Abstract
The retrotrapezoid nucleus (RTN) consists, by definition, of Phox2b-expressing, glutamatergic, non-catecholaminergic, noncholinergic neurons located in the parafacial region of the medulla oblongata. An unknown proportion of RTN neurons are central respiratory chemoreceptors and there is mounting evidence for biochemical diversity among these cells. Here, we used multiplexed in situ hybridization and single-cell RNA-Seq in male and female mice to provide a more comprehensive view of the phenotypic diversity of RTN neurons. We now demonstrate that the RTN of mice can be identified with a single and specific marker, Neuromedin B mRNA (Nmb). Most (∼75%) RTN neurons express low-to-moderate levels of Nmb and display chemoreceptor properties. Namely they are activated by hypercapnia, but not by hypoxia, and express proton sensors, TASK-2 and Gpr4. These Nmb-low RTN neurons also express varying levels of transcripts for Gal, Penk, and Adcyap1, and receptors for substance P, orexin, serotonin, and ATP. A subset of RTN neurons (∼20–25%), typically larger than average, express very high levels of Nmb mRNA. These Nmb-high RTN neurons do not express Fos after hypercapnia and have low-to-undetectable levels of Kcnk5 or Gpr4 transcripts; they also express Adcyap1, but are essentially devoid of Penk and Gal transcripts. In male rats, Nmb is also a marker of the RTN but, unlike in mice, this gene is expressed by other types of nearby neurons located within the ventromedial medulla. In sum, Nmb is a selective marker of the RTN in rodents; Nmb-low neurons, the vast majority, are central respiratory chemoreceptors, whereas Nmb-high neurons likely have other functions.
SIGNIFICANCE STATEMENT Central respiratory chemoreceptors regulate arterial PCO2 by adjusting lung ventilation. Such cells have recently been identified within the retrotrapezoid nucleus (RTN), a brainstem nucleus defined by genetic lineage and a cumbersome combination of markers. Using single-cell RNA-Seq and multiplexed in situ hybridization, we show here that a single marker, Neuromedin B mRNA (Nmb), identifies RTN neurons in rodents. We also suggest that >75% of these Nmb neurons are chemoreceptors because they are strongly activated by hypercapnia and express high levels of proton sensors (Kcnk5 and Gpr4). The other RTN neurons express very high levels of Nmb, but low levels of Kcnk5/Gpr4/pre-pro-galanin/pre-pro-enkephalin, and do not respond to hypercapnia. Their function is unknown.
Keywords: central chemoreceptors, Gpr4, hypercapnia, medulla oblongata, Neuromedin B, TASK-2
Introduction
The term retrotrapezoid nucleus (RTN) was coined in 1989 to describe a region of the reticular formation that lies beneath the facial motor nucleus and provides input to the ventrolateral and dorsomedial medulla oblongata (Smith et al., 1989; Connelly et al., 1990; Ellenberger and Feldman, 1990). This brain area, also called the parafacial region (Onimaru and Homma, 2003; Huckstepp et al., 2015), has long been suspected to contain central respiratory chemoreceptors (Loeschcke, 1982; Liu et al., 2002; Feldman et al., 2003). Since 2004, a substantial set of evidence has accrued to indicate that the parafacial region harbors the hub of the central respiratory chemoreflex, and may be the principal CNS site where CO2 is sensed for the purpose of breathing regulation (for reviews, see Guyenet and Bayliss, 2015; Guyenet et al., 2016). Identifying the phenotype of these chemoreceptors has been an iterative process, guided by the gradual and often fortuitous discovery of biochemical markers expressed by the parafacial neurons that are activated by acidification in slices or by hypercapnia in vivo (Mulkey et al., 2004; Stornetta et al., 2006). It is now clear that these parafacial acid-sensitive neurons express Phox2b, vesicular glutamate transporter-2 (VGlut2), and NK1 receptors from late embryological stages to adulthood; by contrast, they lack tyrosine hydroxylase, choline acetyltransferase, glutamic acid decarboxylases, glycine transporter-2, and tryptophan hydroxylase (Mulkey et al., 2004; Dubreuil et al., 2008; Lazarenko et al., 2009; Thoby-Brisson et al., 2009; Wang et al., 2013b; Onimaru et al., 2014; Kumar et al., 2015; Guyenet et al., 2016). This combination of markers defines a population of 600–800 parafacial neurons in mice (∼2000 in rats) that also have a common developmental lineage (Egr-2, Phox2b, Atoh-1), and which are now referred to specifically as the RTN (Ramanantsoa et al., 2011; Ruffault et al., 2015; Guyenet et al., 2016). RTN neurons innervate the ventrolateral and dorsomedial medulla oblongata, as per the original definition of the term (Smith et al., 1989; Connelly et al., 1990; Ellenberger and Feldman, 1990). They drive breathing in proportion to arterial pH as expected from central respiratory chemoreceptors (Basting et al., 2015). It is now well established that the parafacial CO2-responsive neurons have the combined neurochemical phenotype expected of RTN neurons; conversely, whether every RTN neuron is a respiratory chemoreceptor remains an open question.
Recent evidence suggests that two proton-sensitive membrane proteins—a H+-inhibited background K+ channel, TASK-2 (encoded by Kcnk5), and a H+-activated G-protein-coupled receptor, Gpr4—underlie the intrinsic pH-sensitivity of RTN neurons in vitro and contribute to their central respiratory chemoreceptor properties in vivo (Reyes et al., 1998; Ludwig et al., 2003; Gestreau et al., 2010; Wang et al., 2013b; Kumar et al., 2015). Thus, these proteins could be selective markers of the RTN neurons that have chemoreceptor properties.
RTN neurons are biochemically heterogeneous. Subsets of RTN neurons, including the putative chemoreceptors, express the neuropeptide galanin (Stornetta et al., 2009). The parafacial region also contains Neuromedin B (Nmb)-expressing neurons (Lein et al., 2007) that may control sighing (Li et al., 2016). Given their location, these Nmb neurons could be a subtype of RTN neurons.
Here, we used multiplexed in situ hybridization and single-cell RNA-Seq to provide a more comprehensive view of the phenotypic diversity of RTN neurons. Our primary objectives were to address the following questions. Are the recently identified parafacial Nmb neurons a subset of the neuronal population that we previously defined as RTN? Is there a unique marker of RTN neurons that could simplify their identification? Does the entire population of RTN neurons qualify as central respiratory chemoreceptors? If not, does the presence of the putative proton receptors Gpr4 and or TASK-2 predict their CO2 sensitivity?
Materials and Methods
Animals
JX99 (Phox2b::eGFP BAC transgenic) mice were originally obtained from the Gene Expression Nervous System Atlas Project at Rockefeller University and have been maintained on a mixed genetic background (Lazarenko et al., 2009). C57BL/6J mice were obtained from Jackson Laboratories (RRID:IMSR_JAX:000664). Three male Sprague-Dawley rats were obtained from Taconic (RRID:RGD_1566440). Under the supervision of the University of Virginia veterinary staff, all animals were kept in the Pinn Hall vivarium and allowed ad libitum access to food and water with a 12 h day/night light cycle. All protocols were approved by the University of Virginia Animal Care and Use Committee and complied with NIH animal guidelines.
Immunocytochemistry
Antibodies.
Phox2b::eGFP JX99 mice express eGFP in all Phox2b neurons (Lazarenko et al., 2009). To enhance sensitivity, eGFP was visualized with a chicken antibody to GFP from Aves Labs (catalog #GFP-1020; concentration 1:1000; RRID:AB_10000240) followed by a secondary anti-chicken antibody tagged with Alexa488 (Jackson ImmunoResearch; RRID:SCR_010488).
In situ hybridization.
Mice (N = 37; 20 females, 17 males) were anesthetized with pentobarbital and perfused transcardially with 4% paraformaldehyde. Brains were removed and postfixed in the same fixative for 16–18 h at 4°C. Brains were sectioned and placed in cryoprotectant (30% ethylene glycol, 20% glycerol, 50 mm sodium phosphate buffer, pH 7.4) at −20°C until further processing. Sections were briefly washed in sterile PBS, mounted on charged slides, and dried overnight. All sections for an experimental “run” were mounted and reacted on the same slide and thus experienced the same experimental conditions and solutions. After two rinses in sterile water, sections were incubated with “pretreat 4” from the RNAscope Multiplex Fluorescent Assay kit [Advanced Cell Diagnostics (ACD); RRID:SCR_012481] for 30 min at 40°C. Sections were rinsed twice in sterile water and incubated in RNAscope catalog oligonucleotide probes for Nmb, tyrosine hydroxylase (Th), pre-pro-Galanin (Gal), Pre-pro-enkephalin (Penk), Kcnk5, Gpr4, VGlut2 (Slc17a6), Fos, glutamate decarboxylase 1 (Gad1), Pituitary adenylate cyclase-activating polypeptide (PACAP; Adcyap1), or phenylethanolamine-N-methyltransferase (Pnmt) transcripts as listed in Table 1 for 2 h at 40°C. After incubation with probes, tissue was treated according to the manufacturer's protocol (ACD). When eGFP enhancement was required, sections were first subjected to the in situ hybridization (ISH) protocol and then rinsed 2 × 2 min in blocking buffer (10% horse serum, 0.1% Triton in 100 mm Tris buffer), incubated for 1 h in chicken GFP antibody (1:1000) in blocking buffer. Sections were rinsed 2 × 2 min in Tris buffer, incubated for 30 min in anti-chicken IgG tagged with AlexaFluor 488 (1:500) in Tris buffer, then rinsed and allowed to air dry. Slides were covered with Prolong Gold with DAPI Anti-fade mounting medium (Invitrogen).
Table 1.
Probes used for in situ hybridization
| Probe | ACD Catalog # | No. of pairs | Targeted region (sequence accession no.) |
|---|---|---|---|
| Neuromedin-B (mouse) | 459931(C1); 459931-C3 | 14 | bp 14–685 (NM_001291280.1) |
| Neuromedin-B (rat) | 494791(C1) | 13 | bp 2–651 (NM_001109149.1) |
| Tyrosine Hydroxylase | 317621(C1); 317621-C3 | 20 | bp 483–1603 (NM_009377.1) |
| Galanin | 400961-C3 | 12 | bp 43–652 (NM_010253.3) |
| Pre-pro-Enkephalin | 318761-C2 | 20 | bp 106–1332 (NM_001002927.2) |
| TASK-2 (Kcnk5) | 527951-C3 | 20 | bp 659–1827 (NM_021542.4) |
| Gpr4 | 427941(C1) | 20 | bp 866–1900 (NM_175668.4) |
| VGlut2 (Slc17a6) | 319171 (C1); 319171-C3 | 20 | bp 1986–2998 (NM_080853.3) |
| GAD1 | 400951 (C1) | 15 | bp 62–3113 (NM_008077.4) |
| PACAP (Adcyap1) | 405911-C2 | 20 | bp 702–1839 (NM_009625.2) |
| Fos (FBJ osteosarcoma oncogene) | 316921-C3 | 20 | bp 407–1427 (NM_010234.2) |
| Phox2b | 407861-C2 | 20 | bp 1642–2764 (NM_008888.3) |
| Phenylethanolamine-N-methyltransferase (PNMT) | 426421(C1) | 17 | bp 2–849 (NM_008890.1) |
Effect of hypercapnia or hypoxia on Fos expression by RTN neurons
Mice were placed in chambers designed for whole-body plethysmography (Data Sciences International; Kumar et al., 2015). Mass flow regulators provided quiet, constant, and smooth flow through the chamber (0.5 L/min) and delivered 21% O2 (balance N2) during the habituation period (1 h). Mice were then exposed for 35 min to either normoxia (21% O2; n = 11), hypercapnia (15% FiCO2/21% FiO2/balance N2; n = 7), or hypoxia (8% O2/balance N2; n = 5). Preliminary experiments showed that 8% O2 induced a large (∼4-fold) increase in sigh frequency, whereas less severe hypoxia (10% FiO2) had very little effect (<50% increase). Hypercapnic exposure was under normoxic conditions because preliminary experiments revealed that hyperoxia (30 min; 65% FiO2, no CO2 added; 2 mice), unlike normoxia, caused Fos expression in 10–15% of RTN neurons. In humans, sustained hyperoxia produces a mild hyperventilation and arterial hypocapnia consistent with the possibility that central respiratory chemoreceptors might be activated (Becker et al., 1996). Our evidence suggests that this might be the case in mice as well. After exposure to 35 min of hypercapnia, hypoxia, or normoxia, mice were immediately anesthetized and perfused transcardially with fixative as described above. All experiments, including the histology, were run in pairs (a normoxia control and a mouse exposed to hypercapnia or hypoxia). All plethysmography experiments were conducted between 12:00 and 5:00 P.M.
Neuron mapping and counting
A one-in-three or one-in-six series of 30 μm transverse sections through the brain was examined for each experiment under bright-field and epifluorescence using a Zeiss AxioImager Z.1 or a Zeiss AxioImager M2 microscope (Carl Zeiss Microimaging). Neurons were plotted with the Neurolucida software (Micro Brightfield; RRID:SCR_001775) using a Ludl motor driven microscope stage and a Zeiss MRC or Hamamatsu C11440 Orca-Flash 4.0LT digital camera, after methods previously described (Stornetta et al., 2004). Filter settings for AlexaFluor 488, Atto 550, and Atto 647 fluorophores were as follows: AlexaFluor 488, excitation of 500 nm, emission of 535 nm; Atto 550, excitation of 545, emission of 605 nm; Atto 647, excitation of 640 nm, emission of 690 nm. Only cell profiles that included a nucleus were counted and/or mapped. Sections were matched as closely as possible to brain levels with reference to bregma using the atlas of Paxinos and Franklin (2013). Cells were counted and mapped bilaterally. Most mapping was limited to the ventral half of the brainstem which contains the distinctive and isolated parafacial cluster of Nmb+ neurons.
Photographs were taken with a Hamamatsu C11440 Orca-Flash 4.0LT digital camera (resolution 2048 × 2048 pixels) and the resulting TIFF files were first exported into Fiji (RRID:SCR_002285) and unsharp mask filter and/or brightness/contrast adjusted for clarity and to reflect true rendering as much as possible. Images were not otherwise altered. TIFF images were imported into Canvas v10 (ACD; RRID:SCR_014312) for labeling and final presentation. The neuroanatomical nomenclature is after Paxinos and Franklin (2013).
Single-cell transcriptome of RTN neurons
For single-cell collection, we adapted previously described procedures to harvest eGFP-labeled neurons from brainstem slices obtained from JX99 mice (Kumar et al., 2015; Shi et al., 2016), specifically targeting cells in the region ventrolateral to the facial nucleus to bias our sampling toward obtaining RTN neurons and avoiding other Phox2b-expressing cell types (e.g., C1 neurons, facial motor neurons). Briefly, mice (n = 22 with both sexes represented, P11–P120) were anesthetized with ketamine and xylazine (375 mg/kg and 25 mg/kg, i.m.) and rapidly decapitated; brainstems were immediately removed and slices (300 μm) cut with microslicer (DTK Zero 1; Ted Pella) in an ice-cold, sucrose-substituted Ringer's solution containing the following (in mm): 260 sucrose, 3 KCl, 5 MgCl2, 1 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, 10 glucose, and 1 kynurenic acid. Slices were incubated for 30 min at 37°C, and subsequently at room temperature, in normal Ringer's solution containing the following (in mm): 130 NaCl, 3 KCl, 2 MgCl2, 2 CaCl2, 1.25 NaH2PO4, 26 NaHCO3, and 10 glucose. The cutting and incubation solutions were bubbled with 95% O2 and 5% CO2. Slices were prepared in the early-to-late morning, and used for harvesting neurons from the late morning through the afternoon.
Single RTN neurons were collected for molecular analysis in a recording chamber mounted on a fluorescence microscope (Zeiss Axio Imager FS) in HEPES-based perfusate, containing the following (in mm): 140 NaCl, 3 KCl, 2 MgCl2, 2 CaCl2, 10 HEPES, 10 glucose. Under direct vision (60× objective), viable cells were identified and the entire cell was aspirated into pre-baked pipettes (24 h, 200°C) filled with sterile HEPES-based solution. The pipette was broken into a 0.2 ml RNase-free PCR tube containing ice-cold cell lysis mix before performing RNA reverse transcription and cDNA amplification using the SMART-Seq v4 kit, according to the manufacturer's instructions (Clontech, catalog #634896). Each amplification experiment included 10 pg positive control RNA (from SMART-Seq kit) and a water-only negative control. cDNA concentration was quantified using Qubit dsDNA High-Sensitivity Assay Kit (ThermoFisher Scientific, catalog #Q32854). To select single-cell cDNA samples suitable for library construction, we predetermined expression of Gapdh, VGlut2, Phox2b, Th, and Gad1 in each sample by nested PCR; samples positive for Th and/or Gad1 were deemed to be contaminated with other, non-RTN neurons. We used 0.5–1 ng of the amplified cDNA and the Nextera XT DNA Library kit (Illumina, catalog #FC-131-1096; RRID:SCR_010233) to prepare dual-indexed libraries that were assessed for concentration and library quality using Qubit and the Agilent High Sensitivity D1000 TapeStation. These libraries (12–40 per batch) were pooled in a single lane on an Illumina NextSeq500, and sequenced to generate 75 bp paired-end reads. FASTQ files from five sequencing runs were quasi-mapped to the mouse reference transcriptome (Ensembl GRCm38) as described by Patro et al. (2017), with 3–14 million reads per library (50–80% mapped). Transcript abundance was summarized to the gene level normalized as transcripts per million (TPM), according to Soneson et al. (2015). In two early sequencing runs, 12 cell samples were included per sequencing lane often yielding >10 × 106 reads per cell; because rarefaction analysis of these runs indicated a negligible loss in gene representation when reads were reduced to <1 × 106 reads per cell (<5%), we assayed greater numbers of cells per lane (32–40) at lower sequencing depth (3–5 × 106 reads) in three subsequent runs.
Statistics
Values are presented as means ± SEM, or as medians with ranges, as appropriate. Either one-way or two-way ANOVA or t tests (both parametric or nonparametric) were performed as appropriate after verification of normality assumptions for these tests. TPM data were not normally distributed, and were analyzed using a Kruskal-Wallis ANOVA. All statistical tests were done using PRISM v7 (GraphPad Software).
Results
Distribution of Nmb transcripts in the rostral medulla and pons of mice
By multiplex ISH, we observed a distinctive and isolated cluster of cells containing Nmb transcripts in the parafacial region of the rostral ventrolateral medulla oblongata and pons (Fig. 1A,E). Only two additional groups of Nmb cells (see below) were identified in the pons and rostral medulla oblongata. Both were located in the dorsal third of the brainstem and were clearly separated from the parafacial cluster.
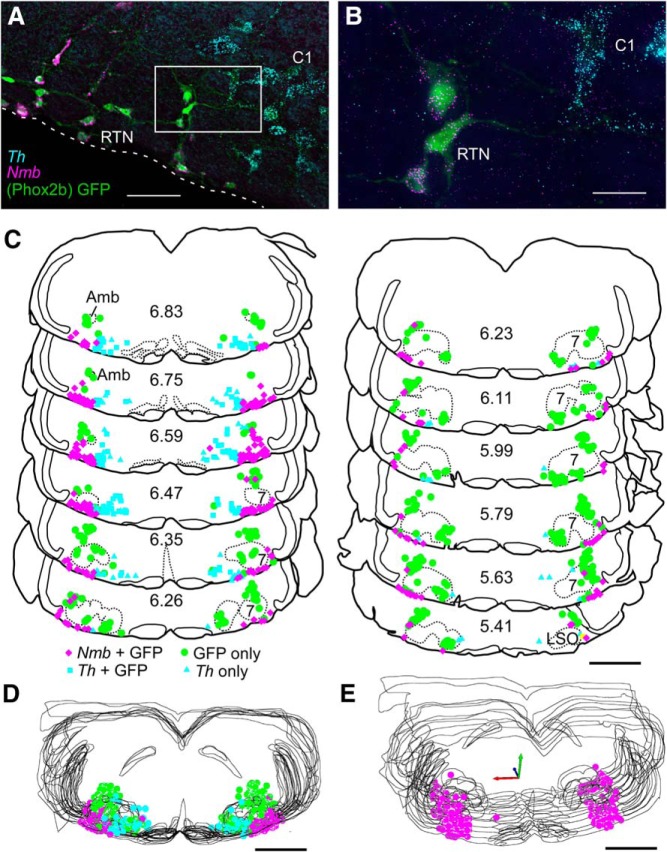
Figure 1.
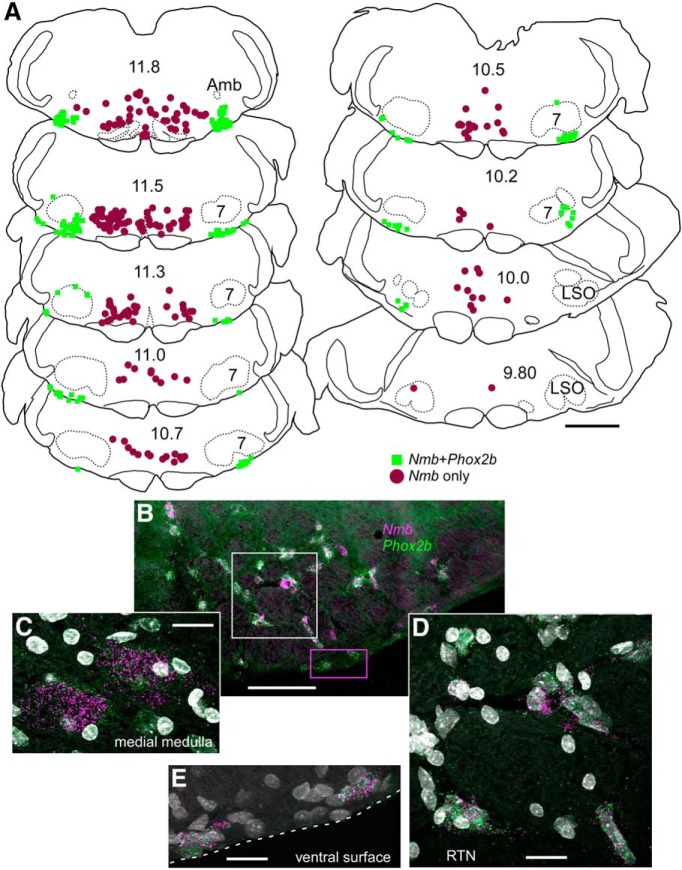
Nmb-positive parafacial neurons express Phox2b but are not catecholaminergic. A, Photomicrograph of a coronal section from a Phox2b::eGFP mouse with eGFP-labeled RTN neurons (green; Phox2b) containing Nmb transcripts (magenta); these neurons do not contain transcripts for Th, unlike the nearby C1 adrenergic cells (Th; blue). Medial to the right, dorsal toward the top. White dashed line represents the ventral medullary surface. B, Enlargement of white boxed area in A. C, Computer stage-assisted drawings of coronal sections showing the distribution of Phox2b-only (green filled circles), Th-only (blue triangles), Th + Phox2b (blue squares), and Nmb + Phox2b neurons (magenta diamonds) in ventral half of the brainstem through the rostral medulla and pons. Approximate millimeters behind bregma (after Paxinos and Franklin, 2013) indicated by numbers in lower center of each section. Abbreviations: 7, Facial motor nucleus; Amb, ambiguus nucleus; LSO, lateral superior olive. D, Sections from C collapsed in a stack to give a general representation of the location of Nmb neurons as ventrolateral to Th as well as Phox2b-only neurons in the RTN region. Nmb in magenta, Th in blue, Phox2b-only in green. E, 3-D representation of Nmb neuron distribution (magenta dots) through the RTN in medulla/pons. Note the bulk of the Nmb neurons are ventral and lateral to the facial motor nucleus. Scale bars: A, 60 μm; B, 20 μm; C–E, 1 mm.
The parafacial group of Nmb+ cells was observed from ∼350 μm caudal to the caudal pole of the facial motor nucleus, extending rostrally to the exit of the seventh nerve (Fig. 1C). Caudal to the facial motor nucleus, Nmb-expressing cells clustered predominantly along the ventral medullary surface but a few were scattered more dorsally within the ventrolateral medulla, approaching the level of nucleus ambiguus. At this caudal level, where C1 adrenergic neurons are also located, most Nmb cells were located ventrolateral to those Th mRNA-expressing neurons, with minimal overlap between the two groups of cell soma (Fig. 1A,C). More rostrally, the bulk of the Nmb cells resided below the facial motor nucleus from its medial border to the medial edge of the trigeminal tract. At these rostral levels, Nmb cells tended to congregate close to the trigeminal tract, with a few of these neurons found around the lateral edge of, and dorsal to, the facial motor nucleus (Fig. 1C).
Using one-in-three or one-in-six series of 30 μm sections from 37 mice (17 males, 20 females), cells containing Nmb transcripts were counted bilaterally within each section. Averages were taken across each bregma level and then added to determine a total average number of cells containing Nmb transcripts (Table 2). Using this method, we counted 266.4 ± 7.2 Nmb cells per mouse averaged across all mice (males: 269.9 ± 10.0; females: 262.9 ± 9.4; Fig. 2; Table 2). The number of Nmb cells and their distribution across the medulla/pons was not significantly different between sexes (Fig. 2; two-way ANOVA; main effect of sex: F(1, 328) = 0.21, p = 0.65; interaction between sex and bregma level: F(12, 328) = 0.92, p = 0.53). Combining data from both sexes, and after Abercrombie correction using an average nuclear width of 6.0 ± 0.2 μm measured from 50 cells in 3 mice (Abercrombie, 1946), we calculate an average total number of Nmb RTN cells in the mouse at 665 cells.
Table 2.
Counts of all cells containing Nmb transcripts in the parafacial area for 37 mice
| Case | Sex | Condition | Caudal to bregma, mm |
Total | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6.74 | 6.65 | 6.56 | 6.47 | 6.38 | 6.29 | 6.2 | 6.11 | 6.02 | 5.93 | 5.84 | 5.75 | 5.66 | ||||
| 22305 | F | Control | 17 | 36 | 63 | 48 | 9 | 11 | 7 | 14 | 7 | 10 | ||||
| 22503 | F | Control | 13 | 21 | 37 | 16 | 14 | 21 | 15 | 15 | 14 | 18 | 24 | 17 | ||
| 22603 | F | Hyperoxic CO2 | 6 | 32 | 38 | 42 | 17 | 17 | ||||||||
| 22609 | F | CO2 | 61 | 33 | 32 | 22 | 8 | 3 | 14 | |||||||
| 23205 | F | CO2 | 13 | 34 | 65 | 36 | 19 | 23 | 11 | 9 | 28 | 32 | 23 | |||
| 26100 | F | CO2 | 5 | 22 | 28 | 58 | 12 | 16 | 18 | 18 | 23 | 16 | 12 | |||
| 26101 | F | Control | 44 | 74 | 45 | 18 | 11 | 9 | 7 | 14 | 9 | 3 | ||||
| C5722203 | F | Control | 14 | 35 | 64 | 69 | 31 | 12 | 11 | 23 | 12 | 16 | 23 | 11 | ||
| C5723204 | F | Control | 36 | 45 | 45 | 31 | 31 | 19 | 22 | 11 | 14 | 15 | 6 | |||
| C5724104 | F | Control | 8 | 21 | 49 | 34 | 20 | 16 | 7 | 9 | 18 | 11 | ||||
| JX99-3407 | F | Control | 20 | 75 | 18 | 14 | 26 | 5 | ||||||||
| JX99-3408 | F | Control | 48 | 13 | 9 | 8 | 9 | 0 | ||||||||
| JX99-3409 | F | CO2 | 59 | 41 | 18 | 12 | 17 | 4 | ||||||||
| JX99-3502 | F | CO2 | 12 | 32 | 37 | 11 | 6 | 2 | ||||||||
| jx992301 | F | Control | 13 | 33 | 13 | 37 | 12 | 16 | 14 | 9 | 37 | 11 | 7 | |||
| jx992303 | F | Control | 4 | 15 | 54 | 75 | 65 | 21 | 19 | 17 | 10 | 6 | ||||
| jx992305 | F | Control | 10 | 32 | 33 | 30 | 13 | 17 | 15 | 11 | 8 | 9 | ||||
| jx993303 | F | Control | 35 | 48 | 19 | 15 | 7 | 6 | 4 | 17 | 12 | 24 | ||||
| jx993305 | F | Control | 20 | 97 | 40 | 23 | 16 | 6 | 11 | 11 | 18 | 22 | 19 | 2 | ||
| jx993306 | F | Control | 20 | 24 | 38 | 16 | 8 | 17 | 18 | 20 | 10 | 8 | 2 | |||
| AVG | F | All | 9.6 | 24.9 | 46.5 | 47.4 | 28.3 | 19.0 | 13.6 | 12.6 | 11.8 | 17.4 | 13.9 | 13.2 | 4.6 | 262.9 |
| SEM | F | All | 1.3 | 2.1 | 4.9 | 4.7 | 3.6 | 1.8 | 1.0 | 1.3 | 1.6 | 2.0 | 1.6 | 2.5 | 1.4 | 9.4 |
| 25100 | M | Hypoxia | 9 | 23 | 31 | 39 | 42 | 18 | 9 | 15 | 20 | 16 | 26 | |||
| 25101 | M | Hypoxia | 28 | 55 | 61 | 31 | 14 | 7 | 11 | 14 | 12 | 15 | 21 | 13 | ||
| 25102 | M | Hypoxia | 43 | 46 | 31 | 20 | 15 | 12 | 16 | 16 | 20 | 31 | 16 | 1 | ||
| 25103 | M | Control | 12 | 30 | 30 | 7 | 10 | 15 | 20 | 17 | 25 | 3 | ||||
| C5700 | M | Control | 0 | 24 | 10 | 5 | 1 | |||||||||
| C5701 | M | CO2 | 24 | 15 | 9 | 18 | 15 | 4 | ||||||||
| C5702 | M | Control | 12 | 31 | 15 | 12 | 6 | 5 | ||||||||
| C5703 | M | CO2 | 3 | 35 | 25 | 16 | 4 | 4 | ||||||||
| C5722502 | M | Control | 6 | 19 | 47 | 52 | 24 | 28 | 16 | 20 | 20 | 7 | 12 | 31 | ||
| JX99-6410 | M | Hypoxia | 6 | 18 | 40 | 50 | 31 | 12 | 8 | 27 | 19 | 12 | 17 | 14 | ||
| JX99-6494 | M | Hypoxia | 27 | 69 | 35 | 45 | 18 | 10 | 15 | 8 | 20 | 15 | 17 | 9 | ||
| JX99-6640 | M | Hypoxia | 43 | 56 | 22 | 18 | 13 | 8 | 20 | 25 | 20 | 24 | ||||
| JX99-7010 | M | Hypoxia | 32 | 57 | 64 | 26 | 14 | 6 | 9 | 19 | 15 | 18 | 33 | 2 | ||
| JX99-7011 | M | Control | 24 | 23 | 32 | 26 | 18 | 2 | 8 | 14 | 12 | 15 | 4 | |||
| jx992401 | M | Control | 5 | 13 | 23 | 64 | 27 | 45 | 35 | 18 | ||||||
| jx992402 | M | Control | 4 | 14 | 48 | 82 | 47 | 21 | 32 | 33 | 21 | 22 | ||||
| jx996031 | M | Control | 19 | 37 | 86 | 41 | 17 | 15 | 12 | 21 | 17 | 29 | 46 | |||
| AVG | M | All | 5.6 | 22.7 | 39.9 | 48.1 | 28.6 | 16.9 | 12.0 | 16.2 | 16.9 | 18.1 | 17.3 | 22.6 | 5.1 | 269.9 |
| SEM | M | All | 1.3 | 2.6 | 3.3 | 5.4 | 2.9 | 1.3 | 1.7 | 2.2 | 2.3 | 1.9 | 2.2 | 4.3 | 1.6 | 10.1 |
| Grand AVG | Both | All | 7.6 | 23.8 | 43.2 | 47.8 | 28.5 | 18.0 | 12.8 | 14.4 | 14.3 | 17.7 | 15.6 | 17.9 | 4.9 | 266.4 |
| Grand SEM | Both | All | 2.0 | 1.1 | 3.3 | 0.3 | 0.1 | 1.0 | 0.8 | 1.8 | 2.5 | 0.3 | 1.7 | 4.7 | 0.3 | 7.2 |
AVG, Average; CO2, 15% CO2; F, female; M, male; hypoxia, 8% O2.
Figure 2.
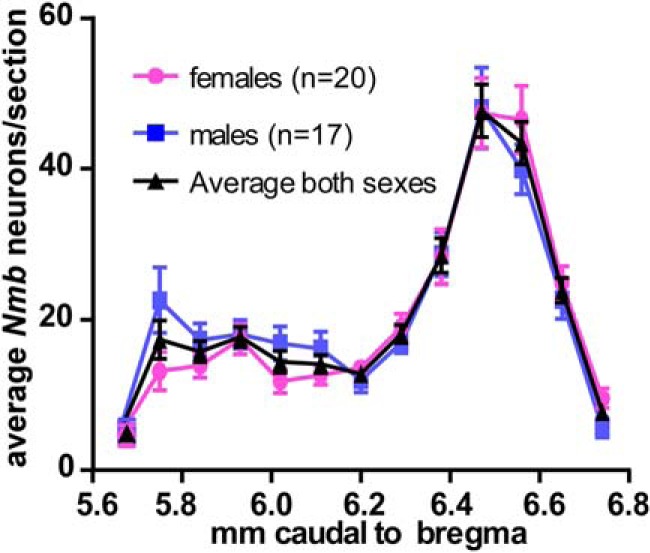
Distribution of parafacial Nmb neurons. Average total numbers of parafacial Nmb-expressing neurons per 30-μm-thick transverse section (SEM indicated by vertical bars). Abscissa indicates the location of the sampled tissue sections (distance from bregma after Paxinos and Franklin, 2013).
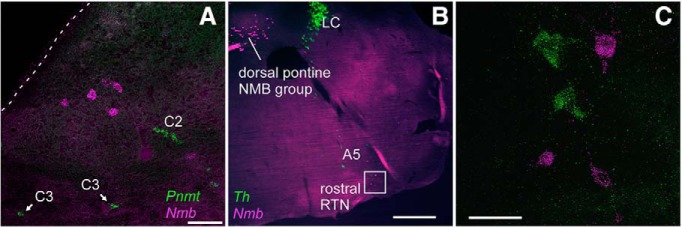
A sparse collection of Nmb cells was present dorsomedial to the prepositus nucleus. These Nmb neurons were located generally medial to the C2 neurons and dorsal to the C3 neurons and, unlike the latter, they did not express Pnmt (Fig. 3A). A second and much larger cluster of Nmb cells was located in the region of the central gray and posterodorsal tegmental nucleus medial to the locus ceruleus (Fig. 3B), from 5.63 to 5.41 mm caudal to bregma (coordinates after the corresponding plates in the atlas of Paxinos and Franklin, 2013) and did not express Th.
Figure 3.
Nmb cells in rostral parafacial region and dorsal pons are not catecholaminergic. A, Transcripts for Pnmt (green) and Nmb (magenta) in dorsal medial medulla at level of C2/C3 adrenergic cell groups. B, Transcripts for Th (green) and Nmb (magenta) in more rostral pontine brain regions. LC, locus coeruleus. C, Enlargement of the white box (rostral RTN) in A. Scale bars: A, 100 μm; B, 500 μm; C, 50 μm.
Parafacial Nmb cells are glutamatergic, non-catecholaminergic neurons that express Phox2b
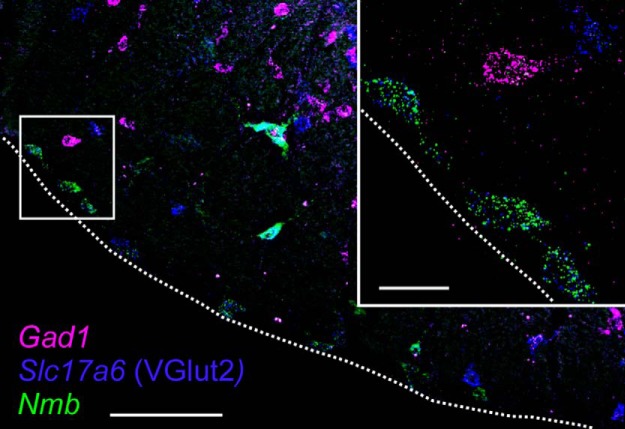
We noted that Nmb and Th cell bodies were non-overlapping in the parafacial region, an observation that was borne out by quantitative analysis of these multiplex in situ hybridization experiments: virtually none of the Nmb neurons expressed Th (only 2/802 cells, counted in 3 mice; Figs. 1, 3). By contrast, essentially all parafacial Nmb neurons expressed Phox2b, as indicated by eGFP staining in JX99 mice (99.6%, 799/802 cells in 3 mice; Fig. 1). Likewise, every Nmb parafacial neuron contained transcripts for VGlut2 (Slc17a6; 613/613 cells, counted in 2 mice), whereas none expressed Gad1 (796/796 cells, counted in 3 mice; Fig. 4). Based on these results, parafacial Nmb neurons meet precisely the criteria used previously to define the RTN cell group; specifically, they are non-catecholaminergic (i.e., TH-negative) neurons located in the parafacial region that coexpress Phox2b and VGlut2 mRNA (Stornetta et al., 2006; Goridis et al., 2010; Ramanantsoa et al., 2011; Ruffault et al., 2015).
Figure 4.
Parafacial Nmb neurons are glutamatergic but not GABAergic. Photomicrograph of coronal section of caudal RTN region with Nmb transcripts in green, VGlut2 transcripts in blue and Gad1 transcripts in magenta. Inset, Enlargement of boxed area with three RTN Nmb neurons along ventral surface that also express VGlut2 transcripts. Note that all neurons that contain Nmb transcripts, including the larger and more dorsal cells that express high levels of Nmb, also contain VGlut2 transcripts; none of the Nmb neurons contain Gad1 transcripts. Dashed white lines represent the ventral medullary surface. Scale bars: larger image, 100 μm; inset, 20 μm.
These ISH data were further validated by RNA-Seq results from 105 single Phox2b-expressing (i.e., GFP-fluorescent) neurons collected from the parafacial region of JX99 mice. From this dataset, we excluded one cell for which the sequencing results were low quality (<650,000 mapped reads). We also excluded six neurons that were clearly C1 neurons, because they expressed transcripts for catecholamine synthesizing enzymes at values much higher than found for those same genes in the remaining sample of 98 non-C1 neurons (median TPM: for Th, 1451 vs 0; for Ddc, 114 vs 33.4; for Dbh, 422 vs 0; and for Pnmt, 326 vs 0); importantly, and consistent with histochemical data, none of the C1 neurons were positive for Nmb (all TPM <20). From the remaining 98 non-C1 neurons, we excluded samples that showed higher than expected levels of either Gad1 or Gad2 (TPM > 30; n = 8 and n = 2). Although it is formally possible that the presence of Gad1/Gad2 represents true expression of these GABA-synthesizing enzymes in a minor subgroup of RTN neurons (Hayes et al., 2017), we think it is more likely to reflect contamination of these particular samples, a common concern in single-cell molecular studies and a conservative interpretation consistent with prior data designating RTN neurons as excitatory rather than inhibitory neurons (Bochorishvili et al., 2012; Holloway et al., 2015). That Gad1 mRNA was undetectable by in situ hybridization, but highly expressed in neighboring Nmb-negative neurons, also supports this view (Fig. 4).
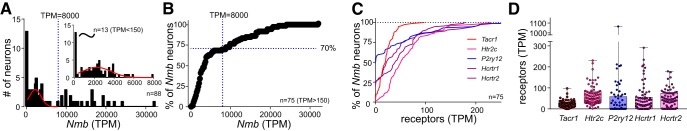
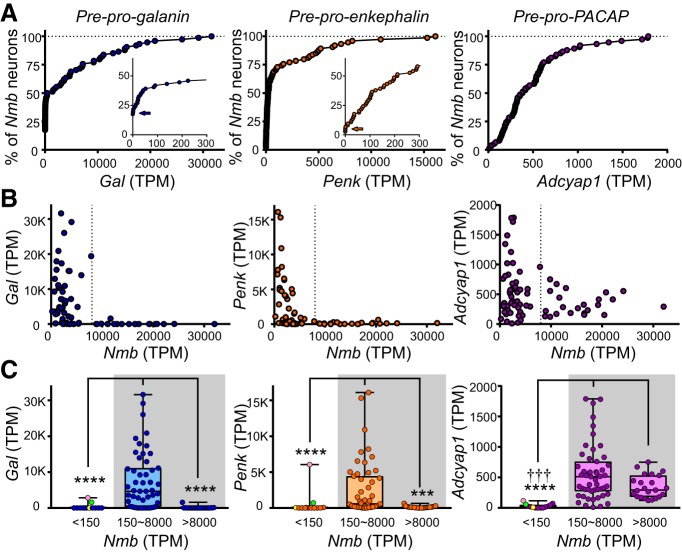
All of the remaining 88 neurons were positive for both Phox2b and VGlut2 mRNA (Slc17a6), and 75 showed appreciable levels of Nmb expression (Fig. 5A; operationally defined as TPM > 150). We expect that the small group of Nmb-negative neurons (n = 13; TPM < 150) present in our GFP-targeted sample likely correspond to the population of Phox2b+, Nmb− neurons encroaching on the dorsal and/or rostral boundaries of the RTN proper (Fig. 1C,D, green circles) because they present with a generally distinct neuropeptide and proton sensor phenotype (see Figs. 7C, 9D). Among the larger group of Nmb-expressing RTN neurons in our sample (n = 75), Nmb transcript levels were widely variable (median: 3,523; range: 224–32,022), and appeared to represent two populations: ∼70% of the neurons were approximately normally distributed with low-to-moderate Nmb transcript levels (150 < TPM < 8,000), whereas a clear inflection point in the cumulative probability plot at TPM ∼8,000 revealed a separate subgroup of ∼30% of RTN neurons that express particularly high levels of Nmb (Fig. 5B).
Figure 5.
Two populations of parafacial Nmb neurons based on expression levels. A, B, Frequency (A) and cumulative probability (B) distributions for Nmb transcript levels assessed by RNA-Seq in individual GFP-expressing neurons from the RTN region (n = 88). A subset of neurons expressed extremely low levels of Nmb (TPM < 150, n = 13). Among the Nmb-expressing neurons (n = 75), a major subgroup of neurons contains moderate levels of Nmb transcripts (∼70%; TPM < 8000), whereas a smaller subgroup of cells express much higher levels of Nmb (∼30%; TPM > 8000). C, Cumulative probability distributions of transcript levels for select G-protein-coupled receptors in Nmb-expressing neurons; the percentage of non-expressing neurons can be estimated from the y-intercept (TPM = 0). D, Expression levels for the indicated receptors in individual Nmb-expressing RTN neurons; in this and other plots of TPM data, the box-and-whiskers represent the median, 25th and 75th percentile, and the range.
Figure 7.
Transcript expression levels for Gal, enkephalin, and pituitary adenylate cyclase-activating peptide in Nmb-expressing RTN neurons. A, Cumulative probability distributions of expression levels for the indicated neuropeptide transcripts in Nmb-expressing neurons; the percentage of non-expressing neurons can be estimated from the y-intercept (TPM = 0; insets for Gal and Penk, arrows). B, Transcript levels for Gal, Penk, and Adcyap1, relative to Nmb, in the same individual RTN neurons. C, Transcript expression levels for Gal, Penk, and Adcyap1 in Nmb-low (150 < TPM < 8000) and Nmb-high (TPM > 8000) RTN neurons. Data from Phox2b-positive neurons that were below the operational threshold for consideration as Nmb-expressing RTN neurons (TPM < 150) are also depicted. Note that Gal and Penk expression is undetectable in the Nmb-high subpopulation of RTN neurons (TPM > 8000); it is also virtually nonexistent in the Phox2b-positive, Nmb-negative neurons (two outliers are marked). ****p < 0.0001, ***p < 0.001 versus Nmb-low RTN neurons (150∼8000); ††† p < 0.001 versus Nmb-negative neurons (TPM < 150) by Kruskal-Wallis ANOVA.
Figure 9.
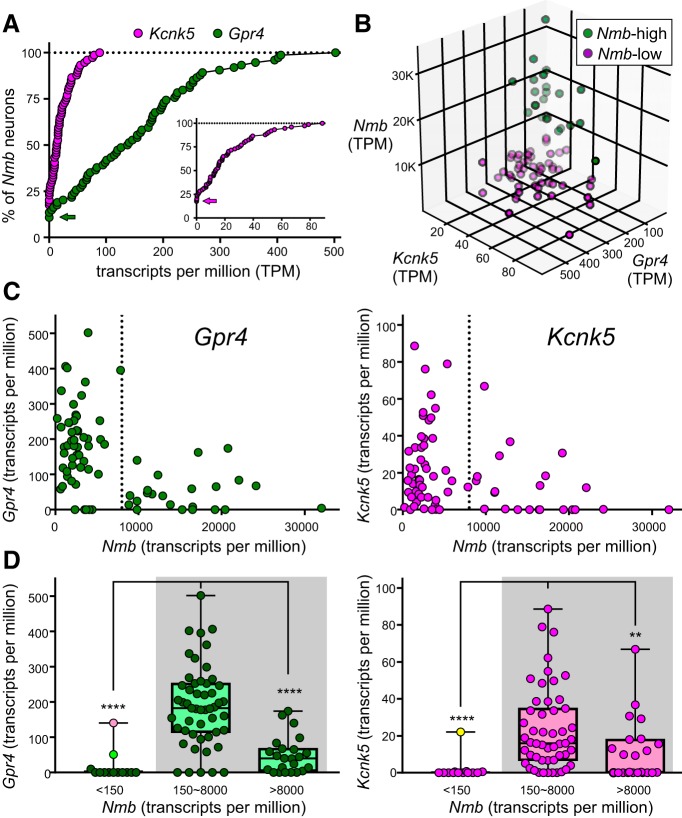
Gpr4/Kcnk5 transcript levels are lower in the Nmb-high subgroup of parafacial neurons. A, Cumulative probability distributions of expression levels for the indicated proton sensor in Nmb-expressing neurons; the percentage of non-expressing neurons can be estimated from the y-intercept (see arrows; inset is expanded for Kcnk5). B, 3-D distribution of Gpr4, Kcnk5, and Nmb expression shows that within both Nmb-high (green) and Nmb-low (purple) populations, a few parafacial neurons predominantly express higher levels of either Gpr4 or Kcnk5. For example, see cells extending outward on the plane of the right rearmost wall (high Kcnk5, low Gpr4) or extending outward on the plane of the left rearmost wall (high Gpr4, low Kcnk5). C, Transcript levels for Gpr4 and Kcnk5 relative to Nmb in the same individual parafacial neurons. D, Transcript expression levels for Gpr4 and Kcnk5 in Nmb-low (150 < TPM < 8000) and Nmb-high (TPM > 8000) RTN neurons. Data from cells that were below the operational threshold for consideration as Nmb-expressing RTN neurons (TPM < 150) are also depicted. Note that Gpr4 and Kcnk5 expression is significantly lower in the Nmb-high subpopulation of RTN neurons (TPM > 8000), by comparison to Nmb-low RTN neurons; it is also virtually nonexistent in the Phox2b-positive, Nmb-negative neurons (outliers are marked, color-coded as in Fig. 7). ****p < 0.0001, **p < 0.01 versus Nmb-low RTN neurons (150∼8000) by Kruskal-Wallis ANOVA.
A number of other transcriptomic features of individual Nmb-expressing parafacial neurons are consistent with those expected for RTN neurons. For example, in developmental genetic analyses, the substance P receptor, NK1R, has been used to define RTN (Thoby-Brisson et al., 2009). Accordingly, we find expression of the cognate gene, Tacr1, in essentially all Nmb-expressing neurons (Fig. 5C,D; in 97.3% of neurons, n = 73/75; TPM median: 19.7; range: 0–98). We also find varying levels of expression for other G-protein-coupled receptors that have been associated previously with RTN neurons (Mulkey et al., 2007; Lazarenko et al., 2011; Barna et al., 2016), as evident in the transcript distributions for the most prominent receptor subtypes in Nmb-positive cells (Fig. 5C,D): the 5-HT2C receptor is universally expressed (Htr2c, TPM median: 51.1; range: 2–231); transcripts for the P2Y12 receptor are found at appreciable levels, albeit in only ∼40–50% of the neurons (P2ry12, TPM median: 0.7; range: 0–1065); and those for each of the orexin/hypocretin receptors are present in ∼75% of cells (Hcrtr1, TPM median: 23.0; range: 0–291; Hcrtr2, TPM median: 40.1; range: 0–179), with only ∼10% of neurons lacking either orexin receptor subtype. Conversely, these Nmb-expressing neurons were virtually devoid of transcripts for the tryptophan hydroxylase enzymes (TPM < 10 for Tph1 and Tph2; n = 75 and n = 74) and the serotonin transporter (TPM < 10 for Slc6a4; n = 75); they were also negative for markers of inhibitory neurons, such as glycine transporter 2, GlyT2, and the GABA vesicular transporter, Vgat (TPM < 10 for Slc6a5 and Slc32a1; n = 73 and n = 75).
In sum, our histochemical and transcriptomic analysis indicates that Phox2b+/Slc17a6+ parafacial neurons that have characteristics of RTN neurons uniformly express Nmb; this was not true of other Phox2b-expressing neurons in the vicinity, such as those located dorsal and/or rostral to the RTN, or the C1 adrenergic neurons. A quantitative analysis of Nmb transcript levels revealed two subgroups of RTN neurons: a larger group (∼70%) with low-to-moderate Nmb expression (henceforth Nmb-low, TPM: 150∼8000) and a smaller group (∼30%) with much higher Nmb expression (Nmb-high, TPM > 8000). We next sought to determine whether these different Nmb-expressing subpopulations were preferentially associated with other genes that had previously been identified in RTN neurons, or perhaps with distinct functional characteristics.
RTN Nmb neurons express additional neuropeptides pre-pro-galanin and pre-pro-enkephalin
Gal could be identified by ISH in 70 ± 2% of Nmb-expressing RTN neurons (n = 4 mice; Fig. 6A,B,E,F). Gal+/Nmb+ neurons were scattered among the Nmb neurons devoid of Gal mRNA without discernible rostrocaudal or mediolateral clustering. The percentage of RTN Gal+ neurons was slightly higher than our previous estimate of 50% in the rat (Stornetta et al., 2009). This could be due to either species differences or the use of the more sensitive RNAscope technique in the current study. When assessed by single-cell RNA-Seq, Gal was detected in 82.7% of the Nmb-expressing neurons (n = 62/75). However, there was marked variability in Gal expression levels in individual neurons (median TPM: 417; range: 0–31,618). Notably, although Gal was not universally expressed in the Nmb-low population of RTN neurons (TPM < 8000), it was always extremely low or absent within the RTN subpopulation expressing the highest levels of Nmb (TPM > 8000; Fig. 7).
Figure 6.
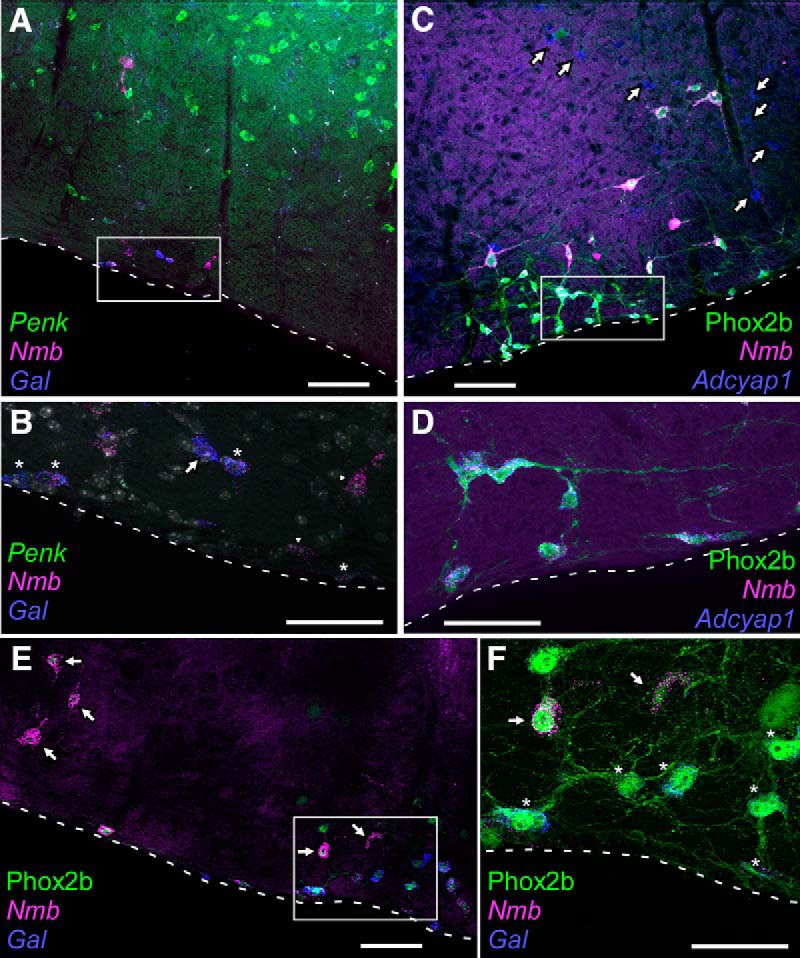
Parafacial Nmb neurons also contain other neuropeptides, including enkephalin, galanin and pituitary adenylate cyclase-activating peptide. A, Photomicrograph of coronal section showing transcripts for Nmb (magenta), Penk (green), and Gal (blue) in the caudal RTN region (level 6.7–6.8 from Fig. 1C). Note the widespread distribution of Penk within the reticular formation dorsal to RTN. Medial to the right, dorsal toward the top. B, Enlargement of the boxed inset from A showing Nmb neurons with both Penk and Gal (stars), an Nmb neuron with Gal and not Penk (arrow) and neurons in which only Nmb was detectable (arrowheads). Nuclei stained with DAPI (white/gray). C, Photomicrograph showing transcripts for Nmb (magenta) and Adcyap1 (blue) in eGFP-immunoreactive (i.e., Phox2b) neurons (green). Note that Adcyap1 transcripts are also expressed outside the RTN (arrows). D, Enlargement of boxed area in C. Note that all Nmb neurons express Adcyap1. Medial to the left, dorsal toward the top. E, Photomicrograph of parafacial regions showing eGFP-immunoreactivity (i.e., Phox2b; green), Nmb transcripts (magenta) and Gal transcripts (blue) neurons. Every neuron is Phox2b. Large neurons with high levels of Nmb transcripts (arrows) do not contain Gal. F, Enlargement of boxed area in E. Nmb-high neurons (arrows) without Gal are generally dorsal to the Nmb-low neurons with Gal (stars). Dashed white lines on all panels represent the ventral medullary surface. Scale bars: A, 100 μm; B, 50 μm; C, 100 μm; D, 50 μm; E, 100 μm; F, 50 μm.
A similar result was obtained with Penk (Fig. 6A,B). By ISH, Penk transcripts were observed in 67 ± 8% of parafacial Nmb neurons throughout the RTN region, with no obvious preferential localization along its rostrocaudal or mediolateral extent (n = 4 mice). By single-cell RNA-Seq, Penk transcripts could be detected in 97.3% of Nmb-expressing neurons (n = 73/75). As with Gal, a broad range of Penk expression was observed across individual Nmb-expressing neurons, although with a lower peak level of expression (median TPM: 212; range: 0–16,089). Also similar to Gal, a striking inverse correlation was observed between Penk and Nmb expression, such that the highest Nmb-expressing neurons were always essentially devoid of Penk transcripts (Fig. 7).
Adcyap1
By ISH, Adcyap1 transcripts were present in a large proportion of the RTN Nmb neurons (88 ± 4%, n = 4), again distributed without discernible clustering throughout the nucleus (Fig. 6C,D). By RNA-Seq, Adcyap1 transcripts could be detected in all Nmb-expressing RTN neurons (n = 75/75). The overall transcript levels were lower than for Gal and Penk (median TPM: 387; range: 12–1787) and, unlike with Gal and Penk, Adcyap1 expression was clearly evident even in cells with the highest levels of Nmb (Fig. 7).
It is also worth mentioning that these neuropeptides were rarely detected in the group of non-C1, Phox2b-positive neurons with Nmb expression below our operational threshold (i.e., with TPM < 150, n = 13), which were judged to represent a distinct population (Fig. 7C). Interestingly, however, the two neurons in this group with appreciable amounts of Gal (TPM: 2846, 1573), Penk (TPM: 6062, 700), and Adcyap1 transcripts (TPM: 117, 57) were also positive for Gpr4 (see below), suggesting that they may indeed be RTN neurons, but at the lower extreme of Nmb expression. Even so, the neuropeptide expression data from these outliers are consistent with the general observation that Gal and Penk expression is limited to the Nmb-low group of RTN neurons.
Collectively, this assessment of neuropeptide expression is consistent with the suggestion that RTN comprises phenotypically distinct subgroups of Nmb-expressing neurons that can be differentiated by either high or low levels of Nmb expression.
Most RTN Nmb neurons express Kcnk5 and Gpr4
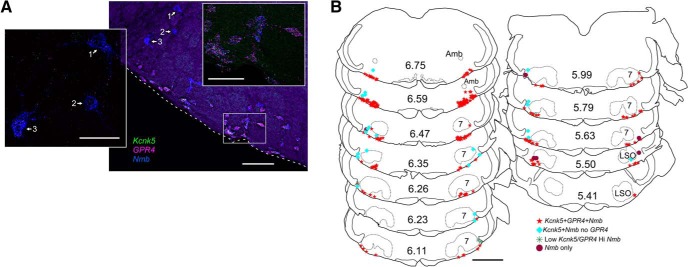
A large but still unspecified proportion of RTN neurons are intrinsically sensitive to CO2/H+; this property is mediated in large part by either (or both) of two distinct pH sensors: TASK-2 (encoded by Kcnk5) and Gpr4. Indeed, by ISH, we found that a major fraction of parafacial Nmb neurons contained transcripts for Kcnk5 (92 ± 3%, n = 6 mice) or Gpr4 (82 ± 1%, n = 25 mice; Fig. 8). However, the neurons that contained high levels of Nmb usually had little or no detectable Gpr4 or Kcnk5 (Fig. 8A). Nmb-high neurons were rarely found on the ventral medullary surface; they were almost always more dorsal (Fig. 8A) and tended to be more lateral to the main group of Nmb cells in more rostral locations (Fig. 8B; especially at levels rostral to 6.35).
Figure 8.
Most parafacial Nmb neurons express proton sensors Gpr4 and TASK-2 (encoded by Kcnk5). A, Photomicrograph of coronal section from caudal parafacial region (left side) showing transcripts for Nmb (blue), Gpr4 (magenta), and Kcnk5 (green). Inset to the left is an enlargement of relatively large cells (numbered 1, 2, and 3) that contain low levels of Kcnk5 and/or Gpr4 transcripts but high levels of Nmb. Inset to the right is an enlargement of the boxed area showing that most parafacial Nmb neurons express transcripts for both Gpr4 and Kcnk5. Note larger cells with high levels of Nmb transcripts and relatively low levels of Kcnk5/Gpr4 are located dorsal and lateral to the other Nmb neurons. Medial to the right, dorsal toward the top. Dashed white lines indicate ventral medullary surface. B, Drawing of coronal sections through medulla/pons showing the distribution of Nmb neurons with Kcnk5 and Gpr4. Note the neurons with either low levels or no detectable Kcnk5/Gpr4 tend to be dorsal and lateral to the Nmb neurons that contain both of these proton sensors. Numbers in center of sections indicate mm behind bregma (after Paxinos and Franklin, 2013). Abbreviations as in Figure 1. Scale bars: A, 100 μm; left inset, 50 μm; right inset, 50 μm; B, 1 mm.
We noticed that many of the Nmb-high expressing neurons appeared to be larger than the Nmb-low cells (compare Figs. 6E, 8A, 10A,C) and measured in 3 JX99 mice the cross sectional soma area of GFP-labeled cells containing Nmb and Gpr4 transcripts (because the eGFP fills the cytoplasm and allows accurate soma tracing). Indeed, the Nmb-high cells had a significantly larger soma area (153.4 μm2; n = 21; range 94.2–390.2) than the Nmb-low cells (74.4 μm2; n = 60; range 35.1–169.5) compared with a one-tailed Mann-Whitney U (U = 48; p < 0.001).
Figure 10.
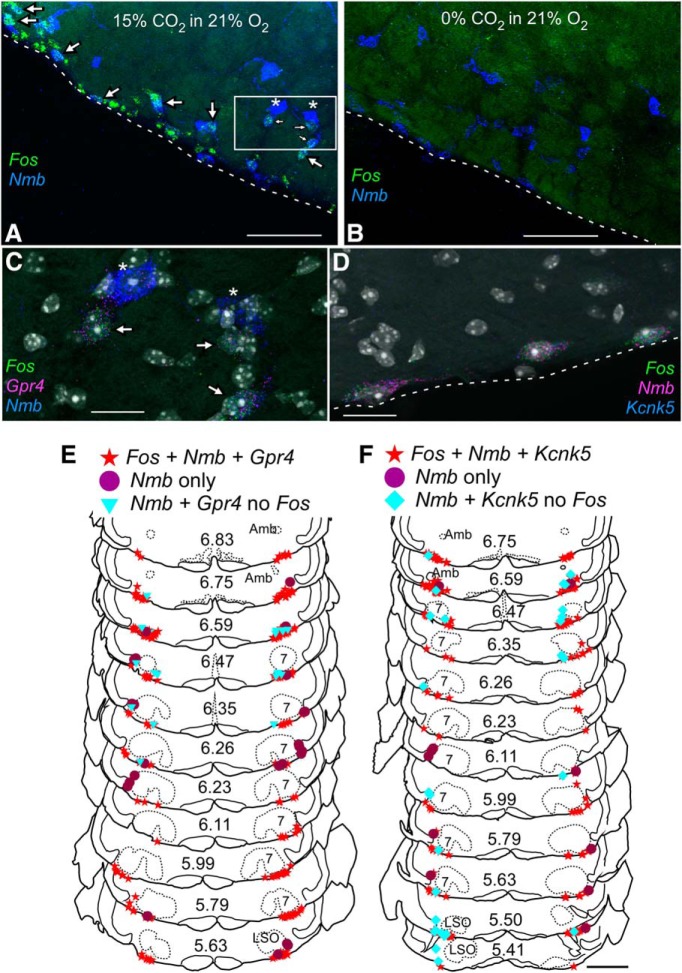
RTN Nmb neurons with proton sensors Gpr4 and TASK-2 express Fos in mice exposed to hypercapnia. A, B, Photomicrographs of coronal section in caudal RTN showing transcripts for Fos (green) and Nmb (blue) in mouse exposed to 15% CO2 in 21% O2 (A) or 21% O2 (B). Many neurons express Fos under hypercapnia (A, arrows). Note that large neurons with high levels of Nmb transcripts in (A) indicated by stars do not express Fos. C, Enlargement of boxed area in A showing neurons expressing transcripts for Nmb (blue), Gpr4 (magenta), and Fos (green). Neurons indicated by stars express high levels of Nmb but low or undetectable levels of Gpr4 and do not express Fos. DAPI stain in white/gray. D, Photomicrograph of neurons expressing transcripts for Nmb (magenta), Kcnk5 (blue), and Fos (green) from mouse exposed to 15% CO2. Dashed lines in A, B, and D represent the ventral medullary surface. E, F, Drawing of transverse sections through the medulla and pons showing the distribution of Nmb neurons with Kcnk5 (E) or Gpr4 (F) and Fos after exposure to 15% CO2. The Nmb neurons that do not express Fos tend to be dorsal and lateral to the Nmb neurons that are activated by hypercapnia. Numbers in center of sections indicate mm behind bregma (Paxinos and Franklin, 2013). Abbreviations as in Figure 1. Scale bars: A, 100 μm; B, 100 μm; C, 20 μm; D, 20 μm; E, 1 mm.
By single-cell RNA-Seq (Fig. 9), we found that Gpr4 was detectable in 89.3% of Nmb-expressing RTN neurons (n = 67/75) and was the highest expressed of all G-protein-coupled receptors in those cells (median TPM: 140.2; range: 0–502; compare with Fig. 5C,D). For Kcnk5, expression could also be detected in a large fraction of Nmb-expressing RTN neurons (82.7%; n = 62/75), although transcript levels were lower overall than for Gpr4 (median TPM for Kcnk5: 14; range: 0–89).
Most RTN neurons expressed both Gpr4 and Kcnk5, but some had high Gpr4 expression and little to no Kcnk5 expression whereas others showed high Kcnk5 expression and little to no Gpr4 expression (Fig. 9B). The neurons that showed a strong preferential expression of either Gpr4 or Kcnk5 likely correspond to the subgroups of previously characterized cells for which one of those genes was required for pH-sensitivity (Wang et al., 2013b; Kumar et al., 2015). In addition, consistent with in situ hybridization results, there was a clear relationship between Gpr4 or Kcnk5 levels and Nmb expression: the subgroup of RTN neurons with the highest levels of Nmb transcripts had correspondingly lower levels of Gpr4 and Kcnk5 (Fig. 9C,D).
As mentioned earlier, the Nmb threshold (TPM < 150) was generally effective for differentiating the RTN population of neurons from other nearby GFP-labeled cells; indeed, most of this other population was also negative for Gpr4 and Kcnk5, with the notable exception of two Gpr4-expressing neurons (also Gal- and Penk-positive; Fig. 7C) and one Kcnk5-expressing neuron. Although these outliers would be at the lower extreme of Nmb expression in this sample of RTN neurons, they nevertheless adhere to the general principle that Nmb-low group of RTN neurons are relatively more likely to express Gpr4 and Kcnk5.
Most RTN Nmb neurons are activated by hypercapnia but unresponsive to hypoxia
We used Fos expression as a readout of neuronal activation to examine whether the quantitative differences in Gpr4 and/or Kcnk5 expression between Nmb-high and Nmb-low subgroups described above might yield corresponding functional differences in effects of CO2 on RTN neurons in vivo. In mice exposed to hypercapnia for 35 min (15% FiCO2, 21% FiO2, balance N2), Fos expression was observed in most RTN Nmb neurons (74 ± 14%, n = 7; Table 3; Fig. 10), whereas in control animals exposed to normoxia (21% FiO2, balance N2) this percentage was very low (1.5 ± 0.5%, n = 7; Table 3; Fig. 10B).
Table 3.
Cell counts for hypercapnia cases
| Sex | Female | Female | Female | Female | Female | Male | Male | Average ± SEM | Average % Total Nmb ± SEM |
|---|---|---|---|---|---|---|---|---|---|
| Strain | C57 | C57 | C57 | JX99 | JX99 | C57 | C57 | ||
| Condition, 15% CO2 | |||||||||
| Case | 22609 | 23205 | 26100 | 3409 | 3502 | J01 | J03 | ||
| Nmb + Fos + Gpr4 | 135 | 234 | 141 | 100 | 73 | 73 | 66 | 117.4 ± 22.5 | 73.5 ± 2.9 |
| Nmb + Gpr4 no Fos | 9 | 20 | 21 | 8 | 15 | 2 | 4 | 11.3 ± 2.8 | 6.8 ± 1.5 |
| Nmb + Gpr4-low no Fos | 13 | 14 | 13 | 5 | 3 | 1 | 4 | 7.6 ± 2.1 | 4.3 ± 0.8 |
| Nmb no Gpr4 no Fos | 28 | 27 | 37 | 38 | 28 | 9 | 12 | 25.6 ± 4.2 | 16.7 ± 2.5 |
| Total Nmb | 185 | 295 | 212 | 151 | 109 | 85 | 86 | 160.4 ± 29.0 | |
| No. of sections | 7 | 11 | 14 | 6 | 6 | 6 | 6 | 8 ± 1.2 | |
| Condition, 21% O2 | |||||||||
| Case | 26101 | 22305 | 22503 | 3407 | 3408 | J02 | J00 | ||
| Nmb + Fos + Gpr4 | 2 | 7 | 8 | 1 | 0 | 2 | 0 | 2.9 ± 1.2 | 1.5 ± 0.6 |
| Nmb + Gpr4 no Fos | 163 | 182 | 182 | 122 | 73 | 57 | 36 | 116.4 ± 23.2 | 78.0 ± 2.2 |
| Nmb + Gpr4-low no Fos | 15 | 14 | 3 | 0 | 1 | 5 | 0 | 5.4 ± 2.4 | 3.0 ± 1.2 |
| Nmb no Gpr4 no Fos | 54 | 33 | 32 | 35 | 13 | 17 | 7 | 27.3 ± 6.1 | 18.0 ± 1.5 |
| Total Nmb | 234 | 226 | 225 | 158 | 87 | 81 | 43 | 150.6 ± 30.4 | |
| No. of sections | 10 | 10 | 12 | 6 | 6 | 6 | 5 | 7.9 ± 1.0 | |
| Condition, 15% CO2 | |||||||||
| Case | 23205 | J01 | |||||||
| Nmb + Fos + TASK-2 | 180 | 213 | 196.5 ± 16.5 | 78.6 ± 3.6 | |||||
| Nmb + TASK-2 no Fos | 29 | 44 | 36.5 ± 7.5 | 14.4 ± 1.1 | |||||
| Nmb no TASK-2 no Fos | 10 | 15 | 12.5 ± 2.5 | 4.9 ± 0.4 | |||||
| Total Nmb | 219 | 284 | 251.5 ± 32.5 | ||||||
| No. of sections | 12 | 12 | 12 |
C57, C57Bl6/J; Gpr4, G-protein-coupled receptor 4; JX99, Phox2b-eGFP BAC transgenic; TASK-2, potassium channel subfamily K member 5. Nmb + Gpr4-low refers to cells with Nmb and very low levels of Gpr4 transcript. No attempt was made to distinguish Nmb-high or Nmb-low cells in these counts.
The vast majority of parafacial neurons that contained both Nmb and moderate levels of Gpr4 transcripts also contained Fos mRNA in mice exposed to hypercapnia (91 ± 2%; n = 7; Table 3; Fig. 10). Notably, however, the Nmb neurons that contained little or no detectable Gpr4 mRNA (Figs. 8A, 9) did not express Fos after CO2 exposure (0/232 Nmb+ Gpr4-low cells positive for Fos mRNA counted across 7 mice; Table 3; Fig. 10A,C). The tissue distribution of the CO2-activated (Fos+) parafacial Nmb neurons was the same as that of the Nmb neurons that contained both Gpr4 and Kcnk5 mRNA (compare Figs. 8B, 10E). By contrast, the CO2-insensitive (Fos−) Nmb neurons were located dorsal and generally lateral to the CO2 responsive cells, with tendency toward being more lateral, particularly in the more rostral areas of RTN. A similar pattern of Fos expression was noted in two of the hypercapnia cases in which alternate sections were hybridized with Kcnk5 probe (i.e., Fos expression was primarily localized to Kcnk5-expressing RTN neurons; Fig. 10D,F; Table 3).
In five male mice exposed to hypoxia (8% FiO2) for 35 min, very few Nmb neurons expressed Fos (3.6 ± 0.7%; Table 4). As in the case for hypercapnia, none of the Nmb-only (i.e., without Kcnk5 or Gpr4) cells expressed Fos in animals exposed to hypoxia (0/259 Nmb-only cells counted in 5 mice). As expected, all mice showed a sharp increase in sighing behavior during hypoxia compared normoxia (21% FiO2: 0.7 ± 1.3 sighs/min, n = 5; 8% FiO2: 33.3 ± 3.5 sighs/min, n = 5; unpaired t = 8.8, p < 0.0001).
Table 4.
Bilateral cell counts for hypoxia cases
| Sex | Male | Male | Male | Male | Male | Average ± SEM | Average % Total Nmb ± SEM |
|---|---|---|---|---|---|---|---|
| Strain | JX99 | JX99 | JX99 | C57 | C57 | ||
| Condition, 8% O2 | |||||||
| Case | 6410 | 7011 | 6640 | 25100 | 25102 | ||
| Nmb + Fos + Gpr4 | 13 | 2 | 13 | 10 | 9 | 9.4 ± 2.0 | 3.6 ± 0.7 |
| Nmb + Gpr4 no Fos | 178 | 156 | 180 | 187 | 195 | 179.2 ± 6.5 | 71.6 ± 1.5 |
| Nmb Gpr4-low no Fos | 11 | 5 | 6 | 14 | 17 | 10.6 ± 2.3 | 4.2 ± 0.8 |
| Nmb only | 52 | 45 | 72 | 46 | 44 | 51.8 ± 5.2 | 20.6 ± 1.7 |
| Total Nmb | 254 | 208 | 271 | 257 | 265 | 251.0 ± 11.2 | |
| No. of sections | 12 | 12 | 10 | 12 | 12 | 11.6 ± 0.4 | |
| Condition, 21% O2 | |||||||
| Case | 7010 | 6494 | 25101 | 25103 | |||
| Nmb + Fos + Gpr4 | 1 | 6 | 6 | 1 | 3.5 ± 1.3 | 1.3 ± 0.4 | |
| Nmb + Gpr4 no Fos | 217 | 229 | 193 | 114 | 188.25 ± 23.1 | 72.3 ± 2.2 | |
| Nmb Gpr4-low no Fos | 18 | 8 | 8 | 11 | 11.3 ± 2.1 | 4.5 ± 0.8 | |
| Nmb only | 59 | 45 | 75 | 44 | 55.8 ± 6.5 | 21.9 ± 2.0 | |
| Total Nmb | 295 | 288 | 282 | 170 | 258.8 ± 26.6 | ||
| No. of sections | 12 | 12 | 12 | 10 | 11.5 ± 0.4 |
C57, C57Bl6/J; Gpr4, G-protein-coupled receptor 4; JX99, Phox2b-eGFP BAC transgenic. Nmb only refers to cells with Nmb transcripts with no Fos and no Gpr4 transcripts.
Parafacial Nmb neurons in the rat
We used a rat-specific probe (Table 1) to examine the distribution of Nmb mRNA in three adult male Sprague Dawley rats, confining our observations to the rostral medulla and pons. As in mice, Nmb cells were abundant in the parafacial region but, unlike in mice, they were also present within the ventromedial medulla oblongata and adjacent portions of the pons (Fig. 11A).
Figure 11.
In rats Nmb transcripts are present both in RTN and in the adjacent medial reticular formation. A, Drawing of coronal sections through medulla and caudal pons of rat showing the distribution of neurons with Nmb transcripts, with or without Phox2b transcripts. As in the mouse, Nmb + Phox2b neurons delineate the RTN. However, a large separate population of Nmb neurons is located more medially in rat. Numbers in center of sections indicate mm behind bregma (after Paxinos and Watson, 2005). Abbreviations as in Fig. 1. B, Photomicrograph of coronal section in caudal RTN showing transcripts for Nmb in magenta and Phox2b in green. DAPI stain is white/gray. Medial to the left, dorsal toward the top. C, Photomicrograph from midline region where Phox2b is not expressed in Nmb neurons. D, Enlargement of white dashed box from B. Note coexpression of Nmb with Phox2b transcripts. E, Enlargement of magenta dashed box in B showing cells expressing both Phox2b and Nmb transcripts. Scale: A, 500 μm. Scale bars: A, 1 mm; B, 100 μm; C–E, 20 μm.
The Nmb neurons located in the parafacial region contained Phox2b transcripts, whereas those located more medially did not (Fig. 11B–E). The two groups were adjoining, but essentially non-overlapping (Fig. 11A). As in mice, none of the Nmb neurons were TH-immunoreactive (data not shown). The distribution of the Nmb+/Phox2b+ neurons was very similar to that of the mouse (compare Figs. 1, 11) and seemed virtually identical to that of the neurons previously defined as RTN in rats based on coexpression of VGlut2 mRNA and Phox2b and absence of TH (Stornetta et al., 2006). Like previously defined RTN neurons, the cluster of Nmb+/Phox2b+ neurons extended from a level ∼500 μm caudal to the facial motor nucleus to the exit point of the seventh nerve (Fig. 11A). Based on cell counts performed in two animals using a one-in-six series of 30 μm sections, and after Abercrombie correction, the rat contains ∼1910 Nmb+/Phox2b+ parafacial neurons, a number virtually identical to the cell population that we previously defined as RTN in this species based on anatomical location, coexpression of Phox2b and VGlut2, and lack of TH.
Discussion
The principal new findings are as follows. A single marker, Nmb mRNA, identifies the RTN, a population of ∼700 parafacial neurons that could be definitively identified previously only by using a combination of several markers (expression of Phox2b, VGlut2, NK1R, and absence of catecholaminergic and other markers). At least 80% of RTN neurons contain transcripts coding for one or both proton receptors, Kcnk5 and Gpr4, and most of these neurons express Fos mRNA in mice exposed to hypercapnia but not hypoxia. We conclude from these observations that RTN consists mostly of CO2-sensitive chemoreceptors, and that Nmb mRNA identifies these chemoreceptors with >75% accuracy. A subset of RTN neurons (∼20–30%) contain extremely high levels of Nmb mRNA, low to undetectable levels of Gpr4 and Kcnk5 transcripts, and typically lack two additional pre-propeptide genes (Penk, Gal) that are variably expressed by other RTN neurons. These Nmb-high RTN neurons are typically larger in size, do not respond to either hypercapnia or hypoxia, and are probably not chemoreceptors. Transcript levels determined by single-cell RNA-seq varied considerably from cell to cell (by 1–2 orders of magnitude). In several instances (Nmb, Gal, Penk, Kcnk5, and Gpr4) this variability could be confirmed by ISH, at least in a semiquantitative sense, suggesting considerable phenotypic variability even in this small population of neurons. Finally, Nmb is also a marker of RTN in rats.
Nmb mRNA: a diagnostic marker of RTN in mice
Phox2b and VGlut2 transcripts were detectable in essentially all Nmb neurons located in the parafacial region of the mouse. These Nmb neurons did not express Th or Gad1/Gad2. In this region, Nmb transcripts are expressed by neurons that have all the phenotypical characteristics that have been used to define the RTN (i.e., presence of Phox2b, VGlut2, and NK1 receptors; widespread expression of Gpr4 and Kcnk5; and absence of Th, Tph1 or Tph2, Gad1 or Gad2, GlyT2, and Vgat; Stornetta et al., 2006; Goridis et al., 2010; Ramanantsoa et al., 2011; Guyenet and Bayliss, 2015; Ruffault et al., 2015). The number of Nmb neurons counted in the present study (∼665) is smaller than our prior estimates of RTN cells based only on eGFP expression (as a proxy for Phox2b) in the parafacial area in Phox2b::eGFP mice (787; Lazarenko et al., 2009) but still within the range of the number of neurons defined as RTN by genetic lineage analysis (∼600:Thoby-Brisson et al., 2009; ∼700–800: Ramanantsoa et al., 2011). In light of these observations, we conclude that Nmb is a reliable single marker of the neuronal population previously defined as RTN based on combinatorial neurochemical and developmental criteria (Dubreuil et al., 2008; Thoby-Brisson et al., 2009; Guyenet et al., 2016).
In rats, Nmb transcripts also identify RTN but, in this species, Nmb is also expressed by a separate group of Phox2b-negative neurons located in the ventromedial medulla oblongata. The two populations are adjacent but overlap very little. Thus, Nmb expression should also serve as a useful marker of RTN in rat, although coexpression of Phox2b and Nmb would provide added precision. Nmb identifies the RTN in both rats and mice suggesting that it could also be a useful marker of the human RTN (Rudzinski and Kapur, 2010).
The majority of RTN neurons are central respiratory chemoreceptors
Although the CO2/H+ sensitivity of RTN neurons is likely enhanced by astrocyte-dependent paracrine mechanisms (Gourine et al., 2005, 2010), it is in large measure a cell-autonomous response to protons (Guyenet et al., 2016). This view is based on two key observations. RTN neurons are activated by CO2/H+ after complete isolation (Wang et al., 2013a) and both their pH sensitivity in vitro and the central respiratory chemoreflex require the expression by RTN neurons of proton sensors Gpr4 and TASK-2 (Gestreau et al., 2010; Wang et al., 2013b; Kumar et al., 2015). We show here that Gpr4 and Kcnk5 are usually coexpressed by RTN neurons. However, we also found cells in which one of those genes was preferentially expressed, likely corresponding to the subset of RTN neurons that only require Gpr4 or TASK-2 for their pH sensitivity (Wang et al., 2013b; Kumar et al., 2015).
We also found that ∼75% of RTN Nmb neurons in mice expressed Fos after exposure to hypercapnia (15% FiCO2). This high level of CO2 stimulus, notwithstanding the limitations of Fos as a measure of neuronal activation, is likely to have revealed close to the true fraction of chemosensitive RTN neurons. Fewer RTN neurons (∼35%) express Fos in mice exposed to 8% FiCO2 (Kumar et al., 2015; Shi et al., 2016). Subsets of RTN neurons, such as those that elicit active expiration (Marina et al., 2010; Burke et al., 2015) probably have a high threshold of activation by CO2. Importantly, all the Fos+ RTN neurons contained Gpr4 and Kcnk5 mRNA whereas these transcripts were generally undetectable in the small subset of RTN neurons that did not respond to CO2. The CO2-activated neurons did not express Fos after hypoxia. Using loss of function optogenetics, we previously demonstrated that the RTN neurons that drive breathing are silenced during hypoxia because of the accompanying respiratory alkalosis (Basting et al., 2015; Guyenet et al., 2016). Activation by hypercapnia and inhibition by hypoxia (as a result of the respiratory alkalosis) are responses that are expected from central respiratory chemoreceptors in intact unanesthetized mammals.
The Nmb neurons that did not detectably respond to hypercapnia typically expressed the highest levels of Nmb. Their cell bodies had a larger cross-sectional area, and tended to be located dorsal and lateral to the main cluster of RTN neurons. The function of these neurons is unknown, but they could be the ∼200 Nmb neurons identified by Li et al. (2016) in the parafacial region of an Nmb-eGFP mouse that were implicated in Nmb-mediated sighing.
RTN peptides
In addition to Nmb, mouse RTN neurons express Gal, Penk, and Adcyap1 transcripts. The concentration of Gal and Penk transcripts was negatively correlated with Nmb levels. In particular, both Gal and Penk mRNA levels were low to undetectable in the Nmb-high neurons. Adycap1 transcripts were also detected in nearby catecholaminergic neurons and facial motor neurons. Most pro-peptides can be processed into several bioactive transmitters. Galanin and Neuromedin B are presumably produced and used as transmitters by RTN neurons because their terminals are immunoreactive for these peptides (Bochorishvili et al., 2012; Li et al., 2016). The other peptides have as yet to be identified in RTN terminals.
Every RTN neuron contains several pro-peptide transcripts. If peptide content is an indication of function, the code is probably combinatorial. The problem is further complicated as transcript levels, and presumably peptide expression, are not all or none but vary over a wide range.
Stochastic gene expression or evidence for multiple functional subsets of RTN neurons?
The transcript level for individual genes examined semiquantitatively by in situ hybridization and quantitatively by RNA-Seq varied considerably from cell to cell (e.g., by 1.5 log units). Clearly, technical issues contribute to some variability for either of these methods, but the general concordance between them suggests that much of this reflects real differences in expression among individual neurons. For example, the generally inverse correlation between Nmb transcripts and mRNA for neuropeptides (Penk or Gal) and proton sensors (Gpr4 or Kcnk5) was detected by both methods, as was the paired and/or preferential expression of Gpr4 and Kcnk5 in individual RTN neurons.
The RTN is a numerically small group of neurons with a unique and well defined developmental genetic lineage (Ruffault et al., 2015). Nonetheless, gene expression in RTN neurons was surprisingly variable, suggesting that this relatively small and select population consists of multiple subtypes of neurons. This interpretation is supported by prior evidence of differential synaptic inputs and firing properties in several preparations (Guyenet et al., 2005; Onimaru et al., 2008; Thoby-Brisson et al., 2009). Subsets of RTN neurons with varying degrees of CO2-sensitivity and distinctive synaptic inputs may differentially regulate various breathing parameters (frequency, amplitude, etc.) and possibly other functions (e.g., arousal, cardiovascular control). Alternatively, the large variability in gene expression that we observed could mean that gene expression is simply stochastic, even among a small cluster of functionally homogeneous neurons.
Conclusions
All RTN neurons express Nmb in mice and rats (Fig. 12 for summary). Nmb also provides a quantitative trait that defines subsets of RTN neurons. Specifically, the cells expressing the highest levels of Nmb never express Gal and Penk, have lower levels of Gpr4 and Kcnk5, and do not express Fos after hypercapnia. The latter seems especially true for the Nmb-high RTN neurons (∼20–25%) that are located at the outer reaches of this nucleus and may subserve a task distinct from the central respiratory chemoreceptor role of the majority of RTN neurons. Finally, based on Fos expression after hypercapnia at least 75% of RTN neurons qualify as potential central chemoreceptors, closely corresponding to the fraction of neurons expressing either Gpr4 or Kcnk5 (80–90%). Since all these neurons express Nmb, this transcript is a marker that, by itself, identifies RTN chemoreceptors with at least 75% fidelity.
Figure 12.
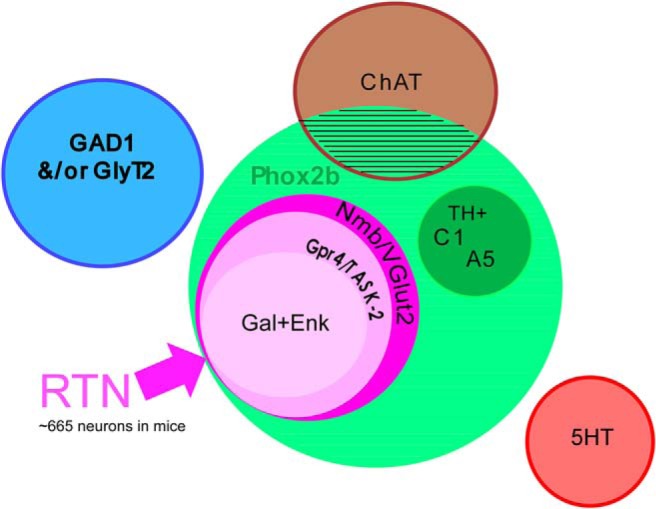
Venn diagram of RTN and other parafacial neurons in mice. The facial/parafacial region contains multiple types of Phox2b neurons (light green circle), which are either cholinergic, noradrenergic or glutamatergic. RTN in mice consists of ∼665 Phox2b+ glutamatergic (VGlut2+) neurons that express Nmb (dark pink circle). Most RTN neurons (80–90%) contain proton sensors TASK-2 and Gpr4 (medium pink circle); these neurons are most probably central respiratory chemoreceptors. A subset of the latter (>70%) express varying levels of Gal and/or Penk mRNA (light pink circle). Parafacial Nmb neurons do not express ChAT, Th, Tph(1 or 2), Gad1, or GlyT2. The size of the circles is not meant to represent relative number of neurons.
Footnotes
This work was supported by HL074011 (P.G.G.), and HL108609 and Pilot Grant Award from the CCHS Family Network (D.A.B.). We thank Drs. Mike McConnell and Ian Burbulis for early advice and support on single-cell sequencing approaches.
The authors declare no competing financial interests.
References
- Abercrombie M. (1946) Estimation of nuclear population from microtome sections. Anat Rec 94:239–247. 10.1002/ar.1090940210 [DOI] [PubMed] [Google Scholar]
- Barna BF, Takakura AC, Mulkey DK, Moreira TS (2016) Purinergic receptor blockade in the retrotrapezoid nucleus attenuates the respiratory chemoreflexes in awake rats. Acta Physiol (Oxf) 217:80–93. 10.1111/apha.12637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basting TM, Burke PG, Kanbar R, Viar KE, Stornetta DS, Stornetta RL, Guyenet PG (2015) Hypoxia silences retrotrapezoid nucleus respiratory chemoreceptors via alkalosis. J Neurosci 35:527–543. 10.1523/JNEUROSCI.2923-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker HF, Polo O, McNamara SG, Berthon-Jones M, Sullivan CE (1996) Effect of different levels of hyperoxia on breathing in healthy subjects. J Appl Physiol 81:1683–1690. [DOI] [PubMed] [Google Scholar]
- Bochorishvili G, Stornetta RL, Coates MB, Guyenet PG (2012) Pre-Bötzinger complex receives glutamatergic innervation from galaninergic and other retrotrapezoid nucleus neurons. J Comp Neurol 520:1047–1061. 10.1002/cne.22769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke PG, Kanbar R, Basting TM, Hodges WM, Viar KE, Stornetta RL, Guyenet PG (2015) State-dependent control of breathing by the retrotrapezoid nucleus. J Physiol 593:2909–2926. 10.1113/JP270053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly CA, Ellenberger HH, Feldman JL (1990) Respiratory activity in retrotrapezoid nucleus in cat. Am J Physiol 258:L33–L44. [DOI] [PubMed] [Google Scholar]
- Dubreuil V, Ramanantsoa N, Trochet D, Vaubourg V, Amiel J, Gallego J, Brunet JF, Goridis C (2008) A human mutation in Phox2b causes lack of CO2 chemosensitivity, fatal central apnoea and specific loss of parafacial neurons. Proc Natl Acad Sci U S A 105:1067–1072. 10.1073/pnas.0709115105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger HH, Feldman JL (1990) Brainstem connections of the rostral ventral respiratory group of the rat. Brain Res 513:35–42. 10.1016/0006-8993(90)91086-V [DOI] [PubMed] [Google Scholar]
- Feldman JL, Mitchell GS, Nattie EE (2003) Breathing: rhythmicity, plasticity, chemosensitivity. Annu Rev Neurosci 26:239–266. 10.1146/annurev.neuro.26.041002.131103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gestreau C, Heitzmann D, Thomas J, Dubreuil V, Bandulik S, Reichold M, Bendahhou S, Pierson P, Sterner C, Peyronnet-Roux J, Benfriha C, Tegtmeier I, Ehnes H, Georgieff M, Lesage F, Brunet JF, Goridis C, Warth R, Barhanin J (2010) Task2 potassium channels set central respiratory CO2 and O2 sensitivity. Proc Natl Acad Sci U S A 107:2325–2330. 10.1073/pnas.0910059107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goridis C, Dubreuil V, Thoby-Brisson M, Fortin G, Brunet JF (2010) Phox2b, congenital central hypoventilation syndrome and the control of respiration. Semin Cell Dev Biol 21:814–822. 10.1016/j.semcdb.2010.07.006 [DOI] [PubMed] [Google Scholar]
- Gourine AV, Llaudet E, Dale N, Spyer KM (2005) ATP is a mediator of chemosensory transduction in the central nervous system. Nature 436:108–111. 10.1038/nature03690 [DOI] [PubMed] [Google Scholar]
- Gourine AV, Kasymov V, Marina N, Tang F, Figueiredo MF, Lane S, Teschemacher AG, Spyer KM, Deisseroth K, Kasparov S (2010) Astrocytes control breathing through pH-dependent release of ATP. Science 329:571–575. 10.1126/science.1190721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyenet PG, Bayliss DA (2015) Neural control of breathing and CO2 homeostasis. Neuron 87:946–961. 10.1016/j.neuron.2015.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyenet PG, Mulkey DK, Stornetta RL, Bayliss DA (2005) Regulation of ventral surface chemoreceptors by the central respiratory pattern generator. J Neurosci 25:8938–8947. 10.1523/JNEUROSCI.2415-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyenet PG, Bayliss DA, Stornetta RL, Ludwig MG, Kumar NN, Shi Y, Burke PG, Kanbar R, Basting TM, Holloway BB, Wenker IC (2016) Proton detection and breathing regulation by the retrotrapezoid nucleus. J Physiol 594:1529–1551. 10.1113/JP271480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JA, Kottick A, Picardo MCD, Halleran AD, Smith RD, Smith GD, Saha MS, Del Negro CA (2017) Transcriptome of neonatal preBötzinger complex neurones in Dbx1 reporter mice. Sci Rep 7:8669. 10.1038/s41598-017-09418-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway BB, Viar KE, Stornetta RL, Guyenet PG (2015) The retrotrapezoid nucleus stimulates breathing by releasing glutamate in adult conscious mice. Eur J Neurosci 42:2271–2282. 10.1111/ejn.12996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckstepp RT, Cardoza KP, Henderson LE, Feldman JL (2015) Role of parafacial nuclei in control of breathing in adult rats. J Neurosci 35:1052–1067. 10.1523/JNEUROSCI.2953-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar NN, Velic A, Soliz J, Shi Y, Li K, Wang S, Weaver JL, Sen J, Abbott SB, Lazarenko RM, Ludwig MG, Perez-Reyes E, Mohebbi N, Bettoni C, Gassmann M, Suply T, Seuwen K, Guyenet PG, Wagner CA, Bayliss DA (2015) Regulation of breathing by CO2 requires the proton-activated receptor GPR4 in retrotrapezoid nucleus neurons. Science 348:1255–1260. 10.1126/science.aaa0922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarenko RM, Milner TA, Depuy SD, Stornetta RL, West GH, Kievits JA, Bayliss DA, Guyenet PG (2009) Acid sensitivity and ultrastructure of the retrotrapezoid nucleus in Phox2b-EGFP transgenic mice. J Comp Neurol 517:69–86. 10.1002/cne.22136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarenko RM, Stornetta RL, Bayliss DA, Guyenet PG (2011) Orexin A activates retrotrapezoid neurons in mice. Respir Physiol Neurobiol 175:283–287. 10.1016/j.resp.2010.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176. 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- Li P, Janczewski WA, Yackle K, Kam K, Pagliardini S, Krasnow MA, Feldman JL (2016) The peptidergic control circuit for sighing. Nature 530:293–297. 10.1038/nature16964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JL, Kulakofsky J, Zucker IH (2002) Exercise training enhances baroreflex control of heart rate by a vagal mechanism in rabbits with heart failure. J Appl Physiol 92:2403–2408. 10.1152/japplphysiol.00039.2002 [DOI] [PubMed] [Google Scholar]
- Loeschcke HH. (1982) Central chemosensitivity and the reaction theory. J Physiol 332:1–24. 10.1113/jphysiol.1982.sp014397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig MG, Vanek M, Guerini D, Gasser JA, Jones CE, Junker U, Hofstetter H, Wolf RM, Seuwen K (2003) Proton-sensing G-protein-coupled receptors. Nature 425:93–98. 10.1038/nature01905 [DOI] [PubMed] [Google Scholar]
- Marina N, Abdala AP, Trapp S, Li A, Nattie EE, Hewinson J, Smith JC, Paton JF, Gourine AV (2010) Essential role of Phox2b-expressing ventrolateral brainstem neurons in the chemosensory control of inspiration and expiration. J Neurosci 30:12466–12473. 10.1523/JNEUROSCI.3141-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulkey DK, Stornetta RL, Weston MC, Simmons JR, Parker A, Bayliss DA, Guyenet PG (2004) Respiratory control by ventral surface chemoreceptor neurons in rats. Nat Neurosci 7:1360–1369. 10.1038/nn1357 [DOI] [PubMed] [Google Scholar]
- Mulkey DK, Rosin DL, West G, Takakura AC, Moreira TS, Bayliss DA, Guyenet PG (2007) Serotonergic neurons activate chemosensitive retrotrapezoid nucleus neurons by a pH-independent mechanism. J Neurosci 27:14128–14138. 10.1523/JNEUROSCI.4167-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onimaru H, Homma I (2003) A novel functional neuron group for respiratory rhythm generation in the ventral medulla. J Neurosci 23:1478–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onimaru H, Ikeda K, Kawakami K (2008) CO2-sensitive preinspiratory neurons of the parafacial respiratory group express Phox2b in the neonatal rat. J Neurosci 28:12845–12850. 10.1523/JNEUROSCI.3625-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onimaru H, Ikeda K, Mariho T, Kawakami K (2014) Cytoarchitecture and CO2 sensitivity of Phox2b-positive parafacial neurons in the newborn rat medulla. Prog Brain Res 209:57–71. 10.1016/B978-0-444-63274-6.00004-7 [DOI] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C (2017) Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ (2013) The mouse brain in stereotaxic coordinates, Ed 4 New York: Academic. [Google Scholar]
- Paxinos G, Watson C (2005) The rat brain in stereotaxic coordinates, Ed 5 San Diego: Elsevier/Academic. [Google Scholar]
- Ramanantsoa N, Hirsch MR, Thoby-Brisson M, Dubreuil V, Bouvier J, Ruffault PL, Matrot B, Fortin G, Brunet JF, Gallego J, Goridis C (2011) Breathing without CO2 chemosensitivity in conditional Phox2b mutants. J Neurosci 31:12880–12888. 10.1523/JNEUROSCI.1721-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes R, Duprat F, Lesage F, Fink M, Salinas M, Farman N, Lazdunski M (1998) Cloning and expression of a novel pH-sensitive two pore domain K+ channel from human kidney. J Biol Chem 273:30863–30869. 10.1074/jbc.273.47.30863 [DOI] [PubMed] [Google Scholar]
- Rudzinski E, Kapur RP (2010) PHOX2B immunolocalization of the candidate human retrotrapezoid nucleus. Pediatr Dev Pathol 13:291–299. 10.2350/09-07-0682-OA.1 [DOI] [PubMed] [Google Scholar]
- Ruffault PL, D'Autreaux F, Hayes JA, Nomaksteinsky M, Autran S, Fujiyama T, Hoshino M, Hagglund M, Kiehn O, Brunet JF, Fortin G, Goridis C (2015) The retrotrapezoid nucleus neurons expressing Atoh1 and Phox2b are essential for the respiratory response to CO2. eLife 4:e07051. 10.7554/eLife.07051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Abe C, Holloway BB, Shu S, Kumar NN, Weaver JL, Sen J, Perez-Reyes E, Stornetta RL, Guyenet PG, Bayliss DA (2016) Nalcn is a “leak” sodium channel that regulates excitability of brainstem chemosensory neurons and breathing. J Neurosci 36:8174–8187. 10.1523/JNEUROSCI.1096-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JC, Morrison DE, Ellenberger HH, Otto MR, Feldman JL (1989) Brainstem projections to the major respiratory neuron populations in the medulla of the cat. J Comp Neurol 281:69–96. 10.1002/cne.902810107 [DOI] [PubMed] [Google Scholar]
- Soneson C, Love MI, Robinson MD (2015) Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res 4:1521. 10.12688/f1000research.7563.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stornetta RL, McQuiston TJ, Guyenet PG (2004) GABAergic and glycinergic presympathetic neurons of rat medulla oblongata identified by retrograde transport of pseudorabies virus and in situ hybridization. J Comp Neurol 479:257–270. 10.1002/cne.20332 [DOI] [PubMed] [Google Scholar]
- Stornetta RL, Moreira TS, Takakura AC, Kang BJ, Chang DA, West GH, Brunet JF, Mulkey DK, Bayliss DA, Guyenet PG (2006) Expression of Phox2b by brainstem neurons involved in chemosensory integration in the adult rat. J Neurosci 26:10305–10314. 10.1523/JNEUROSCI.2917-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stornetta RL, Spirovski D, Moreira TS, Takakura AC, West GH, Gwilt JM, Pilowsky PM, Guyenet PG (2009) Galanin is a selective marker of the retrotrapezoid nucleus in rats. J Comp Neurol 512:373–383. 10.1002/cne.21897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoby-Brisson M, Karlén M, Wu N, Charnay P, Champagnat J, Fortin G (2009) Genetic identification of an embryonic parafacial oscillator coupling to the preBötzinger complex. Nat Neurosci 12:1028–1035. 10.1038/nn.2354 [DOI] [PubMed] [Google Scholar]
- Wang S, Shi Y, Shu S, Guyenet PG, Bayliss DA (2013a) Phox2b-expressing retrotrapezoid neurons are intrinsically responsive to acidification and CO2. J Neurosci 33:7756–7761. 10.1523/JNEUROSCI.5550-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Benamer N, Zanella S, Kumar NN, Shi Y, Bévengut M, Penton D, Guyenet PG, Lesage F, Gestreau C, Barhanin J, Bayliss DA (2013b) TASK-2 channels contribute to pH sensitivity of retrotrapezoid nucleus chemoreceptor neurons. J Neurosci 33:16033–16044. 10.1523/JNEUROSCI.2451-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]