SUMMARY
Chemoreceptors in bacteria detect a variety of signals and feed this information into chemosensory pathways that represent a major mode of signal transduction. The five chemoreceptors from Escherichia coli have served as traditional models in the study of this protein family. Genome analyses revealed that many bacteria contain much larger numbers of chemoreceptors with broader sensory capabilities. Chemoreceptors differ in topology, sensing mode, cellular location, and, above all, the type of ligand binding domain (LBD). Here, we highlight LBD diversity using well-established and emerging model organisms as well as genomic surveys. Nearly a hundred different types of protein domains that are found in chemoreceptor sequences are known or predicted LBDs, but only a few of them are ubiquitous. LBDs of the same class recognize different ligands, and conversely, the same ligand can be recognized by structurally different LBDs; however, recent studies began to reveal common characteristics in signal-LBD relationships. Although signals can stimulate chemoreceptors in a variety of different ways, diverse LBDs appear to employ a universal transmembrane signaling mechanism. Current and future studies aim to establish relationships between LBD types, the nature of signals that they recognize, and the mechanisms of signal recognition and transduction.
KEYWORDS: chemotaxis, receptor-ligand interaction, signal transduction
INTRODUCTION
Bacteria need to constantly adapt to changing environmental conditions to ensure survival. This is achieved through a variety of signal transduction pathways that most commonly include one-component systems (1), two-component systems (2), and chemoreceptor-based signaling cascades, also referred to as chemotaxis or chemosensory systems (3, 4). Chemosensory pathways mediate chemotaxis and type IV pilus-based motility and also regulate other cellular processes (5–7). Central to these systems is the ternary complex formed by chemoreceptors (also known as methyl-accepting chemotaxis proteins [MCPs] [8] and transducer-like proteins or Tlps [9]), the CheA histidine kinase, and the CheW coupling protein. Chemoreceptors recognize various signals and transmit this information to CheA, which ultimately controls the direction of flagellar motor rotation (reviewed in references 3, 8, and 10). Chemoreceptors contain two principal modules: input and output. The input module is usually composed of a single global domain, although in some chemoreceptors, two or more domains comprise the input (11). The output module is a conserved structure, a long dimeric four-helix bundle (4HB) composed of two symmetric antiparallel coiled coils, which comprises the cytoplasmic signaling domain (annotated the MA domain in the SMART database and the MCPsignal domain in the Pfam database) (12, 13). In transmembrane chemoreceptors that form the most common membrane topology class (11), input-output signaling is mediated by the HAMP (found in histidine kinases, adenylate cyclases, methyl-accepting chemotaxis proteins, and phosphatases) domain (14). Chemoreceptors can recognize the signal via their input module in several ways. One common mode of sensing is by the direct binding of chemoeffectors to the ligand binding input domain (LBD) (15, 16). Some input domains contain cofactors, such as heme or flavin adenine dinucleotide (FAD), which enable chemoreceptors to recognize oxygen and changes in redox status (17–19). Chemoreceptor input domains can also bind chemoeffector-loaded periplasmic binding proteins (20–22). Collectively, input domains are often referred to as sensory domains, although the term LBD is also used in a broader sense to depict the input module of chemoreceptors.
Escherichia coli and Salmonella enterica serovar Typhimurium are the classic model organisms to study chemoreceptor-based signaling, but a variety of other species were studied in this respect, primarily during the last decade (23). E. coli has five chemoreceptors that funnel stimuli into a single chemotaxis signaling cascade (8). Four of these receptors, Tar, Tsr, Trg, and Tap, contain a periplasmic 4HB LBD, whereas the signal input of the fifth receptor, Aer, occurs via a PAS (found in Per-Arnt-Sim proteins) sensory domain (24) located in the cytoplasm. Genome analyses of other bacteria have shown that many of them possess more complex chemosensory systems (7, 25, 26). First, many other bacteria possess a much larger number of chemoreceptors: on average, there are 14 chemoreceptor genes per genome (25), but in some species, more than 80 chemoreceptor genes were identified (27). The number of chemoreceptors was found to depend on bacterial lifestyle (28). For example, bacteria that are able to inhabit multiple and variable environments encode approximately five times more chemoreceptors than do those that live in a specific ecological niche (25). Second, many bacterial genomes encode multiple chemosensory pathways (7, 26), which is in marked contrast to the single pathway in E. coli. In this respect, Rhodobacter sphaeroides and Pseudomonas aeruginosa, with three (4) and four (5, 29, 30) chemosensory pathways, respectively, emerged as model organisms to study complex chemosensory networks.
The availability of sequenced genomes from many different chemotactic species revealed that the chemoreceptor family exhibits substantial divergence. While the cytoplasmic signaling domain is conserved in all chemoreceptors (13), there are large differences at the levels of protein topology and LBD type. Although a transmembrane topology with an extracellular LBD is predominant, some chemoreceptors either are entirely cytosolic, lack a LBD, or are membrane anchored and possess an LBD with a cytosolic location (11, 31). A computational analysis performed in 2010 showed that the large majority of chemoreceptors possess an LBD, but its type could be predicted for only a small fraction of them (25).
A central question in the field is how to identify chemoreceptor function, which ultimately depends on which chemoeffector binds to its LBD. However, the different families of LBDs are characterized by enormous sequence divergence (32, 33). This and the fact that chemoeffector specificity can be different in LBDs with similar sequences (34, 35) have largely hampered the functional annotation of chemoreceptors in genomic data sets by extrapolation from experimentally studied homologs in model organisms. Experimental approaches were thus necessary to elucidate chemoreceptor function. In this context, high-throughput ligand screening assays using individual, recombinantly produced LBDs (36–38) become a highly efficient strategy.
Over the last several years, tremendous progress has been made in the field, resulting in (i) the functional characterization of different chemoreceptors in many bacterial species, (ii) the availability of several chemoreceptor LBD structures, and (iii) the development of improved bioinformatics tools enabling computational identification of the LBD type. Here, we review these new developments that allowed us to assess the structural and functional diversity of the chemoreceptor sensory repertoire in greater depth and detail.
CHEMORECEPTORS IN MODEL ORGANISMS
E. coli and S. Typhimurium are classical model organisms to study chemoreceptors. Here, we briefly review the knowledge on these models and then focus on a few other selected organisms for which information about chemoreceptors and their functions became available in recent years, namely, Bacillus subtilis, Helicobacter pylori, Campylobacter jejuni, Pseudomonas aeruginosa, Pseudomonas putida, and Pseudomonas fluorescens. Due to space constraints, it is impossible to review all the information about chemoreceptors in many other species that are subjects of experimental investigation, such as Azospirillum brasilense (39), Comamonas testosteroni (40), Myxococcus xanthus (41), Rhodobacter sphaeroides (42), Sinorhizobium meliloti (43), Vibrio cholerae (44), and others.
E. coli and S. Typhimurium
The enteric bacteria E. coli and S. Typhimurium are ubiquitous colonizers of the intestines of animals. Motility is important for colonization by both commensal and pathogenic E. coli and S. Typhimurium strains (45, 46). Fundamental mechanisms of chemosensory signaling were determined by using these models (reviewed in references 3 and 8). E. coli exhibits chemotaxis toward and away from many chemoeffectors (attractants and repellents) (47–49), and responses to amino acids, sugars, dipeptides, pyrimidine bases, neurotransmitters, phenol, quorum sensing signals, substrates of the phosphotransferase system (PTS), pH, and temperature have been studied in detail (50–60).
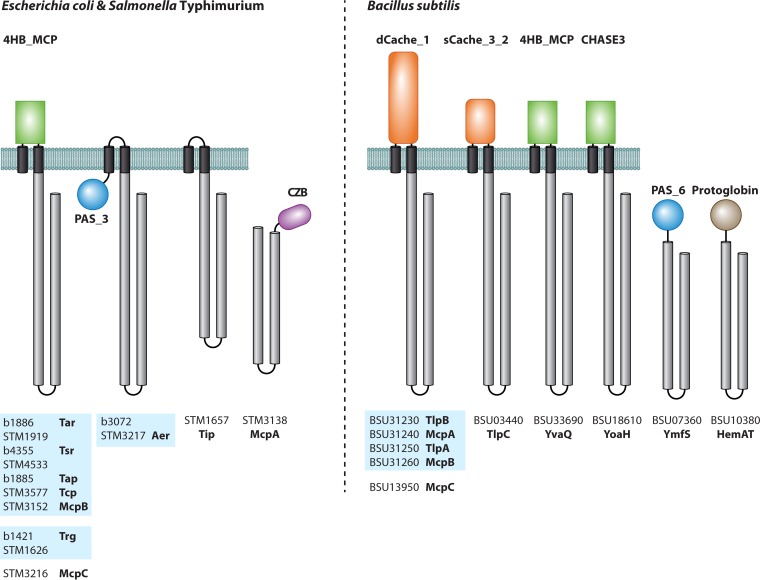
E. coli has five chemoreceptors, four of which (Tar, Tsr, Trg, and Tap) have similar domain architectures and contain the 4HB domain as a sensory module (Fig. 1 and Table 1). Tsr mediates attractant responses to serine (61) and quorum autoinducer 2 (AI-2) (50) as well as responses to oxygen, redox, and oxidizable substrates (18, 19, 62). Serine binds directly to the Tsr LBD (16), whereas AI-2 taxis requires the periplasmic AI-2 binding protein LsrB (50). Tsr was also shown to govern taxis to 3,4-dihydroxymandelic acid, a metabolite of norepinephrine that is produced by human cells (51). Tar mediates attractant responses to aspartate (61) through direct binding to its LBD (63) and to maltose via binding to a maltose binding protein (57, 64). Tar also governs negative responses to metal ions by an as-yet-unidentified mechanism (65). Trg is a chemoreceptor for attractant responses to ribose and galactose (66) that are mediated via its interaction with ribose and galactose binding proteins, respectively. Tap is stimulated by a dipeptide-loaded periplasmic protein (54) and by pyrimidines (52), causing attractant responses in both cases. In contrast to these four receptors, the Aer chemoreceptor contains a cytosolic, redox-sensitive, FAD-containing PAS domain (Fig. 1). The Aer chemoreceptor mediates responses to oxygen as well as energy taxis (18, 19, 62). Many pathogenic E. coli strains lack either Trg, Tap, or both (67).
FIG 1.
The chemoreceptor repertoires of E. coli, S. Typhimurium, and B. subtilis. Receptor topology, names, and locus tags of E. coli K-12, S. Typhimurium strain LT2, and Bacillus subtilis subsp. subtilis strain 168 are shown. The LBDs are colored and their type is annotated according to data in the Pfam database. Orthologous groups of chemoreceptors (including paralogs) are highlighted by blue shading.
TABLE 1.
Functionally characterized chemoreceptors of E. coli, S. Typhimurium, and B. subtilis
| Locus tag (chemoreceptor name) | LBD type | Effector(s) | Binding mode(s) | Reference(s) |
|---|---|---|---|---|
| E. coli | ||||
| b4355 (Tsr) | 4HB | Serine | Direct | 50, 61 |
| AI-2 | Indirect | 50 | ||
| Redox substrates, aerotaxis | Unknown | 19, 62 | ||
| 3,4-Dihydroxymandelic acid | Direct | 51 | ||
| b1886 (Tar) | 4HB | Aspartate | Direct | 61, 63 |
| Maltose | Indirect | 57, 64 | ||
| Metal ions | Unknown | 65 | ||
| b1421 (Trg) | 4HB | Ribose, galactose | Indirect | 66 |
| b1885 (Tap) | 4HB | Dipeptides | Indirect | 54 |
| Pyrimidines | Unknown | 52 | ||
| b3072 (Aer) | PAS | Aerotaxis, energy | Unknown | 18, 19, 62 |
| S. Typhimurium | ||||
| STM1919 (Tar) | 4HB | Cysteine, aspartate | Direct | 70, 208 |
| STM4533 (Tsr) | 4HB | Cysteine | Unknown | 70 |
| STM3577 (Tcp) | 4HB | Citrate | Direct | 209 |
| Phenol | Unknown | |||
| STM3152 (McpB) | 4HB | Cystine | Unknown | 70 |
| STM3216 (McpC) | 4HB | Cystine | Unknown | 70 |
| B. subtilis | ||||
| BSU10380 (HemAT) | Protoglobin | Aerotaxis | Unknown | 17 |
| BSU13950 (McpC) | dCache | 12 amino acids | Direct | 88, 89 |
| PTS substrates | Indirect | 194, 195 | ||
| BSU31240 (McpA) | dCache | Glucose, α-methylglucoside | Unknown | 9 |
| BSU31260 (McpB) | dCache | 4 amino acids | Direct | 9 |
S. Typhimurium LT2 has nine chemoreceptors (Fig. 1 and Table 1), four of which (Tar, Tsr, Trg, and Aer) are orthologs of the corresponding E. coli proteins (68). It lacks the Tap chemoreceptor but has five chemoreceptors that are not present in E. coli. Tcp mediates attractant responses to citrate and repellent responses to phenol (69). McpB and McpC appear to serve as chemoreceptors for repellent responses to cysteine (70). The function of two other chemoreceptors, Tip (71) and McpA (72), is still unknown. At the C-terminal part, McpA contains a CZB (chemoreceptor zinc binding) domain. This domain was first identified in the cytoplasmic chemoreceptor TlpD from Helicobacter pylori (see below) (73). TlpD mediates energy taxis (74) as well as chemotaxis to reactive oxygen species (ROS) (75) and pH (76), and the role of zinc binding in these processes remains to be established. As in the case of E. coli, a majority of chemoreceptors in S. Typhimurium contain a periplasmic 4HB domain (Fig. 1). More detailed information on the chemosensory mechanisms of E. coli and S. Typhimurium was reported previously (8, 10, 77).
Bacillus subtilis
B. subtilis is a member of the phylum Firmicutes, which is often found in soil but is also present in water or associated with plants or inhabits the gastrointestinal tract of ruminants and humans (78). It is an obligate aerobe, which, however, is capable of surviving under anaerobic conditions in the presence of nitrates or glucose. It is the best-characterized Gram-positive bacterium and the first nonenterobacterial model used to study chemotaxis (79). Initial studies of B. subtilis demonstrated that a galactose binding protein was essential for chemotaxis toward this sugar (80, 81), and subsequent studies revealed chemotaxis to many other sugars (82). B. subtilis shows repellent responses to different compounds, including uncouplers of oxidative phosphorylation (79, 83). Chemoattraction was observed for the 20 proteinogenic amino acids (84–86) and oxygen (17, 87).
B. subtilis has 10 chemoreceptors (Fig. 1 and Table 1). In contrast to E. coli and S. Typhimurium, it contains only one chemoreceptor with a 4HB domain, whereas five receptors contain a dCache_1 (double Cache 1) domain. This domain is composed of two α/β subdomains, each homologous to the PAS domain, and a long N-terminal helix, and it was recently shown to be the most ubiquitous extracellular LBD in prokaryotes (33). Two of the dCache_1 domain-containing chemoreceptors, McpB and McpC, mediate chemotaxis toward amino acids. McpC is a broad-range receptor that mediates chemotaxis to all amino acids except l-asparagine (88). Interestingly, McpB was identified as a chemoreceptor for this amino acid (9). McpB has thus evolved to recognize, with high specificity, the only l-amino acid that is not recognized by the broad-range McpC receptor.
Although McpC mediates chemotaxis toward 19 amino acids, only 12 of them bind to its recombinant LBD (89). Using pulldown experiments with the immobilized McpC-LBD, four amino acid binding proteins were identified, suggesting that, similarly to the Tar chemoreceptor in E. coli, McpC recognizes signals both directly by ligand binding and via interactions with ligand binding proteins (89).
Three genes that encode McpB paralogs, namely, TlpA, McpA, and TlpB, are located adjacent to the mcpB gene on the B. subtilis chromosome (9). McpA was shown to mediate chemotaxis toward glucose and α-methylglucoside, whereas chemoeffector screening of TlpA and TlpB mutants failed to provide any information as to receptor function (9). The TlpC chemoreceptor contains the sCache (single Cache) domain, and similarly to TlpA and TlpB, its function remains unknown.
Genome analysis revealed the presence of three other chemoreceptors in B. subtilis, YfmS, YoaH, and YvaQ, that contain 4HB, CHASE (cyclase/histidine kinase-associated sensory extracellular) (90, 91), and PAS domains as their sensory LBDs, respectively (Fig. 1). However, no information as to their function is available. The soluble HemAT receptor (Fig. 1) mediates chemotaxis toward oxygen. However, in contrast to E. coli, where aerotaxis is mediated by the PAS domain-containing Aer chemoreceptor (via redox sensing) and by the 4HB domain-containing Tsr chemoreceptor (via proton motive force sensing), HemAT governs aerotaxis via direct oxygen sensing by a protoglobulin domain, which contains a bound heme (17, 92).
Helicobacter pylori
H. pylori belongs to the class Epsilonproteobacteria. The elevated motility of this pathogen in dilute and viscous media is based on the action of a bundle of unipolar sheathed flagella. It is a microaerophilic organism, and its main habitat is the upper gastrointestinal tract. Bacteria penetrate the gastric mucous layer and cause gastric ulcers, adenocarcinomas, and lymphomas. Its ability to colonize gastric mucosa depends largely on the capacity to produce urease and to neutralize the acidic pH of the stomach by breaking urea into ammonia and bicarbonate (93). Chemotaxis plays a fundamental role in the mechanism of infection (94).
Urea and bicarbonate emanate from the human gastric epithelium, and chemotaxis of H. pylori to both compounds was observed (95–97). Other detected attractants include mucin (98), various amino acids (96, 99), and cholesterol (100), whereas repellent responses were observed for salts (96), AI-2 (101), and metal ions (75, 102). In addition, other behavioral responses, for example, pH (102, 103), ROS (75), and energy taxis (74), were also described. All four chemoreceptors of H. pylori (Fig. 2 and Table 2) were found to play a role in the infection process: mutation of TlpA and TlpC reduced stomach colonization (104), whereas TlpD was required for the proliferation of H. pylori in the gastric antrum, the lowermost part of the stomach (105), or throughout the stomach in different animal models (105, 106). TlpB was shown to be important for persistent colonization in vivo (97, 103), and mutation of TlpB resulted in decreased inflammation (107, 108).
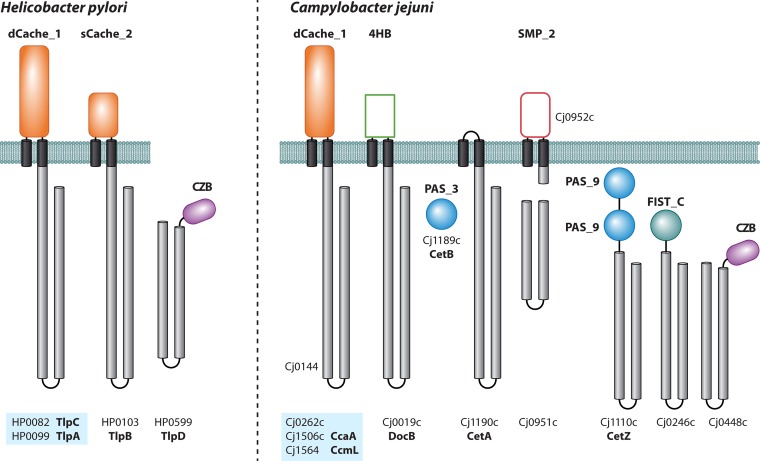
FIG 2.
The chemoreceptor repertoires of H. pylori and C. jejuni. Receptor topology and locus tags of H. pylori 26695 and C. jejuni subsp. jejuni NCTC 11168 are shown. Pfam names for all LBDs are shown, except for 4HB, which is predicted to be a divergent four-helix bundle domain. Domains that are not detected by the Pfam tool HMMER but are recognized by the more sensitive HHpred tool are indicated as empty rectangles. Orthologous groups of chemoreceptors (including paralogs) are highlighted by blue shading.
TABLE 2.
Functionally characterized chemoreceptors of H. pylori and C. jejuni
| Locus tag (chemoreceptor name) | LBD type | Effector(s) | Binding mode | Reference(s) |
|---|---|---|---|---|
| H. pylori | ||||
| HP0099 (TlpA) | dCache | Arginine, bicarbonate | Unknown | 99 |
| HP0103 (TlpB) | sCache | pH | Unknown | 103 |
| AI-2 | Indirect | 110 | ||
| Urea, hydroxyurea, formamide, acetamide | Direct | 97, 109 | ||
| HP0599 (TlpD) | CZB | Energy | Unknown | 74 |
| Hydrogen peroxide, paraquat | Unknown | 75 | ||
| C. jejuni | ||||
| Cj0262c (DocC) | dCache | Deoxycholate | Unknown | 117 |
| Cj1506c (CcaA) | dCache | Aspartate | Direct | 116 |
| Cj1564 (CcmL) | dCache | 3 amino acids, succinate | Direct | 38 |
| Malate, fumarate, purine, thiamine, Ile | 179 | |||
| Cj1189c (CetB) | PAS | Energy | Unknown | 121 |
| Cj1190c (CetA) | None | Energy | Unknown | 121 |
| Cj1110c (CetZ) | PAS | Energy | Unknown | 125 |
| Cj0951c (Tlp7) | None | Formate | Unknown | 124 |
| Cj0952c | Unknown | Formate | Unknown | 124 |
| Tlp11a | dCache | Galactose | Direct | 119 |
Found in only a few isolates.
TlpB is an intriguing chemoreceptor (Fig. 2), because it mediates repellent responses to acidic pH (103, 109) and to the AI-2 quorum sensing signal (101) as well as attractant responses to compounds released by epithelial tissues, such as urea (97). Urea binds to TlpB with high specificity and affinity (97, 109). Strains containing TlpB with single-amino-acid substitutions that prevented urea binding showed responses to AI-2 but lost the pH taxis phenotype (109). A model in which bound urea, acting as a cofactor, modulates the properties of a pH-sensing aspartate residue was proposed. The structure of the urea-containing TlpB LBD has been solved, and it was initially classified as a PAS domain (109). However, a recent study revealed that all extracellular “PAS-like” domains belong to the Cache domain superfamily (33). Accordingly, the TlpB LBD is now classified as an sCache domain. The TlpB LBD does not bind AI-2 directly: AI-2 chemotaxis depends on two periplasmic ligand binding proteins, and consequently, an indirect binding mechanism was proposed (110).
Two other chemoreceptors, TlpA and TlpC, possess dCache domains (Fig. 2). TlpA was identified as a chemoreceptor for arginine and bicarbonate because a corresponding mutant lost chemotaxis to these compounds (111). However, the mode by which these chemoeffectors stimulate the receptor remains unknown. The signals that are recognized by the TlpA paralog TlpC are currently unknown; however, indirect evidence suggests that TlpC can modulate the TlpB-mediated response to acid by an as-yet-unknown mechanism (102).
The fourth receptor in H. pylori, TlpD, is soluble and is composed of an MCP signaling domain and a CZB domain (73) (Fig. 2). TlpD was proposed to serve as an energy taxis receptor that mediates repellent responses away from conditions of reduced electron transport and away from electron transport inhibitors (74). A recent report showed that TlpD mediates a repellent response to agents that promote oxidative stress, such as hydrogen peroxide and paraquat (75). This chemoreceptor appears to function independently of the three transmembrane chemoreceptors, because it forms an autonomous polar signaling complex (75). TlpD localization is affected by metabolic activity (catalase deficiency and lower-energy conditions), which provides clues to its potential mechanism of action (112).
Campylobacter jejuni
Campylobacter species are commensal microorganisms present in the gastrointestinal tracts of different animals and are transmitted to humans primarily by the consumption of infected food. C. jejuni is one of the main causative agents of gastroenteritis but can also lead to more serious diseases such as Guillain-Barre syndrome, Miller Fisher syndrome, and arthritis (113). Mutants with deletions of various chemoreceptor genes showed up to a 10-fold decrease in the ability of bacteria to invade host cells (114). C. jejuni shows attractant responses to amino and organic acids, sugars, and nucleotides (38, 114–119), whereas lysine, arginine, glucosamine, thiamine, and bile constituents act as repellents (38, 115).
The majority of completely sequenced C. jejuni genomes contain 10 chemoreceptor genes, as exemplified by the widely used C. jejuni subsp. jejuni laboratory strain NCTC 11168 (Fig. 2 and Table 2). However, the number of chemoreceptor genes per C. jejuni genome can vary from 5 to 11. Of the 10 typical C. jejuni chemoreceptors (named consecutively Tlp1 to Tlp10 [120]), 5 appear to detect signals in the periplasm, and another 5 appear to detect signals in the cytoplasm. Four of the five transmembrane receptors contain a dCache_1 domain as a single sensory module (Fig. 2). Signal specificity was established for two of them: Tlp1 (renamed CcaA) was identified as an aspartate chemoreceptor (116), and Tlp3 (renamed CcmL) was identified as a chemoreceptor for multiple ligands, which directly binds isoleucine, lysine, arginine, glucosamine, succinate, malate, fumarate, purine, and thiamine (38). CcmL and another chemoreceptor with a similar domain architecture, Tlp4 (renamed DocC), appear to mediate chemotaxis to bile and its major component deoxycholate (117).
C. jejuni also has an unusual bipartite chemoreceptor encoded by two separate genes, cetA and cetB, that are located adjacent to each other (121). Two proteins encoded by these genes work in tandem by forming a fully functional chemoreceptor. The CetA/CetB pair is the first studied example of a “split-gene” chemoreceptor. CetA (or Tlp9) is composed of a chemoreceptor signaling domain and a HAMP domain and is anchored to the membrane by two transmembrane helices (Fig. 2), whereas CetB is a stand-alone PAS domain. The cetA and cetB genes are cotranscribed, and both proteins are membrane associated and present in protein complexes (122). This bipartite chemoreceptor system was shown to govern energy taxis (121), and follow-up studies showed that both components are homologous to energy taxis chemoreceptors in other species, where signal input is achieved through FAD-containing PAS domains, similar to a canonical Aer chemoreceptor in E. coli (39, 123).
More recently, a second bipartite chemoreceptor system was described for C. jejuni (124). Similar to the CetA/CetB pair, Tlp7 is a stand-alone signaling domain that interacts with a protein encoded by the cj0952c gene, which consists of a periplasmic domain flanked by two transmembrane regions and a HAMP domain. This bipartite chemoreceptor system appears to mediate the attractant response to formate and is important for invasion of host cells (124). The soluble CetZ chemoreceptor (Tlp8) has dual PAS domains and was recently shown to counteract the CetA/B system in guiding C. jejuni cells toward optimal energy resources (125).
Roles of other chemoreceptors in C. jejuni have not been established yet; however, their domain composition provides hints as to their putative functions. For example, the soluble Tlp5 chemoreceptor contains a FIST (found in F-box and intracellular signal transduction proteins) domain, a widely distributed sensory module which was suggested to bind primarily amino acids (126). Another soluble chemoreceptor, Tlp6, has a C-terminal CZB domain and is orthologous to the H. pylori TlpD chemoreceptor (73) that mediates a repellent response to conditions that promote oxidative stress (75).
In a recent study, the chemoreceptor Tlp11 was identified, which is present in only a few genomes of invasive C. jejuni species. This receptor binds galactose via its periplasmic dCache_1 domain and mediates positive chemotaxis toward this sugar (119).
Pseudomonas
Figure 3 summarizes the chemoreceptor repertoire of three different species that belong to the genus Pseudomonas, a member of the Gammaproteobacteria. P. aeruginosa is an opportunistic pathogen of humans, animals, and plants and is also present in soil and water. Strain PAO1 is the common reference strain and corresponds to a spontaneous chloramphenicol-resistant mutant of the original PAO strain, which was isolated from a human wound (127). P. putida KT2440 is a Tol plasmid-cured derivative of the mt-2 strain. This strain was isolated from soil, is nutritionally versatile, and has a saprophytic lifestyle (128). P. fluorescens strain Pf0-1 was isolated during a study of bacterial fitness in soil (129). This strain efficiently colonizes plant roots, and chemotaxis was found to be essential for this process (130, 131). In general, Pseudomonas strains have extraordinary metabolic versatility and a large number of chemoreceptors (25). The three model strains analyzed here contain 26 chemoreceptors (PAO1), 27 chemoreceptors (KT2440), and 37 chemoreceptors (Pfl0-1). The chemoeffector repertoire of pseudomonads is very diverse and includes organic and amino acids, aromatic hydrocarbons, sugars, fatty acids, peptides, bivalent metal ions, inorganic anions, herbicides, morphine, as well as purine and pyrimidine bases (132).
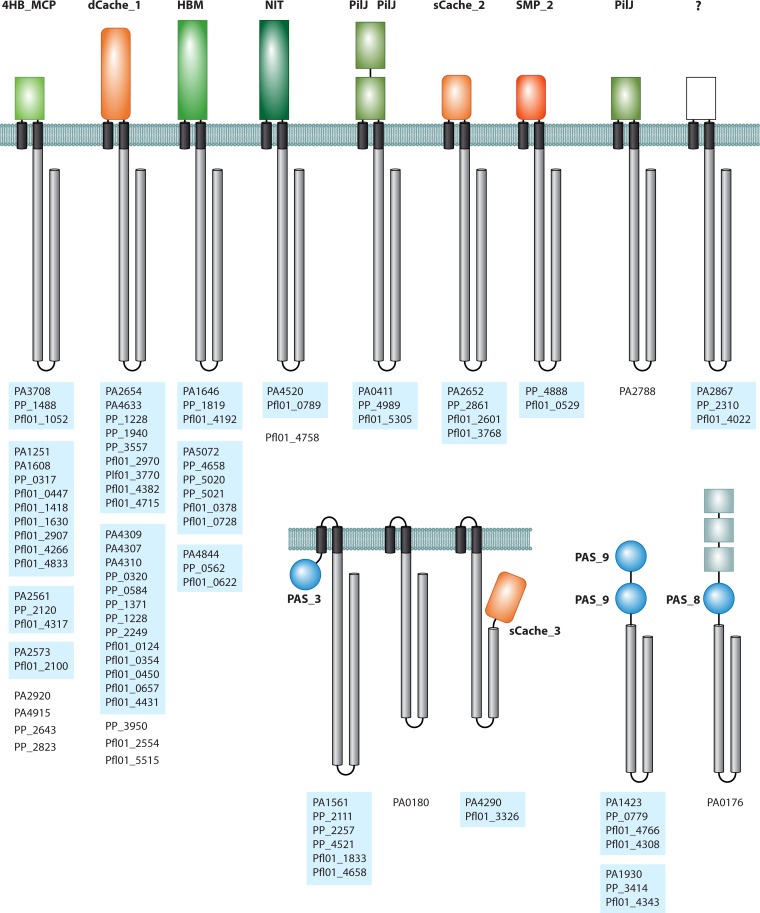
FIG 3.
The chemoreceptor repertoires of three different Pseudomonas strains. Receptor topology and locus tags of P. aeruginosa PAO1, P. putida KT2440, and P. fluorescens Pf0-1 are shown. The LBDs are colored, and their type is annotated. Orthologous groups of chemoreceptors (including paralogs) are highlighted by blue shading. The question mark indicates a domain of an unknown type.
P. aeruginosa.
Many bacteria possess multiple chemosensory pathways, and P. aeruginosa emerged as an important model to study this property. Two of the P. aeruginosa pathways, Che1 and Che2, control flagellum-mediated taxis (30, 133). The Wsp pathway modulates c-di-GMP levels (5), and the Chp pathway is responsible for type IV pilus-mediated motility (29, 134) and for regulating cAMP levels (135).
The P. aeruginosa PAO1 chemoreceptors that mediate chemotaxis to amino acids and inorganic phosphate (Pi) are the best-studied chemoreceptors (Table 3). This strain responds to all 20 proteinogenic amino acids (136), and no amino acid taxis was observed in the PctA/PctB/PctC (PA4309/PA4310/PA4307) triple mutant (Fig. 3) (137). Each of these three paralogous chemoreceptors contains a dCache domain as a single sensory module. Similar to B. subtilis (see above), P. aeruginosa has one broad-spectrum receptor, PctA, which binds most of the amino acids (137, 138). PctB binds several of the PctA ligands but has a strong preference for glutamine, which is one of the two amino acids not recognized by PctA. The signal input in PctA and PctB (represented by the binding constants of individual ligands) correlates with the magnitude of its output, as determined by fluorescence resonance energy transfer (FRET) analyses of a PctA-Tar chimera (139). PctC binds with modest affinity to two of the amino acids that are also recognized by PctA. This receptor, however, has evolved to bind with very high affinity to gamma-aminobutyrate (GABA) and mediates chemotactic responses to low GABA concentrations (34, 138). PctA, PctB, and PctC were also identified as chemoreceptors for trichloroethylene (TCE) (140), but this compound is not recognized directly by their LBDs (138).
TABLE 3.
Functionally characterized chemoreceptors of different pseudomonads
| Locus tag (chemoreceptor name[s]) | LBD type | Effector(s) | Binding mode | Reference(s) |
|---|---|---|---|---|
| P. aeruginosa | ||||
| PA0176 (Aer2, McpB) | PAS | O2, NO, CO, cyanide | Direct | 148, 153, 154, 210 |
| PA0180 (CttP, McpA) | None | Chloroethylenes | Unknown | 140 |
| PA0411 (PilJ) | PilJ | Phosphatidylethanolamine | Unknown | 158 |
| PA1561 (Aer, TlpC) | PAS | Aerotaxis | Unknown | 148, 210 |
| PA2561 (CtpH) | 4HB | Inorganic phosphate | Direct | 141 |
| PA2652 (CtpM) | sCache | Malate | Unknown | 145 |
| PA2654 (TlpQ) | dCache | Ethylene | Unknown | 146 |
| PA3708 (WspA) | 4HB | Growth on solid surfaces | Unknown | 211 |
| Ethanol | Unknown | 212 | ||
| PA4307 (PctC) | dCache | GABA, histidine, proline | Direct | 34, 136–138 |
| Chloroethylenes | Unknown | 213 | ||
| PA4309 (PctA) | dCache | 17 amino acids | Direct | 136–138 |
| Chloroethylenes | Unknown | 213 | ||
| Chloroform | Unknown | |||
| Methylthiocyanate | Unknown | |||
| PA4310 (PctB) | dCache | 5 amino acids | Direct | 136–138 |
| Chloroethylenes | Unknown | 213 | ||
| PA4844 (CtpL) | HBM | Inorganic phosphate | Indirect | 141, 214 |
| Chloroaniline, catechol | Unknown | |||
| PA5072 (McpK) | HBM | α-Ketoglutarate | Direct | 147 |
| P. putida | ||||
| PP0320 (McpH) | dCache | Metabolizable purines | Direct | 162 |
| PP1228 (McpU) | dCache | Polyamines | Direct | 163 |
| PP1371 (McpG) | dCache | GABA | Direct | 34 |
| PP2111 (Aer2) | PAS | Aerotaxis, methylphenols | Unknown | 169 |
| Phenylacetic acid | Unknown | 215 | ||
| PP2249 (McpA) | dCache | 12 amino acids | Direct | 163 |
| PP2257 (Aer1) | PAS | Aerotaxis | Unknown | 216 |
| PP2861 (McpP) | sCache | Acetate, pyruvate, propionate, l-lactate | Direct | 168 |
| PP4658 (McpS) | HBM | TCA intermediates, acetate, butyrate | Direct | 164 |
| PP5020 (McpQ) | HBM | Citrate, citrate/Mg2+ | Direct | 167 |
| Pput_0623 (McpC)a | dCache | Nicotinic acid, cytosine | Unknown | 172, 173 |
| Pput_2149 (PcaY)a | 4HB | Cyclic carboxylic acids | Unknown | 174 |
| Pput_4520 (McfS)a | HBM | Malate | Unknown | 175 |
| Pput_4894 (McfQ)a | HBM | Citrate, fumarate | Unknown | 175 |
| Pput_0339 (McfR)a | 4HB | Succinate, malate, fumarate | Unknown | 175 |
| P. fluorescens | ||||
| Pfl01_0124 (CtaB) | dCache | Amino acids | Unknown | 131 |
| Pfl01_0354 (CtaC) | dCache | Amino acids | Unknown | 131 |
| Pfl01_0728 (McpS) | HBM | Malate, succinate, fumarate | Unknown | 130 |
| Pfl01_3768 (McpT) | sCache | Malate, succinate | Unknown | 130 |
| Pfl01_4431 (CtaA) | dCache | Amino acids | Unknown | 131 |
| NbaYb | sCache | Nitrobenzoate | Unknown | 176 |
P. putida F1.
P. fluorescens KU-7.
Two chemoreceptors in P. aeruginosa respond to changes in Pi concentrations: CtpL (PA4844) detects low Pi concentrations, and CtpH (PA2561) detects high Pi concentrations (141). However, these chemoreceptors differ in their sensory modules: CtpL is predicted to possess an HBM (helical bimodular) domain (142), whereas CtpH has a 4HB domain (Fig. 3). A recent study showed that Pi binds directly to CtpH but not to CtpL (22). CtpL is activated by binding the Pi-loaded PstS periplasmic binding protein that also provides Pi to the Pst transporter (143). Indirect chemoreceptor activation by periplasmic binding proteins was previously shown to mediate chemotaxis to sugars, dipeptides, and AI-2 in E. coli (50, 54, 57, 64, 144). The discovery of a similar mechanism in a different bacterial family, involving a different chemoreceptor type and chemoeffector, suggests that such systems are widespread.
Other chemoreceptors with identified specificity include the malate-specific sCache domain-containing PA2652 chemoreceptor (145), the dCache domain-containing TlpQ (PA2654) receptor for the plant hormone ethylene (146), and the α-ketoglutarate-specific McpK (PA5072) chemoreceptor that harbors an HBM domain (147). The membrane-bound McpA (CttP or PA0180) chemoreceptor mediates positive chemotaxis to TCE. McpA has no LBD (Fig. 3), and the mechanism of its action is unknown.
P. aeruginosa has two chemoreceptors, namely, PA1561 (Aer/TlpC) and PA0176 (Aer-2/McpB/TlpG), which were suggested to mediate aerotaxis (148). While Aer is homologous to that of E. coli, Aer-2/McpB has a very unusual domain architecture. This soluble receptor is composed of three HAMP domains followed by a PAS domain, two HAMP domains, and the signaling domain (149). Similar to the high-abundance chemoreceptors Tar and Tsr in E. coli, Aer-2/McpB is one of the two P. aeruginosa chemoreceptors that contains a pentapeptide tethered via a linker to its C-terminal end. It has been shown that CheR2, and not any other CheR homolog, binds exclusively to this pentapeptide and methylates Aer-2/McpB (150). It was concluded that the presence of this pentapeptide permits the targeting of a specific chemoreceptor with a dedicated CheR homolog, which may be one of the molecular mechanisms to ensure the specificity of the interaction of signaling proteins within a given pathway. This receptor may be associated with pathogenicity, because the mutation of its cognate CheB2 methylesterase causes a dramatic reduction in virulence (151). The Aer-2/McpB PAS domain contains heme, which enables this chemoreceptor to recognize O2, NO, CO, and cyanide. A recent study revealed that oxygen binds to the heme-containing PAS domain with a KD (equilibrium dissociation constant) of 16 μM, which is comparable to the affinities of other PAS-heme O2 sensors (152). This and the fact that amino acid substitutions in the gas binding site affected primarily the binding of O2 and not of other gases led to the proposition that O2 is the natural ligand for this chemoreceptor (152). Comparison of the structures containing ferric, ligand-free heme (153) and cyanide-bound ferric heme (154) also led to a model of receptor function (153). Changes in the heme ligation state cause conformational changes in the PAS domain that shift the PAS monomer-dimer equilibrium or alternatively cause a rearrangement of the PAS dimer. The exact role of Aer-2/McpB remains unclear: the aer-2 (mcpB) mutant shows an aerotaxis phenotype comparable to that of the wild type (133).
BdlA (PA1423) is a soluble chemoreceptor, which is composed of two PAS domains followed by the signaling domain (Fig. 3). This specific domain architecture defines a widely spread chemoreceptor subfamily, several members of which were shown to monitor changes in the redox status of the electron transport system (39). Initial studies showed that the BdlA mutant was deficient in biofilm dispersion and had increased adherence properties and increased intracellular levels of cyclic di-GMP (155). BdlA employs an unorthodox mechanism. The full-length protein appears to be inactive. However, during growth in biofilms, but not during planktonic growth, an amino acid in the segment linking the PAS domains with the signaling domain is phosphorylated by an as-yet-unidentified kinase, and this step permits the proteolytic cleavage of the N-terminal PAS domain. It was proposed that the resulting truncated BdlA chemoreceptor forms efficient signaling complexes (156).
The PilJ chemoreceptor (PA0411) contains dual PilJ domains in its N terminus, and it feeds into the Chp pathway controlling type IV pilus-mediated motility and cAMP levels (29, 135, 157). The pilJ mutant showed a defect in twitching motility toward phosphatidylethanolamine (PE), and it was proposed that PE may be sensed by this receptor (158). PilJ mutant cells have shortened pili, suggesting that PilJ is required for full pilus assembly and/or extension (159). This agrees with data from another study showing that the Chp pathway modulates cAMP levels, which in turn impacts type IV pilus production (135). A more recent study showed that PilA, a subunit that assembles to form type IV pili, directly interacts with PilJ, most likely via its periplasmic LBD (160). The authors of that study suggested that PilJ may act as a mechanosensor that can detect conformational changes induced in stretched type IV pili.
P. putida.
P. putida KT2440 has three chemosensory pathways (161) orthologous to the Che1, Wsp, and Chp pathways in P. aeruginosa; it lacks the Che2 pathway, which is associated with virulence in P. aeruginosa. Eight of its 27 chemoreceptors possess a dCache LBD (Fig. 3). Four of these chemoreceptors, all belonging to the same paralogous group, have been functionally annotated (Table 3). PP0320 (McpH) mediates chemotaxis toward metabolizable purine derivatives such as adenine and guanine, but it does not recognize nonmetabolizable derivatives such as theobromine and theophylline (162). PP1371 (McpG) is a GABA-specific chemoreceptor, and its dissociation constant for this ligand (KD = 175 nM) is one of the highest ever observed for a chemoreceptor. GABA is present in plant root exudates, and bacterial root colonization is delayed in an McpG mutant (34). PP1228 (McpU) was identified as a chemoreceptor that specifically binds three polyamines, putrescine, cadaverine, and spermidine; PP2249 (McpA) responds to 12 different proteinogenic amino acids (163). The common feature of the above-mentioned chemoreceptors is that they recognize their ligands directly and that all attractants are amines that support bacterial growth as sole N sources.
McpS (PP4658) mediates chemotaxis to six tricarboxylic acid (TCA) cycle intermediates, butyrate, and acetate (164, 165), and it is the first characterized chemoreceptor with an HBM domain (142). This domain is composed of 6 helices that are arranged into two stacked helical modules (166). Each module contains a ligand binding site; malate and succinate bind to the membrane-proximal bundle, whereas acetate binds to the membrane-distal bundle. It has been demonstrated that ligand binding to each module causes chemotaxis and that signals are additive (166). In a superimposition of both modules, by translation and 180° rotation, both ligand binding sites overlap. In addition, the Tar LBD can be closely superimposed onto the individual McpS LBD modules, and the Tar aspartate binding site overlaps the binding sites at the McpS modules. McpS thus has a bimodular LBD, and the binding of different ligands to the individual modules was proposed to be a mechanism to fine-tune chemotactic responses (166). The dCache domain is also characterized by a bimodular arrangement, but it remains to be elucidated whether ligands also bind to both modules.
Despite the facts that citrate is abundantly present in plant tissues and root exudates and that it serves as a preferred carbon source, McpS binds citrate with low affinity and does not bind citrate-metal ion complexes (164). The McpS paralog PP5020 (McpQ) was found to specifically bind citrate, either in its free form or in complex with metal ions (167). PP2861 (McpP) is a receptor specific for C-2 and C-3 carboxylic acids and recognizes its ligands directly via the sCache domain (168). In contrast to P. aeruginosa, aerotaxis in P. putida is likely mediated by three paralogous chemoreceptors that all have the exact domain architecture of E. coli Aer, namely, PP2257 (Aer-1), PP2111 (Aer-2), and PP4521 (Aer-3) (169) (Fig. 3).
A characteristic feature of many P. putida strains is that they are able to degrade various aromatic compounds, and many of them serve as chemoattractants (132). So far, two chemoreceptors have been unambiguously identified as mediators of aromatic hydrocarbon taxis: McpT of P. putida DOT-T1E, for taxis to many different mono- and biaromatic hydrocarbons (170), and NahY of P. putida G7, for taxis to naphthalene (171). In contrast to the very large majority of chemoreceptors, McpT and NahY are encoded on plasmids. Another common feature is that both receptors possess 4HB LBDs.
Sensory specificity of several other chemoreceptors was identified in P. putida strain F1. McpC (orthologous to PP0584) (Fig. 3) was identified as a chemoreceptor for cytosine (172) and nicotinic acid (173). Another study led to the identification of a chemoreceptor, termed PcaY, which is an ortholog of PP2643 (Fig. 3); it responds to different aromatic acids such as benzoate and its derivatives (174). McpS of P. putida KT2440 is the dominant chemoreceptor for TCA cycle intermediates, and only very minor responses are observed for the mcpS mutant (164). In contrast, mutation of the McpS ortholog McfS in P. putida F1 (99.5% sequence identity) had almost no effect on chemotaxis toward TCA cycle intermediates (175). Two other chemoreceptors for TCA cycle intermediates, McfQ and McfR, were identified in P. putida F1, and there is evidence for additional, as-yet-unidentified chemoreceptors for TCA cycle intermediates (175). These studies highlight unique evolutionary paths for chemoreceptors, where, in contrast to the general rule, orthologs can respond to different spectra of chemoeffectors, while the same chemoeffector might be recognized by nonhomologous LBDs (e.g., the HBM domain in McfS/McfQ and the 4HB domain in McfR).
P. fluorescens.
Five of the 37 chemoreceptors in P. fluorescens Pf0-1 have been functionally characterized. The response to amino acids was modulated primarily by three chemoreceptors that are orthologous to P. aeruginosa PctA, PctB, and PctC. Consequently, as their P. aeruginosa counterparts, these receptors, Pfl01_4431 (CtaA), Pfl01_0124 (CtaB), and Pfl01_0354 (CtaC) (Fig. 3), contain a dCache domain as a single dedicated sensory module. While CtaA and CtaB are broad-range receptors (each one responds to 16 amino acids), CtaC has a relatively narrow ligand profile and mediates taxis to only 5 amino acids (Cys, Arg, Gly, Met, and Thr) (131).
Chemoreceptors that mediate a strong attractant response to the TCA cycle intermediates succinate, malate, and fumarate in P. fluorescens Pf0-1 have been identified. Both Pfl01_0728 (McpS) and Pfl01_3768 (McpT) respond to malate and succinate, whereas McpS also responds to fumarate in addition to these two compounds (130). The sCache domain-containing chemoreceptor McpT is orthologous to the malate-specific chemoreceptor PA2652 of P. aeruginosa, whereas the HBM-containing chemoreceptor McpS is an ortholog of P. aeruginosa PA5072. Thus, as in the case of P. putida F1, chemotaxis of P. fluorescens to organic acids is mediated by nonhomologous chemoreceptors with different LBDs.
A study of P. fluorescens strain KU-7, which is able to metabolize 2-nitrobenzoate, led to the identification of the NbaY chemoreceptor (176). The nbaY gene was found on the plasmid next to the genes encoding the two initial enzymes of the 2-nitrobenzoate degradation pathway. The proximity of genes involved in degradation and chemotaxis strongly suggests that there is a functional link; indeed, the wild-type strain shows positive chemotaxis to 2-nitrobenzoate, and NahY was identified as a receptor for this response.
The majority of chemoreceptors in Pseudomonas remain functionally uncharacterized. Furthermore, no known LBD type was detected (even when using the most sensitive profile-profile similarity searches) in the P. aeruginosa chemoreceptor PA2867 and its orthologs in two other species (Fig. 3). However, transcript levels of the P. putida KT2440 ortholog (PP2310) were highest among all chemoreceptors in this strain (177), and mutation of the corresponding gene increased biofilm formation (163).
CHEMORECEPTOR LBD DIVERSITY AND ABUNDANCE
Following the analysis of the chemoreceptor repertoire of model organisms, we revisited the issue of chemoreceptor LBD diversity and abundance on the genomic scale. The latest analysis was reported in 2010 (7), and the amount of available genomic information has grown dramatically in recent years. To address this issue, we analyzed the domain architectures of all sequences matching the chemoreceptor signaling domain (MCPsignal; Pfam accession number PF00015), which is the accepted criterion for the chemoreceptor definition (13), that are currently available in the Pfam 31.0 database and its underlying UniProt reference proteome database (178) as of March 2017. This search retrieved 26,530 sequences, 10,658 of which contained either no LBD or a putative LBD that cannot be matched to any known domain model in Pfam. The remaining 15,872 sequences contain at least one known LBD. This set of sequences was used to determine LBD diversity and abundance (Table 4 and Fig. 4).
TABLE 4.
Genomic diversity and abundance of chemoreceptor ligand binding domainsa
| Domain superfamilyb | LBD type | Domain model | No. of domains |
|---|---|---|---|
| 4HB_MCP | Single 4HB | 4HB_MCP_1 | 3,998 |
| TarH | 861 | ||
| CHASE3 | 392 | ||
| Double 4HB | HBM | 231 | |
| Cache-like | Double Cache | dCache_1 | 3,515 |
| Cache_3-Cache_2 | 307 | ||
| dCache_3 | 202 | ||
| dCache_2 | 183 | ||
| Ykul_C | 2 | ||
| Single Cache | sCache_2 | 1,165 | |
| sCache_3_2 | 60 | ||
| sCache_3_3 | 368 | ||
| Diacid_rec | 40 | ||
| CHASE4 | 19 | ||
| CHASE8 | 7 | ||
| PAS | PAS | PAS_3 | 1,930 |
| PAS_9 | 682 | ||
| PAS_4 | 459 | ||
| PAS_8 | 97 | ||
| PAS | 111 | ||
| PAS_7 | 8 | ||
| PAS_6 | 1 | ||
| PAS_10 | 1 | ||
| GAF | GAF | GAF | 463 |
| GAF_2 | 74 | ||
| Protoglobin | Protoglobin | Protoglobin | 552 |
| Globin | 8 | ||
| Bac_globin | 2 | ||
| PBP | Periplasmic binding protein | Phosphonate-bd | 30 |
| SBP_bac_5 | 29 | ||
| SBP_bac_3 | 23 | ||
| SBP_bac_8 | 7 | ||
| Peripla_BP_4 | 16 | ||
| Peripla_BP_5 | 2 | ||
| Peripla_BP_3 | 1 | ||
| Peripla_BP_6 | 1 | ||
| OpuAC | 5 | ||
| DctP | 2 | ||
| NMT1 | 2 | ||
| HNOX-like | Heme and NO binding | HNOB | 75 |
| 4Fe-4S | Iron-sulfur cluster binding | Fer4 | 14 |
| Fe4_10 | 20 | ||
| Fer4_7 | 6 | ||
| Fer4_9 | 4 | ||
| Fer4_6 | 1 | ||
| FeS | 33 | ||
| GPCR_A | GPCR-like | 7TMR-DISM_7TM | 16 |
| NADP_Rossman | NAD binding | GFO_IDH_MocA | 9 |
| Semialdhyde_dh | 1 | ||
| Gx_transp | Transporter | 5TM-5TMR_LYT | 9 |
| Beta_propeller | Extracellular binding | Reg_prop | 8 |
| GBD | Galactose binding | 7TMR-DISMED2 | 6 |
| CBM_4_9 | 2 | ||
| TPR | Protein-protein interactions | TPR_19 | 4 |
| ANAPC3 | 1 | ||
| Cupin | Nucleotide binding | cNMP_binding | 3 |
| HHH | Helix turn helix | HHH_5 | 2 |
| E-set | Sugar binding | Y_Y_Y | 2 |
| Phosphatase | Cyanide binding | Rhodanese | 2 |
| P-loop_NTPase | Nucleotide binding | ABC_tran | 1 |
| 2heme_cytochrom | Heme binding | Ni_hydr_CYTB | 1 |
| ATP-grasp | Glutathione binding | GSH-S_ATP | 1 |
| Flavodoxin | FMN/FAD binding | Falvodoxin_2 | 1 |
| No assigned superfamily | Zinc binding | CZB | 582 |
| PilJ | PilJ | 326 | |
| Nitrate and nitrite binding | NIT | 318 | |
| c-di-GMP binding | PilZ | 137 | |
| Oxygen binding | Hemerythrin | 98 | |
| PocR | 91 | ||
| Domain of unknown function | DUF3365 | 83 | |
| Amino acid binding | FIST | 62 | |
| Hydrogen binding | Fe_hyd_lg_C | 45 | |
| Integral membrane sensor | MHYT | 28 | |
| Adenosyl group binding | CBS | 21 | |
| Ammonium transporter | Ammonium_trans | 16 | |
| Ligand binding | PrpR_N | 9 | |
| Domain of unknown function | DUF4077 | 5 | |
| Integral membrane domain | MASE3 | 5 | |
| Quorum sensing | AI-2E_transport | 1 | |
| Nucleoside binding | Gate | 1 | |
| Phosphate/sugar binding | PTS_EIIC | 1 |
Data from Pfam 31.0 and the underlying UniProt reference proteome databases (178) as of March 2017. A total of 26,530 sequences were retrieved by using the MCPsignal domain model. A total of 17,907 LBDs matching Pfam domain models were identified. GPCR, G-protein-coupled receptor; GBD, galactose-binding domain.
Pfam clan.
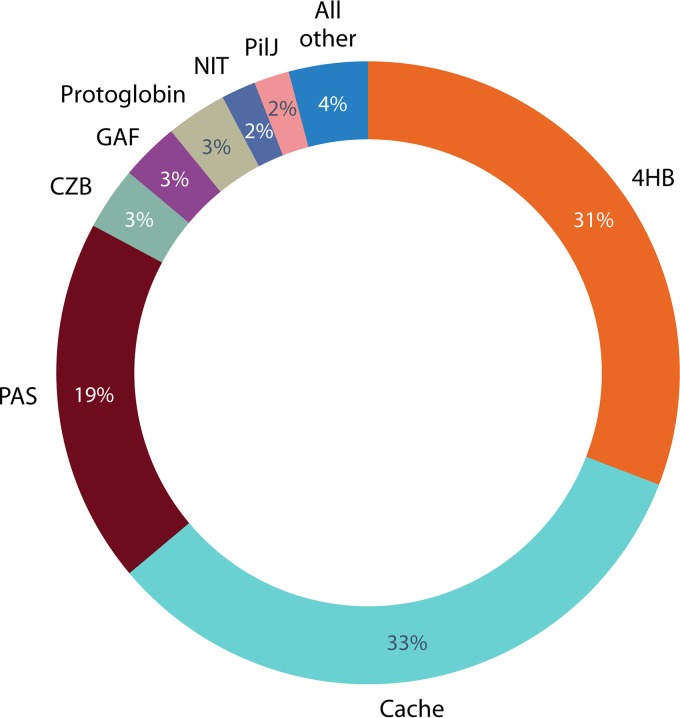
FIG 4.
Relative abundances of different LBD types in chemoreceptors. The analysis includes sequences matching the chemoreceptor signaling domain (MCPsignal; Pfam accession number PF00015) that were available in the Pfam 31.0 database and its underlying UniProt reference proteome database (178) as of March 2017. See Table 4 for details.
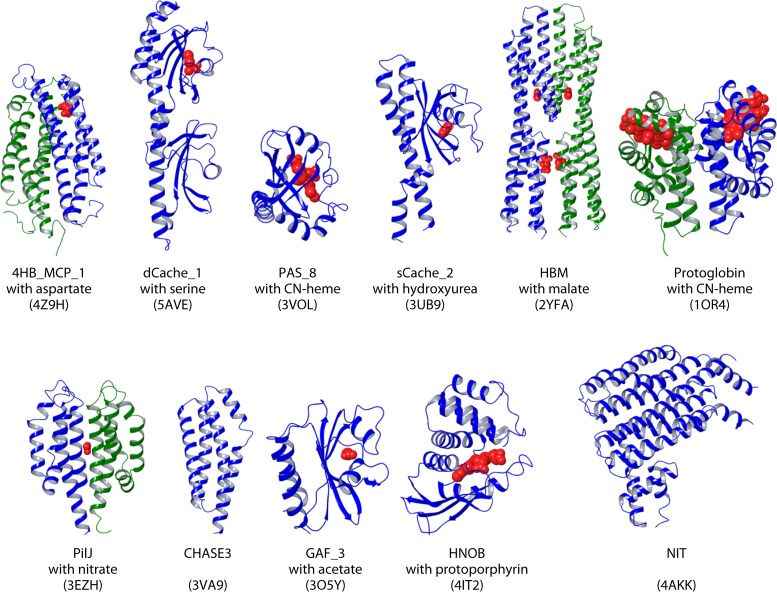
More than 80 different LBD types were found in this data set (Table 4), a number which is at least three times larger than the previously reported number (7); however, their distribution is uneven. Only a few LBD types are abundant, whereas the majority of them are found infrequently. The 4HB domain (defined by Pfam models 4HB_MCP_1 and TarH) is the most abundant LBD in chemoreceptors (Table 4 and Fig. 4 and 5). This domain was previously identified as a ubiquitous extracytoplasmic sensory module found in all major signal transduction families of bacteria and archaea (32). Ligands that are directly recognized by representatives of this domain family are shown in Fig. 6. This domain is best studied for the E. coli chemoreceptors Tar and Tsr, and the mechanism of ligand binding and signal transduction is well understood (reviewed in reference 8).
FIG 5.
Structural diversity of LBDs. Structures of different LBD types that are found in chemoreceptors are shown. Bound ligands are shown in red. Different chain colors indicate that the domain was experimentally shown to be dimeric. Domain definitions were obtained from Pfam. PDB accession numbers are shown in parentheses.
FIG 6.
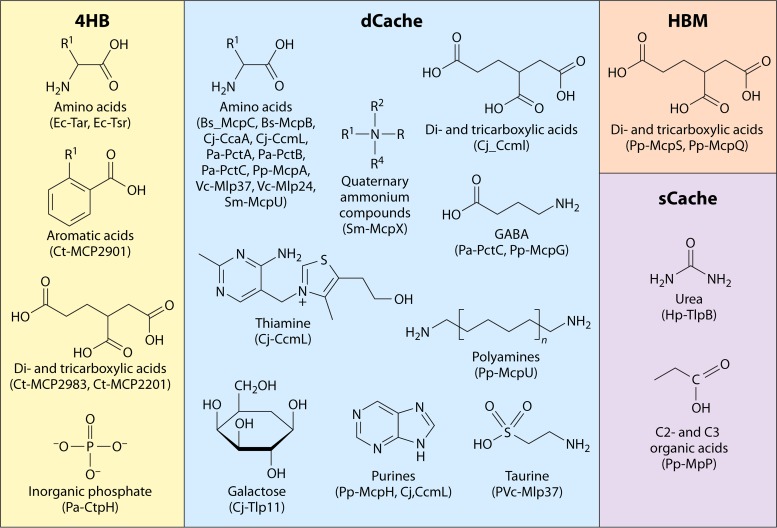
Diversity of ligands recognized by the major classes of chemoreceptor LBDs. The following ligands for which direct binding was observed are shown (along with references for the corresponding evidence): E. coli Tar (Ec-Tar) (63), Ec-Tsr (50), C. testosteroni MCP2901 (Ct-MCP2901) (203), Ct-MCP2983 (204), Ct-MCP2201 (40), P. aeruginosa CtpH (Pa-CtpH) (22, 141), P. putida McpS (Pp-McpS) (164), Pp-McpQ (167), H. pylori TlpB (Hp-TlpB) (97, 109), Pp-McpP (168), B. subtilis McpC (Bs-McpC) (89), Bs-McpB (9, 180), C. jejuni CcaA (Cj-CcaA) (116), Cj-CcmL (38, 179), Pa-PctA (138), Pa-PctB (138), Pp-McpA (163), V. cholerae Mlp37 (Vc-Mlp37) (44), Vc-Mlp24 (205), S. meliloti McpU (Sm-McpU) (206), Cj-Tlp11 (119), Pa-PctC (138), Pp-McpG (34), Sm-McpX (207), Pp-McpU (163), and Pp-McpH (162).
The dCache_1 domain is the second most abundant LBD (Table 4). The dCache domains are composed of two PAS-like modules and a long N-terminal α-helix (Fig. 5). The different subfamilies of dCache domains likely originated from sCache domains that contain only one PAS-like module (33). dCache is one of the two characterized LBDs that have a bimodular arrangement. In all structural and functional studies of dCache domains conducted to date, chemoeffectors were shown to bind to the membrane-distal module (44, 89, 138, 179, 180), and no physiologically relevant ligand interacting with the membrane-proximal module has been identified. The latter module might be involved in signal transmission to the membrane. Different ligands, primarily amines, bind to dCache domains directly (Fig. 6). In contrast to dCache, chemoeffectors bind to both modules of another characterized LBD with a bimodular arrangement, the HBM domain (142). For example, acetate binds to the distal module of the HBM domain in the McpS chemoreceptor of P. putida, whereas malate and succinate bind to the membrane-proximal module (166). Furthermore, ligand binding at both modules is agonistic (166).
PAS domain-containing chemoreceptors represent the third most abundant subfamily (Fig. 4). Although PAS and sCache domains share a similar fold, they are defined and recognized by different sequence signatures. Similar to 4HB and Cache domains, PAS domains are also omnipresent in bacterial signal transduction systems, but in contrast to the 4HB and Cache domains, they are also widely distributed in eukaryotes (24, 181). This domain is found almost exclusively in the cytosol (24, 33), and many family members contain bound heme, FAD, or flavin mononucleotide (FMN) and are involved in oxygen and redox sensing, which govern aerotaxis and related behavioral responses.
The three most abundant LBD superfamilies (4HB, Cache, and PAS) account for more than 80% of chemoreceptors with known LBDs, whereas the CZB, GAF (found in cGMP-specific phosphodiesterases, adenylyl cyclases, and FhlA), protoglobin, NIT (nitrate and nitrite sensing), and PilJ LBDs account for 2 to 3% each (Fig. 4 and Table 4). Representatives of more than 30 other LBD superfamilies and families account for only 4% of all chemoreceptors with known LBDs (Fig. 4 and Table 4). The CZB domain-containing chemoreceptor TlpD mediates energy, ROS, and pH taxis in H. pylori (73, 75, 76), whereas the protoglobin domain of the HemAT chemoreceptor in the archaeon Halobacterium salinarum and in B. subtilis contains a bound heme and mediates taxis to oxygen (17). While chemoreceptors containing representatives of other LBD families have never been studied, putative functions for many of them can be predicted by using comparative genomics. For example, known ligands for GAF domains include cyclic nucleotides (182), whereas the NIT domain was predicted to bind nitrates and nitrites (183). The FIST domain is hypothesized to be associated with amino acid sensing (126), the PilZ domain binds the second messenger c-di-GMP (184), the PocR domain was proposed to be involved in sensing simple hydrocarbon derivatives (185), the HNOB (heme-NO-binding) domain is predicted to function as a heme-dependent sensor for gaseous ligands (186), and TPR (tetratricopeptide repeat) domains typically bind other proteins (187). Many putative LBDs appear to be derivatives of enzymes and transporters with a known or predicted specificity (Table 4). Their lower abundance suggests that many such chemoreceptors evolved recently in some lineages, helping bacteria to adapt to specific environments.
The vast majority of LBDs that have been studied are located in the N-terminal region of chemoreceptors; however, some chemoreceptors, including those from model organisms (Fig. 1 to 3), contain LBDs as their C-terminal domains. Genomic analysis suggests that approximately 5% of LBDs in microbial chemoreceptors are located at the C terminus, and the CZB domain is the most abundant among them. Some of the chemoreceptor-associated LBDs are always located at the very C terminus, for example, the PilZ and hemerythrin domains. Some LBD types are predominantly located at the C terminus but occasionally can be found in the N terminus, for example, domains that belong to the periplasmic binding protein (PBP) superfamily. Finally, many LBD types that are usually located at the N terminus occasionally can be found in the C terminus, for example, domains from the Cache and PAS superfamilies. The molecular mechanism of signal transduction by C-terminally located LBDs remains to be established, but it likely involves a direct interaction with the HAMP domain and/or the signaling domain.
Approximately 14% of bacterial chemoreceptors lack transmembrane regions and are predicted to be located in the cytosol (31). A characteristic feature of this group is the predominant presence of PAS domains that are found in nearly one-half of the cytosolic chemoreceptors (31). In contrast, transmembrane chemoreceptors predominantly contain 4HB and Cache domains as extracellular LBDs.
Current structural information about different LBD types is summarized in Fig. 5, revealing two dominant folds: antiparallel α-helix bundles (4HB, HBM, PilJ, CHASE, and NIT domains) and PAS-like α/β-folds (domains that belong to the PAS, GAF, and Cache superfamilies).
DIVERSITY OF SENSING MECHANISMS AND WAYS TO STIMULATE A CHEMORECEPTOR
The chemoreceptor structural unit is a homodimer. Chemoreceptor homodimers interact to form trimers of dimers that are functional units of signal transduction, and oligomerization appears to be mediated primarily by the signaling domain of chemoreceptors (188). Comprehensive studies of the sensory mechanism were carried out on E. coli chemoreceptors that contain the 4HB LBD. It was shown that aspartate binds only to the dimeric state of the recombinant Tar LBD with a stoichiometry of 1 molecule per dimer and very strong negative cooperativity. Ligand binding was found to stabilize the dimer and to shift the monomer-dimer equilibrium to the dimeric state (189, 190). The analysis of the Tar LBD structure provided the basis for this binding mechanism. The aspartate binding sites are located at the dimer interface, and amino acids from both monomers of the dimer establish contacts with the bound ligand (Fig. 7A), causing dimer stabilization (63).
FIG 7.
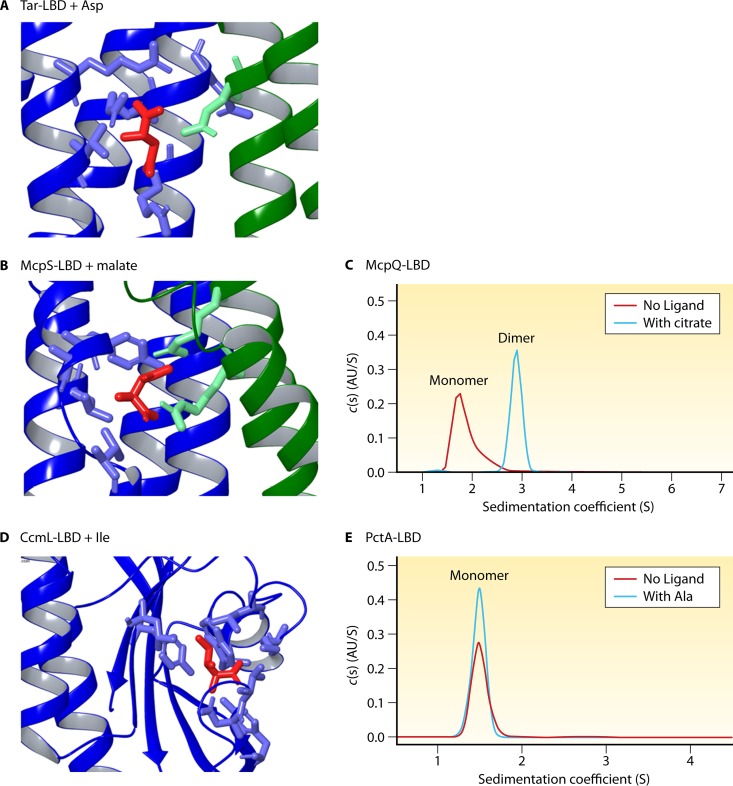
Different sensing mechanisms. (A to C) Sensing mechanism in which ligand binding induces LBD dimerization. (D and E) Sensing mechanism in which ligand binding does not induce LBD dimerization. (A and B) Zoomed-in images of the binding pockets of the Tar LBD (4HB) with bound Asp (PDB accession number 4Z9H) (A) and the McpS LBD (HBM) with malate (PDB accession number 2YFA) (B). The two monomers of the dimer are colored differently. In both cases, the binding site is at the dimer interface, and amino acids from both monomers are involved in ligand binding. (C) Analytical ultracentrifugation studies of the LBD of the McpS homolog McpQ (HBM) in the absence and presence of its ligand citrate. (D) Binding pocket of the dCache_1 LBD of the CcmL receptor (PDB accession number 4XMR) containing bound Ile. Amino acids involved in Ile binding are from the same monomer. (E) Analytical ultracentrifugation data for the PctA LBD (dCache_1) in the absence and presence of Ala. Data were reported previously (63, 138, 166, 167, 179). c(s), sedimentation coefficient distribution; AU, absorbance units; S, Svedberg units.
More recent studies of bimodular HBM domains produced results that are very similar to what was observed for the 4HB domain. In the P. putida McpS LBD, malate binds with a stoichiometry of 1 molecule per dimer, and similar to the Tar LBD structure, the malate binding site in McpS LBD is at the dimer interface; residues from both monomers participate in binding (Fig. 7B). Analytical ultracentrifugation studies showed that the individual LBD of McpS is present in a monomer-dimer equilibrium and that ligand binding shifts this equilibrium entirely to the dimeric state (Fig. 7C) (164). Similar observations were made in studies of the individual LBDs of the McpQ (167) and McpK (147) chemoreceptors. However, new structural information that became available for other LBD types suggests that this binding mode may not be universal. Structures of sCache and dCache domains show that the ligand is buried within the binding cavity of a single protein monomer (Fig. 7D) (44, 109, 179). In agreement with this structural information, the dCache LBDs of the PctA (138) and TlpC (191) chemoreceptors were found to be monomeric, and in the case of the PctA LBD, a saturating ligand concentration did not alter its oligomeric state (Fig. 7E) (138). The data therefore suggest that there are different sensing mechanisms with regard to ligand-induced LBD dimerization.
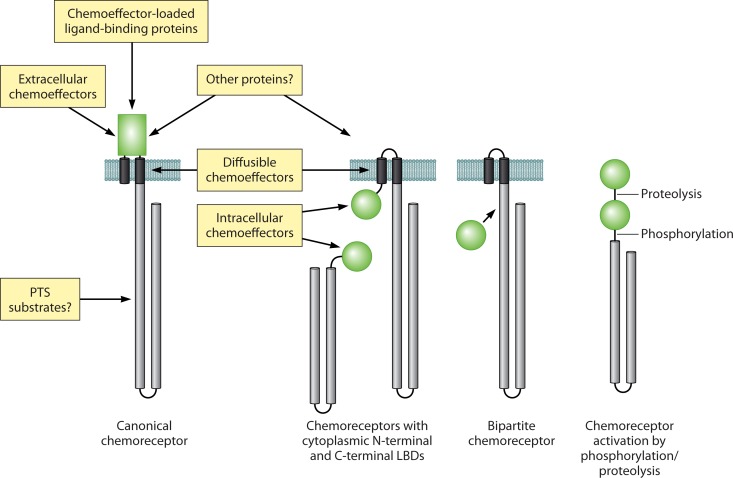
Newly available data also show that there are different ways to stimulate chemoreceptors (Fig. 8). A canonical chemoreceptor such as E. coli Tar can be stimulated either by direct ligand binding or by complexing with a ligand binding protein. The example of the PilJ chemoreceptor suggests that stimulation can also be achieved by binding to other proteins such as PilA, the subunit that forms type IV pili (160). Studies of the mechanism underlying phenol taxis in E. coli, where Tar senses phenol as an attractant and Tsr as a repellent, resulted in the proposition of an alternative mechanism. Diffusible chemoeffectors such as phenol may stimulate a chemoreceptor by perturbing the structural stability or position of the transmembrane bundle helices or other elements, which ultimately causes a modulation of kinase activity (53). Those authors suggested that behavioral responses to cytoplasmic pH and temperature may also involve the detection of alterations in the transmembrane regions, which is in agreement with a significant body of evidence showing that the temperature-sensing histidine kinases employ similar mechanisms (192, 193). However, there is evidence that chemotaxis to diffusible chemoeffectors is not mediated exclusively by the direct action of the effector on the transmembrane helices. One such example is McpT of P. putida DOT-T1E, which mediates chemotaxis to various hydrocarbons (170). Several lines of evidence suggest that this chemoreceptor operates by a canonical mechanism involving specific ligand recognition at the LBD. First, McpT has a broad and well-defined ligand profile and does not mediate responses to many highly hydrophobic and diffusible ligands, and second, a single-amino-acid substitution in its 4HB LBD abolished receptor function. Chemoreceptor stimulation by PTS substrates appears to occur via yet another mechanism (Fig. 8): a model in which the stimuli generated by PTS substrates are transmitted to the cytosolic fragment of the McpC chemoreceptor of B. subtilis was proposed (194, 195). Changes in the redox state of the FAD cofactor is yet another mode of stimulation, which is seen in chemoreceptors that modulate redox and energy taxis (18, 19, 196).
FIG 8.
Diversity in signal recognition and modes of action. LBDs are shown in green.
Approximately 18% of chemoreceptors lack sensory domains (25). Around one-half of these receptors are composed exclusively of a signaling domain, whereas the other half also contains transmembrane regions (25). The function of this receptor family is poorly understood. However, the discovery of bipartite chemoreceptors (122–124) comprised of an individual sensory domain, such as the PAS domain, and a separate sensorless chemoreceptor (Fig. 8) may provide import clues to their potential mechanism of action.
The example of the BdlA chemoreceptor in P. aeruginosa, which requires phosphorylation and, subsequently, the proteolytic cleavage of one of the sensor domains (Fig. 8C) (156), demonstrates that there might be other unorthodox ways of chemoreceptor stimulation. Future research will show whether other receptors employ similar mechanisms and likely reveal other ways to stimulate chemoreceptors.
COMMON MECHANISM FOR TRANSMEMBRANE SIGNALING
Despite the structural diversity of LBDs, there is evidence for a common mechanism of transmembrane signaling. This mechanism has been identified for the E. coli chemoreceptors and consists of chemoeffector-induced, piston-like, and rotational motions of the last α-helix of the 4HB domain, which extends into the second transmembrane helix (197–200). A recent report identified similar types of movements in the sensory domain on a histidine kinase, further strengthening the argument for a common mechanism for transmembrane signaling in bacteria (201). As shown in Fig. 4 and Table 5, chemoreceptors are equipped with a range of structurally diverse LBDs, which in turn raises the question of whether there are different transmembrane signaling mechanisms. Recent studies reported the construction of chimeric receptors in which NIT, HBM, sCache, and dCache LBDs were fused to the cytosolic fragment of the E. coli Tar chemoreceptor (34, 139, 202). All of these constructs were functional and mediated efficient and specific responses to the corresponding chemoeffectors, suggesting that structurally and functionally diverse LBDs in chemoreceptors employ a single universal transmembrane signaling mechanism.
TABLE 5.
Chemoreceptor ligand binding domains with known three-dimensional structures
| Species | Protein | LBD type | Ligand | PDB accession no. | Reference(s) |
|---|---|---|---|---|---|
| E. coli | Tar | 4HB | Aspartate | 4Z9H | 217 |
| V. cholerae | Mlp37 | dCache | Taurine | 5AVF | 44 |
| Serine | 5AVE | ||||
| C. jejuni | Tlp1 | dCache | None | 4WY9 | 218 |
| C. jejuni | CcmL | dCache | Isoleucine | 4XMR | 179 |
| H. pylori | TlpB | sCache | Urea | 3UB6 | 109 |
| Acetamide | 3UB7 | ||||
| Formamide | 3UB8 | ||||
| Hydroxyurea | 3UB9 | ||||
| Vibrio parahaemolyticus | Q87T87_VIBPA | sCache | Pyruvate | 4EXO, 2QHK | 109 |
| P. putida | McpS | HBM | Malate | 2YFA | 166 |
| Succinate | 2YFB | ||||
| E. coli | Tsr | 4HB | Serine | 2D4U, 3ATP | 35 |
| S. Typhimurium | Tar | 4HB | Aspartate | 2LIG, 1WAT, 1JMW | 63, 219, 220 |
| B. subtilis | HemAT | Protolobin | Cyano | 1OR4 | 221 |
| P. aeruginosa | McpB (Aer2) | PAS | Ferric heme | 4HI4 | 153 |
| Cyano | 3VOL | 154 |
CONCLUSIONS
As key components of navigation systems in bacteria and archaea, chemoreceptors enable motile cells to recognize numerous environmental and internal signals that ultimately affect cell behavior and other functions. The types of signals and the mechanisms of signal recognition by chemoreceptors were studied primarily in model organisms such as E. coli and S. enterica as well as B. subtilis, but emerging new models of chemotaxis and the availability of genomic data are changing paradigms and revealing new information about the sensory repertoire of chemoreceptors. Nearly a hundred different types of protein domains are now found in chemoreceptor sequences as known or predicted LBDs, although only a few of them appear to be ubiquitous. Bacteria seem to rely primarily on evolutionarily “old” specialized sensory domains, such as 4HB, Cache, and PAS, that provide countless opportunities to recognize various signals. On the other hand, many bacterial species and strains are “experimenting” with newly evolved versions of chemoreceptors that recruit other domain types as their sensory modules. Consequently, in some species, the chemoreceptor sensory repertoire is extremely limited, whereas in other species, it is very broad. LBDs of the same type that are very similar in sequence might recognize very different ligands, whereas the same ligand can be recognized by sensory domains that belong to fundamentally different protein folds. Although these extremes present a serious challenge to our efforts toward predictive biology, recent experimental and computational advances have begun to reveal certain tendencies. We now know that dCache domains primarily recognize amines and that HBM domains primarily bind TCA cycle intermediates, whereas PAS domains tend to contain redox-sensitive cofactors. The identification of bimodular LBDs, such as dCache_1 and HBM domains, exposed new ways for recognizing multiple signals. On the other hand, signals recognized by the membrane-proximal subdomain of dCache-type domains remain to be identified. The indirect binding of many different ligands emerges as a common mode of signal recognition by chemoreceptors, which might enable the coordination of chemotaxis and uptake, and warrants the development of new strategies and lines of inquiry. Unconventional chemoreceptors that lack LBDs and chemoreceptors with cytoplasmic C-terminal LBDs, for which no mechanistic understanding of signaling is available, add to the overall complexity of chemosensing in bacteria. Future genomics-driven studies will undoubtedly reveal many more relationships between signals and signal-recognizing domains and will broaden and deepen our understanding of the sensory repertoire of chemoreceptors in particular and bacterial signal transduction in general.
ACKNOWLEDGMENTS
We thank members of the BLAST/STiM/ReceptorFest community for many helpful discussions.
This work was supported by FEDER funds and the Fondo Social Europeo through grants from the Junta de Andalucia (grant CVI-7335) and the Spanish Ministry for Economy and Competitiveness (grants BIO2013-42297 and BIO2016-76779-P) to T.K. and National Institutes of Health grant R01 GM072285 to I.B.Z. Á.O. acknowledges a CSIC JAE-Doc contract cofunded by the European Social Fund.
REFERENCES
- 1.Ulrich LE, Koonin EV, Zhulin IB. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 3.Wadhams GH, Armitage JP. 2004. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol 5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 4.Porter SL, Wadhams GH, Armitage JP. 2011. Signal processing in complex chemotaxis pathways. Nat Rev Microbiol 9:153–165. doi: 10.1038/nrmicro2505. [DOI] [PubMed] [Google Scholar]
- 5.Hickman JW, Tifrea DF, Harwood CS. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102:14422–14427. doi: 10.1073/pnas.0507170102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zusman DR, Scott AE, Yang Z, Kirby JR. 2007. Chemosensory pathways, motility and development in Myxococcus xanthus. Nat Rev Microbiol 5:862–872. doi: 10.1038/nrmicro1770. [DOI] [PubMed] [Google Scholar]
- 7.Wuichet K, Zhulin IB. 2010. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal 3:ra50. doi: 10.1126/scisignal.2000724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parkinson JS, Hazelbauer GL, Falke JJ. 30 March 2015. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol doi: 10.1016/j.tim.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanlon DW, Ordal GW. 1994. Cloning and characterization of genes encoding methyl-accepting chemotaxis proteins in Bacillus subtilis. J Biol Chem 269:14038–14046. [PubMed] [Google Scholar]
- 10.Sourjik V, Wingreen NS. 2012. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol 24:262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wuichet K, Alexander RP, Zhulin IB. 2007. Comparative genomic and protein sequence analyses of a complex system controlling bacterial chemotaxis. Methods Enzymol 422:1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KK, Yokota H, Kim SH. 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 13.Alexander RP, Zhulin IB. 2007. Evolutionary genomics reveals conserved structural determinants of signaling and adaptation in microbial chemoreceptors. Proc Natl Acad Sci U S A 104:2885–2890. doi: 10.1073/pnas.0609359104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkinson JS. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol 64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 15.Mowbray SL, Koshland DE Jr. 1990. Mutations in the aspartate receptor of Escherichia coli which affect aspartate binding. J Biol Chem 265:15638–15643. [PubMed] [Google Scholar]
- 16.Lin LN, Li J, Brandts JF, Weis RM. 1994. The serine receptor of bacterial chemotaxis exhibits half-site saturation for serine binding. Biochemistry 33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- 17.Hou S, Larsen RW, Boudko D, Riley CW, Karatan E, Zimmer M, Ordal GW, Alam M. 2000. Myoglobin-like aerotaxis transducers in Archaea and Bacteria. Nature 403:540–544. doi: 10.1038/35000570. [DOI] [PubMed] [Google Scholar]
- 18.Bibikov SI, Biran R, Rudd KE, Parkinson JS. 1997. A signal transducer for aerotaxis in Escherichia coli. J Bacteriol 179:4075–4079. doi: 10.1128/jb.179.12.4075-4079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL. 1997. The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A 94:10541–10546. doi: 10.1073/pnas.94.20.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park C, Hazelbauer GL. 1986. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol 167:101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mowbray SL, Koshland DE Jr. 1987. Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell 50:171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- 22.Rico-Jimenez M, Reyes-Darias JA, Ortega A, Diez Pena AI, Morel B, Krell T. 2016. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci Rep 6:28967. doi: 10.1038/srep28967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bardy SL, Briegel A, Rainville S, Krell T. 8 May 2017. Recent advances and future prospects in bacterial and archaeal locomotion and signal transduction. J Bacteriol doi: 10.1128/JB.00203-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lacal J, Garcia-Fontana C, Munoz-Martinez F, Ramos JL, Krell T. 2010. Sensing of environmental signals: classification of chemoreceptors according to the size of their ligand binding regions. Environ Microbiol 12:2873–2884. doi: 10.1111/j.1462-2920.2010.02325.x. [DOI] [PubMed] [Google Scholar]
- 26.Hamer R, Chen PY, Armitage JP, Reinert G, Deane CM. 2010. Deciphering chemotaxis pathways using cross species comparisons. BMC Syst Biol 4:3. doi: 10.1186/1752-0509-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaneko T, Minamisawa K, Isawa T, Nakatsukasa H, Mitsui H, Kawaharada Y, Nakamura Y, Watanabe A, Kawashima K, Ono A, Shimizu Y, Takahashi C, Minami C, Fujishiro T, Kohara M, Katoh M, Nakazaki N, Nakayama S, Yamada M, Tabata S, Sato S. 2010. Complete genomic structure of the cultivated rice endophyte Azospirillum sp. B510. DNA Res 17:37–50. doi: 10.1093/dnares/dsp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexandre G, Greer-Phillips S, Zhulin IB. 2004. Ecological role of energy taxis in microorganisms. FEMS Microbiol Rev 28:113–126. doi: 10.1016/j.femsre.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Whitchurch CB, Leech AJ, Young MD, Kennedy D, Sargent JL, Bertrand JJ, Semmler AB, Mellick AS, Martin PR, Alm RA, Hobbs M, Beatson SA, Huang B, Nguyen L, Commolli JC, Engel JN, Darzins A, Mattick JS. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol Microbiol 52:873–893. doi: 10.1111/j.1365-2958.2004.04026.x. [DOI] [PubMed] [Google Scholar]
- 30.Ferrandez A, Hawkins AC, Summerfield DT, Harwood CS. 2002. Cluster II che genes from Pseudomonas aeruginosa are required for an optimal chemotactic response. J Bacteriol 184:4374–4383. doi: 10.1128/JB.184.16.4374-4383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collins KD, Lacal J, Ottemann KM. 2014. Internal sense of direction: sensing and signaling from cytoplasmic chemoreceptors. Microbiol Mol Biol Rev 78:672–684. doi: 10.1128/MMBR.00033-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ulrich LE, Zhulin IB. 2005. Four-helix bundle: a ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics 21(Suppl 3):iii45–iii48. doi: 10.1093/bioinformatics/bti1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB. 2016. Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 12:e1004862. doi: 10.1371/journal.pcbi.1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes-Darias JA, Garcia V, Rico-Jimenez M, Corral-Lugo A, Lesouhaitier O, Juarez-Hernandez D, Yang Y, Bi S, Feuilloley M, Munoz-Rojas J, Sourjik V, Krell T. 2015. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol Microbiol 97:488–501. doi: 10.1111/mmi.13045. [DOI] [PubMed] [Google Scholar]
- 35.Tajima H, Imada K, Sakuma M, Hattori F, Nara T, Kamo N, Homma M, Kawagishi I. 2011. Ligand specificity determined by differentially arranged common ligand-binding residues in bacterial amino acid chemoreceptors Tsr and Tar. J Biol Chem 286:42200–42210. doi: 10.1074/jbc.M111.221887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKellar JL, Minnell JJ, Gerth ML. 2015. A high-throughput screen for ligand binding reveals the specificities of three amino acid chemoreceptors from Pseudomonas syringae pv. actinidiae. Mol Microbiol 96:694–707. doi: 10.1111/mmi.12964. [DOI] [PubMed] [Google Scholar]
- 37.Krell T. 2015. Tackling the bottleneck in bacterial signal transduction research: high-throughput identification of signal molecules. Mol Microbiol 96:685–688. doi: 10.1111/mmi.12975. [DOI] [PubMed] [Google Scholar]
- 38.Rahman H, King RM, Shewell LK, Semchenko EA, Hartley-Tassell LE, Wilson JC, Day CJ, Korolik V. 2014. Characterisation of a multi-ligand binding chemoreceptor CcmL (Tlp3) of Campylobacter jejuni. PLoS Pathog 10:e1003822. doi: 10.1371/journal.ppat.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie Z, Ulrich LE, Zhulin IB, Alexandre G. 2010. PAS domain containing chemoreceptor couples dynamic changes in metabolism with chemotaxis. Proc Natl Acad Sci U S A 107:2235–2240. doi: 10.1073/pnas.0910055107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ni B, Huang Z, Fan Z, Jiang CY, Liu SJ. 2013. Comamonas testosteroni uses a chemoreceptor for tricarboxylic acid cycle intermediates to trigger chemotactic responses towards aromatic compounds. Mol Microbiol 90:813–823. doi: 10.1111/mmi.12400. [DOI] [PubMed] [Google Scholar]
- 41.Mauriello EM, Astling DP, Sliusarenko O, Zusman DR. 2009. Localization of a bacterial cytoplasmic receptor is dynamic and changes with cell-cell contacts. Proc Natl Acad Sci U S A 106:4852–4857. doi: 10.1073/pnas.0810583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones CW, Armitage JP. 5 September 2017. Essential role of the cytoplasmic chemoreceptor TlpT in the de novo formation of chemosensory complexes in Rhodobacter sphaeroides. J Bacteriol doi: 10.1128/JB.00366-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb BA, Helm RF, Scharf BE. 2016. Contribution of individual chemoreceptors to Sinorhizobium meliloti chemotaxis towards amino acids of host and nonhost seed exudates. Mol Plant Microbe Interact 29:231–239. doi: 10.1094/MPMI-12-15-0264-R. [DOI] [PubMed] [Google Scholar]
- 44.Nishiyama S, Takahashi Y, Yamamoto K, Suzuki D, Itoh Y, Sumita K, Uchida Y, Homma M, Imada K, Kawagishi I. 2016. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci Rep 6:20866. doi: 10.1038/srep20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lane MC, Lockatell V, Monterosso G, Lamphier D, Weinert J, Hebel JR, Johnson DE, Mobley HL. 2005. Role of motility in the colonization of uropathogenic Escherichia coli in the urinary tract. Infect Immun 73:7644–7656. doi: 10.1128/IAI.73.11.7644-7656.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stecher B, Hapfelmeier S, Muller C, Kremer M, Stallmach T, Hardt WD. 2004. Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect Immun 72:4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adler J. 1966. Chemotaxis in bacteria. Science 153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- 48.Adler J, Hazelbauer GL, Dahl MM. 1973. Chemotaxis toward sugars in Escherichia coli. J Bacteriol 115:824–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mesibov R, Adler J. 1972. Chemotaxis toward amino acids in Escherichia coli. J Bacteriol 112:315–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hegde M, Englert DL, Schrock S, Cohn WB, Vogt C, Wood TK, Manson MD, Jayaraman A. 2011. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J Bacteriol 193:768–773. doi: 10.1128/JB.01196-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pasupuleti S, Sule N, Cohn WB, MacKenzie DS, Jayaraman A, Manson MD. 2014. Chemotaxis of Escherichia coli to norepinephrine (NE) requires conversion of NE to 3,4-dihydroxymandelic acid. J Bacteriol 196:3992–4000. doi: 10.1128/JB.02065-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu X, Parales RE. 2008. Chemotaxis of Escherichia coli to pyrimidines: a new role for the signal transducer tap. J Bacteriol 190:972–979. doi: 10.1128/JB.01590-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pham HT, Parkinson JS. 2011. Phenol sensing by Escherichia coli chemoreceptors: a nonclassical mechanism. J Bacteriol 193:6597–6604. doi: 10.1128/JB.05987-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Manson MD, Blank V, Brade G, Higgins CF. 1986. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 321:253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- 55.Hazelbauer GL, Adler J. 1971. Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose. Nat New Biol 230:101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- 56.Galloway DR, Furlong CE. 1977. The role of ribose-binding protein in transport and chemotaxis in Escherichia coli K12. Arch Biochem Biophys 184:496–504. doi: 10.1016/0003-9861(77)90459-3. [DOI] [PubMed] [Google Scholar]
- 57.Hazelbauer GL. 1975. Maltose chemoreceptor of Escherichia coli. J Bacteriol 122:206–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Somavanshi R, Ghosh B, Sourjik V. 2016. Sugar influx sensing by the phosphotransferase system of Escherichia coli. PLoS Biol 14:e2000074. doi: 10.1371/journal.pbio.2000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang Y, Sourjik V. 2012. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol Microbiol 86:1482–1489. doi: 10.1111/mmi.12070. [DOI] [PubMed] [Google Scholar]
- 60.Paulick A, Jakovljevic V, Zhang S, Erickstad M, Groisman A, Meir Y, Ryu WS, Wingreen NS, Sourjik V. 2017. Mechanism of bidirectional thermotaxis in Escherichia coli. eLife 6:e26607. doi: 10.7554/eLife.26607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Springer MS, Goy MF, Adler J. 1977. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A 74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greer-Phillips SE, Alexandre G, Taylor BL, Zhulin IB. 2003. Aer and Tsr guide Escherichia coli in spatial gradients of oxidizable substrates. Microbiology 149:2661–2667. doi: 10.1099/mic.0.26304-0. [DOI] [PubMed] [Google Scholar]
- 63.Milburn MV, Prive GG, Milligan DL, Scott WG, Yeh J, Jancarik J, Koshland DE Jr, Kim SH. 1991. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science 254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 64.Gardina P, Conway C, Kossman M, Manson M. 1992. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J Bacteriol 174:1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tso WW, Adler J. 1974. Negative chemotaxis in Escherichia coli. J Bacteriol 118:560–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harayama S, Palva ET, Hazelbauer GL. 1979. Transposon-insertion mutants of Escherichia coli K12 defective in a component common to galactose and ribose chemotaxis. Mol Gen Genet 171:193–203. doi: 10.1007/BF00270005. [DOI] [PubMed] [Google Scholar]
- 67.Borziak K, Fleetwood AD, Zhulin IB. 2013. Chemoreceptor gene loss and acquisition via horizontal gene transfer in Escherichia coli. J Bacteriol 195:3596–3602. doi: 10.1128/JB.00421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ortega DR, Zhulin IB. 2016. Evolutionary genomics suggests that CheV is an additional adaptor for accommodating specific chemoreceptors within the chemotaxis signaling complex. PLoS Comput Biol 12:e1004723. doi: 10.1371/journal.pcbi.1004723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yamamoto K, Imae Y. 1993. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc Natl Acad Sci U S A 90:217–221. doi: 10.1073/pnas.90.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lazova MD, Butler MT, Shimizu TS, Harshey RM. 2012. Salmonella chemoreceptors McpB and McpC mediate a repellent response to l-cystine: a potential mechanism to avoid oxidative conditions. Mol Microbiol 84:697–711. doi: 10.1111/j.1365-2958.2012.08051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Russo AF, Koshland DE Jr. 1986. Identification of the tip-encoded receptor in bacterial sensing. J Bacteriol 165:276–282. doi: 10.1128/jb.165.1.276-282.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Frye J, Karlinsey JE, Felise HR, Marzolf B, Dowidar N, McClelland M, Hughes KT. 2006. Identification of new flagellar genes of Salmonella enterica serovar Typhimurium. J Bacteriol 188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Draper J, Karplus K, Ottemann KM. 2011. Identification of a chemoreceptor zinc-binding domain common to cytoplasmic bacterial chemoreceptors. J Bacteriol 193:4338–4345. doi: 10.1128/JB.05140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schweinitzer T, Mizote T, Ishikawa N, Dudnik A, Inatsu S, Schreiber S, Suerbaum S, Aizawa S, Josenhans C. 2008. Functional characterization and mutagenesis of the proposed behavioral sensor TlpD of Helicobacter pylori. J Bacteriol 190:3244–3255. doi: 10.1128/JB.01940-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Collins KD, Andermann TM, Draper J, Sanders L, Williams SM, Araghi C, Ottemann KM. 2016. The Helicobacter pylori CZB cytoplasmic chemoreceptor TlpD forms an autonomous polar chemotaxis signaling complex that mediates a tactic response to oxidative stress. J Bacteriol 198:1563–1575. doi: 10.1128/JB.00071-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huang JY, Goers Sweeney E, Guillemin K, Amieva MR. 2017. Multiple acid sensors control Helicobacter pylori colonization of the stomach. PLoS Pathog 13:e1006118. doi: 10.1371/journal.ppat.1006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hazelbauer GL, Lai WC. 2010. Bacterial chemoreceptors: providing enhanced features to two-component signaling. Curr Opin Microbiol 13:124–132. doi: 10.1016/j.mib.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell SC, Bron S, Brouillet S, Bruschi CV, Caldwell B, Capuano V, Carter NM, Choi SK, Cordani JJ, Connerton IF, Cummings NJ, Daniel RA, Denziot F, Devine KM, Dusterhoft A, Ehrlich SD, Emmerson PT, Entian KD, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim SY, Glaser P, Goffeau A, Golightly EJ, Grandi G, Guiseppi G, Guy BJ, Haga K, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 79.Ordal GW, Goldman DJ. 1975. Chemotaxis away from uncouplers of oxidative phosphorylation in Bacillus subtilis. Science 189:802–805. doi: 10.1126/science.808854. [DOI] [PubMed] [Google Scholar]
- 80.Ordal GW, Adler J. 1974. Properties of mutants in galactose taxis and transport. J Bacteriol 117:517–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ordal GW, Adler J. 1974. Isolation and complementation of mutants in galactose taxis and transport. J Bacteriol 117:509–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ordal GW, Villani DP, Rosendahl MS. 1979. Chemotaxis towards sugars by Bacillus subtilis. J Gen Microbiol 115:167–172. doi: 10.1099/00221287-115-1-167. [DOI] [PubMed] [Google Scholar]
- 83.Ordal GW, Goldman DJ. 1976. Chemotactic repellents of Bacillus subtilis. J Mol Biol 100:103–108. doi: 10.1016/S0022-2836(76)80037-X. [DOI] [PubMed] [Google Scholar]
- 84.Ordal GW, Gibson KJ. 1977. Chemotaxis toward amino acids by Bacillus subtilis. J Bacteriol 129:151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.de Jong MH, van der Drift C, Vogels GD. 1975. Receptors for chemotaxis in Bacillus subtilis. J Bacteriol 123:824–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Van Der Drift C, De Jong MH. 1974. Chemotaxis toward amino acids in Bacillus subtilis. Arch Microbiol 96:83–92. doi: 10.1007/BF00590165. [DOI] [PubMed] [Google Scholar]
- 87.Holscher T, Bartels B, Lin YC, Gallegos-Monterrosa R, Price-Whelan A, Kolter R, Dietrich LE, Kovacs AT. 2015. Motility, chemotaxis and aerotaxis contribute to competitiveness during bacterial pellicle biofilm development. J Mol Biol 427:3695–3708. doi: 10.1016/j.jmb.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Muller J, Schiel S, Ordal GW, Saxild HH. 1997. Functional and genetic characterization of mcpC, which encodes a third methyl-accepting chemotaxis protein in Bacillus subtilis. Microbiology 143(Part 10):3231–3240. doi: 10.1099/00221287-143-10-3231. [DOI] [PubMed] [Google Scholar]
- 89.Glekas GD, Mulhern BJ, Kroc A, Duelfer KA, Lei V, Rao CV, Ordal GW. 2012. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J Biol Chem 287:39412–39418. doi: 10.1074/jbc.M112.413518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Anantharaman V, Aravind L. 2001. The CHASE domain: a predicted ligand-binding module in plant cytokinin receptors and other eukaryotic and bacterial receptors. Trends Biochem Sci 26:579–582. doi: 10.1016/S0968-0004(01)01968-5. [DOI] [PubMed] [Google Scholar]
- 91.Mougel C, Zhulin IB. 2001. CHASE: an extracellular sensing domain common to transmembrane receptors from prokaryotes, lower eukaryotes and plants. Trends Biochem Sci 26:582–584. doi: 10.1016/S0968-0004(01)01969-7. [DOI] [PubMed] [Google Scholar]
- 92.Hou S, Freitas T, Larsen RW, Piatibratov M, Sivozhelezov V, Yamamoto A, Meleshkevitch EA, Zimmer M, Ordal GW, Alam M. 2001. Globin-coupled sensors: a class of heme-containing sensors in Archaea and Bacteria. Proc Natl Acad Sci U S A 98:9353–9358. doi: 10.1073/pnas.161185598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nakamura H, Yoshiyama H, Takeuchi H, Mizote T, Okita K, Nakazawa T. 1998. Urease plays an important role in the chemotactic motility of Helicobacter pylori in a viscous environment. Infect Immun 66:4832–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Keilberg D, Ottemann KM. 2016. How Helicobacter pylori senses, targets and interacts with the gastric epithelium. Environ Microbiol 18:791–806. doi: 10.1111/1462-2920.13222. [DOI] [PubMed] [Google Scholar]
- 95.Mizote T, Yoshiyama H, Nakazawa T. 1997. Urease-independent chemotactic responses of Helicobacter pylori to urea, urease inhibitors, and sodium bicarbonate. Infect Immun 65:1519–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Worku ML, Karim QN, Spencer J, Sidebotham RL. 2004. Chemotactic response of Helicobacter pylori to human plasma and bile. J Med Microbiol 53:807–811. doi: 10.1099/jmm.0.45636-0. [DOI] [PubMed] [Google Scholar]
- 97.Huang JY, Sweeney EG, Sigal M, Zhang HC, Remington SJ, Cantrell MA, Kuo CJ, Guillemin K, Amieva MR. 2015. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe 18:147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yoshiyama H, Nakamura H, Kimoto M, Okita K, Nakazawa T. 1999. Chemotaxis and motility of Helicobacter pylori in a viscous environment. J Gastroenterol 34(Suppl 11):S18–S23. [PubMed] [Google Scholar]
- 99.Cerda O, Rivas A, Toledo H. 2003. Helicobacter pylori strain ATCC700392 encodes a methyl-accepting chemotaxis receptor protein (MCP) for arginine and sodium bicarbonate. FEMS Microbiol Lett 224:175–181. doi: 10.1016/S0378-1097(03)00423-3. [DOI] [PubMed] [Google Scholar]
- 100.Wunder C, Churin Y, Winau F, Warnecke D, Vieth M, Lindner B, Zahringer U, Mollenkopf HJ, Heinz E, Meyer TF. 2006. Cholesterol glucosylation promotes immune evasion by Helicobacter pylori. Nat Med 12:1030–1038. doi: 10.1038/nm1480. [DOI] [PubMed] [Google Scholar]
- 101.Rader BA, Wreden C, Hicks KG, Sweeney EG, Ottemann KM, Guillemin K. 2011. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiology 157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanders L, Andermann TM, Ottemann KM. 2013. A supplemented soft agar chemotaxis assay demonstrates the Helicobacter pylori chemotactic response to zinc and nickel. Microbiology 159:46–57. doi: 10.1099/mic.0.062877-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Croxen MA, Sisson G, Melano R, Hoffman PS. 2006. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J Bacteriol 188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Andermann TM, Chen YT, Ottemann KM. 2002. Two predicted chemoreceptors of Helicobacter pylori promote stomach infection. Infect Immun 70:5877–5881. doi: 10.1128/IAI.70.10.5877-5881.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rolig AS, Shanks J, Carter JE, Ottemann KM. 2012. Helicobacter pylori requires TlpD-driven chemotaxis to proliferate in the antrum. Infect Immun 80:3713–3720. doi: 10.1128/IAI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Behrens W, Schweinitzer T, Bal J, Dorsch M, Bleich A, Kops F, Brenneke B, Didelot X, Suerbaum S, Josenhans C. 2013. Role of energy sensor TlpD of Helicobacter pylori in gerbil colonization and genome analyses after adaptation in the gerbil. Infect Immun 81:3534–3551. doi: 10.1128/IAI.00750-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McGee DJ, Langford ML, Watson EL, Carter JE, Chen YT, Ottemann KM. 2005. Colonization and inflammation deficiencies in Mongolian gerbils infected by Helicobacter pylori chemotaxis mutants. Infect Immun 73:1820–1827. doi: 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Williams SM, Chen YT, Andermann TM, Carter JE, McGee DJ, Ottemann KM. 2007. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect Immun 75:3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Goers Sweeney E, Henderson JN, Goers J, Wreden C, Hicks KG, Foster JK, Parthasarathy R, Remington SJ, Guillemin K. 2012. Structure and proposed mechanism for the pH-sensing Helicobacter pylori chemoreceptor TlpB. Structure 20:1177–1188. doi: 10.1016/j.str.2012.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Anderson JK, Huang JY, Wreden C, Sweeney EG, Goers J, Remington SJ, Guillemin K. 2015. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. mBio 6:e00379-15. doi: 10.1128/mBio.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cerda OA, Nunez-Villena F, Soto SE, Ugalde JM, Lopez-Solis R, Toledo H. 2011. tlpA gene expression is required for arginine and bicarbonate chemotaxis in Helicobacter pylori. Biol Res 44:277–282. doi: 10.4067/S0716-97602011000300009. [DOI] [PubMed] [Google Scholar]
- 112.Behrens W, Schweinitzer T, McMurry JL, Loewen PC, Buettner FF, Menz S, Josenhans C. 2016. Localisation and protein-protein interactions of the Helicobacter pylori taxis sensor TlpD and their connection to metabolic functions. Sci Rep 6:23582. doi: 10.1038/srep23582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S, Lee J, Ha J, Choi Y, Kim S, Lee H, Yoon Y, Choi KH. 2016. Clinical relevance of infections with zoonotic and human oral species of Campylobacter. J Microbiol 54:459–467. doi: 10.1007/s12275-016-6254-x. [DOI] [PubMed] [Google Scholar]
- 114.Vegge CS, Brondsted L, Li YP, Bang DD, Ingmer H. 2009. Energy taxis drives Campylobacter jejuni toward the most favorable conditions for growth. Appl Environ Microbiol 75:5308–5314. doi: 10.1128/AEM.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hugdahl MB, Beery JT, Doyle MP. 1988. Chemotactic behavior of Campylobacter jejuni. Infect Immun 56:1560–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hartley-Tassell LE, Shewell LK, Day CJ, Wilson JC, Sandhu R, Ketley JM, Korolik V. 2010. Identification and characterization of the aspartate chemosensory receptor of Campylobacter jejuni. Mol Microbiol 75:710–730. doi: 10.1111/j.1365-2958.2009.07010.x. [DOI] [PubMed] [Google Scholar]
- 117.Li Z, Lou H, Ojcius DM, Sun A, Sun D, Zhao J, Lin X, Yan J. 2014. Methyl-accepting chemotaxis proteins 3 and 4 are responsible for Campylobacter jejuni chemotaxis and jejuna colonization in mice in response to sodium deoxycholate. J Med Microbiol 63:343–354. doi: 10.1099/jmm.0.068023-0. [DOI] [PubMed] [Google Scholar]
- 118.Dwivedi R, Nothaft H, Garber J, Xin Kin L, Stahl M, Flint A, van Vliet AH, Stintzi A, Szymanski CM. 2016. l-Fucose influences chemotaxis and biofilm formation in Campylobacter jejuni. Mol Microbiol 101:575–589. doi: 10.1111/mmi.13409. [DOI] [PubMed] [Google Scholar]
- 119.Day CJ, King RM, Shewell LK, Tram G, Najnin T, Hartley-Tassell LE, Wilson JC, Fleetwood AD, Zhulin IB, Korolik V. 2016. A direct-sensing galactose chemoreceptor recently evolved in invasive strains of Campylobacter jejuni. Nat Commun 7:13206. doi: 10.1038/ncomms13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Marchant J, Wren B, Ketley J. 2002. Exploiting genome sequence: predictions for mechanisms of Campylobacter chemotaxis. Trends Microbiol 10:155–159. doi: 10.1016/S0966-842X(02)02323-5. [DOI] [PubMed] [Google Scholar]
- 121.Hendrixson DR, DiRita VJ. 2004. Identification of Campylobacter jejuni genes involved in commensal colonization of the chick gastrointestinal tract. Mol Microbiol 52:471–484. doi: 10.1111/j.1365-2958.2004.03988.x. [DOI] [PubMed] [Google Scholar]
- 122.Elliott KT, DiRita VJ. 2008. Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol Microbiol 69:1091–1103. doi: 10.1111/j.1365-2958.2008.06357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Elliott KT, Zhulin IB, Stuckey JA, DiRita VJ. 2009. Conserved residues in the HAMP domain define a new family of proposed bipartite energy taxis receptors. J Bacteriol 191:375–387. doi: 10.1128/JB.00578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Tareen AM, Dasti JI, Zautner AE, Gross U, Lugert R. 2010. Campylobacter jejuni proteins Cj0952c and Cj0951c affect chemotactic behaviour towards formic acid and are important for invasion of host cells. Microbiology 156:3123–3135. doi: 10.1099/mic.0.039438-0. [DOI] [PubMed] [Google Scholar]
- 125.Reuter M, van Vliet AH. 2013. Signal balancing by the CetABC and CetZ chemoreceptors controls energy taxis in Campylobacter jejuni. PLoS One 8:e54390. doi: 10.1371/journal.pone.0054390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Borziak K, Zhulin IB. 2007. FIST: a sensory domain for diverse signal transduction pathways in prokaryotes and ubiquitin signaling in eukaryotes. Bioinformatics 23:2518–2521. doi: 10.1093/bioinformatics/btm384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Klockgether J, Munder A, Neugebauer J, Davenport CF, Stanke F, Larbig KD, Heeb S, Schock U, Pohl TM, Wiehlmann L, Tummler B. 2010. Genome diversity of Pseudomonas aeruginosa PAO1 laboratory strains. J Bacteriol 192:1113–1121. doi: 10.1128/JB.01515-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Regenhardt D, Heuer H, Heim S, Fernandez DU, Strompl C, Moore ER, Timmis KN. 2002. Pedigree and taxonomic credentials of Pseudomonas putida strain KT2440. Environ Microbiol 4:912–915. doi: 10.1046/j.1462-2920.2002.00368.x. [DOI] [PubMed] [Google Scholar]
- 129.Compeau G, Al-Achi BJ, Platsouka E, Levy SB. 1988. Survival of rifampin-resistant mutants of Pseudomonas fluorescens and Pseudomonas putida in soil systems. Appl Environ Microbiol 54:2432–2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Oku S, Komatsu A, Nakashimada Y, Tajima T, Kato J. 2014. Identification of Pseudomonas fluorescens chemotaxis sensory proteins for malate, succinate, and fumarate, and their involvement in root colonization. Microbes Environ 29:413–419. doi: 10.1264/jsme2.ME14128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Oku S, Komatsu A, Tajima T, Nakashimada Y, Kato J. 2012. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ 27:462–469. doi: 10.1264/jsme2.ME12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sampedro I, Parales RE, Krell T, Hill JE. 2015. Pseudomonas chemotaxis. FEMS Microbiol Rev 39:17–46. doi: 10.1111/1574-6976.12081. [DOI] [PubMed] [Google Scholar]
- 133.Guvener ZT, Tifrea DF, Harwood CS. 2006. Two different Pseudomonas aeruginosa chemosensory signal transduction complexes localize to cell poles and form and remould in stationary phase. Mol Microbiol 61:106–118. doi: 10.1111/j.1365-2958.2006.05218.x. [DOI] [PubMed] [Google Scholar]
- 134.Darzins A. 1994. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol 11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 135.Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC. 2010. The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76:889–904. doi: 10.1111/j.1365-2958.2010.07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kuroda A, Kumano T, Taguchi K, Nikata T, Kato J, Ohtake H. 1995. Molecular cloning and characterization of a chemotactic transducer gene in Pseudomonas aeruginosa. J Bacteriol 177:7019–7025. doi: 10.1128/jb.177.24.7019-7025.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Taguchi K, Fukutomi H, Kuroda A, Kato J, Ohtake H. 1997. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 143(Part 10):3223–3229. doi: 10.1099/00221287-143-10-3223. [DOI] [PubMed] [Google Scholar]
- 138.Rico-Jimenez M, Munoz-Martinez F, Garcia-Fontana C, Fernandez M, Morel B, Ortega A, Ramos JL, Krell T. 2013. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol Microbiol 88:1230–1243. doi: 10.1111/mmi.12255. [DOI] [PubMed] [Google Scholar]
- 139.Reyes-Darias JA, Yang Y, Sourjik V, Krell T. 2015. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol Microbiol 96:513–525. doi: 10.1111/mmi.12953. [DOI] [PubMed] [Google Scholar]
- 140.Kim HE, Shitashiro M, Kuroda A, Takiguchi N, Ohtake H, Kato J. 2006. Identification and characterization of the chemotactic transducer in Pseudomonas aeruginosa PAO1 for positive chemotaxis to trichloroethylene. J Bacteriol 188:6700–6702. doi: 10.1128/JB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu H, Kato J, Kuroda A, Ikeda T, Takiguchi N, Ohtake H. 2000. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J Bacteriol 182:3400–3404. doi: 10.1128/JB.182.12.3400-3404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ortega A, Krell T. 2014. The HBM domain: introducing bimodularity to bacterial sensing. Protein Sci 23:332–336. doi: 10.1002/pro.2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Neznansky A, Blus-Kadosh I, Yerushalmi G, Banin E, Opatowsky Y. 2014. The Pseudomonas aeruginosa phosphate transport protein PstS plays a phosphate-independent role in biofilm formation. FASEB J 28:5223–5233. doi: 10.1096/fj.14-258293. [DOI] [PubMed] [Google Scholar]
- 144.Kondoh H, Ball CB, Adler J. 1979. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci U S A 76:260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Alvarez-Ortega C, Harwood CS. 2007. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl Environ Microbiol 73:7793–7795. doi: 10.1128/AEM.01898-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kim HE, Shitashiro M, Kuroda A, Takiguchi N, Kato J. 2007. Ethylene chemotaxis in Pseudomonas aeruginosa and other Pseudomonas species. Microbes Environ 22:186–189. doi: 10.1264/jsme2.22.186. [DOI] [Google Scholar]
- 147.Martin-Mora D, Ortega A, Reyes-Darias JA, García V, López-Farfán D, Matilla MA, Krell T. 2016. Identification of a chemoreceptor in Pseudomonas aeruginosa that specifically mediates chemotaxis towards alpha-ketoglutarate. Front Microbiol 7:1937. doi: 10.3389/fmicb.2016.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hong CS, Shitashiro M, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. 2004. Chemotaxis proteins and transducers for aerotaxis in Pseudomonas aeruginosa. FEMS Microbiol Lett 231:247–252. doi: 10.1016/S0378-1097(04)00009-6. [DOI] [PubMed] [Google Scholar]
- 149.Watts KJ, Taylor BL, Johnson MS. 2011. PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Mol Microbiol 79:686–699. doi: 10.1111/j.1365-2958.2010.07477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Garcia-Fontana C, Corral Lugo A, Krell T. 2014. Specificity of the CheR2 methyltransferase in Pseudomonas aeruginosa is directed by a C-terminal pentapeptide in the McpB chemoreceptor. Sci Signal 7:ra34. doi: 10.1126/scisignal.2004849. [DOI] [PubMed] [Google Scholar]
- 151.Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B, Filloux A. 2009. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5:e1000540. doi: 10.1371/journal.ppat.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Garcia D, Orillard E, Johnson MS, Watts KJ. 22 August 2017. Gas sensing and signaling in the PAS-heme domain of the Pseudomonas aeruginosa Aer2 receptor. J Bacteriol doi: 10.1128/JB.00003-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR. 2013. Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 425:886–901. doi: 10.1016/j.jmb.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sawai H, Sugimoto H, Shiro Y, Ishikawa H, Mizutani Y, Aono S. 2012. Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem Commun (Camb) 48:6523–6525. doi: 10.1039/c2cc32549g. [DOI] [PubMed] [Google Scholar]
- 155.Morgan R, Kohn S, Hwang SH, Hassett DJ, Sauer K. 2006. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Petrova OE, Sauer K. 2012. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc Natl Acad Sci U S A 109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Luo Y, Zhao K, Baker AE, Kuchma SL, Coggan KA, Wolfgang MC, Wong GC, O'Toole GA. 2015. A hierarchical cascade of second messengers regulates Pseudomonas aeruginosa surface behaviors. mBio 6:e02456-14. doi: 10.1128/mBio.02456-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kearns DB, Robinson J, Shimkets LJ. 2001. Pseudomonas aeruginosa exhibits directed twitching motility up phosphatidylethanolamine gradients. J Bacteriol 183:763–767. doi: 10.1128/JB.183.2.763-767.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.DeLange PA, Collins TL, Pierce GE, Robinson JB. 2007. PilJ localizes to cell poles and is required for type IV pilus extension in Pseudomonas aeruginosa. Curr Microbiol 55:389–395. doi: 10.1007/s00284-007-9008-5. [DOI] [PubMed] [Google Scholar]
- 160.Persat A, Inclan YF, Engel JN, Stone HA, Gitai Z. 2015. Type IV pili mechanochemically regulate virulence factors in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A 112:7563–7568. doi: 10.1073/pnas.1502025112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Garcia-Fontana C, Reyes-Darias JA, Munoz-Martinez F, Alfonso C, Morel B, Ramos JL, Krell T. 2013. High specificity in CheR methyltransferase function: CheR2 of Pseudomonas putida is essential for chemotaxis, whereas CheR1 is involved in biofilm formation. J Biol Chem 288:18987–18999. doi: 10.1074/jbc.M113.472605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fernandez M, Morel B, Corral-Lugo A, Krell T. 2016. Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol Microbiol 99:34–42. doi: 10.1111/mmi.13215. [DOI] [PubMed] [Google Scholar]
- 163.Corral-Lugo A, de la Torre J, Matilla MA, Fernandez M, Morel B, Espinosa-Urgel M, Krell T. 2016. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ Microbiol 18:3355–3372. doi: 10.1111/1462-2920.13170. [DOI] [PubMed] [Google Scholar]
- 164.Lacal J, Alfonso C, Liu X, Parales RE, Morel B, Conejero-Lara F, Rivas G, Duque E, Ramos JL, Krell T. 2010. Identification of a chemoreceptor for tricarboxylic acid cycle intermediates: differential chemotactic response towards receptor ligands. J Biol Chem 285:23126–23136. doi: 10.1074/jbc.M110.110403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lacal J, Garcia-Fontana C, Callejo-Garcia C, Ramos JL, Krell T. 2011. Physiologically relevant divalent cations modulate citrate recognition by the McpS chemoreceptor. J Mol Recognit 24:378–385. doi: 10.1002/jmr.1101. [DOI] [PubMed] [Google Scholar]
- 166.Pineda-Molina E, Reyes-Darias JA, Lacal J, Ramos JL, Garcia-Ruiz JM, Gavira JA, Krell T. 2012. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc Natl Acad Sci U S A 109:18926–18931. doi: 10.1073/pnas.1201400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Martin-Mora D, Reyes-Darias JA, Ortega A, Corral-Lugo A, Matilla MA, Krell T. 2016. McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ Microbiol 18:3284–3295. doi: 10.1111/1462-2920.13030. [DOI] [PubMed] [Google Scholar]
- 168.Garcia V, Reyes-Darias JA, Martin-Mora D, Morel B, Matilla MA, Krell T. 2015. Identification of a chemoreceptor for C2 and C3 carboxylic acids. Appl Environ Microbiol 81:5449–5457. doi: 10.1128/AEM.01529-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sarand I, Osterberg S, Holmqvist S, Holmfeldt P, Skarfstad E, Parales RE, Shingler V. 2008. Metabolism-dependent taxis towards (methyl)phenols is coupled through the most abundant of three polar localized Aer-like proteins of Pseudomonas putida. Environ Microbiol 10:1320–1334. doi: 10.1111/j.1462-2920.2007.01546.x. [DOI] [PubMed] [Google Scholar]
- 170.Lacal J, Munoz-Martinez F, Reyes-Darias JA, Duque E, Matilla M, Segura A, Calvo JJ, Jimenez-Sanchez C, Krell T, Ramos JL. 2011. Bacterial chemotaxis towards aromatic hydrocarbons in Pseudomonas. Environ Microbiol 13:1733–1744. doi: 10.1111/j.1462-2920.2011.02493.x. [DOI] [PubMed] [Google Scholar]
- 171.Grimm AC, Harwood CS. 1999. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J Bacteriol 181:3310–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu X, Wood PL, Parales JV, Parales RE. 2009. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J Bacteriol 191:2909–2916. doi: 10.1128/JB.01708-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Parales RE, Nesteryuk V, Hughes JG, Luu RA, Ditty JL. 2014. Cytosine chemoreceptor McpC in Pseudomonas putida F1 also detects nicotinic acid. Microbiology 160:2661–2669. doi: 10.1099/mic.0.081968-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Luu RA, Kootstra JD, Nesteryuk V, Brunton CN, Parales JV, Ditty JL, Parales RE. 2015. Integration of chemotaxis, transport and catabolism in Pseudomonas putida and identification of the aromatic acid chemoreceptor PcaY. Mol Microbiol 96:134–147. doi: 10.1111/mmi.12929. [DOI] [PubMed] [Google Scholar]
- 175.Parales RE, Luu RA, Chen GY, Liu X, Wu V, Lin P, Hughes JG, Nesteryuk V, Parales JV, Ditty JL. 2013. Pseudomonas putida F1 has multiple chemoreceptors with overlapping specificity for organic acids. Microbiology 159:1086–1096. doi: 10.1099/mic.0.065698-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Iwaki H, Muraki T, Ishihara S, Hasegawa Y, Rankin KN, Sulea T, Boyd J, Lau PC. 2007. Characterization of a pseudomonad 2-nitrobenzoate nitroreductase and its catabolic pathway-associated 2-hydroxylaminobenzoate mutase and a chemoreceptor involved in 2-nitrobenzoate chemotaxis. J Bacteriol 189:3502–3514. doi: 10.1128/JB.01098-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.López-Farfán D, Reyes-Darias JA, Krell T. 8 September 2016. The expression of many chemoreceptor genes depends on the cognate chemoeffector as well as on the growth medium and phase. Curr Genet doi: 10.1007/s00294-016-0646-7. [DOI] [PubMed] [Google Scholar]
- 178.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liu YC, Machuca MA, Beckham SA, Gunzburg MJ, Roujeinikova A. 2015. Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr D Biol Crystallogr 71:2127–2136. doi: 10.1107/S139900471501384X. [DOI] [PubMed] [Google Scholar]
- 180.Glekas GD, Foster RM, Cates JR, Estrella JA, Wawrzyniak MJ, Rao CV, Ordal GW. 2010. A PAS domain binds asparagine in the chemotaxis receptor McpB in Bacillus subtilis. J Biol Chem 285:1870–1878. doi: 10.1074/jbc.M109.072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Henry JT, Crosson S. 2011. Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Gross-Langenhoff M, Hofbauer K, Weber J, Schultz A, Schultz JE. 2006. cAMP is a ligand for the tandem GAF domain of human phosphodiesterase 10 and cGMP for the tandem GAF domain of phosphodiesterase 11. J Biol Chem 281:2841–2846. doi: 10.1074/jbc.M511468200. [DOI] [PubMed] [Google Scholar]
- 183.Shu CJ, Ulrich LE, Zhulin IB. 2003. The NIT domain: a predicted nitrate-responsive module in bacterial sensory receptors. Trends Biochem Sci 28:121–124. doi: 10.1016/S0968-0004(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 184.Amikam D, Galperin MY. 2006. PilZ domain is part of the bacterial c-di-GMP binding protein. Bioinformatics 22:3–6. doi: 10.1093/bioinformatics/bti739. [DOI] [PubMed] [Google Scholar]
- 185.Anantharaman V, Aravind L. 2005. MEDS and PocR are novel domains with a predicted role in sensing simple hydrocarbon derivatives in prokaryotic signal transduction systems. Bioinformatics 21:2805–2811. doi: 10.1093/bioinformatics/bti418. [DOI] [PubMed] [Google Scholar]
- 186.Iyer LM, Anantharaman V, Aravind L. 2003. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics 4:5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28:655–662. doi: 10.1016/j.tibs.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 188.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. 2006. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci U S A 103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Milligan DL, Koshland DE Jr. 1993. Purification and characterization of the periplasmic domain of the aspartate chemoreceptor. J Biol Chem 268:19991–19997. [PubMed] [Google Scholar]
- 190.Biemann HP, Koshland DE Jr. 1994. Aspartate receptors of Escherichia coli and Salmonella typhimurium bind ligand with negative and half-of-the-sites cooperativity. Biochemistry 33:629–634. doi: 10.1021/bi00169a002. [DOI] [PubMed] [Google Scholar]
- 191.Liu YC, Roujeinikova A. 2015. Expression, refolding, purification and crystallization of the sensory domain of the TlpC chemoreceptor from Helicobacter pylori for structural studies. Protein Expr Purif 107:29–34. doi: 10.1016/j.pep.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 192.Inda ME, Vandenbranden M, Fernandez A, de Mendoza D, Ruysschaert JM, Cybulski LE. 2014. A lipid-mediated conformational switch modulates the thermosensing activity of DesK. Proc Natl Acad Sci U S A 111:3579–3584. doi: 10.1073/pnas.1317147111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J 20:1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Garrity LF, Schiel SL, Merrill R, Reizer J, Saier MH Jr, Ordal GW. 1998. Unique regulation of carbohydrate chemotaxis in Bacillus subtilis by the phosphoenolpyruvate-dependent phosphotransferase system and the methyl-accepting chemotaxis protein McpC. J Bacteriol 180:4475–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kristich CJ, Glekas GD, Ordal GW. 2003. The conserved cytoplasmic module of the transmembrane chemoreceptor McpC mediates carbohydrate chemotaxis in Bacillus subtilis. Mol Microbiol 47:1353–1366. doi: 10.1046/j.1365-2958.2003.03375.x. [DOI] [PubMed] [Google Scholar]
- 196.Samanta D, Widom J, Borbat PP, Freed JH, Crane BR. 2016. Bacterial energy sensor Aer modulates the activity of the chemotaxis kinase CheA based on the redox state of the flavin cofactor. J Biol Chem 291:25809–25814. doi: 10.1074/jbc.C116.757492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ottemann KM, Xiao W, Shin YK, Koshland DE Jr. 1999. A piston model for transmembrane signaling of the aspartate receptor. Science 285:1751–1754. doi: 10.1126/science.285.5434.1751. [DOI] [PubMed] [Google Scholar]
- 198.Ames P, Hunter S, Parkinson JS. 2016. Evidence for a helix-clutch mechanism of transmembrane signaling in a bacterial chemoreceptor. J Mol Biol 428:3776–3788. doi: 10.1016/j.jmb.2016.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yu D, Ma X, Tu Y, Lai L. 2015. Both piston-like and rotational motions are present in bacterial chemoreceptor signaling. Sci Rep 5:8640. doi: 10.1038/srep08640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Chervitz SA, Falke JJ. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci U S A 93:2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gushchin I, Melnikov I, Polovinkin V, Ishchenko A, Yuzhakova A, Buslaev P, Bourenkov G, Grudinin S, Round E, Balandin T, Borshchevskiy V, Willbold D, Leonard G, Buldt G, Popov A, Gordeliy V. 2017. Mechanism of transmembrane signaling by sensor histidine kinases. Science 356:eaah6345. doi: 10.1126/science.aah6345. [DOI] [PubMed] [Google Scholar]
- 202.Bi S, Pollard AM, Yang Y, Jin F, Sourjik V. 10 June 2016. Engineering hybrid chemotaxis receptors in bacteria. ACS Synth Biol doi: 10.1021/acssynbio.6b00053. [DOI] [PubMed] [Google Scholar]
- 203.Huang Z, Ni B, Jiang CY, Wu YF, He YZ, Parales RE, Liu SJ. 2016. Direct sensing and signal transduction during bacterial chemotaxis toward aromatic compounds in Comamonas testosteroni. Mol Microbiol 101:224–237. doi: 10.1111/mmi.13385. [DOI] [PubMed] [Google Scholar]
- 204.Ni B, Huang Z, Wu YF, Fan Z, Jiang CY, Liu SJ. 2015. A novel chemoreceptor MCP2983 from Comamonas testosteroni specifically binds to cis-aconitate and triggers chemotaxis towards diverse organic compounds. Appl Microbiol Biotechnol 99:2773–2781. doi: 10.1007/s00253-014-6216-3. [DOI] [PubMed] [Google Scholar]
- 205.Nishiyama S, Suzuki D, Itoh Y, Suzuki K, Tajima H, Hyakutake A, Homma M, Butler-Wu SM, Camilli A, Kawagishi I. 2012. Mlp24 (McpX) of Vibrio cholerae implicated in pathogenicity functions as a chemoreceptor for multiple amino acids. Infect Immun 80:3170–3178. doi: 10.1128/IAI.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Webb BA, Hildreth S, Helm RF, Scharf BE. 2014. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl Environ Microbiol 80:3404–3415. doi: 10.1128/AEM.00115-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Webb BA, Compton KK, Castaneda Saldana R, Arapov TD, Ray WK, Helm RF, Scharf BE. 11 November 2016. Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol Microbiol doi: 10.1111/mmi.13561. [DOI] [PubMed] [Google Scholar]
- 208.Yeh JI, Biemann HP, Prive GG, Pandit J, Koshland DE Jr, Kim SH. 1996. High-resolution structures of the ligand binding domain of the wild-type bacterial aspartate receptor. J Mol Biol 262:186–201. doi: 10.1006/jmbi.1996.0507. [DOI] [PubMed] [Google Scholar]
- 209.Iwama T, Ito Y, Aoki H, Sakamoto H, Yamagata S, Kawai K, Kawagishi I. 2006. Differential recognition of citrate and a metal-citrate complex by the bacterial chemoreceptor Tcp. J Biol Chem 281:17727–17735. doi: 10.1074/jbc.M601038200. [DOI] [PubMed] [Google Scholar]
- 210.Hong CS, Kuroda A, Ikeda T, Takiguchi N, Ohtake H, Kato J. 2004. The aerotaxis transducer gene aer, but not aer-2, is transcriptionally regulated by the anaerobic regulator ANR in Pseudomonas aeruginosa. J Biosci Bioeng 97:184–190. doi: 10.1016/S1389-1723(04)70188-7. [DOI] [PubMed] [Google Scholar]
- 211.O'Connor JR, Kuwada NJ, Huangyutitham V, Wiggins PA, Harwood CS. 2012. Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol Microbiol 86:720–729. doi: 10.1111/mmi.12013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S, Ha DG, Willger SD, O'Toole GA, Harwood CS, Dietrich LE, Hogan DA. 2014. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Shitashiro M, Tanaka H, Hong CS, Kuroda A, Takiguchi N, Ohtake H, Kato J. 2005. Identification of chemosensory proteins for trichloroethylene in Pseudomonas aeruginosa. J Biosci Bioeng 99:396–402. doi: 10.1263/jbb.99.396. [DOI] [PubMed] [Google Scholar]
- 214.Vangnai AS, Takeuchi K, Oku S, Kataoka N, Nitisakulkan T, Tajima T, Kato J. 2013. Identification of CtpL as a chromosomally encoded chemoreceptor for 4-chloroaniline and catechol in Pseudomonas aeruginosa PAO1. Appl Environ Microbiol 79:7241–7248. doi: 10.1128/AEM.02428-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Luu RA, Schneider BJ, Ho CC, Nesteryuk V, Ngwesse SE, Liu X, Parales JV, Ditty JL, Parales RE. 2013. Taxis of Pseudomonas putida F1 toward phenylacetic acid is mediated by the energy taxis receptor Aer2. Appl Environ Microbiol 79:2416–2423. doi: 10.1128/AEM.03895-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Nichols NN, Harwood CS. 2000. An aerotaxis transducer gene from Pseudomonas putida. FEMS Microbiol Lett 182:177–183. doi: 10.1111/j.1574-6968.2000.tb08893.x. [DOI] [PubMed] [Google Scholar]
- 217.Mise T. 2016. Structural analysis of the ligand-binding domain of the aspartate receptor Tar from Escherichia coli. Biochemistry 55:3708–3713. doi: 10.1021/acs.biochem.6b00160. [DOI] [PubMed] [Google Scholar]
- 218.Machuca MA, Liu YC, Beckham SA, Gunzburg MJ, Roujeinikova A. 2016. The crystal structure of the tandem-PAS sensing domain of Campylobacter jejuni chemoreceptor Tlp1 suggests indirect mechanism of ligand recognition. J Struct Biol 194:205–213. doi: 10.1016/j.jsb.2016.02.019. [DOI] [PubMed] [Google Scholar]
- 219.Yu EW, Koshland DE Jr. 2001. Propagating conformational changes over long (and short) distances in proteins. Proc Natl Acad Sci U S A 98:9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yeh JI, Biemann HP, Pandit J, Koshland DE, Kim SH. 1993. The three-dimensional structure of the ligand-binding domain of a wild-type bacterial chemotaxis receptor. Structural comparison to the cross-linked mutant forms and conformational changes upon ligand binding. J Biol Chem 268:9787–9792. [PubMed] [Google Scholar]
- 221.Zhang W, Phillips GN Jr. 2003. Structure of the oxygen sensor in Bacillus subtilis: signal transduction of chemotaxis by control of symmetry. Structure 11:1097–1110. doi: 10.1016/S0969-2126(03)00169-2. [DOI] [PubMed] [Google Scholar]