antibiotic-treated, breastfed infant (2, 457). In this case, the low diversity and decrease of bifidobacteria are considered a biomarker for hampered microbiota development and risk for later-life health. The decline of bifidobacterial and Bacteroides sp. counts leads to higher relative abundances of enterobacteria and enterococci. The latter groups contain opportunistic pathogens and increase the risk of infections and disease (456). In prematurely born infants, microbial colonization is hampered, and low microbial richness and the abundance of facultative anaerobic species from the enterobacteria and enterococci are directly linked to an increased risk of NEC development (458).
In early life, low microbial diversity has been described to be correlated with colic and atopic dermatitis (456, 459). In infants suffering from colic, the increase of microbial diversity in the first weeks of life was less marked than that observed in healthy control subjects. This lower microbial diversity in colic infants continued across the first 2 years following birth and was accompanied by a significantly lower abundance of bifidobacteria and lactobacilli in such colic infants compared to healthy controls. The manifestation of colic symptoms is most pronounced in the first 6 weeks after birth; therefore, the reduced diversity and specific microbiota signature observed in infants with colic in the first weeks of life suggest a potential role of microbiota development in the etiology of colic (459).
Microbial diversity is a nonstable factor in early life as it gradually increases toward adulthood. Deviations in this transition, such as lower diversity at certain stages in this transition, might be a measure for risk of certain early-life disease. Specific microbial signatures accompanied with this diversity measure are important and are further discussed below.
Presence and Abundance of Bifidobacteria as a Biomarker for Health
Bifidobacteria colonize the intestine early in life, and bifidobacterial levels become lower yet remain relatively stable in adulthood, tending to decrease as a result of senescence. As discussed above, infants typically possess a microbiota dominated by bifidobacteria. When considering a vaginally delivered, breastfed infant as a healthy reference, formula-fed infants have a more diverse microbiota and infants born preterm or by caesarean section show a delayed colonization by bifidobacteria (83, 123). Bifidobacterial presence and abundance are described to be associated with a positive health status and can therefore be considered a potential microbial biomarker.
There are examples of early-life disease, such as colic and NEC, as well as later-life development of obesity, celiac disease, and autoimmune diseases, that are correlated with diminished levels of bifidobacteria in the infant intestine (460, 461). Malnutrition in children is typically characterized by lower abundances of B. longum and puts these children at increased risk for impaired learning ability and physical stunting in later life (454).
The use of antibiotics shifts the composition of the gut microbiota toward an increased abundance of Proteobacteria by further reducing Bifidobacterium populations. Antibiotic use in the first days of life delays colonization by bifidobacteria in the gut. Antibiotics are routinely administered perinatally during caesarean sections, which is a confounder in analyses where bifidobacteria have been shown to be transmitted vertically with vaginal but not caesarean delivery. The decreased amounts of bifidobacteria and increased amounts of specific Firmicutes, including different Clostridia class groups such as members of the families Ruminococcaceae and Lachnospiraceae, have been associated with the development and onset of allergic diseases (456). Furthermore, in celiac disease and childhood constipation, there is a reduced proportion of Bifidobacterium spp. (279, 462). At the same time, bifidobacteria have been suggested to alleviate gastrointestinal symptoms of adult celiac patients and have been associated with reduced abdominal pain and discomfort in healthy adults, thus opening novel opportunities for nutritional intervention to treat disease with low abundance of bifidobacteria.
The protective ability of bifidobacteria against early-life disease is suggested to work through specific immune stimulation and acidification of the intestinal environment through the production of SCFAs and lactate. It is unclear whether specific strains of bifidobacteria or even a diverse community of different bifidobacteria elicits different effects. The colonization of B. longum subsp. infantis has been associated with normal development of immune tolerance, and the species has been shown to be capable of normalizing the permeability of the intestinal mucosa (463, 464). B. animalis subsp. lactis has been shown to protect against infections in infants (465).
The presence of aberrant bifidobacterial numbers is one of the most frequently observed intestinal microbiota alterations (461) in early-life-associated diseases. This fact suggests an important role for the bifidobacterial population in establishing intestinal homeostasis. Bifidobacteria may therefore be used as a biomarker to assess the intestinal status with regard to a putative dysbiosis. In addition, increasing bifidobacterial levels in the gastrointestinal tract may be considered a target to prevent and/or alleviate microbiota-associated diseases.
The Lachnospiraceae and Ruminococcaceae Families as a Marker for Health
The commonly described microbial fermentation product butyrate is not typically the most abundant SCFA in the early-life intestine, where bifidobacteria dominate. Butyrate-producing bacteria that are present in the adult human gut have been proposed to reduce pain sensation, as pointed out by studies relying on butyrate enemas (466). Butyrate serves as a primary energy source for colonic epithelial cells, and it is also described as beneficial for gut health by causing, for example, decreased intestinal permeability. Signatures of butyrate-producing bacteria are, however, detectable in the infant microbiota and described as markers for health. Colonization by low numbers of butyrogenic microorganisms early in life is thought to help the fast transition of microbes during the process of weaning, which rapidly changes the bifidobacterium-dominated microbiota to a microbiota rich in Bacteroides spp. and Clostridium spp. In this context, it should be noted that a negative association between crying and butyrate-producing bacteria in infants with colic symptoms has been reported (459).
Acidity as a Result of Bifidobacteria and LAB in the Early-Life Intestine
Acetate is the most abundantly produced SCFA in the colons of adults, and its production is a common feature of most gut microbiota members (453). Compared to those in adults, the levels of fecal SCFAs in breastfed infants are characterized by higher relative proportions of acetate, lower proportions of propionate, and an almost complete absence of butyrate. Furthermore, lactate is more commonly detected in the feces of infants, while it is usually found at low levels (<5 mM) in healthy adults. Acetate and lactate are the precursors for butyrate production by certain microbiota members, and these organic acids are therefore rapidly converted in the adult intestine. The high levels of acetate and lactate in breastfed infants reflect the dominance of bifidobacteria and lactobacilli. As such, breastfed infants have a lower fecal pH (average of ∼5.8) than formula-fed infants (average of ∼7.1) (467). It has been suggested that the higher relative abundance of lactic acid in breastfed infants drives this pH difference. The low acidity in the early-life intestine is thought to play a beneficial role, as it is prohibitive to the colonization of pathogens (468).
Microbial Lipids and Proteins as Modulators of Gut Health
As described above, aspects of microbiota maturation, microbial diversity, and abundance of microbial taxa can be indicative of health status in early life. Such characteristics may be used for future diagnostic methods to identify predisease states and states of disease. Apart from the use of the microbiota as a tool, it can be exploited as a target. Stimulation of the right microbes and combinations of microorganisms in the early-life intestine can modulate infant health. The exact mechanisms by which microbiota maturation, diversity, or taxon abundance are able to modulate the onset and severity of various disease states are still lacking. Mechanistic insights into microbial functionality in host-microbe interaction processes are scarce. The role of SCFAs in the intestinal tract and their effects on intestinal health and physiology represent nice examples of the direct effects of the microbiota toward host-microbe interactions. Microbially produced lipids and proteins represent two other major biological compounds that are directly encountered by the host epithelial cells.
Clinicians commonly use the differences in the outer membrane compositions of Gram-negative microorganisms as a first tool to identify pathogenic isolates. The categorization of Gram-positive and Gram-negative bacteria is based on the expression of lipids on the cell surface of bacteria. Gram-positive microorganisms have a thicker peptidoglycan layer and a lower lipid content, whereas in Gram-negative bacteria the outer membrane contains a high lipopolysaccharide (LPS) content and only a thin layer peptidoglycan below it. In the host gut, epithelial cells have evolved separate mechanisms dedicated to sense the presence of these different components. Toll-like receptor 4 (TLR4) is specialized in monitoring LPS from microbiota members, while TLR2 is responsive toward peptidoglycan. LPS is a precursor for host inflammatory responses. This might be related to the fact that Gram-negative bacteria induce a more severe inflammatory response than Gram-positive bacteria (469). A higher relative abundance of Gram-negative bacteria is associated with several inflammatory and metabolic disease states in adults, despite the fact that certain Gram-negative taxa, such as members of the genus Bacteroides, have been shown to exert anti-inflammatory activities. In infants, a higher abundance of Gram-negative bacteria, such as those from the phylum Proteobacteria, is associated with colic symptoms, constipation, and atopic dermatitis (456, 459, 462).
Bifidobacterium spp. and Bacteroides spp. are typically the most abundant microorganisms in the infant microbiota. Both Bifidobacterium spp. and Bacteroides spp. have been reported to have anti-inflammatory properties. They are, for example, rich in TLR9 ligands; TLR9-mediated stimulation is known to both enhance epithelial integrity and direct immune responses, with the ability to direct the cellular and physical maturation of the developing immune system (470). Furthermore, polysaccharide A produced by Bacteroides has been shown to promote immunological tolerance through induction of regulatory T cells, causing suppression of an interleukin-17 (IL-17)-mediated response in mice (404). Bioactive factors secreted by B. longum subsp. infantis have been shown to induce the expression of tight-junction proteins, thus increasing gut barrier function. As described above, the exopolysaccharide of B. breve masks other surface antigens and allows such bacteria to remain immunologically “invisible” (398). The beneficial effects of Bifidobacterium spp. and Bacteroides spp. may be diminished in allergic subjects, who tend to carry reduced numbers of these microorganisms in their gut (456).
There is evidence, however, that species belonging to the Bifidobacterium or Bacteroides genus do matter in terms of immune regulation when present as part of the microbiota. In this context, B. longum subsp. infantis, for example, does not induce production of IL-10 by dendritic cells, while B. bifidum, B. longum subsp. longum, and B. pseudocatenulatum do activate dendritic cells to produce IL-10 (471).
As another example, recent findings indicate that LPS from Bacteroides dorei harbors tetra- and penta-acylated lipid A structures, as opposed to the hexa-acylated lipid A observed in E. coli (472). Children in countries with high susceptibility to autoimmunity more often harbor Bacteroides species in the microbiota that produce the immune-inhibitory LPS. It has been argued that the immune-inhibitory properties of Bacteroides LPS may prevent early education of the mucosal immune system and contribute to development of type 1 diabetes (472).
Altogether, these publications suggest that the host has established many processes to detect and respond to both commensal bacteria and pathogens by differentiating the responses toward the different microbial cell structures on their cell walls. Early in life, the developing immune system is trained to develop immunological tolerance. Examples of both immune stimulation and immune inhibition are reported as beneficial in this process during early life and in the potential effects on autoimmune diseases that may occur in later life. Before microbial cell wall products can be used reliably as biomarkers, more insights are needed to understand the differences between different members of the same genus within the microbiota.
ROLE OF THE INFANT GUT VIROME IN MODULATING THE INFANT GUT HOMEOSTASIS
General Features of the Gut Virome
The gut virome is defined as the portion of the intestinal microbiome representing viruses that target either eukaryotic host cells (eukaryotic virome), bacteria (bacterial virome), or archaea (archaeal virome). It also includes all virus-derived genetic elements which are found integrated in host chromosomes (prophages or endogenous viral elements) (473). It is estimated that the human gut is host to more than 1012 bacterial cells, which in turn are outnumbered by their infecting or associated viral counterparts (bacteriophages or phages) by an estimated ratio of 10:1. The predation of bacterial populations by these phages is believed to play an important role in shaping the bacterial community structure of the gut (Fig. 10) (474).
FIG 10.
Contribution of phages to gut microbiota development through human aging. Putative factors influencing the virome biodiversity from infant to adult are schematically represented as factors of the curve's formula. Phage and bacterial loads are schematically represented to express the concept that while the phage load decreases during aging, the gut microbial population increases in complexity and abundance. The number of bacterial or phage particles schematically represents the number of species and complexity of the population.
An aberrant gut microbiota in early life may cause an imbalance between resident gut bacteria, which in turn may result in incorrect immune system maturation with consequences in later life (60). In this regard, a better understanding of the interactions between the bacterial gut community and its associated virome is necessary in order to devise strategies to positively influence gut microbiota development in a newborn and/or restore an anomalous gut microbiota to a “normal” one.
Compared to bacterial microbiome investigations, the study of the virome is still in its infancy (27). Virome analysis has greatly benefited from modern culture-independent approaches, using next-generation sequencing technologies (e.g., shotgun metagenomics) combined with advanced bioinformatic tools (475). Detection of virus-associated sequences in metagenomic data sets has to overcome the challenge of detecting relationships between extremely divergent sequences, where the majority of genes (up to 90%) identified in viral metagenomic data sets have no known function (476, 477). However, as viral DNA represents only 2 to 5% of the total gut community DNA (478), metagenomic sequencing may not provide the desired depth of data for downstream analysis. Therefore, specific enrichment approaches have been developed to obtain data sets representing deep DNA sequencing (479).
A recent study has established that viruses targeting bacteria are found in abundance in the infant gut (Fig. 10) (480). A hypothesis of transkingdom interactions occurring between viruses and bacteria has been proposed, in which both parties are responsible for modulating their relative composition and impacting the health status of the host (27). This dynamic relationship is exemplified by their progression from early infancy to adulthood (Fig. 10). Immediately following birth and up to 2 to 3 years of age, the infant gut microbiota appears to be extremely dynamic, undergoing a process of rapid expansion and diversification to result in an adult-like microbiota. Very little is known about the factors that drive this early gut microbiota development; however, recent studies have indicated that the gut virome plays a role in this dynamic microbial (re)shaping process (480).
Similar to the bacterial microbiota, the virome appears to be highly dynamic during infant microbiota development, with the highest diversity of bacteriophages, especially Caudovirales (i.e., tailed phages), observed during the first months after birth (27, 480, 481). Subsequently, the bacteriophage virome (or phageome) undergoes a mechanism of contraction and loss of diversity, shifting toward a Microviridae-dominated composition (480, 481). The development and diversification of the gut microbiota appear to occur at the expense of the phage community (27). Interestingly, the aforementioned virome contraction occurs during the same period when the infant microbiota adopts an adult-like composition, which indicates that the resultant reduction in predator numbers facilitates the establishment of a diverse bacterial community in the gastrointestinal tract (27).
In contrast, the eukaryotic virome is observed to be low in diversity during the first days of life (average, 2.6 days), with an increase in richness over a period of 24 months in a study involving four sets of twins. This suggests that eukaryotic viruses are obtained primarily from environmental sources (480). Anelloviridae have repeatedly been identified as the most prevalent eukaryotic DNA virus in infant fecal samples (482). In a longitudinal study, an increase in prevalence of such viruses was observed in fecal samples from infants at 6 and 12 months, yet they were rarely detected before the age of 3 months. It was suggested that the expansion of the annellovirus complement is linked to the immunocompromised state of the infant following reduction of maternal antibodies (480). The virome composition in the gut can be influenced by several factors, among which geography and diet appear to have strongest influence. Interestingly, it has been shown that individuals who follow the same dietary habits have a tendency to harbor a similar virome, likely reflecting a diet-dependent microbiota, allowing the proliferation of phages infecting the more dominant members of this microbiota (483). This becomes even more relevant in the context of early infancy, a crucial period when host immune maturation and various metabolic developments take place.
Particularly relevant information on the virome community has been derived from the study of twins. Comparative analysis of twin-derived viromes has shown that the fecal viromes of corresponding twins are more similar to each other than to those of unrelated individuals, a phenomenon that appears to persist throughout life (477, 480, 481). A longitudinal study following monozygotic and dizygotic twins for 30 months reaffirmed that the viromes of twin pairs (regardless of zygosity) are more similar to each other than to any other group and are distinguishable from those of their mothers or nontwin siblings. Moreover, the viromes of unrelated twins of the same age were found to be more similar to each other than to those of any relative, suggesting that the age of the infant represents a relevant factor in virome composition (481).
Phage-associated sequences of the gut virome may also be detected as integrated genetic elements or prophages within the bacterial chromosome, and as such they constitute part of the bacterial mobilome. Furthermore, their persistence in the host chromosome and subsequent acquisition by lateral transfer between members of a bacterial community make them an important source and catalyst of intraspecies diversity (484).
Integration of temperate phage genomes into the chromosome of their bacterial host(s) may act as a modulating factor of the gut microbiota through lysogenic conversion. Such lysogenic conversion may impart phenotypic alterations on the host cells that provide a competitive advantage (e.g., in the form of phage resistance). For this reason, the carriage of prophages by bacteria may promote their dominance among other competing strains cohabiting the same niche. This phenomenon may explain the apparent lack of “kill-the-winner” phage-host dynamics in the human gut, as phage-resistant lysogenic hosts are allowed to persist (485). Through the analysis of 18 infant fecal samples collected over a time frame of 14 months, an inverse trend was observed between the abundance of a specific taxon (e.g., Bifidobacterium scardovii or B. longum) and bifidobacterium-associated phage (or bifidophage) DNA. This observation indicates that phages impact bifidobacterial establishment and prevalence in the gut and suggests that these phages may enter the lytic cycle, thereby modulating bifidobacterial colonization of the gut and their relative abundance in the gut (486).
These findings therefore support the notion that the virome plays a role in influencing the bacterial microbiome composition and establishment, necessitating the introduction of community-scale genetic approaches for further analyses (487). Therefore, future microbiota engineering methodologies may be aimed at restoring microbiota imbalances by in situ manipulation of bacterial communities using (selected) phages.
PROBIOTIC/PREBIOTIC THERAPIES AS EXTERNAL MODULATORS OF THE INFANT GUT MICROBIOTA
General Features
The recent identification of differences in gut microbiota composition between healthy and diseased individuals makes it a valuable tool that can be exploited as a support for diagnostics and treatment. Disruption of natural development of the infant microbiota may increase the neonate's risk of gastric, metabolic, and immune diseases. The best-known risk factors for differential development of the infant microbiota are caesarean section delivery, pre-/perinatal antibiotic use, and formula feeding. Different strategies are available, and those involving the use of orally supplied pre- and probiotics to influence and direct the microbiota development in these early stage of life are the most common. Another strategy is the transfer of the mother's vaginal and fecal microbiota immediately following birth.
While the literature on probiotic applications is promising, it is unclear how strain specific the beneficial effects are and if combinations of strains might be more efficient. It is also of great importance that safety is taken into account when probiotic trials are performed in infants, especially those infants who are at increased risk for infections, such as prematurely delivered and antibiotic-treated infants. A literature review of studies addressing the safety of probiotics concluded that “there is a lack of assessment and systematic reporting of adverse events in probiotic intervention studies, and interventions are poorly documented” (488). Future research should therefore focus on the different strains and combinations of strains, the timing of administration, safety, and whether or not these probiotics are more efficacious in conjunction with prebiotics.
Probiotic Therapy during Pregnancy or for Infant Nutrition
The fact that infants encounter their first microbiota during the birthing process has led to the development of strategies to modulate the maternal fecal and vaginal microbiota during pregnancy. Treatment of women during the second and third trimesters of pregnancy with L. rhamnosus resulted in maintenance of a vaginal microbiota free of pathogenic microorganisms and helped to maintain a low vaginal pH (489). Probiotic supplementation of the mother during and after pregnancy has also been shown to alter the infant's microbiota. In this context, L. rhamnosus, supplied to women during and after pregnancy, was shown to colonize the intestines of their infants and was correlated with an increase in the abundance of bifidobacteria in the infant gut (490, 491). Noticeably, treatment of mothers before delivery and subsequent supplementation of the infants after delivery with L. rhamnosus GG also positively correlated with the abundance of bifidobacteria and lactobacilli in the infant microbiota.
A culture-based study evaluated a synbiotic treatment of newborns with Lactobacillus plantarum and FOS and found that the synbiotic resulted in rapid microbial colonization of the infant gut. Infants even remained colonized several months after therapy was terminated (492). However, probiotic gut colonization has also been shown to result in lower levels of colonization or in transient colonization (109). The efficacy with which different probiotic microbes are capable of colonizing the gut may underlie this variability.
Preterm-delivered infants are at high risk for the development of gastric infections due to immune immaturity, antibiotic treatments, and delayed microbial colonization. Several publications show that oral probiotics, using either bifidobacteria, lactobacilli, Saccharomyces boulardii, or a combination of different bifidobacteria with Streptococcus thermophilus, reduce the risk of or prevent NEC in preterm-delivered infants (493–495).
Furthermore, the effects of probiotics on pediatric diseases/disorders include effects on allergies, obesity, gastrointestinal infections, or colic (496–498). As such, L. reuteri DSM 17938 has been evaluated for its effects on infantile colic. Average crying and fussing were significantly less in infants from the probiotic group than in infants in the placebo group (499–501). The potential of L. rhamnosus GG in alleviation of colic symptoms has also been reported (502, 503). In contrast, it was recently also shown that L. reuteri DSM 17938 or Lactobacillus salivarius CEC5713 is not effective in protecting newborns from colic (504, 505). While the impacts of these and other data are discussed elsewhere (505), these findings should help to design future clinical approaches for the development of specific therapies for colic prevention and alleviation.
In infants at risk for allergic diseases, the administration of B. longum BB536 and L. rhamnosus GG during the first 6 months of life was not shown to influence the overall composition of the gut microbiota, and the probiotic bacteria did not persist once administration was stopped (506). Based on a systematic literature review, there is evidence suggesting that probiotics can in some cases alleviate allergic symptoms but are usually not effective in modulating microbiota composition and overall are not sufficient for the treatment of allergic diseases in early life (507). However, combinations of bifidobacteria with Streptococcus thermophilus have been shown to be effective in the prevention and treatment of antibiotic-associated diarrhea in children (508).
Proposed mechanisms of probiotic action include enhanced epithelial barrier function, enhanced mucosal IgA responses, direct antagonism against pathogens, competitive exclusion of pathogens, prevention of apoptosis, production of anti-inflammatory cytokines, and downregulation of proinflammatory pathways (109). The exact mechanism of probiotics is probably strain dependent. The overall complexity of the microbiota and its interaction with the immune system make it rather hard to assess. It has been reported, however, that lactobacilli and bifidobacteria exert direct effects on intestinal epithelial barrier function by decreasing intestinal permeability and improving intestinal epithelial resistance (109).
Microbiota Modulation through Prebiotics
The knowledge that bifidobacteria are the main microbes benefitting from the prebiotic HMOs in breast milk has led to the administration of prebiotic bifidogenic fibers to pregnant women and infants. Nondigestible oligosaccharides that are added to formula have shown results similar to those of breastfeeding in reducing the colonic pH and increasing the production of SCFAs and lactate. Prebiotics were also shown to be effective in selectively increasing the abundance of bifidobacteria and lactobacilli in both pregnant women and formula-fed infants (509, 510). The consumption of infant formula containing prebiotics promotes the development of a neonatal gut microbiota similar to that of breastfed infants (108). Nevertheless, such studies evaluated the microbiota composition at the genus level, and thus species-specific effects of probiotics with respect to HMOs still need to be fully elucidated. The main prebiotics that are used in infant formula are GOS, FOS, and polydextrose (PDX). For both GOS and inulin-type fructans, there is strong clinical support that they are beneficial for digestive and immune health (511, 512).
In pregnant women, GOS/long-chain FOS (lcFOS) was shown to significantly increase fecal bifidobacterial levels, with potential benefits for the transmission of microbes to their infant host during birth (513). Feeding with infant formula containing either oligofructose/FOS, GOS/FOS, or standard formula indicated that the bacterial composition in the first two formulas better resembles that of breastfed infants (509, 514). Addition of GOS or a GOS/FOS mixture to infant formula has a positive effect on bifidobacterial abundance (509, 515–517). GOS alone was shown to increase lactobacilli (509, 516, 517). The proportion of bifidobacteria was reported to be higher in a prebiotic group treated with GOS/FOS. Notably, B. breve numbers were higher and those of C. difficile were lower in infants that were fed GOS than in those fed control formula (518). Even though prebiotic administration may cause the microbiota composition to be more similar to that of breastfed infants, this does not mean that these probiotics are also effective for host immune response regulation. In a recent study, the feeding of GOS-containing infant formula did not lead to changes in the incidence of infection or allergic manifestation even though the formula produced a definite prebiotic effect as evident from changes in microbiota composition, stool consistency, and stool frequency (518).
Supplementation of infants with the prebiotic PDX also caused an increased abundance of bifidobacteria (519). It is worth mentioning that the metabolic activity of the prebiotic-stimulated bifidobacteria is probably similar to that exerted by breast milk: a semisynthetic medium containing GOS showed a transcriptional effect similar to that of human milk for B. longum subsp. longum under in vitro conditions (520).
Finally, high-risk, prematurely born infants given FOS-supplemented formula were shown to have a positive response in terms of the (increased) numbers of bifidobacteria present in fecal samples and a corresponding significant reduction of E. coli and enterococci (521). Furthermore, results from a randomized controlled trial showed that oral synbiotics given to preterm babies alter the composition of their gut microbiota and decrease their risk of developing atopic diseases (522), while also reducing the levels of fussing and crying (502).
The use of prebiotics in infant formula is already a common practice in infant feeding systems. There is no clarity about functional differences between the effects of different types of prebiotics, the combination of different prebiotics, or even synbiotics. Usually, the effect of prebiotics is measured as an increase of bifidobacteria in the infant gut. However, information on the type of stimulated bifidobacteria and the direct effects on immune stimulation is still lacking. It is hoped that future research will provide more insights into the mechanisms of pro/prebiotics and their further use in infants.
Mode of Delivery Impacts Microbiota Transfer from Mother to Infant
As described above, the mode of delivery has a major impact on the composition of the microbiota of newborns. In particular, early-colonizing bacteria may commonly be acquired from the infant's mother, while late-colonizing bacteria may be the result of environmental contamination. In order to trace vertical transmission events, several in silico pipelines have recently been developed (45, 523). Vaginally delivered infants harbor bacterial communities resembling those of the maternal vagina, whereas caesarean section-delivered infants are enriched in skin microbiota (70). The deviation of microbiota development is associated with long-term effects on host metabolism and impaired immune development. Restoration of the microbiota development of infants born via caesarean section could be achieved by exposure of the neonate to maternal vaginal and fecal contents. Recent results suggest that the fecal, skin, and oral microbiotas of the exposed neonates more closely resemble those from vaginally born than from caesarean section-born babies (524). Although the long-term health consequences of restoring the microbiota of caesarean section-delivered infants remain unclear, these results demonstrate that vaginal microbes can be partially restored at birth in caesarean section-delivered neonates.
While the risks involved in this procedure should be similar to those during vaginal delivery, research is warranted to optimize the safety and mechanism of such exposure. Given the rapid development of the infant microbiome, early introduction of key founder populations may be crucial in facilitating a more natural microbial ecological succession and host immune and metabolic responses.
CONCLUSIONS
The gut microbiota represents a prime example of how an environment which is considered sterile, or at least poorly colonized, at birth is rapidly occupied by a plethora of microbial communities. Notably, this colonization appears to follow typical trajectories that are governed by stochastic processes and microbe-host coevolution forces. In this context, only specific microorganisms that are maternally inherited are, under natural circumstances, driving the establishment of the infant gut microbiota. The subsequent development of this early gut microbiota is then modulated by specific dietary compounds present in human milk that support selective colonization. It has been shown that the genomes of infant gut commensals, in particular bifidobacteria, are genetically adapted to utilize specific glycan components of this human secretory fluid (232). This represents a very intriguing example of host-microbe coevolution, where both partners are believed to benefit. There is growing evidence that such a mechanism, by which host products act as key agents for the modulation of the gut microbiota, thus acting as natural prebiotics, is a common scheme not limited to humans or other primates but extending to all species across the mammalian tree of life. Notably, other maternally linked forces responsible for the establishment of the very early infant gut microbiota include the mode of delivery, host genetics, gestational age, and maternal diet. There are intriguing indications that the infant gut is not always sterile at delivery and that fetal colonization might sometimes occur, with a concurrent transfer of maternal microbiota to the fetus during pregnancy. In this context, the early stages of life represent a more opportunistic period of human life where the gut microbiota may be more prone to changes by interventions involving probiotics, prebiotics, phages, or combinations of these.
In recent years, a substantial number of publications in the field of microbiology have focused on dissecting microbial (infant) gut communities and their interaction with their human host, being a determining factor in host physiology and metabolic activities. Such studies have highlighted a reduction of microbial diversity and/or an aberrant microbiota composition, also known as dysbiosis, both of which have been causally linked to a number of intestinal diseases in infants, such as NEC, or diseases that manifest themselves at later stages of life, including chronic diseases such as IBD as well as metabolic disorders such as obesity or IBS. Thus, there is preliminary data that suggest that the early human gut microbiota influences risk factors related to both childhood and adult health conditions. However, clinical data about the long-term effects of early dysbiosis in the development of IBD in children and adults are very scarce, most likely because such analyses will require clinical data collected during very long periods of time and ideally during the entire human life span. Thus, future or-already ongoing works (525) involving both animal models and human epidemiological studies must shed light on microbiota-mediated immune and physiological responses that may cause the onset of IBD.
We are only beginning to understand this early-life interaction, which may already be initiated during gestation (206, 526), and its long-term effects on health. Therefore, it may be time to put forward alternatives for favoring the correct development of the microbiota in those cases in which, for various reasons, this process is challenged.
This concept has fueled the development of various strategies based on various nutraceutical products (e.g., probiotics and/or prebiotics) to shape the infant microbiota. This intervention theory is linked to an appropriate evaluation of the composition of the infant gut microbiota and the identification of shifts in the abundance of those microorganisms considered to have a crucial role in the establishment of diseases, i.e., the microbial biomarkers. The identification of microbial biomarkers is very challenging for future prophylactic approaches as well as for early diagnosis of diseases. Currently, substantial efforts have been made to investigate the dominant members of the infant gut microbiota, e.g., bifidobacteria, and to understand their impact on the physiology of the infant gut as well as in the priming of the immune system and the metabolism of the host. Nevertheless, the level of understanding of their interactive behavior with the host as well as with other members of the gut microbiota is still very preliminary and largely restricted to a few species and strains. However, in contrast to the case for other infant gut commensals, in the case of bifidobacteria, some effort is being made beyond the cataloguing of OTUs in order to understand the individuality of strains and species and their impact on the microbiota and their associated host. Notably, the contribution of bifidobacteria to the overall gut microbiota composition displays regional differences much more than other microorganisms typically found in the infant gut. In this context, the presence of bifidobacteria is markedly reduced in the offspring ecosystem of industrialized areas (2). In contrast, surveys of the infant gut microbiota composition from babies born in Africa, as well as Asia, showed a higher occurrence of bifidobacteria (125, 527, 528).
Such findings are also complemented by early studies performed in industrialized countries of Northern Europe, displaying higher a abundance of bifidobacteria in the infant gut (529–531). Thus, comparing data related to bifidobacterial occurrence in infants described in older literature with those in recent publications, one may argue that bifidobacteria represent one of the “missing microbes,” i.e., bacterial taxa that have been lost or are now present in low abundance. This apparent process of extinction of bifidobacteria may have been due to modifications of the human diet, including the widespread use of formula milk instead of breast milk, yet may also be due to or compounded by other factors, such as the use of antibiotics, increased hygiene, and C-sections.
As described in this review, bifidobacteria represent the dominant norm for the infant gut microbiota, and their use as health-promoting microorganisms for infants is already exploited by the food industry. However, most of the probiotic bifidobacterial strains do not fulfill any scientific criteria in terms of ecological origin, i.e., autochthonous origin, as well as knowledge of the molecular basis of their health-promoting effects (532).
Future probiotic interventions directed to prevent and/or counteract gut microbiota dysbiosis may necessarily involve “next-generation” probiotic bacteria, which may include “classical” probiotic bacteria belonging to the Bifidobacterium or the Lactobacillus genus yet may also consist of other microbial groups of the infant core gut microbiota.
It is expected that investigations of the infant gut microbiota will be central to understand human health and disease. Novel experimental approaches available in the coming years will no doubt lead to pivotal discoveries that can be applied in functional food and nutraceutical industries or in medical settings.
ACKNOWLEDGMENTS
This work was funded by the EU Joint Programming Initiative “A Healthy Diet for a Healthy Life” (JPI HDHL, http://www.healthydietforhealthylife.eu/) and MIUR to M.V. and by the Spanish MINECO to M.G. (project PCIN-2015-233). A.M. and S.D.P. are partially funded by grants from the Instituto Danone and by the Spanish Plan Estatal de I+D+I (reference AGL2016-78311-R). D.V.S. is a member of the APC Microbiome Institute funded by Science Foundation Ireland (SFI) through the Irish Government's National Development Plan (grant number SFI/12/RC/2273). J.M.R. is in receipt of AGL2016-75476-R (Ministerio de Economía y Competitividad (Spain). J.M. is in receipt of a Starting Investigator Research Grant (SIRG) (no. 15/SIRG/3430) funded by Science Foundation Ireland (SFI).
Part of this research was conducted using the High Performance Computing (HPC) facility of the University of Parma.
We declare that we have no competing interests.
REFERENCES
- 1.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. 2012. Diversity, stability and resilience of the human gut microbiota. Nature 489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, Lieber AD, Wu F, Perez-Perez GI, Chen Y, Schweizer W, Zheng X, Contreras M, Dominguez-Bello MG, Blaser MJ. 2016. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med 8: 343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yassour M, Vatanen T, Siljander H, Hamalainen AM, Harkonen T, Ryhanen SJ, Franzosa EA, Vlamakis H, Huttenhower C, Gevers D, Lander ES, Knip M, DABIMMUNE Study Group, Xavier RJ. 2016. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci Transl Med 8: 343ra81. doi: 10.1126/scitranslmed.aad0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. 2012. Human gut microbiome viewed across age and geography. Nature 486:222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ventura M, O'Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O'Toole PW. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol 7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- 6.Ventura M, Turroni F, Canchaya C, Vaughan EE, O'Toole PW, van Sinderen D. 2009. Microbial diversity in the human intestine and novel insights from metagenomics. Front Biosci 14:3214–3221. doi: 10.2741/3445. [DOI] [PubMed] [Google Scholar]
- 7.Ursell LK, Clemente JC, Rideout JR, Gevers D, Caporaso JG, Knight R. 2012. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol 129:1204–1208. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. 2015. Cesarean section and chronic immune disorders. Obstet Gynecol Surv 70:303–305. doi: 10.1097/01.ogx.0000466336.81671.9f. [DOI] [PubMed] [Google Scholar]
- 9.Huh SY, Rifas-Shiman SL, Zera CA, Edwards JWR, Oken E, Weiss ST, Gillman MW. 2012. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Arch Dis Child 97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eggesbo M, Botten G, Stigum H, Nafstad P, Magnus P. 2003. Is delivery by cesarean section a risk factor for food allergy? J Allergy Clin Immunol 112:420–426. doi: 10.1067/mai.2003.1610. [DOI] [PubMed] [Google Scholar]
- 11.Relman DA. 2012. The human microbiome: ecosystem resilience and health. Nutr Rev 70(Suppl 1):S2–S9. doi: 10.1111/j.1753-4887.2012.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tamboli CP, Neut C, Desreumaux P, Colombel JF. 2004. Dysbiosis in inflammatory bowel disease. Gut 53:1–4. doi: 10.1136/gut.53.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spor A, Koren O, Ley R. 2011. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol 9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 14.Kim A. 2015. Dysbiosis: a review highlighting obesity and inflammatory bowel disease. J Clin Gastroenterol 49(Suppl 1):S20–S4. doi: 10.1097/MCG.0000000000000356. [DOI] [PubMed] [Google Scholar]
- 15.Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. 2016. The role of Gut microbiota in the development of obesity and diabetes. Lipids Health Dis 15:108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuoka K, Kanai T. 2015. The gut microbiota and inflammatory bowel disease. Semin Immunopathol 37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancabelli L, Milani C, Lugli GA, Turroni F, Mangifesta M, Viappiani A, Ticinesi A, Nouvenne A, Meschi T, van Sinderen D, Ventura M. 2017. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci Rep 7:9879. doi: 10.1038/s41598-017-10663-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vellend M. 2010. Conceptual synthesis in community ecology. Q Rev Biol 85:183–206. doi: 10.1086/652373. [DOI] [PubMed] [Google Scholar]
- 19.Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smillie CS, Smith MB, Friedman J, Cordero OX, David LA, Alm EJ. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 21.Kerr B, Riley MA, Feldman MW, Bohannan BJ. 2002. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature 418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 22.Bordenstein SR, Theis KR. 2015. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol 13:e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bosch TCG, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology 114:185–190. doi: 10.1016/j.zool.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert SF, Sapp J, Tauber AI. 2012. A symbiotic view of life: we have never been individuals. Q Rev Biol 87:325–341. doi: 10.1086/668166. [DOI] [PubMed] [Google Scholar]
- 25.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg E, Sharon G, Atad I, Zilber-Rosenberg I. 2010. The evolution of animals and plants via symbiosis with microorganisms. Environ Microbiol Rep 2:500–506. doi: 10.1111/j.1758-2229.2010.00177.x. [DOI] [PubMed] [Google Scholar]
- 27.Lim ES, Wang D, Holtz LR. 2016. The bacterial microbiome and virome milestones of infant development. Trends Microbiol 24:801–810. doi: 10.1016/j.tim.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Clemente JC, Ursell LK, Parfrey LW, Knight R. 2012. The impact of the gut microbiota on human health: an integrative view. Cell 148:1258–1270. doi: 10.1016/j.cell.2012.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rautava S, Luoto R, Salminen S, Isolauri E. 2012. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol 9:565–576. doi: 10.1038/nrgastro.2012.144. [DOI] [PubMed] [Google Scholar]
- 30.Biteen JS, Blainey PC, Cardon ZG, Chun MY, Church GM, Dorrestein PC, Fraser SE, Gilbert JA, Jansson JK, Knight R, Miller JF, Ozcan A, Prather KA, Quake SR, Ruby EG, Silver PA, Taha S, van den Engh G, Weiss PS, Wong GCL, Wright AT, Young TD. 2016. Tools for the microbiome: nano and beyond. ACS Nano 10:6–37. doi: 10.1021/acsnano.5b07826. [DOI] [PubMed] [Google Scholar]
- 31.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. 2016. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rajilic-Stojanovic M, de Vos WM. 2014. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 38:996–1047. doi: 10.1111/1574-6976.12075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamady M, Knight R. 2009. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res 19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarridge JE. 2004. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 17:840-+. doi: 10.1128/CMR.17.4.840-862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilbert JA, Dupont CL. 2011. Microbial metagenomics: beyond the genome. Annu Rev Mar Sci 3:347–371. doi: 10.1146/annurev-marine-120709-142811. [DOI] [PubMed] [Google Scholar]
- 38.Neefs JM, Vandepeer Y, Derijk P, Chapelle S, Dewachter R. 1993. Compilation of small ribosomal-subunit RNA structures. Nucleic Acids Res 21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sundquist A, Bigdeli S, Jalili R, Druzin ML, Waller S, Pullen KM, El-Sayed YY, Taslimi MM, Batzoglou S, Ronaghi M. 2007. Bacterial flora-typing with targeted, chip-based pyrosequencing. BMC Microbiol 7:108. doi: 10.1186/1471-2180-7-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Claesson MJ, Wang QO, O'Sullivan O, Greene-Diniz R, Cole JR, Ross RP, O'Toole PW. 2010. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res 38:e200. doi: 10.1093/nar/gkq873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. doi: 10.1371/journal.pone.0036957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Milani C, Hevia A, Foroni E, Duranti S, Turroni F, Lugli GA, Sanchez B, Martin R, Gueimonde M, van Sinderen D, Margolles A, Ventura M. 2013. Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS One 8:e68739. doi: 10.1371/journal.pone.0068739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu WT, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 44.Chakravorty S, Helb D, Burday M, Connell N, Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Milani C, Lugli GA, Turroni F, Mancabelli L, Duranti S, Viappiani A, Mangifesta M, Segata N, van Sinderen D, Ventura M. 2014. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol Ecol 90:493–503. doi: 10.1111/1574-6941.12410. [DOI] [PubMed] [Google Scholar]
- 46.Milani C, Turroni F, Duranti S, Lugli GA, Mancabelli L, Ferrario C, van Sinderen D, Ventura M. 2015. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ Microbiol 82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dean FB, Nelson JR, Giesler TL, Lasken RS. 2001. Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res 11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McLean JS, Lombardo MJ, Badger JH, Edlund A, Novotny M, Yee-Greenbaum J, Vyahhi N, Hall AP, Yang Y, Dupont CL, Ziegler MG, Chitsaz H, Allen AE, Yooseph S, Tesler G, Pevzner PA, Friedman RM, Nealson KH, Venter JC, Lasken RS. 2013. Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum. Proc Natl Acad Sci U S A 110:E2390-9. doi: 10.1073/pnas.1219809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fodor AA, DeSantis TZ, Wylie KM, Badger JH, Ye Y, Hepburn T, Hu P, Sodergren E, Liolios K, Huot-Creasy H, Birren BW, Earl AM. 2012. The “most wanted” taxa from the human microbiome for whole genome sequencing. PLoS One 7:e41294. doi: 10.1371/journal.pone.0041294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci U S A 104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lugli GA, Milani C, Mancabelli L, Turroni F, Ferrario C, Duranti S, van Sinderen D, Ventura M. 2017. Ancient bacteria of the Otzi's microbiome: a genomic tale from the Copper Age. Microbiome 5:5. doi: 10.1186/s40168-016-0221-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hugon P, Dufour JC, Colson P, Fournier PE, Sallah K, Raoult D. 2015. A comprehensive repertoire of prokaryotic species identified in human beings. Lancet Infect Dis 15:1211–1219. doi: 10.1016/S1473-3099(15)00293-5. [DOI] [PubMed] [Google Scholar]
- 55.Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, Gordon JI. 2011. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A 108:6252–6257. doi: 10.1073/pnas.1102938108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Walker AW, Duncan SH, Louis P, Flint HJ. 2014. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol 22:267–274. doi: 10.1016/j.tim.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 57.Lagier JC, Khelaifia S, Alou MT, Ndongo S, Dione N, Hugon P, Caputo A, Cadoret F, Traore SI, Seck EH, Dubourg G, Durand G, Mourembou G, Guilhot E, Togo A, Bellali S, Bachar D, Cassir N, Bittar F, Delerce J, Mailhe M, Ricaboni D, Bilen M, Dangui Nieko NP, Dia Badiane NM, Valles C, Mouelhi D, Diop K, Million M, Musso D, Abrahao J, Azhar EI, Bibi F, Yasir M, Diallo A, Sokhna C, Djossou F, Vitton V, Robert C, Rolain JM, La Scola B, Fournier PE, Levasseur A, Raoult D. 2016. Culture of previously uncultured members of the human gut microbiota by culturomics. Nat Microbiol 1:16203. doi: 10.1038/nmicrobiol.2016.203. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, Bittar F, Fournous G, Gimenez G, Maraninchi M, Trape JF, Koonin EV, La Scola B, Raoult D. 2012. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect 18:1185–1193. doi: 10.1111/1469-0691.12023. [DOI] [PubMed] [Google Scholar]
- 59.Browne HP, Forster SC, Anonye BO, Kumar N, Neville BA, Stares MD, Goulding D, Lawley TD. 2016. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature 533:543. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez JM, Murphy K, Stanton C, Ross RP, Kober OI, Juge N, Avershina E, Rudi K, Narbad A, Jenmalm MC, Marchesi JR, Collado MC. 2015. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb Ecol Health Dis 26:26050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jimenez E, Fernandez L, Marin ML, Martin R, Odriozola JM, Nueno-Palop C, Narbad A, Olivares M, Xaus J, Rodriguez JM. 2005. Isolation of commensal bacteria from umbilical cord blood of healthy neonates born by cesarean section. Curr Microbiol 51:270–274. doi: 10.1007/s00284-005-0020-3. [DOI] [PubMed] [Google Scholar]
- 62.DiGiulio DB, Romero R, Amogan HP, Kusanovic JP, Bik EM, Gotsch F, Kim CJ, Erez O, Edwin S, Relman DA. 2008. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: a molecular and culture-based investigation. PLoS One 3:e3056. doi: 10.1371/journal.pone.0003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. 2014. The placenta harbors a unique microbiome. Sci Transl Med 6: 237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lauder AP, Roche AM, Sherrill-Mix S, Bailey A, Laughlin AL, Bittinger K, Leite R, Elovitz MA, Parry S, Bushman FD. 2016. Comparison of placenta samples with contamination controls does not provide evidence for a distinct placenta microbiota. Microbiome 4:29. doi: 10.1186/s40168-016-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, Turner P, Parkhill J, Loman NJ, Walker AW. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perez-Munoz ME, Arrieta MC, Ramer-Tait AE, Walter J. 2017. A critical assessment of the “sterile womb” and “in utero colonization” hypotheses: implications for research on the pioneer infant microbiome. Microbiome 5:48. doi: 10.1186/s40168-017-0268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Avershina E, Lundgard K, Sekelja M, Dotterud C, Storro O, Oien T, Johnsen R, Rudi K. 2016. Transition from infant- to adult-like gut microbiota. Environ Microbiol 18:2226–2236. doi: 10.1111/1462-2920.13248. [DOI] [PubMed] [Google Scholar]
- 68.Avershina E, Storro O, Oien T, Johnsen R, Wilson R, Egeland T, Rudi K. 2013. Bifidobacterial succession and correlation networks in a large unselected cohort of mothers and their children. Appl Environ Microbiol 79:497–507. doi: 10.1128/AEM.02359-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Munyaka PM, Khafipour E, Ghia JE. 2014. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front Pediatr 2:109. doi: 10.3389/fped.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. 2010. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A 107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Biasucci G, Rubini M, Riboni S, Morelli L, Bessi E, Retetangos C. 2010. Mode of delivery affects the bacterial community in the newborn gut. Early Hum Dev 86(Suppl 1):S13–S15. doi: 10.1016/j.earlhumdev.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 72.Fouhy F, Ross RP, Fitzgerald GF, Stanton C, Cotter PD. 2012. Composition of the early intestinal microbiota: knowledge, knowledge gaps and the use of high-throughput sequencing to address these gaps. Gut Microbes 3:203–220. doi: 10.4161/gmic.20169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Backhed F, Roswall J, Peng Y, Feng Q, Jia H, Kovatcheva-Datchary P, Li Y, Xia Y, Xie H, Zhong H, Khan MT, Zhang J, Li J, Xiao L, Al-Aama J, Zhang D, Lee YS, Kotowska D, Colding C, Tremaroli V, Yin Y, Bergman S, Xu X, Madsen L, Kristiansen K, Dahlgren J, Wang J. 2015. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 74.Del Chierico F, Vernocchi P, Petrucca A, Paci P, Fuentes S, Pratico G, Capuani G, Masotti A, Reddel S, Russo A, Vallone C, Salvatori G, Buffone E, Signore F, Rigon G, Dotta A, Miccheli A, de Vos WM, Dallapiccola B, Putignani L. 2015. Phylogenetic and metabolic tracking of gut microbiota during perinatal development. PLoS One 10:e0137347. doi: 10.1371/journal.pone.0137347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Adlerberth I, Strachan DP, Matricardi PM, Ahrne S, Orfei L, Aberg N, Perkin MR, Tripodi S, Hesselmar B, Saalman R, Coates AR, Bonanno CL, Panetta V, Wold AE. 2007. Gut microbiota and development of atopic eczema in 3 European birth cohorts. J Allergy Clin Immunol 120:343–350. doi: 10.1016/j.jaci.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 76.Jakobsson HE, Abrahamsson TR, Jenmalm MC, Harris K, Quince C, Jernberg C, Bjorksten B, Engstrand L, Andersson AF. 2014. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 63:559–566. doi: 10.1136/gutjnl-2012-303249. [DOI] [PubMed] [Google Scholar]
- 77.Neu J, Rushing J. 2011. Cesarean versus vaginal delivery: long-term infant outcomes and the hygiene hypothesis. Clin Perinatol 38:321–331. doi: 10.1016/j.clp.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, van den Brandt PA, Stobberingh EE. 2006. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 79.Hill CJ, Lynch DB, Murphy K, Ulaszewska M, Jeffery IB, O'Shea CA, Watkins C, Dempsey E, Mattivi F, Touhy K, Ross RP, Ryan CA, O'Toole PW, Stanton C. 2017. Evolution of gut microbiota composition from birth to 24 weeks in the INFANTMET cohort. Microbiome 5:4. doi: 10.1186/s40168-016-0213-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Martin R, Makino H, Cetinyurek Yavuz A, Ben-Amor K, Roelofs M, Ishikawa E, Kubota H, Swinkels S, Sakai T, Oishi K, Kushiro A, Knol J. 2016. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One 11:e0158498. doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salminen S, Gibson GR, McCartney AL, Isolauri E. 2004. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 53:1388–1389. doi: 10.1136/gut.2004.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, Kurilshikov A, Bonder MJ, Valles-Colomer M, Vandeputte D, Tito RY, Chaffron S, Rymenans L, Verspecht C, De Sutter L, Lima-Mendez G, D'Hoe K, Jonckheere K, Homola D, Garcia R, Tigchelaar EF, Eeckhaudt L, Fu J, Henckaerts L, Zhernakova A, Wijmenga C, Raes J. 2016. Population-level analysis of gut microbiome variation. Science 352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 83.Chu DM, Ma J, Prince AL, Antony KM, Seferovic MD, Aagaard KM. 23 January 2017. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malamitsi-Puchner A, Protonotariou E, Boutsikou T, Makrakis E, Sarandakou A, Creatsas G. 2005. The influence of the mode of delivery on circulating cytokine concentrations in the perinatal period. Early Hum Dev 81:387–392. doi: 10.1016/j.earlhumdev.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 85.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. 2008. A meta-analysis of the association between Caesarean section and childhood asthma. Clin Exp Allergy 38:629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 86.Bager P, Wohlfahrt J, Westergaard T. 2008. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy 38:634–642. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 87.Cardwell CR, Stene LC, Joner G, Cinek O, Svensson J, Goldacre MJ, Parslow RC, Pozzilli P, Brigis G, Stoyanov D, Urbonaite B, Sipetic S, Schober E, Ionescu-Tirgoviste C, Devoti G, de Beaufort CE, Buschard K, Patterson CC. 2008. Caesarean section is associated with an increased risk of childhood-onset type 1 diabetes mellitus: a meta-analysis of observational studies. Diabetologia 51:726–735. doi: 10.1007/s00125-008-0941-z. [DOI] [PubMed] [Google Scholar]
- 88.Pei Z, Heinrich J, Fuertes E, Flexeder C, Hoffmann B, Lehmann I, Schaaf B, von Berg A, Koletzko S, Influences of Lifestyle-Related Factors on the Immune System and the Development of Allergies in Childhood plus Air Pollution and Genetics (LISAplus) Study Group. 2014. Cesarean delivery and risk of childhood obesity. J Pediatr 164:1068–1073.e2. doi: 10.1016/j.jpeds.2013.12.044. [DOI] [PubMed] [Google Scholar]
- 89.Ruiz L, Moles L, Gueimonde M, Rodriguez JM. 2016. Perinatal microbiomes' influence on preterm birth and preterms' health: influencing factors and modulation strategies. J Pediatr Gastroenterol Nutr 63:e193–e203. doi: 10.1097/MPG.0000000000001196. [DOI] [PubMed] [Google Scholar]
- 90.Rouge C, Goldenberg O, Ferraris L, Berger B, Rochat F, Legrand A, Gobel UB, Vodovar M, Voyer M, Roze JC, Darmaun D, Piloquet H, Butel MJ, de La Cochetiere MF. 2010. Investigation of the intestinal microbiota in preterm infants using different methods. Anaerobe 16:362–370. doi: 10.1016/j.anaerobe.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 91.Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E, Picaud JC. 2011. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr 158:390–396. doi: 10.1016/j.jpeds.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 92.Arboleya S, Binetti A, Salazar N, Fernandez N, Solis G, Hernandez-Barranco A, Margolles A, de los Reyes-Gavilan CG, Gueimonde M. 2012. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiology Ecology 79:763–772. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 93.Arboleya S, Sanchez B, Milani C, Duranti S, Solis G, Fernandez N, de los Reyes-Gavilan CG, Ventura M, Margolles A, Gueimonde M. 2015. Intestinal microbiota development in preterm neonates and effect of perinatal antibiotics. J Pediatr 166:538–544. doi: 10.1016/j.jpeds.2014.09.041. [DOI] [PubMed] [Google Scholar]
- 94.Cong X, Xu W, Janton S, Henderson WA, Matson A, McGrath JM, Maas K, Graf J. 2016. Gut microbiome developmental patterns in early life of preterm infants: impacts of feeding and gender. PLoS One 11:e0152751. doi: 10.1371/journal.pone.0152751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aujoulat F, Roudiere L, Picaud JC, Jacquot A, Filleron A, Neveu D, Baum TP, Marchandin H, Jumas-Bilak E. 2014. Temporal dynamics of the very premature infant gut dominant microbiota. BMC Microbiol 14:325. doi: 10.1186/s12866-014-0325-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.La Rosa PS, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, Stevens HJ, Bennett WE Jr, Shaikh N, Linneman LA, Hoffmann JA, Hamvas A, Deych E, Shands BA, Shannon WD, Tarr PI. 2014. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U S A 111:12522–12527. doi: 10.1073/pnas.1409497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Groer MW, Gregory KE, Louis-Jacques A, Thibeau S, Walker WA. 2015. The very low birth weight infant microbiome and childhood health. Birth Defects Res C Embryo Today 105:252–264. doi: 10.1002/bdrc.21115. [DOI] [PubMed] [Google Scholar]
- 98.Collado MC, Cernada M, Neu J, Perez-Martinez G, Gormaz M, Vento M. 2015. Factors influencing gastrointestinal tract and microbiota immune interaction in preterm infants. Pediatr Res 77:726–731. doi: 10.1038/pr.2015.54. [DOI] [PubMed] [Google Scholar]
- 99.Torrazza RM, Neu J. 2013. The altered gut microbiome and necrotizing enterocolitis. Clin Perinatol 40:93–108. doi: 10.1016/j.clp.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Madan JC, Salari RC, Saxena D, Davidson L, O'Toole GA, Moore JH, Sogin ML, Foster JA, Edwards WH, Palumbo P, Hibberd PL. 2012. Gut microbial colonisation in premature neonates predicts neonatal sepsis. Arch Dis Child Fetal Neonatal Ed 97:F456–F462. doi: 10.1136/fetalneonatal-2011-301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mai V, Torrazza RM, Ukhanova M, Wang X, Sun Y, Li N, Shuster J, Sharma R, Hudak ML, Neu J. 2013. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS One 8:e52876. doi: 10.1371/journal.pone.0052876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cernada M, Bauerl C, Serna E, Collado MC, Martinez GP, Vento M. 2016. Sepsis in preterm infants causes alterations in mucosal gene expression and microbiota profiles compared to non-septic twins. Sci Rep 6:25497. doi: 10.1038/srep25497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Arboleya S, Sanchez B, Solis G, Fernandez N, Suarez M, Hernandez-Barranco AM, Milani C, Margolles A, de Los Reyes-Gavilan CG, Ventura M, Gueimonde M. 2016. Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: a functional inference study. Int J Mol Sci 17:649. doi: 10.3390/ijms17050649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.O'Sullivan A, Farver M, Smilowitz JT. 2015. The influence of early infant-feeding practices on the intestinal microbiome and body composition in infants. Nutr Metab Insights 8:1–9. doi: 10.4137/NMI.S29530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maynard CL, Elson CO, Hatton RD, Weaver CT. 2012. Reciprocal interactions of the intestinal microbiota and immune system. Nature 489:231–241. doi: 10.1038/nature11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bezirtzoglou E, Tsiotsias A, Welling GW. 2011. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH). Anaerobe 17:478–482. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 107.Praveen P, Jordan F, Priami C, Morine MJ. 2015. The role of breast-feeding in infant immune system: a systems perspective on the intestinal microbiome. Microbiome 3:41. doi: 10.1186/s40168-015-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guaraldi F, Salvatori G. 2012. Effect of breast and formula feeding on gut microbiota shaping in newborns. Front Cell Infect Microbiol 2:94. doi: 10.3389/fcimb.2012.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gritz EC, Bhandari V. 2015. The human neonatal gut microbiome: a brief review. Front Pediatr 3:17. doi: 10.3389/fped.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Harmsen HJ, Wildeboer-Veloo AC, Raangs GC, Wagendorp AA, Klijn N, Bindels JG, Welling GW. 2000. Analysis of intestinal flora development in breast-fed and formula-fed infants by using molecular identification and detection methods. J Pediatr Gastroenterol Nutr 30:61–67. doi: 10.1097/00005176-200001000-00019. [DOI] [PubMed] [Google Scholar]
- 111.Le Huerou-Luron I, Blat S, Boudry G. 2010. Breast- v. formula-feeding: impacts on the digestive tract and immediate and long-term health effects. Nutr Res Rev 23:23–36. doi: 10.1017/S0954422410000065. [DOI] [PubMed] [Google Scholar]
- 112.Fallani M, Amarri S, Uusijarvi A, Adam R, Khanna S, Aguilera M, Gil A, Vieites JM, Norin E, Young D, Scott JA, Dore J, Edwards CA, INFABIO Team 2011. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology 157:1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 113.Koenig JE, Spor A, Scalfone N, Fricker AD, Stombaugh J, Knight R, Angenent LT, Ley RE. 2011. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A 108(Suppl 1):S4578–S4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Valles Y, Artacho A, Pascual-Garcia A, Ferrus ML, Gosalbes MJ, Abellan JJ, Francino MP. 2014. Microbial succession in the gut: directional trends of taxonomic and functional change in a birth cohort of Spanish infants. PLoS Genet 10:e1004406. doi: 10.1371/journal.pgen.1004406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Laursen MF, Andersen LB, Michaelsen KF, Molgaard C, Trolle E, Bahl MI, Licht TR. 2016. Infant gut microbiota development is driven by transition to family foods independent of maternal obesity. mSphere 1:e00069-15. doi: 10.1128/mSphere.00069-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bergstrom A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, Molgaard C, Michaelsen KF, Licht TR. 2014. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol 80:2889–2900. doi: 10.1128/AEM.00342-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nuriel-Ohayon M, Neuman H, Koren O. 2016. Microbial changes during pregnancy, birth, and infancy. Front Microbiol 7:1031. doi: 10.3389/fmicb.2016.01031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Levin AM, Sitarik AR, Havstad SL, Fujimura KE, Wegienka G, Cassidy-Bushrow AE, Kim H, Zoratti EM, Lukacs NW, Boushey HA, Ownby DR, Lynch SV, Johnson CC. 2016. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci Rep 6:31775. doi: 10.1038/srep31775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Davis MY, Zhang H, Brannan LE, Carman RJ, Boone JH. 2016. Rapid change of fecal microbiome and disappearance of Clostridium difficile in a colonized infant after transition from breast milk to cow milk. Microbiome 4:53. doi: 10.1186/s40168-016-0198-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Collado MC, Isolauri E, Laitinen K, Salminen S. 2010. Effect of mother's weight on infant's microbiota acquisition, composition, and activity during early infancy: a prospective follow-up study initiated in early pregnancy. Am J Clin Nutr 92:1023–1030. doi: 10.3945/ajcn.2010.29877. [DOI] [PubMed] [Google Scholar]
- 121.Chu DM, Antony KM, Ma J, Prince AL, Showalter L, Moller M, Aagaard KM. 2016. The early infant gut microbiome varies in association with a maternal high-fat diet. Genome Med 8:77. doi: 10.1186/s13073-016-0330-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Laursen MF, Zachariassen G, Bahl MI, Bergstrom A, Host A, Michaelsen KF, Licht TR. 2015. Having older siblings is associated with gut microbiota development during early childhood. BMC Microbiol 15:154. doi: 10.1186/s12866-015-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. 2014. The intestinal microbiome in early life: health and disease. Front Immunol 5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci U S A 107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Grzeskowiak L, Collado MC, Mangani C, Maleta K, Laitinen K, Ashorn P, Isolauri E, Salminen S. 2012. Distinct gut microbiota in southeastern African and northern European infants. J Pediatr Gastroenterol Nutr 54:812–816. doi: 10.1097/MPG.0b013e318249039c. [DOI] [PubMed] [Google Scholar]
- 126.Lin A, Bik EM, Costello EK, Dethlefsen L, Haque R, Relman DA, Singh U. 2013. Distinct distal gut microbiome diversity and composition in healthy children from Bangladesh and the United States. PLoS One 8:e53838. doi: 10.1371/journal.pone.0053838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Echarri PP, Gracia CM, Berruezo GR, Vives I, Ballesta M, Solis G, Morillas IV, Reyes-Gavilan CG, Margolles A, Gueimonde M. 2011. Assessment of intestinal microbiota of full-term breast-fed infants from two different geographical locations. Early Hum Dev 87:511–513. doi: 10.1016/j.earlhumdev.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 128.Stearns JC, Zulyniak MA, de Souza RJ, Campbell NC, Fontes M, Shaikh M, Sears MR, Becker AB, Mandhane PJ, Subbarao P, Turvey SE, Gupta M, Beyene J, Surette MG, Anand SS. 2017. Ethnic and diet-related differences in the healthy infant microbiome. Genome Med 9:32. doi: 10.1186/s13073-017-0421-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Benson AK, Kelly SA, Legge R, Ma F, Low SJ, Kim J, Zhang M, Oh PL, Nehrenberg D, Hua K, Kachman SD, Moriyama EN, Walter J, Peterson DA, Pomp D. 2010. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A 107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Leamy LJ, Kelly SA, Nietfeldt J, Legge RM, Ma F, Hua K, Sinha R, Peterson DA, Walter J, Benson AK, Pomp D. 2014. Host genetics and diet, but not immunoglobulin A expression, converge to shape compositional features of the gut microbiome in an advanced intercross population of mice. Genome Biol 15:552. doi: 10.1186/s13059-014-0552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Turpin W, Espin-Garcia O, Xu W, Silverberg MS, Kevans D, Smith MI, Guttman DS, Griffiths A, Panaccione R, Otley A, Xu L, Shestopaloff K, Moreno-Hagelsieb G, Paterson AD, Croitoru K. 2016. Association of host genome with intestinal microbial composition in a large healthy cohort. Nat Genet 48:1413–1417. doi: 10.1038/ng.3693. [DOI] [PubMed] [Google Scholar]
- 132.Stewart JA, Chadwick VS, Murray A. 2005. Investigations into the influence of host genetics on the predominant eubacteria in the faecal microflora of children. J Med Microbiol 54:1239–1242. doi: 10.1099/jmm.0.46189-0. [DOI] [PubMed] [Google Scholar]
- 133.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Turnbaugh PJ, Quince C, Faith JJ, McHardy AC, Yatsunenko T, Niazi F, Affourtit J, Egholm M, Henrissat B, Knight R, Gordon JI. 2010. Organismal, genetic, and transcriptional variation in the deeply sequenced gut microbiomes of identical twins. Proc Natl Acad Sci U S A 107:7503–7508. doi: 10.1073/pnas.1002355107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Bonder MJ, Kurilshikov A, Tigchelaar EF, Mujagic Z, Imhann F, Vila AV, Deelen P, Vatanen T, Schirmer M, Smeekens SP, Zhernakova DV, Jankipersadsing SA, Jaeger M, Oosting M, Cenit MC, Masclee AA, Swertz MA, Li Y, Kumar V, Joosten L, Harmsen H, Weersma RK, Franke L, Hofker MH, Xavier RJ, Jonkers D, Netea MG, Wijmenga C, Fu J, Zhernakova A. 2016. The effect of host genetics on the gut microbiome. Nat Genet 48:1407–1412. doi: 10.1038/ng.3663. [DOI] [PubMed] [Google Scholar]
- 136.Walker RW, Clemente JC, Peter I, Loos RJF. 2017. The prenatal gut microbiome: are we colonized with bacteria in utero? Pediatr Obes 12(Suppl 1):S3–S17. doi: 10.1111/ijpo.12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Grahn N, Olofsson M, Ellnebo-Svedlund K, Monstein HJ, Jonasson J. 2003. Identification of mixed bacterial DNA contamination in broad-range PCR amplification of 16S rDNA V1 and V3 variable regions by pyrosequencing of cloned amplicons. FEMS Microbiol Lett 219:87–91. doi: 10.1016/S0378-1097(02)01190-4. [DOI] [PubMed] [Google Scholar]
- 138.Muhl H, Kochem AJ, Disque C, Sakka SG. 2010. Activity and DNA contamination of commercial polymerase chain reaction reagents for the universal 16S rDNA real-time polymerase chain reaction detection of bacterial pathogens in blood. Diagn Microbiol Infect Dis 66:41–49. doi: 10.1016/j.diagmicrobio.2008.07.011. [DOI] [PubMed] [Google Scholar]
- 139.Laurence M, Hatzis C, Brash DE. 2014. Common contaminants in next-generation sequencing that hinder discovery of low-abundance microbes. PLoS One 9:e97876. doi: 10.1371/journal.pone.0097876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Parnell LA, Briggs CM, Cao B, Delannoy-Bruno O, Schrieffer AE, Mysorekar IU. 2017. Microbial communities in placentas from term normal pregnancy exhibit spatially variable profiles. Sci Rep 7:11200. doi: 10.1038/s41598-017-11514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hansen R, Scott KP, Khan S, Martin JC, Berry SH, Stevenson M, Okpapi A, Munro MJ, Hold GL. 2015. First-pass meconium samples from healthy term vaginally-delivered neonates: an analysis of the microbiota. PLoS One 10:e0133320. doi: 10.1371/journal.pone.0133320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sekirov I, Russell SL, Antunes LC, Finlay BB. 2010. Gut microbiota in health and disease. Physiol Rev 90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 143.Stout MJ, Conlon B, Landeau M, Lee I, Bower C, Zhao Q, Roehl KA, Nelson DM, Macones GA, Mysorekar IU. 2013. Identification of intracellular bacteria in the basal plate of the human placenta in term and preterm gestations. Am J Obstet Gynecol 208:226.e1–7. doi: 10.1016/j.ajog.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gray J, Oehrle K, Worthen G, Alenghat T, Whitsett J, Deshmukh H. 2017. Intestinal commensal bacteria mediate lung mucosal immunity and promote resistance of newborn mice to infection. Sci Transl Med 9:eaaf9412. doi: 10.1126/scitranslmed.aaf9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Romano-Keeler J, Weitkamp JH. 2015. Maternal influences on fetal microbial colonization and immune development. Pediatr Res 77:189–195. doi: 10.1038/pr.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Macpherson AJ, de Aguero MG, Ganal-Vonarburg SC. 2017. How nutrition and the maternal microbiota shape the neonatal immune system. Nat Rev Immunol 17:508–517. doi: 10.1038/nri.2017.58. [DOI] [PubMed] [Google Scholar]
- 147.Green KA, Zarek SM, Catherino WH. 2015. Gynecologic health and disease in relation to the microbiome of the female reproductive tract. Fertil Steril 104:1351–1357. doi: 10.1016/j.fertnstert.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 148.Solt I. 2015. The human microbiome and the great obstetrical syndromes: a new frontier in maternal-fetal medicine. Best Pract Res Clin Obstet Gynaecol 29:165–175. doi: 10.1016/j.bpobgyn.2014.04.024. [DOI] [PubMed] [Google Scholar]
- 149.Krohn MA, Hillier SL, Nugent RP, Cotch MF, Carey JC, Gibbs RS, Eschenbach DA. 1995. The genital flora of women with intraamniotic infection. Vaginal Infection and Prematurity Study Group. J Infect Dis 171:1475–1480. [DOI] [PubMed] [Google Scholar]
- 150.Newton ER, Piper J, Peairs W. 1997. Bacterial vaginosis and intraamniotic infection. Am J Obstet Gynecol 176:672–677. doi: 10.1016/S0002-9378(97)70568-4. [DOI] [PubMed] [Google Scholar]
- 151.Leitich H, Bodner-Adler B, Brunbauer M, Kaider A, Egarter C, Husslein P. 2003. Bacterial vaginosis as a risk factor for preterm delivery: a meta-analysis. Am J Obstet Gynecol 189:139–147. doi: 10.1067/mob.2003.339. [DOI] [PubMed] [Google Scholar]
- 152.Yudin MH. 2005. Bacterial vaginosis in pregnancy: diagnosis, screening, and management. Clin Perinatol 32:617–627. doi: 10.1016/j.clp.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 153.Eckert LO, Moore DE, Patton DL, Agnew KJ, Eschenbach DA. 2003. Relationship of vaginal bacteria and inflammation with conception and early pregnancy loss following in-vitro fertilization. Infect Dis Obstet Gynecol 11:11–17. doi: 10.1155/S1064744903000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.van Oostrum N, De Sutter P, Meys J, Verstraelen H. 2013. Risks associated with bacterial vaginosis in infertility patients: a systematic review and meta-analysis. Hum Reprod 28:1809–1815. doi: 10.1093/humrep/det096. [DOI] [PubMed] [Google Scholar]
- 155.Reid G, Brigidi P, Burton JP, Contractor N, Duncan S, Fargier E, Hill C, Lebeer S, Martin R, McBain AJ, Mor G, O'Neill C, Rodriguez JM, Swann J, van Hemert S, Ansell J. 2015. Microbes central to human reproduction. Am J Reprod Immunol 73:1–11. doi: 10.1111/aji.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Roos PJ, Malan AF, Woods DL, Botha P, Hyland J, Heese Hde V. 1980. The bacteriological environment of preterm infants. S Afr Med J 57:347–350. [PubMed] [Google Scholar]
- 157.Naeye RL, Peters EC. 1980. Causes and consequences of premature rupture of fetal membranes. Lancet i:192–194. [DOI] [PubMed] [Google Scholar]
- 158.Dong Y, St Clair PJ, Ramzy I, Kagan-Hallet KS, Gibbs RS. 1987. A microbiologic and clinical study of placental inflammation at term. Obstet Gynecol 70:175–182. [PubMed] [Google Scholar]
- 159.Goldenberg RL, Culhane JF, Iams JD, Romero R. 2008. Epidemiology and causes of preterm birth. Lancet 371:75–84. doi: 10.1016/S0140-6736(08)60074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Onderdonk AB, Hecht JL, McElrath TF, Delaney ML, Allred EN, Leviton A. 2008. Colonization of second-trimester placenta parenchyma. Am J Obstet Gynecol 199:52.e1–52.e10. doi: 10.1016/j.ajog.2007.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Payne MS, Bayatibojakhi S. 2014. Exploring preterm birth as a polymicrobial disease: an overview of the uterine microbiome. Front Immunol 5:595. doi: 10.3389/fimmu.2014.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Romero R, Miranda J, Kusanovic JP, Chaiworapongsa T, Chaemsaithong P, Martinez A, Gotsch F, Dong Z, Ahmed AI, Shaman M, Lannaman K, Yoon BH, Hassan SS, Kim CJ, Korzeniewski SJ, Yeo L, Kim YM. 2015. Clinical chorioamnionitis at term I: microbiology of the amniotic cavity using cultivation and molecular techniques. J Perinat Med 43:19–36. doi: 10.1515/jpm-2014-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Vinturache AE, Gyamfi-Bannerman C, Hwang J, Mysorekar IU, Jacobsson B. 2016. Maternal microbiome—a pathway to preterm birth. Semin Fetal Neonatal Med 21:94–99. doi: 10.1016/j.siny.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 164.Mandar R. 2013. Microbiota of male genital tract: impact on the health of man and his partner. Pharmacol Res 69:32–41. doi: 10.1016/j.phrs.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 165.Franasiak JM, Scott RT Jr. 2015. Reproductive tract microbiome in assisted reproductive technologies. Fertil Steril 104:1364–1371. doi: 10.1016/j.fertnstert.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 166.Ansbacher R, Boyson WA, Morris JA. 1967. Sterility of the uterine cavity. Am J Obstet Gynecol 99:394–396. doi: 10.1016/S0002-9378(16)34549-5. [DOI] [PubMed] [Google Scholar]
- 167.Verstraelen H, Vilchez-Vargas R, Desimpel F, Jauregui R, Vankeirsbilck N, Weyers S, Verhelst R, De Sutter P, Pieper DH, Van De Wiele T. 2016. Characterisation of the human uterine microbiome in non-pregnant women through deep sequencing of the V1-2 region of the 16S rRNA gene. PeerJ 4:e1602. doi: 10.7717/peerj.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Franasiak JM, Werner MD, Juneau CR, Tao X, Landis J, Zhan Y, Treff NR, Scott RT. 2016. Endometrial microbiome at the time of embryo transfer: next-generation sequencing of the 16S ribosomal subunit. J Assist Reprod Genet 33:129–136. doi: 10.1007/s10815-015-0614-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Moreno I, Codoner FM, Vilella F, Valbuena D, Martinez-Blanch JF, Jimenez-Almazan J, Alonso R, Alama P, Remohi J, Pellicer A, Ramon D, Simon C. 2016. Evidence that the endometrial microbiota has an effect on implantation success or failure. Am J Obstet Gynecol 215:684–703. doi: 10.1016/j.ajog.2016.09.075. [DOI] [PubMed] [Google Scholar]
- 170.Robertson SA, Chin PY, Glynn DJ, Thompson JG. 2011. Peri-conceptual cytokines—setting the trajectory for embryo implantation, pregnancy and beyond. Am J Reprod Immunol 66(Suppl 1):S2–S10. doi: 10.1111/j.1600-0897.2011.01039.x. [DOI] [PubMed] [Google Scholar]
- 171.Dominguez F, Gadea B, Mercader A, Esteban FJ, Pellicer A, Simon C. 2010. Embryologic outcome and secretome profile of implanted blastocysts obtained after coculture in human endometrial epithelial cells versus the sequential system. Fertil Steril 93:774–782.e1. doi: 10.1016/j.fertnstert.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 172.Pelzer E, Willner D, Huygens F, Buttini M. 2015. Is diversification of the endometrial microbiome significant for reproductive success? Placenta 36:A23–A23. doi: 10.1016/j.placenta.2015.07.253. [DOI] [Google Scholar]
- 173.Khan KN, Fujishita A, Kitajima M, Hiraki K, Nakashima M, Masuzaki H. 2014. Intra-uterine microbial colonization and occurrence of endometritis in women with endometriosis. Hum Reprod 29:2446–2456. doi: 10.1093/humrep/deu222. [DOI] [PubMed] [Google Scholar]
- 174.Fang RL, Chen LX, Shu WS, Yao SZ, Wang SW, Chen YQ. 2016. Barcoded sequencing reveals diverse intrauterine microbiomes in patients suffering with endometrial polyps. Am J Transl Res 8:1581–1592. [PMC free article] [PubMed] [Google Scholar]
- 175.Walther-Antonio MR, Chen J, Multinu F, Hokenstad A, Distad TJ, Cheek EH, Keeney GL, Creedon DJ, Nelson H, Mariani A, Chia N. 2016. Potential contribution of the uterine microbiome in the development of endometrial cancer. Genome Med 8:122. doi: 10.1186/s13073-016-0368-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Giudice LC. 2016. Challenging dogma: the endometrium has a microbiome with functional consequences! Am J Obstet Gynecol 215:682–683. [DOI] [PubMed] [Google Scholar]
- 177.Hillier SL, Martius J, Krohn M, Kiviat N, Holmes KK, Eschenbach DA. 1988. A case-control study of chorioamnionic infection and histologic chorioamnionitis in prematurity. N Engl J Med 319:972–978. doi: 10.1056/NEJM198810133191503. [DOI] [PubMed] [Google Scholar]
- 178.Diaz Heijtz R. 2016. Fetal, neonatal, and infant microbiome: perturbations and subsequent effects on brain development and behavior. Semin Fetal Neonatal Med 21:410–417. doi: 10.1016/j.siny.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 179.Neu J. 2016. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med 21:373–379. doi: 10.1016/j.siny.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 180.Martin R, Langa S, Reviriego C, Jimenez E, Marin ML, Olivares M, Boza J, Jimenez J, Fernandez L, Xaus J, Rodriguez JM. 2004. The commensal microflora of human milk: new perspectives for food bacteriotherapy and probiotics. Trends Food Sci Technol 15:121–127. doi: 10.1016/j.tifs.2003.09.010. [DOI] [Google Scholar]
- 181.Favier CF, Vaughan EE, De Vos WM, Akkermans AD. 2002. Molecular monitoring of succession of bacterial communities in human neonates. Appl Environ Microbiol 68:219–226. doi: 10.1128/AEM.68.1.219-226.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Martin R, Langa S, Reviriego C, Jiminez E, Marin ML, Xaus J, Fernandez L, Rodriguez JM. 2003. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr 143:754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 183.Steel JH, Malatos S, Kennea N, Edwards AD, Miles L, Duggan P, Reynolds PR, Feldman RG, Sullivan MH. 2005. Bacteria and inflammatory cells in fetal membranes do not always cause preterm labor. Pediatr Res 57:404–411. doi: 10.1203/01.PDR.0000153869.96337.90. [DOI] [PubMed] [Google Scholar]
- 184.Queiros da Mota V, Prodhom G, Yan P, Hohlfheld P, Greub G, Rouleau C. 2013. Correlation between placental bacterial culture results and histological chorioamnionitis: a prospective study on 376 placentas. J Clin Pathol 66:243–248. doi: 10.1136/jclinpath-2012-201124. [DOI] [PubMed] [Google Scholar]
- 185.Aagaard K, Petrosino J, Keitel W, Watson M, Katancik J, Garcia N, Patel S, Cutting M, Madden T, Hamilton H, Harris E, Gevers D, Simone G, McInnes P, Versalovic J. 2013. The Human Microbiome Project strategy for comprehensive sampling of the human microbiome and why it matters. FASEB J 27:1012–1022. doi: 10.1096/fj.12-220806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zheng J, Xiao X, Zhang Q, Mao L, Yu M, Xu J. 2015. The placental microbiome varies in association with low birth weight in full-term neonates. Nutrients 7:6924–6937. doi: 10.3390/nu7085315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hauguel-de Mouzon S, Guerre-Millo M. 2006. The placenta cytokine network and inflammatory signals. Placenta 27:794–798. doi: 10.1016/j.placenta.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 188.Stewart FM, Freeman DJ, Ramsay JE, Greer IA, Caslake M, Ferrell WR. 2007. Longitudinal assessment of maternal endothelial function and markers of inflammation and placental function throughout pregnancy in lean and obese mothers. J Clin Endocrinol Metab 92:969–975. doi: 10.1210/jc.2006-2083. [DOI] [PubMed] [Google Scholar]
- 189.Challier JC, Basu S, Bintein T, Minium J, Hotmire K, Catalano PM, Hauguel-de Mouzon S. 2008. Obesity in pregnancy stimulates macrophage accumulation and inflammation in the placenta. Placenta 29:274–281. doi: 10.1016/j.placenta.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Baeten JM, Bukusi EA, Lambe M. 2001. Pregnancy complications and outcomes among overweight and obese nulliparous women. Am J Public Health 91:436–440. doi: 10.2105/AJPH.91.3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.McDonald SD, Han Z, Mulla S, Beyene J. 2010. Overweight and obesity in mothers and risk of preterm birth and low birth weight infants: systematic review and meta-analyses. BMJ 341:c3428. doi: 10.1136/bmj.c3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Azevedo FA, Andrade-Moraes CH, Curado MR, Oliveira-Pinto AV, Guimaraes DM, Szczupak D, Gomes BV, Alho AT, Polichiso L, Tampellini E, Lima L, de Lima DO, da Silva HA, Lent R. 2013. Automatic isotropic fractionation for large-scale quantitative cell analysis of nervous tissue. J Neurosci Methods 212:72–78. doi: 10.1016/j.jneumeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 193.Antony KM, Ma J, Mitchell KB, Racusin DA, Versalovic J, Aagaard K. 2015. The preterm placental microbiome varies in association with excess maternal gestational weight gain. Am J Obstet Gynecol 212:653.e1–e16. doi: 10.1016/j.ajog.2014.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Racicot K, Kwon JY, Aldo P, Silasi M, Mor G. 2014. Understanding the complexity of the immune system during pregnancy. Am J Reprod Immunol 72:107–116. doi: 10.1111/aji.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, Moris F, Rodrigo L, Mira A, Collado MC. 2015. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol 50:167–179. doi: 10.1007/s00535-014-0963-x. [DOI] [PubMed] [Google Scholar]
- 196.Cardenas I, Means RE, Aldo P, Koga K, Lang SM, Booth CJ, Manzur A, Oyarzun E, Romero R, Mor G. 2010. Viral infection of the placenta leads to fetal inflammation and sensitization to bacterial products predisposing to preterm labor. J Immunol 185:1248–1257. doi: 10.4049/jimmunol.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Hitti J, Riley DE, Krohn MA, Hillier SL, Agnew KJ, Krieger JN, Eschenbach DA. 1997. Broad-spectrum bacterial rDNA polymerase chain reaction assay for detecting amniotic fluid infection among women in premature labor. Clin Infect Dis 24:1228–1232. doi: 10.1086/513669. [DOI] [PubMed] [Google Scholar]
- 198.Jimenez E, Marin ML, Martin R, Odriozola JM, Olivares M, Xaus J, Fernandez L, Rodriguez JM. 2008. Is meconium from healthy newborns actually sterile? Res Microbiol 159:187–193. doi: 10.1016/j.resmic.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 199.Dong XD, Li XR, Luan JJ, Liu XF, Peng J, Luo YY, Liu CJ. 2015. Bacterial communities in neonatal feces are similar to mothers' placentae. Can J Infect Dis Med Microbiol 26:90–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Gosalbes MJ, Llop S, Valles Y, Moya A, Ballester F, Francino MP. 2013. Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin Exp Allergy 43:198–211. doi: 10.1111/cea.12063. [DOI] [PubMed] [Google Scholar]
- 201.Moles L, Gomez M, Heilig H, Bustos G, Fuentes S, de Vos W, Fernandez L, Rodriguez JM, Jimenez E. 2013. Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life. PLoS One 8:e66986. doi: 10.1371/journal.pone.0066986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Hu J, Nomura Y, Bashir A, Fernandez-Hernandez H, Itzkowitz S, Pei Z, Stone J, Loudon H, Peter I. 2013. Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One 8:e78257. doi: 10.1371/journal.pone.0078257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ardissone AN, de la Cruz DM, Davis-Richardson AG, Rechcigl KT, Li N, Drew JC, Murgas-Torrazza R, Sharma R, Hudak ML, Triplett EW, Neu J. 2014. Meconium microbiome analysis identifies bacteria correlated with premature birth. PLoS One 9:e90784. doi: 10.1371/journal.pone.0090784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Nagpal R, Tsuji H, Takahashi T, Kawashima K, Nagata S, Nomoto K, Yamashiro Y. 2016. Sensitive quantitative analysis of the meconium bacterial microbiota in healthy term infants born vaginally or by cesarean section. Front Microbiol 7:1997. doi: 10.3389/fmicb.2016.01997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Makino H, Kushiro A, Ishikawa E, Kubota H, Gawad A, Sakai T, Oishi K, Martin R, Ben-Amor K, Knol J, Tanaka R. 2013. Mother-to-infant transmission of intestinal bifidobacterial strains has an impact on the early development of vaginally delivered infant's microbiota. PLoS One 8:e78331. doi: 10.1371/journal.pone.0078331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. 2016. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Mshvildadze M, Neu J, Shuster J, Theriaque D, Li N, Mai V. 2010. Intestinal microbial ecology in premature infants assessed with non-culture-based techniques. J Pediatr 156:20–25. doi: 10.1016/j.jpeds.2009.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Satokari R, Gronroos T, Laitinen K, Salminen S, Isolauri E. 2009. Bifidobacterium and Lactobacillus DNA in the human placenta. Lett Appl Microbiol 48:8–12. doi: 10.1111/j.1472-765X.2008.02475.x. [DOI] [PubMed] [Google Scholar]
- 209.Vanterpool SF, Been JV, Houben ML, Nikkels PG, De Krijger RR, Zimmermann LJ, Kramer BW, Progulske-Fox A, Reyes L. 2016. Porphyromonas gingivalis within placental villous mesenchyme and umbilical cord stroma is associated with adverse pregnancy outcome. PLoS One 11:e0146157. doi: 10.1371/journal.pone.0146157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rodriguez JM. 2014. The origin of human milk bacteria: is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv Nutr 5:779–784. doi: 10.3945/an.114.007229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Han YW, Shen T, Chung P, Buhimschi IA, Buhimschi CS. 2009. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol 47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Han YW, Redline RW, Li M, Yin L, Hill GB, McCormick TS. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun 72:2272–2279. doi: 10.1128/IAI.72.4.2272-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Han YW, Ikegami A, Bissada NF, Herbst M, Redline RW, Ashmead GG. 2006. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J Clin Microbiol 44:1475–1483. doi: 10.1128/JCM.44.4.1475-1483.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Han YW, Fardini Y, Chen C, Iacampo KG, Peraino VA, Shamonki JM, Redline RW. 2010. Term stillbirth caused by oral Fusobacterium nucleatum. Obstet Gynecol 115:442–445. doi: 10.1097/AOG.0b013e3181cb9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Fardini Y, Chung P, Dumm R, Joshi N, Han YW. 2010. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun 78:1789–1796. doi: 10.1128/IAI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Dasanayake AP, Li Y, Wiener H, Ruby JD, Lee MJ. 2005. Salivary Actinomyces naeslundii genospecies 2 and Lactobacillus casei levels predict pregnancy outcomes. J Periodontol 76:171–177. doi: 10.1902/jop.2005.76.2.171. [DOI] [PubMed] [Google Scholar]
- 217.Perez PF, Dore J, Leclerc M, Levenez F, Benyacoub J, Serrant P, Segura-Roggero I, Schiffrin EJ, Donnet-Hughes A. 2007. Bacterial imprinting of the neonatal immune system: lessons from maternal cells? Pediatrics 119:e724–e732. doi: 10.1542/peds.2006-1649. [DOI] [PubMed] [Google Scholar]
- 218.McGuire MK, Meehan CL, McGuire MA, Williams JE, Foster J, Sellen DW, Kamau-Mbuthia EW, Kamundia EW, Mbugua S, Moore SE, Prentice AM, Kvist LJ, Otoo GE, Brooker SL, Price WJ, Shafii B, Placek C, Lackey KA, Robertson B, Manzano S, Ruiz L, Rodriguez JM, Pareja RG, Bode L. 2017. What's normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am J Clin Nutr 105:1086–1100. doi: 10.3945/ajcn.116.139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Kumazaki T, Yoshida A. 1984. Biochemical evidence that secretor gene, Se, is a structural gene encoding a specific fucosyltransferase. Proc Natl Acad Sci U S A 81:4193–4197. doi: 10.1073/pnas.81.13.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Viverge D, Grimmonprez L, Cassanas G, Bardet L, Solere M. 1990. Discriminant carbohydrate components of human milk according to donor secretor types. J Pediatr Gastroenterol Nutr 11:365–370. doi: 10.1097/00005176-199010000-00014. [DOI] [PubMed] [Google Scholar]
- 221.Johnson PH, Watkins WM. 1992. Purification of the Lewis blood-group gene associated alpha-3/4-fucosyltransferase from human milk: an enzyme transferring fucose primarily to type 1 and lactose-based oligosaccharide chains. Glycoconj J 9:241–249. doi: 10.1007/BF00731136. [DOI] [PubMed] [Google Scholar]
- 222.Thurl S, Henker J, Siegel M, Tovar K, Sawatzki G. 1997. Detection of four human milk groups with respect to Lewis blood group dependent oligosaccharides. Glycoconj J 14:795–799. doi: 10.1023/A:1018529703106. [DOI] [PubMed] [Google Scholar]
- 223.Chaturvedi P, Warren CD, Altaye M, Morrow AL, Ruiz-Palacios G, Pickering LK, Newburg DS. 2001. Fucosylated human milk oligosaccharides vary between individuals and over the course of lactation. Glycobiology 11:365–372. doi: 10.1093/glycob/11.5.365. [DOI] [PubMed] [Google Scholar]
- 224.Stahl B, Thurl S, Henker J, Siegel M, Finke B, Sawatzki G. 2001. Detection of four human milk groups with respect to Lewis-blood-group-dependent oligosaccharides by serologic and chromatographic analysis. Adv Exp Med Biol 501:299–306. doi: 10.1007/978-1-4615-1371-1_37. [DOI] [PubMed] [Google Scholar]
- 225.Thurl S, Munzert M, Henker J, Boehm G, Muller-Werner B, Jelinek J, Stahl B. 2010. Variation of human milk oligosaccharides in relation to milk groups and lactational periods. Br J Nutr 104:1261–1271. doi: 10.1017/S0007114510002072. [DOI] [PubMed] [Google Scholar]
- 226.Xu Z, Vo L, Macher BA. 1996. Structure-function analysis of human alpha1,3-fucosyltransferase. Amino acids involved in acceptor substrate specificity. J Biol Chem 271:8818–8823. [DOI] [PubMed] [Google Scholar]
- 227.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. 2006. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol 72:4497–4499. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Ward RE, Ninonuevo M, Mills DA, Lebrilla CB, German JB. 2007. In vitro fermentability of human milk oligosaccharides by several strains of bifidobacteria. Mol Nutr Food Res 51:1398–1405. doi: 10.1002/mnfr.200700150. [DOI] [PubMed] [Google Scholar]
- 229.LoCascio RG, Ninonuevo MR, Freeman SL, Sela DA, Grimm R, Lebrilla CB, Mills DA, German JB. 2007. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem 55:8914–8919. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 230.Locascio RG, Ninonuevo MR, Kronewitter SR, Freeman SL, German JB, Lebrilla CB, Mills DA. 2009. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb Biotechnol 2:333–342. doi: 10.1111/j.1751-7915.2008.00072.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Garrido D, Ruiz-Moyano S, Lemay DG, Sela DA, German JB, Mills DA. 2015. Comparative transcriptomics reveals key differences in the response to milk oligosaccharides of infant gut-associated bifidobacteria. Sci Rep 5:13517. doi: 10.1038/srep13517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, Price NP, Richardson PM, Mills DA. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A 105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Lin AE, Autran CA, Szyszka A, Escajadillo T, Huang M, Godula K, Prudden AR, Boons GJ, Lewis AL, Doran KS, Nizet V, Bode L. 2017. Human milk oligosaccharides inhibit growth of group B Streptococcus. J Biol Chem doi: 10.1074/jbc.M117.789974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Gonia S, Tuepker M, Heisel T, Autran C, Bode L, Gale CA. 2015. Human milk oligosaccharides inhibit Candida albicans invasion of human premature intestinal epithelial cells. J Nutr 145:1992–1998. doi: 10.3945/jn.115.214940. [DOI] [PubMed] [Google Scholar]
- 235.Fridkin SK, Kaufman D, Edwards JR, Shetty S, Horan T. 2006. Changing incidence of Candida bloodstream infections among NICU patients in the United States: 1995-2004. Pediatrics 117:1680–1687. doi: 10.1542/peds.2005-1996. [DOI] [PubMed] [Google Scholar]
- 236.Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. 2003. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem 278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- 237.Morrow AL, Ruiz-Palacios GM, Altaye M, Jiang X, Guerrero ML, Meinzen-Derr JK, Farkas T, Chaturvedi P, Pickering LK, Newburg DS. 2004. Human milk oligosaccharides are associated with protection against diarrhea in breast-fed infants. J Pediatr 145:297–303. doi: 10.1016/j.jpeds.2004.04.054. [DOI] [PubMed] [Google Scholar]
- 238.Manthey CF, Autran CA, Eckmann L, Bode L. 2014. Human milk oligosaccharides protect against enteropathogenic Escherichia coli attachment in vitro and EPEC colonization in suckling mice. J Pediatr Gastroenterol Nutr 58:165–168. doi: 10.1097/MPG.0000000000000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. 2012. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. Br J Nutr 108:1839–1846. doi: 10.1017/S0007114511007392. [DOI] [PubMed] [Google Scholar]
- 240.Eiwegger T, Stahl B, Haidl P, Schmitt J, Boehm G, Dehlink E, Urbanek R, Szepfalusi Z. 2010. Prebiotic oligosaccharides: in vitro evidence for gastrointestinal epithelial transfer and immunomodulatory properties. Pediatr Allergy Immunol 21:1179–1188. doi: 10.1111/j.1399-3038.2010.01062.x. [DOI] [PubMed] [Google Scholar]
- 241.Civardi E, Garofoli F, Tzialla C, Paolillo P, Bollani L, Stronati M. 2013. Microorganisms in human milk: lights and shadows. J Matern Fetal Neonatal Med 26(Suppl 2):S30–S34. doi: 10.3109/14767058.2013.829693. [DOI] [PubMed] [Google Scholar]
- 242.Urbaniak C, Angelini M, Gloor GB, Reid G. 2016. Human milk microbiota profiles in relation to birthing method, gestation and infant gender. Microbiome 4:1. doi: 10.1186/s40168-015-0145-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Hunt KM, Foster JA, Forney LJ, Schutte UM, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. 2011. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS One 6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Jost T, Lacroix C, Braegger C, Chassard C. 2013. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr 110:1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 245.Fernandez L, Langa S, Martin V, Maldonado A, Jimenez E, Martin R, Rodriguez JM. 2013. The human milk microbiota: origin and potential roles in health and disease. Pharmacol Res 69:1–10. doi: 10.1016/j.phrs.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 246.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, van Sinderen D, Ventura M. 2017. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Urashima T, Saito T, Nakamura T, Messer M. 2001. Oligosaccharides of milk and colostrum in non-human mammals. Glycoconj J 18:357–371. doi: 10.1023/A:1014881913541. [DOI] [PubMed] [Google Scholar]
- 248.Saito T, Itoh T, Adachi S, Suzuki T, Usui T. 1981. The chemical structure of neutral and acidic sugar chains obtained from bovine colostrum kappa-casein. Biochim Biophys Acta 678:257–267. doi: 10.1016/0304-4165(81)90215-4. [DOI] [PubMed] [Google Scholar]
- 249.Urashima T, Saito T, Ohmisya K, Shimazaki K. 1991. Structural determination of three neutral oligosaccharides in bovine (Holstein-Friesian) colostrum, including the novel trisaccharide; GalNAc alpha 1-3Gal beta 1-4Glc. Biochim Biophys Acta 1073:225–229. doi: 10.1016/0304-4165(91)90207-W. [DOI] [PubMed] [Google Scholar]
- 250.Martinez-Ferez A, Rudloff S, Guadix A, Henkel CA, Pohlentz G, Boza JJ, Guadix EM, Kunz C. 2006. Goats' milk as a natural source of lactose-derived oligosaccharides: isolation by membrane technology. Int Dairy J 16:173–181. doi: 10.1016/j.idairyj.2005.02.003. [DOI] [Google Scholar]
- 251.Nakajima K, Kinoshita M, Matsushita N, Urashima T, Suzuki M, Suzuki A, Kakehi K. 2006. Capillary affinity electrophoresis using lectins for the analysis of milk oligosaccharide structure and its application to bovine colostrum oligosaccharides. Anal Biochem 348:105–114. doi: 10.1016/j.ab.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 252.Tao N, Ochonicky KL, German JB, Donovan SM, Lebrilla CB. 2010. Structural determination and daily variations of porcine milk oligosaccharides. J Agric Food Chem 58:4653–4659. doi: 10.1021/jf100398u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Goto K, Fukuda K, Senda A, Saito T, Kimura K, Glander KE, Hinde K, Dittus W, Milligan LA, Power ML, Oftedal OT, Urashima T. 2010. Chemical characterization of oligosaccharides in the milk of six species of New and Old World monkeys. Glycoconj J 27:703–715. doi: 10.1007/s10719-010-9315-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tao N, Wu S, Kim J, An HJ, Hinde K, Power ML, Gagneux P, German JB, Lebrilla CB. 2011. Evolutionary glycomics: characterization of milk oligosaccharides in primates. J Proteome Res 10:1548–1557. doi: 10.1021/pr1009367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Gilbert SF. 2014. A holobiont birth narrative: the epigenetic transmission of the human microbiome. Front Genet 5:282. doi: 10.3389/fgene.2014.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, Siuzdak G. 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci U S A 106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, Hentschel U, King N, Kjelleberg S, Knoll AH, Kremer N, Mazmanian SK, Metcalf JL, Nealson K, Pierce NE, Rawls JF, Reid A, Ruby EG, Rumpho M, Sanders JG, Tautz D, Wernegreen JJ. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Funkhouser LJ, Bordenstein SR. 2013. Mom knows best: the universality of maternal microbial transmission. PLoS Biol 11:e1001631. doi: 10.1371/journal.pbio.1001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Moran NA. 2007. Symbiosis as an adaptive process and source of phenotypic complexity. Proc Natl Acad Sci U S A 104:8627–8633. doi: 10.1073/pnas.0611659104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Conroy ME, Shi HN, Walker WA. 2009. The long-term health effects of neonatal microbial flora. Curr Opin Allergy Clin Immunol 9:197–201. doi: 10.1097/ACI.0b013e32832b3f1d. [DOI] [PubMed] [Google Scholar]
- 261.Dotterud CK, Avershina E, Sekelja M, Simpson MR, Rudi K, Storro O, Johnsen R, Oien T. 2015. Does maternal perinatal probiotic supplementation alter the intestinal microbiota of mother and child? J Pediatr Gastroenterol Nutr 61:200–207. doi: 10.1097/MPG.0000000000000781. [DOI] [PubMed] [Google Scholar]
- 262.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, et al. 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Avershina E, Storro O, Oien T, Johnsen R, Pope P, Rudi K. 2014. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol Ecol 87:280–290. doi: 10.1111/1574-6941.12223. [DOI] [PubMed] [Google Scholar]
- 264.Duranti S, Lugli GA, Mancabelli L, Armanini F, Turroni F, James K, Ferretti P, Gorfer V, Ferrario C, Milani C, Mangifesta M, Anzalone R, Zolfo M, Viappiani A, Pasolli E, Bariletti I, Canto R, Clementi R, Cologna M, Crifò T, Cusumano G, Sedi S, Gottardi S, Innamorati C, Masè C, Postai D, Savoi D, Soffiati M, Tateo S, Pedrotti A, Segata N, Van Sinderen D, Ventura M. 2017. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome 5:66. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Milani C, Mangifesta M, Mancabelli L, Lugli GA, James K, Duranti S, Turroni F, Ferrario C, Ossiprandi MC, Van Sinderen D, Ventura M. 22 August 2017. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Cheng J, Ringel-Kulka T, Heikamp-de Jong I, Ringel Y, Carroll I, de Vos WM, Salojarvi J, Satokari R. 2016. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J 10:1002–1014. doi: 10.1038/ismej.2015.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Hollister EB, Riehle K, Luna RA, Weidler EM, Rubio-Gonzales M, Mistretta TA, Raza S, Doddapaneni HV, Metcalf GA, Muzny DM, Gibbs RA, Petrosino JF, Shulman RJ, Versalovic J. 2015. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Ottman N, Smidt H, de Vos WM, Belzer C. 2012. The function of our microbiota: who is out there and what do they do? Front Cell Infect Microbiol 2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Dogra S, Sakwinska O, Soh SE, Ngom-Bru C, Bruck WM, Berger B, Brussow H, Lee YS, Yap F, Chong YS, Godfrey KM, Holbrook JD, GUSTO Study Group . 2015. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. mBio 6:e02419-14. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Fan W, Huo G, Li X, Yang L, Duan C. 2014. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. J Microbiol Biotechnol 24:133–143. doi: 10.4014/jmb.1309.09029. [DOI] [PubMed] [Google Scholar]
- 272.Fan W, Huo G, Li X, Yang L, Duan C, Wang T, Chen J. 2013. Diversity of the intestinal microbiota in different patterns of feeding infants by Illumina high-throughput sequencing. World J Microbiol Biotechnol 29:2365–2372. doi: 10.1007/s11274-013-1404-3. [DOI] [PubMed] [Google Scholar]
- 273.Gensollen T, Iyer SS, Kasper DL, Blumberg RS. 2016. How colonization by microbiota in early life shapes the immune system. Science 352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Hansen CH, Nielsen DS, Kverka M, Zakostelska Z, Klimesova K, Hudcovic T, Tlaskalova-Hogenova H, Hansen AK. 2012. Patterns of early gut colonization shape future immune responses of the host. PLoS One 7:34043. doi: 10.1371/journal.pone.0034043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Sommer F, Backhed F. 2013. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol 11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 276.Clarke G, O'Mahony SM, Dinan TG, Cryan JF. 2014. Priming for health: gut microbiota acquired in early life regulates physiology, brain and behaviour. Acta Paediatr 103:812–819. doi: 10.1111/apa.12674. [DOI] [PubMed] [Google Scholar]
- 277.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Bjorkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. 2011. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci U S A 108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Neuman H, Debelius JW, Knight R, Koren O. 2015. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiol Rev 39:509–521. doi: 10.1093/femsre/fuu010. [DOI] [PubMed] [Google Scholar]
- 279.Nylund L, Satokari R, Salminen S, de Vos WM. 2014. Intestinal microbiota during early life—impact on health and disease. Proc Nutr Soc 73:457–469. doi: 10.1017/S0029665114000627. [DOI] [PubMed] [Google Scholar]
- 280.Moon C, Baldridge MT, Wallace MA, Burnham CA, Virgin HW, Stappenbeck TS. 2015. Vertically transmitted faecal IgA levels determine extra-chromosomal phenotypic variation. Nature 521:90–93. doi: 10.1038/nature14139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, Panzer AR, LaMere B, Rackaityte E, Lukacs NW, Wegienka G, Boushey HA, Ownby DR, Zoratti EM, Levin AM, Johnson CC, Lynch SV. 2016. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 22:1187–1191. doi: 10.1038/nm.4176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Kalliomaki M, Collado MC, Salminen S, Isolauri E. 2008. Early differences in fecal microbiota composition in children may predict overweight. Am J Clin Nutr 87:534–538. [DOI] [PubMed] [Google Scholar]
- 283.Kalliomaki M, Kirjavainen P, Eerola E, Kero P, Salminen S, Isolauri E. 2001. Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J Allergy Clin Immunol 107:129–134. doi: 10.1067/mai.2001.111237. [DOI] [PubMed] [Google Scholar]
- 284.Simonyte Sjodin K, Vidman L, Ryden P, West CE. 2016. Emerging evidence of the role of gut microbiota in the development of allergic diseases. Curr Opin Allergy Clin Immunol 16:390–395. doi: 10.1097/ACI.0000000000000277. [DOI] [PubMed] [Google Scholar]
- 285.Al-Asmakh M, Zadjali F. 2015. Use of germ-free animal models in microbiota-related research. J Microbiol Biotechnol 25:1583–1588. doi: 10.4014/jmb.1501.01039. [DOI] [PubMed] [Google Scholar]
- 286.Olszak T, An D, Zeissig S, Vera MP, Richter J, Franke A, Glickman JN, Siebert R, Baron RM, Kasper DL, Blumberg RS. 2012. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Kristof K, Kocsis E, Nagy K. 2009. Clinical microbiology of early-onset and late-onset neonatal sepsis, particularly among preterm babies. Acta Microbiol Immunol Hung 56:21–51. doi: 10.1556/AMicr.56.2009.1.2. [DOI] [PubMed] [Google Scholar]
- 288.Stewart CJ, Embleton ND, Marrs EC, Smith DP, Nelson A, Abdulkadir B, Skeath T, Petrosino JF, Perry JD, Berrington JE, Cummings SP.. 2016. Temporal bacterial and metabolic development of the preterm gut reveals specific signatures in health and disease. Microbiome 4:67. doi: 10.1186/s40168-016-0216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ward DV, Scholz M, Zolfo M, Taft DH, Schibler KR, Tett A, Segata N, Morrow AL. 2016. Metagenomic sequencing with strain-level resolution implicates uropathogenic E. coli in necrotizing enterocolitis and mortality in preterm infants. Cell Rep 14:2912–2924. doi: 10.1016/j.celrep.2016.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Leach ST, Lui K, Naing Z, Dowd SE, Mitchell HM, Day AS. 2015. Multiple opportunistic pathogens, but not pre-existing inflammation, may be associated with necrotizing enterocolitis. Dig Dis Sci 60:3728–3734. doi: 10.1007/s10620-015-3830-6. [DOI] [PubMed] [Google Scholar]
- 291.Sim K, Shaw AG, Randell P, Cox MJ, McClure ZE, Li MS, Haddad M, Langford PR, Cookson WO, Moffatt MF, Kroll JS. 2015. Dysbiosis anticipating necrotizing enterocolitis in very premature infants. Clin Infect Dis 60:389–397. doi: 10.1093/cid/ciu822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Mai V, Young CM, Ukhanova M, Wang X, Sun Y, Casella G, Theriaque D, Li N, Sharma R, Hudak M, Neu J. 2011. Fecal microbiota in premature infants prior to necrotizing enterocolitis. PLoS One 6:e20647. doi: 10.1371/journal.pone.0020647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Morrow AL, Lagomarcino AJ, Schibler KR, Taft DH, Yu Z, Wang B, Altaye M, Wagner M, Gevers D, Ward DV, Kennedy MA, Huttenhower C, Newburg DS. 2013. Early microbial and metabolomic signatures predict later onset of necrotizing enterocolitis in preterm infants. Microbiome 1:13. doi: 10.1186/2049-2618-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Torrazza RM, Ukhanova M, Wang X, Sharma R, Hudak ML, Neu J, Mai V. 2013. Intestinal microbial ecology and environmental factors affecting necrotizing enterocolitis. PLoS One 8:e83304. doi: 10.1371/journal.pone.0083304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Warner BB, Deych E, Zhou Y, Hall-Moore C, Weinstock GM, Sodergren E, Shaikh N, Hoffmann JA, Linneman LA, Hamvas A, Khanna G, Rouggly-Nickless LC, Ndao IM, Shands BA, Escobedo M, Sullivan JE, Radmacher PG, Shannon WD, Tarr PI. 2016. Gut bacteria dysbiosis and necrotising enterocolitis in very low birthweight infants: a prospective case-control study. Lancet 387:1928–1936. doi: 10.1016/S0140-6736(16)00081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Stewart CJ, Nelson A, Treumann A, Skeath T, Cummings SP, Embleton ND, Berrington JE. 2016. Metabolomic and proteomic analysis of serum from preterm infants with necrotising entercolitis and late-onset sepsis. Pediatr Res 79:425–431. doi: 10.1038/pr.2015.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Gough EK, Stephens DA, Moodie EE, Prendergast AJ, Stoltzfus RJ, Humphrey JH, Manges AR. 2015. Linear growth faltering in infants is associated with Acidaminococcus sp. and community-level changes in the gut microbiota. Microbiome 3:24. doi: 10.1186/s40168-015-0089-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan YM, Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, Gordon JI. 2016. Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351:aad3311. doi: 10.1126/science.aad3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Charbonneau MR, O'Donnell D, Blanton LV, Totten SM, Davis JC, Barratt MJ, Cheng J, Guruge J, Talcott M, Bain JR, Muehlbauer MJ, Ilkayeva O, Wu C, Struckmeyer T, Barile D, Mangani C, Jorgensen J, Fan YM, Maleta K, Dewey KG, Ashorn P, Newgard CB, Lebrilla C, Mills DA, Gordon JI. 2016. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 164:859–871. doi: 10.1016/j.cell.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Schwarzer M, Makki K, Storelli G, Machuca-Gayet I, Srutkova D, Hermanova P, Martino ME, Balmand S, Hudcovic T, Heddi A, Rieusset J, Kozakova H, Vidal H, Leulier F. 2016. Lactobacillus plantarum strain maintains growth of infant mice during chronic undernutrition. Science 351:854–857. doi: 10.1126/science.aad8588. [DOI] [PubMed] [Google Scholar]
- 301.Blanton LV, Barratt MJ, Charbonneau MR, Ahmed T, Gordon JI. 2016. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science 352:1533. doi: 10.1126/science.aad9359. [DOI] [PubMed] [Google Scholar]
- 302.Houghteling PD, Walker WA. 2015. Why is initial bacterial colonization of the intestine important to infants' and children's health? J Pediatr Gastroenterol Nutr 60:294–307. doi: 10.1097/MPG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Johnson CC, Ownby DR. 2017. The infant gut bacterial microbiota and risk of pediatric asthma and allergic diseases. Transl Res 179:60–70. doi: 10.1016/j.trsl.2016.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Johnson CC, Ownby DR. 2016. Allergies and asthma: do atopic disorders result from inadequate immune homeostasis arising from infant gut dysbiosis? Expert Rev Clin Immunol 12:379–388. doi: 10.1586/1744666X.2016.1139452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 306.Bjorksten B, Sepp E, Julge K, Voor T, Mikelsaar M. 2001. Allergy development and the intestinal microflora during the first year of life. J Allergy Clin Immunol 108:516–520. doi: 10.1067/mai.2001.118130. [DOI] [PubMed] [Google Scholar]
- 307.Penders J, Thijs C, van den Brandt PA, Kummeling I, Snijders B, Stelma F, Adams H, van Ree R, Stobberingh EE. 2007. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 56:661–667. doi: 10.1136/gut.2006.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, van den Brandt PA. 2006. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy 36:1602–1608. doi: 10.1111/j.1365-2222.2006.02599.x. [DOI] [PubMed] [Google Scholar]
- 309.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, Martricardi PM, Aberg N, Perkin MR, Tripodi S, Coates AR, Hesselmar B, Saalman R, Molin G, Ahrne S. 2008. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol 121:129–134. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 310.Ismail IH, Oppedisano F, Joseph SJ, Boyle RJ, Licciardi PV, Robins-Browne RM, Tang ML. 2012. Reduced gut microbial diversity in early life is associated with later development of eczema but not atopy in high-risk infants. Pediatr Allergy Immunol 23:674–681. doi: 10.1111/j.1399-3038.2012.01328.x. [DOI] [PubMed] [Google Scholar]
- 311.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. 2012. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol 129:434–440. doi: 10.1016/j.jaci.2011.10.025. [DOI] [PubMed] [Google Scholar]
- 312.Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. 2015. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy 70:241–244. doi: 10.1111/all.12549. [DOI] [PubMed] [Google Scholar]
- 313.Azad MB, Kozyrskyj AL. 2012. Perinatal programming of asthma: the role of gut microbiota. Clin Dev Immunol 2012:932072. doi: 10.1155/2012/932072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Hill DA, Siracusa MC, Abt MC, Kim BS, Kobuley D, Kubo M, Kambayashi T, Larosa DF, Renner ED, Orange JS, Bushman FD, Artis D. 2012. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med 18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Sudo N, Sawamura S, Tanaka K, Aiba Y, Kubo C, Koga Y. 1997. The requirement of intestinal bacterial flora for the development of an IgE production system fully susceptible to oral tolerance induction. J Immunol 159:1739–1745. [PubMed] [Google Scholar]
- 317.Russell SL, Gold MJ, Hartmann M, Willing BP, Thorson L, Wlodarska M, Gill N, Blanchet MR, Mohn WW, McNagny KM, Finlay BB. 2012. Early life antibiotic-driven changes in microbiota enhance susceptibility to allergic asthma. EMBO Rep 13:440–447. doi: 10.1038/embor.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Chu S, Yu H, Chen Y, Chen Q, Wang B, Zhang J. 2015. Periconceptional and gestational exposure to antibiotics and childhood asthma. PLoS One 10:e0140443. doi: 10.1371/journal.pone.0140443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Lapin B, Piorkowski J, Ownby D, Freels S, Chavez N, Hernandez E, Wagner-Cassanova C, Pelzel D, Vergara C, Persky V. 2015. Relationship between prenatal antibiotic use and asthma in at-risk children. Ann Allergy Asthma Immunol 114:203–207. doi: 10.1016/j.anai.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Ong MS, Umetsu DT, Mandl KD. 2014. Consequences of antibiotics and infections in infancy: bugs, drugs, and wheezing. Ann Allergy Asthma Immunol 112:441–445 e1. doi: 10.1016/j.anai.2014.01.022. [DOI] [PubMed] [Google Scholar]
- 321.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Investigators CS, Mohn WW, Turvey SE, Finlay BB. 2015. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med 7: 307ra152. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 322.Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. 2014. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin Exp Allergy 44:842–850. doi: 10.1111/cea.12253. [DOI] [PubMed] [Google Scholar]
- 323.Stiemsma LT, Arrieta MC, Dimitriu PA, Cheng J, Thorson L, Lefebvre DL, Azad MB, Subbarao P, Mandhane P, Becker A, Sears MR, Kollmann TR, Canadian Healthy Infant Longitudinal Development Study Investigators, Mohn WW, Finlay BB, Turvey SE. 2016. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci (Lond) 130:2199–2207. doi: 10.1042/CS20160349. [DOI] [PubMed] [Google Scholar]
- 324.van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, Reijmerink NE, Dompeling E, van den Brandt PA, Ferreira I, Mommers M, Thijs C. 2011. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol 128:948–955.e1–3. doi: 10.1016/j.jaci.2011.07.027. [DOI] [PubMed] [Google Scholar]
- 325.Dzidic M, Abrahamsson TR, Artacho A, Bjorksten B, Collado MC, Mira A, Jenmalm MC. 2017. Aberrant IgA responses to the gut microbiota during infancy precede asthma and allergy development. J Allergy Clin Immunol 139:1017–1025.e14. doi: 10.1016/j.jaci.2016.06.047. [DOI] [PubMed] [Google Scholar]
- 326.Reinhardt C, Reigstad CS, Backhed F. 2009. Intestinal microbiota during infancy and its implications for obesity. J Pediatr Gastroenterol Nutr 48:249–256. doi: 10.1097/MPG.0b013e318183187c. [DOI] [PubMed] [Google Scholar]
- 327.Koleva PT, Bridgman SL, Kozyrskyj AL. 2015. The infant gut microbiome: evidence for obesity risk and dietary intervention. Nutrients 7:2237–2260. doi: 10.3390/nu7042237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Scheepers LE, Penders J, Mbakwa CA, Thijs C, Mommers M, Arts IC. 2015. The intestinal microbiota composition and weight development in children: the KOALA Birth Cohort Study. Int J Obes (Lond) 39:16–25. doi: 10.1038/ijo.2014.178. [DOI] [PubMed] [Google Scholar]
- 329.Kerr CA, Grice DM, Tran CD, Bauer DC, Li D, Hendry P, Hannan GN. 2015. Early life events influence whole-of-life metabolic health via gut microflora and gut permeability. Crit Rev Microbiol 41:326–340. doi: 10.3109/1040841X.2013.837863. [DOI] [PubMed] [Google Scholar]
- 330.Blustein J, Attina T, Liu M, Ryan AM, Cox LM, Blaser MJ, Trasande L. 2013. Association of caesarean delivery with child adiposity from age 6 weeks to 15 years. Int J Obes (Lond) 37:900–906. doi: 10.1038/ijo.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Gohir W, Ratcliffe EM, Sloboda DM. 2015. Of the bugs that shape us: maternal obesity, the gut microbiome, and long-term disease risk. Pediatr Res 77:196–204. doi: 10.1038/pr.2014.169. [DOI] [PubMed] [Google Scholar]
- 332.Chang L, Neu J. 2015. Early factors leading to later obesity: interactions of the microbiome, epigenome, and nutrition. Curr Probl Pediatr Adolesc Health Care 45:134–142. doi: 10.1016/j.cppeds.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 333.Manco M. 2012. Gut microbiota and developmental programming of the brain: from evidence in behavioral endophenotypes to novel perspective in obesity. Front Cell Infect Microbiol 2:109. doi: 10.3389/fcimb.2012.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Mahana D, Trent CM, Kurtz ZD, Bokulich NA, Battaglia T, Chung J, Muller CL, Li H, Bonneau RA, Blaser MJ. 2016. Antibiotic perturbation of the murine gut microbiome enhances the adiposity, insulin resistance, and liver disease associated with high-fat diet. Genome Med 8:48. doi: 10.1186/s13073-016-0297-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, Zarate Rodriguez JG, Rogers AB, Robine N, Loke P, Blaser MJ. 2014. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, Chung J, Sohn J, Barber CM, Goldfarb DS, Raju K, Abubucker S, Zhou Y, Ruiz VE, Li H, Mitreva M, Alekseyenko AV, Weinstock GM, Sodergren E, Blaser MJ. 2015. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Li DK, Chen H, Ferber J, Odouli R. 2017. Infection and antibiotic use in infancy and risk of childhood obesity: a longitudinal birth cohort study. Lancet Diabetes Endocrinol 5:18–25. doi: 10.1016/S2213-8587(16)30281-9. [DOI] [PubMed] [Google Scholar]
- 338.Jess T. 2014. Microbiota, antibiotics, and obesity. N Engl J Med 371:2526–2528. doi: 10.1056/NEJMcibr1409799. [DOI] [PubMed] [Google Scholar]
- 339.Cox LM, Blaser MJ. 2015. Antibiotics in early life and obesity. Nat Rev Endocrinol 11:182–190. doi: 10.1038/nrendo.2014.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Turta O, Rautava S. 2016. Antibiotics, obesity and the link to microbes—what are we doing to our children? BMC Med 14:57. doi: 10.1186/s12916-016-0605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Gulden E, Wong FS, Wen L. 2015. The gut microbiota and type 1 diabetes. Clin Immunol 159:143–153. doi: 10.1016/j.clim.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.de Goffau MC, Fuentes S, van den Bogert B, Honkanen H, de Vos WM, Welling GW, Hyoty H, Harmsen HJ. 2014. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 57:1569–1577. doi: 10.1007/s00125-014-3274-0. [DOI] [PubMed] [Google Scholar]
- 343.Livanos AE, Greiner TU, Vangay P, Pathmasiri W, Stewart D, McRitchie S, Li H, Chung J, Sohn J, Kim S, Gao Z, Barber C, Kim J, Ng S, Rogers AB, Sumner S, Zhang XS, Cadwell K, Knights D, Alekseyenko A, Backhed F, Blaser MJ. 2016. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol 1:16140. doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Paun A, Danska JS. 2016. Modulation of type 1 and type 2 diabetes risk by the intestinal microbiome. Pediatr Diabetes 17:469–477. doi: 10.1111/pedi.12424. [DOI] [PubMed] [Google Scholar]
- 345.Kostic AD, Gevers D, Siljander H, Vatanen T, Hyotylainen T, Hamalainen AM, Peet A, Tillmann V, Poho P, Mattila I, Lahdesmaki H, Franzosa EA, Vaarala O, de Goffau M, Harmsen H, Ilonen J, Virtanen SM, Clish CB, Oresic M, Huttenhower C, Knip M, DABIMMUNE Study Group, Xavier RJ. 2015. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17:260–273. doi: 10.1016/j.chom.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Schirbel A, Fiocchi C. 2011. Targeting the innate immune system in pediatric inflammatory bowel disease. Expert Rev Gastroenterol Hepatol 5:33–41. doi: 10.1586/egh.10.76. [DOI] [PubMed] [Google Scholar]
- 347.Wang F, Kaplan JL, Gold BD, Bhasin MK, Ward NL, Kellermayer R, Kirschner BS, Heyman MB, Dowd SE, Cox SB, Dogan H, Steven B, Ferry GD, Cohen SA, Baldassano RN, Moran CJ, Garnett EA, Drake L, Otu HH, Mirny LA, Libermann TA, Winter HS, Korolev KS. 2016. Detecting microbial dysbiosis associated with pediatric Crohn disease despite the high variability of the gut microbiota. Cell Rep 14:945–955. doi: 10.1016/j.celrep.2015.12.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.O'Mahony SM, Marchesi JR, Scully P, Codling C, Ceolho AM, Quigley EM, Cryan JF, Dinan TG. 2009. Early life stress alters behavior, immunity, and microbiota in rats: implications for irritable bowel syndrome and psychiatric illnesses. Biol Psychiatry 65:263–267. doi: 10.1016/j.biopsych.2008.06.026. [DOI] [PubMed] [Google Scholar]
- 349.O'Mahony SM, Felice VD, Nally K, Savignac HM, Claesson MJ, Scully P, Woznicki J, Hyland NP, Shanahan F, Quigley EM, Marchesi JR, O'Toole PW, Dinan TG, Cryan JF. 2014. Disturbance of the gut microbiota in early-life selectively affects visceral pain in adulthood without impacting cognitive or anxiety-related behaviors in male rats. Neuroscience 277:885–901. doi: 10.1016/j.neuroscience.2014.07.054. [DOI] [PubMed] [Google Scholar]
- 350.Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O'Connor M, Harnedy N, O'Connor K, Henry C, O'Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O'Toole PW. 2011. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 108(Suppl 1):S4586–S4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Roger LC, Costabile A, Holland DT, Hoyles L, McCartney AL. 2010. Examination of faecal Bifidobacterium populations in breast- and formula-fed infants during the first 18 months of life. Microbiology 156:3329–3341. doi: 10.1099/mic.0.043224-0. [DOI] [PubMed] [Google Scholar]
- 352.Turroni F, Serafini F, Mangifesta M, Arioli S, Mora D, van Sinderen D, Ventura M. 2014. Expression of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in response to environmental gut conditions. FEMS Microbiol Lett 357:23–33. doi: 10.1111/1574-6968.12509. [DOI] [PubMed] [Google Scholar]
- 353.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol 20:467–476. doi: 10.1016/j.tim.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 354.Milani C, Mancabelli L, Lugli GA, Duranti S, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Ferretti P, Gorfer V, Tett A, Segata N, van Sinderen D, Ventura M. 2015. Exploring vertical transmission of bifidobacteria from mother to child. Appl Environ Microbiol 81:7078–7087. doi: 10.1128/AEM.02037-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Ventura M, Turroni F, Zomer A, Foroni E, Giubellini V, Bottacini F, Canchaya C, Claesson MJ, He F, Mantzourani M, Mulas L, Ferrarini A, Gao B, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Gupta RS, Zhang Z, Beighton D, Fitzgerald GF, O'Toole PW, van Sinderen D. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet 5:e1000785. doi: 10.1371/journal.pgen.1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Lee JH, O'Sullivan DJ. 2010. Genomic insights into bifidobacteria. Microbiol Mol Biol Rev 74:378–416. doi: 10.1128/MMBR.00004-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol Mol Biol Rev 71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Lugli GA, Milani C, Turroni F, Duranti S, Ferrario C, Viappiani A, Mancabelli L, Mangifesta M, Taminiau B, Delcenserie V, van Sinderen D, Ventura M. 2014. Investigation of the evolutionary development of the genus Bifidobacterium by comparative genomics. Appl Environ Microbiol 80:6383–6394. doi: 10.1128/AEM.02004-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Duranti S, Mangifesta M, Lugli GA, Turroni F, Anzalone R, Milani C, Mancabelli L, Ossiprandi MC, Ventura M. 2017. Bifidobacterium vansinderenii sp. nov., isolated from faeces of emperor tamarin (Saguinus imperator). Int J Syst Evol Microbiol doi: 10.1099/ijsem.0.002243. [DOI] [PubMed] [Google Scholar]
- 360.Lugli GA, Milani C, Turroni F, Duranti S, Mancabelli L, Mangifesta M, Ferrario C, Modesto M, Mattarelli P, Jiri K, van Sinderen D, Ventura M. 2017. Comparative genomic and phylogenomic analyses of the Bifidobacteriaceae family. BMC Genomics 18:568. doi: 10.1186/s12864-017-3955-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Lamendella R, Santo Domingo JW, Kelty C, Oerther DB. 2008. Bifidobacteria in feces and environmental waters. Appl Environ Microbiol 74:575–584. doi: 10.1128/AEM.01221-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Duranti S, Turroni F, Lugli GA, Milani C, Viappiani A, Mangifesta M, Gioiosa L, Palanza P, van Sinderen D, Ventura M. 2014. Genomic characterization and transcriptional studies of the starch-utilizing strain Bifidobacterium adolescentis 22L. Appl Environ Microbiol 80:6080–6090. doi: 10.1128/AEM.01993-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Duranti S, Milani C, Lugli GA, Mancabelli L, Turroni F, Ferrario C, Mangifesta M, Viappiani A, Sanchez B, Margolles A, van Sinderen D, Ventura M. 2016. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci Rep 6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Milani C, Andrea Lugli G, Duranti S, Turroni F, Mancabelli L, Ferrario C, Mangifesta M, Hevia A, Viappiani A, Scholz M, Arioli S, Sanchez B, Lane J, Ward DV, Hickey R, Mora D, Segata N, Margolles A, van Sinderen D, Ventura M. 2015. Bifidobacteria exhibit social behavior through carbohydrate resource sharing in the gut. Sci Rep 5:15782. doi: 10.1038/srep15782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Marcobal A, Barboza M, Sonnenburg ED, Pudlo N, Martens EC, Desai P, Lebrilla CB, Weimer BC, Mills DA, German JB, Sonnenburg JL. 2011. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe 10:507–514. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Bienenstock J, Buck RH, Linke H, Forsythe P, Stanisz AM, Kunze WA. 2013. Fucosylated but not sialylated milk oligosaccharides diminish colon motor contractions. PLoS One 8:e76236. doi: 10.1371/journal.pone.0076236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Lewis ZT, Totten SM, Smilowitz JT, Popovic M, Parker E, Lemay DG, Van Tassell ML, Miller MJ, Jin YS, German JB, Lebrilla CB, Mills DA. 2015. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 3:13. doi: 10.1186/s40168-015-0071-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Wada J, Ando T, Kiyohara M, Ashida H, Kitaoka M, Yamaguchi M, Kumagai H, Katayama T, Yamamoto K. 2008. Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure. Appl Environ Microbiol 74:3996–4004. doi: 10.1128/AEM.00149-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Fujita K, Oura F, Nagamine N, Katayama T, Hiratake J, Sakata K, Kumagai H, Yamamoto K. 2005. Identification and molecular cloning of a novel glycoside hydrolase family of core 1 type O-glycan-specific endo-alpha-N-acetylgalactosaminidase from Bifidobacterium longum. J Biol Chem 280:37415–37422. doi: 10.1074/jbc.M506874200. [DOI] [PubMed] [Google Scholar]
- 371.Ashida H, Miyake A, Kiyohara M, Wada J, Yoshida E, Kumagai H, Katayama T, Yamamoto K. 2009. Two distinct alpha-l-fucosidases from Bifidobacterium bifidum are essential for the utilization of fucosylated milk oligosaccharides and glycoconjugates. Glycobiology 19:1010–1017. doi: 10.1093/glycob/cwp082. [DOI] [PubMed] [Google Scholar]
- 372.Nishimoto M, Kitaoka M. 2007. Identification of N-acetylhexosamine 1-kinase in the complete lacto-N-biose I/galacto-N-biose metabolic pathway in Bifidobacterium longum. Appl Environ Microbiol 73:6444–6449. doi: 10.1128/AEM.01425-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Ruas-Madiedo P, Gueimonde M, Fernandez-Garcia M, de los Reyes-Gavilan CG, Margolles A. 2008. Mucin degradation by Bifidobacterium strains isolated from the human intestinal microbiota. Appl Environ Microbiol 74:1936–1940. doi: 10.1128/AEM.02509-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Ruiz L, Gueimonde M, Coute Y, Salminen S, Sanchez JC, de los Reyes-Gavilan CG, Margolles A. 2011. Evaluation of the ability of Bifidobacterium longum to metabolize human intestinal mucus. FEMS Microbiol Lett 314:125–130. doi: 10.1111/j.1574-6968.2010.02159.x. [DOI] [PubMed] [Google Scholar]
- 375.Turroni F, Bottacini F, Foroni E, Mulder I, Kim JH, Zomer A, Sanchez B, Bidossi A, Ferrarini A, Giubellini V, Delledonne M, Henrissat B, Coutinho P, Oggioni M, Fitzgerald GF, Mills D, Margolles A, Kelly D, van Sinderen D, Ventura M. 2010. Genome analysis of Bifidobacterium bifidum PRL2010 reveals metabolic pathways for host-derived glycan foraging. Proc Natl Acad Sci U S A 107:19514–19519. doi: 10.1073/pnas.1011100107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Turroni F, Milani C, van Sinderen D, Ventura M. 2011. Genetic strategies for mucin metabolism in Bifidobacterium bifidum PRL2010: an example of possible human-microbe co-evolution. Gut Microbes 2:183–189. doi: 10.4161/gmic.2.3.16105. [DOI] [PubMed] [Google Scholar]
- 377.Duranti S, Milani C, Lugli GA, Turroni F, Mancabelli L, Sanchez B, Ferrario C, Viappiani A, Mangifesta M, Mancino W, Gueimonde M, Margolles A, van Sinderen D, Ventura M. 2015. Insights from genomes of representatives of the human gut commensal Bifidobacterium bifidum. Environ Microbiol 17:2515–2531. doi: 10.1111/1462-2920.12743. [DOI] [PubMed] [Google Scholar]
- 378.O'Connell Motherway M, Fitzgerald GF, van Sinderen D. 2011. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb Biotechnol 4:403–416. doi: 10.1111/j.1751-7915.2010.00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6:285–306. doi: 10.1007/s12263-010-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Pokusaeva K, Neves AR, Zomer A, O'Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb Biotechnol 3:311–323. doi: 10.1111/j.1751-7915.2009.00152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Pokusaeva K, O'Connell-Motherway M, Zomer A, Fitzgerald GF, van Sinderen D. 2009. Characterization of two novel alpha-glucosidases from Bifidobacterium breve UCC2003. Appl Environ Microbiol 75:1135–1143. doi: 10.1128/AEM.02391-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Ryan SM, Fitzgerald GF, van Sinderen D. 2006. Screening for and identification of starch-, amylopectin-, and pullulan-degrading activities in bifidobacterial strains. Appl Environ Microbiol 72:5289–5296. doi: 10.1128/AEM.00257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Turroni F, Ozcan E, Milani C, Mancabelli L, Viappiani A, van Sinderen D, Sela DA, Ventura M. 2015. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol 6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Egan M, O'Connell Motherway M, Ventura M, van Sinderen D. 2014. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl Environ Microbiol 80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Egan M, Motherway MO, Kilcoyne M, Kane M, Joshi L, Ventura M, van Sinderen D. 2014. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol 14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Egan M, Jiang H, O'Connell Motherway M, Oscarson S, van Sinderen D. 2016. Glycosulfatase-encoding gene cluster in Bifidobacterium breve UCC2003. Appl Environ Microbiol 82:6611–6623. doi: 10.1128/AEM.02022-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Pande S, Shitut S, Freund L, Westermann M, Bertels F, Colesie C, Bischofs IB, Kost C. 2015. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun 6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 388.Phelan VV, Liu WT, Pogliano K, Dorrestein PC. 2011. Microbial metabolic exchange–the chemotype-to-phenotype link. Nat Chem Biol 8:26–35. doi: 10.1038/nchembio.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. 2013. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 390.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 391.Barcenilla A, Pryde SE, Martin JC, Duncan SH, Stewart CS, Henderson C, Flint HJ. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl Environ Microbiol 66:1654–1661. doi: 10.1128/AEM.66.4.1654-1661.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Morrison DJ, Mackay WG, Edwards CA, Preston T, Dodson B, Weaver LT. 2006. Butyrate production from oligofructose fermentation by the human faecal flora: what is the contribution of extracellular acetate and lactate? Br J Nutr 96:570–577. [PubMed] [Google Scholar]
- 393.Duncan SH, Flint HJ. 2008. Proposal of a neotype strain (A1-86) for Eubacterium rectale. Request for an opinion. Int J Syst Evol Microbiol 58:1735–1736. doi: 10.1099/ijs.0.2008/004580-0. [DOI] [PubMed] [Google Scholar]
- 394.Falony G, Verschaeren A, De Bruycker F, De Preter V, Verbeke K, Leroy F, De Vuyst L. 2009. In vitro kinetics of prebiotic inulin-type fructan fermentation by butyrate-producing colon bacteria: implementation of online gas chromatography for quantitative analysis of carbon dioxide and hydrogen gas production. Appl Environ Microbiol 75:5884–5892. doi: 10.1128/AEM.00876-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Turroni F, Milani C, Duranti S, Mancabelli L, Mangifesta M, Viappiani A, Lugli GA, Ferrario C, Gioiosa L, Ferrarini A, Li J, Palanza P, Delledonne M, van Sinderen D, Ventura M. 2016. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Hidalgo-Cantabrana C, Sanchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. 2014. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol 80:9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Ferrario C, Milani C, Mancabelli L, Lugli GA, Duranti S, Mangifesta M, Viappiani A, Turroni F, Margolles A, Ruas-Madiedo P, van Sinderen D, Ventura M. 2016. Modulation of the eps-ome transcription of bifidobacteria through simulation of human intestinal environment. FEMS Microbiol Ecol 92:fiw056. doi: 10.1093/femsec/fiw056. [DOI] [PubMed] [Google Scholar]
- 398.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A 109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Foroni E, Serafini F, Amidani D, Turroni F, He F, Bottacini F, O'Connell Motherway M, Viappiani A, Zhang Z, Rivetti C, van Sinderen D, Ventura M. 2011. Genetic analysis and morphological identification of pilus-like structures in members of the genus Bifidobacterium. Microb Cell Fact 10(Suppl 1):S16. doi: 10.1186/1475-2859-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sanchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Freitas F, Alves VD, Reis MA. 2011. Advances in bacterial exopolysaccharides: from production to biotechnological applications. Trends Biotechnol 29:388–398. doi: 10.1016/j.tibtech.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 403.Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, Mazmanian SK. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Laws A, Gu Y, Marshall V. 2001. Biosynthesis, characterisation, and design of bacterial exopolysaccharides from lactic acid bacteria. Biotechnol Adv 19:597–625. doi: 10.1016/S0734-9750(01)00084-2. [DOI] [PubMed] [Google Scholar]
- 406.Lopez P, Monteserin DC, Gueimonde M, de los Reyes-Gavilan CG, Margolles A, Suarez A, Ruas-Madiedo P. 2012. Exopolysaccharide-producing Bifidobacterium strains elicit different in vitro responses upon interaction with human cells. Food Res Int 46:99–107. doi: 10.1016/j.foodres.2011.11.020. [DOI] [Google Scholar]
- 407.Hughes KR, Harnisch LC, Alcon-Giner C, Mitra S, Wright CJ, Ketskemety J, van Sinderen D, Watson AJ, Hall LJ. 2017. Bifidobacterium breve reduces apoptotic epithelial cell shedding in an exopolysaccharide and MyD88-dependent manner. Open Biol 7:160155. doi: 10.1098/rsob.160155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Schiavi E, Gleinser M, Molloy E, Groeger D, Frei R, Ferstl R, Rodriguez-Perez N, Ziegler M, Grant R, Moriarty TF, Plattner S, Healy S, O'Connell Motherway M, Akdis CA, Roper J, Altmann F, van Sinderen D, O'Mahony L. 2016. The surface-associated exopolysaccharide of Bifidobacterium longum 35624 plays an essential role in dampening host proinflammatory responses and repressing local TH17 responses. Appl Environ Microbiol 82:7185–7196. doi: 10.1128/AEM.02238-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Altmann F, Kosma P, O'Callaghan A, Leahy S, Bottacini F, Molloy E, Plattner S, Schiavi E, Gleinser M, Groeger D, Grant R, Rodriguez Perez N, Healy S, Svehla E, Windwarder M, Hofinger A, O'Connell Motherway M, Akdis CA, Xu J, Roper J, van Sinderen D, O'Mahony L. 2016. Genome analysis and characterisation of the exopolysaccharide produced by Bifidobacterium longum subsp. longum 35624. PLoS One 11:e0162983. doi: 10.1371/journal.pone.0162983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Bach JF. 2002. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med 347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- 411.Ivanov D, Emonet C, Foata F, Affolter M, Delley M, Fisseha M, Blum-Sperisen S, Kochhar S, Arigoni F. 2006. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J Biol Chem 281:17246–17252. doi: 10.1074/jbc.M601678200. [DOI] [PubMed] [Google Scholar]
- 412.Turroni F, Foroni E, O'Connell Motherway M, Bottacini F, Giubellini V, Zomer A, Ferrarini A, Delledonne M, Zhang Z, van Sinderen D, Ventura M. 2010. Characterization of the serpin-encoding gene of Bifidobacterium breve 210B. Appl Environ Microbiol 76:3206–3219. doi: 10.1128/AEM.02938-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Alvarez-Martin P, Fernandez M, O'Connell-Motherway M, O'Connell KJ, Sauvageot N, Fitzgerald GF, MacSharry J, Zomer A, van Sinderen D. 2012. A conserved two-component signal transduction system controls the response to phosphate starvation in Bifidobacterium breve UCC2003. Appl Environ Microbiol 78:5258–5269. doi: 10.1128/AEM.00804-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Bottacini F, Ventura M, van Sinderen D, O'Connell Motherway M. 2014. Diversity, ecology and intestinal function of bifidobacteria. Microb Cell Fact 13(Suppl 1):S4. doi: 10.1186/1475-2859-13-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Christiaen SE, O'Connell Motherway M, Bottacini F, Lanigan N, Casey PG, Huys G, Nelis HJ, van Sinderen D, Coenye T. 2014. Autoinducer-2 plays a crucial role in gut colonization and probiotic functionality of Bifidobacterium breve UCC2003. PLoS One 9:e98111. doi: 10.1371/journal.pone.0098111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Xavier KB, Bassler BL. 2003. LuxS quorum sensing: more than just a numbers game. Curr Opin Microbiol 6:191–197. doi: 10.1016/S1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 417.Milani C, Lugli GA, Duranti S, Turroni F, Bottacini F, Mangifesta M, Sanchez B, Viappiani A, Mancabelli L, Taminiau B, Delcenserie V, Barrangou R, Margolles A, van Sinderen D, Ventura M. 2014. Genomic encyclopedia of type strains of the genus Bifidobacterium. Appl Environ Microbiol 80:6290–6302. doi: 10.1128/AEM.02308-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Rainey FA. 2015. Clostridia class. nov. In Bergey's manual of systematics of archaea and bacteria. doi: 10.1002/9781118960608.cbm00034. [DOI] [Google Scholar]
- 419.Lawson PA, Citron DM, Tyrrell KL, Finegold SM. 2016. Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe 40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 420.Lakshminarayanan B, Harris HM, Coakley M, O'Sullivan O, Stanton C, Pruteanu M, Shanahan F, O'Toole PW, Ross RP, ELDERMET Consortium. 2013. Prevalence and characterization of Clostridium perfringens from the faecal microbiota of elderly Irish subjects. J Med Microbiol 62:457–466. doi: 10.1099/jmm.0.052258-0. [DOI] [PubMed] [Google Scholar]
- 421.Mevissen-Verhage EA, Marcelis JH, de Vos MN, Harmsen-van Amerongen WC, Verhoef J. 1987. Bifidobacterium, Bacteroides, and Clostridium spp. in fecal samples from breast-fed and bottle-fed infants with and without iron supplement. J Clin Microbiol 25:285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Stackebrandt E, Kramer I, Swiderski J, Hippe H. 1999. Phylogenetic basis for a taxonomic dissection of the genus Clostridium. FEMS Immunol Med Microbiol 24:253–258. doi: 10.1111/j.1574-695X.1999.tb01291.x. [DOI] [PubMed] [Google Scholar]
- 423.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, Kim S, Fritz JV, Wilmes P, Ueha S, Matsushima K, Ohno H, Olle B, Sakaguchi S, Taniguchi T, Morita H, Hattori M, Honda K. 2013. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature 500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 424.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, Cheng G, Yamasaki S, Saito T, Ohba Y, Taniguchi T, Takeda K, Hori S, Ivanov II, Umesaki Y, Itoh K, Honda K. 2011. Induction of colonic regulatory T cells by indigenous Clostridium species. Science 331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. 2004. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 427.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Morotomi M, Nagai F, Sakon H, Tanaka R. 2009. Paraprevotella clara gen. nov., sp. nov. and Paraprevotella xylaniphila sp. nov., members of the family ‘Prevotellaceae’ isolated from human faeces. Int J Syst Evol Microbiol 59:1895–1900. doi: 10.1099/ijs.0.008169-0. [DOI] [PubMed] [Google Scholar]
- 429.Sakamoto M, Ohkuma M. 2012. Bacteroides sartorii is an earlier heterotypic synonym of Bacteroides chinchillae and has priority. Int J Syst Evol Microbiol 62:1241–1244. doi: 10.1099/ijs.0.035659-0. [DOI] [PubMed] [Google Scholar]
- 430.Macfarlane GT, Cummings JH, Allison C. 1986. Protein degradation by human intestinal bacteria. J Gen Microbiol 132:1647–1656. [DOI] [PubMed] [Google Scholar]
- 431.Narushima S, Itoha K, Miyamoto Y, Park SH, Nagata K, Kuruma K, Uchida K. 2006. Deoxycholic acid formation in gnotobiotic mice associated with human intestinal bacteria. Lipids 41:835–843. doi: 10.1007/s11745-006-5038-1. [DOI] [PubMed] [Google Scholar]
- 432.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555–558. doi: 10.1038/35107092. [DOI] [PubMed] [Google Scholar]
- 433.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. 2005. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 434.Liu CH, Lee SM, Vanlare JM, Kasper DL, Mazmanian SK. 2008. Regulation of surface architecture by symbiotic bacteria mediates host colonization. Proc Natl Acad Sci U S A 105:3951–3956. doi: 10.1073/pnas.0709266105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Kuwahara T, Yamashita A, Hirakawa H, Nakayama H, Toh H, Okada N, Kuhara S, Hattori M, Hayashi T, Ohnishi Y. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc Natl Acad Sci U S A 101:14919–14924. doi: 10.1073/pnas.0404172101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 437.Vipperla K, O'Keefe SJ. 2012. The microbiota and its metabolites in colonic mucosal health and cancer risk. Nutr Clin Pract 27:624–635. doi: 10.1177/0884533612452012. [DOI] [PubMed] [Google Scholar]
- 438.Solis G, de Los Reyes-Gavilan CG, Fernandez N, Margolles A, Gueimonde M. 2010. Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16:307–310. doi: 10.1016/j.anaerobe.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 439.Kageyama A, Benno Y, Nakase T. 1999. Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int J Syst Bacteriol 49:557–565. doi: 10.1099/00207713-49-2-557. [DOI] [PubMed] [Google Scholar]
- 440.Nagai F, Watanabe Y, Morotomi M. 2010. Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int J Syst Evol Microbiol 60:2639–2646. doi: 10.1099/ijs.0.017533-0. [DOI] [PubMed] [Google Scholar]
- 441.Durand GA, Cadoret F, Lagier JC, Fournier PE, Raoult D. 2017. Description of ‘Gorbachella massiliensis’ gen. nov., sp. nov., ‘Fenollaria timonensis’ sp. nov., ‘Intestinimonas timonensis’ sp. nov. and ‘Collinsella ihuae’ sp. nov. isolated from healthy fresh stools with culturomics. New Microbes New Infect 16:60–62. doi: 10.1016/j.nmni.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Padmanabhan R, Dubourg G, Jean-Christophe l, Nguyen TT, Couderc C, Rossi-Tamisier M, Caputo A, Raoult D, Fournier PE. 2014. Non-contiguous finished genome sequence and description of Collinsella massiliensis sp. nov. Stand Genomic Sci 9:1144–1158. doi: 10.4056/sigs.5399696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Bidart GN, Rodriguez-Diaz J, Yebra MJ. 2015. The extracellular wall-bound beta-N-acetylglucosaminidase from Lactobacillus casei is involved in the metabolism of the human milk oligosaccharide lacto-N-triose. Appl Environ Microbiol 82:570–577. doi: 10.1128/AEM.02888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Derrien M, Collado MC, Ben-Amor K, Salminen S, de Vos WM. 2008. The mucin degrader Akkermansia muciniphila is an abundant resident of the human intestinal tract. Appl Environ Microbiol 74:1646–1648. doi: 10.1128/AEM.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 446.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KC, Aalvink S, Martinez LO, Dumas ME, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 449.Ottman N, Reunanen J, Meijerink M, Pietila TE, Kainulainen V, Klievink J, Huuskonen L, Aalvink S, Skurnik M, Boeren S, Satokari R, Mercenier A, Palva A, Smidt H, de Vos WM, Belzer C. 2017. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One 12:e0173004. doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Ottman N. 2015. Host immunostimulation and substrate utilization of the gut symbiont Akkermansia muciniphila. PhD thesis. Wageningen University, Wageningen, The Netherlands. [Google Scholar]
- 451.Belzer C, de Vos WM. 2012. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.de Vos WM. 2017. Microbe profile: Akkermansia muciniphila: a conserved intestinal symbiont that acts as the gatekeeper of our mucosa. Microbiology doi: 10.1099/mic.0.000444. [DOI] [PubMed] [Google Scholar]
- 453.Flint HJ, Scott KP, Louis P, Duncan SH. 2012. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol 9:577–589. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 454.Gordon JI, Dewey KG, Mills DA, Medzhitov RM. 2012. The human gut microbiota and undernutrition. Sci Transl Med 4: 137ps12. doi: 10.1126/scitranslmed.3004347. [DOI] [PubMed] [Google Scholar]
- 455.Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jorgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clement K, Dore J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, MetaHIT Consortium, Bork P, Wang J, Ehrlich SD, Pedersen O. 2013. Richness of human gut microbiome correlates with metabolic markers. Nature 500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- 456.Wopereis H, Oozeer R, Knipping K, Belzer C, Knol J. 2014. The first thousand days—intestinal microbiology of early life: establishing a symbiosis. Pediatr Allergy Immunol 25:428–438. doi: 10.1111/pai.12232. [DOI] [PubMed] [Google Scholar]
- 457.Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, de Vos WM. 2016. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun 7:10410. doi: 10.1038/ncomms10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.DiBartolomeo ME, Claud EC. 2016. The developing microbiome of the preterm infant. Clin Ther 38:733–739. doi: 10.1016/j.clinthera.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.de Weerth C, Fuentes S, Puylaert P, de Vos WM. 2013. Intestinal microbiota of infants with colic: development and specific signatures. Pediatrics 131:e550–e558. doi: 10.1542/peds.2012-1449. [DOI] [PubMed] [Google Scholar]
- 460.Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, Kalach N, Leroux B, Dupont C. 2007. Conditions of bifidobacterial colonization in preterm infants: a prospective analysis. J Pediatr Gastroenterol Nutr 44:577–582. doi: 10.1097/MPG.0b013e3180406b20. [DOI] [PubMed] [Google Scholar]
- 461.Tojo R, Suarez A, Clemente MG, de los Reyes-Gavilan CG, Margolles A, Gueimonde M, Ruas-Madiedo P. 2014. Intestinal microbiota in health and disease: role of bifidobacteria in gut homeostasis. World J Gastroenterol 20:15163–15176. doi: 10.3748/wjg.v20.i41.15163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.de Meij TG, de Groot EF, Eck A, Budding AE, Kneepkens CM, Benninga MA, van Bodegraven AA, Savelkoul PH. 2016. Characterization of microbiota in children with chronic functional constipation. PLoS One 11:e0164731. doi: 10.1371/journal.pone.0164731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. 2012. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr 55:321–327. doi: 10.1097/MPG.0b013e31824fb899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 465.Taipale T, Pienihakkinen K, Isolauri E, Larsen C, Brockmann E, Alanen P, Jokela J, Soderling E. 2011. Bifidobacterium animalis subsp. lactis BB-12 in reducing the risk of infections in infancy. Br J Nutr 105:409–416. doi: 10.1017/S0007114510003685. [DOI] [PubMed] [Google Scholar]
- 466.Vanhoutvin SA, Troost FJ, Kilkens TO, Lindsey PJ, Hamer HM, Jonkers DM, Venema K, Brummer RJ. 2009. The effects of butyrate enemas on visceral perception in healthy volunteers. Neurogastroenterol Motil 21:952-e76. doi: 10.1111/j.1365-2982.2009.01324.x. [DOI] [PubMed] [Google Scholar]
- 467.Ogawa K, Ben RA, Pons S, de Paolo MI, Bustos Fernandez L. 1992. Volatile fatty acids, lactic acid, and pH in the stools of breast-fed and bottle-fed infants. J Pediatr Gastroenterol Nutr 15:248–252. doi: 10.1097/00005176-199210000-00004. [DOI] [PubMed] [Google Scholar]
- 468.Haarman M, Knol J. 2006. Quantitative real-time PCR analysis of fecal Lactobacillus species in infants receiving a prebiotic infant formula. Appl Environ Microbiol 72:2359–2365. doi: 10.1128/AEM.72.4.2359-2365.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Abe R, Oda S, Sadahiro T, Nakamura M, Hirayama Y, Tateishi Y, Shinozaki K, Hirasawa H. 2010. Gram-negative bacteremia induces greater magnitude of inflammatory response than Gram-positive bacteremia. Crit Care 14:R27. doi: 10.1186/cc8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 470.Kant R, de Vos WM, Palva A, Satokari R. 2014. Immunostimulatory CpG motifs in the genomes of gut bacteria and their role in human health and disease. J Med Microbiol 63:293–308. doi: 10.1099/jmm.0.064220-0. [DOI] [PubMed] [Google Scholar]
- 471.Young SL, Simon MA, Baird MA, Tannock GW, Bibiloni R, Spencely K, Lane JM, Fitzharris P, Crane J, Town I, Addo-Yobo E, Murray CS, Woodcock A. 2004. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin Diagn Lab Immunol 11:686–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Vatanen T, Kostic AD, d'Hennezel E, Siljander H, Franzosa EA, Yassour M, Kolde R, Vlamakis H, Arthur TD, Hamalainen AM, Peet A, Tillmann V, Uibo R, Mokurov S, Dorshakova N, Ilonen J, Virtanen SM, Szabo SJ, Porter JA, Lahdesmaki H, Huttenhower C, Gevers D, Cullen TW, Knip M, DIABIMMUNE Study Group, Xavier RJ. 2016. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165:842–853. doi: 10.1016/j.cell.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Virgin HW. 2014. The virome in mammalian physiology and disease. Cell 157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Ray K. 2015. Gut microbiota: a ‘friendly’ gut virus? Nat Rev Gastroenterol Hepatol 12:6. doi: 10.1038/nrgastro.2014.220. [DOI] [PubMed] [Google Scholar]
- 475.Hayes S, Mahony J, Nauta A, van Sinderen D. 2017. Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses 9:E127. doi: 10.3390/v9060127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 476.Wommack KE, Bhavsar J, Polson SW, Chen J, Dumas M, Srinivasiah S, Furman M, Jamindar S, Nasko DJ. 2012. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand Genomic Sci 6:421. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, Gordon JI. 2010. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466:334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. 2012. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol 10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. 2016. Healthy human gut phageome. Proc Natl Acad Sci U S A 113:10400–10405. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, Warner BB, Tarr PI, Wang D, Holtz LR. 2015. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med 21:1228–1234. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, Smith MI, Wang D, Virgin HW, Rohwer F, Gordon JI. 2015. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci U S A 112:11941–11946. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Kapusinszky B, Minor P, Delwart E. 2012. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol 50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. 2011. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brussow H. 2003. Phage as agents of lateral gene transfer. Curr Opin Microbiol 6:417–424. doi: 10.1016/S1369-5274(03)00086-9. [DOI] [PubMed] [Google Scholar]
- 485.Ogilvie LA, Jones BV. 2015. The human gut virome: a multifaceted majority. Front Microbiol 6:918. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, Duranti S, Ward DV, Ossiprandi MC, Moineau S, van Sinderen D, Ventura M. 2016. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol 18:2196–2213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 487.Sheth RU, Cabral V, Chen SP, Wang HH. 2016. Manipulating bacterial communities by in situ microbiome engineering. Trends Genet 32:189–200. doi: 10.1016/j.tig.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Hempel S, Newberry S, Ruelaz A, Wang Z, Miles JN, Suttorp MJ, Johnsen B, Shanman R, Slusser W, Fu N, Smith A, Roth B, Polak J, Motala A, Perry T, Shekelle PG. 2011. Safety of probiotics used to reduce risk and prevent or treat disease. Agency for Healthcare Research and Quality, Rockville, MD. [PMC free article] [PubMed] [Google Scholar]
- 489.Stojanovic N, Plecas D, Plesinac S. 2012. Normal vaginal flora, disorders and application of probiotics in pregnancy. Arch Gynecol Obstet 286:325–332. doi: 10.1007/s00404-012-2293-7. [DOI] [PubMed] [Google Scholar]
- 490.Gueimonde M, Sakata S, Kalliomaki M, Isolauri E, Benno Y, Salminen S. 2006. Effect of maternal consumption of lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J Pediatr Gastroenterol Nutr 42:166–170. doi: 10.1097/01.mpg.0000189346.25172.fd. [DOI] [PubMed] [Google Scholar]
- 491.Lahtinen SJ, Boyle RJ, Kivivuori S, Oppedisano F, Smith KR, Robins-Browne R, Salminen SJ, Tang ML. 2009. Prenatal probiotic administration can influence Bifidobacterium microbiota development in infants at high risk of allergy. J Allergy Clin Immunol 123:499–501. doi: 10.1016/j.jaci.2008.11.034. [DOI] [PubMed] [Google Scholar]
- 492.Panigrahi P, Parida S, Pradhan L, Mohapatra SS, Misra PR, Johnson JA, Chaudhry R, Taylor S, Hansen NI, Gewolb IH. 2008. Long-term colonization of a Lactobacillus plantarum synbiotic preparation in the neonatal gut. J Pediatr Gastroenterol Nutr 47:45–53. doi: 10.1097/MPG.0b013e31815a5f2c. [DOI] [PubMed] [Google Scholar]
- 493.AlFaleh K, Anabrees J. 2014. Probiotics for prevention of necrotizing enterocolitis in preterm infants. Cochrane Database Syst Rev doi: 10.1002/14651858.CD005496.pub4. [DOI] [PubMed] [Google Scholar]
- 494.Kajander K, Hatakka K, Poussa T, Farkkila M, Korpela R. 2005. A probiotic mixture alleviates symptoms in irritable bowel syndrome patients: a controlled 6-month intervention. Aliment Pharmacol Ther 22:387–394. doi: 10.1111/j.1365-2036.2005.02579.x. [DOI] [PubMed] [Google Scholar]
- 495.Lin HC, Su BH, Chen AC, Lin TW, Tsai CH, Yeh TF, Oh W. 2005. Oral probiotics reduce the incidence and severity of necrotizing enterocolitis in very low birth weight infants. Pediatrics 115:1–4. doi: 10.1542/peds.2004-1463. [DOI] [PubMed] [Google Scholar]
- 496.Braegger C, Chmielewska A, Decsi T, Kolacek S, Mihatsch W, Moreno L, Piescik M, Puntis J, Shamir R, Szajewska H, Turck D, van Goudoever J, Nutrition ECo. 2011. Supplementation of infant formula with probiotics and/or prebiotics: a systematic review and comment by the ESPGHAN committee on nutrition. J Pediatr Gastroenterol Nutr 52:238–250. doi: 10.1097/MPG.0b013e3181fb9e80. [DOI] [PubMed] [Google Scholar]
- 497.Cuello-Garcia CA, Brozek JL, Fiocchi A, Pawankar R, Yepes-Nunez JJ, Terracciano L, Gandhi S, Agarwal A, Zhang Y, Schunemann HJ. 2015. Probiotics for the prevention of allergy: a systematic review and meta-analysis of randomized controlled trials. J Allergy Clin Immunol 136:952–961. doi: 10.1016/j.jaci.2015.04.031. [DOI] [PubMed] [Google Scholar]
- 498.Bergmann H, Rodriguez JM, Salminen S, Szajewska H. 2014. Probiotics in human milk and probiotic supplementation in infant nutrition: a workshop report. Br J Nutr 112:1119–1128. doi: 10.1017/S0007114514001949. [DOI] [PubMed] [Google Scholar]
- 499.Chau K, Lau E, Greenberg S, Jacobson S, Yazdani-Brojeni P, Verma N, Koren G. 2015. Probiotics for infantile colic: a randomized, double-blind, placebo-controlled trial investigating Lactobacillus reuteri DSM 17938. J Pediatr 166:74–78. doi: 10.1016/j.jpeds.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 500.Szajewska H, Gyrczuk E, Horvath A. 2013. Lactobacillus reuteri DSM 17938 for the management of infantile colic in breastfed infants: a randomized, double-blind, placebo-controlled trial. J Pediatr 162:257–262. doi: 10.1016/j.jpeds.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 501.Savino F, Cordisco L, Tarasco V, Palumeri E, Calabrese R, Oggero R, Roos S, Matteuzzi D. 2010. Lactobacillus reuteri DSM 17938 in infantile colic: a randomized, double-blind, placebo-controlled trial. Pediatrics 126:e526–e533. doi: 10.1542/peds.2010-0433. [DOI] [PubMed] [Google Scholar]
- 502.Partty A, Luoto R, Kalliomaki M, Salminen S, Isolauri E. 2013. Effects of early prebiotic and probiotic supplementation on development of gut microbiota and fussing and crying in preterm infants: a randomized, double-blind, placebo-controlled trial. J Pediatr 163:1272–1277.e1–2. doi: 10.1016/j.jpeds.2013.05.035. [DOI] [PubMed] [Google Scholar]
- 503.Partty A, Isolauri E. 2012. Gut microbiota and infant distress—the association between compositional development of the gut microbiota and fussing and crying in early infancy. Microb Ecol Health Dis doi: 10.3402/mehd.v23i0.18577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Sung V, Hiscock H, Tang ML, Mensah FK, Nation ML, Satzke C, Heine RG, Stock A, Barr RG, Wake M. 2014. Treating infant colic with the probiotic Lactobacillus reuteri: double blind, placebo controlled randomised trial. BMJ 348:g2107. doi: 10.1136/bmj.g2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 505.Skorka A, Piescik-Lech M, Kolodziej M, Szajewska H. 2017. To add or not to add probiotics to infant formulae? An updated systematic review. Benef Microbes doi: 10.3920/BM2016.0233. [DOI] [PubMed] [Google Scholar]
- 506.Mah KW, Chin VI, Wong WS, Lay C, Tannock GW, Shek LP, Aw MM, Chua KY, Wong HB, Panchalingham A, Lee BW. 2007. Effect of a milk formula containing probiotics on the fecal microbiota of Asian infants at risk of atopic diseases. Pediatr Res 62:674–679. doi: 10.1203/PDR.0b013e31815991d5. [DOI] [PubMed] [Google Scholar]
- 507.Boyle RJ, Bath-Hextall FJ, Leonardi-Bee J, Murrell DF, Tang ML. 2008. Probiotics for treating eczema. Cochrane Database Syst Rev doi: 10.1002/14651858.CD006135.pub2. [DOI] [PubMed] [Google Scholar]
- 508.Correa NB, Peret Filho LA, Penna FJ, Lima FM, Nicoli JR. 2005. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic-associated diarrhea in infants. J Clin Gastroenterol 39:385–389. doi: 10.1097/01.mcg.0000159217.47419.5b. [DOI] [PubMed] [Google Scholar]
- 509.Vandenplas Y, Zakharova I, Dmitrieva Y. 2015. Oligosaccharides in infant formula: more evidence to validate the role of prebiotics. Br J Nutr 113:1339–1344. doi: 10.1017/S0007114515000823. [DOI] [PubMed] [Google Scholar]
- 510.Jinno S, Toshimitsu T, Nakamura Y, Kubota T, Igoshi Y, Ozawa N, Suzuki S, Nakano T, Morita Y, Arima T, Yamaide F, Kohno Y, Masuda K, Shimojo N. 2017. Maternal prebiotic ingestion increased the number of fecal bifidobacteria in pregnant women but not in their neonates aged one month. Nutrients 9:E196. doi: 10.3390/nu9030196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Kunz C, Rudloff S, Baier W, Klein N, Strobel S. 2000. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr 20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 512.Roberfroid M, Gibson GR, Hoyles L, McCartney AL, Rastall R, Rowland I, Wolvers D, Watzl B, Szajewska H, Stahl B, Guarner F, Respondek F, Whelan K, Coxam V, Davicco MJ, Leotoing L, Wittrant Y, Delzenne NM, Cani PD, Neyrinck AM, Meheust A. 2010. Prebiotic effects: metabolic and health benefits. Br J Nutr 104(Suppl 2):S1–S63. doi: 10.1017/S0007114510003363. [DOI] [PubMed] [Google Scholar]
- 513.Firmansyah A, Chongviriyaphan N, Dillon DH, Khan NC, Morita T, Tontisirin K, Tuyen LD, Wang W, Bindels J, Deurenberg P, Ong S, Hautvast J, Meyer D, Vaughan EE. 2016. Fructans in the first 1000 days of life and beyond, and for pregnancy. Asia Pac J Clin Nutr 25:652–675. doi: 10.6133/apjcn.092016.02. [DOI] [PubMed] [Google Scholar]
- 514.Veereman-Wauters G, Staelens S, Van de Broek H, Plaskie K, Wesling F, Roger LC, McCartney AL, Assam P. 2011. Physiological and bifidogenic effects of prebiotic supplements in infant formulae. J Pediatr Gastroenterol Nutr 52:763–771. doi: 10.1097/MPG.0b013e3182139f39. [DOI] [PubMed] [Google Scholar]
- 515.Knol J, Scholtens P, Kafka C, Steenbakkers J, Gro S, Helm K, Klarczyk M, Schopfer H, Bockler HM, Wells J. 2005. Colon microflora in infants fed formula with galacto- and fructo-oligosaccharides: more like breast-fed infants. J Pediatr Gastroenterol Nutr 40:36–42. doi: 10.1097/00005176-200501000-00007. [DOI] [PubMed] [Google Scholar]
- 516.Ben XM, Li J, Feng ZT, Shi SY, Lu YD, Chen R, Zhou XY. 2008. Low level of galacto-oligosaccharide in infant formula stimulates growth of intestinal bifidobacteria and lactobacilli. World J Gastroenterol 14:6564–6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 517.Rinne MM, Gueimonde M, Kalliomaki M, Hoppu U, Salminen SJ, Isolauri E. 2005. Similar bifidogenic effects of prebiotic-supplemented partially hydrolyzed infant formula and breastfeeding on infant gut microbiota. FEMS Immunol Med Microbiol 43:59–65. doi: 10.1016/j.femsim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 518.Sierra C, Bernal MJ, Blasco J, Martinez R, Dalmau J, Ortuno I, Espin B, Vasallo MI, Gil D, Vidal ML, Infante D, Leis R, Maldonado J, Moreno JM, Roman E. 2015. Prebiotic effect during the first year of life in healthy infants fed formula containing GOS as the only prebiotic: a multicentre, randomised, double-blind and placebo-controlled trial. Eur J Nutr 54:89–99. doi: 10.1007/s00394-014-0689-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 519.Scalabrin DM, Mitmesser SH, Welling GW, Harris CL, Marunycz JD, Walker DC, Bos NA, Tolkko S, Salminen S, Vanderhoof JA. 2012. New prebiotic blend of polydextrose and galacto-oligosaccharides has a bifidogenic effect in young infants. J Pediatr Gastroenterol Nutr 54:343–352. doi: 10.1097/MPG.0b013e318237ed95. [DOI] [PubMed] [Google Scholar]
- 520.Gonzalez R, Klaassens ES, Malinen E, de Vos WM, Vaughan EE. 2008. Differential transcriptional response of Bifidobacterium longum to human milk, formula milk, and galactooligosaccharide. Appl Environ Microbiol 74:4686–4694. doi: 10.1128/AEM.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Kapiki A, Costalos C, Oikonomidou C, Triantafyllidou A, Loukatou E, Pertrohilou V. 2007. The effect of a fructo-oligosaccharide supplemented formula on gut flora of preterm infants. Early Hum Dev 83:335–339. doi: 10.1016/j.earlhumdev.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 522.Luoto R, Ruuskanen O, Waris M, Kalliomaki M, Salminen S, Isolauri E. 2014. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J Allergy Clin Immunol 133:405–413. doi: 10.1016/j.jaci.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. 2016. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res 26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Dominguez-Bello MG, De Jesus-Laboy KM, Shen N, Cox LM, Amir A, Gonzalez A, Bokulich NA, Song SJ, Hoashi M, Rivera-Vinas JI, Mendez K, Knight R, Clemente JC. 2016. Partial restoration of the microbiota of cesarean-born infants via vaginal microbial transfer. Nat Med 22:250–253. doi: 10.1038/nm.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 525.Dezateux C, Colson D, Brocklehurst P, Elias P. 2016. ‘Life after Life Study’: report of a scientific meeting held at The Royal College of Physicians, 14th January 2016. doi: 10.14324/000.rp.1485681. [DOI] [Google Scholar]
- 526.Gomez-Gallego C, Garcia-Mantrana I, Salminen S, Collado MC. 2016. The human milk microbiome and factors influencing its composition and activity. Semin Fetal Neonatal Med 21:400–405. doi: 10.1016/j.siny.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 527.Jaeggi T, Kortman GA, Moretti D, Chassard C, Holding P, Dostal A, Boekhorst J, Timmerman HM, Swinkels DW, Tjalsma H, Njenga J, Mwangi A, Kvalsvig J, Lacroix C, Zimmermann MB. 2015. Iron fortification adversely affects the gut microbiome, increases pathogen abundance and induces intestinal inflammation in Kenyan infants. Gut 64:731–742. doi: 10.1136/gutjnl-2014-307720. [DOI] [PubMed] [Google Scholar]
- 528.Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, Barratt MJ, VanArendonk LG, Zhang Q, Province MA, Petri WA Jr, Ahmed T, Gordon JI. 2014. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Tissier H. 1900. Recherches sur la flore intestinale normale et pathologique du nourisson. Thesis. University of Paris, Paris, France. [Google Scholar]
- 530.Logan WR. 1913. The intestinal flora of infants and young children. J Pathol Bacteriol 18:527–551. doi: 10.1002/path.1700180154. [DOI] [Google Scholar]
- 531.Norton RC, Shohl AT. 1926. The hydrogen ion concentration of the stools of new-born infants. Am J Dis Child 32:183–191. [Google Scholar]
- 532.Ventura M, Turroni F, van Sinderen D. 2012. Probiogenomics as a tool to obtain genetic insights into adaptation of probiotic bacteria to the human gut. Bioeng Bugs 3:73–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 533.Hemsell DL, Obregon VL, Heard MC, Nobles BJ. 1989. Endometrial bacteria in asymptomatic, nonpregnant women. J Reprod Med 34:872–874. [PubMed] [Google Scholar]