Abstract
Clear cell carcinoma of the endometrium is a rare type of endometrial cancer generally associated with an aggressive clinical behavior. Here we sought to define the repertoire of somatic genetic alterations in endometrial clear cell carcinomas (ECCs) and whether ECCs could be classified into the molecular subtypes described for endometrial endometrioid and serous carcinomas. We performed a rigorous histopathological review, immunohistochemical analysis and massively parallel sequencing targeting 300 cancer-related genes of 32 pure ECCs. Eleven (34%), seven (22%) and six (19%) ECCs displayed abnormal expression patterns of p53, ARID1A and at least one DNA mismatch repair protein, respectively. Targeted sequencing data were obtained from 30 of the 32 ECCs included in this study, which revealed that two ECCs (7%) were ultramutated and harbored mutations affecting the exonuclease domain of POLE. In POLE wild-type ECCs, TP53 (46%), PIK3CA (36%), PPP2R1A (36%), FBXW7 (25%), ARID1A (21%), PIK3R1 (18%) and SPOP (18%) were the genes most commonly affected by mutations, and 18% and 11% harbored CCNE1 and ERBB2 amplifications, respectively, while 11% showed DAXX homozygous deletions. In comparison to non-POLE endometrioid carcinomas from The Cancer Genome Atlas (TCGA), ECCs less frequently harbored mutations affecting CTNNB1 and PTEN but more frequently PPP2R1A and TP53 mutations. Compared to endometrial serous carcinomas (TCGA), ECCs less frequently harbored TP53 mutations. Using a surrogate model for the molecular-based TCGA classification, all molecular subtypes previously identified in endometrial endometrioid and serous carcinomas were present in the ECCs studied, including POLE, MMR-deficient, copy-number high (serous-like)/p53 abnormal and copy-number low (endometrioid)/p53 wild-type, which were significantly associated with disease-free survival in univariate analysis. These findings demonstrate that ECCs are a histologically and genetically heterogeneous group of tumors with varying outcomes. Furthermore, our data suggest that the classification of ECCs as being generally “high-grade” or “type II” tumors may not be warranted.
Keywords: Endometrial clear cell carcinoma, massively parallel sequencing, somatic mutations, copy number alterations, molecular classification
INTRODUCTION
Endometrial clear cell carcinoma (ECC) is rare type of endometrial cancer accounting for <3% of all endometrial cancers [1–3]. ECCs are generally morphologically similar to their ovarian counterparts but it should be noted that diagnosis of these lesions can be challenging and significant inter-observer diagnostic variability has been reported [3–6].
Endometrial carcinoma has been traditionally classified into two groups based on clinical, endocrine, and epidemiological observations, the so-called type I and type II cancers [7,8]. Type I cancers are typically endometrioid carcinomas that tend to show a good prognosis, while type II cancers are generally associated with a poor prognosis and include serous and clear cell histologies [7–10]. There is burgeoning evidence to demonstrate, however, that endometrial cancer is a biologically, clinically and genetically heterogeneous disease and that this dualistic classification may not reflect the actual heterogeneity observed [8]. A study by The Cancer Genome Atlas (TCGA) combining somatic mutations, copy number alterations and microsatellite instability (MSI) data classified endometrial endometrioid and serous carcinomas into four molecular subtypes [11], including i) the POLE (ultramutated) tumors being characterized by extremely high mutation rates and mutations in the exonuclease domain of POLE, ii) the MSI (hypermutated) tumors showing very high mutation rates and few copy number alterations, iii) the copy-number low (endometrioid) tumors being microsatellite stable (MSS), harboring lower mutation frequencies and recurrent CTNNB1 mutations, and iv) the copy-number high (serous-like) group comprising all serous carcinomas and a subset of the grade 3 endometrioid carcinomas, and being characterized by high levels of copy number alterations, low mutation frequencies and recurrent TP53, PPP2R1A and FBXW7 somatic mutations [11].
At variance with endometrial endometrioid and serous carcinomas, there is a paucity of data on the genomic landscape of ECCs, partly due to the rarity of this tumor type. Candidate gene analyses of small series of ECCs have reported mutations affecting PIK3CA, ARID1A, PPP2R1A, TP53, PIK3R1, PTEN and KRAS [4,12,13], and molecular similarities to both serous and endometrioid endometrial cancers have been found [14] but little is known about copy number changes and molecular subtypes of these lesions and whether the genetic alterations correlate with outcome.
To address this gap in our understanding of the genetics of ECCs, we have subjected a series of centrally reviewed ECCs to immunohistochemical and massively parallel sequencing analysis to investigate i) whether ECCs harbor mutations affecting 300 key cancer genes, ii) whether ECCs display a repertoire of somatic mutations that is distinct from endometrial endometrioid and serous carcinomas, and iii) whether ECCs could be classified into the molecular subtypes described for endometrial endometrioid and serous carcinomas.
MATERIAL AND METHODS
Case selection
We selected from the files of the Department of Pathology at Memorial Sloan Kettering Cancer Center (MSKCC), all ECCs (n=45) diagnosed between 1996 and 2013 that had both slides and blocks available. Samples were anonymized prior to analysis. This study was approved by the Institutional Review Board (IRB) of MSKCC, and patient consent was obtained where appropriate. Representative hematoxylin and eosin (H&E)-stained sections of each case diagnosed as ECC were independently reviewed by two specialized gynecologic pathologists with expertise in clear cell morphology (DFD, RAS) [4,5]. For inclusion in the study, the tumors had to show typical clear cell morphology, as defined by the World Health Organization (WHO) [2] and Fadare et al [3]. More specifically, carcinomas with abundant nuclear stratification, diffuse severe pleomorphism, and abundant columnar nuclear shape were excluded. Immunohistochemistry was not used in the classification of these tumors, following Fadare et al [3].
Specific morphologic features were evaluated, including the presence of classic architectural patterns, lymphocytic infiltration, mitotic index (number of mitoses per 10 high powered fields (HPFs)) and nuclear grade using a 3-tiered scale based on nuclear pleomorphism [3] (Supplementary materials and methods). Following this review, we included 32 ECCs. Of the 45 cases reviewed, 13 (29%) were excluded, which were cases where both reviewers diagnosed a tumor other than clear cell carcinoma, cases lacking diagnostic consensus between both pathologists, and/or mixed epithelial carcinoma with a clear cell component. Clinical information, including age, stage, location of metastases at presentation, and follow-up was retrieved from the medical records.
Immunohistochemistry (IHC)
IHC for p53, ARID1A/BAF250a and the DNA mismatch repair (MMR) proteins MSH2, MSH6, MLH1, PMS2 was performed on all cases, as described previously [15–17]; see Supplementary materials and methods.
DNA extraction
Formalin-fixed paraffin-embedded (FFPE) tumor and normal sections were reviewed by a pathologist (DFD). Five-µm sections from tumor samples were manually macro-dissected to ensure >20% neoplastic cells. Normal tissue sections, usually from a benign lymph node, were confirmed to be devoid of any neoplastic cells. Genomic DNA from tumor- and patient-matched normal samples was extracted using the DNeasy Blood & Tissue kit (Qiagen).
Targeted capture massively parallel sequencing
Tumor and normal DNA samples were subjected to targeted massively parallel sequencing using the Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) assay, targeting all exons of 300 key cancer genes (supplementary material, Table S1), as described previously [18,19]. Sequence reads were aligned to the human reference genome GRCh37 using the Burrows-Wheeler Aligner (BWA, v0.7.10) [20], and local realignment, duplicate removal and base quality recalibration were performed using the Genome Analysis Toolkit (GATK, v3.1.1) [21]. Variant calling and copy number analysis was performed as described previously [19,22–27] (Supplementary material and methods). Mutational hotspots were assigned according to Chang et al [28]. Cancer cell fractions of all mutations were inferred using ABSOLUTE (v1.0.6) [29], as described previously [19,22] and the potential functional effect of each mutation was investigated as described previously [19,29–37], see also Supplementary material and methods.
Comparisons of ECCs with endometrial carcinomas from the TCGA dataset
For comparisons of mutational frequencies of ECCs with those of endometrial endometrioid and serous carcinomas, the clinicopathologic data and whole-exome sequencing-derived mutational data from the TCGA data portal (https://tcga-data.nci.nih.gov/docs/publications/ucec_2013/; files “Key Clinical Data”, “UCEC Somatic Mutations”, “Cumulative Data Freeze List”) were retrieved as described previously [19]. We restricted the comparison to the 300 genes targeted by our sequencing panel. All comparisons were performed using Fisher’s exact tests, corrected for multiple comparisons using the Benjamini-Hochberg method.
Molecular classification
To classify the ECCs into the molecular subtypes described for endometrial endometrioid and serous carcinomas by TCGA, we employed a surrogate model described by Talhouk et al. [38] (Supplementary material and methods). As a second approach, hierarchical clustering was performed using Ward’s algorithm and Euclidean distance, using the ECCs and all endometrial endometrioid and serous carcinomas in the TCGA dataset [11] filtered for the 300 genes targeted by our sequencing panel. The stability of the hierarchical cluster analysis was assessed using pvclust [39].
Statistical analysis
The association between molecular subtype and disease-free survival was analyzed, and survival curves were calculated using the Kaplan–Meier method with the log-rank test. Associations between specific clinicopathologic features and molecular subtypes were tested using Fisher’s exact and t-tests. P-values of <0.05 were considered statistically significant.
RESULTS
ECCs are phenotypically heterogeneous
After central review of 45 cases initially classified as ECCs, a final diagnosis of pure ECC was rendered in 32 cases, which were included in this study (see Materials and Methods). The median age of the patients was 65 years (range 33 to 83 years). At presentation, 50% (16/32) of patients were of FIGO stage I, 6% (2/32) stage III, and 44% (14/32) stage IV (Table 1). The median follow-up was 29 months (range 5 to 105 months), and at the end of the follow-up period, 47% (15/32) of patients had died of disease, 41% (13/32) showed no evidence of disease, 9% (3/32) were alive with disease and 3% (1/32) had died of another cause.
Table 1.
Clinicopathologic information of the endometrial clear cell carcinomas included in this study.
| Case ID |
Age at diagnosis (years) |
FIGO stage at presentation |
Architectural pattern (%) | Nuclear grade |
Mitotic index** |
p53 IHC |
ARID1A IHC |
DNA MMR IHC | Follow-up (months, status) |
||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Papillary | Tubulocystic | Solid | |||||||||
| CC01 | 73 | IB | 50 | 50 | 2 (focal 3) | 18 | Null | Retained | Retained | 31, NED*** | |
| CC02 | 74 | IV | 40 | 30 | 30 | 2 (focal 3) | 8 | Positive | Retained | Retained | 23, DOD |
| CC03* | 62 | IV | 40 | 40 | 20 | 2 (focal 3) | 5 | Wild-type | Retained | Retained | 30, DOD |
| CC04 | 51 | IA | 80 | 20 | 2 | 4 | Wild-type | Loss | Retained | 16, DOD | |
| CC05 | 59 | IA | 90 | 10 | 2 | 5 | Wild-type | Loss | MLH1/PMS2 loss | 65, NED | |
| CC06 | 83 | IA | 50 | 50 | 2 | 8 | Wild-type | Loss | Retained | 60, DOD | |
| CC07 | 62 | IV | 40 | 60 | 2 (focal 3) | 16 | Positive | Retained | Retained | 5, DOD | |
| CC08 | 68 | IA | 70 | 30 | 2 | 5 | Wild-type | Retained | Retained | 69, NED | |
| CC10* | 54 | IA | 100 | 2 (focal 3) | 1 | Wild-type | Retained | MSH6 loss | 50, NED | ||
| CC11 | 65 | IA | 100 | 2 | 1 | Wild-type | Loss | MSH6 loss | 83, NED | ||
| CC12 | 61 | IV | 30 | 70 | 2 (focal 3) | 4 | Wild-type | Retained | Retained | 26, DOD | |
| CC13 | 65 | IV | 80 | 20 | 2 (focal 3) | 17 | Positive | Retained | Retained | 8, DOD | |
| CC14 | 70 | IV | 40 | 60 | 2 | 3 | Wild-type | Retained | Retained | 6, DOD | |
| CC15 | 78 | IV | 50 | 50 | 2 | 3 | Positive | Retained | Retained | 53, DOD | |
| CC16 | 63 | IV | 50 | 50 | 2 (focal 3) | 8 | Null | Retained | Retained | 6, DOD | |
| CC17 | 56 | IIIC | 60 | 40 | 2 | 6 | Positive | Retained | Retained | 55, AWD | |
| CC18 | 72 | IA | 50 | 50 | 2 (focal 3) | 2 | Wild-type | Partial loss | Retained | 93, NED | |
| CC19 | 71 | IA | 50 | 50 | 2 | 2 | Wild-type | Retained | Retained | 6, DOC | |
| CC20 | 63 | IIIC | 50 | 50 | 2 | 12 | Wild-type | Loss | MLH1/PMS2/MSH6 loss | 36, NED | |
| CC21 | 66 | IA | 50 | 50 | 2 | 8 | Wild-type | Retained | Retained | 105, AWD | |
| CC22 | 62 | IA | 30 | 70 | 2 (focal 3) | 8 | Wild-type | Retained | Retained | 13, DOD | |
| CC23 | 58 | IV | 30 | 70 | 2 (focal 3) | 2 | Positive | Retained | Retained | 39, AWD | |
| CC24 | 71 | IA | 40 | 40 | 20 | 2 (focal 3) | 4 | Positive | Retained | Retained | 14, NED |
| CC25 | 65 | IA | 70 | 30 | 2 (focal 3) | 9 | Wild-type | Retained | Retained | 21, NED | |
| CC26 | 65 | IA | 20 | 20 | 60 | 2 | 2 | Wild-type | Retained | MSH6 equivocal | 20, NED |
| CC27 | 64 | IA | 50 | 50 | 2 (focal 3) | 8 | Wild-type | Retained | Retained | 61, NED | |
| CC28 | 64 | IV | 100 | 2 (focal 3) | 5 | Positive | Retained | Retained | 8, DOD | ||
| CC29 | 67 | IV | 25 | 25 | 50 | 2 | 2 | Wild-type | Loss | Retained | 65, DOD |
| CC31 | 55 | IA | 10 | 90 | 2 | 9 | Wild-type | Retained | MSH6 loss | 66, NED | |
| CC33 | 33 | IV | 20 | 80 | 2 | 2 | Null | Retained | Retained | 10, DOD | |
| CC34 | 70 | IV | 40 | 30 | 30 | 2 (focal 3) | 3 | Wild-type | Retained | Retained | 27, NED |
| CC35 | 81 | IV | 40 | 30 | 30 | 2 | 8 | Wild-type | Retained | Retained | 14, DOD |
Sequencing data not available;
Mitotic index defined as number of mitoses per 10 high-power fields;
Lost to follow-up
AWD, alive with disease; DOC, died of other cause; DOD, died of disease; IHC, Immunohistochemistry; MMR, mismatch repair; NED, no evidence of disease; Null, null phenotype.
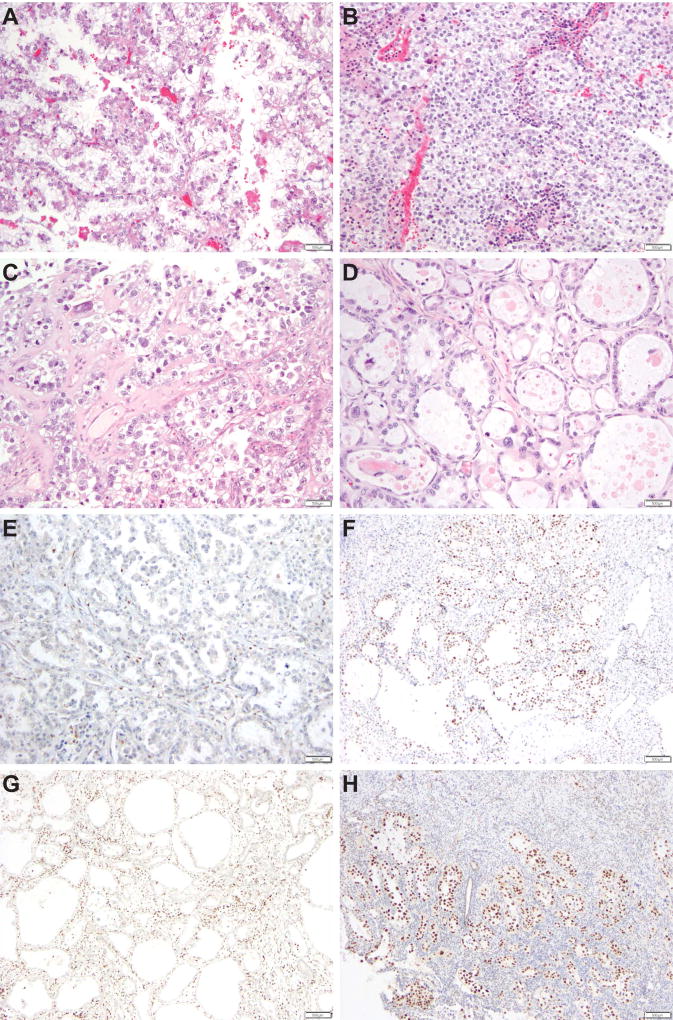
The ECCs included in this study showed varying combinations of the typical morphologic patterns previously described in Mullerian clear cell carcinomas (Table 1, Figure 1), with papillary and tubulocystic combinations (19/32, 60%), and papillary, tubulocystic and solid combinations (7/32, 22%) being the most common. All ECCs predominantly displayed grade 2 nuclei, and in 50% of the cases, focal areas composed of cells with grade 3 nuclei were present (Figure 1). The median mitotic index was five mitotic figures per 10 HPFs (range 1–18).
Figure 1. Histological features of endometrial clear cell carcinomas.
Representative hematoxylin and eosin stained sections displaying endometrial clear cell carcinomas with (A) papillary architectural pattern and grade 2 nuclei, (B) solid architectural pattern and grade 2 nuclei, (C) papillary architectural pattern and focal grade 3 nuclei, and (D) tubulocystic architectural pattern and focal grade 3 nuclei. Immunohistochemical analysis showing endometrial clear cell carcinomas with (E) loss of ARID1A/BAF250a expression, (F) retained nuclear expression of MLH1, (G) loss of MLH6 expression, and (H) retained PMS2 expression. Scale bars, 500µm.
Immunohistochemical analysis revealed that six ECCs (19%) displayed abnormal expression patterns of DNA MMR proteins (Table 1, Figure 1), with MSH6 being the most commonly altered. Of these, case CC11 had a personal history of colorectal cancer and an immediate family member with a Lynch syndrome-related tumor. Case CC10 was not subjected to germline testing, and the remaining four patients were found not to harbor any pathogenic mutations affecting the canonical DNA MMR genes. In addition, CC05 showed MLH1 promoter methylation, whereas CC20 did not. Of the 32 ECCs analyzed, eleven (34%) showed aberrant p53 expression, and six tumors displayed complete loss of ARID1A expression, whereas one ECC showed partial geographic loss for ARID1A. All ECCs with loss of ARID1A expression showed wild-type p53 expression patterns (Table 1). Taken together, the ECCs analyzed here were found to be heterogeneous at the histologic level, and in the expression of DNA MMR markers, p53 and ARID1A.
The repertoire of mutations and gene copy number alterations of ECCs
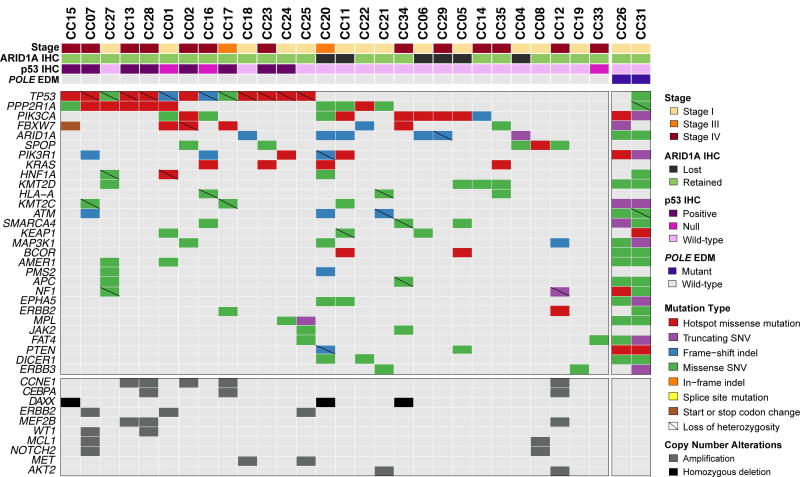
Of the 32 ECCs included in this study, we obtained high-quality targeted massively parallel sequencing data for 30 cases, at a median depth of coverage of 453× (range 156–838x) and 255× (118–540×) for tumor and normal samples, respectively (supplementary material, Table S2). ECCs harbored a median of 5 non-synonymous somatic mutations (range 2–538) in the 300 genes tested. In comparison, endometrial endometrioid and serous carcinomas from the TCGA dataset harbored a median of 8 (2–276) and 4 (1–39) non-synonymous somatic mutations in the 300 genes studied here, respectively, which is statistically significantly different (ECCs vs endometrioid, p=0.0014; ECCs vs serous, p=0.0421, Mann-Whitney U test). Two of the ECCs analyzed here (6%) harbored mutations affecting the exonuclease domain of POLE (CC26, V411L; CC31, P436H), characterized by a very high number of somatic mutations (supplementary material, Table S3). After removing the two POLE ECCs from the analysis, we identified TP53 (46%), PIK3CA (36%), PPP2R1A (36%), followed by PPP2R1A (36%), FBXW7 (25%), ARID1A (21%), PIK3R1 (18%), SPOP (18%) and KRAS (14%), as the most commonly mutated genes (Figure 2; supplementary material, Figure S1 and Table S3). Many of these mutations affected hotspots, including 9/13 (69%) of TP53 mutations, 6/10 (60%) of PPP2R1A mutations, 6/10 (60%) PIK3CA mutations and 4/4 (100%) KRAS mutations (Figure 2), and these were found to be clonal (i.e. bioinformatically inferred to be present in virtually all cancer cells within a tumor) (supplementary material, Figure S2 and Table S3). Noteworthy, a subset of the mutations identified in the non-POLE ECCs affected genes previously reported to be preferentially mutated in endometrial serous carcinomas, including TP53, PPP2R1A, FBXW7 and SPOP, whereas others were previously found to be preferentially mutated in endometrial endometrioid carcinomas, including PIK3R1 and KRAS (Figure 2) [4,8,11,13,14].
Figure 2. Non-synonymous somatic mutations detected by targeted capture massively parallel sequencing in endometrial clear cell carcinomas.
Recurrent (n≥2) non-synonymous somatic mutations (top) and recurrent (n≥2) amplifications and homozygous deletions (bottom) identified in 28 POLE-wild-type endometrial clear cell carcinomas by massively parallel sequencing targeting 300 cancer-related genes. Cases are represented in columns; genes are depicted in rows. Mutation types and gene copy number alterations are color-coded according to the legend. Loss of heterozygosity of the wild-type allele in association with a somatic mutation is depicted by a diagonal bar. Two additional cases, CC26 and CC31, were found to harbor somatic POLE exonuclease domain mutations (EDM), and are shown on the right. The phenobar provides information on stage, POLE status, ARID1A and p53 immunohistochemistry, and is color-coded according to the legend. Indel, small insertion/ deletion; SNV, single nucleotide variant.
We did not identify any correlation between architectural patterns and specific mutations or genes affected by mutations (data not shown), however the presence of focal areas with grade 3 nuclei was significantly higher in non-POLE TP53-mutant ECCs than in TP53 wild-type tumors (n=11 vs n=2; p=0.002, Fisher’s exact test). Thirteen of the 28 non-POLE ECCs subjected to targeted sequencing harbored a mutation in TP53, of which ten (77%) displayed aberrant p53 expression by IHC. CC33 lacked any p53 expression (i.e. null pattern) but no TP53 somatic mutation could be identified (Table 1, supplementary material, Table S3). All six ECCs found to have an ARID1A loss-of-function mutation (i.e. frameshift or stop-gain) lacked ARID1A expression (n=5) or displayed partial geographic loss of expression (n=1) by IHC (Table 1, supplementary material, Table S3). CC05, however, lacked ARID1A expression but was found to be ARID1A wild-type. Finally, three of the four ECCs subjected to sequencing analysis and displaying loss of MSH6 expression (n=3) or equivocal (focal, weak) MSH6 expression (n=1) by IHC also harbored somatic MSH6 loss-of-function mutations, whereas CC11 lacked MSH6 expression but no somatic MSH6 mutation was identified. In CC05, which lacked MLH1 and PMS2 expression, no somatic genetic alterations affecting MLH1 or PMS2 were found (Table 1, supplementary material, Table S3), however MLH1 hypermethylation was identified by clinical genetics testing (data not shown).
At the copy number level, the most recurrent gene copy number changes detected in non-POLE ECCs were amplifications of CCNE1 (18%, 5/28 non-POLE ECCs) and ERBB2 (11%, 3/28) and homozygous deletions of DAXX on 6p21 (11%, 3/28).
These findings demonstrate that ECCs are genetically heterogeneous, and that their repertoire of somatic genetic alterations includes recurrent hotspot mutations in TP53, PIK3CA and PPP2R1A as well as CCNE1 and ERBB2 amplifications. In addition, a subset of ECCs harbored POLE exonuclease domain mutations and displayed an ultramutator phenotype.
Comparison of the mutational repertoire of ECCs with that of endometrioid and serous carcinomas
Given that ECCs displayed mutations in genes previously reported to be either preferentially mutated in endometrial endometrioid or serous carcinomas (see above), we sought to compare the repertoire of mutations affecting the 300 genes analyzed in the 28 non-POLE ECCs included in this study with that of endometrioid and serous carcinomas from the TCGA dataset [11]. In comparison to non-POLE endometrial endometrioid carcinomas (n=183), ECCs harbored significantly less mutations in CTNNB1 (0% ECC vs 37% endometrioid, adjusted p<0.0001, Fisher’s exact test) and PTEN (7% ECC vs 78% endometrioid, adjusted p<0.0001, Fisher’s exact test) but significantly more mutations in PPP2R1A (36% ECC vs 5% endometrioid, adjusted p<0.0001, Fisher’s exact test) and TP53 (46% ECC vs 11% endometrioid, adjusted p=0.0151, Fisher’s exact test)(Supplementary Table S1). By contrast, following corrections for multiple comparisons, the only gene more frequently altered in 44 serous carcinomas as compared to ECCs was TP53 (89% vs 46%, adjusted p<0.0001, Fisher’s exact test, supplementary material, Table S1).
Molecular classification of ECCs and outcome
Given the heterogeneity in the repertoire of somatic mutations identified in ECCs, and the similarities and differences with endometrial endometrioid and serous cancers at the mutational level, we sought to define whether the ECCs could be classified into the molecular subtypes. For this, we employed a surrogate model for the molecular-based TCGA classification of endometrial endometrioid and serous carcinomas as described by Talhouk et al. [38]. This surrogate integrates the POLE mutation status, the IHC-based MSI status and, as a surrogate for ‘copy number’ status, the IHC-based p53 expression status. Two of the 32 EECs harbored a mutation in the POLE exonuclease domain (CC26 and CC31) and were classified as of POLE subtype (Table 2). Of the remaining cases, four ECCs displayed abnormal DNA MMR protein expression and were classified as of MMR-D subtype. Eleven ECCs displayed abnormal p53 expression patterns and were classified as of copy-number high (serous-like) subtype, called p53 abnormal (p53 abn) [38]. The remaining 15 ECCs lacking POLE mutations and showing normal DNA MMR and p53 protein expression were classified as copy-number low (endometrioid), called p53 wild-type (p53 wt) [38] (Table 2, supplementary material, Figure S3).
Table 2.
Molecular-based classification of ECCs.
| Sample ID |
Surrogate assay for molecular-based classification of endometrial cancers [38] | Subtype by hierarchical clustering* | |||
|---|---|---|---|---|---|
| POLE EDM | MMR IHC | p53 IHC | Molecular subtype | ||
| CC26 | V411L | MSH6 equivocal | Wild-type | POLE | POLE |
| CC31 | P436H | MSH6 loss | Wild-type | POLE | POLE |
| CC05 | Wild-type | MLH1/PMS2 loss | Wild-type | MMR-D | CN low (endometrioid)/ MSI |
| CC10 | Wild-type | MSH6 loss | Wild-type | MMR-D | NA |
| CC11 | Wild-type | MSH6 loss | Wild-type | MMR-D | CN low (endometrioid)/ MSI |
| CC20 | Wild-type | MLH1/PMS2/MSH6 loss | Wild-type | MMR-D | CN low (endometrioid)/ MSI |
| CC01 | Wild-type | Retained | Null pattern | p53 abnormal | CN high (serous-like) |
| CC02 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC07 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC13 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC15 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC16 | Wild-type | Retained | Null pattern | p53 abnormal | CN high (serous-like) |
| CC17 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC23 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC24 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC28 | Wild-type | Retained | Positive | p53 abnormal | CN high (serous-like) |
| CC33 | Wild-type | Retained | Null pattern | p53 abnormal | CN high (serous-like) |
| CC03 | Wild-type | Retained | Wild-type | p53 wild-type | NA |
| CC04 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC06 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC08 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC12 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC14 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC18 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC19 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC21 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC22 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC25 | Wild-type | Retained | Wild-type | p53 wild-type | CN low (endometrioid)/ MSI |
| CC27 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC29 | Wild-type | Retained | Wild-type | p53 wild-type | CN low (endometrioid)/ MSI |
| CC34 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
| CC35 | Wild-type | Retained | Wild-type | p53 wild-type | CN high (serous-like) |
CN, copy number; EDM, exonuclease domain mutations; IHC, Immunohistochemistry; MMR, mismatch repair; MMR-D, MMR-deficient; MSI, microsatellite instable; NA, not available as no sequencing data; *Ward's algorithm, Euclidean distance.
Nine of the eleven patients in the p53 abn group presented at advanced stage (82%), whereas only seven patients of the other subtypes presented at advanced stage (33%; p=0.0233, Fisher’s exact test). We also noted that p53 abn ECCs showed a significantly higher rate of dissemination to the peritoneum (8/11, 73%) compared to the other groups (7/22, 32%, p=0.0053, Fisher’s exact test). Of the 15 patients with p53 wt ECCs, nine (60%) presented with early stage and six patients (40%) with stage IV disease. At the time of follow-up, nine patients with p53 wt ECC had died of disease, four were alive with disease, and five had no evidence of disease (Table 1 and Table 2).
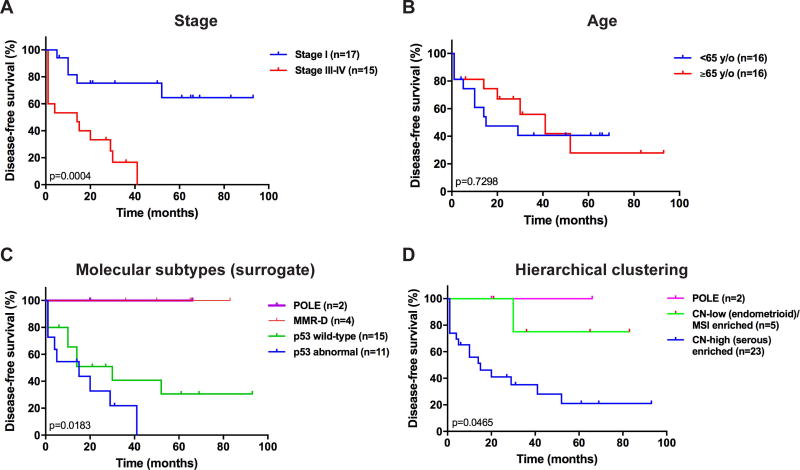
As an exploratory analysis, we assessed whether the molecular subtypes identified using the surrogate model would be associated with outcome. We observed that the molecular subtypes as defined by the surrogate model were significantly associated with disease-free survival (p=0.0183) in univariate analysis (Figure 3). Patients with ECCs of POLE or MMR-D subtype had a favorable outcome (no events) as compared to those with ECCs of p53 wt or p53 abn subtype (Figure 3).
Figure 3. Disease-free survival analyses of endometrial clear cell carcinoma patients stratified according to clinical features and molecular subtypes.
Kaplan–Meier disease-free survival curves for endometrial clear cell carcinoma patients stratified according to (A) stage of disease (stage I vs stage III/IV), (B) age (<65 years vs ≥65 years), (C) molecular subtypes defined using a surrogate model [38] (POLE, MMR-D, p53 wild-type (copy-number low (endometrioid)) and p53 abnormal (copy-number high (serous-like)), and (D) hierarchical clustering (POLE, copy-number high (serous-like) enriched, copy-number low (endometrioid)/ MSI (hypermutated) enriched). P-values of the log-rank test are shown. MMR-D, mismatch repair deficient; MSI, microsatellite instable.
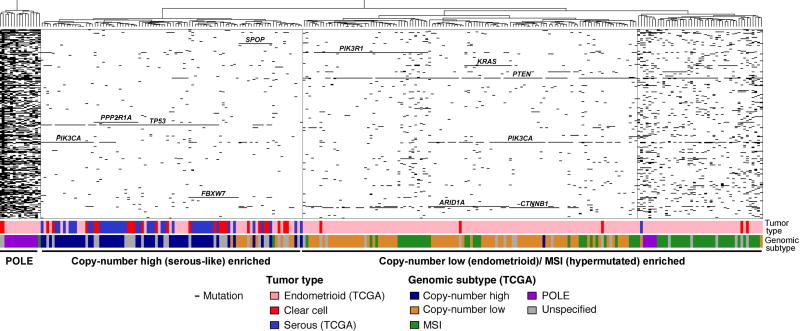
As a hypothesis-generating exploratory aim, we assessed whether unsupervised hierarchical clustering of the mutations identified in the 300 genes studied in the ECCs and all TCGA endometrial endometrioid and serous carcinomas (n=244) would allow a classification of ECCs on the basis of their mutational profiles. This cluster analysis revealed three stable clusters: one enriched for endometrial carcinomas of POLE subtype, one enriched for copy-number high (serous-like) and one encompassing the majority of copy-number low (endometrioid) and MSI (hypermutated) cancers (Figure 4, supplementary material, Figures S4 and S5). The ECCs classified as of POLE, MMR-D, and p53 abn molecular subtypes based on the surrogate IHC assay described above, also clustered with the respective TCGA endometrial cancers classified as POLE, copy-number low (endometrioid)/ MSI and copy-number high (serous-like) in the hierarchical cluster analysis, respectively (Table 2). By contrast, only two of the 14 ECCs classified as of p53 wt subtype by the surrogate assay clustered with the copy-number low (endometrioid)/ MSI endometrial cancers from TCGA; rather, 12/14 of these cases clustered with the copy-number high (serous-like) endometrial cancers based on their mutational profile. We noted that a subset p53 wt ECCs harbored mutations in genes preferentially mutated in serous carcinomas [11], including SPOP, FBXW7 and/ or PPP2R1A (Figure 2), which may drive the clustering. In addition, we observed that a subset of the surrogate assay-defined p53 wt ECCs displayed aberrant copy number profiles (supplementary material, Figure S6). In univariate analysis, the three subgroups identified based on mutational profiles using hierarchical clustering were also significantly associated with outcome (p=0.0465). These data provide further evidence that ECCs are heterogeneous at the genetic level and that all molecular subtypes identified in endometrial endometrioid and serous carcinomas can be found in ECCs.
Figure 4. Hierarchical clustering of endometrial clear cell carcinomas from this study and endometrioid and serous carcinomas from TCGA using somatic mutations identified in 300 cancer genes.
Hierarchical cluster analysis of mutations identified in the 300 cancer genes included in our targeted massively parallel sequencing assay using Euclidean distance metric and Ward’s algorithm, including all endometrial clear cell carcinomas (ECCs) from the current study, and all endometrial endometrioid and serous carcinomas from TCGA. Three stable clusters were identified (supplementary material, Figure S5): a POLE cluster, a cluster enriched for endometrial carcinomas of copy-number high (serous-like) subtype, and a cluster enriched for endometrial cancers of copy-number low (endometrioid) and of MSI (hypermutated) subtypes. The tumor type as well as the molecular subtype of the endometrial endometrioid and serous carcinomas as defined by TCGA are presented in the phenobar below the heatmap, color-coded according to the legend. The majority of ECCs clustered with the serous carcinomas/ copy-number high (serous-like) tumors, however ECCs were also found in the POLE and the copy-number low (endometrioid)/ MSI (hypermutated) enriched clusters.
DISCUSSION
Here we demonstrated that ECCs, a rare type of endometrial cancer, display a heterogeneous repertoire of somatic genetic alterations, affecting cancer genes previously found to be altered preferentially either in endometrioid or in serous endometrial carcinomas [8,11]. Furthermore, we observed that all four molecular subtypes identified in endometrial endometrioid and serous carcinomas in the TCGA dataset are represented in ECCs.
We confirm previous studies showing that TP53/p53 is the most commonly altered gene in ECCs [4,12–14]. Based on immunohistochemical analysis of 21 ECCs with ER, PR, p53 and Ki-67, Lax et al. [40] concluded that there were three types of ECC: typical ECC, serous-like ECC, and endometrioid-like ECC. Hoang et al. [13] suggested that a subset of ECCs with typical clear cell morphology may be biologically and clinically related to serous cancers. In addition, Fadare et al. [3] found 34% of ECCs to harbor aberrant p53 expression by IHC, which was associated with significantly lower progression-free survival by univariate analysis. In our dataset, 62% of the TP53-mutant ECCs studied harbored concomitant mutations in PPP2R1A (6/13 non-POLE TP53-mutant) or SPOP (2/13 non-POLE TP53-mutant), akin to serous/ copy-number high (serous-like) carcinomas [11]. Furthermore, we observed that 34% of ECCs were classified as of p53 abn subtype using a surrogate model, which were associated with poor disease-free survival in univariate analysis and showed a higher rate of peritoneal metastases as compared to the other subtypes. Our findings further support and provide additional evidence that the subgroup of ECCs harboring TP53 mutations may be similar to endometrial serous carcinoma not only in its biological behavior but also at the genetic level.
Importantly, however, not all ECCs are similar to endometrial serous cancers in terms of their molecular profile and clinical behavior. In fact, the entire spectrum of molecular subtypes previously described for endometrial endometrioid and serous carcinomas were identified in the ECCs studied here, which also include POLE, MMR-D and p53 wt/ copy-number low (endometrioid) cancers. Two ECCs were found to harbor POLE exonuclease domain mutations (6%) associated with an extremely high mutational burden (371 and 727 somatic mutations in the 300 genes studied), and both showed abnormal MSH6 expression patterns. In addition, 13% (4/32) of the non-POLE ECCs were classified as MMR-D associated with an increased number of somatic mutations (median 11 mutations (range 8–43) vs non-MMR-D, median 4 mutations (range 2–25), p=0.012, Mann-Whitney U test). Akin to the observations in endometrial endometrioid carcinomas harboring POLE exonuclease domain mutations [11,41–43], both patients with POLE-mutant ECCs are currently alive without evidence of disease. Although a significant association between lost/equivocal expression of MSH6 and POLE exonuclease domain mutations was observed here (p<0.05, Fisher’s exact test), only two ECCs concurrently displayed both alterations (Table 2). Therefore, these findings should be perceived as hypothesis generating, and warrant further studies to define the frequency and the molecular basis of the association between alterations of MSH6 and POLE exonuclease domain mutations. It should be noted that universal Lynch syndrome testing has been recommended for all newly diagnosed endometrial cancers [44]. Importantly, endometrial carcinomas showing a clear cell component have been found to be overrepresented in tumors with DNA MMR abnormalities [45].
MMR-deficient and POLE-mutant EECs have been reported to display distinctive histologic features [15,43]. A detailed histologic review revealed no histologic differences between ECCs with and without abnormal DNA MMR protein expression or POLE mutations other than differences in the pattern of immune infiltrate; 78% (25/32) were found to have a lymphoplasmacytic inflammatory response, a typical histologic feature of ECCs. Importantly, however, 19% (6/32) of cases were found to harbor prominent peri- and/or intra-tumoral infiltrating lymphocytes. Of these six ECCs, four displayed abnormal DNA MMR protein expression, one of which also harbored a POLE exonuclease domain mutation.
The surrogate model for molecular subtype classification employed here has been developed using endometrioid and serous/mixed carcinomas [38], and while the associations with outcome in our ECCs were statistically significant, improvements to this model may be possible. Different types of TP53 mutations (e.g. missense, truncation, frameshift) have been shown to affect the assessment of p53 by IHC [46–48], and not all TP53 mutations identified by massively parallel sequencing showed abnormal IHC patterns. In the model employed, p53 IHC is used as a surrogate for ‘copy number’ status. We observed, however, that a subset of p53/TP53 wild-type ECCs harbored aberrant gene copy number profiles (supplementary material, Figure S6), and based on their mutational profile, most clustered with copy-number high (serous-like) endometrioid and serous carcinomas, suggesting that alterations in genes other than TP53 may lead to a ‘serous-like’ genetic make-up. Similarly, while the immunohistochemical analysis of DNA MMR proteins performed has high sensitivity and specificity for microsatellite instability [49], the correlation is not perfect.
The stratification of ECCs based on their genetic make-up may not only identify subsets with distinct outcomes but in the era of precision medicine may also help guide treatment decision-making in the future. Eleven of the 30 ECCs subjected to targeted sequencing analysis (37%) harbored mutations affecting PIK3CA, PIK3R1 and/ or PTEN, suggesting that akin to endometrioid and serous carcinomas, targeting of the PI3K pathway in a subset of ECCs may constitute a therapeutic strategy [11,50]. Furthermore, we observed that three ECCs (10%) harbored ERBB2 gene amplifications and one (CC12) an ERBB2 hotspot mutation (S310F), providing evidence to suggest that ERBB2 may be a therapeutic target in a subset of ECCs. ARID1A, a member of the SWI/SNF complex, has been previously reported to be important in the pathogenesis of gynecologic clear cell carcinomas, and has been found to be mutated in 46–57% and 13% of ovarian and endometrial clear cell carcinomas, respectively [13,51,52]. Of the ECCs studied here, 22% lacked ARID1A expression, of which all but one had an underlying ARID1A frameshift mutation. It has recently been suggested that pharmacological inhibition of EZH2 may represent a novel treatment strategy for cancers harboring ARID1A mutations [53]. In addition, there is evidence indicating that ARID1A-mutated cancers may also be sensitive to targeting of the PI3K/AKT pathway or DNA damage response [54], and a clinical trial combining olaparib with the AKT inhibitor AZD5363 in ARID1A-mutant advanced solid tumors is currently recruiting participants (ClinicalTrials.gov; NCT02576444). Finally, immunotherapy has recently been added to the repertoire of possible treatments in patients with hyper- and ultramutated cancers [55].
This study has several limitations. First, given that all ECCs were formalin-fixed, paraffin-embedded and that the tumor and normal tissue material was limited, we subjected the ECCs to targeted sequencing focusing on cancer genes rather than to whole-exome sequencing and could not define the prevalence of TAF1 mutations, recently reported in up to 10% of ECCs [14]. Many of the cancer-related genes analyzed in our study, however, are targetable and are recurrently altered in endometrial cancer [11,18]. Second, using MSK-IMPACT, we were unable to find a genetic basis for the differences between ECCs, endometrioid and serous carcinomas. One could hypothesize that a somatic genetic alteration affecting a gene other than those included in MSK-IMPACT could be pathognomonic for ECCs. This is unlikely to be the case, given that a recent whole-exome sequencing analysis of 12 ECCs [14] failed to reveal any somatic mutations pathognomonic for these tumors. Hence, alternative explanations for the distinctive histologic features of ECCs include that ECCs, endometrioid and serous carcinomas may have different cells of origin, or that ECCs differ from the other types of endometrial carcinoma on the basis of distinct cells of origin affected by genetic and/or epigenetic alterations not surveyed with the methods employed in this study. Third, the tumor cell content of a subset of samples was low (i.e. <50%), which may have affected the mutation and/or copy number analysis. The sequencing depth for these samples was high, however (median of 453× for tumors), and the frequency of mutations affecting known genes, including TP53 and ARID1A, similar to that previously described. Fourth, given the low number of somatic mutations in ECCs in the 300 genes analyzed, with the exception of the tumors harboring POLE mutations, mutational signatures could not be determined. Further studies are warranted to define the mutational signatures in ECCs.
Despite the limitations, our data demonstrate that ECCs are a heterogeneous group of tumors in terms of histology, somatic genetic alterations and clinical behavior. Akin to endometrial endometrioid cancers, all molecular subtypes were represented in the ECCs studied, including POLE and MMR-deficient tumors with an excellent prognosis, and copy-number low (endometrioid)/p53 wt and copy-number high (serous-like)/p53 abn with a poor prognosis. Based on these findings, the classification of all ECCs as “high-grade” or “type II” tumor according to Bokhman [7] may not be warranted.
Supplementary Material
Figure S1. Likely pathogenic non-synonymous somatic mutations detected by targeted capture massively parallel sequencing in endometrial clear cell carcinomas
Figure S2. Cancer cell fractions of non-synonymous somatic mutations detected by targeted capture massively parallel sequencing in endometrial clear cell carcinomas
Figure S3. Histological features and copy number profiles of molecular subtypes of endometrial clear cell carcinomas
Figure S4. Hierarchical clustering of endometrial clear cell carcinomas from this study and endometrioid and serous carcinomas from TCGA using somatic mutations identified in 300 cancer genes
Figure S5. Assessment of the stability of the hierarchical cluster analysis
Figure S6. Copy number profiles of endometrial clear cell carcinomas classified as of copy-number low (endometrioid)/ p53 wild-type subtype using a surrogate model
Table S1. List of 300 genes included in the targeted capture massively parallel sequencing assay and frequency of mutations affecting these genes in endometrial clear cell carcinomas from this study and endometrial endometrioid and endometrial serous carcinomas from The Cancer Genome Atlas (TCGA) dataset
Table S2. Sequencing statistics of endometrial clear cell carcinomas subjected to targeted massively parallel sequencing
Table S3. Somatic mutations identified in endometrial clear cell carcinomas by targeted massively parallel sequencing
Acknowledgments
We thank Jorge S Reis-Filho for critically reading the manuscript. Research reported in this paper was supported in part by the Farmer Family Foundation and a Cancer Center Support Grant of the National Institutes of Health/National Cancer Institute (Grant No P30CA008748). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: None to declare
AUTHOR CONTRIBUTIONS STATEMENT
DFD and RAS conceived the study; DFD and RAS reviewed and selected the cases; KAB, PS, RSL, SNS, SM, ASM, DTC and MFB performed bioinformatics analysis; DFD, KAB and BW analyzed and interpreted the data; DFD and BW wrote the first draft of the manuscript. All authors edited and approved the final draft of the manuscript.
This work was presented in part as a platform presentation at the 104th United States and Canadian Academy of Pathology (USCAP) Annual Meeting, Boston, MA, USA, 2015.
References
- 1.Creasman WT, Odicino F, Maisonneuve P, et al. Carcinoma of the corpus uteri. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int J Gynaecol Obstet. 2006;(95 Suppl 1):S105–143. doi: 10.1016/S0020-7292(06)60031-3. [DOI] [PubMed] [Google Scholar]
- 2.Zaino R, Carinelli SG, Ellenson LH, et al. Epithelial tumours and precursors. In: Kurman RJ, Carcangiu M-L, Herrington CS, et al., editors. WHO Classification of Tumours of Female Reproductive Organs. IARC; Lyon: 2014. pp. 125–134. [Google Scholar]
- 3.Fadare O, Zheng W, Crispens MA, et al. Morphologic and other clinicopathologic features of endometrial clear cell carcinoma: a comprehensive analysis of 50 rigorously classified cases. Am J Cancer Res. 2013;3:70–95. [PMC free article] [PubMed] [Google Scholar]
- 4.Han G, Soslow RA, Wethington S, et al. Endometrial Carcinomas With Clear Cells: A Study of a Heterogeneous Group of Tumors Including Interobserver Variability, Mutation Analysis, and Immunohistochemistry With HNF-1beta. Int J Gynecol Pathol. 2015;34:323–333. doi: 10.1097/PGP.0000000000000162. [DOI] [PubMed] [Google Scholar]
- 5.DeLair D, Oliva E, Kobel M, et al. Morphologic spectrum of immunohistochemically characterized clear cell carcinoma of the ovary: a study of 155 cases. Am J Surg Pathol. 2011;35:36–44. doi: 10.1097/PAS.0b013e3181ff400e. [DOI] [PubMed] [Google Scholar]
- 6.Fadare O, Parkash V, Dupont WD, et al. The diagnosis of endometrial carcinomas with clear cells by gynecologic pathologists: an assessment of interobserver variability and associated morphologic features. Am J Surg Pathol. 2012;36:1107–1118. doi: 10.1097/PAS.0b013e31825dd4b3. [DOI] [PubMed] [Google Scholar]
- 7.Bokhman JV. Two pathogenetic types of endometrial carcinoma. Gynecol Oncol. 1983;15:10–17. doi: 10.1016/0090-8258(83)90111-7. [DOI] [PubMed] [Google Scholar]
- 8.Murali R, Soslow RA, Weigelt B. Classification of endometrial carcinoma: more than two types. Lancet Oncol. 2014;15:e268–278. doi: 10.1016/S1470-2045(13)70591-6. [DOI] [PubMed] [Google Scholar]
- 9.Felix AS, Weissfeld JL, Stone RA, et al. Factors associated with Type I and Type II endometrial cancer. Cancer Causes Control. 2010;21:1851–1856. doi: 10.1007/s10552-010-9612-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Plataniotis G, Castiglione M, Group EGW. Endometrial cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;(21 Suppl 5):v41–45. doi: 10.1093/annonc/mdq245. [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.An HJ, Logani S, Isacson C, et al. Molecular characterization of uterine clear cell carcinoma. Mod Pathol. 2004;17:530–537. doi: 10.1038/modpathol.3800057. [DOI] [PubMed] [Google Scholar]
- 13.Hoang LN, McConechy MK, Meng B, et al. Targeted mutation analysis of endometrial clear cell carcinoma. Histopathology. 2015;66:664–674. doi: 10.1111/his.12581. [DOI] [PubMed] [Google Scholar]
- 14.Le Gallo M, Rudd ML, Urick ME, et al. Somatic mutation profiles of clear cell endometrial tumors revealed by whole exome and targeted gene sequencing. Cancer. 2017;123:3261–3268. doi: 10.1002/cncr.30745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garg K, Leitao MM, Jr, Kauff ND, et al. Selection of endometrial carcinomas for DNA mismatch repair protein immunohistochemistry using patient age and tumor morphology enhances detection of mismatch repair abnormalities. Am J Surg Pathol. 2009;33:925–933. doi: 10.1097/PAS.0b013e318197a046. [DOI] [PubMed] [Google Scholar]
- 16.Garg K, Leitao MM, Jr, Wynveen CA, et al. p53 overexpression in morphologically ambiguous endometrial carcinomas correlates with adverse clinical outcomes. Mod Pathol. 2010;23:80–92. doi: 10.1038/modpathol.2009.153. [DOI] [PubMed] [Google Scholar]
- 17.Ye J, Zhou Y, Weiser MR, et al. Immunohistochemical detection of ARID1A in colorectal carcinoma: loss of staining is associated with sporadic microsatellite unstable tumors with medullary histology and high TNM stage. Hum Pathol. 2014;45:2430–2436. doi: 10.1016/j.humpath.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schultheis AM, Ng CK, De Filippo MR, et al. Massively Parallel Sequencing-Based Clonality Analysis of Synchronous Endometrioid Endometrial and Ovarian Carcinomas. J Natl Cancer Inst. 2016;108 doi: 10.1093/jnci/djv427. djv427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piscuoglio S, Ng CK, Murray MP, et al. The genomic landscape of male breast cancers. Clin Cancer Res. 2016;22:4045–4056. doi: 10.1158/1078-0432.CCR-15-2840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen R, Seshan VE. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saunders CT, Wong WS, Swamy S, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 26.Koboldt DC, Zhang Q, Larson DE, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25:1729–1735. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carter SL, Cibulskis K, Helman E, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter H, Chen S, Isik L, et al. Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res. 2009;69:6660–6667. doi: 10.1158/0008-5472.CAN-09-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martelotto LG, Ng CK, De Filippo MR, et al. Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Biol. 2014;15:484. doi: 10.1186/s13059-014-0484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shihab HA, Gough J, Cooper DN, et al. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29:1504–1510. doi: 10.1093/bioinformatics/btt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwarz JM, Rodelsperger C, Schuelke M, et al. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 35.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talhouk A, McConechy MK, Leung S, et al. A clinically applicable molecular-based classification for endometrial cancers. Br J Cancer. 2015;113:299–310. doi: 10.1038/bjc.2015.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 40.Lax SF, Pizer ES, Ronnett BM, et al. Clear cell carcinoma of the endometrium is characterized by a distinctive profile of p53, Ki-67, estrogen, and progesterone receptor expression. Hum Pathol. 1998;29:551–558. doi: 10.1016/s0046-8177(98)80002-6. [DOI] [PubMed] [Google Scholar]
- 41.Billingsley CC, Cohn DE, Mutch DG, et al. Prognostic Significance of POLE Exonuclease Domain Mutations in High-Grade Endometrioid Endometrial Cancer on Survival and Recurrence: A Subanalysis. Int J Gynecol Cancer. 2016;26:933–938. doi: 10.1097/IGC.0000000000000681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Church DN, Stelloo E, Nout RA, et al. Prognostic significance of POLE proofreading mutations in endometrial cancer. J Natl Cancer Inst. 2015;107:402. doi: 10.1093/jnci/dju402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hussein YR, Weigelt B, Levine DA, et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod Pathol. 2015;28:505–514. doi: 10.1038/modpathol.2014.143. [DOI] [PubMed] [Google Scholar]
- 44.Mills AM, Liou S, Ford JM, et al. Lynch syndrome screening should be considered for all patients with newly diagnosed endometrial cancer. Am J Surg Pathol. 2014;38:1501–1509. doi: 10.1097/PAS.0000000000000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kobel M, Tessier-Cloutier B, Leo J, et al. Frequent mismatch repair protein deficiency in mixed endometrioid and clear cell carcinoma of the endometrium. Int J Gynecol Pathol. 2017;36:555–561. doi: 10.1097/PGP.0000000000000369. [DOI] [PubMed] [Google Scholar]
- 46.Hall PA, McCluggage WG. Assessing p53 in clinical contexts: unlearned lessons and new perspectives. J Pathol. 2006;208:1–6. doi: 10.1002/path.1913. [DOI] [PubMed] [Google Scholar]
- 47.Schultheis AM, Martelotto LG, De Filippo MR, et al. TP53 mutational spectrum in endometrioid and serous endometrial cancers. Int J Gynecol Pathol. 2016;35:289–300. doi: 10.1097/PGP.0000000000000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soussi T, Leroy B, Taschner PE. Recommendations for analyzing and reporting TP53 gene variants in the high-throughput sequencing era. Hum Mutat. 2014;35:766–778. doi: 10.1002/humu.22561. [DOI] [PubMed] [Google Scholar]
- 49.Modica I, Soslow RA, Black D, et al. Utility of immunohistochemistry in predicting microsatellite instability in endometrial carcinoma. Am J Surg Pathol. 2007;31:744–751. doi: 10.1097/01.pas.0000213428.61374.06. [DOI] [PubMed] [Google Scholar]
- 50.Myers AP. New strategies in endometrial cancer: targeting the PI3K/mTOR pathway--the devil is in the details. Clin Cancer Res. 2013;19:5264–5274. doi: 10.1158/1078-0432.CCR-13-0615. [DOI] [PubMed] [Google Scholar]
- 51.Jones S, Wang TL, Shih Ie M, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wiegand KC, Shah SP, Al-Agha OM, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bitler BG, Fatkhutdinov N, Zhang R. Potential therapeutic targets in ARID1A-mutated cancers. Expert Opin Ther Targets. 2015;19:1419–1422. doi: 10.1517/14728222.2015.1062879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Piulats JM, Matias-Guiu X. Immunotherapy in endometrial cancer: in the nick of time. Clin Cancer Res. 2016;22:5623–5625. doi: 10.1158/1078-0432.CCR-16-1820. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Likely pathogenic non-synonymous somatic mutations detected by targeted capture massively parallel sequencing in endometrial clear cell carcinomas
Figure S2. Cancer cell fractions of non-synonymous somatic mutations detected by targeted capture massively parallel sequencing in endometrial clear cell carcinomas
Figure S3. Histological features and copy number profiles of molecular subtypes of endometrial clear cell carcinomas
Figure S4. Hierarchical clustering of endometrial clear cell carcinomas from this study and endometrioid and serous carcinomas from TCGA using somatic mutations identified in 300 cancer genes
Figure S5. Assessment of the stability of the hierarchical cluster analysis
Figure S6. Copy number profiles of endometrial clear cell carcinomas classified as of copy-number low (endometrioid)/ p53 wild-type subtype using a surrogate model
Table S1. List of 300 genes included in the targeted capture massively parallel sequencing assay and frequency of mutations affecting these genes in endometrial clear cell carcinomas from this study and endometrial endometrioid and endometrial serous carcinomas from The Cancer Genome Atlas (TCGA) dataset
Table S2. Sequencing statistics of endometrial clear cell carcinomas subjected to targeted massively parallel sequencing
Table S3. Somatic mutations identified in endometrial clear cell carcinomas by targeted massively parallel sequencing