Abstract
Widespread clinical laboratory implementation of next-generation sequencing–based cancer testing has highlighted the importance and potential benefits of standardizing the interpretation and reporting of molecular results among laboratories. A multidisciplinary working group tasked to assess the current status of next-generation sequencing–based cancer testing and establish standardized consensus classification, annotation, interpretation, and reporting conventions for somatic sequence variants was convened by the Association for Molecular Pathology with liaison representation from the American College of Medical Genetics and Genomics, American Society of Clinical Oncology, and College of American Pathologists. On the basis of the results of professional surveys, literature review, and the Working Group's subject matter expert consensus, a four-tiered system to categorize somatic sequence variations based on their clinical significances is proposed: tier I, variants with strong clinical significance; tier II, variants with potential clinical significance; tier III, variants of unknown clinical significance; and tier IV, variants deemed benign or likely benign. Cancer genomics is a rapidly evolving field; therefore, the clinical significance of any variant in therapy, diagnosis, or prognosis should be reevaluated on an ongoing basis. Reporting of genomic variants should follow standard nomenclature, with testing method and limitations clearly described. Clinical recommendations should be concise and correlate with histological and clinical findings.
The sequencing of human DNA for the human genome project has led to the emergence of technologies that identify genomic, transcriptional, proteomic, and epigenetic alterations in patients' tumors. Precision medicine uses concepts of the genetic and environmental basis of disease to individualize prevention, diagnosis, and treatment and integrates tumor molecular data into decision making in medical practice.1, 2, 3, 4 Genomic information–based disease prognosis and the selective use of targeted therapy to target specific genotypic and biological biomarkers, combined with other therapeutic strategies based on the tumor biology of the individual patient, hold the promise of improving clinical outcomes and patient care.
In recent years, assays for single-target detection have been replaced by next-generation sequencing (NGS) or massively parallel sequencing. This technology allows for the simultaneous evaluation of many genes and the generation of millions of short nucleic acid sequences in parallel.5, 6 The NGS high-throughput platform is more efficient and less expensive and provides information that is not provided by single gene-by-gene Sanger DNA sequencing analysis or by gene-specific targeted hot spot mutation assays.7 The vast number of variants identified by NGS in tumor tissue is attributed to the complexity of carcinogenesis, including the multistep process of genetic mutations and tumor heterogeneity (ie, multiple clones of cells with related but distinct molecular signatures within tumors).8, 9 Herein, tumor refers to tissue deriving from either a benign or malignant neoplasm. NGS results obtained using DNA or RNA extracted from tumor tissue frequently demonstrate a complex molecular signature that is different from that of normal tissue for any given patient. The significance of the change relative to tumorigenesis depends on the type of genetic aberration, the location of the variant, and the normal function of the protein. Genetic variants can be germline or somatic. A germline variant is defined as a genetic alteration that occurs within the germ cells (egg or sperm), such that the alteration can be passed to subsequent generations. A somatic variant is defined as a genetic alteration that occurs in any of the cells of the body, except the germ cells, and therefore is not passed on to subsequent generations. Genetic variations may be activating, resulting in a gain of function of the protein, such as a missense mutation in the functional or kinase domain of the protein, allowing for autophosphorylation of the protein, the loss of regulation for downstream signaling, and uncontrolled cell growth and proliferation. Conversely, the genetic alteration may be inactivating, such as nonsense, splice-site, and frameshift insertion/deletion mutations, thereby causing a loss of function of a tumor-suppressor gene. The types of variants observed include single-nucleotide variants (SNVs) that cause a missense, silent, or nonsense amino acid substitution, or a splice site alteration affecting normal splicing of the mRNA transcript. Alternatively, one or more nucleotides may be involved in duplications, deletions, insertions, or even a more complex pattern with a nucleotide(s) deletion coupled with a nucleotide(s) insertion (indels) at a particular location. Also common in the pathogenesis of cancer is a change in copy number of cancer-related genes. Generically identified as copy number variants (CNVs), these include copy number alterations of various types. Examples of CNVs include the common loss (deletion) of the tumor-suppressor RB1 gene in retinoblastoma or the gain (amplification) of the oncogene ERBB2 in invasive breast carcinoma. In addition, structural rearrangements, including chromosome translocations, deletions, duplication, or inversions, are frequently identified in tumor DNA and result in gene fusions and associated fusion proteins with unique cancer-promoting properties, such as the EML4-ALK recurrent inversion mutation that is seen in non-small cell lung cancer.
Molecular profiles obtained on tumor DNA and RNA can guide the clinical management of cancer patients. This information can provide diagnostic or prognostic information, identify a potential treatment regimen or targeted therapy, and determine eligibility for the following: i) a Food and Drug Administration (FDA)–approved medication for that tumor type, ii) a medication available as off-label treatment for the specific molecular alteration in a nonapproved tumor type, or iii) a targeted therapy available in clinical trials with investigational agents based on an identified molecular alteration. In the United States, the Clinical Laboratory Improvement Amendments of 1988 provide regulatory oversight to laboratories performing tumor genomics characterization (US Government Publishing Office, Electronic Code of Federal Regulations, Title 42, §Part 493.1, http://www.ecfr.gov/cgi-bin/text-idx?SID=1248e3189da5e5f936e55315402bc38b&node=pt42.5.493&rgn=div5#se42.5.493_11, last accessed July 6, 2016). Clinical Laboratory Improvement Amendment certification of laboratories, ongoing quality assurance/improvement, and appropriate proficiency testing are required to ensure accurate and reproducible molecular profiling.
Implementaion of NGS identifies large numbers of genetic variations in tumor DNA, which are crucial for optimal patient care, and treatment guidelines are developed based on specific molecular findings; therefore, it is imperative to unify the interpretation and reporting of molecular results among laboratiores performing these tests.10, 11, 12, 13 In the spring of 2015, a clinical laboratory–focused working group was formed to establish recommendations for the interpretation and reporting of sequence variants identified in tumor tissue analogous to the recently published standards and guidelines for the interpretation of sequence variants in genes associated with mendelian disorders.14 The multidisciplinary working group, convened by the Association for Molecular Pathology (AMP), included investigators with expertise in molecular pathology, medical genetics, and clinical oncology and included liaison representation from the American College of Medical Genetics and Genomics (ACMG), American Society of Clinical Oncology, and College of American Pathologists.
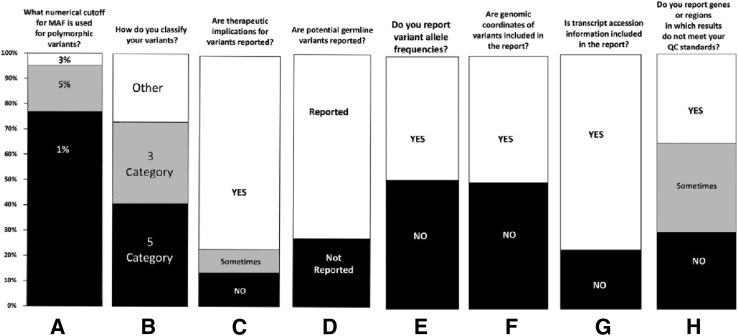
To broadly assess the current practice of NGS testing for tumor tissue, an NGS technical survey and an NGS reporting survey were prepared by the working group and made available online to the AMP membership community for approximately 4 weeks (Figure 1). A representative from each laboratory performing NGS testing was encouraged to participate; 67 responses were received for the technical survey, while 44 participants completed the reporting survey. Participants reported performing NGS testing on both solid tumors and hematological neoplasms, with the number of genes contained within the testing panels ranging from 1 to 10 genes to >100 genes. A minority of respondents reported performing exome (12%) or genome (5%) analysis on tumor tissue. All participants noted the ability to detect SNVs; 95% of participants stated that small indels could be identified by their testing methods, whereas CNVs and gene fusions were interrogated by only 35% and 37% of participating laboratories, respectively.
Figure 1.
Association for Molecular Pathology (AMP) Interpretation of Sequence Variants in Somatic Conditions Working Group technical and reporting survey 1 results. A: Minor allele frequency (MAF). B: Five categories includes pathogenic, likely pathogenic, variant of unknown significance (VUS), likely benign, and benign; three categories includes pathogenic, VUS, and likely benign; other represents participant answers, such as two categories of pathogenic and VUS and no categories. List variants in order of importance for clinical impact, clinical trials, and actionability or predictive of prognosis in that tumor type or in cancer. C: Reported includes therapies approved by Food and Drug Administration (FDA) or contained in professional guidelines, or investigational therapies such as clinical trials for investigational drugs or off-label use of FDA approved drugs. D: Reported includes recommendation of genetic counseling and/or discussion with clinical team providing care to the patient. E: Variant allele frequency (VAF): the proportion (usually shown as %) of variant reads. F: Genomic coordinates of variants: genomic locations of variants denoted by the chromosome number followed by the nucleotide location of the variant (eg, chr17:7674250). G: Transcript accession information includes transcript accession number and version number (eg, NM_000546.5). H: Sometimes includes only if quality control (QC) fails for actionable/targeted variant regions. Survey results obtained anonymously from participating AMP membership regarding reporting of clinical next-generation sequencing results. AMP members are individuals involved in the clinical practice, educational, basic or translational research, economic, and/or regulatory aspects of molecular diagnostics, including but not limited to pathologists, laboratory directors, clinical laboratory scientists, informaticians, technologists, clinicians and other health care personnel, government employees, especially those involved in regulation of the field, and professionals in the in vitro diagnostics industry. Surveys were distributed through AMP's electronic member community [Chat AMP (CHAMP)], and responses were collected between April 7, 2015, and May 1, 2015. There were 44 individual responses for the reporting survey and 67 individual responses for the technical survey. For both surveys, respondents were permitted to select multiple demographic categories describing their occupation. For the technical survey, respondents self-identified as pathologist (39%), laboratory medical director (37%), researcher (15%), bioinformatician (10%), physician (8%), laboratory supervisor (12%), oncologist (3%), and clinical laboratory technologist/technician (3%). For the reporting survey, respondents self-identified as pathologist (50%), laboratory medical director (40%), researcher (13%), bioinformatician (11%), physician (8%), laboratory supervisor (8%), oncologist (3%), and clinical laboratory technologist/technician (3%).
In addition to differences in NGS techniques reported among the participating laboratories, this survey also highlighted significant differences in annotation and reporting of variants (Figure 1). Preliminary findings from this survey and additional public open comment designed to further inform this project were gathered during a workshop held at the 2015 AMP Annual Meeting (Austin, TX). Classification and reporting of variants to health care providers is critical for patient care, including the following: accurate reporting of tumor response to targeted therapy; establishment of national guidelines for patient care; and collaborative institutional clinical trials, thereby supporting the need for standardization among laboratories performing these tests. The goal of these guidelines is to establish standardized classification, annotation, interpretation, and reporting of sequence variants associated with cancer. The guidelines presented herein are based on literature review, empirical data, and the professional judgment of the working group members.
Databases
Genomic Databases
With the publication of an increasing number of large-scale genome sequencing projects for a variety of tumor types, a wealth of genomic information is being generated and consolidated into many public databases globally15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31 (Table 1). For example, the National Cancer Institute's Genome Data Commons contains National Cancer Institute–generated data from some of the largest and most comprehensive cancer genomic data sets, including The Cancer Genome Atlas, Therapeutically Applicable Research to Generate Effective Therapies, and the Cancer Genome Characterization Initiative (https://gdc.cancer.gov, last accessed September 25, 2016). Another public somatic variant database is the Catalog of Somatic Mutations in Cancer (http://cancer.sanger.ac.uk/cosmic, last accessed September 30, 2016), which contains millions of somatic alterations across numerous tumor types. Several other data repositories, such as reference sequence information, population databases, and germline variant databases, that are frequently used in somatic variant analysis are also constantly increasing and improving. The genomic databases provide information that is necessary for accurate annotation and prioritization of somatic variants. As a general rule, clinical laboratories should exercise the following cautionary steps on the use of public databases:
-
1.
Understand the content of the database and how the data are aggregated. The clinical laboratory should review the documentation or published literature relating to a given database to ascertain the source, type, and intent of the database.
-
2.
Pay specific attention to the limitation of each database to avoid overinterpretation of annotation results.
-
3.
Confirm the versions of the human genome assembly as well as mRNA transcript references to ensure appropriate Human Genome Variation Society (HGVS) annotation.
-
4.
Whenever possible, use genomic coordinates, instead of HGVS nomenclature, to unambiguously query genomic databases.
-
5.
Assess the quality of the provided genomic data based on the source, from publications or another database, the number of a specific entry, single or multiple, the depth of the study, the use of appropriate controls, confirmation of a variant's somatic origin, and functional and potential drug response studies.
-
6.
Verify data quality of the pathological diagnosis provided (eg, site, diagnosis, and subtype).
Table 1.
Databases Relevant to Interpretation of Somatic Sequence Variants
| Utility/function | Database | Location (web address) |
|---|---|---|
| Population databases to exclude polymorphisms | 1000 Genomes Project16 | http://browser.1000genomes.org |
| Exome Variant Server | http://evs.gs.washington.edu/EVS | |
| dbSNP17 | http://www.ncbi.nlm.nih.gov/snp | |
| dbVar18 | http://www.ncbi.nlm.nih.gov/dbvar | |
| ExAC | http://exac.broadinstitute.org | |
| Cancer-specific variant databases | Catalog of Somatic Mutations in Cancer19 | http://cancer.sanger.ac.uk/cosmic |
| My Cancer Genome | http://www.mycancergenome.org | |
| Personalized cancer therapy, MD Anderson Cancer Center | https://pct.mdanderson.org | |
| cBioPortal, Memorial Sloan Kettering Cancer Center20 | http://www.cbioportal.org | |
| Intogen21 | https://www.intogen.org/search | |
| ClinicalTrials.gov | https://clinicaltrials.gov | |
| IARC (WHO) TP53 mutation database22 | http://p53.iarc.fr | |
| Pediatric Cancer Genome Project (St. Jude Children's Research Hospital–Washington University) | http://explorepcgp.org | |
| International Cancer Genome Consortium23 | https://dcc.icgc.org | |
| Sequence repositories and data hosts | NCBI Genome | http://www.ncbi.nlm.nih.gov/genome |
| RefSeqGene24 | http://www.ncbi.nlm.nih.gov/refseq/rsg | |
| Locus Reference Genomic25 | http://www.lrg-sequence.org | |
| UCSC table browser26 | https://genome.ucsc.edu/cgi-bin/hgTables | |
| Ensemble BioMart27 | http://useast.ensembl.org/biomart/martview | |
| Other disease/mutation databases useful in the context of variant interpretation for cancer genomics | ClinVar28 | http://www.ncbi.nlm.nih.gov/clinvar |
| Human Gene Mutation Database29 | http://www.hgmd.org | |
| Leiden Open Variation Database30 | http://www.lovd.nl | |
| dbNSFP (compiled database of precomputed in silico prediction scores for nonsynonymous SNVs)31 | https://sites.google.com/site/jpopgen/dbNSFP | |
| Ensemble Variant Effect Predictor15 | http://www.ensembl.org/info/docs/tools/vep/index.html |
These are not comprehensive lists, and inclusion does not represent an organizational endorsement of any individual database or product. All websites last accessed June 7, 2016.
dbSNP, The Database of Short Genetic Variation; ExAC, Exome Aggregation Consortium; IARC, International Agency for Research on Cancer; NCBI, National Center for Biotechnology Information; SNV, single-nucleotide variant; UCSC, University of California, Santa Cruz; WHO, World Health Organization.
Reference Sequence Databases
Reference sequence databases provide information on the version of the human genome assembly and related information, such as genomic coordinates, for unambiguous representation of sequence variants. Additional information, such as accession and version for mRNA transcripts (eg, BRAF NM_004333.4) and exon boundary definition, is critically important for generating correct HGVS nomenclature for variants. Variant location mapping (coding, noncoding, untranslated region, and splice site) and strand representation (positive versus negative) for a gene can be computed from these databases. This also allows for unequivocal representation of the variant in the absence of genomic coordinate information. Some of the commonly used resources include RefSeq (National Center for Biotechnology Information Reference Sequence Database, https://www.ncbi.nlm.nih.gov/refseq, last accessed January 2, 2016), Ensembl (http://www.ensembl.org/index.html, last accessed January 2, 2016), and Locus Reference Genomic (https://www.lrg-sequence.org, last accessed February 2, 2016).
Population Databases
These databases provide comprehensive information about frequencies of alternative (minor) alleles at a given locus in a large cohort of individuals who represent an array of geographically distinct populations. These databases are frequently used to filter out variants that are deemed polymorphic/benign based on an arbitrary cutoff of minor allele frequency (MAF).14, 32 There is no standardized cutoff for MAF to be used for eliminating polymorphic or benign variants. In the absence of paired normal tissue, the work group recommends using 1% (0.01) as a primary cutoff, which is also commonly used across many clinical laboratories. Although the aggregate global MAF is most commonly used, clinical laboratories may consider using ethnicity-specific MAFs based on the ethnic background of the patients. In the context of interpretation of somatic variants, these databases need to be used with caution, because individuals participating in these sequencing studies were assumed to be healthy or free of subclinical diseases at the time of participation in the study. Indeed, several well-known classic cancer-associated and targetable somatic alterations are included as germline variants in population databases. For example, variant NM_004972.3(JAK2):c.1849G>T (c.V617F) is commonly seen as a somatic variant in myeloproliferative neoplasms and can be targeted with FDA-approved Janus kinase (JAK) inhibitors.33 It is also included in multiple population databases, such as The Database of Short Genetic Variation (the National Center for Biotechnology Information database of genetic variation), Exome Variant Server, and Exome Aggregation Consortium (Table 1). Special care should be taken when evaluating possible hematological malignancies because many commonly mutated genes in leukemia and myelodysplastic syndromes may also be somatically mutated in the blood of otherwise healthy individuals and, therefore, may be incorrectly annotated as polymorphisms.34, 35
Cancer-Specific Databases
These databases provide information about the incidence and prevalence of sequence variants across the spectrum of different cancers and subtypes, cross-references to other genomic databases, as well as references to published literature with or without systematic review, cellular pathways, targeted therapies, clinical trials, and outcome data. The prevalence and distribution of sequence variants across different cancers extracted from these databases should be interpreted with caution because of the poor representation of the pathology diagnosis standards, lack of clinical-grade literature curation, and loosely controlled sources of submitted variants (eg, exploratory or discovery studies) in some of these databases. For example, some common germline benign variants are included in these databases, such as NM_000222.2(KIT):c.1621A>C (p.M541L) in the Catalog of Somatic Mutations in Cancer database. Commonly used somatic variant databases are listed in Table 1.
Constitutional Variant Databases
It is common that tumor sequencing with or without matched normal tissues may reveal variants that are of germline origin, such as pathogenic variants in genes associated with cancer predisposition syndromes. Germline mutation databases, such as the Human Gene Mutation Database and other disease- or locus-specific mutation databases, are useful resources for evaluating these variants. These databases can also be used for evaluating somatic variants that have well-studied germline counterparts reported in these databases (eg, certain variants in TP53 and PTEN genes). Another commonly used database is ClinVar (http://www.ncbi.nlm.nih.gov/clinvar, last accessed March 6, 2016). ClinVar addresses all categories of rare germline variants, such as pathogenic and benign, and provides associated clinical and experimental evidence when available. Some of the variants in ClinVar are vetted by expert panels regarding their pathogenicity. At the present time, the database only hosts germline variants and is expected to incorporate somatic variants in the near future.
Internal (Laboratory-Generated) Databases
It is important to emphasize that clinical laboratories should establish a well-annotated in-house database for both tracking variants identified within the laboratory and to provide consistent variant annotations. Such databases are useful to identify potential false-positive calls that may result from sequencing alignment artifacts and to establish the frequency of mutations in cancer types commonly encountered by the laboratory. We strongly encourage somatic variant data sharing and urge clinical laboratories to contribute their well-curated variants to public variant databases to facilitate accurate interpretation of somatic variants. However, such a submission process should be standardized and in compliance with federal privacy regulations, namely, the Health Insurance Portability and Accountability Act and the Health Information Technology for Economic and Clinical Health Act. There are ongoing efforts toward building clinical-grade genomic databases.36
In Silico (Computational) Prediction Algorithms
In silico prediction algorithms are frequently used tools to predict whether a nucleotide change in a gene will change the structure and function of the protein37, 38, 39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 (Table 2). At the onset, the early commonly used algorithms were developed and validated for curating germline variants. Subsequently, their use was extrapolated for interpreting somatic variants. Although the individual algorithms may differ in their core method of risk prediction, they can be binned into two categories: prediction of the impact of a missense variant on protein function and impact of a sequence variant on splicing.
Table 2.
Algorithms for Computational Prediction of Functional Impact of Sequence Variant/Splice Site Changes
| Utility/function | Algorithm/software | Location (web address) |
|---|---|---|
| Missense SNV | PolyPhen237 | http://genetics.bwh.harvard.edu/pph2 |
| SIFT38 | http://sift.jcvi.org | |
| MutationAssessor39 | http://mutationassessor.org | |
| MutationTaster41 | http://www.mutationtaster.org | |
| PROVEAN45 | http://provean.jcvi.org/index.php | |
| Condel46 | http://bg.upf.edu/blog/2012/12/condel-for-prioritization-of-variants-involved-in-hereditary-diseases-and-transfic-for-cancer | |
| CoVEC40 | https://sourceforge.net/projects/covec/files | |
| CADD47 | http://cadd.gs.washington.edu | |
| GERP++48 | http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html | |
| PhyloP and PhastCons49 | http://compgen.bscb.cornell.edu/phast | |
| Splice site prediction | Human Splicing Finder42 | http://www.umd.be/HSF3 |
| MaxEntScan43 | http://genes.mit.edu/burgelab/maxent/Xmaxentscan_scoreseq.html | |
| NetGene244 | http://www.cbs.dtu.dk/services/NetGene2 | |
| NNSplice50 | http://www.fruitfly.org/seq_tools/splice.html | |
| GeneSplicer51 | http://www.cbcb.umd.edu/software/GeneSplicer/gene_spl.shtml |
These are not comprehensive lists, and inclusion does not represent an organizational endorsement of any individual database or product. All websites last accessed June 7, 2016.
SNV, single-nucleotide variant.
The degree of evolutionary conservation of the amino acid or nucleotide residue, the biochemical impact of an amino acid substitution considering the specific physicochemical properties, and the location of the variant in the translated protein are some of the main criteria used by different algorithms to predict functional impact of missense variants. Splice site prediction algorithms use a variety of statistical approaches, such as the Markov model, machine learning (neural networks), and the maximum entropy principle, to predict if a variant will have any impact on splicing.37, 38, 39, 40, 41, 42, 43, 44
In general, missense and splice site prediction tools have a moderate specificity (approximately 60% to 80%) with a tendency of over-predicting deleterious impact.52, 53, 54 The interpretation of these predictions in the context of cancer gene function is usually not straightforward, especially for activating mutations. For example, the canonical BRAF V600E oncogenic mutation is predicted to have conflicting or even benign effect when analyzed across multiple in silico algorithms. This is one of the several scenarios unique to somatic variant interpretation that clinical laboratories should be aware of, and they should exercise caution when interpreting in silico scores. It is recommended that the results of these prediction algorithms should never be used as the sole evidence for variant classification or clinical decision making.
Variant Identification and Annotation
Variant identification is a critical starting point of variant interpretation. There are many variant detection software tools that cater to one specific alteration, such as SNV, indels, structural variants, and CNVs (Supplemental Table S1).55, 56, 57, 58, 59, 60, 61, 62, 63 It is important for clinical laboratories to understand the limitations of these variant detection tools. Appropriate laboratory validation of the bioinformatics pipeline, including commercially purchased bioinformatics packages, is critical to ensure the quality of the results. Certain metrics for called variants are critical for variant interpretation, such as supporting reads (depth of coverage) and variant allele frequency (VAF), and should be included in variant evaluation; the latter is particularly important for somatic variant interpretation in the absence of paired normal and for evaluating tumor clonal diversity.
The results of variant calling are typically represented in one of the several standardized formats, such as the clinical variant call format (VCF), genomic VCF, and general feature format (alias gene-finding format or generic feature format). The VCF is the most widely used schema in the clinical laboratories as of 2016 to represent detected variants (Clinical Variant Call Format, http://vcfclin.org, last accessed September 28, 2016). Required VCF fields include genomic coordinates, reference nucleotide(s), and variant nucleotide(s). However, complex, multinucleotide, and large structural variants are difficult to represent in the current specification of VCF file format version 4.2, despite the ongoing efforts for standardizing variant representation. For further clinical interpretation, additional metadata that add meaningful and readily identifiable information to variants should be included (eg, gene symbol, variant location, variant type, HGVS nomenclature for cDNA sequence changes, and predicted protein sequence alterations). Additional resources, such as cross-references to external databases (cancer-specific and general genomic databases) (Table 1) and precomputed in silico algorithm-based predictions (Table 2), can also be beneficial. This process is formally referred to as variant annotation, and may be automated by software tools (Supplemental Table S2).15, 64, 65, 66, 67
Variant annotation is crucial to accurate interpretation of somatic sequence variants. These multiparametric data of annotated variants form the starting material for variant prioritization and interpretation. One of the challenging aspects of variant annotation is the conversion of genomic coordinates (ie, chromosome and position) to the corresponding cDNA/amino acid coordinate system (c. and p. syntax, respectively) for interpretation. This is specifically problematic for indel variants because of the inherent difference in alignment of variant representation. Although the HGVS system recommends right-aligned (shifting the start position of the variant to the right until it is no longer possible to do so) representation of sequence variants, VCF specifications require left-aligned representation. Currently available annotation solutions only partially address this problem. Lack of standardization of left/right alignment could significantly affect variant localization, leading to incorrect variant nomenclature. It is essential to use correct mRNA transcript accession and version information when multiple alternative transcripts exist to ensure consistent representation of variants, according to HGVS nomenclature, and support accurate interpretation. It is also important that clinical laboratories use the genomic coordinate to store variants in their in-house database to warrant unambiguous references.
Proposed Guideline for Evidence-Based Categorization of Somatic Variants
Somatic variants include SNVs, indels, fusion genes resulting from genomic rearrangements, and CNVs. Unlike interpretation of germline sequence variations, which focuses on pathogenicity of a variant for a specific disease or disease causality, interpretation of somatic variants should be focused on their impact on clinical care. A variant can be considered a biomarker that affects clinical care if it predicts sensitivity, resistance, or toxicity to a specific therapy, alters the function of the gene, which can be targeted by approved or investigational drugs, serves as an inclusion criterion for clinical trials, influences disease prognosis, assists in establishing a diagnosis of a cancer, or warrants implementing surveillance measures for early cancer detection. Clinical impacts should, therefore, include therapeutic, prognostic, diagnostic, and preventive actions. The clinical impact of a given variant should be determined according to currently available evidence. Evidence used for variant categorization can be weighed differently based on its significance in clinical decision making. On the basis of literature review and Working Group consensus, we propose to group clinical and experimental evidence into four levels (Table 3):
-
1.
Level A, biomarkers that predict response or resistance to US FDA-approved therapies for a specific type of tumor or have been included in professional guidelines as therapeutic, diagnostic, and/or prognostic biomarkers for specific types of tumors;
-
2.
Level B, biomarkers that predict response or resistance to a therapy based on well-powered studies with consensus from experts in the field, or have diagnostic and/or prognostic significance of certain diseases based on well-powered studies with expert consensus;
-
3.
Level C, biomarkers that predict response or resistance to therapies approved by FDA or professional societies for a different tumor type (ie, off-label use of a drug), serve as inclusion criteria for clinical trials, or have diagnostic and/or prognostic significance based on the results of multiple small studies;
-
4.
Level D, biomarkers that show plausible therapeutic significance based on preclinical studies, or may assist disease diagnosis and/or prognosis themselves or along with other biomarkers based on small studies or multiple case reports with no consensus.
Table 3.
Categories of Clinical and/or Experimental Evidence
| Category | Therapeutic | Diagnosis | Prognosis |
|---|---|---|---|
| Level A | 1. Biomarkers that predict response or resistance to FDA-approved therapies for a specific type of tumor 2. Biomarkers included in professional guidelines that predict response or resistance to therapies for a specific type of tumor |
Biomarkers included in professional guidelines as diagnostic for a specific type of tumor | Biomarkers included in professional guidelines as prognostic for a specific type of tumor |
| Level B | Biomarkers that predict response or resistance to therapies for a specific type of tumor based on well-powered studies with consensus from experts in the field | Biomarkers of diagnostic significance for a specific type of tumor based on well-powered studies with consensus from experts in the field | Biomarkers of prognostic significance for a specific type of tumor based on well-powered studies with consensus from experts in the field |
| Level C | 1. Biomarkers that predict response or resistance to therapies approved by the FDA or professional societies for a different type of tumor 2. Biomarkers that serve as inclusion criteria for clinical trials |
Biomarkers of diagnostic significance based on the results of multiple small studies | Biomarkers of prognostic significance based on the results of multiple small studies |
| Level D | Biomarkers that show plausible therapeutic significance based on preclinical studies | Biomarkers that may assist disease diagnosis themselves or along with other biomarkers based on small studies or a few case reports | Biomarkers that may assist disease prognosis themselves or along with other biomarkers based on small studies or a few case reports |
FDA, Food and Drug Administration.
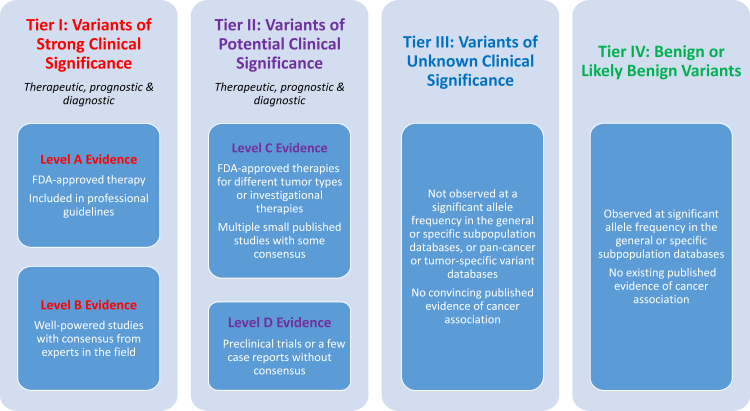
These levels of evidence can be assigned to genomic variants to determine the significance of their clinical impact. We propose to categorize sequence variants in somatic conditions into four categories based on their clinical impact: tier I, variants with strong clinical significance (level A and B evidence); tier II, variants with potential clinical significance (level C or D evidence); tier III, variants with unknown clinical significance; and tier IV, variants that are benign or likely benign (Figure 2).
Figure 2.
Evidence-based variant categorization. Somatic variants are classified into four tiers based on their level of clinical significance in cancer diagnosis, prognosis, and/or therapeutics. Variants in tier I are of strongest clinical significance, and variants in tier IV are benign or likely benign variants. FDA, Food and Drug Administration.
Determination of the pathogenicity or clinical impact of variants in cancer remains a monumental task. Genomic alterations can have a spectrum of clinical utility, including diagnosis, prognosis, therapy selection, and monitoring of therapy, although strong evidence linking genomic alterations to FDA-approved cancer therapies exists only for a few cancers.68 Peer-reviewed literature, clinical practice guidelines, and large-scale cancer mutation databases remain primary resources for evidence needed to effectively assess clinical significance of a particular variant. It is within the scope of the molecular professional's medical practice to use his or her considered judgment regarding the evidence derived from these sources (Table 4, Table 5, Table 6, Table 7).69
Table 4.
Tier I: Variants with Strong Clinical Significance
| Evidence source/type | Available evidence |
|---|---|
| FDA-approved therapies, PG, investigational therapies | Therapeutic: FDA approved or investigational with strong evidence∗ Diagnostic: In PG or reported evidence with consensus Prognostic: In PG or reported evidence with consensus |
| Mutation type | Activating, LOF (missense, nonsense, indel, splicing), CNVs, fusions |
| Variant frequencies | Mostly mosaic |
| Potential germline† | Mostly nonmosaic (VAF approximately 50% or 100%) |
| Population database: ESP, dbSNP, 1000Genome, ExAC | Absent or extremely low MAF |
| Germline database: HGMD, ClinVar | May or may not be present |
| Somatic database: COSMIC, My Cancer Genome, TCGA | Most likely present |
| Predictive software: SIFT, PolyPhen2, MutTaster, CADD | Mostly damaging; information to be used for reference only |
| Pathway involvement | Disease-associated pathways |
| Publications: functional study, population study, other | Therapeutic: reported evidence with consensus Diagnostic: reported evidence with consensus Prognostic: reported evidence with consensus |
Italicized text indicates examples provided within each category; these are not comprehensive lists, and inclusion does not represent an organizational endorsement of any individual database or product.
CNV, copy number variation; COSMIC, Catalog of Somatic Mutations in Cancer; dbSNP, The Database of Short Genetic Variation; ExAC, Exome Aggregation Consortium; FDA, Food and Drug Administration; HGMD, Human Gene Mutation Database; indel, insertion and deletion; LOF, loss of function; MAF, minor allele frequency; PG, professional guideline; TCGA, The Cancer Genome Atlas; VAF, variant allele frequency.
Strong evidence based on well-powered clinical studies with consensus from experts in the field.
Confirmation on normal tissue if tested tumor only and genetic counseling should be recommended.
Table 5.
Tier II: Variants with Potential Clinical Significance
| Evidence source/type | Available evidence |
|---|---|
| FDA-approved therapies, PG, investigational therapies | Therapeutic: FDA approved for different tumor type; investigational therapies with some evidence Diagnostic: not in PG but with convincing published data Prognostic: not in PG but with convincing published data |
| Mutation type | Activating, LOF (missense, nonsense, indel, splicing), CNVs, fusions |
| Variant frequencies | Mostly mosaic |
| Potential germline∗ | Mostly nonmosaic (VAF approximately 50% or 100%) |
| Population database: ESP, dbSNP, 1000Genome, ExAC | Absent or extremely low MAF |
| Germline database: HGMD, ClinVar | May or may not be present |
| Somatic database: COSMIC, My Cancer Genome, TCGA | Likely present |
| Predictive software: SIFT, PolyPhen2, MutTaster, CADD | Mostly damaging; information to be used for reference only |
| Pathway involvement | Involve disease-associated pathways or pathogenic pathways |
| Publications: functional study, population study, other | Therapeutic: evidence of using FDA-approved therapies for different tumor types; phase 2 or 3 clinical trials for investigational therapies Diagnostic: multiple lines of reported evidence without consensus Prognostic: multiple lines of reported evidence without consensus |
Italicized text indicates examples provided within each category; these are not comprehensive lists, and inclusion does not represent an organizational endorsement of any individual database or product.
CNV, copy number variation; COSMIC, Catalog of Somatic Mutations in Cancer; dbSNP, The Database of Short Genetic Variation; ExAC, Exome Aggregation Consortium; FDA, Food and Drug Administration; HGMD, Human Gene Mutation Database; indel, insertion and deletion; LOF, loss of function; MAF, minor allele frequency; PG, professional guideline; TCGA, The Cancer Genome Atlas; VAF, variant allele frequency.
Confirmation on normal tissue if tested tumor only and genetic counseling should be recommended.
Table 6.
Tier III: Variants of Unknown Clinical Significance
| Evidence source/type | Available evidence |
|---|---|
| FDA-approved therapies, PG, investigational therapies | Cancer genes: none Noncancer genes (apply to cancer exome/whole genome sequencing): none |
| Mutation type | Functionally unknown; mostly missense, in-frame indels; less commonly, other types |
| Variant frequencies | Mosaic or nonmosaic |
| Potential germline∗ | Mostly nonmosaic (VAF approximately 50% or 100%) |
| Population database: ESP, dbSNP, 1000Genome, ExAC | Absent or extremely low MAF |
| Germline database: HGMD, ClinVar | Absent or downgraded from pathogenic to VUS |
| Somatic database: COSMIC, My Cancer Genome, TCGA | Absent or present without association to specific tumors (potential germline VUS); present but in very few cases |
| Predictive software: SIFT, PolyPhen2, MutTaster, CADD | Variable; information to be used for reference only |
| Pathway involvement | May or may not involve disease-associated pathways |
| Publications: functional study, population study, other | None or no convincing evidence to determine clinical/biological significance |
Italicized text indicates examples provided within each category; these are not comprehensive lists, and inclusion does not represent an organizational endorsement of any individual database or product.
COSMIC, Catalog of Somatic Mutations in Cancer; dbSNP, The Database of Short Genetic Variation; ExAC, Exome Aggregation Consortium; FDA, Food and Drug Administration; HGMD, Human Gene Mutation Database; indel, insertion and deletion; MAF, minor allele frequency; PG, professional guideline; TCGA, The Cancer Genome Atlas; VAF, variant allele frequency; VUS, variant of unknown clinical significance.
Confirmation on normal tissue if tested tumor only and genetic counseling should be recommended.
Table 7.
Tier IV: Benign/Likely Benign Variants
| Evidence source/type | Available evidence |
|---|---|
| FDA-approved therapies, PG, investigational therapies | None |
| Mutation type | Functionally benign or unknown; mostly missense; less commonly, other types |
| Variant frequencies | Mostly nonmosaic (VAF, approximately 50% or 100%) |
| Potential germline∗ | Mostly nonmosaic (VAF, approximately 50% or 100%) |
| Population database: ESP, dbSNP, 1000Genome, ExAC | MAF ≥ 1% in the general population; or high MAF in some ethnic populations |
| Germline database: HGMD, ClinVar | Absent or present but downgraded to benign/likely benign |
| Somatic database: COSMIC, My Cancer Genome, TCGA | Absent or present without association to specific tumors (potential rare germline polymorphism) |
| Predictive software: SIFT, PolyPhen2, MutTaster, CADD | Mostly benign; information to be used for reference only |
| Pathway involvement | May or may not involve disease-associated pathways |
| Publications: functional study, population study, other | Reported evidence supportive of benign/likely benign; or none |
Italicized text indicates examples provided within each category; these are not comprehensive lists, and inclusion does not represent an organizational endorsement of any individual database or product.
COSMIC, Catalog of Somatic Mutations in Cancer; dbSNP, The Database of Short Genetic Variation; ExAC, Exome Aggregation Consortium; FDA, Food and Drug Administration; HGMD, Human Gene Mutation Database; MAF, minor allele frequency; PG, professional guideline; TCGA, The Cancer Genome Atlas; VAF, variant allele frequency.
Confirmation on normal tissue if tested tumor only and genetic counseling should be recommended.
Tier I Variants: Variants with Strong Clinical Significance (Level A and B Evidence)
Variants with Level A Therapeutic Significance
These variants predict response or resistance to therapies approved by the FDA or included in professional guidelines for specific types of tumors (Table 4). For example, BRAFV600E predicts response to the FDA-approved drug vemurafenib in melanoma (FDA Table of Pharmacogenomic Biomarkers in Drug Labeling, http://www.fda.gov/drugs/scienceresearch/researchareas/pharmacogenetics/ucm083378.htm, last accessed July 2, 2016); KRAS mutations predict resistance to anti–epidermal growth factor receptor monoclonal antibodies in colorectal cancer (National Comprehensive Cancer Network Guidelines—Colon, http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf, last accessed March 6, 2016, registration required).70
Variants with Level A Diagnostic/Prognostic Significance
These variants are included in professional guidelines as diagnostic and/or prognostic biomarkers for specific types of tumors. For instance, PML-RARA fusion is pathognomonic for promyelocytic leukemia, and FLT3 internal tandem duplication variant is associated with poor prognosis in acute myeloid leukemia (National Comprehensive Cancer Network Guidelines in Oncology Acute Myeloid Leukemia, Version 2, 2016, https://www.nccn.org/professionals/physician_gls/PDF/aml.pdf, last accessed July 14, 2016, registration required).71
A biomarker may have significant impact in clinical decision making in multiple areas. PML-RARA fusion is diagnostic for promyelocytic leukemia, and it is also associated with a good prognosis and predicts sensitivity to all-trans retinoic acid or arsenic (National Comprehensive Cancer Network Guidelines—Acute Myelogenous Leukemia, http://www.nccn.org/professionals/physician_gls/pdf/aml.pdf, last accessed July 6, 2016, registration required).72
Variants with Level B Therapeutic Significance
These are biomarkers that predict response or resistance to a therapy based on well-powered studies with expert consensus or smaller studies that are repeatedly confirmed or reproduced by different groups. For example, BRAFV600E, seen in >90% of patients with hairy-cell leukemia, constitutively activates the mitogen-activated protein kinase pathway of hematopoietic stem cells.73, 74, 75 Two recently published multicenter phase 2 clinical trials, in addition to multiple smaller studies, demonstrate that BRAF inhibitor vemurafenib is highly effective (96% response rate) in patients with relapsed or refractory hairy-cell leukemia.76 Another example, multiple recent studies demonstrate strong evidence that mutations of RAS genes or amplification of mutated BRAF gene reactivates the mitogen-activated protein kinase pathway, resulting in acquired resistance to BRAF inhibitor therapy in melanoma.77, 78, 79, 80, 81 It is foreseeable that certain level B evidence may lead to new FDA-approved therapeutic application and/or be adopted into professional guidelines in the near future and become level A evidence.
Variants with Level B Diagnostic/Prognostic Significance
These are diagnostic and/or prognostic biomarkers based on well-powered studies with expert consensus or smaller studies that are repeatedly confirmed or reproduced by different groups. Examples include activating KIT mutations (typically D816V) that are present in virtually all adults (93%) with indolent and aggressive forms of systemic mastocytosis82, 83, 84; a BRAF mutation (typically V600E) that is used as a diagnostic and prognostic marker for papillary thyroid carcinoma in thyroid fine-needle aspiration samples85, 86, 87; KIAA1549-BRAF fusion, which is diagnostic for pilocytic astrocytoma and is associated with a better clinical outcome88, 89; and EWSR1 fusions, mostly EWSR1-FLI1, seen in nearly 100% of the Ewings family of tumors, which have greatly enhanced the ability to differentiate Ewings family of tumors from other small blue round cell tumors.90, 91
Germline Pathogenic Variants Associated with Cancer Predisposition
We recommend reporting germline variants with known evidence of clinical impact. An example of germline variants with therapeutic impact is pathogenic germline variants of BRCA1/2. A multicenter clinical trial showed responses of multiple advanced tumors, including ovarian, breast, pancreatic, and prostate cancers, to poly(adenosine diphosphate-ribose) polymerase inhibitors, which led to the FDA approval of olaparib for treatment of women with BRCA-related advanced ovarian cancer (FDA Olaparib, http://www.fda.gov/drugs/informationondrugs/approveddrugs/ucm427598.htm, last accessed July 6, 2016).92 Germline pathogenic variants leading to the diagnosis of a hereditary cancer syndrome that has an established guideline for clinical surveillance should also be reported with recommendation of cancer genetic counseling. For example, certain pathogenic germline TP53 variants lead to Li-Fraumeni syndrome, a hereditary cancer syndrome with increased risk for a wide spectrum of TP53-associated malignancies, including, but not limited to, breast cancer, sarcomas, brain tumors, and leukemias.93, 94 Management guidelines for TP53 mutation carriers have been established (eviQ Cancer Treatments Online Risk management for Li-Fraumeni syndrome, http://www.eviq.org.au, last accessed July 5, 2016; National Comprehensive Cancer Network Guidelines—Genetic Screening, http://www.nccn.org/professionals/physician_gls/pdf/genetics_screening.pdf, last accessed July 6, 2016, registration required). In the absence of paired normal sequencing data to differentiate somatic variants from constitutional/inherited variants, clinical judgment should be used when reporting potential germline pathogenic variants identified in cancer tissue because many germline pathogenic variants (including those in TP53, PTEN, and BRCA1/2) may also occur as acquired somatic variants in cancer. For definitive classification of germline status, sequencing of nontumor tissue should be performed. Germline variants may also serve as inclusion criteria for clinical trials. For example, multiple clinical studies are currently recruiting children and adults who are carriers of TP53 pathogenic variants to evaluate the clinical significance of implementing more comprehensive surveillance protocols and to investigate the psychological impact of undergoing comprehensive surveillance.93 See the section discussing germline variants identified during cancer testing below.
Tier II Variants: Variants with Potential Clinical Significance (Level C and D Evidence)
Variants with Level C Therapeutic Significance
These are FDA-approved therapies or therapies included in professional guidelines for a different tumor type, or investigative therapies with some clinical evidence (Table 5). As an example, the JAK inhibitor ruxolitinib is an FDA-approved drug for the treatment of myelofibrosis based on the results of clinical trials that showed significant benefits of reducing spleen size and relieving myelofibrosis-related symptoms, and improving overall survival (FDA Ruxolitinib, http://www.fda.gov/AboutFDA/CentersOffices/OfficeofMedicalProductsandTobacco/CDER/ucm280155.htm, last accessed July 6, 2016).95, 96 Recent studies have shown in vivo efficacy of ruxolitinib in JAK-activated Philadelphia chromosome–like acute lymphoblastic leukemia.97 Clinical trials are being developed to test JAK inhibitor therapy in children with acute lymphoblastic leukemia and JAK2 mutations.98 Another group of variants of level C evidence are variants that qualify patients for clinical trials for investigative targeted therapies. For instance, patients with FLT3 ITD mutated acute myeloid leukemia may be included in phase 2/3 studies for FLT3 inhibitors (ClinicalTrials.gov, https://clinicaltrials.gov/ct2/results?term=flt3+INHIBITOR&Search=Search, last accessed May 16, 2016).99
Variants with Level C Diagnostic/Prognostic Significance
These are biomarkers of diagnostic and/or prognostic significance based on the results of multiple small studies. Variants in this category include those that are diagnostic/prognostic for a group of related cancers or variants that are supportive of a diagnosis along with other genomic variants. For instance, somatic variants in genes that encode spliceosome proteins have been recently identified to be associated with certain myeloid malignancies, including myelodysplastic syndromes, acute myeloid leukemia, and myeloproliferative neoplasms.9, 100, 101 Multiple studies have shown that somatic mutations in the SF3B1 gene are associated with a good impact on overall survival and disease progression to acute myeloid leukemia in patients with myelodysplastic syndromes.100, 102
Variants with Level D Therapeutic Significance
These are biomarkers that have been associated with targeted therapies in preclinical trials. TP53 remains the most commonly mutated gene in human cancers. For this reason, restoring normal tumor-suppressor functions of p53 protein has been the center of pharmaceutical research targeting mutated p53. Nutlins, a group of cis-imidazoline small-molecule compounds, have shown selective inhibition of the MDM2:p53 interaction in preclinical studies.103, 104 Nutlins have also been shown in human xenograft models to inhibit wild-type p53 tumor growth in a dose-dependent manner and even exhibited regression in some instances. Phase 1 clinical trials have shown that nutlin RG7112 appeared to be well tolerated and displayed initial evidence of clinical activity in advanced solid tumors, hematological malignancies, and liposarcomas (ClinicalTrials.gov, https://clinicaltrials.gov/ct2/results?term=NCT01164033&Search=Search, last accessed May 6, 2016).105, 106, 107, 108
It is important to recognize that molecular genetics in cancer is a rapidly evolving field; therefore, the level of evidence in therapy, diagnosis, and prognosis should be continuously evaluated based on evolving research data and modified accordingly. As mentioned earlier, level B evidence may become level A evidence if a well-powered clinical trial leads to FDA approval of an investigative therapy. On the other hand, new research may prove that a level C potential prognostic biomarker is of no prognostic significance. Because cancers often contain multiple somatic variants, interpretation of the clinical impact of a somatic variant should consider the effects of other coexisting mutations and the clonal relationship among different variants whenever possible.
Tier III Variants: Variants of Unknown Significance
Variants in this category may include somatic variants in cancer genes reported in the same or different cancer types with unknown clinical significance and variants in cancer genes that have not been reported in any cancers (Table 6). These variants should not have been observed at significant allele frequencies in the general population, such as in the 1000 Genomes Project database, Exome Variant Server, or Exome Aggregation Consortium database. The mutation type of the variants (eg, nonsense versus missense or frameshift versus in-frame deletion or duplication) and the main function of the gene (eg, tumor suppressors versus oncogenes) should be considered when evaluating this group of variants. Multiple in silico algorithms can be used to predict the structure and function of the mutant protein, but the predicted results should be used for reference only (ie, not be used as the only parameter to determine potential clinical significance of a variant). When whole genome or whole exome sequencing is performed on tumor-normal paired samples, somatic variants in noncancer genes may be revealed. These variants should be annotated based on their mutation type, whether present in somatic mutation or normal population databases and their MAFs, in silico algorithms prediction results, and literature search. Most of the noncancer gene somatic variants fall into the category of variant of unknown significance.
Tier IV Variants: Benign or Likely Benign
Variants in this category are most likely rare germline variants (Table 7). They often show VAFs of approximately 50% or 100%, with rare exceptions, and have been observed at significant allele frequencies in the general population or a specific subpopulation. Categorization and interpretation of tier IV variants may refer to recently published ACMG/AMP standards and guidelines for the interpretation of germline sequence variants.14
Germline Variants Identified during Cancer Testing
During the clinical laboratory study of somatic cancer-causing mutations, it is important to distinguish acquired somatic variants from inherited germline variants. Most germline variants are inherited variants that are passed down through generations and have the variant typically in 100% of cells, leading to allelic fractions of 0.5 or 1.0. Somatic variants are acquired after birth and typically result from errors in DNA replication or repair or from environmental insults. Allelic fractions for somatic variants are usually <0.5 because of the presence of contaminating normal tissue, even in apparently pure tumor samples. Laboratories must correctly identify somatic mutations that may be related to diagnosis, prognosis, therapeutic intervention, and/or clinical trial selection, and not misidentify high-frequency somatic mutations as germline, because this could have significant clinical implications. Likewise, laboratories must recognize germline variants that potentially lead to a cancer predisposition syndrome that would have health care implications for the patient and other family members.
To help with classifying variants, some laboratories sequence a normal, matched control DNA sample in parallel testing with the tumor DNA. When a normal, matched control tissue is sequenced along with the tumor, germline variants are typically evident. In this case, the laboratory must have policies that address detection, disclosure/nondisclosure, and interpretation/reporting of germline variants (see section below). When a matched control is not included in the analysis, the laboratory should have criteria that can be used to infer that a variant is somatic or germline. The main criterion for germline designation is the VAF, which should be approximately 50% for a heterozygous variant and 100% for a homozygous variant. Certain germline variants, such as large indels, may lead to preferential amplification (in amplicon-based tests) or capture (in capture-based tests) of normal alleles because of the loss of sequence homology of the variant alleles, resulting in <50% VAF for germline variants. When an apparent germline variant is detected in a known cancer predisposition syndrome gene (eg, TP53 or BRCA1), clinical information on the age of disease onset (young age is associated with higher risk for an inherited germline mutation in a cancer-causing gene), laterality of tumor (bilateral tumors are more likely to be inherited), and family or personal history of cancer can assist in determining the likelihood of a cancer predisposition. Literature review and database queries can also help to determine whether the variant has been reported previously as a recurrent germline variant in patients with a predisposition syndrome. Useful databases for constitutional mutations include the following: Online Mendelian Inheritance in Man (National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/omim, last accessed March 6, 2016), Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php, last accessed March 6, 2016), ClinVar, and locus-specific databases, as described earlier. Interpretation of germline variants should follow ACMG/AMP standards and guidelines for the interpretation of germline sequence variants.14 When a pathogenic germline variant is suspected during tumor-only testing, confirmation of the variant with a normal tissue sample, along with appropriate genetic counseling, should be recommended.
Laboratories should have a policy about testing a germline sample for a genetic variant found in a malignancy to confirm the germline or somatic origin of the variant using a clinically validated germline test after appropriate patient consent is received or per request of a clinician.107, 109 For secondary findings revealed in germline testing, the ACMG recommends disclosure of positive germline results for 53 genes, approximately half of which are associated with cancer predisposition susceptibility genes that will likely be on somatic testing panels.108 Disclosure is recommended even when the germline is only being evaluated as part of a tumor/normal study. By inference, it would seem prudent to also consider the likelihood of germline pathogenic variants in a tumor-only somatic mutation study.110
Interpretation and Reporting
The report is an essential part of any laboratory test and should contain all the information required for the ordering physician and the patient to know what exactly was tested, what results were obtained from the test, and any additional preanalytic, analytic, or postanalytic factors that may influence the clinical interpretation of the results. In this context, what a test does not find (ie, pertinent negatives or suboptimal signal) may be as important, if not more so, than what a test does find. An incomplete or unclear representation of the data can lead to clinical errors and incorrect patient management. Detected variants should be carefully reviewed by appropriately trained and certified molecular diagnostic professionals in the context of each complete case, including histologic findings, and evidence-based variant categorization must be performed before reporting.69, 111 Large panels may need to convey large amounts of information, including technical elements about assay design that are regulatory requirements but may not be of immediate use to all patients and clinical providers; nevertheless, reports should be short, simple, and to the point. All clinically critical information should be at the beginning of the report and formatted in a prominent manner to increase the likelihood that it is seen and understood, with the understanding that data that are on page 2 or beyond stand a high likelihood of being missed by the treating physician. It is desirable to include graphs, charts, and tables to increase the overall clarity of the report, provided that they can be integrated into the medical record.
Tier-Based Reporting
All detected genetic alterations should be classified into a 4-tiered system: tier I, variants of strong clinical significance (Table 4); tier II, variants of potential clinical significance (Table 5); tier III, variants of unknown significance (Table 6); and tier IV, variants of known insignificance (ie, likely benign or benign) (Table 7). Tiers I to III must be reported in descending order of clinical importance. It is NOT recommended to include tier IV or benign/likely benign variants/alterations in the report (see criteria for classification above).
Nomenclature
All detected genetic alterations should be annotated and reported as designated by the HUGO Gene Nomenclature Committee (http://www.genenames.org, last accessed March 6, 2016; HGVS, http://www.hgvs.org, last accessed March 6, 2016). SNVs and indels should be reported using p. and c. notation (eg, BRAF p.V600E, c.1799T>A). Gene fusions should be reported listing both fused gene partners separated by a slash (eg, EML4/ALK fusion).112, 113 CNVs generated from NGS tests should be reported in table format as copy number GAIN or LOSS. Genomic coordinates of gene/genomic locus can be included if applicable. Reporting of numerical copy number changes can be performed when appropriate [eg, EGFR copy number GAIN (copy number ratio 25); CDKN2A copy number LOSS].
The use of standard nomenclature does not outweigh the need for clear and unambiguous communication to the clinical team. Therefore, colloquial nomenclature should be included in addition to the standard nomenclature, as needed, to convey meaning with clarity to the physicians reading the reports and using them to determine treatment. For example, reporting of hTERT promoter variants can be c.1-124C>T (HGVS nomenclature), followed by the colloquial nomenclature in parentheses (TERT C228T). Reporting of genomic variants using HGVS nomenclature allows for unambiguous remapping of the variant to the reference genome, and colloquial nomenclature delivers a clear message to the clinical team.
Other Reporting Elements
In addition to the detected variants, the report should also contain several other elements that may be relevant for more thorough analysis of the results or for comparison with other results obtained from this patient over time, such as the genomic coordinates, the genome build, and the transcript reference sequence (eg, NM_004333.4), provided that this information does not detract from the ability of a patient and clinical provider to interpret the immediately relevant essential elements of the report.112 Along these lines, it may be advisable to include this information in a table format toward the end of the section or in another section with extended description of results, away from the topline results. Allele fraction (VAF) and coverage should be evaluated and included in the report when appropriate (Table 4, Table 5, Table 6, Table 7). The report should include the sequencing coverage cutoff for the NGS assay used. All genes and/or hot spots not meeting the minimal required sequencing coverage criteria should be declared in the report as having failed.
Reports should not be limited to positive findings. Pertinent negatives should be reported, in a disease-specific manner. Pertinent negatives should be included for tier I drug/cancer combinations (eg, the definitive lack of an EGFR mutation in a patient with lung cancer or the definitive lack of a BRAF mutation in a patient with melanoma). Uncertainty, if present, must be communicated in reports; this includes issues of sequence quality, sample adequacy, tumor content, and biomedical knowledge.
Reporting of Germline Variants
Concurrent analysis of a paired germline sample is desirable because it clarifies interpretation.114 However, it is not always practical and should not be required. When a paired germline sample is available, sequencing pipelines may allow separating germline findings from somatic acquired variants. Frequently, only somatic variants are interpreted and reported. If a patient or clinician requests additional analysis for germline findings, germline NGS data can be rereviewed at a later time and reported after appropriate patient consent is received. Consent, and documentation thereof, may be required for paired germline testing, in accordance with local laws and policies. If germline variants are not reported in some of the genes in an NGS panel, the initial report should specifically state that fact.
When paired germline samples are not used, NGS analysis does not distinguish germline and somatic variants, and sequencing results may contain both findings. In this case, findings can be reported with a disclaimer that the NGS test used does not allow definitive differentiation between germline and somatic variants. In certain settings, a germline variant may be suspected (eg, MAF 40% to 60%). However, this interpretation should be made with caution and correlated with tumor cellularity. If a germline variant is suspected, testing of a patient germline sample (eg, blood in patients with solid tumors) can be suggested if clinically indicated after an appropriate patient consent. The reports should include a statement addressing the manner in which the distinction between somatic and germline alterations is made, and indications of remaining uncertainty, where appropriate.
Laboratories that test cancer samples first and paired germline samples at a later date may choose to do the following: i) issue an addendum to the report of the cancer incorporating the findings from the germline sample, ii) issue a separate report of the germline variant with a unifying interpretation statement in the germline report and in an addendum to the initial cancer report, or iii) issue a separate germline report and an additional separate report that integrates the results from the cancer and germline samples. Germline variants should be reported as per ACMG/AMP guidelines.14
If germline testing is ordered for cancer predisposition genes, reporting of germline variants should follow the ACMG/AMP guidelines.14 Genetic counseling and referral to a clinical medical geneticist should be offered. Laboratories should have policies regarding reporting of variants of unknown significance and disclosure of secondary findings, including under what circumstances such findings will or will not be reported.107
Reporting the Clinical Significance of Detected Variants
It is useful to provide an interpretive comment on detected genetic alterations that puts the alteration in clinicopathologic context to inform management decisions. This is essential for mutations in tiers I and II. Detailed analyses of tier III variants must be balanced against the goal of keeping the most critical information in the reports concise, clear, and prominently presented. The comments may include functional, prognostic, or predictive significance of the variant for particular tumor type, impact on biochemical pathway(s), and prevalence in relevant cancers.
Recommendations should be made wherever possible and defensible based on evidence, with appropriate literature citations. However, recommendations should be short and worded carefully, with the understanding that treatment or other patient management decisions are based on many pieces of medical information beyond genetic alterations, many of which are not available to the molecular professional issuing the report. It is important to recognize that suitability for a treatment is based on many factors other than the diagnosis as written on a test requisition and the genotype discovered through testing. Often, these factors are unknown to the molecular professional reporting results (ie, presence of confounding medical conditions, such as glucose intolerance, autoimmune disease, or heart failure), and failure to take these other factors into consideration when recommending a specific therapy can lead to confusion, conflict between patient and oncology team, and anxiety. Treatment suggestions within the NGS laboratory report should be evidence based, should be relevant to the patient's cancer diagnosis, and should contain some kind of language to make it clear that the report contains generalized treatment suggestions incorporating the data points available to the laboratory (ie, diagnosis and genotype), but that additional factors need to be incorporated into crafting a treatment plan for each individual. Recommendations for specific clinical trials should not be made, although general statements about availability of relevant trials or citing results of published trials are acceptable.
Updating Knowledge
Reports should be static, and the date of issue should be clearly presented; they do not need to be automatically recalled and/or reissued when medical knowledge changes. That said, medical knowledge does change rapidly, and laboratories should anticipate being asked to reinterpret previous test results in the light of new knowledge. Consideration should be given to developing a process for updating reports when specifically requested to do so.
Reporting Method
Methodologic details should be presented at the bottom of the reports and should include description of method, assay performance characteristics (especially limit of detection and minimal depth of sequencing coverage), and critical quality metrics for the assay run. The report should include the details of what was actually tested. It is not sufficient for a report to simply list gene names unless the entirety of each of these genes was sequenced or an assay would detect all reported pathogenic mutations in the listed gene. The specific gene loci, exons, or hot spots tested should be listed in the final report. As gene panels become larger, including all of this information into a report can become onerous. Laboratories could post additional information on a website that is available to all of their clients. However, a true stand-alone report is highly preferable.
Integration with Electronic Health Records
Ideally, the report should be in a format that enables integration with an electronic health record. Laboratories, in conjunction with clinical information technology teams, should take Health Level Seven International (http://www.hl7.org/implement/standards, last accessed March 6, 2016) constraints into consideration during technical design of the report format. An aesthetically beautiful report that must be scanned (eg, a printed or PDF file) into a patient's chart is, in the long-term, less valuable for that patient than a report that can be integrated into the structured environment of an electronic health record. Web-based reporting under appropriately secured conditions is an enhanced option, but it is the Working Group's recommendation that it should not be a substitute for a report in the medical record.
Access to germline genetic test results is sometimes restricted to a subset of each patient's health care providers by electronic health record systems; this is not appropriate for somatic genetic test results. Somatic test results should be just as available to privileged health care workers as are complete blood cell counts, chest X-rays, or lymph node biopsy specimens. However, the results of tests performed under research protocols, even when performed in a Clinical Laboratory Improvement Amendment–certified laboratory under clinical-grade protocols, should not be reported into the medical record without specified approval from the responsible institutional review board.
Conclusion
The increasing use of NGS technologies in cancer genomic profiling has raised new challenges for clinical laboratories. One of the key tasks for molecular professionals is to standardize the interpretation and reporting of cancer-associated sequence variants detected during cancer sequencing. The proposed Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer represents the expert consensus opinion of the working group members with input from the stakeholders they represent. The recommendation describes a system for evidence-based variant categorization and the process of variant annotation, classification, and reporting. The report also enlists useful bioinformatics tools and databases commonly used in NGS data analysis. The recommendations should serve as an educational resource for both clinical laboratory professionals and oncologists to aid variant interpretation and clinical decision making. It is our hope that the guidelines presented herein will achieve widespread use in the cancer genomics community and engender significant improvements in the practice of genomic testing and precision care for cancer patients.
Disclaimer
The Association for Molecular Pathology (AMP) Clinical Practice Guidelines and Reports are developed to be of assistance to laboratory and other health care professionals by providing guidance and recommendations for particular areas of practice. The Guidelines or Report should not be considered inclusive of all proper approaches or methods, or exclusive of others. The Guidelines or Report cannot guarantee any specific outcome, nor do they establish a standard of care. The Guidelines or Report is not intended to dictate the treatment of a particular patient. Treatment decisions must be made based on the independent judgment of health care providers and each patient's individual circumstances. AMP makes no warranty, express or implied, regarding the Guidelines or Report and specifically excludes any warranties of merchantability and fitness for a particular use or purpose. AMP shall not be liable for direct, indirect, special, incidental, or consequential damages related to the use of the information contained herein.
Acknowledgments
The Interpretation of Sequence Variants in Somatic Conditions Working Group members thank the many colleagues in the cancer genomics and clinical oncology communities who contributed to the development of this guideline through their responses to surveys and their astute comments and suggestions at workshops; Dr. Robyn Temple-Smolkin for insightful recommendations and careful editing; and Mrudula Pullambhatla for outstanding administrative support to the project.
Footnotes
Supported by the Association for Molecular Pathology.
Disclosures: E.J.D. is the Medical Director for Cofactor Genomics and claims ownership in P&V Licensing, LLC; A.Y. is a consultant for Foundation Medicine; A.M.T. received research funding from Foundation Medicine, EMD Serono, Baxalta, Bayer, and Onyx.
The Interpretation of Sequence Variants in Somatic Conditions Working Group is a working group of the Association for Molecular Pathology Clinical Practice Committee with liaison representation from the American College of Medical Genetics and Genomics (S.K. and D.J.W.), American Society of Clinical Oncology (A.M.T. and A.Y), and College of American Pathologists (M.D. and N.I.L.). The 2015 and 2016 Clinical Practice Committee consisted of Marina Nikiforova (Chair 2015 to 2016), Monica Basehore, Christopher Coldren, Linda Cook, Jennifer Crow, Birgit Funke, Meera Hameed, Larry Jennings, Arivarasan Karunamurthy, Annette Kim, Bryan Krock, Mary Lowery-Nordberg, Melissa Miller, Benjamin Pinsky, Somak Roy, Mark Routbort, Ryan Schmidt, and David Viswanatha.
Standard of practice is not defined by this article and there may be alternatives. See Disclaimer for further details.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2016.10.002.
Supplemental Data
References
- 1.Garraway L.A., Verweij J., Ballman K.V. Precision oncology: an overview. J Clin Oncol. 2013;31:1803–1805. doi: 10.1200/JCO.2013.49.4799. [DOI] [PubMed] [Google Scholar]
- 2.Tsimberidou A.M. Targeted therapy in cancer. Cancer Chemother Pharmacol. 2015;76:1113–1132. doi: 10.1007/s00280-015-2861-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Von Hoff D.D., Stephenson J.J., Rosen P., Loesch D.M., Borad M.J., Anthony S., Jameson G., Brown S., Cantafio N., Richards D.A., Fitch T.R., Wasserman E., Fernandez C., Green S., Sutherland W., Bittner M., Alarcon A., Mallery D., Penny R. Pilot study using molecular profiling of patients' tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 4.Tsimberidou A.-M., Iskander N.G., Hong D.S., Wheler J.J., Falchook G.S., Fu S., Piha-Paul S., Naing A., Janku F., Luthra R., Ye Y., Wen S., Berry D., Kurzrock R. Personalized medicine in a phase I clinical trials program: the MD Anderson Cancer Center initiative. Clin Cancer Res. 2012;18:6373–6383. doi: 10.1158/1078-0432.CCR-12-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shendure J., Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 6.Bentley D.R., Balasubramanian S., Swerdlow H.P., Smith G.P., Milton J., Brown C.G. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabatini L.M., Mathews C., Ptak D., Doshi S., Tynan K., Hegde M.R., Burke T.L., Bossler A.D. Genomic sequencing procedure microcosting analysis and health economic cost-impact analysis: a report of the Association for Molecular Pathology. J Mol Diagn. 2016;18:319–328. doi: 10.1016/j.jmoldx.2015.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kandoth C., McLellan M.D., Vandin F., Ye K., Niu B., Lu C., Xie M., Zhang Q., McMichael J.F., Wyczalkowski M.A., Leiserson M.D., Miller C.A., Welch J.S., Walter M.J., Wendl M.C., Ley T.J., Wilson R.K., Raphael B.J., Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;503:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L., Ley T.J., Larson D.E., Miller C.A., Koboldt D.C., Welch J.S. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481:506–510. doi: 10.1038/nature10738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schilsky R.L. Implementing personalized cancer care. Nat Rev Clin Oncol. 2014;11:432–438. doi: 10.1038/nrclinonc.2014.54. [DOI] [PubMed] [Google Scholar]
- 11.Teutsch S.M., Bradley L.A., Palomaki G.E., Haddow J.E., Piper M., Calonge N., David Dotson W., Douglas M.P., Berg A.O. The Evaluation of Genomic Applications in Practice and Prevention (EGAPP) initiative: methods of the EGAPP Working Group. Genet Med. 2009;11:3–14. doi: 10.1097/GIM.0b013e318184137c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindeman N.I., Cagle P.T., Beasley M.B., Chitale D.A., Dacic S., Giaccone G., Jenkins R.B., Kwiatkowski D.J., Saldivar J.-S., Squire J., Thunnissen E., Ladanyi M. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors. J Mol Diagn. 2013;15:415–453. doi: 10.1016/j.jmoldx.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 13.McShane L.M., Cavenagh M.M., Lively T.G., Eberhard D.A., Bigbee W.L., Williams P.M., Mesirov J.P., Polley M.-Y.C., Kim K.Y., Tricoli J.V., Taylor J.M.G., Shuman D.J., Simon R.M., Doroshow J.H., Conley B.A. Criteria for the use of omics-based predictors in clinical trials: explanation and elaboration. BMC Med. 2013;11:220. doi: 10.1186/1741-7015-11-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., Voelkerding K., Rehm H.L. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Auton A., Abecasis G.R., Altshuler D.M., Durbin R.M., Bentley D.R., Chakravarti A., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lappalainen I., Lopez J., Skipper L., Hefferon T., Spalding J.D., Garner J., Chen C., Maguire M., Corbett M., Zhou G., Paschall J., Ananiev V., Flicek P., Church D.M. DbVar and DGVa: public archives for genomic structural variation. Nucleic Acids Res. 2013;41:D936–D941. doi: 10.1093/nar/gks1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., Ding M., Bamford S., Cole C., Ward S., Kok C.Y., Jia M., De T., Teague J.W., Stratton M.R., McDermott U., Campbell P.J. COSMIC: exploring the world's knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gundem G., Perez-Llamas C., Jene-Sanz A., Kedzierska A., Islam A., Deu-Pons J., Furney S.J., Lopez-Bigas N. IntOGen: integration and data mining of multidimensional oncogenomic data. Nat Methods. 2010;7:92–93. doi: 10.1038/nmeth0210-92. [DOI] [PubMed] [Google Scholar]
- 22.Petitjean A., Mathe E., Kato S., Ishioka C., Tavtigian S.V., Hainaut P., Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Baran J., Cros A., Guberman J.M., Haider S., Hsu J., Liang Y., Rivkin E., Wang J., Whitty B., Wong-Erasmus M., Yao L., Kasprzyk A. International cancer genome consortium data portal: a one-stop shop for cancer genomics data. Database. 2011;2011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pruitt K.D., Brown G.R., Hiatt S.M., Thibaud-Nissen F., Astashyn A., Ermolaeva O., Farrell C.M., Hart J., Landrum M.J., McGarvey K.M., Murphy M.R., O'Leary N.A., Pujar S., Rajput B., Rangwala S.H., Riddick L.D., Shkeda A., Sun H., Tamez P., Tully R.E., Wallin C., Webb D., Weber J., Wu W., Dicuccio M., Kitts P., Maglott D.R., Murphy T.D., Ostell J.M. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–D763. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalgleish R., Flicek P., Cunningham F., Astashyn A., Tully R.E., Proctor G., Chen Y., McLaren W.M., Larsson P., Vaughan B.W., Béroud C., Dobson G., Lehväslaiho H., Taschner P.E., den Dunnen J.T., Devereau A., Birney E., Brookes A.J., Maglott D.R. Locus Reference Genomic sequences: an improved basis for describing human DNA variants. Genome Med. 2010;2:24. doi: 10.1186/gm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smedley D., Haider S., Durinck S., Pandini L., Provero P., Allen J., Arnaiz O., Awedh M., Baldock R. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucleic Acids Res. 2015;43:W589–W598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenson P.D., Ball E.V., Mort M., Phillips A.D., Shaw K., Cooper D.N. The human gene mutation database (HGMD) and its exploitation in the fields of personalized genomics and molecular evolution. Curr Protoc Bioinformatics. 2012;Chapter 1:Unit1.13. doi: 10.1002/0471250953.bi0113s39. [DOI] [PubMed] [Google Scholar]
- 30.Fokkema I.F.A.C., Taschner P.E.M., Schaafsma G.C.P., Celli J., Laros J.F.J., den Dunnen J.T. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 31.Liu X., Wu C., Li C., Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016;37:235–241. doi: 10.1002/humu.22932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cottrell C.E., Al-Kateb H., Bredemeyer A.J., Duncavage E.J., Spencer D.H., Abel H.J., Lockwood C.M., Hagemann I.S., O'Guin S.M., Burcea L.C., Sawyer C.S., Oschwald D.M., Stratman J.L., Sher D.A., Johnson M.R., Brown J.T., Cliften P.F., George B., McIntosh L.D., Shrivastava S., Nguyen T.T., Payton J.E., Watson M.A., Crosby S.D., Head R.D., Mitra R.D., Nagarajan R., Kulkarni S., Seibert K., Virgin I.V.H.W., Milbrandt J., Pfeifer J.D. Validation of a next-generation sequencing assay for clinical molecular oncology. J Mol Diagn. 2014;16:89–105. doi: 10.1016/j.jmoldx.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gotlib J. JAK inhibition in the myeloproliferative neoplasms: lessons learned from the bench and bedside. Hematol Am Soc Hematol Educ Program. 2013;2013:529–537. doi: 10.1182/asheducation-2013.1.529. [DOI] [PubMed] [Google Scholar]
- 34.Jaiswal S., Fontanillas P., Flannick J., Manning A., Grauman P.V., Mar B.G., Lindsley R.C., Mermel C.H., Burtt N., Chavez A., Higgins J.M., Moltchanov V., Kuo F.C., Kluk M.J., Henderson B., Kinnunen L., Koistinen H.A., Ladenvall C., Getz G., Correa A., Banahan B.F., Gabriel S., Kathiresan S., Stringham H.M., McCarthy M.I., Boehnke M., Tuomilehto J., Haiman C., Groop L., Atzmon G., Wilson J.G., Neuberg D., Altshuler D., Ebert B.L. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genovese G., Kähler A.K., Handsaker R.E., Lindberg J., Rose S.A., Bakhoum S.F., Chambert K., Mick E., Neale B.M., Fromer M., Purcell S.M., Svantesson O., Landén M., Höglund M., Lehmann S., Gabriel S.B., Moran J.L., Lander E.S., Sullivan P.F., Sklar P., Grönberg H., Hultman C.M., McCarroll S.A. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yohe S.L., Carter A.B., Pfeifer J.D., Crawford J.M., Cushman-Vokoun A., Caughron S., Leonard D.G.B. Standards for clinical grade genomic databases. Arch Pathol Lab Med. 2015;139:1400–1412. doi: 10.5858/arpa.2014-0568-CP. [DOI] [PubMed] [Google Scholar]
- 37.Adzhubei I., Jordan D.M., Sunyaev S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013;Chapter 7:Unit7.20. doi: 10.1002/0471142905.hg0720s76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sim N.-L., Kumar P., Hu J., Henikoff S., Schneider G., Ng P.C. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457. doi: 10.1093/nar/gks539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frousios K., Iliopoulos C.S., Schlitt T., Simpson M.A. Predicting the functional consequences of non-synonymous DNA sequence variants: evaluation of bioinformatics tools and development of a consensus strategy. Genomics. 2013;102:223–228. doi: 10.1016/j.ygeno.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 41.Schwarz J.M., Cooper D.N., Schuelke M.S.D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 42.Desmet F.O., Hamroun D., Lalande M., Collod-Bëroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeo G., Burge C.B. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 44.Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J Mol Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 45.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:btv195. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzalez-Perez A., Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kircher M., Witten D.M., Jain P., O'Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–121. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reese M., Eeckman F., Kulp D., Haussler D. Improved splice site detection in genie. J Comp Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 51.Pertea M., Lin X., Salzberg S.L. GeneSplicer: a new computational method for splice site prediction. Nucleic Acids Res. 2001;29:1185–1190. doi: 10.1093/nar/29.5.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thusberg J., Olatubosun A., Vihinen M. Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat. 2011;32:358–368. doi: 10.1002/humu.21445. [DOI] [PubMed] [Google Scholar]
- 53.Houdayer C., Caux-Moncoutier V., Krieger S., Barrois M., Bonnet F., Bourdon V., Bronner M., Buisson M., Coulet F., Gaildrat P., Lefol C., Lone M., Mazoyer S., Muller D., Remenieras A., Rvillion F., Rouleau E., Sokolowska J., Vert J.P., Lidereau R., Soubrier F., Sobol H., Sevenet N., Bressac-de Paillerets B., Hardouin A., Tosi M., Sinilnikova O.M., Stoppa-Lyonnet D. Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat. 2012;33:1228–1238. doi: 10.1002/humu.22101. [DOI] [PubMed] [Google Scholar]
- 54.Vreeswijk M.P.G., Kraan J.N., Van Der Klift H.M., Vink G.R., Cornelisse C.J., Wijnen J.T., Bakker E., Van Asperen C.J., Devilee P. Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat. 2009;30:107–114. doi: 10.1002/humu.20811. [DOI] [PubMed] [Google Scholar]
- 55.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Koboldt D.C., Larson D.E., Wilson R.K. Using varscan 2 for germline variant calling and somatic mutation detection. Curr Protoc Bioinformatics. 2013;44:15.4.1–15.4.17. doi: 10.1002/0471250953.bi1504s44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lai Z., Markovets A., Ahdesmaki M., Johnson J. Abstract 4864: VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Cancer Res. 2015;75:4864. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saunders C.T., Wong W.S.W., Swamy S., Becq J., Murray L.J., Cheetham R.K. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 59.Garrison E., Marth G. 2012. Haplotype-based variant detection from short-read sequencing.https://arxiv.org/pdf/1207.3907.pdf arXiv.org. Available at. (accessed November 18, 2016) [Google Scholar]
- 60.Fang H., Grabowska E.A., Arora K., Vacic V., Zody M.C., Iossifov I., ORawe J.A., Wu Y., Barron L.T.J., Rosenbaum J., Ronemus M., Lee Y., Wang Z., Lyon G.J., Wigler M., Schatz M.C., Narzisi G. Indel variant analysis of short-read sequencing data with Scalpel. Nat Protoc. 2016;11:2529–2548. doi: 10.1038/nprot.2016.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye K., Schulz M.H., Long Q., Apweiler R., Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larson D.E., Harris C.C., Chen K., Koboldt D.C., Abbott T.E., Dooling D.J., Ley T.J., Mardis E.R., Wilson R.K., Ding L. Somaticsniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makarov V., O'Grady T., Cai G., Lihm J., Buxbaum J.D., Yoon S. Anntools: a comprehensive and versatile annotation toolkit for genomic variants. Bioinformatics. 2012;28:724–725. doi: 10.1093/bioinformatics/bts032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grant J.R., Arantes A.S., Liao X., Stothard P. In-depth annotation of SNPs arising from resequencing projects using NGS-SNP. Bioinformatics. 2011;27:2300–2301. doi: 10.1093/bioinformatics/btr372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Joseph L., Cankovic M., Caughron S., Chandra P., Emmadi R., Hagenkord J., Hallam S., Jewell K., Klein R., Pratt V., Rothberg P., Temple-Smolkin R.L., Lyon E. The spectrum of clinical utilities in molecular pathology testing procedures for inherited conditions and cancer: a report of the Association for Molecular Pathology. J Mol Diagn. 2016;18:605–619. doi: 10.1016/j.jmoldx.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 69.Schrijver I., Farkas D.H., Gibson J.S., Lyon E. The evolving role of the laboratory professional in the age of genome sequencing: a vision of the Association for Molecular Pathology. J Mol Diagn. 2015;17:335–338. doi: 10.1016/j.jmoldx.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 70.Andreyev H.J., Norman A.R., Cunningham D., Oates J., Dix B.R., Iacopetta B.J. Kirsten ras mutations in patients with colorectal cancer: the “RASCAL II” study. Br J Cancer. 2001;85:692–696. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sinha C., Cunningham L.C., Liu P.P. Core binding factor acute myeloid leukemia: new prognostic categories and therapeutic opportunities. Semin Hematol. 2015;52:215–222. doi: 10.1053/j.seminhematol.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang J.W., Wang J.Y., Chen S.J., Chen Z. Mechanisms of all-trans retinoic acid-induced differentiation of acute promyelocytic leukemia cells. J Biosci. 2000;25:275–284. doi: 10.1007/BF02703936. [DOI] [PubMed] [Google Scholar]
- 73.Dietrich S., Zenz T. BRAF inhibitor therapy in HCL. Best Pract Res Clin Haematol. 2015;28:246–252. doi: 10.1016/j.beha.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 74.Tiacci E., Trifonov V., Schiavoni G., Holmes A., Kern W., Martelli M.P. BRAF mutations in hairy-cell leukemia. N Engl J Med. 2011;364:2305–2315. doi: 10.1056/NEJMoa1014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tiacci E., Schiavoni G., Martelli M.P., Boveri E., Pacini R., Tabarrini A., Zibellini S., Santi A., Pettirossi V., Fortini E., Ascani S., Arcaini L., Inghirami G., Paulli M., Falini B. Constant activation of the RAF-MEK-ERK pathway as a diagnostic and therapeutic target in hairy cell leukemia. Haematologica. 2013;98:635–639. doi: 10.3324/haematol.2012.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tiacci E., Park J.H., De Carolis L., Chung S.S., Broccoli A., Scott S. Targeting mutant BRAF in relapsed or refractory hairy-cell leukemia. N Engl J Med. 2015;373:1733–1747. doi: 10.1056/NEJMoa1506583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilson M.A., Zhao F., Khare S., Roszik J., Woodman S.E., D'Andrea K., Wubbenhorst B., Rimm D.L., Kirkwood J.M., Kluger H.M., Schuchter L.M., Lee S.J., Flaherty K.T., Nathanson K.L. Copy number changes are associated with response to treatment with carboplatin, paclitaxel, and sorafenib in melanoma. Clin Cancer Res. 2015;22:374–382. doi: 10.1158/1078-0432.CCR-15-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi H., Hugo W., Kong X., Hong A., Koya R.C., Moriceau G., Chodon T., Guo R., Johnson D.B., Dahlman K.B., Kelley M.C., Kefford R.F., Chmielowski B., Glaspy J.A., Sosman J.A., Van Baren N., Long G.V., Ribas A., Lo R.S. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shi H., Moriceau G., Kong X., Lee M.-K., Lee H., Koya R.C., Ng C., Chodon T., Scolyer R.A., Dahlman K.B., Sosman J.A., Kefford R.F., Long G.V., Nelson S.F., Ribas A., Lo R.S. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Van Allen E.M., Wagle N., Sucker A., Treacy D.J., Johannessen C.M., Goetz E.M., Place C.S., Taylor-Weiner A., Whittaker S., Kryukov G.V., Hodis E., Rosenberg M., Mckenna A., Cibulskis K., Farlow D., Zimmer L., Hillen U., Gutzmer R., Goldinger S.M., Ugure S., Gogas H.J., Egberts F., Berking C., Trefzer U., Loquai C., Weide B., Hasse J.C., Gabrie S.B., Carter S.L., Getz G., Garraway L.A., Schadendorf D. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wagle N., Van Allen E.M., Treacy D.J., Frederick D.T., Cooper Z.A., Taylor-Weiner A., Rosenberg M., Goetz E.M., Sullivan R.J., Farlow D.N., Friedrich D.C., Anderka K., Perrin D., Johannessen C.M., McKenna A., Cibulskis K., Kryukov G., Hodis E., Lawrence D.P., Fisher S., Getz G., Gabriel S.B., Carter S.L., Flaherty K.T., Wargo J.A., Garraway L.A. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Furitsu T., Tsujimura T., Tono T., Ikeda H., Kitayama H., Koshimizu U., Sugahara H., Butterfield J.H., Ashman L.K., Kanayama Y., Matsuzawa Y., Kitamura Y., Kanakura Y. Identification of mutations in the coding sequence of the proto-oncogene c-kit in a human mast cell leukemia cell line causing ligand-independent activation of c-kit product. J Clin Invest. 1993;92:1736–1744. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nakata Y., Kimura A., Katoh O., Kawaishi K., Hyodo H., Abe K., Kuramoto A., Satow Y. C-kit point mutation of extracellular domain in patients with myeloproliferative disorders. Br J Haematol. 1995;91:661–663. doi: 10.1111/j.1365-2141.1995.tb05364.x. [DOI] [PubMed] [Google Scholar]
- 84.Garcia-Montero A.C., Jara-Acevedo M., Teodosio C., Sanchez M.L., Nunez R., Prados A., Aldanondo I., Sanchez L., Dominguez M., Botana L.M., Sanchez-Jimenez F., Sotlar K., Almeida J., Escribano L., Orfao A. KIT mutation in mast cells and other bone marrow hematopoietic cell lineages in systemic mast cell disorders: a prospective study of the Spanish Network on Mastocytosis (REMA) in a series of 113 patients. Blood. 2006;108:2366–2372. doi: 10.1182/blood-2006-04-015545. [DOI] [PubMed] [Google Scholar]
- 85.Nikiforova M.N., Kimura E.T., Gandhi M., Biddinger P.W., Knauf J.A., Basolo F., Zhu Z., Giannini R., Salvatore G., Fusco A., Santoro M., Fagin J.A., Nikiforov Y.E. BRAF mutations in thyroid tumors are restricted to papillary carcinomas and anaplastic or poorly differentiated carcinomas arising from papillary carcinomas. J Clin Endocrinol Metab. 2003;88:5399–5404. doi: 10.1210/jc.2003-030838. [DOI] [PubMed] [Google Scholar]
- 86.Nikiforova M.N., Wald A.I., Roy S., Durso M.B., Nikiforov Y.E. Targeted next-generation sequencing panel (ThyroSeq) for detection of mutations in thyroid cancer. J Clin Endocrinol Metab. 2013;98:E1852–E1860. doi: 10.1210/jc.2013-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Elisei R., Viola D., Torregrossa L., Giannini R., Romei C., Ugolini C., Molinaro E., Agate L., Biagini A., Lupi C., Valerio L., Materazzi G., Miccoli P., Piaggi P., Pinchera A., Vitti P., Basolo F. The BRAFV600E mutation is an independent, poor prognostic factor for the outcome of patients with low-risk intrathyroid papillary thyroid carcinoma: single-institution results from a large cohort study. J Clin Endocrinol Metab. 2012;97:4390–4398. doi: 10.1210/jc.2012-1775. [DOI] [PubMed] [Google Scholar]
- 88.Collins V.P., Jones D.T.W., Giannini C. Pilocytic astrocytoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:775–788. doi: 10.1007/s00401-015-1410-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones D.T.W., Kocialkowski S., Liu L., Pearson D.M., Bäcklund L.M., Ichimura K., Collins V.P. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–8677. doi: 10.1158/0008-5472.CAN-08-2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gamberi G., Cocchi S., Benini S., Magagnoli G., Morandi L., Kreshak J., Gambarotti M., Picci P., Zanella L., Alberghini M. Molecular diagnosis in ewing family tumors the rizzoli experience: 222 consecutive cases in four years. J Mol Diagn. 2011;13:313–324. doi: 10.1016/j.jmoldx.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Szuhai K., Cleton-Jansen A.M., Hogendoorn P.C., Bovee J.V. Molecular pathology and its diagnostic use in bone tumors. Cancer Genet. 2012;205:193–204. doi: 10.1016/j.cancergen.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 92.Kaufman B., Shapira-Frommer R., Schmutzler R.K., Audeh M.W., Friedlander M., Balmaña J., Mitchell G., Fried G., Stemmer S.M., Hubert A., Rosengarten O., Steiner M., Loman N., Bowen K., Fielding A., Domchek S.M. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33:244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ballinger M.L., Mitchell G.T.D. Surveillance recommendations for patients with germline TP53 mutations. Curr Opin Oncol. 2015;27:332–337. doi: 10.1097/CCO.0000000000000200. [DOI] [PubMed] [Google Scholar]
- 94.Funato M., Kondo N. New knowledge of Li-Fraumeni syndromeGan To Kagaku Ryoho. 2013;40:148–153. Japanese. [PubMed] [Google Scholar]
- 95.Harrison C., Kiladjian J.-J., Al-Ali H.K., Gisslinger H., Waltzman R., Stalbovskaya V., McQuitty M., Hunter D.S., Levy R., Knoops L., Cervantes F., Vannucchi A.M., Barbui T., Barosi G. JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. N Engl J Med. 2012;366:787–798. doi: 10.1056/NEJMoa1110556. [DOI] [PubMed] [Google Scholar]
- 96.Verstovsek S., Mesa R.A., Gotlib J., Levy R.S., Gupta V., DiPersio J.F., Catalano J.V., Deininger M., Miller C., Silver R.T., Talpaz M., Winton E.F., Harvey J.H., Arcasoy M.O., Hexner E., Lyons R.M., Paquette R., Raza A., Vaddi K., Erickson-Viitanen S., Koumenis I.L., Sun W., Sandor V., Kantarjian H.M. A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N Engl J Med. 2012;366:799–807. doi: 10.1056/NEJMoa1110557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maude S.L., Tasian S.K., Vincent T., Hall J.W., Sheen C., Roberts K.G., Seif A.E., Barrett D.M., Chen I.M., Collins J.R., Mullighan C.G., Hunger S.P., Harvey R.C., Willman C.L., Fridman J.S., Loh M.L., Grupp S.A., Teachey D.T. Targeting JAK1/2 and mTOR in murine xenograft models of Ph-like acute lymphoblastic leukemia. Blood. 2012;120:3510–3518. doi: 10.1182/blood-2012-03-415448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Roberts K.G., Li Y., Payne-Turner D., Harvey R.C., Yang Y.-L., Pei D. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–1015. doi: 10.1056/NEJMoa1403088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Starlinger M. Adaptation of the upper gastrointestinal tract following vagotomy. Z Gastroenterol Verh. 1989;24:223–224. [PubMed] [Google Scholar]
- 100.Papaemmanuil E., Cazzola M., Boultwood J., Malcovati L., Vyas P., Bowen D. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med. 2011;365:1384–1395. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshida K., Sanada M., Shiraishi Y., Nowak D., Nagata Y., Yamamoto R., Sato Y., Sato-Otsubo A., Kon A., Nagasaki M., Chalkidis G., Suzuki Y., Shiosaka M., Kawahata R., Yamaguchi T., Otsu M., Obara N., Sakata-Yanagimoto M., Ishiyama K., Mori H., Nolte F., Hofmann W.-K., Miyawaki S., Sugano S., Haferlach C., Koeffler H.P., Shih L.-Y., Haferlach T., Chiba S., Nakauchi H., Miyano S., Ogawa S. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 102.Malcovati L., Papaemmanuil E., Bowen D.T., Boultwood J., Della Porta M.G., Pascutto C., Travaglino E., Groves M.J., Godfrey A.L., Ambaglio I., Gallì A., Da Vià M.C., Conte S., Tauro S., Keenan N., Hyslop A., Hinton J., Mudie L.J., Wainscoat J.S., Futreal P.A., Stratton M.R., Campbell P.J., Hellström-Lindberg E., Cazzola M. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood. 2011;118:6239–6246. doi: 10.1182/blood-2011-09-377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B.T., Qing W., Packman K., Myklebost O., Heimbrook D.C., Vassilev L.T. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tovar C., Graves B., Packman K., Filipovic Z., Xia B.H.M., Tardell C., Garrido R., Lee E., Kolinsky K., To K.H., Linn M., Podlaski F., Wovkulich P., Vu B., Vassilev L.T. MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res. 2013;73:2587–2597. doi: 10.1158/0008-5472.CAN-12-2807. [DOI] [PubMed] [Google Scholar]
- 105.Saha M.N., Qiu L., Chang H. Targeting p53 by small molecules in hematological malignancies. J Hematol Oncol. 2013;6:23. doi: 10.1186/1756-8722-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lehmann S., Bykov V.J.N., Ali D., Andreń O., Cherif H., Tidefelt U., Uggla B., Yachnin J., Juliusson G., Moshfegh A., Paul C., Wiman K.G., Andersson P.O. Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J Clin Oncol. 2012;30:3633–3639. doi: 10.1200/JCO.2011.40.7783. [DOI] [PubMed] [Google Scholar]
- 107.Clinical and Laboratory Standards Institute . ed 2. Clinical Laboratory Standards Institute; Wayne, PA: 2014. Nucleic Acid Sequencing Methods in Diagnostic Laboratory Medicine: Approved Guideline. CLSI document MM09-A2. [Google Scholar]
- 108.Green R.C., Berg J.S., Grody W.W., Kalia S.S., Korf B.R., Martin C.L., McGuire A.L., Nussbaum R.L., O'Daniel J.M., Ormond K.E., Rehm H.L., Watson M.S., Williams M.S., Biesecker L.G. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–574. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hegde M., Bale S., Bayrak-Toydemir P., Gibson J., Bone Jeng L.J., Joseph L., Laser J., Lubin I.M., Miller C.E., Ross L.F., Rothberg P.G., Tanner A.K., Vitazka P., Mao R. Reporting incidental findings in genomic scale clinical sequencing: a clinical laboratory perspective: a report of the Association for Molecular Pathology. J Mol Diagn. 2015;17:107–117. doi: 10.1016/j.jmoldx.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raymond V.M., Gray S.W., Roychowdhury S., Joffe S., Chinnaiyan A.M., Parsons D.W., Plon S.E. Germline findings in tumor-only sequencing: points to consider for clinicians and laboratories: table 1. J Natl Cancer Inst. 2016;108:djv351. doi: 10.1093/jnci/djv351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.MacKinnon A.C., Wang Y.L., Sahota A., Yeung C.C., Weck K.E. Certification in molecular pathology in the United States: an update from the Association for Molecular Pathology training and education committee. J Mol Diagn. 2012;14:541–549. doi: 10.1016/j.jmoldx.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 112.Gulley M.L., Braziel R.M., Halling K.C., Hsi E.D., Kant J.A., Nikiforova M.N., Nowak J.A., Ogino S., Oliveira A., Polesky H.F., Silverman L., Tubbs R.R., Van Deerlin V.M., Vance G.H., Versalovic J. Clinical laboratory reports in molecular pathology. Arch Pathol Lab Med. 2007;131:852–863. doi: 10.5858/2007-131-852-CLRIMP. [DOI] [PubMed] [Google Scholar]
- 113.Ogino S., Gulley M.L., Den Dunnen J.T., Wilson R.B. Standard mutation nomenclature in molecular diagnostics practical and educational challenges. J Mol Diagn. 2007;9:1–6. doi: 10.2353/jmoldx.2007.060081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jones S., Anagnostou V., Lytle K., Parpart-li S., Nesselbush M., Riley D.R., Shukla M., Chesnick B., Kadan M., Papp E., Galens K.G., Murphy D., Zhang T., Kann L., Sausen M., Angiuoli S.V., Jr., Diaz L.A., Velculescu V.E. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.