Abstract
Background
To better assess potential hepatotoxicity of nanomaterials, human liver HepG2 cells were exposed for 3 days to five different CeO2 (either 30 or 100 μg/ml), 3 SiO2 based (30 μg/ml) or 1 CuO (3 μg/ml) nanomaterials with dry primary particle sizes ranging from 15 to 213 nm. Metabolomic assessment of exposed cells was then performed using four mass spectroscopy dependent platforms (LC and GC), finding 344 biochemicals.
Results
Four CeO2, 1 SiO2 and 1 CuO nanomaterials increased hepatocyte concentrations of many lipids, particularly free fatty acids and monoacylglycerols but only CuO elevated lysolipids and sphingolipids. In respect to structure-activity, we now know that five out of six tested CeO2, and both SiO2 and CuO, but zero out of four TiO2 nanomaterials have caused this elevated lipids effect in HepG2 cells. Observed decreases in UDP-glucuronate (by CeO2) and S-adenosylmethionine (by CeO2 and CuO) and increased S-adenosylhomocysteine (by CuO and some CeO2) suggest that a nanomaterial exposure increases transmethylation reactions and depletes hepatic methylation and glucuronidation capacity. Our metabolomics data suggests increased free radical attack on nucleotides. There was a clear pattern of nanomaterial-induced decreased nucleotide concentrations coupled with increased concentrations of nucleic acid degradation products. Purine and pyrimidine alterations included concentration increases for hypoxanthine, xanthine, allantoin, urate, inosine, adenosine 3′,5′-diphosphate, cytidine and thymidine while decreases were seen for uridine 5′-diphosphate, UDP-glucuronate, uridine 5′-monophosphate, adenosine 5′-diphosphate, adenosine 5′-monophophate, cytidine 5′-monophosphate and cytidine 3′-monophosphate. Observed depletions of both 6-phosphogluconate, NADPH and NADH (all by CeO2) suggest that the HepG2 cells may be deficient in reducing equivalents and thus in a state of oxidative stress.
Conclusions
Metal oxide nanomaterial exposure may compromise the methylation, glucuronidation and reduced glutathione conjugation systems; thus Phase II conjugational capacity of hepatocytes may be decreased. This metabolomics study of the effects of nine different nanomaterials has not only confirmed some observations of the prior 2014 study (lipid elevations caused by one CeO2 nanomaterial) but also found some entirely new effects (both SiO2 and CuO nanomaterials also increased the concentrations of several lipid classes, nanomaterial induced decreases in S-adenosylmethionine, UDP-glucuronate, dipeptides, 6-phosphogluconate, NADPH and NADH).
Electronic supplementary material
The online version of this article (10.1186/s12989-017-0230-4) contains supplementary material, which is available to authorized users.
Keywords: Nanomaterial, SiO2, CeO2, CuO, Metabolomics, Fatty acids, HepG2
Background
Metal oxide nanomaterials have many uses including: coatings, grinding, ceramics, catalysis, electronics, biomedical, energy and fuel additives (for CeO2); biocides, sensor applications, catalysis and electronics (for CuO); and additives for rubber and plastics, composites for concrete and other construction materials and biomedical applications such as drug delivery and theranostics (for SiO2). It is difficult to evaluate nanomaterials to determine their degree and type of toxicity [1]. For nanomaterials a major determinant of their biological action may be their surface properties, particularly their ability to donate or accept electrons [2] and/or to generate free radicals and to form reactive oxygen species (ROS) [3].
After the development of the genomics and proteomics technologies, metabolomics has more recently been developed and used as an analytical tool in general biological research [4] and toxicological studies (Kitchin et al. [5]). The analytical platforms most commonly used to determine cellular metabolites are liquid chromatography tandem mass spectroscopy (LC-MS/MS), LC-MS/MS with hydrophilic interaction liquid chromatography (HILIC), gas chromatography-mass spectroscopy (GC-MS) and nuclear magnetic resonance (NMR). Metabolomics offers environmental and toxicological researchers the opportunity to determine the concentrations of many important cellular biochemicals in one experiment and provide complimentary information to traditional toxicological tests and other modern ‘omics approaches to biological questions.
In the nanotoxicology world, functional assays have recently been proposed as a way to better predict and connect the physical-chemical properties of nanomaterials and their potential adverse health outcomes [6]. Metabolomics based determinations of the altered concentrations of many important cellular biochemicals offer many good possible functional assays as intermediates in the long causal chain between physical-chemical properties of nanomaterials and eventual toxicity.
This study partnered with the Metabolon Inc. (Durham, NC) which used four analytical platforms to measure as many HepG2 (human liver) metabolites as possible – liquid chromatography-tandem mass spectroscopy with positive ionization (LC-MS/MS+), liquid chromatography-tandem mass spectroscopy with negative ionization (LC-MS/MS−), HILIC LC-MS/MS with negative ionization and gas chromatography mass spectroscopy (GC-MS) (with positive ionization via electron impact ionization). With metabolomics tools such as these, cellular biochemicals from different metabolic classes can be determined – lipids, energy molecules, amino acids, peptides, carbohydrates, purines, pyrimidines and nucleotides etc. A prior metabolomics study had discovered several interesting biochemical changes in TiO2 and CeO2 exposed HepG2 cells – a large number of lipid increases, particularly of fatty acids and many decreases in glutathione-related biochemicals and increased asymmetric dimethylarginine by two CeO2 nanomaterials [5]. Because of strong interest in the prior CeO2 nanomaterial induced effects, five new CeO2 nanomaterials were selected for the current study (labelled W4, X5, Y6, Z7 and Q) (Table 1). CeO2 based materials offer the possibility of Ce+4 <--> Ce+3 redox cycling [7] and the generation of ROS. Additionally, atomic layer deposition (ALD) using tris(isopropylcyclopentadienyl)cerium was attempted in an effort to produce a CeO2 coated SiO2 nanoparticle with a large amount of Ce+3 on the surface (nanomaterials labelled SiO2 K1 and SiO2 N2). Finally, a CuO nanomaterial was included because of interest in the toxicity of soluble copper ions and the oxidative stress theory of nanomaterial toxicity (all treatment nanomaterials are summarized in Table 1).
Table 1.
Physical-chemical characterization of CeO2, SiO2 and CuO particles
| ID | Chemical | Vendor | Cat No. | Lot number | Primary particle size (nm) | TEM particle size (nm) | SEM aggregate Size (um) | Surface area (m2/g) | Diameter by BET (nm) | FTIR | Elements by SEM-EDX | Elements by TEM-EDX | Form by XRD | Assayer |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CeO2 W4 | CeO2 | Nano-oxides | 10-025 | 68740 | 15 by BET | 55 | 15 | Nano-oxides | ||||||
| 20–50 | 1–3 | 52.8 | 14.9 | -OH, Ce-O | Ce, O, Al | Ce, O, Al, Ti, Si | crystalline | University of Kentucky | ||||||
| CeO2 X5 | CeO2 | Nano-oxides | 10-025-3 | 67722 | 200 by BET | 5–9 | 200 | Nano-oxides | ||||||
| 5–20 | 1–5 | 20.8 | 38.1 | -OH, Ce-O | Ce, O | Ce, O | crystalline | University of Kentucky | ||||||
| CeO2 Y6 | CeO2 | Aldrich | 544841 | 67722 | <25 by BET | Aldrich | ||||||||
| 5–20 | 1–20 | 40.3 | 19.5 | -OH, Ce-O | Ce, O | Ce, O | crystalline | University of Kentucky | ||||||
| CeO2 Z7 | CeO2 | Alfa aesar | 44960 | J06 U027 | 15–30 | 30–50 | Alfa aesar | |||||||
| 5–20 | 1–5 | 57.0 | 13.8 | -OH, Ce-O | Ce, O | Ce, O | crystalline | University of Kentucky | ||||||
| CeO2 Q | CeO2 | Sigma Aldrich | 211575 | NM-213 | <5000 | >500 | 0.615 | 3.73 | 213 | Geraets et al. [26] | ||||
| NDa | ND | ND | ND | ND | ND | ND | ND | University of Kentucky | ||||||
| SiO2 J0 | SiO2 | US Research Nanomaterials | US3438 | None | 20–30 | US Research Nanomaterials | ||||||||
| 10–30 | 1–10 | 137.4 | 16.5 | -OH, Si-O | Si, O | Si, O | amorphorus | University of Kentucky | ||||||
| SiO2 K1 | SiO2 | ALDb | NAc | None | 20–30 | US Research Nanomaterials | ||||||||
| 10–30 | 1–10 | 128.8 | 17.6 | -OH, Si-O | Si, O | Si, O | amorphorus | University of Kentucky | ||||||
| SiO2 N2 | SiO2 | ALDb | NAc | None | 20–30 | US Research Nanomaterials | ||||||||
| 10–30 | 1–10 | 120.5 | 18.8 | -OH, Si-O | Si, O | Si, O | amorphorus | University of Kentucky | ||||||
| CuO | CuO | Nanostruct-ured and Amorphous Materials | 2110FY | US3438 | 47 | Nanostructured and Amorphous Materials | ||||||||
| 20–80 | 1–3 | 10.8 | 88. | -OH | Cu, O | Cu, O | crystalline | University of Kentucky |
Abbreviations: TEM transmission electron microscopy, SEM scanning electron microscopy, BET surface area/porosity determination by the Brunauer, Emmett, Teller test method, FTIR Fourier transform infrared spectroscopy, EDX energy-dispersive x-ray analysis, XRD X-ray diffraction
aNot done
bAtomic layer deposition on SiO2
cNot available
In vitro toxicity testing allows us to link molecular, biochemical and cellular functions to physicochemical properties of nanomaterials, adverse biological outcomes and better predict risk. The specific major goals of this metabolomics study was to replicate and/or further explore: 1) the findings of lipid elevations (e. g. fatty acids) caused by one CeO2 nanomaterial, 2) the depletion of glutathione and gamma-glutamyl amino acids by several metal oxide nanomaterials (both CeO2 and TiO2), 3) elevations in asymmetric dimethylarginine found with 2 CeO2 nanomaterials and 4) to explore the metabolomics effects of two new metal oxide nanomaterials based on SiO2 and CuO and 5) to discover possible functional assays. Overall, functional assays can link individual experimental data with proposed mechanisms of action to inform adverse outcome pathway model development in support of regulatory decisions.
To assess potential hepatotoxicity issues from oral and/or inhalation exposure routes, 72 h exposures were conducted in human liver HepG2 cells. Thus, human liver HepG2 cells were exposed for 3 days to five different CeO2 (either 30 or 100 μg/ml), 3 SiO2 based (30 μg/ml) or 1 CuO (3 μg/ml) nanomaterials with dry primary particle sizes ranging from 15 to 213 nm. Nanomaterial-exposed cells were examined for their ability to cause cellular toxicity and effects on the concentrations of cellular metabolites in HepG2 cells (Table 1, from 15 to 213 nm dry size). In our study 344 cellular metabolites were found and relatively quantified. This metabolomics study included sufficient biochemicals to examine the biochemical components of several major cellular systems – lipid homeostasis, cellular energetics, hepatic conjugation and excretion, urea cycle, polyamines, purines and pyrimidines. These metabolomics experimental results are discussed in the context of systems biology and the toxicology of nanomaterials.
Methods
Nanomaterials and their characterization and dispersion via ultrasound
The nine nanomaterials used in this study (Table 1) were selected to further determine the biological properties of various forms of CeO2 nanomaterials as well as some other metal oxide based nanomaterials (SiO2 and CuO). These nine nanomaterials are being used by three research laboratories at the US EPA in a coordinated research effort with many different scientific disciplines and experimental techniques.
Physical-chemical characterization of these nanomaterials was conducted by a variety of techniques for dry primary particle size, range of particle size, surface area and percent purity mostly by their manufacturer (Table 1). The nanomaterials were obtained from six different vendors (Alfa Aesar, Aldrich, Sigma, Nanoxides, US Research Nanomaterials and Nanostructured and Amorphous Materials). When given, the chemical purity was high (>99.5%). The primary dry particle sizes ranged from 15 to 213 nm. All nine nanomaterials in Table 1 have been physical-chemical characterized by nine different techniques by a University of Kentucky group led by Dr. Eric Grulke and the results will be published elsewhere.
For dispersion prior to cell culture, measured amounts of bovine serum albumin (BSA, Sigma-Aldrich, product A7906) at 200 mg/ml and phosphate buffered saline (PBS) were added to the dry nanomaterials in a glass vial. The general protein coating recipe of Dale Porter [8] was followed with the mass ratio of the nanomaterial to BSA of 1/0.6. For example, in preparation of CeO2 “Z7” for study, 16.04 mg nanomaterial CeO2 Z7, 9.624 mg BSA and 4.95 ml of PBS were combined. Sonication occurred at a nanomaterial concentration of 3.21 mg/ml and 5.0 ml of volume. Sonication was done at room temperature with a S-4000 Misonix Ultrasonic Liquid Processor with a 2.5 in. cup horn (part #431-A, Farmington, NY) for two 10 min cycles of 13 s on, 7 s off with a total power of about 131 watts and a total energy of 166,120 joules. Excess unbound albumin was removed by pelleting (9300 × g for 5 min) the nanomaterials and resuspending them in cell culture media without any sonication of the cell culture media.
After nanomaterial dispersion, the degree of agglomeration was determined by dynamic light scattering at 35o C at each treatment concentration used for metabolomics study and sometimes one lower concentration. Size and zeta potential measurements were made both just after sonication and 72 h later at the end of treatment period with a Malvern Model Zen3600 Zetasizer (data in Additional file 1: Table S1).
Chemicals and cell culture methods
The chemicals and suppliers used in this study were: BSA (Sigma) and fetal bovine serum, GlutaMAX™, sodium pyruvate, fetal bovine serum, Dulbecco’s Phosphate-Buffered Saline and phosphate buffered saline (all from Invitrogen). Human Hepatocellular Carcinoma Cells, designation HepG2 (ATCC catalog number HB-8065), were obtained and expanded through passage seven using Basal Medium Eagle (Gibco) containing 2 mM GlutaMAX™, 1 mM sodium pyruvate and 10% fetal bovine serum and then frozen in liquid nitrogen. This combined cell culture media is called Eagle’s mimimum essential medium (EMEM). Cells were subsequently carefully thawed and expanded before experimentation at passages 10 and 11. Cultures were maintained in a humidified incubator at 37 °C and 95% air/5% CO2 during the study. Cells were plated at 80,000 cells/cm2 in vented T-25 flasks (Corning) for 48 h prior to nanomaterial exposure. After sonifcation, centrifugation and resuspension, working stocks of each nanomaterial were prepared at 1.0 mg per mL and diluted using culture medium. Individual flasks were dosed with 200 uL per cm2 of the appropriate nanomaterial dilution to achieve either 100 μg/ml (CeO2 Q), 30 μg/ml (7 other nanomaterials) or 3 μg/ml (CuO) exposure concentrations. Cultures were then incubated for 72 h prior to harvesting. At 72 h, the media was vacuum aspirated and the flasks rinsed with warm Dulbecco’s Phosphate-Buffered Saline (DPBS). The DPBS was aspirated and cells were scraped free of the flask and collected in labeled 15 mL tubes using 1 mL of warm DPBS by micropipette. The cells were then centrifuged at room temperature at 100 × g for 5 min. The supernatant was carefully removed via vacuum aspiration and the cellular pellet was flash frozen on dry ice before transfer to −80o C freezer for storage prior to metabolomic analysis.
Cytotoxicity assays and kits
Many common cytotoxicity assays [MTT (3-[4,5-dimethyl-2-thiazol]-2,5-diphenyl-2H-tetrazolium bromide), MTS (4-[5-[3-(carboxymethoxy)phenyl]-3-(4,5-dimethyl-1,3-thiazol-2-yl)tetrazol-3-ium-2-yl]benzenesulfonate), alamar blue (resazurin), neutral red (3-amino-7-dimethylamino-2 methylphenazine hydrochloride), ATP and simple visual examination of the cells] have been used by our laboratory seeking to avoid or minimize interferences from the nanomaterials themselves. After 72 h of culture with various nanomaterials, cytotoxicity assays based on MTT (Sigma-Aldrich, St Louis, MO), MTS (Promega, Madison, WI) and alamar blue (Cell Tier-Blue, Promega, Madison, WI) were performed in accordance with the enclosed kit directions. Alamar blue and MTS were used for all nanomaterial cytotoxicity experiments except for CeO2 Q (MTT only was used). A PerkinElmer 1420 Multilabel Counter Victor3V plate reader was used for all cytotoxicity assays. Cytotoxicity assays results were always checked with each other and versus visual assessment of the cells to ensure the cytotoxicity assays were functioning properly.
Study design
For metabolomics study, three different exposure concentrations (3, 30 or 100 μg/ml) were used for the nanomaterials. Only CuO at 3 μg/ml and CeO2 Q at 100 μg/ml were not run at 30 μg/ml. The intent was (a) to give approximately equally cytotoxic concentrations of the nine different nanomaterials and (b) if feasible to compare CeO2 nanomaterials at 30 μg/ml for better comparison to a prior study of our group that used this exposure dose for two prior CeO2 nanomaterials [5]. The number of samples per group is either five for treatments or six for controls. Two different days were used for HepG2 culturing. On day 1 most of the CeO2 (W4, X5, Z7 and Q) and the CuO treatment groups were run. On day 2 nanomaterials J0, K1 and N2 (the 3 SiO2 based nanomaterials) and CeO2 Y6 were run together.
Statistical analysis
Biochemical ion signals were processed by normalization to Bradford protein concentration, log transformation and imputation of missing values, if any, with the minimum observed value for each compound. Biochemicals that were detected in all samples from one or more groups, but not in samples from other groups, were assumed to be near the lower limit of detection in the groups in which they were not detected. In this case, the lowest detected level of these biochemicals was imputed for samples in which that biochemical was not detected. Then, Welch’s two-sample t-test was used to identify biochemicals that differed significantly between experimental groups [9]. In modern gene array work, using the False Discovery Rate (FDR) is a common method of controlling false positive (Type I) error rates. Thus, to account for multiple comparisons in this metabolomics testing, false-discovery rates were computed for each comparison via the Q-value method [10]. P values and Q value false discovery rate-values for all statistical comparisons are reported in Additional file 2: Table S2.
Pathways were assigned for each metabolite, allowing examination of overrepresented pathways. The degree of statistical significance presented in this study is both the common P < 0.05 level used if this 0.05 criteria is met by both P and Q statistics and the more lenient standard of 0.10 if both P and Q are <0.10, because this more lenient standard is less likely to miss some true biological effects. Tables 3, 4, 5, 6 and 7 and Additional file 2: Table S2 have color high lighting to graphically display these P < 0.05 and <0.10 significance levels. The text of the paper uses the P < 0.05 level of claimed statistical significance with the P < 0.10 level mentioned only for NADPH.
Table 3.
Nanomaterial effects on responsive lipids
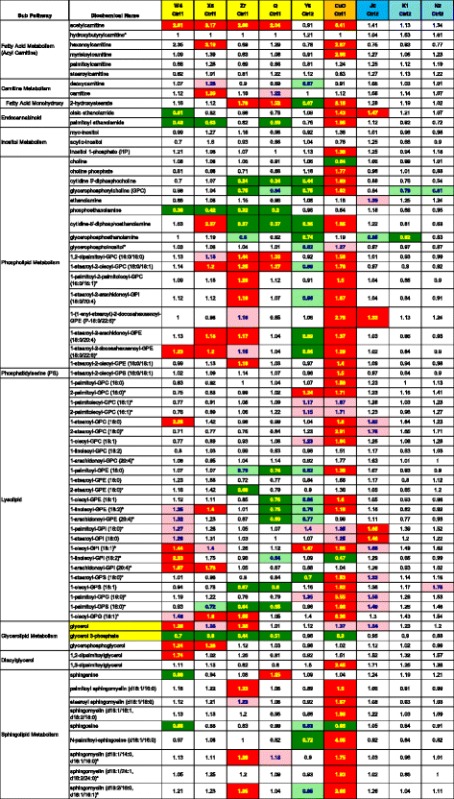
Darker shading (red for increases, green for decreases) means P and Q are both <0.05; Lighter shading means P and Q are both <0.10
The numbers are the ratio of the treated mean divided by the control mean
Table 4.
Nanomaterial effects on less responsive lipids
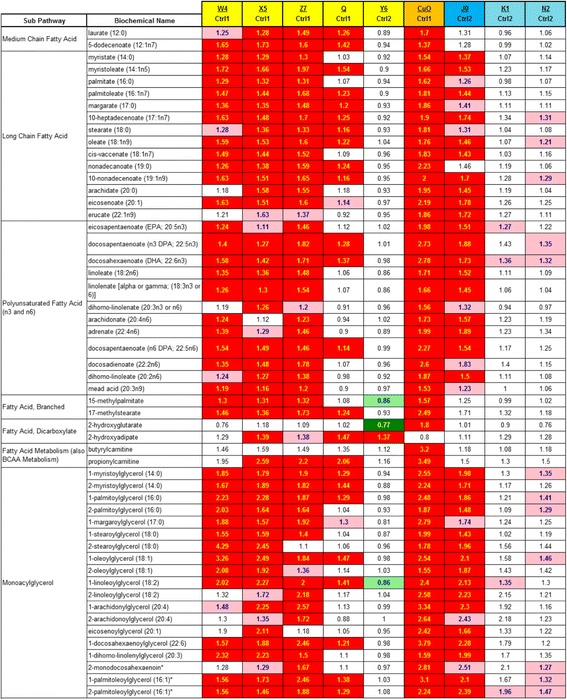
Darker shading (red for increases, green for decreases) means P and Q are both <0.05; Lighter shading means P and Q are both <0.10
The numbers are the ratio of the treated mean divided by the control mean
Table 5.
Nanomaterial effects on SAM, SAH, glutathione-related and nucleotide sugar metabolites
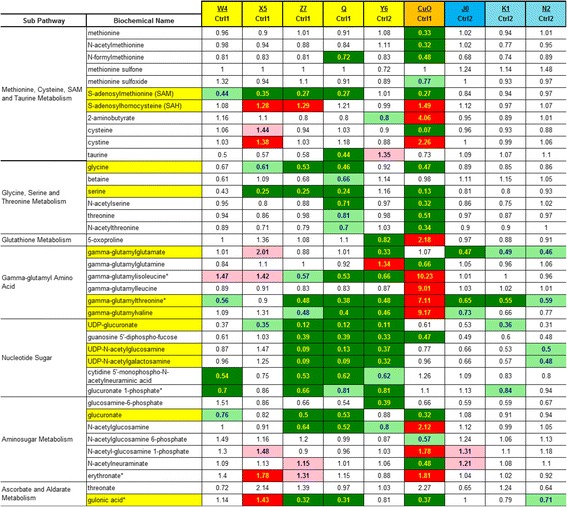
Darker shading (red for increases, green for decreases) means P and Q are both <0.05; Lighter shading means P and Q are both <0.10
The numbers are the ratio of the treated mean divided by the control mean
Table 6.
Nanomaterial effects on maltotriose, 6-phosphogluconate, nicotinamide metabolites and dipeptides
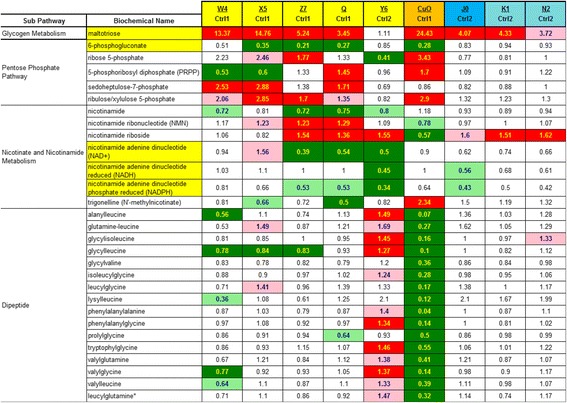
Darker shading (red for increases, green for decreases) means P and Q are both <0.05; Lighter shading means P and Q are both <0.10
The numbers are the ratio of the treated mean divided by the control mean
Table 7.
Nanomaterial effects on urea cycle, polyamines, purine and pyrimidine metabolites
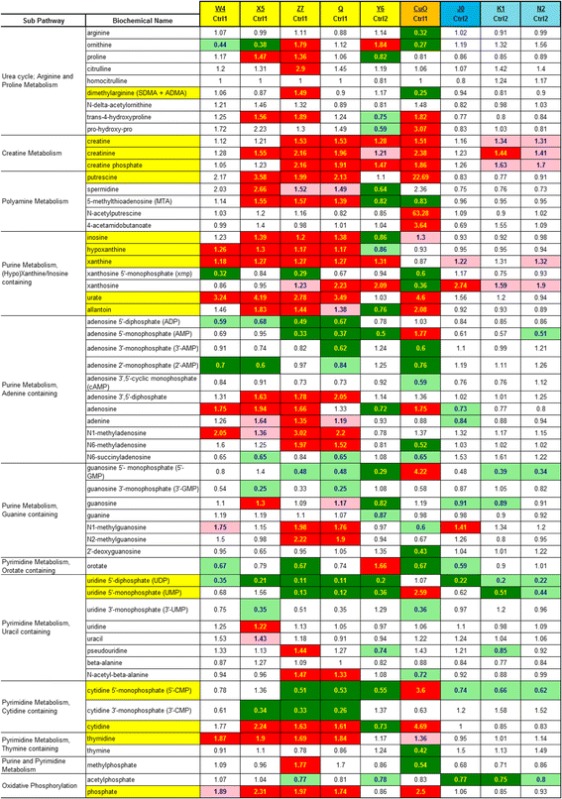
Darker shading (red for increases, green for decreases) means P and Q are both <0.05; Lighter shading means P and Q are both <0.10
The numbers are the ratio of the treated mean divided by the control mean
Results
Dispersion and agglomeration of nanomaterials (size and zeta potential)
By dynamic light scattering, these sonicated nanomaterial samples displayed a fairly large hydrodynamic diameter in both water based cell culture media (EMEM with 10% fetal bovine serum) and PBS (Additional file 1: Table S1). In cell culture media the mean sizes by peak intensity ranged between 154 to 540 nm for CeO2, 312 to 554 nm for SiO2 and 148 to 188 nm for CuO (Additional file 1: Table S1). These hydrodynamic sizes are much larger than the dry primary particle sizes of 15, 22.5, 25, 200 and 213 nm for the five different forms of CeO2 studied. In cell culture media the mean zeta potentials ranged between −4.4 to −10.3 mV for CeO2, −4.7 to −10.5 for CuO and −4.7 to −8.7 for SiO2 (Additional file 1: Table S1).
The coating of SiO2 K1 and SiO2 N2 and ICP-MS results
Our attempt to use atomic layer deposition to put a thin layer of CeO2 on the J0 SiO2 based particles failed. By ICP-OES analysis performed at both Missouri University of Science and Technology and the US EPA, almost zero Ce was found in nanomaterials SiO2 K1 and SiO2 N2 (Additional file 3: Table S3).
Cytotoxicity results
The exposure concentrations used in this metabolomics study (3, 30 or 100 μg/ml) were below concentrations which produced a full degree of cytotoxicity in HepG2 cells via common colorimetric and fluorimetric assays (Table 2). At the administered dose, no sign of cytotoxicity was observed for CeO2 W4, CeO2 X5 and CeO2 Y6; a low degree of cytotoxicity for CeO2 Z7, CeO2 Q, SiO2 K1 and SiO2 N2; and a medium degree of cytotoxicity for SiO2 J0 and CuO (Table 2).
Table 2.
Cytotoxicity of the CeO2, SiO2 and CuO nanomaterials in HepG2 cells
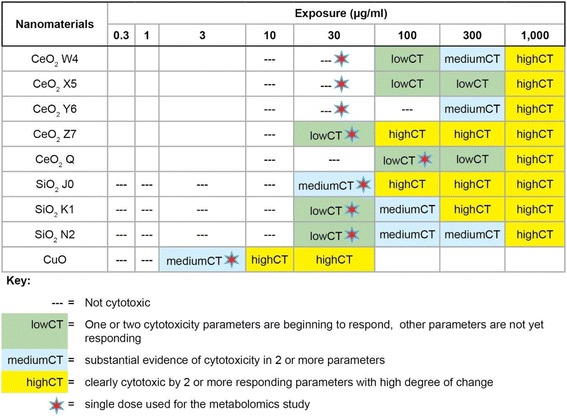
Both the number and degree of response was considered for each of the eight parameters germane to “cytotoxicity”
The eight cytotoxicity parameters are visual microscopic cellular appearance, alamar blue, MTS, cellular protein and microalbumin concentrations and release of lactate dehydrogenase, alanine aminotransferase and aspartate aminotransferase
Metabolomic results
For the metabolomics results the nanomaterial exposure concentrations were 3 μg/ml for CuO, 30 μg/ml for CeO2 W4, CeO2 X5, CeO2 Y6, CeO2 Z7, SiO2 J0, SiO2 K1 and SiO2 N2 and 100 μg/ml for CeO2 Q. Additional file 4: Table S4 presents the number and direction of statistically significant metabolite concentration alterations following nanomaterial treatments. Overall, the number of P < 0.05 total metabolite concentration changes, increased and decreased biochemical concentrations versus concurrent controls were: 75, 59 and 16 for CeO2 W4; 117, 99 and 18 for CeO2 X5; 67, 19 and 48 for CeO2 Y6; 157, 115 and 42 for CeO2 Z7; 124, 70 and 54 for CeO2 Q; 52, 43 and 9 for SiO2 J0; 9, 3 and 6 for SiO2 K1; 1, 1 and 0 for SiO2 N2; and 226, 145 and 81 for CuO, respectively. With the exception of CuO (226 altered metabolite concentrations at a medium degree of cytotoxicity), the number of significantly changed metabolite concentrations did not correlate with degree of cytotoxicity observed for the other eight nanomaterials.
Altered lipids
In Tables 3, 4, 5, 6 and 7, the displayed numbers are the ratio of the treatment metabolite concentration mean divided by the concurrent control metabolite concentration mean. Increased concentrations of medium and long chain fatty acids, polyunsaturated fatty acid (n3 and n6), fatty acid branched, fatty acid dicarboxylate and monoacylglycerols were observed after treatment with several CeO2 (W4, X5, Z7 and Q), SiO2 (J0 only) and CuO nanomaterials (Tables 3 and 4). In this study far fewer increases were noted with fatty acid metabolites, lysolipids, carnitine, inositol metabolites, phospholipid metabolites, phospholipidserine, diacylglycerol and sphingolipid metabolites, showing the selectivity of this lipid effect (Tables 3 and 4). CuO was the only nanomaterial to induce many increases in these classes of less responsive lipids (Tables 3 and 4). The most active lipid-elevating nanomaterials were W4, X5, Z7 (all are CeO2), SiO2 J0 and CuO. CeO2 Y6 and the two ALD coated SiO2 based nanoparticles (K1 and N2) did not elevate as many lipid metabolite concentrations. P and Q numbers are tabulated for all 344 biochemicals for every nanomaterial treatment comparison with concurrent controls in Additional file 2: Table S2.
Hepatic conjugation systems (methylation, glucuronidation and glutathione)
Treatment of HepG2 cells with nanoparticles from the day-1 set (CeO2 X5, CeO2 Z7, CeO2 Q and CuO) resulted in declines in S-adenosylmethionine (SAM) and several increases in S-adenosylhomocysteine (SAH) (by CeO2 X5 and CeO2 Z7) (Table 5), though methionine levels were largely unchanged. In the liver methylation capacity is required to support Phase II methylation of xenobiotics to facilitate clearance. The lower SAM levels were accompanied by a sharp decline in serine (by CeO2 X5, CeO2 Z7, CeO2 Q and CuO), in day-1 nanomaterial treated cells. Serine is consumed in the regeneration of methionine from homocysteine, in the one-carbon metabolism pathway. Most of the day-1 nanoparticle-treated samples had SAM below the limit of detection, however 5 of 6 day-1 control cell samples had SAM levels above the lower limit of measurement. SAM levels were relatively unchanged with exposure to the day-2 nanoparticles (CeO2 Y6, SiO2 J0, SiO2 K1 and SiO2 N2) and declines in serine were also limited and not statistically significant.
The three observed UDP-glucuronate fold decreases were rather large, 0.12 (CeO2 Z7), 0.12 (CeO2 Q), and 0.11 (CeO2 Y6) of concurrent control values (Table 5). Glucuronate itself was significantly decreased by nanomaterials CeO2 Z7, CeO2 Q and CuO (Table 5). Uridine diphosphate (UDP) is an important metabolite for cellular glycogen synthesis, protein glycosylation and glucuronidation. After treatment with several nanoparticles, a decreases in UDP as well as the measured UDP-sugars UDP-glucuronate, UDP-N-acetylgalactosamine and UDP-N-acetylglucosamine were also observed (Table 5).
It is quite surprising that reduced glutathione (GSH) levels were below detection limit in most control and treated samples in this study (some GSH was detected in three of our samples). Similar to prior results with 4 TiO2 and 2 CeO2 nanomaterials [5], there were decreases observed in gamma–glutamyl amino acids with several CeO2 and SiO2 based nanomaterials (Table 5). Most effected were gamma–glutamylthreonine, gamma–glutamylvaline and gamma–glutamylgluatamate. In contrast, the CuO nanomaterial caused large fold increases in four gamma–glutamyl-amino acid compounds –leucine (9.0 fold increase), –isoleucine (10.2), –threonine (7.1) and –valine (9.2) but not –glutamine (0.66) or –glutamate (1.07) (Table 5).
Cellular energetics, reducing capacity and oxidative stress (maltotriose, 6-phosphogluconate, NADPH, NADH and NAD+ and dipeptides)
Seven out of nine nanomaterial treatment groups (only CeO2 Y6 and SiO2 N2 did not) increased maltotriose concentrations ranging from 3.45 to 24.4 fold of concurrent control values. Three increases were above 10 fold increases (13.4 by CeO2 W4, 14.8 by CeO2 X5 and 24.4 by CuO). Maltotriose levels can represent a measure of glycogen degradation, from which maltotriose is derived. The first step in conversion of glucose 6-phosphate to 6-phosphogluconate generates NADPH. 6-phosphogluconate was significantly depleted by four of the 5 day-1 set of nanoparticles (Table 6). NADPH concentrations were numerically decreased in all nine nanoparticle treatments (range 0.34 to 0.81) (Table 6), achieving statistical significance for nanoparticle CeO2 Y6 at the P < 0.05 level, while the CeO2 Z7, CeO2 Q and SiO2 J0 particles were statistically significant at the lower P < 0.10 level, relative to controls. NADH concentration was significantly decreased (P < 0.05) by CeO2 Y6 (0.45). No significant elevations were seen for NADH or NADPH. Both nicotinamide (2 decreases) and NAD+ were significantly decreased by three nano CeO2 treatments (CeO2 Z7, CeO2 Q and CeO2 Y6) (Table 6). Nicotinamide riboside (a NAD+ precursor) was significantly elevated in all three cases where NAD+ was depleted (CeO2 Z7, CeO2 Q and CeO2 Y6) (Table 6).
CuO nanomaterial exposure decreased (P < 0.05) the concentrations of all 16 dipeptides ranging from 0.07 to 0.55 fold change. With the exception of CeO2 W4, CeO2 X5 and CeO2 Z7 induced decreases in the dipeptide glycylleucine, few other dipeptides were decreased by CeO2, or SiO2 based nanomaterials. CuO was also the only nanomaterial that caused a large decrease in the concentration of cysteine (0.07) while elevating cystine (2.26) (Table 5). This cysteine-cystine redox perturbation suggests oxidative stress caused by CuO exposure.
Cellular effects (urea cycle, polyamines, purine and pyrimidine metabolism, nucleotide sugars)
Several urea cycle, creatinine and polyamine pathway biochemicals were significantly increased by nanomaterial treatment, such as creatine (4 increases), creatinine (5 increases), creatine phosphate (4 increases), putrescine (4 increases) and 5-methylthioadenosine (5 changes with 3 increases) (Table 7). Levels of putrescine, spermidine and 5-methylthioadenosine were significantly elevated for many of the CeO2 nanoparticles in the day-1 set, but these biochemical were not elevated in the day-2 nanomaterials (Table 7). CuO exposure increased putrescine 22.7 fold and N-acetylputrescine 63.3 fold, among the highest elevations observed in this data set. Following CuO exposure, high putrescine concentration (22.7 fold) coupled with low ornithine concentration (0.27 fold) suggest that the enzyme activity of the rate limiting step of polyamine synthesis, ornithine decarboxylase, may have been increased. To a much lesser extent this pattern also occurred with CeO2 X5 (putrescine (3.58) and ornithine (0.38)) CeO2 exposures.
In the general area of purine and pyrimidine metabolism, there were many nanomaterial induced changes with both increases and decreases in concentrations observed. Phosphate ion concentration was significantly increased in four of the nine comparisons (3 with nano CeO2 and 1 with CuO). Nanomaterial exposures often decreased nucleotide concentrations: adenosine 5′-diphosphate (ADP) (2 decreases), adenosine 5′-monophophate (AMP) (3 decreases), uridine 5′-diphosphate (UDP) (5 decreases), uridine 5′monophosphate (UMP) (4 decreases), cytidine 5′monophosphate (5′-CMP) (3 decreases) and cytidine 3′-monophophate (3′-CMP) (3 decreases).
However, there were many examples of increased nucleic acid degradation products: inosine (4 changes with 3 increases), hypoxanthine (4 increases), xanthine (5 increases), urate (5 increases) and allantoin (4 changes, 3 increases). Thus, the overall purine and pyrimidine pattern is one of decreased nucleotides and increased nucleic acid degradation products.
In the six component nucleotide sugar biochemical sub pathway (Table 5), all six members of the group showed statistically significant (P < 0.05) decreases in 3 or more of the nine treatment groups (often following CeO2 Z7, CeO2 Q, CeO2 Y6, SiO2 K1 and SiO2 N2 exposure). The nucleotide sugars are important in Phase II glucuronidation and glycation reactions. Most active nanomaterials were CeO2 Z7, CeO2 Q and CeO2 Y6; least active were CeO2 X5, SiO2 J0, SiO2 K1, SiO2 N2 and CuO. There is a major data imbalance here with no significant increases and 19 significant decreases observed in 54 nucleotide sugar observations (Table 5). Moreover, some of the treated-to-control ratios were quite low for three nucleotide sugars – between 0.09 and 0.13 for UDP-glucuronate (by CeO2 Z7, CeO2 Q and CeO2 Y6), UDP-N-acetylglucosamine (by CeO2 Z7 and CeO2 Q) and UDP-N-acetylgalactosamine (by CeO2 Z7 and CeO2 Q).
Discussion
Altered lipids
Comparison of the results of this study with prior results from one CeO2 nanomaterial (M from Nanoamour, dry size 8 nm) [5] shows that the results of the two studies are similar in respect to CeO2 nanomaterial-induced elevations in fatty acids and monoacylglycerols. There were additional elevations in lysolipids, diacylglycerols and sphingolipids caused by CuO (this study) and by CeO2 M [5], but in the current study the other five CeO2 nanomaterials did not cause these particular lipid elevations. Possible explanations of the lipid increases seen with 3 CeO2, 1 SiO2 and 1 CuO nanomaterial include: a) increases in lipolysis of complex lipids, b) increased synthesis of fatty acids, c) decreased utilization in β-oxidation or complex lipid assembly or d) greater uptake of lipids from the cell culture media containing 10% fetal bovine serum because of nanoparticle uptake through endocytosis or nanomaterial induced cell membrane leakage. The major fatty acids of fetal bovine serum are palmitic, stearic and oleic [11]. However, these fatty acids were not particularly elevated over other fatty acids, thus arguing somewhat against the “greater uptake of lipids” interpretation.
A literature search showed elevated free fatty acids mentioned as a biomarker in ozone toxicity studies and ethanol-induced liver injury. Free fatty acids have been proposed as an “emerging biomarker” of nonalcoholic steatohepatitis [12]. From 1 to 48 h after exposure to hepatic irradiation, rat hepatic fatty acid concentrations were elevated [13]. Ozone exposures to both rats [14] and humans [15] elevated serum fatty acid concentrations. In addition, rat serum, brain and liver fatty acid concentrations were elevated by ethanol-induced liver injury [16]. In one in vitro study, exposure to quantum dots caused the down-regulation of beta-oxidation of fatty acids in PC12 cells (rat pheochromocytoma) [17]. In both PC12 cells and primary mouse hypothalamic cell culture, Zn-S coated quantum dots induced the accumulation of lipid droplets [17].
Glycerol levels were higher in several of nanoparticle-treated cells relative to controls (Tables 3 and 4). Reduced glycerol 3-phosphate concentration was observed with each of the day-1 nanoparticles that elevated lipid concentrations (Tables 3 and 4). Glycerol 3-phosphate is utilized in the assembly of free fatty acids into triacylglycerides. A decline in glycerol 3-phosphate concentrations may be an indication of increased complex lipid assembly for storage [18]. Alternatively, a partial blockage in the transformation of glycerol into glycerol 3-phosphate might reduce the synthesis of triglycerides and thus elevated free fatty acids, exactly what is observed in many cases (Tables 3 and 4).
Hepatic conjugation systems (methylation, glucuronidation and glutathione)
An important role of the liver is to conjugate various molecules with methyl, glucuronic acid or glutathione groups often as part of Phase II “drug metabolism” pathways [19]. Nanoparticle exposure may result in an increase in trans-methylation reactions and thus explain the observed SAM depletion.
One potentially important consequence of an insufficient supply of hepatocyte UDP-glucuronate would be a lack of glucuronidation capacity for Phase II metabolism of xenobiotic substances. Thus, even if nanoparticle clearance does not require glucuronidation per se, nanoparticle-induced UDP-glucuronate depletion may impair glucuronidation and clearance of other medicinal or toxic substances. Thus, with declines in both UDP-glucuronate (Table 5) and SAM (Table 5), hepatocytes may have a diminished capacity to methylate, glucuronidate and excrete xenobiotics. In many animals, but not humans or guinea pigs, UDP-glucuronate is also a synthetic intermediate in the biosynthesis of ascorbic acid, an important cellular antioxidant. Gulonic acid, another biochemical intermediate in ascorbic acid biosynthesis was also decreased by prior administration of nanomaterials CeO2 Z7, CeO2 Q, and CuO (Table 5).
In this study, no useful GSH concentrations information was obtained because the measured GSH concentrations were often below the quantitation limit. In the sample preparation for metabolomics profiling, there was no added acid, chelators or deoxygenation of solutions, all well established factors that preserve GSH in the reduced oxidation state [20]. The size of the cell pellet was about 1/3 of that in our previous study so the factor of small cell pellet size also probably contributed to GSH being below the lower limit of measurement in most samples. It seems that the LC-MS/MS parts of the analytical procedure were working properly because other cell based studies run the following day and 2 days previous to our study measured GSH at typical levels for a cell based assay.
Cellular energetics, reducing capacity and oxidative stress (maltotriose, 6-phosphogluconate, NADPH, NADH and NAD+ and dipeptides)
Maltotriose, a trisaccharide consisting of three glucose moieties with alpha 1–>4 glycosidic bonds between them is not known to be connected to toxicology or environmental health in any major way. However, maltotriose might be valuable as a biomarker of exposure for some metal oxide nanomaterials (e.g. 24.4 fold elevation by CuO). In yeast, exposure to either H2O2 or CuSO4 leads to increased maltotriose concentrations (https://www.wikipathways.org/index.php/Pathway:WP478).
Most nano forms of copper give off Cu+ and/or Cu++ ions [21]. The single peptide bond of all dipeptides is capable of reducing Cu++ to Cu+ (the biuret reaction). In the presence of H2O2 and Cu+, hydroxyl radical can be generated (the Fenton reaction) [22]. Such hydroxyl radicals are capable of destroying molecules within a short diffusional distance, such as the dipeptides binding site at which the Cu+ may have been generated. This could explain why all 16 dipeptide concentrations were decreased (0.07 to 0.55 fold) by CuO nanomaterial administration. Neither CeO2, SiO2 (Table 6) or TiO2 [5] nanoparticles caused large numbers of decreases in the dipeptide concentrations. After CuO exposure, 17 out of 20 single amino acids also exhibited decreases in concentration but not to as large an extent as observed for dipeptides (Additional file 2: Table S2). It does not seem as if CuO administration causes selective reductions of primary amine or carboxy group containing biochemical concentrations as there is substantial evidence against this possibility. For example, two primary amines containing biochemicals are significantly increased by CuO nanomaterial administration, namely putrescine (22.7 fold) and N-acetyl putrescine (63.3) (Additional file 2: Table S2). Three carboxy group containing biochemicals were also significantly increased by CuO nanomaterial treatment namely trans-4-hydroxyproline (1.8 fold), 4-acetamidobutanoate (3.6) and pro-hydroxy-pro (proline-hydroxyproline, CAS 18684-24-7) (3.1 fold) (Additional file 2: Table S2).
Thus, CuO nanomaterials produced three effects at very high frequency of occurrence – elevation of certain lipids (Tables 3 and 4), decrease of most dipeptides (Table 6) and decreases in many single amino acids (Additional file 2: Table S2). Thus, even if dissolution of CuO to copper ions produces hydroxy radicals, dipeptides and single amino acids are showing the large, consistently decreased cellular concentrations while other similar biochemicals are not showing decreases. An alternative explanation of the observed dipeptide decreases would be that protein breakdown was decreased.
Cellular effects (urea cycle, polyamines, purine and pyrimidine metabolism, nucleotide sugar)
Among the CeO2 nanoparticles from the day-1 set, CeO2 Z7 stood out for its elevation of citrulline, ornithine and dimethylarginine, relative to controls and the other CeO2 nanoparticles in the set. The higher levels of citrulline and ornithine in CeO2 Z7-treated cells were not accompanied by a decrease in arginine, relative to control or the other CeO2 nanoparticles. Dimethylarginine (both asymmetric and symmetric dimethylarginine were quantified together) were highest in CeO2 Z7 treated cells and, given the inhibitory properties of asymmetric dimethylarginine towards iNOS, it is possible that less arginine gets converted directly to citrulline through iNOS and instead is converted to ornithine. There were fewer dimethylarginine increases observed in this data set than in the preceding metabolomics study in which 2 CeO2 nanomaterials increased asymmetric dimethylarginine [5]. In addition, this study determined asymmetric and symmetric dimethylarginine together (Table 7) so this might have masked some asymmetric dimethylarginine increases.
Changes in urea cycle metabolites were also observed in the prior study with two forms of CeO2 [5], with changes being more pronounced in the current study. The levels of creatine were correlated with creatinine and creatine phosphate (Table 7). Glycine is consumed in the synthesis of creatine. Glycine levels are decreased with several nanoparticle exposures (CeO2 Z7, CeO2 Q, and CuO) (Table 5).
Among the day-1 nanomaterials, CuO caused the greatest amount of purine nucleotide degradation, as judged by the urate and allantoin levels. Metabolites connected with pyrimidine nucleotide degradation, such as thymidine and cytidine were elevated with several day-1 nanoparticle treatments (Table 7). Other purine nucleotide degradation metabolites were also increased. Hypoxanthine (4 increases) oxidation to xanthine (5 increases) and subsequent xanthine oxidation to urate (5 increases) by the enzyme xanthine oxidoreductase can produce superoxide or hydrogen peroxide, under some conditions. This can result in redox stress if sufficient anti-oxidants such as glutathione are not present.
Our first study with TiO2 and CeO2 and this current study with CeO2 and SiO2 agree in respect to the metabolite identity and direction of changes (increase or decrease) for several biochemicals notably NAD+, 6-phosphogluconate, UDP-glucuronate, UDP-acetylglucosamine, UDP-galactosamine and gamma-glutamlyglutamate. In summarizing the results, there does not appear to be a single, obvious cause of some of the metabolomics effects observed (Additional file 5: Table S5). The single CuO nanomaterial studied was quite different in number and some types of metabolomics effects it caused. This could be because of the different nanomaterial elemental composition (Cu rather than Ce or Si), higher degree of cytotoxicity observed with 3 μg/ml of CuO and the ability to form toxic copper ions via dissolution.
Pattern of significant effects within biochemical pathways
Table 8 presents a summary of the treatment effects of the CeO2, SiO2 and CuO particles for 13 of the more important altered biochemical pathways. Table 8 shows the direction of significant changes (up or down) for some of the altered biochemicals in each pathway. The number of significant changes observed per biochemical pathway was one in the glycogen pathway (maltotriose), two in the ascorbic acid synthesis pathway (gulonic acid and UDP-glucuronate), six in the glucuronidation-related pathway (glucoronate, UDP-N-acetylgalactosamine, UDP-N-acetylglucosamine, UDP-glucuronate, uridine 5′-diphosphate (UDP), and uridine 5′-monophosphate (UMP)) and over 40 in the lipid pathways (e. g. oleate, sterate and palmitate).
Table 8.
Overview of the direction of observed metabolomic effects in various biochemical pathways following HepG2 exposures to CeO2, CuO and SiO2 particles
| Biochemical Pathway | Lipids fatty acids monoacyl-glycerols | Lipids Phospholipids Sphingolipids Lysolipids | Ascorbic acid synthesis | Glucuronidation-related | Methylation-related | Glutathione-related | Dipeptides | NAD-(P)H system | Glycogen | Creatinine metabolism | Polyamines | Nucleotides | Nucleic acid degrad-ation products |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CeO2 | Up | – | Down | Down | Down | Down | – | Down | Up | Up | Up | Down | Up |
| CuO | Up | Up | Down | – | Down | Up | Down | – | Up | Up | Up | Up | Up |
| SiO2 | Up | – | Down | Down | – | Down | – | – | Up | Up | – | Down | – |
| Example biochemical in pathway | oleate (18:1n9) | choline phosphate | gulonic acid | UDP-glucuronate | S-adenosyl-methionine (SAM) | gamma-glutamyl-threonine | phenylalanyl-alanine | NADPH | maltotriose | creatinine | putrescine | Cytidine 5′- monophos-phate (5′-CMP) | urate |
| Data in Table number | Tables 3 and 4 | Tables 3 and 4 | Table 5 | Table 5 | Table 5 | Table 5 | Table 6 | Table 6 | Table 6 | Table 7 | Table 7 | Table 7 | Table 7 |
Key: Up = several increases observed; Down = several decreases observed; − = No obvious pattern observed
Dosimetry
In in vitro nanomaterial toxicology there are large numbers of complex factors involved in the pharmacokinetics and dosimetry between administered dose (expressed as μg/ml in this study) and internalized dose to the cultured HepG2 cell. Some of the major factors that determine in vitro intracellular dose of nanomaterials include particle dose, shape, surface chemistry, size, charge, density, binding of molecules to the particle surface (protein corona), agglomeration, diffusion and gravitational settling [23–25]. In our nanomaterial studies we have collected ICP-OES data on Ce and Cu cellular concentrations from CeO2 and CuO exposed HepG2 cells. Eventually this cellular Ce and Cu dosimetry data may be useful in more deeply understanding the complex relationship between administered dose, internal cellular dose and various biological effects.
Conclusions
Altered lipids
This study confirms and extends the prior observation that a single CeO2 nanomaterial (M) caused concentration increases in large numbers of several classes of lipids in HepG2 cells (most notably fatty acids and monoacylglycerols) [5]. In this study 4 CeO2, 1 SiO2 and 1CuO nanomaterials were also shown to have this property of increasing lipid concentrations (Tables 3 and 4). In respect to structure-activity, we know that five out of six tested CeO2, and both SiO2 and CuO, but zero out of 4 TiO2 nanomaterials have caused this elevated concentration of lipids effect (Tables 3 and 4 and [5]). Thus, cellular lipid concentration increases may be a general property of exposure to many metal oxide nanomaterials and may impact hepatocyte and systemic lipid homeostatis.
Hepatic conjugation systems (methylation, glucuronidation and glutathione)
Metal oxide nanomaterial exposure may compromise the methylation, glucuronidation (Table 5) and glutathione conjugation systems (GSH data of [5]). The large number of metabolomics findings of decreased SAM coupled with increased SAH suggest an increase in transmethylation reactions and a depletion of SAM capacity. This shortage of methyl groups could have profound and adverse effects on cells in respect to DNA methylation and drug metabolism. From gamma-glutamyl amino acid decreases data (Table 5), there was a degree of indirect confirmation of glutathione depletion and oxidative stress observed in our prior study with TiO2 and CeO2 nanomaterials [5].
Cellular energetics, reducing capacity and oxidative stress (maltotriose, 6-phosphogluconate, NADPH, NADH and NAD+ and dipeptides)
Increases in the concentration of maltotriose occurred in the prior metabolomics study (1.76 fold increase by CeO2 M) [5] and also in this current study where the observed increases were much larger (a range of from 3.45 to 24.4-fold). To date, maltotriose concentrations have been significantly elevated by four out of six tested CeO2, along with both CuO and SiO2, but zero out of 4 TiO2 nanomaterials (Table 6 and [5]).
Observed depletions of both 6-phosphogluconate, NADPH and NADH suggest that the HepG2 cells may be out of redox equilibrium (not enough reducing equivalents) and thus in a state of oxidative stress. The unexpected pattern of CuO nanomaterial decreasing all 16 quantified dipeptides (Table 6) can be explained by the dissolution of CuO to ionic copper, peptide bond binding of Cu++, and the eventual free radical attack of hydroxyl radical on the dipeptides.
Cellular effects (urea cycle, polyamines, purine and pyrimidine metabolism, nucleotide sugar)
Cellular metabolism related to amino groups was strongly perturbed by these metal oxide nanomaterials. In HepG2 cells, the urea cycle and the metabolism of proline, creatine and polyamines were strongly effected by nanomaterial exposures. Both increases and decreases were seen with ornithine and proline concentrations. All significant findings were elevations for creatine, creatinine and creatine phosphate, molecules important in cellular energetics. Polyamines, one of the few positively charged cellular modulators, were usually increased by nanomaterial exposure, particularly by putrescine.
Because there was a clear pattern of nanomaterial-induced decreased nucleotide concentrations coupled with increased concentrations of nucleic acid degradation products, this study supports the interpretation of either increased free radical attack on nucleotides or increased turnover of important purines and pyrimidine biomolecules.
This metabolomics study of the effects of nine different nanomaterials has not only confirmed some observations of the prior 2014 study (lipid elevations caused by one CeO2 nanomaterial) but also found some entirely new effects (both SiO2 and CuO nanomaterials also increased the concentrations of several lipid classes, nanomaterial induced declines in SAM, UDP-glucuronate, dipeptides, 6-phosphogluconate, NADPH and NADH).
Additional files
Nanomaterial size and & zeta potential from dynamic light scattering (DLS). (DOC 184 kb)
Large excel Metabolon data sheet (heat map) with all results. (XLSX 533 kb)
Characterization of atomic layer deposition coated SiO2 nanoparticles at Missouri University of Science and Technology. (DOC 79 kb)
The number and direction of statistically significant metabolite concentration alterations after nanomaterial treatments. (DOC 109 kb)
Summary of some physical-chemical properties and biological effects of nanomaterial exposures. (DOC 86 kb)
Possible Metabolomic Functional Assays. (DOC 82 kb)
TEM images of SiO2 nanoparticles. (DOC 6894 kb)
Atomic layer deposition. (DOC 74 kb)
Metabolomic sample assessioning, preparation, chromatography, QA/QC, data extraction and compound identification. (DOC 77 kb)
Acknowledgements
We are grateful for the participation of many individuals of the Metabolon Inc. group who performed the analysis of the HepG2 cells, the statistical data reduction and some of the figures and tables. The elemental analysis of SiO2 J0, SiO2 K1 and SiO2 N2 (ICP-MS and/or OES at US EPA) were ably done by John McGee and Drs. Patrick Pancras and Mark Higuchi. We thank Drs. Urmila Kodavanti and Michael F. Hughes for reviewing this manuscript as part of EPA clearance procedures.
Funding
This work was supported by the U.S. Environmental Protection Agency.
Availability of data and materials
The metabolomic data set and Additional files 1, 2, 3, 4, 5 and 6: Tables S1–S6, Additional file 7: Figure S1, Additional files 8 and 9 are all available at https://intranet.ord.epa.gov/science/sciencehub The identification number is ScID: A-2rbs.
Disclaimer
The information in this document has been funded wholly by the U. S. Environmental Protection Agency. It has been subjected to review by the National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents necessarily reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Abbreviations
- 3′-CMP
Cytidine 3′-monophosphate
- 5′-CMP
Cytidine 5′monophosphate
- ADP
Adenosine 5′-diphosphate
- ALD
Atomic layer deposition
- AMP
Adenosine 5′-monophosphate
- ATP
Adenosine 5′-triphosphate
- BSA
Bovine serum albumin
- DPBS
Dulbecco’s Phosphate-Buffered Saline
- EMEM
Eagle’s minimum essential medium
- FDR
False Discovery Rate
- GC-MS
Gas chromatography-mass spectroscopy
- GSH
Reduced glutathione
- HepG2
Human Hepatocellular Carcinoma Cells, ATCC catalog number HB-8065
- HILIC
Hydrophilic interaction liquid chromatography based LC-MS-MS
- ICP-MS
Inductively coupled plasma mass spectroscopy
- ICP-OES
Inductively coupled plasma optical emission spectroscopy
- LC-MS/MS
Liquid chromatography tandem mass spectroscopy
- MTS
4-[5-[3-(carboxymethoxy)phenyl]-3-(4,5-dimethyl-1,3-thiazol-2-yl)tetrazol-3-ium-2-yl]benzenesulfonate
- MTT
3-[4,5-dimethyl-2-thiazol]-2,5-diphenyl-2H-tetrazolium bromide
- NMR
Nuclear magnetic resonance
- PBS
Phosphate buffered saline
- ROS
Reactive oxygen species
- SAH
S-adenosylhomocysteine
- SAM
S-adenosylmethionine
- UDP
Uridine 5′-diphosphate
- UMP
Uridine 5′-monophosphate
Authors’ contributions
KTK conceived of the idea and coordinated the research team. SS first interpreted the metabolomics data and wrote a summary report of this study’s findings. BLR performed cell culture and cytotoxicity experiments. BTC sized the nanomaterial dispersions. XL performed the atomic layer deposition. KTK and SS drafted the paper with input and critical revision from all authors. All authors have read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12989-017-0230-4) contains supplementary material, which is available to authorized users.
Contributor Information
Kirk T. Kitchin, Phone: 919-541-7502, Email: kitchin.kirk@epa.gov
Steve Stirdivant, Email: sstirdivant@metabolon.com.
Brian L. Robinette, Email: robinette.brian@epa.gov
Benjamin T. Castellon, Email: ben.castellon@gmail.com
Xinhua Liang, Email: liangxin@mist.edu.
References
- 1.Warheit DB, Borm PJ, Hennes C, Lademann J. Testing strategies to establish the safety of nanomaterials: conclusions of an ECETOC workshop. Inhal Toxicol. 2007;19(8):631–643. doi: 10.1080/08958370701353080. [DOI] [PubMed] [Google Scholar]
- 2.Thompson TL, Yates JT., Jr Surface science studies of the photoactivation of TiO2--new photochemical processes. Chem Rev. 2006;106(10):4428–4453. doi: 10.1021/cr050172k. [DOI] [PubMed] [Google Scholar]
- 3.Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311(5761):622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- 4.Jeong S-H, Cho W-S, Kim J-E, Cho M-H. Systems toxicological approach to the risk assessment of Nanomaterials. In: Casciano DA, Saura C, editors. Handbook of systems toxicology, vol. 15. SEP 2011 ed. Hoboken: Wiley; 2011. p. 871–89.
- 5.Kitchin K, Grulke E, Robinette BL, Castellon BT. Metabolomic effects in HepG2 cells exposed to four TiO2 and two CeO2 nanomaterials. Environ Sci Nano. 2014;1:466–477. doi: 10.1039/C4EN00096J. [DOI] [Google Scholar]
- 6.Hendren CO, Lowry GV, Unrine JM, Wiesner MR. A functional assay-based strategy for nanomaterial risk forecasting. Sci Total Environ. 2015;536:1029–1037. doi: 10.1016/j.scitotenv.2015.06.100. [DOI] [PubMed] [Google Scholar]
- 7.Celardo I, Traversa E, Ghibelli L. Cerium oxide nanoparticles: a promise for applications in therapy. J Exp Ther Oncol. 2011;9(1):47–51. [PubMed] [Google Scholar]
- 8.Porter D, Shiram K, Wolfarth M, Jefferson A, Schwegler-Berry D, Andrew M, Castranova V. A biocompatible medium for nanoparticle dispersion. Nanotoxicology 2008, 2(3):144-154.
- 9.Welch BL. The generalisation of student’s problems when several different population variances are involved. Biometrika. 1947;34(1–2):28–35. doi: 10.1093/biomet/34.1-2.28. [DOI] [PubMed] [Google Scholar]
- 10.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoll LL, Spector AA. Changes in serum influence the fatty acid composition of established cell lines. In vitro. 1984;20(9):732–738. doi: 10.1007/BF02618879. [DOI] [PubMed] [Google Scholar]
- 12.Neuman MG, Cohen LB, Nanau RM. Biomarkers in nonalcoholic fatty liver disease. Can J Gastroenterol Hepatol. 2014;28(11):607–618. doi: 10.1155/2014/757929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martius G, Alwahsh SM, Rave-Frank M, Hess CF, Christiansen H, Ramadori G, Malik IA. Hepatic fat accumulation and regulation of FAT/CD36: an effect of hepatic irradiation. Int J Clin Exp Pathol. 2014;7(8):5379–5392. [PMC free article] [PubMed] [Google Scholar]
- 14.Miller DB, Karoly ED, Jones JC, Ward WO, Vallanat BD, Andrews DL, Schladweiler MC, Snow SJ, Bass VL, Richards JE, et al. Inhaled ozone (O3)-induces changes in serum metabolomic and liver transcriptomic profiles in rats. Toxicol Appl Pharmacol. 2015;286(2):65–79. doi: 10.1016/j.taap.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller DB, Ghio AJ, Karoly ED, Bell LN, Snow SJ, Madden MC, Soukup J, Cascio WE, Gilmour MI, Kodavanti UP. Ozone exposure increases circulating stress hormones and lipid metabolites in humans. Am J Respir Crit Care Med. 2016;193(12):1382-91. doi:10.1164/rccm.201508-1599OC. PMID: 26745856. [DOI] [PMC free article] [PubMed]
- 16.Senthilkumar R, Viswanathan P, Nalini N. Glycine modulates hepatic lipid accumulation in alcohol-induced liver injury. Pol J Pharmacol. 2003;55(4):603–611. [PubMed] [Google Scholar]
- 17.Przybytkowski E, Behrendt M, Dubois D, Maysinger D. Nanoparticles can induce changes in the intracellular metabolism of lipids without compromising cellular viability. FEBS J. 2009;276(21):6204–6217. doi: 10.1111/j.1742-4658.2009.07324.x. [DOI] [PubMed] [Google Scholar]
- 18.Henriksen BS, Curtis ME, Fillmore N, Cardon BR, Thomson DM, Hancock CR. The effects of chronic AMPK activation on hepatic triglyceride accumulation and glycerol 3-phosphate acyltransferase activity with high fat feeding. Diabetol Metab Syndr. 2013;5:29. doi: 10.1186/1758-5996-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Testa B, Kramer SD. The biochemistry of drug metabolism--an introduction: part 4. Reactions of conjugation and their enzymes. Chem Biodivers. 2008;5(11):2171–2336. doi: 10.1002/cbdv.200890199. [DOI] [PubMed] [Google Scholar]
- 20.Hissin PJ, Hilf R. A fluorometric method for determination of oxidized and reduced glutathione in tissues. Anal Biochem. 1976;74(1):214–226. doi: 10.1016/0003-2697(76)90326-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z, von dem Bussche A, Kabadi PK, Kane AB, Hurt RH. Biological and environmental transformations of copper-based nanomaterials. ACS Nano. 2013;7(10):8715–8727. doi: 10.1021/nn403080y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kehrer JP, Klotz LO. Free radicals and related reactive species as mediators of tissue injury and disease: implications for health. Crit Rev Toxicol. 2015;45(9):765–798. doi: 10.3109/10408444.2015.1074159. [DOI] [PubMed] [Google Scholar]
- 23.Teeguarden JG, Hinderliter PM, Orr G, Thrall BD, Pounds JG. Particokinetics in vitro: dosimetry considerations for in vitro nanoparticle toxicity assessments. Toxicol Sci. 2007;95(2):300–312. doi: 10.1093/toxsci/kfl165. [DOI] [PubMed] [Google Scholar]
- 24.Pal AK, Bello D, Cohen J, Demokritou P. Implications of in vitro dosimetry on toxicological ranking of low aspect ratio engineered nanomaterials. Nanotoxicology. 2015;9(7):871–885. doi: 10.3109/17435390.2014.986670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen JM, DeLoid GM, Demokritou P. A critical review of in vitro dosimetry for engineered nanomaterials. Nanomedicine (Lond) 2015;10(19):3015–3032. doi: 10.2217/nnm.15.129. [DOI] [PubMed] [Google Scholar]
- 26.Geraets L, Oomen AG, Schroeter JD, Coleman VA, Cassee, FR. Tissue distribution of inhaled microand nano-sized cerium oxide particles in rats: results from a 28-day exposure study. Toxicol Sci. 2012;127(2):463–473. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nanomaterial size and & zeta potential from dynamic light scattering (DLS). (DOC 184 kb)
Large excel Metabolon data sheet (heat map) with all results. (XLSX 533 kb)
Characterization of atomic layer deposition coated SiO2 nanoparticles at Missouri University of Science and Technology. (DOC 79 kb)
The number and direction of statistically significant metabolite concentration alterations after nanomaterial treatments. (DOC 109 kb)
Summary of some physical-chemical properties and biological effects of nanomaterial exposures. (DOC 86 kb)
Possible Metabolomic Functional Assays. (DOC 82 kb)
TEM images of SiO2 nanoparticles. (DOC 6894 kb)
Atomic layer deposition. (DOC 74 kb)
Metabolomic sample assessioning, preparation, chromatography, QA/QC, data extraction and compound identification. (DOC 77 kb)
Data Availability Statement
The metabolomic data set and Additional files 1, 2, 3, 4, 5 and 6: Tables S1–S6, Additional file 7: Figure S1, Additional files 8 and 9 are all available at https://intranet.ord.epa.gov/science/sciencehub The identification number is ScID: A-2rbs.


