Abstract
Induction of the cell cycle is emerging as an intervention to treat heart failure. Here, we tested the hypothesis that enhanced cardiomyocyte renewal in transgenic mice expressing cyclin D2 would be beneficial during hemodynamic overload. We induced pressure overload by transthoracic aortic constriction (TAC) or volume overload by aortocaval shunt in cyclin D2–expressing and WT mice. Although cyclin D2 expression dramatically improved survival following TAC, it did not confer a survival advantage to mice following aortocaval shunt. Cardiac function decreased following TAC in WT mice, but was preserved in cyclin D2–expressing mice. On the other hand, cardiac structure and function were compromised in response to aortocaval shunt in both WT and cyclin D2–expressing mice. The preserved function and improved survival in cyclin D2–expressing mice after TAC was associated with an approximately 50% increase in cardiomyocyte number and exaggerated cardiac hypertrophy, as indicated by increased septum thickness. Aortocaval shunt did not further impact cardiomyocyte number in mice expressing cyclin D2. Following TAC, cyclin D2 expression attenuated cardiomyocyte hypertrophy, reduced cardiomyocyte apoptosis, fibrosis, calcium/calmodulin–dependent protein kinase IIδ phosphorylation, brain natriuretic peptide expression, and sustained capillarization. Thus, we show that cyclin D2–induced cardiomyocyte renewal reduced myocardial remodeling and dysfunction after pressure overload but not after volume overload.
Keywords: Cardiology, Cell Biology
Keywords: Cell cycle, Heart failure
Introduction
In structural heart diseases, increased ventricular load is a key trigger mechanism for altered gene expression and cardiac remodeling (1). Increased ventricular load translates into increased wall stress for the individual cardiomyocyte, which is a major determinant of myocardial energy consumption. The heart may adapt to elevated load by increasing wall thickness, which, according to the law of Laplace, reduces wall stress, or by improving its energy efficiency. The latter may occur at 3 distinct levels. First, at the level of the acto-myosin crossbridge, economy may change due to switches in isoforms of the myosin heavy chain gene, or thin filament regulatory proteins, or due to altered phosphorylation of regulatory proteins (2). Secondly, energy efficiency may change at the level of the excitation-contraction-coupling system due to alterations in calcium sensitivity (3). Thirdly, energy efficiency may change at the level of metabolism due to a switch from fatty acids to glucose consumption (4). Furthermore, a number of genetically modified mouse models seem to accomplish load adaptation without increasing wall thickness (5, 6).
An increase in wall thickness may occur through cardiomyocyte hypertrophy or cardiomyocyte proliferation. Because intrinsic cardiomyocyte renewal is infrequent in the adult mammalian heart (7, 8), the former is the dominant mechanism for increasing wall thickness. Accordingly, the development of therapeutic interventions aimed at enhancing cardiomyocyte renewal is a challenging consideration (9). We have previously shown that targeted expression of cyclin D1 was sufficient to activate cardiomyocyte DNA synthesis in transgenic mice following myocardial infarction (10). Moreover, cardiomyocyte cell cycle activity resulting from the targeted expression of cyclin D2 (CCND2) was sufficient to improve cardiac architecture (11) and function (12) following myocardial infarction in adult animals. These mice (designated D2 mice) used the α cardiac myosin heavy chain (αMHC) promoter to target constitutive CCND2 expression.
In infarction models, both pre- and afterload are increased in the surviving left ventricular cardiomyocytes. The present study was undertaken to determine if cardiomyocyte cell cycle activation is similarly beneficial in models with distinct hemodynamic phenotypes of increased afterload (transversal aortic constriction, TAC) versus increased preload (aortocaval shunt, Shunt). In a previous study we showed that these models, at comparable levels of increased load, exhibit significantly different gene expression signatures resulting in detrimental maladaptive remodeling in TAC and more favorable remodeling with better prognosis in Shunt (13). The present data show that TAC enhances the level of transgene-induced cardiomyocyte cell cycle activity in D2 mice, resulting in increased cardiomyocyte number and increased wall thickness despite attenuated hypertrophy at the cellular level. This in turn prevented heart failure and improved survival. In contrast, Shunt did not enhance the level of transgene-induced cardiomyocyte renewal, and there was no improvement in either phenotypic or prognostic effects in the D2 mice.
Results
D2 mice exhibit improved survival in TAC but not in Shunt.
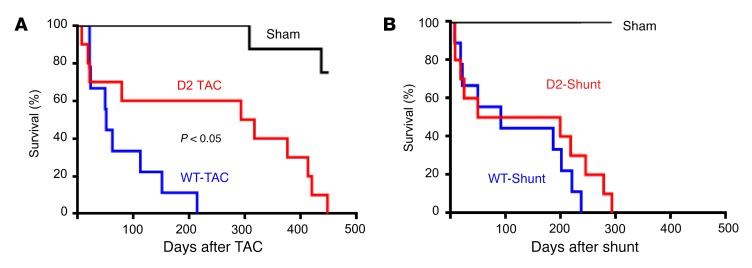
Following TAC surgery, survival was greatly enhanced in D2 as compared with wild-type (WT) mice (Figure 1A); transgene expression resulted in a 6-fold increase in median survival after TAC, with some animals surviving for more than 450 days after imposition of pressure overload. In contrast, cardiomyocyte CCND2 expression did not affect survival following shunt surgery (Figure 1B).
Figure 1. Enhanced survival of D2 mice following TAC but not shunt surgery.
(A) Survival plot of WT-TAC (blue line, n = 9), D2-TAC (red line, n = 10), and sham mice (black line, n = 9); animals that died acutely (i.e., within 24 hours of surgery) are not included in the plot. (B) Survival plot of WT-Shunt (blue line, n = 9), D2-Shunt (red line, n = 10), and sham mice (black line, n = 8); animals that died acutely (i.e., within 24 hours of surgery) are not included in the plot. Statistical analysis: log-rank (Mantel-Cox) test.
D2 mice are resistant to heart failure in TAC but not in Shunt.
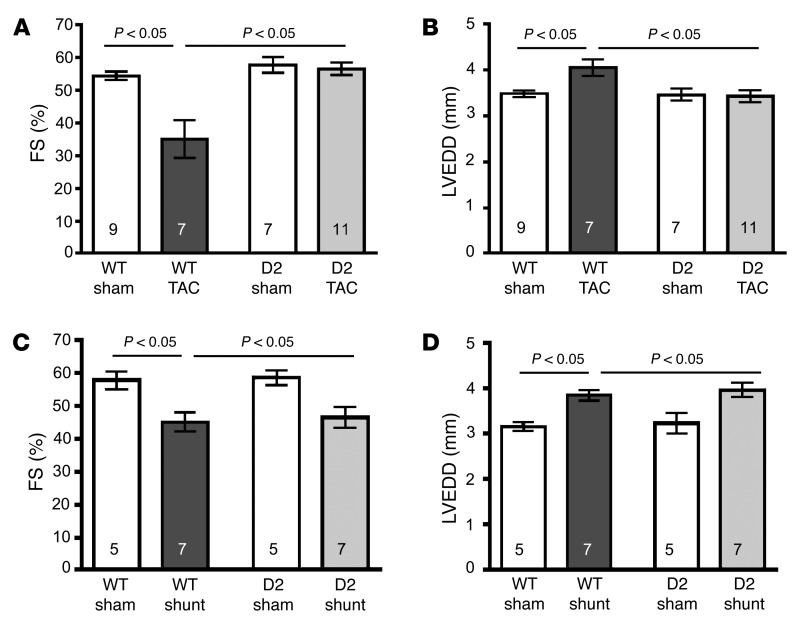
Cardiac function was monitored at 1, 3, and 10 weeks after TAC and shunt surgery. Cardiac function, as evidenced via fractional shortening (FS), was reduced at 3 weeks after TAC in the WT mice and continued to decline with time (Figure 2A and Supplemental Figures 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/JCI81870DS1). Decreased function in the WT-TAC mice was associated with left ventricular dilation (Figure 2B). In contrast, cardiac function and chamber dimensions were preserved in the D2 mice over the same period (Figure 2, A and B). In Shunt, transgenic CCND2 expression did not affect the decrease in FS and left ventricular dilatation as determined at 10 weeks after surgery (Figure 2, C and D).
Figure 2. Preserved cardiac function in D2 mice following TAC but not shunt surgery.
(A) Fractional shortening (FS) of WT and D2 mice 10 weeks after either sham or TAC surgery. (B) Left ventricular end-diastolic dimension (LVEDD) of WT and D2 mice 10 weeks after either sham or TAC surgery. (C) FS of WT and D2 mice 10 weeks after either sham or shunt surgery. (D) LVEDD of WT and D2 mice 10 weeks after either sham or shunt surgery. Statistical analysis: 2-way ANOVA followed by the Bonferroni procedure. Numbers within bars are the numbers of animals examined.
TAC but not Shunt results in further increases in cardiomyocyte number and wall thickness in D2 mice.
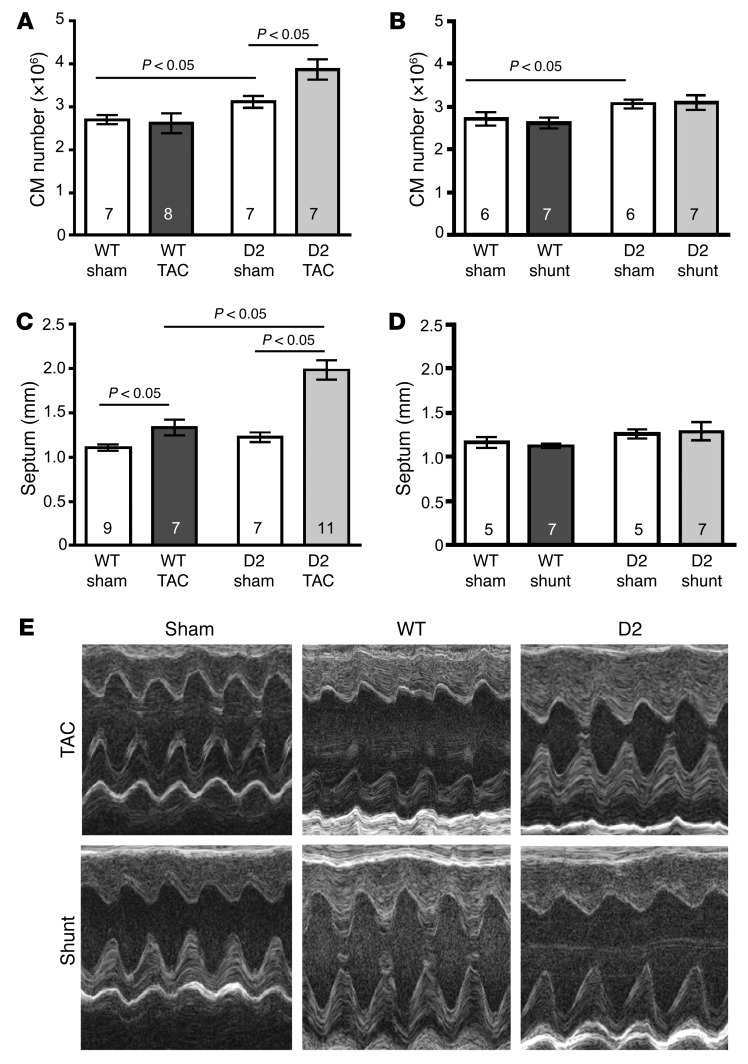
To investigate if the transgenic expression of CCND2 resulted in an increase in cardiomyocyte number, hearts from WT and D2 mice were harvested 10 weeks after sham or TAC and shunt surgery. Following retrograde collagenase perfusion, the number of cardiomyocytes in the resulting dispersed cell preparations was quantified. Cardiomyocyte number was similar in sham versus TAC WT mice (Figure 3A). Sham-operated D2 mice had a 15% increase in the number of cardiomyocytes as compared with the WT mice, consistent with the presence of transgene-induced postnatal cardiomyocyte cell cycle activity. Interestingly, TAC resulted in a further 24% increase in cardiomyocyte number in the D2 mice. In contrast, cardiomyocyte number was not increased in D2-sham versus D2-Shunt mice (Figure 3B).
Figure 3. Increased cardiomyocyte number and wall thickness in D2 mice following TAC but not shunt surgery.
(A and B) Total cardiomyocyte (CM) number for WT and D2 mice at 10 weeks following sham, TAC (A), or shunt (B) surgery. (C and D) Heart septum thickness of WT and D2 mice 10 weeks following sham, TAC (C), or shunt (D) surgery. (E) Representative echocardiographic recordings from a sham-operated, a WT, and a D2 mouse 10 weeks after TAC or shunt surgery. Statistical analysis: 2-way ANOVA followed by the Bonferroni procedure. Numbers within bars are the numbers of animals examined.
TAC resulted in cardiac hypertrophy in the WT mice, as evidenced by the presence of increased septum thickness in the echocardiographic recordings at the 10-week time point (Figure 3C). Interestingly, TAC-induced septum thickening was exaggerated in the D2 mice as compared with their WT littermates (62% vs. 21% increase, respectively). Septum thickness was not significantly different between WT and D2-Shunt mice (Figure 3D). Figure 3E shows representative echocardiographic recordings from WT and D2 mice.
TAC enhances cardiomyocyte cell cycle activity in D2 mice.
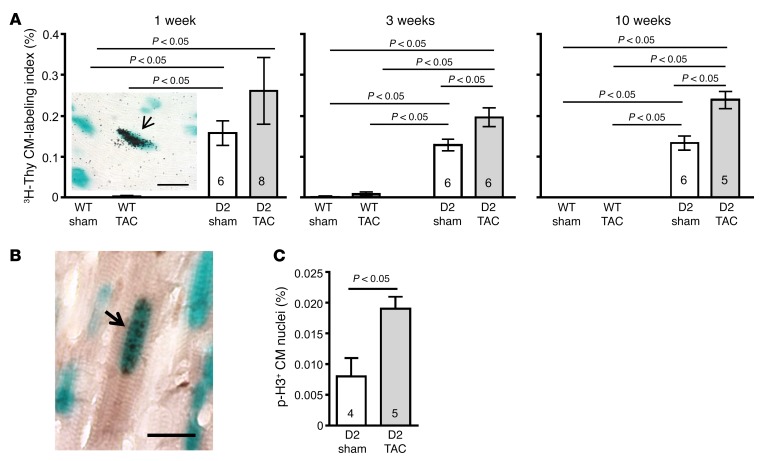
Tritiated thymidine (3H-Thy) incorporation analyses were used to quantify the number of S-phase cardiomyocytes; these studies utilized the αMHC-nLAC reporter transgene, which directs expression of a nucleus-localized β-galactosidase reporter exclusively to cardiomyocytes (14). At 1, 3, or 10 weeks following sham or TAC surgery, D2/αMHC-nLAC double-transgenic and αMHC-nLAC single-transgenic mice received a single injection of 3H-Thy and 4 hours later their hearts were harvested, sectioned, reacted with 5-bromo-4-chloro-3-indolyl-β-D-galactoside (X-GAL), and processed for autoradiography. Cardiomyocyte nuclei undergoing DNA synthesis were identified by the presence of silver grains over the blue X-GAL reaction product (Figure 4A shows a representative example of an S-phase cardiomyocyte). TAC resulted in significantly increased levels of cardiomyocyte DNA synthesis in the D2/αMHC-nLAC mice as compared with the D2/αMHC-nLAC sham group at 3 and 10 weeks after surgery. The level of cardiomyocyte DNA synthesis in sham- and TAC-operated WT mice was extremely low.
Figure 4. Increased cardiomyocyte cell cycle activity in D2 mice following TAC surgery.
(A) Cardiomyocyte (CM) tritiated thymidine (3H-Thy) labeling index (%) in WT and D2 mice at 1, 3, and 10 weeks following sham or TAC surgery (n = 4–8). Insert shows a representative example of an S-phase cardiomyocyte nucleus (arrow), as evidenced by 3H-Thy incorporation (blue signal is X-GAL reaction product; black signal is silver grains in the autoradiograph). Scale bar: 25 μm. (B) Phosphorylated histone H3 (p-H3) immunoreactivity 10 weeks after surgery. Representative example of a mitotic cardiomyocyte nucleus (arrow), as evidence by the presence of p-H3 immunoreactivity (blue signal is X-GAL reaction product; black signal is p-H3 immunoreactivity). Scale bar: 25 μm. (C) Quantification of p-H3 in sham versus TAC. Statistical analyses: 2-way ANOVA followed by the Bonferroni procedure or unpaired Student’s t test. Numbers within bars are the numbers of animals examined.
Sections from sham- and TAC-operated D2 hearts were also evaluated for the presence of phosphorylated histone H3 (p-H3) immunoreactivity; phosphorylation of histone H3 at serine residue 10 is a marker for mitosis (15). Once again, the X-GAL reaction was used to identify cardiomyocyte nuclei, and overlay of blue X-GAL reaction product and black/brown p-H3 immune signal was quantified in the D2 hearts (Figure 4B). Ten weeks after TAC surgery, D2 mice exhibited increased levels of cardiomyocyte p-H3 immunoreactivity as compared with sham-operated D2 mice (0.019 ± 0.002 vs. 0.008 ± 0.003, respectively, P < 0.05, Figure 4C). Collectively, these data suggest that the increased cardiomyocyte number in TAC- versus sham-operated D2 mice results from enhanced cardiomyocyte cell cycle activity.
D2-TAC mice exhibit reduced maladaptive remodeling.
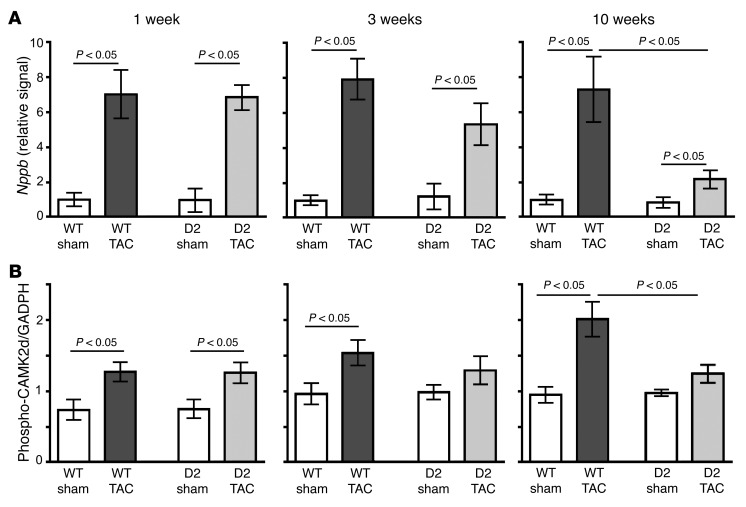
Quantitative reverse transcription PCR (qRT-PCR) analyses were performed to measure brain natriuretic peptide (BNP) levels following sham and TAC surgery; BNP is induced in response to cardiomyocyte load and thus provides a molecular marker for myocardial strain. BNP levels were similar in WT and D2 mice at 1 week after TAC (Figure 5A), implying a similar degree of TAC-induced myocardial stress in the 2 groups. BNP levels remained elevated in the WT mice at 3 and 10 weeks after TAC, consistent with the persistent cardiomyocyte load in these animals. In contrast, BNP levels dropped in the TAC D2 mice at 3 and 10 weeks after surgery, suggesting that transgene-induced cardiomyocyte proliferation provided a compensatory mechanism.
Figure 5. Normalization of heart failure molecular characteristics in D2 mice following TAC surgery.
(A) Quantitative RT-PCR analysis of Nppb expression in sham- and TAC-operated WT and D2 hearts at 1, 3, and 10 weeks after surgery (n = 5 in each group). (B) Quantitative analysis of Ca2+-calmodulin kinase 2δ (CaMK2d) phosphorylation in sham- and TAC-operated WT and D2 hearts at 1, 3, and 10 weeks after surgery (n = 5–6 per group). Statistical analysis: 2-way ANOVA followed by the Bonferroni procedure.
Activation of calcium/calmodulin–dependent protein kinase II (CaMKII) was shown to be a key pathological mechanism in TAC heart failure (5). Although CaMKII phosphorylation was increased comparably in WT and D2 mice at 1 week after TAC (Figure 5B and Supplemental Figure 3), CaMKII phosphorylation increased further in WT mice at 3 and 10 weeks after TAC, consistent with maladaptive signaling. In contrast, CaMKII phosphorylation returned to sham levels in D2-TAC mice at 3 and 10 weeks after surgery.
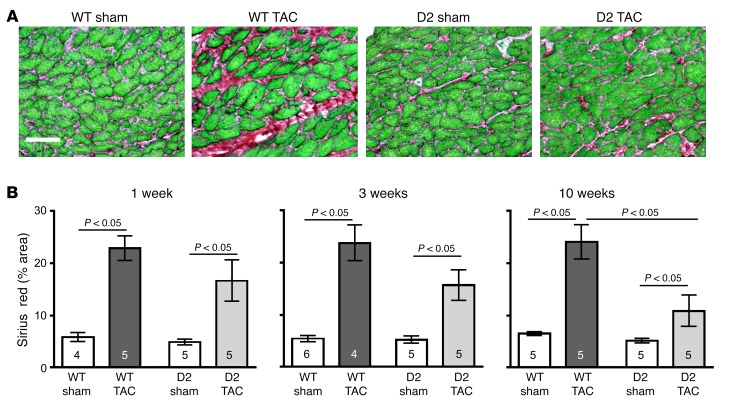
Sirius red/fast green histochemistry was employed to monitor interstitial cardiac fibrosis; representative images of hearts harvested at 10 weeks after TAC are shown in Figure 6A. Image analysis of the red pixel content in the Sirius red/fast green–stained sections revealed elevated levels of fibrosis in TAC mice as compared with their respective sham-operated controls at the 1-, 3-, and 10-week time points (Figure 6B). Interestingly, the level of fibrosis in the D2-TAC mice was significantly lower than in the WT-TAC mice at 10 weeks after surgery. There was no overt difference in perivascular fibrosis in D2 versus WT mice subjected to TAC (Supplemental Figure 4).
Figure 6. Reduced myocardial fibrosis in D2 mice after TAC surgery.
(A) Representative heart sections stained with Sirius red/fast green from a WT and D2 mouse at 10 weeks following sham or TAC surgery. Scale bar: 50 μm. (B) Quantification of myocardial fibrosis in WT and D2 hearts at 1, 3, and 10 weeks following sham or TAC surgery (peri-vascular regions were excluded from the analyses). Statistical analysis: 2-way ANOVA followed by the Bonferroni procedure. Numbers within bars are the numbers of animals examined.
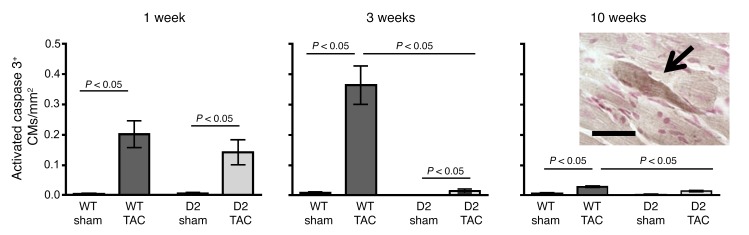
We have previously shown that TAC-induced interstitial fibrosis is accompanied by a marked increase in the level of cardiomyocyte apoptosis (13). Accordingly, apoptosis levels were approximated by screening for the presence of activated caspase 3 immunoreactivity. Since activated caspase 3 immunoreactivity is present in the cytoplasm, cardiomyocytes at early stages of apoptosis can be identified in histologic sections based on cell size and shape (Figure 7, insert) (16). Similar levels of cardiomyocyte activated caspase 3 immunoreactivity were observed in WT and D2 mice at 1 week after TAC. The immunoreactive cardiomyocytes were distributed throughout the left ventricle and septum with no apparent preferential anatomical localization. At 3 weeks after TAC, activated caspase 3 immunoreactivity was markedly reduced in the D2 mice as compared with the WT animals (TUNEL analyses confirmed distinct levels of apoptosis at this time point: 1.1% ± 0.10 vs. 0.2% ± 0.01, in WT-TAC animals vs. D2-TAC, respectively; n = 4 mice, P < 0.05). At the 10-week time point, the level of caspase 3 immunoreactivity remained higher in WT than in the D2 mice.
Figure 7. Reduced cardiomyocyte apoptosis in D2 mice following TAC surgery.
Heart sections from WT and D2 mice at 1, 3, and 10 weeks following sham or TAC surgery were processed for anti–activated caspase 3 immunoreactivity (n = 4–6 per group). The number of immunoreactive activated caspase 3 cardiomyocytes (CMs) per mm2 myocardium is shown. Insert shows a representative activated caspase 3 immunoreactive cardiomyocyte (arrow). Scale bar: 25 μm. Statistical analysis: 2-way ANOVA followed by the Bonferroni procedure.
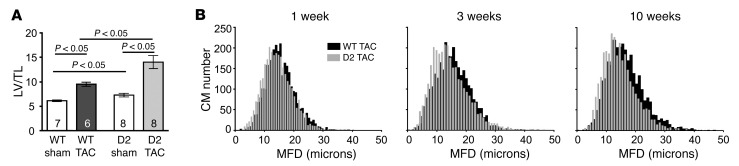
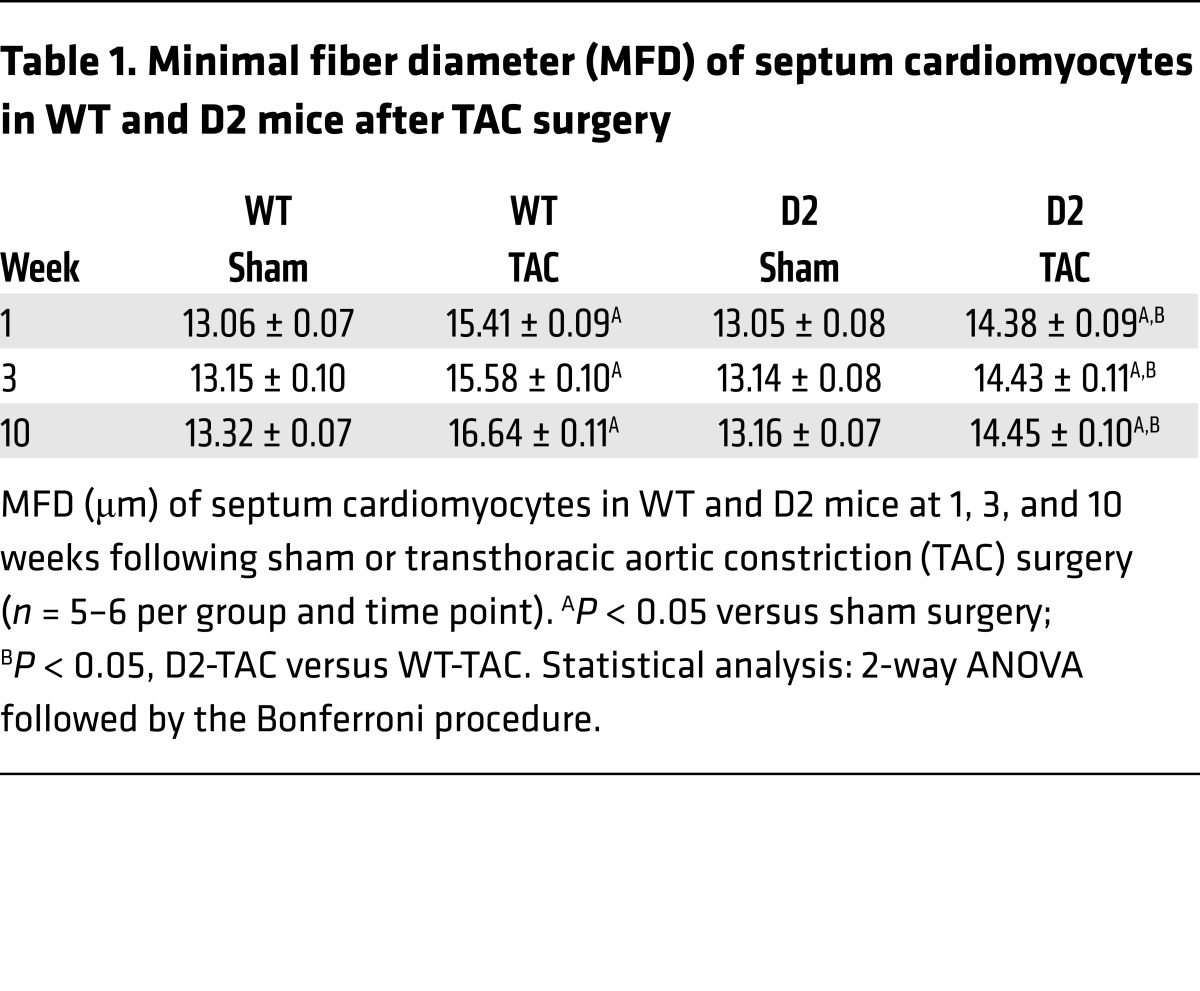
To further elucidate how transgene expression mitigates TAC-mediated heart failure, the hypertrophic phenotype in D2 and WT mice was analyzed in depth. Left ventricular weight/tibia length analyses confirmed that the D2-TAC mice exhibited exaggerated heart hypertrophy as compared with their WT siblings (Figure 8A). To characterize TAC-induced hypertrophy at the cellular level, the minimal fiber diameter (MFD) of septum cardiomyocytes was determined. Cardiomyocyte MFD in the sham-operated groups was similar in the WT and D2 mice at all time points. As expected, TAC resulted in an increase in MFD. However, the average increase in MFD was attenuated in the D2-TAC as compared with WT mice (Figure 8B and Table 1). Previous studies indicated a decrease in capillary density (measured as the number of CD31+ cells/cardiomyocyte) following prolonged pressure overload; this decrease is thought to contribute to the development of heart failure (17). Decreased capillary density was observed in WT but not in D2 mice following 10 weeks of TAC (Supplemental Figure 5).
Figure 8. Exaggerated heart hypertrophy, but reduced cardiomyocyte hypertrophy, in D2 mice following TAC surgery.
(A) Left ventricle weight/tibia length ratio (LV/TL) for WT and D2 mice at 10 weeks following sham or TAC surgery. (B) Minimal fiber diameter (MFD) distribution of WT and D2 septum cardiomyocytes (black and gray columns, respectively) at 1, 3, and 10 weeks following TAC surgery. Statistical analysis: 2-way ANOVA followed by the Bonferroni procedure.
Table 1. Minimal fiber diameter (MFD) of septum cardiomyocytes in WT and D2 mice after TAC surgery.
Cardiomyocyte proliferation signaling pathways in D2-TAC mice.
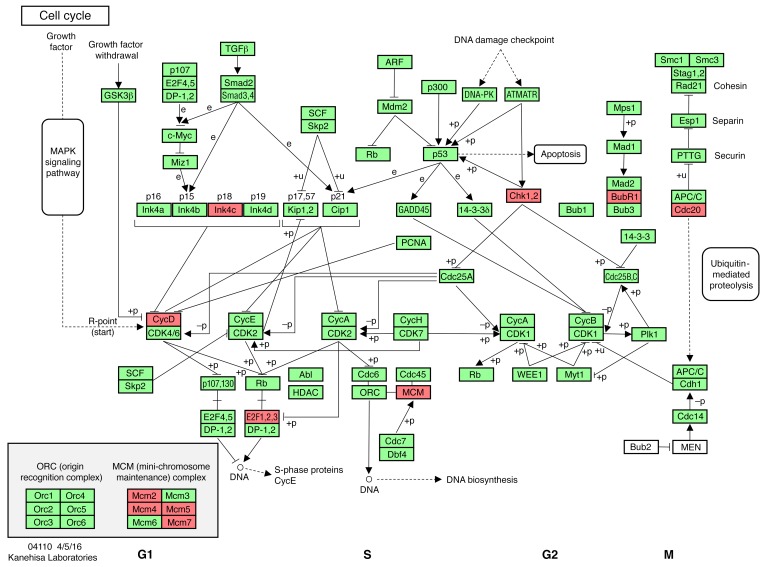
Unbiased analysis of RNA-sequencing (RNA-seq) data sets using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was employed to compare the transcriptomes of WT and D2 hearts under various conditions. Comparison of KEGG cell cycle pathway regulators in RNA-seq data sets from sham WT and sham D2 hearts 3 weeks after surgery identified 11 differentially regulated genes (Figure 9 and Supplemental Table 1). The data set was also interrogated for genes previously implicated in regulating cardiomyocyte cell cycle activity, including HIPPO pathway members (i.e., YAP and TAZ) (18), ALMS1 (Alstroem’s 1) (19), FGF1 (20), FGF2 (21), WNT/β-catenin (22), neuregulin (23), ERBB2 (a neuregulin coreceptor) (24), PITX2 (25), Meis1 (26), Notch/Jagged1 (27), periostin (28), agrin (29), follistatin-like 1 (FSTL1) (30), and TERT (31). Of these, only PITX2 expression was observed to be increased in D2-sham hearts versus WT-sham hearts (Supplemental Table 1).
Figure 9. Differentially regulated cell cycle genes in D2 versus WT mice.
Regulated genes (red) in the cell cycle KEGG pathway from RNA-seq analyses at 3 weeks after surgery (n = 5 hearts each) in WT-sham versus D2-sham hearts. Adapted with permission from Kanehisa Laboratories.
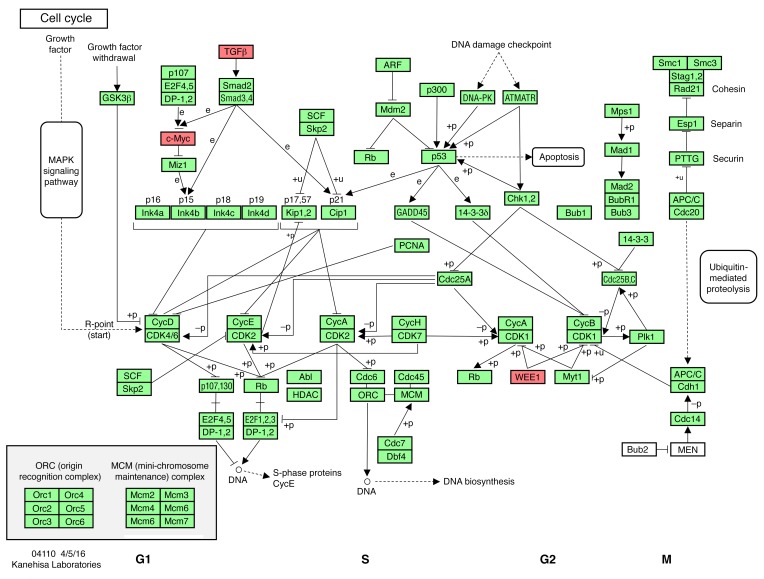
Given that D2 mice show a significant increase in cardiomyocyte number after chronically elevatsed afterload, but not after chronically elevated preload, the RNA-seq data sets from D2-TAC hearts versus D2-Shunt hearts were compared for changes in gene expression in the KEGG cell cycle pathway (Figure 10 and Supplemental Table 1), as well as in the cardiomyocyte cell cycle regulators listed above. In D2 hearts, TAC resulted in decreased WEE1 expression and increased TGFB2 expression, whereas Shunt did not impact their expression. Finally, additional unbiased analysis of the next-generation sequencing (NGS) data sets were performed to identify other differentially regulated genes in D2-TAC hearts versus D2-Shunt hearts. Differences were observed in 93 unique molecular pathways (Supplemental Figure 11 and Supplemental Table 2). Not surprisingly, changes in cell cycle, fibrosis, and extracellular matrix pathways were observed, as well as changes in the Akt and MAPK signaling pathways. Analysis of the top 100 differentially regulated genes also showed differences in fibrotic (COL1a1, TGFB2) and signaling (RCAN1) pathways (Supplemental Table 3).
Figure 10. Differentially regulated cell cycle genes in D2-TAC versus D2-Shunt.
Regulated genes (red) in the cell cycle KEGG pathway from RNA-seq analyses at 3 weeks after surgery in D2-TAC (n = 3) versus D2-Shunt (n = 5) hearts. Adapted with permission from Kanehisa Laboratories.
Discussion
The present study gives potentially new insight into adaptive mechanisms in structural heart diseases with increased load. Specifically, (a) pressure overload enhances CCND2-induced cardiomyocyte cell cycle activity, resulting in increased cardiomyocyte number (hyperplasia) in TAC- versus sham-operated animals; in contrast, volume overload does not enhance hyperplasia in CCND2-expressing mice. (b) Enhanced hyperplasia is associated with rescue from pressure overload–induced heart failure and improved survival. (c) Enhanced hyperplasia is associated with attenuated cardiomyocyte hypertrophy and sustained capillary density, and correlates with reversal of maladaptive gene expression and reverse myocardial remodeling during pressure overload. (d) Expression analysis by RNA-seq identified downregulation of the kinase WEE1 as a possible regulator of cardiomyocyte proliferation in D2-TAC mice. (e) Cell cycle induction may be of therapeutic value in structural heart disease with increased afterload.
Pressure overload, but not volume overload, results in increased cardiomyocyte numbers in D2 mice.
The D2 mice with increased afterload exhibited enhanced cardiomyocyte cell cycle activity as evidenced by increased levels of DNA synthesis and p-H3 immunoreactivity. This translated into an increase in cardiomyocyte number. These observations suggest that enhanced heart hypertrophy in the D2-TAC model is dependent upon ongoing cardiomyocyte renewal. Although the assays employed strongly suggest that the enhanced renewal results from proliferation of preexisting cardiomyocytes, they do not rule out the potential contribution of stem cell–mediated de novo cardiomyogenesis. However, studies claiming adult cardiomyogenic activity have indicated that the resulting cardiomyocytes are small and somewhat immature (32). In contrast, the MFDs of 3H-Thy–positive cardiomyocytes in the current study were not different from their neighboring 3H-Thy–negative cells (16.8 ± 0.54 vs. 16.6 ± 0.45 μm, respectively; P > 0.05, n = 58 cells), arguing against the presence of cardiomyogenic stem cell activity. The ultimate origin of the proliferating cardiomyocytes does not alter the main conclusion of the study, namely that sustained cardiomyocyte renewal has a profound impact on survival and function in response to chronic pressure overload.
In contrast, no increase in cardiomyocyte number was observed in Shunt- versus sham-operated D2 mice, suggesting that volume overload does not enhance cardiomyocyte renewal rates in mice expressing CCND2. In agreement with this, cardiomyocyte p-H3 immunoreactivity was significantly lower in Shunt compared with TAC D2 mice (0.009 ± 0.002 vs. 0.019 ± 0.003, respectively; P < 0.05, n = 5 mice per group; Supplemental Figure 6). Also, no difference in cardiomyocyte hypertrophy was detected in WT versus D2 mice with volume overload. Shunt increased cardiomyocyte width and length comparably (20% vs. 18%, and 24% vs. 25%, respectively) in WT versus D2 mice (Supplemental Figure 6). The degree of fibrosis, the expression of BNP, and the activation of signaling pathways are quite different in the Shunt model compared with the TAC model (13, 33). Although shunt surgery is known to induce significant volume overload, CCND2 expression did not impact either cardiac function or survival.
Unbiased analysis of the NGS data sets for KEGG cell cycle regulatory pathways provides mechanistic insight into the differential cardiomyocyte cell cycle activity observed in D2 mice with chronic afterload versus chronic preload. Of note, TAC was associated with decreased levels of WEE1 and increased levels of myc and TGFB2, whereas shunt surgery did not impact the expression of these genes. WEE1 is a negative regulator of cardiomyocyte cell cycle activity (34), while myc and TGF signaling have been implicated in regulating proliferation of fetal (35) and left atrial (36) cardiomyocytes, respectively. Decreased expression of negative and increased expression of positive regulators may contribute to the enhanced cell cycle activity observed in D2 mice subjected to chronically elevated afterload but not preload. Unbiased analysis of the NGS data sets from WT-sham and D2-sham hearts for KEGG cell cycle pathway regulators was also informative. As expected, expression of a number of cell cycle regulatory genes was observed to be altered in the transgenic hearts, consistent with the presence of sustained, transgene-induced cardiomyocyte cell cycle activity. Candidate gene analysis of the NGS data sets revealed that only 1 gene shown to modulate cardiomyocyte cell cycle activity in other studies, PitX2, was differentially expressed in WT-sham versus D2-sham hearts, suggesting that CCND2-mediated cardiomyocyte proliferation occurs largely independently of these pathways.
Increased cardiomyocyte number is cardioprotective during chronic pressure overload.
Morphological comparison of WT-sham and D2-sham hearts suggest that most increases in cardiomyocyte number in the D2 mice in the absence of injury occur after 9 weeks of age (Supplemental Figure 7). Although there was a tendency towards increased cardiac mass in D2 mice prior to TAC surgery, this did not provide intrinsic protection against pressure overload at 1 week after TAC, as evidenced by the levels of cardiomyocyte apoptosis, Nppb expression, CaMK2d phosphorylation, and interstitial myocardial fibrosis. In contrast, in D2-TAC mice cardiomyocyte number increased ultimately by 48%, while BNP expression and CaMKII phosphorylation declined. These data are consistent with the hypothesis that increased levels of cardiomyocyte renewal is cardioprotective in the context of chronic pressure overload.
Examination of the literature suggests that at least some of the cardioprotective activity in D2 mice with chronic pressure overload likely results from the presence of ongoing cardiomyocyte cell cycle activity. DUSP6-null mice exhibit a modest degree of cardiomyocyte hyperplasia during development and early neonatal life (but not in adult life), which results in an increase in cardiomyocyte number (37). When subjected to TAC, these animals exhibit lower levels of cardiomyocyte apoptosis and myocardial fibrosis as compared with WT animals. Nonetheless, there is a decline in cardiac function in the DUSP6-null mice as early as 4 weeks following TAC surgery and there is no overt improvement in viability as compared with WT control mice. In contrast, the D2 mice were protected from TAC-induced heart failure and had remarkably increased survival as compared with WT control mice. These observations support the notion that the presence of cardiomyocyte cell cycle activity in the D2 mice is critical for the rescue of heart failure and enhanced survival observed following the imposition of chronically increased afterload, whereas reduced levels of cardiomyocyte apoptosis and fibrosis is likely due to the presence of increased cardiomyocyte numbers at the time of TAC surgery (since these latter attributes are present in both the D2 and DUSP6-null mice). Although preliminary data using a conditional expression system indicate that de novo CCND2 expression is sufficient to initiate cardiomyocyte cell cycle activity in the adult heart (L. Field, unpublished observation), additional validation studies are needed before the model can be used to definitively address the contribution of ongoing cell cycle activity to the cardioprotection observed in the current study.
Exaggerated organ hypertrophy, preserved cardiac function, and enhanced survival are associated with sustained cardiomyocyte renewal and blunted cardiomyocyte hypertrophy in D2 mice.
A striking feature of the current study is the large degree of heart hypertrophy observed in the D2 mice. Analysis of echocardiography recordings revealed that the increase in septum thickness following 10 weeks of pressure overload was 2.9-fold greater in D2 mice as compared with that in WT mice (62% increase vs. 21% increase, respectively). Similarly, TAC resulted in a 1.7-fold greater increase in left ventricle mass in the D2 mice as compared with that in their WT littermates (92% increase vs. 55% increase, respectively). In contrast, TAC-induced hypertrophy at the level of individual cardiomyocytes was less in D2 as compared with the WT mice at all times examined. Thus, the pronounced hypertrophy in TAC mice results exclusively from the increased number of cardiomyocytes in the D2 hearts, as evidenced by the 48% increase in cardiomyocyte number (albeit with a concomitant 15% decrease in cardiomyocyte MFD).
A priori, one does not associate an exaggerated hypertrophic response with a beneficial functional outcome. Indeed, progressive heart failure is often associated with increased cardiac hypertrophy accompanied by myocardial fibrosis, cardiomyocyte apoptosis, and other indicators of adverse remodeling (7, 38, 39). The simplest interpretation of the data is that ongoing cardiomyocyte renewal led to sustained cardiac function culminating in enhanced viability. We speculate that cardiomyocyte renewal leads to exaggerated wall thickness, thereby reducing wall stress and hypertrophy of the individual cardiomyocyte. This is consistent with the notion that excessive cardiomyocyte hypertrophy is the critical event leading to apoptosis and fibrosis. Moreover, the fact that capillary density (CD31-positive cells per cardiomyocyte) was reduced in WT-TAC but unaltered in D2-TAC despite reduced cardiomyocyte hypertrophy may suggest that improved energy supply contributes to preserved function and improved survival in the D2-TAC mice. While we cannot rule out the potential impact of cell cycle–independent CCND2 activities (40), similarities in adverse gene expression, myocardial fibrosis, and cardiomyocyte apoptosis in D2 versus WT mice at 1 week after TAC suggest that this was likely not a major contributor to the observed long-term cardioprotection and enhanced survival.
The data presented suggest that cardiomyocyte cell cycle induction has a beneficial impact in the setting of chronic pressure overload. Therefore, it would be logical to develop cell cycle–based interventions to treat structural heart diseases with increased afterload. In particular, the finding that load-induced adverse remodeling can be reversed by cardiomyocyte proliferation seems promising. Additional efforts are nonetheless warranted to develop a better understanding of the molecular mechanisms that regulate the cardiomyocyte cell cycle, with the goal of developing more efficient interventions to enhance cell cycle–based regenerative growth.
Methods
Mice.
D2 mice (which were previously designated MHC-cycD2 mice) express a cDNA encoding CCND2 under the regulation of the cardiomyocyte-restricted αMHC (41). αMHC-nLAC mice express a nucleus-localized β-galactosidase reporter under the regulation of the αMHC promoter (14). All experimental mice were in a (DBA/2J × C3HeB/FeJ) F1 genetic background. C3H mice were obtained from Charles River.
TAC surgery.
Surgery was done using a minimally invasive approach as described previously (13, 42), with the exception that the degree of aortic constriction was reduced to yield comparable survival in WT mice with TAC versus Shunt. Briefly, 8-week-old mice were anesthetized using i.p. injections of ketamine (100 mg/kg, Pfizer Pharma) and xylazine (5 mg/kg, Riemser). A horizontal incision at the jugulum was used to display the transverse aorta. A needle was tied against the aorta using a 5-0 nonabsorbable suture. After removal of the needle, the skin was closed and the mice were kept on a heating plate until they recovered from anesthesia. Sham animals underwent the same procedure except banding of the aorta. (DBA/2J × C3HeB/FeJ) F1 mice exhibit a pronounced sex-based difference in body weight at 8 weeks of age (29.6 ± 0.8 vs. 23.3 ± 0.4, males vs. females, n = 10 mice per sex). Because aortic dimension correlates with body weight (43, 44), and because aortic dimensions influence the gradient, 25-gauge needles were used to induce TAC in males and 26-gauge needles were used in females. The resulting pressure gradients were measured by echocardiography, and comparable gradients were observed in male and female mice (Supplemental Figures 8 and 9).
Aortocaval shunt.
Surgery was done as described previously (45). Briefly, 8-week-old mice were anesthetized using isoflurane (AbbVie) insufflation. A longitudinal abdominal incision was made and the vessels prepared. The aorta was clamped and punctured with a needle (23-gauge) through the vena cava inferior. After removing the needle the external hole in the aorta was closed using cyanoacrylate glue (Pattex). A successful shunt operation was immediately seen by inflow of oxygenated blood from the abdominal aorta into the vena cava. The abdomen was then closed and the mice were kept on a heating plate until full recovery from anesthesia. Sham animals underwent the same procedure except for the puncture of the vessels. In a previous analysis we showed that a decline of left ventricular end-diastolic dimension (LVEDD) after an initial increase is associated with a decrease in right ventricular oxygen saturation as an indicator of shunt closure (Supplemental Figure 10). In the present study all animals after shunt surgery showed a persistent increase in LVEDD.
Echocardiography.
Two-dimensional (2-D) guided M-mode echocardiography was performed using a VS-VEVO 660/230 High Resolution Imaging System (Visualsonics). Mice were lightly anesthetized with 2.5% 2,2,2-tribromoethanol (Avertin, 0.01 ml/g i.p.) and were allowed to breathe spontaneously. LVEDD was measured from original tracings using the leading-edge convention of the American Society of Echocardiography. Left ventricular percentage FS and septum thickness were calculated as previously described (46). Pressure gradients were measured using a Vevo 2100 (VisualSonics) system with a 30-MHz respiration- and ECG-controlled probe under anesthesia with 1.5% isoflurane. The transverse aorta was also visualized with 2-D and color flow imaging. The distal transverse aortic flow velocity (distal to constriction in TAC mice) was measured by pulsed wave (PW) Doppler to assess the pressure gradient, which was estimated using the modified Bernoulli equation (pressure gradient = 4 × velocity2). The investigator was blinded with respect to genotype and treatment.
qRT-PCR.
RNA was extracted from frozen left ventricular tissues using TRIzol (Invitrogen), treated with RNA-free DNase (Sigma-Aldrich), and reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories). qRT-PCR was performed in triplicate using IQ SYBR Green Supermix and the iCycler iQ Detection System (Bio-Rad Laboratories). Primer sequences were as follows: natriuretic peptide type B (Nppb) 5′-CAGCTCTTGAAGGACCAAGG-3′ (forward), and 5′-ACTTCAGTGCGTTACAGCCC-3′ (reverse); hypoxanthine guanine phosphoribosyl transferase (Hprt) 5′-AGCCCCAAAATGGTTAAGGTTGC-3′ (forward) and 5′-TTGCAGATTCAACTTGCGCTCAT-3′ (reverse). Nppb gene expression was quantified using the 2-δδ-CT method (47) normalized to the constitutively expressed housekeeping gene Hprt. qRT-PCR analysis data are shown as the mean ± standard error of the mean (SEM).
Western immunoblot analysis.
Left ventricle samples were homogenized as described previously (13). Protein (20 μg) was resolved by SDS-PAGE using standard protocols (48). Western blotting was carried out according to standard protocols (49) using antibodies against CaMK2d (a gift from Don M. Bers, UC Davis, Davis, California, USA), p-CaMKII (MA1-047, Affinity BioReagents), and GAPDH (BT46-9995-55, BioTrend). For quantification an enhanced chemoluminescence detection system (GE Life Sciences) was used according to the manufacturer’s instructions.
Histology.
Hearts were harvested, cryoprotected in 30% sucrose, embedded, and sectioned at 10 μm using standard techniques (50). Sirius red/fast green staining was performed and quantified on sections postfixed in Bouin’s solution as described previously (11, 36). To quantify cardiomyocyte MFD, images from Sirius red/fast green–stained sections were captured, digitized, and analyzed with NIH ImageJ 1.36b software as described previously (16). At least 400 randomly selected cardiomyocytes from the interventricular septum of each animal were analyzed. To quantify cardiomyocyte apoptosis, 6 coronal sections from each heart were postfixed in 4% paraformaldehyde and screened for anti–activated caspase 3 immunoreactivity (catalog G7481, Promega), followed by a horseradish peroxidase–conjugated secondary antibody; signal was visualized with a diaminobenzidine reaction as described previously (16). TUNEL analysis was performed on adjacent sections using the ApopTag Apoptosis Detection kit according to the manufacturer’s protocol (Chemicon International). Cardiac endothelial cells were visualized using monoclonal antibodies against mouse CD31 (BD Pharmingen, clone MEC 13.3) and their number per cardiomyocyte manually counted on at least 4 optical fields per cross section. For p-H3 immunoreactivity, sections were postfixed with 4% paraformaldehyde and cardiomyocyte nuclei were visualized by reaction with X-GAL as described previously (7). Sections were then reacted with rabbit anti–p-H3 antibody (catalog 06-570, EMD Millipore) and signal was developed with a horseradish peroxidase–conjugated secondary antibody (ABC kit, Vector Laboratories) as described previously (16).
Quantification of cardiomyocyte number and dimensions.
Cardiomyocyte isolation was performed as described previously (51). In brief, mice were anesthetized with isoflurane, hearts were excised and mounted on a Langendorff perfusion system and retrogradely perfused with a nominally Ca-free Tyrodes’ solution containing (in mM) 113 NaCl, 4.7 KCl, 0.6 KH2PO4, 0.6 Na2HPO4 × 2 H2O, 1.2 MgSO4 × 7 H2O, 12 NaHCO3, 10 KHCO3, 10 HEPES, 30 taurine, 10 2,3-butanedione monoxime (BDM), 5.5 glucose, and 0.032 phenol red for 4 minutes at 37°C and pH 7.4. Thereafter, perfusion solution was switched to the same solution containing 7.5 mg/ml Liberase 1 (Roche diagnostics), 0.6% trypsin, and 0.125 mM CaCl2, and perfusion was continued for about 3 minutes until the heart became flaccid. Ventricular tissue was removed, cut into small pieces, and dispersed until no solid cardiac tissue was left. The total volume was increased to 12 ml and cardiomyocytes were counted 5 times in an improved Neubauer counting chamber (Karl Hecht GmbH). Cardiomyocyte dimensions were measured with ImageJ software. Width and length of at least 200 cardiomyocytes per heart were analyzed.
Cardiomyocyte DNA synthesis assay.
Cardiomyocyte DNA synthesis was monitored in experimental animals carrying the αMHC-nLAC reporter transgene. Cardiomyocyte nuclei in αMHC-nLAC mice are readily identified in tissue sections by incubation with the chromogenic β-galactosidase substrate X-GAL, which gives rise to a blue reaction product. For 3H-Thy incorporation (7), mice received a single injection of radiolabeled nucleotide (200 μCi i.p. at 20 Ci/mM, Perkin Elmer) and were sacrificed 4 hours later. Hearts were harvested, cryoprotected in 30% sucrose, embedded, and sectioned at 10 μm using standard histologic techniques (50). Sections were postfixed in 50 mM cacodylic acid/1% paraformaldehyde and reacted with X-GAL (1 mg/ml in 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, and 2 mM magnesium chloride in PBS). The sections were counterstained with Hoechst 33342 (Molecular Probes), and autoradiographic emulsion was applied and processed as described previously (7).
RNA-seq.
RNA-seq was carried out for WT (n = 5), D2-sham (n = 5), D2-TAC (n = 3), and D2-Shunt (n = 5) mice. Total RNA was isolated from left ventricular tissue using the RNeasy Fibrous Tissue Mini Kit (Qiagen) according to the manufacturer’s protocol. NGS was performed at the Transcriptome and Genome Analysis Laboratory (TAL) core facility in Goettingen. Library preparation was conducted according to the instructions of the TruSeq RNA Sample Preparation v2 Kit (catalog RS-122-2002, Illumina) using 1 μg total RNA as starting material. For accurate quantification of cDNA libraries, the fluorometry-based QuantiFluor dsDNA System from Promega was used. The size of the final cDNA libraries was determined using the DNA 1000 chip (280 bp) on the Bioanalyzer 2100 (Agilent). cDNA libraries were amplified and sequenced (single read; 1 × 50 bp; ca. 30 Mio reads per sample) using the cBot and HiSeq2000 (Illumina). Sequence images were transformed to bcl files with the software BaseCaller (Illumina), and then demultiplexed to fastq files with CASAVA v1.8.2. Quality checking was done via fastqc. Sequences were aligned to the genome reference sequence of Mus musculus (GRCm38/mm10). The alignment was performed using STAR alignment software (version 2.3.0e) allowing for 2 mismatches within 50 bases. Subsequently, conversion of resulting SAM files to sorted BAM files, filtering of unique hits, and counting were conducted with SAMtools (version 0.1.19) and HTSeq (version 0.6.1p1). Data were preprocessed and analyzed in the R/Bioconductor environment (www.bioconductor.org) using the DESeq2 package (version 1.8). Specifically, the data were normalized and tested for differentially expressed genes based on a generalized linear model likelihood ratio test assuming negative binomial data distribution. Candidate genes were filtered to a minimum of log2(fold change) greater than 1/–1 and a false discovery rate–corrected P value less than 0.05. Gene annotation was performed using Mus musculus entries from Ensembl (www.ensembl.org) via the biomaRt package (version 2.18.0). Raw RNA-seq data have been made available through the NCBI’s Gene Expression Omnibus (GEO GSE103545).
Pathway analysis of cell cycle genes.
Pathway analysis was conducted using significantly regulated genes (cutoff log2[fold change] > 1/–1; adjusted P value < 0.05). The analysis is based on KEGG (52) pathway annotations (KEGG pathway mmu04110, mmu04310) and was carried out using the KEGG Mapper – Search&Color Pathway tool (http://www.genome.jp/kegg/tool/map_pathway2.html).
Gene ontology (GO) and pathway enrichment analysis.
GO and pathway enrichment analysis was conducted using Cytoscape (53) and ClueGO (54) and significantly regulated genes (cutoff adjusted P value < 0.05) of the transcriptomes of D2-TAC mice compared with D2-Shunt mice. The analysis is based on the following databases: GO biological function, GO cellular component, GO immune system process, GO molecular function, KEGG, REACTOME, and WikiPathways using GO Term Fusion and showing pathways with a P value of less than 0.05, a Kappa score of 0.4, and a minimum of 20 genes representing at least 5% of all genes of the pathway.
Statistics.
Results are expressed as mean ± SEM. Survival was tested with the log-rank (Mantel-Cox) test. Multiple group comparisons were performed by 2-way analysis of variance (ANOVA) followed by the Bonferroni procedure for comparison of means. Comparisons between 2 groups were performed using the unpaired 2-tailed Student’s t test. All tests were 2-sided. Data were considered statistically significant at P less than 0.05.
Study approval.
The investigation conformed to the Guide for the Care and Use of Laboratory Animals (NIH publication number 85–23, revised 1996). The study was approved by the Lower Saxony State Office for Consumer Protection and Food Safety (G64/08 and 15/1860).
Author contributions
KT and LJF designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. WZ, MH, BAM, SK, SPR, and KS conducted experiments, acquired data, and analyzed data. DR conducted experiments. SE provided reagents. GH designed research studies, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (HL132927 to L.J.F.) and the German Research Foundation (DFG: SFB 1002 to K.T. and G.H.). We are grateful to Sabrina Koszewa, Joanna Jakubiczka-Smorag, and the service project of SFB1002 (Sarah Zaffar, Beate Knocke, Roland Blume, Marcel Zoremba) for excellent technical assistance.
Version 1. 10/30/2017
Electronic publication
Version 2. 12/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(12):4285–4296.https://doi.org/10.1172/JCI81870.
Contributor Information
Karl Toischer, Email: ktoischer@med.uni-goettingen.de.
Mark Hünlich, Email: huenlich@med.uni-goettingen.de.
Sara Khadjeh, Email: Sara.Khadjeh@med.uni-goettingen.de.
Katrin Schäfer, Email: kschaefer@med.uni-goettingen.de.
Deepak Ramanujam, Email: deepak.ramanujam@tum.de.
Stefan Engelhardt, Email: stefan.engelhardt@tum.de.
Loren J. Field, Email: ljfield@iupui.edu.
Gerd Hasenfuss, Email: hasenfus@med.uni-goettingen.de.
References
- 1.Grossman W, Jones D, McLaurin LP. Wall stress and patterns of hypertrophy in the human left ventricle. J Clin Invest. 1975;56(1):56–64. doi: 10.1172/JCI108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasenfuss G, et al. Energetics of isometric force development in control and volume-overload human myocardium. Comparison with animal species. Circ Res. 1991;68(3):836–846. doi: 10.1161/01.RES.68.3.836. [DOI] [PubMed] [Google Scholar]
- 3.Huke S, et al. Focal energy deprivation underlies arrhythmia susceptibility in mice with calcium-sensitized myofilaments. Circ Res. 2013;112(10):1334–1344. doi: 10.1161/CIRCRESAHA.113.301055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belke DD, et al. Insulin signaling coordinately regulates cardiac size, metabolism, and contractile protein isoform expression. J Clin Invest. 2002;109(5):629–639. doi: 10.1172/JCI13946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backs J, et al. The delta isoform of CaM kinase II is required for pathological cardiac hypertrophy and remodeling after pressure overload. Proc Natl Acad Sci U S A. 2009;106(7):2342–2347. doi: 10.1073/pnas.0813013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buitrago M, et al. The transcriptional repressor Nab1 is a specific regulator of pathological cardiac hypertrophy. Nat Med. 2005;11(8):837–844. doi: 10.1038/nm1272. [DOI] [PubMed] [Google Scholar]
- 7.Soonpaa MH, Field LJ. Assessment of cardiomyocyte DNA synthesis in normal and injured adult mouse hearts. Am J Physiol. 1997;272(1 Pt 2):H220–H226. doi: 10.1152/ajpheart.1997.272.1.H220. [DOI] [PubMed] [Google Scholar]
- 8.Bergmann O, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324(5923):98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473(7347):326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soonpaa MH, et al. Cyclin D1 overexpression promotes cardiomyocyte DNA synthesis and multinucleation in transgenic mice. J Clin Invest. 1997;99(11):2644–2654. doi: 10.1172/JCI119453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasumarthi KB, Nakajima H, Nakajima HO, Soonpaa MH, Field LJ. Targeted expression of cyclin D2 results in cardiomyocyte DNA synthesis and infarct regression in transgenic mice. Circ Res. 2005;96(1):110–118. doi: 10.1161/01.RES.0000152326.91223.4F. [DOI] [PubMed] [Google Scholar]
- 12.Hassink RJ, et al. Cardiomyocyte cell cycle activation improves cardiac function after myocardial infarction. Cardiovasc Res. 2008;78(1):18–25. doi: 10.1093/cvr/cvm101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toischer K, et al. Differential cardiac remodeling in preload versus afterload. Circulation. 2010;122(10):993–1003. doi: 10.1161/CIRCULATIONAHA.110.943431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soonpaa MH, Koh GY, Klug MG, Field LJ. Formation of nascent intercalated disks between grafted fetal cardiomyocytes and host myocardium. Science. 1994;264(5155):98–101. doi: 10.1126/science.8140423. [DOI] [PubMed] [Google Scholar]
- 15.Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci U S A. 1998;95(13):7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakajima H, Nakajima HO, Tsai SC, Field LJ. Expression of mutant p193 and p53 permits cardiomyocyte cell cycle reentry after myocardial infarction in transgenic mice. Circ Res. 2004;94(12):1606–1614. doi: 10.1161/01.RES.0000132279.99249.f4. [DOI] [PubMed] [Google Scholar]
- 17.Sano M, et al. p53-induced inhibition of Hif-1 causes cardiac dysfunction during pressure overload. Nature. 2007;446(7134):444–448. doi: 10.1038/nature05602. [DOI] [PubMed] [Google Scholar]
- 18.Xin M, et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110(34):13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shenje LT, et al. Mutations in Alström protein impair terminal differentiation of cardiomyocytes. Nat Commun. 2014;5:3416. doi: 10.1038/ncomms4416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Formiga FR, et al. Controlled delivery of fibroblast growth factor-1 and neuregulin-1 from biodegradable microparticles promotes cardiac repair in a rat myocardial infarction model through activation of endogenous regeneration. J Control Release. 2014;173:132–139. doi: 10.1016/j.jconrel.2013.10.034. [DOI] [PubMed] [Google Scholar]
- 21.Sheikh F, et al. Inhibition of TGFbeta signaling potentiates the FGF-2-induced stimulation of cardiomyocyte DNA synthesis. Cardiovasc Res. 2004;64(3):516–525. doi: 10.1016/j.cardiores.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notch1/beta-catenin/Isl1 determines cardiac progenitor cell fate. Nat Cell Biol. 2009;11(8):951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bersell K, Arab S, Haring B, Kühn B. Neuregulin1/ErbB4 signaling induces cardiomyocyte proliferation and repair of heart injury. Cell. 2009;138(2):257–270. doi: 10.1016/j.cell.2009.04.060. [DOI] [PubMed] [Google Scholar]
- 24.D’Uva G, et al. ERBB2 triggers mammalian heart regeneration by promoting cardiomyocyte dedifferentiation and proliferation. Nat Cell Biol. 2015;17(5):627–638. doi: 10.1038/ncb3149. [DOI] [PubMed] [Google Scholar]
- 25.Tao G, et al. Pitx2 promotes heart repair by activating the antioxidant response after cardiac injury. Nature. 2016;534(7605):119–123. doi: 10.1038/nature17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahmoud AI, et al. Meis1 regulates postnatal cardiomyocyte cell cycle arrest. Nature. 2013;497(7448):249–253. doi: 10.1038/nature12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nemir M, et al. The Notch pathway controls fibrotic and regenerative repair in the adult heart. Eur Heart J. 2014;35(32):2174–2185. doi: 10.1093/eurheartj/ehs269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kühn B, et al. Periostin induces proliferation of differentiated cardiomyocytes and promotes cardiac repair. Nat Med. 2007;13(8):962–969. doi: 10.1038/nm1619. [DOI] [PubMed] [Google Scholar]
- 29.Bassat E, et al. The extracellular matrix protein agrin promotes heart regeneration in mice. Nature. 2017;547(7662):179–184. doi: 10.1038/nature22978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei K, et al. Epicardial FSTL1 reconstitution regenerates the adult mammalian heart. Nature. 2015;525(7570):479–485. doi: 10.1038/nature15372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh H, et al. Telomerase reverse transcriptase promotes cardiac muscle cell proliferation, hypertrophy, and survival. Proc Natl Acad Sci U S A. 2001;98(18):10308–10313. doi: 10.1073/pnas.191169098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beltrami AP, et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell. 2003;114(6):763–776. doi: 10.1016/S0092-8674(03)00687-1. [DOI] [PubMed] [Google Scholar]
- 33.Mohamed BA, et al. Molecular and structural transition mechanisms in long-term volume overload. Eur J Heart Fail. 2016;18(4):362–371. doi: 10.1002/ejhf.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puente BN, et al. The oxygen-rich postnatal environment induces cardiomyocyte cell-cycle arrest through DNA damage response. Cell. 2014;157(3):565–579. doi: 10.1016/j.cell.2014.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson T, Allard MF, Sreenan CM, Doss LK, Bishop SP, Swain JL. The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol Cell Biol. 1990;10(7):3709–3716. doi: 10.1128/MCB.10.7.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakajima H, Nakajima HO, Dembowsky K, Pasumarthi KB, Field LJ. Cardiomyocyte cell cycle activation ameliorates fibrosis in the atrium. Circ Res. 2006;98(1):141–148. doi: 10.1161/01.RES.0000197783.70106.4a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maillet M, Purcell NH, Sargent MA, York AJ, Bueno OF, Molkentin JD. DUSP6 (MKP3) null mice show enhanced ERK1/2 phosphorylation at baseline and increased myocyte proliferation in the heart affecting disease susceptibility. J Biol Chem. 2008;283(45):31246–31255. doi: 10.1074/jbc.M806085200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Senyo SE, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493(7432):433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swynghedauw B. Molecular mechanisms of myocardial remodeling. Physiol Rev. 1999;79(1):215–262. doi: 10.1152/physrev.1999.79.1.215. [DOI] [PubMed] [Google Scholar]
- 40.Angelis E, et al. A cyclin D2-Rb pathway regulates cardiac myocyte size and RNA polymerase III after biomechanical stress in adult myocardium. Circ Res. 2008;102(10):1222–1229. doi: 10.1161/CIRCRESAHA.107.163550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pasumarthi KB, Nakajima H, Nakajima HO, Jing S, Field LJ. Enhanced cardiomyocyte DNA synthesis during myocardial hypertrophy in mice expressing a modified TSC2 transgene. Circ Res. 2000;86(10):1069–1077. doi: 10.1161/01.RES.86.10.1069. [DOI] [PubMed] [Google Scholar]
- 42.Rockman HA, et al. Segregation of atrial-specific and inducible expression of an atrial natriuretic factor transgene in an in vivo murine model of cardiac hypertrophy. Proc Natl Acad Sci U S A. 1991;88(18):8277–8281. doi: 10.1073/pnas.88.18.8277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hinton RB, et al. Mouse heart valve structure and function: echocardiographic and morphometric analyses from the fetus through the aged adult. Am J Physiol Heart Circ Physiol. 2008;294(6):H2480–H2488. doi: 10.1152/ajpheart.91431.2007. [DOI] [PubMed] [Google Scholar]
- 44.Stypmann J, et al. Age and gender related reference values for transthoracic Doppler-echocardiography in the anesthetized CD1 mouse. Int J Cardiovasc Imaging. 2006;22(3-4):353–362. doi: 10.1007/s10554-005-9052-9. [DOI] [PubMed] [Google Scholar]
- 45.Tanaka N, et al. Transthoracic echocardiography in models of cardiac disease in the mouse. Circulation. 1996;94(5):1109–1117. doi: 10.1161/01.CIR.94.5.1109. [DOI] [PubMed] [Google Scholar]
- 46.Schmidt AG, et al. Cardiac-specific overexpression of calsequestrin results in left ventricular hypertrophy, depressed force-frequency relation and pulsus alternans in vivo. J Mol Cell Cardiol. 2000;32(9):1735–1744. doi: 10.1006/jmcc.2000.1209. [DOI] [PubMed] [Google Scholar]
- 47.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 49.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carneiro LC, Kelley J, Junqueira RO. Basic histology. 7th ed Norwalk, CT(4): Appleton & Lange;1992. [Google Scholar]
- 51.Maier LS, Zhang T, Chen L, DeSantiago J, Brown JH, Bers DM. Transgenic CaMKIIdeltaC overexpression uniquely alters cardiac myocyte Ca2+ handling: reduced SR Ca2+ load and activated SR Ca2+ release. Circ Res. 2003;92(8):904–911. doi: 10.1161/01.RES.0000069685.20258.F1. [DOI] [PubMed] [Google Scholar]
- 52.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bindea G, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.