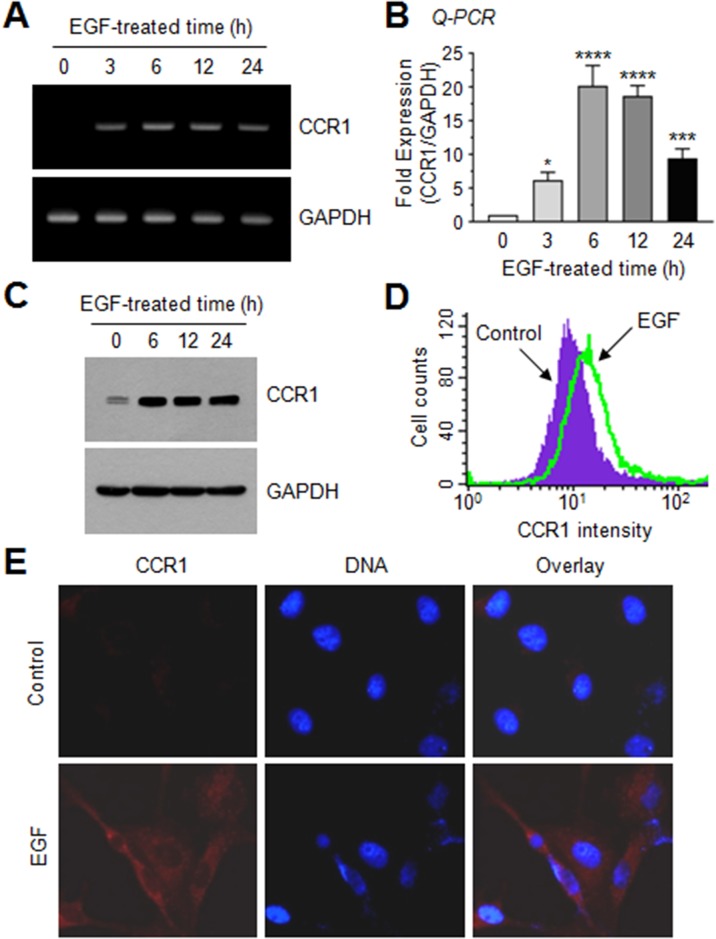
Figure 3. EGF enhances CCR1 expression in MDA-MB-231 breast cancer cells.
(A) MDA-MB-231 cells were serum-depleted overnight and treated with EGF (50 ng/mL) for 0–24 h, and total RNA was extracted. CCR1 mRNA levels were examined by RT-PCR. GAPDH was used as an internal control. (B) Serum-starved MDA-MB-231 cells were treated as in A, and CCR1 mRNA levels were assessed by quantitative real-time PCR (Q-PCR). Values were normalized to GAPDH mRNA levels. Data are mean ± SD (n = 3). *, P = 0.0247; ***, P < 0.0009; ****, P < 0.0001 (compared to untreated control; by Sidak’s multiple comparisons test). (C) Serum-starved MDA-MB-231 cells were treated as in A, and CCR1 protein levels were assessed by immunoblotting. GAPDH was used as an internal control. (D) Flow cytometry. After serum-starved MDA-MB-231 cells were treated with EGF (50 ng/mL) for 12 h, CCR1 protein expression on the cell surface was measured using flow cytometry. (E) MDA-MB-231 cells were treated with EGF (50 ng/mL) for 12 h, and then incubated with an antibody against CCR1, and a AlexaFluor 555-conjugated (red signal) secondary antibody for 30 min. Nuclear DNA was stained with 0.1 μg/mL Hoechst 33258 for 10 min (blue signal). Fluorescence-positive cells were examined under an EVOSf1® fluorescence microscope.