Abstract
Primary immunodeficiencies are often monogenic disorders characterized by vulnerability to specific infectious pathogens. Here, we performed whole-exome sequencing of a patient with disseminated Mycobacterium abscessus, Streptococcus viridians bacteremia, and cytomegalovirus (CMV) viremia and identified mutations in 2 genes that regulate distinct IFN pathways. The patient had a homozygous frameshift deletion in IFNGR2, which encodes the signal transducing chain of the IFN-γ receptor, that resulted in minimal protein expression and abolished downstream signaling. The patient also harbored a homozygous deletion in IFNAR1 (IFNAR1*557Gluext*46), which encodes the IFN-α receptor signaling subunit. The IFNAR1*557Gluext*46 resulted in replacement of the stop codon with 46 additional codons at the C-terminus. The level of IFNAR1*557Gluext*46 mutant protein expressed in patient fibroblasts was comparable to levels of WT IFNAR1 in control fibroblasts. IFN-α–induced signaling was impaired in the patient fibroblasts, as evidenced by decreased STAT1/STAT2 phosphorylation, nuclear translocation of STAT1, and expression of IFN-α–stimulated genes critical for CMV immunity. Pretreatment with IFN-α failed to suppress CMV protein expression in patient fibroblasts, whereas expression of WT IFNAR1 restored IFN-α–mediated suppression of CMV. This study identifies a human IFNAR1 mutation and describes a digenic immunodeficiency specific to type I and type II IFNs.
Keywords: Immunology, Virology
Keywords: Innate immunity
Introduction
A paradigm of host immunity is that an individual’s susceptibility to pathogens indicates a specific molecular defect. Yet, there are apparent exceptions to this paradigm: individuals who develop infections from a spectrum of organisms either broader or narrower than that predicted by the identified molecular defect. Several cases of human mutations affecting the interferon (IFN) pathways exemplify such exceptions. There are 3 families of IFNs: type I (IFN-α, IFN-β, and IFN-ω), type II (IFN-γ), and type III (IFN-λ1, IFN-λ2, and IFN-λ3) (1). Each IFN family has its specific receptor (IFNAR, IFNGR, IFNLR, respectively) and unique signaling pathways, although crosstalk occurs among these pathways (1). Type I IFNs induce the expression of MHC class I and stimulate NK cell– and CD8+ T cell–mediated cytotoxicity to promote the killing of cells infected with virus (1). Only 2 types of defects in the type I IFN pathway have been reported to date: one kindred with a truncation mutation in IFNAR2 and patients deficient in STAT2, a transcription factor downstream of IFNAR activation (2–4). These patients presented with disseminated live vaccine–strain viral infections, indicating the contribution of IFNAR signaling to the induction of IFN-stimulated genes (ISGs) important for inhibiting viral replication and infection (2–4). However, despite the known importance of type I IFNs in controlling viral pathogens, both the IFNAR2- and STAT2-deficient patients had either absent or mild symptoms after infection with childhood viruses, including cytomegalovirus (CMV), Epstein-Barr virus, varicella zoster virus (VZV), and respiratory viruses (2, 3). This suggests that the type I IFNs have an essential, but narrow, role in host immunity against viral pathogens.
The contributions of the type II IFN pathway to host immunity have been illustrated in patients with mutations in the genes encoding the IFNGR, 2 downstream transcription factors (STAT1 and IRF8), and the proteins important for IFN-γ secretion, such as IL-12, IL-12R, and ISG15 (5, 6). Due to the role of IFN-γ in macrophage activation, these patients are susceptible to intracellular organisms, most commonly mycobacteria and Salmonella, but also Listeria monocytogenes and Nocardia (5, 7). A few patients have been reported to have concomitant infections with additional pathogens, including CMV, respiratory syncytial virus, VZV, klebsiella, histoplasmosis, and paracoccidioidomycosis (7–10). It is not known if these atypical infections resulted from differences in environmental exposures or additional mutations affecting other mechanisms of host immunity.
We present a patient with disseminated mycobacteria, Streptococcus viridans bacteremia, and CMV viremia. We identified, for the first time to our knowledge, a digenic innate primary immunodeficiency leading to defective type I and type II IFN signaling that illustrates the diverse contributions of these pathways to host defense against specific pathogens.
Results and Discussion
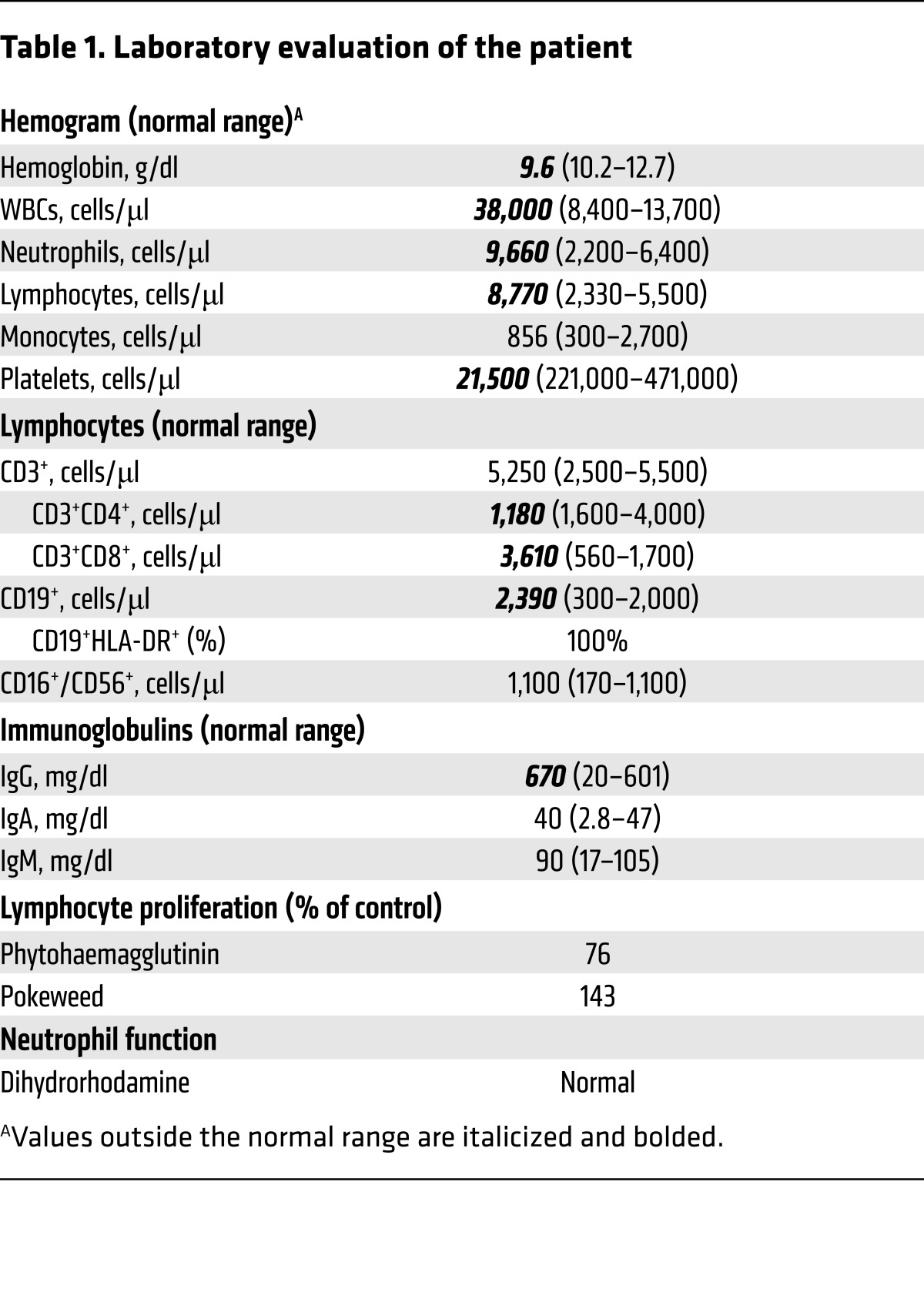
The proband is the only child of consanguineous Saudi Arabian parents (Figure 1A). He was healthy until 2 months of age, when he developed a high fever and a blood culture positive for Streptococcus viridans, and severe CMV viremia (4,000,000 copies/ml). Within a few days, he developed anemia, thrombocytopenia, splenomegaly, and lymphadenopathy. He had laboratory findings consistent with hemophagocytic lymphohistiocytosis (HLH): elevated ferritin (4,000 μg/l), increased triglycerides (7.5 mmol/l), high soluble CD25 (6,500 U/ml), and hemophagocytosis in his bone marrow. His bone marrow was positive for Mycobacterium abscessus and CMV. His laboratory evaluation revealed elevated numbers of white blood cells and total lymphocytes, a mildly decreased CD4+ T cell count, and increased numbers of CD8+ T cells (Table 1). His HIV PCR was negative. Targeted Sanger sequencing identified no mutations in genes associated with familial HLH (PRF1, UNC13D, STX11, STXBP2). He was treated with dexamethasone and cyclosporine for HLH, gancyclovir for the CMV viremia, and isoniazid, rifampin, clarithromycin, ciprofloxacin, and amikacin for his mycobacterial infection. He recovered from this episode and has been treated with prophylactic antimycobacterial medications without any subsequent significant infections. He has received attenuated measles, mumps, and rubella virus vaccines without adverse sequelae. Due to his history of disseminated infections, the proband is now scheduled to undergo a hematopoietic stem cell transplant.
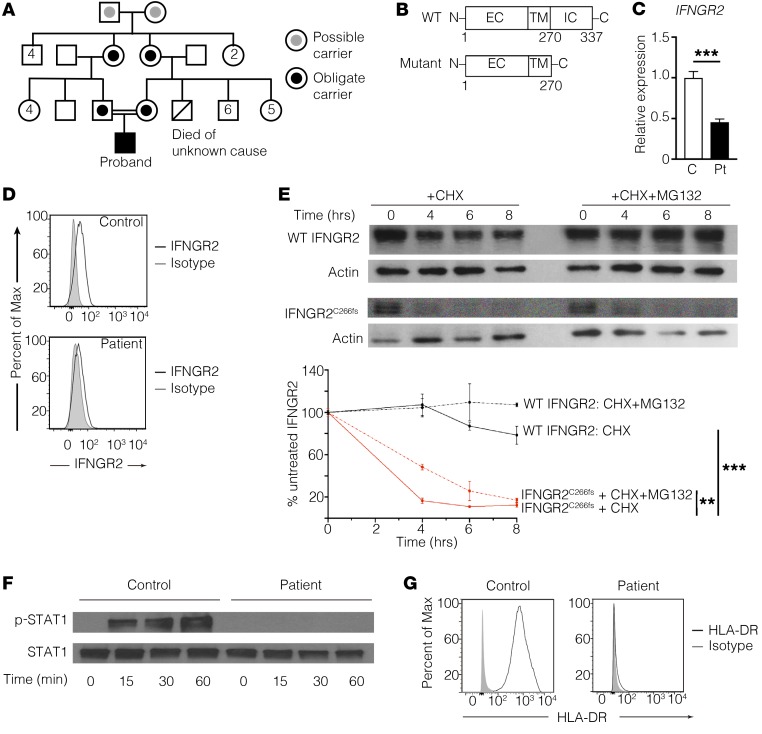
Figure 1. IFNGR2C266fs has minimal protein expression and abrogates downstream signaling.
(A) Pedigree of the proband’s family. No one in the extended family has recurrent infections. (B) Schematic of wild-type (WT) and the mutant (IFNGR2C266fs) IFNGR2. EC, extracellular domain; TM, transmembrane domain; IC, intracellular domain. (C) Quantitative PCR of IFNGR2 mRNA from control and proband (Pt) fibroblasts, normalized to GAPDH as a housekeeping gene and expressed relative to the control. (D) Flow cytometric analysis of the wild-type (WT IFNGR2) and mutant (IFNGR2C266fs) protein expression on CD14+ cells from a control and the proband, respectively. (E) Decay curves of WT and mutant IFNGR2 transfected into 293T cells, treated with the protein inhibitor cycloheximide (CHX) with or without the proteasome inhibitor MG132. **P < 0.01, ***P < 0.001, 2-way ANOVA. Results are pooled from 3 experiments. (F) Immunoblot of STAT1 phosphorylation after IFN-γ stimulation of fibroblasts from the proband and a control. (G) Flow cytometric analysis of HLA-DR expression on patient or control fibroblasts stimulated with IFN-γ. All data are representative of 3 independent experiments using 2 controls.
Table 1. Laboratory evaluation of the patient.
The proband’s consanguineous ancestry suggested an autosomal recessive immunodeficiency (Figure 1A). Whole-exome sequencing (WES) of the proband identified 2 potentially novel variants affecting distinct IFN pathways, in addition to 13 rare homozygous variants (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI93486DS1). The first is a homozygous single nucleotide deletion in IFNGR2 (IFNGR2C266fs, NM_005534: c.798delT: p.C266fs), which encodes the IFNGR signal transducing chain. Sanger sequencing of genomic as well as cDNA from the proband’s fibroblasts demonstrated that this mutation results in a frameshift at residue 266, followed by premature truncation of the protein at residue 270 upstream of the residues essential for JAK2 binding (Figure 1B and Supplemental Figure 1, A and B). The mutant IFNGR2 mRNA was expressed at a significantly lower level in the patient’s fibroblasts than the WT IFNGR2 mRNA in control fibroblasts, consistent with possible nonsense-mediated decay of the mutant mRNA (Figure 1C). Flow cytometry using an antibody specific for the N-terminus of IFNGR2 demonstrated that the patient’s monocytes had nearly absent IFNGR2C266fs protein expression (Figure 1D). To investigate the turnover of IFNGR2C266fs, we transfected HEK293T cells with cDNA encoding WT or mutant IFNGR2, in the absence or presence of the protein synthesis inhibitor cycloheximide (CHX). IFNGR2C266fs had impaired expression at baseline and had a shorter half-life than the WT protein (Figure 1E). Misfolded proteins in the endoplasmic reticulum are cleared by endoplasmic reticulum–associated protein degradation (ERAD), a process that is blocked by the proteasomal inhibitor MG132. The addition of MG132 partially prevented the degradation of IFNGR2C266fs, demonstrating that ERAD contributes to the accelerated degradation of the mutant (Figure 1E). To determine if IFNGR2C266fs retained any residual activity, we stimulated the patient’s skin-derived fibroblasts with IFN-γ and assessed STAT1 phosphorylation and upregulation of the MHC class II protein HLA-DR. Ligation of IFNGR2C266fs with IFN-γ failed to result in STAT1 phosphorylation or upregulation of HLA-DR after stimulation (Figure 1, F and G). The minimal expression of IFNGR2C266fs and its lack of activity were consistent with the patient’s early-onset disseminated mycobacterial disease, a phenotype associated with autosomal recessive complete IFN-γR2 deficiency.
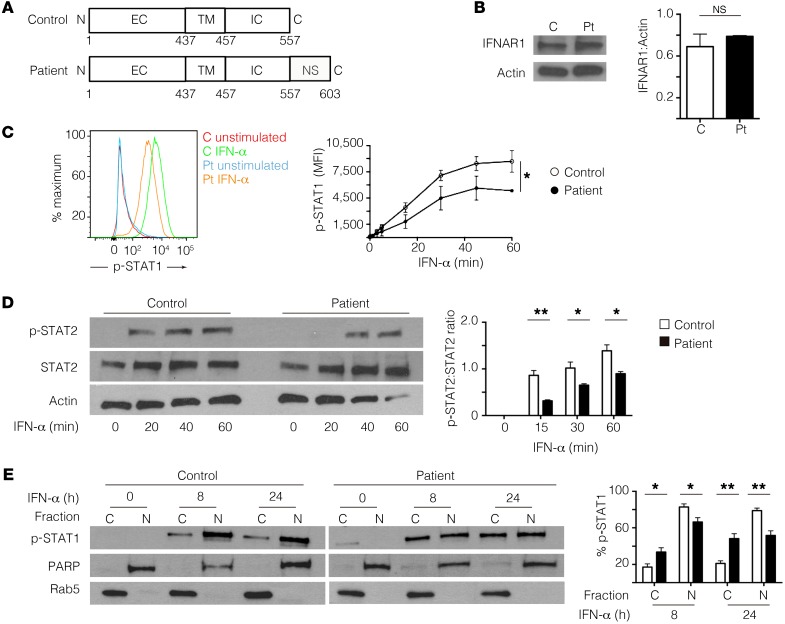
The proband’s history of disseminated CMV is atypical for a monogenic defect in the type II IFN axis because very few patients with IFNGR2 deficiency or autoantibodies against IFN-γ have been previously reported to have CMV viremia (9–11). In our patient, WES also identified a potentially novel homozygous deletion in the IFNAR1 gene (IFNAR1*557Gluext*46, NM_000629: c.1671_1821del: p.*557Gluext*46), which encodes the signaling chain of the IFNAR. Sanger sequencing of genomic DNA confirmed that the mutation was homozygous in the proband and heterozygous in his parents (Supplemental Figure 2A). The IFNAR1*557Gluext*46 mutation is predicted to replace the gene’s stop codon with 46 novel C-terminal codons, which was confirmed by Sanger sequencing of cDNA from the patient’s fibroblasts (Figure 2A and Supplemental Figure 2B). The additional C-terminal amino acids were predicted to add 5 kDa to the protein’s molecular weight. Immunoblotting of fibroblast lysates from the patient using an antibody directed against residues 450–500 revealed protein expression of IFNAR1*557Gluext*46 at a level comparable with that of WT IFNAR1 in control fibroblasts (Figure 2B). The small difference in molecular weight between WT IFNAR1 (130 kDa) and IFNAR1*557Gluext*46 (135 kDa) was not evident on immunoblotting due to the relatively high molecular weight of both proteins. IFNAR1 serves as the signaling chain of the IFNAR and is required for responsiveness to type I IFNs (1, 12). Binding of IFN-α to the IFNAR complex leads to the phosphorylation of STAT1 and STAT2 followed by assembly of the p-STAT1/p-STAT2 heterodimer. The p-STAT1/p-STAT2 heterodimer is released from the IFNAR complex, translocates to the nucleus, and induces expression of ISGs necessary for controlling viral replication and spread (12). IFN-α stimulation of the patient’s fibroblasts resulted in significantly decreased STAT1 and STAT2 phosphorylation compared with control fibroblasts (Figure 2, C and D). Additionally, the nuclear translocation of p-STAT1 was significantly impaired in the patient’s fibroblasts 8 and 24 hours after IFN-α stimulation (Figure 2E). Although the mutant IFNAR1*557Gluext*46 protein retains the docking sites required for downstream intracellular signaling events, these results indicated that the addition of the potentially novel 46 amino acids partially disrupts the receptor’s downstream signaling. Unlike the IFNAR2- or STAT2-deficient patients, who developed fatal vaccine strain measles encephalopathy (2, 3), our proband had no complications after MMR vaccination. The residual signaling and ISG expression downstream of IFNAR1*557Gluext*46 in our proband may have been sufficient for providing immunity against attenuated, but not WT viruses.
Figure 2. The mutant IFNAR1*557next*46 protein retains normal expression, but has impaired signaling.
(A) Schematic of wild-type IFNAR1 and the mutant IFNAR1*557next*46 proteins. EC, extracellular domain; TM, transmembrane domain; IC, intracellular domain; NS, previously uncharacterized sequence. (B) Immunoblot of IFNAR1 using lysates from control (C) and patient (Pt) skin-derived fibroblasts. (C) Representative flow cytometric plot (left) and quantification (right) of STAT1 phosphorylation after IFN-α stimulation of patient and control fibroblasts. MFI, mean fluorescence intensity. (D) Representative immunoblot (left) and quantification (right) of STAT2 phosphorylation after IFN-α stimulation of control and patient fibroblasts. *P < 0.05, 2-way ANOVA. (E) Representative immunoblots (left) and percentage of p-STAT1 in the cytoplasmic (C) and nuclear (N) fractions (right) after IFN-α stimulation of control and patient fibroblasts. *P < 0.05, **P < 0.01, Student’s t test. Data shown are from 3 independent experiments, using the proband and 2 controls. NS, not significant.
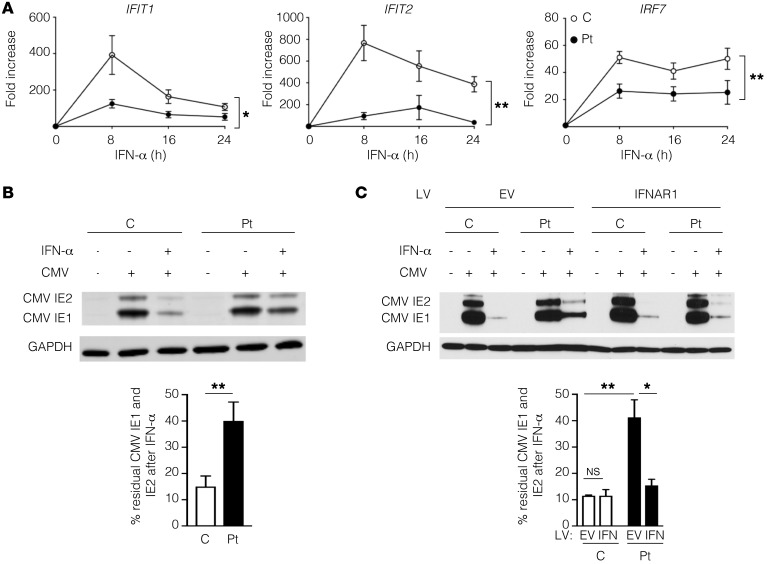
We next assessed the antiviral state induced by IFN-α stimulation of patient and control fibroblasts. Since fibroblasts do not secrete IFN-γ, a cytokine produced exclusively by hematopoietic cells (13), we used patient-derived fibroblasts to specifically determine the impact of the IFNAR1*557Gluext*46 mutant protein on the induction of ISGs and CMV susceptibility independent of IFN-γ. IFN-α stimulation resulted in significantly impaired induction of IRF7, a transcription factor upregulated by activation of the IFNAR, as well as IFIT1 and IFIT2, two ISGs regulated by type I IFNs (12) and important for the inhibition of CMV replication (Figure 3A). We directly tested the ability of IFN to control human CMV (HCMV) infection in the patient and control fibroblasts. Patient and control fibroblasts were incubated in media with or without IFN-α for 17 hours, followed by infection with HCMV for 6 hours. At this early time point, which occurs prior to the induction of maximal endogenous type I IFNs, there was no difference in the expression of CMV IE1 and IE2 in the untreated patient and control fibroblasts (Figure 3B). IFN-α pretreatment of normal fibroblasts reduced CMV IE1 and IE2 expression to 15% of the level in untreated cells (Figure 3B). IFN-α pretreatment of the patient’s fibroblasts resulted in a significantly higher residual CMV IE1 and IE2 expression that was 40% of the level in untreated patient cells (Figure 3B). The partial inhibition of CMV protein expression in the patient’s fibroblasts by IFN-α was concordant with the residual signaling downstream of the mutant IFNAR1*557Gluext*46 (Figure 2, C and D, and Figure 3A). To definitively prove the contribution of the mutant IFNAR1*557Gluext*46 to the CMV susceptibility seen in the patient’s cells, we examined whether the expression of WT IFNAR1 in the patient’s cells would restore the ability to control CMV infection. Patient and control fibroblasts were transduced with lentivirus containing either empty vector (EV) or IFNAR1, then treated with IFN-α for 17 hours, and subsequently infected with HCMV for 6 hours. The introduction of WT IFNAR1 had no effect on the residual CMV IE1 or IE2 expression in control fibroblasts pretreated with IFN-α (Figure 3C). In contrast, introduction of WT IFNAR1 reduced CMV IE1 and IE2 expression to 15%, which is significantly less than the 40% residual expression observed in EV-transduced patient cells (Figure 3C). Notably, the expression of WT IFNAR1 in patient cells restored IFN-α–mediated suppression of CMV IE1 and IE2 expression to a level comparable to that observed in control IFN-α–treated cells (Figure 3C). Collectively, these results demonstrated that the impaired signaling downstream of IFNAR1*557Gluext*46 is insufficient for controlling CMV protein expression.
Figure 3. IFNAR1*557next*46 demonstrates impaired induction of ISGs and inhibition of CMV protein expression.
(A) Quantitative PCR of IFIT1, IFIT2, and IRF7 expression in control (C) and patient (Pt) fibroblasts after stimulation with IFN-α. *P < 0.05; **P < 0.01, 2-way ANOVA. (B) Representative immunoblot (left) and residual CMV IE1 and IE2 protein (right) in control and patient fibroblasts, with and without IFN-α pretreatment. GAPDH was used as a loading control. (C) Representative immunoblot (left) and residual CMV IE1 and IE2 protein (right) in control and patient fibroblasts transduced with lentivirus (LV) containing empty vector (EV) or wild-type IFNAR1, with or without IFN-α pretreatment prior to CMV infection. Three independent experiments were performed. *P < 0.05; **P < 0.01, Student’s t test. NS, not significant.
The unbiased approach inherent in next-generation sequencing has shown that approximately 5% of sequenced patients have disease-causing multiloci variants (14). Multigenic mutations have been shown to shape the phenotype of patients with autoimmunity, severe congenital neutropenia, and familial HLH (15–17). This is the first report, to our knowledge, of a digenic human primary immunodeficiency caused by defects specific to IFNAR1 and IFNGR2. This is unique from previously published reports of patients with selective susceptibility to mycobacteria and viruses due to mutations in genes encoding proteins important for signaling via multiple IFNs and other cytokines. These include STAT1, TYK-2, and NEMO, all of which are downstream of all 3 types of IFNs as well as multiple cytokines. IL-27 signals via STAT1, IL-6 and IL-12 signal via TYK2, and multiple cytokines signal through the NEMO-dependent NF-κB classical pathway (7). Therefore, our patient’s susceptibility to multiple types of pathogens, specifically mycobacteria, extracellular bacteria, and CMV, illustrates the specific contributions of the IFNAR and IFNGR to host immunity in vivo.
Our patient had disseminated CMV characterized by an exceptionally high viral load, a striking phenotype not seen in a previously reported IFNAR2-deficient patient (2). We have shown an essential contribution of IFNAR1 signaling to IFN-α–mediated control of HCMV IE1 and IE2 protein expression. In addition, our patient’s mutation in IFNGR2 abrogates the contribution of IFN-γ to host defense against CMV. IFNGR2 and IFNAR1 form heterodimers (18). The binding of IFN-γ to IFNGR2/IFNAR1 heterodimers results in IFNAR1 phosphorylation, STAT1/STAT2 recruitment to the IFNAR1 subunit of the heterodimer, and ultimately, assembly of ISG factor 3 (ISGF3) for inhibition of viral activity (18). Additionally, IFN-γ induces ISGs through activation of IFNGR2/IFNAR1 heterodimers on the caveolar membrane (18). The importance of IFNAR1/IFNGR2 heterodimers to the antiviral state is seen in IFNAR1–/– mouse embryonic fibroblasts, in which IFN-γ fails to mount protection against the cytopathic effect of encephalomyocarditis virus (18). The immunocompetence against CMV observed in a previously reported IFNAR2-deficient patient may have been due to the preserved IFNAR1/IFNGR2 heterodimers, as well as an intact response to IFN-γ, as evidenced by the ability of IFNAR2-deficient fibroblasts to control viral infection after stimulation with IFN-γ, but not IFN-α (2). The dual contribution of the type I and type II IFNs to susceptibility to viral infection is illustrated by the IFNAR/IFNGR double-knockout mice, which are more susceptible to lymphocytic choriomeningitis virus than either the IFNR or IFNGR single-knockout mice (19). Both type I and II IFNs are important for upregulating the phagosomal proteins critical for defense against extracellular bacteria (20, 21). The dual defect in type I and II IFNs in our patient may have contributed to the disseminated infection with Streptococcus viridans, a weakly virulent, extracellular organism. The proband’s history of disseminated infections with opportunistic pathogens underscores his need for curative therapy. Although drugs that enable translational read-through of nonsense mutations have been used to treat cystic fibrosis and muscular dystrophy, this approach cannot correct defects caused by frameshift mutations. Hematopoietic stem cell transplant is the only cure at this time for this digenic disease.
Primary immunodeficiencies have been classically considered monogenic diseases, but WES/whole-genome sequencing has begun to identify phenotypes shaped by multiple mutations (15, 22, 23). This study demonstrates how a disease previously ascribed to a monogenic cause may arise from potentially novel multigenic mutations, indicating the utility of an unbiased approach for variant identification. Cases with atypical phenotypes studied before the widespread use of WES may thus have had secondary, unknown pathogenic mutations. Additionally, this study illustrates the burden of proof needed to determine the effect of novel secondary mutations, because the IFNAR1*557Gluext*46 mutant allele retained all of the protein’s coding amino acids and protein expression, but led to impaired downstream signaling and defective inhibition of viral replication. The dissociation of predictable genotype-phenotype relationships and the approaches needed to prove new mechanisms of disease will define the practice of genomic medicine.
Methods
The full description of methods is in the supplemental material.
Statistics.
All data are presented as mean ± SEM. Unpaired Student’s t test for single comparisons or 2-way ANOVA were used for multiple comparisons. P less than 0.05 was considered significant.
Study approval.
The study was approved by the Boston Children’s Hospital Institutional Review Board. Written informed consent was obtained from all participants.
Author contributions
RHB, JC, CNS, AB, WB, and M. Angelova performed experiments. EAI, MKH, HAA, FA, MAS, NE, MAE, and M. AlZahrani provided clinical data and gave critical advice. RHB, JC, DMK, M. AlZahrani, and RSG designed and coordinated the investigations. RHB, JC, DMK, and RSG wrote the manuscript. The final version of the manuscript was approved by all authors.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants P01AI-076210 and R21-AI124101 (to RSG) AI-106934 (to DMK), and K08AI116979-01 (to JC); a grant from the Perkin Fund (to RSG); a Jeffrey Modell Translational Research Award (to JC); and the Chilean Ministry of Education by the CONICYT PAI/INDUSTRIA 79090016 (to RHB).
Version 1. 11/06/2017
Electronic publication
Version 2. 11/10/2017
Error corrected in second sentence of abstract.
Version 3. 12/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(12):4415–4420.https://doi.org/10.1172/JCI93486.
Contributor Information
Rodrigo Hoyos-Bachiloglu, Email: rahoyosbachiloglu@gmail.com.
Janet Chou, Email: Janet.Chou@childrens.harvard.edu.
Abdallah Beano, Email: Abdallah.Beano@childrens.harvard.edu.
Wayne Bainter, Email: wayne.bainter@childrens.harvard.edu.
Magdalena Angelova, Email: magangelova@gmail.com.
Fahd Almanjomi, Email: Falmanjomi@kfmc.med.sa.
Mohamed Al Shehri, Email: malshehri@kfmc.med.sa.
Nagi Elsidig, Email: nagiamin16@hotmail.com.
Mofareh AlZahrani, Email: msalzahrani@kfmc.med.sa.
References
- 1.Zhang S-Y, et al. Inborn errors of interferon (IFN)-mediated immunity in humans: insights into the respective roles of IFN-α/β, IFN-γ, and IFN-γ in host defense. Immunol Rev. 2008;226(1):29–40. doi: 10.1111/j.1600-065X.2008.00698.x. [DOI] [PubMed] [Google Scholar]
- 2.Duncan CJA, et al. Human IFNAR2 deficiency: Lessons for antiviral immunity. Sci Transl Med. 2015;7(307):307ra154–307ra154. doi: 10.1126/scitranslmed.aac4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hambleton S, et al. STAT2 deficiency and susceptibility to viral illness in humans. Proc Natl Acad Sci U S A. 2013;110(8):3053–3058. doi: 10.1073/pnas.1220098110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shahni R, et al. Signal transducer and activator of transcription 2 deficiency is a novel disorder of mitochondrial fission. Brain J Neurol. 2015;138(Pt 10):2834–2846. doi: 10.1093/brain/awv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Muhsen S, Casanova J-L. The genetic heterogeneity of mendelian susceptibility to mycobacterial diseases. J Allergy Clin Immunol. 2008;122(6):1043–1051. doi: 10.1016/j.jaci.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 6.Bogunovic D, et al. Mycobacterial disease and impaired IFN-γ immunity in humans with inherited ISG15 deficiency. Science. 2012;337(6102):1684–1688. doi: 10.1126/science.1224026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustamante J, Boisson-Dupuis S, Abel L, Casanova J-L. Mendelian susceptibility to mycobacterial disease: genetic, immunological, and clinical features of inborn errors of IFN-γ immunity. Semin Immunol. 2014;26(6):454–470. doi: 10.1016/j.smim.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sampaio EP, et al. Signal transducer and activator of transcription 1 (STAT1) gain-of-function mutations and disseminated coccidioidomycosis and histoplasmosis. J Allergy Clin Immunol. 2013;131(6):1624–1634. doi: 10.1016/j.jaci.2013.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorman SE, et al. Viral infections in interferon-γ receptor deficiency. J Pediatr. 1999;135(5):640–643. doi: 10.1016/S0022-3476(99)70064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorman SE, et al. Clinical features of dominant and recessive interferon γ receptor 1 deficiencies. Lancet. 2004;364(9451):2113–2121. doi: 10.1016/S0140-6736(04)17552-1. [DOI] [PubMed] [Google Scholar]
- 11.Poulin S, et al. Fatal Mycobacterium colombiense/cytomegalovirus coinfection associated with acquired immunodeficiency due to autoantibodies against interferon gamma: a case report. BMC Infect Dis. 2013;13(1):1. doi: 10.1186/1471-2334-13-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchieri G. Type I interferon: friend or foe? J Exp Med. 2010;207(10):2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75(2):163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 14.Posey JE, et al. Resolution of disease phenotypes resulting from multilocus genomic variation. N Engl J Med. 2017;376(1):21–31. doi: 10.1056/NEJMoa1516767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Massaad MJ, et al. Deficiency of base excision repair enzyme NEIL3 drives increased predisposition to autoimmunity. J Clin Invest. 2016;126(11):4219–4236. doi: 10.1172/JCI85647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Germeshausen M, et al. Digenic mutations in severe congenital neutropenia. Haematologica. 2010;95(7):1207–1210. doi: 10.3324/haematol.2009.017665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang K, et al. Synergistic defects of different molecules in the cytotoxic pathway lead to clinical familial hemophagocytic lymphohistiocytosis. Blood. 2014;124(8):1331–1334. doi: 10.1182/blood-2014-05-573105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takaoka A, et al. Cross talk between interferon-gamma and -alpha/beta signaling components in caveolar membrane domains. Science. 2000;288(5475):2357–2360. doi: 10.1126/science.288.5475.2357. [DOI] [PubMed] [Google Scholar]
- 19.Ou R, Zhou S, Huang L, Moskophidis D. Critical role for alpha/beta and gamma interferons in persistence of lymphocytic choriomeningitis virus by clonal exhaustion of cytotoxic T cells. J Virol. 2001;75(18):8407–8423. doi: 10.1128/JVI.75.18.8407-8423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jutras I, et al. Modulation of the phagosome proteome by interferon-gamma. Mol Cell Proteomics. 2007;7(4):697–715. doi: 10.1074/mcp.M700267-MCP200. [DOI] [PubMed] [Google Scholar]
- 21.Charrel-Dennis M, et al. TLR-independent type I interferon induction in response to an extracellular bacterial pathogen via intracellular recognition of its DNA. Cell Host Microbe. 2008;4(6):543–554. doi: 10.1016/j.chom.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tangye SG. Genetic cause of immune dysregulation — one gene or two? J Clin Invest. 2016;126(11):4065–4067. doi: 10.1172/JCI90831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casanova J-L, Fieschi C, Zhang S-Y, Abel L. Revisiting human primary immunodeficiencies. J Intern Med. 2008;264(2):115–127. doi: 10.1111/j.1365-2796.2008.01971.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.