Abstract
Objectives
P450 oxidoreductase (POR) donates electrons to all microsomal cytochrome P450s, including drug-metabolizing and steroidogenic enzymes. Severe POR mutations cause skeletal malformations and disordered steroidogenesis. The POR polymorphism A503V is found on ~28% of human alleles and decreases activities of CYP3A4 and steroidogenic CYP17, but not the activities of steroidogenic CYP21 or drug-metabolizing CYP1A2 and CYP2C19. CYP2D6 metabolizes about 25% of clinically used drugs; we assessed the capacity of POR variants to support the activities of human CYP2D6.
Methods
N-27 forms of wild type (WT), Q153R, A287P, R457H and A503V POR, and WT CYP2D6 were expressed in E.coli. POR proteins in bacterial membranes were reconstituted with purified CYP2D6. Support of CYP2D6 was measured by metabolism of EOMCC (2H-1-benzopyran-3-carbonitrile,7-(ethoxy-methoxy)-2-oxo-(9Cl)), dextromethorphan and bufuralol. Km and Vmax were determined in three triplicate experiments for each reaction; catalytic efficiency is expressed as Vmax/Km.
Results
Compared to WT POR, disease-causing POR mutants A287P and R457H supported no detectable CYP2D6 activity with EOMCC, but A287P supported ~ 25% activity with dextromethorphan and bufuralol. Q153R had increased function with CYP2D6 (128% with EOMCC; 198% with dextromethorphan; 153% with bufuralol). A503V supported decreased CYP2D6 activity: 85% with EOMCC, 62% with dextromethorphan and 53% with bufuralol.
Conclusions
POR variants have different effects depending on the substrate metabolized. Disease-causing POR mutations R457H and A287P had poor activities, suggesting that diminished drug metabolism should be considered in affected patients. The common A503V polymorphism impaired CYP2D6 activities with two commonly-used drugs by 40–50%; potentially explaining some genetic variation in drug metabolism.
Keywords: Drug metabolism, cytochrome P450, dextromethorphan, bufuralol, pharmacogenetics, electron transfer, enzyme kinetics
Introduction
Cytochrome P450 (P450) enzymes are heme-containing proteins that catalyze the metabolism of drugs and xenobiotics and participate in the biosynthesis of endogenous small molecules. The human genome contains 57 genes encoding P450s [1]. Seven of these encode Type I P450s found in mitochondria and fifty encode type II P450s, found in the endoplasmic reticulum. Among the 50 Type II P450 enzymes, approximately 20 are involved in the biosynthesis of steroids, sterols, fatty acids, cholesterol and eicosanoids, 15 participate in xenobiotic and drug metabolism, and the remaining 15 are “orphan” enzymes whose activities have not been identified [2]. Type I P450s receive electrons form NADPH via two mitochondrial proteins, ferredoxin reductase and ferredoxin [3]. Type II P450 enzymes receive electrons from NADPH via microsomal P450 oxidoreductase (POR). The single human POR gene on chromosome 7 consists of 15 protein-coding exons that encode 680 amino acids, plus an untranslated exon 38.8 kb upstream that initiates POR transcription [4]. POR is a 78 kDa membrane-bound protein that contains two distinct domains separated by a flexible ‘hinge’; one domain binds NADPH and FAD, the other domain binds FMN [5–7]. Electrons from NADPH pass to the FAD, eliciting a conformational change that permits the electrons to pass from the FAD to the FMN, following which the POR protein can then interact with the redox partner binding site of the P450 enzymes so that electrons reach the heme iron of the P450 to permit catalysis.
The crucial role of POR and type II P450 enzymes in so many essential processes suggests that POR defects would have global effects, and POR gene deletions in mice are lethal at embryonic day 9.5 or 13.5, depending on how the knockout vector is constructed [8, 9]. This phenotype is due to loss of function of extra-hepatic P450 enzyme activities, as liver-specific knockout of POR results in phenotypically normal mice with normal reproductive capacity and grossly diminished capacity for hepatic drug metabolism [10–12]. While the embryonic lethality of POR-knockout mice suggested that human POR mutations would have devastating consequences, patients with POR deficiency were described in 2004 having craniosynostosis, radio-ulnar synostosis, midface hypoplasia, disordered steroidogenesis and genital ambiguity [13], a phenotype known as ‘Antley-Bixler syndrome’ (ABS) [14]. An indistinguishable ABS skeletal phenotype is also caused by gain-of-function mutations in fibroblast growth factor receptor 2, but those patients have normal steroidogenesis and normal genital development [15].
Substantial research in pharmacogenetics has concerned genetic variations in drug metabolizing P450 enzymes [16]. For example, the activity of CYP2D6, which metabolizes about 25% of clinically-used drugs [17, 18] ranges from complete deficiency to ultra-fast metabolism, depending on up to 78 known alleles (www.cypalleles.ki.se/cyp2d6.htm). However, CYP2D6 variants do not explain all variation in CYP2D6-mediated drug metabolism. In a study of over 500 Europeans given dextromethorphan, an individual’s CYP2D6 alleles correlated with their urinary ratio of dextromethorphan to its metabolite dextrorphan, but even among individuals homozygous for particular CYP2D6 alleles, there was a large variation in the urinary ratio of dextromethorphan to dextrorphan [19]. Similarly, when the urinary ratio of debrisoquine to 4-hydroxydebrisoquine was examined, large inter-individual differences were again observed in individuals who were homozygous for particular CYP2D6 alleles[20–22]. Thus genetic variation in CYP2D6 alone cannot account for all the variation in metabolism of CYP2D6 substrates.
In addition to the human POR mutants that cause genetic disease [15], sequencing the POR genes of 842 healthy individuals belonging to four different ethnic groups identified the coding sequence polymorphism A503V on ~28% of alleles [23]. POR A503V, encoded by the POR*28 allele [24], reduced the 17α-hydroxylase and 17,20 lyase activities of steroidogenic P450c17 (CYP17) to 68% and 58%, respectively [15], but had no impact on catalysis by steroidogenic P450c21 (CYP21) [25]. Similarly, POR A503V supports normal activity by CYP1A2 and CYP2C19 [26], but supported 61–97% of activities of CYP3A4, depending on the substrate metabolized [27]. The POR mutant A287P, which is the most common cause of ABS in persons of European heritage, dramatically reduces the activity of P450c17 [13, 15] but has no impact on catalysis by P450aro (aromatase, CYP19) [28]. Thus the activity of a particular POR variant to support catalysis by one P450 cannot predict its ability to support catalysis by another P450, and the recent work with CYP3A4 indicates that the effects may vary with the specific substrate metabolized by a single P450 [27]. Thus each POR/P450 combination requires separate study. To determine the potential effects of POR sequence variants on catalysis by CYP2D6, we examined the metabolism of three substrates by bacterially-expressed human CYP2D6 supported by five different variants of human POR.
Methods
Chemicals and reagents
Dextromethorphan, levallorphan, metoprolol and bovine serum albumin (BSA) were purchased from Sigma-Aldrich (St. Louis, Missouri, USA). NADP, glucose-6-phosphate (G6P) and glucose-6-phosphate dehydrogenase (G6PD) were purchased from EMD Biosciences (La Jolla California, USA). Vivid EOMCC (2H-1-benzopyran-3-carbonitrile,7-(ethoxy-methoxy)-2-oxo-(9Cl)) substrate and its corresponding fluorescent dye standard (3-cyano-7-hydroxy-coumarin) were purchased from Invitrogen (Madison, Wisconsin, USA). Dextrorphan, bufuralol and 1-hydroxybufuralol were purchased from BD Biosciences (San Jose, California, USA).
Preparation and quantitation of P450 oxidoreductase (POR)
The construction of vectors for human WT-POR and its mutants lacking 27 N-terminal residues cloned in pET22b has been described [15]. The expression of POR in E. coli C41(DE3) (Lucigen Corp., Middleton, Wisconsin, USA) and the preparation of bacterial membrane proteins have been described [15, 26]. POR variants were quantitated by Western blotting as described [15, 26], and equimolar amounts of each isoform were used. Total membrane-bound proteins were separated by electrophoresis on sodium dodecyl sulfate (SDS) 10% polyacrylamide gel, and transferred to Immobilon-FL transfer membrane (Millipore, Bedford, Massachusetts, USA). Membranes were then incubated in blocking buffer (5% skimmed milk and 5% horse serum in Tris-buffered saline (TBS) at pH 8.0) for 1 hr and then incubated for 24 hr at 4°C with rabbit polyclonal antibody directed against rat POR (Stressgen, Ann Arbor, Michigan, USA) at 1:2000 dilution. After washing in TBS containing 0.1% Tween-20, membranes were incubated with blocking buffer containing goat anti-rabbit infrared-labeled secondary antibody (Li-COR Bioscience, Lincoln, Nebraska, USA) at a dilution of 1:10,000 at room temperature for 1 hr. Membranes were washed three times in TBS containing 0.1% Tween-20, and protein bands were detected in the green fluorescent channel (700 nm) on an Odyssey Infrared Imaging System (LI-COR Bioscience) and analyzed with Odyssey software. Membrane-bound POR protein content was extrapolated from linear regression of the POR standards.
Expression and purification of CYP2D6
CYP2D6 cDNA with amino-terminal modifications and codon selections that facilitate expression in E. coli [29] was synthesized and inserted into plasmid pCW containing an Amp marker (Genscript, Inc. Piscataway, New Jersey, USA). Plasmid pCW expressing CYP2D6 and plasmid pGro7 (Takara Bio, Shiga, Japan) containing an Cm marker and expressing GroES/EL, were then co-transformed into E.coli JM109 and spread on Luria-Bertani (LB) plates containing ampicillin and chloramphenicol. A single colony was inoculated into 10 ml of LB medium containing 100 µg/ml ampicillin and 20 µg/ml chloramphenicol, and grown overnight at 37°C with shaking at 225 rpm. This starter culture was inoculated to 500 ml of modified Terrific broth containing 100 µg/ml ampicillin and 20 µg/ml chloramphenicol, 1 mmol/l thiamine, 125 µl of 4000× stock trace elements and cultivated 4–6 h at 37°C with shaking. The 4000× stock contains: 2.7 g FeCl3.6H2O, 0.2 g ZnCl2.4H2O, 0.2 g CoCl2.6H2O, 0.2 g Na2MoO4.H2O, 0.1 g CaCl2.6H2O, 0.1 g CuCl2, 0.05 g H3BO3, 10 ml HCl in a final volume of 100 ml [30]. At OD600 0.4–0.6, 0.5 ml 100× BME vitamin stock (Sigma-Aldrich) and δ–aminolevulinic acid (final concentration 0.5 mmol/l) were added to the culture. Isopropyl β-d-thiogalactopyranose (IPTG) was added to a final concentration of 1 mmol/l to induce expression of CYP2D6, and arabinose (Acros Organics, Geel, Belgium) was added to a final concentration of 0.5 mg/ml to induce expression of GroES/EL. Growth was continued at 28°C while shaking at 175 rpm for 24–36 hr. Bacteria were harvested by centrifugation at 5000g for 5 min, resuspended in 20 ml storage buffer (50 mmol/l potassium phosphate, 20% glycerol, 1 mmol/l EDTA, and 1 mmol/l NaCl), transferred to a pre-chilled 50 ml Falcon tube, centrifuged at 5,000g for 30 min and the pellet was stored at −80°C.
Stored E.coli were resuspended in 25 ml of B-PER reagent (Thermo Scientific, Rockford, Illinois, USA) containing 15% glycerol, 0.5 mol/l NaCl, 20 mmol/l imidazole, 0.5% sodium cholate, 2% Tergitol NP-11 (Dow Chemical, Midland, Michigan, USA), 160 mmol/l β-mercaptoethanol, 20 mg lyzozyme, 1mg DNase, 1 ml protease inhibitor cocktail (P8849, Sigma-Aldrich), 0.2 mmol/l phenylmethylsulfonyl fluoride (PMSF). Bacteria were kept on ice and stirred until fully suspended, then lysed with a French press (American Instrument Co., Silver Spring, Maryland, USA) at 16,000 psi twice and centrifuged at 100,000g for one hour at 4°C. The supernatant was collected and filtered with a Millipore 5µm Millex-SV Syringe-Driven Filter.
A 5 ml HisTrap column (HP-GE Healthcare, Piscataway, New Jersey, USA) was equilibrated with 50 ml of binding buffer (100 mmol/l potassium phosphate pH 7.4, 15% glycerol, 40 mmol/l imidazole, 2% cholate, 0.3 mol/l NaCl, 0.2 mmol/l PMSF). The E. coli lysate containing CYP2D6 was loaded slowly onto the column; the flow through was monitored for P450 content by carbon monoxide reduced difference spectra [31] to ensure proper binding. The column was washed with 50–70 ml of washing buffer (100 mmol/l potassium phosphate pH 7.4, 15% glycerol, 40 mmol/l imidazole, 2% cholate, 0.3 mol/l NaCl, 0.2 mmol/l PMSF, 50 mmol/l glycine) with monitoring the flow through for P450 content. The column was then eluted with 100 mmol/l potassium phosphate pH 7.4, 15% glycerol, 500 mmol/l imidazole, 2% cholate, 0.3 mol/l NaCl, 0.2 mmol/l PMSF, 0.5% Tergitol NP-11 and fractions were collected until the flow through was clear of yellowish-brown color. An aliquot was assayed for P450 content and analyzed on SDS-PAGE.
Batch chromatography on hydroxyapatite resin was used to remove detergent. Three grams of acro-Prep Ceramic Hydroxyapatite Type1 40 µm (Bio-Rad) were combined with 20 ml of 50 mmol/l potassium phosphate pH 7.4, 20% glycerol, mixed and centrifuged at 5000g, 4°C, for 5 minutes and the supernatant was discarded. The protein sample was then added to the resin, mixed well and centrifuged as above, and the supernatant was monitored for P450 content to ensure that the protein had bound to the resin. The resin was washed three times with 5–10 ml 50 mmol/l potassium phosphate pH 7.4, 20% glycerol, 0.5% cholate, centrifuged after each wash and the supernatant was monitored for absent P450 content. After washing, the bound P450 was eluted with 5–10 ml 300 mmol/l potassium phosphate pH 7.4, 20% glycerol. The tube was centrifuged as above, the yellowish-pink supernatant was collected, and the P450 content was measured. The purified CYP2D6 was dialyzed overnight twice against 50 mmol/l potassium phosphate pH 7.4, 20% glycerol, 0.5 mmol/l EDTA, 1 mmol/l NaCl, the final Bradford protein concentration was measured and the CYP2D6 content was determined by carbon monoxide reduced difference spectra [31].
CYP2D6 protein at 14 µmol/l was diluted either ten-fold or 100-fold in 50 mmol/l potassium phosphate buffer. Dithiothreitol and SDS were added to final concentrations of 40 mmol/l and 1%, respectively. The solutions were heated for five min at 100°C and a 30 µl aliquot of each was loaded on 12% SDS-PAGE gels (Pierce, St Louis, Missouri, USA) with a protein ladder standard (Bio-Rad). After electrophoresis for one hour at 115V, gels were equilibrated in distilled water and stained using GelCode Blue Stain Reagent per manufacturer's instructions (Pierce).
CYP2D6 catalysis with vivid EOMCC as substrate
Optimal conditions for CYP2D6-mediated catalysis of vivid EOMCC were determined by comparing activities with ratios of CYP2D6 to WT POR set at 1:10, 1:20 and 1:30 with EOMCC substrate concentrations of 10, 20, 30, 50, 100, 150 and 200 µmol/l in a final reaction volume of 100 µl. EOMCC and fluorescent dye standard (3-cyano-7-hydroxy-coumarin) were dissolved in acetonitrile, which did not exceed 1% of final volume. Reactions were performed in 96-well flat bottom black walled plates (Griener, Friekenhausen, Germany) at 37°C as described [26] with minor modifications. Three pmol of CYP2D6 was mixed with 30 or 60 or 90 pmol of membrane-bound POR and incubated on ice for 5 min then 10 µg of DLPC (1,2-didodecanoyl-sn-glycero-3-phosphocholine) was added and incubated on ice for 5 min. 50 mmol/l of phosphate buffer pH 7.4 was added to 70 µl final volume. Each substrate concentration (10, 20, 30, 50, 100, 150 and 200 µmol/l) was then added to the reaction and incubated at 37°C for 5 min. Catalysis was initiated by adding 30 µl of NADPH regeneration mix (3.3 mmol/l glucose-6-phosphate, 1 mmol/l NADP, 0.8 U/ml glucose-6-phosphate dehydrogenase and 50 mmol/l HEPES (N-2-hydroxyl piperazine-N´-2-ethane sulfonic acid) buffer (pH 7.4)) [32, 33]. After incubation at 37°C for 20 min, the fluorescent signal was read in a Molecular Dynamics Spectramax M2 plate reader with excitation at 409 nm and emission at 460 nm. Velocity was measured using a standard curve and presented as nmol of product per pmol of P450 per min.
CYP2D6 catalysis with dextromethorphan and bufuralol as substrates
Optimal conditions for CYP2D6-mediated catalysis of dextromethorphan and bufuralol were determined by comparing activities with ratios of CYP2D6 to POR set at 1:10, 1:20 and 1:30 with dextromethorphan substrate concentrations from 0.625–15 µmol/l or bufuralol concentrations from 0.325–30 µmol/l in a final reaction volume of 100 µl. Dextromethorphan and bufuralol were dissolved in DMSO, which did not exceed 1% in the final incubation. Reactions (total volume 100 µl) were performed in 1.5 ml microcentrifuge tubes (USA Scientific, Orlando, Florida, USA) at 37°C. One pmol of CYP2D6 was mixed with 10, 20 or 30 pmol of membrane-bound POR, incubated on ice for 5 min, then 10 µg of DLPC was added and incubated on ice for 5 min. Each concentration of substrate was added in 70 µl of 50 mmol/l phosphate buffer, pH 7.4, incubated at 37°C for 5 min, and catalysis was initiated by adding 30 µl of NADPH regeneration mix. After incubation at 37°C for 30 min, the reactions were stopped by adding 1 ml of methylene chloride containing 10 µmol/l levallorphan as internal standard for dextromethorphan and 1.5 µmol/l metoprolol as internal standard for bufuralol. Dextromethorphan and its metabolite (dextrorphan) or bufuralol and its metabolite (1-hydroxy bufuralol) were extracted by vortexing for 1 minute and centrifuging at 15,000×g for 10 min. After centrifugation, the organic phase was transferred into new tube and dried under nitrogen. The pellet was dissolved in 200 µl of nanopure water, and the products were measured by a liquid chromatography/mass spectrometry (LC-MS/MS) system. The amounts of dextromethorphan, dextrorphan, bufuralol and 1-hydroxy bufuralol were calculated using area peak in comparison with their standard curves.
LC-MS/MS analysis
Quantitative analysis of dextromethorphan, dextrorphan, bufuralol and 1-hydroxy-bufuralol were determined by LC-MS/MS. The instrumentation consisted of LC pumps (Shimadzu, Kyoto, Japan) and an API 4000 mass spectrometer (Applied Biosystems, Foster City, California, USA). Liquid-liquid extraction with methylene chloride was used to clean the samples. Samples (20 µl) were analyzed using a ZORBAX XBD-C8 column (3.5 µm, 4.6×50mm; Agilent Technologies, Santa Clara, California, USA) over a linear gradient from 50% acetonitrile to 100% acetonitrile in 0.1% formic acid for 4 min (dextromethorphan/dextrorphan) or 5 min (bufuralol/1-hydroxy-bufuralol) at a flow rate of 0.4 ml/min. Analytes and internal standards were detected under positive-ion, electrospray ionization conditions and identified via multiple reaction monitoring using the transitions m/z 273.6→216.2 (dextromethorphan), 258.2→157.0 (dextrorphan), 213.6→199.1 (levallorphan), 263.0→189.1 (bufuralol), 278.8→187.2 (1-hydroxybufuralol) and 268.0→74.0 (metoprolol). The rate of metabolite formation was determined using a standard curve for each metabolite.
Data Analysis
The Michaelis constant (Km, µmol/l) and maximum velocity (Vmax, nmol product/pmol P450/min) were determined using the Michaelis-Menten equation (rate of metabolite formation vs. substrate concentration) using PRISM 3 software (GraphPad Software, San Diego, California, USA). Km and Vmax data are the mean ± SEM of three independent experiments, each performed in triplicate. Analysis of variance (ANOVA) followed by post hoc test was used in the statistical analysis. Differences are considered significant if P < 0.05.
Results
Preparation of human CYP2D6
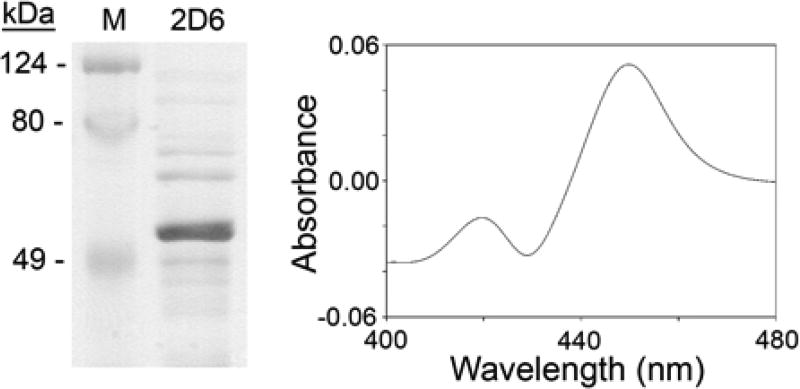
CYP2D6 was expressed in E. coli, purified nearly to homogeneity (Fig. 1, left panel) and quantitated by carbon monoxide reduced difference spectra at 450 nm [31] (Fig. 1, right panel). CYP2D6 expression and purification protocols have been published using yeast or insect cell systems [34–36]; E. coli systems offer the advantages of a more easily maintained cell system and greater yields of enzyme when one modifies the N-terminus to facilitate expression in E. coli [29]. The modified N-terminus, which interacts with the membrane, does not alter catalytic activity [29]. High expression levels in E. coli can yield misfolded, inactive protein; this was circumvented by co-expressing CYP2D6 with the chaperone protein GroES/EL, facilitating proper folding of CYP2D6.
Fig. 1. Expression of human CYP2D6.
Left, Coomassie blue staining of expressed, purified CYP2D6 (prominent band at ~50 kDa). Right, spectrum of the expressed, purified protein; CYP2D6 content was determined from the carbon-monoxide difference spectrum and the molar absorption coefficient for cytochrome P450.
Experimental design and assay optimization
POR is required for steroidogenesis and for drug metabolism. Determining the effects of POR variants on the activities of hepatic CYP enzymes may explain some of the genetic variation in drug metabolism. The forms of POR studied were: WT (encoded by POR*1); Q153R (POR*13), a rare mutant associated with both loss-of-function and gain-of-function in vitro [15, 26]; A287P (POR*5) and R457H (POR*2), which cause most cases of ABS [15, 37]; and A503V (POR*28), the common variant found on 28% of human alleles [23]. Because the ability of an individual POR variant to support catalysis by a specific cytochrome P450 enzyme can vary with both the particular P450 being assayed [15, 26], and with the substrate metabolized by the P450 [27], we assayed the abilities of several POR variants to support CYP2D6 using three different substrates: EOMCC, dextromethorphan and bufuralol. EOMCC was chosen because it is easily assayed and gave excellent activities with CYP1A2 and CYP2C19 in similar assays [26]. Dextromethorphan and bufuralol were chosen as they are selective CYP2D6 substrates that undergo different reactions; dextromethorphan is demethylated whereas bufuralol is hydroxylated. The FDA also recommends that metabolism of dextromethorphan and bufuralol be assessed in in vitro studies of CYP2D6-catalyzed drug disposition (http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm081177.htm#inVitro).
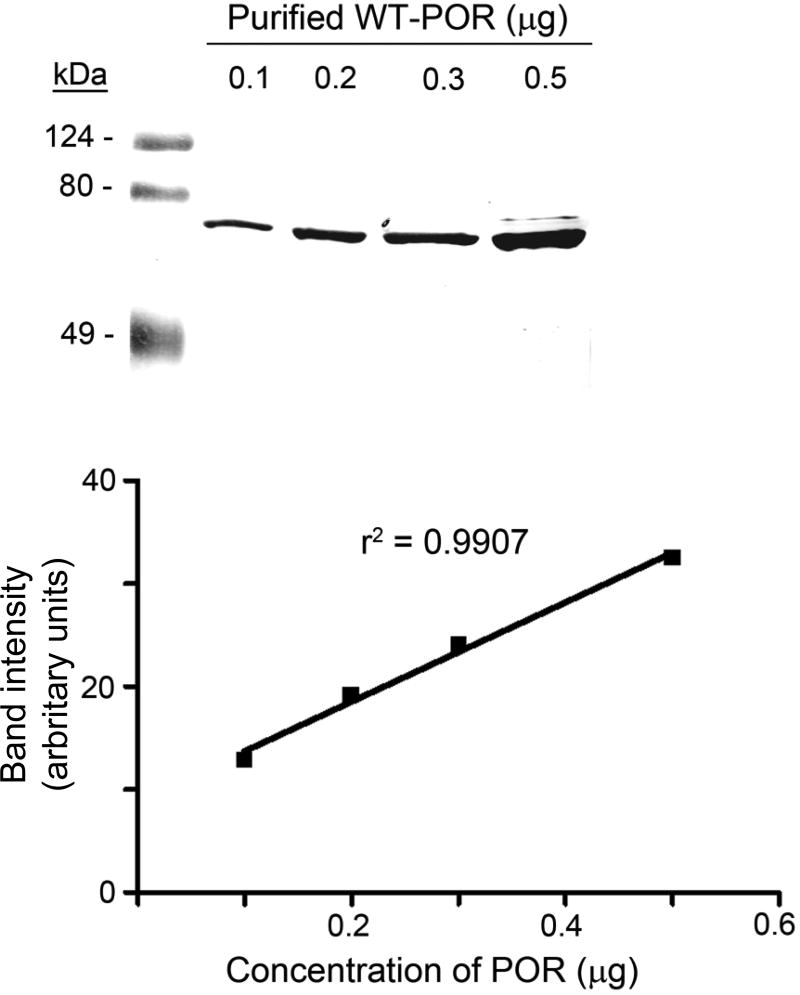
To examine the effects of POR variants on CYP2D6 activities, bacterially-expressed POR and its variants were combined with bacterially expressed CYP2D6 and catalysis of the three substrates was measured. Each POR variant was expressed lacking 27 N-terminal amino acids, as this form facilitates bacterial expression without affecting catalytic activity [15, 23, 26, 38, 39]. Although POR can easily be quantitated by absorbance at 446 nm (the absorbance wavelength for the flavins) [40], some POR sequence variants will affect flavin binding [15] rendering this mode of quantitation unreliable for POR mutants. Therefore, we quantitated our bacterially-expressed POR sequence variants in E.coli membranes by immunoblotting compared to a standard curve of immunoblotted WT POR, as described previously [26] (Fig. 2).
Fig. 2. Quantitation of POR proteins.
The upper panel shows a western blot of 0.1–0.5 µg of purified WT POR displayed on an SDS 10% polyacrylamide gel. The lower panel shows a standard curve plotting the known amount of WT POR protein against band intensity, in arbitrary units. POR mutants run on gels were then quantitated by comparison with this curve.
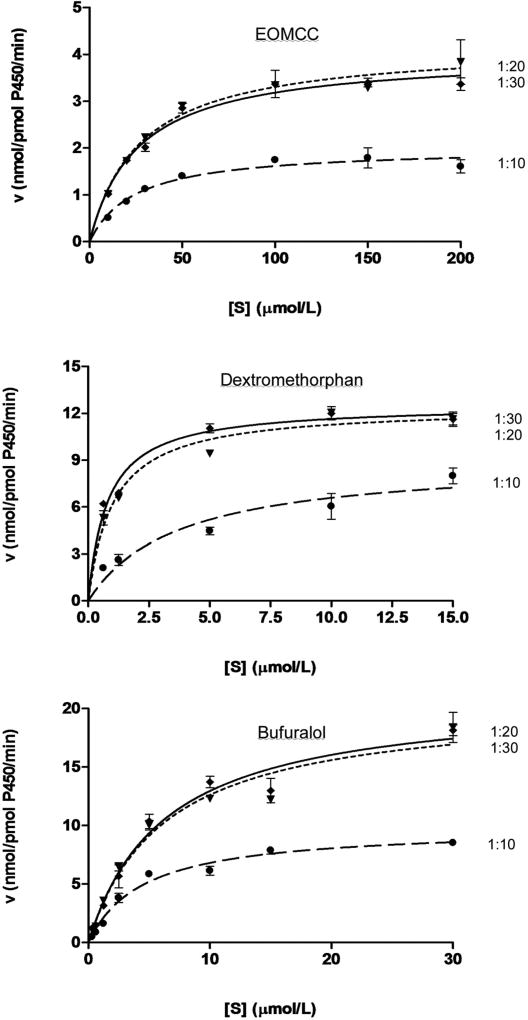
To assess the influence of sequence variants on the ability of POR to support catalysis by CYP2D6, the amount of WT POR should not be limiting; therefore, the concentration of WT POR that will saturate CYP2D6 activity was examined by incubating various amounts of POR with CYP2D6 and examining catalysis (Fig. 3). Irrespective of whether CYP2D6 catalysis was assayed with EOMCC, dextromethorphan or bufuralol as substrate, the reaction was saturated at a CYP:POR molar ratio of 1:20. Therefore, this ratio was used for all subsequent enzymatic assays.
Fig. 3. Optimizing the ratio of CYP:POR.
CYP2D6 was incubated with WT POR at ratios of 1:10, 1:20 and 1:30, and catalysis was examined with EOMCC, dextromethorphan or bufuralol as substrate. Substrate concentrations were plotted against the velocity (nmol/pmol P450/min). Each experiment was done three times in duplicate, and data are depicted as mean ± SEM.
POR support of CYP2D6-mediated metabolism of EOMCC
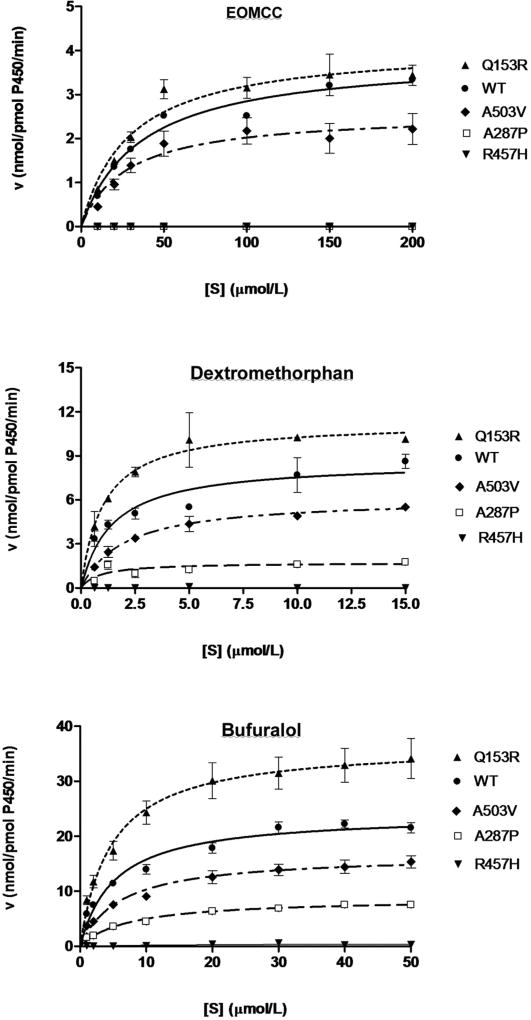
We measured the conversion of EOMCC to 3-cyano-7-hydroxy coumarin by fluorescence spectroscopy. The data from three independent experiments, each performed in triplicate, are displayed as Michaelis-Menten plots of the velocity of metabolite formation against substrate concentrations (Fig. 4, upper panel). The Michaelis constant (Km), maximal velocity (Vmax), catalytic efficiency (Vmax/Km) and catalytic efficiency compared to the efficiency with WT POR were calculated for each reaction, and are summarized in Table 1. Although easily assayable, EOMCC was a poor substrate for CYP2D6, as indicated by its high Km value (35 µmol/l). Compared to WT POR, the A287P and R457H POR mutants had no detectable capacity to support the metabolism of EOMCC by CYP2D6. By contrast, the A503V polymorphism reduced CYP2D6 activity modestly (85%), while the Q153R mutant had activity slightly higher (128%) than WT POR (P<0.05). The Michaelis-Menten plot, which emphasizes the Vmax, suggests that, compared to WT POR, the negative deviation of the A503V mutant is greater than the positive deviation of the Q153R mutant, whereas the calculated Vmax/Km values imply the opposite. This probably reflects the poor binding of EOMCC to CYP2D6, as indicated by the high Km values, so that the impact of the variation in Km is magnified.
Fig. 4. Kinetics of CYP2D6 metabolism of EOMCC, dextromethorphan and bufuralol.
The activities of wild type (WT) and the indicated mutants of POR to support CYP2D6 catalysis was measured with the indicated substrates and are displayed as Michaelis-Menten plots of substrate concentration [S] versus enzymatic velocity, V. Each experiment was done three times in triplicate, and the data are shown as mean ± SEM.
Table 1.
Kinetics of CYP2D6 supported by POR variants, using EOMCC, dextromethorphan and bufuralol as substrates
| EOMCC | ||||
|---|---|---|---|---|
|
| ||||
| POR | Vmax ± SEM | Km ± SEM | Vmax / Km | % WT |
| WT | 3.8 ± 0.50 | 35.1 ± 2.11 | 0.111 | 100 |
| Q153R | 4.2 ± 0.04 | 29.6 ± 3.68 | 0.142 | 128* |
| A503V | 2.6 ± 0.51 | 27.4 ± 3.40 | 0.095 | 85* |
| A287P | NC | NC | NC | NC |
| R457H | NC | NC | NC | NC |
| Dextromethorphan | ||||
|---|---|---|---|---|
|
| ||||
| POR | Vmax ± SEM | Km ± SEM | Vmax / Km | % WT |
| WT | 9.3 ± 0.35 | 1.6 ± 0.67 | 5.813 | 100 |
| Q153R | 13.8 ± 2.27 | 1.2 ± 0.56 | 11.500 | 198* |
| A503V | 6.5 ± 0.86 | 1.8 ± 0.34 | 3.611 | 62* |
| A287P | 2.2 ± 0.41 | 1.4 ± 0.10 | 1.571 | 27* |
| R457H | NC | NC | NC | NC |
| Bufuralol | ||||
|---|---|---|---|---|
|
| ||||
| POR | Vmax ± SEM | Km ± SEM | Vmax / Km | % WT |
| WT | 24.4 ± 1.82 | 5.0 ± 0.26 | 4.880 | 100 |
| Q153R | 35.0 ± 6.18 | 4.7 ± 0.43 | 7.447 | 153* |
| A503V | 16.4 ± 2.29 | 6.4 ± 1.16 | 2.563 | 53* |
| A287P | 8.7 ± 0.46 | 7.5 ± 0.86 | 1.160 | 24* |
| R457H | NC | NC | NC | NC |
Note: The asterisk (*) indicates P < 0.05 for comparisons of WT and variant POR using ANOVA followed by post hoc test. NC = data could not be calculated.
POR support of CYP2D6-mediated metabolism of dextromethorphan
The ability of POR variants to support the conversion of dextromethorphan to dextrorphan by CYP2D6 was assayed by LC-MS/MS (Fig 4, middle panel). Dextromethorphan was an excellent substrate for CYP2D6, with a Km of 1.6 µmol/l. One of the two ABS mutants, R457H, supported no measurable activity, but the ABS mutant A287P supported 27% of WT activity. The A503V polymorphism supported 62% of WT POR activity, and the Q153R mutant showed a clear gain of function, to 198% of WT POR. The catalytic efficiencies of all mutants (Table 1) were statistically different from WT (P<0.05).
POR support of CYP2D6-mediated metabolism of bufuralol
The ability of POR variants to support the conversion of bufuralol to 1-hydroxybufuralol by CYP2D6 was assayed by LC-MS/MS (Fig 4, lower panel). Bufuralol was a good substrate for CYP2D6, with a Km of 5.0 µmol/l, intermediate between that of dextromethorphan and EOMCC. The pattern of POR activities to support the metabolism of bufuralol was essentially indistinguishable from that with dextromethorphan. R457H supported no measurable activity while A287P supported 24% of WT activity. The A503V polymorphism again showed diminished activity, to 53%, and the Q153R mutant again had a gain of activity, to 153%. The catalytic efficiencies of all mutants (Table 1) were statistically different from WT (P<0.05).
Discussion
Pharmacogenetics correlates variations in drug responses with genetic polymorphisms. Pharmacogenetic variation may be at the levels of drug absorption, distribution, metabolism and excretion; the most important of these is metabolism. Approximately 80% of drug metabolism is catalyzed by cytochrome P450 enzymes, especially CYP3A4, CYP2D6 and CYP2C9, which oxidize approximately 70% of currently prescribed drugs [17, 18]. However, the genetic variations in hepatic cytochrome P450 enzymes do not account for all the observed variations in the metabolism of their substrates. For example, CYP3A4 polymorphisms are found in <1% of people, which fails to explain the wide variation in CYP3A4-catalyzed drug metabolism [41, 42]. Although genome-wide association studies of variation in response to clopidogrel show statistically significant influences of genetic variants in CYP2C19, such variants do not account for all of the variation in response to this highly prescribed drug [43]. Because all drug-metabolizing P450 enzymes require POR, polymorphisms in POR are an attractive potential basis for genetic variation in drug metabolism.
To study the potential effects of POR sequence variants, we initially characterized the activities of 35 naturally occurring POR mutants. Each was expressed in E.coli and assayed for activities to oxidize NADPH, reduce cytochrome c, to support the 17α-hydroxylase and 17,20 lyase activities of P450c17 (CYP17), and to support the metabolism of EOMCC by CYP1A2 and CYP2C19 [15, 23, 26]. These studies showed that the activity of a specific POR variant to support the activity of one P450 could not predict its ability to support the activity of a different P450. In our recent studies examining the ability of selected POR mutants to support catalysis by CYP3A4, we found that the activity of a specific POR variant would also vary with the size and geometry of the CYP3A4 substrate [27]. Therefore, the impact of a specific POR mutant must be assayed individually with both the specific P450 of interest and with the specific P450 substrate of interest.
The POR mutation A287P is the most common disease-causing mutation in the European population [15] and R457H is the most common disease-causing mutation in the Japanese population [15, 37]. The phenotypic manifestations in patients homozygous for these mutations are similar [15], although patients homozygous for R457H are more severely affected, as A287P has minimal effects on CYP21 [44] and no effect on CYP19 [28]. In our assays of the ability of these two mutants to support catalysis by CYP2D6, A287P had 24–27% of WT activity to support the metabolism of dextromethorphan and bufuralol, whereas R457H had no detectable activity, and neither mutant had detectable activity in the assay based on EOMCC. In prior assays examining oxidation of NADPH and reduction of cytochrome c, A287P retained 9–16% of WT activity whereas R457H retained no detectable activity [15]; in assays examining the 17α-hydroxylase activity and 17,20 lyase activity of human P450c17, A287P retained 21–40% of WT activities whereas R457H retained <3% activity [15]; in assays examining the activation of EOMCC by CYP1A2 and CYP2C19, neither A287P nor R457H supported detectable activity [26]; in assays of CYP3A4-mediated hydroxylation of testosterone and midazolam, A287P supported 14–17% of WT activity whereas R457H supported no detectable activity [27] and in assays of CYP3A4 mediated metabolism of quinidine and erythromycin, neither mutant had significant activity [27]. Thus both of these POR mutants have lost substantial activity, and the degree of loss varies with the electron recipient assayed, but the R457H mutant is consistently more severely affected than the A287P mutant, consistent with the clinical observations. Thus, while clinically-apparent defects in drug metabolism have not yet been reported in patients carrying these mutations, physicians caring for these patients should consider that their ability to dispose of many drugs may be substantially impaired.
The differences in the activities of A287P and R457H are clarified by their inferred structures. R457 is located in the highly-conserved FAD binding domain (residues 453–495) and forms a hydrogen bond with the pyrophosphate group of FAD [5]. By contrast, A287 is located below the FAD-binding region, and may affect FAD binding and possibly influence the interaction of POR with some P450s. A287 is located at the start of a loop that interacts with the C-terminal loop of P450c17. The change from alanine to proline breaks the POR loop so that it will no longer interact with P450c17 [28]. By contrast, A287P does not effect the interaction of POR with P450aro because that interaction involves different POR residues [28]. Alternatively, it has been suggested that A287P is located adjacent to a motif rich in proline and phenylalanine residues at residues 227–286 that also contains many charged residues and hence interacts with negative charges on phospholipid head groups of the endoplasmic reticulum [44]. This model suggests that this region is involved in membrane attachment of POR, and that A287P would induce a conformational change in 227–286, which would disorder the attachment of POR to the endoplasmic reticulum. The altered membrane attachment could alter the access of some enzymes to CYP docking site of POR. Present data do not permit one to discriminate between these two models.
The Q153R mutant was studied because it is the only known POR gain-of-function mutant. Q153R was initially found in a patient with Antley-Bixler Syndrome and disordered steroidogenesis; consistent with this, Q153R had only ~10% of WT activity in assays of NADPH oxidation and cytochrome c reduction, and 27–31% of WT activity in the two assays of P450c17 activity [13, 15]. However in the assays of CYP1A2 and CYP2C19 to metabolize EOMCC it had 144% and 284% of WT activity, respectively [26] and in assays of CYP3A4 activities it supported 76–150% of WT activity, depending on the CYP3A4 substrate being assayed [27]. Similarly to its activities with CYP1A2, 2C19 and 3A4, we found that Q153R supported 128–198% of WT activity with CYP2D6. Models of human POR indicate that Q153 is surface-exposed in the electron-donating domain, a highly conserved region near the FMN binding site [15, 45]. The Q153R mutation changes the electrostatic charge, possibly affecting electron donation to CYP enzyme. Thus there are great differences in the activities of some POR mutants depending on the electron recipient assayed. Further structural studies will be needed to determine why Q153R exhibits such different behavior with different P450s.
A503V is a common variant found in the POR alleles of 19.1% of African-Americans, 26.4% of Caucasians, 31% of Mexican-Americans, and 36.7% of Chinese-Americans [23]. A503V has moderately decreased function for NADPH oxidation, cytochrome c reduction, and the 17α-hydroxylase and 17,20 lyase activities of P450c17 [15, 23], but no significant influence on the 21-hydroxylase activities of P450c21 (CYP21) [25], or on the activities of CYP1A2 and CYP2C19 to metabolize EOMCC [26]. A503V reduced the ability of CYP3A4 to hydroxylate testosterone and midazolam to 61–74% of WT, but had little effect on the CYP3A4-mediated metabolism of quinidine or erythromycin [27]. We now find that A503V inhibits the ability of POR to support the metabolism of dextromethorphan and bufuralol by CYP2D6 to 62% and 53% of WT activity, respectively, while reducing the metabolism of EOMCC modestly to 85% of WT. The mechanism by which A503V inhibits POR activity with some P450 enzymes is not known. Modeling of human POR indicates that A503 is in the FAD-binding domain [15, 45], but the change of an alanine residue to a valine is very conservative. Crystallographic studies of human WT and A503V POR will be needed to determine how this common variant affects POR structure and function.
Our results confirm that the activity of a POR variant with one P450 may not predict its activity with another P450. Therefore, the impact of a particular POR mutant needs to be tested individually with each P450. Moreover, the A503V polymorphism has moderately impaired activity with CYP2D6 and CYP3A4, possibly contributing to genetic variability in drug and xenobiotic metabolism. CYP variants that have not lost function when tested with wild-type POR may have reduced activity with A503V POR. CYP2D6, CYP2C9 and CYP2C19 are polymorphic, having high frequency variants, including several coding region variants, with reduced activities [18, 46], and both metabolize many clinically-used drugs [17, 46]; hence it will be important to study the effects of A503V POR on these CYP variants. There is an imperfect correlation between the hypomorphic genotypes of these CYP enzymes and in vivo drug metabolism; genetic variants of POR, such as A503V, are good candidates for modifier genes, which contribute to variations in metabolism.
Acknowledgments
This work was supported by NIH grant R01 GM073020 to WLM and R01 GM069753 to TST.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nelson DR. Comparison of P450s from human and fugu: 420 million years of vertebrate P450 evolution. Arch Biochem Biophys. 2003;409:18–24. doi: 10.1016/s0003-9861(02)00553-2. [DOI] [PubMed] [Google Scholar]
- 2.Guengerich FP. Cytochrome P450: what have we learned and what are the future issues? Drug Metab Rev. 2004;36:159–197. doi: 10.1081/dmr-120033996. [DOI] [PubMed] [Google Scholar]
- 3.Miller WL. Minireview: regulation of steroidogenesis by electron transfer. Endocrinology. 2005;146:2544–2550. doi: 10.1210/en.2005-0096. [DOI] [PubMed] [Google Scholar]
- 4.Scott RR, Gomes LG, Huang N, Van Vliet G, Miller WL. Apparent manifesting heterozygosity in P450 oxidoreductase deficiency and its effect on coexisting 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2007;92:2318–2322. doi: 10.1210/jc.2006-2345. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Roberts DL, Paschke R, Shea TM, Masters BS, Kim JJ. Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes. Proc Natl Acad Sci U S A. 1997;94:8411–8416. doi: 10.1073/pnas.94.16.8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard PA, Shen AL, Paschke R, Kasper CB, Kim JJ. NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer. J Biol Chem. 2001;276:29163–29170. doi: 10.1074/jbc.M101731200. [DOI] [PubMed] [Google Scholar]
- 7.Ellis J, Gutierrez A, Barsukov IL, Huang WC, Grossmann JG, Roberts GC. Domain motion in cytochrome P450 reductase: conformational equilibria revealed by NMR and small-angle x-ray scattering. J Biol Chem. 2009;284:36628–36637. doi: 10.1074/jbc.M109.054304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen AL, O'Leary KA, Kasper CB. Association of multiple developmental defects and embryonic lethality with loss of microsomal NADPH-cytochrome P450 oxidoreductase. J Biol Chem. 2002;277:6536–6541. doi: 10.1074/jbc.M111408200. [DOI] [PubMed] [Google Scholar]
- 9.Otto DM, Henderson CJ, Carrie D, Davey M, Gundersen TE, Blomhoff R, et al. Identification of novel roles of the cytochrome P450 system in early embryogenesis: effects on vasculogenesis and retinoic Acid homeostasis. Mol Cell Biol. 2003;23:6103–6116. doi: 10.1128/MCB.23.17.6103-6116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, et al. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem. 2003;278:13480–13486. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- 11.Gu J, Weng Y, Zhang QY, Cui H, Behr M, Wu L, et al. Liver-specific deletion of the NADPH-cytochrome P450 reductase gene: impact on plasma cholesterol homeostasis and the function and regulation of microsomal cytochrome P450 and heme oxygenase. J Biol Chem. 2003;278:25895–25901. doi: 10.1074/jbc.M303125200. [DOI] [PubMed] [Google Scholar]
- 12.Finn RD, McLaren AW, Carrie D, Henderson CJ, Wolf CR. Conditional deletion of cytochrome P450 oxidoreductase in the liver and gastrointestinal tract: a new model for studying the functions of the P450 system. J Pharmacol Exp Ther. 2007;322:40–47. doi: 10.1124/jpet.107.121780. [DOI] [PubMed] [Google Scholar]
- 13.Fluck CE, Tajima T, Pandey AV, Arlt W, Okuhara K, Verge CF, et al. Mutant P450 oxidoreductase causes disordered steroidogenesis with and without Antley-Bixler syndrome. Nat Genet. 2004;36:228–230. doi: 10.1038/ng1300. [DOI] [PubMed] [Google Scholar]
- 14.Antley R, Bixler D. Trapezoidocephaly, midfacial hypoplasia and cartilage abnormalities with multiple synostoses and skeletal fractures. Birth Defects Orig Artic Ser. 1975;11:397–401. [PubMed] [Google Scholar]
- 15.Huang N, Pandey AV, Agrawal V, Reardon W, Lapunzina PD, Mowat D, et al. Diversity and function of mutations in P450 oxidoreductase in patients with Antley-Bixler syndrome and disordered steroidogenesis. Am J Hum Genet. 2005;76:729–749. doi: 10.1086/429417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertz RJ, Granneman GR. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokinet. 1997;32:210–258. doi: 10.2165/00003088-199732030-00004. [DOI] [PubMed] [Google Scholar]
- 18.Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- 19.Sachse C, Brockmoller J, Bauer S, Roots I. Cytochrome P450 2D6 variants in a Caucasian population: allele frequencies and phenotypic consequences. Am J Hum Genet. 1997;60:284–295. [PMC free article] [PubMed] [Google Scholar]
- 20.Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith RL. Polymorphic hydroxylation of Debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- 21.Dalen P, Dahl ML, Eichelbaum M, Bertilsson L, Wilkinson GR. Disposition of debrisoquine in Caucasians with different CYP2D6-genotypes including those with multiple genes. Pharmacogenetics. 1999;9:697–706. doi: 10.1097/01213011-199912000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Dorado P, Berecz R, Caceres MC, Gonzalez I, Cobaleda J, Llerena A. Determination of debrisoquine and 4-hydroxydebrisoquine by high-performance liquid chromatography: application to the evaluation of CYP2D6 genotype and debrisoquine metabolic ratio relationship. Clin Chem Lab Med. 2005;43:275–279. doi: 10.1515/CCLM.2005.046. [DOI] [PubMed] [Google Scholar]
- 23.Huang N, Agrawal V, Giacomini KM, Miller WL. Genetics of P450 oxidoreductase: sequence variation in 842 individuals of four ethnicities and activities of 15 missense mutations. Proc Natl Acad Sci U S A. 2008;105:1733–1738. doi: 10.1073/pnas.0711621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sim SC, Miller WL, Zhong XB, Arlt W, Ogata T, Ding X, et al. Nomenclature for alleles of the cytochrome P450 oxidoreductase gene. Pharmacogenet Genomics. 2009;19:565–566. doi: 10.1097/FPC.0b013e32832af5b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomes LG, Huang N, Agrawal V, Mendonca BB, Bachega TA, Miller WL. The common P450 oxidoreductase variant A503V is not a modifier gene for 21-hydroxylase deficiency. J Clin Endocrinol Metab. 2008;93:2913–2916. doi: 10.1210/jc.2008-0304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Agrawal V, Huang N, Miller WL. Pharmacogenetics of P450 oxidoreductase: effect of sequence variants on activities of CYP1A2 and CYP2C19. Pharmacogenet Genomics. 2008;18:569–576. doi: 10.1097/FPC.0b013e32830054ac. [DOI] [PubMed] [Google Scholar]
- 27.Agrawal V Choi JH, Giacomini KM, Miller WL. Substrate-specific modulation of CYP3A4 activity by genetic variants of cytochrome P450 oxidoreductase (POR) Pharmacogenet Genomics. 2010 doi: 10.1097/FPC.0b013e328330cb5. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pandey AV, Kempna P, Hofer G, Mullis PE, Flück CE. Modulation of human CYP19A1 activity by mutant NADPH P450 oxidoreductase. Mol Endocrinol. 2007;21:2579–2595. doi: 10.1210/me.2007-0245. [DOI] [PubMed] [Google Scholar]
- 29.Barnes HJ, Arlotto MP, Waterman MR. Expression and enzymatic activity of recombinant cytochrome P450 17 α-hydroxylase in Escherichia coli. Proc Natl Acad Sci U S A. 1991;88:5597–5601. doi: 10.1073/pnas.88.13.5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Halkier BA, Nielsen HL, Koch B, Moller BL. Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch Biochem Biophys. 1995;322:369–377. doi: 10.1006/abbi.1995.1477. [DOI] [PubMed] [Google Scholar]
- 31.Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 32.Marks BD, Smith RW, Braun HA, Goossens TA, Christenson M, Ozers MS, et al. A high throughput screening assay to screen for CYP2E1 metabolism and inhibition using a fluorogenic vivid P450 substrate. Assay Drug Dev Technol. 2002;1:73–81. doi: 10.1089/154065802761001329. [DOI] [PubMed] [Google Scholar]
- 33.Backes WL, Kelley RW. Organization of multiple cytochrome P450s with NADPH-cytochrome P450 reductase in membranes. Pharmacol Ther. 2003;98:221–233. doi: 10.1016/s0163-7258(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 34.Deeni YY, Paine MJ, Ayrton AD, Clarke SE, Chenery R, Wolf CR. Expression, purification, and biochemical characterization of a human cytochrome P450 CYP2D6-NADPH cytochrome P450 reductase fusion protein. Arch Biochem Biophys. 2001;396:16–24. doi: 10.1006/abbi.2001.2585. [DOI] [PubMed] [Google Scholar]
- 35.Yu A, Kneller BM, Rettie AE, Haining RL. Expression, purification, biochemical characterization, and comparative function of human cytochrome P450 2D6.1, 2D6.2, 2D6.10, and 2D6.17 allelic isoforms. J Pharmacol Exp Ther. 2002;303:1291–1300. doi: 10.1124/jpet.102.039891. [DOI] [PubMed] [Google Scholar]
- 36.Peters FT, Dragan CA, Kauffels A, Schwaninger AE, Zapp J, Bureik M, et al. Biotechnological synthesis of the designer drug metabolite 4'-hydroxymethyl-α-pyrrolidinohexanophenone in fission yeast heterologously expressing human cytochrome P450 2D6--a versatile alternative to multistep chemical synthesis. J Anal Toxicol. 2009;33:190–197. doi: 10.1093/jat/33.4.190. [DOI] [PubMed] [Google Scholar]
- 37.Fukami M, Horikawa R, Nagai T, Tanaka T, Naiki Y, Sato N, et al. Cytochrome P450 oxidoreductase gene mutations and Antley-Bixler syndrome with abnormal genitalia and/or impaired steroidogenesis: molecular and clinical studies in 10 patients. J Clin Endocrinol Metab. 2005;90:414–426. doi: 10.1210/jc.2004-0810. [DOI] [PubMed] [Google Scholar]
- 38.Dierks EA, Davis SC, Ortiz de Montellano PR. Glu-320 and Asp-323 are determinants of the CYP4A1 hydroxylation regiospecificity and resistance to inactivation by 1-aminobenzotriazole. Biochemistry. 1998;37:1839–1847. doi: 10.1021/bi972458s. [DOI] [PubMed] [Google Scholar]
- 39.Nicolo C, Flück CE, Mullis PE, Pandey AV. Restoration of mutant cytochrome P450 reductase activity by external flavin. Mol Cell Endocrinol. 2010;321:245–252. doi: 10.1016/j.mce.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 40.Hinkson JW. Azotobacter free-radical flavoprotein. Preparation and properties of the apoprotein. Biochemistry. 1968;7:2666–2672. doi: 10.1021/bi00847a033. [DOI] [PubMed] [Google Scholar]
- 41.Hirth J, Watkins PB, Strawderman M, Schott A, Bruno R, Baker LH. The effect of an individual's cytochrome CYP3A4 activity on docetaxel clearance. Clin Cancer Res. 2000;6:1255–1258. [PubMed] [Google Scholar]
- 42.Mizutani T. PM frequencies of major CYPs in Asians and Caucasians. Drug Metab Rev. 2003;35:99–106. doi: 10.1081/dmr-120023681. [DOI] [PubMed] [Google Scholar]
- 43.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dhir V, Ivison HE, Krone N, Shackleton CH, Doherty AJ, Stewart PM, et al. Differential inhibition of CYP17A1 and CYP21A2 activities by the P450 oxidoreductase mutant A287P. Mol Endocrinol. 2007;21:1958–1968. doi: 10.1210/me.2007-0066. [DOI] [PubMed] [Google Scholar]
- 45.Fluck CE, Mullis PE, Pandey AV. Modeling of human P450 oxidoreductase structure by in silico mutagenesis and MD simulation. Mol Cell Endocrinol. 2009;313:17–22. doi: 10.1016/j.mce.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003;348:538–549. doi: 10.1056/NEJMra020526. [DOI] [PubMed] [Google Scholar]