The Rab GTPase Ypt1 recruits the Golgi Arf-GEFs Gea1, Gea2, and Sec7 to distinct parts of the Golgi. Early Golgi localized Gea1 and Gea2 prefer neutral membranes, whereas late Golgi localized Sec7 prefers anionic membranes. The C-terminal domains of Gea1 and Gea2 regulate their localization and activity and are required for recruitment by Ypt1.
Abstract
At the Golgi complex, the biosynthetic sorting center of the cell, the Arf GTPases are responsible for coordinating vesicle formation. The Arf-GEFs activate Arf GTPases and are therefore the key molecular decision-makers for trafficking from the Golgi. In Saccharomyces cerevisiae, three conserved Arf-GEFs function at the Golgi: Sec7, Gea1, and Gea2. Our group has described the regulation of Sec7, the trans-Golgi Arf-GEF, through autoinhibition, positive feedback, dimerization, and interactions with a suite of small GTPases. However, we lack a clear understanding of the regulation of the early Golgi Arf-GEFs Gea1 and Gea2. Here we demonstrate that Gea1 and Gea2 prefer neutral over anionic membrane surfaces in vitro, consistent with their localization to the early Golgi. We illustrate a requirement for a critical mass of either Gea1 or Gea2 for cell growth under stress conditions. We show that the C-terminal domains of Gea1 and Gea2 toggle roles in the cytosol and at the membrane surface, preventing membrane binding in the absence of a recruiting interaction but promoting maximum catalytic activity once recruited. We also identify the small GTPase Ypt1 as a recruiter for Gea1 and Gea2. Our findings illuminate core regulatory mechanisms unique to the early Golgi Arf-GEFs.
INTRODUCTION
Intracellular membrane trafficking is an essential and intricately coordinated process in eukaryotes. Membrane-bound vesicles transport synthesized proteins and lipids to compartments where modifications occur, deliver them to their final destinations, and shuttle them between organelles and the plasma membrane as needed. The Golgi complex is the central sorting compartment for intracellular membrane trafficking, and vesicular traffic out of the Golgi is both tightly regulated and highly conserved to ensure that cargo only leaves the Golgi at the appropriate place and time.
A key regulator of vesicle formation throughout the Golgi is the small GTPase Arf1 and its paralogs (Stearns et al., 1990; Donaldson and Honda, 2005). As with other small GTPases, Arf1 functions as a molecular switch. When GDP-bound, it is inactive and cytoplasmic. On GTP binding, Arf1 inserts its myristoylated N-terminal amphipathic helix into the membrane and changes conformation to recruit cargoes, cargo adapters, and coat proteins to generate a vesicle (Antonny et al., 1997; Goldberg, 1998). Thus, the decision to switch Arf1 “on” through nucleotide exchange is a pivotal regulatory event in Golgi membrane trafficking.
Nucleotide exchange on Arf1 is carried out by the Sec7 family of guanine nucleotide exchange factors (Arf-GEFs) (Jackson and Casanova, 2000; Casanova, 2007; Gillingham and Munro, 2007). Two highly conserved subfamilies of Arf-GEFs function at the Golgi complex: Gea/GBF and Sec7/BIG in Saccharomyces cerevisiae/humans, respectively (Achstetter et al., 1988; Peyroche et al., 1996). Outside the highly conserved catalytic GEF domain, these Golgi Arf-GEFs differ greatly from the rest of the Sec7 family to which they belong (Mouratou et al., 2005; Bui et al., 2009). They are considerably larger and share no sequence homology with any known domains in other proteins, including canonical membrane-targeting domains, outside the GEF domain. Yet understanding how these Golgi Arf-GEFs are regulated and recruited to the correct membrane surface is essential to understanding the regulation of Arf1 activation and subsequent vesicle formation.
The Sec7/BIG subfamily of Golgi Arf-GEFs functions at the trans-Golgi network (TGN), activating Arf1 to form secretory vesicles and vesicles that traffic to endosomes and lysosomes. Our group has shown that Sec7 is regulated through strong autoinhibition, a positive feedback loop with Arf1, interactions with several other small GTPases, and dimerization through its C-terminal HDS4 domain (Richardson et al., 2012, 2016; McDonold and Fromme, 2014). Sec7, Gea1, and Gea2 share predicted domain architecture (Bui et al., 2009), which suggests shared regulation. However, Gea1 and Gea2, which function in intra-Golgi and Golgi-ER retrograde traffic, lack similar positive feedback and dimerization regulatory mechanisms. Evidence of protein interactors has been presented by other groups (Chantalat et al., 2003, 2004; Monetta et al., 2007; Deng et al., 2009; Christis and Munro, 2012; Tsai et al., 2013), but to date no clear picture of the regulation of Gea/GBF has been established.
Here we present evidence for a model in which the C-terminal domains of Gea1 and Gea2 serve both inhibitory and stimulatory functions in regulation. We also show that the nature of the membrane surface is important for function of Gea1 and Gea2 and that the Rab GTPase Ypt1 recruits Gea1 and Gea2 to membranes in a manner dependent on the C-terminus of Gea2. Our findings define several important mechanistic differences between the Sec7/BIG and Gea/GBF1 families and indicate that Arf1-dependent trafficking can be regulated independently at early versus late Golgi compartments.
RESULTS
Gea1 and Gea2 localize differently relative to early and late Golgi markers
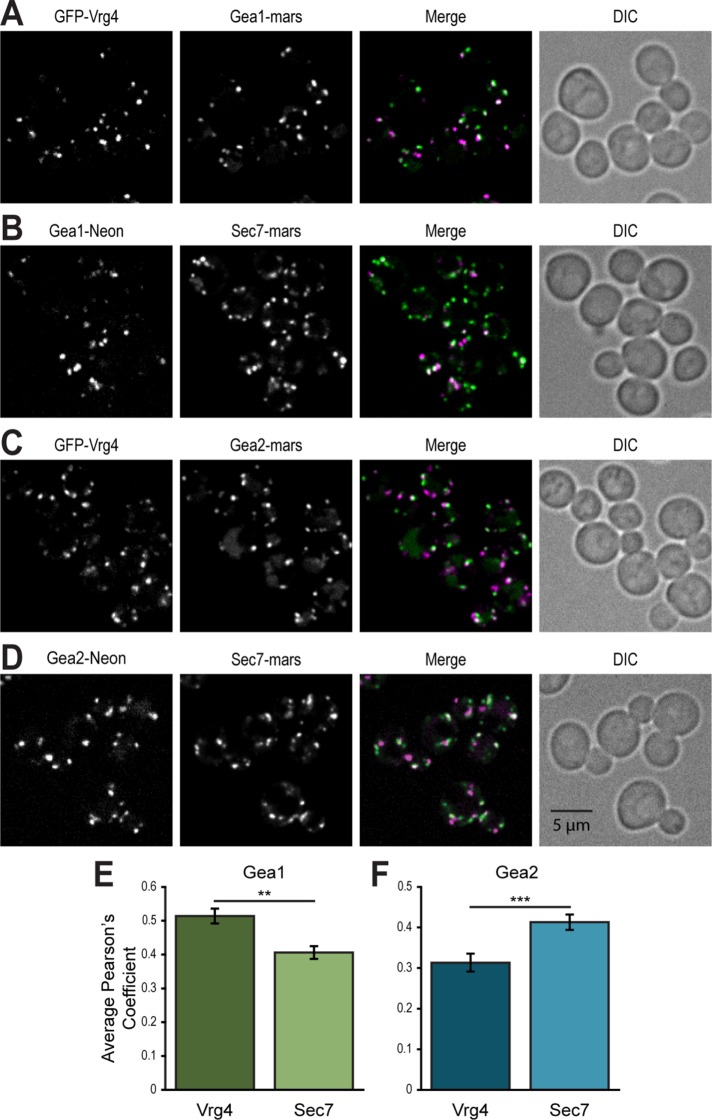
Sec7 is well established as localizing to the TGN, where it activates Arf1 to initiate formation of secretory vesicles and vesicles that traffic to endosomes and the lysosome/vacuole. In contrast, Gea1 and Gea2 have been shown to function in COPI vesicle-mediated intra-Golgi and Golgi-ER traffic and to fractionate with early Golgi markers (Spang et al., 2001; Deng et al., 2009). To confirm the distribution of large Arf-GEFs at the Golgi in live cells, we generated strains that coexpressed endogenously-tagged Gea1 or Gea2 with either the early Golgi marker Vrg4 or with Sec7 as a marker for the TGN (Losev et al., 2006; Matsuura-Tokita et al., 2006). Gea1 shows greater colocalization with Vrg4-labeled Golgi compartments (Figure 1A) than with Sec7 compartments (Figure 1, B and E), while Gea2 colocalizes more with Sec7 (Figure 1C) than Vrg4 (Figure 1, D and F). Neither Gea1 nor Gea2 shows perfect colocalization or anticorrelation with either marker, suggesting that the Gea Arf-GEFs occupy intermediate or hybrid compartments and reflecting the dynamic nature of the Golgi. These data indicate that Gea1 occupies earlier Golgi compartments than Gea2 (Figure 2A), hinting at differences in their roles and regulatory mechanisms.
FIGURE 1:
Gea1 and Gea2 localize differently relative to early and late Golgi markers. Subcellular localization (A) of GFP-Vrg4, an early Golgi marker, and Gea1-3xmRFPmars and (B) of Gea1-mNeonGreen and mRFPmars-Sec7, a late Golgi marker. Subcellular localization (C) of GFP-Vrg4 and Gea2-3xmRFPmars and (D) of Gea2-mNeonGreen and mRFPmars-Sec7. Quantification of colocalization of Gea1 (E) or Gea2 (F) with Vrg4 or Sec7 at puncta. Error bars represent 95% CIs for n = 76 (Gea1 vs. Vrg4), n = 84 (Gea1 vs. Sec7) cells, n = 59 (Gea2 vs. Vrg4), or n = 58 (Gea2 vs. Sec7) cells. In all Merge panels, the GFP channel is shown in green, the RFP channel in magenta, and areas of overlap in white. Differential interference contrast (DIC) panels show cells in the field. **p < 0.01; ***p < 0.0001.
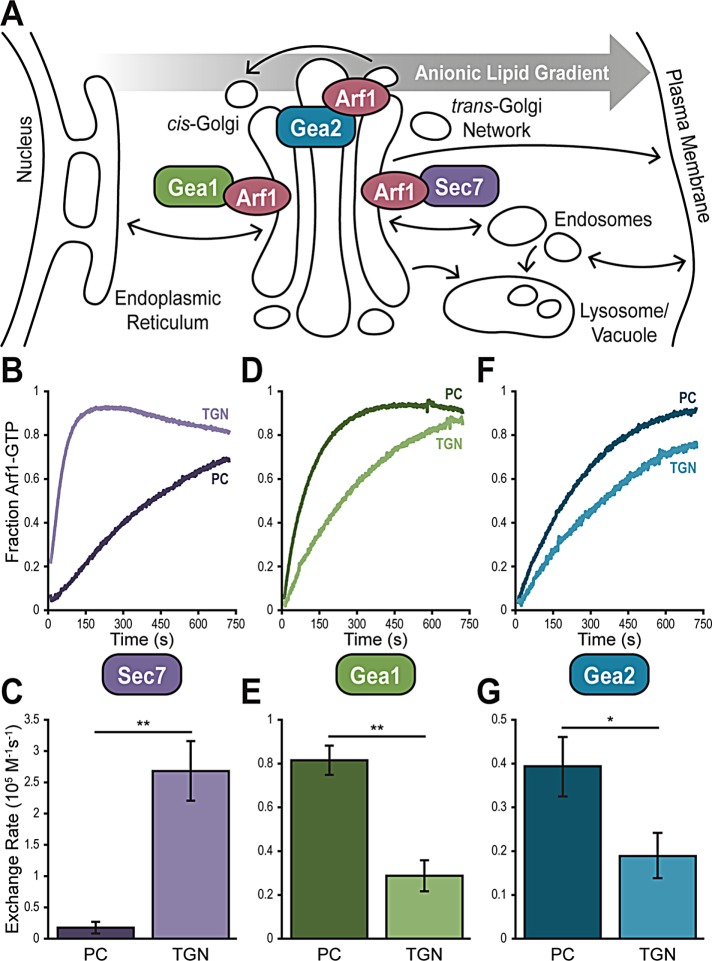
FIGURE 2:
The in vitro membrane preferences of Gea1, Gea2, and Sec7 correspond to their sub-Golgi localization in vivo. (A) Cartoon of the secretory pathway, showing relative sub-Golgi localization of Gea1, Gea2, and Sec7 as well as the gradient of anionic lipids on the cytosolic membrane surface from the ER to the plasma membrane. Representative normalized traces showing activation of Arf1 by Sec7f (B), Gea1 (D), and Gea2 (F) on synthetic PC and TGN liposomes. Rates of Arf1 activation determined from full sets of traces for Sec7f (C), Gea1 (E), and Gea2 (G). Error bars represent 95% CIs for n = 3 reactions. *p < 0.05; **p < 0.01.
The in vitro membrane preferences of Gea1, Gea2, and Sec7 correspond to their sub-Golgi localization in vivo
One feature that distinguishes the early Golgi from the TGN is the net charge of the cytosolic membrane surface. There is a well-established gradient of anionic phosphatidylserine (PS) across the secretory pathway, with very little PS exposed to the cytosol at the endoplasmic reticulum (ER) and as much as 10% or more PS in the cytosolic lipid content of the plasma membrane (Figure 2A) (van Meer et al., 2008; Leventis and Grinstein, 2010; Bigay and Antonny, 2012). This gradient is achieved by lipid flippases including Drs2, which flips PS from the lumen to the cytosol at the TGN (Natarajan et al., 2004). Additionally, the phosphatidylinositol 4-kinase Pik1 phosphorylates PI to PI4P at the TGN (Walch-Solimena and Novick, 1999; Strahl et al., 2005). Both PI4P and PS lend negative charges to the membrane surface of the TGN that are absent at earlier Golgi compartments. We hypothesized that the nature of the lipid environment in which each of the Golgi Arf-GEFs functions would impact their regulation, so we tested the membrane preferences of each Arf-GEF using in vitro catalytic assays.
To measure the catalytic activity of Arf-GEFs, we employed a well-established assay that measures native tryptophan fluorescence to monitor GEF-catalyzed Arf1 nucleotide exchange in real time (Higashijima et al., 1987; Richardson and Fromme, 2015). These experiments were performed at approximate physiological concentrations of GEF (100 nM) and Arf1 (600 nM) (Ghaemmaghami et al., 2003) in the presence of artificial liposomes.
As we reported previously, our group has purified a functional recombinant construct of Sec7 (Sec7f) for use in in vitro studies (Richardson et al., 2012; McDonold and Fromme, 2014). Similar biochemical studies of Gea1 and Gea2 have been precluded by the difficulty in purifying stable Arf-GEFs in sufficient quantities for study. Therefore, we established protocols for purifying full-length recombinant Gea1 and Gea2, allowing us to make biochemical inquiries into the mechanisms of Gea1 and Gea2 regulation (Supplemental Figure 1A).
We benefit from a detailed model for TGN lipid composition (Klemm et al., 2009; Richardson et al., 2012), which we used to generate liposomes that mimic the TGN’s lipid environment. We lack similar data describing the precise lipid composition of the earlier Golgi compartments, so we used a simplistic model of neutral phosphatidylcholine (PC) to simulate the lipid environment of the early Golgi.
When we measured catalysis of Arf1 activation by Sec7, we observed a 10-fold higher rate on TGN than on PC liposomes (Figure 2, B and C). This parallels the inherent preference of Arf1 for TGN liposomes when intrinsic exchange was stimulated by incubation with EDTA (Supplemental Figure 2) and matches the known localization of Sec7 to the anionic TGN. Notably, Gea1 and Gea2 both display the opposite preference: Gea1 demonstrated a 3-fold and Gea2 a 2-fold higher rate of Arf1 exchange on PC over TGN liposomes (Figure 2, D–G). This is contrary to the intrinsic preference of Arf1, indicating that the Gea GEFs themselves prefer PC over TGN lipids. This finding is consistent with the localization of Gea1 and Gea2 to earlier Golgi compartments, which lack exposed anionic lipids such as PS and PI4P.
Cells require a critical mass of either Gea1 or Gea2 for growth
Gea1 and Gea2 are genetically redundant under normal growth conditions (Peyroche et al., 1996). However, although an arf1Δgea1Δ mutant (sustained by wild-type Arf2, which is expressed at 10-fold lower cellular concentrations than Arf1 [ Stearns et al., 1990]) is viable, an arf1Δgea2Δ mutant is not (Spang et al., 2001). One possible explanation for this phenotype is some separation of function between Gea1 and Gea2, with Gea2 serving a function that is essential under stress conditions. Another possibility is a simple difference in expression levels: Gea2 is expressed at ∼5-fold higher levels than Gea1 (Ghaemmaghami et al., 2003). In light of our observation that Gea1 and Gea2 show different localization patterns, we tested the simpler of these two possibilities by creating promoter swaps to invert expression levels of the two Arf-GEFs.
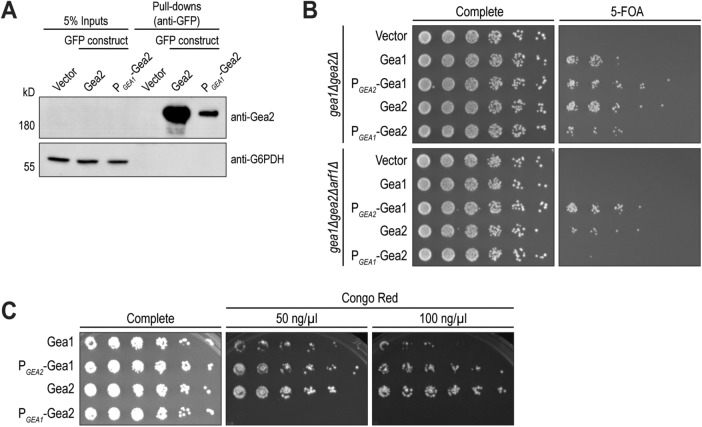
Using a plasmid shuffling strain (gea1Δgea2Δ) maintained by GEA2 on a URA3 plasmid, we compared LEU2 plasmids harboring GFP-tagged wild-type GEA1 or GEA2 to the promoter swaps PGEA2-GEA1 and PGEA1-GEA2. Western blotting of GFP-immunoprecipitation samples confirmed that the promoter swap had the expected effect, significantly reducing cellular expression levels of Gea2 (Figure 3A).
FIGURE 3:
Cells require a critical mass of either Gea1 or Gea2 for growth. (A) Protein expression levels of Gea2 expressed through its endogenous promoter (Gea2) and through Gea1’s promoter (PGEA1-Gea2) were visualized by Western blot after GFP pull downs. G6PDH serves as a loading control. (B) Gea1 and Gea2, expressed through their endogenous promoters or with swapped promoters, were expressed on CEN plasmids in gea1Δgea2Δ and gea1Δgea2Δarf1Δ cells and selected for with 5-FOA. Relative growth of colonies on 5-FOA media reflects sufficiency of each construct to support growth as the sole early Arf-GEF in the cell. (C) Cells from the top right panel of B were cultured and plated onto media containing 50 and 100 ng/µl Congo Red to test for sensitivity to this compound. Data shown represent ≥3 independent experiments.
We observed that yeast growth was subtly impaired in cells harboring only wild-type Gea1, compared with only wild-type Gea2 (Figure 3B). Yeast harboring the promoter-swapped PGEA2-Gea1 grew as well as those with wild-type Gea2, while promoter swapped PGEA1-Gea2 yielded a slight growth impairment similar to wild-type Gea1 (Figure 3B). Furthermore, in a shuffling strain stressed by reduced levels of Arf at the Golgi (gea1Δgea2Δarf1Δ), the phenotype was more stark: cells harboring Gea1 or Gea2 expressed via the GEA2 promoter grew, while cells with the GEA1 promoter-driven Gea1 or Gea2 constructs failed to grow.
Another previously observed difference between Gea1 and Gea2 is the sensitivity of gea2Δ cells to Congo Red, a dye that interferes with cell wall integrity by binding nascent β-glucan chains. gea2Δ cells, but not gea1Δ cells, are hypersensitive to Congo Red (Tsai et al., 2013), again suggesting some function of Gea2 that Gea1 cannot complement. However, when we tested the promoter swap constructs, the Congo Red phenotype was revealed to also depend on expression levels (Figure 3C). Cells with only wild-type Gea1 were more sensitive to Congo Red than cells with only wild-type Gea2. Cells harboring PGEA2-Gea1, on the other hand, grew as well as cells with wild-type Gea2, while cells with PGEA1-Gea2 did not grow.
These results demonstrate that the sensitivity of gea2Δ and survival of gea1Δ cells under stress conditions can be attributed to differential expression levels of Gea1 and Gea2, rather than to an essential function specific to Gea2. They also highlight the requirement for a critical mass of either Gea1 or Gea2 under stress conditions.
The HDS1 and HDS2 domains of Gea1 and Gea2 are required for localization and essential in vivo, while the HDS3 domain is dispensable
Studies of human GBF1 have revealed roles for its N-terminal domains in homodimerization and for its HDS1 domain in membrane targeting (Ramaen et al., 2007; Bouvet et al., 2013; Bhatt et al., 2016). The roles of the HDS2 and HDS3 domains remain unresolved, and Gea1 and Gea2 may have evolved separate regulatory mechanisms from GBF1 after the whole genome duplication event in yeast. Therefore, we pursued more information regarding the roles of the C-terminal domains of Gea1 and Gea2.
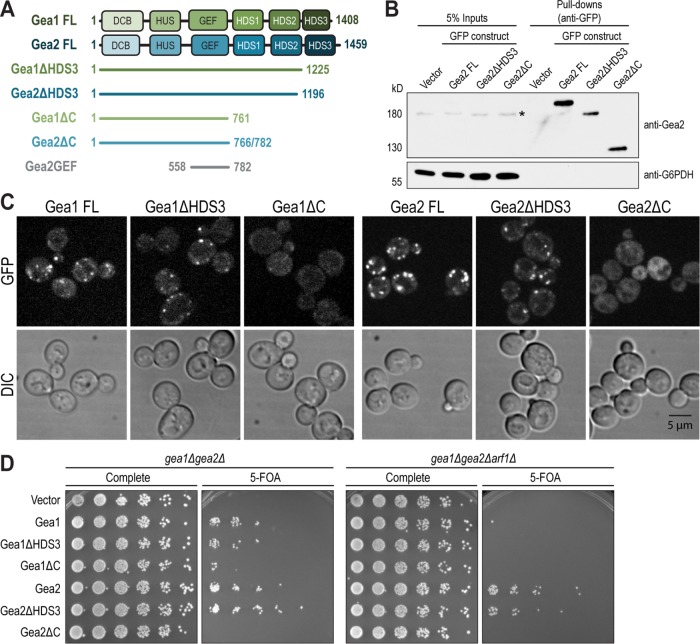
In addition to full-length (FL) Gea1 (1-1408) and Gea2 (1-1459), we generated C-terminal truncations of Gea1 and Gea2 harboring C-terminal mNeonGreen tags: Gea1ΔHDS3 (1-1225), Gea1ΔC (1-774), Gea2ΔHDS3 (1-1234), and Gea2ΔC (1-782) (Figure 4A). We found that constructs lacking both the HDS2 and HDS3 domains were unstable. The stable constructs were introduced into wild-type yeast and expressed under the native GEA1 and GEA2 promoters. We verified by Western blotting that the truncations do not dramatically diminish expression levels under these conditions (Figure 4B).
FIGURE 4:
The HDS1 and HDS2 domains of Gea1 and Gea2 are required for localization and essential in vivo, while the HDS3 domain is dispensable. (A) Diagram of full length and truncated constructs of Gea1 and Gea2 employed in this study. DCB, dimerization and cyclophilin binding; HUS, homology upstream of Sec7; GEF, guanine nucleotide exchange factor (catalytic, aka “Sec7” domain); HDS, homology downstream of Sec7. Note that the HDS1, 2, and 3 domains are not homologous to one another. (B) Full-length Gea2 (Gea2 FL), Gea2 lacking the HDS3 domain (Gea2ΔHDS3), and Gea2 lacking all domains downstream of the GEF domain (Gea2ΔC) were expressed with GFP tags through Gea2’s endogenous promoter on CEN plasmids in wild-type cells. After pull-down with GFP, expression levels of each construct were assessed through Western blot. *, signal from endogenous Gea2 in the whole cell extract before pull downs. G6PDH serves as a loading control. (C) Gea1 and Gea2 constructs (FL, ΔHDS3, and ΔC) were tagged with 3xGFP and visualized in wild-type cells. DIC panels show cells in the field. (D) The same constructs of Gea1 and Gea2 were expressed in gea1Δgea2Δ and gea1Δgea2Δarf1Δ cells as in Figure 3B. Data shown represent ≥3 independent experiments.
Fluorescence microscopy revealed that, for both Gea1 and Gea2, the FL and ΔHDS3 constructs localized normally to Golgi puncta (Figure 4C). The ΔHDS3 signal was weaker than the FL, likely reflecting the slight reduction in expression shown in Figure 4B. Strikingly, Gea1ΔC and Gea2ΔC were completely mislocalized to the cytoplasm, showing no punctate signal despite expression comparable to that of the ΔHDS3 construct.
We next tested the ability of these C-terminal truncations to provide essential Gea function and observed that for both Gea1 and Gea2, the FL and ΔHDS3 constructs supported growth in the gea1Δgea2Δ strain (Figure 4D). The growth of cells harboring only Gea1ΔC is considerably impaired, while cells harboring only Gea2ΔC do not grow at all. As expected, none of the Gea1 constructs supported growth in gea1Δgea2Δarf1Δ strain, while the relative growth of cells harboring the Gea2 truncation constructs remained the same.
Together, these results indicate that the HDS3 domains of Gea1 and Gea2 are dispensable for subcellular localization and essential function. In contrast the HDS1 and HDS2 domains are required for localization to Golgi cisternae and for cell survival.
The C-terminus of Gea2 both inhibits membrane binding in vitro and contributes to Arf1 nucleotide exchange
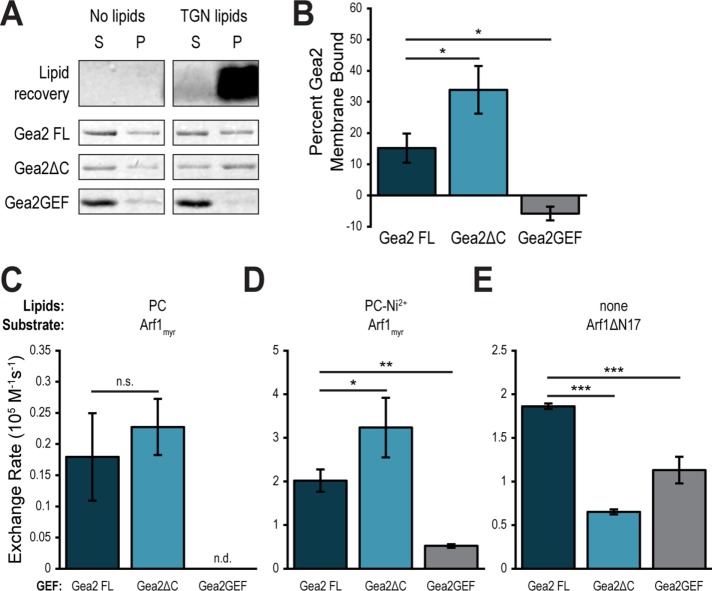
To try to understand the reason for mislocalization of Gea1ΔC and Gea2ΔC, we purified recombinant Gea2ΔC as well as a construct comprising the GEF domain of Gea2 (558–782) (Figure 4A). We then tested membrane binding of each construct in membrane pelleting assays (Paczkowski and Fromme, 2016). As PC liposomes do not pellet efficiently in this assay, we incubated each construct with TGN liposomes. After subjecting each binding reaction to ultracentrifugation, we isolated the membrane pellet, including any membrane-associated proteins, and quantified the amount of each Gea2 construct in the pellet and the supernatant by measuring band intensity after SDS–PAGE. We controlled for background pelleting of Gea2 by carrying out parallel experiments in the absence of liposomes (Figure 5A). After normalizing for background pelleting, we found ∼15% of Gea2 FL bound to membranes (Figure 5B). Unexpectedly, considering the in vivo results, Gea2ΔC showed higher affinity for membranes, with around 30% of that construct pelleting. Finally, the Gea2GEF construct showed no affinity for membranes above background.
FIGURE 5:
The C-terminus of Gea2 both inhibits membrane binding and contributes to Arf1 nucleotide exchange. (A) Purified full length Gea2 (Gea2 FL), Gea2 lacking its C-terminal domains (Gea2ΔC), and the isolated catalytic domain (Gea2GEF) were incubated with or without TGN liposomes before ultracentrifugation. Supernatant (S) and pellet (P) were separated and lipids and proteins in each fraction were visualized by SDS–PAGE. (B) Quantification of band intensity represented as percent Gea2 in pellet after subtracting background, for n = 3 (Gea2FL and Gea2ΔC) and n = 2 (Gea2GEF) independent assays. Rates of Arf1 activation by Gea2 FL, Gea2ΔC, and Gea2GEF on PC (C) or PC-Ni2+ (D) liposomes or in the absence of liposomes using the soluble mutant Arf1ΔN17 (E). n = 3. n.d., not detectable (the Arf1 was not activated by the GEF, and exponential functions could not be fit to experimental curves); n.s., not significant; *p < 0.05; **p < 0.01; ***p < 0.001.
As the C-terminal domains of Gea2 are essential in vivo but not required for membrane binding in vitro, we tested whether the C-terminus might play a role in the nucleotide exchange function of Gea2. First, we tested Arf1 nucleotide exchange by Gea2 FL, Gea2ΔC, and Gea2GEF in the presence of PC liposomes. Gea2FL and Gea2ΔC showed no significant difference in exchange rates on these liposomes, while the GEF domain displayed no measurable exchange activity (Figure 5C).
Considering the very slow rates observed on PC liposomes, we hoped to tease out subtle differences in activity by observing reactions under different conditions. We have observed that Gea1 and Gea2 show a significant preference for liposomes containing the artificial lipid Ni2+-dioleoyl-glycero-succinate (DOGS), used in assaying potential recruiting interactions with poly-histidine–anchored proteins. Despite lacking histidine tags (Supplemental Figure 1, B–D), full-length Gea1 and Gea2 display higher reaction rates on PC-Ni2+ liposomes than on PC liposomes, and Arf1 itself shows a subtle preference for PC-Ni2+ over PC liposomes when activated by EDTA (Supplemental Figure 2). A similar affinity for Ni2+-DOGS lipids has been observed previously for other GEFs (Thomas and Fromme, 2016). While we lack a physiological explanation for the phenomenon, we reasoned that faster rates overall would amplify differences in catalytic rates. Therefore, we tested the catalytic activity of Gea2ΔC and Gea2GEF on PC-Ni2+ liposomes.
Interestingly, Gea2ΔC catalyzed exchange on Arf1 at a higher rate than Gea2 FL on PC-Ni2+ liposomes (Figure 5D). The GEF domain remained inefficient at catalysis in the absence of Gea2’s other domains but catalyzed low-level, measurable exchange on PC-Ni2+ liposomes. These relative exchange reaction rates correlated with the relative membrane affinities of these constructs (Figure 5, A and B).
To examine how the C-terminus contributes to catalysis without the confounding factor of membrane interaction, we employed the mutant Arf1ΔN17, which is missing its amphipathic membrane-inserting helix and can therefore be activated in solution. On the basis of the results in Figures 5, C and D, we expected one of two outcomes when testing Arf1 exchange by Gea2 constructs in solution. If the difference in rates observed on membranes was due to an allosteric effect of removing the HDS domains, then the activity of Gea2ΔC would be higher than the activity of Gea2 FL in solution. If the increased catalytic activity of Gea2ΔC was due to its increased membrane binding, and therefore higher likelihood of successful catalytic events, then Gea2ΔC and Gea2 FL would activate Arf1 at similar rates in solution. Surprisingly, a third possibility proved true for this experiment: in these soluble exchange reactions, Gea2 FL showed the highest catalytic rate on Arf1ΔN17, followed by Gea2GEF (Figure 5E). Gea2ΔC showed the slowest catalytic rate on Arf1ΔN17 in the absence of liposomes.
Taken together, the results of these membrane binding and catalytic assays suggest a complex role for the C-terminus of Gea2 in regulating activation of Arf1.
Gea1 and Gea2 are recruited to membranes by the small GTPase Ypt1
As shown in Figure 5A, ∼15% of Gea2 FL is membrane bound in vitro in the absence of other factors. Furthermore, in vivo, both N-terminal and C-terminal domains of Gea1 and Gea2 are needed for Golgi localization. This suggests that the intrinsic membrane affinity of Gea1 and Gea2 is likely not the only factor regulating their recruitment to Golgi membranes. Sec7 has been shown to be recruited to membranes and is regulated by interactions with several GTPases, so an analogous mechanism may regulate Gea1 and Gea2. Although Gea1 lacks the positive feedback interaction observed between Sec7 and Arf1 (Richardson et al., 2012), the Arf-like GTPase Arl1 interacts with Gea2 (Tsai et al., 2013) and human Rab1b (yeast Ypt1) GTPase interacts with the N-terminus of human GBF1 (yeast Gea1/Gea2) (Monetta et al., 2007).
To explore physical regulatory interactions between Golgi GTPases and Gea1/2, we carried out Gea1 and Gea2 membrane binding assays on PC-Ni2+ liposomes preloaded with activated (GTP-bound) myristoylated Arf1, myristoylated Arl1, Ypt1-His7, or the Rab GTPase Ypt6-His7. These assays confirmed that Arf1 does not recruit either Gea1 or Gea2 to membranes (Figure 6, A–C). Furthermore, neither Arl1 nor Ypt6 increased membrane binding of Gea1 or Gea2 (Figure 6A). The only Golgi small GTPase that increased membrane binding was Ypt1 (Figure 6, A–C).
FIGURE 6:
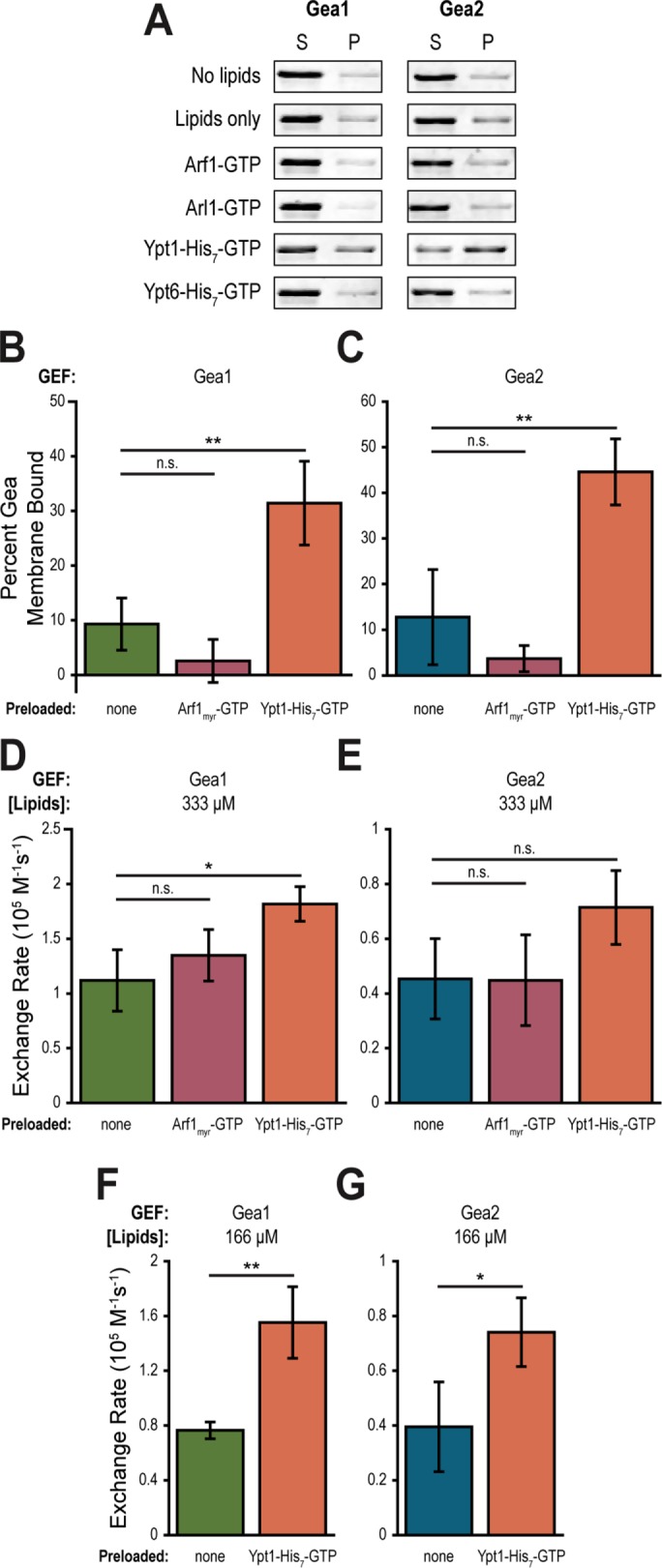
Gea1 and Gea2 are recruited to membranes by the small GTPase Ypt1. (A) Purified Gea1 and Gea2 were incubated with or without PC-Ni2+ liposomes and with no recruiter or GTP-bound Arf1, Arl1, Ypt1-His7, or Ypt6-His7 before ultracentrifugation. Supernatant (S) and pellet (P) were separated and proteins in each fraction were visualized by SDS–PAGE. Graphs show percent of Gea1 (B) or Gea2 (C) bound to membranes after incubation with PC-Ni2+ liposomes alone, preloaded with Arf1-GTP, or preloaded with Ypt1-His7-GTP. n = 3. Rates of Arf1 activation by Gea1 (D) and Gea2 (E) on 333 µM PC-Ni2+ liposomes alone, preloaded with Arf1-GTP, or preloaded with Ypt1-His7-GTP. n = 3. Rates of Arf1 activation by Gea1 (F) and Gea2 (G) with 166 µM PC-Ni2+ liposomes alone or preloaded with Ypt1-His7-GTP. n = 3. n.s., not significant; *p < 0.05; **p < 0.01.
We hypothesized that this recruitment of Gea1 and Gea2 to membranes by Ypt1 would increase their catalytic rates on Arf1 by concentrating the GEFs at the membrane surface where activation of Arf1 must occur. To test this hypothesis, we performed catalytic assays using liposomes alone, liposomes preloaded with activated Arf1, and liposomes preloaded with activated Ypt1-His7. As expected from the membrane binding results, Arf1 conferred no improvement of catalytic activity for Gea1 or Gea2 (Figure 6, D and E). Surprisingly, recruitment by Ypt1 under these conditions yielded only a subtle increase in catalytic rate for Gea1 and a statistically insignificant increase for Gea2. To dissect this apparent discrepancy between our expectations and results, we halved the concentration of liposomes in the catalytic assays. If recruitment is important for activity, increasing the scarcity of membranes in the reaction should resolve a difference between intrinsic, weak membrane binding of Gea1 or Gea2 and active recruitment by Ypt1. Indeed, in catalytic assays with reduced liposome concentrations, both Gea1 and Gea2 showed higher catalytic rates on Ypt1-preloaded liposomes compared with liposomes alone (Figure 6, F and G).
These results indicate that Ypt1 increases Gea1 and Gea2 GEF activity by increasing membrane recruitment of the GEF, thereby increasing the likelihood of productive catalytic interactions between the GEF and Arf1 at the membrane surface.
The C-terminus of Gea2 is required for recruitment by Ypt1
Finally, we set out to understand how regulation by the domains of Gea2 is coordinated with regulation through recruitment by Ypt1. To add a further degree of biological relevance, we carried out these assays using recombinant prenylated Ypt1 (prenyl-Ypt1). We used a protocol for purifying and modifying Ypt1 in vitro, yielding a complex of prenyl-Ypt1 and its stabilizing protein, GDP dissociation inhibitor (GDI) (Thomas and Fromme, 2016). Using prenyl-Ypt1 enabled us to test Gea2 membrane binding and catalytic activity on membranes without any confounding effect from Ni2+-DOGS.
First, we observed that prenyl-Ypt1 can recruit Gea2 FL to TGN liposomes (Figure 7A). While the increase in membrane binding was more subtle with prenyl-Ypt1 and TGN liposomes than with Ypt1-His7 and PC-Ni2+ liposomes, the trend endured under the more physiological conditions. Despite the documented interaction between Rab1b and GBF1’s N-terminus (Monetta et al., 2007), prenyl-Ypt1 did not increase membrane binding of Gea2ΔC on TGN liposomes (Figure 7B). The observed slight reduction in membrane binding of Gea2ΔC may reflect crowding of the membrane surface by prenyl-Ypt1 that cannot effectively recruit the truncated GEF. This result indicates that the C-terminal domains of Gea2 are required to stabilize the Ypt1-Gea2 interaction on membranes.
FIGURE 7:
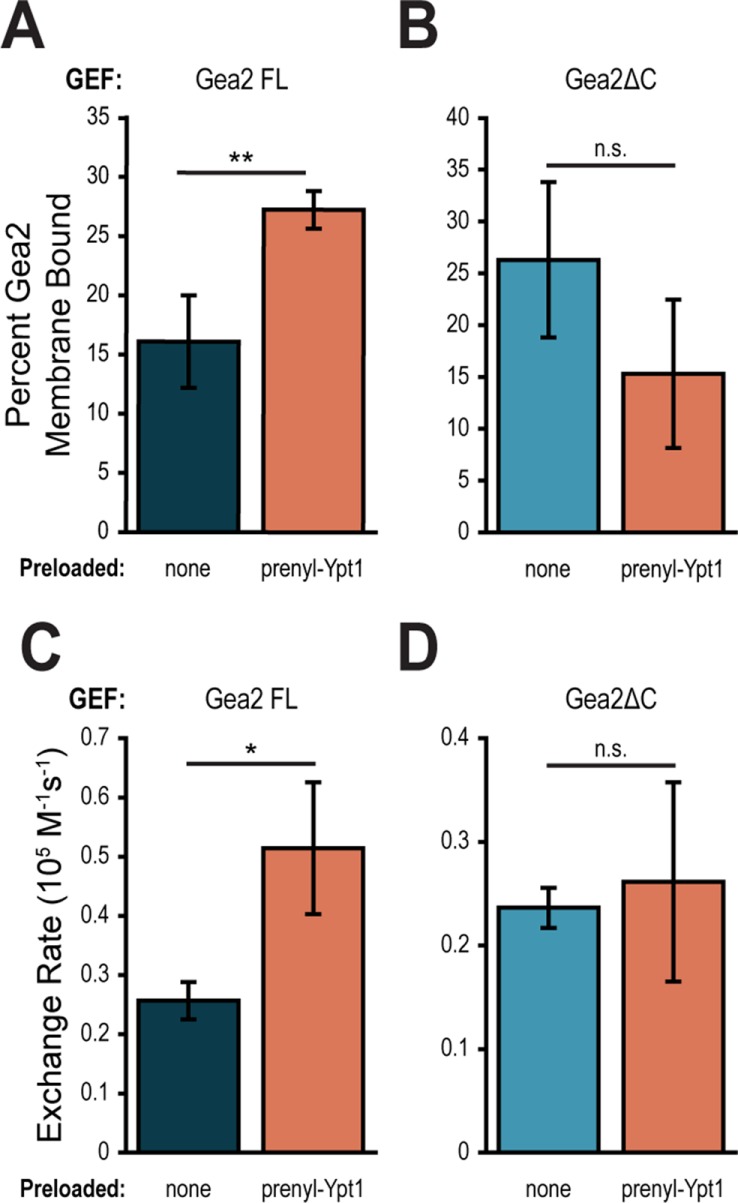
The C-terminus of Gea2 is required for recruitment by Ypt1. Percent of Gea2 FL (A) or Gea2ΔC (B) in membrane pellet after incubation with either TGN liposomes alone or with prenylated Ypt1-GTP (prenyl-Ypt1). n ≥ 3. Rates of Arf1 activation by Gea2 FL (C) or Gea2ΔC (D) on PC liposomes alone or preloaded with prenylated Ypt1-GTP. n = 3. n.s., not significant; *p < 0.05; **p < 0.01.
We next tested whether the failure of prenyl-Ypt1 to recruit Gea2ΔC to membranes coincided with an inability to improve catalytic activity. Gea2 FL showed an approximately twofold increase in catalytic rate on PC liposomes preloaded with prenyl-Ypt1 (Figure 7C), similarly to that observed on PC-Ni2+ liposomes preloaded with Ypt1-His7 (Figure 6G). However, Gea2ΔC catalyzed Arf1 exchange at nearly the same rate on PC liposomes alone as on PC liposomes preloaded with prenyl-Ypt1 (Figure 7D), indicating that the C-terminus of Gea2 is required for effective membrane recruitment by Ypt1, as well as for the positive effect on catalysis conferred by Ypt1 recruitment.
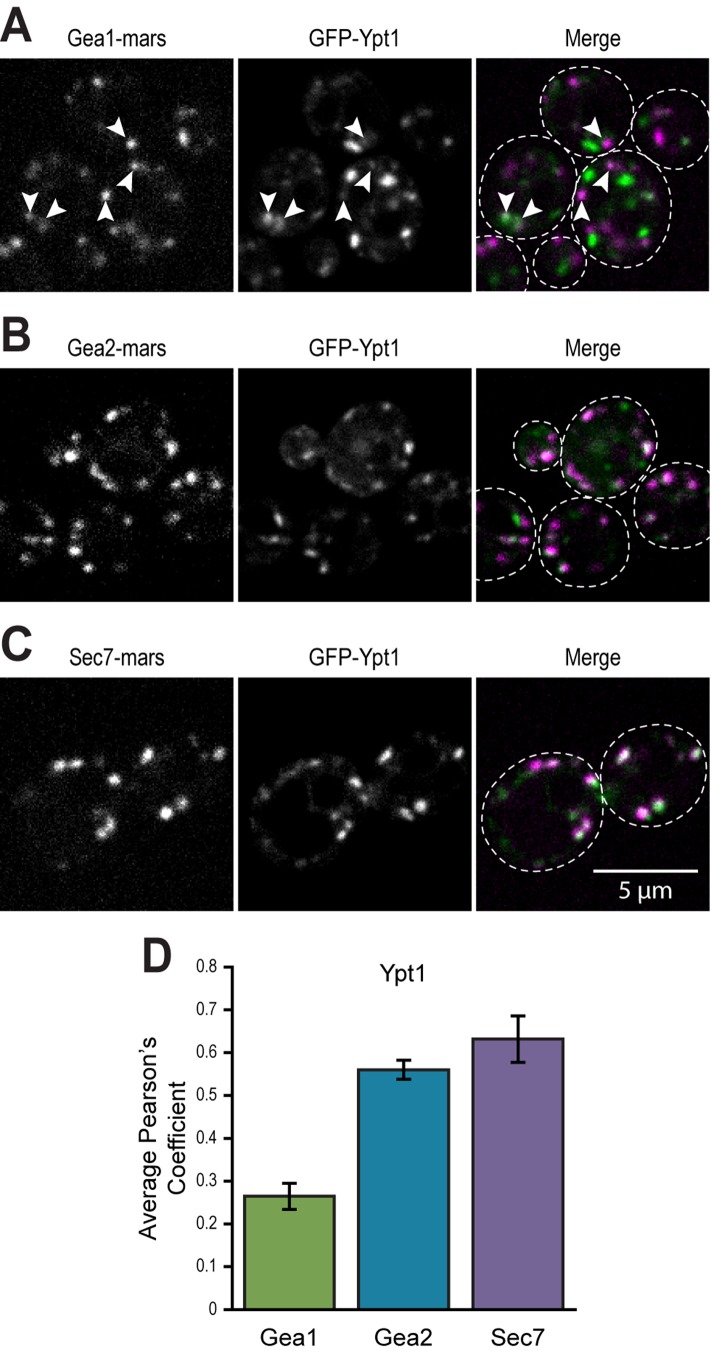
Ypt1 colocalizes well with Gea2 and Sec7 and to a lesser degree with Gea1
To assess the potential relevance of in vitro recruitment by Ypt1 of Gea1 and Gea2 to membranes, we compared the subcellular localization of Gea1, Gea2, and Sec7 to that of Ypt1. While Gea1 colocalized poorly with Ypt1, Ypt1 colocalized well with Gea2 and Sec7 (Figure 8, A–D). We observed instances of overlap between faintly labeled compartments Gea1 and Ypt1 compartments (Figure 8A). The fact that all three Arf-GEFs exhibit partial colocalization with Ypt1 yet occupy different regions of the Golgi indicates that factors other than Ypt1, such as lipids and other proteins, provide sub-compartment specificity.
FIGURE 8:
Ypt1 colocalizes well with Gea2 and Sec7 and to a lesser degree with Gea1. (A) Subcellular localization of GFP-Ypt1 relative to (A) Gea1-mRFPmars, (B) Gea2-mRFPmars, and (C) mRFPmars-Sec7. (D) Quantification of colocalization of Gea1, Gea2, and Sec7 with Ypt1 at puncta. Error bars represent 95% CIs for n = 31 (Ypt1 vs. Gea1), n = 33 (Ypt1 vs. Gea2) cells, or n = 22 (Ypt1 vs. Sec7). In all Merge panels, the GFP channel is shown in green, the RFP channel in magenta, and areas of overlap in white. Arrowheads in A label instances of faint overlap. Cells boundaries are outlined in Merge panels.
DISCUSSION
Arf1 and its homologues are the central coordinators for Golgi vesicle formation in eukaryotic cells, where cargoes are sorted for transport to the plasma membrane, the lysosome, and the endosome, as well as for retrograde traffic within the Golgi and to the ER. Thus, the activation of Arf GTPases at the Golgi represents a critical and conserved point for regulation of Golgi membrane trafficking. In yeast, the activation of Arf1/2 is carried out at the late Golgi by Sec7 and at earlier Golgi compartments by Gea1 and Gea2, implicating these Arf-GEFs as the key decision-makers in initiation of vesicle formation at the Golgi. Despite the essential role of Gea1 and Gea2 in retrograde transport, the mechanisms that govern regulation of these Arf-GEFs have remained elusive.
We have relatively little structural information about these large proteins, beyond structures of the GEF domain (Renault et al., 2002) and of a portion of the N-terminus of Thielavia terrestris Sec7 (Richardson et al., 2016), leaving any structure/function clues in the C-terminus obscured. Temperature-sensitive mutants have proven useful in describing whole-Golgi or whole cell phenotypes for Gea1 and Gea2 mutations (Spang et al., 2001; Park et al., 2005), but the precise reasons for these phenotypes are difficult to infer. Descriptions of genetic and physical interactions with Rab1b, COPI coat proteins, Drs2, Arl1, and Gmh1 allude to the complex environment in which Gea/GBF function (Chantalat et al., 2003, 2004; Monetta et al., 2007; Deng et al., 2009; Christis and Munro, 2012; Tsai et al., 2013) but fail to separate recruiting from effector interactions, leaving the order of events of these interactions and the identity of recruiting partners unknown. The question of how these interactions fit into the overall tasks of Golgi membrane trafficking is unclear, and an exact picture of detailed regulatory mechanisms for Gea/GBF recruitment and function has yet to emerge.
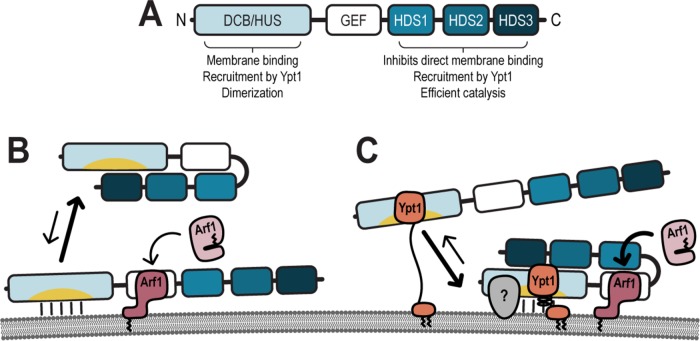
In this study, we demonstrate that Gea1 and Gea2 share their essential function and several regulatory features (Figure 9A). We show that while the HDS3 domain of Gea1 and Gea2 is dispensable for both localization and function in vivo, the HDS1 and HDS2 domains are required for localization. Our findings also reveal that the C-terminus of Gea2 is required for the interaction between Gea2 and Ypt1, which recruits Gea2 to membranes in vitro. Together with a previously reported role for the GBF1 HDS1 domain in targeting to lipid droplets (Bouvet et al., 2013), our results indicate a general role of the C-terminal domains in organelle targeting.
FIGURE 9:
Model of Gea regulation. (A) Assignment of functions and interactions to domains of Gea1/Gea2. Membrane binding and the behavior of the C-terminus of Gea1/Gea2 in the absence (B) and presence (C) of active recruiting interactions. Note that the membrane diagramed represents a neutral membrane, lacking anionic lipids. Gray (?) represents a potential interactor which confers specificity of localization to Gea1 and Gea2.
Our colocalization studies show Gea1 and Gea2 to have different colocalization patterns relative to the early and late Golgi markers Vrg4 and Sec7. This is consistent with a model in which Gea1 and Gea2 occupy intermediate compartments within the Golgi, overlapping at cis and trans compartments with Vrg4 and Sec7. This model posits a continuum of Arf-GEFs across the Golgi (Figure 2A) and is consistent with evidence that GBF1 (Gea1/Gea2) provides the seed Arf-GTP to recruit BIG1/BIG2 (Sec7) to the TGN (Richardson et al., 2012; Lowery et al., 2013). In addition, Gea1 occupies earlier Golgi compartments than Gea2, which must require distinct recruitment mechanisms and implies different roles for the two GEFs. Further study is needed to understand how and why Gea1 and Gea2 show different colocalization patterns.
It appears that any specialized roles for Gea1 or Gea2 are not essential, however, as we show here that Gea1 and Gea2 are equally functional in stressed cells when expressed with the higher copy GEA2 promoter. This confirms a redundant essential function and highlights the requirement for a critical cellular concentration of Gea under stress conditions.
We observed that Gea1 and Gea2 display a preference for neutral over anionic membranes in catalytic assays. Sec7, on the other hand, prefers anionic membranes. These preferences correlate with the localization patterns of the GEFs, with Gea1 and Gea2 occupying earlier Golgi compartments than Sec7, which functions at the late Golgi. Intriguingly, a recent study showed that recruitment of Caenorhabditis elegans GBF1 to membranes was reduced in a small interfering RNA knockdown of the rate-limiting enzyme for PC synthesis (Smulan et al., 2016), suggesting a physiological role for Golgi lipid composition in recruitment of GBF1/Gea family Arf-GEFs. Together with our previous observation that another late Golgi GEF, TRAPPII, prefers anionic membranes (Thomas and Fromme, 2016), these results suggest a general mechanism for regulating the membrane specificity of Golgi GEFs. As these GEFs lack traditional membrane binding domains, the increasing negative charge of the late Golgi and TGN may help exclude early Golgi GEFs and recruit late Golgi GEFs to the TGN.
We found that the HDS3 domain of Gea1 and Gea2 is dispensable for in vivo localization to the Golgi and for the essential function of Gea. In biochemical studies (not shown), we found that Gea2ΔHDS3 showed a milder version of each phenotype observed for Gea2ΔC. Future investigations may reveal whether this conserved domain plays a regulatory role separate from the HDS1 and HDS2 domains.
Unlike Sec7, whose C-terminal domains function in both strong catalytic autoinhibition and the protein interactions that relieve that autoinhibition, Gea1 and Gea2 are not catalytically autoinhibited. Instead, we show here that the C-terminus of Gea2 inhibits membrane binding, perhaps by masking the intrinsic affinity of the Gea2 N-terminus for membranes. Removal of the C-terminal domains of Gea2 increased both membrane binding and the rate of Arf1 exchange on membranes, yet the C-terminus of Gea2 was required for Golgi localization and full exchange function in the absence of membranes.
Our results combine to evoke a model for carefully calibrated regulation of Gea2 by its C-terminal domains: in the absence of active recruitment to membranes, the C-terminus functions to keep Gea2 in the cytosol (Figure 9B). Once this membrane-binding inhibition is relieved by active recruitment, the C-terminus switches functions to promote efficient catalysis of Arf1 exchange (Figure 9C).
We have identified Ypt1 as one of the active recruiters for Gea1 and Gea2. Ypt1 recruits Gea1 and Gea2 to membranes in vitro, and this recruiting interaction requires the C-terminal domains, despite previous evidence of a direct interaction between the N-terminus of GBF1 and Rab1b (Figure 9C) (Monetta et al., 2007). It is possible that the C-terminus stabilizes the interaction with Ypt1 or that intrinsic membrane binding in the HDS1 domain of Gea1 and Gea2 complements the Ypt1 recruitment. As both the Ypt1 interaction and intrinsic membrane binding seem weak or transient, it is likely that concomitant interactions cooperate to fully recruit Gea1 and Gea2 to the membrane surface (Figure 9C).
Interestingly, while Gea1 is recruited by Ypt1 in vitro, our in vivo colocalization analysis revealed very little overlap between the two. One possibility is that an interaction between Gea1 and Ypt1 occurs only very briefly at the early Golgi. Further studies are required to determine the physiological role of the Ypt1/Gea1 interaction.
Our results clearly distinguish the regulation of Gea1 and Gea2 from that of Sec7. Gea1 and Gea2 do not participate in a positive feedback loop with Arf1 (Richardson et al., 2012); Arf1 cannot recruit either Gea1 or Gea2 to membranes, and it does not stimulate GEF activity in vitro. This characteristic represents an important regulatory divergence from Sec7, as does the absence of apparent catalytic autoinhibition and the failure of Arl1 to have a recruiting or stimulating effect on Gea1 or Gea2. This lack of autoinhibition resembles more distant members of the Sec7 family of Arf-GEFs, such as BRAG, which functions in endocytosis (Aizel et al., 2013). Additionally, Gea1 and Gea2 lack a fourth C-terminal domain, HDS4, which is conserved within the Sec7/BIG subfamily. We recently described the role of this domain in homodimerization of Sec7 (Richardson et al., 2016). However, Gea2 is dimeric in the absence of the HDS4 domain, reinforcing previous observations that the N-terminal domains of Gea/GBF1 are responsible for dimerization (Grebe et al., 2000; Ramaen et al., 2007; Bhatt et al., 2016) and further differentiating regulation of the early Golgi Arf-GEFs from that of the late.
One regulatory feature that Gea1 and Gea2 share with Sec7 is the interaction with Ypt1. Ypt1 may serve as a general Golgi recruiter for Arf-GEFs. However, the specific sub-Golgi localization of Gea1, Gea2, and Sec7 must require additional specific recruiting interactions for each Gea1 and Gea2, either with proteins or with the membrane surface (Figure 9C). Our results imply that membrane lipid character likely plays an important role in Golgi Arf-GEF targeting and GEF activity. The comprehensive identification and characterization of protein–protein and protein–membrane interactions will be crucial to a full understanding of how vesicular membrane trafficking at the Golgi is coordinated.
MATERIALS AND METHODS
Strains and plasmids
Yeast strains used in this study are detailed in Supplemental Table 1 and plasmids in Supplemental Table 2. All new plasmids used in this study were verified by sequencing.
Antibodies and immunoprecipitations
The anti-Gea2 rabbit polyclonal antibody was generated using purified Gea2 GEF domain (Covance) and used at a 1:500 dilution. This antibody cross-reacts to some extent with purified recombinant Gea1 but fails to detect Gea1 in yeast extracts. The anti-G6PDH rabbit polyclonal antibody was purchased from Sigma and used at a 1:30,000 dilution. The anti-His6 mouse monoclonal antibody (mAb) used to verify cleavage of His tags was purchased from Covance and used at a 1:500 dilution.
The GFP nanobody resin used for pull downs in Figures 3 and 4 was made using purified GFP nanobody and NHS-activated Sepharose Fast Flow (GE Healthcare) (Kirchhofer et al., 2010). Twenty-five ODs of cells were suspended in lysis buffer (50 mM Tris–HCl, pH 7.5, 0.2% NP40 substitute, 150 mM NaCl, 1 mM EDTA, 1× complete protease inhibitor cocktail [Roche] and 1 mM phenylmethylsulfonyl fluoride [PMSF]) before lysis by mechanical disruption. After incubation, resin was washed three times with lysis buffer, and proteins were eluted in SDS sample buffer.
Yeast growth assays
Yeast plasmid shuffling assays were used to assess in vivo sufficiency of Gea1 and Gea2 mutants (Gea2ΔC is residues 1-766). Double-deletion strains (gea1Δgea2Δ and gea1Δgea2Δarf1Δ) were maintained by a copy of GEA2 on a URA3 plasmid. LEU2 plasmids carrying the mutant constructs were introduced, and cells were cultured overnight in –Leu media. Cells were plated at threefold dilutions onto synthetic complete media or media with 5-fluoroorotic acid (5-FOA) and incubated at 30°C for two days before imaging.
To test for sensitivity to the drug Congo Red, shuffled cells were plated at threefold dilutions onto synthetic complete media containing either 50 or 100 ng/µl Congo Red (Sigma) and incubated at 30°C for 2 d before imaging.
Protein purification
Full-length Gea1 and Gea2, as well as Gea2ΔC (1–782), were expressed with an N-terminal His6 tag in Rosetta2 cells, with expression induced overnight with 250 µM IPTG at 15°C. Pelleted cultures were resuspended in 25 ml/l of culture of lysis buffer (40 mM Tris, pH 8.0, 300 mM NaCl, 10% glycerol, 10 mM imidazole, 0.25× Roche complete protease inhibitor, 1 mM PMSF, 10 mM β-mercaptoethanol) before lysis by sonication. After the lysate was clarified by centrifugation, protein constructs were purified via nickel affinity (Ni-NTA resin; Qiagen) in batch, followed by anion exchange (MonoQ, GE Healthcare), overnight cleavage of the His6 tag by TEV protease at 4°C, and gel filtration (Superdex 200; GE Healthcare), with a final buffer containing 20 mM Tris, pH 8, 150 mM NaCl, and 1 mM dithiothreitol. The Gea2GEF construct was expressed and purified similarly through the nickel affinity step, and then incubated at room temperature overnight with TEV, before an additional round nickel binding and elution to remove any uncleaved protein.
Purification of Arf and Rab GTPases has previously been described in detail (Ha et al., 2005; McDonold and Fromme, 2014; Richardson and Fromme, 2015; Thomas and Fromme, 2016).
Liposome preparation
Lipid composition of PC, PC-Ni2+, TGN, and TGN-Ni2+ liposomes is described in Supplemental Table 3. Liposomes were prepared in HK buffer (20 mM HEPES, pH 7.5, 150 mM KOAc) as described previously (Paczkowski and Fromme, 2016), with 100-nm filters used to extrude liposomes used for GEF activity assays and 400-nm filters used to extrude liposomes used for membrane pelleting assays.
GEF activity assays
GEF activity was measured as described previously (Richardson and Fromme, 2015). All assays were carried out in HKM buffer (HK buffer plus 2 mM MgCl2) at 30°C. Arf1 exchange was measured after sequentially adding 200 µM GTP and 100 nM GEF to HKM buffer with 333 µM liposomes (or 166 µM liposomes, for Figure 6, F and G). After equilibration for 5 min, Arf1 was added and native tryptophan fluorescence was measured for 12 min. For assays testing the effect of preloaded GTPases, 500 nM Arf1, Arl1, Ypt1-His7, or Ypt6-His7 or 400 nM prenylated Ypt1 was added to HKM buffer with liposomes, followed by 200 µM GTP and 2 mM EDTA. After incubation for 12 min, 4 mM MgCl2 was added, followed by 100 nM GEF. After 5 min of equilibration, 500 nM Arf1 was added and monitored for an additional 12 min. Each assay was carried out in triplicate for statistical analysis.
Liposome pelleting assays
Liposome pelleting assays were carried out as previously described (Paczkowski and Fromme, 2016). Liposomes (500 µM) were mixed with HKM buffer. To test intrinsic membrane binding of GEF constructs, 4 µg of each construct was incubated with liposomes for 10 min at room temperature before ultracentrifugation, separation of supernatant from pellet, and PAGE analysis. For recruiting assays, 2 µg of each GTPase was activated by 15 min of EDTA-mediated GTP exchange at 30°C before the addition of extra MgCl2 and finally 4 µg of GEF. Parallel reactions lacking liposomes were run with all sets of binding reactions to account for background pelleting of GEF constructs.
PAGE gels were stained with Bio-Safe Coomassie (Bio-Rad) and imaged using a LI-COR Odyssey system. Band intensities were quantified in ImageJ. Percentage GEF in pellet was calculated after subtraction of background pelleting for each construct. Each set of reactions was performed in at least triplicate for statistical analysis.
Microscopy
Cells were cultured in either synthetic complete media (Figure 1) or synthetic dropout media (Figure 4) at 30°C and imaged in log phase. Note that Gea2ΔC corresponds to residues 1–766.
Images were captured a CSU-X spinning disk confocal microscope system (Intelligent Imaging Innovations) using a DMI600B microscope (Leica Biosystems), 100 ×/1.46 NA objective, and a QuantEM EMCCD camera (Photometrics). Images were acquired, leveled, and analyzed using Slidebook 5.0 software (Intelligent Imaging Innovations). Single confocal sections are shown. After background subtraction to limit analysis to Golgi puncta and selection of individual cells, Pearson’s analysis was performed in Slidebook 5.0 to quantify colocalization of fluorescently tagged proteins for Figure 1.
Statistical tests
Significance for Figures 1, 2, 6, F and G, and 7 was determined using an unpaired t test with a Welch’s correction. Significance for all other figures was determined by a one-way analysis of variance with a Tukey post-test. Error bars represent 95% confidence intervals.
Supplementary Material
Acknowledgments
We acknowledge C. McDonold for performing preliminary experiments and construction of plasmids. We thank Y. F. Chen for plasmid and strain construction. We thank the lab of Y. Mao for advice on generating GFP nanobody resin. We acknowledge the labs of T. Bretscher and S. Emr for strains, plasmids, and use of equipment. We are grateful to C. Diefenderfer and L. Thomas for advice on the manuscript. We thank B. Richardson for training and support. This work was supported by National Institutes of Health (NIH) Grant No. R01GM098621. M.A.G. was supported by NIH training Grant No. T32GM007273 and by the Harry and Samuel Mann Outstanding Graduate Student Award.
Abbreviations used:
- DCB
dimerization and cyclophilin binding
- ER
endoplasmic reticulum
- GDI
GDP dissociation inhibitor
- GEF
guanine nucleotide exchange factor
- HDS
homology downstream of Sec7
- HUS
homology upstream of Sec7
- PC
phosphatidylcholine
- PS
phosphatidylserine
- TGN
trans-Golgi network
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E17-06-0370) on October 4, 2017.
REFERENCES
- Achstetter T, Franzusoff A, Field C, Schekman R. Sec7 encodes an unusual, high molecular weight protein required for membrane traffic from the yeast Golgi apparatus. J Biol Chem. 1988;263:11711–11717. [PubMed] [Google Scholar]
- Aizel K, Biou V, Navaza J, Duarte LV, Campanacci V, Cherfils J, Zeghouf M. Integrated conformational and lipid-sensing regulation of endosomal ArfGEF BRAG2. PLoS Biol. 2013;11:e1001652. doi: 10.1371/journal.pbio.1001652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonny B, Huber I, Paris S, Chabre M, Cassel D. Activation of ADP-ribosylation factor 1 GTPase-activating protein by phosphatidylcholine-derived diacylglycerols. J Biol Chem. 1997;272:30848–30851. doi: 10.1074/jbc.272.49.30848. [DOI] [PubMed] [Google Scholar]
- Bhatt JM, Viktorova EG, Busby T, Wyrozumska P, Newman LE, Lin H, Lee E, Wright J, Belov GA, Kahn RA, et al. Oligomerization of the Sec7 domain Arf guanine nucleotide exchange factor GBF1 is dispensable for Golgi localization and function but regulates degradation. Am J Physiol Cell Physiol. 2016;310:C456–C469. doi: 10.1152/ajpcell.00185.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Bouvet S, Golinelli-Cohen M-P, Contremoulins V, Jackson CL. Targeting of the Arf-GEF GBF1 to lipid droplets and Golgi membranes. J Cell Sci. 2013;126:4794–4805. doi: 10.1242/jcs.134254. [DOI] [PubMed] [Google Scholar]
- Bui QT, Golinelli-Cohen M-P, Jackson CL. Large Arf1 guanine nucleotide exchange factors: evolution, domain structure, and roles in membrane trafficking and human disease. Mol Genet Genomics. 2009;282:329–350. doi: 10.1007/s00438-009-0473-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova JE. Regulation of arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- Chantalat S, Courbeyrette R, Senic-Matuglia F, Jackson CL, Goud B, Peyroche A. A novel golgi membrane protein is a partner of the ARF exchange factors Gea1p and Gea2p. Mol Biol Cell. 2003;14:2357–2371. doi: 10.1091/mbc.E02-10-0693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantalat S, Park S-K, Hua Z, Liu K, Gobin R, Peyroche A, Rambourg A, Graham T, Jackson CL. The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J Cell Sci. 2004;117:711–722. doi: 10.1242/jcs.00896. [DOI] [PubMed] [Google Scholar]
- Christis C, Munro S. The small G protein Arl1 directs the trans-Golgi-specific targeting of the Arf1 exchange factors BIG1 and BIG2. J Cell Biol. 2012;196:327–335. doi: 10.1083/jcb.201107115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Golinelli-Cohen M-P, Smirnova E, Jackson CL. A COPI coat subunit interacts directly with an early-Golgi localized Arf exchange factor. EMBO Rep. 2009;10:58–64. doi: 10.1038/embor.2008.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem Soc Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. Identification of a guanine nucleotide exchange factor for Arf3, the yeast orthologue of mammalian Arf6. PLoS One. 2007;2:e842. doi: 10.1371/journal.pone.0000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld J-U, Salchert K, Koncz C, Jürgens G. A conserved domain of the arabidopsis GNOM protein mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha V, Thomas G, Stauffer S, Randazzo P. Preparation of myristoylated Arf1 and Arf6. Methods Enzymol. 2005;404:164–174. doi: 10.1016/S0076-6879(05)04016-4. [DOI] [PubMed] [Google Scholar]
- Higashijima T, Ferguson K, Sternweis P, Ross E, Smigel M, Gilman A. The effect of activating ligands on the intrinsic fluorescence of guanine nucleotide-binding regulatory proteins. J Biol Chem. 1987;262:752–756. [PubMed] [Google Scholar]
- Jackson CL, Casanova JE. Turning on ARF: the Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- Kirchhofer A, Helma J, Schmidthals K, Frauer C, Cui S, Karcher A, Pellis M, Muyldermans S, Casas-Delucchi CS, Cardoso MC, et al. Modulation of protein properties in living cells using nanobodies. Nat Struct Mol Biol. 2010;17:133–138. doi: 10.1038/nsmb.1727. [DOI] [PubMed] [Google Scholar]
- Klemm R, Ejsing C, Surma M, Kaiser H-J, Gerl M, Sampaio J, Robillard Q, Ferguson C, Proszynski T, Shevchenko A, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185:601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventis PA, Grinstein S. The distribution and function of phosphatidylserine in cellular membranes. Annu Rev Biophys. 2010;39:407–427. doi: 10.1146/annurev.biophys.093008.131234. [DOI] [PubMed] [Google Scholar]
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS. Golgi maturation visualized in living yeast. Nature. 2006;441:1002–1006. doi: 10.1038/nature04717. [DOI] [PubMed] [Google Scholar]
- Lowery J, Szul T, Styers M, Holloway Z, Oorschot V, Klumperman J, Sztul E. The Sec7 guanine nucleotide exchange factor GBF1 regulates membrane recruitment of BIG1 and BIG2 guanine nucleotide exchange factors to the trans-Golgi network (TGN) J Biol Chem. 2013;288:11532–11545. doi: 10.1074/jbc.M112.438481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K, Takeuchi M, Ichihara A, Mikuriya K, Nakano A. Live imaging of yeast Golgi cisternal maturation. Nature. 2006;441:1007–1010. doi: 10.1038/nature04737. [DOI] [PubMed] [Google Scholar]
- McDonold CM, Fromme JC. Four GTPases differentially regulate the Sec7 Arf-GEF to direct traffic at the trans-Golgi network. Dev Cell. 2014;30:759–767. doi: 10.1016/j.devcel.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol Biol Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouratou B, Biou V, Joubert A, Cohen J, Shields D, Geldner N, Jürgens G, Melançon P, Cherfils J. The domain architecture of large guanine nucleotide exchange factors for the small GTP-binding protein Arf. BMC Genomics. 2005;6:20. doi: 10.1186/1471-2164-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan P, Wang J, Hua Z, Graham TR. Drs2p-coupled aminophospholipid translocase activity in yeast Golgi membranes and relationship to in vivo function. Proc Natl Acad Sci USA. 2004;101:10614–10619. doi: 10.1073/pnas.0404146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paczkowski JE, Fromme JC. Analysis of Arf1 GTPase-dependent membrane binding and remodeling using the exomer secretory vesicle cargo adaptor. In: Brown WJ, editor. The Golgi Complex: Methods and Protocols. New York: Springer; 2016. pp. 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-K, Hartnell L, Jackson CL. Mutations in a highly conserved region of the Arf1p activator GEA2 block anterograde Golgi transport but not COPI recruitment to membranes. Mol Biol Cell. 2005;16:3786–3799. doi: 10.1091/mbc.E05-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Ramaen O, Joubert A, Simister P, Belgareh-Touzé N, Olivares-Sanchez MC, Zeeh J-C, Chantalat S, Golinelli-Cohen M-P, Jackson CL, Biou V, et al. Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J Biol Chem. 2007;282:28834–28842. doi: 10.1074/jbc.M705525200. [DOI] [PubMed] [Google Scholar]
- Renault L, Christova P, Guibert B, Pasqualato S, Cherfils J. Mechanism of domain closure of Sec7 domains and role in BFA sensitivity. Biochemistry. 2002;41:3605–3612. doi: 10.1021/bi012123h. [DOI] [PubMed] [Google Scholar]
- Richardson BC, Fromme JC. Biochemical methods for studying kinetic regulation of Arf1 activation by Sec7. Methods Cell Biol. 2015;130:101–126. doi: 10.1016/bs.mcb.2015.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC, Halaby SL, Gustafson MA, Fromme JC. The Sec7 N-terminal regulatory domains facilitate membrane-proximal activation of the Arf1 GTPase. Elife. 2016;5:1–20. doi: 10.7554/eLife.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson BC, McDonold CM, Fromme JC. The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell. 2012;22:799–810. doi: 10.1016/j.devcel.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smulan LJ, Ding W, Freinkman E, Gujja S, Edwards YJK, Walker AK. Cholesterol-independent SREBP-1 maturation is linked to ARF1 inactivation. Cell Rep. 2016;16:9–18. doi: 10.1016/j.celrep.2016.05.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Herrmann JM, Hamamoto S, Schekman R. The ADP ribosylation factor-nucleotide exchange factors Gea1p and Gea2p have overlapping, but not redundant functions in retrograde transport from the Golgi to the endoplasmic reticulum. Mol Biol Cell. 2001;12:1035–1045. doi: 10.1091/mbc.12.4.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T, Kahn RA, Botstein D, Hoyt MA. ADP ribosylation factor is an essential protein in saccharomyces cerevisiae and is encoded by two genes. Mol Cell Biol. 1990;10:6690–6699. doi: 10.1128/mcb.10.12.6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Hama H, DeWald DB, Thorner J. Yeast phosphatidylinositol 4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J Cell Biol. 2005;171:967–979. doi: 10.1083/jcb.200504104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LL, Fromme JC. GTPase cross talk regulates TRAPPII activation of Rab11 homologues during vesicle biogenesis. J Cell Biol. 2016;215:499–513. doi: 10.1083/jcb.201608123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai P-C, Hsu J-W, Liu Y-W, Chen K-Y, Lee F-JS. Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc Natl Acad Sci USA. 2013;110:E668–E677. doi: 10.1073/pnas.1221484110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase Pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.