Abstract
Objectives
Anti-epidermal growth factor receptor (EGFR) therapy has been found to be more effective against left-sided colorectal cancer (LCRC) than right-sided colorectal cancer (RCRC). We hypothesized that RCRC is more likely to harbor genetic alterations associated with resistance to anti-EGFR therapy and tested this using comprehensive genomic sequencing.
Materials and methods
A total of 201 patients with either primary RCRC or LCRC were analyzed. We investigated tumors for genetic alterations using a 415-gene panel, which included alterations associated with resistance to anti-EGFR therapy: TK receptors (ERBB2, MET, EGFR, FGFR1, and PDGFRA), RAS pathway (KRAS, NRAS, HRAS, BRAF, and MAPK2K1), and PI3K pathway (PTEN and PIK3CA). Patients whose tumors had no alterations in these 12 genes, theoretically considered to respond to anti-EGFR therapy, were defined as “all wild-type”, while remaining patients were defined as “mutant-type”.
Results
Fifty-six patients (28%) and 145 patients (72%) had RCRC and LCRC, respectively. Regarding genetic alterations associated with anti-EGFR therapy, only 6 of 56 patients (11%) with RCRC were “all wild-type” compared with 41 of 145 patients (28%) with LCRC (P = 0.009). Among the 49 patients who received anti-EGFR therapy, RCRC showed significantly worse progression-free survival (PFS) than LCRC (P = 0.022), and “mutant-type” RCRC showed significantly worse PFS compared with “all wild-type” LCRC (P = 0.004).
Conclusions
RCRC is more likely to harbor genetic alterations associated with resistance to anti-EGFR therapy compared with LCRC. Furthermore, our data shows primary tumor sidedness is a surrogate for the non-random distribution of genetic alterations in CRC.
Keywords: colorectal cancer, right-sided, anti-EGFR therapy, next-generation sequencing, comprehensive genomic sequencing
INTRODUCTION
The colon is an embryological derivative of the midgut and hindgut separately, and thus the right-sided colon, the left-sided colon, and the rectum each have different anatomical and physiological features. Evidence shows that tumors arising from right colon have distinct clinical and biological characteristics compared with tumors of the left colon or rectum [1-6]. Right-sided colorectal cancer (RCRC) is generally characterized by being more common in women, and associated with Lynch syndrome, the serrated pathway, Mitogen-activated protein kinase signaling, microsatellite instability-high (MSI-H), deficiency of mismatch repair genes, CpG island methylation, and KRAS and BRAF mutations [7-10]. Left-sided colorectal cancer (LCRC) is more common in men, and associated with familial adenomatous polyposis syndrome, Wnt and EGFR signaling, chromosomal instability, ERBB1 and ERBB2 amplifications, and APC, p53, and NRAS mutations [10, 11]. Based on these molecular differences, sidedness of CRC is thought to be associated with efficacy of chemotherapy and targeted therapy.
The monoclonal antibodies cetuximab and panitumumab are epidermal growth factor receptor (EGFR) inhibitors that block downstream signaling of the EGFR pathway. Randomized phase III clinical trials have shown a survival benefit of these anti-EGFR monoclonal antibodies in RAS wild-type metastatic CRC [12-14]; however, tumor location has not traditionally been included as a stratification criterion in clinical trials. Recently, several retrospective, unplanned analyses examined primary tumor sidedness and revealed that anti-EGFR therapy clearly benefitted patients with LCRC, whereas patients with RCRC derived limited benefit [15-17]. Consequently, while these analyses were limited by low numbers of RCRC patients, the related imbalance between groups, and no randomization; primary tumor sidedness of CRC has emerged as new predictive marker for efficacy of anti-EGFR therapy.
The mechanism of resistance to anti-EGFR therapy in patients with RCRC has not been fully elucidated. Although RAS mutations are established biomarkers of efficacy to anti-EGFR therapy, anti-EGFR therapy is not effective for all patients with a RAS wild-type tumor [18-21]. Genetic alterations in tyrosine kinase (TK) receptors, the RAS pathway (other than KRAS and NRAS mutations), and the PI3K pathway are other possible mechanisms of resistance to anti-EGFR therapy [22, 23]. While the most clinically important alterations, such as KRAS, NRAS, and BRAF mutations, have been widely analyzed among patients with metastatic CRC, the other alterations have not been widely studied.
Next-generation sequencing projects, such as The Cancer Genome Atlas, have profiled genomic changes in many cancers including CRC [24]. We have similarly reported a genomic analysis of Japanese CRC patients using comprehensive genomic sequencing (CGS) [25, 26]. CGS detects gene mutations and copy number alterations in TK receptors, and the RAS and PI3K pathway in a single assay. In the present analysis, we hypothesized that RCRC more frequently harbors genetic alterations associated with resistance to anti-EGFR therapy compared with LCRC. To test this hypothesis, we investigated these genetic alterations using CGS.
RESULTS
Association between primary tumor sidedness and clinicopathological characteristics
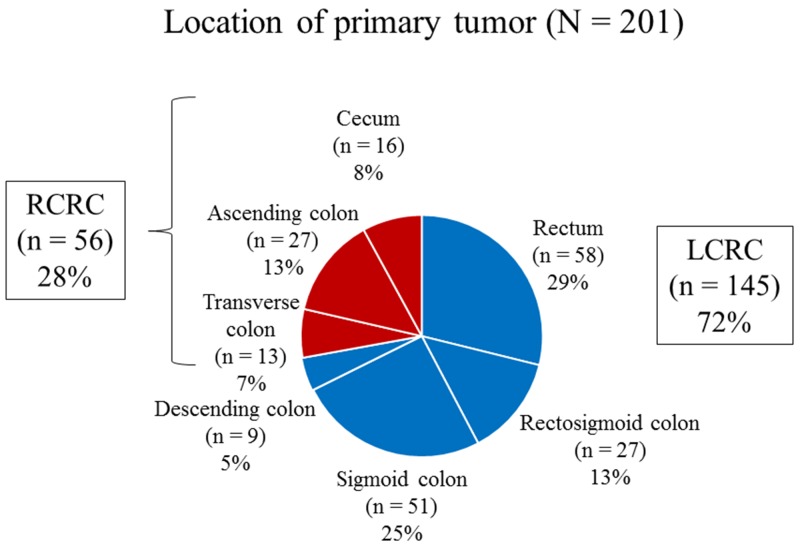
Fifty-six patients (28%) and 145 patients (72%) had RCRC and LCRC, respectively (Figure 1). Histopathological grade 3 was significantly associated with RCRC (P < 0.001; Table 1). Medullary type, mucinous type, and MLH1/MSH2 status were significantly associated with RCRC (P = 0.022, P = 0.007, and P = 0.024, respectively; Table 2).
Figure 1. Primary tumor locations in right-sided colorectal cancer and left-sided colorectal cancer.
RCRC, right-sided colorectal cancer; LCRC, left-sided colorectal cancer.
Table 1. Association between primary tumor sidedness and clinicopathological characteristics (N = 201).
| Primary tumor sidedness | P-value | ||
|---|---|---|---|
| Variable | Right (n = 56) | Left (n = 145) | |
| Age (years) | |||
| < 65 | 22 | 78 | 0.065 |
| ≥ 65 | 34 | 67 | |
| Sex | |||
| Male | 29 | 55 | 0.074 |
| Female | 27 | 90 | |
| Tumor size (mm) | |||
| < 50 | 23 | 65 | 0.630 |
| ≥ 50 | 33 | 80 | |
| T category | |||
| T1, 2 | 6 | 18 | 0.739 |
| T3, 4 | 50 | 127 | |
| Histopathological grading | |||
| G1, 2 | 31 | 116 | < 0.001 |
| G3 | 25 | 29 | |
| Lymphatic invasion | |||
| Absence | 18 | 61 | 0.196 |
| Presence | 38 | 84 | |
| Venous invasion | |||
| Absence | 10 | 38 | 0.213 |
| Presence | 46 | 107 | |
| N category | |||
| N0 | 13 | 46 | 0.235 |
| N1, 2 | 43 | 99 | |
| M category | |||
| M0 | 22 | 68 | 0.331 |
| M1 | 34 | 77 | |
Table 2. Primary tumor sidedness and pathological and genetic characteristics related with deficiency of mismatch repair genes (N = 201).
| Primary tumor sidedness | P-value | ||
|---|---|---|---|
| Variable | Right (n = 56) | Left (n = 145) | |
| Medullary type | |||
| Yes | 52 | 144 | 0.022 |
| No | 4 | 1 | |
| Mucinous type | |||
| Yes | 47 | 139 | 0.007 |
| No | 9 | 6 | |
| Signet ring type | |||
| Yes | 55 | 143 | 0.999 |
| No | 1 | 2 | |
| Tumor infiltrating lymphocytesa | |||
| Yes | 13 | 23 | 0.223 |
| No | 43 | 122 | |
| MLH1/MSH2 status | |||
| Normal | 22 | 69 | 0.013 |
| Abnormal | 10 | 9 | |
| Hypermutated phenotype | |||
| Hypermutated | 10 | 7 | 0.008 |
| Non-hypermutated | 46 | 138 | |
a Cut-off value = 10 lymphocytes/5 high power fields.
Association between primary tumor sidedness and genetic alterations evaluated using CGS
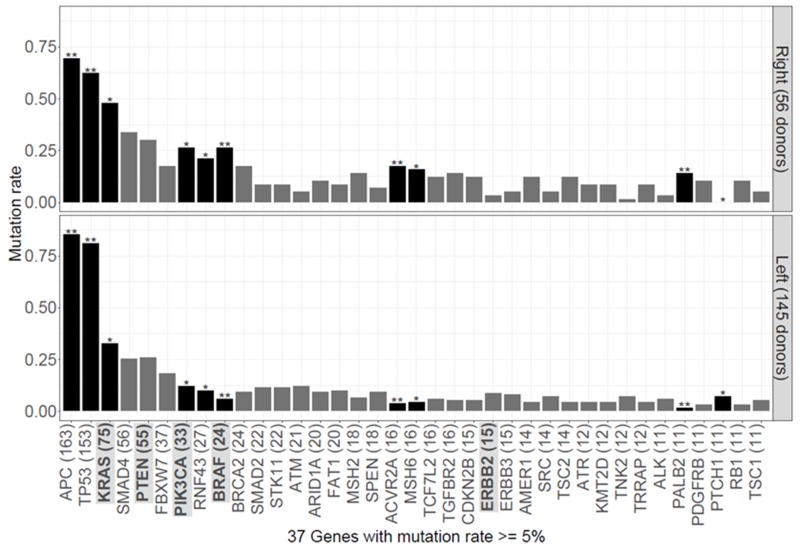
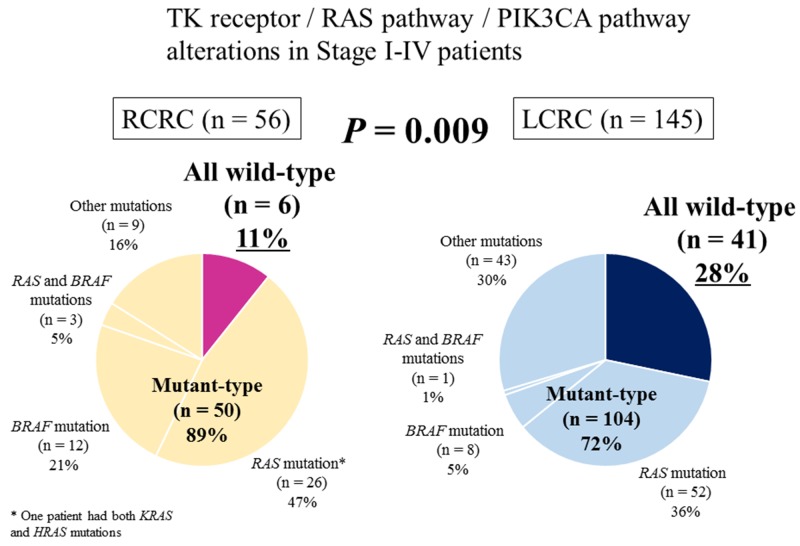
CGS of the 415-gene panel in our cohort of 201 patients detected genetic alterations in 268 genes (Supplementary Table 1). Mutations in KRAS, PIK3CA, RNF43, BRAF, ACVR2A, MSH6, and PALB2 were significantly associated with RCRC (P = 0.047, P = 0.014, P = 0.039, P < 0.001, P = 0.003, P = 0.016, and P = 0.001, respectively; Figure 2), and mutations in APC, TP53 and PTCH1 were significantly associated with LCRC (P = 0.010, P = 0.005, and P = 0.036, respectively; Figure 2). The hypermutated tumor was significantly associated with RCRC (P = 0.008; Table 2). Genetic alterations found in the 12 genes associated with resistance to anti-EGFR therapy (TK receptors: ERBB2, MET, EGFR, FGFR1, and PDGFRA; RAS pathway: KRAS, NRAS, HRAS, BRAF, and MAPK2K1; and PI3K pathway: PTEN and PIK3CA) are detailed in Supplementary Table 2. Of the 201 patients, 154 harbored one or more genetic alterations in these 12 genes with 80, 63, 8, 2, and 1 patients had 1, 2, 3, 4, and 5 gene alterations, respectively. Of the 56 RCRC patients, 6 (11%) were wild-type in all 12 genes (termed “all wild-type”); while 41 of 145 patients (28%) of LCRC were “all wild-type” (P = 0.009; Figure 3, Table 3).
Figure 2. Distribution of genetic alterations in right-sided and left-sided colorectal cancer.
Dark bars indicate genes with a significant difference (P < 0.05, two-tailed Fisher’s exact test or Chi-squared test) in the frequency of genetic alterations compared with other-sided donors. Light bars indicate genes that are not significantly different (*, P < 0.05; **, P < 0.01). The genes associated with anti-EGFR resistance were highlighted.
Figure 3. Percentage of genetic alterations associated with resistance to anti-EGFR therapy in right-sided colorectal cancer and left-sided colorectal cancer.
Genetic alterations in TK receptors (ERBB2, MET, EGFR, FGFR1, and PDGFRA), RAS pathway (KRAS, NRAS, HRAS, BRAF, and MAPK2K1), and PI3K pathway (PTEN and PIK3CA) were evaluated using comprehensive genomic sequencing of the 415-gene panel. Patients who had no alterations in all 12 genes were defined as “all wild-type”. RCRC, right-sided colorectal cancer; LCRC, left-sided colorectal cancer.
Table 3. Association between primary tumor sidedness and gene alterations of TK receptors/RAS pathway/PI3K pathway (N = 201).
| Primary tumor sidedness | P-value | ||
|---|---|---|---|
| Variable | Right (n = 56) | Left (n = 145) | |
| ERBB2 status | |||
| Wild-type | 54 | 132 | 0.243 |
| Mutanta | 2 | 13 | |
| MET status | |||
| Wild-type | 55 | 139 | 0.676 |
| Mutanta | 1 | 6 | |
| EGFR status | |||
| Wild-type | 56 | 141 | 0.578 |
| Mutant | 0 | 4 | |
| FGFR1 status | |||
| Wild-type | 56 | 135 | 0.065 |
| Mutant | 0 | 10 | |
| PDGFRA status | |||
| Wild-type | 55 | 144 | 0.481 |
| Mutant a | 1 | 1 | |
| KRAS status | |||
| Wild-type | 29 | 97 | 0.047 |
| Mutant | 27 | 48 | |
| NRAS status | |||
| Wild-type | 54 | 142 | 0.620 |
| Mutant | 2 | 3 | |
| HRAS status | |||
| Wild-type | 55 | 143 | 0.999 |
| Mutant | 1 | 2 | |
| BRAF status | |||
| Wild-type | 41 | 136 | < 0.001 |
| Mutant | 15 | 9 | |
| MAPK2K1 status | |||
| Wild-type | 54 | 137 | 0.729 |
| Mutant | 2 | 8 | |
| PTEN status | |||
| Wild-type | 39 | 107 | 0.554 |
| Mutant b | 17 | 38 | |
| PIK3CA status | |||
| Wild-type | 41 | 127 | 0.014 |
| Mutant | 15 | 18 | |
| Alterations in TK receptors/RAS pathway/PI3K pathway | |||
| 0 | 6 | 41 | 0.024 |
| 1 | 24 | 56 | |
| 2 or more | 26 | 48 | |
| All wild-type | 6 | 41 | 0.009 |
| Mutant-type | 50 | 104 | |
a Including mutation and amplification.
b Including mutation and deletion
Efficacy of anti-EGFR therapy according to primary tumor sidedness and genetic alterations associated with resistance to anti-EGFR therapy
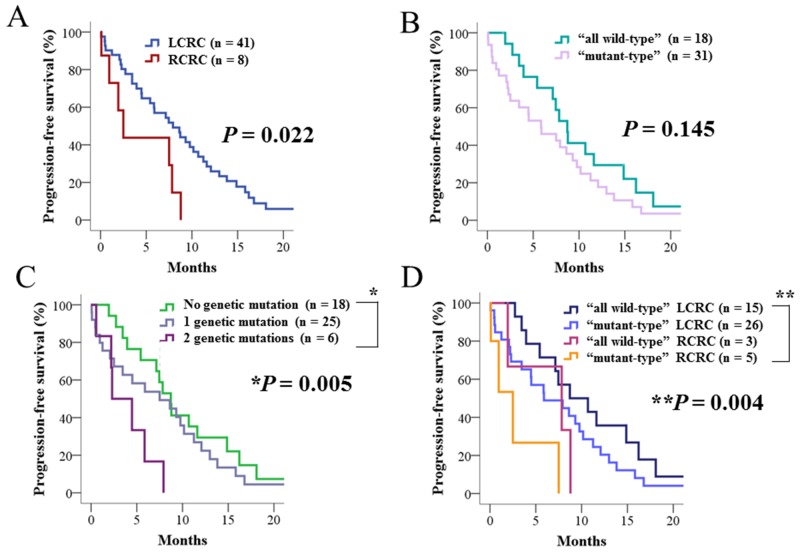
Among the 49 patients treated with anti-EGFR therapy in addition to cytotoxic chemotherapy, patients with RCRC showed significantly worse progression-free survival (PFS) than patients with LCRC (P = 0.022; Figure 4A). Regarding the 12 genes associated with anti-EGFR therapy resistance in these 49 patients, 18, 25, and 6 patients had 0, 1 and 2 genetic alterations, respectively. No significant difference was observed in PFS between “all wild-type” and “mutant-type” (Figure 4B), but the six patients with two genetic alterations showed significantly worse PFS than patients with no genetic mutations (P = 0.005; Figure 4C). After stratification by primary tumor sidedness, we found BRAF mutations were significantly associated with RCRC (P = 0.047; Table 4). When the 49 patients were classified into four groups according to primary tumor sidedness and genetic alterations associated with anti-EGFR therapy resistance, “mutant-type” RCRC showed a significantly worse PFS compared with “all wild-type” LCRC (P = 0.004; Figure 4D).
Figure 4. Progression-free survival of patients who received anti-EGFR therapy in addition to cytotoxic chemotherapy.
(A) Progression-free survival stratified by primary tumor sidedness. (B) Progression-free survival stratified by genetic alterations associated with resistance to anti-EGFR therapy. (C) Progression-free survival stratified by the number of genetic alterations associated with resistance to anti-EGFR therapy. (D) Progression-free survival stratified by primary tumor sidedness and genetic alterations. “All wild-type” indicates patients without any genetic alterations associated with resistance to anti-EGFR therapy, and “mutant-type” indicates those with one or more genetic alterations. RCRC, right-sided colorectal cancer; LCRC, left-sided colorectal cancer.
Table 4. Association between primary tumor sidedness and clinicopathological characteristics in 49 patients with anti-EGFR therapy in addition to cytotoxic chemotherapy.
| Primary tumor sidedness | P-value | ||
|---|---|---|---|
| Variable | Right (n = 8) | Left (n = 41) | |
| Age (years) | |||
| < 65 | 4 | 27 | 0.443 |
| ≥ 65 | 4 | 14 | |
| Sex | |||
| Male | 4 | 27 | 0.443 |
| Female | 4 | 14 | |
| Tumor size (mm) | |||
| < 50 | 5 | 19 | 0.463 |
| ≥ 50 | 3 | 22 | |
| T category | |||
| T2, 3 | 4 | 18 | 0.999 |
| T4 | 4 | 23 | |
| Histopathological grading | |||
| G1, 2 | 3 | 32 | 0.033 |
| G3 | 5 | 9 | |
| Lymphatic invasion | |||
| Absence | 1 | 14 | 0.406 |
| Presence | 7 | 27 | |
| Venous invasion | |||
| Absence | 2 | 6 | 0.601 |
| Presence | 6 | 35 | |
| N category | |||
| N0 | 1 | 5 | 0.999 |
| N1, 2 | 7 | 36 | |
| M category | |||
| M0 | 1 | 1 | 0.421 |
| M1a | 4 | 23 | |
| M1b | 3 | 17 | |
| KRAS status | |||
| Wild-type | 7 | 39 | 0.421 |
| Mutant | 1 | 2 | |
| BRAF status | |||
| Wild-type | 5 | 38 | 0.047 |
| Mutant | 3 | 3 | |
| Alterations in TK receptors/RAS pathway/PI3K pathway excluding KRAS and BRAF mutations | |||
| Absence | 7 | 20 | 0.059 |
| Presence | 1 | 21 | |
| Alterations in TK receptors/RAS pathway/PI3K pathway | |||
| 0 | 3 | 15 | 0.492 |
| 1 | 5 | 20 | |
| 2 | 0 | 6 | |
| All wild-type | 3 | 15 | 0.999 |
| Mutant-type | 5 | 26 | |
| Anti-EGFR drug | |||
| Cetuximab | 2 | 9 | 0.999 |
| Panitumumab | 6 | 32 | |
| Anti-EGFR therapy | |||
| Initial therapy | 3 | 9 | 0.386 |
| Subsequent therapy | 5 | 32 | |
| Chemotherapy added to anti-EGFR therapy | |||
| Oxaliplatin-based | 3 | 7 | 0.195 |
| Irinotecan-based | 3 | 29 | |
| Anti-EGFR drug only | 2 | 5 | |
DISCUSSION
CGS analysis of genetic alterations in 201 primary CRCs revealed important genetic differences in relation to tumor sidedness: that there are genetic alterations in RCRC that are distinct from LCRC, and that CRCs wild-type in TK receptors and the RAS and PI3K pathways (termed “all wild-type” tumors and theoretically more likely to respond to anti-EGFR therapy), were significantly less common amongst RCRC. These genetic differences likely drive the inherent resistance of RCRCs to anti-EGFR therapy.
Primary tumor sidedness of CRC has prognostic importance and relates to response to targeted therapy [15-18]. Recent meta-analyses reported that RCRC was a negative prognostic variable independent of Tumor-Node-Metastasis (TNM) stage [16]. Furthermore, patients with RAS wild-type LCRC had significantly greater survival benefit from anti-EGFR therapy compared with anti-vascular endothelial growth factor (VEGF) therapy; and, conversely, RCRC had poor benefit from standard therapies including anti-EGFR therapy, but was associated with longer survival with anti-VEGF therapy [17, 18]. The National Comprehensive Cancer Network (NCCN) guidelines noted that cetuximab and panitumumab confer little, if any, benefit to patients with metastatic CRC if the primary tumor originated on the right side, and primary tumor sidedness is a surrogate for the non-random distribution of molecular subtypes [18]. However, the molecular background of RCRC and LCRC has not been fully elucidated, and it is still unclear why anti-EGFR therapy is less efficacious in patients with RCRC compared with patients with LCRC. Hence, we investigated genomic differences between RCRC and LCRC using CGS, focusing on identifying the mechanism driving the observed difference in response to anti-EGFR therapy.
CGS has been shown to detect numerous genetic alterations, including driver mutations, in many solid cancers [24, 25]. Mutations in the RAS pathway, such as KRAS, NRAS, and BRAF, are benchmarks to determine treatment strategies for patients with metastatic CRC. The NCCN guidelines state that all patients with metastatic CRC should have tumor tissue genotyped for KRAS, NRAS, and BRAF mutations, and patients with any known KRAS or NRAS mutation should not be treated with anti-EGFR therapy such as cetuximab and panitumumab [18]. In the present analysis, we assumed that genetic alterations in TK receptors, the RAS pathway, or the PI3K pathway are possible mechanisms underlying resistance to anti-EGFR therapy [22, 23]. We successfully detected genetic alterations, not only in the RAS pathway, but also among TK receptors and the PI3K pathway that may be associated with resistance to anti-EGFR therapy. Furthermore, patients with RCRC showed a significantly worse PFS than those with LCRC.
Cancer genome profiling seeks to enable precision medicine, modifying therapies based on the unique genomic changes inherent in the individual tumor of each patient. In the present analysis, we showed an association between tumor sidedness and gene mutations, which may explain the difference in efficacy of anti-EGFR therapy in RCRC compared with LCRC. The genomic background of RCRC as revealed by CGS is consistent with the results of previous meta-analyses [15-17] and the NCCN guidelines regarding the relevance of tumor sidedness [18]. However, we also demonstrated that approximately 10% of RCRC patients had the “all wild-type” phenotype with no mutations detected in TK receptors or the RAS or PI3K pathways, and therefore, theoretically, these patients would be considered as responders to anti-EGFR therapy despite having RCRC. As such, while we showed that RCRC commonly demonstrates a genomic profile associated with resistance to anti-EGFR therapy, we propose future analyses should focus on individual tumors rather than primary tumor sidedness to best facilitate precision medicine.
CGS has ability to detect numerous actionable mutations that can guide new treatment strategies. In this analysis, a novel finding was PALB2 mutations occurring significantly more frequent in RCRC than LCRC. PALB2 is a DNA maintenance gene, where the encoded protein binds to and colocalizes with BRCA2 in nuclear foci, and plays a role of tumor suppression [27]. In CRC, the significance of PALB2 mutations has not been elucidated, and this is the first report regarding PALB2 mutations in relation to CRC sidedness. PALB2 mutations are considered to be actionable, and are a biomarker for response to Poly (ADP-ribose) polymerase (PARP) inhibitors in pancreatic (ClinicalTrials.gov Identifier: NCT03140670) and prostatic (ClinicalTrials.gov Identifier: NCT02952534) cancers. In this analysis, we found 8 of 56 (14%) RCRCs had PALB2 mutations compared with 3 of 145 (2%) LCRCs. Thus, targeting PALB2 may represent a future treatment strategy for RCRC.
To the best of our knowledge, this is the first report describing a genomic overview of RCRC and LCRC using CGS. However, this analysis has several limitations. First, it was a retrospective analysis performed at two institutions and included a relatively small number of patients. Second, the selection of genomic biomarkers of resistance outside of RAS is not yet well supported by prospective studies. Third, as patients who received anti-EGFR therapy were analyzed retrospectively, we could not definitively associate primary tumor sidedness with response to anti-EGFR therapy. Fourth, as the number of RCRC patients who received anti-EGFR therapy was small, we need for increasing the number of RCRC patients in future. However, we did demonstrate that RCRC was significantly associated with genetic alterations associated with resistance to anti-EGFR therapy, which provides a plausible mechanism of resistance to anti-EGFR therapy in patients with RCRC.
In conclusion, we show RCRC is more likely to harbor genetic alterations associated with resistance to anti-EGFR therapy compared with LCRC, and primary tumor sidedness is a surrogate for a non-random distribution of genetic alterations in CRC.
MATERIALS AND METHODS
Patients
This retrospective analysis was performed in accordance with the Helsinki Declaration and the protocol was approved by the Ethics Committee of the School of Medicine, Niigata University. We randomly selected and enrolled 201 patients diagnosed with stage I - IV CRC based on the 7th edition of the American Joint Committee on Cancer staging manual [28] who had a primary tumor resection between 2009 and 2015 at Niigata University Medical and Dental Hospital or Niigata Cancer Center Hospital. In this analysis, we included the 201 independent individuals, all unrelated, confirmed from our database and medical charts. Patients with familial adenomatous polyposis or inflammatory bowel disease were excluded.
Primary tumor sidedness and clinicopathological characteristics
Primary tumor location was determined by operative findings. Cancer in the cecum, ascending colon, hepatic flexure, or transverse colon was classified as right-sided; and cancer in the splenic flexure, descending colon, sigmoid colon, rectosigmoid, or rectum was classified as left-sided [29, 30]. Histopathological features associated with RCRC, such as medullary type, mucinous type, signet ring type, and tumor-infiltrating lymphocytes were analyzed by a previously reported method [31]. MutL homologue 1 (MLH1)/MutS homologue 2 (MSH2) status was evaluated in 110 of the 201 patients by immunohistochemistry with anti-MLH1 (1:50; BD Biosciences PharMingen, San Diego, CA) and anti-MSH2 (1:50; Leica Microbiosystems, Tokyo, Japan) antibodies. Hypermutation was defined as a tumor with MSI-H and/or high tumor mutation burden (TMB), as described previously [25], using CGS. TMB was calculated as the number of non-synonymous mutations per megabase of sequence in the panel (panel size = 1.3 Mb). To be classified as hypermutated, the threshold of TMB was set as the lowest TMB observed in tumors with MSI-H. Tumors with mutations in POLE or other DNA repair genes can have very high TMB but not show MSI-H [25].
CGS analysis of primary tumors
Archival tissue in the form of formalin-fixed, paraffin-embedded (FFPE) tumor or unstained tissue sections obtained during primary tumor resection were used for CGS. An independent pathologist evaluated tumor content in each sample using hematoxylin and eosin-stained slides to ensure > 50% tumor content was present. Where applicable, unstained slides were macro-dissected to enrich for tumor content and DNA was extracted using a BioStic FFPE Tissue DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA). All sample preparation, CGS, and analytics were performed in a CLIA/CAP-accredited laboratory (KEW Inc., Cambridge, MA). DNA fragment (50–150 ng) libraries were prepared and enriched for the CancerPlex 415-gene panel (KEW Inc.) [25, 26], a large clinically validated panel of 415 genes enriched for coding regions and selected introns of known cancer-related genes. Sequencing was performed on Illumina MiSeq and NextSeq platforms with an average 500× sequencing depth. Genomic data were then processed through a proprietary bioinformatics platform and knowledgebase to identify multiple classes of genomic abnormalities including single nucleotide substitutions, small insertions/deletions, copy number variations, and translocations. Single nucleotide variant (SNV) and insertion or deletion (indel) calling were only performed in genomic regions intended to be captured by the assay (region of interest). We set a standard threshold of 10% allelic fraction for calling SNVs and indels to focus on primary truncal driver mutations and avoid subclonal events. Copy number variants were called for exons as well as globally. We segmented regions using a Fused-Lasso method and export the results to a VCF file. The threshold for gain was > 2.5 fold and for loss was < 0.5 fold. Variants were filtered or flagged according to technical quality (e.g. coverage, allelic fraction, number of supporting reads), presence in previously characterized normal samples, or presence/absence in the following databases: dbSNP, ExAC, COSMIC, ClinVar, and KEW. SNVs and indels in VCF format were annotated using SnpEff and the output was adapted according to HGVS recommendations [25, 26].
Genetic alterations in TK receptors and the RAS and PI3K pathways in RCRC and LCRC
Genetic alterations of TK receptors (ERBB2, MET, EGFR, FGFR1, and PDGFRA), RAS pathway (KRAS, NRAS, HRAS, BRAF, and MAPK2K1), and PI3K pathway (PTEN and PIK3CA) were analyzed using CGS of the 415-gene panel. We defined patients who had no alterations in all 12 genes as “all wild-type”; theoretically, these patients should respond to anti-EGFR therapy [22, 23]. We defined the remaining patients with genetic alterations as “mutant-type”. We also estimated the incidence of “all wild-type” for RCRC and LCRC. In this analysis of 201 patients, 49 received anti-EGFR therapy. In these 49 patients, we investigated the efficacy of anti-EGFR therapy according to primary tumor sidedness and genetic alterations associated with resistance to anti-EGFR therapy.
Statistical analysis
Statistical analyses were performed with IBM SPSS Statistics 22 (IBM Japan, Inc., Tokyo, Japan). A Fisher’s exact test or Chi-squared test was used to evaluate associations between primary tumor sidedness and clinicopathological characteristics, and primary tumor sidedness and genetic alterations were evaluated with CGS. The association between primary tumor sidedness and genetic alterations associated with resistance to anti-EGFR therapy was examined by Fisher’s exact test or Chi-squared test. PFS rates in patients treated with anti-EGFR therapy (cetuximab or panitumumab) in addition to cytotoxic chemotherapy were estimated using Kaplan-Meier analysis. A log-rank test was used to assess for a significant difference between right-sided and left-sided tumors. P-values < 0.05 were considered statistically significant.
SUPPLEMENTARY MATERIAL TABLES
Author contributions
Yoshifumi Shimada: Study conception, acquisition of data, analysis and interpretation of data, and drafting of manuscript. Hitoshi Kameyama: Acquisition and interpretation of data. Masayuki Nagahashi: Acquisition and interpretation of data. Hiroshi Ichikawa: Acquisition and interpretation of data. Yusuke Muneoka: Acquisition and interpretation of data. Ryoma Yagi: Acquisition and interpretation of data. Yosuke Tajima: Acquisition and interpretation of data. Takuma Okamura: Acquisition and interpretation of data. Masato Nakano: Acquisition and interpretation of data. Jun Sakata: Acquisition and interpretation of data. Takashi Kobayashi: Acquisition and interpretation of data. Hitoshi Nogami: Acquisition of data and critical revision of manuscript. Satoshi Maruyama: Acquisition of data and critical revision of manuscript. Yasumasa Takii: Acquisition of data and critical revision of manuscript. Tetsu Hayashida: Interpretation of data and critical revision of manuscript. Hiromasa Takaishi: Interpretation of data and critical revision of manuscript. Yuko Kitagawa: Interpretation of data and critical revision of manuscript. Eiji Oki: Interpretation of data and critical revision of manuscript. Tsuyoshi Konishi: Interpretation of data and critical revision of manuscript. Fumio Ishida: Interpretation of data and critical revision of manuscript. Shin-ei Kudo: Interpretation of data and critical revision of manuscript. Jennifer E. Ring: Acquisition of data and critical revision of manuscript. Alexei Protopopov: Acquisition of data and critical revision of manuscript. Stephen Lyle: Acquisition of data and critical revision of manuscript. Yiwei Ling: Analysis of data and critical revision of manuscript. Shujiro Okuda: Analysis of data and critical revision of manuscript. Takashi Ishikawa: Analysis of data and critical revision of manuscript. Kohei Akazawa: Analysis of data and critical revision of manuscript. Kazuaki Takabe: Interpretation of data and critical revision of manuscript. Toshifumi Wakai: Study conception, acquisition of data, analysis and interpretation of data, critical revision of manuscript. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST
Jennifer E. Ring, Alexei Protopopov, and Stephen Lyle are employees of KEW Inc., who have been granted stock options by KEW Inc. The remaining authors declare that they have no competing interests.
FUNDING
This project was supported by Denka Co., Ltd. Tokyo, Japan and in part by JSPS KAKENHI Grant Number JP15K10130.
REFERENCES
- 1.Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, Maus MK, Antoniotti C, Langer C, Scherer SJ, Müller T, Hurwitz HI, Saltz L, et al. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst. 2015;107 doi: 10.1093/jnci/dju427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss JM, Pfau PR, O’Connor ES, King J, LoConte N, Kennedy G, Smith MA. Mortality by stage for right- versus left-sided colon cancer: analysis of surveillance, epidemiology, and end results--Medicare data. J Clin Oncol. 2011;29:4401–4409. doi: 10.1200/JCO.2011.36.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishihara S, Watanabe T, Akahane T, Shimada R, Horiuchi A, Shibuya H, Hayama T, Yamada H, Nozawa K, Matsuda K, Maeda K, Sugihara K. Tumor location is a prognostic factor in poorly differentiated adenocarcinoma, mucinous adenocarcinoma, and signet-ring cell carcinoma of the colon. Int J Colorectal Dis. 2012;27:371–379. doi: 10.1007/s00384-011-1343-0. [DOI] [PubMed] [Google Scholar]
- 4.Lee MS, McGuffey EJ, Morris JS, Manyam G, Baladandayuthapani V, Wei W, Morris VK, Overman MJ, Maru DM, Jiang ZQ, Hamilton SR, Kopetz S. Association of CpG island methylator phenotype and EREG/AREG methylation and expression in colorectal cancer. Br J Cancer. 2016;114:1352–1361. doi: 10.1038/bjc.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Powell AG, Wallace R, McKee RF, Anderson JH, Going JJ, Edwards J, Horgan PG. The relationship between tumour site, clinicopathological characteristics and cancer-specific survival in patients undergoing surgery for colorectal cancer. Colorectal Dis. 2012;14:1493–1499. doi: 10.1111/j.1463-1318.2012.03048.x. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi Y, Sugai T, Habano W, Ishida K, Eizuka M, Otsuka K, Sasaki A, Takayuki Matsumoto, Morikawa T, Unno M, Suzuki H. Molecular differences in the microsatellite stable phenotype between left-sided and right-sided colorectal cancer. Int J Cancer. 2016;139:2493–2501. doi: 10.1002/ijc.30377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, Agarwal A, Maru DM, Sieber O, Desai J. Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer. 2011;117:4623–4632. doi: 10.1002/cncr.26086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iacopetta B. Are there two sides to colorectal cancer? Int J Cancer. 2002;101:403–408. doi: 10.1002/ijc.10635. [DOI] [PubMed] [Google Scholar]
- 9.Hutchins G, Southward K, Handley K, Magill L, Beaumont C, Stahlschmidt J, Richman S, Chambers P, Seymour M, Kerr D, Gray R, Quirke P. Value of mismatch repair, KRAS, and BRAF mutations in predicting recurrence and benefits from chemotherapy in colorectal cancer. J Clin Oncol. 2011;29:1261–1270. doi: 10.1200/JCO.2010.30.1366. [DOI] [PubMed] [Google Scholar]
- 10.Shen H, Yang J, Huang Q, Jiang MJ, Tan YN, Fu JF, Zhu LZ, Fang XF, Yuan Y. Different treatment strategies and molecular features between right-sided and left-sided colon cancers. World J Gastroenterol. 2015;21:6470–6478. doi: 10.3748/wjg.v21.i21.6470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breivik J, Lothe RA, Meling GI, Rognum TO, Børresen-Dale AL, Gaudernack G. Different genetic pathways to proximal and distal colorectal cancer influenced by sex-related factors. Int J Cancer. 1997;74:664–669. doi: 10.1002/(sici)1097-0215(19971219)74:6<664::aid-ijc18>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 12.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 13.Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 2010;28:4697–4705. doi: 10.1200/JCO.2009.27.4860. [DOI] [PubMed] [Google Scholar]
- 14.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocákova I, Ruff P, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 15.Holch JW, Ricard I, Stintzing S, Modest DP, Heinemann V. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. doi: 10.1016/j.ejca.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 16.Petrelli F, Tomasello G, Borgonovo K, Ghidini M, Turati L, Dallera P, Passalacqua R, Sgroi G, Barni S. Prognostic Survival Associated With Left-Sided vs Right-Sided Colon Cancer: A Systematic Review and Meta-analysis. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.4227. Oct 27. https://doi.org/10.1001/jamaoncol.2016.4227. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Tejpar S, Stintzing S, Ciardiello F, Tabernero J, Van Cutsem E, Beier F, Esser R, Lenz HJ, Heinemann V. Prognostic and Predictive Relevance of Primary Tumor Location in Patients With RAS Wild-Type Metastatic Colorectal Cancer: Retrospective Analyses of the CRYSTAL and FIRE-3 Trials. JAMA Oncol. 2016 doi: 10.1001/jamaoncol.2016.3797. Oct 10. https://doi.org/10.1001/jamaoncol.2016.3797. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Comprehensive Cancer Network NCCN clinical practice guidelines in oncology-rectal cancer (version 2. 2017) doi: 10.6004/jnccn.2017.0146. Available: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf[accessed April 3, 2017] [DOI] [PMC free article] [PubMed]
- 19.Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, Aranda Aguilar E, Bardelli A, Benson A, Bodoky G, Ciardiello F, D’Hoore A, Diaz-Rubio E, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016;27:1386–1422. doi: 10.1093/annonc/mdw235. [DOI] [PubMed] [Google Scholar]
- 20.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 21.Saltz LB, Meropol NJ, Loehrer PJ, Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 22.Bertotti A, Papp E, Jones S, Adleff V, Anagnostou V, Lupo B, Sausen M, Phallen J, Hruban CA, Tokheim C, Niknafs N, Nesselbush M, Lytle K, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526:263–267. doi: 10.1038/nature14969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertotti A, Migliardi G, Galimi F, Sassi F, Torti D, Isella C, Corà D, Di Nicolantonio F, Buscarino M, Petti C, Ribero D, Russolillo N, Muratore A, et al. A molecularly annotated platform of patient-derived xenografts (“xenopatients”) identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov. 2011;1:508–523. doi: 10.1158/2159-8290.CD-11-0109. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagahashi M, Wakai T, Shimada Y, Ichikawa H, Kameyama H, Kobayashi T, Sakata J, Yagi R, Sato N, Kitagawa Y, Uetake H, Yoshida K, Oki E, et al. Genomic landscape of colorectal cancer in Japan: clinical implications of comprehensive genomic sequencing for precision medicine. Genome Med. 2016;8:136. doi: 10.1186/s13073-016-0387-8. https://doi.org/10.1186/s13073-016-0387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimada Y, Yagi R, Kameyama H, Nagahashi M, Ichikawa H, Tajima Y, Okamura T, Nakano M, Nakano M, Sato Y, Matsuzawa T, Sakata J, Kobayashi T, et al. Utility of comprehensive genomic sequencing for detecting HER2-positive colorectal cancer. Hum Pathol. 2017 doi: 10.1016/j.humpath.2017.02.004. Feb 21. https://doi.org/10.1016/10.1016/j.humpath.2017.02.004. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, Quinn MC, Robertson AJ, Fadlullah MZ, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature. 2015;518:495–501. doi: 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, editors. AJCC Cancer Staging Manual. 7th edition. Springer; New York: 2010. [Google Scholar]
- 29.Lee GH, Malietzis G, Askari A, Bernardo D, Al-Hassi HO, Clark SK. Is right-sided colon cancer different to left-sided colorectal cancer? — a systematic review. Eur J Surg Oncol. 2015;41:300–308. doi: 10.1016/j.ejso.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 30.Li FY, Lai MD. Colorectal cancer, one entity or three. J Zhejiang Univ Sci B. 2009;10:219–229. doi: 10.1631/jzus.B0820273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumours of the Digestive System. 4th edition. IARC; Lyon: 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.