Abstract
Angiocrine factors, such as Notch ligands, supplied by the specialized endothelial cells (ECs) within the bone marrow and splenic vascular niche play an essential role in modulating the physiology of adult hematopoietic stem and progenitor cells (HSPCs). However, the relative contribution of various Notch ligands, specifically jagged-2, to the homeostasis of HSPCs is unknown. Here, we show that under steady state, jagged-2 is differentially expressed in tissue-specific vascular beds, but its expression is induced in hematopoietic vascular niches after myelosuppressive injury. We used mice with EC-specific deletion of the gene encoding jagged-2 (Jag2) to demonstrate that while EC-derived jagged-2 was dispensable for maintaining the capacity of HSPCs to repopulate under steady-state conditions, by activating Notch2 it did contribute to the recovery of HSPCs in response to myelosuppressive conditions. Engraftment and/or expansion of HSPCs was dependent on the expression of endothelial-derived jagged-2 following myeloablation. Additionally, jagged-2 expressed in bone marrow ECs regulated HSPC cell cycle and quiescence during regeneration. Endothelial-deployed jagged-2 triggered Notch2/Hey1, while tempering Notch2/Hes1 signaling in HSPCs. Collectively, these data demonstrate that EC-derived jagged-2 activates Notch2 signaling in HSPCs to promote hematopoietic recovery and has potential as a therapeutic target to accelerate balanced hematopoietic reconstitution after myelosuppression.
Keywords: Hematology, Vascular Biology
Keywords: Adult stem cells, Bone marrow, endothelial cells
Introduction
Defining the cellular and molecular components of the bone marrow (BM) niche that regulate the quiescence, self-renewal, and lineage differentiation of hematopoietic stem and progenitor cells (HSPCs) can facilitate targeted interventions toward improved hematopoietic recovery after injury and the expansion of sufficient numbers of HSPCs for transplantation (1, 2). At the core of this dynamic cell-cell interaction is the instructive vascular niche provided by bone marrow endothelial cells (BMECs), which deploy angiocrine signals such as jagged-1 (1–3), Kit ligand (KitL) (1, 2, 4, 5), and CXCL12 (3, 6), to regulate hematopoietic stem cell (HSC) self-renewal and lineage differentiation under both homeostatic and regenerative conditions.
In mammals, endothelial cells (ECs) express 4 types of Notch ligands, jagged-1 (Jag1), jagged-2 (Jag2), Delta-like 1 (Dll1), and Delta-like 4 (Dll4) (7). In mice, endothelial-specific knockout of Jag1 (3) results in the reduction of the number and repopulating capacity of HSPCs at steady state, as well as diminished hematopoietic recovery following myelosuppressive stress. In these mice, the residual Notch activity within HSPCs suggested complementary mechanisms of Notch activation, likely contributed by other Notch ligands, including jagged-2, DLL1, and DLL4. DLL4 haploinsufficiency results in defects in arterial and yolk sac vascular development (8–12). DLL1 was shown to regulate fetal artery development (13). This suggested that DLL1 or DLL4 regulates vascular development in part in a cell-autonomous manner. DLL4 has also been shown to regulate adult hematopoiesis (14). Nonetheless, accumulating evidence demonstrates that signaling afforded by expression of various Notch ligands might perform collectively to induce Notch activation in a dose-dependent manner (15, 16). In this paradigm, the dose of each ligand consummates to induce the level of physiological Notch signaling that ultimately dictates HSPC function. Thus, we hypothesized that the stoichiometry of other Notch ligands, specifically jagged-2 supplied by ECs, might participate in HSPC maintenance by modulating the degree of Notch signaling and HSPC recovery.
To this end, we first characterized the expression of Jag2 mRNA among adult vascular ECs from different tissue types. In the BM, Jag2 mRNA and jagged-2 protein are enriched in BMECs compared with non-BMECs. During hematopoietic regeneration, the expression of jagged-2 in BMECs is increased compared with that in homeostatic conditions. Next, using a transgenic mouse line that expresses a Cre recombinase under a Cdh5 promoter (17), we deleted exons 1–2 of the Jag2 gene specifically in ECs (18). This deletion generated a truncated Jag2 mRNA and truncated jagged-2 protein in ECs. Under steady state, there were minor changes in the hematopoietic indexes in the peripheral blood and in the BM. However, in a more defined EC-HSPC coculture model that mimics HSPC regeneration following myeloablative injury, jagged-2 expressed in ECs was required to promote the HSPC in vitro expansion. Following in vivo myelosuppressive injuries, endothelial jagged-2 preserves the survival rate of mice via maintenance of the HSPCs at both early and later stages of HSPC regeneration. Previous publications suggested that jagged-2 was expressed in both hematopoietic progenitor cells and ECs (19, 20); using transplantation studies, we demonstrated that the engraftment and/or expansion of HSPCs requires endothelial jagged-2. Mechanistically, endothelial jagged-2 induced Notch2/Hey1 signaling and repressed Notch2/Hes1 signaling in HSPCs. The differential requirement of jagged-2 for HSPC function under homeostatic compared with myelosuppressive conditions correlated with the level of jagged-2 expression under these conditions. Indeed, when Jag2 was deleted from both ECs and hematopoietic cells, there was a more profound alteration of repopulating capacity of HSPCs under steady state conditions. Therefore, jagged-2 serves as an activating module in Notch signaling to promote hematopoietic recovery.
Results
Heterogeneity of Jag2 mRNA expression in organotypic ECs.
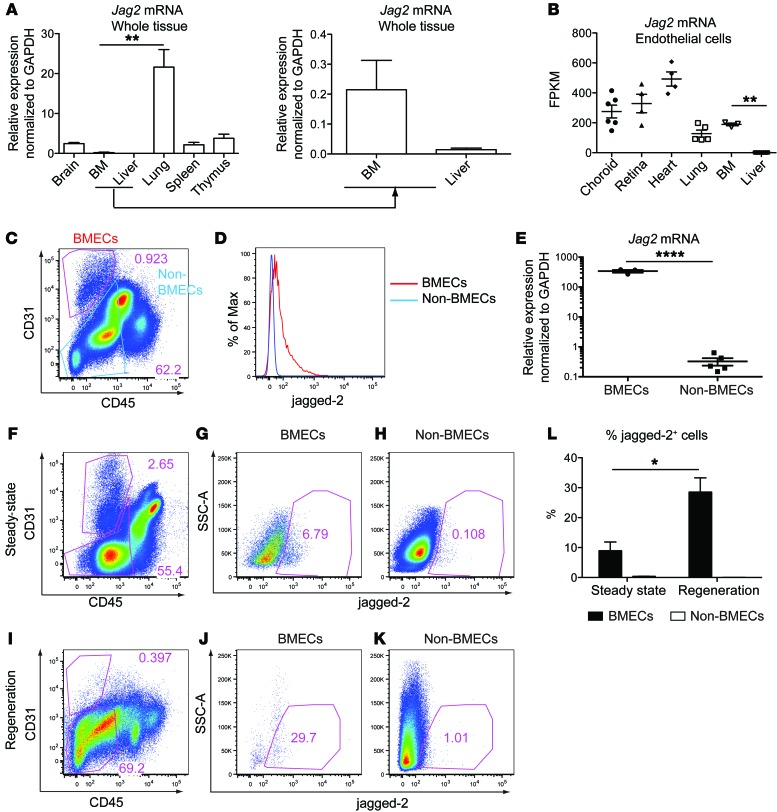
To systematically examine Jag2 mRNA expression, we carried out reverse transcriptase quantitative PCR (RT–qPCR) in whole tissue lysate from various organs (Figure 1A). Jag2 mRNA is abundantly expressed in lung, modestly expressed in spleen, thymus, and brain, and expressed at lower levels in BM and liver. Using a previously established protocol (21), we isolated CD45–CD31+VE-cadherin+ primary vascular ECs from various mouse organs and subjected them to RNA sequencing analysis. The expression of Jag2 mRNA (Figure 1B) was comparable between freshly isolated ECs from lung and BM, suggesting the relative enrichment of Jag2 expression in BMECs compared with other cell types in the BM. Examination of jagged-2 protein expression pattern in BM via flow cytometry revealed higher expression of jagged-2 in CD31+CD45– BMECs than in CD31–CD45– non-BMECs (Figure 1, C, D, and F–H). The level of Jag2 mRNA in sorted BMECs was significantly higher than in non-BMECs (Figure 1E).
Figure 1. Jagged-2 is dynamically expressed in BMECs.
(A) The expression level of Jag2 mRNA in different mouse whole organs (n = 3). The mRNA expression is calculated using GAPDH as internal control. (B) The FPKM (fragments per kilobase of exon per million fragments mapped) value for Jag2 mRNA in primary ECs from various organs. The number of dots indicates the number of biological replicates. (C) Representative flow cytometric plots for the gating of CD31+CD45– BMECs and CD31–CD45– non-BMECs (n = 4). (D) Histogram of jagged-2 expression on BMECs and non-BMECs. (E) qPCR quantification of Jag2 mRNA from sorted BMECs (n = 3) and non-BMECs (n = 5). The RNA expression level is calculated using GAPDH as internal control. (F–H) Representative flow plots for jagged-2 expression in BMECs and non-BMECs (n = 4) under homeostatic conditions. (I–K) Jagged-2 expression within BMECs and non-BMECs at 2 weeks after 650 cGy sublethal irradiation (n = 5). (L) Comparison of percentage of jagged-2+ cells among BMECs under steady state and during regeneration after myeloablative injuries. Error bars indicate the SEM. *P < 0.05, **P < 0.01, and ****P < 0.0001, by 2-tailed unpaired t test. The numbers in the flow plots represent percentages of cells.
When comparing the Notch ligand profile of primary microvascular ECs with cultured ECs, Jag2 mRNA expression (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI92309DS1) was significantly reduced in the cultured ECs compared with primary isolated choroid ECs, retinal ECs, and lung ECs, but not in BMECs, suggesting that the in vivo microenvironment selectively sustains the expression level of Jag2 in ECs outside the BM. In contrast, the expression level of Jag1 mRNA showed either no significant change or increased expression in cultured ECs compared with primary isolated ECs (Supplemental Figure 1B), except for retinal ECs. The expression of Dll4, Dll1, Notch1, and Notch4 (Supplemental Figure 1, C–F) was also changed in cultured ECs compared with freshly isolated ECs, suggesting an overall change of Notch signaling profiles in cultured versus primary isolated ECs. However, as compared with primary isolated BMECs, the cultured BMECs retained the molecular signatures of Jag1, Jag2, and Dll4 expression, with reduction of Notch1 expression.
In vivo, the level of Jag2 mRNA expression in ECs also dynamically changed from steady state to myeloablative conditions (Supplemental Figure 1, G–L). During the recovery phase after myelosuppression, the level of Jag2 and Dll1 mRNA expression was significantly increased in liver ECs in comparison with steady state (Supplemental Figure 1, G and J). The level of Dll4 mRNA was significantly decreased, whereas the level of Jag1, Notch4, and Notch1 expression did not change, at steady state and following myelosuppression (Supplemental Figure 1, H–L). These data correlate with the previous published microarray data demonstrating the dynamic changes of Notch ligand Jag2 expression in ECs under steady state and following myeloablative stress (21). In the BM, following myeloablative injuries, the percentage of BMECs expressing jagged-2 was increased at 2 weeks after sublethal irradiation compared with that in homeostatic conditions (Figure 1, I–L), suggesting differential requirement of jagged-2 under steady state and hematopoietic regeneration.
In summary, both Jag2 mRNA and jagged-2 protein are enriched in BMECs compared with non-BMECs. The dynamic changes of jagged-2 level in BMECs after myelosuppression prompted us to examine the physiological role of endothelial jagged-2 in regulating HSPC homeostasis and regeneration. To this end, we used a conditional knockout mouse model in which the exon 1–2 region of the Jag2 gene is selectively deleted from ECs, and examined the HSPC behavior under homeostasis and during regeneration.
Normal angiogenesis in mice with endothelial deletion of Jag2.
To conditionally delete Jag2 from ECs, we crossed transgenic mice carrying VE-cadherin promoter–driven Cre recombinase (17) with mice carrying Jag2fl allele (18) that flanked the first 2 exons of Jag2 gene (Supplemental Figure 2A). We denote the mice with EC-specific deletion of Jag2 as Jag2ECKO mice. We next used the reporter analysis (Supplemental Figure 2, B–D) and qPCR analysis of primary isolated ECs (Supplemental Figure 2, E–J) to confirm the deletion efficiency of the VE-cadherin-Cre transgenic line (17) in ECs from various vascular beds. The percentage of td-tomato+ cells among BMECs was about 89.5%, indicating efficient deletion of VE-cadherin-Cre. Via qPCR analysis, we observed that in both BMECs and lung microvascular ECs, the expression level of Jag2 at the indicated exon regions was reduced by more than 90%; the expression level of the other 3 Notch ligands Jag1, Dll1, and Dll4 was not significantly changed (Supplemental Figure 2, H and I). In BMECs, the Notch targets Hes1 and Hey1 were reduced when Jag2 was deleted from BMECs (Supplemental Figure 2J). In lung ECs, the Notch targets Hes1 and Hey1 were not significantly changed.
From the RNA sequencing results of primary lung microvascular ECs, we observed that truncated Jag2 mRNA was generated from Jag2ECKO mice. The truncated Jag2 mRNA retained the expression from exons 6–26 (Supplemental Figure 2K). Using 2 antibodies that recognize the N- or C-terminus of jagged-2 protein, respectively, we confirmed that truncated jagged-2 protein was generated in ECs from Jag2ECKO mice, which lacks the conserved Notch receptor binding DSL and Notch N-terminal (MNNL) domains (ref. 22 and Supplemental Figure 2, L–Q). Most important, we noted that under steady-state conditions there were no apparent defects in the angiogenic profile and vascular perfusion of the Jag2ECKO mice in the key hematopoietic organs (Supplemental Figure 3). These data indicate that in contrast to Dll4 deficiency, in which there is a significant defect in angiogenesis, lack of Jag2 is dispensable for the maintenance of adult organotypic ECs.
Endothelial jagged-2 is not required for HSPC homeostasis.
We next examined HSPC function under steady-state conditions by quantifying the lineage distributions of hematopoietic cells and the number of phenotypic HSPCs in the BM, followed by competitive transplantation studies. Lack of jagged-2 in the ECs minimally altered the total number of hematopoietic cells in the BM (Figure 2A). The total number of BM mononucleated cells (BMMNCs) was increased in the Jag2ECKO group (Figure 2A) as compared with Jag2fl/fl mice. In the peripheral blood, there were no differences in the WBC number in Jag2ECKO mice compared with Jag2fl/fl mice (Figure 2C). Closer inspection of the different lineage contributions did not reveal any significant differences in Gr-1+/CD11b+ myeloid cells, CD3+ T cells, or B220+ B cells in the BM (Figure 2B) or in the peripheral blood (Figure 2D).
Figure 2. Endothelial-supplied jagged-2 is not required to maintain HSPC function under homeostatic conditions.
(A–D) The total number of BMMNCs (A) and the WBC count (C) were quantified in adult Jag2fl/fl or Jag2ECKO mice. Lineage distribution of hematopoietic cells within the BM (B) and peripheral blood (D) was quantified in Jag2fl/fl or Jag2ECKO mice. For A and B, n = 3. For C, the number of dots indicates the number of biological replicates. For D, n = 9 for Jag2fl/fl, n = 8 for Jag2ECKO. (E) Representative flow cytometric gating of phenotypic Lin–c-Kit+Sca1+CD150+CD48– long-term HSCs (LT-HSCs). (F) Quantification of the number of phenotypic LT-HSCs per million BMMNCs in the femur of Jag2fl/fl or Jag2ECKO (n = 8 for each group). (G) Schematic view of competitive repopulating assay. (H) The percentage of CD45.2+ hematopoietic cells in the peripheral blood of CD45.1+ mice at 15.5 weeks after transplantation (n = 6 CD45.1 recipients for Jag2fl/fl and n = 8 CD45.1 recipients for Jag2ECKO mice). (I) Representative flow cytometric plots showing the multilineage engraftment. The CD45.2+ cells were further gated to reveal donor-derived CD3+ T cells and CD19+ B cells. (J) Quantification of multilineage engraftment within the CD45.2+ cells. Error bars indicate the SEM. *P < 0.05 and **P < 0.01, by unpaired 2-tailed t test. The numbers in the flow plots represent percentages of cells.
Next, we examined the number and function of the more primitive populations of long-term hematopoietic stem cells (LT-HSCs) defined as Lin–c-Kit+Sca1+CD150+CD48– cells (KLS-SLAM) (Figure 2E). There was no difference in the total number of phenotypic LT-HSCs within Jag2ECKO mice compared with Jag2fl/fl mice (Figure 2F), suggesting that jagged-2 expressed on ECs was not required to maintain the cell number of primitive LT-HSCs under steady-state conditions.
The repopulating capacity of HSPCs was determined by competitive repopulating assay (Figure 2G). At 15.5 weeks after transplantation (Figure 2H), there were no apparent differences in the percentage of CD45.2+ cells in the peripheral blood of CD45.1 mice, demonstrating that BMMNCs from Jag2ECKO mice contributed to peripheral blood engraftment at a similar level to that of control mice. Multilineage engraftment analysis of the CD45.2+ cells revealed a significant reduction in the percentage of T cells contributed from Jag2ECKO BM cells (Figure 2, I and J) compared with that of the control mice, suggesting the role of endothelial jagged-2 in regulating lymphoid-bias potential of HSPCs (23).
Collectively, the quantification of phenotypic LT-HSCs and functional validation of HSPCs from Jag2ECKO mice have demonstrated a minimal requirement of EC–jagged-2 in maintaining HSPC number and repopulating capacity under steady state. The lack of obvious HSPC phenotype under steady state suggested that other Notch ligands on ECs or jagged-2 expressed by stromal cells, hematopoietic cells, or other cytokines expressed by the ECs (Supplemental Figure 4) compensate for the loss of function of Jag2 on ECs. To further reveal the role of endothelial jagged-2 in HSPC function, a more defined in vitro coculture system using BMECs and HSPCs was carried out to test the hypothesis that EC–jagged-2 is necessary for the in vitro expansion of HSPCs. Subsequently, in vivo myelosuppressive models were used to reveal the role of endothelial jagged-2 in promoting HSPC regeneration following myelosuppressive stress.
Jagged-2 expressed on BMECs is required for in vitro expansion of HSPCs.
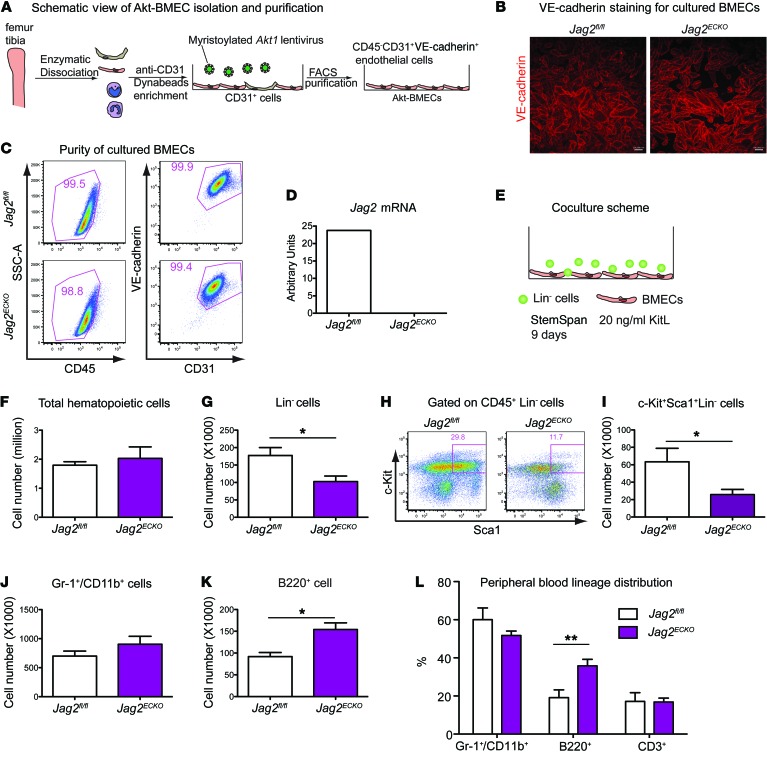
A more defined in vitro coculture assay that modeled hematopoietic regeneration was used to dissect the role of jagged-2 on BMECs in promoting the expansion of HSPCs (Supplemental Figure 5A). To this end, we enriched BMECs from Jag2fl/fl mice or Jag2ECKO mice (3, 17). Dynabead-based cell enrichment (24) was carried out using an mAb directed to mouse CD31 (Figure 3A). After cell separation, lentivirus-encoding myristoylated Akt1 gene was transduced into the primary cells, and we term these cells Akt-BMECs. Following a minimum of 5 cell passages and additional CD31+VE-Cadherin+ cell sorting to ensure purification, Akt-BMECs from both control and Jag2ECKO mice displayed cobblestone pattern with contact inhibition and robust VE-cadherin expression (Figure 3B). The survival and proliferation of BMECs from Jag2ECKO mice were not significantly different from those of the control mice (data not shown). After several passages, the identity and purity of BMECs were maintained (Figure 3C); more than 97% of the cultured BMECs were CD45–CD31+VE-cadherin+ ECs. RT–qPCR analysis using primers flanking exons 1–2 of Jag2 mRNA demonstrated that Jag2 mRNA was abundantly expressed in Jag2fl/fl, and its expression was almost undetectable in BMECs from Jag2ECKO mice (Figure 3D). Western blot confirmed loss of the N-terminus of jagged-2 protein in the in vitro cultured BMECs (Supplemental Figure 2N).
Figure 3. Jagged-2 expressed by BMECs promotes in vitro expansion of HSPCs.
(A) Schematic view of the method to enrich BMECs from Jag2fl/fl or Jag2ECKO mice. (B) The cultured BMECs were stained with anti–VE-cadherin antibody. Scale bars: 100 μm. (C) After passages, the purity of BMECs from Jag2fl/fl or Jag2ECKO mice was confirmed by flow cytometric staining of CD45, CD31, and VE-cadherin (n = 5). The percentage of CD45–CD31+VE-cadherin+ ECs is shown. (D) Using primers flanking the first 2 exons of Jag2 mRNA, the expression level of Jag2 was quantified via real-time qPCR (n = 3). (E) Schematic view of the coculture setup using BMECs and lineage-negative (Lin–) hematopoietic cells (n = 3 biological replicates of Lin– cells were used for the coculture; 1 Jag2fl/fl BMEC and 2 lines of Jag2ECKO BMECs were used as feeders). (F–I) At day 9 after coculture, the total number of CD45+ cells (F), Lin– cells (G), and KLS cells (I) within the culture was summarized. The representative flow cytometric plots of KLS HSPCs are shown in H. (J and K) At day 9 after coculture, the total number of Gr-1+/CD11b+ myeloid cells (J) and B220+ cells (K) was quantified. (L) Quantification of peripheral blood multilineage distribution in 18-month-old Jag2fl/fl or Jag2ECKO mice (n = 7 for Jag2fl/fl, n = 8 for Jag2ECKO). Error bars indicate the SEM. *P < 0.05 and **P < 0.01, by 2-tailed t test. The numbers in the flow plots represent percentages of cells.
Following the coculture of Lin– cells on BMECs derived from Jag2fl/fl or Jag2ECKO mice (Figure 3E), the total numbers of CD45+ hematopoietic cells, Lin– cells, and HSPCs were quantified. Though there were no significant differences between the numbers of total expanded CD45+ hematopoietic cells (Figure 3F), there was a significant reduction in the number of expanded Lin– cells (Figure 3G) and KLS HSPCs (Figure 3, H and I, and Supplemental Figure 5, B–D) when Lin– cells were cocultured on BMECs from Jag2ECKO compared with Jag2fl/fl mice. Loss of jagged-2 on BMECs reduced HSPC expansion to 40% of that expanded on control BMECs. To determine whether jagged-2 on BMECs modulates HSPC differentiation, the total of expanded cells was examined by antibody staining with Gr-1 and CD11b for myeloid cells and B220 for B cells. The number of expanded Gr-1+/CD11b+ myeloid cells was not significantly different when Lin– cells were cocultured on BMECs from Jag2fl/fl or Jag2ECKO mice (Figure 3J). The total number of B220+ cells was significantly increased when Lin– cells were cultured on BMECs derived from Jag2ECKO mice, compared with BMECs from Jag2fl/fl mice (Figure 3K and Supplemental Figure 5E). Thus, jagged-2 on BMECs prevented the differentiation of Lin– cells or HSPCs into B220+ cells. Indeed, in a long-term analysis to reveal the role of endothelial jagged-2 in regulating HSPC differentiation, we observed that within the peripheral blood of 18-month-old Jag2ECKO mice, the percentage of B220+ B cells was significantly increased compared with that in Jag2fl/fl mice (Figure 3L and Supplemental Figure 5F), suggesting increased differentiation of HSPCs into B cells (25). Taken together, these results suggest that jagged-2 supplied by BMECs maintains HSPCs by inhibiting the differentiation of Lin– cells or HSPCs into B220+ cells.
Endothelial jagged-2 promotes recovery of HSPCs after myelosuppression.
The coculture model demonstrates the requirement of jagged-2 to expand HSPCs in vitro. To test the role of endothelial jagged-2 in protecting hematopoietic reconstitution after myelosuppression, 2 regimens were used to cause myeloablative injuries to mice. One involves weekly injection of the chemotherapy reagent 5-fluorouracil; the other is γ-irradiation administered at a sublethal dose of 650 cGy. Eight- to ten-week-old Jag2fl/fl mice or Jag2ECKO mice were subjected to weekly 5-flurouracil injections. The survival rate of the mice was monitored weekly (Figure 4A). The whole cohort of Jag2ECKO mice died faster and earlier than the cohort of control mice. There was a reduced survival rate of Jag2ECKO mice compared with Jag2fl/fl mice (Figure 4A). This result underscores the protective role of endothelial jagged-2 in preventing myelosuppressive damage.
Figure 4. Endothelial jagged-2 ensures proper HSPC recovery after myelosuppression.
(A) Jag2fl/fl or Jag2ECKO mice were subjected to weekly 5-fluorouracil (5-FU) injections at the dose of 150 mg/kg. Kaplan-Meier survival curve was generated after monitoring of the survival rate of the mice. n = 12 for Jag2fl/fl, n = 9 for Jag2ECKO mice. (B) Jag2fl/fl or Jag2ECKO mice were subjected to sublethal irradiation at 650 cGy, and their survival rate was monitored. This experiment was performed 4 times. Each time, n = 4 mice were used for each genotype. The data are combined and shown in B. (C–H) To monitor the kinetics of hematopoiesis regeneration following irradiation, the quantification of KLS cells and LT-HSCs at day 10 (C and D) (n = 4 for each genotype), day 16 (E and F) (n = 5 for each group), and day 29 (G and H) (n = 5 for each group) after sublethal irradiation is further shown. Error bars indicate the SEM. *P < 0.05 and **P < 0.01, by unpaired 2-tailed t test.
We further tested whether endothelial jagged-2 could protect the mice from sublethal irradiation (26). Following 650 cGy sublethal irradiation, WT mice undergo a regeneration phase that lasts about 4–6 weeks (24). In a combination of 4 batches of mice (a total of 16 mice were used for each genotype), more Jag2ECKO mice died than control mice (Figure 4B). There was a trend toward reduction of survival rate of Jag2ECKO mice compared with that of the control mice. Because the death of mice was detected around day 12 after irradiation in the Jag2ECKO mice, and on the basis of the regeneration kinetics observed in previous publications (3, 26), we examined the hematopoietic recovery in the BM on day 10, day 16, and day 29 after irradiation. During the recovery of the hematopoietic reconstitution, there was a slight decrease in the number of RBC and platelets in the Jag2ECKO mice compared with Jag2fl/fl control mice, yet the WBC count did not show significant differences between the 2 groups (Supplemental Figure 6, A–E). At day 10, the total number of hematopoietic cells within the BM of Jag2fl/fl or Jag2ECKO was similar; however, the more primitive KLS cells (Figure 4C) showed a trend toward reduction in Jag2ECKO mice compared with Jag2fl/fl mice. The analysis of LT-HSCs revealed a significant decrease in the number of LT-HSCs at day 10 after irradiation in Jag2ECKO mice compared with Jag2fl/fl mice (Figure 4D).
At day 16 after irradiation, the total numbers of hematopoietic cells, Lin– cells, KLS cells, and LT-HSCs were the lowest among the 3 time points examined, correlating with the lowest number of WBC, RBC, and platelets in the peripheral blood (Supplemental Figure 6, A–C) and the need for rapid regeneration of KLS cells at this stage of recovery. At day 16 after irradiation, there were no significant differences between the KLS numbers (Figure 4E and Supplemental Figure 6, F and G) or between the LT-HSC numbers (Figure 4F) in Jag2fl/fl and Jag2ECKO mice. At day 29, the number of LT-HSCs showed a trend toward increase in Jag2ECKO mice compared with Jag2fl/fl mice (Figure 4, G and H).
As Notch signaling regulates vascular sprouting angiogenesis and arterial maintenance (16), we examined whether the reduced number of phenotypic LT-HSCs in Jag2ECKO mice was due to defective vasculature functions. At day 10 after 650 cGy irradiation, hematopoietic organs including spleen (Figure 5A), liver (Figure 5B), and BM (Figure 5C) were examined for vascular perfusion function. Vasculature was well perfused in the spleen and liver in Jag2ECKO mice compared with Jag2fl/fl mice, suggesting that there were no vascular patterning defects or perfusion abnormalities in those organs. In the BM, VE-cadherin staining indicated preservation of perfusion functions of BM of Jag2ECKO mice and Jag2fl/fl mice. In nonhematopoietic organs such as lung (Figure 5D), there were slight alterations of lung alveolar structures, with staining of VE-cadherin in Jag2ECKO mice compared with control mice.
Figure 5. Vascular perfusion function is preserved in Jag2ECKO mice following myelosuppression.
At day 10 after 650 cGy irradiation, the perfusion function of spleen vasculature (A), liver vasculature (B), BM vasculature (C), and lung vasculature (D) was preserved in Jag2ECKO mice, as demonstrated by fluorescence-labeled VE-cadherin antibodies perfused into the vasculature (n = 4 for each group). Scale bars: 50 μm.
We next examined the cell cycle status of the KLS cells at day 16 and day 29 after irradiation. On day 16, when the least number of KLS cells and LT-HSCs resided in the BM, only 45.2% of KLS cells remained at the Ki67–G0 quiescent stage in Jag2fl/fl mice (Figure 6, A and B), compared with 70% at G0 stage at day 29 after 650 cGy irradiation (Figure 6D). At day 16, there was significant reduction of percentage (27.42%) of Ki67– G0 stage quiescent KLS cells in Jag2ECKO mice compared with Jag2fl/fl (45.2%) mice (Figure 6, A and B), suggesting the requirement of endothelial jagged-2 to maintain the fine balance between quiescence and proliferation of KLS cells. The apoptosis rate of KLS cells at day 14 after irradiation was similar between the 2 groups (Figure 6C and Supplemental Figure 6, H and I). At day 21, there was a significant increase in both the forward scatter and the side scatter (Supplemental Figure 6, J and K) of KLS cells from Jag2ECKO mice compared with Jag2fl/fl mice, suggesting the altered morphology of KLS cells during regeneration when jagged-2 is deleted from ECs. At day 29, the difference between the cell cycle status of KLS cells was diminished between Jag2ECKO and Jag2fl/fl mice (Figure 6D).
Figure 6. Endothelial jagged-2 modulates the cell-cycle status of HSPCs during regeneration.
(A) On day 16 after irradiation, the flow cytometric gating for KLS, CD150–CD48– short-term KLS (ST-HSCs), and CD150–CD48+ KLS multipotent progenitor cells is shown. (B) At day 16 after irradiation, the percentage of Ki67– G0 cells among KLS cells was quantified (n = 5 for each group). (C) At day 14 after irradiation, the percentage of apoptotic cells among Lin– cells, cKit+Lin– cells, KLS cells, and Sca1+Lin– cells was quantified. (D) On day 29 after 650 cGy irradiation, the percentage of Ki67– G0 cells among Lin– cells and KLS cells was quantified (n = 5 for each group). Error bars indicate the SEM. *P < 0.05, by unpaired 2-tailed t test. The numbers in the flow plots represent percentages of cells.
Taken together, the kinetics of total hematopoietic cells, Lin– cells, KLS cells, and LT-HSCs after sublethal irradiation indicated that although the number of total hematopoietic cells did not reduce to almost undetectable at day 16 after irradiation, the more primitive cells such as KLS cells and LT-HSCs had undergone a rapid exhaustion (day 0 to day 16). This was followed by a decision of KLS cells to delicately balance between their quiescent stem cell identity and rapid proliferation to replenish the progenitor cell pool (day 10 to day 16), and they finally returned to a state similar to steady-state conditions (~day 29 and onward). Therefore, the decrease of LT-HSC number in the BM of Jag2ECKO mice at day 10 indicates that loss of jagged-2 correlates with reduced HSPC maintenance at the early stage of myelosuppression. At the mid-phase that precedes the rapid regeneration, loss of endothelial jagged-2 causes rapid proliferation of KLS cells, which leads to the upward trend in LT-HSCs at the final stage of hematopoietic regeneration.
Previous studies of the microvascular EC gene expression profiles before and after 650 cGy sublethal irradiation have revealed significant upregulation of Jag2 mRNA in BMECs at day 10 after irradiation compared with steady state levels. However, the level of Jag2 expression gradually fell to steady-state level, as indicated by reduced Jag2 mRNA expression in liver ECs and BMECs at day 28 after irradiation compared with day 10 after irradiation (21). These data supported the critical role of endothelial jagged-2 in mediating hematopoietic recovery after irradiation, and the notion that the biological outcome of the loss of function of Jag2 correlated with its dose of expression in specific biological contexts.
Engraftment of HSPCs is not dependent on the Notch2/Hes1 signaling axis in HSPCs.
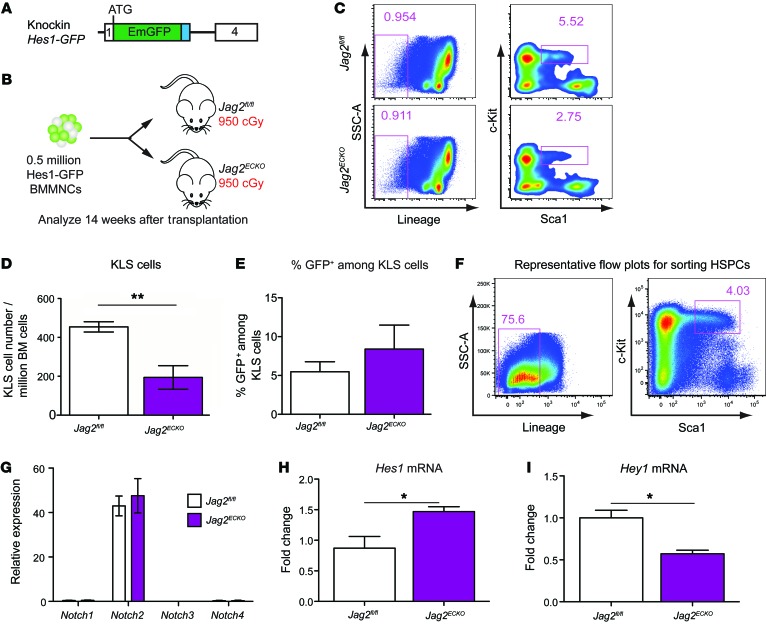
Jagged-2 is expressed on both ECs and hematopoietic cells (19). To test whether endothelial jagged-2 is necessary for engraftment and/or expansion of HSPCs, we used an experimental model in which an equal number of BMMNCs was transplanted into lethally irradiated Jag2fl/fl or Jag2ECKO mice (Figure 7B). Specifically, we took advantage of Hes1-GFP knockin reporter (27, 28) mice to visualize the activation status of Hes1 via flow cytometry (Figure 7A). Fourteen weeks after the transplantation of Hes1-GFP BM mononuclear cells into Jag2fl/fl or Jag2ECKO mice, the number of HSPCs per million total hematopoietic cells in the BM of Jag2ECKO mice was significantly reduced compared with that in control mice (Figure 7, C and D), suggesting the requirement of endothelial jagged-2 in maintaining HSPC number after transplantation. Loss of jagged-2 on ECs caused reduction of c-Kit+Sca1+Lin– HSPCs by 58% in comparison with Jag2fl/fl mice. There was no significant difference in the percentage of Hes1+ cells within KLS cells that were transplanted into Jag2ECKO mice compared with Jag2flfl mice (Figure 7E).
Figure 7. Endothelial jagged-2 induces Notch2/Hey1 signaling in HSPCs, enhancing engraftment.
(A and B) Schematic view of the knockin Hes1-GFP reporter mouse line (A) and the transplantation assays (B) used to test the role of BMEC jagged-2 in promoting engraftment/expansion of HSPCs. n = 4 for Jag2fl/fl group, n = 3 for Jag2ECKO group. (C) Representative flow plots for the gating strategies for KLS cells in the Jag2fl/fl and Jag2ECKO mice. (D) The number of HSPCs per million BM cells was quantified. (E) The percentage of GFP+ cells among the gated KLS cells is summarized. The data were collected following the transplantation experiment listed in B. (F) Representative flow cytometric plots showing the sorting strategies for KLS cells from Jag2fl/fl or Jag2ECKO mice. (G–I) Real-time qPCR analysis of the sorted HSPCs from Jag2fl/fl or Jag2ECKO mice was carried out for Notch receptors (G) and Notch targets (H and I). For G and I, n = 3 biological replicates for each group. For H, n = 5 for each group. Error bars indicate the SEM. *P < 0.05 and **P < 0.01, by unpaired 2-tailed t test. The numbers in the flow plots represent percentages of cells.
We next aimed to delineate the Notch signaling pathways involved in angiocrine communication between vascular ECs and HSPCs (3) by identifying the Notch receptors and Notch downstream targets expressed in HSPCs in both Jag2flfl and Jag2ECKO mice. To this end, we sorted out KLS HSPCs (Figure 7F) from Jag2fl/fl mice and Jag2ECKO mice under steady state. Via RT–qPCR analysis, we confirmed that the Notch receptors expressed in HSPCs from control mice were mainly Notch2, with undetectable expression of Notch1, Notch3, or Notch4 (Figure 7G). These data correlated with the recent publication using Notch receptor promoter–driven Cre to map out the Notch receptor expression in the hematopoietic system (27). There were no changes of Notch receptor expression levels in Jag2ECKO mice compared with the control mice. For the Notch downstream targets, there was a significant reduction of the Notch target Hey1 in sorted HSPCs in Jag2ECKO mice compared with Jag2fl/fl mice (Figure 7I), suggesting that endothelial jagged-2 activated the Notch2/Hey1 axis in HSPCs. Notably, loss of jagged-2 in ECs caused upregulation of Hes1 level in the HSPCs both under steady state (Figure 7H) and during hematopoietic regeneration following irradiation (Supplemental Figure 6L), suggesting the role of endothelial jagged-2 in curbing the expression of Hes1 in HSPCs.
Collectively, these data demonstrated the non–cell-autonomous role of endothelial jagged-2 in maintaining HSPC cell number following myelosuppressive regimens. For the downstream signaling events, endothelial jagged-2 activates the Notch2/Hey1 signaling axis in HSPCs, and balances the upward trend of Notch2/Hes1 signals in HSPCs. The unique dichotomy of Hey1 and Hes1 signals in HSPCs might lead to as-yet unrecognized specific changes in HSPC behaviors, which will manifest under various physiological or stress situations.
Endothelial and hematopoietic cell–derived jagged-2 maintains HSPC number.
It was previously reported that jagged-2 is also expressed in HSPCs (refs. 19, 29, and Figure 8A). To investigate the consequence of compound deletion of Jag2 from both ECs and hematopoietic cells, we used another VE-cadherin-Cre transgenic line (30), which was shown to recombine in ECs of various organs and hematopoietic cells (Figure 8, B and C). We denote the resulting VE-cadherin-Cre+/– Jag2fl/fl mice as Jag2KO mice, to distinguish them from the Jag2ECKO mice (Supplemental Figure 2) used for the majority of this study.
Figure 8. Jagged-2 supplied by ECs and hematopoietic cells maintains HSC number.
(A) Summary of mRNA expression of Notch ligands Jag1, Jag2, Dll1, Dll3, and Dll4 in HSPCs. Data are from the published RNA sequencing data of SP-KLS-CD150+ cells (29). VE-cadherin-Cre mice (30) were crossed with Jag2fl/fl mice to delete exons 1 and 2 of the Jag2 gene from ECs and hematopoietic cells. (B and C) After crossing of VE-cadherin-Cre mice with Rosa26CAG<stop>tdtomato mice, the deletion efficiency of VE-cadherin-Cre in ECs (B) and hematopoietic cells within the BM (C) was quantified. (D–F) The platelets (D), WBC (E), and RBC (F) were monitored after sublethal irradiation with 650 cGy at the indicated time points. (G) The survival curve of Jag2fl/fl or Jag2KO mice was plotted. (H and I) Competitive repopulating assay was carried out using BMMNCs from Jag2fl/fl or Jag2KO mice (n = 7 for each group). For B and C, n = 3 biological replicates for each group. For D–G, n = 5 for Jag2fl/fl, n = 4 for Jag2KO mice. For I, n = 7 each for Jag2fl/fl and Jag2KO mice. Error bars indicate SEM. For D–F, at each individual time point, the difference between Jag2fl/fl and Jag2KO mice was compared using a 2-tailed t test. The resulting P value is shown. For D–F and J, the overall differences of the 2 curves were also compared using 2-way ANOVA, and the P values of the observed variance based on the genotype are as follows: for D, P = 0.0025; for E, P < 0.0001; for F, P = 0.0373; for I, P = 0.0016. *P < 0.05; **P < 0.01 (**P value was determined by 2-way ANOVA).
The deletion of Jag2 from ECs and hematopoietic cells led to increased WBC count following myelosuppressive injury (Figure 8, D–F) with a trend of reduced survival for the Jag2KO mice compared with Jag2fl/fl mice (Figure 8G). When an equal number of BMMNCs from Jag2KO mice or Jag2fl/fl mice were transplanted into lethally irradiated CD45.1 mice, BMMNCs from Jag2KO mice resulted in elevated peripheral blood chimerism into CD45.1 host mice (Figure 8, H and I), suggesting the role of jagged-2 in regulating HSPC repopulating capacity. This suggested a dose effect of jagged-2 as Notch ligand; when Jag2 was deleted from both hematopoietic cells and ECs (Figure 8G), the resulting interference with marrow reconstitution was more profound than when Jag2 was deleted only from ECs (Figure 4B).
Discussion
Balanced self-renewal and fine-tuned differentiation of HSPCs are essential for the proper reconstitution of hematopoiesis. While numerous niche-derived pathways have been shown to choreograph HSPC regeneration, the precise mechanism(s) by which niche signals drive long-term regeneration of the HSPCs is being actively scrutinized and refined. Proper activation of Wnt, BMP, and Notch signaling pathways conveyed by the niche cells, such as the vascular niche, orchestrates the hematopoietic reconstitution. Among these pathways, the Notch signaling pathway plays a seminal role in mandating HSPC self-renewal at the expense of differentiation, thereby safeguarding long-term hematopoiesis. However, the mechanism by which Notch pathways mastermind hematopoietic recovery is complex and depends on coordinated and tissue-specific angiocrine expression of Notch ligands such as jagged-2. Here, we have shown that jagged-2 represents an essential arm of Notch signaling that ensures proper HSPC recovery after myeloablative injury.
Contribution of Notch signaling to hematopoietic recovery.
Notch signaling was shown to mediate the regeneration of long-term repopulating HSPCs following myelosuppressive conditions such as 5-fluorouracil treatment (31). Notch signaling enhanced the expansion of murine HSPCs by ECs (32, 33). Therefore, the Notch signaling was required for HSPC regeneration and in vitro expansion.
The differential contribution of Notch ligands expressed by ECs to HSPCs during in vitro expansion and regeneration is not well studied. Mouse ECs express 4 types of Notch ligands, jagged-1, jagged-2, DLL1, and DLL4. It was previously shown that immobilized Notch ligand DLL1 in combination with hematopoietic cytokines expands the absolute number of HSPCs, and that those expanded hematopoietic progenitor cells engraft into BM of immunodeficient NOD-SCID mice (34). EC-specific deletion of Dll4 increased tip cell formation in retinal angiogenesis, suggesting autocrine function of DLL4 in regulating angiogenesis and vascular remodeling (8, 16). Similarly, Dll1 is expressed in embryonic arteries and is required to induce VEGF-A expression in arteries (13). Postnatally, expression of DLL1 on ECs ensures arteriogenesis in hind limb ischemia models. Therefore, DLL4 and DLL1 might primarily be engaged in neoangiogenesis.
Because of the apparent vascular defects in Dll4- or Dll1-knockout mice, we used conditional knockout mouse models (Jag1ECKO or Jag2ECKO) in which the perfusion and vascular patterning were not severely affected to examine the perfusion-independent role of ECs in HSPC homeostasis and reconstitution. Indeed, in Jag1ECKO mice (3), vascular patterning was changed minimally compared with that in control mice. However, the number of HSPCs was reduced under steady state and hematopoietic regeneration was delayed following myelosuppressive stresses. Furthermore, residual HSPC function and Notch activity within HSPCs in the Jag1 conditional knockout mice suggested a complementary mechanism of Notch activation. To this end, we hypothesize that the other member of the mammalian serrate Notch ligands expressed on ECs, jagged-2, complements jagged-1 and maintains HSPCs during homeostasis and reconstitution.
Jagged-2/Notch regulation of steady-state and stressed hematopoiesis.
Our results indicate that jagged-2 is differentially expressed among vascular beds. In the BM, Jag2 mRNA and jagged-2 protein are enriched in ECs. Jagged-2 supplied by ECs is not required for HSPC maintenance under steady state. Here, we show that jagged-2 is required for HSPC expansion in BMEC-mediated in vitro coculture experiments. Moreover, jagged-2 expression on ECs maintains HSPC reconstitution following myelosuppressive conditions. Although hematopoietic cells also express jagged-2, the maintenance and engraftment of HSPCs into irradiated BM is dependent on endothelial jagged-2. Finally, jagged-2 triggers Notch2/Hey1 signaling in HSPCs to maintain HSPC function during recovery following myelosuppressive stress.
These lines of evidence indicate that endothelial jagged-2 performs as a functional Notch ligand and induces Notch signaling between ECs and HSPCs. Since under steady state, HSPCs undergo self-renewal and differentiation at a physiological rate; it is plausible that upon loss of jagged-2, other Notch ligands, jagged-1, DLL1, or DLL4, are compensating for Notch activation. It is also conceivable that jagged-2 expressed by BM stromal cells or hematopoietic cells (Figure 8A) or other pro-HSPC angiocrine factors in the BM niche (Supplemental Figure 4) compensates for loss of function of jagged-2 on ECs. In contrast, the in vitro EC-HSPC coculture model and HSPC regeneration following myelosuppressive stresses represent processes where rapid HSPC recovery accompanied by HSPC self-renewal, expansion, and differentiation takes places within a narrow time window, and concerted actions of a plethora of angiocrine factors are required to maintain the fine balance between HSPC self-renewal and HSPC exhaustion. In line with the requirement of Notch signaling in maintaining HSPC reconstitution, the expression of Notch ligands in key hematopoietic organs such as BMECs and liver ECs was increased in the time window corresponding to rapid HSPC recovery following myelosuppressive stress (ref. 21, Figure 1L, and Supplemental Figure 1G). This can also explain the more dramatic HSPC phenotype observed following myelosuppressive stress compared with steady state. Alternatively, it is possible that during regenerative processes, the role of Notch ligands in angiogenesis was more apparent, and the observed lethality phenotype associated with loss of the Notch ligand jagged-1 or jagged-2 on ECs might be due to a vascular defect or perfusion impairment. To identify the key event leading to defective HSPC recovery, we quantified HSPC number during the recovery phase (Figure 4, C–H), and monitored vascular perfusion function via i.v. injection of fluorescence-labeled VE-cadherin antibodies (Figure 5, A–D). Histological sections showed minor changes in vascular structures and preservation of vascular perfusion function as demonstrated by VE-cadherin staining of key organs involved in HSPC recovery. Nonetheless, the number of LT-HSCs was significantly reduced at the early phase of myelosuppressive responses (Figure 4D), and during the mid-recovery phase, the KLS cells failed to go back to quiescence as efficiently when jagged-2 was deleted from ECs (Figure 6B).
The observed HSPC phenotype might be also due to the differential regulation of downstream Notch targets in HSPCs induced by jagged-2 or jagged-1 expression on ECs. Notably, jagged-1 induced both Hey1 and Hes1 expression in HSPCs, and jagged-2 induced Hey1 activation in HSPCs (Figure 7I). The ability to influence multiple Notch downstream targets by EC–jagged-1 (3) would likely result in more drastic perturbation of HSPC function when Jag1 was deleted from ECs, consistent with the observation that under steady state, HSPC number was reduced in Jag1ECKO mice but remained unchanged in Jag2ECKO mice. Hes1 expression in HSPCs of Jag2ECKO mice is increased in comparison with control mice. Given the recent findings that increased Notch activity caused rapid proliferation of premature exhaustion of bona fide HSPCs and increased lymphoid differentiation (35), it is conceivable that jagged-2 expression on ECs curbs the expression of Hes1 on HSPCs to ensure proper balance of downstream Notch targets in HSPCs and maintains HSPC self-renewal and differentiation.
We note that because Jag2fl/fl mice were maintained on a B6/129 background, the studies using Jag2ECKO and Jag2KO mice bear the caveat that, regardless of absence of rejection, strain inconsistencies might induce phenotypes that could be responsible for the differences attributed to the specific deletion discussed above (Figures 2 and 4–8). We have repeated these experiments several times to diminish potential bias introduced by minor strain differences.
In summary, our data set forth the new finding that jagged-2 is a functional Notch ligand in which its angiocrine expression by the vascular niche promotes HSPC reconstitution under myelosuppressive conditions without altering vascular perfusion. Moreover, angiocrine expression of jagged-2 by triggering Notch2/Hey1, augments engraftment and expansion of HSPCs. Future experimental studies are needed to dissect the role of Notch2/Hey1 activation induced by EC–jagged-2 in promoting HSPC maintenance. Collectively, these data lay the foundation for employing jagged-2 expression as a therapeutic modality to accelerate balanced hematopoietic recovery after myelosuppression.
Methods
Mice.
VE-cadherin-Cre mice were obtained from the laboratories of M.L. Iruela-Arispe (Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, CA, USA) and N.A. Speck (Abramson Family Cancer Research Institute and Department of Cell and Developmental Biology, University of Pennsylvania, Philadelphia, PA, USA) and were maintained on a B6 background. Jag2fl/fl mice were obtained from the laboratory of T. Gridley (Center for Molecular Medicine, Maine Medical Center Research Institute, Scarborough, ME, USA) and were of B6/129 background.
Cell lines.
Cultured choroid ECs, retinal ECs, lung ECs, and liver ECs were from Angiocrine Biosciences. The ECs were authenticated via flow cytometric staining of CD45–CD31+VE-cadherin+ markers. The cells were tested free of mycoplasma.
Generation of BMEC lines.
Ten microliters per animal of Dynabeads (Dynabeads Sheep Anti-Rat IgG; Invitrogen; 11035) was prepared 1 day before the EC isolation. The proper amount of Dynabeads was taken out and washed in MACS buffer (PBS + 2 mM EDTA + 0.1% BSA + 1% penicillin/streptomycin/antimitotic). Dynabeads were then incubated with CD31 antibody for more than 1 hour to overnight. Age- and sex-matched adult Jag2fl/fl or Jag2ECKO mice were used. Two femurs and 2 tibiae from hind limb were dissected out. The attached muscle was cleared first by scissors and then by Kimwipes (Kimberly-Clark Worldwide, Inc.). The femurs and tibiae were then grounded using sterile mortars and pestles. The crude BM solution was then digested using collagenase/dispase at a working concentration of 2.5 mg/ml (Roche 11088793001 for collagenase A, 04942078001 for Dispase II) for 15 minutes at 37°C on an orbital shaker. Following the enzymatic dissociation, the reaction was stopped by addition of DMEM plus 10% FBS. The cells were then filtered through a 40-μm cell strainer to obtain single-cell suspension. The cells were then incubated with anti-CD31–coated Dynabeads for 45 minutes at 4°C on a rotation shaker. After the incubation, the Dynabeads-bound cells were washed 5 times in MACS buffer. The cells were then grown on a fibronectin-coated 12-well plate using mouse EC complete medium. Three days later or when the cobblestone structure of mouse ECs appeared, lentivirus-encoding myristoylated Akt1 was transduced into ECs, and cells were grown to full confluence, which was achieved within a month. Following passages and expansion, BMECs were further purified using FACS sorting. Frozen stocks of early passages of BMECs were prepared for future use. The staining of cultured BMECs using VE-cadherin antibody (R&D Systems) was carried out to verify the purity and morphology of BMECs. The VE-cadherin staining was repeated at least 6 times to verify the ECs.
Cell culture.
BMECs were grown in mouse EC complete medium. To prepare 500 ml of mouse EC complete medium, 200 ml of F-12 medium (Corning, Cellgro; 10-080-CV) and 200 ml DMEM low-glucose medium (Corning, Cellgro; 10-014-CV) were mixed with 100 ml heat-inactivated FBS. The medium was then supplemented with 5 ml nonessential amino acid (Corning, Cellgro; 25-025-CI), 5 ml of penicillin/streptomycin/amphotericin (Corning, Cellgro; 30-004-CI), 10 ml of 1 M HEPES (Corning, Cellgro, 25-060-CI), 5 ml of 10 mg/ml heparin stock (Sigma-Aldrich; H3149-100KU), and 5 ml of 7.5 mg/ml EC growth supplement (Alfa Aesar J64516). Cells were passed every 3 days or whenever cells reached confluence. Freezing medium was prepared by mixing of 4:1 volume of heat-inactivated FBS and DMSO (Sigma-Aldrich). Cells were pelleted and resuspended into mouse EC complete medium. An equal volume of freezing medium was added into the cell suspension in a dropwise manner. To thaw cells, a frozen tube was immediately put into a 37°C incubator. After transfer of the cells into a 15-ml Falcon tube, mouse EC complete medium was added dropwise into the cell suspension. The same procedure was repeated to remove residual DMSO. Cells were then plated on tissue culture flasks.
In vitro BMEC-HSPC coculture assays.
BMECs isolated from Jag2fl/fl control mice or Jag2ECKO mice were seeded into a 12-well plate and grown into confluence in mouse EC complete medium. Lin– hematopoietic cells were isolated from BM of adult mice. The same number of BM Lin– cells were added into each well on top of BMECs and cultured in defined medium StemSpan (Stemcell Technologies) supplemented with knockout serum replacement and 20 μg/ml mouse Kit-ligand (stem cell factor) (PeproTech). Every other day 1 ml of fresh medium was added into the coculture well; following the fourth day, the floating hematopoietic cells were collected and spun down and transferred into a new confluent layer of BMECs. Fresh medium was added to the old well of BMECs with attached hematopoietic cells. Following the split event, fresh medium was added to both the new well and old wells every other day. At 9 days after coculture, all the cells in the wells were collected after enzymatic dissociation. The cells were then separated into CD45hi hematopoietic cells and CD45dim hematopoietic or ECs. For each cell population, cells were stained with the hematopoietic marker CD45, lineage antibodies, and the HSPC markers c-Kit and Sca1. Cell counting beads were added before flow cytometric analysis. The total number of hematopoietic cells, Lin– cells, and HSPCs was quantified by normalizing to the number of counting beads and the gating frequencies. The total numbers of hematopoietic cells, Lin– cells, and KLS cells were added from both the CD45dim and CD45hi populations.
Competitive transplantation assays.
Ten- to twelve-week-old CD45.1 mice were purchased from The Jackson Laboratory. The CD45.1 mice were lethally irradiated at 950 cGy 1 day before transplantation. On the day of transplantation, 1 femur was dissected out from Jag2fl/fl or Jag2ECKO mice. Muscles were detached from the femur. After cutting of the 2 ends of the femur, BM was flushed out using a 26G needle infused with sterile PBS. The BM cell suspension was filtered through a 40-μm cell strainer. After cell counting, 0.5 million CD45.2+ BMMNCs were combined with 0.5 million CD45.1+ BMMNCs and were transplanted into lethally irradiated CD45.1 mice. At the indicated time points after transplantation (Figure 2H and Figure 8I), peripheral blood was examined by flow cytometric staining to check the percentage of CD45.2 engraftment; lineage antibodies were also stained to check the multilineage engraftment and lineage differentiation potential of the engrafted HSPCs.
Flow cytometric analysis to analyze competitive repopulating assay.
To draw blood, mice were anesthetized using an isoflurane chamber at a constant flow rate of vaporized isoflurane 3-5% for induction and adjust the oxygen to the flow rate of 0.5-1.0L/ml. Two caliber tubes of blood were drawn from retro-orbital veins of mice and added into 200 ml of 10 mM EDTA/PBS buffer. After brief vortexing, the sample was put on ice until all the blood samples were collected. RBC were then lysed using 5 ml of RBC Lysis Buffer (BioLegend; 420301) on ice. The cells were then transferred into an Eppendorf tube for staining. Cells were incubated with mouse FcR block in 50 μl MACS buffer for 10 minutes. Antibodies for Ter119, CD45.1, CD45.2, CD3, B220, and Gr-1/CD11b were then mixed to generate a master mix and added into each tube. The information for the antibodies is listed in Supplemental Table 1. For all the experiments, unstained sample or fluorescence-minus-one (FMO) rule was used for gating of positive populations. Unstained control and single-stained cells or ultracompensation beads (eBioscience; 01-2222-42) were used to calculate the fluorescence spillover from all the channels. Compensation was calculated either manually or automatically and applied to all the samples.
Flow cytometric analysis to quantify LT-HSC number and cell cycle status.
Age- and sex-matched adult Jag2fl/fl or Jag2ECKO mice were used for this quantification. Femurs and tibiae from mice were dissected. Following muscle cleanup, the femurs and tibiae were ground using mortars and pestles in MACS buffer. The cells were then filtered through a 40-μm cell strainer; the remaining bones were ground twice more following the same procedure until the bones appeared white. The total number of filtered cells was counted as the total hematopoietic cells (excluding mature RBC) in the BM. Two million total hematopoietic cells were then stained with lineage antibodies and the SLAM-HSPC markers c-Kit, Sca1, CD150, and CD48. The total number of SLAM-HSPCs was calculated using the total number of hematopoietic cells and the frequencies of Lin– cells and of c-Kit+Sca1+ and CD150+CD48– gating. For steady-state HSPC quantification, this same method was used. For quantification of HSPC recovery kinetics following myelosuppression, total hematopoietic cells were counted. Lineage depletion was then carried out. The Lin– cells were stained with SLAM-HSPC markers. The total number of SLAM-HSPCs was quantified using the Lin– cell number and the frequencies of c-Kit+Sca1+ and CD150+CD48– gating. For cell cycle analysis of the SLAM-HSPCs, following surface staining of c-Kit, Sca1, CD150, and CD48, the cells were washed in MACS buffer and fixed in 1% paraformaldehyde (PFA) for 3 minutes. Following intracellular permeabilization (BD Biosciences; Cytofix/Cytoperm), the cells were stained with Ki67. Finally, the cells were washed of excessive Ki67 staining and incubated with DAPI overnight. WT Lin– cells that had undergone the same surface staining and intracellular permeabilization were used as FMO gating control. Ultracompensation beads were stained with each antibody for compensation purposes.
Perfusion function of vasculature at steady-state conditions or after 650 cGy irradiation.
Monoclonal antibody that targets mouse VE-cadherin (BioLegend; clone BV13) was conjugated with Alexa Fluor 647 dye (Life Technologies, Thermo Fisher Scientific) in the laboratory. At day 10 after sublethal irradiation (650 cGy), 25 μg conjugated VE-cadherin antibodies was retro-orbitally injected into Jag2fl/fl control or Jag2ECKO mice. Ten minutes after antibody injection, the mice were euthanized with CO2. Organs such as liver, lung, heart, BM, and spleen were dissected out and washed in PBS. After fixation in 4% PFA, tissues were dehydrated in 30% sucrose, embedded in OCT compound, and snap-frozen. Tissue sections (10-μm-thick) were then prepared using a cryostat machine. The tissue sections were counterstained with DAPI to reveal the nuclei. Pictures were taken using a Zeiss LSM 710 confocal microscope. For the imaging of mouse BM vasculature, more than 3 images were taken for the vasculature near the growth plate; more than 5 images were taken for the metaphysis region. For image analysis, the indicated numbers of fields were randomly chosen. The data from the different image fields were then averaged as the vessel density of the organ of interest. The investigators were unaware of the genotype of mice when recording the images.
FACS sorting of primary ECs.
Monoclonal antibody that targets mouse VE-cadherin (Biolegend, clone BV13) was conjugated with Alexa Fluor 647 dye (Life Technologies) in the laboratory. Age- and sex-matched Jag2fl/fl or Jag2ECKO mice were used for EC isolation and RNA preparation. Twenty-five micrograms of conjugated VE-cadherin antibodies was retro-orbitally injected into Jag2fl/fl control or Jag2ECKO mice. Ten minutes after antibody injection, the mice were euthanized with CO2. For BMEC isolation, femurs and tibiae were dissected, then ground with mortars and pestles, and then enzymatically digested with collagenase/dispase mixture at 37°C for 15 minutes. Following retrieval of single-cell suspension, lineage depletion was carried out using a lineage cell depletion kit (Miltenyi Biotec). Lineage-negative cells were then stained with the hematopoietic and EC surface markers CD45 and CD31. DAPI was stained lastly in the buffer of PBS plus 2 mM EDTA. BMECs were then sorted as DAPI–CD45–CD31+VE-cadherin+ cells.
For isolation of ECs from choroid, retina, lungs, and livers, 25 μg isolectin and 25 μg VE-cadherin were injected into mice. After tissue dissection, the tissues were first cut into small pieces using razor blades in a sterile Petri dish. The small tissue pieces were then enzymatically digested using collagenase/dispase mixture at 37°C for 30 minutes on an orbital shaker. Following enzymatic digestion, the cells were then further broken into homogenous single-cell suspension via titration using a 21-gauge needle attached to a syringe. The enzymatic reaction was then stopped via neutralization by FBS. The RBC within the single-cell suspension were lysed using RBC lysis buffer (BioLegend). The cells were then resuspended in MACS buffer and stained with the EC marker CD31. Microvascular ECs were sorted also as CD45–isolectin+VE-cadherin+ cells.
Quantitative real-time PCR analysis and sample preparation of RNA sequencing analysis.
For whole-organ Jag2 mRNA expression analysis, a piece of tissue from lung, liver, or BM was homogenized in RNA lysis buffer, RLT buffer (Qiagen) with 1% β-mercaptoethanol. The RLT solution was then cleared of undigested tissue using a QIAthredder column (Qiagen). RNA was then prepared following the RNeasy protocol (QIAGEN). DNA was digested using DNase I (QIAGEN) during the RNA extraction processes. The obtained RNA was measured for concentration using NanoDrop (Thermo Fisher Scientific). RNA (200 ng) was used for cDNA conversion using qScript Super Mix (Quanta Biosciences). After cDNA dilution with double-distilled H2O at a 1:5 ratio, the qPCR reaction was set up by mixing the gene-specific primers and SYBR Green qPCR Reaction Mix (KAPA Biosystems). qPCR was run and detected using a ViiA-7 qPCR machine (Life Technologies, Thermo Fisher Scientific). Please refer to Supplemental Table 2 for the exact primer sequence(s) for indicated gene. For RNA extraction of abundant cells, a QIAGEN RNeasy kit was used. For RNA extraction of low numbers of cells following FACS sorting, the cells were pelleted down and lysed using extraction buffer from PicoPure RNA Isolation Kit (Life Technologies, Thermo Fisher Scientific). DNA was removed during RNA extraction processed with DNase I (Qiagen). For RNA sequencing analysis, the extracted RNA was submitted to the Genomic Core Facility within Weill Cornell Medicine. The RNA concentration and integrity were measured using a NanoDrop and Agilent 2100 Bioanalyzer (Agilent), respectively. The integrity of RNA was indicated by the RNA integrity number (RIN). RNA samples with sufficient concentration and RIN greater than 8.0 were further prepared for cDNA library preparation and subsequent sequencing. Refer to GSE95835 (Gene Expression Omnibus, NCBI) for RNA sequencing results of primary choroid and retinal ECs.
Western blot.
For Western blotting of tissue of Jag2fl/fl or Jag2ECKO mice, small pieces of tissue were homogenized using automatic tissue homogenizer (Qiagen) in RIPA buffer that contained protease inhibitor, phosphate inhibitor, and sodium orthovanadate. Tissues were lysed in this buffer on ice for 1 hour. After sonication, supernatant was collected for protein concentration measurement and Western blot analysis. Protein concentration was measured using a BCA assay kit (Thermo Fisher Scientific). Protein sample was then mixed with loading buffer and reduced using 5% β-mercaptoethanol. The protein sample was boiled at 95°C for 10 minutes before being loaded onto SDS-PAGE gel. Membrane transfer was done using wet transfer technique. Primary antibody was incubated with 5% milk in PBST (PBS+0.2% Tween-20) buffer overnight at the dilution of 1:1,000. Secondary antibodies were incubated at room temperature for 1 hour. Signals were amplified and detected using an ECL kit using x-ray films. For the Western blotting of lung tissues, more than 2 separate batches of experiments were performed (each time, n = 2 for Jag2fl/fl or Jag2ECKO mice).
Immunofluorescent staining of cultured cells.
For immunofluorescent staining of cultured cells, cells were removed from growth medium and washed with PBS once. The cells were then fixed in 4% PFA for 10 minutes at room temperature. After blocking with blocking buffer, cells were incubated with primary antibody overnight. Secondary antibodies were stained the next day. Finally, cells were counterstained using Hoechst 33328 nuclear dye. After washes with PBS, cells were then incubated with PBS before images were taken using a Zeiss LSM 710 confocal microscope.
Statistics.
The differences in the phenotypic analysis of experimental parameters were statistically compared using a 2-tailed t test. For measurements that were made across different time points between the 2 genotypes, 2-way ANOVA was carried out to measure the statistical significance of the observed data for the 2 factors, time and genotype; the P value of the observed phenotype variance based on genotype was reported and considered for statistical significance. P less than 0.05 was considered to be statistically significant; *P < 0.05, **P < 0.01, and ***P < 0.001. Experiments were repeated at least 2–3 times. The variance was similar between the groups being statistically compared. If higher variability was found, we increased the sample size to fully confirm statistical significance. No statistical method was used to predetermine sample size for a given effect size. We included all tested animals for quantification to analyze statistical difference.
Study approval.
All animal experiments were performed under the approval of Weill Cornell Medicine Institutional Animal Care and Use Committee, New York, NY. The breeding and maintenance of animal colonies abided by the guidelines of the IACUC of Weill Cornell Medical College, New York, New York, USA. All experimental procedures followed the IACUC guidelines. Genotyping was carried out in the laboratory or the tails were sent to Transnetyx (transnetyx.com). To compare the phenotypes between different mouse genotypes, sex- and weight-matched littermates were used. Mice at 20–28 g body weight were used. Investigators who performed the experiments and recorded the data were blinded to genotypes of individual mice.
Author contributions
PG designed the study, performed the experiments, interpreted the results, and wrote the paper. MGP, BP, CRB, RL, BK, and BSD performed the experiments and analyzed the data. BP, CRB, and BSD performed mouse experiments and collected and analyzed the data. BSD, SYR, KS, and JMB helped to formulate the hypothesis, interpreted the results, and edited the manuscript. PG and SR conceived the project, analyzed the data, and wrote the paper.
Supplementary Material
Acknowledgments
We are grateful to M.L. Iruela-Arispe and N.A. Speck for the VE-cadherin-Cre transgenic mice and T. Gridley for Jag2fl/fl mice. We are indebted to I. Aifantis for providing the Hes1-GFP reporter mice. RL is supported by the Ansary Stem Cell Institute, the Starr Foundation Tri-Institutional Stem Cell core project, and the Qatar National Priorities Research Program (NPRP 8-1898-3-392). BK is supported by NIH T32 HD060600. BSD is supported by the Ansary Stem Cell Institute and a National Scientist Development Grant from the American Heart Association (12SDG1213004). SYR is supported by the Ansary Stem Cell Institute and a BioTime Stem Cell Research grant. KS is supported by the Ansary Stem Cell Institute, the Starr Foundation Tri-Institutional Stem Cell core project, the Empire State Stem Cell Board, and New York State Department of Health grants. JMB and MGP are supported by the Ansary Stem Cell Institute, the Tri-Institutional Stem Cell Initiative (TRI-SCI 2013-022 and 2014-004), Leukemia & Lymphoma Society grant 0859-15, and the American Federation for Aging Research Research Grant for Junior Faculty. SR is supported by the Ansary Stem Cell Institute, the Starr Foundation Tri-Institutional Stem Cell core project, the Tri-Institutional Stem Cell Initiative (TRI-SCI 2013-032, 2014-023, 2016-013), the Empire State Stem Cell Board, and New York State Department of Health grants, and by grants from the NIH R01 (DK095039, HL119872, HL128158, HL115128, HL099997) and U54 CA163167.
Version 1. 10/23/2017
Electronic publication
Version 2. 12/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: This work is licensed under the Creative Commons Attribution 4.0 International License. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reference information: J Clin Invest. 2017;127(12):4242–4256.https://doi.org/10.1172/JCI92309.
Contributor Information
Peipei Guo, Email: peg2005@med.cornell.edu.
Brisa Palikuqi, Email: brp2008@med.cornell.edu.
Raphael Lis, Email: ral2020@med.cornell.edu.
Balvir Kunar, Email: bak2010@med.cornell.edu.
Bi-Sen Ding, Email: bid2004@med.cornell.edu.
Koji Shido, Email: kos2001@med.cornell.edu.
Shahin Rafii, Email: srafii@med.cornell.edu.
References
- 1.Mendelson A, Frenette PS. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat Med. 2014;20(8):833–846. doi: 10.1038/nm.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramalingam P, Poulos MG, Butler JM. Regulation of the hematopoietic stem cell lifecycle by the endothelial niche. Curr Opin Hematol. 2017;24(4):289–299. doi: 10.1097/MOH.0000000000000350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poulos MG, et al. Endothelial Jagged-1 is necessary for homeostatic and regenerative hematopoiesis. Cell Rep. 2013;4(5):1022–1034. doi: 10.1016/j.celrep.2013.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heissig B, et al. Recruitment of stem and progenitor cells from the bone marrow niche requires MMP-9 mediated release of kit-ligand. Cell. 2002;109(5):625–637. doi: 10.1016/S0092-8674(02)00754-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rafii S, Butler JM, Ding BS. Angiocrine functions of organ-specific endothelial cells. Nature. 2016;529(7586):316–325. doi: 10.1038/nature17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi H, et al. Angiocrine factors from Akt-activated endothelial cells balance self-renewal and differentiation of haematopoietic stem cells. Nat Cell Biol. 2010;12(11):1046–1056. doi: 10.1038/ncb2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hofmann JJ, Iruela-Arispe ML. Notch signaling in blood vessels: who is talking to whom about what? Circ Res. 2007;100(11):1556–1568. doi: 10.1161/01.RES.0000266408.42939.e4. [DOI] [PubMed] [Google Scholar]
- 8.Gale NW, et al. Haploinsufficiency of delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc Natl Acad Sci U S A. 2004;101(45):15949–15954. doi: 10.1073/pnas.0407290101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benedito R, et al. Loss of Notch signalling induced by Dll4 causes arterial calibre reduction by increasing endothelial cell response to angiogenic stimuli. BMC Dev Biol. 2008;8:117. doi: 10.1186/1471-213X-8-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leslie JD, Ariza-McNaughton L, Bermange AL, McAdow R, Johnson SL, Lewis J. Endothelial signalling by the Notch ligand Delta-like 4 restricts angiogenesis. Development. 2007;134(5):839–844. doi: 10.1242/dev.003244. [DOI] [PubMed] [Google Scholar]
- 11.Lobov IB, et al. Delta-like ligand 4 (Dll4) is induced by VEGF as a negative regulator of angiogenic sprouting. Proc Natl Acad Sci U S A. 2007;104(9):3219–3224. doi: 10.1073/pnas.0611206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445(7129):776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 13.Sörensen I, Adams RH, Gossler A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 2009;113(22):5680–5688. doi: 10.1182/blood-2008-08-174508. [DOI] [PubMed] [Google Scholar]
- 14.Lee SU, et al. LRF-mediated Dll4 repression in erythroblasts is necessary for hematopoietic stem cell maintenance. Blood. 2013;121(6):918–929. doi: 10.1182/blood-2012-03-418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delaney C, Varnum-Finney B, Aoyama K, Brashem-Stein C, Bernstein ID. Dose-dependent effects of the Notch ligand Delta1 on ex vivo differentiation and in vivo marrow repopulating ability of cord blood cells. Blood. 2005;106(8):2693–2699. doi: 10.1182/blood-2005-03-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benedito R, et al. The notch ligands Dll4 and Jagged1 have opposing effects on angiogenesis. Cell. 2009;137(6):1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 17.Alva JA, et al. VE-cadherin-Cre-recombinase transgenic mouse: a tool for lineage analysis and gene deletion in endothelial cells. Dev Dyn. 2006;235(3):759–767. doi: 10.1002/dvdy.20643. [DOI] [PubMed] [Google Scholar]
- 18.Xu J, Krebs LT, Gridley T. Generation of mice with a conditional null allele of the Jagged2 gene. Genesis. 2010;48(6):390–393. doi: 10.1002/dvg.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsai S, Fero J, Bartelmez S. Mouse Jagged2 is differentially expressed in hematopoietic progenitors and endothelial cells and promotes the survival and proliferation of hematopoietic progenitors by direct cell-to-cell contact. Blood. 2000;96(3):950–957. [PubMed] [Google Scholar]
- 20.Kared H, et al. Jagged2-expressing hematopoietic progenitors promote regulatory T cell expansion in the periphery through notch signaling. Immunity. 2006;25(5):823–834. doi: 10.1016/j.immuni.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Nolan DJ, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26(2):204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luca VC, Jude KM, Pierce NW, Nachury MV, Fischer S, Garcia KC. Structural biology. Structural basis for Notch1 engagement of Delta-like 4. Science. 2015;347(6224):847–853. doi: 10.1126/science.1261093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6(3):265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulos MG, et al. Vascular platform to define hematopoietic stem cell factors and enhance regenerative hematopoiesis. Stem Cell Reports. 2015;5(5):881–894. doi: 10.1016/j.stemcr.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13(6):427–437. doi: 10.1038/nri3445. [DOI] [PubMed] [Google Scholar]
- 26.Hooper AT, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell Stem Cell. 2009;4(3):263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oh P, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13(2):190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fre S, et al. Notch lineages and activity in intestinal stem cells determined by a new set of knock-in mice. PLoS One. 2011;6(10):e25785. doi: 10.1371/journal.pone.0025785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun D, et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell Stem Cell. 2014;14(5):673–688. doi: 10.1016/j.stem.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457(7231):887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Varnum-Finney B, Halasz LM, Sun M, Gridley T, Radtke F, Bernstein ID. Notch2 governs the rate of generation of mouse long- and short-term repopulating stem cells. J Clin Invest. 2011;121(3):1207–1216. doi: 10.1172/JCI43868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Butler JM, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell Stem Cell. 2010;6(3):251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benveniste P, et al. Notch signals are required for in vitro but not in vivo maintenance of human hematopoietic stem cells and delay the appearance of multipotent progenitors. Blood. 2014;123(8):1167–1177. doi: 10.1182/blood-2013-07-505099. [DOI] [PubMed] [Google Scholar]
- 34.Delaney C, Heimfeld S, Brashem-Stein C, Voorhies H, Manger RL, Bernstein ID. Notch-mediated expansion of human cord blood progenitor cells capable of rapid myeloid reconstitution. Nat Med. 2010;16(2):232–236. doi: 10.1038/nm.2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang MY, Shestova O, Xu L, Aster JC, Pear WS. Divergent effects of supraphysiologic Notch signals on leukemia stem cells and hematopoietic stem cells. Blood. 2013;121(6):905–917. doi: 10.1182/blood-2012-03-416503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.