Abstract
To identify potential reservoirs/vectors of Mycobacterium ulcerans in northern Queensland, Australia, we analyzed environmental samples collected from the Daintree River catchment area, to which Buruli ulcer is endemic, and adjacent coastal lowlands by species-specific PCR. We detected M. ulcerans DNA in soil, mosquitoes, and excreta of bandicoots, which are small terrestrial marsupials.
Keywords: Buruli ulcer, animal reservoir, Mycobacterium ulcerans, bacteria, DNA, insertion sequence element IS2404, tuberculosis and other mycobacteria, mycolactone-producing mycobacteria, bandicoot, excreta, zoonoses, Far North Queensland, Queensland, Australia, Switzerland
Mycobacterium ulcerans infections, which cause the chronic, necrotizing skin disease Buruli ulcer (BU), occur mainly in focalized areas in West Africa but have also been reported in Australia, Asia, and the Americas (1). Much of the pathology of this debilitating disease is caused by mycolactone, a macrolide toxin (2) unique for members of the species M. ulcerans, which are also referred to as mycolactone-producing mycobacteria (3).
Although the definite route of infection with M. ulcerans remains obscure, in Victoria, Australia, small arboreal marsupials (possums) have been implicated as reservoirs of the pathogen, and mosquitoes have been implicated as vectors of the pathogen (4,5). In a second BU-endemic area of Australia, in Far North Queensland (6), a similar animal reservoir has not been identified. In this region, outbreaks of BU occur in waves, separated by several years, and are believed to be associated with environmental changes caused by heavy rainfall.
The major region in Far North Queensland to which BU is endemic is a rim of valleys and lowlands surrounding the Dagmar Range and extending from Daintree and Forest Creek in the northern region to Mossman in the southern region (6). In this area, a BU outbreak with 64 reported cases occurred in 2011 after the exceptionally long and wet rainy season during 2010–2011, which led to flooding of the Daintree River basin (7). An additional 23 cases were identified in 2012, but numbers subsequently reported have been sporadic.
Although proximity to stagnant water bodies and hydromorphologic alterations are well-established risk factors for emergence of M. ulcerans infection foci (8), definite ecologic factors leading to focal emergence of BU in humans have not been determined. The aim of this study was to identify potential reservoirs and vectors of M. ulcerans in the BU-endemic area of Far North Queensland by analyzing environmental samples for the presence of species-specific DNA sequences.
The Study
We collected 102 environmental samples (55 from soil/mud/vegetation, 35 from insects or small insect pools, and 12 from animal excreta) in September 2013 from different locations within the Daintree River basin, in which BU cases were reported during the outbreak in 2011. Global positioning system coordinates were recorded for all sampling locations (Figure 1). Specimens were collected in sterile plastic containers and shipped to the Victorian Infectious Disease Reference Laboratory (Melbourne, Victoria, Australia) for PCR analysis.
Figure 1.
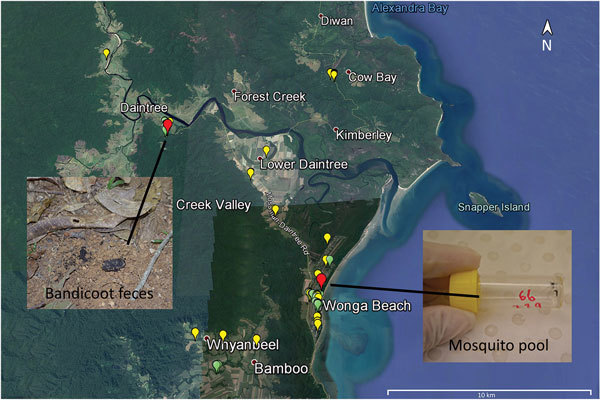
Sample collection for detection of Mycobacterium ulcerans DNA in Buruli ulcer–endemic area, northern Queensland, Australia. Environmental samples were collected in the Daintree River basin during September (yellow) and October (green) 2013. Red indicates locations where Bandicoot feces and mosquito pool samples with positive results by real-time PCR for all 3 M. ulcerans targets were collected. Inset shows specimens from bandicoots and mosquitoes. Map created by using Google Earth (https://www.google.com/earth/).
We extracted DNA by using the FastPrep Instrument (MP Biomedicals, Solon, OH, USA) as described (4). We used the FastDNA Kit (MP Biomedicals) for insect samples and the FastDNA Spin Kit (MP Biomedicals) for soil, mud, vegetation, and feces samples. We analyzed DNA extracts by using semiquantitative real-time PCRs optimized for detection of M. ulcerans in environmental samples (9). We first screened all extracted DNA samples for M. ulcerans insertion sequence element IS2404. Subsequently, we analyzed IS2404-positive samples in a second real-time PCR to detect 2 additional regions in the genome of M. ulcerans: IS2606 and a sequence encoding the ketoreductase B domain of the mycolactone polyketide synthase genes.
Of the 102 samples, 5 (1 soil specimen, 2 bandicoot [Isoodon macrourus] feces samples, 1 sample of an individual mosquito, and 1 pool of 2 mosquitoes) were positive for IS2404. Although 3 of the 5 specimens did not contain sufficient amounts of DNA to identify IS2606 and the ketoreductase B domain of the mycolactone polyketide synthase genes, as indicated by the high cycle threshold (Ct) values for multicopy IS2404 (Table), these markers were detectable in the other 2 samples (bandicoot feces and the pool of 2 mosquitoes) (Figure 1). The IS2404–positive soil sample and the 2 bandicoot feces specimens were collected at the same location (Figure 1), close to a small pond (Figure 2, panel A), and 2 positive mosquito samples were collected at Wonga Beach (Figure 1).
Table. Molecular genetic analysis of environmental samples for Mycobacterium ulcerans DNA in Buruli ulcer–endemic area, northern Queensland, Australia*.
| Sample | Real-time PCR analysis |
|||
|---|---|---|---|---|
| IS2404 | IS2606 | IS2606–IS2404 | KR | |
| Bandicoot feces 1† | 37.3 | ND | ND | ND |
| Bandicoot feces 2† | 27.8 | 29.1 | 1.3 | 28.9 |
| Soil | 36.3 | ND | ND | ND |
| Mosquito | 39.2 | ND | ND | ND |
| Mosquito pool | 31.0 | 38.6 | 7.6 | 31.9 |
| Bandicoot feces‡ | 31.4 | 33.9 | 2.5 | 33.9 |
*Values are mean cycle thresholds of duplicate tests. IS, insertion sequence; KR, ketoreductase B domain of mycolactone polyketide synthase genes; ND, not detected. †Collected in September 2013. ‡Collected in October 2013.
Figure 2.
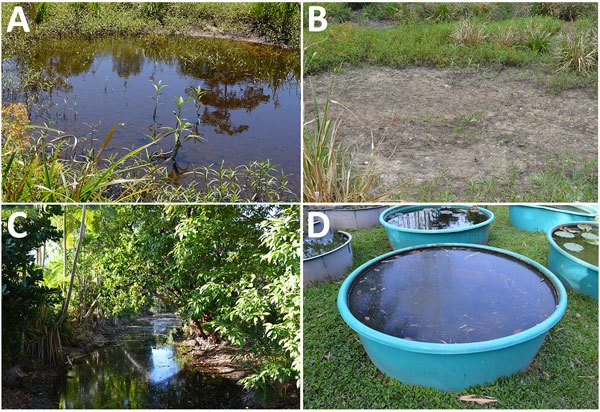
Sample locations for detection of Mycobacterium ulcerans DNA in Buruli ulcer–endemic area, northern Queensland, Australia. A) Pond where IS2404-positive soil sample was collected at first sampling time in September 2013; B) same location dried out at the end of the dry season in October. C, D) Other water bodies suspected to be linked to M. ulcerans infections, such as creeks (C) or water surfaces near houses (D), showed negative results for IS2404. IS, insertion sequence.
As reported (9), analysis of difference in real-time PCR cycle thresholds between IS2606 and IS2404 (ΔCt [IS2606 – IS2404]) enables differentiation between strains of M. ulcerans known to cause BU in humans and other mycolactone-producing mycobacteria strains that contain IS2404 but have fewer copy numbers of IS2606 and are not known to cause disease in endotherms. Thus, DNA in bandicoot feces could be attributed to the M. ulcerans genotype known to cause BU (Table). Conversely, the mosquito pool contained DNA of a closely related M. ulcerans subspecies that had a low copy number for IS2606 (Table), which is usually not associated with disease in endotherms.
The location at which the bandicoot specimen containing M. ulcerans DNA was collected was situated in a nature refuge. In October 2013, we performed a followup environmental study that focused on environmental samples, predominantly animal excreta, collected in this refuge. Of 18 soil/mud/vegetation specimens, 12 insects/insect pools, and 74 animal excreta samples collected, only 1 sample, a bandicoot feces specimen found at almost the same location as the M. ulcerans–positive bandicoot feces specimens collected during the first sampling (Figure 1), was also positive for all M. ulcerans DNA markers tested (Table).
Conclusions
Motivated by increased evidence for the role of M. ulcerans–infected possums in the ecology of human BU in southeastern Australia (4,5), we searched for a similar animal reservoir of M. ulcerans in a BU-endemic area of northern Queensland. Detection of M. ulcerans DNA in bandicoot feces at 2 sampling time points separated by 4 weeks suggests that small mammals might be a potential reservoir for this pathogen. Our data provide a basis for future investigations, which should include a survey of the local animal population for lesions of BU. Similar studies have not identified a mammalian animal reservoir in BU-endemic regions of West Africa (10). Therefore, investigations should be conducted to determine whether at least some marsupial species are more susceptible to M. ulcerans infection than other mammals.
In Far North Queensland, the Daintree River obtains its waters from the mountainous rainforest region northwest of the small town of Mossman and flows into the sea at Cape Tribulation. The wet season is November/December–April, and the dry season is May–October/November. We encountered different environmental conditions at the 2 sampling time points. In September, water bodies, including creeks, small lakes, and swamps, were filled with water (Figure 2, panel A). However, water levels were comparatively low at the end of the dry season in October; some water bodies had even dried up (Figure 2, panel B). Samples from other water bodies, such as creeks (Figure 2, panel C) or biotopes near houses (Figure 2, panel D) in which the presence of M. ulcerans was suspected, all showed negative results for IS2404.
Because outbreaks of BU in Far North Queensland were historically related to heavy rainfalls and flooding, future studies in this region should be performed during or shortly after the rainy season. Although mosquitoes seem to be involved in the ecology of BU in Victoria (5,11,12), the mosquito sample positive for M. ulcerans in this study did not have a bacterial genotype that is known to commonly cause disease in endotherms.
Acknowledgments
We thank Hendrik John Weimar, Barbara Maslen, and Allen Sheather for facilitating collection of environmental samples and Samuel Yaw Aboagye for assisting with testing of collected samples.
Biography
Dr. Röltgen is a scientist at the Swiss Tropical and Public Health Institute, Basel, Switzerland. Her research interests are molecular and seroepidemiologic aspects of M. ulcerans infection and development of point-of-care diagnostics for Buruli ulcer.
Footnotes
Suggested citation for this article: Röltgen K, Pluschke G, Johnson PDR, Fyfe J. Mycobacterium ulcerans DNA in bandicoot excreta in Buruli ulcer–endemic area, northern Queensland, Australia. Emerg Infect Dis. 2017 Dec [date cited]. https://doi.org/10.3201/eid2312.170780
References
- 1.Röltgen K, Pluschke G. Epidemiology and disease burden of Buruli ulcer: a review. Rearch and Reports in Tropical Medicine. 2015;2015:59–73.
- 2.Hong H, Coutanceau E, Leclerc M, Caleechurn L, Leadlay PF, Demangel C. Mycolactone diffuses from Mycobacterium ulcerans-infected tissues and targets mononuclear cells in peripheral blood and lymphoid organs. PLoS Negl Trop Dis. 2008;2:e325. 10.1371/journal.pntd.0000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pidot SJ, Asiedu K, Käser M, Fyfe JA, Stinear TP. Mycobacterium ulcerans and other mycolactone-producing mycobacteria should be considered a single species. PLoS Negl Trop Dis. 2010;4:e663. 10.1371/journal.pntd.0000663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fyfe JA, Lavender CJ, Handasyde KA, Legione AR, O’Brien CR, Stinear TP, et al. A major role for mammals in the ecology of Mycobacterium ulcerans. PLoS Negl Trop Dis. 2010;4:e791. 10.1371/journal.pntd.0000791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson PD, Lavender CJ. Correlation between Buruli ulcer and vector-borne notifiable diseases, Victoria, Australia. Emerg Infect Dis. 2009;15:614–5. 10.3201/eid1504.081162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Steffen CM, Smith M, McBride WJ. Mycobacterium ulcerans infection in North Queensland: the ‘Daintree ulcer’. ANZ J Surg. 2010;80:732–6. 10.1111/j.1445-2197.2010.05338.x [DOI] [PubMed] [Google Scholar]
- 7.Steffen CM, Freeborn H. Mycobacterium ulcerans in the Daintree 2009-2015 and the mini-epidemic of 2011. ANZ J Surg. 2016. 10.1111/ans.13817 [DOI] [PubMed] [Google Scholar]
- 8.Merritt RW, Walker ED, Small PL, Wallace JR, Johnson PD, Benbow ME, et al. Ecology and transmission of Buruli ulcer disease: a systematic review. PLoS Negl Trop Dis. 2010;4:e911. 10.1371/journal.pntd.0000911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fyfe JA, Lavender CJ, Johnson PD, Globan M, Sievers A, Azuolas J, et al. Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl Environ Microbiol. 2007;73:4733–40. 10.1128/AEM.02971-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Röltgen K, Pluschke G. Mycobacterium ulcerans disease (Buruli ulcer): potential reservoirs and vectors. Curr Clin Microbiol Rep. 2015;2:35–43. 10.1007/s40588-015-0013-3 [DOI] [Google Scholar]
- 11.Johnson PD, Azuolas J, Lavender CJ, Wishart E, Stinear TP, Hayman JA, et al. Mycobacterium ulcerans in mosquitoes captured during outbreak of Buruli ulcer, southeastern Australia. Emerg Infect Dis. 2007;13:1653–60. 10.3201/eid1311.061369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lavender CJ, Fyfe JA, Azuolas J, Brown K, Evans RN, Ray LR, et al. Risk of Buruli ulcer and detection of Mycobacterium ulcerans in mosquitoes in southeastern Australia. PLoS Negl Trop Dis. 2011;5:e1305. 10.1371/journal.pntd.0001305 [DOI] [PMC free article] [PubMed] [Google Scholar]


