Abstract
Steroid-resistant nephrotic syndrome (SRNS) is a frequent cause of chronic kidney disease. Here, we identified recessive mutations in the gene encoding the actin-binding protein advillin (AVIL) in 3 unrelated families with SRNS. While all AVIL mutations resulted in a marked loss of its actin-bundling ability, truncation of AVIL also disrupted colocalization with F-actin, thereby leading to impaired actin binding and severing. Additionally, AVIL colocalized and interacted with the phospholipase enzyme PLCE1 and with the ARP2/3 actin-modulating complex. Knockdown of AVIL in human podocytes reduced actin stress fibers at the cell periphery, prevented recruitment of PLCE1 to the ARP3-rich lamellipodia, blocked EGF-induced generation of diacylglycerol (DAG) by PLCE1, and attenuated the podocyte migration rate (PMR). These effects were reversed by overexpression of WT AVIL but not by overexpression of any of the 3 patient-derived AVIL mutants. The PMR was increased by overexpression of WT Avil or PLCE1, or by EGF stimulation; however, this increased PMR was ameliorated by inhibition of the ARP2/3 complex, indicating that ARP-dependent lamellipodia formation occurs downstream of AVIL and PLCE1 function. Together, these results delineate a comprehensive pathogenic axis of SRNS that integrates loss of AVIL function with alterations in the action of PLCE1, an established SRNS protein.
Keywords: Nephrology
Keywords: Monogenic diseases
Introduction
Nephrotic syndrome (NS) is characterized by proteinuria caused by disruption of the glomerular filtration barrier (1). It is the second most frequent cause of chronic kidney disease before the age of 25 years and has an incidence of approximately 2 per 100,000 individuals (2). Steroid-resistant nephrotic syndrome (SRNS), which manifests histologically as focal segmental glomerulosclerosis (FSGS), inescapably results in end-stage renal disease (ESRD) through the progressive loss of the filtration barrier. It remains one of the most intractable kidney diseases (3). The identification of single-gene causes of SRNS and subsequent research on the function of those genes have helped to assemble the essential components of glomerular podocyte function (4–6). SRNS is genetically heterogeneous, and more than 50 monogenic genes have been discovered to cause podocyte dysfunction if mutated. These findings revealed podocytes as the critical site of pathogenesis in SRNS (5). Fascinatingly, the proteins encoded by the genes that are mutated in monogenic causes of disease have started to coalesce to protein interaction complexes that, through their loss of function, define the pathomechanisms of nephrotic syndrome (5).
One of the protein-protein interaction clusters is involved in the dynamic control of the actin cytoskeleton, which is essential in maintaining the shape and movement of the podocyte foot processes, as well as in maintaining cell-cell contacts. More than one hundred proteins are involved in the regulation of actin, enabling processes such as polymerization and depolymerization, which are necessary for foot process formation and lamellipodia formation in cell culture (7). Interestingly, 15 of the 40 disease-causing genes encode proteins that regulate the small Rho-like GTPases RhoA, Rac1,and Cdc42, which are the master regulators of actin cytoskeleton rearrangement including the Rho GDP disassociation inhibitor ARHGDIA (8). In addition, actin remodeling has also been implicated in NS (9). Defects in actin remodeling may be part of the reason why a defect in the podocyte migration rate (PMR) in cell culture has been established as a reliable intermediate phenotype in monogenic forms of SRNS (8, 10–12).
We have recently shown in a cohort of 1,780 families with SRNS that 30% of all cases are caused by mutations in 1 of the 26 genes known at the time to cause SRNS (13). However, this indicated that additional single-gene causes of SRNS must exist that are yet to be discovered. Therefore, to identify monogenic causes of SRNS and to further understand its pathogenesis, we applied homozygosity mapping (HM) and whole-exome sequencing (WES) to individuals with SRNS and identified mutations in AVIL as a cause of NS and characterized the related pathogenesis (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI94138DS1).
Results
Mutations of AVIL cause SRNS.
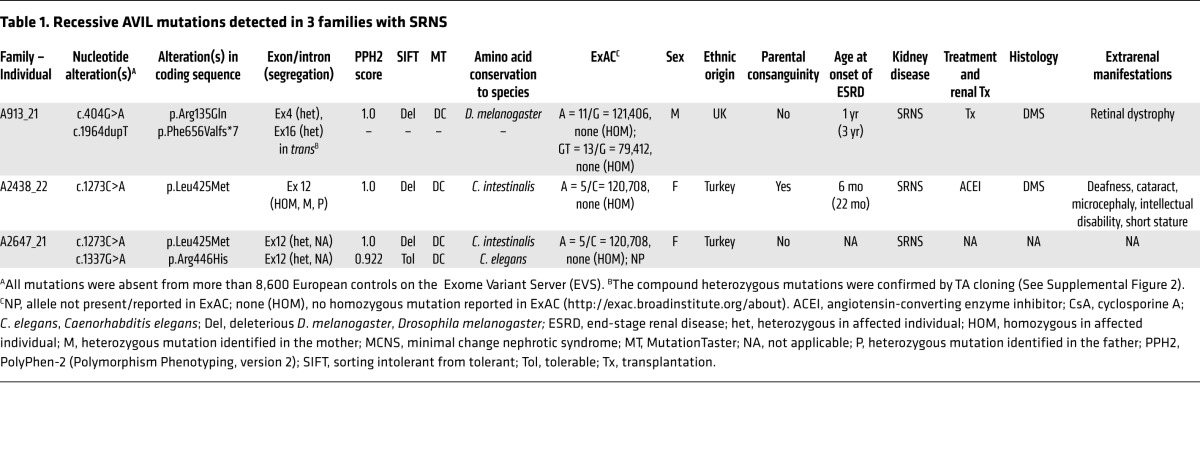
To identify monogenic causes of SRNS and further understand its pathogenesis, we applied HM (14) and WES (15) to individuals with SRNS. HM in 1 of the affected siblings with SRNS from a consanguineous Turkish family (A2438_22) (Figure 1, A and B) yielded 14 candidate regions of homozygosity by descent, with a cumulative genomic length of approximately 94 Mb (Figure 1C). Using WES, we detected a homozygous missense mutation (c.1273C>A; p.Leu425Met, conserved to Ciona intestinalis) in the advillin gene (AVIL) (RefSeq accession number NM 006576.3) (Table 1, Figure 2, A–D, and Supplemental Table 1). The mutation segregated with the affected status in this family and was not present homozygously in the Exome Aggregation Consortium (ExAC) data set (Table 1). Direct inspection of sequence alignments from whole-exome data did not yield a mutation in any of the 40 known NS genes.
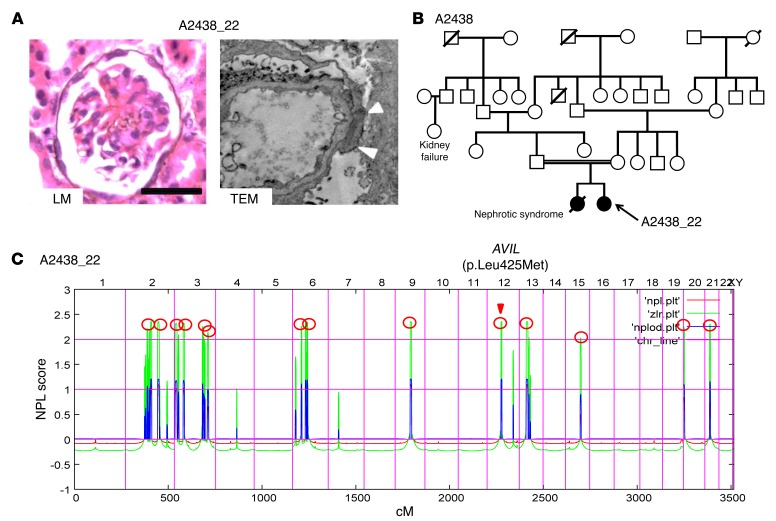
Figure 1. Renal histology, pedigree analysis and homozygosity mapping in family A2438 diagnosed with SRNS.
(A) Renal histology of individuals with AVIL mutation. Renal histology by light microscopy (LM) from individual A2438_22 with PAS staining showed DMS (scale bars: 50 μm). Transmission electron microscopy (TEM) image from individual A2438_22 showed diffuse foot process effacement (arrowheads). Original magnification, ×7,000. (B) Pedigree for index family A2438. Squares represent males, circles females, black symbols indicate affected persons, and white symbols designate unaffected persons. The double line indicates the consanguineous family. (C) Homozygosity mapping identified recessive candidate loci. In individual A2438_22 with SRNS, nonparametric lod (NPL) scores were calculated and plotted across the human genome. The x axis plots Affymetrix 250K StyI array SNP positions on human chromosomes concatenated from p-ter (left) to q-ter (right). Genetic distance is given in centimorgans (cM). Maximum NPL peaks (red circles) indicate 14 candidate regions of homozygosity by descent as recessive candidate loci. The AVIL locus (arrowhead) is positioned within one of the maximum NPL peaks on chromosome 12q.
Table 1. Recessive AVIL mutations detected in 3 families with SRNS.
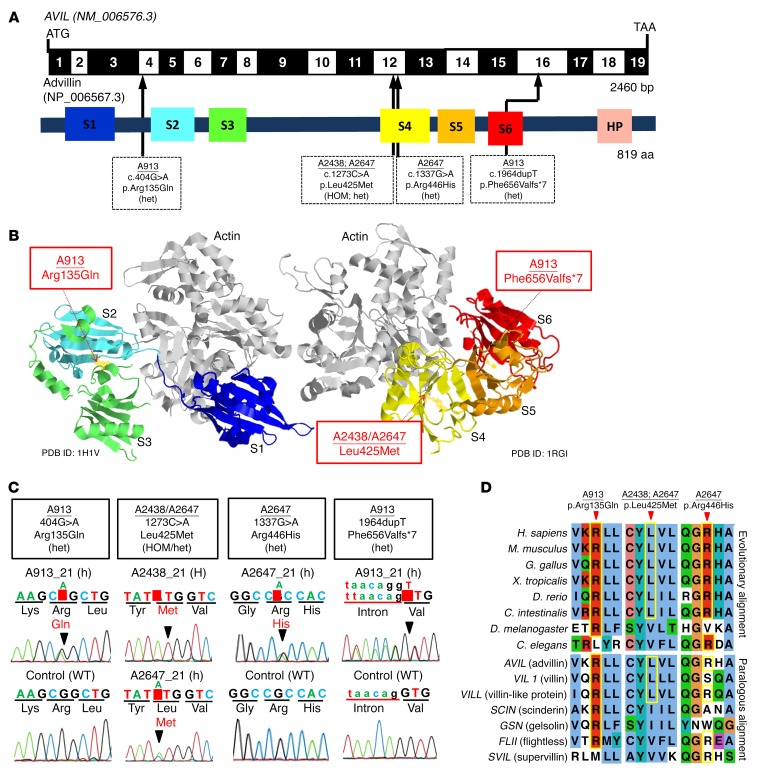
Figure 2. WES identifies recessive mutations of AVIL in 3 families with SRNS.
(A) Exon structure and functional domains of human AVIL cDNA. Positions of the start codon (ATG) and the stop codon (TAA) are indicated. Exons are marked on a black or white background. The length of protein domains is indicated by colored boxes. The positions of mutations (family numbers are underlined) are lined up by black arrows in relation to exons and protein domains (see also Table 1). (B) Ribbon diagram of the atomic structure of advillin. The gelsolin domains labeled S1 to S6 are color-coded blue (S1), aqua (S2), green (S3), yellow (S4), orange (S5), and red (S6), with actin shown in gray (Protein Data Bank [PDB] references: 1H1V and 1RGI). (C) Chromatograms of AVIL variants identified in individuals with SRNS. Sequence traces are shown for the variants above normal controls. Arrowheads denote altered nucleotides. (D) Multiple amino acid sequence alignment of AVIL throughout evolution and multiple sequence alignment of the human AVIL paralogs VIL1, VILL, SCIN, GSN, FLII, and SVIL using the Clustal_O program.
In order to investigate whether AVIL mutations occur in additional individuals with NS, we examined a worldwide cohort of approximately 800 individuals with SRNS by exon resequencing of all 19 AVIL-coding exons (Figure 2A) using an approach of multiplex PCR followed by next-generation sequencing (NGS), which we developed (16). In an individual with SRNS from the United Kingdom (A913_21), we detected 2 compound heterozygous mutations in AVIL: c.404G>A (p.Arg135Gln) and c.1964dupT (p.Phe656Valfs*7) (Table 1 and Figure 2, A–D). The 2 heterozygous mutations were sequenced after subcloning into a vector plasmid by TA cloning, and it was found that the compound heterozygous mutations were in trans. (Supplemental Figure 2). In an SRNS patient of Turkish descent (A2647_21), we detected 2 compound heterozygous mutations in AVIL: c.1337G>A (p.Arg446His) and c.1273C>A (p.Leu425Met) (Table 1 and Figure 2, A–D). We further confirmed the rarity of the genetic variants identified in the Turkish SRNS patients A2433_22 and A2647_21 by examining and confirming their absence from the high-throughput exon-sequencing data from approximately 200 SRNS patients of Turkish descent.
The renal histological analyses for individuals A2438_22 and A913_21 revealed diffuse mesangial sclerosis (DMS) (Table 1 and Figure 1A). The 2 individuals manifested signs of NS before 1 year of age and progressed to ESRD before 3 years of age (Table 1). DMS is a distinct clinicopathologic entity of severe NS (17) that shares a pathogenic spectrum with focal segmental glomerulosclerosis (FSGS) (18). It is characterized clinically by early-onset SRNS and rapid progression to ESRD. Individual A2438_22 had severe SRNS combined with deafness, cataracts, microcephaly, and intellectual disability. Individual A913_21 had severe NS combined with retinal dystrophy and pervasive refusal syndrome. A spectrum ranging from severe early-onset NS with DMS to a later-onset form with FSGSwas observed here, as has also been described in phospholipase C ϵ1 (PLCE1) mutations (19).
In total, we detected 4 different homozygous or compound heterozygous mutations of AVIL in 3 families affected by NS (Table 1, Figure 1, A–D, and Supplemental Figure 1, A–C). Copy number variation (CNV) analysis of these affected individuals revealed no deleterious CNV. Mutation segregation was consistent with recessive inheritance when parental DNA was available (Table 1). We thereby identified recessive mutations in AVIL as a cause of recessive NS and introduce the term “NPHS25” for this variant of monogenic SRNS.
Location of and impact on structural flexibility of the mutations.
Advillin (human Avil-1 [hAvil-1]) is a member of the gelsolin/villin family of proteins and is most similar in sequence to villin. It shares the 6 gelsolin homology (GH) domains (Figure 2, A and B) with a number of other mammalian gelsolin/villin family members (Figure 2D), as well as the carboxyl terminal headpiece domain of villin. The 2 missense mutations (Arg135Gln and Leu425Met) are located in the interior of the GH2 and GH4 domains between the β-sheet and the long α-helix involved in actin binding (Figure 3A). In order to assess the impact on the structure and flexibility of the resulting mutant domains, we performed classical and scaled molecular dynamic (MD) simulations. The comparison of descriptors reflecting flexibility during the scaled MD simulations was indicative of dynamic changes between the WT and the mutated GH2 (Supplemental Figure 3, A–D) and GH4 domains (Supplemental Figure 3, E–H). These regions are mapped down on the overlay of the structures averaged over the trajectories of the WT domain and that of the mutant domains (Figure 3A). The Arg135Gln mutation in GH2 results in displacement of the loop in between the first and second β strand. As the latter contains residues involved in the coordination of the calcium ion at the type 2 site, this may in turn influence the ability of hAvil-1 to adopt its active, actin-binding state. The Arg135Gln mutation replaces a long, positively charged side chain with the shorter, polar but uncharged one of glutamine, with a propensity for the formation of hydrogen bonds rather than salt bridges (Figure 3A). According to scaled MD simulations, the Leu425Met mutation in GH4 correlates with a displacement of the C-terminal end of the actin-binding α helix. Since this α-helix on the GH4 domain is involved in binding the actin filament, the mutation could therefore potentially affect the ability of hAvil-1 to carry out its biological function. The Leu425Met mutation involves a change for leucine with a bulky, hydrophobic side chain to methionine, which is similarly long but less bulky and more polar in nature (Figure 3A).
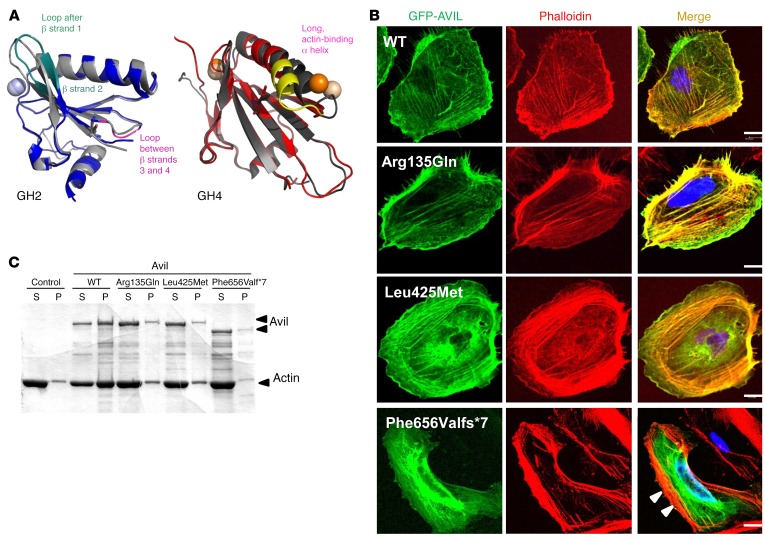
Figure 3. Mutations in AVIL affect podocyte cytoskeleton architecture and inhibit the actin-bundling function of AVIL.
(A) Locations of mutations in gelsolin homology domains of advillin and their effect on conformational flexibility. Overlay of averaged structures of WT (dark gray) and mutated GH2 (blue) and GH4 (red) domains from MD studies of human AVIL (PDB references: 1RGI and 1H1V). Averaging was carried out over the whole scaled MD trajectory. A homology model for hAvil-1 was built using gelsolin as a template based on the crystal structures of actin-bound gelsolin 1H1V4 (S4–S6 or GH4–GH6 domains) (57) and 1RGI (S1–S3 or GH1–GH3 domains) (58). The models built served as starting structures for all MD studies. Structural elements with increased conformational flexibility are highlighted in teal and magenta for the GH2 domain and in yellow for the GH4 domain. Mutated residues are depicted with sticks, and spheres denote calcium ions. (B) Colocalization of AVIL with F-actin using cDNA clones representing WT and mutations detected in individuals with SRNS (Table 1). Human podocytes were transfected with GFP-tagged WT or the mutant AVIL (Arg135Gln, Leu425Met, or Phe656Valfs*7) and were stained for F-actin with phalloidin (red). Colocalization of AVIL and phalloidin-labeled F-actin resulted in yellow fluorescence. Note that podocytes with overexpression of the truncating mutant Phe656Valfs*7 lost the costaining pattern with phalloidin-labeled F-actin at the cell periphery (white arrowheads). Cell nuclei were stained with DAPI (blue). Scale bars: 10 μm. (C) For the actin-bundling assay, WT or mutant (Arg135Gln, Leu425Met, or Phe656Valfs*7) Avil GST proteins (1 μM) were incubated with F-actin. To determine actin bundling, samples were centrifuged at 10,000 g for 15 minutes, and actin distribution in supernatant (S) and pellet (P) fractions was analyzed by 10% SDS-PAGE and GelCode Blue staining. Control refers to F-actin filaments in the absence of Avil GST protein.
The AVIL-truncating mutant lacks colocalization with F-actin.
AVIL is known to be involved in neurite outgrowth and morphogenesis (20), but there is no information on its role in the kidney or podocytes. To study AVIL protein localization, we characterized 2 different polyclonal antibodies directed against the N-terminus (α-AVIL-91606) and C-terminus (α-AVIL-80312) of AVIL (Supplemental Figure 4, A and B). We demonstrated the presence of AVIL in protein extracts from cultured human podocytes (Supplemental Figure 4, A and B). Using immunoblotting, we found that α-AVIL-80312 recognized a band of 92 kDa (Supplemental Figure 4B). To investigate subcellular localization of AVIL in podocytes, we performed immunofluorescence (IF) studies in a human podocyte cell line upon overexpression of GFP-tagged AVIL (Supplemental Figure 4, C and D). We found that AVIL was present in the F-actin–enriched cell periphery labeled with phalloidin that generates lamellipodia and focal adhesions (FAs) (Figure 3B). However, the truncating mutant construct of AVIL (Phe656Valfs*7) failed to localize with F-actin in podocytes (Figure 3B). To explore the deleteriousness of the AVIL mutations detected in individuals with SRNS, we generated the AVIL-truncating mutant constructs AVIL-Δ628-819 (deletion of the C-terminal S6 and HP domains) and AVIL-Δ135-143 (lacking one of the possible PIP2-binding motifs) (Supplemental Figure 5A). We found that GFP-tagged AVIL-Δ628-819 lacked localization with F-actin in podocytes, which is an equivalent to the mutant construct of AVIL (Phe656Valfs*7) (Supplemental Figure 5B).
Endogenous expression of AVIL in rat kidney glomeruli.
We performed immunofluorescence staining in 2-day-old rat kidney glomeruli, which showed that AVIL localized to the cytoplasm of podocytes in rat glomeruli (Supplemental Figure 6). We observed AVIL staining in cells positive for the podocyte marker WT1 in later stages of development of rat kidney glomeruli.
Mutations in AVIL inhibit its actin-regulatory functions.
Like villin, gelsolin, and other members of actin-binding proteins, advillin has been described as playing an important role in F-actin polymerization (21). To further characterize the pathogenic effects of AVIL mutations detected in patients with SRNS, we studied the role of AVIL for actin-bundling,- binding, and -severing properties. Actin bundling was measured by low-speed centrifugation and sedimentation of bundled actin filaments. We found that WT AVIL protein bundled actin and that the majority of the actin filaments appeared in the pellet (P) fraction (Figure 3C). However, in all 3 AVIL mutants (Arg135Gln, Leu425Met, and Phe656Valfs*7) that represented the mutations found in patients with SRNS, a majority of the actin filaments now appeared in the supernatant (S) fraction, demonstrating a severe defect in actin bundling.
We also tested whether the actin-binding and -severing functions of AVIL were altered in its mutants. We examined the binding properties of AVIL by using a cosedimentation assay and found that only the truncating mutant of AVIL (Phe656Valfs*7) that lacks its F-actin–binding C-terminal domain showed a decrease in the binding affinity of AVIL for actin (Supplemental Figure 7A). Further, we investigated the effect of mutant AVIL proteins on the actin-severing activity of advillin. Whereas the WT AVIL and 2 mutants representing missense alleles showed normal actin-severing function, the truncating mutant exhibited a complete loss of the actin-severing ability of AVIL (Supplemental Figure 7B).
AVIL interacts with PLCE1 and the actin-related protein 2/3 complex.
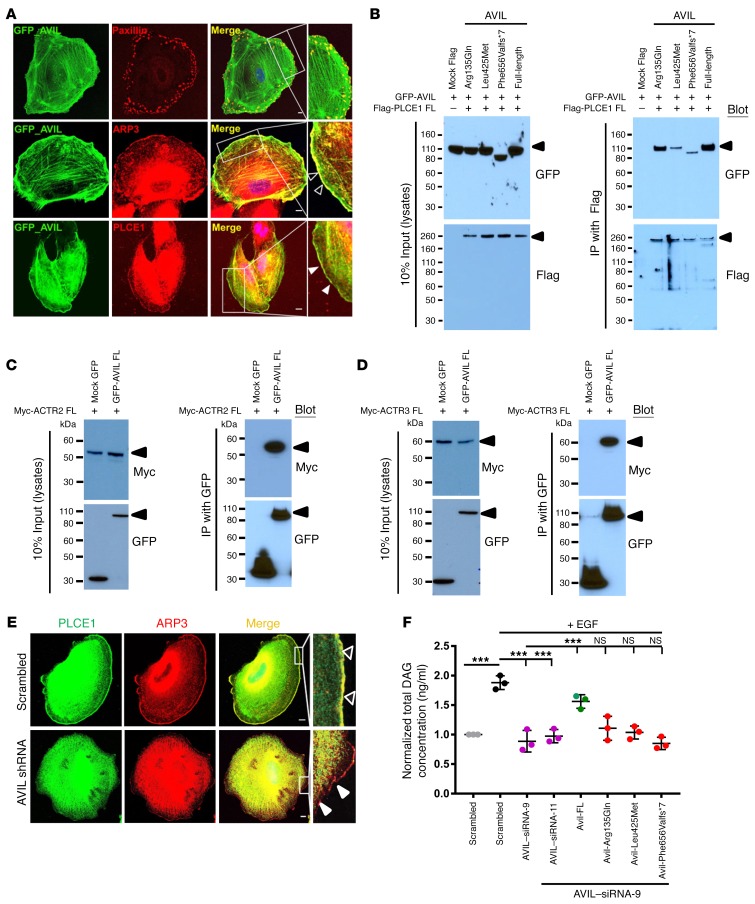
We then delineated a comprehensive pathogenic axis of SRNS that integrates PLCE1 loss of function as follows: Immunofluorescence staining of AVIL and paxillin, as a marker for FA (22), showed stronger AVIL localization at lamellipodia than at FAs (Figure 4A). It is well known that villin, a paralog of advillin (Figure 2D), interacts with phospholipase C γ1 (PLC-γ1) and regulates PLC-γ1 catalytic activity, thus modifying phosphatidylinositol signaling pathways (23). We previously demonstrated that PLCE1 mutations cause a form of SRNS that involves podocyte dysfunction (19). We therefore hypothesized that PLCE1, rather than PLCG1, may represent the phospholipase C isoform relevant for podocyte function and that advillin, rather than villin, may be activated by PLCE1 in podocytes (Supplemental Figure 1, A–C). Immunostaining for AVIL and PLCE1 revealed colocalization of both proteins at the cell periphery of human podocytes (Figure 4A). This tight subcellular colocalization is consistent with defects of AVIL or PLCE1 causing monogenic recessive SRNS. Because of the colocalization of AVIL with F-actin in podocytes (Figure 3B), and because of the known role of AVIL in actin severing, which is a prerequisite for lamellipodia formation, we looked for AVIL colocalization with the actin-nucleating complex member actin-related protein 2/3 (ARP2/3), which has been shown to play a major role in dynamic actin assembly, especially in lamellipodia (24). AVIL and ARP3 colocalized at the podocyte periphery (Figure 4A), as we observed with F-actin (Figure 3B).
Figure 4. AVIL colocalizes with and interacts with PLCE1 and the ARP2/3 complex.
(A) Human podocytes transfected with a GFP-tagged, full-length (FL) mouse Avil construct were immunostained with mouse anti-paxillin (red), mouse anti-ARP3 (red), and rabbit anti-PLCE1 (red) antibodies, respectively. Note the colocalization of AVIL and ARP3 at podocyte lamellipodia (empty white arrowheads) and the colocalization of AVIL and PLCE1 at lamellipodia (solid white arrowheads). AVIL also overlapped with paxillin at FAs. Scale bars: 10 μm. Inset scale bars: 25 μm. (B) AVIL interacted with PLCE1 upon co-overexpression in HEK293T cells. GFP-tagged AVIL was co-overexpressed with Flag-tagged PLCE1 in HEK293T cells. Co-IP using Flag showed that GFP-tagged AVIL interacted with Flag-tagged PLCE1. (C and D) Upon co-overexpression in HEK293T cells, Myc-tagged ACTR2 (ARP2 complex) (C) and ACTR3 (ARP3 complex) (D) interacted with GFP-tagged AVIL. (E) Under EGF stimulation (100 ng/ml) in human cultured podocytes, PLCE1 (green) translocated to cell membrane ruffles, where it colocalized with ARP3 (red, solid white arrowheads). Cells depleted of AVIL by shRNA failed to recruit PLCE1 to the ARP-marked lamellipodia (empty white arrowheads) upon EGF stimulation. Scale bars: 10 μm. Inset scale bars: 25 μm. (F) Human podocytes were transfected with scrambled siRNA, siRNA-9, or siRNA-11 for AVIL. EGF stimulation increased the concentration of DAG in scrambled siRNA–treated control cells (black circles). siRNA-mediated knockdown of AVIL decreased active DAG levels (pink circles). The overexpression of WT Avil rescued this effect (green circles), while the 3 mutant clones (Arg135Gln, Leu425Met, and Phe656Valfs*7) from patients with SRNS did not (red circles). Individual data points were derived from 3 independent experiments and are displayed as the mean and SD. Statistical analysis was performed using a 2-tailed, 1-way ANOVA [F (7,16) = 92.09, P < 0.0001]. ***P < 0.0001, by Sidak’s multiple comparisons test.
The observation that AVIL colocalized to the cell periphery with PLCE1 and ARP3 in a human podocyte cell line led us to perform co-IP studies to test for molecular interaction between AVIL and PLCE1 or ARP2/3. We found that AVIL coimmunoprecipitated with PLCE1 following cotransfection of Flag-tagged PLCE1 and GFP-tagged AVIL into HEK293T cells. We showed that the mutants Leu425Met and Phe656Valfs*7, reflecting the alleles found in the patients with NS, weakened this interaction (Figure 4B). We next tested the interaction of AVIL with Myc-tagged ATCR2 (member of the ARP2 complex) (Figure 4C) and Myc-tagged ATCR3 (member of the ARP3 complex) (Figure 4D) by co-IP in HEK293T cells and observed that AVIL can form a protein complex with ARP2/3. We thereby implicated AVIL interaction with PLCE1 and the ARP2/3 complex in podocyte function (Supplemental Figure 1, C–G).
Our findings integrate the pathogenic mechanisms of AVIL mutations with the function of PLCE1, F-actin, and the ARP complex, all of which play a central role in podocyte lamellipodia formation. To further determine the role of AVIL in podocyte function, we studied whether depletion of AVIL by shRNA knockdown would disrupt the actin cytoskeleton and podocyte morphology. We found that depletion of AVIL from human podocytes did not result in a significant change in podocyte morphology (Supplemental Figure 8 and Figure 4E), and FA still formed, as shown by α-actinin-4 (labeling the actin structure) and paxillin antibodies, respectively (Supplemental Figure 8, A and C). However, there were fewer formations of actin stress fibers and less abundance of paxillin at the podocyte lamellipodia following shRNA-mediated knockdown of AVIL compared with the scrambled shRNA negative control (Supplemental Figure 8, A and C, and Figure 4E). To confirm that the effect on actin stress fibers was specific to knockdown of AVIL, we performed siRNA-mediated knockdown of NUP107, a known monogenic cause of SRNS (25), in human podocytes and observed no effect on stress fibers compared with the scrambled siRNA–knockdown control (Supplemental Figure 8B). Upon stimulation with EGF, PLCE1 translocated to the lamellipodia (as described in ref. 26), where it colocalized with ARP3 (Figure 4E and Supplemental Figure 8D). Interestingly, depletion of AVIL from podocytes failed to recruit PLCE1 to the ARP3-rich region, which marks the formation of lamellipodia upon EGF stimulation (Figure 4E and Supplemental Figure 8D). We confirmed the efficiency of knockdown by shRNA in podocytes by immunoblotting using an anti-advillin antibody (Supplemental Figure 4B). The results indicated that AVIL plays a role in lamellipodia formation through the ARP2/3 complex (Supplemental Figure 1, F and G).
EGF-induced diacylglycerol generation by phospholipase C ϵ1 activity is blocked by AVIL knockdown.
PLCE1 is a member of the phospholipase family of enzymes that catalyzes the hydrolysis of polyphosphoinositides to generate the second messengers inositol-1,4,5 trisphosphate (IP3) and diacylglycerol (DAG) (19). Our group has previously demonstrated that PLCE1 causes SRNS (19). In addition, it is well known that villin, a paralog of advillin (Figure 2D), interacts with PLC-γ1 and regulates PLC-γ1 catalytic activity, thus modifying phosphatidylinositol signaling pathways (23). We therefore hypothesized that PLCE1, rather than PLCG1, may represent the PLC isoform relevant for podocyte function and that advillin, rather than villin, may be the relevant activator of podocytes. We tested and confirmed the interaction of AVIL with PLCE1 and delineated a pathogenic axis in SRNS that integrates AVIL loss of function with the established role of the SRNS protein PLCE1 (Supplemental Figure 1). Next, we measured the effect of AVIL knockdown on the levels of EGF-induced DAG generated by PLCE1 in human cultured podocytes and found that upon knockdown of AVIL, DAG levels where reduced, even after stimulation with EGF. This effect was found to be reversed when WT mouse Avil was transfected into human podocytes (Figure 4F). In contrast, all 3 AVIL mutants (Arg135Gln, Leu425Met, and Phe656Valfs*7), detected in patients with NS, failed to fully rescue the decreased DAG levels (Figure 4F), thereby connecting the pathogenesis of AVIL mutations to the loss of function of DAG generation by the SRNS-related protein PLCE1 (Supplemental Figure 1, D and E).
AVIL mutations lack modulation of the PMR.
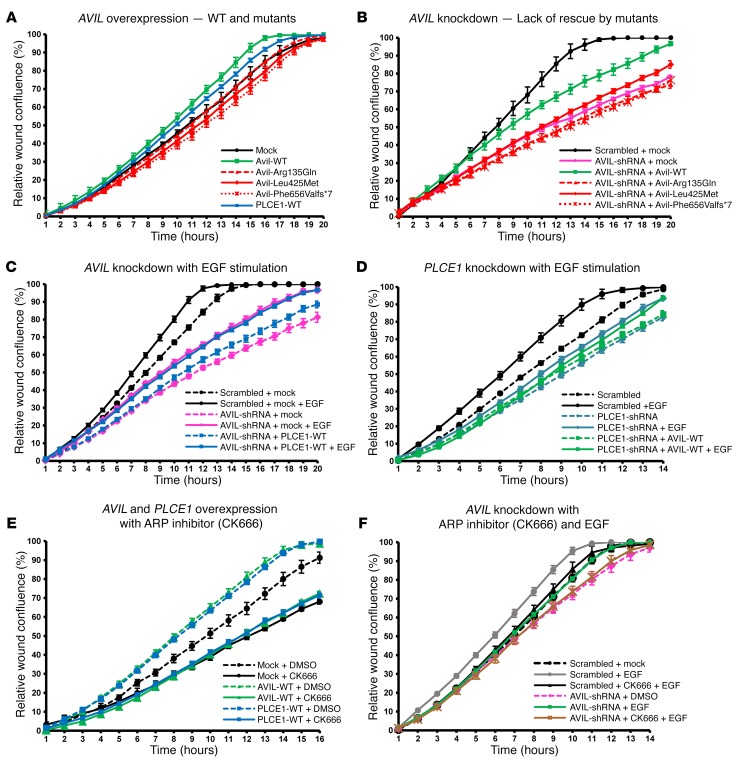
We next examined the effect of AVIL on the PMR, which represents a relevant intermediate cellular phenotype in many forms of monogenic SRNS (8, 10, 12). We used the IncuCyte ZOOM video microscopy system, which monitors cell migration in real time. We observed an increase in the PMR upon overexpression of mouse Avil or human PLCE1 in human podocytes (Figure 5A). In contrast, the AVIL construct representing mutations detected in individuals with SRNS failed to increase the PMR (Figure 5A). Furthermore, we observed a PMR decrease after shRNA-mediated knockdown of AVIL in cultured human podocytes (Figure 5B). Interestingly, the PMR decrease upon knockdown of AVIL was reversed by overexpression of WT mouse Avil. In contrast, overexpression of Avil-mutant constructs that represented the mutations found in patients with SRNS (Arg135Gln, Leu425Met, and Phe656Valfs*7) failed to rescue the reduction in the PMR caused by AVIL knockdown (Figure 5B). We thereby connected AVIL loss of function to altered PMR as an established intermediate phenotype of SRNS (Supplemental Figure 1, H and I).
Figure 5. The ARP complex is an effector for podocyte migration downstream of EGF, AVIL, and PLCE1.
(A) Human podocytes transfected with WT Avil (green) or PLCE1 (blue) showed a significant increase in the PMR compared with mock (black). Overexpression of Avil mutants failed to increase the PMR. Mutations: Arg135Gln (red triangle dotted line), Leu425Met (red rhombus solid line), and Phe656Valfs*7 (red cross dotted line). (B) Knockdown of AVIL (pink) showed a strongly reduced PMR compared with scrambled shRNA (black), which was partially rescued by overexpression of WT Avil (green) but not by mutants. (C) EGF stimulation increased the PMR in scrambled shRNA (black solid line). The PMR reduction upon AVIL knockdown (pink dotted line) was partially rescued by EGF treatment (pink solid line), but overexpression of PLCE1 (blue dotted line) showed no additional rescue effect over EGF (blue solid line). (D) The PMR reduction upon PLCE1 knockdown (turquoise dotted line) was partially rescued by EGF (turquoise solid line). Overexpression of AVIL (green dotted line) failed to rescue the PMR reduction in PLCE1-knockdown cells (turquoise dotted line). Overexpression of Avil showed no additional rescue effect over EGF treatment (green solid line). (E) Human podocytes transfected with the scrambled control showed a decreased PMR upon treatment with the ARP inhibitor CK666 (black solid line) compared with the scrambled control without CK666 treatment (black dotted line). The PMR increase upon overexpression of WT Avil (green dotted line) or PLCE1 (blue dotted line) was blocked by the addition of CK666 (green or blue solid lines, respectively). (F) Human podocytes were pretreated with CK666 for 45 minutes before the wound scratch was performed. Migration assays with scrambled shRNA as a negative control were conducted with (gray solid line) or without (black dotted line) EGF stimulation following the wound scratch in podocytes. The reduction in PMR upon AVIL knockdown (pink dotted line) was rescued by EGF (green solid line), but was not rescued in the presence of CK666 (brown solid line). The relative migration rate and wound density (percentage) were calculated by IncuCyte assay. Multiple scratch wounds were made in confluent cells. Scratch wounds were allowed to heal for 24 hours, and the PMR was plotted on graphs.
Regulation of podocyte migration by interaction of AVIL with PLCE1.
Next, we tested the mutual functional interaction of AVIL and PLCE1 with regard to PMR modulation. Both genes cause SRNS if mutated, and both proteins colocalized to lamellipodia in cultured human podocytes (Figure 4A). Interestingly, we found an increase in the PMR upon overexpression of human PLCE1 in human podocytes (Figure 5A). Furthermore, we observed that the decreased PMR upon knockdown of AVIL was rescued by overexpression of human PLCE1 (Figure 5C). In contrast, we found that the decreased PMR upon knockdown of PLCE1 was not rescued by overexpression of human AVIL (Figure 5D), indicating that PLCE1 is downstream of AVIL in terms of PMR modulation, again connecting the SRNS-related pathogenesis of AVIL loss of function to PLCE1 (Supplemental Figure 1, H and I).
We then explored the effect of EGF on PMR modulation under AVIL or PLCE1 knockdown in human podocytes. EGF stimulation led to a marked increase in podocyte migration (Figure 5, C and D). The overexpression of human PLCE1 in AVIL-knockdown podocytes showed no additional effect after EGF stimulation (Figure 5C). This indicates that AVIL and PLCE1 are necessary for EGF-induced migration and are thus both downstream of PMR modulation by EGF (Supplemental Figure 1, C–I).
The ARP complex is a PMR effector downstream of EGF, AVIL, and PLCE1.
As we found that AVIL colocalized and interacted with the ARP2/3 complex in actin-rich lamellipodia of the podocytes described above (Figure 4, B–D), we next assessed the effects of ARP2/3 complex inhibition on podocyte migration using the small-molecule inhibitor CK666, which binds at the interface of ARP2 and ARP3, thereby blocking their conversion into the active conformation (27). We found that CK666 treatment led to a marked PMR decrease (Figure 5E). Interestingly, the increase in the PMR upon AVIL or PLCE1 overexpression was abolished upon treatment with CK666 (Figure 5E). This strongly suggests that the PMR increase induced by AVIL or PLCE1 is dependent on ARP2/3 complex activity. Furthermore, CK666 treatment blocked EGF-induced migration in AVIL-knockdown cells (Figure 5F), demonstrating that ARP2/3 function is downstream of PMR modulation by EGF, AVIL, and PLCE1 (Supplemental Figure 1, F–I).
Discussion
In this study, we have identified recessive mutations in advillin (AVIL) as a single-gene cause of SRNS. We describe how AVIL mutations result in a defective PMR as an intermediate phenotype of SRNS and elucidate the role of EGF signaling, PLCE1 recruitment by AVIL, IP3 and DAG generation by PLCE1, actin bundling by AVIL, and assembly of the ARP2/3 complex to form podocyte lamellipodia (Supplemental Figure 1).
Advillin is similar to other members of the gelsolin/villin family and is most similar to villin in terms of sequence. It shares the 6-domain gelsolin structure (Figure 2, A and B) with a number of other mammalian gelsolin/villin family members (Figure 2D), as well as the carboxyl terminal headpiece domain of villin. Gelsolin/villin proteins have been shown to serve important functions in regulating the actin organization. Here, we studied and characterized the role of AVIL for actin-bundling, -binding, and -severing properties. We demonstrate the involvement and role of AVIL in actin dynamics and further characterize the pathogenic effects of AVIL mutations that were detected in individuals with SRNS. Interestingly, we found that, while all AVIL mutants showed a significant loss of its actin-bundling ability, the AVIL-truncating mutation also disrupted colocalization with F-actin and showed impaired actin-binding and -severing function.
PLCE1, a known NS-causing gene if mutated, is a member of the phospholipase family of enzymes that catalyzes the hydrolysis of polyphosphoinositides to generate the second messengers IP3 and DAG (Supplemental Figure 1). DAG regulates the release of calcium from intracellular stores (19), thereby initiating actin severing, which is a prerequisite for actin nucleation by the ARP2/3 complex and a prerequisite for lamellipodia formation and podocyte migration (Supplemental Figure 1, D–I). PLCE1, linked with second messengers and the small Rho-like GTPases, has been helpful in identifying the pathogenic pathway in SRNS (Supplemental Figure 1). We confirmed that AVIL interacts and colocalizes with PLCE1 in human podocytes. Furthermore, we also found that AVIL interacts with ARP2/3 and colocalizes with it at the podocyte periphery.
We explored the effect of AVIL on the PMR, which represents a relevant intermediate cellular phenotype in many forms of monogenic SRNS. Interestingly, we found that EGF stimulation increased the PMR, which was partially diminished by knockdown of the SRNS-causing gene AVIL or PLCE1. In addition, the PMR increase caused by overexpression of AVIL or PLCE1 or by EGF stimulation was abolished by the ARP2/3 complex inhibitor CK666, indicating that ARP-dependent lamellipodia formation occurs downstream of AVIL or PLCE1 function.
To test whether the knockdown of Avil in zebrafish results in a renal phenotype, we also generated a CRISPR model of Avil knockdown in zebrafish, but our initial experiments did not show a larval edema phenotype. However, virtually the entire literature on zebrafish models for NS has been based on morpholino oligonucleotide (MO) knockdown in zebrafish larvae. This includes the genes PLCE1 (19), ADCK4 (28), ARHGDIA (8), and EMP2 (29). And it is of note that the approach of morpholino-based knockdown of genes in zebrafish disease models has been recently questioned in a major way, because these data were not reproducible in mutant stable lines (30). Therefore, we currently do not expect to be able to easily generate a zebrafish model for NS.
In summary, we have delineated a complete pathogenic axis in SRNS that integrates AVIL loss of function with the role of the established SRNS protein PLCE1. Multiple components of this pathogenic axis may represent drug targets for SRNS, for which no efficient treatment exists.
Methods
Research subjects.
We obtained blood samples and pedigrees after obtaining informed consent from individuals with NS. The diagnoses of NS and SRNS were based on published clinical criteria and renal biopsies criteria evaluated by renal pathologists (31). Clinical data were obtained using a standardized questionnaire (http://www.renalgenes.org).
Linkage analysis.
For genome-wide HM, the GeneChip Human Mapping 250k StyI Array from Affymetrix was used. Nonparametric lod scores were calculated using a modified version of the program GENEHUNTER 2.1 (32, 33) with stepwise use of a sliding window with sets of 110 SNPs and the program ALLEGRO (33) to identify regions of homozygosity, as described previously (14, 34), using a disease allele frequency of 0.0001 and Caucasian marker allele frequencies.
WES and mutation calling.
WES and variant burden analysis were performed as described previously (35).
High-throughput mutation analysis using array-based multiplex PCR and NGS.
We used PCR-based 48.48 Access Array microfluidic technology (Fluidigm) with consecutive NGS. A 12-fold primer multiplexing approach was applied, allowing PCR-based amplification for 48 DNA samples simultaneously in 576 amplicons (16). A total of 800 individuals with SRNS were analyzed. The primer sequences for targeted exon sequencing are provided in Supplemental Table 2.
cDNA cloning.
A mouse Avil full-length cDNA was subcloned by PCR from mouse full-length cDNA (clone 40129468, catalog MMM1013-211691434; Thermo Fisher Scientific). Human full-length cDNA was subcloned from a full-length human AVIL cDNA clone purchased from Open Biosystems (BC111730). Three variations to the human reference sequence (NM_009635.3) were corrected by mutagenesis using the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies). A human PLCE1 construct was created as described previously (19). Expression constructs (pGEX4T-3, pRK5-N-Myc, pCDNA6.2-N-GFP, pCS-Dest2, and pSirenRetroQ) were produced using LR Clonase (Invitrogen, Thermo Fisher Scientific) following the manufacturer’s instructions.
TA cloning.
In the case of A913_21, two SNPs (c.940-88G>T and c.1672-1161T>G) were found between the two mutations c.404G>A and c.1964dupT in the AVIL gene. By designing the primers for nested PCR, 3 products were amplified from genomic DNA, gel-purified, ligated by means of a TA ligation method into the TA-cloning vector pGEM-T (Promega), and subcloned into E. coli DH5α–competent cells (Invitrogen, Thermo Fisher Scientific) using standard procedures. The plasmid inserts were sequenced using the forward T7 and SP6 primers. DNA from 10 to 15 independent clones was sequenced. WT and mutated sequences were compared manually. The primer sequences for nested PCR were as follows: P1.1, AGTGAGGCTGCATCTGTGTAATT; P1.2, GATGTTCCCTTAGCTACAAGCAGG; P1.3, CATTCCCAAAGGCAGGTCAGT; P1.4, ATCGTTGACGGTCTCCACATT; P2.1, CACCGCAAGCCAATAAACAGAT; P2.2, CTTGGTCCTTAGACAAAAGTGAGTTAGA; P2.3, CAACATGGCGAAACCTCGTCT; P2.4, CAGGAGTAGGATTGCTGGATCA; P3.1, CATCATTAGTCACTAGGGAGATGCA; P3.2, TGACAACATAAAATATGAGCAAGGATGC; 3.3, GTGCAGGTACTGCTGTGCT; and P3.4, CTGTGAAGATGGGAGGCTCAAAC.
Computational methods for MD.
The automatic model-building search for a structural template was carried out against the SWISS-MODEL template library (SMTL, update: 2015-04.01) using BLAST (36) and HHbit (37) for evolutionarily related structures matching the target sequence. The template was selected on the basis of quality assessment using the QMEAN scoring function (38). Multiple sequence alignment was performed using Promod-II16. The template selected was the x-ray structure of gelsolin solved at 2.80 Å resolution (2FGH, chain A). The sequence covered 88% of the modeled structure, with 43% sequence similarity and 46.11% sequence identity.
Model building.
Manual multiple sequence alignment was carried out using ClustalX (39). Homology models were built with Modeller 9.16 (40) using the Chimera interface (41). The sequence identity was calculated in the Multiple Alignment Viewer of Chimera. For modeling of the gelsolin domain–containing part of hAvil-1 (without the headpiece), the crystal structures of 1H1V, 1RGI, and 3FFK were used. 1RGI and 3FFK covered the GH1-3 part of hAvil-1 (48.82% and 48.40% sequence identity, respectively), while 1H1V covered the GH4-GH6 part (48.62% sequence identity). No attempt was made to model the headpiece domain. The 1H1V chain G structure was used as a template to build the model of the individual GH4 domain. The sequence of 1H1V covered 100% of the modeled GH4 domain, with 48.63% sequence identity and 23.35% sequence similarity. The 1RGI crystal structure was used as a template for the GH2 domain model. The sequence of 1RGI covered the whole GH2 domain, with 46.24% sequence identity and 23.6% sequence similarity. For modeling of the GH4 domain, the sequence between residues 391 and 504 of the built model was used, producing a 114-residue-long construct. Two calcium ions were positioned at the same coordinates as in 1H1V.pdb. For the GH2 domain model, the construct contained residues between 134 and 226. The GH2 domain contains 1 calcium ion, which was positioned at the same location as in 1RGI.
MD.
All MD simulations were performed using AMBER version 14, implemented for GPUs (42). The AMBER ff99SB force field (43) for the peptides and the TIP3P model (44) for water were used. The cutoff used for nonbonded interactions was 8 Å. The particle-mesh Ewald (45) procedure was used to describe long-range electrostatic interactions with maximal grid spacing of 1 Å. Periodic boundary conditions were applied using truncated octahedron geometry. The SHAKE (46) algorithm was used to keep the bond lengths of hydrogen atoms rigid, thus allowing a time step of 2 fs to be used. The protocol for constant temperature and constant pressure (NPT) simulations started with minimization in 2,000 steps, switching from the steepest descent to conjugate the gradient algorithm after 1,000 steps. After minimization, a constant energy/constant volume (NVE ensemble) MD was carried out for 50 ps while increasing the temperature from 0°K to 50°K. The system was further relaxed in a 1-ns-long MD simulation using NPT ensemble while increasing the temperature from 50°K to 310°K over a 500-ps period and keeping it at this temperature for 500 ps. During both relaxation steps, backbone atoms were restrained with a 4 kcal/mol/Å force constant. Subsequently, a third relaxation step was conducted at 310°K and 1 bar pressure using an NPT ensemble for 1 ns without restraints. A 200-ns classical MD-producing canonical NPT ensemble was carried out at 310°K and 1 bar. Both NPT ensembles were performed using temperature regulation with Langevin dynamics, with a collision frequency of 1 ps–1 and application of isotropic pressure scaling with a pressure relaxation time of 1 ps. Protocols identical to those for scaled MD (47) were used for the NPT simulations to produce a 100-ns-long trajectory with a scaling factor of 0.7. Analysis 50000 coordinate snapshots were saved for analysis in all cases. Order parameters for the NH bond vectors of each residue were extracted using Isotropic Reorientational Eigenmode Dynamics (iRED) analysis (48). The trajectories were analyzed with cpptraj29 (49) and visualized in VMD (50). Visualization and analysis of the molecules were done in the PyMOL Molecular Graphics System, version 1.8 (Schrodinger, LLC).
Measurement of actin-bundling, -binding, and -severing activities of advillin.
Human WT AVIL and AVIL mutants were cloned into a pGEX-4T-3 GST vector, and GST-tagged (recombinant) protein was generated and purified (51) (Supplemental Figure 9). Actin-bundling, -binding, and -severing assays were performed as described previously (52, 53). Fluorescence measurements were performed at 25°C using the FluoroMax 3 spectrofluorometer (Horiba Scientific).
Cell lines.
The experiments described here were performed using HEK293T cells and immortalized human podocytes. HEK293T cells were purchased from the ATCC Biological Resource Center. Human immortalized podocytes were a gift of Moin Saleem (University of Bristol, Bristol, United Kingdom) and were cultured as previously described (54).
siRNA and shRNA transfection.
For some knockdown experiments, cells were transfected with ON-TARGET siRNA against AVIL (GE Dharmacon) at a final concentration of 100 nM using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturers’ instructions. Experiments were performed 48 hours after transfection. Additionally, shRNA against human AVIL and PLCE1 was subcloned into pSIREN RetroQ for retroviral transduction using HEK293T cells. Forty-eight hours after transduction, puromycin at a final concentration of 4 μg/ml was added to the medium for selection of transduced cells. See Supplemental Table 2 for the list of target sequences.
Reagents and antibodies.
For immunofluorescence experiments, the following primary antibodies were used: rabbit anti-AVIL (NBP-80312; Novus Biologicals); rabbit anti-PLCE1 (CS117) (19); mouse anti–α–actinin-4 (sc393495; Santa Cruz Biotechnology); mouse anti-paxillin (610619; BD); mouse anti-Arp3 (ab49671; Abcam); and anti-WT1 (sc-7385; Santa Cruz Biotechnology). Donkey anti-goat Alexa 488– and Alexa 594–conjugated secondary antibodies and DAPI were obtained from Invitrogen (Thermo Fisher Scientific). For immunoblotting, the following primary antibodies were used: rabbit anti-AVIL (NBP-91606; Novus Biologicals); rabbit anti-AVIL (NBP-80312; Novus Biologicals); rabbit anti-PLCE1 (LS-C152751; Cell Signaling Technology); and rabbit anti-PLCE1 (CS117), which has been described previously (19). HRP-labeled secondary antibodies were purchased from Santa Cruz Biotechnology. For cell treatment, the following reagents were used: EGF recombinant human protein (Life Technologies, Thermo Fisher Scientific); ROCK inhibitor (Y-27632; Cytoskeleton); and ARP2/3 complex inhibitor I (CK666; Calbiochem).
Immunofluorescence and confocal microscopy in cell lines and tissue sections.
For immuno-staining, human immortalized podocytes were seeded on coverglasses and grown at a permissive temperature. For overexpression studies, human podocytes were transiently transfected using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. Experiments were performed 24–48 hours after transfection. Cells were fixed and permeabilized for 10 minutes using 4% paraformaldehyde and 0.01% SDS. After blocking, cells were incubated overnight at 4°C. The cells were incubated in secondary antibodies for 90 minutes at room temperature, followed by 5 minutes of staining with 1× DAPI and PBS. For immunostaining of paraffin-embedded tissue sections (rat kidney, day 2 postpartum), permeabilization was performed using 0.025% Triton-X 100. Confocal imaging was performed using the Leica SP5X system with an upright DM6000 microscope, and images were processed with the Leica AF software suite.
Co-IP.
Co-IP experiments were performed as described previously (55). Briefly, cell lysates were precleared with protein G or A beads. Next, cell lysates were mixed with the appropriate antibodies and incubated overnight at 4°C in lysis buffer containing the complete protease inhibitor mixture. Immune complexes were collected by binding to mixed protein G or A beads and washed 4 times with lysis buffer prior to immunoblotting.
DAG activation assay.
Cells were transfected in 6-well plates with siRNA against AVIL or a scrambled control using Lipofectamine RNAiMAX. Transfected cells were incubated in RPMI (human podocytes) with 10% FBS for approximately 36 hours and then stimulated with EGF for 30 hours. DAG activity was determined using a DAG ELISA kit (LifeSpan BioSciences) according to the manufacturer’s instructions.
Podocyte migration assay.
Briefly, the podocyte migration assays were performed using the IncuCyte ZOOM System (Essen BioScience) according to the manufacturer’s instructions (56).
Bioinformatics.
Genetic location information is according to the February 2009 Human Genome Browser data, hg19 assembly (http://www.genome.ucsc.edu).
Accession numbers.
Human AVIL full-length protein: GenBank accession number NM_006576.3; mouse AVIL full-length protein: GenBank accession number NM_009635.3; human PLCE1 full-length isoform-1: GenBank accession number NM_016341.3.
Statistics.
Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software). Results from unpaired, 1-way ANOVA with 2-tailed P values were combined with post-hoc multivariate analysis according to Sidak’s method to calculate statistical significance (F values and P values, as indicated in the figure legends). A standard CI of 95% was applied in the analyses. The figures show the mean values of all technical replicates for each of the independent experiments (displayed as single data points). Black lines indicate the mean values of all independent experiments. Error bars represent the SD. A P value of less than 0.05 was considered statistically significant.
Study approval.
Approval for human subjects research was granted by the IRBs of the University of Michigan and Boston Children’s Hospital.
Author contributions
JR, SA, WT, ATvdV, HYG, DAB, WIC, TJS, RS, JMS, EW, JKW, TH, DS, SL, SS, AD, S. Mane, RPL, and FH generated total genome linkage, performed exome capture and massively parallel sequencing, and performed whole-exome evaluation and mutation analyses. JR, SA, SPG, AE, and SK performed cDNA cloning, protein purification and immunofluorescence, and subcellular localization studies in cell lines by confocal microscopy. JR performed migration assays in immortalized human podocytes. SA performed co-IP experiments and measurement of DAG levels. KF, JCM, and MAJ performed and analyzed MD simulation experiments. SPG, AE, and SK performed actin-bundling, -binding, and-severing assays. AN, MS, YT, MB, UH, S. Mir, A. Berdeli, JAK, SED, NAS, A. Bagga, and FH recruited patients and gathered detailed clinical information for the study. FH conceived the study. JR, SA, and FH wrote the manuscript, which was critically reviewed by all the authors.
Supplementary Material
Acknowledgments
We are grateful to the families and study participants for their contributions. We thank the Yale Center for Mendelian Genomics (U54HG006504) for WES analysis. FH is a William E. Harmon Professor of Pediatrics. This research was supported by the NIH (DK076683, to FH); the Young Scholars Program of Children’s Hospital of Fudan University (to JR); Basic Science Research Program through the National Research Foundation of Korea 2015R1D1A1A01056685 (to HYG); DFG fellowships (VE 196/1-1, to ATvdV; Jo 1324/1-1, to TJS; and HE 7456/1-1, to TH); the German National Academy of Sciences Leopoldina (LPDS-2015-07, to EW); the Egyptian Group for Orphan Renal Diseases (EGORD) (to NAS); the Department of Science and Technology, Government of India (DST-SERB, to MAJ); the National Institute of Diabetes and Digestive and Kidney Diseases (DK-98120, to SK); and the Public Health Service (DK-56338, to SK).
Version 1. 10/23/2017
Electronic publication
Version 2. 12/01/2017
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2017;127(12):4257–4269.https://doi.org/10.1172/JCI94138.
Contributor Information
Jia Rao, Email: amayraojia@gmail.com.
Shazia Ashraf, Email: Shazia.Ashraf@childrens.harvard.edu.
Weizhen Tan, Email: Weizhen.Tan@childrens.harvard.edu.
Heon Yung Gee, Email: hygee@yuhs.ac.
Daniela A. Braun, Email: daniela.braun@childrens.harvard.edu.
Krisztina Fehér, Email: Krisztina.Feher@ugent.be.
Amin Esmaeilniakooshkghazi, Email: aesmaeil@Central.UH.EDU.
Tilman Jobst-Schwan, Email: Tilman.Jobst-Schwan@childrens.harvard.edu.
Ronen Schneider, Email: Ronen.Schneider@childrens.harvard.edu.
Johanna Magdalena Schmidt, Email: Magdalena-schmidt@uni-bonn.de.
Eugen Widmeier, Email: Eugen.Widmeier@childrens.harvard.edu.
Jillian K. Warejko, Email: Jillian.Warejko@childrens.harvard.edu.
Tobias Hermle, Email: Tobias.Hermle@childrens.harvard.edu.
David Schapiro, Email: David.Schapiro@childrens.harvard.edu.
Svjetlana Lovric, Email: svjetlanalovric@googlemail.com.
Shirlee Shril, Email: Shirlee.Shril@childrens.harvard.edu.
Ankana Daga, Email: Ankana.Daga@childrens.harvard.edu.
Ahmet Nayir, Email: nayira@istanbul.edu.tr.
Mohan Shenoy, Email: Mohan.Shenoy@cmft.nhs.uk.
Yincent Tse, Email: yincent1@gmail.com.
Martin Bald, Email: m.bald@klinikum-stuttgart.de.
Udo Helmchen, Email: helmchen-hh@t-online.de.
Sevgi Mir, Email: sevgi.mir@ege.edu.tr.
Afig Berdeli, Email: afig.berdeli@ege.edu.tr.
Jameela A. Kari, Email: jkari@doctors.org.uk.
Sherif El Desoky, Email: sherifd68@hotmail.com.
Neveen A. Soliman, Email: neveenase@yahoo.com.
Arvind Bagga, Email: arvindbagga@hotmail.com.
Shrikant Mane, Email: shrikant.mane@yale.edu.
Mohamad A. Jairajpuri, Email: mjairajpuri@jmi.ac.in.
Richard P. Lifton, Email: rickl@mail.rockefeller.edu.
Seema Khurana, Email: skhurana@uh.edu.
Jose C. Martins, Email: jose.martins@ugent.be.
Friedhelm Hildebrandt, Email: friedhelm.hildebrandt@childrens.harvard.edu.
References
- 1.Wiggins RC. The spectrum of podocytopathies: a unifying view of glomerular diseases. Kidney Int. 2007;71(12):1205–1214. doi: 10.1038/sj.ki.5002222. [DOI] [PubMed] [Google Scholar]
- 2.Tryggvason K, Patrakka J, Wartiovaara J. Hereditary proteinuria syndromes and mechanisms of proteinuria. N Engl J Med. 2006;354(13):1387–1401. doi: 10.1056/NEJMra052131. [DOI] [PubMed] [Google Scholar]
- 3.Hildebrandt F. Genetic kidney diseases. Lancet. 2010;375(9722):1287–1295. doi: 10.1016/S0140-6736(10)60236-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Somlo S, Mundel P. Getting a foothold in nephrotic syndrome. Nat Genet. 2000;24(4):333–335. doi: 10.1038/74139. [DOI] [PubMed] [Google Scholar]
- 5.Lovric S, Ashraf S, Tan W, Hildebrandt F. Genetic testing in steroid-resistant nephrotic syndrome: when and how? Nephrol Dial Transplant. 2016;31(11):1802–1813. doi: 10.1093/ndt/gfv355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hildebrandt F. Decade in review--genetics of kidney diseases: Genetic dissection of kidney disorders. Nat Rev Nephrol. 2015;11(11):635–636. doi: 10.1038/nrneph.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Insall RH, Machesky LM. Actin dynamics at the leading edge: from simple machinery to complex networks. Dev Cell. 2009;17(3):310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Gee HY, et al. ARHGDIA mutations cause nephrotic syndrome via defective RHO GTPase signaling. J Clin Invest. 2013;123(8):3243–3253. doi: 10.1172/JCI69134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh J, Reiser J, Mundel P. Dynamic (re)organization of the podocyte actin cytoskeleton in the nephrotic syndrome. Pediatr Nephrol. 2004;19(2):130–137. doi: 10.1007/s00467-003-1367-y. [DOI] [PubMed] [Google Scholar]
- 10.Gee HY, et al. FAT1 mutations cause a glomerulotubular nephropathy. Nat Commun. 2016;7:10822. doi: 10.1038/ncomms10822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun DA, et al. Mutations in nuclear pore genes NUP93, NUP205 and XPO5 cause steroid-resistant nephrotic syndrome. Nat Genet. 2016;48(4):457–465. doi: 10.1038/ng.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gee HY, et al. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest. 2015;125(6):2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sadowski CE, et al. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26(6):1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hildebrandt F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5(1):e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otto EA, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42(10):840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halbritter J, et al. High-throughput mutation analysis in patients with a nephronophthisis-associated ciliopathy applying multiplexed barcoded array-based PCR amplification and next-generation sequencing. J Med Genet. 2012;49(12):756–767. doi: 10.1136/jmedgenet-2012-100973. [DOI] [PubMed] [Google Scholar]
- 17.Habib R, Gubler MC, Antignac C, Gagnadoux MF. Diffuse mesangial sclerosis: a congenital glomerulopathy with nephrotic syndrome. Adv Nephrol Necker Hosp. 1993;22:43–57. [PubMed] [Google Scholar]
- 18.Hildebrandt F, Heeringa SF. Specific podocin mutations determine age of onset of nephrotic syndrome all the way into adult life. Kidney Int. 2009;75(7):669–671. doi: 10.1038/ki.2008.693. [DOI] [PubMed] [Google Scholar]
- 19.Hinkes B, et al. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38(12):1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 20.Marks PW, Arai M, Bandura JL, Kwiatkowski DJ. Advillin (p92): a new member of the gelsolin/villin family of actin regulatory proteins. J Cell Sci. 1998;111(Pt 15):2129–2136. doi: 10.1242/jcs.111.15.2129. [DOI] [PubMed] [Google Scholar]
- 21.Vermeulen W, et al. Solution structures of the C-terminal headpiece subdomains of human villin and advillin, evaluation of headpiece F-actin-binding requirements. Protein Sci. 2004;13(5):1276–1287. doi: 10.1110/ps.03518104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown MC, Perrotta JA, Turner CE. Identification of LIM3 as the principal determinant of paxillin focal adhesion localization and characterization of a novel motif on paxillin directing vinculin and focal adhesion kinase binding. J Cell Biol. 1996;135(4):1109–1123. doi: 10.1083/jcb.135.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tomar A, et al. Regulation of cell motility by tyrosine phosphorylated villin. Mol Biol Cell. 2004;15(11):4807–4817. doi: 10.1091/mbc.E04-05-0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7(10):713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 25.Miyake N, et al. Biallelic mutations in nuclear pore complex subunit NUP107 cause aarly-childhood-onset steroid-resistant nephrotic syndrome. Am J Hum Genet. 2015;97(4):555–566. doi: 10.1016/j.ajhg.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song C, et al. Regulation of a novel human phospholipase C, PLCepsilon, through membrane targeting by Ras. J Biol Chem. 2001;276(4):2752–2757. doi: 10.1074/jbc.M008324200. [DOI] [PubMed] [Google Scholar]
- 27.Nolen BJ, et al. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature. 2009;460(7258):1031–1034. doi: 10.1038/nature08231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashraf S, et al. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest. 2013;123(12):5179–5189. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wan X, Chen Z, Choi WI, Gee HY, Hildebrandt F, Zhou W. Loss of epithelial membrane protein 2 aggravates podocyte injury via upregulation of caveolin-1. J Am Soc Nephrol. 2016;27(4):1066–1075. doi: 10.1681/ASN.2014121197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kok FO, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell. 2015;32(1):97–108. doi: 10.1016/j.devcel.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.[No authors listed] Primary nephrotic syndrome in children: clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int. 1981;20(6):765–771. doi: 10.1038/ki.1981.209. [DOI] [PubMed] [Google Scholar]
- 32.Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet. 1996;58(6):1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 33.Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP. Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet. 2000;66(6):1945–1957. doi: 10.1086/302911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sayer JA, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38(6):674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 35.Boyden LM, et al. Mutations in kelch-like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature. 2012;482(7383):98–102. doi: 10.1038/nature10814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Remmert M, Biegert A, Hauser A, Söding J. HHblits: lightning-fast iterative protein sequence searching by HMM-HMM alignment. Nat Methods. 2011;9(2):173–175. doi: 10.1038/nmeth.1818. [DOI] [PubMed] [Google Scholar]
- 38.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27(3):343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 40.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 41.Yang Z, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179(3):269–278. doi: 10.1016/j.jsb.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salomon-Ferrer R, Götz AW, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh Ewald. J Chem Theory Comput. 2013;9(9):3878–3888. doi: 10.1021/ct400314y. [DOI] [PubMed] [Google Scholar]
- 43.Lindorff-Larsen K, et al. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Chandreskhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 45.Darden T. Particle mesh Ewald—an n·log(n) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 46.Miyamoto S, Kollman PA. Molecular dynamics studies of calixspherand complexes with alkali metal cations: calculation of the absolute and relative free energies of binding of cations to a calixspherand. J Am Chem Soc. 1992;114:3668–3674. doi: 10.1021/ja00036a015. [DOI] [Google Scholar]
- 47.Sinko W, Miao Y, de Oliveira CA, McCammon JA. Population based reweighting of scaled molecular dynamics. J Phys Chem B. 2013;117(42):12759–12768. doi: 10.1021/jp401587e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prompers JJ, Brüschweiler R. General framework for studying the dynamics of folded and nonfolded proteins by NMR relaxation spectroscopy and MD simulation. J Am Chem Soc. 2002;124(16):4522–4534. doi: 10.1021/ja012750u. [DOI] [PubMed] [Google Scholar]
- 49.Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9(7):3084–3095. doi: 10.1021/ct400341p. [DOI] [PubMed] [Google Scholar]
- 50.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 51.Panebra A, Ma SX, Zhai LW, Wang XT, Rhee SG, Khurana S. Regulation of phospholipase C-gamma(1) by the actin-regulatory protein villin. Am J Physiol, Cell Physiol. 2001;281(3):C1046–C1058. doi: 10.1152/ajpcell.2001.281.3.C1046. [DOI] [PubMed] [Google Scholar]
- 52.Kumar N, Zhao P, Tomar A, Galea CA, Khurana S. Association of villin with phosphatidylinositol 4,5-bisphosphate regulates the actin cytoskeleton. J Biol Chem. 2004;279(4):3096–3110. doi: 10.1074/jbc.M308878200. [DOI] [PubMed] [Google Scholar]
- 53.George SP, Wang Y, Mathew S, Srinivasan K, Khurana S. Dimerization and actin-bundling properties of villin and its role in the assembly of epithelial cell brush borders. J Biol Chem. 2007;282(36):26528–26541. doi: 10.1074/jbc.M703617200. [DOI] [PubMed] [Google Scholar]
- 54.Saleem MA, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol. 2002;13(3):630–638. doi: 10.1681/ASN.V133630. [DOI] [PubMed] [Google Scholar]
- 55.Zariwala MA, et al. ZMYND10 is mutated in primary ciliary dyskinesia and interacts with LRRC6. Am J Hum Genet. 2013;93(2):336–345. doi: 10.1016/j.ajhg.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price ST, Beckham TH, Cheng JC, Lu P, Liu X, Norris JS. Sphingosine 1-phosphate receptor 2 regulates the migration, proliferation, and differentiation of mesenchymal stem cells. Int J Stem Cell Res Ther. 2015;2(2):014. doi: 10.23937/2469-570x/1410014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choe H, Burtnick LD, Mejillano M, Yin HL, Robinson RC, Choe S. The calcium activation of gelsolin: insights from the 3A structure of the G4-G6/actin complex. J Mol Biol. 2002;324(4):691–702. doi: 10.1016/S0022-2836(02)01131-2. [DOI] [PubMed] [Google Scholar]
- 58.Burtnick LD, Urosev D, Irobi E, Narayan K, Robinson RC. Structure of the N-terminal half of gelsolin bound to actin: roles in severing, apoptosis and FAF. EMBO J. 2004;23(14):2713–2722. doi: 10.1038/sj.emboj.7600280. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.