Abstract
Prostate cancer is the most common cancer among men. The prospective discrimination of aggressive and clinically insignificant tumors still poses a significant and, as yet, unsolved problem. PITX2 DNA methylation is a strong prognostic biomarker in prostate cancer. Recently, a diagnostic microarray for prostate cancer prognosis based on PITX2 methylation has been developed and validated. Because this microarray requires nonstandard laboratory equipment, its use in a diagnostic setting is limited. This study aimed to develop and validate an alternative quantitative real-time PCR assay for measuring PITX2 methylation that can easily be established in clinical laboratories, thereby facilitating the implementation of this biomarker in clinical practice. A methylation cut-off for patient stratification was established in a training cohort (n = 157) and validated in an independent test set (n = 523) of men treated with radical prostatectomy. In univariate Cox proportional hazards analysis, PITX2 hypermethylation was a significant predictor for biochemical recurrence (P < 0.001, hazard ratio = 2.614). Moreover, PITX2 hypermethylation added significant prognostic information (P = 0.003, hazard ratio = 1.814) to the Gleason score, pathological T stage, prostate-specific antigen, and surgical margins in a multivariate analysis. The clinical performance was particularly high in patients at intermediate risk (Gleason score of 7) and in samples containing high tumor cell content. This assay might aid in risk stratification and support the decision-making process when determining whether a patient might benefit from adjuvant treatment after radical prostatectomy.
In the western world, prostate cancer is by far the most common cancer and also one of the leading causes of cancer-related deaths among men.1 However, because of the relatively good prognosis of prostate cancer, only a relatively low percentage of men diagnosed as having localized prostate cancer will die of it. Prostate-specific antigen (PSA) screening for prostate cancer allows for early detection of prostate cancer and, regardless of the limitations of PSA and the problem of overdiagnosis, saves lives.2 However, PSA testing often results in men being diagnosed as having clinically insignificant prostate cancer that may pose little or no threat to their life if left untreated.3, 4 Currently, most patients with biopsy-confirmed localized prostate cancer undergo some form of treatment, such as radical prostatectomy, even though a large fraction of these patients are likely to be overtreated. On the other hand, despite the overall good prognosis of the disease and the problem of overdiagnosis, 15% to 30% of patients receiving radical prostatectomy will experience a biochemical relapse5 and might benefit from an adjuvant therapy after radical prostatectomy. Prognostic biomarkers, which allow for the identification of patients with an aggressive tumor, will therefore help to improve prostate cancer management. These biomarkers might be useful in two distinct clinical situations in which a clinician has to reach a treatment decision. First, a prognostic biomarker for outcome prediction in patients who received radical prostatectomy might be considered in the future to personalize adjuvant therapy. Patients at high risk for biochemical recurrence (BCR) might benefit from an adjuvant endocrine therapy, chemotherapy, or radiotherapy. Second, a prognostic biomarker, which works in biopsy samples from patients with prostate cancer, might allow for the identification of the subgroup of patients who do not need a radical prostatectomy and could instead be monitored by active surveillance. Therefore, such an assay might help combat the issue of overtreatment in prostate cancer patients.
DNA methylation is an epigenetic mechanism that plays an important role in fundamental biological processes, such as cell differentiation and development.6, 7 In addition, aberrant DNA methylation is a hallmark of malignant tumors and plays a major role during carcinogenesis, suggesting that aberrant methylated loci may be a valuable source for cancer biomarkers.8, 9 DNA methylation of the PITX2 gene locus has been established in several studies as a prognostic biomarker in various cancers. Methylation of PITX2 has been reported to be strongly associated with disease outcome in lung cancer patients10 and in hormone receptor–positive breast cancer patients.11, 12, 13, 14 In addition, PITX2 methylation was found to be strongly associated with BCR in prostate cancer patients who received radical prostatectomy.15, 16, 17 In multivariate analysis, PITX2 methylation added significant independent prognostic information to established clinicopathologic parameters (age, pathological T stage, Gleason score, and presurgical PSA). In particular, PITX2 methylation was able to provide independent information on the risk of BCR in patients at intermediate risk (Gleason score of 7), which represents a high portion of all individuals with prostate cancer treated with radical prostatectomy and for whom the assessment of disease prognosis is particularly challenging.15, 17
PITX2 is a member of the homeobox gene family, which has been characterized extensively as a transcriptional factor involved in pattern formation in both invertebrate and vertebrate species. The protein encoded by PITX2 is involved in the development of several organs, including the heart, lungs, pituitary gland, and teeth,18 and in the determination of left-right asymmetry during development.19 Mutations in PITX2 lead to Axenfeld-Rieger syndrome, a genetic disease affecting multiple organ systems.20 However, the functional role of PITX2 during carcinogenesis is still largely unknown. Interestingly, PITX2 DNA methylation has recently been identified as an upstream regulator of the insulin-like growth factor 1 receptor and androgen receptor in prostate cancer,21 suggesting its important role in prostate carcinogenesis and progression.
Because of the high clinical performance of the PITX2 DNA methylation biomarker in prostate cancer patients, a robust and reliable diagnostic test for assessing the methylation status of PITX2 was developed, enabling an improved outcome prediction in prostate cancer patients after radical prostatectomy.16 This microarray-based test (EpiChip PITX2; Epigenomics AG, Berlin, Germany) was developed for use with a validated diagnostic platform, the Affymetrix GeneChip System (Affymetrix, Santa Clara, CA). The clinical performance of the test was further successfully validated in a large retrospective cohort study17 of >500 patients. However, the Affymetrix GeneChip System is nonstandard laboratory equipment, which limits the utility of this test. In addition, the test requires several whole formalin-fixed, paraffin-embedded (FFPE) tumor sections and, therefore, cannot be used in biopsy samples, where such a prognostic test might be of particular value.
The goal of this study was to develop a real-time PCR-based assay for accurate and reliable assessment of the methylation level of PITX2. The assay was designed to probe the identical CpG sites analyzed in the previous studies using different methodological approaches.15, 16, 17 A methylation cut-off for patient stratification was established and verified using leftover bisulfite-converted DNA from a cohort of 157 patients analyzed in a previous study.16 The success of the assay development and cut-off transfer was further verified by comparing the results directly to the results of the EpiChip PITX2 obtained from the same samples. The clinical performance of the assay was then successfully validated using leftover DNA samples from the cohort, which were previously used to validate the microarray-based EpiChip PITX2.17 The developed real-time PCR assay might be successfully applied to biopsy specimens in the future and therefore support the decision-making process when determining whether a prostate cancer patient will benefit from a radical prostatectomy or should be left untreated.
Materials and Methods
Preparation of a Calibrator Sample and Model DNAs with Defined Methylation Levels
Sperm DNA was extracted as previously described.16 Bisulfite conversion of artificially methylated human DNA and DNA from human sperm was conducted according to the ammonium bisulfite protocol.10 Bisulfite-converted, artificially (fully) methylated human DNA was used as a calibrator sample. For analytical performance evaluation, model DNA samples with defined methylation levels (0%, 0.25%, 0.5%, 1%, 2%, 4%, 8%, 16%, 32%, 64%, and 100%) were prepared by mixing bisulfite-converted, artificially methylated human DNA and DNA from sperm. The concentration of bisulfite-converted DNA was measured by means of UV spectrophotometry using a Nanodrop ND-1000 spectral photometer (Nanodrop Technologies, Wilmington, DE).
Cohorts and Patient Samples
Leftover bisulfite-converted DNA prepared from tumor samples from FFPE radical prostatectomy tissue blocks in the course of two previous studies was used.16, 17 For cut-off establishment and verification, leftover bisulfite samples from 157 patient samples were taken from a previous study in which the cut-off was already transferred to the microarray-based EpiChip PITX2 microarray assay.16 This cohort was obtained from the Virginia Mason Medical Center (Seattle, WA) and included patient samples from patients who were treated between 1996 and 2000 and were analyzed according to the protocol approved by the institutional review board. This cohort is described in more detail elsewhere.15, 16
For the validation of the prognostic power of the developed assay, leftover bisulfite-converted DNA from the EpiChip PITX2 validation study was used.17 During that previous study, cohorts were recruited retrospectively from four sites under institutional review board approval. Samples were obtained from patients who underwent radical prostatectomy between 1995 and 2001 at Baylor College of Medicine, Duke University, Durham Veterans Affairs Medical Center, and Erasmus Medical Center. Patients who received any treatment other than radical prostatectomy before BCR were excluded from the study. BCR was defined as the clinical end point of this study as described previously.17 In brief, two consecutive PSA measurements ≥0.2 ng/mL were considered as BCR. Patient eligibility included confirmed prostate adenocarcinoma (stage pT2 or pT3), no evidence of lymph node or distant metastasis, and a postoperative PSA decrease to <0.1 ng/mL within 6 months of radical prostatectomy.17 The characteristics of the patients in the validation study are listed in Table 1.
Table 1.
Characteristics of the Cohort Composed of 523 Prostate Cancer Patients Who Received Radical Prostatectomy as Primary Treatment
| Characteristic | No. (%) of patients |
|---|---|
| All patients | 523 (100) |
| BCR | |
| Yes | 109 (21) |
| No | 414 (79) |
| Clinical center | |
| Baylor College of Medicine | 98 (19) |
| Duke University | 198 (38) |
| Durham Veterans Affairs Medical Center | 88 (17) |
| Erasmus Medical Center | 139 (27) |
| Ethnicity | |
| African American | 95 (18) |
| White | 282 (54) |
| Other or unknown | 146 (28) |
| Age (years) | |
| 40–49 | 7 (1) |
| 50–59 | 158 (30) |
| 60–69 | 288 (55) |
| 70–79 | 70 (13) |
| Pathological stage | |
| pT2 (organ confined) | 368 (70) |
| pT3a (extraprostatic extension) | 125 (24) |
| pT3b (seminal vesicle invasion) | 29 (6) |
| Unknown | 1 (0.2) |
| Preoperative PSA (ng/mL) | |
| ≤4.0 | 100 (19) |
| 4.1–10 | 311 (59) |
| 10.1–20 | 86 (16) |
| >20 | 26 (5) |
| Surgical margin | |
| R1 | 157 (30) |
| R0 | 362 (69) |
| Unknown | 4 (0.8) |
| Tumor content in sample | |
| 1%–9% | 40 (8) |
| 10%–24% | 85 (16) |
| 25%–49% | 136 (26) |
| 50%–74% | 144 (28) |
| 75%–100% | 118 (23) |
| Pathological Gleason score | |
| 4 | 1 (0.2) |
| 5 | 55 (11) |
| 6 | 173 (33) |
| 7 | 253 (48) |
| 8 | 22 (4) |
| 9 | 17 (3) |
| 10 | 2 (0.4) |
All patients had a localized disease (N0, M0) with a postoperative decrease of PSA serum levels <0.1 ng/mL.
In contrast to the previous validation study,17 in the current study, all samples were included and no patients were excluded because of low tumor cell content. Specifically, 40 men who were excluded from the prior validation study were included in the current study.
Real-Time PCR Quantification of PITX2 DNA Methylation
Relative DNA methylation of the PITX2 locus (PITX2CH3), compared with total DNA as determined with a methylation unspecific PITX2 assay (PITX2Total), was analyzed via quantitative, duplex real-time PCR. Twenty μL PCR reactions were performed with the following composition: 35 mmol/L Tris-HCl, pH 8.4, 6 mmol/L MgCl2, 50 mmol/L KCl, 5% glycerol, dNTPs [0.25 mmol/L dCTP, dGTP, and dATP each and 0.50 mmol/L dUTP (Bioline, London, UK)], 3 U of FastStart TaqDNA polymerase (Roche Applied Science, Penzberg, Germany), 0.004 mL of ROX/DMSO solution (prepared as previously described10), primers (0.3 mmol/L 5′-GGAGGGAAGTAGATGTTA-3′, 0.3 mmol/L 5′-CCAAATCCCCTCTCCTTTC-3′, 0.5 mmol/L 5′-TTGGTGATTAATTTAAAGGAGTTAT-3′, 0.5 mmol/L 5′-AATTACCTAAAAACCAAACCTAA-3′), 1 mmol/L blocker (5′-CCTTTCACTCTCCCAACTCCAACTCCCAA-SpacerC3-3′), 0.1 mmol/L each detection probe (PITX2CH3: 5′-6-FAM-TCGGAGTCGGGAGAGC-DABCYL-3′, PITX2Total: 5′-R6G-TTAAA-GAAATGGTGAGAGTTTGGTAT-BHQ-1-3′), and 2.5 μL of DNA. PCR was performed using the 7500 Fast Real-Time PCR System (Life Technologies, Carlsbad, CA) with the following temperature profile: 15 minutes at 95°C and 50 cycles 75 seconds at 57°C (at 100% ramp rate) and 15 seconds at 95°C (at 75% ramp rate).
For each valid sample, a relative methylation value was determined using the ΔΔCT method as previously described10: ΔΔCTSample = ΔCTSample – ΔCTCalibrator, where ΔCTSample = CTPITX2_CH3[Sample] – CTPITX2_Total[Sample] and ΔCTCalibrator = CTPITX2_CH3[Calibrator] – CTPITX2_Total[Calibrator]. ΔΔCT were measured in triplicate, and median average was computed. Percentage methylation was calculated using the following formula: Methylationsample = 100% · 2−ΔΔCTSample.
Statistical Analysis
Associations between PITX2 DNA methylation and clinicopathologic variables were analyzed using the Kendall τ, Spearman, and Pearson tests. The independent t-test was used to test for a DNA methylation difference between races. BCR-free survival was calculated using the Kaplan-Meier method, and survival time differences were compared using the log-rank test. DNA methylation was also examined within univariate and multivariate Cox proportional hazards regression models. P values refer to the Wald test. P < 0.05 was considered statistically significant. All analyses were performed using the SPSS software version 20 (IBM, Armonk, NY).
Results
Analytical Assay Performance
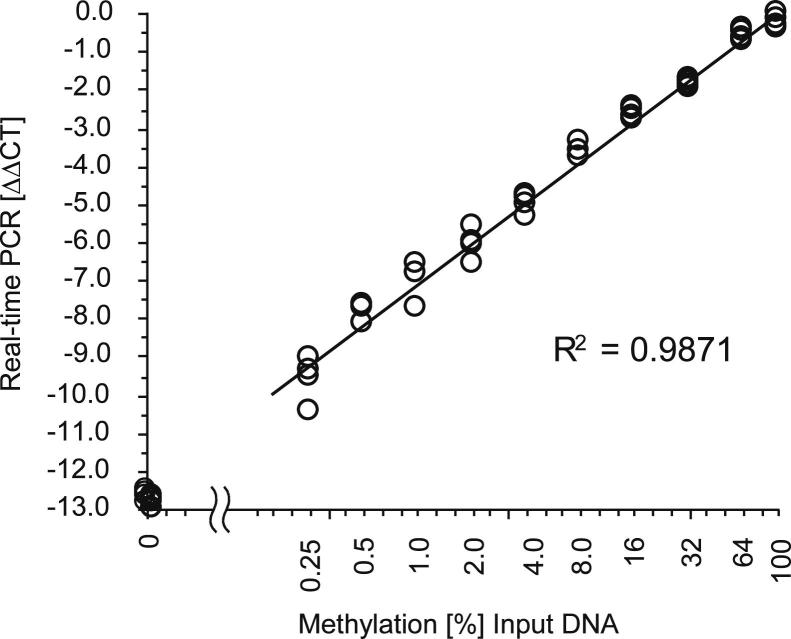
A quantitative real-time PCR (qPCR) assay to accurately determine the methylation levels of the PITX2 promoter was developed. This qPCR assay combined HeavyMethyl22 and TaqMan technologies together with the ΔΔCT method10, 23, 24, 25 for the determination of the relative amount of methylated PITX2 in a sample. The PCR is a duplex reaction for the simultaneous detection of methylated PITX2 gene copies and the total number of PITX2 gene copies irrespective of their methylation in a single tube. The PCR for assaying the DNA methylation of PITX2 is a methylation-specific HeavyMethyl PCR. The assay for measuring the total number of PITX2 gene copies is located in a region of the gene, which contains no CpG and therefore leads to an amplification of the total DNA. As shown in Figure 1, the developed assay allowed for the highly accurate determination of PITX2 DNA methylation over the whole spectrum of possible methylation levels (0% to 100%). Each measurement in Figure 1 reflects a single PCR result. To further increase the accuracy of the assay result, the patient samples were measured in triplicate.
Figure 1.
Analytical performance of the developed real-time PCR assay to measure the DNA methylation of the PITX2 gene locus. DNA samples with defined levels of DNA methylation were prepared by mixing artificially methylated DNA and DNA from sperm. Each mixture was analyzed in quadruplicate, and each measurement reflects the result from a single PCR.
Establishment and Verification of a Methylation Cut-Off for Patient Stratification
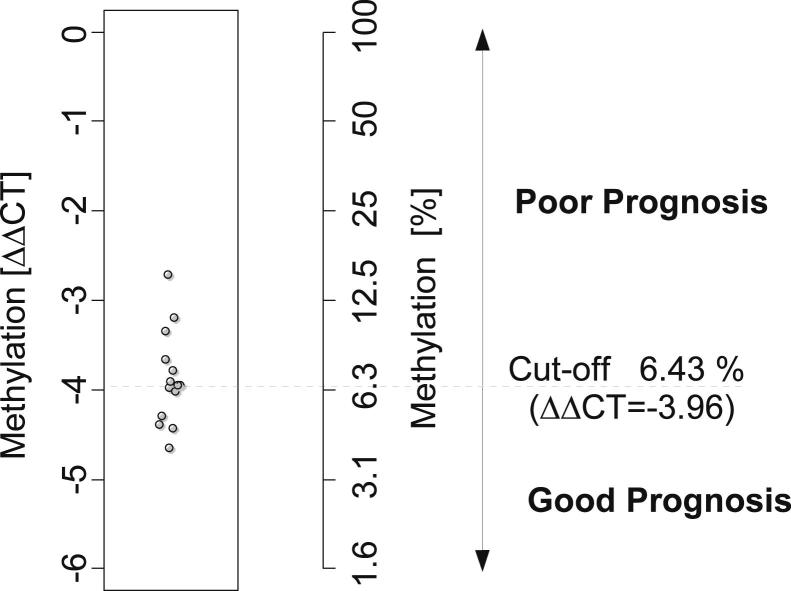
PITX2 DNA methylation has previously been reported to be a prognostic marker for predicting BCR in patients who have undergone radical prostatectomy.15 The previous study revealed that methylation at different levels could be found in most samples, and patient stratification into a high-risk and a low-risk group necessitated the use of a methylation cut-off. The cut-off had been defined with regard to an optimized clinical performance of the assay.15 To transfer this cut-off to the developed real-time PCR assay, samples from 14 patients were selected that revealed methylation levels close to the cut-off in the former study.15 Seven patient samples were close to but above and seven samples were close to but below the cut-off from the previous study. Leftover bisulfite-converted DNA from these patient samples was taken from a study in which this cut-off had already been transferred to a microarray-based EpiChip PITX2 assay.16 The median methylation level of these patient samples was defined as the cut-off for the developed real-time PCR assay. Figure 2 shows the result of the analysis of the 14 samples. A ΔΔCT of −3.96 (refers to 6.43% methylation) was determined and established as the cut-off for the new assay. According to this clinical cut-off, samples with PITX2 methylation levels <6.43% methylation are rated as having a “good prognosis,” whereas samples with PITX2 methylation level values >6.43% are relegated to the “poor prognosis” group.
Figure 2.
Establishment of a methylation cut-off for patient stratification into groups of patients at high risk and low risk of BCR. Leftover DNAs from 14 patient samples, which reflected the cut-off in a previous sample cohort,15 were measured using the developed PITX2 real-time PCR assay. The resulting cut-off was defined as the median of the 14 patient samples. Resulting cut-off: ΔΔCT = −3.96 (refers to 6.43% methylation).
Concordance Evaluation
To evaluate the concordance of the results derived from the developed real-time PCR assay to the results obtained from the microarray measurement, the full cohort from the Virginia Mason Medical Center, used in the previous studies,15, 16 was reanalyzed. Leftover bisulfite-converted DNA from 157 patient samples as prepared by Schatz et al16 was analyzed with the new PCR assay. The results were compared with results generated with the EpiChip PITX2 within the same cohort.16 Valid results with both assays were obtained from 153 patients. The classification of the patients into two groups with low and high methylation status was used to provide estimates of the probability of BCR after prostatectomy.
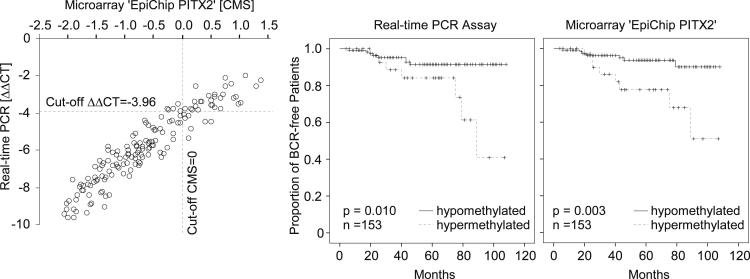
Figure 3 displays the analytical and clinical concordance of the developed real-time PCR assay and the results from the EpiChip PITX2 assay as generated earlier.16 Obviously, the classification is highly similar because the corresponding pairs of Kaplan-Meier survival curves are similar for both assays. In addition, both assays separate the cohort into two groups of good and poor prognosis (log rank test: P = 0.010 for real-time PCR assay and P = 0.003 for EpiChip PITX2). In univariate analyses, estimates of the hazard ratio (HR) to quantify the prognostic value of dichotomized PITX2 methylation levels are highly similar and significantly larger than 1 (EpiChip PITX2: HR = 4.124; 95% CI = 1.493 to 11.395; real-time PCR: HR = 3.479; 95% CI = 1.259 to 9.614). These results verified that the cut-off for patient stratification was successfully transferred to the new assay format.
Figure 3.
Left panel: Concordance of results obtained using real-time PCR and EpiChip PITX2 assays, respectively. EpiChip PITX2 measurements were taken from a previous study.16 Kaplan-Meier analysis of BCR-free survival in 153 prostate cancer patients stratified by the DNA methylation status of PITX2 as determined by real-time PCR (middle panel) and microarray (right panel). Methylation cut-off for patient stratification was applied as established above. CMS, calibrated methylation score according to Schatz et al.16
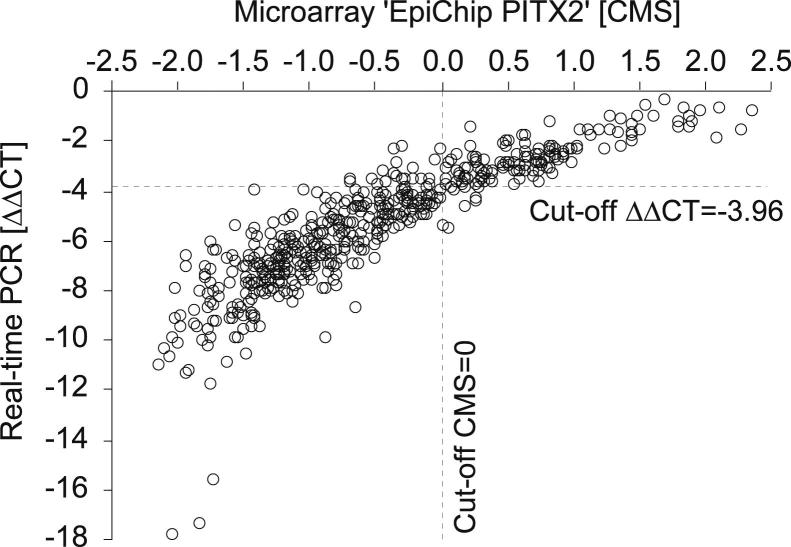
The developed real-time PCR assay for PITX2 DNA methylation was further used to measure leftover bisulfite-converted DNA samples from a study in which the clinical performance of the EpiChip PITX2 assay had been validated.17 Figure 4 shows the concordance between the measurement of the real-time PCR assay and the EpiChip PITX2 measurements as previously generated.17 Both assays gave highly concordant results (Spearman’s ρ = 0.922, P < 0.001).
Figure 4.
Concordance of the results as derived by real-time PCR and EpiChip PITX2 DNA methylation analyses. EpiChip PITX2 measurements were taken from a previous study.17 Valid results with both assays were obtained from 523 patients. CMS, calibrated methylation score according to Schatz et al.16
Clinical Performance Evaluation
The leftover samples from the validation study performed by Bañez and colleagues17 were further used to investigate the prognostic power of the developed real-time PCR. First, before dichotomization, the correlation of PITX2 DNA methylation level as a continuous variable with clinicopathologic variables (age, tumor cell content, Gleason score, surgical margin, pathological T stage, and preoperative PSA) was investigated. Parametric and nonparametric correlation models were applied. Table 2 indicates that when using different correlation methods (Kendall τ, Pearson, and Spearman tests) the PITX2 DNA methylation level significantly and strongly correlates with tumor cell content, Gleason score, pathological T stage, preoperative PSA, and surgical margin (P < 0.001 for all). In addition, PITX2 DNA methylation correlated with the age of the patients (Table 2). The independent t-test revealed no DNA methylation difference among tumors from white and African American patients (P = 0.28).
Table 2.
Associations of PITX2 DNA Methylation with Clinicopathologic Variables
| Variable | Kendall τ test | P | Pearson test | P | Spearman test | P |
|---|---|---|---|---|---|---|
| Age | 0.088 | 0.003 | 0.140 | 0.001 | 0.131 | 0.003 |
| Tumor cell content | 0.349 | <0.001 | 0.482 | <0.001 | 0.489 | <0.001 |
| Gleason score | 0.212 | <0.001 | 0.247 | <0.001 | 0.271 | <0.001 |
| Surgical margin | 0.155 | <0.001 | 0.184 | <0.001 | 0.190 | <0.001 |
| Pathological stage (pT) | 0.232 | <0.001 | 0.285 | <0.001 | 0.284 | <0.001 |
| Preoperative PSA | 0.128 | <0.001 | 0.130 | 0.003 | 0.185 | <0.001 |
P values and correlation coefficients (Kendall τ, Pearson, and Spearman tests) are listed in the table. PITX2 DNA methylation is analyzed as a continuous variable. Higher PITX2 methylation was found in older patients.
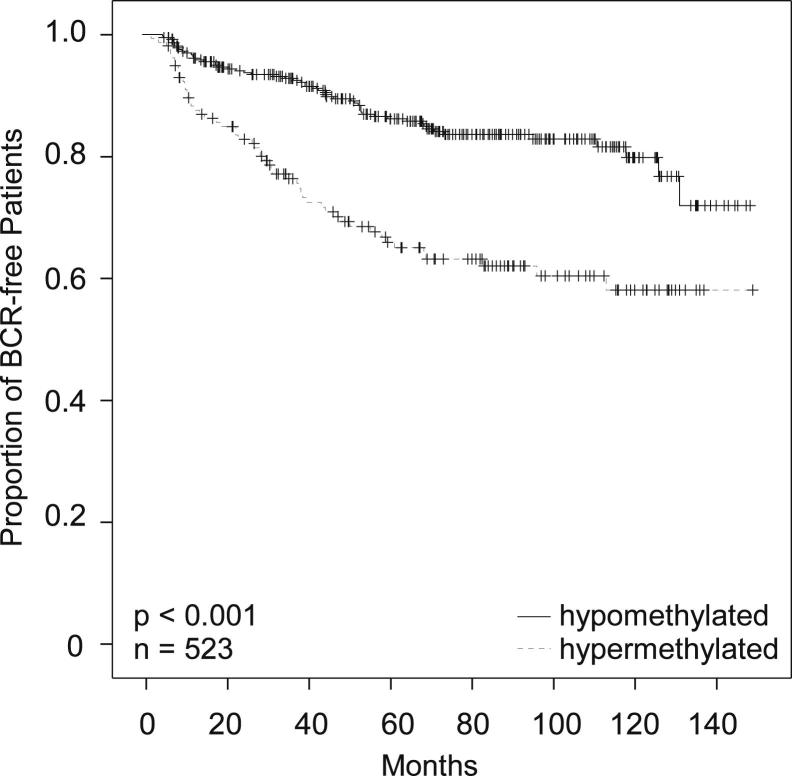
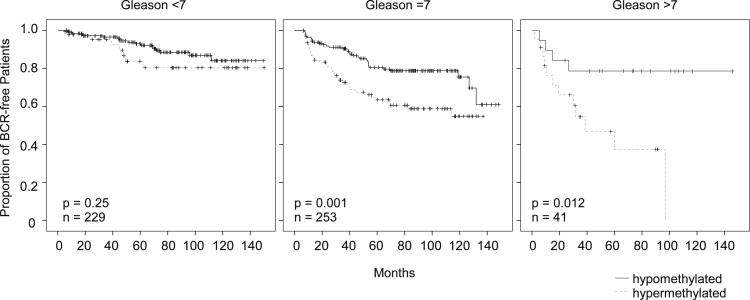
PITX2 DNA methylation levels were dichotomized using the cut-off as established. The prognostic power of the assay was studied by means of Kaplan-Meier analysis. Figure 5 shows the results of the Kaplan-Meier analysis in the whole cohort of 523 prostate cancer patients who were stratified by dichotomized PITX2 DNA methylation levels. The developed real-time PCR assay for measuring PITX2 DNA methylation allowed for a highly significant prediction of BCR in the analyzed cohort (P < 0.001). The clinical performance of the assay was shown to be particularly high in patients at intermediate risk (Gleason score of 7) who require further risk stratification (Figure 6).
Figure 5.
Kaplan-Meier analysis of BCR-free survival in 523 prostate cancer patients stratified by the DNA methylation status of PITX2. Methylation cut-off for patient stratification was applied as established above.
Figure 6.
Kaplan-Meier analysis of BCR-free survival in prostate cancer patients stratified by the DNA methylation status of PITX2 with respect to Gleason score.
In univariate Cox proportional hazards model analysis (Table 3), PITX2 DNA hypermethylation was a significant and strong predictor of BCR (P < 0.001; HR = 2.614; 95% CI = 1.795 to 3.807). It is comparable to the results obtained by reanalyzing the data generated during the previous microarray study.17 In the same patient group, including patient samples with a <10% tumor cell content (n = 523), the HRs were highly similar (P < 0.001; HR = 2.757; 95% CI = 1.891 to 4.018), and the 95% CIs were largely overlapping.
Table 3.
Univariate and Multivariate Cox Proportional Hazard Model Analyses on BCR-Free Survival for PITX2 DNA Methylation, Gleason Score, Pathological Stage, Preoperative PSA Level, and Surgical Margin
| Variable | Univariate Cox analysis |
Multivariate Cox analysis |
||
|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | |
| Pathological stage (pT2 reference) | ||||
| pT3 | 3.165 (2.171–4.615) | <0.001 | 2.255 (1.530–3.325) | <0.001 |
| Gleason score (7 reference) | ||||
| <7 | 0.388 (0.245–0.614) | <0.001 | 0.569 (0.356–0.911) | 0.019 |
| >7 | 1.910 (1.107–3.297) | 0.020 | 1.766 (1.006–3.100) | 0.048 |
| Surgical margin (R0 reference) | ||||
| R1 | 3.203 (2.196–4.671) | <0.001 | 2.224 (1.507–3.283) | <0.001 |
| PITX2 methylation (hypomethylated reference) | ||||
| Hypermethylated | 2.614 (1.795–3.807) | <0.001 | 1.814 (1.232–2.673) | 0.003 |
| Preoperative PSA | 1.012 (1.002–1.022) | 0.016 | 1.011 (0.995–1.028) | 0.177 |
Gleason score was analyzed as categorized (<7, 7, and >7) variable. Preoperative PSA level was analyzed as continuous variable.
In addition, known prognostic factors (pathological T stage, Gleason score, preoperative PSA, and surgical margin) were confirmed to be prognostic in this cohort (Table 3). To test whether PITX2 DNA methylation measured with the real-time PCR assay added independent information to these known prognostic clinicopathologic parameters, multivariate Cox proportional hazard model analysis was performed. In multivariate analysis, PITX2 hypermethylation added significant independent prognostic information (P = 0.003; HR = 1.814; 95% CI = 1.232 to 2.673) to Gleason score, pathological T stage, preoperative PSA, and surgical margin (Table 3).
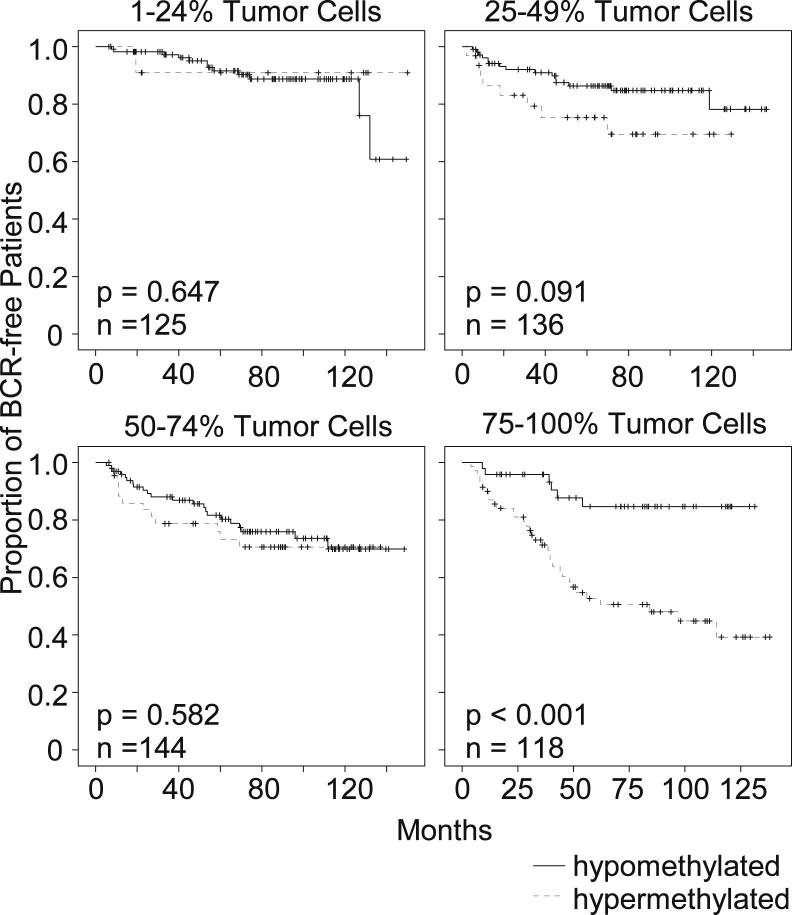
Table 2 indicates that PITX2 hypermethylation is strongly associated with the tumor cell content. In addition, tumor cell content is a strong prognostic factor in univariate Cox proportional hazards analysis (P < 0.001). These findings trigger the assumption that PITX2 DNA methylation might in fact be more of a surrogate marker for the tumor cell content and therefore the tumor size than a true prognostic biomarker. However, the multivariate Cox analysis adjusting for tumor cell content and pT stage, together with PITX2 methylation, clearly revealed that PITX2 hypermethylation added significant prognostic information (P = 0.002; HR = 1.889; 95% CI = 1.259 to 2.832). Therefore, it can be hypothesized that the prognostic value of the biomarker might be increased when analyzing a highly enriched tumor cell population without any contaminating nontumor cells. Therefore, an analysis of the prognostic power with respect to tumor cell content was performed. Patient samples were divided into four groups according to the tumor cell content: low content (<25%), low-medium (25% to 49%), medium-high (50% to 74%), and high tumor cell content (≥75%). Figure 7 shows the result of a Kaplan-Meier analysis in these four groups. The stratification of the patients by PITX2 DNA methylation allows for a significant discrimination between patients at high and low risk of BCR in the group in which samples with high tumor cell content were analyzed. Statistically significant risk stratification by PITX2 DNA methylation in the groups with tumor cell content <75% in the analyzed samples was not possible.
Figure 7.
Clinical assay performance with regard to the tumor cell content. Kaplan-Meier analysis of BCR-free survival in prostate cancer patients stratified by the DNA methylation status of PITX2 as measured with the developed real-time PCR assay.
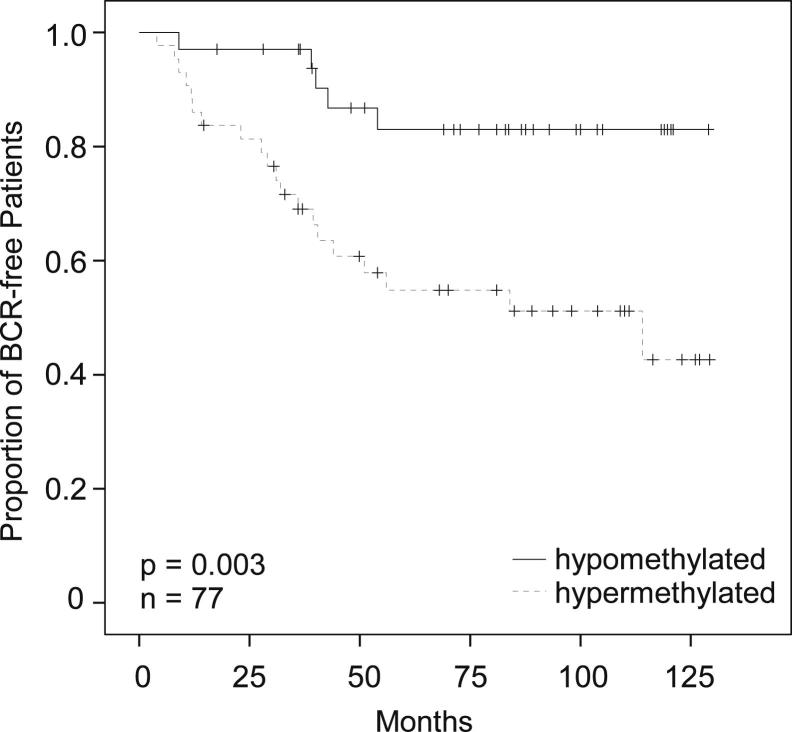
A biomarker for risk stratification in prostate cancer patients is particularly valuable in patients with Gleason scores of 7 (intermediate-risk patients) for whom the risk of recurrence is increased but not a forgone conclusion. To get an idea of how the biomarker might perform when analyzing biopsy material from patients with Gleason scores of 7, which is enriched with tumor cells by macrodissection, the group of patients with Gleason scores of 7 and ≥75% tumor cell content was analyzed. The result of a Kaplan-Meier analysis of these patients (n = 77) stratified by PITX2 DNA methylation is shown in Figure 8. PITX2 DNA methylation is a strong prognostic biomarker in patients with Gleason scores of 7 when analyzing samples with a high tumor cell content (P = 0.003).
Figure 8.
Clinical performance of the developed PITX2 real-time PCR assay in the subgroup of patients with Gleason score 7 tumors and ≥75% tumor cell content in the sample. The figure shows a Kaplan-Meier analysis of BCR-free survival in patients stratified by PITX2 DNA methylation.
Discussion
In 2012, prostate cancer alone will account for 29% (n = 241,740) of incident cases in men1 in the United States. However, at the same time, prostate cancer will account for only 9% of cancer-related deaths among men.1 PSA screening has been reported to save lives; however, it has also led to the problem of overdiagnosis because many clinically insignificant cases that would otherwise not impair patient life are being detected. The huge imbalance between incidence and mortality in the context of increasingly more sensitive screening methods necessitates the proper management of the disease. Therefore, prognostic biomarkers allowing for the discrimination between clinically insignificant cancers and aggressive tumors are of particular clinical value in the field of prostate cancer.
Many biomarker candidates have been evaluated in the context of prostate cancer prognosis.26 In addition, there are numerous studies on DNA methylation biomarkers for prostate cancer prognosis.27 However, none of the biomarkers has been successfully implemented into routine clinical practice. The implementation of DNA methylation analysis in routine clinical practice requires highly precise and accurate determination of the methylation in a sample from each patient. In addition, high interlaboratory and intralaboratory reproducibility and robustness are mandatory. DNA methylation in a heterogeneous tissue sample is not dichotomous, and a prognosis is usually not indicated by the presence or absence of methylation but by the relative abundance of methylated alleles. Thus, a classification into good and poor prognosis patients has to be based on quantitative rather than qualitative measure. This quantitative result has to be converted into a semiquantitative result using a certain methylation level as a cut-off. A high level of accuracy and a precise measurement are mandatory, in particular for samples that have methylation close to the cut point, to avoid misclassification of patients. The transfer of such a cut-off value into other laboratories represents an additional challenge.
Recently, a robust and reliable diagnostic microarray (EpiChip PITX2) for assessing the methylation status of PITX2, enabling an improved outcome prediction in cancer patients after radical prostatectomy, has been developed and validated.16, 17 The analytical performance of the assay has been verified by means of daily-use reproducibility, cut-off verification, and repeatability,16 indicating the general feasibility of introducing such a biomarker test into clinical routine. However, the utility of the test is limited for two specific reasons. First, the test is based on a customized microarray and the Affymetrix GeneChip System, which is nonstandard laboratory equipment. Second, up until now, no therapy option has been linked to a prognostic test predicting BCR in patients who have received radical prostatectomy. The latter is due to the lack of approved treatment options in such scenarios. However, with the development of new drugs, new treatment options in this setting are likely to become available in the future.
In this study, a qPCR for assessing the methylation status of the PITX2 promoter region has been successfully developed. This qPCR assay is a duplex reaction for the simultaneous detection of methylated PITX2 gene copies and the total number of PITX2 genes. Methylation-specific amplification of methylated PITX2 copies is achieved using HeavyMethyl technology.22 TaqMan probes are used for real-time quantification of the PCR product. CT values are translated into a quantitative measure using the ΔΔCT method 10, 23, 24, 25 for the determination of the relative amount of methylated PITX2 in a sample. A real-time PCR assay for measuring SHOX2 DNA methylation in bronchial aspirates from lung cancer patients, based on the same principal and a similar assay composition, has recently been developed as a CE (Conformité Européenne) marked in vitro diagnostic test in Europe.25 Its development proved the general suitability of the technology for routine diagnostic use. The developed assay for measuring PITX2 DNA methylation is optimized to be used together with the 7500 Fast Real-Time PCR System. This and similar instruments are now considered standard laboratory equipment at pathology institutes. Moreover, the Food and Drug Administration–approved Applied Biosystems 7500 Fast Dx Real-Time PCR Instrument (for in vitro diagnostic use) recently became available in the United States. Therefore, the development of a Food and Drug Administration–approved in vitro diagnostic test seems feasible. The developed real-time PCR assay can be performed with standard laboratory equipment, allowing for its smooth implementation into clinical routine.
Overall, this study has found that the clinical performance of the PITX2 real-time PCR assay gave highly concordant results with a diagnostic microarray (EpiChip PITX2) validated in the same patient cohort.17 However, a marginally worse clinical performance of the real-time PCR could have been anticipated because of two factors. The limited availability of leftover DNA from the previous studies resulted in a lower input amount of template DNA in the PCR, which might have led to higher intra-assay variability. In addition, the storage of the bisulfite-converted DNA for >2 years at −20°C might have caused a degradation of the single-stranded, bisulfite-converted DNA. This degradation might have resulted in an additional source of variability. Further studies are needed to elucidate the effect of DNA degradation and PCR template amount on assay variability and clinical performance.
A prognostic test that works in biopsy specimens from patients with primary-diagnosed prostate cancer would be of particular clinical value. Such a prognostic test could contribute to the decision-making process when determining whether a patient diagnosed as having prostate cancer might benefit from a radical prostatectomy or could instead be monitored by active surveillance. The analysis of biopsy specimens represents one major technological challenge. The amount of tissue is very low, which hampers the quantitative DNA methylation analysis. The EpiChip PITX2, for example, works with whole FFPE tissue sections as an input sample. Samples yielding too little DNA are excluded from the analysis because it is anticipated that the result will be unreliable.16 A real-time PCR allows for the quantification of the total DNA content with the simultaneous quantification of the DNA methylation. Thus, such an assay is much more suited for analyzing samples that contain only minute DNA amounts because the results can be interpreted in the context of the total DNA content. On the other hand, the analysis of biopsy material also offers a major advantage. Highly enriched tumor cell population can be obtained by macrodissecting or microdissecting microscopically identified areas of interest from tissue sections mounted on glass slides. Therefore, the resulting sample is only miniscule and the expected DNA yield low; however, it represents highly enriched tumor cell content. In this study, it has been found that the clinical performance of the developed real-time PCR assay was particularly high in samples with high tumor cell content. A quantitative biomarker, which is only expressed in tumor cells, can be expected to exhibit the highest clinical performance when analyzing a highly enriched tumor cell population. Such an analysis of highly enriched tumor cells would avoid a dilution effect that results from contamination with nontumoral cells. This dilution of the biomarker signal caused by contamination with nontumor DNA would lead to an underestimation of the true biomarker level. Further studies using microdissected cells are needed to investigate this issue. However, high performance in highly enriched tumor cells qualifies the assay for further studies with macrodissected biopsy samples. It is possible that clinical performance is highest when analyzing only a few ducts selected and macrodissected or microdissected by a pathologist. This obviously represents a challenge for any assay from a technological point of view. However, an earlier study has found that quantification of DNA methylation biomarkers, even in <100 cells laser microdissected from FFPE tumor sections, is feasible.28 However, the limit of quantification of the presented assay needs to be assessed. A study comparing the clinical assay performance in biopsy samples taken before radical prostatectomy and matched prostatectomy samples might allow for further evaluation of the suitability of the assay for diagnostic purposes.
The clinical performance of the assay was found to be particularly high in patients at intermediate risk (Gleason score of 7) who require further risk stratification. An assay that works in microdissected or macrodissected tumor cells from FFPE biopsy specimens may support the decision-making process when determining whether a patient diagnosed as having prostate cancer at intermediate risk might benefit from a radical prostatectomy or could instead be monitored by active surveillance. However, the clinical performance of the assay in a suited patient cohort (ie, an active surveillance cohort) needs to be evaluated in further studies.
Footnotes
Supported by Epigenomics AG, Berlin, Germany.
Disclosures: D.D., O.H. and P.S. are or have been employees and/or stockholders of Epigenomics AG, a company that aims to commercialize DNA methylation markers (eg, PITX2).
A guest editor acted as editor-in-chief for this manuscript. No person at the Department of Veterans Affairs was involved in the peer review process or final disposition for this article.
References
- 1.Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Schröder F.H., Hugosson J., Roobol M.J., Tammela T.L., Ciatto S., Nelen V., Kwiatkowski M., Lujan M., Lilja H., Zappa M., Denis L.J., Recker F., Berenguer A., Määttänen L., Bangma C.H., Aus G., Villers A., Rebillard X., van der Kwast T., Blijenberg B.G., Moss S.M., de Koning H.J., Auvinen A., ERSPC Investigators Screening and prostate-cancer mortality in a randomized European study. N Engl J Med. 2009;360:1320–1328. doi: 10.1056/NEJMoa0810084. [DOI] [PubMed] [Google Scholar]
- 3.Klotz L. Active surveillance for prostate cancer: a review. Arch Esp Urol. 2011;64:806–814. [PubMed] [Google Scholar]
- 4.Klotz L. Cancer overdiagnosis and overtreatment. Curr Opin Urol. 2012;22:203–209. doi: 10.1097/MOU.0b013e32835259aa. [DOI] [PubMed] [Google Scholar]
- 5.Kattan M.W., Eastham J.A., Stapleton A.M., Wheeler T.M., Scardino P.T. A preoperative nomogram for disease recurrence following radical prostatectomy for prostate cancer. J Natl Cancer Inst. 1998;90:766–771. doi: 10.1093/jnci/90.10.766. [DOI] [PubMed] [Google Scholar]
- 6.De Carvalho D.D., You J.S., Jones P.A. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiman T.M., Muegge K. DNA methylation in early development. Mol Reprod Dev. 2010;77:105–113. doi: 10.1002/mrd.21118. [DOI] [PubMed] [Google Scholar]
- 8.Sinčić N., Herceg Z. DNA methylation and cancer: ghosts and angels above the genes. Curr Opin Oncol. 2011;23:69–76. doi: 10.1097/CCO.0b013e3283412eb4. [DOI] [PubMed] [Google Scholar]
- 9.Kulis M., Esteller M. DNA methylation and cancer. Adv Genet. 2010;70:27–56. doi: 10.1016/B978-0-12-380866-0.60002-2. [DOI] [PubMed] [Google Scholar]
- 10.Dietrich D., Hasinger O., Liebenberg V., Field J.K., Kristiansen G., Soltermann A. DNA methylation of the homeobox genes PITX2 and SHOX2 predicts outcome in non-small-cell lung cancer patients. Diagn Mol Pathol. 2012;21:93–104. doi: 10.1097/PDM.0b013e318240503b. [DOI] [PubMed] [Google Scholar]
- 11.Maier S., Nimmrich I., Koenig T., Eppenberger-Castori S., Bohlmann I., Paradiso A., Spyratos F., Thomssen C., Mueller V., Nährig J., Schittulli F., Kates R., Lesche R., Schwope I., Kluth A., Marx A., Martens J.W., Foekens J.A., Schmitt M., Harbeck N., European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group DNA-methylation of the homeodomain transcription factor PITX2 reliably predicts risk of distant disease recurrence in tamoxifen-treated, node-negative breast cancer patients–Technical and clinical validation in a multi-centre setting in collaboration with the European Organisation for Research and Treatment of Cancer (EORTC) PathoBiology group. Eur J Cancer. 2007;43:1679–1686. doi: 10.1016/j.ejca.2007.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Nimmrich I., Sieuwerts A.M., Meijer-van Gelder M.E., Schwope I., Bolt-de Vries J., Harbeck N., Koenig T., Hartmann O., Kluth A., Dietrich D., Magdolen V., Portengen H., Look M.P., Klijn J.G., Lesche R., Schmitt M., Maier S., Foekens J.A., Martens J.W. DNA hypermethylation of PITX2 is a marker of poor prognosis in untreated lymph node-negative hormone receptor-positive breast cancer patients. Breast Cancer Res Treat. 2008;111:429–437. doi: 10.1007/s10549-007-9800-8. [DOI] [PubMed] [Google Scholar]
- 13.Harbeck N., Nimmrich I., Hartmann A., Ross J.S., Cufer T., Grützmann R., Kristiansen G., Paradiso A., Hartmann O., Margossian A., Martens J., Schwope I., Lukas A., Müller V., Milde-Langosch K., Nährig J., Foekens J., Maier S., Schmitt M., Lesche R. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26:5036–5042. doi: 10.1200/JCO.2007.14.1697. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann O., Spyratos F., Harbeck N., Dietrich D., Fassbender A., Schmitt M., Eppenberger-Castori S., Vuaroqueaux V., Lerebours F., Welzel K., Maier S., Plum A., Niemann S., Foekens J.A., Lesche R., Martens J.W. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009;15:315–323. doi: 10.1158/1078-0432.CCR-08-0166. [DOI] [PubMed] [Google Scholar]
- 15.Weiss G., Cottrell S., Distler J., Schatz P., Kristiansen G., Ittmann M., Haefliger C., Lesche R., Hartmann A., Corman J., Wheeler T. DNA methylation of the PITX2 gene promoter region is a strong independent prognostic marker of biochemical recurrence in patients with prostate cancer after radical prostatectomy. J Urol. 2009;181:1678–1685. doi: 10.1016/j.juro.2008.11.120. [DOI] [PubMed] [Google Scholar]
- 16.Schatz P., Dietrich D., Koenig T., Burger M., Lukas A., Fuhrmann I., Kristiansen G., Stoehr R., Schuster M., Lesche R., Weiss G., Corman J., Hartmann A. Development of a diagnostic microarray assay to assess the risk of recurrence of prostate cancer based on PITX2 DNA methylation. J Mol Diagn. 2010;12:345–353. doi: 10.2353/jmoldx.2010.090088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bañez L.L., Sun L., van Leenders G.J., Wheeler T.M., Bangma C.H., Freedland S.J., Ittmann M.M., Lark A.L., Madden J.F., Hartman A., Weiss G., Castaños-Vélez E. Multicenter clinical validation of PITX2 methylation as a prostate specific antigen recurrence predictor in patients with post-radical prostatectomy prostate cancer. J Urol. 2010;184:149–156. doi: 10.1016/j.juro.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 18.Lin C.R., Kioussi C., O’Connell S., Briata P., Szeto D., Liu F., Izpisúa-Belmonte J.C., Rosenfeld M.G. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- 19.Kato Y. The multiple roles of Notch signaling during left-right patterning. Cell Mol Life Sci. 2011;68:2555–2567. doi: 10.1007/s00018-011-0695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang T.C., Summers C.G., Schimmenti L.A., Grajewski A.L. Axenfeld-Rieger syndrome: new perspectives. Br J Ophthalmol. 2012;96:318–322. doi: 10.1136/bjophthalmol-2011-300801. [DOI] [PubMed] [Google Scholar]
- 21.Schayek H., Bentov I., Jacob-Hirsch J., Yeung C., Khanna C., Helman L.J., Plymate S.R., Werner H. Global methylation analysis identifies PITX2 as an upstream regulator of the androgen receptor and IGF-I receptor genes in prostate cancer. Horm Metab Res. 2012;44:511–519. doi: 10.1055/s-0032-1311566. [DOI] [PubMed] [Google Scholar]
- 22.Cottrell S.E., Distler J., Goodman N.S., Mooney S.H., Kluth A., Olek A., Schwope I., Tetzner R., Ziebarth H., Berlin K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004;32:e10. doi: 10.1093/nar/gnh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Winer J., Jung C.K., Shackel I., Williams P.M. Development and validation of real-time quantitative reverse transcriptase-polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem. 1999;270:41–49. doi: 10.1006/abio.1999.4085. [DOI] [PubMed] [Google Scholar]
- 25.Dietrich D., Kneip C., Raji O., Liloglou T., Seegebarth A., Schlegel T., Flemming N., Rausch S., Distler J., Fleischhacker M., Schmidt B., Giles T., Walshaw M., Warburton C., Liebenberg V., Field J.K. Performance evaluation of the DNA methylation biomarker SHOX2 for the aid in diagnosis of lung cancer based on the analysis of bronchial aspirates. Int J Oncol. 2012;40:825–832. doi: 10.3892/ijo.2011.1264. [DOI] [PubMed] [Google Scholar]
- 26.Kristiansen G. Diagnostic and prognostic molecular biomarkers for prostate cancer. Histopathology. 2012;60:125–141. doi: 10.1111/j.1365-2559.2011.04083.x. [DOI] [PubMed] [Google Scholar]
- 27.Costa V.L., Henrique R., Jerónimo C. Epigenetic markers for molecular detection of prostate cancer. Dis Markers. 2007;23:31–41. doi: 10.1155/2007/356742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dietrich D., Lesche R., Tetzner R., Krispin M., Dietrich J., Haedicke W., Schuster M., Kristiansen G. Analysis of DNA methylation of multiple genes in microdissected cells from formalin-fixed and paraffin-embedded tissues. J Histochem Cytochem. 2009;57:477–489. doi: 10.1369/jhc.2009.953026. [DOI] [PMC free article] [PubMed] [Google Scholar]