Abstract
Objective
This study aims to demonstrate the effect of oral doxycycline on fecal microbiota of mice. Doxycycline is a common effector for control of gene expression using the tet-inducible system in transgenic mice. The effect of oral doxycycline on murine gut microbiota has not been reported. We evaluated the effect of doxycycline treatment by sequencing the V4 hypervariable region of the 16S rRNA gene from fecal samples collected during a 4 week course of treatment at a dose of 2 mg/ml in the drinking water.
Results
The fecal microbiota of treated animals were distinct from control animals; the decreased richness and diversity were characterized primarily by Bacteroides sp. enrichment. These effects persisted when the treatment was temporarily discontinued for 1 week. These data suggest that doxycycline treatment can induce significant dysbiosis, and its effects should be considered when used in animal models that are or maybe sensitive to perturbation of the gut microbiota.
Electronic supplementary material
The online version of this article (10.1186/s13104-017-2960-7) contains supplementary material, which is available to authorized users.
Keywords: Doxycycline, Tet-on, Tet-off, Microbiome, Microbiota, Mouse, Animal model
Introduction
Altered gut microbiota (GM) has been associated with a growing list of human conditions (e.g. colorectal cancer [1, 2], inflammatory bowel disease [3, 4], rheumatoid arthritis [5]) and animal models (e.g. type 2 diabetes [6–8], multiple sclerosis [9], anxiety [10], colon cancer [11], arthritis [12], atherosclerosis [13, 14]). Furthermore, the GM plays an important role in many mammalian physiologic processes such as energy balance and metabolism [15], immune function [16–18], angiogenesis [19, 20], and brain development and behavior [21–24]. This suggests that the GM of mouse models should be considered as a variable in animal experiments.
Tetracycline-inducible models have become increasing popular since their creation in the 1990s, though doxycycline’s efficacy has made it the preferred effector over tetracycline [25]. A PubMed search for “mouse” and “doxycycline” yields over 2000 articles describing tet-inducible mouse models of heart failure [26], memory and reversal learning [27], mammary tumors [28], type-1 diabetes mellitus [29], colorectal cancer [30], intestinal inflammation [31] and others. This crude metric demonstrates the pervasiveness of doxycycline use in many disciplines. Oral amoxicillin, metronidazole, vancomycin have already been shown to induce significant and sometimes long-lasting GM perturbations in mice [32, 33] but the effect of oral doxycycline on the GM has not been described.
In light of these findings, the authors believe it is important to determine if and how oral doxycycline affects the GM of mice. Demonstrating and characterizing any effect may enable investigators to control for or eliminate microbiota-induced variability in their experimental design.
We hypothesized that oral doxycycline would alter the composition of the GM (as evidenced by changes in the fecal microbiota [FM]) in female C57BL/6NCrl mice, and that the GM would return to baseline when the drug was temporarily discontinued. The C57BL/6 strain was chosen because it is most commonly used to generate transgenic animals. Females were used to allow for group housing with the intention of enhancing animal welfare. The dose of 2 mg/ml is the most commonly used dose at the authors’ institution, and is the approximate median of published doses which range from 0.2 to 7.5 mg/ml [34, 35]. The collection of fecal pellets allowed for longitudinal collection as opposed to collection of ceca or cecal contents, which are terminal procedures.
Main text
Methods
Animal models
All studies were performed in accordance with the recommendations put forth in the Guide for the Care and Use of Laboratory Animals and were approved by the University of Michigan Institutional Animal Care and Use Committee. Detailed descriptions of animals and husbandry, welfare assessments and interventions, doxycycline administration and sample collection as recommended by the ARRIVE Guidelines are included in Additional file 1.
Experimental design
On day 0, 20 female C57BL/6NCrl mice were divided into experimental (DOX, n = 10) and control (n = 10) groups. No formal randomization was performed. DOX animals were administered doxycycline in the drinking water at a concentration of 2 mg/ml during weeks 1, 2, and 4; they received distilled water during week 3. Animals in the control group received distilled water for the duration of the study. Feces were collected on days 0, 7, 14, 21, and 28 and stored at − 80 °C until analysis. No other experimental manipulations were performed on the animals. At the end of the 4 week study, animals were transferred to another protocol.
DNA extraction, quantification and assessment of purity
DNA extraction was performed as previously described [36] (see Additional file 2 for details). DNA concentrations were determined fluorometrically (Qubit dsDNA BR assay, Life Technologies, Carlsbad, CA) and purity was assessed via 260/280 and 260/230 absorbance ratios, as determined via spectrophotometry (Nanodrop 1000 Spectrophotometer, Thermo Fisher Scientific, Waltham, MA). Samples were stored at − 20 °C until sequencing.
Library construction and 16S rRNA sequencing
Library construction and sequencing was performed at the University of Missouri DNA Core facility. DNA concentration of samples was determined fluorometrically and all samples were normalized to 3.51 ng/µL for PCR amplification. Bacterial 16S rRNA amplicons were generated via amplification of the V4 hypervariable region of the 16S rRNA gene using single-indexed universal primers (U515F/806R) flanked by Illumina standard adapter sequences and the following parameters: 98 °C (3:00) + [98 °C (0:15) + 50 °C (0:30) + 72 °C (0:30)] × 25 cycles +72 °C (7:00). Amplicons were then pooled for sequencing using the Illumina MiSeq platform and V2 chemistry with 2 × 250 bp paired-end reads, as previously described [36]. Samples returning greater than 10,000 reads were deemed to have successful amplification. All samples were sequenced on two separate plates and the data was concatenated during the informatics stage. All treatment groups and time-points were split as evenly as possible between plates to account for any minor plate effect during sequencing.
Informatics analysis
Assembly, binning, and annotation of DNA sequences were performed at the MU Informatics Research Core Facility. Briefly, contiguous DNA sequences were assembled using FLASH software [37] and culled if found to be short after trimming for a base quality less than 31. Qiime v1.8 [38] software was used to perform de novo and reference-based chimera detection and removal, and remaining contiguous sequences were assigned to operational taxonomic units (OTUs) via de novo OTU clustering and a criterion of 97% nucleotide identity. Taxonomy was assigned to selected OTUs using BLAST [39] against the Greengenes database [40] of 16S rRNA sequences and taxonomy. Principal component analyses were performed using ¼ root-transformed OTU relative abundance data via a non-linear iterative partial least squares (NIPALS) algorithm, using an open access Excel macro available from the Riken Institute (http://prime.psc.riken.jp/Metabolomics_Software/StatisticalAnalysisOnMicrosoftExcel/index.html).
Statistical methods
Testing for significant differences between groups and time-points in richness and α-diversity was performed using SigmaPlot 13.0. Briefly, data were tested for normality and equal variance. Once confirmed, main effects of treatment and time-point, and interactions between independent variables, were tested via two-way ANOVA. Main effects of treatment and time-point (and interactions) on β-diversity were tested using non-transformed data via two-way PERMANOVA using Past 3.13 [41]. One-way PERMANOVA was performed post hoc to determine pair-wise differences.
Results
Sequencing at a mean depth of 75407 sequences per sample, between 4 and 146,082 total unique sequences were detected. Rarefaction analysis indicates the number of OTUs detected per sample was independent of sequencing depth above approximately 20,000 sequences indicating the sequencing depth was adequate to detect rare taxa. Twelve samples, all in the DOX group on days 7 and 14, yielded approximately 10,000 reads or fewer (range = 4–10,048) and were omitted from further analysis. A total of 88 samples were included.
Doxycycline treatment decreases richness and diversity
Assembly and binning of the raw sequence data from the 88 samples resulted in 23–50 distinct OTUs represented per sample at each time point. The control group contained 40-50 OTUs per sample per time point, with an overall average of 44.2 OTUs per sample. DOX samples collected after treatment range from 23 to 41 OTUs per animal per time point with an overall average of 30 OTUs per sample (Additional file 3). Rarefaction analysis indicated an apparent difference in microbial richness between treatment groups with control animals at all time points clustering above doxycycline-treated animals at all post-treatment time points. DOX samples on day 0 clustered with the control animals (Fig. 1). Comparison of Chao1 and Shannon indices between treatment groups indicates significant decreases in richness and diversity in the DOX animals post-treatment, with significant main effects of time, treatment, and a significant time × treatment interaction (p < 0.001, 2-way ANOVA, Additional file 3).
Fig. 1.
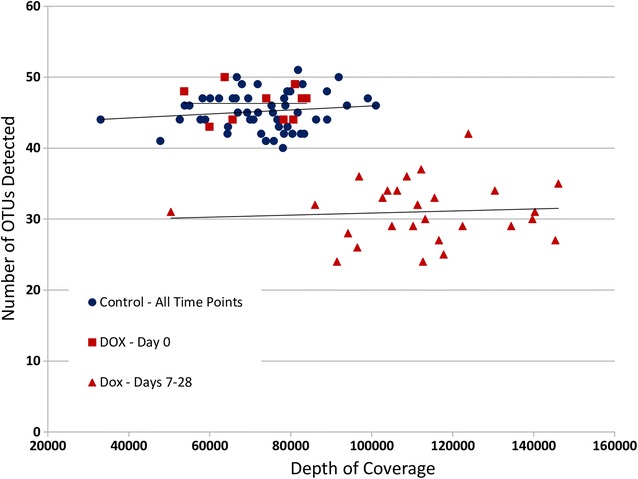
Rarefaction of sequencing data. Rarefaction analysis comparing the number of detected OTUs to the total number of sequences obtained for each individual sample at all time points. Data points are colored to indicate experimental group: Control, blue; DOX, red
Doxycycline treatment alters relative abundance
The OTUs from all samples were annotated to nine phyla: Actinobacteria, Bacteroidetes, Cyanobacteria, Deferribacteres, Firmicutes, Proteobacteria, Tenericutes, TM7, and Verrucomicrobia (Fig. 2). As expected, the FM was composed primarily of Firmicutes and Bacteroidetes in both groups on day 0, the relative abundance of these and other phyla was stable over time in the control animals. Conversely, the relative abundance of these phyla was markedly altered in the DOX samples on day 28, characterized primarily by enrichment of the phylum Bacteroidetes. Firmicutes were decreased, and samples from DOX cage 2 demonstrate relative enrichment of Proteobacteria.
Fig. 2.
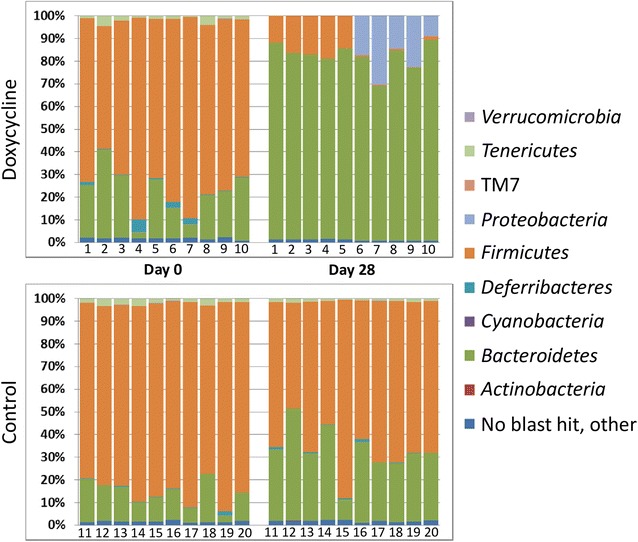
Relative abundance at taxonomic level of phylum. Bar charts showing the bacterial composition of the same control and doxycycline-treated mice on days 0 and 28 annotated to the taxonomic level of phylum. Legend of phyla is shown at right. Each bar represents an individual animal (DOX animals numbered 1–10, control animals numbered 11–20)
Relative abundance at the OTU level is shown for all time points in Additional file 4. The predominant Firmicutes OTU in control animals at all time points, and DOX animals on day 0 was annotated to unclassified (UC) order Clostridiales, with an average relative abundance of 37.3% in the control animals at all points and 40.1% in the DOX animals on day 0. At the same time points, most Bacteroidetes species were annotated to Bacteroides and fewer Parabacteroides. Conversely, Bacteroides species dominated the DOX samples on days 7–28, accounting for an average of 75.0% of all reads in post-treatment samples. In these same samples, the relative abundance of UC order Clostridiales dropped from the day 0 average of 40.1% to 3.4%. In these samples, the marked decrease in UC order Clostridiales was accompanied by an increase in UC family Enterobacteriaceae.
Doxycycline-treated animals have distinct populations from control mice
Control animals at all time points and DOX animals at day 0 cluster tightly on principal component analysis (PCA). Post-treatment DOX animals form a looser but distinct cluster (Fig. 3). Testing via two-way PERMANOVA for significant differences in β-diversity confirmed that both treatment and time were significant factors (p < 0.0001, F = 95.15 and p < 0.0001, F = 8.91, respectively). One-way PERMANOVA, performed to determine pair-wise differences, detected significant differences between the later three time-points of DOX-treated mice and all control time-points (p = 0.0135 to 0.0045, F = 47.03 to 208.2). Conversely, within-group differences in DOX-treated mice were limited to differences between the starting time-point and the last three time-points (p = 0.0045, F = 59.47 to 103.6). Significant within-group differences in the control group were found between time-points 1 and both 3 (p = 0.018, F = 13.44) and 5 (p = 0.014, F = 11).
Fig. 3.
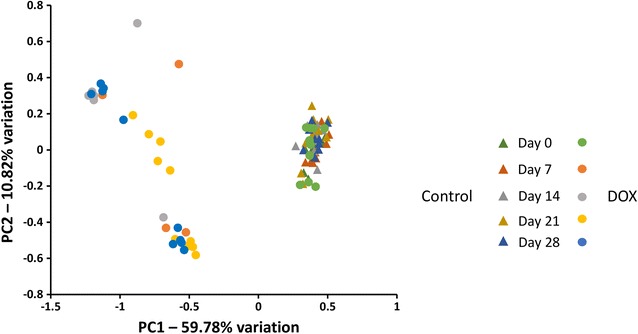
Principal component analysis. Principal component analysis of the fecal microbiota in DOX (circles) and control (triangles) animals. The DOX animals at day 0, and the control animals at all time points cluster tightly
Discussion
These data indicate that the FM of female C57BL/6NCrl mice is significantly altered by administration of doxycycline at a concentration of 2 mg/ml in the drinking water. This was evidenced by decreased richness and evenness, and disparate populations observed after treatment. They also suggest that after 2 weeks of treatment, effects persist for greater than 1 week, as samples taken at the end of week 3 still exhibited these changes.
The results reported here are not surprising when one considers doxycycline’s spectrum of activity and intestinal route of elimination [42–45]. Though unsurprising, the results are not underwhelming, as they illustrate the occult malleability of animals’ baseline condition. These findings suggest that investigators utilizing tet-on or tet-off models should consider the GM as a variable to be minimized or controlled for in their experiments.
To our knowledge, there are no published recommendations or guidelines to aid investigators, thus we propose use of appropriate controls: ideal control animals are littermates or animals of the same strain and individual history (including source, number of backcrosses if any, sex, age, husbandry management, etc.). Controls must be unresponsive to doxycycline (which requires het X wild type breeding to yield transgene negative animals) and must be given the same dose of doxycycline as experimental animals. Finally, any experimental outcomes that are or may be responsive to changes in the GM must be interpreted with care, especially when comparing the results with other models that do not require doxycycline such as knockouts and Cre/lox models. Alternatively, non-antibiotic analogs of doxycycline could be considered [46, 47].
In summary, the common gene-regulating dose of doxycycline causes a significant dysbiosis in female C57BL/6NCrl mice. This altered microbial community may present challenges or confounds in experimental animals.
Limitations
Impact limitation
Results are/may not be broadly applicable to other study populations e.g. mice of different age, sex, strain, vendor source, diet, disease state, etc.
Study limitation
Results do not account for different doses, durations, or routes of administration of doxycycline.
Results do not include effects of doxycycline on other microbial communities e.g. vaginal, skin etc.
Study does not control for stochastic influences.
Additional files
Additional file 1. Detailed descriptions of animals and husbandry, welfare assessments and interventions, doxycycline administration, and sample collection that were omitted from the primary manuscript due to length restrictions.
Additional file 2. Detailed description of DNA extraction that was omitted from the primary manuscript due to length restrictions.
Additional file 3. Fecal microbial community parameter comparisons by treatment and time.This table includes fecal microbial community parameters (mean OTUs per sample, Shannon diversity index, and Chao1) for each sample collected.
Additional file 4. Relative abundance at taxonomic level of OTU. Bar charts showing the bacterial composition of the same control and doxycycline-treated mice at all time points annotated to the taxonomic level of OTU. Legend of prominent OTUs is shown below. Each bar represents an individual animal (DOX animals numbered 1–10, control animals numbered 11–20).
Authors’ contributions
JEW and MU designed the research; FB, MU, and MD performed the research; AE analyzed the data; and FB, JEW, and AE wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank Dr. Ingrid Bergin for her mentorship and assistance during this project; Lisa Burlingame and Jennie Jones for their exceptional care of the animals; and Chris Fry for his assistance with laboratory procedures.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data supporting the conclusions of this article will be provided by the corresponding author on reasonable request.
Consent for publication
Not applicable.
Ethics approval and consent to participate
This study was approved by the Institutional Animal Care and Use Committee at the University of Michigan.
Funding
The authors received no specific funding for this work.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- GM
gut microbiota
- FM
fecal microbiota
- DOX
doxycycline-treated
- OTU
operational taxonomic unit
- UC
unclassified
- PCA
principal component analysis
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s13104-017-2960-7) contains supplementary material, which is available to authorized users.
Contributor Information
Felicia D. Duke Boynton, Email: fduke@umn.edu.
Aaron C. Ericsson, Email: ericssona@missouri.edu
Mayu Uchihashi, Email: mayu.uchihashi@medtronic.com.
Misha L. Dunbar, Email: mldunbar@umn.edu
J. Erby Wilkinson, Email: jerby@med.umich.edu.
References
- 1.Blaut M, Clavel T. Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr. 2007;137(3):751S–755S. doi: 10.1093/jn/137.3.751S. [DOI] [PubMed] [Google Scholar]
- 2.Scanlan PD, Shanahan F, Clune Y, Collins JK, O’Sullivan GC, O’Riorda M, et al. Culture-independent analysis of the gut microbiota in colorectal cancer and polyposis. Environ Microbiol. 2008;10(3):789–798. doi: 10.1111/j.1462-2920.2007.01503.x. [DOI] [PubMed] [Google Scholar]
- 3.Lupp C, Roberston ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2(2):119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, et al. Salmonella enterica serovar Typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS Biol. 2007;5(10):e244. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. 2008;35(8):1500–1505. [PubMed] [Google Scholar]
- 6.Ellekilde M, Krych L, Hansen CH, Hufeldt MR, Dahl K, Hansen LH, et al. Characterization of the gut microbiota in leptin deficient obese mice—correlation to inflammatory and diabetic parameters. Res Vet Sci. 2014;96:241–250. doi: 10.1016/j.rvsc.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 7.Membrez M, Blancher F, Jaquet M, Bibiloni R, Cani PD, Burtcelin RG, et al. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. FASEB J. 2008;22:2416–2426. doi: 10.1096/fj.07-102723. [DOI] [PubMed] [Google Scholar]
- 8.Rune I, Hansen CH, Ellekilde M, Nielsen DS, Skovgaard K, Rolin BC, et al. Ampicillin-improved glucose tolerance in diet-induced obese C57BL/6NTac mice is age dependent. J Diabetes Res. 2013 doi: 10.1155/2013/319321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berer K, Mues M, Koutrolos M, Rasbi ZA, Boziki M, Johner C, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 10.Pyndt Jorgeson B, Hansen JT, Krych L, Larsen C, Klein AB, Nielsen DS, et al. A possible link between food and mood: dietary impact on gut microbiota and behavior in BALB/c mice. PLoS ONE. 2014;9(8):e103398. doi: 10.1371/journal.pone.0103398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zackular JP, Baxter NT, Iverson KD, Sadler WD, Petrosino JF, Chen GY, et al. The gut microbiome modulates colon tumorigenesis. MBio. 2013;4(6):e00692. doi: 10.1128/mBio.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorozynska I, Majewska-Szczepanik M, Marcinska K, Szczepanik M. Partial depletion of the natural gut flora by antibiotic aggravates collagen induced arthritis (CIA) in mice. Pharmacol Rep. 2014;66:250–255. doi: 10.1016/j.pharep.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Martinez I, Wallace G, Zhang CM, Legge R, Benson AK, Carr TP, et al. Diet-induced metabolic improvements in hamster model of hypercholesterolemia are strongly linked to alterations of the gut microbiota. Appl Environ Microbiol. 2009;75:4175–4184. doi: 10.1128/AEM.00380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon HS, Ju JH, Kim HN, Park HJ, Ji Y, Lee JE, et al. Reduction in cholesterol absorption in Caco-2 cells through the down-regulation of Niemann-Pick C1-like 1 by the putative probiotic strains Lactobacillus rhamnosus BFE5264 and Lactobacillus plantarum NR74 from fermented foods. Int J Food Sci Nutr. 2013;64:44–52. doi: 10.3109/09637486.2012.706598. [DOI] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;2006(444):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 16.Garrett WS, Gordon JI, Glimcher LH. Homeostasis and inflammation in the intestine. Cell. 2010;140:859–870. doi: 10.1016/j.cell.2010.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Littman DR, Pamer EG. Role of the commensal microbiota in normal and pathogenic host immune responses. Cell Host Microbe. 2011;10:311–323. doi: 10.1016/j.chom.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;2012(336):1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci. 2002;99:15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reinhardt C, Bergentall M, Greiner TU, Schaffner F, Östergren-Lundén G, Petersen LC, Ruf W, Bäckhed F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature. 2012;483:627–631. doi: 10.1038/nature10893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sudo N, Chida Y, Aiba Y, Sonoda J, Oyama N, Yu X-N, Kubo C, Koga Y. Postnatal microbial colonization programs the hypothalamic–pituitary–adrenal system for stress response in mice. J Physiol. 2004;558:263–275. doi: 10.1113/jphysiol.2004.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bravo JA, Forsythe P, Chew MV, Escaravage E, Savignac HM, Dinan TG, Bienenstock J, Cryan JF. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diaz Heijtz R, Wang S, Anuar F, Qian Y, Björkholm B, Samuelsson A, Hibberd ML, Forssberg H, Pettersson S. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci. 2011;108:3047–3052. doi: 10.1073/pnas.1010529108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ezenwa VO, Gerardo NM, Inouye DW, Medina M, Xavier JB. Animal behavior and the microbiome. Science. 2012;338:198–199. doi: 10.1126/science.1227412. [DOI] [PubMed] [Google Scholar]
- 25.Baron U, Bujard H. Tet repressor-based system for regulated gene expression in eukaryotic calls: principles and advances. Methods Enzymol. 2000;327:401–421. doi: 10.1016/S0076-6879(00)27292-3. [DOI] [PubMed] [Google Scholar]
- 26.Ujihara Y, Iwasaki K, Takatsu S, Hashimoto K, Naruse K, Mohri S, et al. Induced NCX1 overexpression attenuates pressure overload-induced pathological cardiac remodelling. Cardiovasc Res. 2016;111(4):348–361. doi: 10.1093/cvr/cvw113. [DOI] [PubMed] [Google Scholar]
- 27.Kesby JP, Markou A. Semenova S; TMARC Group. Effects of HIV/TAT protein expression and chronic selegiline treatment on spatial memory, reversal learning and neurotransmitter levels in mice. Behav Brain Res. 2016;311:131–140. doi: 10.1016/j.bbr.2016.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boulay PL, Mitchell L, Turpin J, Huot-Marchand JÉ, Lavoie C, Sanguin-Gendreau V, et al. Rab11-FIP1C is a critical negative regulator in ErbB2-mediated mammary tumor progression. Cancer Res. 2016;76(9):2662–2674. doi: 10.1158/0008-5472.CAN-15-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mellado-Gil JM, Jiménez-Moreno CM, Martin-Montalvo A, Alvarez-Mercado AI, Fuente-Martin E, Cobo-Vuilleumier N, et al. PAX4 preserves endoplasmic reticulum integrity preventing beta cell degeneration in a mouse model of type 1 diabetes mellitus. Diabetologia. 2016;59(4):755–765. doi: 10.1007/s00125-016-3864-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dow LE, O’Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161(7):1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kucharzik T, Williams IR. Neutrophil migration across the intestinal epithelial barrier–summary of in vitro data and description of a new transgenic mouse model with doxycycline-inducible interleukin-8 expression in intestinal epithelial cells. Pathobiology. 2002;70(3):143–149. doi: 10.1159/000068146. [DOI] [PubMed] [Google Scholar]
- 32.Antonopoulos DA, Huse SM, Morrison HG, Schmidt TM, Sogin ML, Young VB. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77(6):2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson CJ, Young VB. Antibiotic administration alters the community structure of the gastrointestinal microbiota. Gut Microbes. 2010;1(4):279–284. doi: 10.4161/gmic.1.4.12614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saunders TL. Inducible transgenic mouse models. In: Hofker MH, van Deursen J, editors. Transgenic mouse methods and protocols. Methods in molecular biology (Methods and Protocols), vol 693. Humana Press; 2011. p. 103–15. [DOI] [PubMed]
- 35.Eger K, Hermes M, Uhlemann K, Rodewals S, Ortwein J, Brulport M, et al. Epidoxycycline: an alternative to doxycycline to control gene expression in conditional mouse models. Biochem Biophys Res Commun. 2004;323(3):979–986. doi: 10.1016/j.bbrc.2004.08.187. [DOI] [PubMed] [Google Scholar]
- 36.Ericsson AC, Davis JW, Spollen W, Bivens N, Givan S, Hagan C, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS ONE. 2015;10(2):e0116704. doi: 10.1371/journal.pone.0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magoc T, Salzberg SL. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, Knight R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Microbiol. 2012 doi: 10.1002/9780471729259.mc01e05s27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hammer Ø, Harper DAT, Ryan PD. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- 42.Klein NC, Cunha BA. Tetracyclines. Med Clin North Am. 1995;79(4):789. doi: 10.1016/S0025-7125(16)30039-6. [DOI] [PubMed] [Google Scholar]
- 43.Sutter VL, Finegold SM. Susceptibility of anaerobic bacteria to 23 antimicrobial agents. Antimicrobial Agents Chemother. 1976;1976(10):736–752. doi: 10.1128/AAC.10.4.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chow AW, Patten V, Guze LB. Comparative susceptibility of aerobic bacteria to minocycline, doxycycline, and tetracycline. Antimicrob Agents Chemother. 1975;1975(7):46–49. doi: 10.1128/AAC.7.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saivin S, Houin G. Clinical pharmacokinetics of doxycycline and minocycline. 1988. Clin Pharmacokinet. 1988;15(6):355. doi: 10.2165/00003088-198815060-00001. [DOI] [PubMed] [Google Scholar]
- 46.Croubels S, Baert K, De Busser J, De Backer P. Residue study of doxycycline and 4-epidoxycycline in pigs medicated via-drinking water. Analyst. 1998;123(12):2733–2736. doi: 10.1039/a804936j. [DOI] [PubMed] [Google Scholar]
- 47.Kertscher HP, Habermann A, Pohle R, Tauchert H, Hennig L, Eger K. Preparation and characterization of 4-epidoxycycline. Sci Pharm. 1998;66:287–296. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Detailed descriptions of animals and husbandry, welfare assessments and interventions, doxycycline administration, and sample collection that were omitted from the primary manuscript due to length restrictions.
Additional file 2. Detailed description of DNA extraction that was omitted from the primary manuscript due to length restrictions.
Additional file 3. Fecal microbial community parameter comparisons by treatment and time.This table includes fecal microbial community parameters (mean OTUs per sample, Shannon diversity index, and Chao1) for each sample collected.
Additional file 4. Relative abundance at taxonomic level of OTU. Bar charts showing the bacterial composition of the same control and doxycycline-treated mice at all time points annotated to the taxonomic level of OTU. Legend of prominent OTUs is shown below. Each bar represents an individual animal (DOX animals numbered 1–10, control animals numbered 11–20).
Data Availability Statement
The data supporting the conclusions of this article will be provided by the corresponding author on reasonable request.


