Abstract
In the course of our investigations into the toxicity of tungstate, we discovered that cellular exposure resulted in the loss of the histone demethylase protein. We specifically investigated the loss of two histone demethylase dioxygenases, JARID1A and JMJD1A. Both of these proteins were degraded in the presence of tungstate and this resulted in increased global levels of H3K4me3 and H3K9me2, the substrates of JARID1A and JMJD1A respectively. Treatment with MG132 completely inhibited the loss of the demethylase proteins induced by tungstate treatment, suggesting that tungstate activated the proteasomal degradation of these proteins. The changes in global histone marks and loss of histone demethylase protein persisted for at least 48 hours after removing sodium tungstate from the culture. The increase in global histone methylation remained when cells were cultured in methionine-free media, indicating that the increased histone methylation did not depend upon any de novo methylation process, but rather was due to the loss of the demethylase protein. Similar increases of H3K4me3 and H3K9me2 were observed in the livers of the mice that were acutely exposed to tungstate via their drinking water. Taken together, our results indicated that tungstate exposure specifically reduced histone demethylase JARID1A and JMJD1A via proteasomal degradation, leading to increased histone methylation.
Keywords: Tungsten, Cancer, BEAS-2B, In Vitro, Lung Cancer, Histone Demethylase, Histone Methylation
Introduction
Over the past few decades, we have learned that metallic species may exert their carcinogenic potential via epigenetic mechanisms [1]. Exposure to the IARC class I carcinogenic metals—arsenic, nickel, cadmium, and chromium(VI)—have all displayed significant epigenetic alterations that likely drive the cancer phenotype [2]. These metals have been widely studied in cancer research, with a large body of literature establishing them as agents that alter the cellular epigenetic program. Therefore, it is likely that other metal carcinogens may also alter the epigenetic program of cells, which may drive carcinogenesis. Recently, we have reported that the oxyanion vanadate may have tumorigenic potential based on its ability to induce transformation of BEAS-2B cells in tissue culture [3]. This research may have generated other studies investigating vanadium’s ability to induce cancer [4, 5]. Tungsten is another metal that has not been explored extensively in toxicology studies. We aim to investigate the carcinogenic potential of tungsten compounds and uncover possible epigenetic mechanisms that drive its carcinogenicity.
Tungsten has unique chemical properties; at just over 3,000 degrees Celsius, tungsten has the highest melting point of all metals [6]. Elemental tungsten is a dense transition metal, which exhibits oxidation states from −2 to +6. When exposed to atmospheric conditions, it readily forms oxides. The most common oxide is WO3. In particle form, tungsten exists as clusters of WO3 molecules that can adopt either crystalline or amorphous structures connected by non-covalent bonds. In alkaline conditions, WO3 particulate powder dissolves in aqueous solutions to form WO42− (orthotungstate) ions. Adding tungsten to steel significantly increases its toughness. Coating machining tools with tungsten carbide improves hardness and increases melting temperature, enabling the machining of stainless steels. When WO3 intercalates within alkali metals it produces bronzes. Tungsten oxides are widely used in machining applications, which often produce fine particulates [7–10].
Human exposure to tungsten containing material is widespread. Also, since tungsten is commonly used in creating tools for machining steel, machine shop workers regularly inhale particulate tungsten and its alloys. Tungsten is also commonly used in munitions manufacturing, which causes exposure in the manufacture of munitions, as well as those injured by their deployment. While tungsten compounds are considered relatively non-toxic and cells can tolerate mM levels of exposure, the effects of exposure at the mM level are important to explore given the numerous routes by which human tungsten exposure can occur due to its widespread industrial utility.
Alterations in histone modifications, on both a global and gene-specific level, represent emerging mechanisms by which many metals alter gene expression, which can be epigenetic if inherited. A study by Arita et al., 2012 evaluated peripheral blood mononuclear cells (PBMCs) of Chinese nickel refinery workers occupationally exposed to nickel for changes in histone methylation. The investigation found that nickel altered the genome-wide levels of H3K9me2 and H3K4me3 [11]. Nickel has been shown to inhibit the activity of the major class of histone demethylases known as dioxygenases by readily displacing the Fe from the active site of these enzymes [12]. A hallmark property of cancer cells is global hypoacetylation [2]. Metals have been shown to cause hypoacetylation [1, 6] in cells and nickel also inhibits histone acetyltransferases (HATs) [13].
Tungsten’s unique chemical properties make it attractive to industry. Therefore, occupational exposures to tungsten raise significant toxicity concerns. Previously, we reported on tungsten's ability to induce, following in vitro exposure of BEAS-2B cells, cancer-related endpoints including cell transformation, increased migration, xenograft growth in nude mice, and the activation of multiple cancer-related pathways [18]. Histone modifications, specifically alterations in histone acetylation and methylation, have been altered following exposure of a number other carcinogenic metals [1, 19]. Here, we show perturbations in the levels of global histone methylation that occur after tungstate exposure and suggest unusual mechanisms that mediate this effect involving the ability of tungstate to selectively degrade the histone demethylase dioxygenases.
Materials and Methods
Cell culture
Human bronchial epithelial cells (BEAS-2B, ATCC, Manassas, VA) were adapted to serum growth after their purchase and have been carefully maintained. They were recently tested through Genetica DNA Laboratories against a reference BEAS-2B cell line and were 100% matched to prove authenticity (Burlington, NC). BEAS-2B were cultured in 1 × Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA) and 1% Pen Strep (GIBCO, Grand Island, NY). Adenocarcinomic human alveolar basal epithelial cells (A549, ATCC) were cultured in F-12K media (Invitrogen) supplemented with 10% FBS and 1% Pen Strep.
The cells were maintained in 10 cm2 polystyrene tissue culture dishes in an incubator at 37 °C with 5% CO2. Media were changed every four days as well as passaged using 0.05% trypsin-EDTA (GIBCO) as described previously [12]. Cells were split and seeded at the same concentration, 3 × 105, in the presence of Na2WO4 (Sigma-Aldrich, St. Louis, MO), dissolved in distilled water, with concentrations ranging from 1 mM to 5 mM. Cells were treated with Na2WO4 for 48-hours, and then in absence of tungstate for 48-hour, for a washout. Histones and nuclear extracts were extracted.
For BEAS-2B treated in methionine-free media, cells were seeded at 3 × 105. After 24 hours 1 × DMEM media was removed and cells were rinsed with PBS. Cells were incubated in methionine-free media for four hours. Cells were treated with concentrations ranging from 1 mM to 5 mM Na2WO4 for 24 hours. Subsequently, histones and nuclear extracts were extracted.
BEAS-2B cells were co-treated with 0.5 µM MG132 (Sigma-Aldrich) and 1 mM to 5 mM Na2WO4 for 24 hours, and nuclear extracts were prepared.
Plating Efficiency
To evaluate the cell toxicity of BEAS-2B with Na2WO4, cells were seeded at 3 × 105 and treated with 0, 1, 2.5 and 5 mM Na2WO4 for 48 hours. After the exposure, the cells were rinsed with PBS (Thermo Fisher Scientific, Waltham, MA) and then counted and seeded at 500 cells per dish in fresh media. Cells were cultured for two weeks and then fixed and stained with Giemsa. Plating efficiency was calculated by the number of colonies divided by the number of cells plated multiplied by 100%. Three replicate dishes were evaluated per each dose and the results are expressed as mean±standard deviation.
Silver stain
BEAS-2B cells treated acutely with 0, 1, 2.5 and 5 mM Na2WO4 were lysed using boiling buffer (1% SDS, 1mM Na3VO4, 10 mM Tris-Cl, pH 7.4). Briefly, the preheated boiling buffer was added to each 10 cm cell culture plate at 70% confluency after PBS wash. The cell lysate was denatured at 100°C for 5 minutes and sonicated using Diagenode Biorupter (Denville, NJ) at a maximum setting for 10 minutes. Next the samples were centrifuged at 4°C for 15 minutes for 21,000 rpm. Protein was quantified using the Bio-Rad DC colorimetric Protein Assay (Hercules, CA), using bovine serum albumin (Sigma-Aldrich) as the protein standard. Next, 500 µg of proteins were separated by 12% (w/v) gels and the Pierce Silver Stain protocol (Thermo Fisher Scientific) was followed provided by the manufacturer.
Inductively coupled plasma mass spectrometry (ICP-MS)
BEAS-2B were cultured as described above and treated with Na2WO4 in concentrations ranging from 1 mM to 5 mM for 48 hours. The cells were trypsinized and counted to determine cell number. There was equal number of cells in each sample pellet. Each sample pellet was treated with HNO3 (70%), and then incubated at 80°C for 3 hours. The digested material was utilized to measure the levels of tungsten by ICP-MS (Perkin Elmer, Warsaw, Poland).
Mouse experiment with tungstate treated drinking water
Twelve 6-week-old male AJ mice (body weight 22–25g) were obtained from Taconic Farms (Germantown, NY) and housed in our AAALAC accredited housing facility in Tuxedo, NY. Approval was obtained by IACUC for the use of animals in experimental studies. Tungstate was added to the drinking water at 0, 5 and 10 g/L concentrations and the mice were given access to the drinking water ad libitum for 14 days. Four mice were treated with 5g/L of tungstate, the low-dose group. Four mice were treated with 10g/L of tungstate, the high-dose group. All procedures were conducted in compliance with New York University’s guidelines for ethical animal research and the Declaration of Helsinki.
Preparation of histones and nuclear extracts
At 80–90% confluency of cells, histones or nuclear extracts were collected. Livers from mice were collected post-sacrifice, processed and the histones were collected. Histones were collected from cells as previously described [20, 21]. Briefly, ice-cold radioimmunoprecipitation assay buffer (50 mM Tris-HCl at pH 7.4, 1% NP-40, 0.25% Na-deoxycholate, 150 mM NaCl and 1 mM ethylenediaminetetraacetic acid) (Santa Cruz Biotechnology) supplemented with a protease inhibitor mixer (Roche Applied Sciences) was used to extract histones from cells and livers. Nuclear extracts were collected using CelLytic NuCLEAR extract kit (Sigma-Aldrich). The histone concentration and nuclear extract protein concentration was determined using Bio-Rad DC colorimetric Protein Assay using bovine serum albumin as the protein standard.
Western blotting
The protein concentration was measured using Bio-Rad DC protein assay. For histones, 2.5 µg were separated by any kD SDS-PAGE gels and transferred to polyvinylidene difluoride membranes, PVDF (Bio-Rad). Immunoblotting was performed using trimethyl H3K4 (1:1000; Abcam) primary antibody and horseradish peroxidase-conjugated anti-rabbit secondary antibody (Santa Cruz Biotechnology). Immunoblotting was performed using dimethyl H3K9 (1:2000; Abcam) primary antibody and horseradish peroxidase-conjugated anti-mouse secondary (Santa Cruz Biotechnology). In order to obtain relative intensities, loading of histones was attained using H3 (1;30,000; Abcam).
For nuclear extracts containing 25 µg of proteins were separated by 6% (w/v) gels and transferred to PVDF membranes. Immunoblotting was performed using JARID1A (1:1000; Bethyl) primary antibody and horseradish peroxidase-conjugated anti-rabbit secondary antibody. Immunoblotting was performed using JMJD1A (1:1000; Bethyl) primary antibody and horseradish peroxidase-conjugated anti-rabbit secondary antibody. In order to obtain relative intensities, the loading of nuclear extracts was attained using Lamin A (1:500; Santa Cruz).
Detection was accomplished using Pierce ECL Western Blotting Substrate (Thermo Scientific). The relative intensities of the bands were quantified by ImageJ and normalized to the control.
Real-Time Quantitative PCR
Total RNA was extracted by TRIzol (Invitrogen), and then purified using RNeasy Plus Micro Kit (Qiagen). 250 ng of RNA that was extracted and purified was converted into single-strand cDNA using Superscript III (Invitrogen). Next, real-time qPCR was conducted on 20 ng of cDNA using SYBR Green PCR Master Mix (Applied Biosystems) on an ABI PRISM 7900HT. The specific primers are indicated below: JARID1A, 5’-GCTTGGCAATGGGAACAAAA-3’ (forward) and 5’-CGTTGTCTCATTTGCATGTTAA-3’ (reverse); JMJD1A, 5’-TCTGTTCCACAAGCATTGACTGG-3’ (forward), and 5’-AGGGCTATCACTATTGACACCTGC-3’ (reverse); β-actin, 5’-ATCGTCCACCGCAAATGCTTCTA-3’ (forward) and 5’-AGCCATGCCAATCTCATCTTGTT-3’ (reverse). All PCR was performed in triplicates, and the relative gene expression levels were normalized to β-actin expression. The results are represented as a fold change to the untreated control cell expression levels.
Results
Tungstate exposure results in little cell death at highest dose
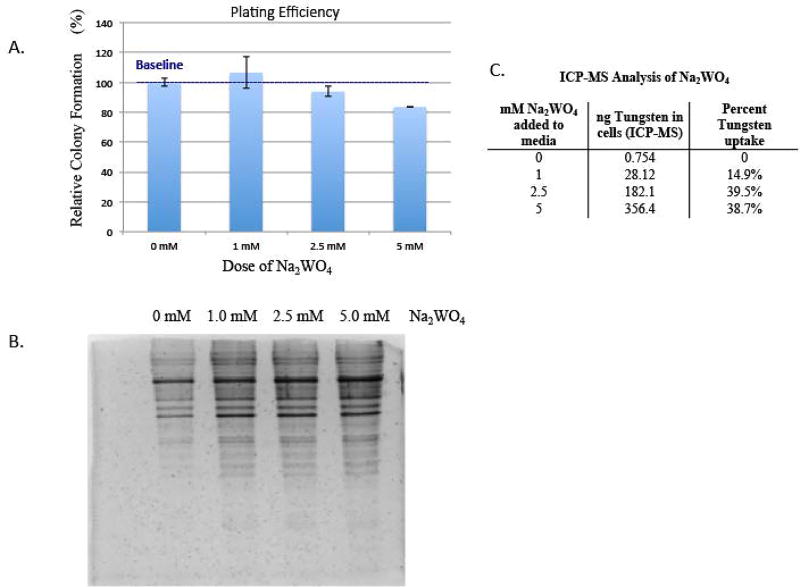
BEAS-2B cells were treated with 0, 1, 2.5 and 5 mM Na2WO4 for 48 hours and then seeded at 500 cells per dish and cultured for two weeks (figure 1a). The lowest dose, 1 mM Na2WO4, resulted in more cell survival than the parental BEAS-2B. The 2.5 mM Na2WO4 dose resulted in 5% cell death. The highest dose, 5 mM Na2WO4, resulted in 16.4% cell death. Plating efficiency was carried out to find appropriate doses of tungstate for treatment. The assay was performed with three biological replicates. These results clearly illustrate the low cellular toxicity of tungstate.
Figure 1. The effect of tungstate on cell viability, its effect on protein degradation and its uptake into cells.
A, BEAS-2B cells were treated with 0, 1, 2.5 and 5 mM Na2WO4 for 48-hours, they were seeded at 500 cells per dish and cultured for two weeks. Cells were fixed then counted for cell survival. Toxicity was determined using plating efficiency. Values are presented as the number of colonies divided by the number of control colonies (0 mM) multiplied by 100%.
B, BEAS-2B cells were acutely treated with 0, 1, 2.5 or 5 mM of Na2WO4, then lysed and tested for protein degradation via silver stain as described in Materials and Methods.
C, ICP-MS analysis of BEAS-2B cells after 48-hour treatment of 0, 1, 2.5 and 5 mM Na2WO4.
Silver stain gel shows no protein degradation after acute tungstate-treatment
BEAS-2B cells were treated with 0, 1, 2.5 and 5 mM Na2WO4 for 48 hours and then collected and assessed for protein degradation using silver staining. In order to evaluate the protein degradation of the tungstate-treated cells, the protein was separated on a 12% (w/v) polyacrylamide gel and then treated with silver stain to visualize the protein (figure 1b). There is no protein degradation evident after 48-hour treatment of tungstate at any dose. These results clearly demonstrate that tungstate does not cause generalized cell-wide protein degradation.
Tungstate exposure results in 15–40% cellular uptake
BEAS-2B cells treated with 1, 2.5 and 5 mM of Na2WO4 for 48 hours were collected and the uptake of tungsten was measured by ICP-MS. The higher doses of tungstate, 2.5 and 5 mM, showed a larger percent uptake (~40%) compared to the lower dose, 1 mM (figure 1c), where the uptake was only ~15%. These results illustrate that tungstate is capable of crossing cell membranes and entering cells, yet has a low level of toxicity.
Acute exposure to tungstate results in increased global level of H3K9me2 and H3K4me3
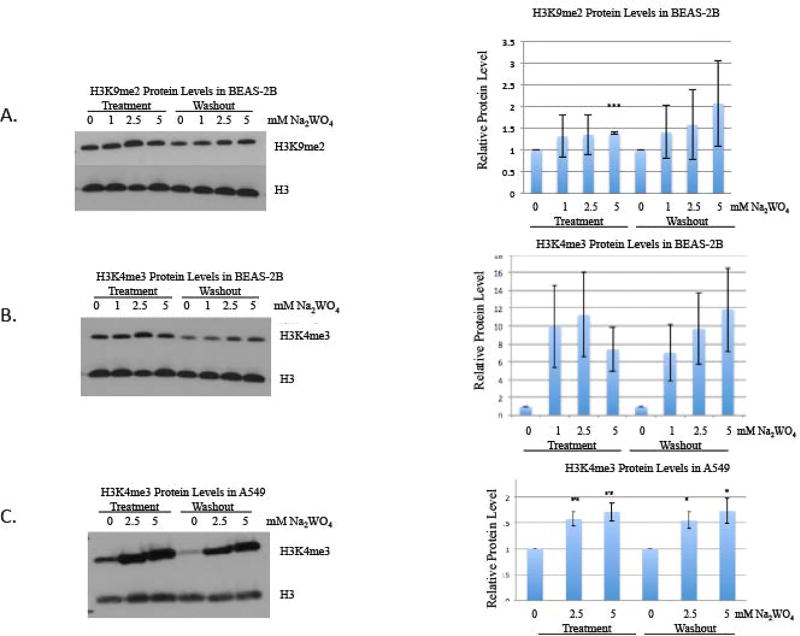
BEAS-2B cells were treated with 0, 1, 2.5, 5 and 10 mM Na2WO4 for 48 hours. The cells were either harvested or continued to grow for 48 hours in the absence of tungstate (washout). After tungstate exposure, histones and nuclear extracts were prepared. Global levels of H3K4me3 and H3K9me2 were assessed by western blots with antibodies specific to these modifications. Global levels of H3K4me3 were significantly increased after 48-hour acute exposure when compared to the untreated controls. In addition, global levels of H3K9me2 were also significantly increased after acute treatment when compared to the untreated controls. The global increase in both H3K4me3 and H3K9me2 persisted, even after a 48-hour washout period (figure 2a, 2b). Global levels of H3K4me3 were significantly increased after 48-hour acute exposure to 2.5 and 5 mM using a different human lung cell line, an adenocarcinomic human alveolar basal epithelial cell line (A549), and these changes persisted after a 48-hour washout (figure 2c).
Figure 2. The effect of tungstate on H3K9me2 and H3K4me3 in BEAS-2B and A549 cells.
BEAS-2B cells were exposed to 0, 1, 2.5 and 5 mM of Na2WO4 for 48-hours, and then cells were allowed to proliferate in the absence of tungstate for 48 hours, for a washout. Histones were extracted after treatment and washout.
A, H3K9me2 was detected using Western blotting as described in Materials and Methods. The same membrane was stripped and reprobed with H3 antibody as loading control.
B, H3K4me3 was detected using Western blotting. The same membrane was stripped and reprobed using H3 antibody as loading control.
C, A549 cells were exposed to 0, 2.5 and 5 mM of Na2WO4 for 48-hours, and then cells were allowed to proliferate in the absence of tungsten for 48 hours, for a washout. Histones were extracted after treatment and washout. H3K4me3 was detected using Western and the same membrane was stripped and reprobed using H3 antibody as loading control.
The results were repeated in three independent experiments; one representative blot is shown here. Graphical representations of relative intensities are calculated by ImageJ.
Statistical significance was calculated using an unpaired, two-tailed t test
* p-value < 0.05
** p-value < 0.01
*** p-value < 0.001
Error bars represent Standard Deviation.
Increased histone methylation is due to depletion of histone demethylases
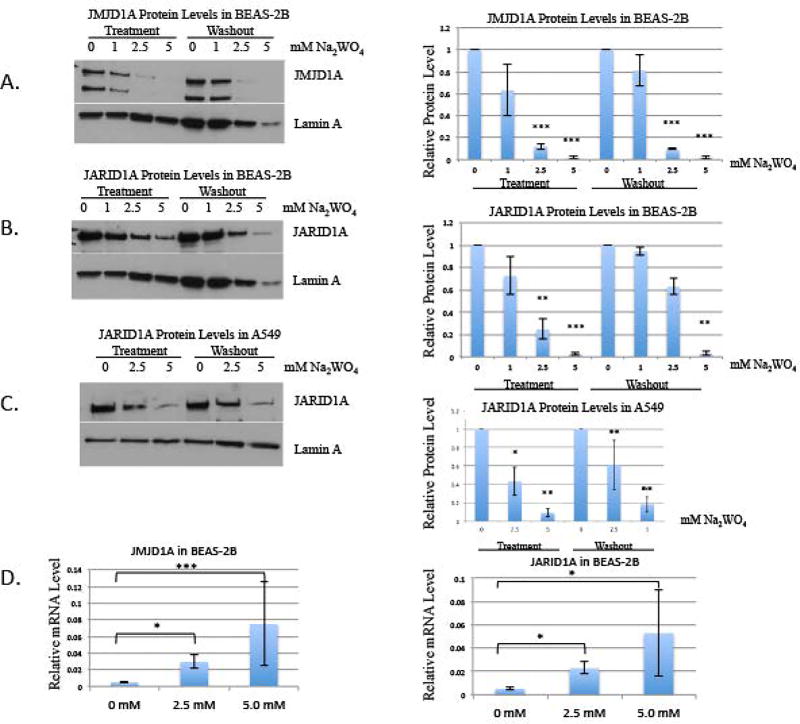
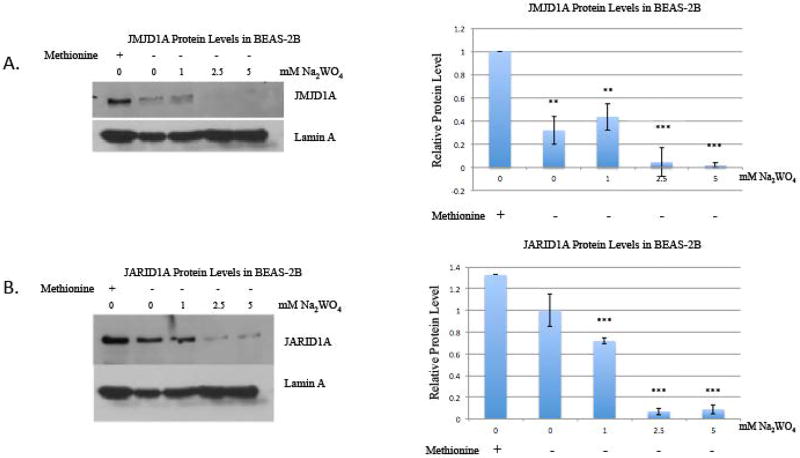
We investigated whether tungstate affecting the protein levels of the histone demethylases could be a possible mechanism that would have caused the increased methylation. BEAS-2B cells were treated with 0, 1, 2.5, and 5 mM Na2WO4 for 48 hours and allowed to proliferate in the absence of tungstate for 48 hours. JMJD1A (a histone demethylase for H3K9me2) levels were depleted by tungstate treatment when compared to the untreated control. The loss of this demethylase protein likely explains the increased H3K9me2 by tungstate (figure 3a).
Figure 3. The effect of tungstate on JMJD1A and JARID1A protein levels in BEAS-2B and A549 cells, and the effect of tungstate on relative mRNA levels of JMJD1A and JARID1A in BEAS-2B.
BEAS-2B cells were exposed to 0, 1, 2.5 and 5 mM of Na2WO4 for 48-hours, and then cells were allowed to proliferate in the absence of tungstate for 48 hours, for a washout. Nuclear extracts were extracted after treatment and washout.
A, JMJD1A was detected using Western blotting. The same membrane was re-probed with Lamin A antibody as loading control.
The results were repeated in three independent experiments; one representative blot is shown here. Graphical representations of relative intensities are calculated by ImageJ.
B, JARID1A was detected using Western blotting. The same membrane was re-probed using Lamin A antibody as loading control.
The results were repeated in three independent experiments; one representative blot is shown here. Graphical representations of relative intensities are calculated by ImageJ.
C, A549 cells were exposed to 0, 2.5 and 5 mM of Na2WO4 for 48-hours, and then cells were allowed to proliferate in the absence of tungsten for 48 hours, for a washout. Nuclear extracts were extracted after treatment and washout. JARID1A was detected using Western blotting and the same membrane was re-probed using Lamin A antibody as loading control.
The results were repeated in three independent experiments; one representative blot is shown here. Graphical representations of relative intensities are calculated by ImageJ.
D, After 48-hour acute treatment of 2.5 and 5 mM Na2WO4, RT-qPCR of histone demethylases, JMJD1A and JARID1A, was evaluated.
Statistical significance was calculated using an unpaired, two-tailed t test
* p-value < 0.05
** p-value < 0.01
*** p-value < 0.001
Error bars represent Standard Deviation.
Subsequently, we evaluated JARID1A, which demethylates H3K4me3. JARID1A levels were depleted by tungstate treatment, suggesting that JARID1A apparent protein loss contributed to the increased H3K4me2. Although there are four known histone demethylases targeting H3K4me3, we have previously shown that JARID1A is the major demethylase expressed in BEAS-2B cells [16]. We assessed if the decrease persisted after tungstate was removed and found that both histone demethylases were still decreased after the washout (figure 3b). In a parallel experiment, A549 cells were treated with 0, 2.5, and 5 mM Na2WO4 for 48 hours. JARID1A was persistently decreased by Na2WO4 in these A549 cells (figure 3c).
We investigated the mRNA levels of JMJD1A and JARID1A using qPCR. BEAS-2B cells were treated with 2.5 and 5 mM Na2WO4 for 48 hours and the mRNA levels coding for these enzymes was determined. We found that there was an increase mRNA levels of JMJD1A and JARID1A in tungstate treated cells compared to control BEAS-2B (figure 3d). This was likely due to the striking depletion of the demethylase protein and compensatory attempts by the cells to restore the protein to normal levels.
Tungstate-induced increase in histone methylation was still observed in cells with depleted SAM levels
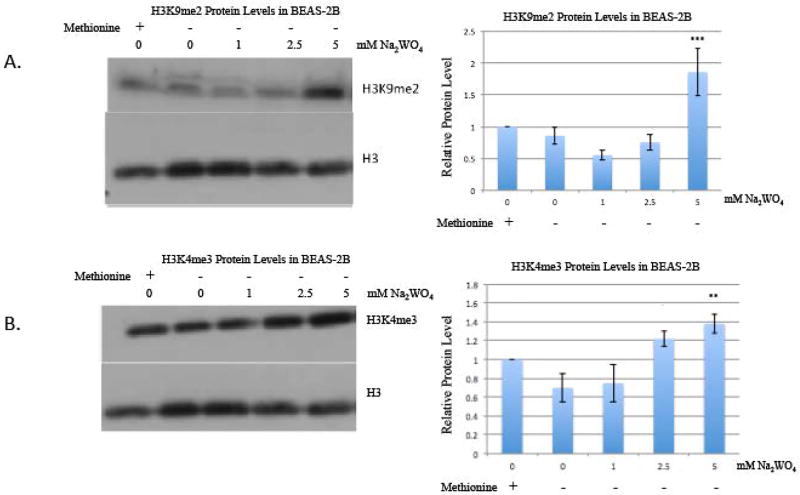
We investigated whether the increased histone methylation was due to a de novo methylation process. When BEAS-2B cells were placed into methionine-free media, there was still an increase in global methylation and a decrease in their histone demethylases after tungstate treatment, suggesting the absence of such activity. BEAS-2B were incubated in methionine-free media for four hours because of the short intracellular half-life of S-adenosyl-methionine (SAM) [16]. SAM is the only known source of methyl groups for all enzymatic reactions involving methylation. SAM is the methyl donor for the histone methyltransferases (HMTs) that methylates the lysine residues on the histone tail [22]. Methionine-deficient medium decreases intracellular SAM pools and inhibits methyl transfer reactions. After four hours incubation, cells were treated with 0, 1, 2.5, and 5 mM of tungstate. The same trend in a global increase of H3K9me2 and H3K4me3 was evident, even in methionine-free media (figure 4a and figure 4b). Additionally, the histone demethylase proteins also decreased with tungstate treatment in methionine-free media (figure 5a and figure 5b), and moreover the effect of tungstate appears to be enhanced in methionine-free media.
Figure 4. The effect of methionine depletion on H3K4me3 and H3K9me2 in BEAS-2B cells treated with tungstate.
BEAS-2B cells were seeded with DMEM complete medium. The following day, cells were pre-incubated for 4 hours in methionine-deficient DMEM, and then cells were exposed to either 0, 1, 2.5 or 5 mM Na2WO4 for 24-hours. Histones were extracted after treatment.
A, Histones were immunoblotted with H3K9me2 antibody and then stripped and reblotted with H3 antibody as loading control.
B, Histones were immunoblotted with H3K4me3 antibody and then stripped and reblotted with H3 antibody as loading control.
The results were repeated in three independent experiments; one representative blot is shown here. The intensity of the bands was quantified using ImageJ, and values were normalized to the each loading control and plotted in the graph. Error bars represent SD.
Statistical significance was calculated using an unpaired, two-tailed t test
* p-value < 0.05
** p-value < 0.01
*** p-value < 0.001
Figure 5. The effect of methionine depletion on JMJD1A and JARID1A protein levels in BEAS-2B cells treated with tungstate.
BEAS-2B cells were seeded with DMEM complete medium. The following day, cells were pre-incubated for 4 hours in methionine-deficient DMEM, and then cells were exposed to either 0, 1, 2.5 or 5 mM Na2WO4 for 24-hours. Nuclear extracts were extracted after treatment.
A, JMJD1A was detected using Western blotting. The same membrane was reprobed with Lamin A antibody as loading control.
B, JARID1A was detected using Western blotting. The same membrane was reprobed using Lamin A antibody as loading control
The results were repeated in three independent experiments; one representative blot is shown here. The intensity of the bands was quantified using ImageJ, and values were normalized to the each loading control and plotted in the graph. Error bars represent SD.
Statistical significance was calculated using an unpaired, two-tailed t test
* p-value < 0.05
** p-value < 0.01
*** p-value < 0.001
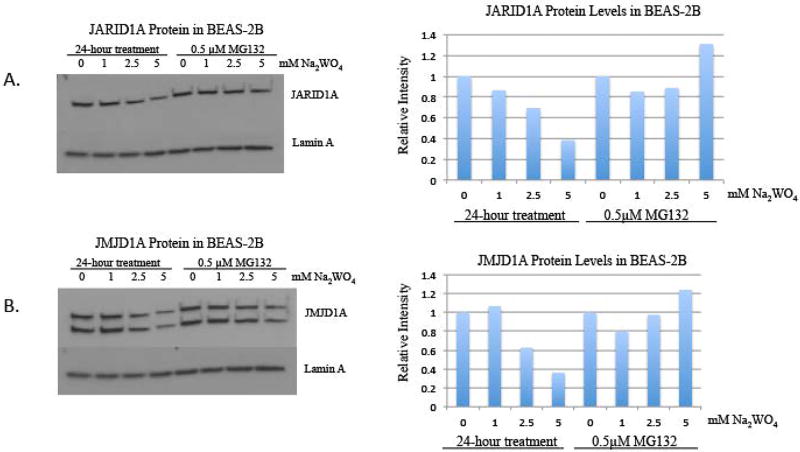
Proteasomal inhibition with MG132 prevented the loss of demethylase protein induced by tungstate
We investigated if the decrease in the protein level of the histone demethylases was due to enhanced proteasomal degradation induced by tungstate. We co-treated the cells with the proteasomal degradation inhibitor, MG132, and tungstate for 24 hours. Acute treatment of tungstate alone decreased the demethylase levels, but when the cells were co-treated with tungstate and MG132, the protein levels were not significantly decreased (figure 6). These findings suggest that tungstate lowers the demethylase protein levels by enhancing proteasomal degradation.
Figure 6. The effect of proteasonmal degradation inhibitor, MG132, on JMJD1A and JARID1A protein levels in BEAS-2B cells treated with tungstate.
BEAS-2B cells were co-treated with a proteasomal degradation inhibitor, MG132, and varying doses of Na2WO4 for 24 hours. Nuclear extracts were extracted after the experiment.
A, JARID1A was detected using Western blotting. The same membrane was reprobed with Lamin A antibody as loading control.
B, JMJD1A was detected using Western blotting. The same membrane was reprobed using Lamin A antibody as loading control
One representative blot is shown here, the experiment was repeated in two independent experiments. The intensity of the bands was quantified using ImageJ, and values were normalized to the each loading control and plotted in the graph.
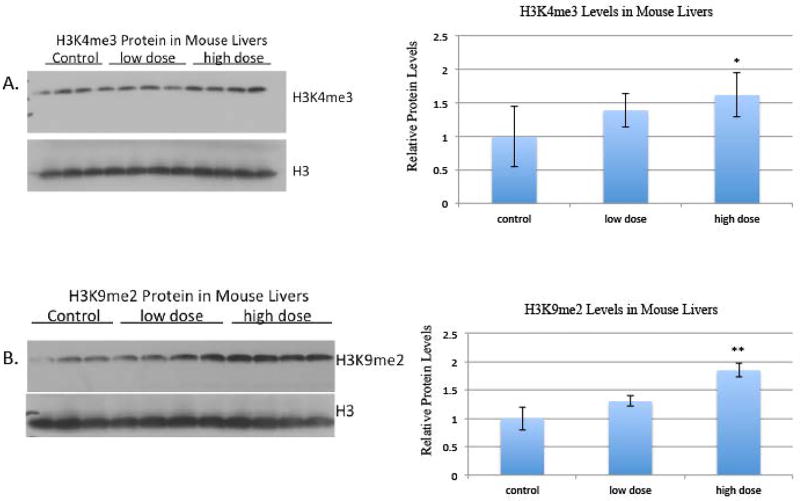
Mice acutely exposed to tungstate in drinking water exhibited increased global level of H3K9me2 and H3K4me3
Each treatment group cohort consisted of four mice. Mice were given access to drinking water with 0, 5 or 10 g/L tungstate ad libitum for two weeks. Over the course of the two-week exposure, the four mice in the 5 g/L tungstate group drank 90 mL cumulatively equating to an average daily exposure of 8 mg of tungstate per mouse. The mice in the 10 g/L cohort drank 60 mL cumulatively, equating to an average daily exposure of 11 mg of tungstate per mouse. Based on average water consumption rates, the 5 g/L tungstate group drank approximately half of the expected water consumption and the 10 g/L tungstate group drank approximately one-third of the expected consumption [23]. After two weeks of exposure, the mice were sacrificed and their livers were harvested.
Based on the equivalent surface area dosage conversion factors from the National Cancer Institute, the amount of tungstate that the mice were exposed to in the low-dose group is equivalent to 28 mg W/kg human per day, calculated by weighing the amount of water they drank. The amount of tungstate that the mice were exposed to in the high-dose group is equivalent to 39 mg W/kg human per day [24]. Global levels of H3K4me3 and H3K9me2 were assessed by western blots with antibodies specific to these modifications. Both global levels of H3K4me3 and H3K9me2 were increased in the mice treated with low-dose tungstate (5 g/L) and high-dose tungstate (10 g/L) drinking water compared to the control group (0 g/L) (figure 7).
Figure 7. The effect of tungstate drinking water exposure on H3K4me3 and H3K9me2 in mouse liver.
Mice were treated with 0g/L tungsten (control), 5g/L tungsten (low-dose), 10g/L tungsten (high-dose) in their drinking water for 2 weeks. Post sacrifice, organs were collected and histones were extracted.
A, Histones were immunoblotted with H3K4me3 antibody and then stripped and reblotted with H3 antibody as loading control.
B, Histones were immunoblotted with H3K9me2 antibody and then stripped and reblotted with H3 antibody as loading control.
Histones were extracted from parts of the liver three separate times; one representative blot is shown here. The intensity of the bands was quantified using ImageJ, and values were normalized to the each loading control and plotted in the graph. Error bars represent SD.
Statistical significance was calculated using an unpaired, two-tailed t test
* p-value < 0.05
** p-value < 0.01
*** p-value < 0.001
Discussion
Previous in vitro and epidemiological studies have shown that transition metals can induce changes in the levels of histone methylation in the cell [1, 11, 12, 20]. We aimed to investigate if tungstate could similarly affect histone methylation. As our previous study on tungstate established that it interferes with genetic pathways and could lead to lung cancer development, we chose to expose BEAS-2B cells, a lung bronchial tissue cell line, to tungstate. We treated BEAS-2B cells with relatively nontoxic doses and observed a striking global increase in H3K4me3 and H3K9me2; notably, the increase persisted after cells were allowed to proliferate for 48 hours through several cell divisions in tungstate-free media. These findings were confirmed after repeating the experiment in another lung cell line, A549 cells, where all the effects seen in BEAS-2B cells were also observed in A549 cells. H3K4me3 is an activating histone mark found near the transcriptional start sites of active genes. This histone mark has been shown to increase globally after metal exposure in humans and this global increase could be associated with activating genes that drive diseases such as cancer. H3K9me2 is a silencing mark that can be found in the promoter region of repressed genes. Methylated H3K9 prevents formation of acetylated H3K9, which promotes gene transcription [2]. A recent study by Teneng et al. found that two genes often repressed in lung cancer, MAL and OLIG2, are silenced due to enrichment of H3K9me2 [25].
Tungstate exposure caused increases in histone marks because it appeared to have induced the proteasomal degradation of histone demethylases. Exposure to proteasomal degradation inhibitor, MG132, suppressed the tungstate-induced loss of the histone demethylase protein induced by tungstate, indicating that the proteasome pathway was activated by tungstate and played a major role in this effect. These changes in histone modifications persisted even after tungstate was removed indicating that the effects were epigenetic. Tungstate is remarkably nontoxic to cells and this is not due to a lack of uptake (figure 1C). In addition, it does not induce a general degradation of cellular proteins as shown in figure 1B, but appears to specifically target the histone demethylases. We have studied other dioxygenases such as prolyl hydroxylase or Tet 2 protein, but did not find any similar degradation (data not shown). Currently, no mechanism has been identified to explain how tungstate targets the histone demethylases for degradation. However, the consequences of tungstate exposure were the increase in histone marks, which are normally removed by these enzymes, and an apparent compensatory increase in the mRNA coding for the lost histone demethylase enzymes.
There are very few studies conducted on tungstate, yet there is ample human exposure to warrant an understanding of what effects it may have in humans. Given its low level of toxicity and selective effects on the histone demethylase, it is possible that its ability to target degradation of histone demethylases by the proteasome may have future therapeutic value especially in the treatment of certain cancers. The mechanisms shown here clearly illustrate loss of protein via an MG132-sensitive proteasomal degradation and not metal inhibition of enzymatic activity. In fact, an oxyanion such as tungstate or chromate would not displace the Fe or Zn that are found on these enzymes but they may affect the oxidation state of Fe, which could lead to loss of enzyme activity if Fe is oxidized to the ferric state. An increase in oxidative stress can lead to inhibition of histone demethylases, since these enzymes are dioxygenases and require iron, ascorbate, and oxygen as co-factors. Chromate, in comparison, can inhibit these enzymes by oxidation of the ferrous to the ferric, Fe2+ to Fe3+ [17]. However ascorbate, which is required for the enzymatic reaction, is present to reduce the Fe back to the active ferric state. Although ascorbate is not present in high concentrations in tissue culture systems, it is present at high levels in the livers of mice, which were similarly affected when the animals were exposed to aqueous tungstate in their drinking water.
Tungstate, like chromate, which we have studied extensively, is hexavalent and is an oxyanion [14–16]. Due to the similarities in their valence, tungsten chemistry is likely to resemble chromate intracellularly and extracellularly. Humans are exposed to oxidized oxyanions of tungstate, which is similar to the oxidation state of chromate. Tungstate likely enters the cell via oxyanion transporters. Ions that resemble phosphate are able to cross into the cells via these transporters. Hexavalent chromium, Cr(VI), enters the cells via the phosphate ion transporter and is reduced inside the cell to chromium(III), Cr(III). Cr(VI) is a well-documented carcinogen and can form Cr(III) adducts with DNA to cause mutations [17]. Although not as well studied, a similar scenario would be expected with tungstate entering the cell like Cr(VI).
A possible mechanism by which metals change the epigenome is by increasing oxidative stress intracellularly. An increase of oxidative stress can lead to inhibition of histone demethylases, since these enzymes are dioxygenases and require iron, ascorbate, and oxygen as co-factors. Chromate, in comparison, can inhibit these enzymes by oxidation of the Fe2+ to Fe3+ [17]. While S-adenosyl methionine (SAM) acts as the universal methyl donor for all cellular methylation activities. SAM acquires its methyl group from methionine. Methionine is an essential amino acid that must be provided by the environment [30–32]. We examined if depletion of cellular SAM would prevent the increase in histone methylation observed after tungstate exposure. Tungstate-treated cells were grown in methionine-free media for four hours in order to deplete cellular SAM levels and JARID1A and JMJD1A protein levels were assessed. We found that even after SAM depletion, the tungstate-treated cells still displayed an increase in both H3K9me2 and H3K4me3, clearly indicating that there is not a gain in methylating capacity that is responsible for the observed increased in these histone marks.
Histones are methylated by histone methyltransferases (HMTs), which use SAM as a coenzyme. An initial theory was that the observed tungstate-induced increase in histone methylation levels may have been secondary to an increase in HMT activity. However, we determined otherwise, since SAM was not available to donate a methyl group to HMTs in methionine-deficient media and the histone marks were increased by tungstate (figure 4). Moreover, there was a decrease in the histone demethylase proteins (figure 5).
We aimed to investigate if animals exposed to drinking water with tungstate displayed any alteration to histone methylation in vivo. Since we found that cells treated with Na2WO4 exhibited increased levels of H3K9me2 and H3K4me3, our next aim was to investigate if tungstate exposure via drinking water affected histone methylation in the liver. Livers from mice exposed to tungstate in the drinking water exhibited an increase in the levels for H3K9me2 and H3K4me3. Future investigations should examine chronic low-dose exposure as well to simulate drinking water with mild contaminations that persist for long periods of time.
Based on the evidence provided in this study and our previous investigations [18], tungsten exposure should be a significant concern for human health despite its low cellular toxicity. Moreover, these findings should generate concern among regulatory agencies and industry to more seriously consider tungsten as a carcinogen. Human exposure arises from the natural occurrence of tungsten compounds in the environment and the use of tungsten in many occupational settings including machining, munitions manufacturing and hydraulic fracturing. Considering the low mutagenicity of metallic species, significant attention should be given to epigenetic mechanisms in order to more fully understand tungsten’s carcinogenic potential.
Summary.
Following acute tungstate exposure, lung cells exhibited increased global histone methylation and the loss of corresponding histone demethylase proteins. Mice acutely exposed to tungstate via their drinking water displayed similar increased global histone methylation patterns in their liver tissues.
Citations
- 1.Chervona Y, et al. Carcinogenic metals and the epigenome: understanding the effect of nickel, arsenic, and chromium. Metallomics. 2012;4(7):619–27. doi: 10.1039/c2mt20033c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brocato J, et al. 10th NTES Conference: Nickel and arsenic compounds alter the epigenome of peripheral blood mononuclear cells. J Trace Elem Med Biol. 2015;31:209–13. doi: 10.1016/j.jtemb.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Passantino L, et al. Sodium metavanadate exhibits carcinogenic tendencies in vitro in immortalized human bronchial epithelial cells. Metallomics. 2013;5(10):1357–67. doi: 10.1039/c3mt00149k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banda M, et al. Quantification of Kras mutant fraction in the lung DNA of mice exposed to aerosolized particulate vanadium pentoxide by inhalation. Mutat Res Genet Toxicol Environ Mutagen. 2015;789–790:53–60. doi: 10.1016/j.mrgentox.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 5.Black MB, et al. Using gene expression profiling to evaluate cellular responses in mouse lungs exposed to V2O5 and a group of other mouse lung tumorigens and non-tumorigens. Regul Toxicol Pharmacol. 2015;73(1):339–47. doi: 10.1016/j.yrtph.2015.07.017. [DOI] [PubMed] [Google Scholar]
- 6.Desoize B. Metals and metal compounds in carcinogenesis. In Vivo. 2003;17(6):529–39. [PubMed] [Google Scholar]
- 7.Lemus R, et al. An update to the toxicological profile for water-soluble and sparingly soluble tungsten substances. Crit Rev Toxicol. 2015;45(5):388–411. doi: 10.3109/10408444.2014.1003422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schrock RR. Metathesis by Molybdenum and Tungsten Catalysts. Chimia (Aarau) 2015;69(7–8):388–92. doi: 10.2533/chimia.2015.388. [DOI] [PubMed] [Google Scholar]
- 9.Wicaksana Y, et al. Tungsten trioxide as a visible light photocatalyst for volatile organic carbon removal. Molecules. 2014;19(11):17747–62. doi: 10.3390/molecules191117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou P, et al. Fabrication of Two-Dimensional Lateral Heterostructures of WS /WO H O Through Selective Oxidation of Monolayer WS. Angew Chem Int Ed Engl. 2015 doi: 10.1002/anie.201508216. [DOI] [PubMed] [Google Scholar]
- 11.Arita A, et al. Global levels of histone modifications in peripheral blood mononuclear cells of subjects with exposure to nickel. Environ Health Perspect. 2012;120(2):198–203. doi: 10.1289/ehp.1104140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, et al. Hypoxia and nickel inhibit histone demethylase JMJD1A and repress Spry2 expression in human bronchial epithelial BEAS-2B cells. Carcinogenesis. 2010;31(12):2136–44. doi: 10.1093/carcin/bgq197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Golebiowski F, et al. Inhibition of core histones acetylation by carcinogenic nickel(II) Mol Cell Biochem. 2005;279(1–2):133–9. doi: 10.1007/s11010-005-8285-1. [DOI] [PubMed] [Google Scholar]
- 14.Clancy HA, et al. Gene expression changes in human lung cells exposed to arsenic, chromium, nickel or vanadium indicate the first steps in cancer. Metallomics. 2012;4(8):784–93. doi: 10.1039/c2mt20074k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun H, et al. Comparison of gene expression profiles in chromate transformed BEAS-2B cells. PLoS One. 2011;6(3):e17982. doi: 10.1371/journal.pone.0017982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X, et al. Hypoxia induces trimethylated H3 lysine 4 by inhibition of JARID1A demethylase. Cancer Res. 2010;10:4214–4221. doi: 10.1158/0008-5472.CAN-09-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun H, et al. Oral Chromium Exposure and Toxicity. Curr Environ Health Rep. 2015;2(3):295–303. doi: 10.1007/s40572-015-0054-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laulicht F, et al. Tungsten-induced carcinogenesis in human bronchial epithelial cells. Toxicol Appl Pharmacol. 2015;288(1):33–9. doi: 10.1016/j.taap.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yao Y, et al. Toxicogenomic effect of nickel and beyond. Arch Toxicol. 2014;88(9):1645–50. doi: 10.1007/s00204-014-1313-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H, et al. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol Cell Biol. 2006;26(10):3728–37. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou X, et al. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008;29(9):1831–6. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoyama A, et al. Regulated histone methyltransferase and demethylase complexes in the control of genes by nuclear receptors. Cold Spring Harb Symp Quant Biol. 2011;76:165–173. doi: 10.1101/sqb.2011.76.010736. [DOI] [PubMed] [Google Scholar]
- 23.Animal Care and Use Committee. http://web.jhu.edu/animalcare/procedures/mouse.html.
- 24.Equivalent Surface Area Dosage Conversion Factors. 2015 https://ncifrederick.cancer.gov/lasp/acuc/frederick/Media/Documents/ACUC42.pdf.
- 25.Teneng I, et al. Global identification of genes targeted by DNMT3b for epigenetic silencing in lung cancer. Oncogene. 2015;34:621–640. doi: 10.1038/onc.2013.580. [DOI] [PubMed] [Google Scholar]
- 26.Loh YH, et al. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao C, et al. Cadmium induces histone H3 lysine methylation by inhibiting histone demethylase activity. Toxicol Sci. 2015;145:80–89. doi: 10.1093/toxsci/kfv019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itoh Y, et al. Identification of Jumonji AT-Rich Interactive Domain 1A Inhibitors and Their Effect on Cancer Cells. ACS Med Chem Lett. 2015;6:665–670. doi: 10.1021/acsmedchemlett.5b00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watanabe K, et al. Histone methylation-mediated silencing of miR-139 enhances invasion of non-small-cell lung cancer. Cancer Med. 2015;4:1573–1582. doi: 10.1002/cam4.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrer A, et al. Metabolism and epigenetics: a link cancer cells exploit. Curr Opin Biotechnol. 2015;34:23–9. doi: 10.1016/j.copbio.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jahan S, et al. Protein arginine methyltransferases (PRMTs): role in chromatin organization. Adv Biol Regul. 2015;57:173–84. doi: 10.1016/j.jbior.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 32.Nordgren KK, et al. The deep end of the metabolite pool: influences on epigenetic regulatory mechanisms in cancer. Eur J Clin Invest. 2015;45(Suppl 1):9–15. doi: 10.1111/eci.12361. [DOI] [PubMed] [Google Scholar]