Abstract
Background
One of the major barriers to the development of lignocellulosic feedstocks is the recalcitrance of plant cell walls to deconstruction and saccharification. Recalcitrance can be reduced by targeting genes involved in cell wall biosynthesis, but this can have unintended consequences that compromise the agronomic performance of the trees under field conditions. Here we report the results of a field trial of fourteen distinct transgenic Populus deltoides lines that had previously demonstrated reduced recalcitrance without yield penalties under greenhouse conditions.
Results
Survival and productivity of the trial were excellent in the first year, and there was little evidence for reduced performance of the transgenic lines with modified target gene expression. Surprisingly, the most striking phenotypic effects in this trial were for two empty-vector control lines that had modified bud set and bud flush. This is most likely due to somaclonal variation or insertional mutagenesis. Traits related to yield, crown architecture, herbivory, pathogen response, and frost damage showed few significant differences between target gene transgenics and empty vector controls. However, there were a few interesting exceptions. Lines overexpressing the DUF231 gene, a putative O-acetyltransferase, showed early bud flush and marginally increased height growth. Lines overexpressing the DUF266 gene, a putative glycosyltransferase, had significantly decreased stem internode length and slightly higher volume index. Finally, lines overexpressing the PFD2 gene, a putative member of the prefoldin complex, had a slightly reduced volume index.
Conclusions
This field trial demonstrates that these cell wall modifications, which decreased cell wall recalcitrance under laboratory conditions, did not seriously compromise first-year performance in the field, despite substantial challenges, including an outbreak of a stem boring insect (Gypsonoma haimbachiana), attack by a leaf rust pathogen (Melampsora spp.), and a late frost event. This bodes well for the potential utility of these lines as advanced biofuels feedstocks.
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-017-0934-6) contains supplementary material, which is available to authorized users.
Keywords: Populus, Transgenic, Field trial, Insects, Frost, Yield, Phenology, Cell wall, Biofuel
Background
The considerable energy contained in plant cell walls is an attractive target for the biofuels industry. Cell walls contain approximately 70% of the carbon fixed by plants globally, and constitute a relatively untapped global energy resource [1]. One of the main barriers for the utilization of lignocellulosic biomass for biofuel production is the recalcitrance of plant cell walls to chemical and enzymatic deconstruction, which is a necessary step to release sugars for subsequent conversion to fuels. Recalcitrance is primarily a consequence of the plant packaging carbohydrates in forms that are inaccessible to degradation by chemical and biological agents. Recalcitrance can be a feature of the cellulose polymer itself, which is packaged in tightly interconnected fibers that can be organized into crystalline sheets that themselves are relatively inaccessible to cellulolytic enzymes [1, 2]. These fibers occur within a largely hydrophobic matrix of lignin, which also contributes to recalcitrance. Cellulose, a polymer of 6-carbon glucose molecules (C6) is also entwined with and bound to hemicelluloses, principally xylans in angiosperms, which are mainly comprised of 5-carbon sugars (C5) that are not as readily converted to fuel as the 6-carbon sugars like the glucose monomers that make up the cellulose chains [1–3]. The hemicelluloses and other non-cellulosic cell wall polymers may also contribute to recalcitrance. This structural complexity of the wall makes bioconversion of lignocellulosic biomass to liquid fuels challenging and expensive.
Release of sugars for subsequent fermentation to fuels can be achieved by a series of separate steps aimed at (1) physically reducing the size of the biomass to maximize surface-to-volume and/or weight-to-volume (density) ratio; (2) pretreatment with heat and chemicals such as dilute acids to enhance porosity; (3) treatment with biocatalysts to break down the cross-linkages between cellulose microfibrils and the cell wall matrix; and (4) subsequent hydrolysis with industrial enzymes such as cellulases to produce the sugars [4, 5]. These processes are expensive due to the large energy requirements and the cost of the enzymes. An attractive alternative is consolidated bioprocessing (CBP), which ideally involves minimal pretreatment, and integrates the production of the hydrolytic enzymes with the fermentation step [6]. Major technological advances are however needed to enable CBP. Ideally the process would involve microbes that can hydrolyze cellulose and hemicellulose from minimally processed biomass feedstock and utilize both C5 and C6 sugars in fermentation under harsh conditions and with minimal inhibition from the fermentation products [7, 8]. Major advances have been achieved in recent years, such as with recent breakthroughs in optimizing organisms such as Clostridium thermocellum [9] and Caldicellulosiruptor bescii [10] for CBP utilization.
Another potential component of efficient biofuel production is the development of biomass feedstocks with cell walls that can be readily deconstructed to yield fermentable sugars [4, 11, 12]. One way to achieve this is to manipulate the expression of genes involved in the biosynthesis of cell walls using genetic transformation. Major phenotypic targets to reduce recalcitrance include: (1) altering cellulose biosynthesis to increase cellulose content and reduce crystallinity; (2) altering hemicellulose composition to decrease H bonding with cellulose; (3) altering enzymes in the phenylpropanoid pathway to reduce lignin content or composition to reduce covalent cross-linkages; and (4) altering the structural proteins in the cell wall or and/or cortical microtubules [1, 3, 5]. To this end, the Department of Energy’s Bioenergy Science Center (BESC) has targeted over 500 distinct genes for overexpression and/or knockdown using Agrobacterium-mediated transformation of Populus deltoides. These transformants have been intensively screened using high-throughput assays to evaluate cell wall composition [13] and sugar release from wood with minimal pretreatment [14]. This evaluation has resulted in the identification of 14 genes that, when overexpressed or knocked down, result in biomass with reduced recalcitrance and no yield penalty based on greenhouse and growth chamber trials (Table 1). The selected genes fall into seven categories, based on the pathways or characteristics that they are expected to affect: (1) phenylpropanoid biosynthesis (CAD, EPSPS); (2) cellulose biosynthesis (IQD10); (3) noncellulosic cell wall polysaccharide biosynthesis (GAUT12); (4) cell wall glycoproteins (EXT1,EXT2); (5) cell wall modifiers (DUF231, DUF266, P4HA1, RWA2,SHMT); (6) cortical microtubule formation (PFD2); and (7) transcription factors controlling enzymes involved in cell wall biosynthesis (HB3,VND6).
Table 1.
Description of genes targeted in this study
| Name | Genea | Description | Typeb | Sugar Releaseb | Yieldb | Rametsc |
|---|---|---|---|---|---|---|
| CAD | Potri.009G095800 | Cinnamyl alcohol dehydrogenase, catalyzes the formation of coniferyl or coumaryl alcohol from their respective aldehydes [58] | KD | + | + | 12(C), 12(T) |
| DUF231 | Potri.009G072800 | Domain of unknown function, a member of the Trichome Birefringence-Like (TBL) gene family, possibly responsible for O-acetylation of hemicelluloses [49] | OE | + | + | 12(C), 12(T) |
| DUF266 | Potri.011G009500 | Domain of unknown function, possibly acting as a glycosyltransferase [53] | OE | + | + | 12(C), 5(C), 5(T) |
| EPSPS | Potri.002G146400 | 5-enolpyruvylshikimate-3-phosphate synthase, key enzyme in biosynthesis of aromatic amino acids [59] | OE | + | = | 12(C), 12(T) |
| EXT1;EXT2 |
Potri.001G020100
Potri.005G190100 |
Extensin, a basic hydroxyproline-rich glycoprotein localized to the cell wall [60] | KD | + | + | 12(C), 12(T); EXT2: 12(T) |
| GAUT12 | Potri.001G416800 | Galacturonosyltransferase targeted to Golgi and involved in xylan and homogalacturonan biosynthesis [61] | KD | + | + | 12(C), 12(C), 12(T) |
| HB3 | Potri.011G098300 | Transcription factor belonging to the HDZIPIII family, with high expression in xylem tissue [62] | KD | + | + | 12(C), 14(T) |
| IQD10 | Potri.001G375700 | Calmodulin-binding protein with IQ amino acid-rich region with high expression in tension wood [63] | KD | + | + | 11(C), 13(T) |
| P4HA1 | Potri.017G075300 | Prolyl 4-hydroxylase alpha subunit, hydroxylation of proline residues, potentially in hydroxyproline-rich glycoproteins in the cell wall [64] | OE | + | = | 12(C), 12(T) |
| PFD2 | Potri.008G153900 | Probable Prefoldin 2 protein. A heterohexameric chaperon protein that binds to actin, tubulin, and other proteins, possibly affecting the cortical spindle [55] | OE | + | + | 12(C), 13(T) |
| RWA2 | Potri.010G148500 | Reduced wall acetylation 2, catalyzes O-acetylation of cell wall polysaccharides [50] | OE | + | + | 13(C), 12(T) |
| SHMT | Potri.001G320400 | Serine hydroxymethyltransferase, reversible conversion of Ser and tetrahydrofolate to Gly and 5,10-methylene tetrahydrofolate, providing a major 1-carbon source for the cell [65] | OE | + | + | 12(C), 12(T) |
| VND6 | Potri.015G127400 | Vascular-related NAC-domain Protein 6, transcription factors involved in xylem vessel differentiation [66] | OE | - | - | 12(C), 13(T) |
KD knockdown of gene expression using RNAi, OE overexpression of target gene using a constitutive promoter (UBIQUITIN3)
aGene model name based on phytozome [67] version 3.0 of the Populus trichocarpa genome
bSugar release [14] and yield refer to performance relative to controls in greenhouse trials prior to the field trial
cThe number of replicates (ramets) included in the statistical analyses for TOP (T) and comparator (C) lines. For DUF266 and GAUT12 there were two comparator lines, which are listed separately
While demonstration of enhanced performance under greenhouse conditions is a significant achievement, it is essential to evaluate the performance of these lines in replicated field trials under realistic field conditions, where results are often qualitatively different [15]. This is particularly important in the case of traits that affect cell wall structure and composition, as the cell wall plays a crucial role in resisting the pervasive biotic and abiotic stresses that predominate under field conditions [11, 16, 17]. Furthermore, although there is ample evidence that transgene expression can be stable over many years and through multiple rounds of vegetative propagation [18–20], there are also many examples of differential performance of transgenic trees under field and laboratory conditions [16].
One illustrative example is the case of the 4-hydroxycinnamoyl-CoA Ligase (4CL) gene in Populus. This enzyme catalyzes a key step in the lignin biosynthetic pathway, responsible for the conversion of p-coumaric acid to p-coumaroyl CoA [21]. Knocking down expression of this gene in Populus tremuloides led to reduced lignin and enhanced growth under greenhouse conditions [22]. Although the lignin reduction has mostly been consistent in subsequent field trials in this and other genetic backgrounds, growth has typically been reduced relative to wild-type under most field conditions [23, 24]. This impaired performance was apparently due to problems with vessel collapse under water stress and partial occlusion of vessels by tyloses and phenylpropanoid deposition in the transgenics [23, 25]. Clearly evaluation of transgenics with altered cell wall properties under field conditions is essential, and should include evaluation of growth as well as responses to biotic and abiotic stressors [16].
Here we describe the results of a field trial of 36 transgenic lines of Populus deltoides representing modification of 14 genes that previously satisfied an intensive screening process under greenhouse and growth chamber conditions. We show that, by and large, the transgenic lines perform equally well as controls in terms of biomass productivity, crown form, and biotic and abiotic stress tolerance during the first year. This is an important milestone in the development of these improved biofuel feedstocks.
Methods
Generation of transgenic lines
Gene targets (Table 1) were initially identified using a combination of data mining approaches [26], expression studies of tissues undergoing enhanced cellulose synthesis [27–29], analysis of activation-tagged lines with altered cell wall characteristics [30], and association genetics analyses of wild populations of P. trichocarpa [31]. Agrobacterium tumefaciens-mediated transformation was performed in Populus deltoides clone WV94 from Issaquena County, MS by Arborgen, LLC as described previously [32]. For overexpression (OE) constructs, full-length transcripts were amplified from either P. deltoides or P. trichocarpa and inserted 3′ of a constitutive promoter (UBQ3 from Arabidopsis thaliana) and 5′ of the NOS terminator from Agrobacterium tumefaciens. In the case of knockdown (KD) constructs, a unique fragment of the coding sequence of the target gene was cloned as an inverted repeat separated by an intron cloned from the CHALCONE SYNTHASE gene of Petunia hybrida, with the same promoter and terminator as described above. Empty vector controls (seven independent lines) were produced simultaneously using identical methods and vectors, minus the transgenes. These plants were propagated from tissue culture and subsequently from greenwood cuttings, together with non-transformed ramets of clone WV94 that had not been through tissue culture (wild type controls). The plants were propagated in a greenhouse at Oak Ridge National Laboratory (Oak Ridge, TN) at 25 °C and 16 h day length. All lines were evaluated in the greenhouse for growth and form, and analyzed for lignin content, syringyl:guaiacyl (S:G) ratio, and sugar release using methods described previously [13, 33]. The top-performing line (referred to below as the TOP line) and at least one transgenic comparator line were selected for each target gene, except for EXT2, for which only the TOP line was available.
Field trial establishment and design
The field trial was established near Morgantown, WV under USDA APHIS permit 15-047-101. The site has mildly sloped topography and had mostly been under hay cultivation for at least a decade prior to the trial. Site preparation was conducted during the spring and summer of 2015 and included treating with herbicide (Glyphosate and Clopyralid (Stinger®, Dow AgroSciences)), grading, plowing, and tilling. The site was then left fallow for a year, with repeated herbicide sprays to exhaust the seed bank. The site was then tilled again in the spring of 2016 prior to transplanting the rooted cuttings for all P. deltoides lines, comparators, and controls.
Rooted cuttings were planted on June 20, 2016, consisting of 512 ramets in the WV94 background. All lines had at least 11 clonal replicates, with the exception of two of the DUF266 lines, which only had 5 replicates. At the time of establishment the plants averaged 76.4 ± 10 cm (SD; range 45–99 cm) tall and had been maintained at tight spacing in Leach Tubes (3.8 cm in diameter, 14.0 cm deep). The trees were planted at a spacing of 1.2 m within rows and 3 m between columns, with columns in an approximately North–South orientation. There were 16 trees per column and 32 columns. Trees were randomized within blocks, which corresponded to approximately 2.5 columns each. The plantation was surrounded by a single border row consisting of extra transgenic and nontransgenic trees from the same background. Each tree was planted in the center of a 91 × 91 cm porous mat to control weed competition (VisPore® Tree Mats, Forestry Suppliers, MS, USA), staked and encircled by a 45-cm plastic tree collar to protect from rodents (Protex® Tree Collars, Forestry Suppliers, MS, USA). All trees were supported by a 1 m bamboo stake to prevent lodging due to high wind. The entire trial was surrounded by an electric fence to exclude large mammals.
All trees received irrigation using a T-tape drip irrigation system with 20-cm spacing between emitters (Aqua-Traxx). Trees were irrigated for 2 h per night for the first 2 months after establishment. This was reduced to 1 h on August 30 and to 30 min on September 5. Plants were fertilized twice with approximately 5 g of 19:19:19 N:P:K fertilizer (ca. 50 kg/ha) on July 30 and again on August 15. Granules were poured directly into the tree collars. Weeds were controlled by periodic sprays of Glyphosate and Clopyralid around the porous mats and by manual removal within the tree tubes, as needed.
Phenotyping and trait measurements
In order to evaluate the field performance of the 37 transgenic lines, 17 phenotypes were measured. These traits were selected to account for (1) yield and growth, (2) crown architecture, (3) vegetative phenology, and (4) response to an array of biotic and abiotic stressors (Table 2).
Table 2.
Phenotypes measured in the field trial
| Phenotypes | Units | Mean | SDa | r 2 b | P-valuec |
|---|---|---|---|---|---|
| Growth and yield | |||||
| Total height | cm | 207 | 16 | 0.558 | 2.23E−12 |
| Internode length | cm | 14.8 | 1.8 | 0.373 | 6.92E−03 |
| Height growth | cm | 130 | 17 | 0.317 | 3.28E−06 |
| Quadratic mean diameter | mm | 23.2 | 2.9 | 0.602 | 5.86E−05 |
| Volume index | m3 | 0.362 | 0.108 | 0.670 | 1.87E−05 |
| Crown architecture | |||||
| Height to first branch | cm | 117 | 15 | 0.297 | 5.03E−06 |
| Number of branches | Counts | 13.3 | 4.0 | 0.286 | 6.85E−09 |
| Stem sinuosity | Score: 0–4 | 1.21 | 0.86 | 0.028 | 3.89E−02 |
| Stem length-height ratio | NA | 0.988 | 0.019 | 0.187 | 4.70E−02 |
| Apical index | NA | 1.40 | 0.13 | 0.257 | 1.65E−02 |
| Trunk section eccentricity | NA | 0.215 | 0.097 | 0.069 | 6.00E−01 |
| Vegetative phenology | |||||
| Bud set | Score: 1–6 | 3.04 | 0.17 | 0.007 | 1.39E−74 |
| Bud flush | Score: 1–6 | 5.18 | 0.35 | 0.189 | 2.55E−09 |
| Stress response | |||||
| Frost damage | Score: 0–3 | 1.93 | 0.26 | 0.385 | 1.50E−01 |
| Melampsora severity | Score: 0–4 | 2.99 | 0.12 | 0.064 | 3.51E−01 |
| Overall herbivory | Score: 1–10 | 2.41 | 1.69 | 0.100 | 8.56E−01 |
| Twig borer incidence | Counts | 2.79 | 1.86 | 0.096 | 6.18E−01 |
NA not applicable for dimensionless trait
aThe standard deviation (SD) of observed values
bCoefficient of determination, r 2, between the observed values and the values predicted by the TPS models, an indicator of the degree of spatial-dependent variation in the trait
cSignificance level, P value, of the one-way ANOVAs for all lines (k = 37; including the seven empty-vector control lines and the wild type)
All measurements were performed on November 12–13, 2016 after all trees had become dormant, except as noted. Yield was estimated by (1) total height: the perpendicular distance between the ground and the apical bud; (2) relative height growth: the difference between the total height and the height of the plants at establishment; (3) quadratic mean diameter: the quadratic mean of the largest trunk transverse section axis and its perpendicular axis; (4) the volume index: the volume of a virtual cylinder with dimensions of total height and quadratic mean diameter; and (5) internode length: the total length of four internodes on the dominant stem leader. The four internodes were selected from the middle portion of the current year growth, where the size of the internodes was more uniform than at the beginning and end of the growing season.
To depict tree crown architecture, we measured (1) height to the first branch: the perpendicular distance between ground and the lowest branch on the tree; (2) number of branches: the number of primary branches on the stem; (3) stem sinuosity: a perceptual score from 0 (straight trunk) to 4 (heavily sinuous trunk); (4) stem length-height ratio: the ratio between the actual trunk length and the total height (defined as above); (5) the apical index: the ratio between the diameter of the apical stem, and of the mean of six lateral branch twig diameters, measured at the base of the 2017 new growth; and (6) trunk section eccentricity: the mathematical first eccentricity of the virtual ellipse created by the largest trunk transverse section axis and its perpendicular axis, as measured above.
Vegetative phenology was appraised by means of (1) bud set stage of the apical bud on October 11, 2016 using a visual scale ranging from 1 (actively growing) to 6 (bud completely set) [34]; and (2) bud flush stage on April 12, 2017, scoring from 1 (bud still dormant) to 6 (actively growing with fully developed leaves).
Finally, response to biotic and abiotic stress was evaluated by quantifying the incidence of three pervasive stressors in the field trial and a general estimation of arthropod grazing pressure. Frost damage was estimated on May 18, 2017 after an episode of late frost, using a visual scale of damage in the apical shoot from 0 (no necrosis) to 3 (apical meristem macroscopically detrimentally affected). Melampsora spp. severity was also measured with a visual score from 0 (no macroscopic symptoms) to 4 (> 50% canopy defoliation). Overall insect herbivory was scored from 0 to 10 based on the proportion of leaf area affected by feeding. Finally, incidence of the cottonwood twig borer Gypsonoma haimbachiana was assessed by counting the total number of larval holes made in six lateral branches plus the apical stem.
Statistical analyses
Although the experiment was designed to minimize environmental sources of variance, most of the traits studied are very influenced by microsite heterogeneity. To account for this, we modeled spatial variation of each trait using a thin plate spline (TPS) algorithm, using the R package module ‘fields’ [35]. The residuals of the models were retrieved and rescaled to the overall trait means to generate trait estimates with minimized spatial variation.
We performed an overall one-way ANOVA for each trait (k = 37), using transgenic line as factor. This analysis included the nine empty-vector control lines as well as the untransformed wild type WV94. To test for non-target effects of transformation, we performed one-way ANOVA for each trait using just the wild type and the empty-vector control lines as factors (k = 8). Finally, to test the actual effects of the transgenes in the WV94 background, we performed specific contrasts between the empty vector control lines and the lines containing the target gene constructs, as follows. First, to avoid an unbalanced contrast, we randomly selected a subset of 15 individuals of the empty-vector lines to be used as controls. We excluded lines EV1 and EV9 because these lines had clear evidence of somaclonal variation (see Results). Second, we tested for trait mean significant differences (one-way ANOVA) for all lines per construct together with the empty-vector control subset (k = 2–4, depending on the construct). Finally, whenever the ANOVA was significant, we conducted a Tukey’s HSD test to identify the pairs of lines that were significantly different. To account for false positive rate due to multi-testing, we restricted the significance threshold using Bonferroni correction.
Results and discussion
Trial establishment
The establishment of the trial was excellent (Fig. 1a). The survival rate was 100% and the trees had negligible transplant shock prior to resumption of growth. The overall productivity was high, with an average uncorrected height of 207 ± 16 cm (SD) and volume indexes of 0.362 ± 0.108 m3 (SD) at the end of the growing season (Fig. 1b). This growth was comparable to that reported in a transgenic trial in Belgium in the Populus tremula × alba cv. “717–1B4” background, but our trial had substantially lower variance [36]. The Belgian trial was also for a single growing season, and heights were approximately 225 ± 25 cm (SD). Stem dry weight, which should be proportional to volume index, was approximately 60 ± 20 g (SD) within genotypes. An Anglo-French study revealed similar net growth and slightly higher estimates of SD [37] than herein. Therefore, given the high productivity of poplar trials, 1 year analyses are highly informative of relative performance of transgenic lines [36].
Fig. 1.

Pictures of the trial a immediately after establishment on June 20, 2016, and b on October 3, 2016
Thin plate spline correction
TPS models reduced spatial variation in nearly all measured traits, though to different extents. Traits related to yield and growth had relatively high (predicted-vs-observed r 2 ranging 0.50–0.70; Fig. 2a, b) or moderate (r 2 = 0.15–0.40) spatial variability (Table 2). Crown architecture traits were mostly moderately affected by position, except trunk sinuosity and eccentricity, which were lightly affected (r 2 ≤ 0.10). Regarding vegetative phenology, bud flush was moderately affected by position whereas bud set was the trait with the lowest r 2 (almost negligible), as expected, given that it is primarily driven by day length rather than temperature [34, 38]. Finally, none of the biotic stressors showed strong position-dependence (r 2 ≤ 0.10; Fig. 2c, d; Additional file 1), but abiotic stress in the form of frost damage was moderately influenced by position in the field (r 2 = 0.385; Table 2).
Fig. 2.
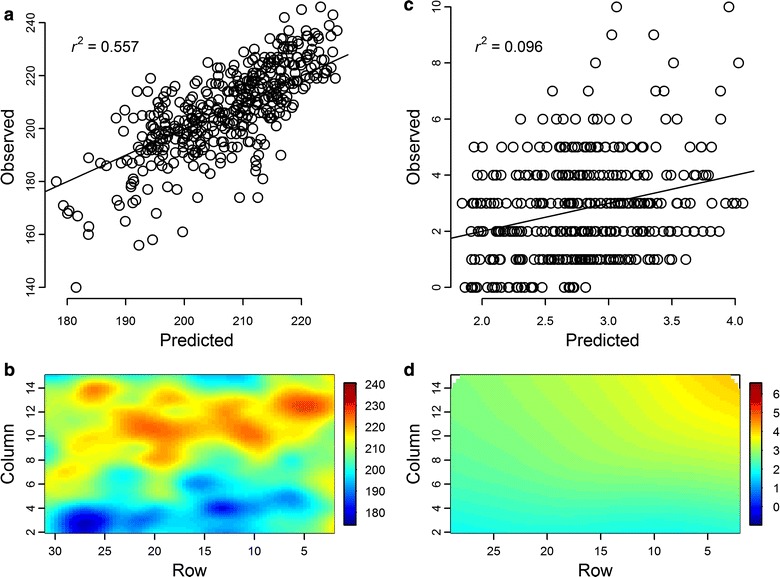
Thin-plate spline correction models. a Scatter plot of the total height values predicted by the model versus the observed values with the coefficient of determination (r 2). Also shown is the 1:1 line. b Heatmap of the trial layout with the total height predicted values by coordinate. Note that the color scale ranges from twice the standard deviation over the mean of the observed values to twice the standard deviation below the mean, to reflect the proportion of trait variance accounted for by the model. c, d Same plots for twig borer incidence
Trait variance across lines
The overall significance of the trait differences among lines was tested using a one-way ANOVA with k = 37 groups (i.e. lines) (Table 2). Interestingly, none of the traits reflecting direct responses to environmental stressors showed significant differences across the line means (ANOVA P > 0.15; Table 2). This is despite the fact that there was a serious outbreak of Melampsora leaf rust that affected 100% of the trees, attack by the cottonwood stem borer (Gypsonoma haimbachiana) that affected 94.2% of the trees, and a late frost event in May 2017 that caused visible damage on 99.9% of the trees.
Conversely, vegetative phenology showed strong differences among lines for both bud flush and bud set (ANOVA P < 1E−08). Within crown architecture traits, tests on height to first (highest) branch and number of branches were strongly significant (ANOVA P < 1E−05), whereas all other crown architecture traits were marginally or not significant (ANOVA P > 0.01). In general, yield trait tests were very significant (ANOVA P < 1E−05), with the sole exception of internode length, which was marginally significant (ANOVA P = 0.007) (Table 2).
It is worth noting that most of the traits with reduced spatial variation (estimated by the TPS predicted-vs-observed r 2) also displayed non-significant one way ANOVAs (Table 2). This indicates either that the inter-individual variance was very high (i.e. they are traits with high phenotypic plasticity in the background WV94) or it was very low (i.e. all the individuals have almost the same value). Only bud set did not follow the pattern, with very low spatial dependence but enormous inter-line variance.
Empty-vector controls
Empty vector control lines showed highly significant differences among lines for bud set, total height, height growth, and volume index and moderately significant differences for height to the first branch, number of branches, trunk diameter, and bud flush (Fig. 3). Post hoc pairwise contrasts between individual lines and the wild type control (Tukey’s HSD) revealed a lack of pairwise significant differences for most traits (Fig. 4a–d). However, lines EV2, EV4 and EV7 had significantly greater height than the wild type, though the differences were not dramatic, amounting to an approximately 6% increase in average height (Fig. 4a). More strikingly, bud set for line EV1 was markedly earlier than for the wild type control line and all of the other empty-vector lines (Fig. 4d). The resulting reduction in growing season ostensibly affected other traits like total height and number of branches as well (Fig. 4a, b). In contrast, line EV5 flushed significantly earlier than four other empty-vector lines, but not than the wild type (Fig. 4d).
Fig. 3.
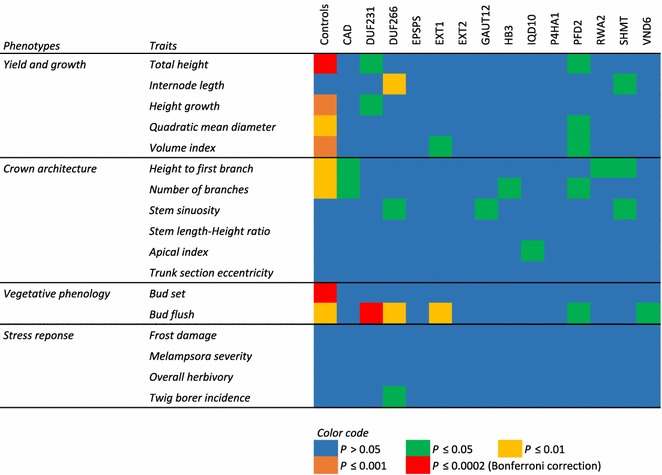
Heatmap of the level of significance of one-way ANOVAs per gene and per trait. Number of groups (k) varies as a function of the number of lines per gene: Control tests include the wild type and the seven empty-vector controls (k = 8) whereas the target gene tests include the random subsample of empty-vector trees plus the transgenic lines of each gene (from one to three; thus, k = 2–4)
Fig. 4.
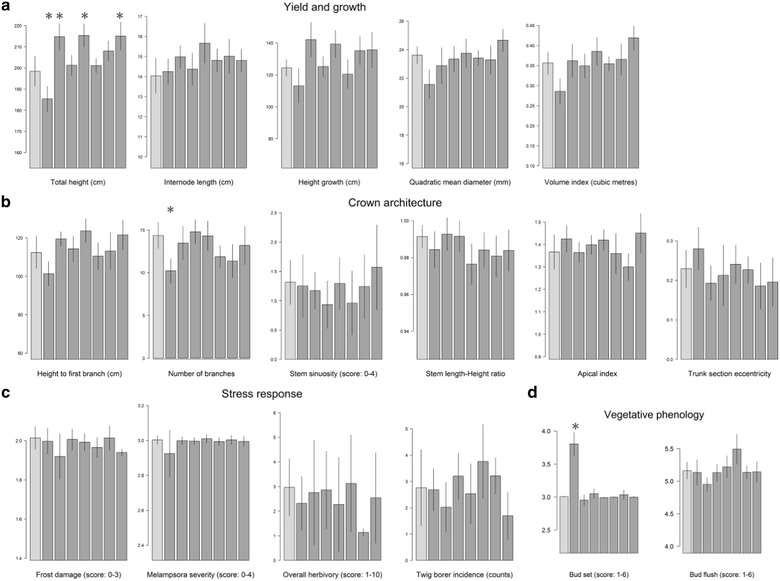
Bar plots of the eight control lines for the measured traits after TPS correction. Traits are indicative of a growth and yield, b crown architecture, c responses to stressors, and d vegetative phenology. Wild type WV94 is represented as the light grey bar and the empty-vector controls as the dark grey bars, ordered from EV1 to EV7. Error bars represent 95% confidence intervals. Asterisks indicate Tukey’s HSD mean difference significance (α = 0.05) between the marked empty-vector line and the wild type
Comparing the one-way ANOVAs between the control lines (WT and EVs) and the target gene lines (EV subsample plus the Comparator and TOP lines), significance was in general much larger within the control lines (Fig. 3), reflecting greater inter-line variance for the empty-vector controls compared to the gene vectors. It is well known that tissue culture and organogenesis can generate genetic instability due to cytosine methylation, repeat-induced point mutations, gross chromosomal rearrangements, and retrotransposon activation [39–43]. This somaclonal variation is apparently driven by oxidative stress cascades triggered by tissue culture conditions [44]. Furthermore, the T-DNA insertions of empty vectors could disrupt coding sequences or regulatory elements, thereby causing genetic changes and sometimes observable phenotypic modifications [45]. This process, called insertional mutagenesis, has been well characterized and widely used in functional genomics of model organisms, including plants [46, 47]. Furthermore, the promoters within the empty vectors could activate nearby genes, a fact that has been exploited previously in activation tagging efforts in Populus [30, 48]. Which of these different possible phenomena underlie our case remains to be explored further. However, this finding highlights the importance of including several independent empty-vector controls in transgenic filed trials to adequately estimate the background phenotypic variance generated solely by tissue culture and vector insertion and, therefore, appropriately calculate the significance of transgenic gains.
Transgenic TOP lines
The effects of the target genes on the measured traits were weak in general, estimated through one-way ANOVAs per gene and trait (number of tests = 238), with lines as groups including the empty-vector random subset as a negative control (k from 2 to 4). The main general trend observed was the lack of effect on the four traits related with stress responses (Additional file 2). Only the lines targeting the DUF266 gene seemed to be slightly affected, with a marginal ANOVA P value that could be an artifact of multiple testing (Fig. 3). Trunk section eccentricity was also not affected by any of the transgenes. Likewise, there were no significant differences from the controls for target genes EPSPS, EXT2 and P4HA1, and only weak (P > 0.01) effects for CAD, GAUT12, HB3, IQD10, RWA2, SHMT and VND6 (Fig. 3). None of these lines were significantly different from controls based on the Tukey’s HSD tests (Fig. 5).
Fig. 5.
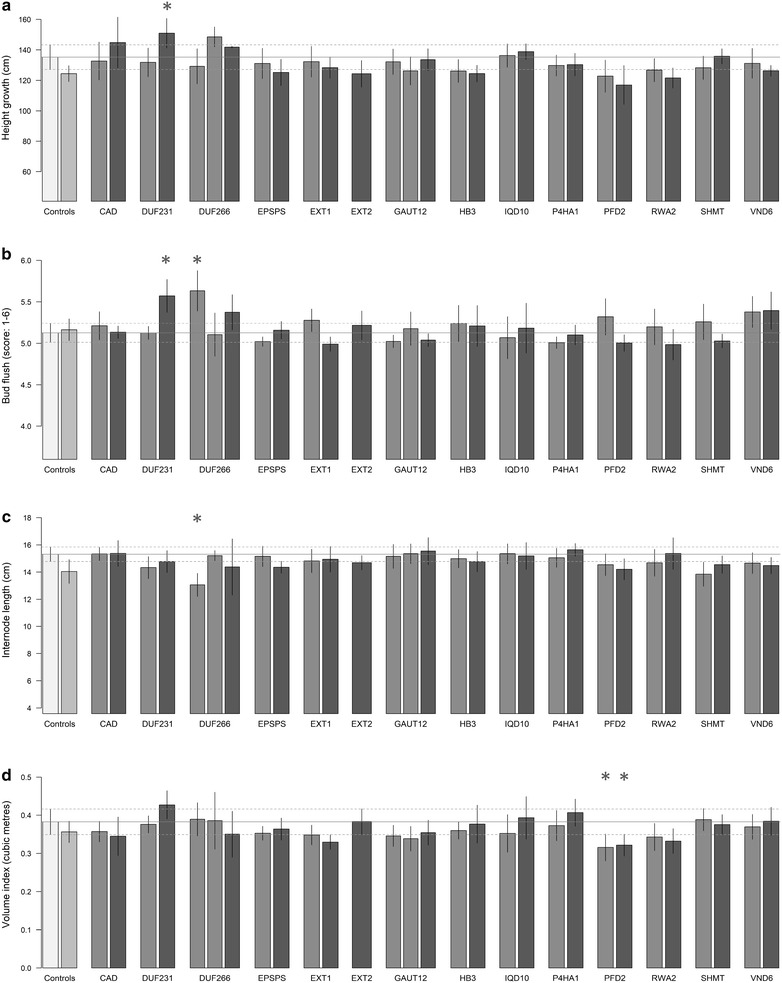
Bar plots of several measured traits after TPS correction for the random subsample of empty-vector trees (very light grey), the wild type (light grey), and the 29 trans-lines grouped by genes. Dark grey indicates Comparator lines and very dark grey indicates TOP lines. Error bars represent 95% confidence intervals. Asterisks indicate Tukey’s HSD mean difference significance (α = 0.05) between the marked transgenic line and the empty-vector control. Traits depicted are a height growth, b bud flush, c internode length, and d volume index
The DUF231 TOP line flushed significantly earlier and had also increased height growth compared to controls (Fig. 5a, b). This gene belongs to the Trichome Birefringence-Like (TBL) gene family [49]. Members of the TBL family are responsible for O-acetylation of hemicelluloses in Arabidopsis thaliana, and knockouts of these genes show altered cell wall phenotypes, including reduced cellulose crystallinity and decreased esterification [50]. Although the mechanisms of early bud flush remain to be determined, one might speculate that increased cell wall permeability in the DUF231 overexpression line facilitates diffusion of growth-promoting signals such as the FT1 protein into the dormant bud to promote resumption of growth, a scenario that is consistent with the central role of glucan hydrolases in releasing dormancy in Populus [51, 52]. Other target genes also showed a trend toward early bud flush, including DUF266, EXT1, PFD2, and VND6 (Figs. 3, 5b). Each of these could also have impacts on cell wall permeability, so a similar explanation for this trend could apply in each of these cases.
One of the comparator lines of the DUF266 target gene also showed significantly decreased internode length in addition to early bud flush (Fig. 5b, c). Interestingly, total height was barely reduced and volume index was slightly higher than the controls, due to an increase in stem diameter. It is worth noting that the bud flush phenotype could not have had a direct effect on the yield and growth values for this specific study, since it was measured in 2017 and the growth reported here occurred prior to this. Therefore, early bud flush could not have compensated for the observed reduction in internode length. This gene is a putative glycosyltransferase with direct impacts on cellulose biosynthesis. The proportion of cellulose and cellulose polymerization were both substantially elevated in stems of these transgenic lines in greenhouse studies [53]. However, the molecular mechanisms underlying these phenotypic effects have yet to be determined, so the reduced internode length and enhanced stem diameter remain to be explained.
Overexpression lines of PFD2 showed marginal significance for five traits, related to biomass and bud flush, pointing at a possible subtle trend (Fig. 3). Indeed, the two lines showed a significantly reduced volume index compared to the controls (Fig. 5d). The closest ortholog of this gene in Arabidopsis thaliana is AT3G22480 [54], which is part of the heterohexameric prefoldin complex, comprised of PFD1-6. Other members of this complex, specifically PFD3 and PFD5, bind to the DELLA protein, which mediates their levels in the cytosol, where the prefoldin complex is responsible for proper cortical microtubule formation [55]. DELLA proteins are diurnally regulated by gibberellin (GA) phytohormones, and their interactions with the prefoldin complex provides a possible mechanism for regulating cell wall expansion and anisotropic growth based on the formation and orientation of cortical microtubules [55, 56]. Overexpression of one member of the prefoldin complex may have disrupted this regulation, leading to reduced volume growth in the field. It is unclear why the opposite effect was seen in greenhouse studies (Table 1; unpublished observations), but since DELLA proteins are responsible for mediating photomorphogenesis, light quality (e.g., the red:far-red ratio), could be a factor [15, 57].
Conclusions
Overall the results of this trial reflect well upon the transgenic lines that have emerged from the intensive screening process conducted by the BESC. More than 500 gene targets have been evaluated in numerous greenhouse and growth chamber trials to identify genes with positive effects on sugar release in a high-throughput assay using thermochemical pretreatment and enzymatic hydrolysis [14]. Most lines have not shown any significant reductions in growth or tolerance of biotic or abiotic stresses in this field trial, despite several substantial challenges, including large outbreaks of the cottonwood twig borer and Melampsora leaf rust, as well as a late frost event. This is in contrast to some previous field studies of Populus trees with modified cell wall characteristics that show reduced yield in the field, including down-regulation of 4CL [23, 24], and down-regulation of cinnamoyl-CoA reductase [36]. One note of caution is that these trees have not yet experienced substantial drought stress due to irrigation in the first year, and mechanical stresses were mitigated by the use of tree collars and stakes during the establishment period. Irrigation and fertilization has been discontinued and the stakes have been removed, so it will be interesting to see if there are differential responses to drought, insects, and pathogens under more stressful conditions. It will also be important to determine if cell wall characteristics and enhanced saccharification efficiency persist in the field. Finally, a replicate trial is underway in Georgia, so there will be an opportunity to evaluate genotype-by-environment interactions for these lines, which have proven to be important for other cell wall modifications, such as 4CL down-regulation [24]. Nevertheless, this first year performance is a positive step toward the development of feedstocks that are optimized for consolidated bioprocessing for biofuel production.
Additional files
Additional file 1. Heatmaps for trait predicted values on each of the coordinates of the trial using a thin-plate spline correction model. Note that the color scale ranges from twice the standard deviation over the mean of the trait observed values to twice the standard deviation below the mean, to reflect the proportion of trait variance accounted for by the model. Traits represented are (A) internode length, (B) height growth, (C) quadratic mean diameter, (D) volume index, (E) height to first branch, (F) number of branches, (G) stem sinuosity, (H) stem length-height ratio, (I) apical index, (J) trunk section eccentricity, (K) bud set, (L) bud flush, (M) frost damage, (N) Melampsora severity, and (O) overall herbivory.
Additional file 2. Bar plots of several measured traits after TPS correction. Bars correspond to a random subsample of empty-vector trees (very light grey), the wild type (light grey), and the 29 transgenic lines grouped by genes. Dark grey indicates Comparator lines and very dark grey indicates TOP lines. Error bars represent 95% confidence intervals. Asterisks indicate Tukey’s HSD mean difference significance (α = 0.05) between the marked transgenic line and the empty-vector control. Traits depicted are (A) total height, (B) quadratic mean diameter, (C) height to first branch, (D) number of branches, (E) stem sinuosity, (F) stem length-height ratio, (G) apical index, (H) trunk section eccentricity, (I) bud set, (J) frost damage, (K) Melampsora severity, (L) overall herbivory, and (M) twig borer incidence.
Authors’ contributions
DMS performed phenotyping and statistical analyses and wrote the manuscript; J‐GC, UK, DM, WM, and TJT led the work on individual genes; LEG, ACB, RP, MX, YY, JZ, and AKB performed laboratory and greenhouse analyses of the transgenic lines; DMS, SJS, SPD, J-GC, LEG, WM, and JZ performed phenotypic measurements in the field trial; SJS performed insect and pathogen surveys and statistical analyses; GAT conceived the study and led the work on characterizing the transgenic lines; SPD led the field study and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
We thank Will Rottmann, Cassandra Collins, and Les Pearson of Arborgen LLC for producing the transgenic trees used in this trial, and the administration and staff of the West Virginia University Davis College of Agriculture, Natural Resources, and Design for assistance in establishing the trial. Field assistance was also provided by Roshan Abeyratne, Hari Chhetri, Luke Evans, Julianne Grady, Nesatalu Hiese, Dayane Lima, Sunita Mahat, Jacob Miller, Lucas Stover, Patrick Whitehouse, and Ran Zhou.
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the US Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
Competing interests
The strategy to produce improved biomass feedstock described in this paper has been included in a patent application. Patent applications have also been submitted or are in progress for several of the gene targets described herein.
Availability of data and materials
All raw data and plant materials are available from the corresponding author upon request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
This research was supported, in part, by funding from US Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science and by ORNL. ORNL is managed by UT-Battelle, LLC, for the US Department of Energy under Contract DE-AC05-00OR22725.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- BESC
BioEnergy Science Center
- ANOVA
analysis of variance
- SD
standard deviation
- TPS
thin-plate spline
- Tukey’s HSD
Tukey’s honest significant difference
- 4CL
4-hydroxycinnamoyl-CoA Ligase
- CAD
cinnamyl alcohol dehydrogenase
- DUF231
domain of unknown function 231
- DUF266
domain of unknown function 266
- EPSPS
5-enolpyruvylshikimate-3-phosphate synthase
- EXT1
extensin 1
- EXT2
extensin 2
- GAUT12
galacturonosyltransferase 12
- HB3
HOMEOBOX 3
- IQD10
isoleucine/glutamine (IQ) 67 domain 10
- P4HA1
prolyl 4-hydroxylase alpha subunit
- PFD2
prefoldin domain protein 2
- RWA2
reduced wall acetylation 2
- SHMT
serine hydroxymethyltransferase
- VND6
vascular-related NAC-domain protein 6
Footnotes
Electronic supplementary material
The online version of this article (doi:10.1186/s13068-017-0934-6) contains supplementary material, which is available to authorized users.
Contributor Information
David Macaya-Sanz, Email: david.macayasanz@mail.wvu.edu.
Jin‐Gui Chen, Email: chenj@ornl.gov.
Udaya C. Kalluri, Email: kalluriudayc@ornl.gov
Wellington Muchero, Email: mucherow@ornl.gov.
Timothy J. Tschaplinski, Email: tschaplinstj@ornl.gov
Lee E. Gunter, Email: gunterle@ornl.gov
Sandra J. Simon, Email: ssimon4@mix.wvu.edu
Ajaya K. Biswal, Email: biswalajaya@ccrc.uga.edu
Anthony C. Bryan, Email: acbryan11@gmail.com
Raja Payyavula, Email: rajasekhar78@yahoo.com.
Meng Xie, Email: xiem@ornl.gov.
Yongil Yang, Email: yi_yang@ornl.gov.
Jin Zhang, Email: zhangj1@ornl.gov.
Debra Mohnen, Email: dmohnen@ccrc.uga.edu.
Gerald A. Tuskan, Email: tuskanga@ornl.gov
Stephen P. DiFazio, Email: spdifazio@mail.wvu.edu
References
- 1.Pauly M, Keegstra K. Cell-wall carbohydrates and their modification as a resource for biofuels. Plant J. 2008;54:559–568. doi: 10.1111/j.1365-313X.2008.03463.x. [DOI] [PubMed] [Google Scholar]
- 2.Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 3.Zhao X, Zhang L, Liu D. Biomass recalcitrance. Part I: the chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels Bioprod Biorefining. 2012;6:465–482. doi: 10.1002/bbb.1331. [DOI] [Google Scholar]
- 4.Carroll A, Somerville C. Cellulosic biofuels. Annu Rev Plant Biol. 2009;60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- 5.Chundawat SPS, Beckham GT, Himmel ME, Dale BE. Deconstruction of lignocellulosic biomass to fuels and chemicals. Annu Rev Chem Biomol Eng. 2011;2:121–145. doi: 10.1146/annurev-chembioeng-061010-114205. [DOI] [PubMed] [Google Scholar]
- 6.Lynd L, van Zyl W, McBride J, Laser M. Consolidated bioprocessing of cellulosic biomass: an update. Curr Opin Biotechnol. 2005;16:577–583. doi: 10.1016/j.copbio.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Olson DG, McBride JE, Joe Shaw A, Lynd LR. Recent progress in consolidated bioprocessing. Curr Opin Biotechnol. 2012;23:396–405. doi: 10.1016/j.copbio.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 8.Parisutham V, Kim TH, Lee SK. Feasibilities of consolidated bioprocessing microbes: from pretreatment to biofuel production. Bioresour Technol. 2014;161:431–440. doi: 10.1016/j.biortech.2014.03.114. [DOI] [PubMed] [Google Scholar]
- 9.Papanek B, Biswas R, Rydzak T, Guss AM. Elimination of metabolic pathways to all traditional fermentation products increases ethanol yields in Clostridium thermocellum. Metab Eng. 2015;32:49–54. doi: 10.1016/j.ymben.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 10.Chung D, Cha M, Guss AM, Westpheling J. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc Natl Acad Sci USA. 2014;111:8931–8936. doi: 10.1073/pnas.1402210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polle A, Janz D, Teichmann T, Lipka V. Poplar genetic engineering: promoting desirable wood characteristics and pest resistance. Appl Microbiol Biotechnol. 2013;97:5669–5679. doi: 10.1007/s00253-013-4940-8. [DOI] [PubMed] [Google Scholar]
- 12.Kalluri UC, Keller M. Bioenergy research: a new paradigm in multidisciplinary research. J R Soc Interface. 2010;7:1391–1401. doi: 10.1098/rsif.2009.0564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuskan G, West D, Bradshaw HD, Neale D, Sewell M, Wheeler N, et al. Two high-throughput techniques for determining wood properties as part of a molecular genetics analysis of hybrid poplar and loblolly pine. Appl Biochem Biotechnol. 1999;77:55–66. doi: 10.1385/ABAB:77:1-3:55. [DOI] [Google Scholar]
- 14.Selig MJ, Tucker MP, Sykes RW, Reichel KL, Brunecky R, Himmel ME, et al. Original research: lignocellulose recalcitrance screening by integrated high-throughput hydrothermal pretreatment and enzymatic saccharification. Ind Biotechnol. 2010;6:104–111. doi: 10.1089/ind.2010.0009. [DOI] [Google Scholar]
- 15.Poorter H, Fiorani F, Pieruschka R, Wojciechowski T, van der Putten WH, Kleyer M, et al. Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol. 2016;212:838–855. doi: 10.1111/nph.14243. [DOI] [PubMed] [Google Scholar]
- 16.Pilate G, Dejardin A, Leple J-C. Field trials with lignin-modified transgenic trees. Adv Bot Res. 2012;61:1–36. doi: 10.1016/B978-0-12-416023-1.00001-X. [DOI] [Google Scholar]
- 17.Cabane M, Afif D, Hawkins S. Lignins and abiotic stresses. Adv Bot Res. 2012;61:219–262. doi: 10.1016/B978-0-12-416023-1.00007-0. [DOI] [Google Scholar]
- 18.Li J, Brunner AM, Meilan R, Strauss SH. Stability of transgenes in trees: expression of two reporter genes in poplar over three field seasons. Tree Physiol. 2008;29:299–312. doi: 10.1093/treephys/tpn028. [DOI] [PubMed] [Google Scholar]
- 19.Walter C, Fladung M, Boerjan W. The 20-year environmental safety record of GM trees. Nat Biotechnol. 2010;28:656–658. doi: 10.1038/nbt0710-656. [DOI] [PubMed] [Google Scholar]
- 20.Li JL, Meilan R, Ma C, Barish M, Strauss SH. Stability of herbicide resistance over 8 years of coppice in field-grown, genetically engineered poplars. West J Appl For. 2008;23:89–93. [Google Scholar]
- 21.Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet. 2010;44:337–363. doi: 10.1146/annurev-genet-102209-163508. [DOI] [PubMed] [Google Scholar]
- 22.Hu WJ, Harding S, Lung J, Popko JL, Ralph J, Stokke DD, et al. Repression of lignin biosynthesis promotes cellulose accumulation and growth in transgenic trees. Nat Biotechnol. 1999;17:808–812. doi: 10.1038/11758. [DOI] [PubMed] [Google Scholar]
- 23.Voelker SL, Lachenbruch B, Meinzer FC, Kitin P, Strauss SH. Transgenic poplars with reduced lignin show impaired xylem conductivity, growth efficiency and survival. Plant Cell Environ. 2011;34:655–668. doi: 10.1111/j.1365-3040.2010.02270.x. [DOI] [PubMed] [Google Scholar]
- 24.Stout AT, Davis AA, Domec JC, Yang C, Shi R, King JS. Growth under field conditions affects lignin content and productivity in transgenic Populus trichocarpa with altered lignin biosynthesis. Biomass Bioenerg. 2014;68:228–239. doi: 10.1016/j.biombioe.2014.06.008. [DOI] [Google Scholar]
- 25.Kitin P, Voelker SL, Meinzer FC, Beeckman H, Strauss SH, Lachenbruch B. Tyloses and phenolic deposits in xylem vessels impede water transport in low-lignin transgenic poplars: a study by cryo-fluorescence microscopy. Plant Physiol. 2010;154:887–898. doi: 10.1104/pp.110.156224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ranjan P, Yin T, Zhang X, Kalluri UC, Yang X, Jawdy S, et al. Bioinformatics-based identification of candidate genes from QTLs associated with cell wall traits in Populus. BioEnergy Res. 2010;3:172–182. doi: 10.1007/s12155-009-9060-z. [DOI] [Google Scholar]
- 27.Kalluri UC, Hurst GB, Lankford PK, Ranjan P, Pelletier DA. Shotgun proteome profile of Populus developing xylem. Proteomics. 2009;9:4871–4880. doi: 10.1002/pmic.200800854. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Ye C-Y, Bisaria A, Tuskan G, Kalluri UC. Identification of candidate genes in Arabidopsis and Populus cell wall biosynthesis using text-mining, co-expression network analysis and comparative genomics. Plant Sci. 2011;181:675–687. doi: 10.1016/j.plantsci.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 29.Abraham P, Adams R, Giannone RJ, Kalluri U, Ranjan P, Erickson B, et al. Defining the boundaries and characterizing the landscape of functional genome expression in vascular tissues of Populus using shotgun proteomics. J Proteome Res. 2012;11:449–460. doi: 10.1021/pr200851y. [DOI] [PubMed] [Google Scholar]
- 30.Busov V, Yordanov Y, Gou J, Meilan R, Ma C, Regan S, et al. Activation tagging is an effective gene tagging system in Populus. Tree Genet Genom. 2011;7:91–101. doi: 10.1007/s11295-010-0317-7. [DOI] [Google Scholar]
- 31.Evans LM, Slavov GT, Rodgers-Melnick E, Martin J, Ranjan P, Muchero W, et al. Population genomics of Populus trichocarpa identifies signatures of selection and adaptive trait associations. Nat Genet. 2014;46:1089–1096. doi: 10.1038/ng.3075. [DOI] [PubMed] [Google Scholar]
- 32.Karve AA, Jawdy SS, Gunter LE, Allen SM, Yang X, Tuskan GA, et al. Initial characterization of shade avoidance response suggests functional diversity between Populus phytochrome B genes. New Phytol. 2012;196:726–737. doi: 10.1111/j.1469-8137.2012.04288.x. [DOI] [PubMed] [Google Scholar]
- 33.Studer MH, DeMartini JD, Davis MF, Sykes RW, Davison B, Keller M, et al. Lignin content in natural Populus variants affects sugar release. Proc Natl Acad Sci USA. 2011;108:6300–6305. doi: 10.1073/pnas.1009252108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohde A, Storme V, Jorge V, Gaudet M, Vitacolonna N, Fabbrini F, et al. Bud set in poplar-genetic dissection of a complex trait in natural and hybrid populations. New Phytol. 2011;189:106–121. doi: 10.1111/j.1469-8137.2010.03469.x. [DOI] [PubMed] [Google Scholar]
- 35.Nychka D, Furrer R, Paige J, Sain S. Fields: Tools for spatial data. 2015. http://www.image.ucar.edu/fields. Accessed 25 June 2017.
- 36.Van Acker R, Leple J-C, Aerts D, Storme V, Goeminne G, Ivens B, et al. Improved saccharification and ethanol yield from field-grown transgenic poplar deficient in cinnamoyl-CoA reductase. Proc Natl Acad Sci USA. 2014;111:845–850. doi: 10.1073/pnas.1321673111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilate G, Guiney E, Holt K, Petit-Conil M, Lapierre C, Leple J-C, et al. Field and pulping performances of transgenic trees with altered lignification. Nat Biotechnol. 2002;20:607–612. doi: 10.1038/nbt0602-607. [DOI] [PubMed] [Google Scholar]
- 38.Singh RK, Svystun T, AlDahmash B, Jönsson AM, Bhalerao RP. Photoperiod- and temperature-mediated control of phenology in trees—a molecular perspective. New Phytol. 2017;213:511–524. doi: 10.1111/nph.14346. [DOI] [PubMed] [Google Scholar]
- 39.Phillips RL, Kaeppler SM, Olhoft P. Genetic instability of plant tissue cultures: breakdown of normal controls. Proc Natl Acad Sci USA. 1994;91:5222–5226. doi: 10.1073/pnas.91.12.5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaeppler SM, Phillips RL. Tissue culture-induced DNA methylation variation in maize. Proc Natl Acad Sci USA. 1993;90:8773–8776. doi: 10.1073/pnas.90.19.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hirochika H, Sugimoto K, Otsuki Y, Tsugawa H, Kanda M. Retrotransposons of rice involved in mutations induced by tissue culture. Proc Natl Acad Sci USA. 1996;93:7783–7788. doi: 10.1073/pnas.93.15.7783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin PJ, Scowcroft WR. Somaclonal variation—a novel source of variability from cell cultures for plant improvement. Theor Appl Genet. 1981;60:197–214. doi: 10.1007/BF02342540. [DOI] [PubMed] [Google Scholar]
- 43.Lee M, Phillips RL. The chromosomal basis of somaclonal variation. Annu Rev Plant Physiol Plant Mol Biol. 1988;39:413–437. doi: 10.1146/annurev.pp.39.060188.002213. [DOI] [Google Scholar]
- 44.Cassells AC, Curry RF. Oxidative stress and physiological, epigenetic and genetic variability in plant tissue culture: Implications for micropropagators and genetic engineers. Plant Cell Tissue Organ Cult. 2001;64:145–157. doi: 10.1023/A:1010692104861. [DOI] [Google Scholar]
- 45.Stanford WL, Cohn JB, Cordes SP. Mouse genomic technologies: gene-trap mutagenesis: past, present and beyond. Nat Rev Genet. 2001;2:756–768. doi: 10.1038/35093548. [DOI] [PubMed] [Google Scholar]
- 46.Jeon J-S, Lee S, Jung K-H, Jun S-H, Jeong D-H, Lee J, et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- 47.Tadege M, Wen J, He J, Tu H, Kwak Y, Eschstruth A, et al. Large-scale insertional mutagenesis using the Tnt1 retrotransposon in the model legume Medicago truncatula. Plant J. 2008;54:335–347. doi: 10.1111/j.1365-313X.2008.03418.x. [DOI] [PubMed] [Google Scholar]
- 48.Harrison EJ, Bush M, Plett JM, McPhee DP, Vitez R, O’Malley B, et al. Diverse developmental mutants revealed in an activation-tagged population of poplar. Can J Bot. 2007;85:1071–1081. doi: 10.1139/B07-063. [DOI] [Google Scholar]
- 49.Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, et al. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 2010;153:590–602. doi: 10.1104/pp.110.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Manabe Y, Verhertbruggen Y, Gille S, Harholt J, Chong S-L, Pawar PM-A, et al. Reduced wall acetylation proteins play vital and distinct roles in cell wall O-acetylation in Arabidopsis. Plant Physiol. 2013;163:1107–1117. doi: 10.1104/pp.113.225193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rinne PLH, Welling A, Vahala J, Ripel L, Ruonala R, Kangasjärvi J, et al. Chilling of dormant buds hyperinduces FLOWERING LOCUS T and recruits GA-inducible 1,3-beta-glucanases to reopen signal conduits and release dormancy in Populus. Plant Cell. 2011;23:130–146. doi: 10.1105/tpc.110.081307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brunner AM, Evans LM, Hsu C-Y, Sheng X. Vernalization and the chilling requirement to exit bud dormancy: shared or separate regulation? Front Plant Sci. 2014;5:1–7. doi: 10.3389/fpls.2014.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang Y, Yoo CG, Guo H-B, Rottmann W, Winkeler KA, Collins CM, et al. Overexpression of a Domain of Unknown Function 266-containing protein results in high cellulose content, reduced recalcitrance, and enhanced plant growth in the bioenergy crop Populus. Biotechnol Biofuels. 2017;10:74. doi: 10.1186/s13068-017-0760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cao J. Analysis of the prefoldin gene family in 14 plant species. Front Plant Sci. 2016;7:317. doi: 10.3389/fpls.2016.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Locascio A, Blázquez MA, Alabadí D. Dynamic regulation of cortical microtubule organization through prefoldin-DELLA interaction. Curr Biol. 2013;23:804–809. doi: 10.1016/j.cub.2013.03.053. [DOI] [PubMed] [Google Scholar]
- 56.Oda Y. Cortical microtubule rearrangements and cell wall patterning. Front Plant Sci. 2015;6:236. doi: 10.3389/fpls.2015.00236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Achard P, Liao L, Jiang C, Desnos T, Bartlett J, Fu X, et al. DELLAs contribute to plant photomorphogenesis. Plant Physiol. 2007;143:1163–1172. doi: 10.1104/pp.106.092254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barakat A, Bagniewska-Zadworna A, Choi A, Plakkat U, DiLoreto DS, Yellanki P, et al. The cinnamyl alcohol dehydrogenase gene family in Populus: phylogeny, organization, and expression. BMC Plant Biol. 2009;9:26. doi: 10.1186/1471-2229-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maeda H, Dudareva N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu Rev Plant Biol. 2012;63:73–105. doi: 10.1146/annurev-arplant-042811-105439. [DOI] [PubMed] [Google Scholar]
- 60.Lamport DTA, Kieliszewski MJ, Chen Y, Cannon MC. Role of the extensin superfamily in primary cell wall architecture. Plant Physiol. 2011;156:11–19. doi: 10.1104/pp.110.169011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Biswal AK, Hao Z, Pattathil S, Yang X, Winkeler K, Collins C, et al. Downregulation of GAUT12 in Populus deltoides by RNA silencing results in reduced recalcitrance, increased growth and reduced xylan and pectin in a woody biofuel feedstock. Biotechnol Biofuels. 2015;8:41. doi: 10.1186/s13068-015-0218-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du J, Miura E, Robischon M, Martinez C, Groover A. The Populus class III HD ZIP transcription factor popcorona affects cell differentiation during secondary growth of woody stems. PLoS One. 2011;6:e17458. doi: 10.1371/journal.pone.0017458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Abel S, Bürstenbinder K, Müller J. The emerging function of IQD proteins as scaffolds in cellular signaling and trafficking. Plant Signal Behav. 2013;8:e24369. doi: 10.4161/psb.24369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Keskiaho K, Hieta R, Sormunen R, Myllyharju J. Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly. Plant Cell. 2007;19:256–269. doi: 10.1105/tpc.106.042739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prabhu V, Chatson KB, Abrams GD, King J. 13C nuclear magnetic resonance detection of interactions of serine hydroxymethyltransferase with C1-tetrahydrofolate synthase and glycine decarboxylase complex activities in Arabidopsis. Plant Physiol. 1996;112:207–216. doi: 10.1104/pp.112.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhong R, McCarthy RL, Lee C, Ye Z-H. Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar. Plant Physiol. 2011;157:1452–1468. doi: 10.1104/pp.111.181354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Heatmaps for trait predicted values on each of the coordinates of the trial using a thin-plate spline correction model. Note that the color scale ranges from twice the standard deviation over the mean of the trait observed values to twice the standard deviation below the mean, to reflect the proportion of trait variance accounted for by the model. Traits represented are (A) internode length, (B) height growth, (C) quadratic mean diameter, (D) volume index, (E) height to first branch, (F) number of branches, (G) stem sinuosity, (H) stem length-height ratio, (I) apical index, (J) trunk section eccentricity, (K) bud set, (L) bud flush, (M) frost damage, (N) Melampsora severity, and (O) overall herbivory.
Additional file 2. Bar plots of several measured traits after TPS correction. Bars correspond to a random subsample of empty-vector trees (very light grey), the wild type (light grey), and the 29 transgenic lines grouped by genes. Dark grey indicates Comparator lines and very dark grey indicates TOP lines. Error bars represent 95% confidence intervals. Asterisks indicate Tukey’s HSD mean difference significance (α = 0.05) between the marked transgenic line and the empty-vector control. Traits depicted are (A) total height, (B) quadratic mean diameter, (C) height to first branch, (D) number of branches, (E) stem sinuosity, (F) stem length-height ratio, (G) apical index, (H) trunk section eccentricity, (I) bud set, (J) frost damage, (K) Melampsora severity, (L) overall herbivory, and (M) twig borer incidence.
Data Availability Statement
All raw data and plant materials are available from the corresponding author upon request.


