Abstract
Glycosylation is one of the most common and essential protein modifications. Glycans conjugated to biomolecules modulate the function of such molecules through both direct recognition of glycan structures and indirect mechanisms that involve the control of protein turnover rates, stability, and conformation. The biological attributes of glycans in numerous biological processes and implications in a number of diseases highlight the necessity for comprehensive characterization of protein glycosylation. This chapter review cutting-edge-methods and tools developed to facilitated quantitative glycomics. The chapter highlight the different methods employed for the release and purification of glycans from biological samples. The most effective labeling methods developed for sensitive quantitative glycomics are also described and discussed. The chromatographic approaches that have been used effectively in glycomics are also highlighted.
Keywords: Glycomics, Release of Glycans, Purification, Chromatographic Methods, Labeling of Glycans, Biomedical Applications
14.1 Protein Glycosylations
Glycosylation is one of the most common and essential protein modifications. More than half of proteins are glycosylated (Apweiler, Hermjakob, & Sharon, 1999). Glycans conjugated to biomolecules modulate the function of such molecules through both direct recognition of glycan structures and indirect mechanisms that involve the control of protein turnover rates, stability, and conformation. The biological attributes of glycans in numerous biological processes and implications in a number of diseases highlight the necessity for comprehensive characterization of protein glycosylation.
14.1.1 Definition of Glycosylation
Proteins are a biomolecule biosynthesized utilizing 20 natural amino acids. Shortly after or during the biosynthesis of proteins, several modifications may occur to amino acid residues. These modifications are referred to as posttranslational modification (PTM). PTM chemically or enzymatically alters specific amino acid residues, the properties, and functions of proteins are altered as a consequence. Among more than 200 types of PTMs, acetylation, methylation, phosphorylation, ubiquitination, lipidation, proteolysis and glycosylation occur most commonly. Glycosylation refers to modifications that commonly add carbohydrate molecules to asparagine and serine or threonine in the polypeptide backbone of proteins. This process is distinct from other common PTMs because of two facts, first is the molecular weight shift by glycosylation is much larger (usually greater than 800 Dalton) than other PTMs (phosphorylation adds 80 Dalton, acetylation adds 42 Dalton and methylation adds 14 Dalton to the modified amino acid); the second is the uncertainty of the added carbohydrate structure, since this process is a template-free process. This structural heterogeneity contributes to the diverse biofunctions of glycoconjugates, while simultaneously increasing the difficulty of achieving a comprehensive characterization of such protein modification.
14.1.2 Classification of Protein Glycosylation
Based on the occurrence site and core structure, glycosylation can be classified as N-linked glycosylation or O-linked glycosylation. N-linked glycosylation occurs on asparagine in a sub-sequence of Asn-Xxx-Ser/Thr, where Xxx refers to any amino acid except proline. There are several steps in the biosynthesis of N-glycans. N-glycosylation is initiated on the cytosolic surface of the endoplasmic reticulum (ER) membrane, where a lipid precursor acts as the carrier for immature glycans (Kornfeld & Kornfeld, 1985). In this process, glycans composed of two N-acetylglucosamine and five mannose residues are synthesized and anchored to the lipid precursor. After which, the oligosaccharide is flipped to the luminal side of the membrane, where four mannose and three glucose units are added to the glycan structure. The formation of this structure indicates that the glycan is ready to be transferred from the lipid precursor to the asparagine on the polypeptide chain by oligosaccharyltransferase (Lizak, Gerber, Numao, Aebi, & Locher, 2011). Terminal glucose and mannose are subsequently trimmed in the ER (Helenius & Aebi, 2004). When the glycoprotein reaches the Golgi complex, glycans are remodeled into diverse structures by different glycosidases and glycosyltransferases, while the core structure of the N-glycan that consists of two N-acetylglucosamine and three mannose units is always kept untouched (Rabouille, Hui, Hunte, Kieckbusch, Berger, Warren et al., 1995; Stanley, 2011). Monosaccharides that commonly compose N-glycans are N-acetylglucosamine, N-acetylgalactosamine, mannose, galactose, fucose and neuraminic acids. The biosynthesis of O-linked glycans is less complex than that of N-glycans. Synthesis starts by direct modification of polypeptide chain Ser/Thr residues in the Golgi apparatus (Clausen & Bennett, 1996; Ten Hagen, Fritz, & Tabak, 2003). However, O-glycan structures are also highly differentiated because of trimming and addition by various glycosidases and glycosyltransferases (Gill, Clausen, & Bard, 2011).
N-glycans can be classified into high mannose, complex and hybrid types according to their structures. High mannose refers to glycans that only have mannose residues beyond the core structure, while the complex type has other monosaccharides. Hybrid glycans have bi-antennary structures with characteristics of high mannose and complex type on each branch. Due to the unique biological functions of fucose and sialic acid, glycans can also be classified as fucosylated and sialylated types when they contain corresponding monosaccharides.
14.1.3 Glycan Biofunctions and Relationships to Diseases
Glycosylation is a type of PTM that adds significant mass to the polypeptide backbone, sometimes glycans cover a major portion of the protein surface. Hence, it is not surprising that glycosylation has significant impacts on protein structure and properties. The functions of protein glycosylation can be observed in a variety of ways. First, glycans may interfere with protein folding (Freeze, Esko, & Parodi, 2009; Molinari, 2007); during the protein folding process, glycans act as quality control and checkpoints for protein folding and determine whether the protein can be folded in the luminal side of the ER or be degraded in the cytosolic side (Helenius et al., 2004; Lederkremer, 2009). Second, glycans influence the stability and solubility of folded proteins; glycans play roles in protecting polypeptides from proteolysis by hindering the accessibility of proteases to peptide bonds (Ellgaard, Molinari, & Helenius, 1999; Means & Desrosiers, 2000; Trombetta & Helenius, 1998). Moreover, glycans mediate protein-protein interactions. Glycans with certain structures may also act as binding targets for pathogens and microbes.
Although only 1 to 2% of the genome is responsible for encoding glycan-related enzymes (Lowe & Marth, 2003), glycan patterns and abundance are altered significantly in various human diseases such as skin diseases (Vabres, Sevin, Amoric, Odievre, Saudubray, & de Prost, 1998), diabetes (Miura & Endo, 2016), rheumatoid arthritis (Gornik & Lauc, 2008), liver diseases (Mehta, Herrera, & Block, 2015) and numerous cancers (Cheresh, Reisfeld, & Varki, 1984; Dall’Olio & Chiricolo, 2001; Kohla, Stockfleth, & Schauer, 2002; Laubli, Stevenson, Varki, Varki, & Borsig, 2006; Seales, Jurado, Brunson, Wakefield, Frost, & Bellis, 2005; Stevenson, Choi, & Varki, 2005; N. M. Varki & Varki, 2002). Glycans have been found to play roles in cell recognition and adhesion (Bucior, Scheuring, Engel, & Burger, 2004), these capabilities are responsible for cancer progression and migration. Various studies have been conducted to investigate the relationships between glycan expression and cancer progression, and several structures of glycans have been identified as putative biomarkers for tumor development (Varki, Kannagi, & Toole, 2009).
The importance of obtaining a comprehensive understanding of protein glycosylation can be concluded from the examples discussed above. Hence, developing sensitive and reliable strategies for glycomic analysis is decidedly necessary. In the following paragraphs, different glycomic platforms will be reviewed and compared.
14.2 Sample handling in Glycomics
Glycans that are linked to proteins are generally oriented such that they either extend outward from the protein surface or are embraced inside of the folded protein. Regardless of their placement on the protein molecule, sample treatments are always required for their analysis. In the field of glycomics, glycans are typically released from heat denatured proteins or peptides prior to analysis. In the sections that follow, a comprehensive list of the methods utilized to release glycans is provided.
14.2.1 Glycan release
Although glycans are analyzed together with the peptide backbone in glycoproteomics, studies to acquire site specific glycosylation information are not common, largely because they require sophisticated tandem mass spectrometers and complex data processing efforts. Releasing glycans from glycoproteins benefits their analysis by increasing sensitivity and providing simplicity. Glycan releasing methods differ between N-linked and O-linked glycans, and can be classified into chemical and enzymatic based reactions.
The most commonly utilized N-glycan releasing approach is cleavage of glycans from asparagine by enzymatic digestion with peptide N-glycosidase F (PNGase F), which can be isolated from Flavobacterium meningosepticum (Plummer & Tarentino, 1991). PNGase F is capable of selectively and efficiently releasing all N-linked glycans from peptide backbones except for those containing core α 1-3 linked fucose, which usually do not exist in mammalian glycans. In the case of plants and insects, PNGase A should be employed to release glycans with core α 1-3 linked fucose (Takahashi & Nishibe, 1978; Tretter, Altmann, & Marz, 1991). However, PNGase A is not sensitive to sialylated glycans. After PNGase F treatment, asparagine in the peptide backbone is converted into aspartic acid, making identification of N-glycosylation sites possible. Additional enzymes, such as endo -F1, -F2, -F3 and –H, are also capable of releasing N-glycans (Trimble & Tarentino, 1991). Unlike PNGase F, the endo F series and endo H only cleave N-glycans containing particular structures and leave one N-acetylglucosamine on the peptide backbone. These specific N-glycan releasing enzymes assist addressing specific scientific questions, such as selecting or depleting high mannose glycans in complex samples. N-glycans can also be released by chemical reactions; however, chemical cleavage relies on high temperature and pH, which tends to prompt glycan cleavages and formation of by-products.
There are no universal and efficient enzymes for releasing O-linked glycans. For example, endo-α-N-acetylgalactosaminidase only cleaves O-glycans with a core 1 structure (Huang & Aminoff, 1972). Therefore, in the case of O-glycans, chemical release methods are more efficient. Reductive β-elimination is the most common method of releasing O-glycans (Carlson, 1968). This reaction can be achieved by mixing a weak base (e.g. ammonium hydroxide) with an O-glycoprotein prior to an overnight incubation at 60 °C in a water bath. However, side reactions during this procedure are inevitable, and site information is lost. Several studies have sought to optimize the reaction conditions for β elimination. For example, in one recent study, pyrazolone analogues (BEP) were utilized, and the reaction was accelerated by microwave radiation (Furukawa, Piao, Yoshida, Okada, Yokota, Higashino et al., 2015). BEP improved reaction selectivity and reduced the frequency of peeling reactions commonly observed in conventional β elimination methods. The assistance of a microwave also allowed reaction time to be reduced from 16 hours to 2 hours. The different methods available for releasing glycans are summarized in Table 14.1.
Table 14.1.
Release of glycans from glycoproteins is attained through different methods.
| N-glycan | Chemical Release | |
| Using anhydrous hydrazine at 95 °C | (Takasaki, Mizuochi, & Kobata, 1982) | |
| Oxidation using sodium hypochlorite | (Song, Ju, Lasanajak, Kudelka, Smith, & Cummings, 2016) | |
| Enzymatic Release | ||
| PNGase F purified from Flavobacterium meningosepticum | (Takahashi et al., 1978; Tarentino, Gomez, & Plummer, 1985) | |
| PNGase A purified from almond Prunus amygdalus var. dulcis | (Takahashi et al., 1978; Tretter et al., 1991) | |
| Endoglycosidase F1 purified from Elizabethkingia miricola | (Trimble et al., 1991) | |
| Endoglycosidase F2 purified from Elizabethkingia miricola | (Trimble et al., 1991) | |
| Endoglycosidase F3 purified from Elizabethkingia miricola | (Trimble et al., 1991) | |
| Endoglycosidase H purified from Streptomyces plicatus | (Tarentino & Maley, 1974; Tarentino, Plummer, & Maley, 1974) | |
| Endo-β-N-acetylglucosaminidase (Endo-M) purified from Mucor hiemalis | (Kadowaki, Yamamoto, Fujisaki, Izumi, Tochikura, & Yokoyama, 1990) | |
| O-glycan | Chemical Release | |
| Using anhydrous hydrazine at 60 °C | (Patel, Bruce, Merry, Bigge, Wormald, Jaques et al., 1993) | |
| Alkaline-β-elimination | (Carlson, 1968) | |
| Non-reductive β-elimination | (Hulsmeier, Gehrig, Geyer, Sack, Gottstein, Deplazes et al., 2002) | |
| Oxidation using sodium hypochlorite | (Song et al., 2016) | |
| Enzymatic Release | ||
| Endo-α-N-acetylgalactosaminidase | (Huang et al., 1972) | |
| Enzymatic/Chemical Release | ||
| Pronase followed by solid phase permethylation | (Goetz, Novotny, & Mechref, 2009) | |
It is worth mentioning that although most glycosidase enzymes work with intact native protein, enzymatic digestion efficiency is much lower compared to protocols in which glycoproteins are denatured prior to digestion. For example, the effectiveness of PNGase F digestion is 100 times higher when denaturation is conducted to pre-treat protein samples. The benefits of effective protein denaturation include accelerating enzymatic digestion, reducing enzyme consumption and ensuring complete release of all glycans, especially when the analyte is a complex biological sample such as serum or tissue. Thermal denaturation and detergent denaturation are two commonly utilized strategies. Although detergents improve protein extraction from complex biological samples and ensure the disruption of 3-D protein structures, most detergents are not MS-friendly and are difficult to remove. Hence, thermal denaturation or MS-compatible detergents such as RapiGest™ are more widely used in MS-based glycomics.
14.2.2 Glycan Purification
After enzymatic digestion, glycans are in solutions containing a mix of proteins, peptides, salts and possibly detergents. Therefore, purification is necessary to eliminate inference from the sample matrix in the derivatization and analytical procedures that follow. Protein precipitation is commonly utilized to remove proteins from sample mixtures; high concentration methanol, ethanol or aqueous acetone solutions are added to samples and incubation at −80 °C follows, the precipitated proteins can then be isolated by high speed centrifugation. Dialysis or filtration methods, based on the molecular weight difference between glycans and their contaminants, can be used to separate glycans from buffer salts or detergents.
Solid phase extraction (SPE) is another efficient method for extracting glycans from digested mixtures according to differences in hydrophobicity. Commonly used SPE materials are C18, and charcoal, both of which have high selectivity for glycans. The affinity of charcoal for carbohydrate structures enables the separation of different types of glycans, which has been demonstrated by its use for glycan fractionation in some studies. However, glycans are not completely trapped by or eluted out of SPE stationary phase materials which results in sample loss, the major drawback of SPE purification.
14.2.3 Glycan Derivatization
Most glycomic analysis strategies require derivatization of released glycans. The advantages of derivatizing glycans can be reflected in the following aspects. Unlike proteins or peptides, native glycans are not suitable for optical detection methods, labeling glycans with tags containing chromophores or fluorophores can make glycans detectable by UV or laser induced fluorescence (LIF). Native glycans, especially acidic species, are hydrophilic and have low proton affinity, resulting in low ionization efficiency in positive mode MS analysis. Increased hydrophobicity through derivatization improves the detection sensitivity of glycans in MS analysis. Furthermore, native sialylated and fucosylated glycans are not stable during ionization and MS analysis. Hence, derivatization is necessary in order to stabilize these glycan structures. A variety of derivatization methods that are employed to simultaneously improve glycan detection and separation efficiency in capillary electrophoresis (CE) and various types of LC will be discussed in the sections that follow.
14.2.3.1 Derivatization through Reductive Amination
The unique chemical properties of the reducing end of glycans make it an ideal site for the attachment of labeling tags. The most commonly utilized reducing end labeling reaction is reductive amination. The reaction occurs between an aldehyde on the glycan reducing end and an amine from the labeling tag (Figure 14.1A). The intermediate imine is quickly reduced in the presence of a reducing reagent in the reaction mixture. The reaction is usually accomplished in dimethyl sulfoxide (Bigge, Patel, Bruce, Goulding, Charles, & Parekh, 1995), tetrahydrofuran (Evangelista, Guttman, & Chen, 1996) or methanol (Anumula, 1994) with the presence of acids such as acetic acid and citric acid (Evangelista et al., 1996; Szabo, Guttman, Rejtar, & Karger, 2010). Sodium cyanoborohydride is most frequently used as the reducing reagent in this reaction, although recently 2-picoline-borane was reported as a non- toxic reducing reagent for glycan labeling (Ruhaak, Steenvoorden, Koeleman, Deelder, & Wuhrer, 2010). Common labeling reagents are anthranilic acid (2-AA) (Anumula, 1994), 2‑aminobenzamide (2-AB) (Bigge et al., 1995) and 2-aminopyridine (2-AP) (Hase, Ikenaka, & Matsushima, 1978), all of which have fluorophores enabling LIF detection after HPLC separation. 9-Aminopyrene-1, 4, 6-trisulfonic acid (APTS) (Guttman, Chen, & Evangelista, 1996) and 8-Aminonaphthalene-1, 3, 6-trisulfonic acid (ANTS) (Chiesa & Horvath, 1993) are regularly utilized to induce fixed charges to glycans so that they can be separated by CE. An additional function of these labeling reagents is to improve ionization efficiency and glycan stability in both matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI). Several other labeling reagents have also been reported for quantitative glycomics, including 2-aminoacridone (AMAC) (Charlwood, Birrell, Organ, & Camilleri, 1999), 4-aminobenzonitrile (ABN) (Schwaiger, Oefner, Huber, Grill, & Bonn, 1994), 3-aminobenzoic acid (3-AA) (Nakajima, Oda, Kinoshita, Masuko, & Kakehi, 2002) and 3-aminobenzamide (3-AB) (Kakehi, Funakubo, Suzuki, Oda, & Kitada, 1999).
Figure 14.1.
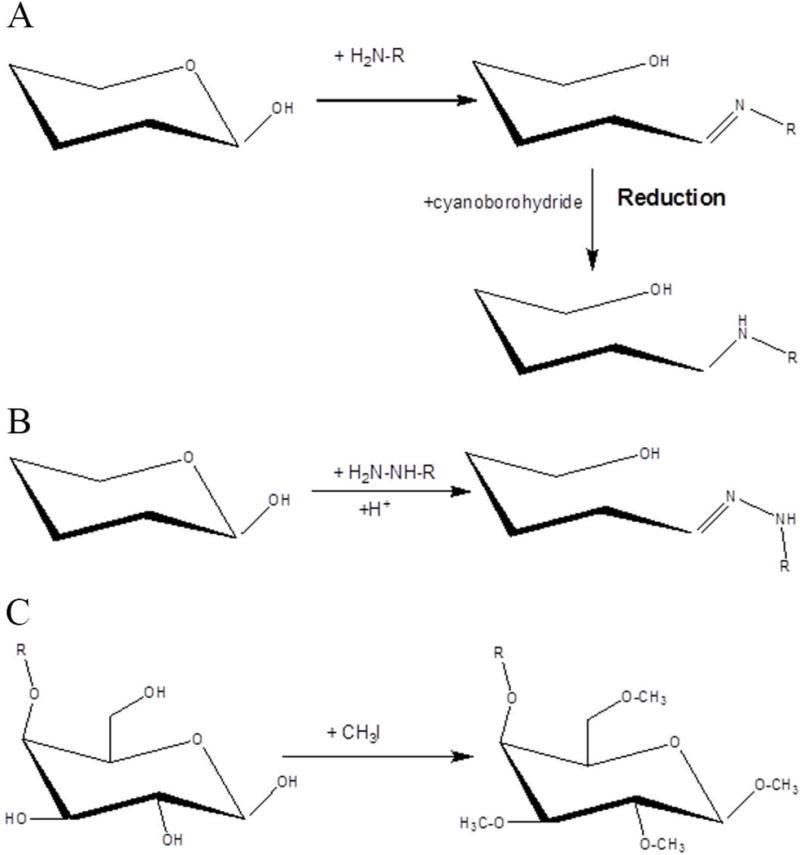
Derivatization by (A) reductive amination, (B) hydrazide derivatization and (C) permethylation.
Reductive amination requires excess labeling and reducing reagents, and a clean-up step is necessary to purify labeled glycans. Commonly utilized purification strategies are SPE with different stationary phase materials (e.g. C18, HILIC, and PGC), paper chromatography (Bigge et al., 1995), filtration (Hase, 1994; Prater, Anumula, & Hutchins, 2007), dialysis and precipitation (Pabst, Kolarich, Poltl, Dalik, Lubec, Hofinger et al., 2009). Unfortunately, the introduction of an additional purification step reduces sample preparation throughput and creates the potential for sample loss.
14.2.3.2 Derivatization through the Hydrazide Reaction
The hydrazide reaction is another widely applied glycan reducing end labeling method. Hydrazide on labeling reagents interacts with the reducing end aldehyde of glycans, after which the resulting CN double bond is left in place. A positive aspect of this reaction is that the employment of toxic reducing reagents is avoided (Figure 14.1B), this renders a post-labeling purification step optional and makes hydrazide labeling more convenient in comparison with reductive amination. 2,4-Dinitrophenylhydrazine (Karamanos, Tsegenidis, & Antonopoulos, 1987), pyrenebutyric acid hydrazide (Sugahara, Amano, & Irimura, 2003) and 1-Alkoxyamino-1-deoxy alditols (Vondeyn, York, Albersheim, & Darvill, 1990) have been reported to label glycans such that their derivatives are compatible with optical detection methods. Additionally, research involving phenylhydrazine has exhibited a 100% labeling yield (Lattova & Perreault, 2003a), demonstrating the efficiency and reliability of hydrazide derivatization for glycan analysis.
Despite the potential for optical detection, most researchers utilizing hydrazide glycan derivatization are employing MS for glycomic profiling. The function of hydrazide in these cases is to induce a permanent charge so that both neutral and acidic glycans can be ionization with similar efficiency in MALDI and ESI. 2-hydrazino-N,N,N-trimethyl-2-oxo-ethanaminium chloride (Girard’s T reagent) is a well-established reagent for improvement of ionization efficiency in positive mode (Gil, Kim, & Kim, 2008; Gouw, Burgers, Trikoupis, & Terlouw, 2002; Naven & Harvey, 1998). An essential characteristics for evaluating hydrazide reagents is their ability to boost sensitivity. Walker and co-workers, in the laboratory of David C. Muddiman, have made comparisons among five labeling reagents, 5-phenylpetane hydrazide, 3-phenylpropane hydrazide, 2-phenylacethydrazide, 4-phenylbutane hydrazide and 4-phenylbenzohydrazide (Phenyl2-GPN), in order to evaluate their ionization efficiency enhancement capability. The results indicated a linear relationship between ion abundance and tag hydrophobicity with Phenyl2-GPN, the most hydrophobic of the labels, being the most effective with a 10% higher sensitivity (Figure 14.2). Other reported hydrazide labeling reagents for glycomics include Biotinyl-L-3-(2-naphthyl)-alanine hydrazide (Walker, Lilley, Enamorado, Comins, & Muddiman, 2011; Walker, Papas, Comins, & Muddiman, 2010), keratan sulfates (Y. Zhang, Cai, Zhao, & Luo, 2008), cyanine dyes (Kameyama, Kaneda, Yamanaka, Yoshimine, Narimatsu, & Shinohara, 2004) and 4-phenyl pyridine(Bereman, Comins, & Muddiman, 2010).
Figure 14.2.
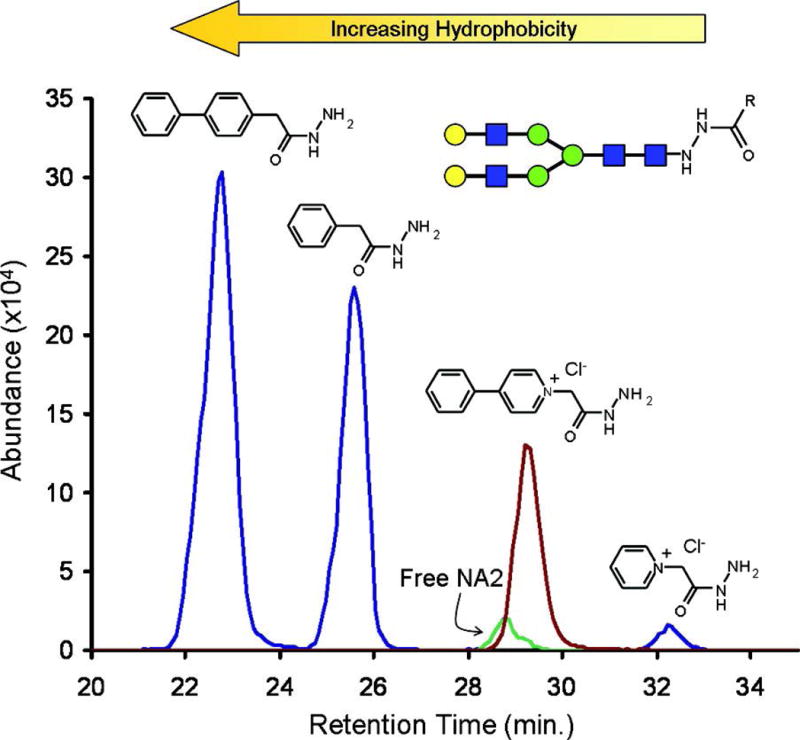
Extracted ion chromatogram of the NA2 glycan labeled with hydrazide tags, demonstrating the relationship between hydrophobicity and ionization efficiency. Reprinted and modified from Walker, S.H., et al. (Walker et al., 2010), with permission. Copyright (2010) American Chemical Society.
Hydrazide glycan interactions have also been utilized for recognizing and capturing glycans (Leteux, Childs, Chai, Stoll, Kogelberg, & Feizi, 1998; Yang & Zhang, 2012). A solid-phase glycan isolation method using hydrazide beads was reported (Yang et al., 2012) to achieve more specific glycan binding than conventional glycan isolation methods, which rely on the hydrophobicity difference between glycan and sample matrix. The formed hydrazone bond can be cleaved in an acidic elution solution so that glycans can be recovered from the hydrazide beads. A negative aspect of the hydrazide labeling approach is that hydrazide derivatized glycans are less stable than reductive amination labeled glycans, due to the lability of the hydrazone bond in acidic or basic solutions.
14.2.3.3 Derivatization of Glycan Sialic Acids
Unlike the two reducing end labeling methods discussed above, the derivatization method discussed in this section is focused on the sialic acids of glycans, which are commonly observed as terminal residues. Because sialic acid is the most labile monosaccharide found in glycans and is commonly found to exhibit significant changes in disease progression, it has attracted attention from a large number of researchers. 4-(4,6-Dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium (DMT-MM) is often utilized to stabilize sialic acids by amidation, especially in MALDI-MS where in source decay regularly occurs (Sekiya, Wada, & Tanaka, 2005). Moreover, DMT-MM can act as a catalyst to differentiate α 2-3 linked and α 2-6 linked sialic acids when proper reaction conditions are employed (Wheeler, Domann, & Harvey, 2009). In this reaction, α 2-3 linked acids are converted into lactones while α 2-6 linked sialic acids are converted into methyl esters, resulting in a 13 Dalton mass difference. These mass differences can be resolved using MS to reveal the number of differently linked sialic acids in a glycan (W. R. Alley, Jr. & Novotny, 2010; Harvey, Royle, Radcliffe, Rudd, & Dwek, 2008; Montesino, Calvo, Vallin, Rudd, Harvey, & Cremata, 2012; Tousi, Bones, Hancock, & Hincapie, 2013). To date, this is still one of the most efficient sialic acid linkage isomer identification methods.
14.2.3.4 Permethylation
Permethylation is another commonly used glycomic derivatization method (Figure 14.1C). It was specifically developed for MS-based glycomics because permethylation only improves glycan stability and ionization efficiency during MS analysis, no chromophores or fluorophores are introduced by this derivatization method. Another difference between permethylation and the reducing end labeling methods described above is that permethylation is a multiple site modification method. Hence, the proton affinity improvement by permethylation is much higher than single site derivatization methods. However, multiple site modification brings with it the challenge of managing derivatization efficiency. The overall incomplete derivatization ratio increases exponentially with the number of modification sites. Thus, selecting a proper permethylation protocol is critical for achieving efficient permethylation. Glycan permethylation was first reported by Ciccanu and Kerek in 1984 (Ciucanu I, 1984), DMSO, solid sodium hydroxide, and iodomethane were utilized in this protocol. The procedure was optimized by the same group in 2003 in order to reduce oxidative degradation in the reaction (Ciucanu & Costello, 2003). In 2005, Kang and Mechref proposed a solid-phase permethylation protocol. In this protocol, conventional in solution reaction was replaced by a reaction in spin columns of capillaries packed with sodium hydroxide beads (P. Kang, Mechref, Klouckova, & Novotny, 2005; P. Kang, Mechref, & Novotny, 2008). This solid-phase permethylation protocol reduced reaction time, improved derivatization efficiency for sialylated glycans and exhibited higher throughput compared with in solution protocols (Figure 14.3).
Figure 14.3.
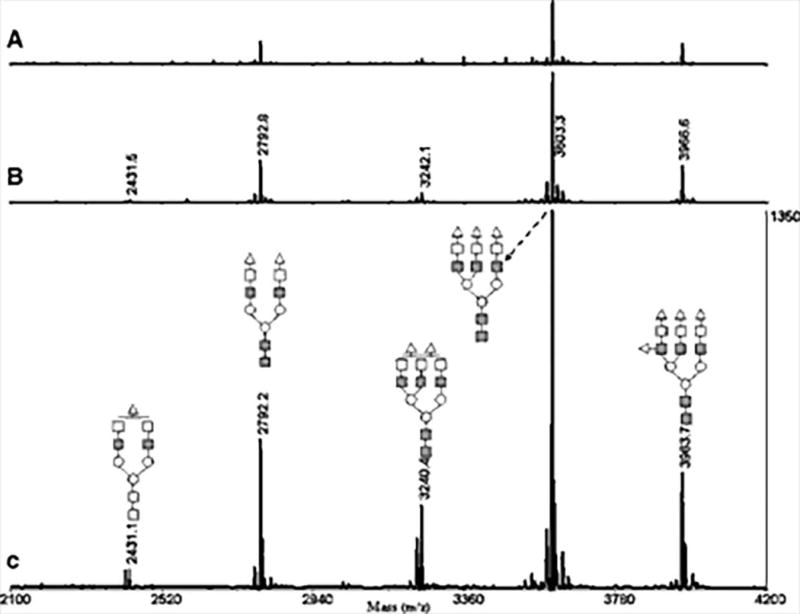
MALDI-TOF-MS spectra of permethylated N-glycans from fetuin, demonstrating the advantage of solid phase permethylation utilizing a microspin column (B) or fused-silica capillary (C) compared to the conventional in-solution method (A). Reprinted and modified from Kang, P., et al., 2005 (P. Kang et al., 2005), with permission. Copyright (2005) John Wiley and Sons.
14.2.3.5 Glycosylamine Derivatization
Recently reagents for the rapid labeling of released N-glycans have been investigated. In contrast to traditional reductive amination approaches, which tag the aldehyde termini of glycans after they are hydrolyzed from their glycosylamine forms, these reagents quickly label glycans as glycosylamines. Cook et al., 2012, initially reported a labeling method based on a 2-AB analog where glycans were tagged prior to the elimination of water from the reducing end by a urea linked aminobenzamide (Cook, Bullock, & Sullivan, 2012). ProZyme (Hayward, CA) has since marketed a kit termed GlykoPrep® Rapid-Reductive-Amination™ 2-AB Labeling Module that utilizes this strategy. The product is reported to provide highly fluorescent labeling, with no loss of sialic acid, with an efficiency of greater than 85%. Although this technique improved the speed of the labeling procedure, its application was limited to fluorescent detection. The Waters Corp. (Milford, MA) has since introduced a labeling reagent termed RapiFluor-MS™ that imparts both fluorescent properties and enhanced ionization efficiency for mass spectrometric analysis (Lauber, Brousmiche, Hua, Koza, Guthrie, Magnelli et al., 2015). The RapiFluor-MS™ reagent is composed of a quinolone fluorophore, an N-hydroxysuccinimide carbamate group, for tagging, and a basic tertiary amine that provides improved ionization efficiency. The reagent is sold in a kit that reports the preparation of glycans for analysis in less than 30 minutes through the use of a rapid PNGase F deglycosylation protocol involving RapiGest SF surfactant and a HILIC μElution solid phase extraction purification step. This method was recently utilized for the comparative glycomic analysis of two biosimilar monoclonal antibodies and the highly sensitive detection of released N-glycans by mass spectrometry was reported (Fang, Doneanu, Alley, Yu, Beck, & Chen, 2016).
14.3 Quantitative Glycomics Platforms
Reliable quantitative analysis of glycans is essential for understanding glycan biofunctions and discovering potential biomarkers for diseases. Numerous strategies for quantitative glycomics have been developed in recent decades. While glycomics is marching towards high sensitivity, high reliability, and high throughput, the remarkable complexity of the glycome still presents a number of challenges that must be addressed. In the sections that follow the strategies currently employed in the field of quantitative glycomics are discussed.
14.3.1 MS-Based Glycomics
Since the invention of soft ionization techniques, such as MALDI and ESI, mass spectrometry has become one of the most powerful bio-analytical platforms available. The rapid development of high resolution mass spectrometry (HRMS) in recent decades has enabled precise glycan identification through molecular weight determination. Tandem MS assists glycan structure elucidation. Direct analysis by MS skips the glycan separation step, significantly accelerating the analysis speed, allowing quantitative glycomic profiling to be accomplished in seconds. However, competitive ionization is not negligible when analyte composition is complex. Therefore, direct MS analysis is most often utilized for relatively simple glycan samples or glycan structural elucidation studies.
MALDI-MS is one of the direct MS glycomic analysis strategies. Native or derivatized glycans are mixed with matrix and then ionized by a laser beam. The selection of matrix is a critical factor that influences mass spectrum quality. 2, 5-dihydroxybenzoic acid (2, 5-DHB) is currently the most suitable matrix for glycan analysis (Stahl, Steup, Karas, & Hillenkamp, 1991). Sodium salts are often spiked into the matrix to prompt the formation of singly charged sodium adducts. Compared with ESI, the most pronounced advantage of MALDI is its capability to generate ions with a simplified charge state and adduct form, thus concentrating ion species while avoiding complicated spectra.
Many studies have reported improved sensitivity when co-matrix chemicals have been spiked into solution with 2, 5-DHB. For example, 2-hydroxy-5-methoxybenzoic acid (Karas, Ehring, Nordhoff, Stahl, Strupat, Hillenkamp et al., 1993), 1-hydroxyisoquinoline (Mohr, Bornsen, & Widmer, 1995), base spermine (Y. Mechref & Novotny, 1998) and aniline (Snovida, Chen, & Perreault, 2006) are all reported as efficient co-matrices for stabilizing glycans and improving ionization efficiency. The derivatization methods discussed above are necessary for MALDI-MS glycan analysis due to the fact that native glycans are fragile during ionization.
MALDI-MS has been utilized for the discovery of many glycomic biomarkers for a variety of diseases, such as hepatocellular carcinoma (Ressom, Varghese, Goldman, An, Loffredo, Abdel-Hamid et al., 2008), breast cancer (Kyselova, Mechref, Kang, Goetz, Dobrolecki, Sledge et al., 2008), prostate cancer (de Leoz, An, Kronewitter, Kim, Beecroft, Vinall et al., 2008) and ovarian cancer (Barkauskas, An, Kronewitter, de Leoz, Chew, White et al., 2009) as well as the therapeutic antibody trastuzumab (Matsumoto, Shimizu, Arao, Andoh, Katsumata, Kohno et al., 2009). Glycan analytes derived from serum, tissue or cell lines are all compatible with MALDI-MS analyses. Therefore, this technique is well suited for the identification of putative glycan biomarkers of disease.
ESI-MS is another major direct MS analysis strategy. It can be utilized in both positive and negative ionization modes for glycomic analysis. However, the performance of this type of analysis, when applied to complex biological samples, is limited due to the high dynamic range of glycans. Direct infusion ESI-MS is most commonly used for glycan structural elucidation studies, where only one or a few glycans are subjected to analysis (Harvey, 2005a, 2005b; Pfenninger, Karas, Finke, & Stahl, 2002a, 2002b).
14.3.2 LC-MS-Based Glycomics
Although direct MS analysis facilitates precise glycan identification and reliable quantification for glycans in high abundance, separation techniques are necessary when only optical detection devices are available, or minor glycans are masked due to competitive ionization in direct MS analysis. Furthermore, deconvolution of the fragmentation spectra of isomeric structures is greatly simplified by the elimination of co-elution through chromatographic separation. Various offline and online separation techniques, for both native and derivatized glycans, have been developed and applied in quantitative glycomics and are discussed herein.
14.3.2.1 Offline separation and fractionation
Several early studies combined LC separation with MALDI-MS analysis (Catala, Howe, Hucko, Rose, & Thannhauser, 2011; Lochnit & Geyer, 2004; Sparbier, Asperger, Kessler, Koch, Shi, Wenzel et al., 2006), where LC fractions were spotted on MALDI plates and subjected to MS analysis. In this method, a complex mixture of glycans is fractionated to reduce competitive ionization, which facilitates the identification of minor glycan species. In a recent study, MALDI-MS, LC-MALDI-MS, and LC-ESI-MS were compared side by side for the analysis of N-glycans derived from human blood serum (Hu & Mechref, 2012). According to the results, the LC-MALDI-MS method identified 12 more glycans than direct MALDI-MS, indicating the necessity of utilizing a separation technique prior to MS analysis. However, LC-ESI-MS provided the highest glycan identification number and sensitivity, demonstrating the advantage of coupling online separation to MS.
Offline glycan fractionation can also be achieved using SPE spin columns (Addeo, Soulier, Pelissier, Chobert, Mercier, & Ribadeaudumas, 1978; Packer, Lawson, Jardine, & Redmond, 1998). Lebrilla and coworkers have been using PGC SPE pre-fractionation in their sample preparation protocol for years. In this method, neutral glycans can be separated from acidic glycans to minimize suppression effects (Chu, Ninonuevo, Clowers, Perkins, An, Yin et al., 2009; Ninonuevo, Park, Yin, Zhang, Ward, Clowers et al., 2006; Ruhaak, Miyamoto, & Lebrilla, 2013).
14.3.2.2 Glycan Separation by Hydrophilic Interaction Chromatography (HILIC)
Native glycans are highly hydrophilic molecules that contain hydroxyl groups capable of hydrogen bonding. HILIC-LC provides highly efficient separation of native and reducing end labeled glycans. The retention mechanism of this chromatographic technique is primarily based on the formation of hydrogen bonds with the hydrophilic stationary phase; ionic interactions also play a role in the separation (Figure 14.4). Commonly used HILIC materials for glycan separation are amide coated silica, amine coated silica, and zwitterionic (ZIC) HILIC phases (Alpert, Shukla, Shukla, Zieske, Yuen, Ferguson et al., 1994; Takegawa, Deguchi, Ito, Keira, Nakagawa, & Nishimura, 2006; Wuhrer, de Boer, & Deelder, 2009). LC elution solutions are most commonly composed of water and acetonitrile with the addition of ammonium formate (5 to 50 mM) to increase ionic strength. The gradients generally begin with a high organic concentration and native or reducing end labeled glycans are eluted as the percentage of water increases. Smaller and neutral glycans elute first, while larger and sialylated glycans are retained by the column for a longer period of time. HILIC-LC is a separation technique that is sensitive to structural isomerism, enabling the partial separation of compositional or linkage glycan isomers.
Figure 14.4.
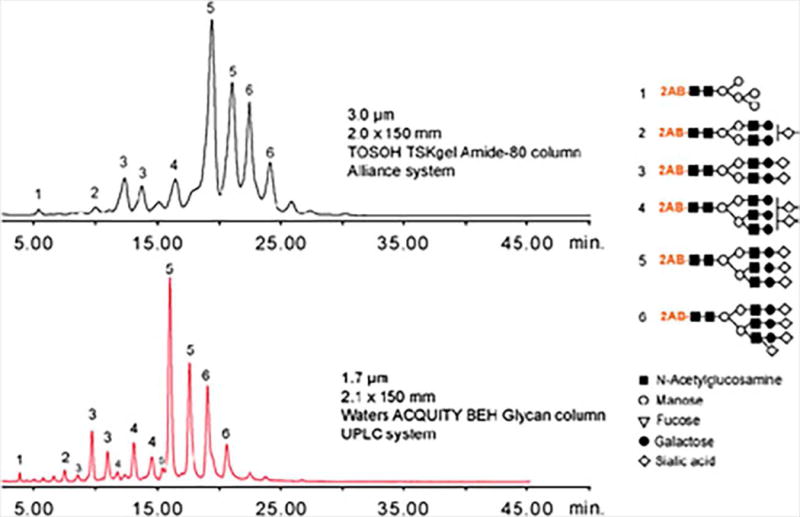
HILIC separation of 2-AB labeled glycans. Reprinted and modified from Zauner Deelder and Wuhrer, 2011, (Zauner, Deelder, & Wuhrer, 2011) with permission.
HILIC-LC can be coupled with UV or fluorescence detection for glycomic analysis. As mentioned above, this strategy requires native glycans to be derivatized with chromophore or fluorophore containing tags. 2-AA (Benet & Austin, 2011; Lonardi, Deelder, Wuhrer, & Balog, 2012; Royle, Campbell, Radcliffe, White, Harvey, Abrahams et al., 2008), 2-AB (Kalay, Ambrosini, van Berkel, Parren, van Kooyk, & Vallejo, 2012; Kozak, Royle, Gardner, Fernandes, & Wuhrer, 2012; Mauko, Lacher, Pelzing, Nordborg, Haddad, & Hilder, 2012) and PA (Takahashi, 1996) are frequently used in HILIC-fluorescence (FL) glycomic profiling. In contrast to MS detection strategies, optical detection methods are not capable of identifying the sequence of glycans; glycan identification is therefore based solely on retention time. Due to retention times varying over time and between instruments, the introduction of an internal standard is necessary for retention time calibration. Dextrin and dextran, which are composed of a series of repeating glucose units, are often used as retention time references with the concept of glycan units (GUs) employed. Hence, the glycan retention times in HILIC-LC can be represented as GUs rather than a precise measure of time. The most comprehensive HILIC-FL glycomic strategy was developed by Rudd and coworkers, who established the GlycoBase and autoGU databases (Campbell, Royle, Radcliffe, Dwek, & Rudd, 2008; Royle et al., 2008) in which more than 350 N-glycan structures can be found. Similarly, databases for O-glycans have also been developed (Royle, Mattu, Hart, Langridge, Merry, Murphy et al., 2002).
Automatic data processing systems are available for HILIC-FL profiling of 2-AB labeled glycans with the assistance of databases, making rapid and high-throughput glycomic analysis for scores of samples possible. This platform has been widely used for biomarker discovery studies involving diseases such as ovarian (Saldova, Royle, Radcliffe, Abd Hamid, Evans, Arnold et al., 2007), lung (Arnold, Saldova, Galligan, Murphy, Mimura-Kimura, Telford et al., 2011) and breast cancer (Abd Hamid, Royle, Saldova, Radcliffe, Harvey, Storr et al., 2008; R. Saldova, Reuben, Abd Hamid, Rudd, & Cristofanilli, 2011).
Common flow rates and mobile phases of HILIC-LC permit convenient coupling of HILIC separation to MS analysis through ESI. As a more informative detection platform, MS can provide precise glycan identification and detailed glycan structural information can be obtained using tandem MS (Wuhrer et al., 2009). Additionally, native or reduced glycans can be subjected to HILIC-MS analysis without the need for fluorescence labeling. Due to the isomeric separation capability of HILIC, it has been coupled to tandem MS for the structural analysis of glycans. Efforts were made to minimize the physical size of the HILIC separation platform, to increase its sensitivity, leading to the development of nanoLC (Clarke, Harmon, & DeFelippis, 2009; Wuhrer, Koeleman, Hokke, & Deelder, 2004) and chip-based (Staples, Bowman, Costello, Hitchcock, Lau, Leymarie et al., 2009) HILIC systems. The detection limit of these miniaturized systems can be as low as femtomoles (Wuhrer, Koeleman, Deelder, & Hokke, 2004; Wuhrer, Koeleman, Hokke, & Deelder, 2004).
14.3.2.3 Glycan Separation by Reverse Phase Liquid Chromatography (RPLC)
One of the most obvious advantages of utilizing RPLC to separate glycans is that RPLC is the separation technique of choice for proteomics and metabolomics. Therefore no adjustment is required for chromatography columns or mobile phases when a laboratory has multiple research projects. The retention mechanism on RPLC based C18 stationary phase columns relies on the hydrophobicity of analyte molecules. Hence, derivatization is required in order to achieve sufficient retention of glycans (Wuhrer, Deelder, & Hokke, 2005). Choices for reducing end labeling tags for glycomic RPLC separation include 2-AB (X. Chen & Flynn, 2007; Wing, Garner, Hunnam, Reinkensmeier, Andersson, Harvey et al., 2001), PA (Yamada, Tsukamoto, Sasaki, Yagyu, & Takahashi, 1997), 1-Phenyl-3-methyl-5-pyrazolone (PMP) (Saba, Shen, Jamieson, & Perreault, 1999, 2001), phenylhydrazine (Lattova & Perreault, 2003b), 2-amino-5-bromopyridine (Li & Kinzer, 2003) and ANTS (Saba et al., 2001). After derivatization, the tags and the glycan itself influence retention time in concert. When the hydrophobicity of a tag is greater than that of the glycan, retention time decreases with increased numbers of monosaccharides in the glycan; this relationship reverses when the hydrophobicity of a tag is less than the glycan (Pabst et al., 2009).
Permethylation is another widely used derivatization method for RPLC glycan separation. After permethylation, glycan hydrophobicity increases significantly. Additionally, all hydrophobic groups are directly related to the glycan structure, thus making the retention of permethylated glycans on C18 stationary phase more predictable. Isomeric separations can be achieved using RPLC for permethylated O-glycans (Figure 14.5) (Hanisch & Muller, 2009). However, when the analyte is an N-glycan, which is larger on average, the isomeric separation capability of RPLC decreases (Figure 14.6) (Dong, Zhou, & Mechref, 2016). Furthermore, peak broadening can often be observed for highly branched N-glycan structures. In this case, high separation temperatures may be employed to increase peak resolution. Nano scale RPLC systems have also been developed for increased sensitivity. Online purification may be achieved by utilizing a C18 trap in a 10 port valve system (Desantos-Garcia, Khalil, Hussein, Hu, & Mechref, 2011). Studies have demonstrated improved sensitivity when offline liquid-liquid extraction is replaced by online purification. This strategy can be utilized for quantitative glycomic profiling of serum, the surface of tissues and cell lines to study a variety of diseases (W. R. J. Alley, Vasseur, Goetz, Svoboda, Mann, Matei et al., 2012; Y. Hu, Zhou, Khalil, Renteria, & Mechref, 2013; Y. Mechref, Hu, Garcia, Zhou, Desantos-Garcia, & Hussein, 2012; Tsai, Wang, Di Poto, Hu, Zhou, Zhao et al., 2014).
Figure 14.5.
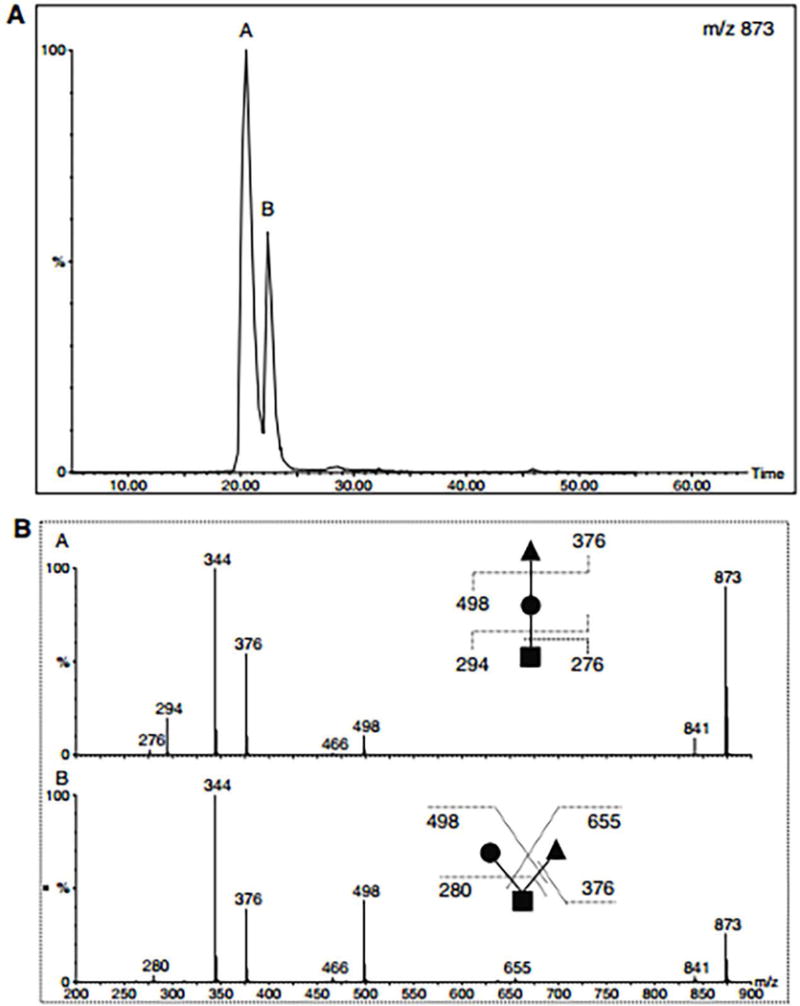
(A) RPLC separation of permethylated isomeric O-glycan trisaccharides. (B) MS/MS spectra of isomers in peaks A and B (upper panel). Reprinted and modified from Hanisch and Muller, 2009, (Hanisch et al., 2009) with permission.
Figure 14.6.
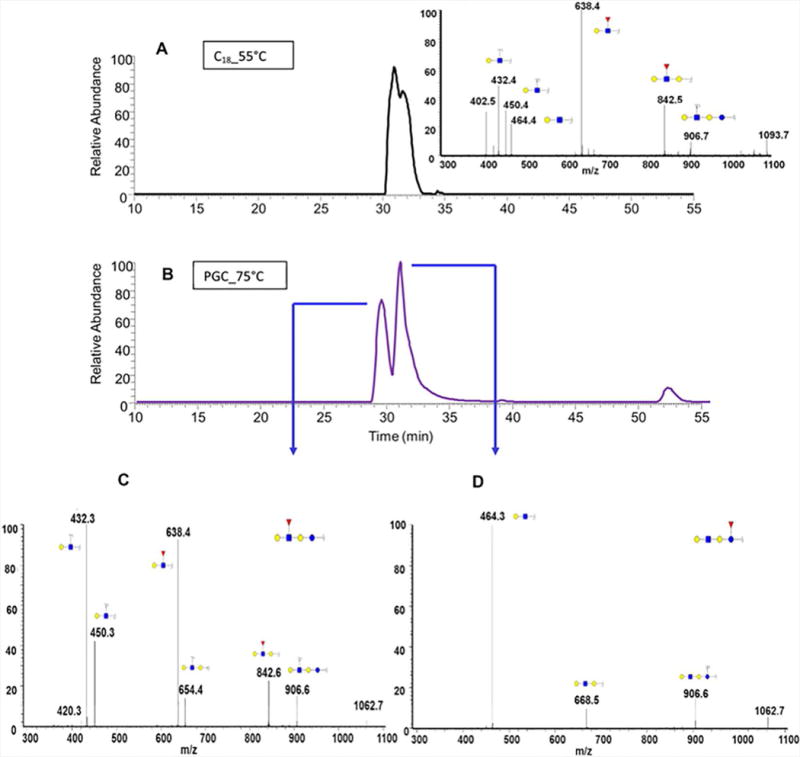
EIC of fucosylated Gal-GlcNAc-lactose isomers determined in human milk using (A) C18 and (B) PGC columns. Tandem MS of the two isomers associated with the free oligosaccharide structures are depicted in (C) and (D). Reprinted and modified from Dong, Zhou and Mechref, 2016, (Dong et al., 2016) with permission.
14.3.2.4 Glycan Separation by Porous Graphitized Carbon (PGC) Liquid Chromatography
PGC was first utilized as an SPE material for glycan purification in 1998 (Packer et al., 1998), and was subsequently applied to LC-ESI-MS in 1999 (Thomsson, Karlsson, & Hansson, 1999). Among currently utilized LC stationary phases, PGC exhibits the greatest capacity for isomeric separation, despite the separation mechanism not being completely understood. Currently, retention is believed to be caused by a combination of hydrophobic and ionic interactions (Ruhaak, Deelder, & Wuhrer, 2009). Native and reducing end labeled glycans are most commonly subjected to PGC separation. However, the elution of acidic glycans on PGC may be problematic. Pabst and coworkers have systematically investigated the separation of acidic glycans on PGC. The results indicated that elution buffer ionic strength is critical for elution of acidic glycans (Pabst & Altmann, 2008).
PGC separation coupled with UV detection or MS analysis has been widely applied for quantitative glycomic analysis. For example, a chip based PGC nanoLC-ESI-MS technique was developed by the Lebrilla group. The method stability was investigated, and the results indicated that analyses repeated on different days had a coefficient of variation of 4%, calculated from log10 transformed peak intensities (Ruhaak, Taylor, Miyamoto, Kelly, Leiserowitz, Gandara et al., 2013). This strategy has was successfully applied to the analysis of human and bovine milk (Nwosu, Aldredge, Lee, Lerno, Zivkovic, German et al., 2012; Wu, Grimm, German, & Lebrilla, 2011), dry blood spots (Ruhaak, Miyamoto, Kelly, & Lebrilla, 2012), plasma from lung cancer patients (Ruhaak, Nguyen, Stroble, Taylor, Taguchi, Hanash et al., 2013), prostate cancer and lymphoproliferative cancer cell lines (Hua, Saunders, Dimapasoc, Jeong, Kim, Kim et al., 2014).
In 2007, permethylated glycans were also reported to be separated by PGC-LC (Costello, Contado-Miller, & Cipollo, 2007). In this study, isomeric separations of high mannose glycans were observed, and detailed structures were interpreted from MS2 spectra. However, peak shapes were not smooth, and baseline resolution was not achieved. Moreover, sialylated glycans were not efficiently separated by the method employed. In a study more recently conducted, in our laboratory, the optimal conditions for PGC separation of permethylated glycans were determined. Improved resolution, similar to that attained for native glycans, was achieved while more informative MS2 spectra were acquired, prompted by glycan permethylation (Figure 14.6) (Dong et al., 2016).
14.3.2.5 Glycan Separation by Capillary Electrophoresis (CE)
CE has been described in-depth in recent reviews (Campa, Coslovi, Flamigni, & Rossi, 2006; Y. Mechref, Hu, Desantos-Garcia, Hussein, & Tang, 2013; S. Suzuki, 2013; Zamfir & Peter-Katalinic, 2004). The method involves the use of capillaries with 50 to 100 μm bore sizes in high electric fields (750 V/cm). Short separation times and small injection volumes are typical. Thus, highly sensitive detection methods must be employed. One such method is LIF following reductive amination labeling with fluorophores such as an APTS (Chen & Evangelista, 1998; Evangelista et al., 1996), utilizing reducing agents such as cyanoborohydride (Evangelista et al., 1996) or 2-picoline-borane (Ruhaak et al., 2010). CE coupled to LIF has been successfully applied for the analysis of APTS labeled N-glycans derived from a number of purified glycoproteins (Chen et al., 1998; Evangelista et al., 1996; Guttman et al., 1996; Guttman & Pritchett, 1995; Ma & Nashabeh, 1999; Mechref, Muzikar, & Novotny, 2005; Suzuki, Muller, Guttman, & Karger, 1997) as well as human serum from chronic hepatitis C patients (Callewaert, Geysens, Molemans, & Contreras, 2001; Callewaert, Van Vlierberghe, Van Hecke, Laroy, Delanghe, & Contreras, 2004; Vanderschaeghe, Debruyne, Van Vlierberghe, Callewaert, & Delanghe, 2009; Vanderschaeghe, Laroy, Sablon, Halfon, Van Hecke, Delanghe et al., 2009). In addition to LIF, CE has also been successfully conjugated to tandem MS for the quantification of glycans, which has been discussed in previous reviews (Campa et al., 2006; Mechref et al., 2013; Zamfir et al., 2004). Both on-line and off-line coupling of CE to MS instrumentation has been utilized. In the case of off-line coupling, MALDI-MS was initially employed to analyze APTS labeled N-glycans from ribonuclease B (Suzuki et al., 1997). On-line coupling entails the use of CE-ESI-MS systems, which have been utilized for the analysis of mannooligosaccharide caps in mycobacterial lipoarabinomannan (Monsarrat, Brando, Condouret, Nigou, & Puzo, 1999) and phosphorylated APTS-labeled N-glycans from cellobiohydrolase I (Sandra, Devreese, Van Beeumen, Stals, & Claeyssens, 2004).
14.3.2.6 Glycan Separation by Ion Mobility Spectroscopy-Mass Spectrometry (IMS-MS)
IMS separates gas phase ions, typically generated by ESI, based on their collisional cross-sections which are approximately proportionate to molecular shape and size (Clowers, Dwivedi, Steiner, Hill, & Bendiak, 2005; Clowers & Hill, 2005). Due to the isomeric separation capabilities of IMS, the technique has been gaining popularity in the field of glycan analysis. The advantages of having an additional gas phase separation during IMS-MS have been demonstrated by the characterization of N-glycan isomers from RNase B, fetuin and gp120 (Fenn & McLean, 2009; Harvey, Scarff, Crispin, Scanlan, Bonomelli, & Scrivens, 2012; Harvey, Scarff, Edgeworth, Crispin, Scanlan, Sobott et al., 2013; Zhu, Lee, Valentine, Reilly, & Clemmer, 2012). IMS has also been applied to the analysis of glycans in complex biological matrices; for example, the profiling of total serum glycans from individuals with liver cancer or cirrhosis of the liver has been reported (Isailovic, Kurulugama, Plasencia, Stokes, Kyselova, Goldman et al., 2008), as well as the comparison of N-glycans from the serum of patients with Barrett’s esophagus, high-grade dysplasia and esophageal adenocarcinoma with healthy control individuals (Gaye, Valentine, Hu, Mirjankar, Hammoud, Mechref et al., 2012). The applications of IMS for the analysis of glycans in the context of high throughput and high resolution studies have recently been reviewed (Gray, Thomas, Upton, Migas, Eyers, Barran et al., 2016).
14.3.3 MS-based Quantitation Methods
Although MS is a powerful glycan identification and structure elucidation tool, its performance in glycomic quantitation is not as stable as optical detection methods. This is due to the nature of ionization and detection of MS instruments. First, both MALDI and ESI do not ionize all analyte molecules; the ionization efficiency is influenced by many factors. For MALDI, the uncertainty of crystal quality, spot layer thickness, and salt concentration in the sample results in the fluctuation of ionization efficiency. For ESI, the needle inner diameter, sheath gas flow, and analyte solution composition all influence the stability of ionization. Especially in the case of nanoESI, which is the common interface between nanoLC or chip/capillary based separation techniques and MS, ionization fluctuation can be as high as 30%.
A multi-institute glycomic profiling study was proposed by the Human Proteome Organization (HUPO) and the Human Disease Glycomics/Proteome Initiative (HGPI) in 2007 (Wada, Azadi, Costello, Dell, Dwek, Geyer et al., 2007). While the final conclusion, of the study, was that MS-based analysis appears to be an efficient method for identification and quantitation of oligosaccharides in glycomic studies. Variation was also observed in the data, indicating the necessity of efforts to improve the reliability of MS quantitation. Currently, spiking internal standards and multiplexing through the use of stable isotope labeling represent two major approaches for increasing the reliability of analyses.
14.3.3.1 Internal Standard
The necessity of spiking internal standards is well recognized in MALDI-MS glycomic profiling. An internal standard can effectively normalize results from different runs. Maltoheptaose and other carbohydrates with repeating units of glucose have been utilized as internal standards (Bereman et al., 2010; Snovida, Rak-Banville, & Perreault, 2008; Szabo et al., 2010; Walker, Budhathoki-Uprety, Novak, & Muddiman, 2011). The selection criteria for internal standards are that their molecular weight should be close to the average molecular weight of the glycans being analyzed and they should have chemical properties similar to glycans so they can be derivatized using the same methods applied to the glycans of interest. Recently, iGlycoMab was reported as an internal standard for normalizing variation observed in glycan release, sample preparation and MS analysis. iGlycoMab is a purified murine IgG2b monoclonal antibody that has all glycan nitrogen atoms replaced by the stable isotope 15N. Hence, the glycan internal standard does not have a molecular weight that overlaps with analyte glycans (~4 Dalton mass difference is induced by 15N on N-acetylglucosamine). The coefficient of variation of model glycoprotein and cell line glycan analysis was found to decrease when results were normalized by iGlycoMab (Figure 14.7), demonstrating its utility (Zhou, Tello, Harvey, Boyes, Orlando, & Mechref, 2016).
Figure 14.7.
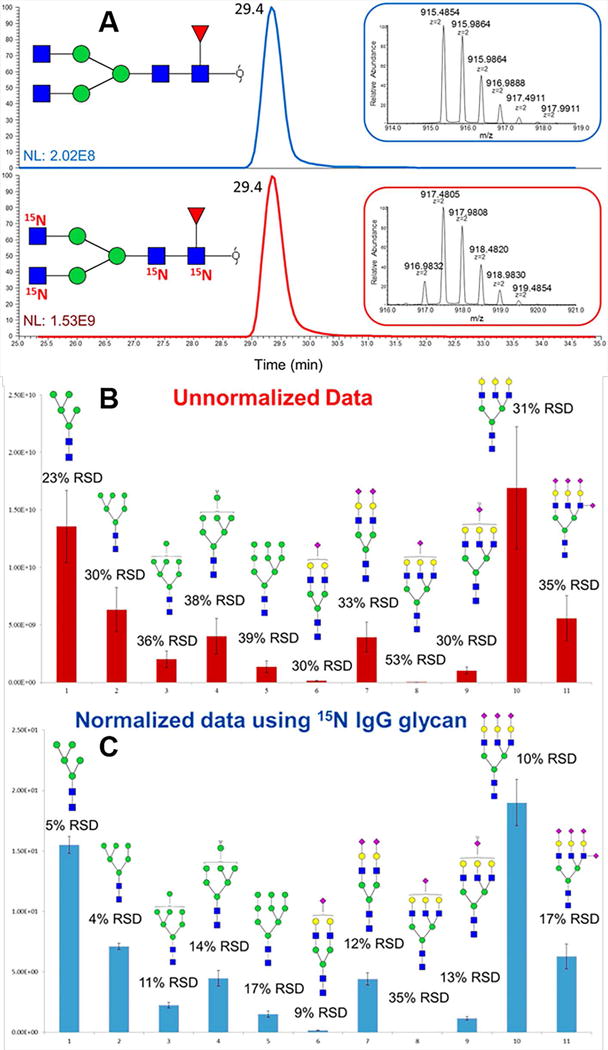
(A) EIC and MS spectra of N-glycans released from unlabeled IgG standard and 15N iGlycoMab. Bar graphs representing intensity variation of biological (n = 3) analyses of N-glycans released from model glycoprotein mixtures (B) without normalization and (C) with normalization using 15N-glycans derived from iGlycoMab. Reprinted and modified from Zhou, S., et al., 2016, (Zhou, Tello, et al., 2016) with permission.
14.3.3.2 Multiplexing Through Derivatization
As mentioned above, variation between different runs is the primary source of uncertainty when reliable analysis for multiple samples is required, for example, biomarker discovery. Hence, combining multiple samples in one MS analysis functions to eliminate a great deal of uncertainty. Multiplexed MS analysis also provides the additional advantage of saving time. Total analysis time can be reduced by a factor of the number of multiplexed samples. In order to resolve identical glycan structures derived from different multiplexed samples in a single MS analysis, molecular weight differences must be generated. The most convenient way to introduce different masses to glycans is through derivatization. Atoms on the derivatization reagents can be replaced with stable isotopes such as deuterium (D), 13C or 15N. Glycans labeled with standard reagents are commonly referred to as “light” and those labeled with the staple isotope reagents are termed “heavy”. It is worth mentioning that the mass difference between light- and heavy-labeled glycans should be larger than 4 Dalton. Otherwise, the overlapping natural isotopic distribution of light-labeled glycans will interfere with the intensity of heavy-labeled glycans.
Reductive amination is a single site derivatization method, therefore the mass difference between heavy and light reagents utilized should be larger than 4 Dalton. Commonly used stable isotope replacement patterns include introducing 4 deuteriums or 6 13C atoms into an aromatic ring. These patterns are widely used for 2-AA (Hitchcock, Costello, & Zaia, 2006; Hitchcock, Yates, Shortkroff, Costello, & Zaia, 2006), aniline (Lawrence, Olson, Steele, Wang, Warrior, Cummings et al., 2008; Xia, Feasley, Sachdev, Smith, & Cummings, 2009) and PA (Yuan, Hashii, Kawasaki, Itoh, Kawanishi, & Hayakawa, 2005). However, due to the limitation of the labeling reagent size, these strategies only allow for the simultaneous analysis of two samples per run. To overcome this, larger tags have been developed to enable the inclusion of more glycan samples per analysis. Bowman and Zaia introduced a tetraplex stable-isotope coded tagging strategy (Bowman & Zaia, 2007), where four tags (d0, d4, d8, and d12) with a 4 Dalton mass difference between them were utilized to analyze glycans derived from model glycoproteins mixed in a predetermined ratio. The results demonstrated the reliability of glycomic quantitation using this type of tag. The tag strategy was then applied for the analysis of glycans derived from human, ovine, bovine and baboon samples (Bowman & Zaia, 2010).
Heavy/light labeling can also be achieved through permethylation. Because permethylation is a multiple site derivatization method, the rule that the mass difference between tags should be equal to or greater than 4 Dalton can be ignored. The first report of multiplexing through permethylation, introduced by Mechref, Novotny and coworkers in 2007, was termed Comparative glycomic mapping (C-GlycoMAP) (Kang, Mechref, Kyselova, Goetz, & Novotny, 2007). The permethylation reagents CH3I and CD3I were used for labeling glycans released from RNaseB and the spectra generated by MALDI-MS exhibited reliable quantitation ratios from 0.125 to 6. This strategy was subsequently extended by introducing an additional iodomethane derivative, CH2DI, allowing triplex analysis to be achieved (Hu, Desantos-Garcia, & Mechref, 2013). In this study, glycans released from human blood serum of patients with esophageal cancer, Barret’s disease and high grade dysplasia were analyzed. Quantitation by isobaric labeling (QUIBL) represents an additional type of permethylation that incorporates multiplexing (Atwood, Cheng, Alvarez-Manilla, Warren, York, & Orlando, 2008). 13CH3I and 12CH2DI are utilized for the isobaric labeling of glycans, resulting in a mass difference of 0.002922 Dalton between each permethylation site. The use of a high resolution mass spectrometer is required for the isolation of such similar masses. When utilized for the analysis of N-glycans from a model glycoprotein and human serum, a dynamic range of 2 orders of magnitude was exhibited. O-glycans from mucin were also quantified using this strategy (Botelho, Atwood III, Alvarez-Manilla, York, & Orlando, 2008). When applying a permethylation-based multiplexing strategy on an RPLC-MS platform, it is important to note that retention times are modified when deuterium iodomethane is used. This occurs as a result of the hydrophobicity of hydrogen being greater than deuterium, the relationship of retention times is CH3 > CH2D > CHD2 > CD3 on a C18 column. This phenomenon does not extend to carbon, as no difference in RPLC retention time is observed between carbon and 13C iodomethane derivatized glycans. Although multiplexing techniques reduce variation, ESI efficiency changes throughout an LC-MS analysis. This temporal variation may not be eliminated when multiplexed samples have significantly different retention times. The isobaric permethylation reagents, 13CH3I and 12CH3I, have been utilized for LC-MS analysis of N-glycans released from embryos of Drosophila melanogaster at various temporal stages of development (Aoki, Perlman, Lim, Cantu, Wells, & Tiemeyer, 2007).
14.3.3.3 Metabolic Isotopic Labeling
Metabolic labeling presents an alternative to derivatization reagents for the incorporation of stable isotopes into glycan structures. This strategy allows for simplified sample preparation and selection of analytical platforms due to the elimination of potential issues stemming from derivatization reagents. In this case, even native glycans can be subjected to multiplex analysis. Isotopic Detection of Aminosugars With Glutamine (IDAWG) is a metabolic glycan labeling strategy introduced by Orlando and coworkers (Orlando, Lim, Atwood, Angel, Fang, Aoki et al., 2009). The labeling mechanism employed in the strategy is as follows; 15N-Gln was spiked into cell culture media to act as the nitrogen source for the biosynthesis of UDP-GlcNAc, which could be further converted to UDP-GalNAc and CMP-Neu5Ac. As a result, all GlcNAc, Neu5Ac, and GalNAc in cell glycans were 15N labeled. The nitrogen replacement efficiency of this method was demonstrated to be higher than 95%. Another metabolic labeling strategy was proposed by Breidenbach and coworkers in 2010 (Breidenbacha, Gallagherb, Kingc, Smarta, Wua, & Bertozzia, 2010). In their approach, 13C6-GlcNAc was introduced to cell secreted glycoproteins by an exogenous GlcNAc salvage pathway.
Stable isotopes can also be introduced into glycans through PNGase F digestion in 18O-water; this method is known as glycan reducing-end 18O labeling (GREOL) (Zhang, Wang, Tang, & Yang, 2011). During the procedure, oxygen atoms of glycan reducing ends are replaced by 18O and a 2 Dalton mass difference is generated. The quantitative capability of this method was verified using ovalbumin glycans; it was additionally applied to glycans released from the serum of patients with hepatocellular carcinoma (HCC). The linear response range of this method was determined to be 2 orders of magnitude or higher.
14.3.3.4 Multiplexing using a Reporter Ion in Tandem MS
Full MS based multiplexing always relies on the criteria of a mass difference of at least 4 Dalton between heavy and light labeled molecules. This can significantly limit the maximum number of samples that can be analyzed together. Additionally, full MS based multiplexing can be complicated by tags resulting in different retention times and isotopic labeling resulting in identical glycan structures having different m/z values. In order to overcome these drawbacks, a multiplex method employing reporter ions in MS/MS was developed. This strategy was accomplished by utilizing a series of reducing end labeling tags with reporter ion portions that differed by mass. A mass balancer, or normalizer, was included in the tags to ensure they were isobaric. Hence, the same glycans with different multiplex tags were identical during LC separation and full MS analysis; this reduced the complexity of LC-MS data collected during the analysis of multiple samples. Due to the fact that the reporter ions were small, m/z =100 - 200 Dalton, the influence of isotopic peaks was negligible. In this strategy, the mass gap between adjacent reporter ions can be 1 Dalton or less, which contributes to its potential capacity for multiplex analysis.
AminoxyTMT (Hahne, Neubert, Kuhn, Etienne, Bomgarden, Rogers et al., 2012; Zhong, Chen, Snovida, Liu, Rogers, & Li, 2015), iART (Yang, Yuan, Yang, Zhou, Harlan, Edwards et al., 2013) and QUANTITY (Yang, Wang, Chen, Yin, Song, Turko et al., 2015) are all examples of multiplexing reagents for quantitative glycomics. AminoxyTMT is a derivative of the TMT reagent developed for proteomics. A six-plex kit of aminoxyTMT is commercially available. However, reporter ion yield has been reported to be an issue for aminoxyTMT because glycosidic bonds are weaker than the bonds linking the reporter ions. This issue can be overcome by conducting MS3 on Y1 ions or by modifying fragmentation patterns through sodium adducts (Figure 14.8) (Zhou, Hu, Veillon, Snovida, Rogers, Saba et al., 2016). Quantitation error was reported to be controlled, within 10%, when an optimized LC-MS/MS method was utilized to facilitate the formation of sodium adducts. The iART tetra-plex reagent is reported to have issues with low reporter ion yield, similar to what has been observed with aminoxyTMT. On the other hand, QUANTITY (Figure 14.9) is a tetra-plex reagent that results in high reporter ion yield without the help of sodium or metal ion adducts. This strategy was applied to the quantitative analysis of glycans released from three bioengineered Chinese Hamster Ovary (CHO) cell lines (Yang et al., 2015).
Figure 14.8.
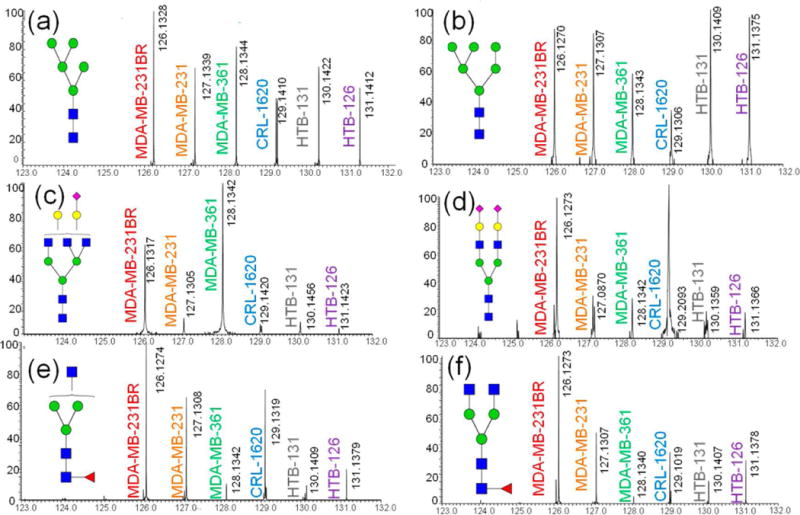
Tandem mass spectra of different aminoxyTMT labeled glycans derived from various cell lines. Tandem mass spectra of several glycan structures are shown in (a)–(f). Reprinted and modified from Zhou, S., et al., 2016, (Zhou, Hu, et al., 2016) with permission.
Figure 14.9.
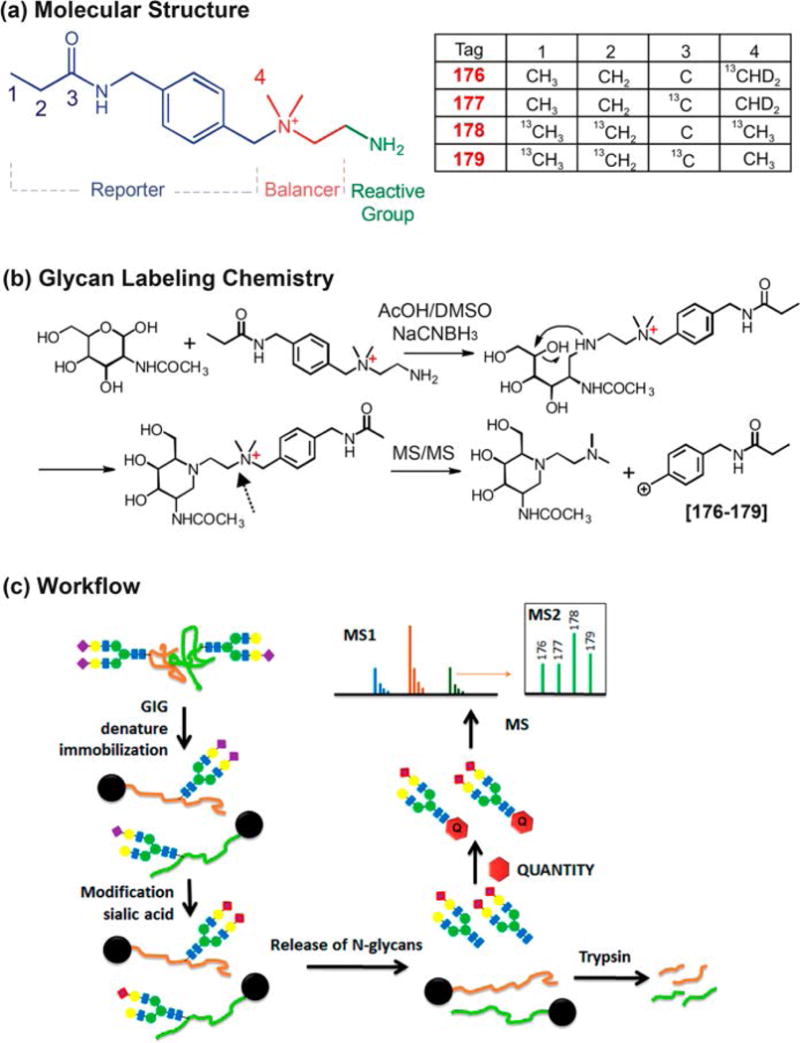
QUANTITY isobaric tandem mass tags for glycan labeling and quantitation. Reprinted and modified from Yang, S., et al., 2015, (Yang et al., 2015) with permission.
14.3.4 Data Processing Software for Glycomics
Omics analyses are always based on the analysis of a vast number of samples, therefore reliable and high throughput data processing software is necessary. While this also the case for glycomics, data processing software for glycomics is relatively under developed compared with proteomics software. MultiGlycan is software that was specifically developed for LC-MS analysis of permethylated glycans, where an algorithm was employed for deconvolution in order to reduce false negatives (Figure 14.10) (Hu, Zhou, Yu, Tang, & Mechref, 2015). Additionally, MultiGlycan is capable of analyzing permethylated glycan data with the functionality of enabling the normalization of reagent purity and isotopic distribution differences.
Figure 14.10.

Workflow of MultiGlycan-ESI-based automatic quantitation strategy: (a) permethylated N-glycan preparation workflow and (b) software annotation and quantitation method. Reprinted and modified from Hu, Y., et al., 2014, (Y. Hu et al., 2015) with permission.
SimGlycan (Apte & Meitei, 2010a; Meitei, Apte, Snovida, Rogers, & Saba, 2015) (Figure 14.11) is an additional software package that allows the identification and quantitation of glycans derivatized using several methods, in addition to permethylation, including aminoxyTMT labeling. Furthermore, a library has been built for the automated analysis of analytes separated on different LC columns. It represents one of the most powerful software solutions for quantitative glycan data analysis and structural elucidation based on MS/MS spectra.
Figure 14.11.
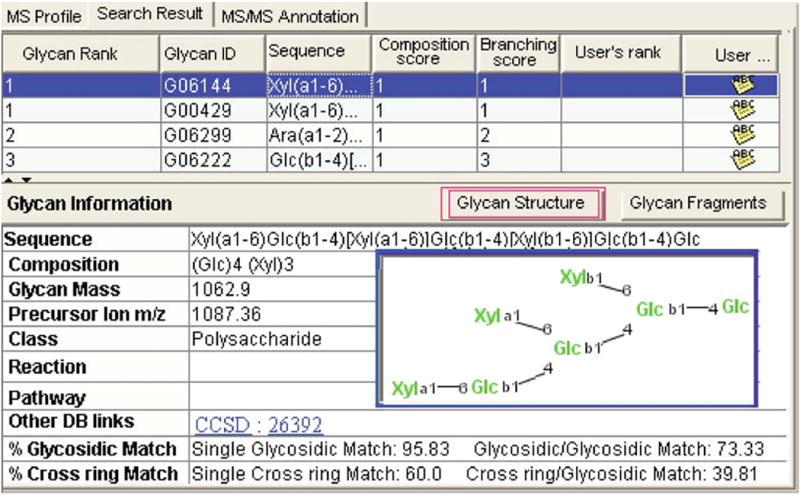
Glycan identified through SimGlycan MS/MS database search. Reprinted and modified from ref (Apte & Meitei, 2010b) with permission.
GlycoWorkbench (Ceroni, Maass, Geyer, Geyer, Dell, & Haslam, 2008; Damerell, Ceroni, Maass, Ranzinger, Dell, & Haslam, 2012, 2015) possesses the function of predicting fragment ions for glycans; Cartoonist (Goldberg, Sutton-Smith, Paulson, & Dell, 2005) provides automated annotation of MALDI-MS analyzed glycans; software like Glycomod (Cooper, Gasteiger, & Packer, 2001), GlycoQuest, Glyco-Peakfinder (Maass, Ranzinger, Geyer, von der Lieth, & Geyer, 2007) and SysBioWare (Vakhrushev, Dadimov, & Peter-Katalinic, 2009) are also employed in glycomic data processing.
Acknowledgments
This work was supported by an NIH grant (1R01GM112490-01) and a grant from the Cancer Prevention and Research Institute of Texas (RP130624).
References
- Abd Hamid UM, Royle L, Saldova R, Radcliffe CM, Harvey DJ, Storr SJ, Rudd PM. A strategy to reveal potential glycan markers from serum glycoproteins associated with breast cancer progression. Glycobiology. 2008;18(12):1105–1118. doi: 10.1093/glycob/cwn095. [DOI] [PubMed] [Google Scholar]
- Addeo F, Soulier S, Pelissier JP, Chobert JM, Mercier JC, Ribadeaudumas B. Preparation and Fractionation of Goat Kappa-Casein - Analysis of Glycan and Peptide Components. Journal of Dairy Research. 1978;45(2):191–196. doi: 10.1017/s0022029900016368. [DOI] [PubMed] [Google Scholar]
- Alley WR, Jr, Novotny MV. Glycomic analysis of sialic acid linkages in glycans derived from blood serum glycoproteins. J Proteome Res. 2010;9(6):3062–3072. doi: 10.1021/pr901210r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley WRJ, Vasseur JA, Goetz JA, Svoboda M, Mann BF, Matei DE, Novotny MV. N-linked glycan structures and their expressions change in the blood sera of ovarian cancer patients. J Proteome Res. 2012;11:2282–2300. doi: 10.1021/pr201070k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert AJ, Shukla M, Shukla AK, Zieske LR, Yuen SW, Ferguson MAJ, Orlando R. Hydrophilic-Interaction Chromatography of Complex Carbohydrates. Journal of Chromatography A. 1994;676(1):191–202. doi: 10.1016/0021-9673(94)00467-6. [DOI] [PubMed] [Google Scholar]
- Anumula KR. Quantitative determination of monosaccharides in glycoproteins by high-performance liquid chromatography with highly sensitive fluorescence detection. Anal Biochem. 1994;220(2):275–283. doi: 10.1006/abio.1994.1338. [DOI] [PubMed] [Google Scholar]
- Aoki K, Perlman M, Lim JM, Cantu R, Wells L, Tiemeyer M. Dynamic Developmental Elaboration of N-Linked Glycan Complexity in the Drosophila melanogaster Embryo. J Biol Chem. 2007;282:9127–9142. doi: 10.1074/jbc.M606711200. [DOI] [PubMed] [Google Scholar]
- Apte A, Meitei NS. Bioinformatics in glycomics: glycan characterization with mass spectrometric data using SimGlycan. Carbohydrate Microarrays: Methods and Protocols. 2010a;600:269–281. doi: 10.1007/978-1-60761-454-8_19. [DOI] [PubMed] [Google Scholar]
- Apte A, Meitei NS. Bioinformatics in Glycomics: Glycan Characterization with Mass Spectrometric Data Using SimGlycan (TM) Functional Glycomics: Methods and Protocols. 2010b;600:269–281. doi: 10.1007/978-1-60761-454-8_19. [DOI] [PubMed] [Google Scholar]
- Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473(1):4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- Arnold JN, Saldova R, Galligan MC, Murphy TB, Mimura-Kimura Y, Telford JE, Rudd PM. Novel Glycan Biomarkers for the Detection of Lung Cancer. J Proteome Res. 2011;10(4):1755–1764. doi: 10.1021/pr101034t. [DOI] [PubMed] [Google Scholar]
- Atwood JA, Cheng L, Alvarez-Manilla G, Warren NL, York WS, Orlando R. Quantitation by isobaric labeling: Applications to glycomics. J Proteome Res. 2008;7(1):367–374. doi: 10.1021/pr070476i. [DOI] [PubMed] [Google Scholar]
- Barkauskas DA, An HJ, Kronewitter SR, de Leoz ML, Chew HK, White RWD, Rocke DM. Detecting glycan cancer biomarkers in serum samples using MALDI FT-ICR mass spectrometry data. Bioinformatics. 2009;25(2):251–257. doi: 10.1093/bioinformatics/btn610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benet T, Austin S. On-line cleanup for 2-aminobenzamide-labeled oligosaccharides. Anal Biochem. 2011;414(1):166–168. doi: 10.1016/j.ab.2011.03.002. [DOI] [PubMed] [Google Scholar]
- Bereman MS, Comins DL, Muddiman DC. Increasing the hydrophobicity and electrospray response of glycans through derivatization with novel cationic hydrazides. Chemical Communications. 2010;46(2):237–239. doi: 10.1039/B915589a. [DOI] [PubMed] [Google Scholar]
- Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230(2):229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- Botelho JC, Atwood JA, III, L C, Alvarez-Manilla G, York WS, Orlando R. Quantification by isobaric labeling (QUIBL) for the comparative glycomic study of O-linked glycans. Int J Mass Spectrom. 2008;287:137–142. [Google Scholar]
- Bowman MJ, Zaia J. Tags for the stable isotopic labeling of carbohydrates and quantitative analysis by mass spectrometry. Anal Chem. 2007;79(15):5777–5784. doi: 10.1021/ac070581b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman MJ, Zaia J. Comparative Glycomics Using a Tetraplex Stable-Isotope Coded Tag. Anal Chem. 2010;82(7):3023–3031. doi: 10.1021/ac100108w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breidenbacha MA, Gallagherb JEG, Kingc DS, Smarta BP, Wua P, Bertozzia CR. Targeted metabolic labeling of yeast N-glycans with unnatural sugars. Proc Natl Acad Sci USA. 2010;107:3988–3993. doi: 10.1073/pnas.0911247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucior I, Scheuring S, Engel A, Burger MM. Carbohydrate-carbohydrate interaction provides adhesion force and specificity for cellular recognition. J Cell Biol. 2004;165(4):529–537. doi: 10.1083/jcb.200309005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callewaert N, Geysens S, Molemans F, Contreras R. Ultrasensitive profiling and sequencing of N-linked oligosaccharides using standard DNA-sequencing equipment. Glycobiology. 2001;11(4):275–281. doi: 10.1093/glycob/11.4.275. [DOI] [PubMed] [Google Scholar]
- Callewaert N, Van Vlierberghe H, Van Hecke A, Laroy W, Delanghe J, Contreras R. Noninvasive diagnosis of liver cirrhosis using DNA sequencer-based total serum protein glycomics. Nat Med. 2004;10(4):429–434. doi: 10.1038/nm1006. [DOI] [PubMed] [Google Scholar]
- Campa C, Coslovi A, Flamigni A, Rossi M. Overview on advances in capillary electrophoresis-mass spectrometry of carbohydrates: a tabulated review. Electrophoresis. 2006;27(11):2027–2050. doi: 10.1002/elps.200500960. [DOI] [PubMed] [Google Scholar]
- Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics. 2008;24(9):1214–1216. doi: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- Carlson DM. Structures and immunochemical properties of oligosaccharides isolated from pig submaxillary mucins. J Biol Chem. 1968;243(3):616–626. [PubMed] [Google Scholar]
- Catala C, Howe KJ, Hucko S, Rose JKC, Thannhauser TW. Towards characterization of the glycoproteome of tomato (Solanum lycopersicum) fruit using Concanavalin A lectin affinity chromatography and LC-MALDI-MS/MS analysis. Proteomics. 2011;11(8):1530–1544. doi: 10.1002/pmic.201000424. [DOI] [PubMed] [Google Scholar]
- Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM. GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res. 2008;7(4):1650–1659. doi: 10.1021/pr7008252. [DOI] [PubMed] [Google Scholar]
- Charlwood J, Birrell H, Organ A, Camilleri P. A chromatographic and mass spectrometric strategy for the analysis of oligosaccharides: determination of the glycan structures in porcine thyroglobulin. Rapid Commun Mass Spectrom. 1999;13(8):716–723. doi: 10.1002/(sici)1097-0231(19990430)13:8<716::aid-rcm547>3.0.co;2-c. doi:10.1002/(SICI)1097-0231(19990430)13:8<716::AID-RCM547>3.0.CO;2-C10.1002/(SICI)109 7-0231(19990430)13:8<716::AID-RCM547>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Chen FT, Evangelista RA. Profiling glycoprotein n-linked oligosaccharide by capillary electrophoresis. Electrophoresis. 1998;19(15):2639–2644. doi: 10.1002/elps.1150191512. [DOI] [PubMed] [Google Scholar]
- Chen X, Flynn GC. Analysis of N-glycans from recombinant immunoglobulin G by on-line reversed-phase high-performance liquid chromatography/mass spectrometry. Anal Biochem. 2007;370(2):147–161. doi: 10.1016/j.ab.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Cheresh DA, Reisfeld RA, Varki AP. O-acetylation of disialoganglioside GD3 by human melanoma cells creates a unique antigenic determinant. Science. 1984;225(4664):844–846. doi: 10.1126/science.6206564. [DOI] [PubMed] [Google Scholar]
- Chiesa C, Horvath C. Capillary zone electrophoresis of malto-oligosaccharides derivatized with 8-aminonaphthalene-1,3,6-trisulfonic acid. J Chromatogr. 1993;645(2):337–352. doi: 10.1016/0021-9673(93)83394-8. [DOI] [PubMed] [Google Scholar]
- Chu CS, Ninonuevo MR, Clowers BH, Perkins PD, An HJ, Yin HF, Lebrilla CB. Profile of native N-linked glycan structures from human serum using high performance liquid chromatography on a microfluidic chip and time-of-flight mass spectrometry. Proteomics. 2009;9(7):1939–1951. doi: 10.1002/pmic.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciucanu I, Costello CE. Elimination of oxidative degradation during the per-O-methylation of carbohydrates. J Am Chem Soc. 2003;125(52):16213–16219. doi: 10.1021/ja035660t. [DOI] [PubMed] [Google Scholar]
- Ciucanu I, K F. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 1984;131(2):209–217. [Google Scholar]
- Clarke A, Harmon B, DeFelippis MR. Analysis of 3-(acetylamino)-6-aminoacridine-derivatized oligosaccharides from recombinant monoclonal antibodies by liquid chromatography-mass spectrometry. Anal Biochem. 2009;390:209–211. doi: 10.1016/j.ab.2009.04.022. [DOI] [PubMed] [Google Scholar]
- Clausen H, Bennett EP. A family of UDP-GalNAc: polypeptide N-acetylgalactosaminyl-transferases control the initiation of mucin-type O-linked glycosylation. Glycobiology. 1996;6(6):635–646. doi: 10.1093/glycob/6.6.635. [DOI] [PubMed] [Google Scholar]
- Clowers BH, Dwivedi P, Steiner WE, Hill HH, Jr, Bendiak B. Separation of sodiated isobaric disaccharides and trisaccharides using electrospray ionization-atmospheric pressure ion mobility-time of flight mass spectrometry. J Am Soc Mass Spectrom. 2005;16(5):660–669. doi: 10.1016/j.jasms.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Clowers BH, Hill HH., Jr Mass analysis of mobility-selected ion populations using dual gate, ion mobility, quadrupole ion trap mass spectrometry. Anal Chem. 2005;77(18):5877–5885. doi: 10.1021/ac050700s. [DOI] [PubMed] [Google Scholar]
- Cook KS, Bullock K, Sullivan T. Development and qualification of an antibody rapid deglycosylation method. Biologicals. 2012;40(2):109–117. doi: 10.1016/j.biologicals.2011.12.010. [DOI] [PubMed] [Google Scholar]
- Cooper CA, Gasteiger E, Packer NH. GlycoMod–a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics. 2001;1(2):340–349. doi: 10.1002/1615-9861(200102)1:2<340::AID-PROT340>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Costello CE, Contado-Miller JM, Cipollo JF. A glycomics platform for the analysis of permethylated oligosaccharide alditols. J Am Soc Mass Spectrom. 2007;18(10):1799–1812. doi: 10.1016/j.jasms.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall’Olio F, Chiricolo M. Sialyltransferases in cancer. Glycoconj J. 2001;18(11-12):841–850. doi: 10.1023/a:1022288022969. [DOI] [PubMed] [Google Scholar]
- Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. The GlycanBuilder and GlycoWorkbench glycoinformatics tools: updates and new developments. Biol Chem. 2012;393(11):1357–1362. doi: 10.1515/hsz-2012-0135. [DOI] [PubMed] [Google Scholar]
- Damerell D, Ceroni A, Maass K, Ranzinger R, Dell A, Haslam SM. Annotation of glycomics MS and MS/MS spectra using the GlycoWorkbench software tool. Carbohydrate Microarrays: Methods and Protocols. 2015;1273:3–15. doi: 10.1007/978-1-4939-2343-4_1. [DOI] [PubMed] [Google Scholar]
- de Leoz MLA, An HJ, Kronewitter S, Kim J, Beecroft S, Vinall R, Lebrilla C. Glycomic approach for potential biomarkers on prostate cancer: Profiling of N-linked glycans in human sera and pRNS cell lines. Disease Markers. 2008;25(4-5):243–258. doi: 10.1155/2008/515318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desantos-Garcia JL, Khalil SI, Hussein A, Hu Y, Mechref Y. Enhanced sensitivity of LC-MS analysis of permethylated N-glycans through online purification. Electrophoresis. 2011;32(24):3516–3525. doi: 10.1002/elps.201100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Zhou S, Mechref Y. LC-MS/MS analysis of permethylated free oligosaccharides and N-glycans derived from human, bovine, and goat milk samples. Electrophoresis. 2016;37(11):1532–1548. doi: 10.1002/elps.201500561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: quality control in the secretory pathway. Science. 1999;286(5446):1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Evangelista RA, Guttman A, Chen FT. Acid-catalyzed reductive amination of aldoses with 8-aminopyrene-1,3,6-trisulfonate. Electrophoresis. 1996;17(2):347–351. doi: 10.1002/elps.1150170210. [DOI] [PubMed] [Google Scholar]
- Fang J, Doneanu C, Alley WR, Jr, Yu YQ, Beck A, Chen W. Advanced assessment of the physicochemical characteristics of Remicade(R) and Inflectra(R) by sensitive LC/MS techniques. MAbs. 2016;8(6):1021–1034. doi: 10.1080/19420862.2016.1193661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn LS, McLean JA. Simultaneous glycoproteomics on the basis of structure using ion mobility-mass spectrometry. Mol Biosyst. 2009;5(11):1298–1302. doi: 10.1039/b909745g. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Esko JD, Parodi AJ. Glycans in Glycoprotein Quality Control. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd. Cold Spring Harbor; NY: 2009. [PubMed] [Google Scholar]
- Furukawa J, Piao J, Yoshida Y, Okada K, Yokota I, Higashino K, Shinohara Y. Quantitative O-Glycomics by Microwave-Assisted beta-Elimination in the Presence of Pyrazolone Analogues. Anal Chem. 2015;87(15):7524–7528. doi: 10.1021/acs.analchem.5b02155. [DOI] [PubMed] [Google Scholar]
- Gaye MM, Valentine SJ, Hu Y, Mirjankar N, Hammoud ZT, Mechref Y, Clemmer DE. Ion Mobility-Mass Spectrometry Analysis of Serum N-linked Glycans from Esophageal Adenocarcinoma Phenotypes. J Proteome Res. 2012;11(12):6102–6110. doi: 10.1021/pr300756e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil GC, Kim YG, Kim BG. A relative and absolute quantification of neutral N-linked oligosaccharides using modification with carboxymethyl trimethylammonium hydrazide and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Biochem. 2008;379(1):45–59. doi: 10.1016/j.ab.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Gill DJ, Clausen H, Bard F. Location, location, location: new insights into O-GalNAc protein glycosylation. Trends Cell Biol. 2011;21(3):149–158. doi: 10.1016/j.tcb.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Goetz JA, Novotny MV, Mechref Y. Enzymatic/chemical release of O-glycans allowing MS analysis at high sensitivity. Anal Chem. 2009;81(23):9546–9552. doi: 10.1021/ac901363h. [DOI] [PubMed] [Google Scholar]
- Goldberg D, Sutton-Smith M, Paulson J, Dell A. Automatic annotation of matrix-assisted laser desorption/ionization N-glycan spectra. Proteomics. 2005;5(4):865–875. doi: 10.1002/pmic.200401071. [DOI] [PubMed] [Google Scholar]
- Gornik O, Lauc G. Glycosylation of serum proteins in inflammatory diseases. Dis Markers. 2008;25(4-5):267–278. doi: 10.1155/2008/493289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw JW, Burgers PC, Trikoupis MA, Terlouw JK. Derivatization of small oligosaccharides prior to analysis by matrix-assisted laser desorption/ionization using glycidyltrimethylammonium chloride and Girard’s reagent T. Rapid Communications in Mass Spectrometry. 2002;16(10):905–912. doi: 10.1002/Rcm.654. [DOI] [PubMed] [Google Scholar]
- Gray CJ, Thomas B, Upton R, Migas LG, Eyers CE, Barran PE, Flitsch SL. Applications of ion mobility mass spectrometry for high throughput, high resolution glycan analysis. Biochim Biophys Acta. 2016;1860(8):1688–1709. doi: 10.1016/j.bbagen.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Guttman A, Chen FT, Evangelista RA. Separation of 1-aminopyrene-3,6,8-trisulfonate-labeled asparagine-linked fetuin glycans by capillary gel electrophoresis. Electrophoresis. 1996;17(2):412–417. doi: 10.1002/elps.1150170221. [DOI] [PubMed] [Google Scholar]
- Guttman A, Pritchett T. Capillary gel electrophoresis separation of high-mannose type oligosaccharides derivatized by 1-aminopyrene-3,6,8-trisulfonic acid. Electrophoresis. 1995;16(10):1906–1911. doi: 10.1002/elps.11501601314. [DOI] [PubMed] [Google Scholar]
- Hahne H, Neubert P, Kuhn K, Etienne C, Bomgarden R, Rogers JC, Kuster B. Carbonyl-Reactive Tandem Mass Tags for the Proteome-Wide Quantification of N-Linked Glycans. Anal Chem. 2012;84(8):3716–3724. doi: 10.1021/ac300197c. [DOI] [PubMed] [Google Scholar]
- Hanisch FG, Muller S. Analysis of methylated O-glycan alditols by reversed-phase NanoLC coupled CAD-ESI mass spectrometry. Carbohydrate Microarrays: Methods and Protocols. 2009;534:107–115. doi: 10.1007/978-1-59745-022-5_8. [DOI] [PubMed] [Google Scholar]
- Harvey DJ. Fragmentation of negative ions from carbohydrates: part 1. Use of nitrate and other anionic adducts for the production of negative ion electrospray spectra from N-linked carbohydrates. J Am Soc Mass Spectrom. 2005a;16(5):622–630. doi: 10.1016/j.jasms.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Harvey DJ. Fragmentation of negative ions from carbohydrates: part 3. Fragmentation of hybrid and complex N-linked glycans. J Am Soc Mass Spectrom. 2005b;16(5):647–659. doi: 10.1016/j.jasms.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Harvey DJ, Royle L, Radcliffe CM, Rudd PM, Dwek RA. Structural and quantitative analysis of N-linked glycans by matrix-assisted laser desorption ionization and negative ion nanospray mass spectrometry. Anal Biochem. 2008;376(1):44–60. doi: 10.1016/j.ab.2008.01.025. [DOI] [PubMed] [Google Scholar]
- Harvey DJ, Scarff CA, Crispin M, Scanlan CN, Bonomelli C, Scrivens JH. MALDI-MS/MS with traveling wave ion mobility for the structural analysis of N-linked glycans. J Am Soc Mass Spectrom. 2012;23(11):1955–1966. doi: 10.1007/s13361-012-0425-8. [DOI] [PubMed] [Google Scholar]
- Harvey DJ, Scarff CA, Edgeworth M, Crispin M, Scanlan CN, Sobott F, Scrivens JH. Travelling wave ion mobility and negative ion fragmentation for the structural determination of N-linked glycans. Electrophoresis. 2013;34(16):2368–2378. doi: 10.1002/elps.201200669. [DOI] [PubMed] [Google Scholar]
- Hase S. High-performance liquid chromatography of pyridylaminated saccharides. Methods Enzymol. 1994;230:225–237. doi: 10.1016/0076-6879(94)30015-1. [DOI] [PubMed] [Google Scholar]
- Hase S, Ikenaka T, Matsushima Y. Structure analyses of oligosaccharides by tagging of the reducing end sugars with a fluorescent compound. Biochem Biophys Res Commun. 1978;85(1):257–263. doi: 10.1016/s0006-291x(78)80037-0. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hitchcock AM, Costello CE, Zaia J. Glycoform quantification of chondroitin/dermatan sulfate using an LC/MS/MS platform. Biochemistry. 2006;45:2350–2361. doi: 10.1021/bi052100t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock AM, Yates KE, Shortkroff S, Costello CE, Zaia J. Optimized extraction of glycosaminoglycans from normal and osteoarthritic cartilage for glycomics profiling. Glycobiology. 2006;17:25–35. doi: 10.1093/glycob/cwl046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Desantos-Garcia JL, Mechref Y. Comparative glycomic profiling of isotopically permethylated N-glycans by LC-ESI-MS. Rapid Commun Mass Spectrom. 2013;27:865–877. doi: 10.1002/rcm.6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhou S, Khalil SI, Renteria CL, Mechref Y. Glycomic profiling of tissue sections by LC-MS. Anal Chem. 2013;85(8):4074–4079. doi: 10.1021/ac400106x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Zhou S, Yu CY, Tang H, Mechref Y. Automated annotation and quantitation of glycans by liquid chromatography/electrospray ionization mass spectrometric analysis using the MultiGlycan-ESI computational tool. Rapid Commun Mass Spectrom. 2015;29(1):135–142. doi: 10.1002/rcm.7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YL, Mechref Y. Comparing MALDI-MS, RP-LC-MALDI-MS and RP-LC-ESI-MS glycomic profiles of permethylated N-glycans derived from model glycoproteins and human blood serum. Electrophoresis. 2012;33(12):1768–1777. doi: 10.1002/elps.201100703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua S, Saunders M, Dimapasoc LM, Jeong SH, Kim BJ, Kim S, An HJ. Differentiation of cancer cell origin and molecular subtype by plasma membrane N-glycan profiling. J Proteome Res. 2014;13(2):961–968. doi: 10.1021/pr400987f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Aminoff D. Enzymes that destroy blood group specificity. V. The oligosaccharase of Clostridium perfringens. J Biol Chem. 1972;247(21):6737–6742. [PubMed] [Google Scholar]
- Hulsmeier AJ, Gehrig PM, Geyer R, Sack R, Gottstein B, Deplazes P, Kohler P. A major Echinococcus multilocularis antigen is a mucin-type glycoprotein. J Biol Chem. 2002;277(8):5742–5748. doi: 10.1074/jbc.M107161200. [DOI] [PubMed] [Google Scholar]
- Isailovic D, Kurulugama RT, Plasencia MD, Stokes ST, Kyselova Z, Goldman R, Clemmer DE. Profiling of human serum glycans associated with liver cancer and cirrhosis by IMS-MS. J Proteome Res. 2008;7(3):1109–1117. doi: 10.1021/pr700702r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki S, Yamamoto K, Fujisaki M, Izumi K, Tochikura T, Yokoyama T. Purification and characterization of a novel fungal endo-beta-N-acetylglucosaminidase acting on complex oligosaccharides of glycoproteins. Agric Biol Chem. 1990;54(1):97–106. [PubMed] [Google Scholar]
- Kakehi K, Funakubo T, Suzuki S, Oda Y, Kitada Y. 3-Aminobenzamide and 3-aminobenzoic acid, tags for capillary electrophoresis of complex carbohydrates with laser-induced fluorescent detection. J Chromatogr A. 1999;863(2):205–218. doi: 10.1016/s0021-9673(99)00978-4. [DOI] [PubMed] [Google Scholar]
- Kalay H, Ambrosini M, van Berkel PHC, Parren PWHI, van Kooyk Y, Vallejo JJG. Online nanoliquid chromatography-mass spectrometry and nanofluorescence detection for high-resolution quantitative N-glycan analysis. Anal Biochem. 2012;423(1):153–162. doi: 10.1016/j.ab.2012.01.015. [DOI] [PubMed] [Google Scholar]
- Kameyama A, Kaneda Y, Yamanaka H, Yoshimine H, Narimatsu H, Shinohara Y. Detection of oligosaccharides labeled with cyanine dyes using matrix-assisted laser desorption/ionization mass spectrometry. Anal Chem. 2004;76(15):4537–4542. doi: 10.1021/Ac049897z. [DOI] [PubMed] [Google Scholar]
- Kang P, Mechref Y, Klouckova I, Novotny MV. Solid-phase permethylation of glycans for mass spectrometric analysis. Rapid Commun Mass Spectrom. 2005;19(23):3421–3428. doi: 10.1002/rcm.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P, Mechref Y, Kyselova Z, Goetz JA, Novotny MV. Comparative Glycomic Mapping through Quantitative Permethylation and Stable-Isotope Labeling. Anal Chem. 2007;79:6064–6073. doi: 10.1021/ac062098r. [DOI] [PubMed] [Google Scholar]
- Kang P, Mechref Y, Novotny MV. High-throughput solid-phase permethylation of glycans prior to mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:721–734. doi: 10.1002/rcm.3395. [DOI] [PubMed] [Google Scholar]
- Karamanos NK, Tsegenidis T, Antonopoulos CA. Analysis of Neutral Sugars as Dinitrophenyl-Hydrazones by High-Performance Liquid-Chromatography. J Chromatogr. 1987;405:221–228. doi: 10.1016/S0021-9673(01)81764-7. [DOI] [Google Scholar]
- Karas M, Ehring H, Nordhoff E, Stahl B, Strupat K, Hillenkamp F, Krebs B. Matrix-Assisted Laser-Desorption Ionization Mass-Spectrometry with Additives to 2,5-Dihydroxybenzoic Acid. Organic Mass Spectrometry. 1993;28(12):1476–1481. doi: 10.1002/oms.1210281219. [DOI] [Google Scholar]
- Kohla G, Stockfleth E, Schauer R. Gangliosides with O-acetylated sialic acids in tumors of neuroectodermal origin. Neurochem Res. 2002;27(7-8):583–592. doi: 10.1023/a:1020211714104. [DOI] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Kozak RP, Royle L, Gardner RA, Fernandes DL, Wuhrer M. Suppression of peeling during the release of O-glycans by hydrazinolysis. Anal Biochem. 2012;423(1):119–128. doi: 10.1016/j.ab.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Kyselova Z, Mechref Y, Kang P, Goetz JA, Dobrolecki LE, Sledge GW, Novotny MV. Breast cancer diagnosis and prognosis through quantitative measurements of serum glycan profiles. Clinical Chemistry. 2008;54(7):1166–1175. doi: 10.1373/clinchem.2007.087148. [DOI] [PubMed] [Google Scholar]
- Lattova E, Perreault H. Labelling saccharides with phenylhydrazine for electrospray and matrix-assisted laser desorption-ionization mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2003a;793(1):167–179. doi: 10.1016/s1570-0232(03)00374-x. [DOI] [PubMed] [Google Scholar]
- Lattova E, Perreault H. Profiling of N-linked oligosaccharides using phenylhydrazine derivatization and mass spectrometry. J Chromatogr A. 2003b;1016(1):71–87. doi: 10.1016/s0021-9673(03)01297-4. [DOI] [PubMed] [Google Scholar]
- Lauber MA, Brousmiche DW, Hua Z, Koza SM, Guthrie E, Magnelli P, Fountain KJ. Rapid Preparation of Released N-Glycans for HILIC Analysis Using a Novel Fluorescence and MS-Active Labeling Reagent [Application Note] Waters Corp.; Milford, MA, U.S.A.: 2015. Retrieved from. [DOI] [PubMed] [Google Scholar]
- Laubli H, Stevenson JL, Varki A, Varki NM, Borsig L. L-selectin facilitation of metastasis involves temporal induction of Fut7-dependent ligands at sites of tumor cell arrest. Cancer Res. 2006;66(3):1536–1542. doi: 10.1158/0008-5472.can-05-3121. [DOI] [PubMed] [Google Scholar]
- Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. Evolutionary Differences in Glycosaminoglycan Fine Structure Detected by Quantitative Glycan Reductive Isotope Labeling. J Biol Chem. 2008;283:33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer GZ. Glycoprotein folding, quality control and ER-associated degradation. Curr Opin Struct Biol. 2009;19(5):515–523. doi: 10.1016/j.sbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Leteux C, Childs RA, Chai WG, Stoll MS, Kogelberg H, Feizi T. Biotinyl-L-3-(2-naphthyl)-alanine hydrazide derivatives of N-glycans: versatile solid-phase probes for carbohydrate-recognition studies. Glycobiology. 1998;8(3):227–236. doi: 10.1093/glycob/8.3.227. [DOI] [PubMed] [Google Scholar]
- Li M, Kinzer JA. Structural analysis of oligosaccharides by a combination of electrospray mass spectrometry and bromine isotope tagging of reducing-end sugars with 2-amino-5-bromopyridine. Rapid Commun Mass Spectrom. 2003;17(13):1462–1466. doi: 10.1002/rcm.1064. [DOI] [PubMed] [Google Scholar]
- Lizak C, Gerber S, Numao S, Aebi M, Locher KP. X-ray structure of a bacterial oligosaccharyltransferase. Nature. 2011;474(7351):350–355. doi: 10.1038/nature10151. [DOI] [PubMed] [Google Scholar]
- Lochnit G, Geyer R. An optimized protocol for nano-LC-MALDI-TOF-MS coupling for the analysis of proteolytic digests of glycoproteins. Biomedical Chromatography. 2004;18(10):841–848. doi: 10.1002/bmc.399. [DOI] [PubMed] [Google Scholar]
- Lonardi E, Deelder AM, Wuhrer M, Balog CIA. Microarray Technology Using Glycans Extracted from Natural Sources for Serum Antibody Fluorescent Detection. Carbohydrate Microarrays: Methods and Protocols. 2012;808:285–302. doi: 10.1007/978-1-61779-373-8_20. [DOI] [PubMed] [Google Scholar]
- Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- Ma S, Nashabeh W. Carbohydrate analysis of a chimeric recombinant monoclonal antibody by capillary electrophoresis with laser-induced fluorescence detection. Anal Chem. 1999;71(22):5185–5192. doi: 10.1021/ac990376z. [DOI] [PubMed] [Google Scholar]
- Maass K, Ranzinger R, Geyer H, von der Lieth CW, Geyer R. “Glyco-peakfinder”–de novo composition analysis of glycoconjugates. Proteomics. 2007;7(24):4435–4444. doi: 10.1002/pmic.200700253. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Shimizu C, Arao T, Andoh M, Katsumata N, Kohno T, Fujiwara Y. Identification of Predictive Biomarkers for Response to Trastuzumab Using Plasma FUCA Activity and N-Glycan Identified by MALDI-TOF-MS. J Proteome Res. 2009;8(2):457–462. doi: 10.1021/pr800655p. [DOI] [PubMed] [Google Scholar]
- Mauko L, Lacher NA, Pelzing M, Nordborg A, Haddad PR, Hilder EF. Comparison of ZIC-HILIC and graphitized carbon-based analytical approaches combined with exoglycosidase digestions for analysis of glycans from monoclonal antibodies. Journal of Chromatography B-Analytical Technologies in the Biomedical and Life Sciences. 2012;911:93–104. doi: 10.1016/j.jchromb.2012.10.043. [DOI] [PubMed] [Google Scholar]
- Means RE, Desrosiers RC. Resistance of native, oligomeric envelope on simian immunodeficiency virus to digestion by glycosidases. J Virol. 2000;74(23):11181–11190. doi: 10.1128/jvi.74.23.11181-11190.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechref Y, Hu Y, Desantos-Garcia JL, Hussein A, Tang H. Quantitative glycomics strategies. Mol Cell Proteomics. 2013;12(4):874–884. doi: 10.1074/mcp.R112.026310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechref Y, Hu Y, Garcia A, Zhou S, Desantos-Garcia JL, Hussein A. Defining putative glycan cancer biomarkers by MS. Bioanalysis. 2012;4:2457–2469. doi: 10.4155/bio.12.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechref Y, Muzikar J, Novotny MV. Comprehensive assessment of N-glycans derived from a murine monoclonal antibody: a case for multimethodological approach. Electrophoresis. 2005;26(10):2034–2046. doi: 10.1002/elps.200410345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechref Y, Novotny MV. Matrix-assisted laser desorption ionization mass spectrometry of acidic glycoconjugates facilitated by the use of spermine as a co-matrix. Journal of the American Society for Mass Spectrometry. 1998;9(12):1293–1302. doi: 10.1016/S1044-0305(98)00106-8. [DOI] [PubMed] [Google Scholar]
- Mehta A, Herrera H, Block T. Glycosylation and liver cancer. Adv Cancer Res. 2015;126:257–279. doi: 10.1016/bs.acr.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitei NS, Apte A, Snovida SI, Rogers JC, Saba J. Automating mass spectrometry-based quantitative glycomics using aminoxy tandem mass tag reagents with SimGlycan. J Proteomics. 2015;127(Pt A):211–222. doi: 10.1016/j.jprot.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Miura Y, Endo T. Glycomics and glycoproteomics focused on aging and age-related diseases–Glycans as a potential biomarker for physiological alterations. Biochim Biophys Acta. 2016;1860(8):1608–1614. doi: 10.1016/j.bbagen.2016.01.013. [DOI] [PubMed] [Google Scholar]
- Mohr MD, Bornsen KO, Widmer HM. Matrix-Assisted Laser-Desorption Ionization Mass-Spectrometry - Improved Matrix for Oligosaccharides. Rapid Communications in Mass Spectrometry. 1995;9(9):809–814. doi: 10.1002/rcm.1290090919. [DOI] [PubMed] [Google Scholar]
- Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3(6):313–320. doi: 10.1038/nchembio880. [DOI] [PubMed] [Google Scholar]
- Monsarrat B, Brando T, Condouret P, Nigou J, Puzo G. Characterization of mannooligosaccharide caps in mycobacterial lipoarabinomannan by capillary electrophoresis/electrospray mass spectrometry. Glycobiology. 1999;9(4):335–342. doi: 10.1093/glycob/9.4.335. [DOI] [PubMed] [Google Scholar]
- Montesino R, Calvo L, Vallin A, Rudd PM, Harvey DJ, Cremata JA. Structural characterization of N-linked oligosaccharides on monoclonal antibody Nimotuzumab through process development. Biologicals. 2012;40(4):288–298. doi: 10.1016/j.biologicals.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Nakajima K, Oda Y, Kinoshita M, Masuko T, Kakehi K. Time-resolved fluorometric analysis of carbohydrates labeled with amino-aromatic compounds by reductive amination. Analyst. 2002;127(7):972–976. doi: 10.1039/b202950b. [DOI] [PubMed] [Google Scholar]
- Naven TJP, Harvey DJ. Cationic Derivatization of Oligosaccharides with Girard’s T Reagent for Improved Performance in Matrix-assisted Laser Desorption/Ionization and Electrospray Mass Spectrometry. Rapid Commun Mass Spectrom. 1998;10(7):829–834. [Google Scholar]
- Ninonuevo MR, Park Y, Yin HF, Zhang JH, Ward RE, Clowers BH, Lebrilla CB. A strategy for annotating the human milk glycome. Journal of Agricultural and Food Chemistry. 2006;54(20):7471–7480. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- Nwosu CC, Aldredge DL, Lee H, Lerno LA, Zivkovic AM, German JB, Lebrilla CB. Comparison of the human and bovine milk N-glycome via high-performance microfluidic chip liquid chromatography and tandem mass spectrometry. J Proteome Res. 2012;11(5):2912–2924. doi: 10.1021/pr300008u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando R, Lim JM, Atwood JA, 3rd, Angel PM, Fang M, Aoki K, Wells L. IDAWG: Metabolic incorporation of stable isotope labels for quantitative glycomics of cultured cells. J Proteome Res. 2009;8(8):3816–3823. doi: 10.1021/pr8010028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst M, Altmann F. Influence of electrosorption, solvent, temperature, and ion polarity on the performance of LC-ESI-MS using graphitic carbon for acidic oligosaccharides. Anal Chem. 2008;80(19):7534–7542. doi: 10.1021/ac801024r. [DOI] [PubMed] [Google Scholar]
- Pabst M, Kolarich D, Poltl G, Dalik T, Lubec G, Hofinger A, Altmann F. Comparison of fluorescent labels for oligosaccharides and introduction of a new postlabeling purification method. Anal Biochem. 2009;384(2):263–273. doi: 10.1016/j.ab.2008.09.041. [DOI] [PubMed] [Google Scholar]
- Packer NH, Lawson MA, Jardine DR, Redmond JW. A general approach to desalting oligosaccharides released from glycoproteins. Glycoconjugate Journal. 1998;15(8):737–747. doi: 10.1023/A:1006983125913. [DOI] [PubMed] [Google Scholar]
- Patel T, Bruce J, Merry A, Bigge C, Wormald M, Jaques A, Parekh R. Use of hydrazine to release in intact and unreduced form both N- and O-linked oligosaccharides from glycoproteins. Biochemistry. 1993;32(2):679–693. doi: 10.1021/bi00053a037. [DOI] [PubMed] [Google Scholar]
- Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 1: methodology) J Am Soc Mass Spectrom. 2002a;13(11):1331–1340. doi: 10.1016/S1044-0305(02)00645-1. [DOI] [PubMed] [Google Scholar]
- Pfenninger A, Karas M, Finke B, Stahl B. Structural analysis of underivatized neutral human milk oligosaccharides in the negative ion mode by nano-electrospray MS(n) (part 2: application to isomeric mixtures) J Am Soc Mass Spectrom. 2002b;13(11):1341–1348. doi: 10.1016/S1044-0305(02)00646-3. [DOI] [PubMed] [Google Scholar]
- Plummer TH, Jr, Tarentino AL. Purification of the oligosaccharide-cleaving enzymes of Flavobacterium meningosepticum. Glycobiology. 1991;1(3):257–263. doi: 10.1093/glycob/1.3.257. [DOI] [PubMed] [Google Scholar]
- Prater BD, Anumula KR, Hutchins JT. Automated sample preparation facilitated by PhyNexus MEA purification system for oligosaccharide mapping of glycoproteins. Anal Biochem. 2007;369(2):202–209. doi: 10.1016/j.ab.2007.07.011. [DOI] [PubMed] [Google Scholar]
- Rabouille C, Hui N, Hunte F, Kieckbusch R, Berger EG, Warren G, Nilsson T. Mapping the distribution of Golgi enzymes involved in the construction of complex oligosaccharides. J Cell Sci. 1995;108(Pt 4):1617–1627. doi: 10.1242/jcs.108.4.1617. [DOI] [PubMed] [Google Scholar]
- Ressom HW, Varghese RS, Goldman L, An Y, Loffredo CA, Abdel-Hamid M, Goldman R. Analysis of MALDI-TOF mass spectrometry data for discovery of peptide and glycan biomarkers of hepatocellular carcinoma. J Proteome Res. 2008;7(2):603–610. doi: 10.1021/pr0705237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Dwek RA. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem. 2008;376(1):1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Royle L, Mattu TS, Hart E, Langridge JI, Merry AH, Murphy N, Rudd PM. An analytical and structural database provides a strategy for Sequencing O-glycans from microgram quantities of glycoproteins. Anal Biochem. 2002;304(1):70–90. doi: 10.1006/abio.2002.5619. [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Deelder AM, Wuhrer M. Oligosaccharide analysis by graphitized carbon liquid chromatography-mass spectrometry. Anal Bioanal Chem. 2009;394(1):163–174. doi: 10.1007/s00216-009-2664-5. [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Miyamoto S, Kelly K, Lebrilla CB. N-Glycan profiling of dried blood spots. Anal Chem. 2012;84(1):396–402. doi: 10.1021/ac202775t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Miyamoto S, Lebrilla CB. Developments in the Identification of Glycan Biomarkers for the Detection of Cancer. Molecular & Cellular Proteomics. 2013;12(4):846–855. doi: 10.1074/mcp.R112.026799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Nguyen UT, Stroble C, Taylor SL, Taguchi A, Hanash SM, Miyamoto S. Enrichment strategies in glycomics-based lung cancer biomarker development. Proteomics Clin Appl. 2013;7(9-10):664–676. doi: 10.1002/prca.201200131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhaak LR, Steenvoorden E, Koeleman CA, Deelder AM, Wuhrer M. 2-picoline-borane: a non-toxic reducing agent for oligosaccharide labeling by reductive amination. Proteomics. 2010;10(12):2330–2336. doi: 10.1002/pmic.200900804. [DOI] [PubMed] [Google Scholar]
- Ruhaak LR, Taylor SL, Miyamoto S, Kelly K, Leiserowitz GS, Gandara D, Kim K. Chip-based nLC-TOF-MS is a highly stable technology for large-scale high-throughput analyses. Anal Bioanal Chem. 2013;405(14):4953–4958. doi: 10.1007/s00216-013-6908-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saba JA, Shen X, Jamieson JC, Perreault H. Effect of 1-phenyl-3-methyl-5-pyrazolone labeling on the fragmentation behavior of asialo and sialylated N-linked glycans under electrospray ionization conditions. Rapid Commun Mass Spectrom. 1999;13(8):704–711. doi: 10.1002/(sici)1097-0231(19990430)13:8<704::aid-rcm543>3.0.co;2-v. doi:10.1002/(SICI)1097-0231(19990430)13:8<704::AID-RCM543>3.0.CO;2-V10.1002/(SICI)109 7-0231(19990430)13:8<704::AID-RCM543>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Saba JA, Shen X, Jamieson JC, Perreault H. Investigation of different combinations of derivatization, separation methods and electrospray ionization mass spectrometry for standard oligosaccharides and glycans from ovalbumin. J Mass Spectrom. 2001;36(5):563–574. doi: 10.1002/jms.158. [DOI] [PubMed] [Google Scholar]
- Saldova R, Reuben JM, Abd Hamid UM, Rudd PM, Cristofanilli M. Levels of specific serum N-glycans identify breast cancer patients with higher circulating tumor cell counts. Ann Oncol. 2011;22(5):1113–1119. doi: 10.1093/annonc/mdq570. [DOI] [PubMed] [Google Scholar]
- Saldova R, Royle L, Radcliffe CM, Abd Hamid UM, Evans R, Arnold JN, Rudd PM. Ovarian cancer is associated with changes in glycosylation in both acute-phase proteins and IgG. Glycobiology. 2007;17(12):1344–1356. doi: 10.1093/glycob/cwm100. [DOI] [PubMed] [Google Scholar]
- Sandra K, Devreese B, Van Beeumen J, Stals I, Claeyssens M. The Q-Trap mass spectrometer, a novel tool in the study of protein glycosylation. J Am Soc Mass Spectrom. 2004;15(3):413–423. doi: 10.1016/j.jasms.2003.11.003. [DOI] [PubMed] [Google Scholar]
- Schwaiger H, Oefner PJ, Huber C, Grill E, Bonn GK. Capillary zone electrophoresis and micellar electrokinetic chromatography of 4-aminobenzonitrile carbohydrate derivatives. Electrophoresis. 1994;15(7):941–952. doi: 10.1002/elps.11501501138. [DOI] [PubMed] [Google Scholar]
- Seales EC, Jurado GA, Brunson BA, Wakefield JK, Frost AR, Bellis SL. Hypersialylation of beta1 integrins, observed in colon adenocarcinoma, may contribute to cancer progression by up-regulating cell motility. Cancer Res. 2005;65(11):4645–4652. doi: 10.1158/0008-5472.can-04-3117. [DOI] [PubMed] [Google Scholar]
- Sekiya S, Wada Y, Tanaka K. Derivatization for stabilizing sialic acids in MALDI-MS. Anal Chem. 2005;77(15):4962–4968. doi: 10.1021/ac050287o. [DOI] [PubMed] [Google Scholar]
- Snovida SI, Chen VC, Perreault H. Use of a 2,5-dihydroxybenzoic acid/aniline MALDI matrix for improved detection and on-target derivatization of glycans: A preliminary report. Anal Chem. 2006;78(24):8561–8568. doi: 10.1021/ac061375r. [DOI] [PubMed] [Google Scholar]
- Snovida SI, Rak-Banville JM, Perreault H. On the use of DHB/aniline and DHB/N,N-dimethylaniline matrices for improved detection of carbohydrates: automated identification of oligosaccharides and quantitative analysis of sialylated glycans by MALDI-TOF mass spectrometry. J Am Soc Mass Spectrom. 2008;19(8):1138–1146. doi: 10.1016/j.jasms.2008.04.033. [DOI] [PubMed] [Google Scholar]
- Song X, Ju H, Lasanajak Y, Kudelka MR, Smith DF, Cummings RD. Oxidative release of natural glycans for functional glycomics. Nat Methods. 2016;13(6):528–534. doi: 10.1038/nmeth.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparbier K, Asperger A, Kessler I, Koch S, Shi G, Wenzel T, Kostrzewa M. Identification and characterization of glycoproteins in human serum by means of glyco-specific magnetic bead separation and LCMALDI analysis. Molecular & Cellular Proteomics. 2006;5(10):S134–S134. [Google Scholar]
- Stahl B, Steup M, Karas M, Hillenkamp F. Analysis of Neutral Oligosaccharides by Matrix-Assisted Laser Desorption - Ionization Mass-Spectrometry. Anal Chem. 1991;63(14):1463–1466. doi: 10.1021/Ac00014a022. [DOI] [Google Scholar]
- Stanley P. Golgi glycosylation. Cold Spring Harb Perspect Biol. 2011;3(4) doi: 10.1101/cshperspect.a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staples GO, Bowman MJ, Costello CE, Hitchcock AM, Lau JM, Leymarie N, Zaia J. A chip-based amide-HILIC LC/MS platform for glycosaminoglycan glycomics profiling. Proteomics. 2009;9(3):686–695. doi: 10.1002/pmic.200701008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson JL, Choi SH, Varki A. Differential metastasis inhibition by clinically relevant levels of heparins–correlation with selectin inhibition, not antithrombotic activity. Clin Cancer Res. 2005;11(19 Pt 1):7003–7011. doi: 10.1158/1078-0432.ccr-05-1131. [DOI] [PubMed] [Google Scholar]
- Sugahara D, Amano J, Irimura T. Fluorescence labeling of oligosaccharides useful in the determination of molecular interactions. Analytical Sciences. 2003;19(1):167–169. doi: 10.2116/analsci.19.167. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Muller O, Guttman A, Karger BL. Analysis of 1-aminopyrene-3,6,8-trisulfonate-derivatized oligosaccharides by capillary electrophoresis with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal Chem. 1997;69(22):4554–4559. doi: 10.1021/ac970090z. [DOI] [PubMed] [Google Scholar]
- Suzuki S. Recent developments in liquid chromatography and capillary electrophoresis for the analysis of glycoprotein glycans. Anal Sci. 2013;29(12):1117–1128. doi: 10.2116/analsci.29.1117. [DOI] [PubMed] [Google Scholar]
- Szabo Z, Guttman A, Rejtar T, Karger BL. Improved sample preparation method for glycan analysis of glycoproteins by CE-LIF and CE-MS. Electrophoresis. 2010;31(8):1389–1395. doi: 10.1002/elps.201000037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N. Three-dimensional mapping of N-linked oligosaccharides using anion-exchange, hydrophobic and hydrophilic interaction modes of high-performance liquid chromatography. Journal of Chromatography A. 1996;720(1-2):217–225. doi: 10.1016/0021-9673(95)00328-2. [DOI] [PubMed] [Google Scholar]
- Takahashi N, Nishibe H. Some characteristics of a new glycopeptidase acting on aspartylglycosylamine linkages. J Biochem. 1978;84(6):1467–1473. doi: 10.1093/oxfordjournals.jbchem.a132270. [DOI] [PubMed] [Google Scholar]
- Takasaki S, Mizuochi T, Kobata A. Hydrazinolysis of asparagine-linked sugar chains to produce free oligosaccharides. Methods Enzymol. 1982;83:263–268. doi: 10.1016/0076-6879(82)83019-x. [DOI] [PubMed] [Google Scholar]
- Takegawa Y, Deguchi K, Ito H, Keira T, Nakagawa H, Nishimura SI. Simple separation of isomeric sialylated N-glycopeptides by a zwitterionic type of hydrophilic interaction chromatography. J Sep Sci. 2006;29(16):2533–2540. doi: 10.1002/jssc.200600133. [DOI] [PubMed] [Google Scholar]
- Tarentino AL, Gomez CM, Plummer TH., Jr Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry. 1985;24(17):4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- Tarentino AL, Maley F. Purification and properties of an endo-beta-N-acetylglucosaminidase from Streptomyces griseus. J Biol Chem. 1974;249(3):811–817. [PubMed] [Google Scholar]
- Tarentino AL, Plummer TH, Jr, Maley F. The release of intact oligosaccharides from specific glycoproteins by endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1974;249(3):818–824. [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA. All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13(1):1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Thomsson KA, Karlsson NG, Hansson GC. Liquid chromatography-electrospray mass spectrometry as a tool for the analysis of sulfated oligosaccharides from mucin glycoproteins. J Chromatogr A. 1999;854(1-2):131–139. doi: 10.1016/s0021-9673(99)00625-1. [DOI] [PubMed] [Google Scholar]
- Tousi F, Bones J, Hancock WS, Hincapie M. Differential chemical derivatization integrated with chromatographic separation for analysis of isomeric sialylated N-glycans: a nano-hydrophilic interaction liquid chromatography-MS platform. Anal Chem. 2013;85(17):8421–8428. doi: 10.1021/ac4018007. [DOI] [PubMed] [Google Scholar]
- Tretter V, Altmann F, Marz L. Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached alpha 1—3 to the asparagine-linked N-acetylglucosamine residue. Eur J Biochem. 1991;199(3):647–652. doi: 10.1111/j.1432-1033.1991.tb16166.x. [DOI] [PubMed] [Google Scholar]
- Trimble RB, Tarentino AL. Identification of distinct endoglycosidase (endo) activities in Flavobacterium meningosepticum: endo F1, endo F2, and endo F3. Endo F1 and endo H hydrolyze only high mannose and hybrid glycans. J Biol Chem. 1991;266(3):1646–1651. [PubMed] [Google Scholar]
- Trombetta ES, Helenius A. Lectins as chaperones in glycoprotein folding. Curr Opin Struct Biol. 1998;8(5):587–592. doi: 10.1016/s0959-440x(98)80148-6. [DOI] [PubMed] [Google Scholar]
- Tsai TH, Wang M, Di Poto C, Hu Y, Zhou S, Zhao Y, Ressom HW. LC-MS profiling of N-Glycans derived from human serum samples for biomarker discovery in hepatocellular carcinoma. J Proteome Res. 2014;13(11):4859–4868. doi: 10.1021/pr500460k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabres P, Sevin C, Amoric JC, Odievre MH, Saudubray JM, de Prost Y. Skin manifestations of protein glycosylation deficiency, the CDG (carbohydrate deficient glycoprotein) type 1 syndrome. Ann Dermatol Venereol. 1998;125(10):715–716. [PubMed] [Google Scholar]
- Vakhrushev SY, Dadimov D, Peter-Katalinic J. Software platform for high-throughput glycomics. Anal Chem. 2009;81(9):3252–3260. doi: 10.1021/ac802408f. [DOI] [PubMed] [Google Scholar]
- Vanderschaeghe D, Debruyne E, Van Vlierberghe H, Callewaert N, Delanghe J. Analysis of gamma-globulin mobility on routine clinical CE equipment: exploring its molecular basis and potential clinical utility. Electrophoresis. 2009;30(15):2617–2623. doi: 10.1002/elps.200900054. [DOI] [PubMed] [Google Scholar]
- Vanderschaeghe D, Laroy W, Sablon E, Halfon P, Van Hecke A, Delanghe J, Callewaert N. GlycoFibroTest is a highly performant liver fibrosis biomarker derived from DNA sequencer-based serum protein glycomics. Mol Cell Proteomics. 2009;8(5):986–994. doi: 10.1074/mcp.M800470-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Kannagi R, Toole BP. Glycosylation Changes in Cancer. In: Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd. Cold Spring Harbor; NY: 2009. [PubMed] [Google Scholar]
- Varki NM, Varki A. Heparin inhibition of selectin-mediated interactions during the hematogenous phase of carcinoma metastasis: rationale for clinical studies in humans. Semin Thromb Hemost. 2002;28(1):53–66. doi: 10.1055/s-2002-20564. [DOI] [PubMed] [Google Scholar]
- Vondeyn W, York WS, Albersheim P, Darvill AG. 1-Alkoxyamino-1-Deoxy Alditols, Useful Uv-Absorbing Derivatives of Neutral and Acidic Oligosaccharides. Carbohydrate Research. 1990;201(1):135–144. doi: 10.1016/0008-6215(90)84230-R. [DOI] [PubMed] [Google Scholar]
- Wada Y, Azadi P, Costello CE, Dell A, Dwek RA, Geyer H, Taniguchi N. Comparison of the methods for profiling glycoprotein glycans–HUPO Human Disease Glycomics/Proteome Initiative multi-institutional study. Glycobiology. 2007;17(4):411–422. doi: 10.1093/glycob/cwl086. [DOI] [PubMed] [Google Scholar]
- Walker SH, Budhathoki-Uprety J, Novak BM, Muddiman DC. Stable-Isotope Labeled Hydrophobic Hydrazide Reagents for the Relative Quantification of N-Linked Glycans by Electrospray Ionization Mass Spectrometry. Anal Chem. 2011;83(17):6738–6745. doi: 10.1021/ac201376q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SH, Lilley LM, Enamorado MF, Comins DL, Muddiman DC. Hydrophobic Derivatization of N-linked Glycans for Increased Ion Abundance in Electrospray Ionization Mass Spectrometry. Journal of the American Society for Mass Spectrometry. 2011;22(8):1309–1317. doi: 10.1007/s13361-011-0140-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SH, Papas BN, Comins DL, Muddiman DC. Interplay of Permanent Charge and Hydrophobicity in the Electrospray Ionization of Glycans. Anal Chem. 2010;82(15):6636–6642. doi: 10.1021/Ac101227a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler SF, Domann P, Harvey DJ. Derivatization of sialic acids for stabilization in matrix-assisted laser desorption/ionization mass spectrometry and concomitant differentiation of alpha(2 –> 3)- and alpha(2 –> 6)-isomers. Rapid Commun Mass Spectrom. 2009;23(2):303–312. doi: 10.1002/rcm.3867. [DOI] [PubMed] [Google Scholar]
- Wing DR, Garner B, Hunnam V, Reinkensmeier G, Andersson U, Harvey DJ, Butters TD. High-performance liquid chromatography analysis of ganglioside carbohydrates at the picomole level after ceramide glycanase digestion and fluorescent labeling with 2-aminobenzamide. Anal Biochem. 2001;298(2):207–217. doi: 10.1006/abio.2001.5393. [DOI] [PubMed] [Google Scholar]
- Wu S, Grimm R, German JB, Lebrilla CB. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10(2):856–868. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M, de Boer AR, Deelder AM. Structural Glycomics Using Hydrophilic Interaction Chromatography (Hilic) with Mass Spectrometry. Mass Spectrometry Reviews. 2009;28(2):192–206. doi: 10.1002/mas.20195. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Deelder AM, Hokke CH. Protein glycosylation analysis by liquid chromatography-mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;825(2):124–133. doi: 10.1016/j.jchromb.2005.01.030. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Koeleman CA, Deelder AM, Hokke CH. Normal-phase nanoscale liquid chromatography-mass spectrometry of underivatized oligosaccharides at low-femtomole sensitivity. Anal Chem. 2004;76(3):833–838. doi: 10.1021/ac034936c. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Koeleman CA, Hokke CH, Deelder AM. Nano-scale liquid chromatography-mass spectrometry of 2-aminobenzamide-labeled oligosaccharides at low femtomole sensitivity. Int J Mass Spectrom. 2004;232:51–57. doi: 10.1021/ac034936c. [DOI] [PubMed] [Google Scholar]
- Wuhrer M, Koeleman CAM, Hokke CH, Deelder AM. Nano-scale liquid chroamtgraphy-mass spectrometry of 2-aminobenzamide-labeled oligosaccharides at low femtomole sensitivity. Int J Mass Spectrom. 2004;232:51–57. [Google Scholar]
- Xia B, Feasley CL, Sachdev GP, Smith DF, Cummings RD. Glycan reductive isotope labeling for quantitative glycomics. Anal Biochem. 2009;387:162–170. doi: 10.1016/j.ab.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada E, Tsukamoto Y, Sasaki R, Yagyu K, Takahashi N. Structural changes of immunoglobulin G oligosaccharides with age in healthy human serum. Glycoconj J. 1997;14(3):401–405. doi: 10.1023/a:1018582930906. [DOI] [PubMed] [Google Scholar]
- Yang S, Wang MY, Chen LJ, Yin BJ, Song GQ, Turko IV, Li SW. QUANTITY: An Isobaric Tag for Quantitative Glycomics. Scientific Reports. 2015;5 doi: 10.1038/srep17585. doi:Artn 17585 10.1038/Srep17585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yuan W, Yang WM, Zhou JY, Harlan R, Edwards J, Zhang H. Glycan Analysis by Isobaric Aldehyde Reactive Tags and Mass Spectrometry. Anal Chem. 2013;85(17):8188–8195. doi: 10.1021/ac401226d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Zhang H. Solid-phase glycan isolation for glycomics analysis. Proteomics Clin Appl. 2012;6(11-12):596–608. doi: 10.1002/prca.201200045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Hashii N, Kawasaki N, Itoh S, Kawanishi T, Hayakawa T. Isotope tag method for quantitative analysis of carbohydrates by liquid chromatography–mass spectrometry. J Chromatogr A. 2005;1067:145–152. doi: 10.1016/j.chroma.2004.11.070. [DOI] [PubMed] [Google Scholar]
- Zamfir A, Peter-Katalinic J. Capillary electrophoresis-mass spectrometry for glycoscreening in biomedical research. Electrophoresis. 2004;25(13):1949–1963. doi: 10.1002/elps.200405825. [DOI] [PubMed] [Google Scholar]
- Zauner G, Deelder AM, Wuhrer M. Recent advances in hydrophilic interaction liquid chromatography (HILIC) for structural glycomics. Electrophoresis. 2011;32(24):3456–3466. doi: 10.1002/elps.201100247. [DOI] [PubMed] [Google Scholar]
- Zhang W, Wang H, Tang H, Yang P. Endoglycosidase-mediated incorporation of 18O into glycans for relative glycan quantitation. Anal Chem. 2011;83(12):4975–4981. doi: 10.1021/ac200753e. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cai MF, Zhao YY, Luo P. Mining of coal seam under mined out space and foundation stability of transmission tower. Boundaries of Rock Mechanics: Recent Advances and Challenges for the 21st Century. 2008:765–768. [Google Scholar]
- Zhong XF, Chen ZW, Snovida S, Liu Y, Rogers JC, Li LJ. Capillary Electrophoresis-Electrospray Ionization-Mass Spectrometry for Quantitative Analysis of Glycans Labeled with Multiplex Carbonyl-Reactive Tandem Mass Tags. Anal Chem. 2015;87(13):6527–6534. doi: 10.1021/acs.analchem.5b01835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Hu Y, Veillon L, Snovida SI, Rogers JC, Saba J, Mechref Y. Quantitative LC-MS/MS Glycomic Analysis of Biological Samples Using AminoxyTMT. Anal Chem. 2016;88(15):7515–7522. doi: 10.1021/acs.analchem.6b00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S, Tello N, Harvey A, Boyes B, Orlando R, Mechref Y. Reliable LC-MS quantitative glycomics using iGlycoMab stable isotope labeled glycans as internal standards. Electrophoresis. 2016;37(11):1489–1497. doi: 10.1002/elps.201600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Lee S, Valentine SJ, Reilly JP, Clemmer DE. Mannose7 glycan isomer characterization by IMS-MS/MS analysis. J Am Soc Mass Spectrom. 2012;23(12):2158–2166. doi: 10.1007/s13361-012-0491-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


