Abstract
Disorders resulting from HBB (β-globin) gene mutations, mainly sickle cell disease (SCD) and β-thalassemia, become symptomatic postnatally as fetal γ-globin expression from two parologous genes HBG1 and HBG2 falls and adult β-globin increases, thereby shifting red blood cell (RBC) hemoglobin from the fetal (HbF, α2γ2) to adult form (HbA, α2β2). These disorders are alleviated when postnatal expression of fetal γ-globin is maintained. For example, in hereditary persistence of fetal hemoglobin (HPFH), a benign genetic condition, mutations attenuate γ-to-β switching, causing high-level HbF expression throughout life. Co-inheritance of HPFH with β-thalassemia or SCD mutations alleviates their clinical manifestations. Here we performed CRISPR-Cas9-mediated genome editing of human blood progenitors to mutate a 13-nucleotide HBG1/HBG2 promoter sequence, thereby recapitulating a naturally occurring HPFH-associated mutation. Edited progenitors produced RBCs with increased HbF levels that were sufficient to inhibit pathological hypoxia-induced RBC morphology of SCD. Our findings identify a potential DNA target for genome editing-mediated therapy of β–hemoglobinopathies.
Typical HPFH mutations include heterozygous deletions or nucleotide (nt) substitutions within the extended β-globin locus1. For example, the −175 T>C HBG1 substitution de-represses γ-globin by creating a binding site for the transcriptional activator TAL12. Other forms of HPFH may be associated with loss of cis elements that recruit γ-globin repressor proteins. We focused on a 13-nt deletion within the HBG1 (Aγ) promoter (−102 to −114, Fig. 1a)3. The deleted region contains a CCAAT box and direct repeat (DR), both of which recruit transcriptional repressor proteins4–8. In heterozygous individuals, HbF expression is pancellular and comprises approximately 30% of all Hb protein (compared to < 1% for normal individuals lacking the mutation), which is potentially therapeutic for hemoglobinopathies, as SCD patients with this level of HbF are asymptomatic9. We reasoned that a DNA break induced by site-directed CRISPR/Cas9 mutagenesis, followed by error-prone non-homologous end joining (NHEJ) might recapitulate effects of the 13-nt-deleted HPFH mutation10. We used lentivirus to express mCherry, Cas9 and 2 different guide RNAs (gRNAs) targeting this region in the erythroblast cell line HUDEP-2, which expresses mainly HbA11 (Fig. 1a). HbF protein levels were undetectable in mock infected and Cas9 only expressing control cultures and rose to approximately 17% and 3% in cells expressing mCherry, Cas9 and gRNA-1 or gRNA-2, respectively (Supplementary Fig. 1a). Cells staining positive for HbF (“F-cells”) increased from 2% to 46% with gRNA-1, and to 26% with gRNA-2 (Fig. 1b). The %HBG1/2 (γ-globin) mRNA [γ/(γ+β)] was increased by both gRNAs; HBB (β-globin) mRNA expression was decreased by gRNA-1 (Supplementary Fig. 1b–c). Induction of HbF by two adjacent non-overlapping gRNAs suggests that the effect is due to on-target mutations rather than to off-target mutations.
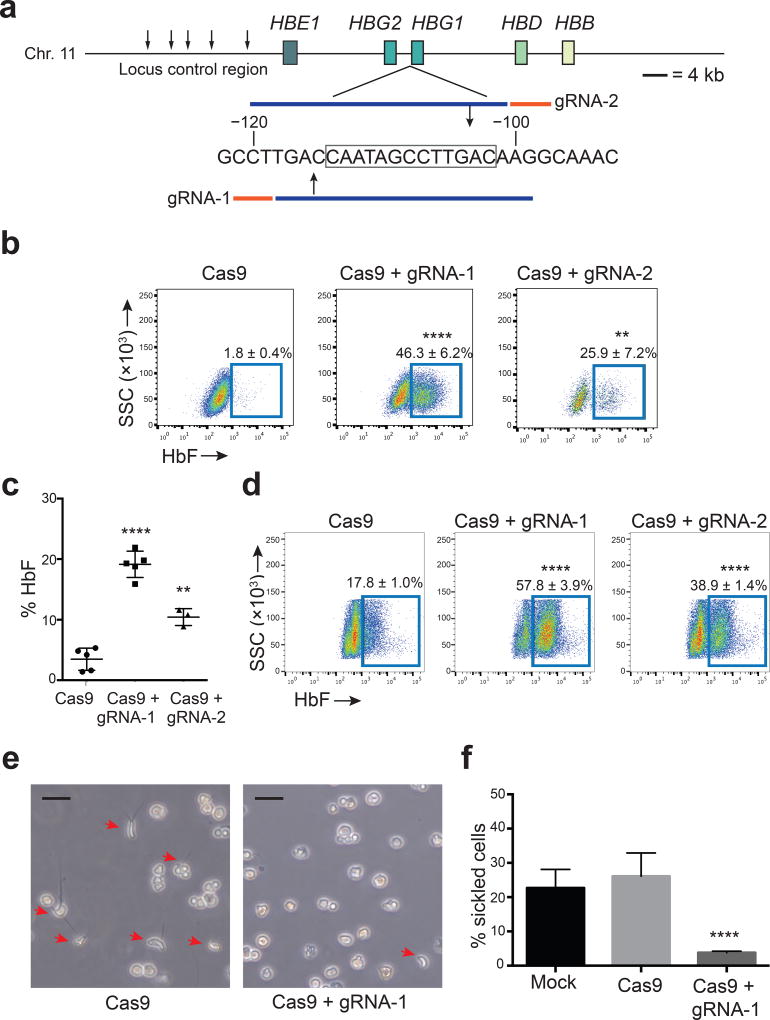
Figure 1. Genome editing of the HBG1 and HBG2 promoters increases erythroid fetal hemoglobin (HbF) levels.
(a) Extended β-globin locus showing β-like genes as boxes. Small arrows mark DNase hypersensitive sites within the locus control region, an upstream enhancer. A region of the HBG1 promoter is shown numbered according to position upstream of the transcription start with the 13-nt HPFH deletion boxed. Guide RNA spacer sequences are blue and PAM motifs (NGG) are orange; gRNA-1 and gRNA-2 are complementary to the sense and antisense strands, respectively. Large arrows show predicted Cas9 cleavage sites.
(b) Representative flow cytometry plots showing HbF+ immunostaining HUDEP-2 cells 5d after transduction with Cas9 ± gRNA-1 or gRNA-2 lentivirus. Numbers indicate mean ± standard error (SE) from four independent experiments.
(c) Normal human CD34+ cells transduced with lentivirus encoding Cas9 ± gRNA-1 or gRNA-2 were cultured for 21d in erythroid cytokines, then analyzed for hemoglobin (Hb) protein by HPLC. %HbF = [HbF/(HbA + HbF) × 100]. Each dot represents a separate experiment performed with CD34+ cells from the same donor. On-target editing rates of HBG1/HBG2 in three experiments were 56%, 65% and 77%.
(d) HbF+ erythroblasts, derived as described for panel (c). Numbers indicate mean ± SE from three experiments.
(e) CD34+ cells from an SCD (HbSS) patient were transduced with lentivirus expressing Cas9 ± gRNA-1, differentiated into RBCs, and cultured in 2% O2.Red arrows denote cells with sickle-like morphology. Original magnification 200×. Size bars indicate 20 µm.
(f) Quantification of hypoxia-induced sickled cells depicted in panel (e). Mean ± SE from three experiments using CD34+ cells from three different SCD donors (> 1,000 cells scored per experiment).
The unpaired t-test was used to analyze data in panels b-d and f. **** P < 0.0001, ** P < 0.01.
Next, we used the same mCherry/Cas9/gRNA-encoding lentiviral vectors to edit peripheral blood CD34+ hematopoietic stem/progenitor cells (HPSCs) from two healthy adults. Transduced HPSCs expressing mCherry were enriched by cell sorting and then induced to undergo erythroid differentiation. HbF protein rose from approximately 5% to 20% (Fig. 1c), and F-cells increased from 18% to 58% in erythroid progeny of Cas9/gRNA-1-expressing CD34+ cells (Fig. 1d); gRNA-2 produced similar effects of lesser magnitude. Gene editing did not alter the expression of erythroid stage-specific maturation markers (Supplementary Fig. 2a–d). In FACS-purified α4-integrin+/Band3+ late basophilic erythroblasts, gRNA-1 increased %HBG1/2 mRNA from 4% to 35% and decreased absolute HBB mRNA by about 50% (Supplementary Fig. 2e–f). Increased γ-globin with decreased β-globin expression indicates reversal of the γ-to-β switch, which is controlled by competition of the corresponding genes for an upstream enhancer, termed locus control region (LCR)12.
To test potential therapeutic effects of this genome-editing approach, we edited patient-derived CD34+ HSPCs from three SCD patients, induced erythroid differentiation, and examined the RBC progeny. Deoxygenated HbS forms rigid polymers that underlie the characteristic RBC morphology and pathophysiology of SCD, while increased levels of HbF inhibit this process9. To test the effects of genome editing on HbS polymerization, we cultured SCD CD34+ cell-derived RBCs under hypoxia (2% O2). Approximately 30–40% of control and gene-edited cells matured to reticulocytes, late stage anucleate RBC precursors (Supplementary Fig. 3a). Consistent with previous reports, control (mock transduced or Cas9 alone) cultures contained relatively high percentages of F-cells (approximately 65%)13,14, although 25% of the cells within the entire populations exhibited sickle morphology, presumably because HbF was absent in those cells or expressed at insufficient levels to prevent HbS polymerization (Supplementary Fig. 3b). In contrast, Cas9 + gRNA-1 expression increased F-cells to 90% and reduced sickle morphology to 4% (Fig. 1e–f). Thus, targeting the −102 to −114 HPFH region inhibited HbS polymerization in cultured reticulocytes. Residual sickling in Cas9/gRNA-1-expressing RBCs could occur from lack of editing or from mutations that do not induce HbF.
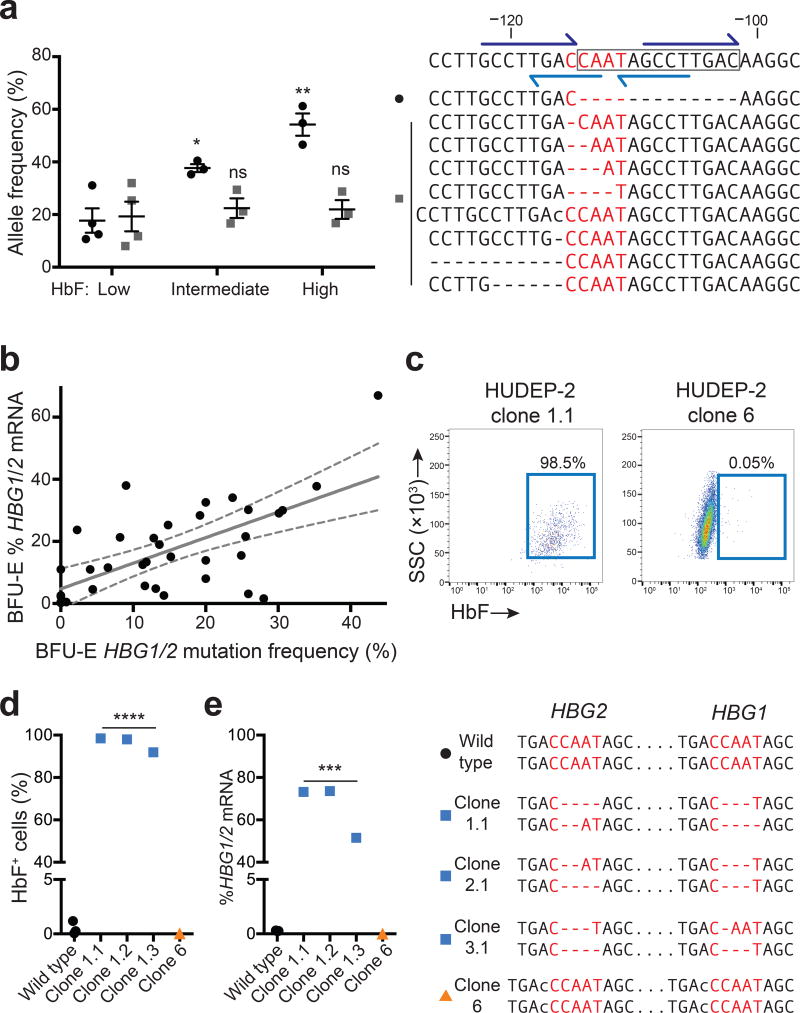
CRISPR/Cas9 creates targeted double-strand DNA breaks that are repaired via error-prone NHEJ, causing nt insertions and deletions (indels). γ-globin is expressed from tandem homologous genes, HBG2 and HBG1 (Supplementary Fig. 4a). While the naturally occurring 13-nt HPFH deletion alters HBG1, HBG2 contains an identical region that is mutated in different HPFH individuals1,15. To characterize the mutations induced by Cas9/gRNA-1-expressing lentivirus in CD34+ HSPCs, we PCR-amplified a 431-nt region surrounding the predicted cleavage sites in HBG2 or HBG1 and deep-sequenced the products. In three biological replicate experiments, the indel incidence in all transduced cells was 56, 65 and 77%, with equal mutation rates in HBG1 and HBG2 (not shown). Approximately half of the mutations were identical to the −102 to −114 HPFH deletion (not shown). Most likely, the 13-nt deletion predominates because the Cas9 cleavage site is flanked by 8-nt tandem repeats that facilitate microhomology-mediated end-joining (Fig. 2a)16. To identify mutations associated with γ-globin induction, we purified low-, intermediate-, and high-HbF erythroblasts and analyzed the promoter regions by deep sequencing (Supplementary Fig. 4b). In four independent experiments, the 13-nt HPFH deletion was enriched in HbF-high cells (P < 0.05) (Fig. 2a and Supplementary Fig. 4c).
Figure 2. Spectrum of γ-globin-inducing mutations caused by Cas9 and gRNA-1.
(a) Normal adult CD34+ cells were edited with Cas9/gRNA-1 lentivirus, differentiated into RBCs, FACS-purified according to HbF immunostaining intensity (low, intermediate or high) and analyzed for on-target mutations. The wild type sequence is shown on the top left with the 13-nt HPFH deletion boxed and the CCAAT box in red. Dark blue half arrows show flanking 8-nt repeats; light blue show the DR element. The top nine mutant alleles (of more than 40 total indels identified) are shown below. Dashes indicate nucleotide deletions and lower-case letters insertions. In the graph at right, black dots denote the 13-nt HPFH deletion, which occurred at the highest frequency; gray squares show the combined frequencies of the eight next common mutations. Each symbol represents an independent experiment. * P < 0.05, ** P < 0.01 by unpaired t-test.
(b) CD34+ cells were electroporated with Cas9/gRNA-1/GFP expression plasmids. GFP+ cells were FACS purified and seeded into methylcellulose. Burst forming unit-erythroid (BFU-E) colonies were analyzed for globin mRNAs and HBG1/HBG2 mutations. All colonies were mosaic for mutations; the total HBG [(HBG1 + HBG2)/2 × 100] mutation frequency for each colony is plotted against %HBG1/2 mRNA [γ / (γ+β)]. Regression analysis shows best-fit line as solid gray (y = 0.82x + 4.8, r2 = 0.41, P < 0.0001, n = 35 colonies from three experiments) and 95% confidence intervals as dashed gray. See also Supplementary Figs. 4a and 5.
(c) HUDEP-2 cells were electroporated with Cas9/gRNA-1/GFP plasmid and cloned. HbF immunostaining is shown for two representative clones with different mutations (see also panel (d) and Supplementary Fig. 6).
(d–e) Characterization of genome edited HUDEP-2 clones. The HBG1 and HBG2 genotypes corresponding to each clone is shown on the right, according to the convention used in panel (a); all clones are homogenous for the indicated mutations. *** P < 0.001, **** P < 0.0001, ns = not significant by unpaired t-test.
Transduction of CD34+ cells with Cas9/gRNA-1 lentivirus produced over 40 different indels smaller than 13 nt (Fig. 2a and Supplementary Fig. 4c), although their low individual frequencies impeded efforts to determine the effects on HbF expression. Thus, we performed clonal studies by expressing gRNA-1/Cas9 in CD34+ cells transiently via DNA electroporation followed by methylcellulose culture to analyze single cell-derived burst-forming unit-erythroid (BFU-E) colonies (Fig. 2b and Supplementary Fig. 5). Of 344 colonies screened from three experiments, 30 had on-target mutations, with 16 containing at least one 13-nt HPFH deletion (Supplementary Fig. 5). All BFU-E colonies analyzed were mosaic for HBG1/2 mutations, reflecting editing over several progenitor divisions. HBG1 and HBG2 were mutated equally. The mutation frequencies correlated moderately with %HBG1/2 mRNA levels in each colony (Fig. 2b) (r2 = 0.41, P < 0.0001). Several small indels within the CCAAT box and/or DR were associated with elevated γ-globin expression in one or more colonies (Supplementary Fig. 5). We also derived HUDEP-2 cell clones after transient Cas9/gRNA-1 expression, including three lines with small (1–4 nt) bi-allelic HBG1/HBG2 CCAAT box mutations and strong γ–globin induction (Fig. 2c–e, Supplementary Fig. 6). HUDEP clone 6 (Fig. 2e) and BFU-E colony 4 (Supplementary Fig. 5) contained single-nt HBG1 and HBG2 insertions that preserved the core CCAAT box element and altered the DR, but was not associated with elevated γ-globin expression.
Simultaneous double-stranded DNA cleavage at gRNA-1 recognition sites in the HBG2 and HBG1 promoters could result in NHEJ-mediated joining of the two ends with loss of the intervening 5.2 kb of genomic DNA (Supplementary Fig. 7a). Indeed, we identified this deletion in several genome edited HUDEP-2 clones (Supplementary Figs. 6–7). We determined the frequency of this event in transiently edited cells using quantitative PCR (qPCR) and fluorescence in situ hybridization (FISH) (Supplementary Fig. 7a). The 5.2-kb deletion was not detected in HUDEP-2 cells after electroporation of 2 µg Cas9/gRNA-1 DNA plasmid, but occurred in about 20% of HBG1/2 alleles after 10 µg DNA transfection (Supplementary Fig. 7b–c). The deletion was not detected by qPCR in human CD34+ HSPCs electroporated with 10 µg Cas9/gRNA-1 plasmid (Supplementary Fig. 7d). Lastly, deep sequencing of CD34+ cells after lentiviral delivery of Cas9/gRNA-1 showed no indels at the top 15 bioinformatically predicted off-target sites (Supplementary Table 1).
The −114 γ-globin promoter CCAAT box and overlapping DR element likely mediate postnatal γ-to-β globin switching by recruiting transcriptional repressors in a developmentally regulated fashion1,15. Candidate CCAAT box binding proteins include COUP-TFII (NR2F2)4, NF-Y (CP-1, CBF)4,5, NF-E36,8, CDP7,8, and C/EBP7; the DR element, binds nuclear hormone receptors TR2 and TR417. Although the molecular triggers of γ-to-β globin switching are not fully defined, the current study shows that disruption of the −114 HBG1/HBG2 CCAAT box/DR region via gene editing partially reverses the switch in adult-type erythroid cells and delineates further the cis elements involved. Importantly, we show that γ-globin induction can occur via small (< 13 nt) NHEJ-associated mutations caused by transient editing and independent of microhomology-mediated end-joining, which is preferentially utilized during S-phase and may not occur efficiently in hematopoietic stem cells16.
Genome editing technologies to manipulate hematopoietic stem cells have fueled innovative strategies for treating β thalassemia and SCD, including correction of the SCD mutation by homology-directed DNA repair18, reactivation of γ-globin via forced promoter-LCR looping19 or disruption of the repressor gene BCL11A via NHEJ20. These approaches are untested in patients. Here we present an additional approach; CRISPR-Cas9-mediated disruption of an HBG1/2 region associated with a benign human condition (HPFH) induces HbF expression to potentially therapeutic levels, similar to those achieved via forced DNA looping or Cas9/gRNA-mediated disruption of a BCL11A erythroid enhancer. Future studies are now required to optimize editing of the γ-globin CCAAT box/DR site in human hematopoietic stem cells and minimize potentially harmful off-target mutations. Overall, our study provides proof-of-principle for a potential approach to treat common b-hemoglobinopathies by genome editing.
Methods
gRNAs and Constructs
gRNA-1: GCTTGTCAAGGCTATTGGTCA
gRNA-2: GTGTCAAGGCTATTGGTCAAG
gRNA sequences were selected using a CRISPR design tool (www.crispr.mit.edu), generated as oligonucleotides, and cloned into plasmids using BbsI. The all-in-one expression plasmid spCas9(BB)-2A-GFP (PX458) was a gift from Feng Zhang (Addgene plasmid #48138). AIO-mCherry vector was cloned by replacing T2A-GFP with IRES-mCherry. The lentiCRISPRv2 was a gift from Feng Zhang (Addgene plasmid #52961). Lentiviral CRISPR-GFP and –mCherry plasmids used in this study were generated by replacing the puromycin resistance gene with IRES-GFP or IRES-mCherry.
Erythroid differentiation of human peripheral blood CD34+ cells
Circulating G-CSF-mobilized human CD34+ cells were obtained from two deidentified healthy donors (Key Biologics, Lifeblood) and enriched by immunomagnetic bead selection using an AutoMACS instrument (Miltenyi Biotec).
CD34+ cells were cultured in a three-phase erythroid differentiation protocol consisting of IMDM (Gibco) supplemented with 2% human AB plasma, 3% human AB serum, 1% penicillin/streptomycin, 3 units/mL heparin, 10 µg/mL insulin, and 3 units/mL erythropoietin (EPO) (Amgen). Phase I (days 1–7) also included 200 µg/mL Holo-Transferrin (Sigma-Aldrich), 10 ng/mL stem cell factor (SCF) (PeproTech, Inc.) and 1 ng/mL IL-3 (PeproTech, Inc.). Phase II (days 8–12) included the same cytokines, except that IL-3 was withdrawn. During phase III (days 13 and beyond) Holo-Transferrin was increased to 1 mg/mL, and SCF was removed. Erythroid differentiation and maturation were monitored by flow cytometry using anti-CD71-PE (BD Biosciences, clone M-A712), anti-CD235-FITC (BD Biosciences, clone GA-R2), anti-Band3-APC (gift from Xiuli An, NY Blood Center), anti-α4-integrin-VioBlue (Miltenyi, clone MZ18-24A9).
HUDEP Cell culture
HUDEP clone 2 (HUDEP-2) cells were cultured as previously described. Cells were expanded in StemSpan SFEM (Stem Cell Technologies) supplemented with 1 µM dexamethasone, 1 µg/mL doxycycline, 50 ng/mL human SCF, 3 units/mL EPO, and 1% penicillin/streptomycin. HUDEP-2 cells were differentiated in the Phase III medium used for CD34+ erythroid cultures. These cells tested negative for mycoplasma.
Generation of genomic deletions
CD34+ cells were transduced by centrifuging with lentivirus (multiplicity of infection 40) at 2,800 rpm, 37°C for 90 min with 8 µg/mL polybrene in Phase I erythroid differentiation medium (CD34+) cells or HUDEP-2 expansion medium (for HUDEP-2 cells). The cells were incubated overnight with virus and were switched to fresh medium the following morning. For transient editing, the Amaxa 2b (Lonza; program U-008) was used to electroporate 1–5 million CD34+ or HUDEP-2 cells with DNA plasmid containing gRNA, Cas9, and GFP (2 or 10 µg).
To generate edited HUDEP-2 clones, cells were electroporated with 10 µg DNA plasmid encoding gRNA-1, Cas9, and GFP. Single GFP+ cells were then sorted into 96-well plates after 24 hours and expanded for 14–21 days. Genomic DNA was isolated using a DNA extraction buffer (100 mM Tris HCl, pH 8.3; 200 mM NaCl; 5 mM EDTA; 1% Triton X-100; 200 µg/mL) proteinase K) followed by incubation at 50°C for 1 hour, then 85°C for 30 minutes.
qRT-PCR
RNA from 0.5–1 million cells was extracted with RNeasy kits (Invitrogen). Reverse transcription reactions were performed with random hexamers using iScript (Bio-Rad). HBG1/2 and HBB mRNAs were quantified by SYBRGreen qPCR, as described16.
HbF protein analysis
Erythroid cells derived from CD34+ cells were fixed with 0.05% glutaraldehyde and permeabilized with 0.1% TritonX-100. Cells were stained with anti-HbF-APC antibody (Invitrogen, HBF-1 clone) and analyzed by flow cytometry. HPLC quantification of HbF was performed using a cation-exchange column (Primus Diagnostics).
In vitro sickling assay
Human SCD CD34+ HSPCs (genotype HbSS) were purified from deidentified, discarded whole blood from partial exchange RBC transfusions (considered “not human subject research” by the St. Jude Children’s Hospital Institutional Review Board), differentiated into erythroblasts and cultured in 2% oxygen levels between days 14 and 17 of culture1. Phase contrast microscopy (CKX41 microscope, Olympus) was performed within 10 minutes of exposure to atmospheric oxygen and analyzed by manual counting of sickled cells, with blinding to sample genotype. The cell permeable nuclear stain Hoechst 33342 (Sigma) was used to quantify nucleated cells by flow cytometry.
Methylcellulose colony assays
Human CD34+ cells were electroporated with GFP, Cas9 and gRNA expression plasmid using an Amaxa Nucleofector 2b (Lonza), program U-008 and cultured for 24 hours. Live GFP+ cells were purified by fluorescent activated cell sorting (FACS) for GFP expression and seeded at 300 cells/ml into cytokine-free human methylcellulose (Stemcell) (300 cells/3 cm dish) supplemented with 2 U/mL EPO, 10 ng/mL SCF, 1 ng/mL IL-3, and 1% penicillin/streptomycin. Individual BFU-E colonies were picked after two weeks of culture.
Deep sequencing of genome modifications
DNA was extracted from cells using Blood and Tissue DNA Extraction Kits (Qiagen). PCR amplification using CloneAmp HiFi Premix was performed with primers including Nextera adapters. An additional PCR was performed to individually index each sample, followed by sequencing on a MiSeq platform (Illumina) with 250bp, paired-end reads. The sequence alignment and mutation detection were performed using CLC Genomics Workbench (CLC Bio). HBG1/2 PCR primers used are as follows:
HBG1-specific Fwd: CGCTGAAACTGTGGTCTTTATGAAAATT
HBG2-specific Fwd: GCACTGAAACTGTTGCTTTATAGGAT
HBG common Rev1 (with HBG-specific Fwd): GGCGTCTGGACTAGGAGCTTATTG
HBG-nonspecific Fwd: ATAACCTCAGACGTTCCAGAAGCGAGTGTG
HBG common Rev2 (with HBG-nonspecific Fwd): AGAAGTCCTGGTATCCTCTATGATGGGAG
qPCR detection of 5.2-kb deletion alleles
A primer and probe set was designed to detect amplification of a HBG1 promoter-specific sequence. TaqMan qPCR was performed on genomic DNA samples from HUDEP-2 and CD34+ cells using Universal TaqMan Mix (Thermo Fisher Scientific) for quantification of triplicates for each sample. ΔΔCt values were calculated based on amplification of RNaseP (Thermo Fisher Scientific) for copy number reference.
5.2kb Fwd: ACGGATAAGTAGATATTGAGGTAAGC
5.2kb Rev: GTCTCTTTCAGTTAGCAGTGG
Taqman probe (FAM): ACTGCGCTGAAACTGTGGTCTTTATGA
Fluorescence in situ hybridization (FISH)
A 5.2-kb probe encompassing the intervening region between gRNA-1 cleavage sites in HBG2 and HBG1 was generated by PCR amplification and cloned using TA vector (Promega). Nick translation was used to label purified DNA with red-dUTP (Alexa-Fluor-594, Molecular Probes), and a control HBB probe (RP11-1205H24) independently with green-dUTP (Alexa-Fluor-488, Molecular Probes). The probes were hybridized simultaneously with interphase and metaphase cells in 50% formamide, 10% dextran sulfate, and 2× SCC. Metaphase cells were stained with DAPI and scored for signals representing the potentially deleted region (red) and HBB (green).
Statistical analyses
Pairwise comparisons were assessed using an unpaired two-tailed Student's t-test. Results were considered significant when P - value < 0.05. Linear regression analysis was performed assess potential correlation between BFU-E colony mutation frequencies and γ-globin ratio. Tests were performed and graphed using Prism software (GraphPad).
Supplementary Material
Acknowledgments
We are grateful to P. Mead, G. Neale, S. Olsen, C. Sherr and Y. Yasui for valuable discussions and technical expertise. DNA sequencing studies were performed at the Hartwell Center, St. Jude Children’s Research Hospital (SJCRH). FISH studies were performed by the SJCRH Cytogenetics Resource. Lentivirus was produced by the SJCRH Hematology Department Vector Core Facility. We thank David Phillips for his support and encouragement. E.A.T. was supported by predoctoral fellowship F30DK102291 (NIDDK). We thank our SCD patients who contributed samples for this study.
Footnotes
Contributions
E.A.T., Y.Y., and M.J.W. designed experiments, analyzed data, and wrote the manuscript. Y.-D.W. performed mutational analyses. K.J.W. performed off-target site analysis. R.K. and Y.N. provided HUDEP-2 cell lines. J.R.H., R.C.H, G.A.B., and C.L. provided conceptual advice and technical expertise. M.J.W. supervised the study. All of the authors discussed the results and assisted in the preparation of the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Forget BG. Molecular basis of hereditary persistence of fetal hemoglobin. Ann. N. Y. Acad. Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 2.Wienert B, et al. Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat Commun. 2015;6:7085. doi: 10.1038/ncomms8085. [DOI] [PubMed] [Google Scholar]
- 3.Gilman JG, et al. Distal CCAAT box deletion in the A gamma globin gene of two black adolescents with elevated fetal A gamma globin. Nucleic Acids Res. 1988;16:10635–10642. doi: 10.1093/nar/16.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liberati C, et al. Cooperation and competition between the binding of COUP-TFII and NF-Y on human epsilon- and gamma-globin gene promoters. J Biol Chem. 2001;276:41700–41709. doi: 10.1074/jbc.M102987200. [DOI] [PubMed] [Google Scholar]
- 5.Zhu X, et al. NF-Y recruits both transcription activator and repressor to modulate tissue- and developmental stage-specific expression of human γ-globin gene. PLoS ONE. 2012;7:e47175. doi: 10.1371/journal.pone.0047175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronchi AE, Bottardi S, Mazzucchelli C, Ottolenghi S, Santoro C. Differential binding of the NFE3 and CP1/NFY transcription factors to the human gamma- and epsilon-globin CCAAT boxes. J Biol Chem. 1995;270:21934–21941. doi: 10.1074/jbc.270.37.21934. [DOI] [PubMed] [Google Scholar]
- 7.Superti-Furga G, Barberis A, Schaffner G, Busslinger M. The-117 mutation in Greek HPFH affects the binding of three nuclear factors to the CCAAT region of the gamma-globin gene. The EMBO journal. 1988;7:3099–3107. doi: 10.1002/j.1460-2075.1988.tb03176.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mantovani R, Superti-Furga G, Gilman J, Ottolenghi S. The deletion of the distal CCAAT box region of the A gamma-globin gene in black HPFH abolishes the binding of the erythroid specific protein NFE3 and of the CCAAT displacement protein. Nucleic Acids Res. 1989;17:6681–6691. doi: 10.1093/nar/17.16.6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoban MD, Orkin SH, Bauer DE. Genetic treatment of a molecular disorder: gene therapy approaches to sickle cell disease. Blood. 2016;127:839–848. doi: 10.1182/blood-2015-09-618587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 11.Kurita R, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS ONE. 2013;8:e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palstra RJ, de Laat W, Grosveld F. Beta-globin regulation and long-range interactions. Adv. Genet. 2008;61:107–142. doi: 10.1016/S0065-2660(07)00004-1. [DOI] [PubMed] [Google Scholar]
- 13.Akinsheye I, et al. Fetal hemoglobin in sickle cell anemia. Blood. 2011;118:19–27. doi: 10.1182/blood-2011-03-325258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidoguchi K, Ogawa M, Karam JD, Martin AG. Augmentation of fetal hemoglobin (HbF) synthesis in culture by human erythropoietic precursors in the marrow and peripheral blood: studies in sickle cell anemia and nonhemoglobinopathic adults. Blood. 1978;52:1115–1124. [PubMed] [Google Scholar]
- 15.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Truong LN, et al. Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proceedings of the National Academy of Sciences. 2013;110:7720–7725. doi: 10.1073/pnas.1213431110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanabe O, et al. An embryonic/fetal beta-type globin gene repressor contains a nuclear receptor TR2/TR4 heterodimer. The EMBO journal. 2002;21:3434–3442. doi: 10.1093/emboj/cdf340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoban MD, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125:2597–2604. doi: 10.1182/blood-2014-12-615948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng W, et al. Reactivation of developmentally silenced globin genes by forced chromatin looping. Cell. 2014;158:849–860. doi: 10.1016/j.cell.2014.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1.de Vasconcellos JF, et al. LIN28A expression reduces sickling of cultured human erythrocytes. PLoS ONE. 2014;9:e106924. doi: 10.1371/journal.pone.0106924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.