Abstract
Targeted next-generation sequencing panels to identify genetic alterations in cancers are increasingly becoming an integral part of clinical practice. We report here the design, validation, and implementation of a comprehensive 95-gene next-generation sequencing panel targeted for hematologic malignancies that we named rapid heme panel. Rapid heme panel is amplicon based and covers hotspot regions of oncogenes and most of the coding regions of tumor suppressor genes. It is composed of 1330 amplicons and covers 175 kb of genomic sequence in total. Rapid heme panel's average coverage is 1500× with <5% of the amplicons with <50× coverage, and it reproducibly detects single nucleotide variants and small insertions/deletions at allele frequencies of ≥5%. Comparison with a capture-based next-generation sequencing assay showed that there is >95% concordance among a wide array of variants across a range of allele frequencies. Read count analyses that used rapid heme panel showed high concordance with karyotypic results when tumor content was >30%. The average turnaround time was 7 days over a 6-month span with an average volume of ≥40 specimens per week and a low sample fail rate (<1%), demonstrating its suitability for clinical application.
Advances in nucleic acid sequencing methods [eg, next-generation sequencing (NGS)]1 have significantly increased sequencing capacity while reducing costs. These technical advances, together with the ever-improving accuracy and reliability of sequencing instrumentation and associated informatics pipelines, have permitted sequencing to move from the realm of research to the clinical laboratory at many institutions. Over the past several years, sequencing of hematologic malignancies has expanded our understanding of the molecular abnormalities that underlie these disorders and has identified a number of genes that bear driver mutations with diagnostic, prognostic, and/or therapeutic implications.2, 3, 4, 5, 6, 7, 8, 9, 10
Given the potential clinical implications of these mutations and diagnostic difficulties that are often presented by patients with peripheral blood cytopenias or cytoses, we established a custom NGS assay to evaluate patients with known or suspected hematologic malignancies, principally acute myeloid leukemia, myelodysplasia, and myeloproliferative neoplasms. Other strategies for integrating new molecular testing approaches into the workup of myeloid neoplasms were proposed or implemented,11 including targeted NGS panels.12, 13, 14 Our goal was to be comprehensive, covering all of the recurrently mutated genes, and to use a platform with the potential of a turnaround time of less than a week. The assay relies on an Illumina Truseq Custom Amplicon (TSCA) (San Diego, CA) kit and identifies single nucleotide variants (SNVs) and insertions/deletions (indels) in genes that are recurrently mutated in myeloid disorders and sequence variants in certain genes that are commonly mutated in lymphoid leukemias, such as NOTCH1, NOTCH2, STAT3, and MYD88.15, 16, 17, 18 Genes covered by the assay encode a diverse collection of transcription factors, epigenetic regulators,19 cohesin family members,20 splicing factors,21 cell surface receptors, and downstream signaling components. In this report, we describe the design, analysis pipeline, validation, strategy for annotation, and reporting of this NGS panel [referred to as rapid heme panel (RHP)] and present the comparison of results from this amplicon-based panel with results obtained with a capture-based NGS panel. Our results show that RHP has similar sensitivity and specificity across a wide range of alterations, including SNV, indel, FLT3 internal tandem duplications (FLT3-ITDs), and selected copy number variations (CNVs) and that results can be delivered in as few as 4 days, permitting integration of the molecular results with the other pathologic findings (morphologic characteristics, flow cytometry, cytogenetics, RT-PCR, etc.), leading to a comprehensive pathologic report.
Materials and Methods
Design
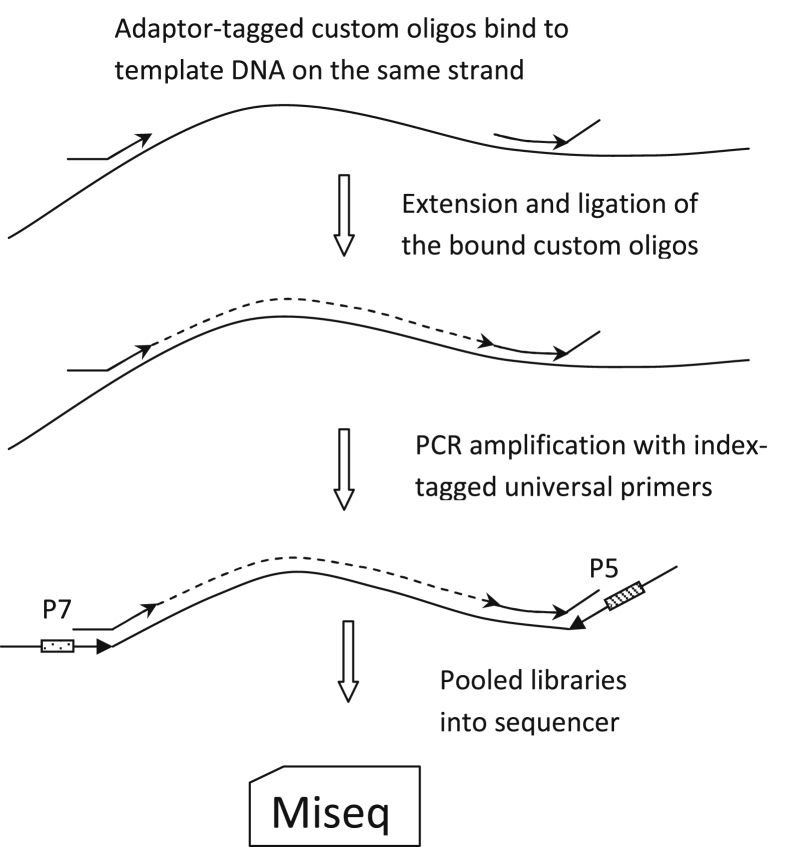
RHP is based on the Illumina TSCA version 1.0 platform (Figure 1). Amplicons (approximately 250 bp) were designed with the Illumina DesignStudio and target 95 genes (Supplemental Table S1), including mutational hotspots in oncogenic driver genes and the coding sequence of tumor suppressor genes. A seven-person working group of molecular diagnosticians, hematopathologists, hematologists, oncologists, and researchers was assembled to curate from the published literature and meeting proceedings a comprehensive list of genes more frequently mutated in myeloid malignancies, acute lymphoid leukemias, and lymphoid neoplasms that commonly involve bone marrow (chronic lymphocytic leukemia, Waldenström macroglobulinemia, hairy cell leukemia, etc.). The initial list was >120 genes. The criteria used to determine whether a gene is included in the final panel were i) importance for classification according to World Health Organization; ii) risk stratification with National Comprehensive Cancer Network category 2B or above of evidence; iii) frequency of mutations; iv) germline predisposition of myeloid neoplasms; and v) primer compatibility in a multiplex panel. The amplicons (N = 1330) span 757 coding exons and 175 kb of genomic sequence in aggregate. The final design had a success rate of 98.5% and a total gap of <1% of the targeted genomic region. Approximately 50% of the target regions are covered in both directions. Difficult to design areas are mainly high-GC sequences, such as regions of CEBPA (Supplemental Table S2).
Figure 1.
TruSeq Custom Amplicon assay workflow diagram.
Sample Processing and Sequencing
Genomic DNA was isolated manually from fresh blood or marrow aspirate with the use of the QIAamp DNA mini kit (Qiagen, Valencia, CA). Batch library preparation was performed according to Illumina TSCA's standard procedure. Briefly, the custom primer pool was annealed to genomic DNA, and adjacent pairs of primers were joined by extension-ligation. The target template was amplified with Illumina TruSeq adaptors and indexes, and the resulting PCR products were isolated on AMPure XP beads (Agencourt; Beckman Coulter, Brea, CA). The DNA concentration of the library from each sample was measured fluorometrically with Qubit (Qubit double-stranded DNA, high sensitivity assay kit, Q32851; Life Technologies, Inc., Carlsbad, CA). Fifty picograms of each library prepared from 16 samples are pooled (800 pg total, approximately 6 pmole) and run on MiSeq with the use of version 2.2 chemistry, with a targeted cluster density of 1100 k/mm2 (actual 800 to 1450 k/mm2) and a cluster passing filter rate of >80%. Tumor samples were sequenced alone without a matched normal sample. DNA isolated from other fresh/frozen and/or alcohol-fixed sources such as coverslips of bone marrow aspirate, fine needle aspirate (both stained and unstained), and cerebrospinal fluid have also yielded satisfactory results. DNA from formalin-fixed, paraffin-embedded was not tested in this panel.
Data Analysis
The analysis pipeline is outlined in Supplemental Figure S1. Paired-end reads (150 bp) were de-multiplexed into individual BAM files and aligned to Hg19 by the onboard Illumina Reporter software version 2.5.1.3 with the use of the TruSeq Amplicon Workflow with one modification of the default variables (maximum indel size, 300). The genome vcf file was scanned for any nucleotide position with a variant read count >9 regardless of variant allele frequency (VAF) or a position with variant read count of 5 to 9 and a VAF >33%. The candidate list was then filtered for the presence of missense, nonsense, frameshift, splice-site, insertion, and deletion mutations. Recurrent assay-specific false positives (those found in the run control) were filtered out. The resulting candidate list was loaded into a custom database and was reviewed with a web-based interface with links that allow autoloading of BAM files and viewing of each variant in Integrated Genome Viewer (Broad Institute, Cambridge, MA). A variant was classified as pathogenic if it was on a curated list of known pathogenic variants for an oncogene, or if it was a frameshift or nonsense mutation in known functional domains of tumor suppressor genes. The final diagnostic report (Supplemental Appendix S1) included Human Genome Variation Society gene name; VAF; Human Genome Variation Society coding nucleotide, and amino acid level annotation for any variants; read depth; and an interpretive comment from a custom in-house knowledge database that relies on information culled from the published literature. The genomic vcf file was also scanned against a must-call list of known clinically relevant codons (Supplemental Table S3), and any non-reference reads on these codons were separately recorded in the database for review. Findings in the must-call review, case-specific amplicon failures (defined as <50× coverage), and clinically relevant gaps in coverage of the assay were all included in each diagnostic report (Supplemental Appendix S1).
FLT3-ITD Identification
Aligned BAM files were filtered for reads in which one side maps to exon 13, 14, or 15 of FLT3 and the other side is unmapped. These one-sided mapped reads were then scanned for the presence of duplications of ≥10 bp. Reads that contain duplications were re-aligned among themselves to make a final determination for FLT3-ITD.
Read Count Analysis for Copy Number Evaluation
Fractional read count for each amplicon (amplicon read count/total read count) was calculated and normalized to the corresponding amplicon fractional read count of the normal control in the same run. The log2 of the sample/normal ratio (log2 ratio) was centered to achieve a median of 0 across each amplicon to reduce batch effects and was then used to generate a read count plot, which was evaluated for copy number gains or losses through visual inspection. A log2 ratio of ±0.5 was used as a cutoff for calling gain and loss.
Results
A typical RHP assay yielded approximately two million reads per specimen with the following characteristics: average read depth approximately 1500×; approximately 90% of amplicons >200× read depth; and <5% of amplicons with <50× read depth (Supplemental Figure S2). Twenty DNA samples isolated from peripheral blood of normal control subjects were first run to generate a baseline for performance of individual amplicons and to assemble a list of platform/assay-specific sequencing noise, which were then used as filters in the analysis pipeline. Validation studies (described in the next section) included method comparison (including deletions of ≤52 bp in CALR and tandem duplications in FLT3 of ≤69 bp), evaluation of analytical sensitivity, evaluation of low-level false positives, assessment of inter- and intra-run reproducibility, and detection of single copy loss of genes on 7q, 17q, and 20q by read count analysis.
Method Comparison
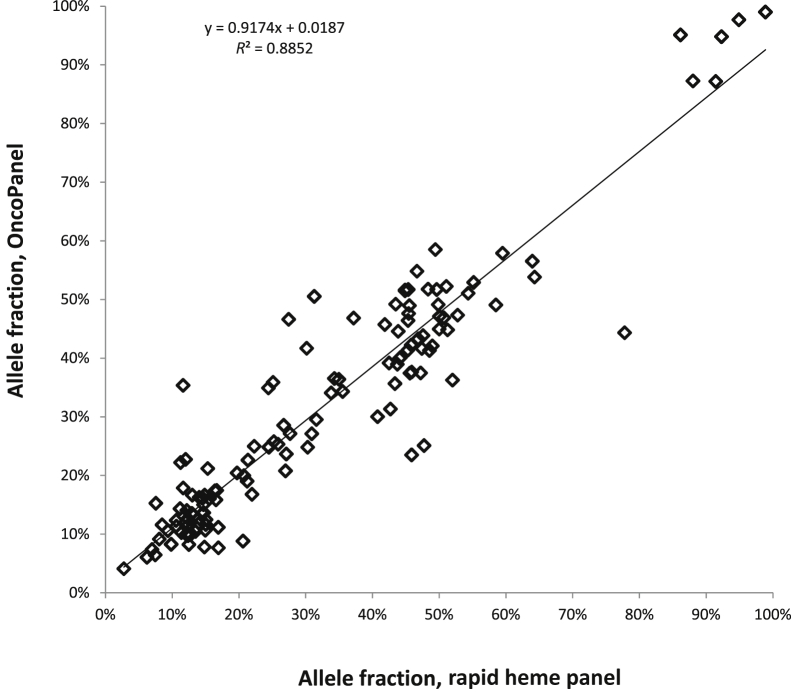
We compared RHP with a previously validated capture-based NGS assay that we launched >1 year ago (OncoPanel version 1). RHP was performed on genomic DNA from 24 peripheral blood and bone marrow aspirate specimens that had been previously analyzed by OncoPanel. Among the 24 samples, there were 130 SNVs and indels identified by the OncoPanel assay with VAF, ranging from 4% to 98%, distributed across 46 different genes that are shared between the two panels. The concordance rate for detection of these SNVs and indels in RHP was 99% (129 of 130). The only discrepancy was a variant (STAG2 c.436C>T, 19% VAF) detected by OncoPanel that was poorly covered in the RHP assay (only one read present for that sample). Furthermore, allele frequencies (AFs) between the two assays were highly concordant (correlation coefficient, R2 = 0.89) (Figure 2). Additional validation was performed for specific disease-associated variants in which known positive samples or cell lines were available, including 30 samples with a variety of SNVs and indels (Supplemental Table S4), with VAF of 3% to 80% that were previously identified by alternate methods. All of these variants were detected by RHP. We found that the default variable in the Illumina pipeline (maximum indel size = 25 bp) would not identify indels >25 bp in size. This could be overcome by changing the variable to 300 bp without a significant increase in execution time as stated in the Illumina manual. We found that most reads containing FLT3-ITD were not mapped, whereas their read-mates were. Our customized algorithm thus collects these one-side-mapped-reads whose mates were mapped to FLT3 exon 13 to 15 and re-aligned them to identify the ITD. All nine samples with FLT3-ITDs that ranged in size from 18 to 69 bp identified by OncoPanel or reference laboratory were also successfully detected by RHP with this approach (data not shown).
Figure 2.
Comparison of allele frequencies determined by amplification- and reference hybrid capture–based next-generation sequencing assays. One hundred twenty-nine variants in 24 samples are identified by both methods, and the variant allele frequencies as determined by both assays across a wide range of variant allele frequencies are highly concordant (correlation coefficient, R2 = 0.89).
Analytical Sensitivity
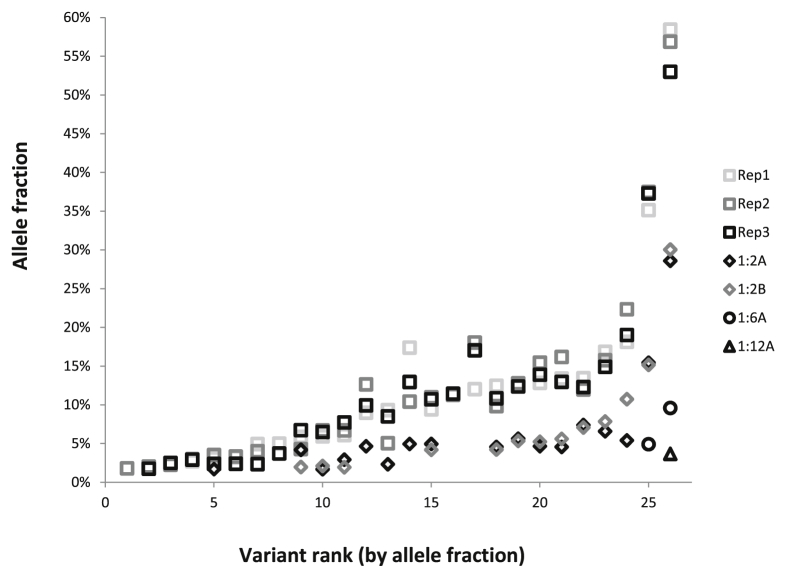
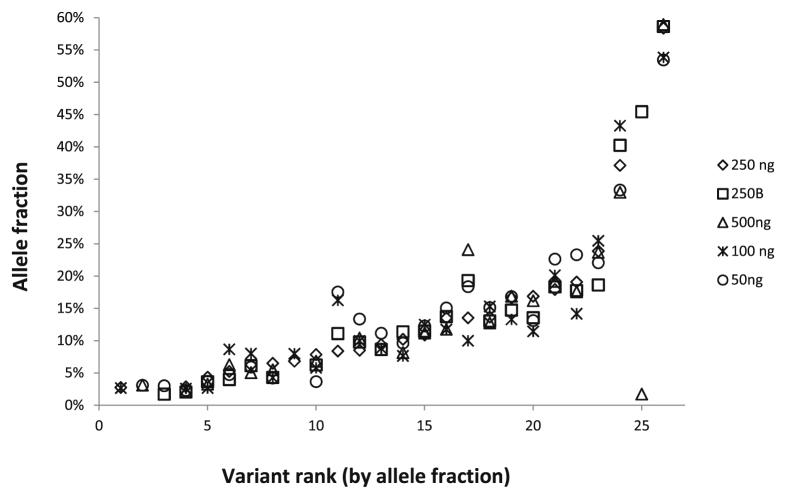
The analytical sensitivity of RHP was evaluated by mixing a positive control sample with normal control DNA. Specifically, genomic DNA from five different cell lines with 22 different SNVs or indels in 19 different genes (Supplemental Table S5) at VAF, ranging from 3% to 60% (before mixing), was used to prepare a 1:1:1:1:1 mixture. This mixed-positive sample was then serially diluted into normal control DNA at dilutions of 1:2, 1:6, and 1:12. The undiluted mixed-positive sample was run in triplicate, and the 1:2 diluted samples were run in duplicate. Known variants were reproducibly detected down to a 5% AF threshold (Figure 3). Below this level, the reproducibility of detection was variant dependent. In addition, the mixed-positive sample was used to evaluate the effect of varying input DNA amounts from 50 to 500 ng. Input of as little as 50 ng of DNA was sufficient for detection of all tested variants across all AFs down to the 5% threshold (Figure 4). However, more false-positive variants [defined as variants with frequencies above the pipeline cutoff (>9 reads or, 5 to 9 reads and >33% AF, plus Illumina Q score > 30) that were not reproducible and that were not detected by other reference assays] were seen with small amounts of input DNA. For example, at an input of 250 ng of DNA, only one variant was determined to be a false positive, whereas 100 ng of input DNA produced four false-positive variants and 50 ng of input DNA produced 11 false variants. The AFs of false-positive variants were generally <5% when they were identified in >10 reads, and the most frequent false-positive mutations were transitions (C>T or T>C). Given these findings, the input DNA amount used in this assay was set at 250 ng, consistent with the manufacturer's recommendation for this platform.
Figure 3.
Determination of analytical sensitivity. DNA from the mixed positive sample (five different cell lines comprising 26 different allele variants; allele frequencies ranging from 3% to 60%) was tested, undiluted, by rapid heme panel in triplicate (Rep1, 2, 3, squares) and was serially diluted into normal DNA at dilutions of 1:2 (A and B in duplicate, diamonds), 1:6 (circles), and 1:12 (triangle). On the x axis, the variants are sorted by increasing allele frequency and numbered 1 to 26.
Figure 4.
Evaluation of optimal DNA input amount. DNA from the mixed positive cell line sample was tested, undiluted, using different amounts of input DNA for library preparation. Two hundred fifty nanograms was run in duplicate, and the other amounts were run once. On the x axis, the variants are sorted by increasing allele frequency and numbered 1 to 26.
Inter- and Intra-Run Reproducibility
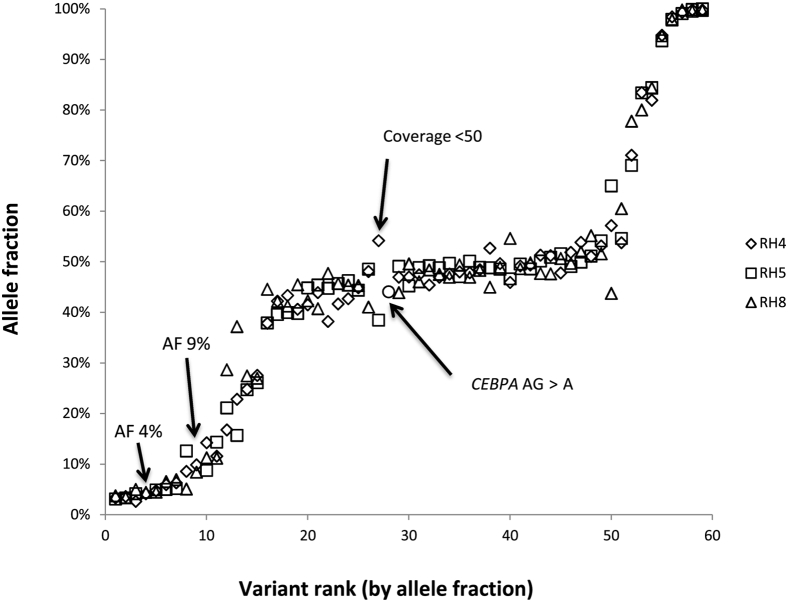
For evaluation of inter-run reproducibility, eight DNA samples that contained 59 SNVs and indels with AFs that ranged from 3% to 100% were tested in three separate runs; among these variants, 55 of 59 (93%) were detected in all three runs (Figure 5). To assess intra-run reproducibility a mixed-positive control DNA sample that contained 20 different SNVs and indels with AFs that ranged from 2% to 56% were tested in triplicate on two separate runs. Among the 40 total variants (20 variants × 2 runs), 88% (35 of 40) were detected in all three replicates; four of the five variants that were not detected in every replicate had AFs <5% (range, 2% to 4%), consistent with limit of analytical sensitivity of this assay previously determined as described in the previous section.
Figure 5.
Inter-run reproducibility. Eight samples comprising 59 single nucleotide variants and insertions/deletions with allele frequencies that ranged from 3% to 100% were tested in three separate runs; among these variants, 55 of 59 (93%) were detected in all three runs. Arrows point to four variants not detected in all three runs. Two variants were in regions with poor coverage in the runs where they were not identified (CEBPA variant was adequately covered in one of three runs and the other had <50× coverage in one run). The two low AF variants (average 4% and 9%) were not identified in a third replicate despite adequate coverage (>200×). Review of the BAM file in Integrated Genome Viewer shows that there were reads of the variant sequences in the third run, but below the pipeline cutoff. AF, allele frequency.
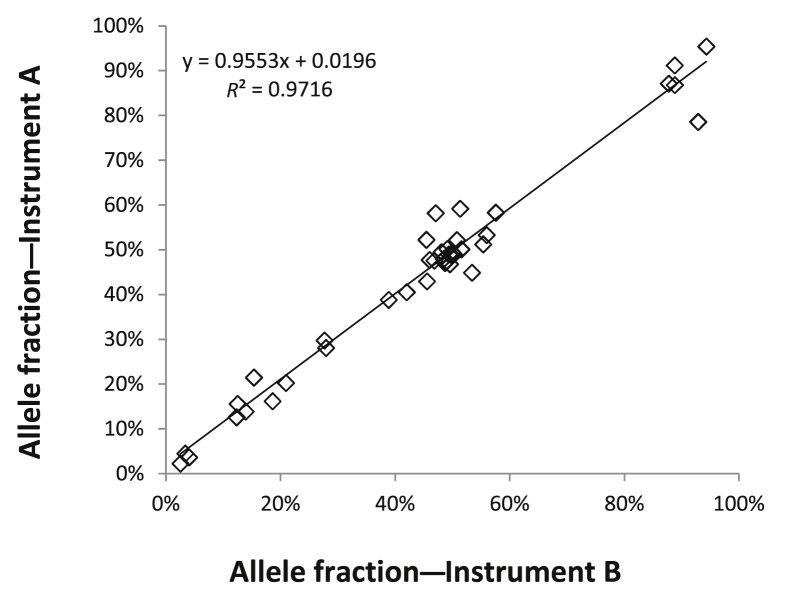
Inter-Instrument Comparison
Twelve DNA samples that contained 41 different SNVs and indels with AFs that ranged from <5% to >90% were tested on two separate MiSeq instruments in two different locations at our institution. The concordance rate for detection of the variants was 98% (40 of 41). In addition, the AFs of the different variants were highly reproducible between both instruments across the full range of AFs (correlation coefficient, R2 = 0.97) (Figure 6).
Figure 6.
Reproducibility between two MiSeq instruments. Twelve samples comprising 41 different single nucleotide variants and insertions/deletions with allele frequencies that ranged from <5% to >90% were tested on two separate MiSeq instruments. The allele frequencies for the different variants were highly concordant between both instruments across the full range of allele frequencies (correlation coefficient, R2 = 0.97).
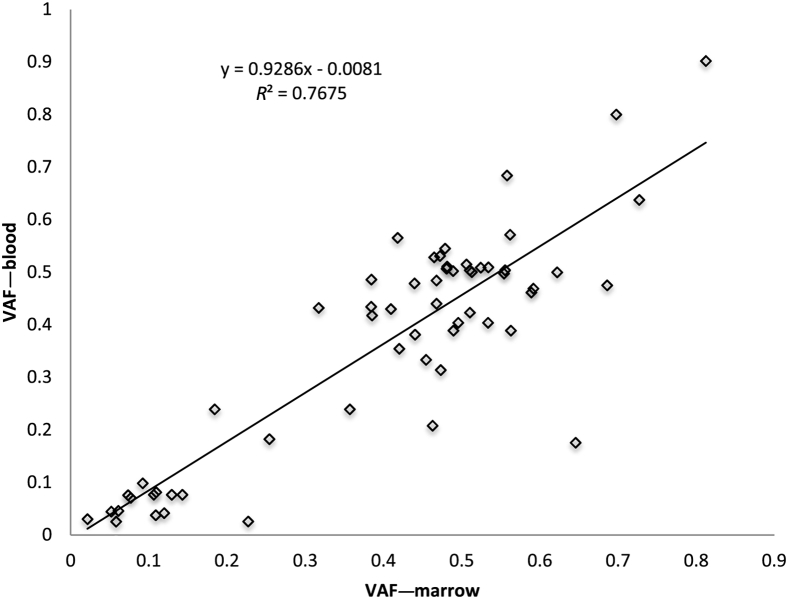
Peripheral Blood and Bone Marrow Comparison
Myeloid neoplasms are diseases of the hematopoietic stem/progenitor cells.22 Mature neutrophils, monocytes, eosinophils, but typically not lymphocytes, in the peripheral blood are expected to harbor the same genomic alterations that are found in myeloid progenitors in the bone marrow. We therefore tested 18 paired DNA samples isolated from peripheral blood and bone marrow of the same individuals on the same day. Fifty-nine variants were identified in both paired samples (100% concordance) with allele frequencies that ranged from 2.5% to 90% (correlation coefficient, R2 = 0.77) (Figure 7). As expected, because lymphocytes make up a higher fraction of nucleated cells in the peripheral blood than in the marrow, there is a trend toward a slightly higher VAF in bone marrow.
Figure 7.
Comparison between peripheral blood and bone marrow samples. Eighteen pairs of peripheral blood and bone marrow collected at the same time were analyzed by rapid heme panel, and the variants were sorted by increasing allele frequency and plotted to show the concordance. Fifty-nine variants were found in both samples with good correlation. The allele frequencies are lower in peripheral blood when the percentage of lymphocytes is >20%. VAF, variant allele frequency.
Read Count Analysis for CNV
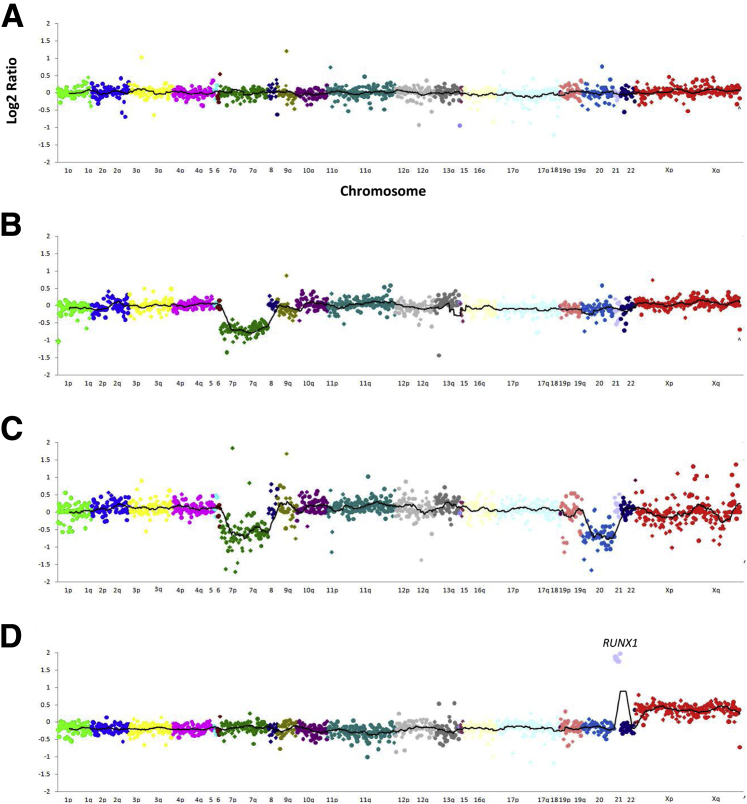
Several CNVs are common in myeloid neoplasms. These include 5q-, 7q-, 20q-, and trisomy 8, among others. We therefore evaluated the use of relative read counts to determine CNV by RHP (see Materials and Methods). Fifteen samples with normal karyotypes and 34 samples with karyotypic abnormalities were analyzed by RHP, and log2 ratio plots were visually inspected for deviation from normal (log2 ratio of ± −0.5 or greater) and scored for the presence of increased or decreased read counts (Figure 8). The results were then compared with the previously determined karyotypic findings. Forty of the 41 karyotypic abnormalities were identified by RHP read count analysis (Table 1). A few single gene deletions not included in the table (in TET2 and ETV6) were identified only by RHP, probably because of the presence of small deletions that are below the resolution of karyotypic analysis. By diluting samples with known CNV alterations into normal control DNA, we determined that identification of copy number gains and losses by read count analysis requires approximately 30% tumor content (data not shown).
Figure 8.
Examples of copy number variant analysis by rapid heme panel. Log2 ratio plots of normalized read count across the 1300 amplicons in rapid heme panel sorted by chromosomal coordinates. One dot represents one amplicon. Different chromosomes are represented by different colors. The line represents 30-amplicon sliding average. The y axis indicates log2 ratio; x axis, chromosomes. A: A representative female sample with no gain or loss of genes identified. B: A sample from a female with loss of all amplicons on chromosome 7. C: A male sample with loss of all amplicons on chromosomes 7 and 20. D: A female sample with amplification of RUNX1 on chromosome 21 and gain of chromosome X.
Table 1.
Comparison between Read Count Analysis of RHP and Karyotypic Findings
| Sample | del(20q) | del(7q)/7- | 8+ | i(17)/17q- | 4+ | i(X) | 10+ | 21+ | 9+ | 19+ | 12+ | del(11q) | Karyotype |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Y | Y | 46,XY,del(20)(q11.2)[8]/45,XY,-7,del(20)[10]/46,XY[2] | ||||||||||
| 2 | Y | Y | 45,XY,-7,del(20)(q11.2)[cp17]/46,XY[3] | ||||||||||
| 3 | Y | 47,XY,+8[20] | |||||||||||
| 4 | Y | Y | 45,XY,-7[7]/46,XY,i(17)(q10{4]46,XY[cp9] | ||||||||||
| 5 | Y | 45,XY,-7[14]/45,idem,?r(6)(p22q27)[6] | |||||||||||
| 6 | Y | 47,X,-Y,del(6)(q15q21),+8,+14[17]/47,idem,t(1;9)(q23;q34)[3] | |||||||||||
| 7 | Y | 46,XY,del(20)(q11.2)[cp18]/45,idem,-7[1]/46,XY[1] | |||||||||||
| 8 | Y | 46,XY,i(17)(q10)[cp20] | |||||||||||
| 9 | Y | 47,XY,+8,t(15;17)(q24;q21)[18]/46,XY[2] | |||||||||||
| 10 | Y | Y | 48,XY,+8,+10[cp20] | ||||||||||
| 11 | Y | Y | 48,XX,+4,+8[17]/46,XX[3] | ||||||||||
| 12 | Y | Y | 45,XY,-7[8]/46,XY,-7,+21[cp12] | ||||||||||
| 13 | Y | 46,XX,del(7)(q22q36)[17]//46,XY[3] | |||||||||||
| 14 | Y | 45,XY,i(7)(q10),del(12)(p12),-15[7]/46,XY[13] | |||||||||||
| 15 | N | Y | 47,XX,+9,del(20)(q11.2)[5] | ||||||||||
| 16 | Y | 47,XX,+21[20] | |||||||||||
| 17 | Y | 47,XY,t(1;17)(q42;q25),+19[20] | |||||||||||
| 18 | Y | 46,XX,del(7)(q21q31),t(15;17)(q24;q21)[cp18]/46,XX[2] | |||||||||||
| 19 | Y | 47,XY,+21[20] | |||||||||||
| 20 | Y | 47,XX,+8[20] | |||||||||||
| 21 | Y | 47,XX,+10[17]/46,XX[cp3] | |||||||||||
| 22 | Y | 46,XY,r(7)(p2?2?q11.2)[6]/46,XY[14] | |||||||||||
| 23 | Y | 45,XY,-7[19]/46,XY,t(17;22)(q21;q13)[1] | |||||||||||
| 24 | Y | 47,XY,+i(X)(q10)[20] | |||||||||||
| 25 | Y | 47,XX,+8[cp4]/46,XX[7] | |||||||||||
| 26 | Y | 47,XX,+12[2]/46,XX[7] | |||||||||||
| 27 | Y | 45,XY,-7[6]/46,XY[10] | |||||||||||
| 28 | Y | 45,XX,-7[19]/46,XX[1] | |||||||||||
| 29 | Y | 46,XY,del(11)(q13q23)[14]/46,XY[6] | |||||||||||
| 30 | Y | 45,X,-X,del(6)(q21),add(16)(p13),del(17)(p11.2)[cp8] | |||||||||||
| 31 | Y | 46,XX,del(11)(q13q23)[cp15]/46,XX[5] | |||||||||||
| 32 | Y | 45,XX,add(3)(q26.2),-7[20] | |||||||||||
| 33 | Y | 46,XY,del(20)(q11.2)[3]/46,XY[12] | |||||||||||
| 34 | Y | 47,XY,+i(X)(q10)[20] | |||||||||||
| Count | 5 | 13 | 7 | 3 | 1 | 2 | 2 | 3 | 1 | 1 | 1 | 2 |
Del, deletion; i, isochromosome; N, no; RHP, rapid heme panel; Y, yes.
Discussion
In this report, we summarize the design, validation, and implementation of a 95-gene panel test on the basis of NGS that is targeted to hematologic malignancies. It is one of the most comprehensive, up-to-date panels of its kind and includes many recently discovered genes (eg, CALR,23, 24 GNB1,25 and STAG220, 26) and most oncogenes and tumor suppressor genes frequently mutated in hematologic malignancies. We chose the Illumina TSCA platform for its capacity (≤1500 amplicons), relative ease of use, and uniform performance across amplicons (<5% amplicons are poor performing). Similar to other amplicon-based NGS platforms, we found that variants present at fractions of >5% are reproducibly identified at read depths of approximately 1500. Because myeloid neoplasms are stem/progenitor cell disorders, mutations are present in early progenitors and differentiated myeloid cells. Except in the setting of extreme neutropenia/monocytopenia, peripheral blood can substitute for bone marrow in evaluating the presence of myeloid-specific alterations (Figure 7). For patients who present with cytopenia or cytosis, panel tests such as RHP on peripheral blood may provide valuable information for the physicians to decide whether to proceed with a bone marrow biopsy and aspirate. For patients with a diagnosis of myelodysplastic/myeloproliferative neoplasms, RHP can establish a personalized baseline of genetic alterations for that patient and inform therapeutic options on disease progression.
Below the 5% VAF level, it is not easy to reliably differentiate between false-positive results and real variants present at low frequencies without additional time-consuming and costly measures such as ultra-deep sequencing or subcloning which are not practical in clinical laboratory. Because of the high concordance rate between the RHP assay and a previously validated capture-based NGS assay (OncoPanel) and known positive samples of specific variants, we did not perform Sanger sequencing or pyrosequencing for further validation of previously unknown variants. We observed increased numbers of false-positive calls when the amount of input DNA was reduced in this platform. Presumably, fewer available starting templates led to reduced complexity of the library, and errors in earlier rounds of PCR became over-represented in the library and on sequencing led to false-positive calls.
We unexpectedly found excellent correlation between VAFs by RHP and OncoPanel, a capture-based assay. In dilution studies, we also found that effect on VAFs was proportional to the expected dilutional effect. These findings suggest that the multiplex reaction in the TSCA protocol amplifies multiple amplicons with reproducible efficiency. Although the best-performing amplicons produce >100-fold more reads than the worst-performing amplicons, the behavior of an individual amplicon is sufficiently consistent across different specimens, including controls, over many different runs such that normalized fractional read counts can be used to deduce the amount of DNA present in the input sample and in turn infer CNVs. Not surprisingly, there are variations among individual amplicons, especially those with low absolute read counts and the sensitivity of detection of CN variants (30% of tumor content) is lower than the sensitivity of detection of SNVs (5% allele frequency, or 10% tumor content for a heterozygous locus). These findings are consistent with those reported by other investigators who have also recently demonstrated reliable detection of focal CNV loss or gain with the use of amplification-based sequencing data.27, 28
For annotation, we elected to limit the classification of variants as either pathogenic or variant of unknown significance. We used a strategy of treating oncogenes and tumor suppressor genes in two different ways in assigning pathogenicity. For oncogenes, a list of curated, known activating pathogenic variants was first assembled. Newly encountered variants are initially classified as variant of unknown significance and later added to the pathogenic list when a consensus is reached at the weekly review meetings. For tumor suppressor genes, structural domains in which mutations were shown to lead to loss of function are defined and used to assign significance to a variant. This strategy allows assignment of variants as pathogenic to be driven by an algorithm after the presence of a variant is affirmed. This in turn allows reports to be generated through automation. The resulting reports have a standard format and uniformly contain all required elements (Supplemental Appendix S1).
After its launch as a clinical test, the RHP had an average turnaround time of 7 days (including weekend and holidays) over a 6-month span, with a re-run rate of <5% and a fail rate of <1% with ≥1000 specimens tested (average of ≥40 samples per week). These numbers demonstrate that RHP is a robust test that can scale to meet the needs of an academic medical center with a busy hematologic malignancy service in a timely fashion.
Since the introduction of molecular markers for diagnosis and classification of hematologic malignancies in the 2008 World Health Organization classification, our knowledge of genetic alterations in hematologic malignancies has expanded exponentially. The clinical significance, both in therapeutic and prognostic terms, of sequence variants is an area of active investigation, and the next iteration of the World Health Organization classification will contain a number of new entities that are defined in part by the presence of particular sequence variants. An increasing number of targeted therapies are also under development in myeloid malignancies (such as IDH1/IDH2 and FLT3 inhibitors29, 30, 31) that may rely on genetic findings for patient selection. On the basis of these developments, it can be anticipated that focused NGS tests will become the standard of care over the next few years.32 Amplicon-based NGS panels such as the one described here have the advantage of being relatively easy to refine (by adding or removing genes or amplicons) on the basis of new findings in a rapidly moving field. With the caveat that they cannot detect gene rearrangements/fusion, NGS panels such as those described here are well positioned to fill an important gap in current clinical cancer diagnostic testing.
Footnotes
Supported by internal funds dedicated for test validation in the laboratory through the Department of Pathology at Brigham and Women's Hospital for assay development.
Disclosures: None declared.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.02.003.
Supplemental Data
Data analysis pipeline for rapid heme panel. AF, allele frequency; FLT3-ITD, FLT3 internal tandem repeat; SNP, single nucleotide polymorphism.
The amplicons are ranked by average depth of coverage for six normal control DNA (from lowest to highest) and plotted with their %GC contents. Red bars indicate %GC content; green bars, average coverage; blue bars, SD of coverage of six control DNA; x axis, amplicon rank; left y axis, average coverage; right y axis, GC%. Average coverage is not correlated with %GC content of the amplicon.
References
- 1.Metzker M.L. Sequencing technologies — the next generation. Nat Rev Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 2.Cazzola M., Della Porta M.G., Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood. 2013;122:4021–4034. doi: 10.1182/blood-2013-09-381665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindsley R.C., Ebert B.L. The biology and clinical impact of genetic lesions in myeloid malignancies. Blood. 2013;122:3741–3748. doi: 10.1182/blood-2013-06-460295. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lundberg P., Karow A., Nienhold R., Looser R., Hao-Shen H., Nissen I., Girsberger S., Lehmann T., Passweg J., Stern M., Beisel C., Kralovics R., Skoda R.C. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–2228. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- 6.Matynia A.P., Szankasi P., Shen W., Kelley T.W. Molecular genetic biomarkers in myeloid malignancies. Arch Pathol Lab Med. 2015;139:594–601. doi: 10.5858/arpa.2014-0096-RA. [DOI] [PubMed] [Google Scholar]
- 7.Meyer S.C., Levine R.L. Translational implications of somatic genomics in acute myeloid leukaemia. Lancet Oncol. 2014;15:e382–e394. doi: 10.1016/S1470-2045(14)70008-7. [DOI] [PubMed] [Google Scholar]
- 8.Zoi K., Cross N.C. Molecular pathogenesis of atypical CML, CMML and MDS/MPN-unclassifiable. Int J Hematol. 2015;101:229–242. doi: 10.1007/s12185-014-1670-3. [DOI] [PubMed] [Google Scholar]
- 9.Lindsley R.C., Mar B.G., Mazzola E., Grauman P.V., Shareef S., Allen S.L., Pigneux A., Wetzler M., Stuart R.K., Erba H.P., Damon L.E., Powell B.L., Lindeman N., Steensma D.P., Wadleigh M., DeAngelo D.J., Neuberg D., Stone R.M., Ebert B.L. Acute myeloid leukemia ontogeny is defined by distinct somatic mutations. Blood. 2015;125:1367–1376. doi: 10.1182/blood-2014-11-610543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang L., Padron E., Lancet J. The molecular basis and clinical significance of genetic mutations identified in myelodysplastic syndromes. Leuk Res. 2015;39:6–17. doi: 10.1016/j.leukres.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Kohlmann A., Bacher U., Schnittger S., Haferlach T. Perspective on how to approach molecular diagnostics in acute myeloid leukemia and myelodysplastic syndromes in the era of next-generation sequencing. Leuk Lymphoma. 2014;55:1725–1734. doi: 10.3109/10428194.2013.856427. [DOI] [PubMed] [Google Scholar]
- 12.Duncavage E.J., Abel H.J., Szankasi P., Kelley T.W., Pfeifer J.D. Targeted next generation sequencing of clinically significant gene mutations and translocations in leukemia. Mod Pathol. 2012;25:795–804. doi: 10.1038/modpathol.2012.29. [DOI] [PubMed] [Google Scholar]
- 13.Cheng D.T., Cheng J., Mitchell T.N., Syed A., Zehir A., Mensah N.Y., Oultache A., Nafa K., Levine R.L., Arcila M.E., Berger M.F., Hedvat C.V. Detection of mutations in myeloid malignancies through paired-sample analysis of microdroplet-PCR deep sequencing data. J Mol Diagn. 2014;16:504–518. doi: 10.1016/j.jmoldx.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luthra R., Patel K.P., Reddy N.G., Haghshenas V., Routbort M.J., Harmon M.A., Barkoh B.A., Kanagal-Shamanna R., Ravandi F., Cortes J.E., Kantarjian H.M., Medeiros L.J., Singh R.R. Next-generation sequencing-based multigene mutational screening for acute myeloid leukemia using MiSeq: applicability for diagnostics and disease monitoring. Haematologica. 2014;99:465–473. doi: 10.3324/haematol.2013.093765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koskela H.L.M., Eldfors S., Ellonen P., van Adrichem A.J., Kuusanmäki H., Andersson E.I., Lagström S., Clemente M.J., Olson T., Jalkanen S.E., Majumder M.M., Almusa H., Edgren H., Lepistö M., Mattila P., Guinta K., Koistinen P., Kuittinen T., Penttinen K., Parsons A., Knowles J., Saarela J., Wennerberg K., Kallioniemi O., Porkka K., Loughran T.P., Jr., Heckman C.A., Maciejewski J.P., Mustjoki S. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366:1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treon S.P., Xu L., Yang G., Zhou Y., Liu X., Cao Y., Sheehy P., Manning R.J., Patterson C.J., Tripsas C., Arcaini L., Pinkus G.S., Rodig S.J., Sohani A.R., Harris N.L., Laramie J.M., Skifter D.A., Lincoln S.E., Hunter Z.R. MYD88 L265P somatic mutation in Waldenström's macroglobulinemia. N Engl J Med. 2012;367:826–833. doi: 10.1056/NEJMoa1200710. [DOI] [PubMed] [Google Scholar]
- 17.Weng A.P., Ferrando A.A., Lee W., Morris J.P.I.V., Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 18.Rossi D., Trifonov V., Fangazio M., Bruscaggin A., Rasi S., Spina V. The coding genome of splenic marginal zone lymphoma: activation of NOTCH2 and other pathways regulating marginal zone development. J Exp Med. 2012;209:1537–1551. doi: 10.1084/jem.20120904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kon A., Shih L.Y., Minamino M., Sanada M., Shiraishi Y., Nagata Y. Recurrent mutations in multiple components of the cohesin complex in myeloid neoplasms. Nat Genet. 2013;45:1232–1237. doi: 10.1038/ng.2731. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., Chalkidis G., Suzuki Y., Shiosaka M., Kawahata R., Yamaguchi T., Otsu M., Obara N., Sakata-Yanagimoto M., Ishiyama K., Mori H., Nolte F., Hofmann W.K., Miyawaki S., Sugano S., Haferlach C., Koeffler H.P., Shih L.Y., Haferlach T., Chiba S., Nakauchi H., Miyano S., Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 22.Bonnet D., Dick J.E. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 23.Nangalia J., Massie C.E., Baxter E.J., Nice F.L., Gundem G., Wedge D.C. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med. 2013;369:2391–2405. doi: 10.1056/NEJMoa1312542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klampfl T., Gisslinger H., Harutyunyan A.S., Nivarthi H., Rumi E., Milosevic J.D., Them N.C.C., Berg T., Gisslinger B., Pietra D., Chen D., Vladimer G.I., Bagienski K., Milanesi C., Casetti I.C., Sant'Antonio E., Ferretti V., Elena C., Schischlik F., Cleary C., Six M., Schalling M., Schönegger A., Bock C., Malcovati L., Pascutto C., Superti-Furga G., Cazzola M., Kralovics R. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med. 2013;369:2379–2390. doi: 10.1056/NEJMoa1311347. [DOI] [PubMed] [Google Scholar]
- 25.Gambacorti-Passerini C., Donadoni C., Parmiani A., Pirola A., Redaelli S., Signore G., Piazza V., Malcovati L., Fontana D., Spinelli R., Magistroni V., Gaipa G., Peronaci M., Morotti A., Panuzzo C., Saglio G., Usala E., Kim D., Rea D., Zervakis K., Viniou N., Symeonidis A., Becker H., Boutlwood J., Campiotti L., Carrabba M., Elli E., Bignell G.R., Papaemmanuil E., Campbell P.J., Cazzola M., Piazza R. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood. 2015;125:499–503. doi: 10.1182/blood-2014-06-579466. [DOI] [PubMed] [Google Scholar]
- 26.Thol F., Bollin R., Gehlhaar M., Walter C., Dugas M., Suchanek K.J., Kirchner A., Huang L., Chaturvedi A., Wichmann M., Wiehlmann L., Shahswar R., Damm F., Göhring G., Schlegelberger B., Schlenk R., Döhner K., Döhner H., Krauter J., Ganser A., Heuser M. Mutations in the cohesin complex in acute myeloid leukemia: clinical and prognostic implications. Blood. 2014;123:914–920. doi: 10.1182/blood-2013-07-518746. [DOI] [PubMed] [Google Scholar]
- 27.Grasso C., Butler T., Rhodes K., Quist M., Neff T.L., Moore S., Tomlins S.A., Reinig E., Beadling C., Andersen M., Corless C.L. Assessing copy number alterations in targeted, amplicon-based next-generation sequencing data. J Mol Diagn. 2015;17:53–63. doi: 10.1016/j.jmoldx.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoogstraat M., Hinrichs J.W.J., Besselink N.J.M., Radersma-van Loon J.H., de Voijs C.M.A., Peeters T., Nijman I.J., de Weger R.A., Voest E.E., Willems S.M., Cuppen E., Koudijs M.J. Simultaneous detection of clinically relevant mutations and amplifications for routine cancer pathology. J Mol Diagn. 2015;17:10–18. doi: 10.1016/j.jmoldx.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Kampa-Schittenhelm K.M., Heinrich M.C., Akmut F., Döhner H., Döhner K., Schittenhelm M.M. Quizartinib (AC220) is a potent second generation class III tyrosine kinase inhibitor that displays a distinct inhibition profile against mutant-FLT3, -PDGFRA and -KIT isoforms. Mol Cancer. 2013;12:19. doi: 10.1186/1476-4598-12-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang F., Travins J., DeLaBarre B., Penard-Lacronique V., Schalm S., Hansen E., Straley K., Kernytsky A., Liu W., Gliser C., Yang H., Gross S., Artin E., Saada V., Mylonas E., Quivoron C., Popovici-Muller J., Saunders J.O., Salituro F.G., Yan S., Murray S., Wei W., Gao Y., Dang L., Dorsch M., Agresta S., Schenkein D.P., Biller S.A., Su S.M., de Botton S., Yen K.E. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 31.Hatzimichael E., Georgiou G., Benetatos L., Briasoulis E. Gene mutations and molecularly targeted therapies in acute myeloid leukemia. Am J Blood Res. 2013;3:29–51. [PMC free article] [PubMed] [Google Scholar]
- 32.Kuo F.C., Dong F. Next-generation sequencing-based panel testing for myeloid neoplasms. Curr Hematol Malig Rep. 2015;10:104–111. doi: 10.1007/s11899-015-0256-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data analysis pipeline for rapid heme panel. AF, allele frequency; FLT3-ITD, FLT3 internal tandem repeat; SNP, single nucleotide polymorphism.
The amplicons are ranked by average depth of coverage for six normal control DNA (from lowest to highest) and plotted with their %GC contents. Red bars indicate %GC content; green bars, average coverage; blue bars, SD of coverage of six control DNA; x axis, amplicon rank; left y axis, average coverage; right y axis, GC%. Average coverage is not correlated with %GC content of the amplicon.