Abstract
The copper-mediated coupling between alkynes to generate a structurally rigid, linear 1,3-diyne linkage has been known for over a century. However, the mechanistic requirement to simultaneously maintain Cu(I) and an oxidant has limited its practical utility, especially for complex functional molecules in aqueous solution. We find that addition of a specific bpy-diol ligand protects unprotected peptides from Cu(II) mediated oxidative damage through formation of an insoluble Cu(II) gel which solves the critical challenge of applying the Glaser coupling to substrates that are degraded by Cu(II). The generality of this method is illustrated through conjugation of a series of polar and nonpolar labels onto a fully unprotected GLP-1R agonist through a linear 7 Å diynyl linker.
Keywords: aqueous bioconjugation, Glaser, unprotected peptide, copper acetylide, chemoselective
Table of Contents
Diyne to Meet: A specific water-soluble bis-diol bipyridine (bpy-diol) ligand is shown to be optimal in the Cu(I)-catalyzed cross-Glaser coupling of unprotected peptides and functional molecules in aqueous media. This bifunctional ligand acts to both stabilize obligate Cu(I/II) species along the productive path to the desired 1,3-diyne linkage and sequester off-cycle, peptide-degrading Cu(II) species as an insoluble gel. The resulting linear diyne linker is well suited to the task of separating molecular tags from the appended biological macromolecule.
The development of chemoselective reactions that generate novel skeletal linkages between unprotected biomolecules lies at the center of chemical biology.[1–3] Synthetic bioconjugates often contain diverse functionality that allow them to be used as biomarker-specific imaging agents, tagged or immobilized constructs for a variety of affinity assays, as well as selective drug delivery systems.[3–7] However, the demands of reaction rate and chemoselectivity often require compromises on the covalent nature of the linkage which is often large and/or hydrophobic. As a result, conjugation chemistries that yield a structurally desirable linkage while maintaining chemoselectivity have enhanced utility.
A seminal example of such a reaction is the copper-catalyzed azide-alkyne cycloaddition (CuAAC), developed independently by Sharpless[8] and Meldal[9] in 2002. While this reaction tolerates a variety of pendant functionality and proceeds quickly at room temperature to join molecules of interest through a compact 1,4-disubstituted triazole linkage, significant amounts of oxidative coupling products between both acetylenic starting materials and in-situ-generated Cu-triazolides are commonly observed.[10] For example, bistriazoles can be obtained in high yield by performing the CuAAC under oxidative conditions that favor the formation of basic Cu(II)-species.[11] Taking inspiration from these reports as well as our own experience obtaining off-cycle coupling products (alkyne homodimers) during application of CuAAC, and in light of the commercial ubiquity of functionalized acetylenes, we endeavored to optimize the oxidative coupling of terminal alkynes and render it useful in the context of bioconjugation. The formation of a linear, aliphatic 1,3-diyne linkage represents a valuable complement to existing techniques used to label and constrain macromolecules.[12–19]
The Glaser coupling is the oldest known acetylenic coupling, first reported by Carl Glaser in 1869.[20,21] The method describes the head-to-head homocoupling of terminal acetylenes to forge symmetrical 1,3-diynes under Cu(I)-catalysis. Despite a number of useful modifications having been developed in the 150 years since its inception,[22–25] the Glaser coupling remains a tenuous reaction with no broadly applicable reaction conditions. The lack of robust protocols has limited the incorporation of the 1,3-diyne motif in complex molecules as the commonly employed “witches-brew” of Cu(I/II/III) salts, strong amine bases, oxidants, high temperatures, and arbitrary additives results in degradation of starting material. Consequently, the 1,3-diyne linkage is somewhat obscure and underexplored as a linker in bioconjugates – an unfortunate fact given its unique rigidity, polarity, and potential as a functionalizable handle. Recent attempts to incorporate it into biological macromolecules have had limited success.
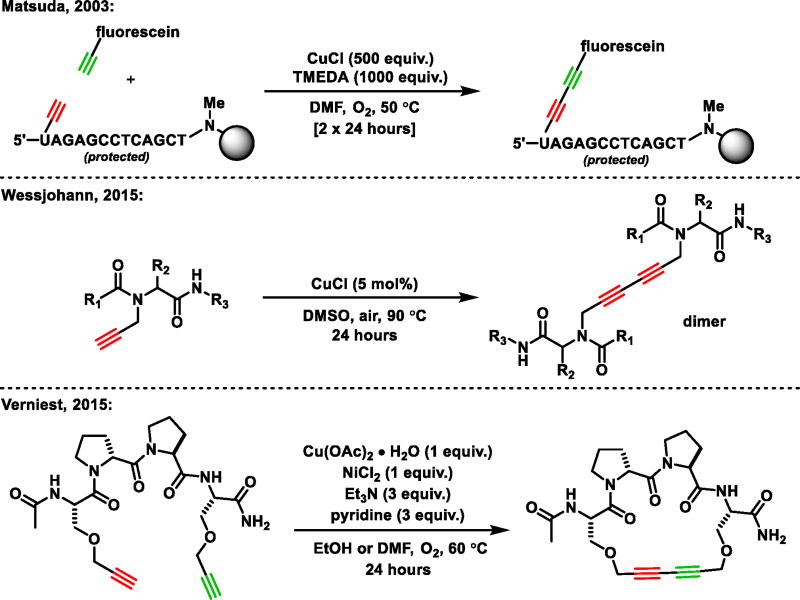
The first example of a cross-Glaser on biologically relevant molecules was reported by Matsuda and coworkers in 2003 and showcased the ability of complexed CuCl to attach functional probes to protected oligodeoxynucleotides (ODNs), Figure 1.[26] This method requires long reaction times (48 hours), an oxygen atmosphere, and protected ODNs on a CPG support to achieve satisfactory yields of the cross-Glaser product. In addition, while homocoupling of a nucleoside could be realized off resin, the rate in aqueous solution remained sluggish. Similar reaction conditions were applied to peptoid homodimerization, the utility of which was limited by harsh reaction conditions (90 °C in DMSO for 24 hours).[27] Intramolecular macrocyclization of peptides was achieved by using a dual Cu(OAc)2 and NiCl2 catalytic system.[28] This approach suffers limitations, requiring N-terminal acetylation, protected residues, as well as the incorporation of a heterochiral D-Pro-L-Pro β-turn motif in the backbone to correctly preorganize the alkynes. The reaction has been applied to the highly stable β-barrel protein GFP,[29,30] facilitating AlexaFluor conjugation.[31] However, increases in aqueous O2 content and long reaction times lead to significant protein degradation – likely a consequence of reactive Cu(II) species and uncontrolled basic pH. These studies highlight the practical challenges associated with applying classic synthetic reactions to biological macromolecules.[34]
Figure 1.
Prior work employing the Glaser coupling on biologically relevant molecules.
Herein, we report the development of reliable and relatively benign conditions for the cross-Glaser coupling between unprotected alkynylated peptides and small molecule alkynes. Taking previously employed conditions (CuCl and a multidentate ligand; Figure 1) as a starting point, we focused our optimization on the conjugation of propargyl benzamide to a model peptide, H-Leu-Tyr-Arg-Ala-Pra-Gly-NH2 (Pra = propargyl glycine). After screening a variety of Cu(I) sources, ligands, aqueous buffers, temperatures, and pHs we developed conditions that delivered heterocoupled product in 60% yield after 24 hours at 40 °C without requiring adjustment of reaction acidity, which remained at a constant pH of 8.8, Figure 2. This reaction is highly pH dependent; increasing the pH from 9.5 to 11 results in a greater than 5-fold increase in reaction rate. At pH 11 the reaction is greater than 75% complete after only 30 minutes at 40 °C. While this pH dependence may indicate that base-mediated Cu-acetylide formation is rate-determining, the possibility that a higher pH simply favors a copper species that is more reactive cannot be ruled out.
Figure 2.
A. Cross-Glaser coupling of propargyl benzamide and H-Leu-Tyr-Arg-Ala-Pra-Gly-NH2. B. pH-dependent rate for the conversion of 1a to 1b.
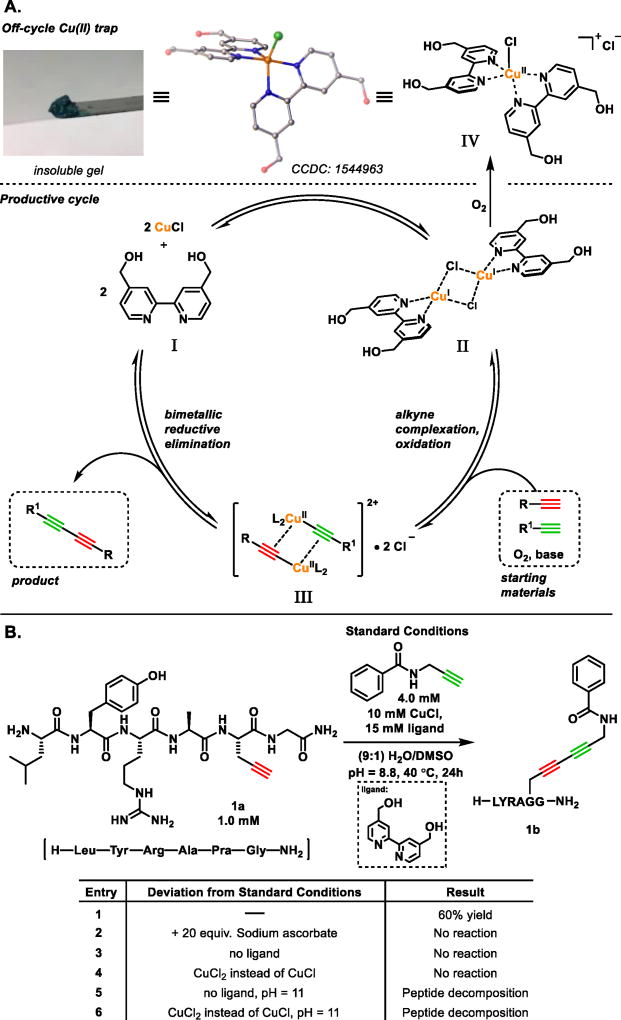
Ligand choice proved absolutely crucial in maintaining solubilized and reactive Cu(I) over the course of the reaction. Specifically, 4,4’-bis(hydroxymethyl)-2,2’-bipyridine (bpy-diol) was found to be unique in its capacity to maintain catalytically competent Cu(I) by inhibiting the formation of inert Cu(I)-acetylide polynucleates known to act as thermodynamic copper sinks in the CuAAC,[35,36] and preventing peptide-mediated copper complexation.[37–40] For applications in biomolecule conjugation, this ligand has the additional advantage of sequestering off-cycle Cu(II) species as an insoluble, hygroscopic blue–green gel, Figure 3A. In contrast, no product is observed when using bipyridine and only minimal product when water-soluble bidentate tetramethylethylenediamine (TMEDA) is employed. Polydentate tris(triazole)methylamines such as THPTA are routinely employed for solubilizing and stabilizing copper in aqueous CuAAC systems.[41–45] However, in the context of cross-Glaser couplings, this ligand displayed similarly low activity, never producing greater than 5% product.
Figure 3.
A. Proposed mechanism of the cross-Glaser and off-cycle, insoluble Cu(II)-complex formation. Chloride counterion, water, and hydrogen atoms omitted for clarity. B. Effect of sodium ascorbate and CuCl2 on model reaction between H-Leu-Tyr-Arg-Ala-Pra-Gly-NH2 and propargyl benzamide.
The proposed catalytic cycle, Figure 3A, begins with the complexation of two equivalents of CuCl by two equivalents of bpy-diol I to yield dimeric Cu(I) intermediate II – a complex previously substantiated by EXAFS spectroscopy in related Glaser homocouplings.[46] This species may then proceed down one of two paths. Along the productive route, dimer II engages two different alkynes and undergoes O2-mediated oxidation to yield III, the existence of which has been verified in analogous systems.[47–51] Indeed, preventing the formation of a high-oxidation intermediate such as III by adding an excess of sodium ascorbate shuts down the reaction completely, Figure 3B; entry 2. Bimetallic reduction of III produces the desired 1,3-diyne and regenerates CuCl/bpy-diol I, thereby completing the catalytic cycle.
Glaser couplings are complicated by the requirement for Cu(I) and O2 to co-exist throughout the course of the reaction. As a result, direct oxidation of II by O2 can lead to basic, off-cycle Cu(II) complexes that are known to cause protein degradation.[31,32,33]
However, a unique property of bpy diol I is that the cupric ion is sequestered as an insoluble pentacoordinate Cu(II) gel, IV. Crystallization of this gel from hexafluoroisopropanol (HFIP) revealed that IV formed a 3:2 co-crystal with bpy diol (see Supplemental Information). The rapid degradation of biological macromolecules mediated by Cu(II) has been well established.[32–33] Indeed, we find that employing CuCl or CuCl2 under the required basic, oxidative conditions necessary to perform this reaction (see Figure 4) leads to significant degradation of more complex peptide starting materials and no recoverable product, Figure 3B; entries 5,6. Critically, the funneling of unproductive Cu(II) species into an insoluble reservoir bpy-diol acts to protect peptide material from degradation.
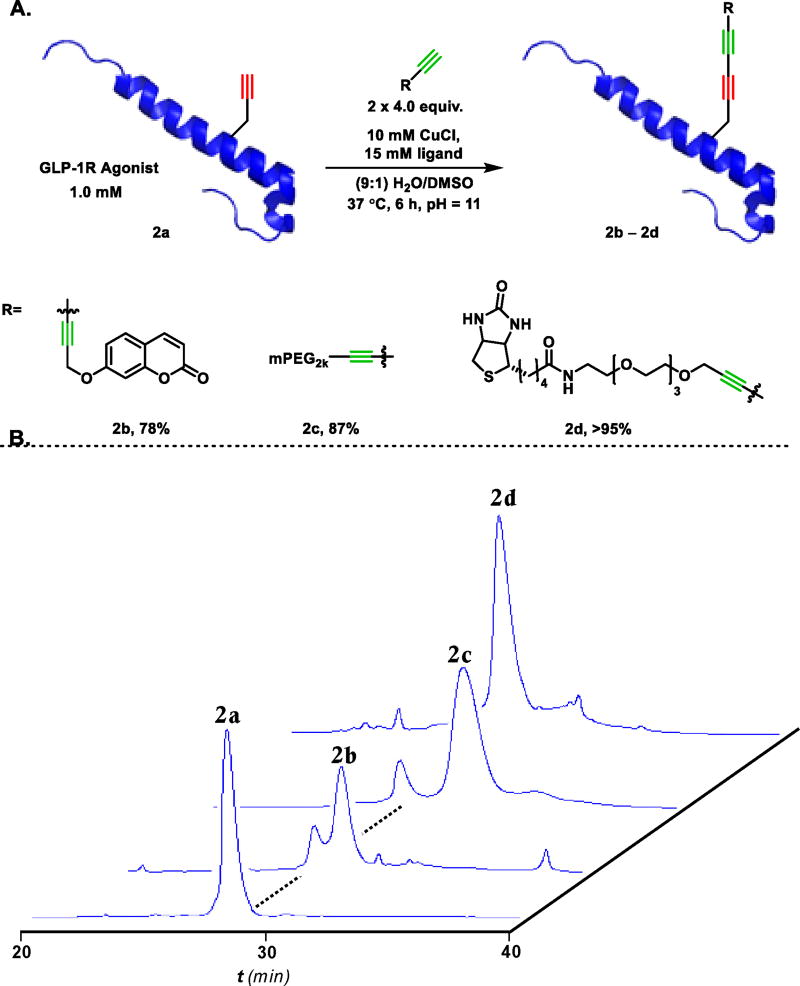
Figure 4.
A. Cross-Glaser coupling of GLP-1R agonist 2a and alkynylated tags (2b–2d). As no structure of 2a exists, the structure of the closely related peptide Ex-4 (PDB accession code: 1JRJ)[50] is used for illustration. Only a single addition of 4 equivalents of tag was used for the 6 hour reaction to form 2b. Two additions of tag at t=0 and t=3 hours were used for the synthesis of 2c and 2d. B. HPLC profile of pure 2a and crude reaction traces forming bioconjugates 2b–2d.
With optimized conditions for the functionalization of our model peptide in hand we focused on expanding our methodology to a larger, more complicated peptide. Exendin-4 (Ex-4) is a 39 amino acid peptide agonist of the Glucagon-like peptide 1 (GLP-1) receptor, whose binding stimulates insulin secretion to control blood sugar concentrations.[52,53] In contrast, the peptide Ex9–39 (Ex-4 lacking the first 8 amino acids) behaves as a competitive antagonist of the receptor.[54] From this unique structure-function relationship, a full-length biased agonist capable of affecting hypoglycemia in mice via selective signaling at the GLP-1R was discovered from a combinatorial peptide library.[55] This peptide agonist, of sequence: H-ELVDNAVGGDLSKQMEEEAVRLFIEWLKNGGPSSGAPPPS-NH2, was selected as a suitable target for modification with a variety of functional tags. The requisite propargyl glycine residue (Pra21) was substituted for Arg21, a site known to be amenable to half-life improving modifications in the related GLP-1 hormones liraglutide and semaglutide.[56] We selected three alkynylated tag molecules spanning a range of solubility, size, and functionality to showcase the versatility of cross-Glaser bioconjugation. Using propargyl umbelliferone, we identified reaction conditions that are uniquely suited to the modification of complex peptides. The addition of 4 equivalents of non-polar propargyl umbelliferone in DMSO to a 1.0 mM aqueous solution of protein, 2a, at 37 °C, and subsequent adjustment to pH 11, cleanly delivered 2b in 78% HPLC yield after only 6 hours, Figure 4B. For the conjugation of more soluble alkyne tags two additions of 4 equivalents (one at t = 0 and one at t = 3 hours) were necessary as some homocoupling of the small molecule was observed. This double addition technique was effective at minimizing tag dimerization while also maintaining an excess of tag relative to protein throughout the course of reaction. In this way, we efficiently PEGylated and biotinylated our GLP-1R agonist to generate bioconjugates 2c and 2d in 87% and >95% by HPLC integration. The use of elevated pH in the manipulation of unprotected peptides in aqueous buffers must be approached with caution due to the potential for asparagine residues proximal to glycine to be converted to aspartate residues via aspartimide formation. However, high resolution mass spectral data obtained for compound 2d confirmed that despite these basic conditions, Asn29 did not undergo deamination via base-mediated cyclization with Gly30. This stability could result from conformational effects propagating from the proline-rich C-terminus.[57–60]
In summary, we have refined the historic cross-Glaser coupling into a robust bioconjugation method that combines the efficiency and chemoselectivity of the CuAAC and delivers a structurally privileged, linear 1,3-diyne linkage between biological macromolecules and functional probes. In addition, the diynyl linkage has the potential to be selectively elaborated following conjugation. The method benefits from a short reaction time at ambient temperature with air as the sole oxidant. The high chemoselectivity facilitates its application to fully deprotected peptides at low concentrations in aqueous solution. The commercial availability of CuCl catalyst, bis-diol bypyridine ligand, numerous functionalized acetylenes, and the well established routes to alkynylated proteins via synthetic and recombinant methods, are expected to facilitate the use of this method in the synthesis of complex bioconjugates.
Supplementary Material
Acknowledgments
This work was funded by the National Institutes of Health (NIH R01 AI13867). We are grateful to A. L. Rheingold, C. E. Moore, and M. Gembicky (University of San Diego) for X-ray crystallographic analysis.
References
- 1.Kalia J, Raines RT. Curr. Org. Chem. 2010;14:138–147. doi: 10.2174/138527210790069839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hackenberger CPR, Schwarzer D. Angew. Chem. Int. Ed. 2008;47:10030–10074. doi: 10.1002/anie.200801313. [DOI] [PubMed] [Google Scholar]
- 3.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 4.Algar WR, Prasuhn DE, Stewart MH, Jennings TL, Blanco-Canosa JB, Dawson PE, Medintz IL. Bioconjug. Chem. 2011;22:825–858. doi: 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- 5.Ulbrich K, Holá K, Šubr V, Bakandritsos A, Tuček J, Zbořil R. Chem. Rev. 2016;116:5338–5431. doi: 10.1021/acs.chemrev.5b00589. [DOI] [PubMed] [Google Scholar]
- 6.Lu Z-R, Shiah J-G, Sakuma S, Kopečková P, Kopeček J. Proc. Tenth Int. Symp. Recent Adv. Drug Deliv. Syst. 2002;78:165–173. [Google Scholar]
- 7.Srivastava A, O’Connor IB, Pandit A, Gerard Wall J. Top. Issue Biorelated Polym. 2014;39:308–329. [Google Scholar]
- 8.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. Angew. Chem. Int. Ed. 2002;41:2596. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Tornøe CW, Christensen C, Meldal M. J. Org. Chem. 2002;67:3057–3064. doi: 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- 10.Hein JE, Fokin VV. Chem. Soc. Rev. 2010;39:1302–1315. doi: 10.1039/b904091a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angell Y, Burgess K. Angew. Chem. Int. Ed. 2007;46:3649–3651. doi: 10.1002/anie.200700399. [DOI] [PubMed] [Google Scholar]
- 12.Blackwell HE, Grubbs RH. Angew. Chem. Int. Ed. 1998;37:3281. doi: 10.1002/(SICI)1521-3773(19981217)37:23<3281::AID-ANIE3281>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y-W, Grossmann TN, Verdine GL. Nat Protoc. 2011;6:761–771. doi: 10.1038/nprot.2011.324. [DOI] [PubMed] [Google Scholar]
- 14.Lin YA, Chalker JM, Davis BG. Chem Bio Chem. 2009;10:959–969. doi: 10.1002/cbic.200900002. [DOI] [PubMed] [Google Scholar]
- 15.Lin YA, Chalker JM, Davis BG. J. Am. Chem. Soc. 2010;132:16805–16811. doi: 10.1021/ja104994d. [DOI] [PubMed] [Google Scholar]
- 16.Vinogradov AA, Choo Z-N, Totaro KA, Pentelute BL. Org. Lett. 2016;18:1226–1229. doi: 10.1021/acs.orglett.5b03626. [DOI] [PubMed] [Google Scholar]
- 17.Assem N, Ferreira DJ, Wolan DW, Dawson PE. Angew. Chem. Int. Ed. 2015;54:8665–8668. doi: 10.1002/anie.201502607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cromm PM, Schaubach S, Spiegel J, Fürstner A, Grossmann TN, Waldmann H. Nat. Commun. 2016;7:11300. doi: 10.1038/ncomms11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lautrette G, Touti F, Lee HG, Dai P, Pentelute BL. J. Am. Chem. Soc. 2016;138:8340–8343. doi: 10.1021/jacs.6b03757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glaser C. Berichte Dtsch. Chem. Ges. 1869;2:422–424. [Google Scholar]
- 21.Glaser C. Justus Liebigs Ann. Chem. 1870;154:137–171. [Google Scholar]
- 22.Eglinton G, Galbraith AR. J. Chem. Soc. Resumed. 1959:889–896. [Google Scholar]
- 23.Hay AS. J. Org. Chem. 1962;27:3320–3321. [Google Scholar]
- 24.Chodkiewicz W. Ann Chim Paris. 1957;2:819–869. [Google Scholar]
- 25.Cadiot P, Chodkiewicz W. Marcel Dekker; New York: 1969. pp. 597–647. [Google Scholar]
- 26.Minakawa N, Ono Y, Matsuda A. J. Am. Chem. Soc. 2003;125:11545–11552. doi: 10.1021/ja036055t. [DOI] [PubMed] [Google Scholar]
- 27.Brauer MCN, Neves Filho RAW, Westermann B, Heinke R, Wessjohann LA. Beilstein J. Org. Chem. 2015;11:25–30. doi: 10.3762/bjoc.11.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verlinden S, Geudens N, Martins JC, Tourwe D, Ballet S, Verniest G. Org. Biomol. Chem. 2015;13:9398–9404. doi: 10.1039/c5ob01153a. [DOI] [PubMed] [Google Scholar]
- 29.Yang F, Moss LG, Phillips GN. Nat Biotech. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 30.Ward WW. Green Fluorescent Protein: Properties, Applications and Protocols. John Wiley & Sons; 2005. pp. 48–83. [Google Scholar]
- 31.Lampkowski JS, Villa JK, Young TS, Young DD. Angew. Chem. Int. Ed. 2015;54:9343–9346. doi: 10.1002/anie.201502676. [DOI] [PubMed] [Google Scholar]
- 32.Kumar A, Li K, Cai C. Chem. Comm. 2011;47:3186–3188. doi: 10.1039/c0cc05376g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong V, Presolski SI, Ma C, Finn MG. Angew. Chem. Int. Ed. 2009;48:9879–9883. doi: 10.1002/anie.200905087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tiefenbrunn TK, Dawson PE. Pept. Sci. 2010;94:95–106. doi: 10.1002/bip.21337. [DOI] [PubMed] [Google Scholar]
- 35.Mykhalichko MB, Temkin ON, Mys’kiv MG. Russ. Chem. Rev. 2000;69:957–984. [Google Scholar]
- 36.Presolski SI, Hong V, Cho S-H, Finn MG. J. Am. Chem. Soc. 2010;132:14570–14576. doi: 10.1021/ja105743g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sigel H, Martin RB. Chem. Rev. 1982;82:385–426. [Google Scholar]
- 38.Kozłowski H, Kowalik-Jankowska T, Jeżowska-Bojczuk M. Coord. Chem. Poland J Ziolkowski. 2005;249:2323–2334. [Google Scholar]
- 39.Fragoso A, Delgado R, Iranzo O. Dalton Trans. 2013;42:6182–6192. doi: 10.1039/c3dt32384f. [DOI] [PubMed] [Google Scholar]
- 40.Park GY, Lee JY, Himes RA, Thomas GS, Blackburn NJ, Karlin KD. J. Am. Chem. Soc. 2014;136:12532–12535. doi: 10.1021/ja505098v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Org. Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 42.Hong V, Steinmetz NF, Manchester M, Finn MG. Bioconjug. Chem. 2010;21:1912–1916. doi: 10.1021/bc100272z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soriano del Amo D, Wang W, Jiang H, Besanceney C, Yan AC, Levy M, Liu Y, Marlow FL, Wu P. J. Am. Chem. Soc. 2010;132:16893–16899. doi: 10.1021/ja106553e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Besanceney-Webler C, Jiang H, Zheng T, Feng L, Soriano del Amo D, Wang W, Klivansky LM, Marlow FL, Liu Y, Wu P. Angew. Chem. Int. Ed. 2011;50:8051–8056. doi: 10.1002/anie.201101817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKay CS, Finn MG. Chem. Biol. 2014;21:1075–1101. doi: 10.1016/j.chembiol.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang G, Yi H, Zhang G, Deng Y, Bai R, Zhang H, Miller JT, Kropf AJ, Bunel EE, Lei A. J. Am. Chem. Soc. 2014;136:924–926. doi: 10.1021/ja410756b. [DOI] [PubMed] [Google Scholar]
- 47.Bohlmann F, Schönowsky H, Inhoffen E, Grau G. Chem. Ber. 1964;97:794–800. [Google Scholar]
- 48.Siemsen P, Livingston RC, Diederich F. Angew. Chem. Int. Ed. 2000;39:2632–2657. [PubMed] [Google Scholar]
- 49.Wendlandt AE, Suess AM, Stahl SS. Angew. Chem. Int. Ed. 2011;50:11062–11087. doi: 10.1002/anie.201103945. [DOI] [PubMed] [Google Scholar]
- 50.Bai R, Zhang G, Yi H, Huang Z, Qi X, Liu C, Miller JT, Kropf AJ, Bunel EE, Lan Y, et al. J. Am. Chem. Soc. 2014;136:16760–16763. doi: 10.1021/ja5097489. [DOI] [PubMed] [Google Scholar]
- 51.Su L, Dong J, Liu L, Sun M, Qiu R, Zhou Y, Yin S-F. J. Am. Chem. Soc. 2016;138:12348–12351. doi: 10.1021/jacs.6b07984. [DOI] [PubMed] [Google Scholar]
- 52.Neidigh JW, Fesinmeyer RM, Prickett KS, Andersen NH. Biochemistry (Mosc.) 2001;40:13188–13200. doi: 10.1021/bi010902s. [DOI] [PubMed] [Google Scholar]
- 53.Willard FS, Sloop KW. Exp. Diabetes Res. 2012;2012:470851. doi: 10.1155/2012/470851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montrose-Rafizadeh C, Yang H, Rodgers BD, Beday A, Pritchette LA, Eng J. J. Biol. Chem. 1997;272:21201–21206. doi: 10.1074/jbc.272.34.21201. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H, Sturchler E, Zhu J, Nieto A, Cistrone PA, Xie J, He L, Yea K, Jones T, Turn R, et al. Nat. Commun. 2015;6:8918. doi: 10.1038/ncomms9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lau J, Bloch P, Schäffer L, Pettersson I, Spetzler J, Kofoed J, Madsen K, Knudsen LB, McGuire J, Steensgaard DB, et al. J. Med. Chem. 2015;58:7370–7380. doi: 10.1021/acs.jmedchem.5b00726. [DOI] [PubMed] [Google Scholar]
- 57.Coin I, Beyermann M, Bienert M. Nat Protoc. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]
- 58.Yee CS, Seyedsayamdost MR, Chang MCY, Nocera DG, Stubbe J. Biochemistry (Mosc.) 2003;42:14541–14552. doi: 10.1021/bi0352365. [DOI] [PubMed] [Google Scholar]
- 59.Wang P, Aussedat B, Vohra Y, Danishefsky SJ. Angew. Chem. Int. Ed Engl. 2012;51:11571–11575. doi: 10.1002/anie.201205038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Abdel-Aal A-BM, Papageorgiou G, Raz R, Quibell M, Burlina F, Offer J. J. Pept. Sci. 2016;22:360–367. doi: 10.1002/psc.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.