Abstract
Angiosarcoma is an aggressive vascular sarcoma with an extremely poor prognosis. Due to the relative rarity of this disease, its molecular drivers and optimal treatment strategies are obscure. DICER1 is an RNase III endoribonuclease central to microRNA biogenesis, and germline DICER1 mutations result in a cancer predisposition syndrome, associated with an increased risk of many tumor types. Here we show that biallelic Dicer1 deletion with aP2-Cre drives aggressive and metastatic angiosarcoma independent of other genetically engineered oncogenes or tumor suppressor loss. Angiosarcomas in aP2-Cre;Dicer1Flox/−mice histologically and genetically resemble human angiosarcoma. MicroRNA-23 target genes including the oncogenes Ccnd1 as well as Adam19, Plau, and Wsb1 that promote invasiveness and metastasis were enriched in mouse and human angiosarcoma. These studies illustrate that Dicer1 can function as a traditional loss-of-function tumor suppressor gene, and they provide a fully penetrant animal model for the study of angiosarcoma development and metastasis.
Keywords: Dicer1, microRNA, angiosarcoma, sarcoma
Introduction
Angiosarcoma is a rare vascular tumor composed of proliferating and invasive endothelial cells. The prognosis for patients is extremely poor with a 5-year survival rate of only 30–40% (1). Patients are treated aggressively with surgery, radiation, and chemotherapy and unfortunately 50% die within one year of diagnosis. Angiosarcoma occurs as two distinct entities either spontaneously (primary) or more commonly secondary to ionizing radiation or chronic lymphedema. Recently, recurrent mutations in genes involved in angiogenesis, PTPRB and PLCG1, have been identified in secondary angiosarcomas while the drivers of spontaneous angiosarcoma are less clear (2), highlighting the need for genetic, in vivo animal models to further understand the molecular mechanisms of angiosarcoma development and assess novel therapeutics.
MicroRNAs are short non-coding RNAs that regulate target gene expression through translational repression or transcript degradation. MicroRNAs are involved in essentially every cellular process and their deregulation has been linked to numerous pathologic processes including cancer. DICER1 is a ribonuclease III enzyme that processes pre-microRNAs into mature 20–23 nucleotide microRNAs and thus is required for canonical microRNA biogenesis (3). DICER1 cleavage of pre-microRNAs results in the separation of the hairpin loop and production of two mature microRNA products, a 5-prime (5p) microRNA derived from the 5’ end of the stemloop and a 3-prime (3p) microRNA from the 3’ end of the stemloop. The 5p and 3p microRNAs from a single pre-microRNA have different base pair sequences and therefore have distinct target genes. Due to indispensable functions of microRNAs in development, global Dicer1 loss causes embryonic lethality in mice (4). Numerous studies have described Dicer1 dysregulation in cancer. In several mouse models, Dicer1 functions as a haploinsufficient tumor suppressor where loss of a single allele promotes tumorigenesis yet loss of both alleles abrogates tumor formation (5).
Heterozygous germline DICER1 loss-of-function mutations in humans is associated with a distinct cancer predisposition syndrome in which affected kindreds present with rare malignant and benign neoplasms including pleuropulmonary blastoma (PPB), pineoblastoma, embryonal rhabdomyosarcoma (ERMS), and others (6). Interestingly, some of these tumors have recently been shown to have secondary somatic missense mutations in the remaining DICER1 allele in exons 24 and 25 encoding the RNAse IIIb domain. These DICER1 RNAse IIIb domain missense mutations specifically block 5p microRNA processing while leaving 3p microRNA processing intact (7, 8). In addition, complete DICER1 loss has been shown to contribute to human pineoblastoma as well as mouse PPB in cell-autonomous and non-cell-autonomous mechanisms respectively (9, 10).
Previously, we described a genetically engineered mouse model of ERMS driven by activation of an oncogenic Smoothened allele, SmoM2, with adipocyte protein 2 (aP2)-Cre (11). The aP2 promoter drives expression of Cre recombinase in adipose tissue as well as other tissues including endothelium but not in skeletal muscle (12, 13). The cell of origin in the aP2-Cre;SmoM2 ERMS model remains to be determined. Given the occurrence of DICER1 mutations in both sporadic ERMS and DICER1 syndrome associated ERMS (8, 14), we sought to further interrogate the role of Dicer1 in ERMS oncogenesis utilizing mice with conditional deletion of Dicer1 with aP2-Cre. Here we describe our findings illustrating Dicer1 deletion in aP2 expressing cells results in angiosarcoma.
Materials and Methods
Mouse strains
Mice were maintained in mixed genetic backgrounds thus littermate controls were used for all comparisons. All strains have been described previously: aP2-Cre (12), Dicer1Flox/Flox (15), and Rosa26LSL-tdTomato (16). Global Dicer1+/− animals were generated by breeding Dicer1Flox/Flox to CAG-Cre (17). Primary angiosarcoma tumors were transplanted into immunocompromised mice as described previously (18). All mouse studies were reviewed and approved by the Institutional Animal Care and Use Committee at St. Jude Children’s Research Hospital.
Immunohistochemistry and mRNA in situ hybridization
Histology and IHC performed following standard protocols (19) using antibodies detailed in Supplementary Table S2. RNAScope mRNA in situ hybridization performed following manufacturer’s protocol using the RNAscope 2.5 VS Reagent kit–Red and Dicer1 probes (456799, Advanced Cell Diagnostics, Inc).
RNA and gene expression
Total RNA was prepared from tissue using the miRNeasy Mini Kit (217004, Qiagen) after homogenizing the tissue using the TissueLyser LT (Qiagen) according to manufacturer’s instruction. qRT-PCR was performed with Taqman probes (Applied Biosystems) and SYBR primers detailed in Supplementary Table S3. Relative expression by qRT-PCR was quantified using the ΔΔCT method normalized to 18S ribosomal RNA or β-Actin and expressed relative to WT tissue. Mature microRNA expression was normalized to snRNA U6 and also expressed relative to WT tissue.
Gene Microarray Analysis
The GeneChip Mouse Gene 2.0 ST Microarray (902463, Affymetrix) was utilized for gene expression analysis of mouse angiosarcoma tumors (n = 7) and normal tissue from WT adult mice including quadriceps femoris (n = 4), sternocleidomastoid (SCM, n = 4), and aorta (n = 4). A previously described cohort of primary human angiosarcoma (n = 7) (20)) and cohort of normal human tissue from GSE7307 (skeletal muscle n = 5, deltoid muscle n = 6, aorta n = 4, and coronary artery n = 3) were utilized for cross species comparison and human aortas from GSE26155 were used for comparison to human angiosarcoma. Normalized signal data was log2 transformed in STATA/MP 14.1 (College Station, TX) and the transformed data was batch corrected and compared. Each probeset was compared by unequal variance t-test comparing mouse angiosarcomas to mouse quadriceps (Partek Genomics Suite 6.6 St. Louis, MO, USA). Mouse and human data were deduplicated with respect to gene, keeping only the most highly expressed probeset for each species. Next the median absolute difference (MAD) was calculated for each probe set across all mouse samples. Next the top 1,000 by MAD scoring genes were selected. This data was joined to the human set and rank transformed in STATA/MP 14.1. The ranks were then imported in Partek 6.6 Genomics Suite for visualization. Unsupervised hierarchical cluster analysis was performed using average linked Euclidean distance metrics of z-transformed ranks. Principle component analysis (PCA) was then performed on the top 1,000 genes MAD score. Separately the maximum orthologous probesets per gene symbol were compared cross species resulting in 13,267 log2 ratios of angiosarcoma to quadriceps (mouse) and angiosarcoma to skeletal muscle (human) STATA/MP 14.1. We compared mouse angiosarcomas to a panel of human soft tissue sarcomas, including angiosarcoma (20), fusion-negative and fusion-positive RMS (21), Ewing Sarcoma (GSE37371), osteosarcoma (22), dedifferentiated liposarcoma, differentiated liposarcoma, leiomyosarcoma, myxofibrosarcoma, and undifferentiated sarcoma (GSE71121)(23). Again the mouse and human probesets were de-duplicated with respect to gene, only the highest for each probeset was retained for each gene. Mouse and human genes were then merged by gene symbol resulting in 12,993 genes. The mean of each disease was found and ranked. The mouse and human sets were joined and ranked according to the mean for each disease and the global mean of the ranks was calculated to generate a synthetic reference for comparison. First, unsupervised hierarchical cluster analysis was performed using average linked Euclidean distance metrics of z-transformed ranks, represented as a dendogram. Finally, the difference in ranked mean expression for each sample type versus the global mean of the ranks was calculated. The difference in the rank of each data set verses the mean of all ranked data was calculated and correlated. Expression of microRNA targets were analyzed using Enrichr (24) using the overexpressed genes in comparing angiosarcoma tumors to normal aorta with a P value < 0.05 and log ratio > 2.5. Enrichr analyzes gene sets for enrichment of computationally predicted microRNA target genes identified by the TargetScan algorithm for every known microRNA. Array data deposited in GEO database (GSE85834).
DNA and genomic PCR
Genomic DNA was isolated from tissue using the DNeasy blood and tissue kit (Qiagen 69504). Dicer1 genotypes were determined using primers as described previously (15). Microdissection was performed on H&E stained sections (Arcturus Paradise Plus LCM Staining Kit, Thermo Fisher KIT0312-S) of mouse angiosarcoma samples followed by genomic DNA extraction using the PicoPure DNA extraction kit (KIT0103 Thermo Fisher).
Immunoblots
Immunoblots were performed on total cell lysates prepared in RIPA buffer as described previously (25). Blots were probed with antibodies detailed in Supplementary Table S4.
Transmission electron microscopy
Angiosarcoma tumors were fixed in 2.5% glutaraldehyde, 2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4) and post-fixed in 2% osmium tetroxide in 0.1 M cacodylate buffer with 0.15% potassium ferrocyanide. The sample was dehydrated in a graded series of ethanol to propylene oxide, and embedded overnight at 70° C in epoxy resin. Ultrathin sections (80 nm) were imaged using a JEOL 1200EX TEM with an AMT XR111 camera or Tecnai TF20 TEM with an AMT XR41 camera.
Results
aP2-Cre;Dicer1Flox/Flox mice are viable
We initially sought to interrogate if Dicer1 loss in aP2-Cre expressing cells cooperates with SmoM2 in our genetically engineered mouse model of rhabdomyosarcoma and phenocopies DICER1 syndrome rhabdomyosarcoma in humans. Our studies used the aP2-Cre mouse generated by Tang et al. (12) and the Dicer1Flox mouse generated by Murchison et al. (15). We first bred compound mutant aP2-Cre;Dicer1Flox/+ (ADFlox/+) to Dicer1Flox/+ (DFlox/+) mice to determine the viability of aP2-Cre;Dicer1Flox/Flox (ADFlox/Flox) mice. Multiple independent aP2-Cre transgenic mouse lines have been generated utilizing the same 5.4-kb aP2 promoter/enhancer directing Cre recombinase expression (12, 26–28). Interestingly, ADFlox/Flox mice derived from independently generated aP2-Cre transgenic mice (created by the laboratories of Barbara Kahn (27) and Ronald Evans (28)) and different conditional Dicer1Flox alleles (generated by Stephen Jones (29) and Clifford Tabin (30)) were previously found to die shortly after birth (13, 31).
In contrast, ADFlox/Flox mice in our studies using the aP2-Cre generated by Jonathan Graff (12) and Dicer1Flox allele generated by Gregory Hannon with loxP sites flanking regions of the RNaseIII domain (15) were born at normal Mendelian ratios and were phenotypically indistinguishable from AD+/+ and ADFlox/+ littermates. In addition, ADFlox/Flox animals maintained normal adipose weight and histology (Supplementary Fig. S1A and S1B). Thus, no early postnatal death or lack of WAT was observed in our studies using the aP2-Cre generated by Tang et al. (12). Dicer1 expression decreased although was still detectable in both BAT and WAT from ADFlox/+ and ADFlox/Flox animals suggesting incomplete Cre-mediated recombination of the Dicer1Flox allele (Supplementary Fig. S1C). To assess Cre-mediated recombination efficiency of Dicer1Flox allele we performed genomic PCR from DNA isolated from BAT and WAT with oligonucleotides to detect the wild-type, non-recombined and recombined Dicer1Flox allele (Supplementary Fig. S1D). We detected retention of the Dicer1Flox allele in both the BAT and WAT (Supplementary Fig. S1E and S1F). This incomplete Cre-mediated excision and resulting Dicer1 expression may allow viability of the animals. Therefore, the phenotypic distinction in our findings from previous studies is likely related to recombination efficiency variability associated with the independent Cre drivers, although we cannot exclude differing positional effects from random transgene insertion.
Angiosarcomas develop in aP2-Cre;DicerFlox/− mice
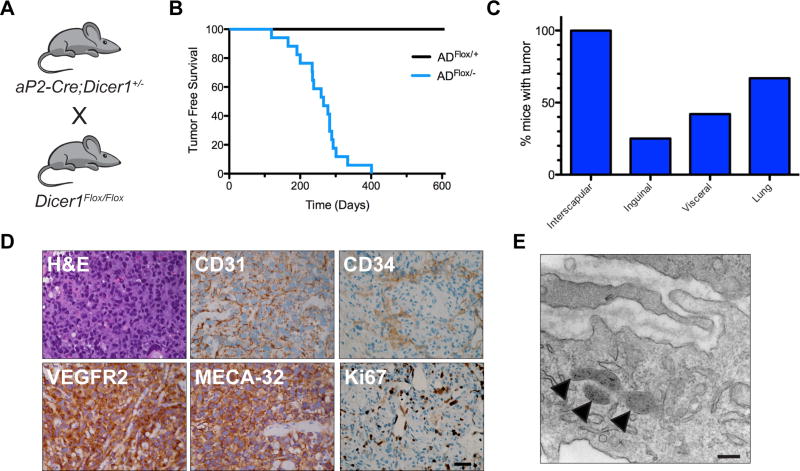
Given that ADFlox/Flox animals were viable and exhibited incomplete Dicer1Flox Cre-mediated recombination in adipose tissue, we generated mice with global heterozygous Dicer1 deletion (Dicer1+/−) by breeding Dicer1Flox/Flox mice to mice with ubiquitous Cre expression under control of the cytomegalovirus immediate early enhancer and chicken β-actin promoter (CAG-Cre). Then, aP2-Cre;Dicer1+/− mice were bred to DicerFlox/Flox (Fig. 1A). We then observed mice with AD+/+, ADFlox/+, AD+/−, and ADFlox/− genotypes to assess phenotypes with both global heterozygous and conditional loss of Dicer1. Surprisingly, 100% of ADFlox/− mice developed large multifocal hemorrhagic blood-filled masses consistent with angiosarcomas with a median onset of 266 days (Fig. 1B). Angiosarcomas were never observed in ADFlox/+, AD+/− or AD+/+ compound mutant mice (Fig. 1B). The hemorrhagic tumors in ADFlox/− mice occurred at various locations while primarily developing in the interscapular region of the back (75%) invading both the adjacent interscapular BAT and skeletal muscle. Angiosarcomas occurred in the inguinal region adjacent to and invading the inguinal WAT and skeletal muscle. Small tumors were also evident in the abdominal visceral fat, and lungs (Fig. 1C). Both the locations and histologic appearance of the tumors were consistent with human angiosarcoma of soft tissue with epithelioid appearance composed of sheets of rounded endothelial cells with pleomorphic nuclei and prominent nucleoli and IHC demonstrating strong expression of CD31, CD34, VEGFR2, MECA-32, and Ki67 consistent with highly proliferative angiosarcoma (Fig. 1D). Ultrastructural analysis with transmission electron microscopy revealed tumor cells contain storage granules consistent with Weibel-Palade bodies in endothelial cells that store von Willebrand factor, P-selectin, and chemokines thus suggesting endothelial tumor origins (Fig. 1E). The recent findings that aP2 is expressed in endothelial cells further suggests an endothelial cell origin of these tumors (13, 32, 33). Lung angiosarcomas were observed to similarly express VEGFR2 and CD31 (Supplementary Fig. S2). Although the lung is the most common metastatic site of angiosarcoma, it is unclear if the lung tumors are metastases or independent primary tumors.
Figure 1. Angiosarcomas develop in aP2-Cre;DicerFlox/− mice.
(A) Breeding scheme to generate ADFlox/− mice that develop angiosarcomas. (B) Kaplan-Meier tumor free survival comparing ADFlox/+ (black line n = 9) to ADFlox/− (blue line, n = 17, median tumor free survival of 266 days, with 100% penetrance), Log rank P = 0.0001. (C) Percentage of mice with tumors at anatomic locations. (D) Representative histology of tumors from ADFlox/− mice with H&E and IHC for angiosarcoma markers CD31, VEGFR2, MECA-32, CD34, and Ki67. Scale bar, 25 µm. (E) Transmission electron microscopy of angiosarcoma tumor cell highlighting presence of Weibel-Palade bodies (arrowheads). Scale bar, 500 nm.
Angiosarcomas in aP2-Cre;Dicer1Flox/− mice are autonomous and metastatic
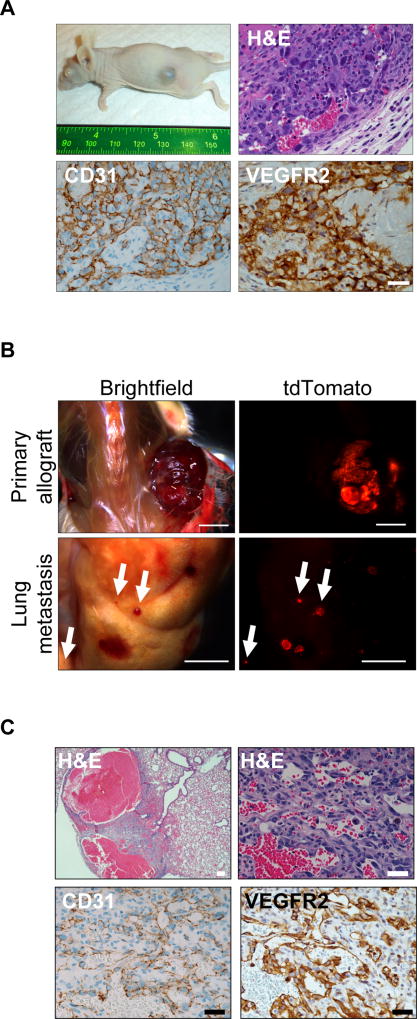
To further assess the tumorigenicity of the ADFlox/− angiosarcomas, primary tumors were transplanted into the flank of immunocompromised mice, and 5 of 5 allografts formed subcutaneous tumors that recapitulated the primary tumors histology and expression of CD31 and VEGFR2 (Fig. 2A). In order to indelibly label tumor cells we bred a conditional tdTomato reporter, R26LSL-tdTom (16), into the tumor model to generate aP2-Cre;R26LSL-tdTom/+;Dicer1Flox/− (ATDFlox/−) compound mutant mice that fluorescently label aP2-Cre derived tumor cells. ATDFlox/− allografts metastasized to the lungs highlighting the aggressive potential of the tumors (Fig.2B and 2C). Although aP2-Cre;Dicer1Flox/− angiosarcoma tumors were successfully serially transplanted, we were unable to establish cell lines from either primary or transplanted tumors. This difficulty is consistent with previous angiosarcoma models (34). Taken together, Dicer1 loss is sufficient to drive angiosarcoma development, leading to highly proliferative, invasive and metastatic angiosarcomas.
Figure 2. aP2-Cre;DicerFlox/− angiosarcomas form allografts and metastasize.
(A) Tumor formation in subcutaneous allograft of ADFlox/− angiosarcoma tumor with H&E and IHC for CD31 and VEGFR2. Scale bar, 25 µm. (B) R26LSL-tdTom/+;ADFlox/− primary allograft and lung metastasis visualized with tdTomato fluorescence. (C) H&E and IHC for CD31 and VEGFR2 in lung metastasis from (B). Scale bar, upper left 100 µm all others 25 µm.
Biallelic loss of Dicer1 drives angiosarcoma
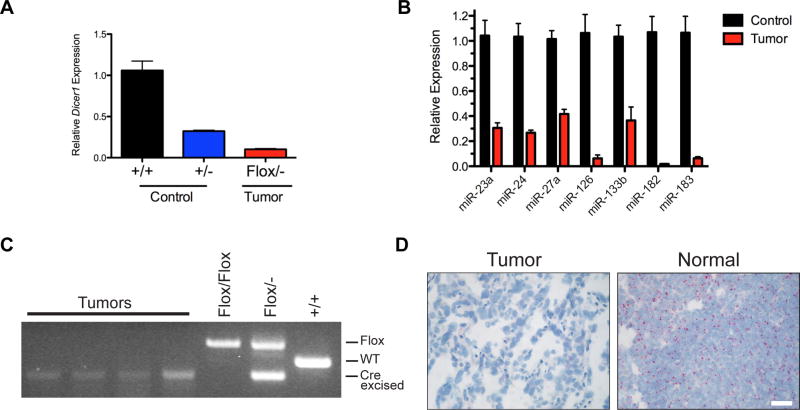
In the ADFlox/− angiosarcoma model, genetic evidence suggested that biallelic Dicer1 loss alone led to angiosarcoma development. To interrogate Dicer1 loss, expression was first assessed in tumors by qRT-PCR. Despite likely contamination of normal stroma and blood in tumor samples, Dicer1 expression was significantly reduced in tumors compared to Dicer1+/+ and Dicer1+/− heterozygous control aortas (Fig. 3A). In addition, mature microRNA expression of microRNAs expressed in endothelial cells (35) was significantly reduced in the tumors (Fig. 3B). Further evidence for biallelic Dicer1 loss in the tumors was observed by genomic PCR of microscopically dissected tumor cells from H&E stained tumor sections (Fig. 3C). Genomic DNA from angiosarcomas of ADFlox/− mice only amplified the Cre-excised Dicer1− allele without detecting the presence of the non-recombined Dicer1Flox allele, indicating complete Cre-mediated recombination of Dicer1Flox and resulting Dicer1 deletion in angiosarcomas. Finally, in situ hybridization using RNAscope probe pairs directed to exons 22–23 (region flanked by LoxP sites in Dicer1Flox mice) demonstrates Dicer1 mRNA levels were significantly reduced or absent in tumor cells compared to adjacent normal tissue (Fig. 3D). Thus, qRT-PCR, genomic PCR, mRNA in situ hybridization, and genetic evidence suggest biallelic Dicer1 loss results in angiosarcoma development independent of genetically engineered cooperating oncogenes or tumor suppressor loss. These data suggest that Dicer1 loss and resulting reduction of microRNAs drives the development of angiosarcoma in aP2-Cre expressing cells.
Figure 3. Biallelic Dicer1 loss in angiosarcomas.
(A) Relative expression of Dicer1 by qRT-PCR in ADFlox/− angiosarcoma tumors (n = 3) compared to normal aorta from wild type, AD+/+, (n = 3) and heterozygous, AD+/−, (n = 3), mice. P < 0.0001 for all comparisons of control to tumors. (B) Mature miRNA expression in ADFlox/− angiosarcoma and control AD+/+ aortas from (A), P < 0.01 for all except miR-133b P = 0.0207. (C) Genomic PCR for Dicer1 in microscopic dissected tumors with tail DNA as controls. (D) Dicer1 mRNA in situ hybridization (red) in angiosarcoma tumor and adjacent normal tissue. Scale bar, 25 µm.
ADFlox/− tumors display genetic signature and signaling pathway activation of angiosarcoma
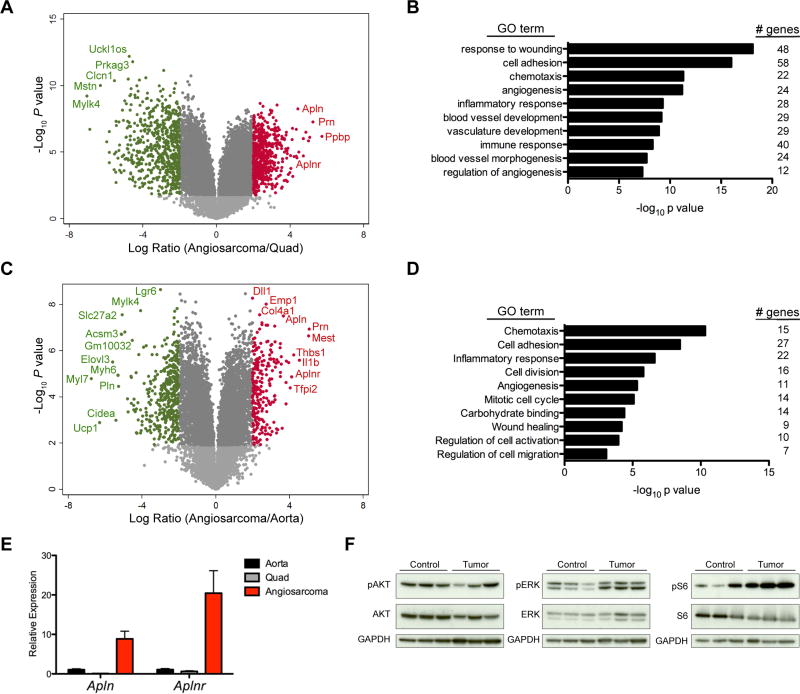
To evaluate whether these were true angiosarcomas we sought to determine if gene sets related to endothelial cells and blood vessels were specifically altered in the tumors from the ADFlox/− mice. First, we used mRNA expression profiling to compare the gene expression of the tumors to a normal non-endothelial tissue, quadriceps femoris skeletal muscle, to determine if tumors were enriched for endothelial genes (Fig. 4A). Gene ontology analysis of the top 578 differentially expressed genes with a P value less than 0.05 and 4-fold increased expression in angiosarcoma compared to skeletal muscle identified 5 of the top 10 gene ontology (GO) terms involved in angiogenesis and blood vessel development (Fig. 4B). With the tumors enriched in endothelial and blood vessels genes, we sought to determine what genes are enriched in the tumors compared to normal blood vessels (Figure 4C). Gene ontology analysis of the top 248 differentially expressed genes with P value less than 0.05 and 4-fold increased expression in angiosarcoma compared to normal aorta enriched for terms involved in angiogenesis, migration and cell proliferation (Figure 4D). Furthermore, the ligand Apelin (Apln) and the Apelin Receptor (Aplnr) were among the top up-regulated genes in both the comparison to skeletal muscle and aorta and were validated by qRT-PCR (Fig. 4A, 4C, and 4E). APLNR and APLN are a known G protein-coupled receptor and its cognate ligand respectively involved in blood vessel development and angiogenesis (36). APLNR signaling can lead to downstream activation of the RAS-ERK1/2 pathway and or PI3K/AKT pathway, both potentially resulting in activation of mTOR and p70S6K. Immunoblots of angiosarcoma tumor lysates indicate hyperphosphorylation of p-ERK and p-S6 (Fig. 4F). Activation of the mTOR pathway is consistent with many mouse models of angiosarcoma as well as human vascular disease (37, 38). Furthermore, a lack of elevated AKT phosphorylation is also consistent with recent findings in an mTOR driven mouse model of lymphangiosarcoma (39).
Figure 4. ADFlox/− tumors display genetic signature of angiosarcomas.
(A) Volcano plot of the Log10 of the P value versus the log ratio fold difference in mRNA expression in ADFlox/− angiosarcomas (n = 7) versus normal quadriceps femoris skeletal muscle (n = 4). Genes with P < 0.01 and log ratio > 2 (red) or log ratio < −2 (green) and genes of interest are labeled. (B) Gene ontology analysis with significantly enriched pathways in genes upregulated in tissue from (A) with P value < 0.05 and > 4-fold increased expression in angiosarcoma (578 genes). (C) Volcano plot of the Log10 of the P value versus the log ratio fold difference in mRNA expression in ADFlox/− angiosarcomas (n = 7) versus normal WT aortas (n = 4). Genes with P < 0.01 and log ratio > 2 (red) or log ratio < −2 (green) and genes of interest are labeled. (D) Gene ontology analysis with significantly enriched pathways in genes upregulated in tissue from (C) P value < 0.05 and > 4-fold increased expression in angiosarcoma (248 genes). (E) Expression of Apln and Aplnr by qRT-PCR in ADFlox/− angiosarcomas (n = 3), normal aorta (n = 3), and normal quadriceps (n = 3), P < 0.05 for all comparisons of angiosarcoma versus normal tissue. (F) Immunoblot analysis of lysates from ADFlox/− angiosarcomas and control normal aortas. Antibodies used are shown to the left.
Mouse angiosarcomas are similar to human angiosarcoma
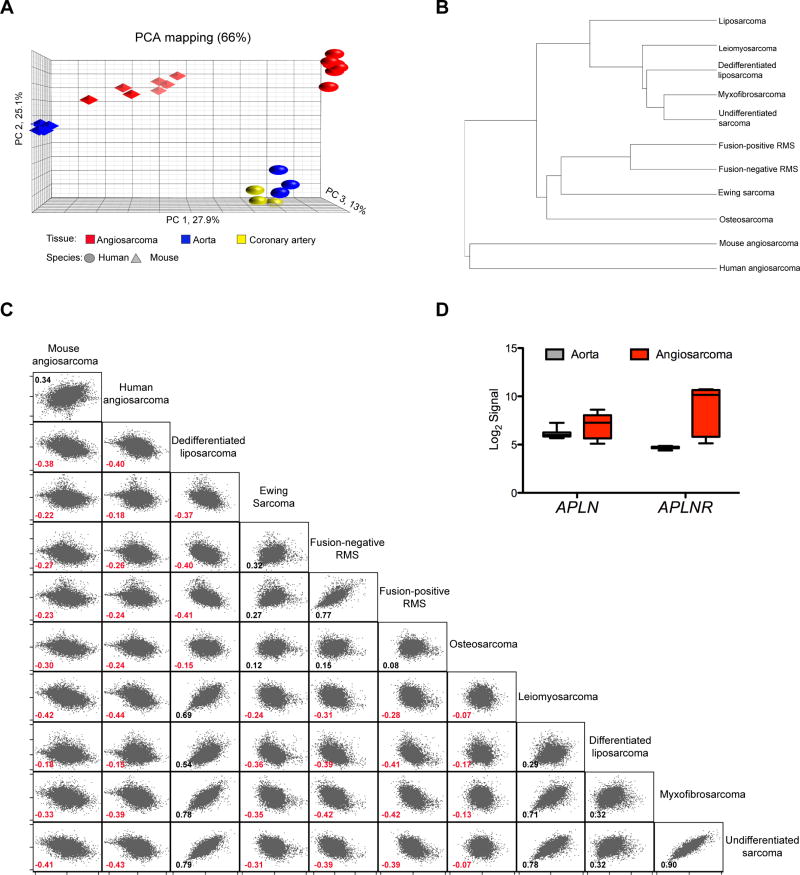
We hypothesized that the mouse angiosarcomas would be related to human angiosarcomas. Therefore, we compared mouse angiosarcomas to a human cohort of 7 primary angiosarcomas (20) and a collection of normal human (GSE7307) and mouse tissues (Supplementary Fig. S3A). Principle component analysis (PCA) revealed that the mouse and human angiosarcomas clustered distinctly and more closely to each other than the normal mouse aorta clustered to the human aorta and coronary artery (Fig. 5A). We further illustrate that the ADFlox/− angiosarcomas gene expression profile more significantly resembles human angiosarcoma than a broad panel of other human sarcomas (Fig.5B and 5C). Interestingly, the human angiosarcoma only has a positive correlation with the mouse angiosarcoma and none of the 9 other sarcoma types. To account for species specific gene expression variability, we further corroborated the association of the mouse and human angiosarcomas with 13,267 orthologous gene pairs normalized to skeletal muscle in a cross-species comparison and observed 62% agreement in genes expression between mouse and human angiosarcomas with a modest 0.49 Pearson’s correlation (Supplementary Fig. S3B). In addition APLNR is also overexpressed in human angiosarcomas as in the mouse model (Fig. 5D). Thus, the mouse angiosarcomas closely resemble human angiosarcoma by gene expression analysis and demonstrate an angiogenic and migratory signature with activation of cell signaling pathways known to be activated in human angiosarcoma.
Figure 5. Mouse angiosarcomas gene expression consistent with human angiosarcoma.
(A) Principle component analysis (PCA) of samples from human (sphere) and mouse (polyhedron) tissue; angiosarcomas (red), coronary artery (yellow) and aorta (blue). (B) Dendogram of panel of soft tissue sarcomas including human angiosarcoma and ADFlox/− mouse angiosarcomas showing ranked correlation clustering. Spearman correlation between mouse and human angiosarcoma = 0.75. (C) Ranked-correlation matrix among 12,993 ortholog gene pairs in ADFlox/− mouse angiosarcomas and panel of human soft tissue sarcomas. Spearman correlation coefficients shown for each comparison. (D) APLN and APLNR expression in human angiosarcoma (n = 7) compared to human normal aorta (GSE26155, n = 13) from deposited microarray analysis expressed as Log2 signal intensity. P value = 0.067 for APLN and P value < 0.001 for APLNR.
MicroRNA-23 target genes enriched in angiosarcoma
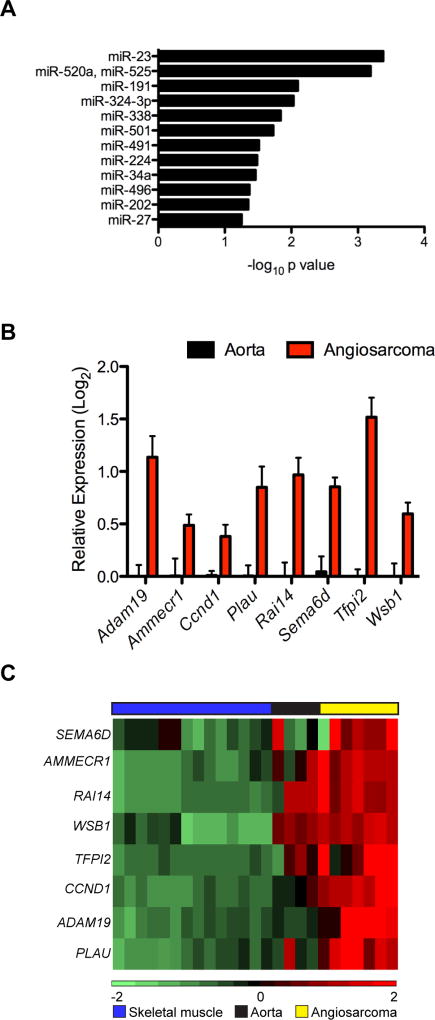
The deletion of Dicer1 with aP2-Cre is sufficient to drive angiosarcoma formation and results in decreased mature microRNA expression. Therefore, we hypothesized that expression of target genes of the key microRNAs contributing to angiosarcoma development in the ADFlox/− mice would be enriched in tumors compared to normal blood vessels (Fig. 4C). Using predicted microRNA target gene sets for each microRNA from TargetScan, we performed gene set enrichment analysis with Enrichr to identify the microRNAs with target genes enriched in mouse angiosarcomas compared to normal aorta with a Log2 ratio > 2.5 (99 genes). Target genes for miR-23 and miR-520a/miR-525 were significantly enriched in the most up-regulated genes; however, miR-520 and miR-525 are not conserved in the mouse shifting our focus to miR-23 (Fig. 6A, and Supplementary Table S1). MiR-23 is expressed in a polycistronic cluster with miR-27 and miR-24 with two clusters encoded in both the mouse and human genomes (Supplementary Fig. S4A and S4B). Mature miR-23a, 27a, and 24 expression is decreased in the ADFlox/− tumors (Fig. 3B), but only miR-23 target genes were enriched in the ADFlox/− angiosarcomas, although miR-27 is trending in the top 12 microRNAs (Fig. 6A). Eight of the 99 genes increased in ADFlox/− tumors with Log2 ratio > 2.5 are predicted targets of miR-23 (Ammerc1, Adam19, Ccnd1, Plau, Rai14, Sema6d, Tfpi2, Wsb1) and these were validated by qRT-PCR in independent tumor samples (Fig. 6B, and Supplementary Fig. S4C). Importantly, these eight miR-23 target genes are also enriched in human angiosarcoma (Fig. 6C). Thus, ADFlox/− mice develop angiosarcoma driven by biallelic Dicer1 loss resulting in decreased mature microRNA expression.
Figure 6. miR-23 targets enriched in mouse and human angiosarcomas.
(A) Gene set enrichment analysis for microRNA target genes among genes overexpressed in angiosarcoma compared to normal aorta (from Fig 4C) with a 2.5 or greater log ratio (99 genes). MicroRNAs with enriched target genes on left represented as Log10 of P value. (B) Validation of enriched miR-23 target genes by qRT-PCR in independent samples, expressed as Log2 transformed relative expression of miR-23 targets in ADFlox/− angiosarcomas (n = 3) compared to normal aortas (n = 3), P < 0.05 for all comparisons of angiosarcoma to normal aorta. (C) Heatmap showing gene expression profiles of miR-23 gene targets in human angiosarcoma (yellow), normal aorta (black) and skeletal muscle (blue).
Discussion
Our results illustrate that biallelic Dicer1 loss in cells expressing aP2-Cre drives angiosarcoma development providing a fully penetrant and metastatic model that mimics human angiosarcoma by histology and gene expression. Angiosarcoma is a rare and highly lethal vascular tumor accounting for 2–3% of all soft tissue sarcomas (1). Because of the rarity of the disease, most treatment decisions are based on retrospective, anecdotal studies consisting of relatively small numbers of cases, highlighting the need for more basic and clinical research to more accurately model the disease. Some mouse models of angiosarcoma exist, but are restricted to either hepatic angiosarcomas or specific to lymphangiosarcomas associated with lymphedema, exhibit variable penetrance and incidence, or complicated with the occurrence of other tumor types (34, 39, 40). The ADFlox/− genetic murine model of primary angiosarcoma is simple involving only two alleles, 100% penetrant, and specific with angiosarcomas being the only gross lesions detected.
Insights from DICER1 syndrome patient tumors illustrate that DICER1 functions as a tumor suppressor but with a non-traditional mechanism of action (3). DICER1 syndrome patients harbor germline nonsense or frameshift mutations resulting in DICER1 protein truncation and loss of enzymatic activity. In most cases, second hit somatic mutations do not exhibit loss of heterozygosity (LOH) of DICER1 but rather missense mutations in the RNase IIIb domain (exons 24– 25). The RNase IIIb domain is primarily responsible for generating 5p microRNAs, thus mutations in this domain affecting the metal-ion binding residues result in deficient pre-microRNA processing for mature 5p microRNAs while retaining expression of mature 3p microRNAs (7, 8). It is unclear if the loss of 5p microRNAs or the disruption in the balance of 3p to 5p microRNAs drive tumorigenesis.
An exception occurs in human pineoblastomas that display LOH of DICER1 and thus bialleic loss (41). This initially appeared to conflict with experiments suggesting Dicer1 was required for cell survival. However several groups have recently reported studies in cells with complete deletion of Dicer1 (42, 43). In these studies, survival of Dicer1 null cells was contingent on cooperating oncogenes such as KrasG12D and/or loss of tumor suppressors such as p53. Interestingly, Chen et al. found that Dicer1 deletion in non-small cell lung cancer led to an angiogenic phenotype (42). In this context, loss of Dicer1 relieved repression of the HIF inhibitor FIH1 leading to a non-cell autonomous reduction in tumor angiogenesis through down-regulation of Hif1a and Vegf. In contrast, we found that Dicer1 deletion directly in endothelial cells leads to cell-autonomous tumorigenesis with no significant changes in Hif1a and Vegfa expression. Our aP2-Cre;Dicer1Flox/− angiosarcoma model represents the first in vivo evidence that biallelic Dicer1 loss functions as a cell-autonomous cancer driver. However, given the prolonged latency of angiosarcoma onset in ADFlox/− mice we cannot exclude the acquisitions of secondary “hits.” These results suggest that Dicer1 loss and resulting loss of microRNA function is sufficient for the formation of angiosarcoma.
Dicer1 deletion drives angiosarcoma formation in our ADFlox/− model suggesting that abolishing microRNA regulation of target genes is central to tumorigenesis. Apelin (Apln) ligand and its receptor Aplnr were among the most overexpressed genes in the ADFlox/− angiosarcoma tumors and correlated with downstream mTOR signaling pathway activation in tumors. In addition, miR~23~27~24 cluster expression decreased and miR-23 target genes were significantly enriched in the ADFlox/− angiosarcoma tumors. The miR~23~27~24 clusters are expressed in endothelial cells and are highly involved in angiogenesis (44). Interestingly, Apln and Aplnr 3’UTRs contain recognition sites in both human and mouse for microRNAs in the miR~23~27~24 cluster further highlighting potential regulation of APLN/APLNR signaling (Supplementary Fig. S4D). Furthermore, deletion of Dicer1 and subsequent increased expression of miR-23 target genes likely participate in the pathogenesis of angiosarcoma through facilitating the G1/S transition with increased CCND1 and resulting CDK4/6 activity. As well miR-23 targets Adam19, Plau and Wsb1 may contribute to the invasiveness and metastasis in the ADFlox/− model as has been shown for miR-23b and PLAU in breast cancer (45). Furthermore, WSB1 can promote metastasis through promoting the degradation of RhoGDI2 leading to activation of RAC1 in osteosarcoma (46), a cancer where miR-23 functions as a tumor suppressor (47). Interestingly SEMA6D can function as an oncogene by activating VEGFR2 independent of ligand VEGF through facilitating interactions between PLEXINA1 and VEGFR2 (48). Although only miR-23 targets were enriched in the most up-regulated genes, we do not expect that angiosarcoma in the ADFlox/− model results from decreased miR-23 function alone but from the combination of global loss of microRNA regulation.
In summary, we have identified a pivotal role of DICER1 in angiosarcoma development and illustrated biallelic Dicer1 loss is sufficient to drive tumorigenesis. Further interrogation of the essential microRNAs and respective target genes as well as the contribution of microRNAs to the tumor microenvironment could provide novel insights into the core dependencies in angiosarcoma. Mechanistic insights from these studies could offer novel therapeutic approaches with microRNA replacement therapy in angiosarcoma. Since a single microRNA targets many mRNAs, manipulating microRNAs therapeutically could prove more efficacious than directed agents against a single molecule. This study highlights the crucial and distinct roles of Dicer1 and microRNAs in sarcoma and suggests microRNA-based therapeutics as a potential novel strategy in sarcoma.
Supplementary Material
Acknowledgments
Financial support: This work was supported by NIH grants NCI-R01CA216344 (M. E. Hatley) and K08CA151649 (M. E. Hatley), V Foundation for Cancer Research (M. E. Hatley), and all authors supported by St. Jude Cancer Center support grant P30 CA021765 and American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
The authors thank Jonathan Graff for aP2-Cre mice and Greg Hannon for Dicer1Flox/Flox mice as well as the St. Jude Cancer Center Shared Resources Cell and Tissue Imaging Resource, VPC Resource for IHC and ISH services, the Hartwell Center for microarray services, and the Flow Cytometry and Cell sorting Resource. We are grateful to Eric N. Olson, Charles W.M. Roberts, Suzanne J. Baker and Mitch J. Weiss for helpful discussions and review of manuscript.
References
- 1.D'Angelo SP, Munhoz RR, Kuk D, Landa J, Hartley EW, Bonafede M, et al. Outcomes of Systemic Therapy for Patients with Metastatic Angiosarcoma. Oncology. 2015;89:205–14. doi: 10.1159/000381917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behjati S, Tarpey PS, Sheldon H, Martincorena I, Van Loo P, Gundem G, et al. Recurrent PTPRB and PLCG1 mutations in angiosarcoma. Nat Genet. 2014;46:376–9. doi: 10.1038/ng.2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat Rev Cancer. 2014;14:662–72. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 5.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325:965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rakheja D, Chen KS, Liu Y, Shukla AA, Schmid V, Chang TC, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun. 2014;2:4802. doi: 10.1038/ncomms5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, et al. Recurrent somatic DICER1 mutations in nonepithelial ovarian cancers. N Engl J Med. 2012;366:234–42. doi: 10.1056/NEJMoa1102903. [DOI] [PubMed] [Google Scholar]
- 9.de Kock L, Sabbaghian N, Druker H, Weber E, Hamel N, Miller S, et al. Germ-line and somatic DICER1 mutations in pineoblastoma. Acta Neuropathol. 2014;128:583–95. doi: 10.1007/s00401-014-1318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y, Castro AM, Hoekstra M, Yan TJ, Kanakamedala AC, Dehner LP, et al. Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links DICER1 Loss to the Pathogenesis of Pleuropulmonary Blastoma. PLoS Genet. 2015;11:e1005242. doi: 10.1371/journal.pgen.1005242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatley ME, Tang W, Garcia MR, Finkelstein D, Millay DP, Liu N, et al. A mouse model of rhabdomyosarcoma originating from the adipocyte lineage. Cancer cell. 2012;22:536–46. doi: 10.1016/j.ccr.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang W, Zeve D, Suh JM, Bosnakovski D, Kyba M, Hammer RE, et al. White fat progenitor cells reside in the adipose vasculature. Science. 2008;322:583–6. doi: 10.1126/science.1156232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, et al. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62:864–74. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doros L, Yang J, Dehner L, Rossi CT, Skiver K, Jarzembowski JA, et al. DICER1 mutations in embryonal rhabdomyosarcomas from children with and without familial PPB-tumor predisposition syndrome. Pediatr Blood Cancer. 2012;59:558–60. doi: 10.1002/pbc.24020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–40. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–40. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–24. doi: 10.1006/bbrc.1997.7111. [DOI] [PubMed] [Google Scholar]
- 18.Morton CL, Houghton PJ. Establishment of human tumor xenografts in immunodeficient mice. Nat Protoc. 2007;2:247–50. doi: 10.1038/nprot.2007.25. [DOI] [PubMed] [Google Scholar]
- 19.Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R, van Rooij E, et al. Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell. 2010;18:282–93. doi: 10.1016/j.ccr.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadj-Hamou NS, Lae M, Almeida A, de la Grange P, Kirova Y, Sastre-Garau X, et al. A transcriptome signature of endothelial lymphatic cells coexists with the chronic oxidative stress signature in radiation-induced post-radiotherapy breast angiosarcomas. Carcinogenesis. 2012;33:1399–405. doi: 10.1093/carcin/bgs155. [DOI] [PubMed] [Google Scholar]
- 21.Williamson D, Missiaglia E, de Reynies A, Pierron G, Thuille B, Palenzuela G, et al. Fusion gene-negative alveolar rhabdomyosarcoma is clinically and molecularly indistinguishable from embryonal rhabdomyosarcoma. J Clin Oncol. 2010;28:2151–8. doi: 10.1200/JCO.2009.26.3814. [DOI] [PubMed] [Google Scholar]
- 22.Stewart E, Federico S, Karlstrom A, Shelat A, Sablauer A, Pappo A, et al. The Childhood Solid Tumor Network: A new resource for the developmental biology and oncology research communities. Dev Biol. 2016;411:287–93. doi: 10.1016/j.ydbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesluyes T, Perot G, Largeau MR, Brulard C, Lagarde P, Dapremont V, et al. RNA sequencing validation of the Complexity INdex in SARComas prognostic signature. Eur J Cancer. 2016;57:104–11. doi: 10.1016/j.ejca.2015.12.027. [DOI] [PubMed] [Google Scholar]
- 24.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–7. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hanna JA, Garcia MR, Go JC, Finkelstein D, Kodali K, Pagala V, et al. PAX7 is a required target for microRNA-206-induced differentiation of fusion-negative rhabdomyosarcoma. Cell Death Dis. 2016;7:e2256. doi: 10.1038/cddis.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barlow C, Schroeder M, Lekstrom-Himes J, Kylefjord H, Deng CX, Wynshaw-Boris A, et al. Targeted expression of Cre recombinase to adipose tissue of transgenic mice directs adipose-specific excision of loxP-flanked gene segments. Nucleic acids research. 1997;25:2543–5. doi: 10.1093/nar/25.12.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abel ED, Peroni O, Kim JK, Kim YB, Boss O, Hadro E, et al. Adipose-selective targeting of the GLUT4 gene impairs insulin action in muscle and liver. Nature. 2001;409:729–33. doi: 10.1038/35055575. [DOI] [PubMed] [Google Scholar]
- 28.He W, Barak Y, Hevener A, Olson P, Liao D, Le J, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci U S A. 2003;100:15712–7. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mudhasani R, Zhu Z, Hutvagner G, Eischen CM, Lyle S, Hall LL, et al. Loss of miRNA biogenesis induces p19Arf-p53 signaling and senescence in primary cells. J Cell Biol. 2008;181:1055–63. doi: 10.1083/jcb.200802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mudhasani R, Puri V, Hoover K, Czech MP, Imbalzano AN, Jones SN. Dicer is required for the formation of white but not brown adipose tissue. Journal of cellular physiology. 2011;226:1399–406. doi: 10.1002/jcp.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elmasri H, Karaaslan C, Teper Y, Ghelfi E, Weng M, Ince TA, et al. Fatty acid binding protein 4 is a target of VEGF and a regulator of cell proliferation in endothelial cells. FASEB J. 2009;23:3865–73. doi: 10.1096/fj.09-134882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Dyck F, Scroyen I, Declercq J, Sciot R, Kahn B, Lijnen R, et al. aP2-Cre-mediated expression activation of an oncogenic PLAG1 transgene results in cavernous angiomatosis in mice. Int J Oncol. 2008;32:33–40. [PubMed] [Google Scholar]
- 34.Zindy F, Nilsson LM, Nguyen L, Meunier C, Smeyne RJ, Rehg JE, et al. Hemangiosarcomas, medulloblastomas, and other tumors in Ink4c/p53-null mice. Cancer Res. 2003;63:5420–7. [PubMed] [Google Scholar]
- 35.Santoro MM, Nicoli S. miRNAs in endothelial cell signaling: the endomiRNAs. Exp Cell Res. 2013;319:1324–30. doi: 10.1016/j.yexcr.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papangeli I, Kim J, Maier I, Park S, Lee A, Kang Y, et al. MicroRNA 139-5p coordinates APLNR-CXCR4 crosstalk during vascular maturation. Nat Commun. 2016;7:11268. doi: 10.1038/ncomms11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hammill AM, Wentzel M, Gupta A, Nelson S, Lucky A, Elluru R, et al. Sirolimus for the treatment of complicated vascular anomalies in children. Pediatr Blood Cancer. 2011;57:1018–24. doi: 10.1002/pbc.23124. [DOI] [PubMed] [Google Scholar]
- 38.Italiano A, Chen CL, Thomas R, Breen M, Bonnet F, Sevenet N, et al. Alterations of the p53 and PIK3CA/AKT/mTOR pathways in angiosarcomas: a pattern distinct from other sarcomas with complex genomics. Cancer. 2012;118:5878–87. doi: 10.1002/cncr.27614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun S, Chen S, Liu F, Wu H, McHugh J, Bergin IL, et al. Constitutive Activation of mTORC1 in Endothelial Cells Leads to the Development and Progression of Lymphangiosarcoma through VEGF Autocrine Signaling. Cancer Cell. 2015;28:758–72. doi: 10.1016/j.ccell.2015.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharpless NE, Alson S, Chan S, Silver DP, Castrillon DH, DePinho RA. p16(INK4a) and p53 deficiency cooperate in tumorigenesis. Cancer Res. 2002;62:2761–5. [PubMed] [Google Scholar]
- 41.Sabbaghian N, Hamel N, Srivastava A, Albrecht S, Priest JR, Foulkes WD. Germline DICER1 mutation and associated loss of heterozygosity in a pineoblastoma. J Med Genet. 2012;49:417–9. doi: 10.1136/jmedgenet-2012-100898. [DOI] [PubMed] [Google Scholar]
- 42.Chen S, Xue Y, Wu X, Le C, Bhutkar A, Bell EL, et al. Global microRNA depletion suppresses tumor angiogenesis. Genes Dev. 2014;28:1054–67. doi: 10.1101/gad.239681.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ravi A, Gurtan AM, Kumar MS, Bhutkar A, Chin C, Lu V, et al. Proliferation and tumorigenesis of a murine sarcoma cell line in the absence of DICER1. Cancer Cell. 2012;21:848–55. doi: 10.1016/j.ccr.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou Q, Gallagher R, Ufret-Vincenty R, Li X, Olson EN, Wang S. Regulation of angiogenesis and choroidal neovascularization by members of microRNA-23~27~24 clusters. Proc Natl Acad Sci U S A. 2011;108:8287–92. doi: 10.1073/pnas.1105254108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pellegrino L, Stebbing J, Braga VM, Frampton AE, Jacob J, Buluwela L, et al. miR-23b regulates cytoskeletal remodeling, motility and metastasis by directly targeting multiple transcripts. Nucleic Acids Res. 2013;41:5400–12. doi: 10.1093/nar/gkt245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cao J, Wang Y, Dong R, Lin G, Zhang N, Wang J, et al. Hypoxia-Induced WSB1 Promotes the Metastatic Potential of Osteosarcoma Cells. Cancer Res. 2015;75:4839–51. doi: 10.1158/0008-5472.CAN-15-0711. [DOI] [PubMed] [Google Scholar]
- 47.He Y, Meng C, Shao Z, Wang H, Yang S. MiR-23a functions as a tumor suppressor in osteosarcoma. Cell Physiol Biochem. 2014;34:1485–96. doi: 10.1159/000366353. [DOI] [PubMed] [Google Scholar]
- 48.Catalano A, Lazzarini R, Di Nuzzo S, Orciari S, Procopio A. The plexin-A1 receptor activates vascular endothelial growth factor-receptor 2 and nuclear factor-kappaB to mediate survival and anchorage-independent growth of malignant mesothelioma cells. Cancer Res. 2009;69:1485–93. doi: 10.1158/0008-5472.CAN-08-3659. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.