Abstract
The contributions of cytochrome P450 3A5 to the metabolic clearance of marketed drugs is unclear, but its probable role is to augment the metabolism of several drugs that are largely cleared by P450 3A4. Selective metabolism by 3A4 is often a concern in drug development owing to potential drug-drug interactions and the variability of 3A4 and 3A5 expression. The contribution of P450 3A5 to these clearance pathways varies between individuals owing to genetic differences and similarities and differences in the metabolic properties of 3A5 compared with 3A4. To better understand the structural differences between P450s 3A4 and 3A5, the structure of 3A5 complexed with ritonavir was determined by X-ray crystallography to a limiting resolution of 2.91 Å. The secondary and tertiary structures of 3A5 and 3A4 are similar, but the architectures of their active sites differ. The 3A5 active site is taller and narrower than that of 3A4. As a result, ritonavir adopts a distinctly different conformation to fit into the cavity of 3A5 than seen for 3A4. These structural changes reflect amino acid differences that alter the conformation of the helix F through helix G region in the upper portion of the cavity and ionic interactions between residues in the beta-sheet domain that reduce the width of the cavity. The structural differences exhibited by 3A4 and 3A5 suggest that the overlap of catalytic activities may reflect molecular flexibility that determines how alternative conformers fit into the different active site architectures of the two enzymes.
Introduction
The cytochrome P450 3A family contributes significantly to the metabolic clearance of marketed drugs (Zanger et al., 2008) and to the underlying reaction pathways (Rendic and Guengerich, 2015). P450 3A4 is prominently expressed in adult liver, but the contributions of P450s 3A7 and 3A43 to drug clearance in adults is negligible owing to their low expression in liver (Stevens, 2006; Ince et al., 2013). In contrast, P450 3A7 rather than 3A4 is prominently expressed in liver during the fetal and perinatal periods. P450 3A5 is expressed in both fetal and adult liver, but the functional CYP3A5*1 allele has a low frequency of occurrence that varies between ethnic populations (Kuehl et al., 2001). P450 3A5*1 expression is highest in African ethnic groups, where the frequency of the CYP3A5*1 allele is greater than 50%. In contrast, most Caucasians and Asians do not express detectable amounts of hepatic P450 3A5 owing to low CYP3A5*1 allele frequencies of roughly 5% and 30%, respectively. This low or absent expression of P450 3A5 is the result of a high frequency of CYP3A5*3 allele in these ethnic groups, which has a splicing defect that leads to premature termination of protein translation (Kuehl et al., 2001; Westlind-Johnsson et al., 2003).
An overlap for metabolic clearance by P450s 3A4 and 3A5 is evident for many drugs (Koudriakova et al., 1998; Williams et al., 2002; Yamazaki et al., 2010; Tseng et al., 2014). Recent development of a P450 3A4 selective inhibitor (Walsky et al., 2012) and subsequent in vitro drug metabolism studies with 3A5 *1/*1 liver microsomes prepared from genotyped livers indicated that P450 3A5 contributes most highly to metabolism of atazanavir, vincristine, midazolam, vardenafil, otenabant, verapamil, and tacrolimus, whereas a negligible contribution was observed for metabolism of 17 of 32 drugs that are predominantly cleared by P450 3A4 (Tseng et al., 2014), suggesting differences in their respective active site features. Furthermore, organ transplant patients with the 3A5*1 allele would probably benefit from individualized adjustments for the initial dosing of tacrolimus to establish immunosuppression quickly, owing to increased metabolic clearance of the drug (Birdwell et al., 2015). Additionally, these two enzymes can catalyze reactions that form different metabolites from the same substrate. For instance, P450 3A5 produces a unique N-oxide metabolite of the phosphodiesterase inhibitor T-1032 (Li et al., 2014) and catalyzes oxygenation of the dichlorocyclohexane ring of maraviroc, whereas 3A4 oxygenates other sites (Lu et al., 2012).
The structure of P450 3A4 exhibits significant differences of secondary and tertiary structure from other drug-metabolizing P450s in families 1 and 2, and these contribute to a large active site cavity (Williams et al., 2004; Yano et al., 2004; Johnson and Stout, 2013). The capacity of this active site cavity to bind two or more molecules potentially underlies the complex kinetic properties displayed by 3A4. Additionally, structures of 3A4 indicate that the binding of substrates and inhibitors can expand the active site cavity (Ekroos and Sjögren, 2006; Sevrioukova and Poulos, 2010, 2012b) or trigger contractions (Sevrioukova and Poulos, 2017) that are enabled by the flexibility of the upper surface of the cavity opposite from the heme prosthetic group. The flexibility of the upper surface reflects a lack of secondary structure in the helical F through G region that is typically seen in other drug-metabolizing P450s (Williams et al., 2004; Yano et al., 2004; Johnson and Stout, 2013). The amino acid sequences of 3A4 and 3A5 exhibit 83% identity, with most of the differences residing in the helical F through G region that forms the roof of the active site cavity and in the N-terminal region of the catalytic domain that forms one side of the cavity in the 3A4 structure. To determine whether these distinctive features are conserved in 3A5 and to identify structural features that may contribute to the distinctive metabolic properties of 3A5, we crystallized 3A5 complexed with ritonavir (a human immunodeficiency virus type I protease inhibitor) and determined the structure of the complex by X-ray crystallography. Although the secondary and tertiary structures of P450 3A5 and 3A4 complexes with ritonavir are similar, the two enzymes exhibit distinctly different active site architectures. The 3A5 active site is taller and narrower than that of 3A4. The differences in the active site structures reflect amino acid differences that alter the conformation of the helix F through helix G region in the upper portion of the cavity and change ionic interactions between residues in the β-sheet domain opposite to helix I that increase the height but reduce the width of the cavity in the structure of the 3A5 ritonavir complex.
Materials and Methods
Chemicals and Reagents.
Isopropyl β-d-1-thiogalactopyranoside and C12E9 were obtained from Anatrace (Maumee, OH), lysozyme was from Worthington (Lakewood, NJ), nickel-nitrotriloacetate agarose (Ni-NTA agarose) was from Qiagen (Valencia, CA), carboxymethyl Sepharose Fast Flow was from GE Healthcare Biosciences (Pittsburgh, PA), and HA ultragel was from Pall Life Sciences (Port Washington, NY). Restriction enzymes, NdeI, XbaI, and HindIII were obtained from New England BioLabs (Ipswich, MA), and PfuUltra II fusion HS DNA polymerase was from Agilent Technologies (Santa Clara, CA). The following chemicals were purchased from Sigma-Aldrich (St Louis, MO): 5-aminolevulinic acid hydrochloride, l-arabinose, sodium cholate, l-histidine, DNase I, phenylmethylsulfonyl fluoride (PMSF), ritonavir, and Sephacryl S-200 HR. Pierce EDTA-free protease inhibitor tablets and all other chemicals were purchased from Thermo Fisher Scientific (Waltham, MA).
Construction of Expression Plasmids.
For expression of P450 3A5 in Escherichia coli for crystallization, plasmids utilizing pCWori vector were generated with sequences encoding modified and truncated N- and C-termini of the protein (Supplemental Fig. S1) using a strategy similar to the one used for construction of the expression plasmid for P450 3A4dH (Yano et al., 2004). The polymerase chain reaction (PCR) template was a CYP3A5*1 cDNA clone (ID: 5188554) purchased from Thermo Fisher Scientific–Open Biosystems. DNA sequencing confirmed the correctness of the allele. Primers were used to generate modified cDNA with deleted portions of the transmembrane helix and linker region. The modified cDNA also contained restriction sites for insertion into the pCWori vector as described in Supplemental Fig. S1. The C-terminal modification contained the additional codons encoding four histidine residues to facilitate protein purification. PfuUltra II fusion HS DNA polymerase was used to generate the PCR fragments using the following cycling condition: one cycle 2 minutes 95°C, 30 cycles of 20 seconds 95°C, 20 seconds 58°C and 1 minute 72°C, and final extension at 72°C for 3 minutes. The PCR products were digested with NdeI/XbaI or NdeI/HindIII, and the fragments were subcloned into pCWori+ vector (Gegner and Dahlquist, 1991). The authenticity of the expression constructs was confirmed by DNA sequencing. Expression plasmids were cotransformed with pGro7 (TakaRa Bio, Kusatsu/Shiga, Japan) into E. coli strain DH5α for protein expression.
Expression and Purification.
A single colony was grown in super broth at 37°C overnight. The bacterial culture was centrifuged at 2500g for 10 minutes, and the pellet was suspended in Terrific Broth. The culture was grown at 200 rpm at 37°C until the optical density of 600 nm reached 0.4–0.5. The shaking rate and temperature were then reduced to 170 rpm and 28°C. After 30 minutes, the expression of the modified P450 3A5 and GroEL and GroES was induced by adding 1 mM isopropyl β-d-1-thiogalactopyranoside, 0.5 mM 5-aminolevulinic acid hydrochloride, and 4 g/l arabinose. Cells were harvested 44–48 hours later. Spheroplasts were generated and suspended in 500 mM potassium phosphate buffer, pH 7.4, containing 20% glycerol, 0.5 mM PMSF, and protease inhibitor tablet. The spheroplasts were lysed by sonication and cleared by centrifugation at 3100g for 15 minutes. CHAPS (8 mM) was added to the cleared lysate, and after a 1 hour incubation at 4°C, the mixture was centrifuged at 100,000g for 60 minutes. The supernatant was then incubated with nickel-nitrilotriacetate agarose at a ratio of 90 nmol P450 per milliliter of resin overnight at 4°C on a bottle roller. The 3A5-nickel-nitrotriloacetate agarose slurry was loaded onto a column and sequentially washed with five column volumes of buffer A (500 mM potassium phosphate buffer, pH 7.4, containing 20% glycerol, 8 mM CHAPS, 10 mM β-mercaptoethanol, and 0.5 mM PMSF), and then buffer A containing 5 mM ATP, 10 mM MgCl2, and 100 mM KCl to remove E. coli chaperone proteins. This was followed by washes with five column volumes of buffer A containing 3 mM histidine. The enzyme was eluted with buffer A containing 40 mM histidine, and pooled fractions were diluted 25-fold with buffer C [100 mM sodium Hepes, pH 7.4, containing 20% glycerol, 8 mM CHAPS, 0.5 mM EDTA, 10 mM β-mercaptoethanol, 20 mM potassium acetate (KOAc), pH 7.4, and 0.5 mM PMSF], and incubated with carboxymethyl Sepharose at a ratio of 30 nmol P450 per milliliter of resin for 3 hours at 4°C on a bottle roller. The slurry was transferred to a chromatography column and washed with five column volumes of buffer C containing 20 mM potassium phosphate (pH 7.4), which was followed by 10 column volumes of buffer D (100 mM sodium Hepes, pH 7.4, containing 20% glycerol, 0.5 mM EDTA, 10 mM β-mercaptoethanol, and 0.5 mM PMSF) with 60 mM KOAc, pH 7.4. The protein was eluted with buffer D containing 250 mM KOAc, pH 7.4. Selected fractions were pooled, concentrated, and loaded onto a Sephacryl S-200 HR. The protein was eluted with buffer D containing 50 mM KOAc, pH 7.4 (buffer E). Selected fractions were pooled and concentrated. The concentration of the enzyme was determined from the intensity of the Soret band of the reduced carbon monoxide complex using visible absorption difference spectroscopy and an extinction coefficient of 0.091 μM−1 cm−1 (Omura and Sato, 1964).
Ligand Binding Studies.
The binding of ritonavir to 3A5 in buffer E was monitored by the reduction in the intensity of the absorption spectrum at 417 nm and the increase of intensity at 421 nm owing to coordination of the thiazole group of ritonavir with the ferric iron of the heme cofactor (Sevrioukova and Poulos, 2010). After each addition of ritonavir (in methanol), the spectra were recorded after a 3-minute incubation at room temperature. The total volume of the solvent added was <2% (v/v). After subtracting the spectrum of the enzyme obtained in the absence of the ligand from each of the spectra recorded following the addition of the ligand, the concentration-dependent changes in the trough-to-peak absorbance for the difference spectra were fit with the quadratic form of the binding equation by nonlinear regression using Slide Write Plus as described (Jung et al., 1998). The binding of ritonavir to P450 3A4dH was determined using the same procedure.
Crystallization and Structure Determination.
Owing to the limited solubility of ritonavir, the P450 3A5-ritonavir complex was prepared from ligand-free purified P450 3A5 at low protein concentrations in a stoichiometric excess of ritonavir. The 3A5-ritonavir protein complex was then concentrated in the presence of excess ritonavir. The wavelength maximum of the Soret band was used to monitor ritonavir binding along with the reduction in the intensity of the α-band relative to the β-band (Supplemental Fig. S2). If necessary, dilution and concentration of the 3A5-ritonavir complex in the presence of ritonavir was repeated to achieve values seen for saturation of the enzyme. Ritonavir-bound P450 3A5 was crystalized by a sitting drop vapor diffusion in a 24-well Cryschem Plate (Hampton Research, Aliso Viejo, CA). One microliter of the ritonavir-bound 3A5 protein (450 μM) in buffer E was mixed with 0.1 μl 10% (w/v) C12E9 and 1 μl of the precipitant containing 0.2 M sodium N-(2-acetamido)iminodiacetate, pH 6.5, 1 mM zinc sulfate, 5% polyethylene glycol (PEG) 300, and 28% PEG 3350. The drop was set to equilibrate against 0.5 ml of precipitant solution in the reservoir at 24°C. Crystals appeared and grew within several days. Prior to crystal collection, 2 μl of cryoprotection solution was added into the drop. Cryoprotection solution is a 1:1 (v/v) mixture of PEG300 and mother liquor [protein buffer: precipitant solution = 1:1 (v/v)]. In addition, 10 mM ritonavir stock was added into the cryoprotectant to final concentration of 0.1 mM. Crystals were harvested and flash frozen in liquid nitrogen prior to shipment to the Stanford Synchrotron Radiation Lightsource. X-ray diffraction data used for structure determination were collected from a single crystal at 100 K on Stanford Synchrotron Radiation Lightsource beam line 7–1. The X-ray diffraction reflections were integrated using XDS (Kabsch, 2010) and merged and scaled using AIMLESS (Evans and Murshudov, 2013) to a limiting resolution of 2.91 Å in the P 21 21 21 space group. Initial phases were obtained by molecular replacement, using the Phaser Crystallographic Software 2.5.6 (McCoy et al., 2007) as implemented in PHENIX (Lawrence Berkeley Laboratory, Berkeley, CA) and chain A of the structure of the 3A4-ritonavir complex [Protein Data Bank (PDB):3NXU] as the search model. Phaser version 2.5.6 identified 12 chains in the unit cell. The Crystallographic Object-Oriented Toolkit Coot (Emsley et al., 2010) was used for model building, and PHENIX was used for model refinement. Chain A was built initially, and the remaining chains were generated by noncrystallographic symmetry (NCS) operators using PHENIX. Adjustments were made to individual chains followed by refinement initially with torsion angle NCS restraints in PHENIX. The NCS restraints were removed in final phases of model building. The refinement protocols were optimized in PHENIX to lower R and free R and retain differences between R and free R to <4.5%. The model-to-data fit for the final structure exhibits an R work of 0.214 and free R value of 0.258 with a high resolution limit of 2.91 Å. Statistics for data processing, the fit of the model to the data, and the quality of the model are provided in Table 1. Graphic images were generated using PyMOL (Schrödinger, Cambridge, MA) (DeLano, 2005), and VOIDOO (Uppsala Software Factory, Uppsala, Sweden) (Kleywegt and Jones, 1994) was used to generate surfaces of the substrate binding cavities on the basis of the outer surface accessibility to a 1.4 Å probe. The atomic coordinates and structure factors (code: 5VEU) have been deposited with the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
TABLE 1.
Data reduction and refinement statistics
| Data reduction | Space group | P 21 21 21 |
| Unit cell dimensions | ||
| a, b, c (Å) | 148.99, 198.38, 234.88 | |
| α, β, γ (°) | 90, 90, 90 | |
| Nominal resolution range | 39.16–2.91 (2.96–2.91)* | |
| Unique reflections | 152,197 (7276) | |
| Rmerge | 0.040 (0.427) | |
| Rpim | 0.023 (0.277) | |
| CC(1/2) | 0.999 (0.848) | |
| Mean (I/σ(I)) | 17.0 (1.7) | |
| Completeness (%) | 99.9 (96.9) | |
| Multiplicity | 3.9 (3.1) | |
| Wilson B factor (Å2) (Xtriage) | 70.0 | |
| Protein chains in ASU | 12 | |
| Model refinement | Resolution range | 39.155–2.910 (2.943–2.910) |
| Unique reflections | 152,082 (4548) | |
| Completeness | 99.66 | |
| Test set (%) | 4.96 | |
| R work (%) | 21.37 (31.03) | |
| R test (%) | 25.76 (37.81) | |
| Heavy atoms | 44,746 | |
| Mean B-factors (Å2) | ||
| Protein | 68.5 | |
| Heme | 49.0 | |
| Ritonavir (A, B, and H chains) | 83.2 | |
| Model quality | RMSD bond lengths (Å) | 0.005 |
| RMSD bond angle (°) | 1.066 | |
| Ramachandran plot (Molprobity) | ||
| Favored (%) | 95.92 | |
| Allowed (%) | 4.03 | |
| Outliers (%) | 0.06 | |
| Rotamer outliers | 0.36% | |
| All-atom clash score | 5.37 |
Values in parentheses are for the highest resolution shell; ASU, asymmetric unit; CC(1/2), correlation coefficient between intensities from random half-datasets.
Results
Optimization of P450 3A5 Expression for Crystallization and Structure Determination.
The initial expression construct for crystallization, 3A5dH, incorporated an N-terminal truncation of 3A5 (residues 3 through 23) encompassing a transmembrane helix to reduce protein aggregation and a C-terminal four-histidine tag to facilitate purification, as described previously for the 3A4dH expression plasmid used to crystallize 3A4 in our laboratory (Yano et al., 2004). Five amino acids at the 3A5 C-terminus were deleted from 3A5dH with retention of the His-tag in a second construct, 3A5C2. The deleted residues extend outside of the catalytic domain and are disordered in the 1TQN structure of 3A4. An additional three residues, 24, 25, and 26, were removed from 3A5C2 construct to generate a third construct (3A5-N2C2) with a shorter N-terminus (Supplemental Fig. S1). Among the three CYP3A5 expression constructs, the C2 construct exhibited the highest expression level (average expression: dH, 700; C2, 1100; and N2C2, 550 nmol/l of culture). The 3A5C2 protein behaved well during purification and storage, and it was selected for crystallization screens.
Structure Determination.
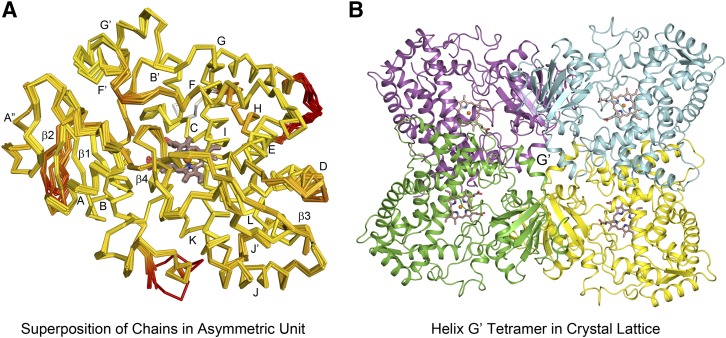
X-ray diffraction data collected from a single crystal of P450 3A5C2 was used for structure determination. Molecular replacement provided initial phasing and identified 12 protein chains in the asymmetric unit using the structure for P450 3A4-ritonavir complex as the search model, PDB:3NXU, chain A. Cα traces of the 12 chains are highly similar when superimposed (Fig. 1A). Chains A, B, D, E, F, G, H, J, K, and L exhibited pairwise RMSD values for main chain atoms of 0.51 ± 0.05 Å. Chains I and C exhibited a pairwise RMSD value 0.39 Å but exhibited larger differences from the other chains by 0.62–1.11 Å. The most divergent regions for the 12 chains when changes in side chain rotamers were included in the comparison are shaded red in Fig. 1A, and in most cases, these regions are associated with external loops. Additionally, amino acid residues 282–287 in the helix H-I loop were modeled for chains C and I, but not, for other chains. Residues 421–424 in another external loop were only modeled into the chains A and C, but the two chains exhibit different conformations for this surface loop. The conformation in this region for chain A probably reflects an interaction with chain C, whereas neighboring interactions for the 421–424 region are not evident for chain C. P450 3A5 is missing a residue corresponding to Asn-423 of P450 3A4, and the numbering of the residues in the C-terminal region of P450 3A4 differ by +1 from those in P450 3A5. Residues 263–280 in the external helix G-H loop could not be modeled for chains E and L, which exhibit higher overall B values. The 12 chains differ in their mean B-values, with mean B-values for chains E, F, and L being higher (99, 82, 106 Å2) than the average for all chains of 68.54 Å2 and the Wilson B-value of 69.99 Å2 determined by Xtriage for the diffraction data, whereas the average B-values for the remaining chains were lower (49–67 Å2) than the Wilson B-value.
Fig. 1.
(A) Overlay of the 12 chains in the asymmetric unit computed using ProSmart (CCP4) are depicted by Cα traces for each chain. Yellow-to-red color changes indicate increased divergence for both the backbone and side-chain rotamers. Helices are indicated by letters and β-sheets by numbers. (B) One of the three tetrameric substructures of the asymmetric unit, centered on lattice interactions of G′, helices is illustrated for chains A (yellow), D (cyan), E (magenta), and I (green). The heme and Fe are depicted as sticks and spheres, respectively. The carbons of the heme are colored brown, oxygen red, nitrogen blue, and iron orange.
The 12 molecules are organized in the crystal lattice as three dimers of dimers composed of chains A–E and D–I (Fig. 1B); B–F and H–L; and C–J and G–K. The dimers are formed by a roughly antiparallel hydrophobic interaction of helix G′ for two monomers (Fig. 1B). The second dimer is oriented with its axis perpendicular to the first dimer and the outer hydrophobic surface of helix G′ positioned above the helix F′-G′ surfaces of the other dimer (Fig. 1B). Differences between the 12 chains for the helix F′-G′ region and helix A′′–β1 turn probably reflect differences in the protein interactions for the two dimers in the three tetramers of asymmetric unit.
Ritonavir Binding to P450 3A5.
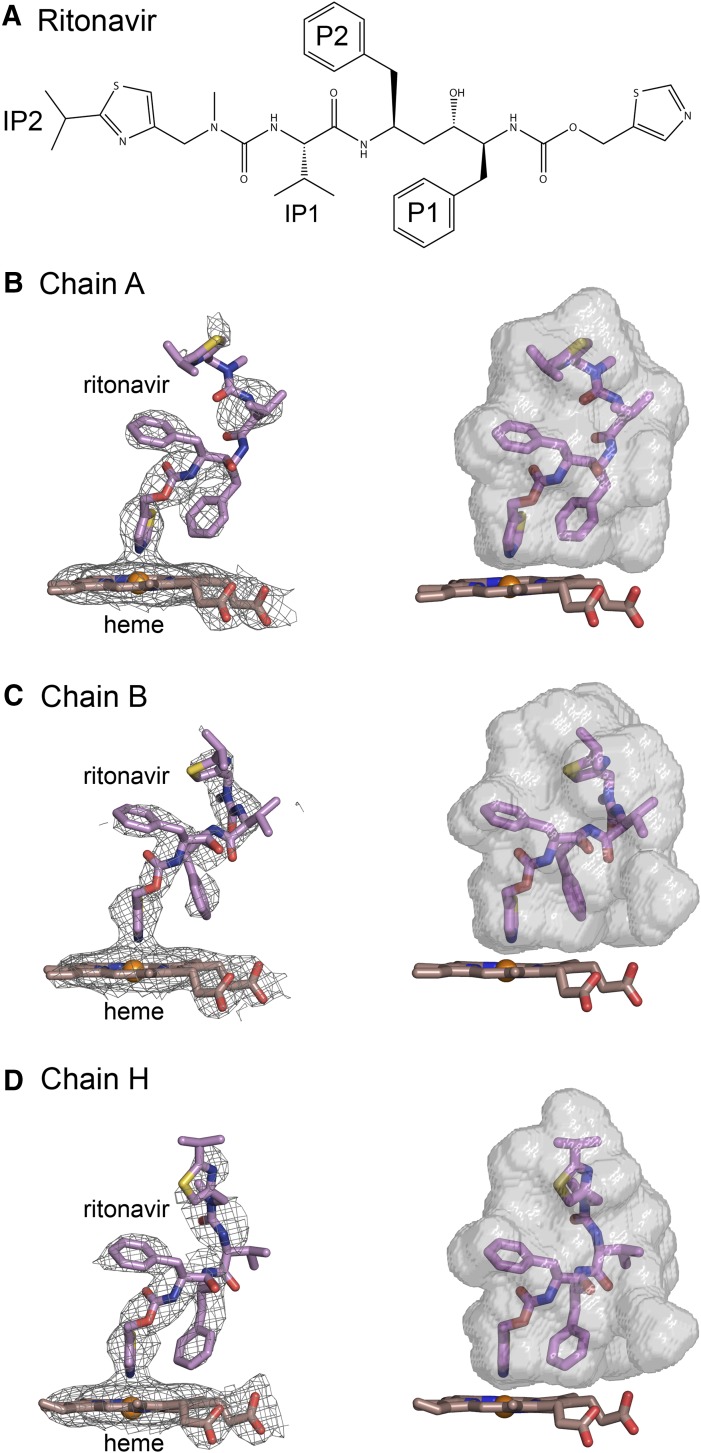
Binding of ritonavir to the enzyme was monitored by changes in the visible absorption spectrum of the heme. The nitrogen of the thiazole moiety of ritonavir donates a lone pair of electrons to the ferric heme iron leading to a change in the visible absorption spectrum of the heme prosthetic group, Supplemental Fig. S2. Using concentration-dependent changes in the UV-visible spectrum, 3A5C2 exhibited a dissociation constant for ritonavir of 51 ± 5 nM, Supplemental Fig. S2, which is similar to that estimated for the binding of ritonavir to 3A4dH, 47 ± 8 nM in our laboratory, and to a reported value of 51 ± 10 nM for 3A4dH (Sevrioukova and Poulos, 2012a). Despite the high binding affinity, electron density for ritonavir was not complete (Fig. 2, B, C, and D, left panels), and the completeness varied between chains. Models for ritonavir in chains A, B, and H were retained in the final structure, but ritonavir models were removed from other chains because of their lower real-space R-values and real-space density correlation coefficients. The nitrogen of the terminal thiazole moiety is located 2.20, 2.16, and 2.27 Å above the heme iron in chains A, B, and H, respectively. This distance is consistent with coordinate covalent binding to the heme iron and the observed spectral changes when bound.
Fig. 2.
(A) Structure of ritonavir. (B–D) Composite omit 2mFo-DFc maps contoured at 1 σ around the heme and ritonavir are shown on the left side, and surfaces for the substrate binding cavity are rendered in relation to the heme and ritonavir on the right for chains A, B, and H, respectively. The conformations of ritonavir and the shapes of the cavities vary between chains. The nitrogen of the thiazole group is coordinated to the heme with bond distances of 2.20, 2.16 and 2.27 Å, respectively for chains A, B, and H. Carbons are colored violet for ritonavir and brown for the heme. Sulfur is colored yellow. Other atom colors are defined in the legend to Fig. 1. Fc, calculated structure factor; Fo, observed structure factor.
We suspect that the incomplete density for the ligand reflects conformational heterogeneity and higher B-values in some chains. The presence of ritonavir in other chains is supported by observed density for the thiazole group bound to the heme in each chain, but electron density for the distal portions of ritonavir was diminished. The exception is chain L, which has the highest average B-values. Additionally, all chains in the asymmetric unit exhibit a conformation of helix I observed for the 3A4-ritonavir complex (Sevrioukova and Poulos, 2010) that reflects a displacement of helix I outward by the presence of the thiazole group bound to the heme, and an adoption of an alternative side-chain rotamer for Phe-304, compared with the substrate-free structures (1TQN and 1WOE) of 3A4 (Supplemental Fig. S3). Additionally, the absolute spectrum of 3A5-ritonavir protein preparation used for crystallization displays a shift of the Soret band of the heme from 416 to 421 nm and a significantly diminished intensity for the Q-band at 565 nm (Supplemental Fig. S2) as reported for 3A4-ritonavair complex relative to ligand free protein (Sevrioukova and Poulos, 2010). The varied degrees of ritonavir disorder in the different chains are similar to variation in disorder seen for different analogs of ritonavir in structures of their complexes with 3A4 (Sevrioukova and Poulos, 2013a,b).
Differences are apparent for the conformations of ritonavir among chains A, B, and H, Fig. 2, B–D. The similarity is greatest near the heme and diverges toward the distal ends of the molecules. The right panel for each chain depicts the conformation of ritonavir in relation to a computed surface of the substrate binding cavity of each chain. The differences in shapes of the upper portions of the cavity correlate with differences in the conformation of ritonavir evident from the electron density maps. The differences in the shapes of the upper portion of the cavity probably reflect the asymmetric interactions in the tetrameric, crystal lattice interface involving the roof of the cavity, Fig. 1B.
Differences between the Substrate Binding Cavities of 3A4 and 3A5.
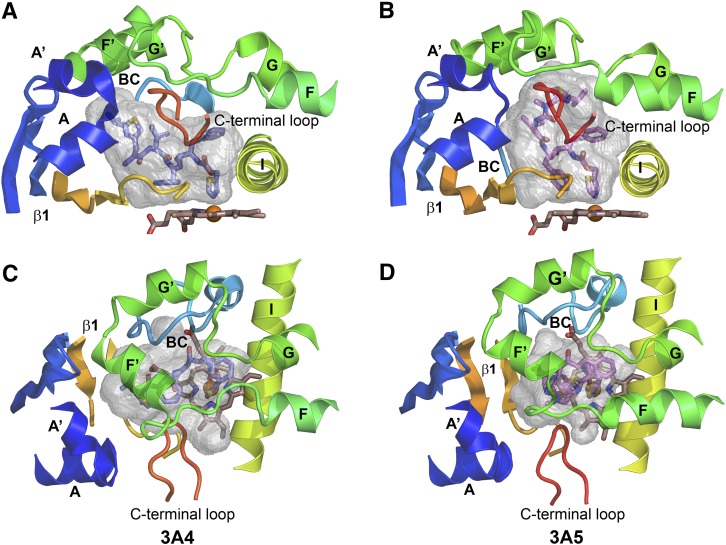
The shape of the active site cavity observed for 3A5 differs significantly from that of the 3A4 ritonavir complex, which exhibits a shorter but wider cavity, Fig. 3, A and B. The wider cavity allows the isopropyl thiazole moiety of ritonavir (IP2, Fig. 2A) to extend above sheet β-1 and under helix F′, whereas this space is not accessible in 3A5, Fig. 3B versus 3A. In contrast, the isopropyl thiazole group of ritonavir engages the upper portion of the cavity in 3A5 defined by the helix F through helix G region. This region forms a loop-like structure with four helices that begins and ends in close proximity as helices F and G pass over helix I, Fig. 3, C and D. The helix F through F′ region differs between 3A4 and 3A5 in the lengths of the F and F′ helices and the conformations of the connecting loop, Fig. 3, C and D.
Fig. 3.
The substrate binding cavity of the 3NXU structure of the 3A4 ritonavir complex (A) is shorter and wider than that of 3A5 (B). The cartoon structures are colored from blue at the N-terminus to red at the C terminus with helices and beta sheets labeled. The A chains of each structure are depicted, and only portions of the structure that surround the cavity are shown. The carbons of ritonavir are colored slate blue in 3A4 (A and C) and violet for 3A5 (B and D). The cavities are rendered as gray semitransparent surfaces. (A and B) Views of the side of the cavities from the C-terminal loop side. (C and D) Views from the helix F through G side above the cavity. Note the longer F and shorter F′ helices for 3A5 in (D) compared with 3A4 in (C). The opening between F-F′ and G-G′ connecters is larger in 3A5. Additionally, the N-terminal region (blue) pushes upward and inward in 3A5 (B) relative to 3A4 (A). These changes are associated with the taller and narrower active site cavity of 3A5.
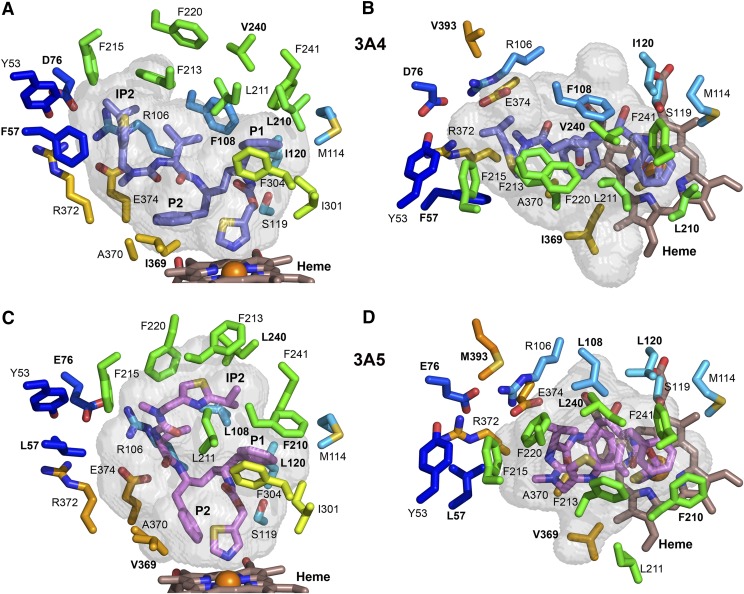
Amino acid differences between the two proteins probably influence the conformation of the helix F-G region and alter the active site architecture. One such difference is evident for residue 210. This residue contributes to a hydrophobic pocket that is formed by residues 108, 210, 211, 241, 301, and 304 in the 3A4 ritonavir complex, where the ritonavir P1 phenyl group (Fig. 2A) is sequestered (Fig. 4, A and B). Residue 210 is located on the extended portion of helix F in 3A5, and the Phe-210 of 3A5 is longer than Leu-210 of 3A4 (Fig. 4, C and D). This longer residue, which contacts ritonavir P1, pushes the C-terminus of helix F away from the P1 moiety of ritonavir and from Phe-241 located on the loop between helices G′ and G. As a result, Leu-211 (which forms part of the P1 hydrophobic pocket in 3A4) is displaced outward where it can no longer contact P1, comparing Fig. 4A versus 4C and 4B versus 4D. This change increases the size of the upper region of the substrate binding cavity in 3A5 by increasing the space between the helix F-F′ and helix G′-G connectors, Fig. 3C versus 3D. This space is further enlarged by the replacement of Phe-108, which contacts P1, in 3A4 with a shorter Leu-108 in 3A5 (Fig. 4B vs. 4D), and Leu-108 is further displaced by the outward bow of the helix B-B′ loop (Fig. 5A).
Fig. 4.
Differences in the identity and positioning of selected, conserved residues that shape the substrate binding cavities of chain A of 3A4 (A and B) and of 3A5 (C and D). Amino acids that differ between 3A4 and 3A5 are labeled with bold text. The longer F210 pushes the protein chain of 3A5 outward and displaces residue 211 away from the P1 group of ritonavir, which enlarges the upper portion of the cavity relative to 3A4. Additionally, the shorter L108 together with outward positioning of L108 Cα contribute to the larger upper cavity of 3A5. P2 of ritonavir displaces residues 369 and 370 on β1-strand 4 outward in 3A4. The smaller V369 does not contact the thiazole group of ritonavir or P2 in 3A5, which adopts a different orientation owing to the upward extension of ritonavir. The color scheme for carbons and heteroatoms is defined in earlier figure legends, and the views correspond to those in Fig. 3.
Fig. 5.
(A) Overlay of 3A5 (yellow carbons) and 3A4 (cyan carbons) protein chains viewed from the helix B′ side with ritonavir rendered as a space filling model. The inward positions of the helices A′′ and A neighboring regions is evident for the 3A5 ritonavir complex and are related to changes in the position of residue 106 and the accommodation of longer residues 76 and 393 in 3A5. A difference in the conformation of the helix B-B′ loop contributes to the positioning of residues 108 and 106. Residue 107 is a proline in 3A4 and a serine in 3A5, and the rigidity and conformation of the proline probably contribute to the positioning of residues 108 and 106. Changes in coulombic and H-bonding interactions that form a wall of the cavity are depicted for (B) 3A4 and (C) 3A5. (D) An overlay of the 1TQN substrate-free structure of 3A4 (magenta) with that of the 3A4 (cyan) and 3A5 (yellow) ritonavir complexes. The presence of ritonavir in 3A4 and 3A5 displaces the central portion of helix I, and ritonavir also displaces other regions of 3A4 relative to the 1TQN structure.
Another amino acid difference that affects the substrate binding cavity is seen for residue 369. Ile-369 of 3A4 forms a hydrophobic contact with the thiazole and P2 groups of ritonavir (Fig. 4A). In the 3A4-ritonavir complex, the plane of the P2 phenyl group is oriented parallel to the plane of the heme creating a stacking interaction with heme and additional hydrophobic interactions with Ile-369 and Ala-370. In 3A5, residue 369 is a smaller valine (Fig. 4C) that does not contact the thiazole group of ritonavir or the P2 moiety, which exhibits a more perpendicular orientation to the plane of the heme and only contacts Ala-370 of 3A5. The orientation of the P2 group in 3A4 displaces the loop preceding the strand of β-sheet 1, Figs. 3 and 5. Interestingly, the PDB:4I4H structure of 3A4 complexed with an analog (GS3) of ritonavir indicates that the P2 group of the analog exhibits a more perpendicular orientation relative the heme, and the loop preceding the strand of β-sheet 1 moves inward to adopt a conformation similar to the substrate-free 3A4 structure (Sevrioukova and Poulos, 2013b) and to that seen for 3A5 ritonavir complex. There are two modifications in the GS3 analog. The thiazole was replaced by a 3-pyridyl group that coordinates to the heme, and the hydroxyl group of ritonavir (Fig. 2A) was removed, disrupting a hydrogen bonding interaction with Ser-119 (Fig. 4A). Interestingly, the carbamoyl group adjacent to P1 flips in the 3A4 complex with GS3 so that its carbonyl accepts an H-bond from Ser-119. In contrast, the conformation adopted by ritonavir in 3A5 is not sufficiently close to Ser-119 for hydrogen bond formation. Additionally, the neighboring residue in 3A5, Leu-120, is longer than Ile-120 in 3A4. As a consequence of these differences and the lack of access to the extended width of 3A4 substrate binding cavity, there are fewer protein contacts with ritonavir in 3A5, which may contribute to the conformational disorder of ritonavir in 3A5. A similar pattern of disorder was observed for the GS3 analog relative to ritonavir in the complexes with 3A4 (Sevrioukova and Poulos, 2013b).
Overall, these differences contribute to the expansion of the upper portion of the cavity in 3A5 and lead to the taller cavity relative to 3A4. Although differences in the upper portions of the 3A5 substrate cavities were observed between chains in the asymmetric unit (Fig. 1A), these differences are small compared with the differences between 3A5 and 3A4. Most structurally characterized mammalian P450s exhibit longer F and G helices that pass over their active site cavities. In contrast, the flexible connectors between helices F and F′ and helices G′ and G form the roof of the active site cavity in both 3A4 and 3A5. Ritonavir passes under this region and extends across β-sheet 1 in 3A4. This pocket is blocked, however, in 3A5, and the terminal isopropyl thiazole moiety engages the center of the loop formed by the helix F-G region between the F-F′ and G′-G connectors, Fig. 3, C and D.
Differences in the Width of the Cavity Reflect Changes in Ionic Interactions.
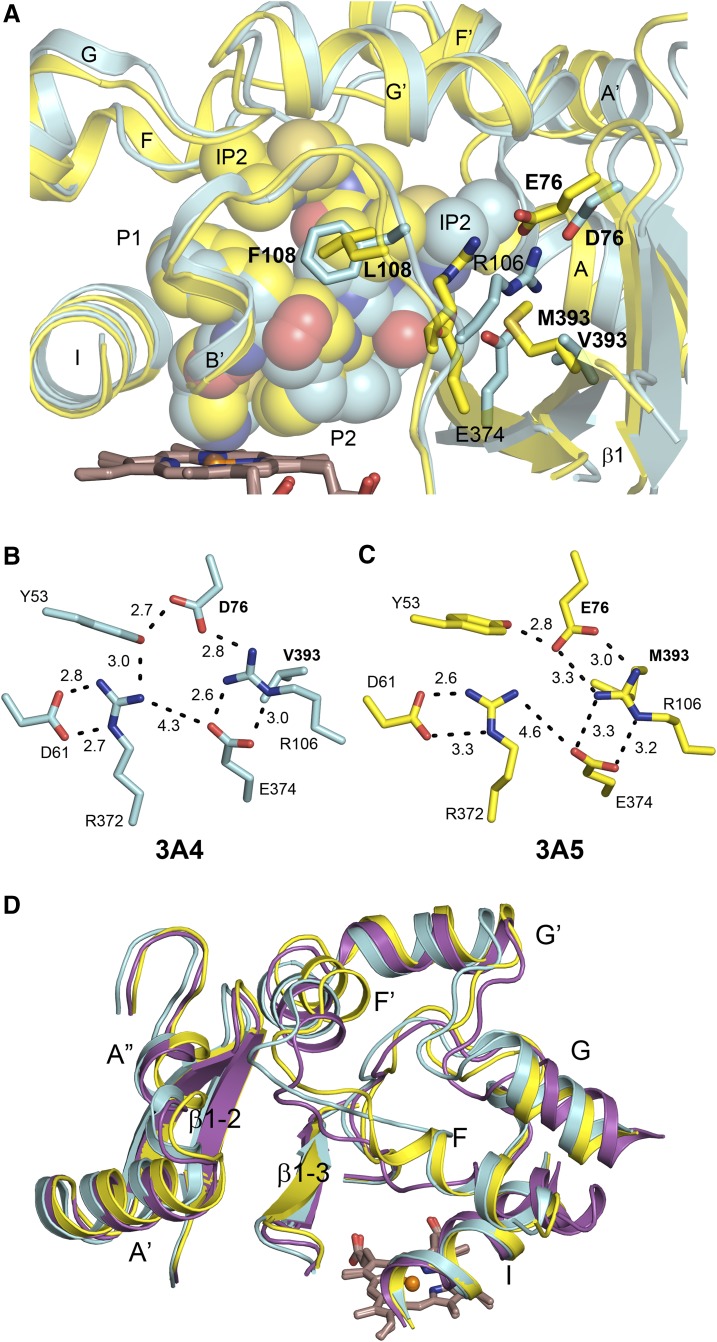
Amino acid differences also contribute the narrower active site cavity that redirects the isopropyl thiazole group to engage the helix F-G region in 3A5. In 3A4, the isopropyl thiazole group of ritonavir extends into a pocket above β-sheet 1 and below helix F′ (Figs. 3A, 4A, and 5B) that is formed in part by ionic interactions between Arg-106 on the helix B-B′ connector, with Glu-374 on strand 3 of β-sheet 1, and Asp-76 on strand 1 of β-sheet 1, as well as by hydrophobic side chains. Glu-76 in 3A5 is longer than Asp-76 in 3A4, and the ionic residues move inward to fill the pocket occupied by the IP2 group of ritonavir in 3A4, and this inward movement is reinforced by the larger Met-393 of 3A5, which fills in the space occupied by the R106 in 3A4 (Fig. 4B vs. 4D and Fig. 5A).
These differences are associated with a significant inward shift of the N-terminal region of 3A5 that includes helices A′ and A, Fig. 5A, and a reorganization of the ionic and hydrogen bonding interactions among the ionic residues that form part of the cavity wall, Fig. 5, B and C. In addition to its hydrogen bond with Arg-106, residue 76 accepts an H-bond with Tyr-53 on the loop between helices A′ and A in both 3A4 and 3A5. In 3A4, Arg-372 is also H-bonded to Tyr-53. Arg-372 forms H-bonded charge-stabilized interactions with Asp-61 on Helix A in both 3A4 and 3A5. Although Arg-372 is a neighbor to Glu-374, they are positioned too far apart to form an H-bond, but they probably exhibit a strong coulombic attraction because they are too close for an intervening water molecule to shield the electrostatic interaction. Additionally, Arg-106 has a hydrophobic interaction with residue 393 on strand 4 of β-sheet 1 which resides outside of the cavity (Fig. 5, A, B, and C). Residues 76 and 393 differ from 3A4 in 3A5. First, residue 393 is a methionine that is longer than valine, and the increased length of Met-393 reinforces the positioning Arg-106 in the pocket, where the isopropyl thiazole group of ritonavir resides in 3A4 (Fig. 5A). Additionally, residue 76 is a longer glutamic acid residue in 3A5 that forms a charge-stabilized H-bond with Arg-106 in 3A5 (Fig. 5C). Tyr-53 remains hydrogen-bonded to Glu-76 but is no longer hydrogen-bonded to Arg-372, Fig. 5C. As a result, the pocket occupied by the isopropyl thiazole group of ritonavir in 3A4 is no longer available in 3A5, and the isopropyl thiazole group of ritonavir is more easily accommodated in the center of the loop-like structure formed by the helix F-G region where the phenylalanine residues Phe-213 and Phe-220 rotate outward rather than stack inward to form the roof of the cavity as seen in 3A4, Figs. 4A versus 4C and Fig. 4B versus 4D. This helix F-G region of 3A4 collapses inward toward the heme in the absence of ritonavir (Fig. 5D), and the N-terminal region of the substrate-free structure of 3A4 more closely resembles that of the 3A5 ritonavir complex than the 3A4-ritonavir complex, Fig. 5D.
Discussion
Determination of the atomic structure of 3A5 was sought to address whether characteristic structural features that distinguish 3A4 from other human drug-metabolizing P450s were conserved in 3A5 and to better understand structural differences between 3A4 and 3A5 that underlie divergent metabolic properties of the two enzymes. The F and G helices of most P450s cross above the heme to form the upper boundary of the substrate binding cavity, but in both 3A4 and 3A5 the roof of the substrate binding site is formed by more flexible connections between helices F-F′ and helices G′-G. The F′ and G′ helices are also longer and positioned differently in 3A4 and 3A5 than seen in other human drug-metabolizing P450s. The hydrophobic exterior surfaces of the F′ and G′ helices are thought to contribute to interactions of the catalytic domain with the hydrophobic portion of the membrane (Baylon et al., 2013; Berka et al., 2013; Treuheit et al., 2016). Although differences in the conformation of the helix F-G region are evident between 3A4 and 3A5, the basic architecture is retained in 3A5, and membrane binding interactions are also probably similar to those exhibited by 3A4. Compared with families 1 and 2 human drug-metabolizing P450s, a larger portion of the heme surface is accessible to substrates in 3A4 (Johnson and Stout, 2013; Johnson et al., 2014), and this is also evident in the 3A5 structure. When coupled with the flexible F-F′ and G′-G connectors, access of substrates to approach the reactive intermediate formed by reduction of oxygen bound to the iron of the heme cofactor is less restricted than is seen for human families 1 and 2 drug-metabolizing P450s (Johnson and Stout, 2013; Johnson et al., 2014). This active site architecture contributes to the capacities of 3A4 and 3A5 to catalyze reactions that produce multiple alternative products from a single substrate and enables them to participate in metabolic clearance of a larger portion of clinically used drug. The predominance of hydrophobic interactions between substrates and the protein also contributes to the energetic equivalency of different binding orientations owing to the omnidirectional nature of the interactions. Additionally, broad access to the heme contributes to utilization of sites of metabolism that are not evident for the families 1 and 2 enzymes as shown recently for the binding of aflatoxin B1 to 3A4 and 1A2 (Bonomo et al., 2017).
Ritonavir was chosen for cocrystallization with 3A5 owing to its high binding affinity and to enable a direct comparison between 3A5 and 3A4 complexed with the same ligand. Ritonavir exhibits higher configurational entropy in the 3A5 complex compared with the 3A4 complex. The 18 rotatable bonds of ritonavir are largely restrained in the complex with 3A4, but these degrees of freedom also enable ritonavir to adapt to the different shape of the 3A5 active site and to retain of greater configurational entropy. The high binding affinities of ritonavir to 3A4 and 3A5 probably reflect the strength of the interaction of the thiazole group to the heme iron as well as ritonavir’s high lipophilicity and sparse aqueous solubility rather than a lock and key type of fit with the two proteins. The strong interaction of the thiazole with the heme iron restrains one end of ritonavir in the active site of both enzymes with the distal end of ritonavir sequestered in a small hydrophobic pocket under helix F′ and above β-sheet 1 in 3A4. The higher configurational entropy of ritonavir in 3A5 probably reflects the binding of the distal end of ritonavir in the most flexible portion of the active site, the helix F-G region.
Although the secondary and tertiary structures of 3A5 and 3A4 are similar, the 3A5 active site is taller and narrower than that of 3A4. Compared with the structure of substrate-free 3A4, the active site cavity widens and increases in height in the structure of the 3A4-ritonavir complex (Fig. 5D). The narrower active site of 3A5 appears to resist this displacement of the N-terminal region. As a consequence, the distal portion of ritonavir is redirected to occupy an expanded upper portion of the active site cavity above the heme. These observations probably reflect amino acid differences between the two proteins for residues 76 and 393 that reinforce the inward movement of Arg-106 and provide increased resistance to the deformation seen for the 3A4-ritonavir complex (Fig. 5, B and C). Interestingly, compensating for the inward movement of Arg-106 is the longer Glu-76 of 3A5, which maintains ionic interactions with Arg-106, whereas the shorter Asp-76 of 3A4 is consistent with the larger width of the 3A4 active site. These differences in the ionic interaction of the side chains reduce the width of the substrate binding cavity in 3A5 and contribute to decreased plasticity of 3A5 N-terminal region. Interestingly, a docking study indicates that the narrow cavity and positions of Arg-106 and Glu-374 probably contribute to the higher selectivity of 3A5 over 3A4 for the O-demethylation of schisantherin E. The 3A5 selectivity relative to related compounds is dependent on a hydroxyl group and a benzoate ester moiety (Wu et al., 2017). When docked into the active site in position for O-demethylation, the critical hydroxyl group H-bonds with Arg-106 and Glu-374, and the benzoate ester group resides in a hydrophobic pocket near F210, Supplemental Fig. S4.
Conformational differences are also evident for the helix F-G region that increase the height of the 3A5 active site. The differences between 3A4 and 3A5 ritonavir complexes in this region reflect in part adaptations of this flexible region to accommodate ritonavir in 3A5 and amino acid differences between the two proteins that appear to facilitate adoption of the more open conformation in the center of this region of 3A5. There are 18 amino acid differences between 3A4 and 3A5 in helix F-G region beginning with N206S and ending with T246I. A difference in the length of one divergent pair, L210F, is associated with the significant divergence of the polypeptide backbones (Supplemental Fig. S5). Introduction of the L210F mutation in 3A4 has been shown to reduce activity toward aflatoxin (Wang et al., 1998), and the reciprocal mutation F210L in 3A5 reduced catalytic activity for the 3A5 selective hydroxylation of the dichlorocyclohexane moiety of maraviroc (Lu et al., 2012) and for the O-demethylation of schisantherin E (Wu et al., 2017).
Other divergent amino acid pairs exhibit packing interactions that are associated with conformational differences in the helix F-G region and interactions of the F-G region with the C-terminal loop and helices A′ and A that indirectly alter the substrate binding cavity (Supplemental Fig. S5). Moreover, when more than one pair of divergent residues interact directly, the effects can be amplified or be compensatory. An example of the latter is the interaction of the divergent F219L and L233F pairs (Supplemental Fig. S5). Mutations that exchange one residue of an interacting pair probably produce an effect that is not representative of either 3A4 or 3A5. For instance, a G214D substitution on outer surface of 3A5 was shown to decrease oxygenation of the dichlorocyclohexane moiety of maraviroc (Lu et al., 2012). The longer aspartate residue probably conflicts with Q479 of 3A5 on the C-terminal loop leading to an indirect effect on substrate binding. The corresponding G480 of 3A4 is more accommodating for D214 of 3A4, and the native G214 of 3A5 is more accommodating for Q479 of 3A5. Other differences in the F-G region create a stronger interaction between helices F and G in 3A5. T245 donates an H-bond to the carbonyl of S206, whereas Val245 of 3A4 exhibits a nonpolar interaction with the carbonyl. T245 also contacts F210, but Val245 does not interact with L210 in 3A4. Additionally, D244 of 3A5 forms a salt-bridge with K209, whereas crowding prevents this interaction between K209 and the longer E244 of 3A4. Nevertheless, examination of the structures of 3A5 and 3A4 do not reveal a structural basis for the reduced activity seen for the N206S mutant of 3A4 (Wang et al., 1998) or the modestly enhanced activity of the 3A5 S206N mutant (Lu et al., 2012). The numerous indirect contributions of divergent residues to active site differences between 3A4 and 3A5 probably underlie difficulties in interchanging 3A4 or 3A5 phenotypes by way of a small number of reciprocal amino acid substitutions (Wang et al., 1998; Xue et al., 2001; Khan et al., 2002; Lu et al., 2012).
The differential plasticity in width and height of the 3A4 and 3A5 active sites probably underlies differences in substrate dynamics and the degree of accessibility of the site of metabolism to the iron-bound reactive intermediate. These differences lead to alternative metabolites for the two enzymes or that differentially modulate rates of oxygenation for shared reaction pathways. Comparisons of different X-ray crystal structures of 3A4 cocrystallized with substrates of different sizes and shapes indicate that the flexible F-F′ and G′-G connections enable large changes in volume and shape of the substrate binding cavity (Ekroos and Sjogren, 2006; Sevrioukova and Poulos, 2010, 2012b, 2013b, 2017). Additional structures of 3A5 and 3A4 complexed with the same ligand would better define the differential plasticity exhibited by these two enzymes that leads to differences in their metabolite profiles. These insights could aid in the design of selective inhibitors or targeted drug design to eliminate undesirable reaction pathways catalyzed by one or both enzymes.
Acknowledgments
The authors thank Mariam Salib for technical support.
Abbreviations
- NCS
noncrystallographic symmetry
- P450
cytochrome P450 enzyme
- PCR
polymerase chain reaction
- PDB
Protein Data Bank
- PEG
polyethylene glycol
- PMSF
phenylmethylsulfonyl fluoride
Authorship Contributions
Participated in research design: Hsu, Savas, Johnson.
Conducted experiments: Hsu, Savas.
Contributed new reagents or analytic tools: Hsu, Savas, Johnson.
Performed data analysis: Hsu, Savas, Johnson.
Wrote or contributed to the writing of the manuscript: Hsu, Savas, Johnson.
Footnotes
This work was supported by the National Institutes of Health, National Institute of General Medical Sciences [Grant R01GM031001]. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the NIH, NIGMS [including Grant P41GM103393]. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. The authors declare that they have no conflicts of interest with the contents of this article.
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. (2013) Characterizing the membrane-bound state of cytochrome P450 3A4: structure, depth of insertion, and orientation. J Am Chem Soc 135:8542–8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berka K, Paloncýová M, Anzenbacher P, Otyepka M. (2013) Behavior of human cytochromes P450 on lipid membranes. J Phys Chem B 117:11556–11564. [DOI] [PubMed] [Google Scholar]
- Birdwell KA, Decker B, Barbarino JM, Peterson JF, Stein CM, Sadee W, Wang D, Vinks AA, He Y, Swen JJ, et al. (2015) Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP3A5 genotype and tacrolimus dosing. Clin Pharmacol Ther 98:19–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonomo S, Jørgensen FS, Olsen L. (2017) Dissecting the cytochrome P450 1A2- and 3A4-mediated metabolism of aflatoxin B1 in ligand and protein contributions. Chemistry 23:2884–2893. [DOI] [PubMed] [Google Scholar]
- DeLano WL. (2005) The case for open-source software in drug discovery. Drug Discov Today 10:213–217. [DOI] [PubMed] [Google Scholar]
- Ekroos M, Sjögren T. (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA 103:13682–13687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. (2010) Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR, Murshudov GN. (2013) How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner JA, Dahlquist FW. (1991) Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci USA 88:750–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince I, Knibbe CA, Danhof M, de Wildt SN. (2013) Developmental changes in the expression and function of cytochrome P450 3A isoforms: evidence from in vitro and in vivo investigations. Clin Pharmacokinet 52:333–345. [DOI] [PubMed] [Google Scholar]
- Johnson EF, Connick JP, Reed JR, Backes WL, Desai MC, Xu L, Estrada DF, Laurence JS, Scott EE. (2014) Correlating structure and function of drug-metabolizing enzymes: progress and ongoing challenges. Drug Metab Dispos 42:9–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EF, Stout CD. (2013) Structural diversity of eukaryotic membrane cytochrome p450s. J Biol Chem 288:17082–17090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung F, Griffin KJ, Song W, Richardson TH, Yang M, Johnson EF. (1998) Identification of amino acid substitutions that confer a high affinity for sulfaphenazole binding and a high catalytic efficiency for warfarin metabolism to P450 2C19. Biochemistry 37:16270–16279. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (2010) XDS. Acta Crystallogr D Biol Crystallogr 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KK, He YQ, Correia MA, Halpert JR. (2002) Differential oxidation of mifepristone by cytochromes P450 3A4 and 3A5: selective inactivation of P450 3A4. Drug Metab Dispos 30:985–990. [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr 50:178–185. [DOI] [PubMed] [Google Scholar]
- Koudriakova T, Iatsimirskaia E, Utkin I, Gangl E, Vouros P, Storozhuk E, Orza D, Marinina J, Gerber N. (1998) Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab Dispos 26:552–561. [PubMed] [Google Scholar]
- Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, et al. (2001) Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet 27:383–391. [DOI] [PubMed] [Google Scholar]
- Li X, Jeso V, Heyward S, Walker GS, Sharma R, Micalizio GC, Cameron MD. (2014) Characterization of T-5 N-oxide formation as the first highly selective measure of CYP3A5 activity. Drug Metab Dispos 42:334–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Hendrix CW, Bumpus NN. (2012) Cytochrome P450 3A5 plays a prominent role in the oxidative metabolism of the anti-human immunodeficiency virus drug maraviroc. Drug Metab Dispos 40:2221–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omura T, Sato R. (1964) The carbon monoxide-binding pigment of liver microsomes. II. Solubilization, purification, and properties. J Biol Chem 239:2379–2385. [PubMed] [Google Scholar]
- Rendic S, Guengerich FP. (2015) Survey of human oxidoreductases and cytochrome P450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol 28:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2010) Structure and mechanism of the complex between cytochrome P4503A4 and ritonavir. Proc Natl Acad Sci USA 107:18422–18427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2012a) Interaction of human cytochrome P4503A4 with ritonavir analogs. Arch Biochem Biophys 520:108–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2012b) Structural and mechanistic insights into the interaction of cytochrome P4503A4 with bromoergocryptine, a type I ligand. J Biol Chem 287:3510–3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2013a) Dissecting cytochrome P450 3A4-ligand interactions using ritonavir analogues. Biochemistry 52:4474–4481. [DOI] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2013b) Pyridine-substituted desoxyritonavir is a more potent inhibitor of cytochrome P450 3A4 than ritonavir. J Med Chem 56:3733–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevrioukova IF, Poulos TL. (2017) Structural basis for regiospecific midazolam oxidation by human cytochrome P450 3A4. Proc Natl Acad Sci USA 114:486–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JC. (2006) New perspectives on the impact of cytochrome P450 3A expression for pediatric pharmacology. Drug Discov Today 11:440–445. [DOI] [PubMed] [Google Scholar]
- Treuheit NA, Redhair M, Kwon H, McClary WD, Guttman M, Sumida JP, Atkins WM. (2016) Membrane interactions, ligand-dependent dynamics, and stability of cytochrome P4503A4 in lipid nanodiscs. Biochemistry 55:1058–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng E, Walsky RL, Luzietti RA, Jr, Harris JJ, Kosa RE, Goosen TC, Zientek MA, Obach RS. (2014) Relative contributions of cytochrome CYP3A4 versus CYP3A5 for CYP3A-cleared drugs assessed in vitro using a CYP3A4-selective inactivator (CYP3cide). Drug Metab Dispos 42:1163–1173. [DOI] [PubMed] [Google Scholar]
- Walsky RL, Obach RS, Hyland R, Kang P, Zhou S, West M, Geoghegan KF, Helal CJ, Walker GS, Goosen TC, et al. (2012) Selective mechanism-based inactivation of CYP3A4 by CYP3cide (PF-04981517) and its utility as an in vitro tool for delineating the relative roles of CYP3A4 versus CYP3A5 in the metabolism of drugs. Drug Metab Dispos 40:1686–1697. [DOI] [PubMed] [Google Scholar]
- Wang H, Dick R, Yin H, Licad-Coles E, Kroetz DL, Szklarz G, Harlow G, Halpert JR, Correia MA. (1998) Structure-function relationships of human liver cytochromes P450 3A: aflatoxin B1 metabolism as a probe. Biochemistry 37:12536–12545. [DOI] [PubMed] [Google Scholar]
- Westlind-Johnsson A, Malmebo S, Johansson A, Otter C, Andersson TB, Johansson I, Edwards RJ, Boobis AR, Ingelman-Sundberg M. (2003) Comparative analysis of CYP3A expression in human liver suggests only a minor role for CYP3A5 in drug metabolism. Drug Metab Dispos 31:755–761. [DOI] [PubMed] [Google Scholar]
- Williams JA, Ring BJ, Cantrell VE, Jones DR, Eckstein J, Ruterbories K, Hamman MA, Hall SD, Wrighton SA. (2002) Comparative metabolic capabilities of CYP3A4, CYP3A5, and CYP3A7. Drug Metab Dispos 30:883–891. [DOI] [PubMed] [Google Scholar]
- Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. (2004) Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science 305:683–686. [DOI] [PubMed] [Google Scholar]
- Wu JJ, Cao YF, Feng L, He YQ, Hong JY, Dou TY, Wang P, Hao DC, Ge GB, Yang L. (2017) A naturally occurring isoform-specific probe for highly selective and sensitive detection of human cytochrome P450 3A5. J Med Chem 60:3804–3813. [DOI] [PubMed] [Google Scholar]
- Xue L, Wang HF, Wang Q, Szklarz GD, Domanski TL, Halpert JR, Correia MA. (2001) Influence of P450 3A4 SRS-2 residues on cooperativity and/or regioselectivity of aflatoxin B(1) oxidation. Chem Res Toxicol 14:483–491. [DOI] [PubMed] [Google Scholar]
- Yamazaki H, Nakamoto M, Shimizu M, Murayama N, Niwa T. (2010) Potential impact of cytochrome P450 3A5 in human liver on drug interactions with triazoles. Br J Clin Pharmacol 69:593–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. (2004) The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem 279:38091–38094. [DOI] [PubMed] [Google Scholar]
- Zanger UM, Turpeinen M, Klein K, Schwab M. (2008) Functional pharmacogenetics/genomics of human cytochromes P450 involved in drug biotransformation. Anal Bioanal Chem 392:1093–1108. [DOI] [PubMed] [Google Scholar]