 Ice-binding proteins (IBP) facilitate survival under extreme conditions in diverse life forms. Successful translation of this natural cryoprotective ability into man-made materials would open up new avenues in biomedicine, agrifood and materials science. This review covers recent advances in the field of IBPs and their synthetic analogues, focusing on fundamental insights of biological and technological relevance.
Ice-binding proteins (IBP) facilitate survival under extreme conditions in diverse life forms. Successful translation of this natural cryoprotective ability into man-made materials would open up new avenues in biomedicine, agrifood and materials science. This review covers recent advances in the field of IBPs and their synthetic analogues, focusing on fundamental insights of biological and technological relevance.
Abstract
Ice-binding proteins (IBP) facilitate survival under extreme conditions in diverse life forms. IBPs in polar fishes block further growth of internalized environmental ice and inhibit ice recrystallization of accumulated internal crystals. Algae use IBPs to structure ice, while ice adhesion is critical for the Antarctic bacterium Marinomonas primoryensis. Successful translation of this natural cryoprotective ability into man-made materials holds great promise but is still in its infancy. This review covers recent advances in the field of ice-binding proteins and their synthetic analogues, highlighting fundamental insights into IBP functioning as a foundation for the knowledge-based development of cheap, bio-inspired mimics through scalable production routes. Recent advances in the utilisation of IBPs and their analogues to e.g. improve cryopreservation, ice-templating strategies, gas hydrate inhibition and other technologies are presented.
1. Introduction
Polar fish produce ice-binding proteins to cope with the extremes of their natural habitat: ice-laden ocean waters with temperatures as low as –1.9 °C.1 Inspired by these cryoprotective biopolymers, chemists set out to develop efficient routes to prepare synthetic analogues in high yield, high purity, and low cost, for scientific research and potential application in e.g. food technology,2,3 agriculture,4,5 fishery,6,7 cryopreservation,8,9 petroleum industry,10–12 coating technology,13,14 and ice-templating.15
This review discusses the recent literature on native ice-binding proteins, including ice-nucleating proteins (INPs),16 as well as analogues thereof, focusing in particular on the outstanding questions of fundamental interest and the development of artificial, non-colligative antifreezes for a broad spectrum of innovative technologies. The umbrella term ice-binding proteins (IBPs) is utilized to denote all proteins that bind ice. Those IBPs that depress the hysteresis freezing point hFP below the hysteresis melting point hMP create a so-called ‘thermal hysteresis gap’ TH = hMP – hFP.17 These are better known as antifreeze proteins (AFPs), antifreeze glycoproteins (AFGPs), or thermal hysteresis proteins (THP). Ice-nucleating proteins (INPs) reduce supercooling and induce freezing. This IBP subclass is thought to organize its surface waters into an ice-like template that favours ice nucleation as temperature decreases.16,18
2. Structure and biological functions of ice-binding proteins
Diverse life forms produce ice-binding proteins for protection against freezing.19,20 Early studies focused on AF(G)Ps isolated from natural sources,1,21,22 mostly fishes, purified by chromatography or (ice-)affinity purification.23 Nowadays most laboratories produce proteins and peptides either by recombinant expression in a suitable host like E. coli24,25 or solid-phase peptide synthesis.26,27 An important advantage of these methods is the possibility to introduce site-specific mutations and engineer fusion proteins.25,28 This facilitates the introduction of fluorescent tags and proteins25 and studies on structure–property relationships, such as the composition of the ice-binding site.29,30 For example, the ice-binding sites of type I and III AFPs have been systematically altered to probe the relative importance of specific amino acids for ice-binding and activity.28,31 A delicate interplay of IBP/interactions seems likely, involving contributions of van der Waals interactions,32 hydrogen bonding,33 hydrophobic forces,24,26,32 and water structuring.34–36
2.1. IBP classification
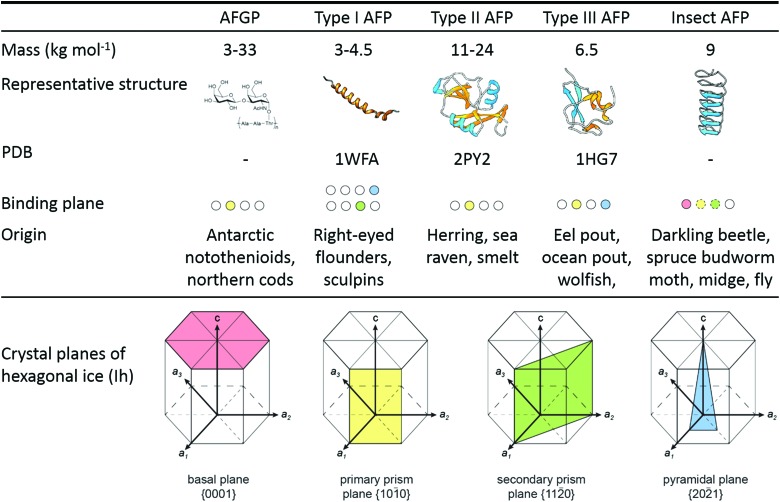
Fishes, like Antarctic notothenioids,1,21 ocean pout,28,32,37 and winter flounder,26,38 express antifreeze glycoproteins (AFGPs)1,21 and type I–III antifreeze proteins (Fig. 1). AFGPs are categorized into 8 classes of descending size from AFGP1 with Mw = 33.7 kDa to AFGP8 with Mw = 2.6 kDa.21,39,40 AFGPs contain 4 to 50 tripeptide repeats of Ala–Ala–Thr (occasionally substituted by Pro–Ala–Thr in AFGPs 6–8) with the disaccharide galactose-N-acetylgalactosamine attached to each Thr. Type I AFPs are 3.3–4.5 kDa alanine-rich α-helices found flounders,27,33 sculpins,24,33 and Alaskan plaice.33 An unusually large and potent type I AFP (‘Maxi’) coordinating over 400 waters was recently discovered in winter flounder.34 Its solution and XRD crystal structure were found to be in good agreement.41 The winter flounder and Alaskan plaice AFPs bind pyramidal planes of ice, the shorthorn sculpin AFP binds secondary prism planes, while Maxi adsorbs to multiple planes including the basal plane (Fig. 1).33,34,42 Type II AFPs from American herring,43 sea raven,44 poacher,45 and smelt46 are globular, cysteine-rich lectin-like proteins of 11–24 kDa that bind nonbasal planes.19,43 Type III AFPs from eel pout,47 ocean pout,28,32,37 and wolffish6 are small, globular proteins of approx. 6–7 kDa without cysteins. Ocean pout type III AFP binds both the primary prism plane and a pyramidal plane of ice.28 Fish type IV AFP found in the longhorn sculpin does not appear to be used as IBP in its host.48
Fig. 1. Antifreeze glycoproteins (AFGPs) from Antarctic Notothenioid fishes and antifreeze proteins (AFPs) from winter flounder (type I wfAFP, PDB: ; 1WFA), herring (type II hAFP, PDB: ; 2PY2), ocean pout (type III opAFP, ; 1HG7), and the beetle Dendroides canadensis (DAFP-1). Secondary structural elements are color-coded as follows: α-helix (cyan), β-sheet (orange), and coil (gray). The ice crystal planes are color-coded: basal plane (pink), primary prism plane (yellow), secondary prism plane (green), and pyramidal plane (blue). Adapted from Oude Vrielink et al., Biointerphases 2016, 11, 018906 and Olijve et al., Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 3740.
IBPs from bacteria,36 insects (Fig. 1),10 diatoms,49 plants,4,50 and fungi51 are often rich in β-solenoids and silk-like solenoids.19 By contrast, the sfAFP from the snow flea contains six antiparallel polyproline type II coils.52 Recently, several xylomannan-based antifreeze glycolipids (AFGLs) were discovered in the darkling beetle Upis ceramboides.53 INPs are found in the hemolymph of insects54 and on bacterial membranes.3 The ice-nucleating protein (INP) from Pseudomonas syringae is modelled as a large β-helical 120–180 kDa protein anchored to the outer membrane with a highly repetitive central domain flanked by non-repetitive domains on either side.55 Other ice nucleation active Gram-negative bacteria are Pseudomonas fluorescens, Erwinia ananas, Erwinia uredovora, Erwinia herbicola, and Xanthomonas campestris.3
2.2. Biological roles of IBPs
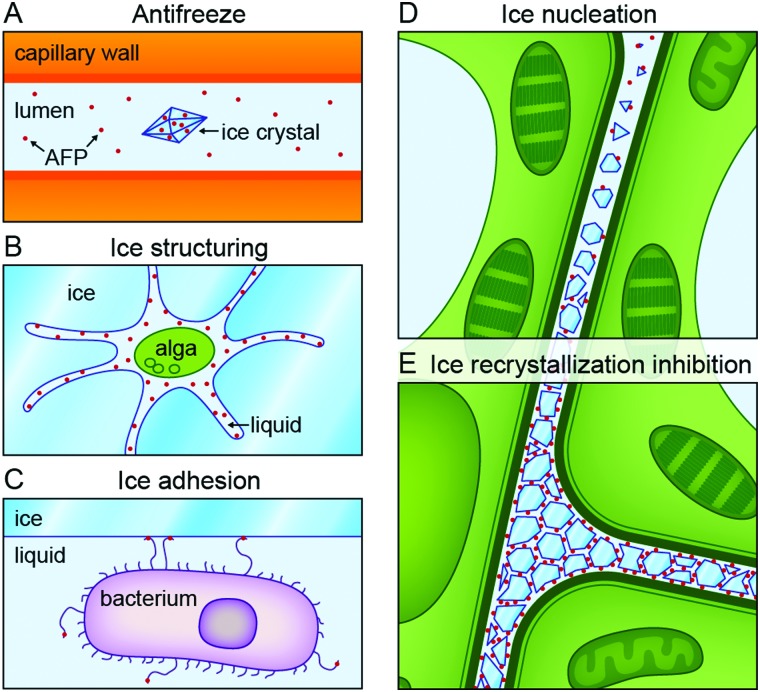
Five distinct functional roles have been attributed to proteins that modulate ice nucleation and growth (Fig. 2).19,20,30 DeVries and others attributed 30% of the serum freezing point depression in Antarctic notothenioids to non-colligative effects arising from circulating AFGPs.1 This ‘thermal hysteresis activity’ of AF(G)Ps (Fig. 2A) is essential for the survival of ‘freeze-avoidant’ polar fish living in shallow waters. Within the TH gap, growth or melting of internalized ice crystals is prevented.17 These accumulate in organs like the spleen, and do not melt during the entire lifespan of the fish.17 Freeze-tolerant species are able to survive freezing. These use IBPs to inhibit ice recrystallization processes (Fig. 2E), during which ice crystals merge (accretive recrystallization),2 change shape (isomass recrystallization),2 or grow at the expense of smaller ones (migratory recrystallization).56 Various microorganisms likely produce IBPs to gain access to nutrient-rich environments. Algae29,57 and diatoms49 live in liquid-like cavities within sea ice created by secreted IBPs (Fig. 2B).49,57 The Antarctic bacterium Marinomonas primoryensis accesses oxygen and nutrients in the upper regions of sea lakes by binding to surface ice through an ice-binding domain at the tip of a MDa-large surface-bound adhesin (Fig. 2C).58,59 There is a growing interest in ice-nucleating proteins (INPs, Fig. 2D) from e.g. Pseudomonas syringae.3,60 INPs induce ice nucleation at undercoolings ranging from –5 to –12 °C in plants.60 This enables pathogens to cause injuries in the epithelia of fruits and plants to gain food access. On the contrary, INPs can increase freeze-tolerance in e.g. bacteria and plants by protective extracellular freezing and utilisation of the released heat of fusion as heat source.60,61 In freeze-avoidant insects, ice-nucleating agents are removed or inactivated by complexation with IBPs.54,60,62
Fig. 2. Biological functions of ice-binding proteins (IBPs). (a) Antifreeze (glyco)proteins in ‘freeze-avoiding’ species lower the freezing point of bodily fluids to halt further growth of internalized ice crystals. (b) Secreted IBPs from microorganisms like algae create a liquid habitat for their host in sea ice. (c) The terminal ice-binding domain of adhesins on the surface of the bacterium Marinomas primoryensis facilitates attachment to ice on Antarctic lakes for access to oxygen and nutrients. (d) Ice nucleation proteins in freeze-tolerant species of e.g. plants locally trigger ice nucleation at high subzero temperatures. (e) Many IBPs inhibit ice recrystallization during which ice crystals grow, merge, or change shape. Reproduced from Oude Vrielink et al., Biointerphases, 2016, 11, 018906.
3. IBP activity
Various assays of hallmark features of IBPs have been developed to evaluate whether a newly discovered protein belongs to the class of IBPs and if so, measure its potency. Similarly, these are used to measure the effectiveness of IBP analogues. Quantitative assays and image analysis routines have been developed to accurately measure thermal hysteresis (TH) activity,63–66 ice recrystallization inhibition (IRI) activity,56,67,68 dynamic ice-shaping (DIS),69 and ice nucleation (IN) activity.70 Furthermore, ice-binding can be detected directly, albeit in a qualitative fashion, by ice-etching33 and fluorescently based ice-plane affinity (FIPA)28 if IBPs are tagged with fluorescent proteins or dyes.33,71
3.1. Antifreeze activity
Antifreeze proteins are a subclass of IBPs that depress the hysteresis freezing point hFP below the hysteresis melting point hMP to create a so-called ‘thermal hysteresis gap’ TH = hMP – hFP.17 This TH gap comprises two hysteresis intervals: a melting hysteresis interval (MH) between hMP and the equilibrium freezing/melting point eqFMP,72,73 and a freezing hysteresis interval (FH) between hFP and eqFMP. Freezing point depression by colligative antifreezes, like ethanol and ethylene glycol, scales linearly with the solution osmolality. Using ΔTf = KF × b × i, with the freezing point depression, ΔTf, the cryoscopic constant, Kf (= 1.853 K kg mol–1 for water), the molality, b, and the van't Hoff factor, i (= 2 for NaCl), we obtain ΔTf ∼ 0.004 °C for 1 mM NaCl in water. AF(G)Ps lower hFP in a non-colligative manner, which is much more effective. For example, TH = 0.6 °C for type I wfAFP and type III rQAE at 5 mg mL–1 and 3 mg mL–1,66 while the colligative effect is not more than ΔTf ∼ 0.002 °C.
Thermal hysteresis (TH) activity is traditionally evaluated by nanolitre cryoscopy (also known as nanolitre osmometry)21,74 under conditions of very slow cooling of single ice crystals.75,76 Innovative microfluidic designs63,64 greatly facilitate sample handling and experimentation compared to traditional osmometers. An alternative method to determine TH activity termed ‘sonocrystallization’ affords high-throughput measurements of both the freezing and melting temperatures of IBP solutions.65,66 It utilizes ultrasound to trigger ice nucleation at a significant undercooling of typically –6 °C corresponding to fast ice growth conditions. As TH activity is concentration dependent,77ζ = (hMP – hFP)/√CAF(G)P has been introduced as a quantitative measure for TH activity.66,78 Maximal TH activities measured by cryoscopy range from 0.1–0.5 °C in apoplastic extracts from overwintering plants4,61 and ∼2 °C for 30–40 g L–1 protein in fish serum21,74 to ∼2 to 13 °C for 10-times lower concentrations in insect hemolymph.54 TH activity can be improved upon addition of (sub)molar concentrations of small molecular solutes79 and ions,20,80 conjugation81 to or complexation82 with potentiating proteins82 or other ice-binding proteins81 and polymers,83,84 and IBS enlargement.85 However, salts like sodium borate can also inactivate AFGPs.74,86
3.2. Ice recrystallization inhibition activity
Ice recrystallization is a thermodynamically driven process, during which the ice grain boundary area per unit volume decreases. As this lowers the free energy of the system, it occurs spontaneously. There are three types of recrystallization processes: isomass, accretive, and migratory recrystallization.2 During isomass recrystallization, ice crystals change shape or internal structure, as irregular grain surfaces are rounded-off and ice crystal defects are reduced. During accretive recrystallization, two or more neighbouring crystals merge into one. During migratory recrystallization, also known as Ostwald ripening, large crystals grow at the expense of small ones with high radii of curvature at constant ice volume fraction, temperature, and pressure. IBPs already inhibit recrystallization at very low, micromolar concentrations.66 The IRI activity can be enhanced further by specific ions87 and hydrocolloids.88
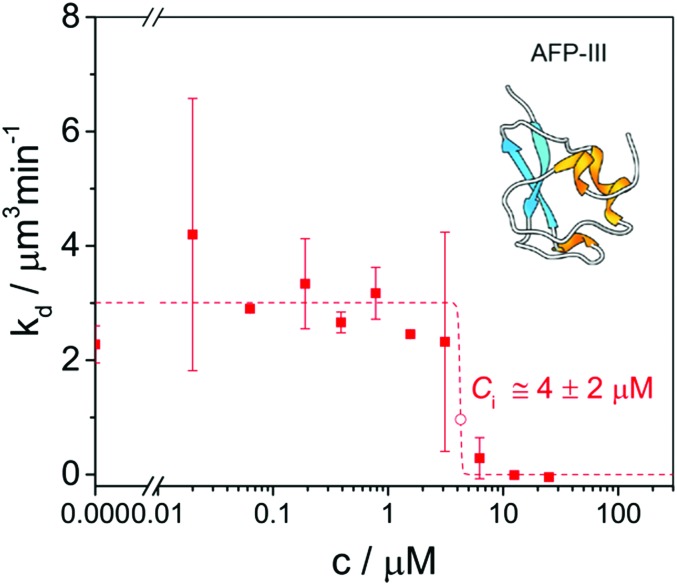
Ice recrystallization inhibition (IRI) activity is determined by polarized optical microscopy (POM) in a so-called ‘splat-assay’89 or ‘sandwich assay’,67 which probe the rate and extent of ice recrystallization in thin wafers of ice. Splat assays are typically performed in the presence of >2 mM NaCl90 or 1–100 mM phosphate-buffered saline (PBS) buffer68 and sandwich assays in the presence of 18–45% sucrose.56,91 In a splat assay quantification of efficacy is based on measurements of the (time-evolution of the) mean (largest) grain size (M(L)GS).92 In a sandwich assay (Fig. 3),93 the inhibitory concentration Ci, is taken as a quantitative measure for IRI activity. It demarcates the boundary between a high recrystallization rate kd at low IBP concentration CIBP and a very low kd at high CIBP.67
Fig. 3. A ‘sandwich assay’ of ice recrystallization inhibition (IRI) activity of type III opAFP in 30% sucrose at –7 °C yields an inhibitory concentration Ci = 4 ± 2 μM. Reproduced (adapted) with permission from Olijve et al., Cryst. Growth Des., 2016, 16. Copyright (2014) American Chemical Society.
A direct comparison between the results of the two IRI assays is unfortunately hampered by differences in sample composition and assay conditions, such as nucleation temperature, annealing temperature, and cooling rate. Care must be taken when identifying antifreezes or inferring ice-binding from ice-structuring (vide infra) and IRI assays. Prism facets may also appear in the melt under mechanical pressure94 and inactive antifreezes also exhibit IRI when too little intercrystalline liquid is present.90 This is because virtually all impurities block ice recrystallization when forced to reside on ice grain boundaries. To exclude false positives, it is thus essential to employ a significant amount of osmolytes in an IRI assay to ensure a sufficiently high liquid volume fraction.90
3.3. Ice plane affinity & ice crystal morphology
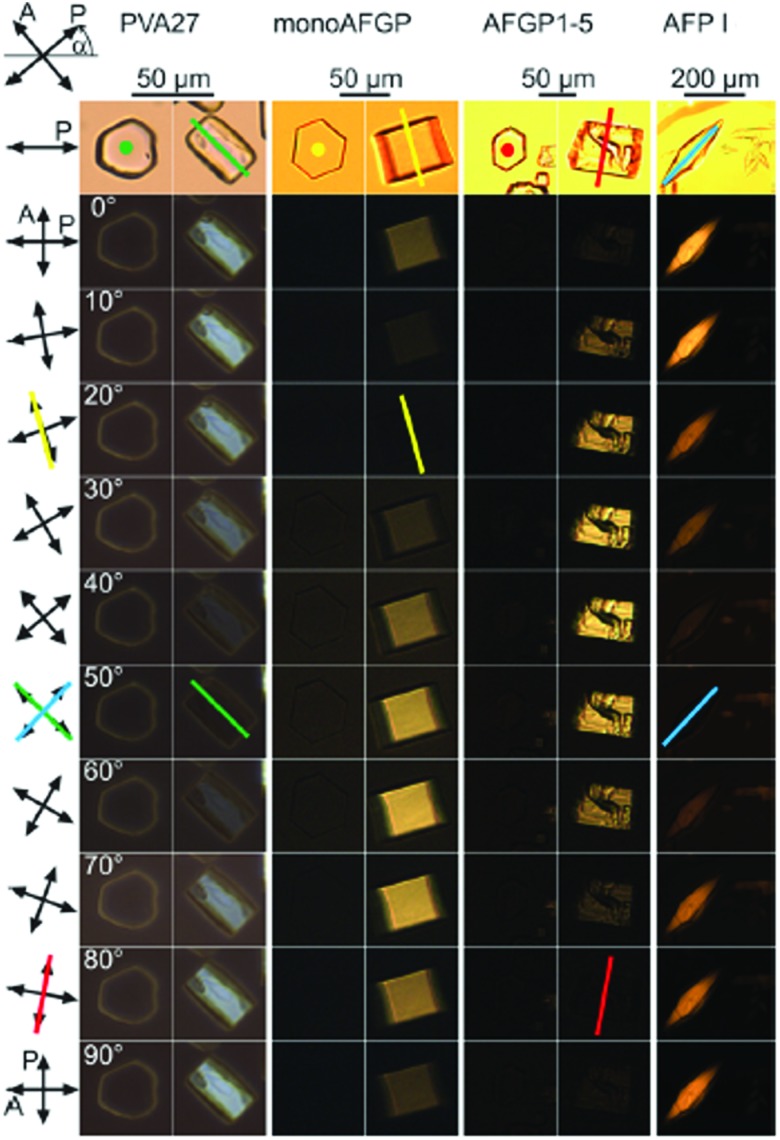
The morphology of ice crystals grown in IBP solutions differs from the flat disks of ice observed in pure water. Dynamic ice shaping (DIS) or ice faceting is typically probed under an optical microscope (Fig. 4)95 or fluorescence microscope69 under conditions of slow ice growth or melting in (microfluidic) chambers utilized for TH assays by cryoscopy. The differential affinity of IBPs for the various crystal planes (prism, basal, pyramidal, see Fig. 1) leads to distinct (bipyramidal) structures within the thermal hysteresis gap when T > hFP (Fig. 4, c-axis normal to the paper plane).95 These undergo burst growth along the c-axis (fish),75 perpendicular to the c-axis (insects),75 or along both the c- and a-axis (plants)19 below the hysteresis freezing point hFP. Although simulations are able to reproduce these growth and burst morphologies,96 the molecular origin of these differences in affinity are not entirely understood. They may well be related to differences in the ice-binding sites (IBS) of AFPs,97 which vary in amino acid composition, structure98 and hydration.34,35 Some IBS display highly ordered arrays of specific amino acids,99 often threonines.20 A recombinant 96-residue polypeptide corresponding to the P. syringae INP segment Tyr176–Gly273 shaped ice crystals into hexagonal-bipyramids and exhibited a dose-dependent thermal hysteresis activity.100
Fig. 4. Ice crystal habits in the presence of poly(vinyl alcohol) PVA27, a monosaccharide bearing synthetic AFGP analogue monoAFGP, natural AFGP1-5, and type I AFP from winter flounder AFP I. Bright field images without the analyser are shown in the top row. Subsequent rows shows ice crystals under crossed polarizers (A, P). The counterclockwise angle α of the polarizers relative to three o'clock is indicated. The colored lines (respectively dots), in the bright field and darkest polarized images indicate the direction of the c-axis (normal to the paper plane). Reproduced (adapted) with permission from Budke et al., Cryst. Growth Des., 2014, 14. Copyright (2014) American Chemical Society.
3.4. Ice nucleation activity
An energy barrier, also called the nucleation barrier, must be overcome to form a small, critical nucleus of ice in an aqueous solution that is supercooled below its equilibrium melting point (which equals its equilibrium freezing point). The metastable liquid state remains until the so-called ‘supercooling point’ or ‘nucleation temperature’ is reached. At this point, sufficiently large ice-like clusters form that grow into ice crystals. Ice-nucleating proteins limit supercooling and trigger ice nucleation at elevated subzero temperatures. This is probably because the ice-binding surface of INPs templates the organization of water molecules into an ice-like lattice which induces nucleation.
While the impact of IBPs on ice crystal growth is well-documented, much less is known about heterogeneous ice nucleation by INPs3,54,60 and AFPs.101 Attempts to recombinantly express and subsequently purify membrane-associated INPs for structure elucidation have failed thus far, since these large membrane proteins have a tendency to misfold and aggregate.60 Furthermore, the quantification of nucleation events is challenging experimentally, due to e.g. their stochastic nature.102 To ensure reasonable statistics, ice nucleation activity is probed in ensembles of droplets prepared by emulsification,3,103 or otherwise,70 by monitoring changes in scattering70,103 or heat effects104 upon ice formation.
3.5. Comparison between TH and IRI activity
A recent study compared TH and IRI activity measured by nanolitre cryoscopy, sonocrystallization, and POM for six different types of AF(G)Ps from different structural classes. No correlation was found between TH and IRI activity expressed in ζ and Ci.66 IBPs with high IRI activity may thus have low TH activity. Furthermore, TH activity determined by cryoscopy and sonocrystallization may differ by as much as 25-fold.66 These variations must be related to differences in the operating conditions of the assays, the ice nucleation and growth characteristics (e.g., single crystal vs. polycrystalline), as well as the adsorption characteristics of the AF(G)Ps, like ice-plane affinity, accumulation rates63 etcetera. Indeed, previous experimental studies already showed a number of parameters including crystal size,47,105 ice volume fraction,106 annealing time,47,107 holding temperature,47 and cooling rate107 impact TH activity studied by cryoscopy. Furthermore, discrepancies were previously observed between TH values determined by different types of cryoscopy like nanoliter osmometry and capillary methods.54,108 Clearly, systematic studies under standardized conditions are required to elucidate precisely how AF(G)Ps work and IBP mimics should be tailored. Furthermore, experimental work should be complemented by computational studies to e.g. gain insight into IBS hydration109,110 and the interplay between various driving forces for ice-binding.111
4. Physical underpinning of IBP activity
4.1. Adsorption-inhibition model
In 1977, Raymond and DeVries introduced an adsorption-inhibition model to explain how AF(G)Ps block ice crystal growth in a non-colligative fashion.112 This step-pinning model proposed that AFPs bind irreversibly onto nascent ice crystals in the path of a growth step. The bound proteins thus force ice to advance between the adsorption sites. Above a critical surface density, this raises the curvature of the growing ice front and thus lowers the freezing point via the Gibbs–Thomson or Kelvin effect.77 Consequentially, a temperature interval known as the thermal hysteresis (TH) gap appears wherein steps are immobile and further ice growth is halted.112 Monte Carlo simulations on 3D ice crystals in the presence of TmAFP and type I wfAFP showed inhibition of ordered step growth.96 The step-pinning mechanism results in a square-root dependence of TH activity on IBP concentration,77 which is in line with numerous experiments.66
The original model was later refined by various others to take into account e.g. the anisotropic surface energy of ice,113 non-stepwise growth on rough prism planes,114,115 and the polymeric nature of IBPs.116 Future revisions are anticipated to discriminate between various inhibition scenarios96 and to achieve quantitative agreement between experiments117,118 and theory.119
4.2. Molecular origin of ice-binding
Research aiming to elucidate the mechanistic details of how IBPs work recently concentrated on two focal points: how do IBPs bind ice and how long do IBPs remain bound? Hydrogen-bonding,33 ice-binding site (IBS) planarity22 (facilitating van der Waals contacts32), structural match with ice-lattice,22 inclusion of methyl functionalities within the ice lattice,32 and ice-like waters34–36 have all been implicated as essential for docking of IBPs to ice. A combination of these factors is likely decisive, with variations in relative importance per IBP (class).
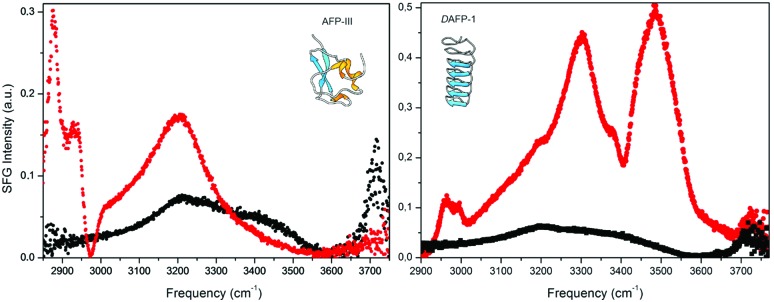
A number of experimental34,35 and computational109,110 studies have revealed clathrate-like or ice-like waters at the surface of IBPs. These are thought to mediate ice adhesion.36 An extensive hydrogen-bond network of bound waters was visible in the X-ray diffraction structure of a large, α-helical type I antifreeze-protein (Maxi),34 not to be mistaken for the smaller α-helical type I antifreeze-protein (wfAFP-1) from the same species. Interestingly, the structure suggests that the active dimer must bind via this network of organized waters, which emanates from the core of the dimer beyond the proteins' surface. The IBS of both dimerized monomers is buried in the interior and seemingly ‘glued’ together by the bound waters. Direct experimental evidence for IBP associated ice-like waters was obtained in a sum-frequency-generation (SFG) experiment performed at room temperature on type III antifreeze proteins from ocean pout (opAFP-3) adsorbed at the air–water interface (Fig. 5, left panel).35 An SFG spectrum reminiscent of ice was observed for wild-type opAFP-3 within a temperature interval ranging from –2.5 °C up to room temperature. This characteristic ice-like signal was absent from spectra of an inactive T18N mutant and other non-ice-binding protein controls like bovine serum albumin (BSA). Interestingly, such ice-like waters were not visible either in the SFG spectra of the Dendroides Canadensis AFP (Fig. 5, right panel).120 This raises the intriguing questions: Which IBPs utilize ice-like surface waters for docking to ice? How does this work precisely? And which IBPs bind ice through a different mechanism? These questions will undoubtedly be subject of future work.
Fig. 5. Vibrational sum-frequency-generation (VSFG) spectra of pure water (right/left panel, black) and two AFP solutions: a 93 μM type III AFP solution at pH 7.8 (left panel, red) and a 20 μM the Dendroides Canadensis AFP (DAFP-1) solution at pH 7.5 (right panel, red) at the air–water interface. (left) The strong and relatively narrow peak at 3.200 cm–1 for type III AFP (red) is attributed to ice-like waters and the spectral features at <3.000 cm–1 to CH vibrations. (right) Contributions belonging to CH-, NH-, and OH-stretching vibrations are visible for DAFP-1 (red) including sharp bands at ∼3300 and ∼3485 cm–1. Reproduced (adapted) from Meister et al., Proc. Natl. Acad. Sci. U. S. A., 2014, 111(50), 17732 and reprinted (adapted) with permission from Meister et al., J. Phys. Chem. Lett., 2015, 1162. Copyright (2015) American Chemical Society.
4.3. Binding characteristics of IBPs
A central premise in the adsorption-inhibition theory is irreversible binding of IBPs to ice. Although experimental evidence for the contrary has been reported,118,121 recent studies by fluorescence microscopy consistently demonstrated quasi-permanent attachment for TmAFP-GFP122,123 and type III AFP-GFP.25,123 On first sight, this seems contradictory to the concentration dependence of thermal hysteresis activity77 and the differences in activity of various types of AF(G)Ps measured under identical operating conditions and concentrations.66 Consequently, irreversible attachment has been a subject of debate. To reconcile the apparently conflicting findings, two-step adsorption mechanisms were invoked,118 involving e.g. a surface-solution equilibrium close to the melting temperature and irreversible attachment at lower temperatures.77 But, experimental evidence of superheating for AFGPs17,72 and AFPs like MpAFP and TmAFP73 contests this explanation. Sander and Tkachenko developed a ‘kinetic pinning theory’,115 which predicted a vanishing growth velocity as a function of the square root of the AFP concentration at a particular temperature. This is in line with the majority of experimental observations. Knight and DeVries also put forth kinetic arguments to rationalize the concentration-dependence and irreversible attachment of non-basal plane binding AFPs.124 The authors proposed that the concentration-dependence is a result of AFPs in solution that bind irreversibly onto newly exposed prism planes. These emanate from growing layers of ice, which appear due to secondary nucleation on the ‘bare’ basal planes. Below a critical size of these ‘sensitive’ planes, which is temperature-dependent, secondary nucleation on the basal plane ceases and crystal growth stops altogether. This argumentation is in agreement with recent fluorescence microscopy experiments demonstrating that AFPs in solution are essential to prevent growth of ice crystals grown from solutions of non-basal plane binding AFPs, while it is not required for insect AFPs that bind to both prism and basal planes.123 This suggests that insect AFPs cover all exposed ice crystal planes in a semi-permanent fashion, as demonstrated previously.25,122 Therefore, the AFP solution could be buffer-exchanged without loss of thermal hysteresis.123 Based on these findings, Braslavsky and co-workers propose that the TH activity of type III AFPs is related to their binding rate and solution concentration, while it is instead the ice surface concentration for TmAFP.64,123
5. Synthetic analogues of ice-binding proteins
An IBP can be discriminated from other proteins in the protein databank by a computational algorithm that recognises ice-binding sites.99 Notwithstanding, it is currently impossible to engineer a synthetic IBP analogue with an artificial, fully functional, IBS. De novo design of IBP mimics with high TH activity thus lies in the distant future. (Nonprotein) IRI active compounds, however, have been made successfully.68 The advantage of synthetic antifreezes is that these may be manufactured at lower cost, in higher yield, with higher purity and long-term stability.
5.1. Small molecular IBP analogues
Inspired by the high efficacy of AFGPs (Fig. 1) as ice recrystallization inhibitors, various groups set out to develop versatile strategies for the production of native glycopeptides125,126 and effective AFGPs analogues.126,127 These include various solution and solid phase approaches, like sequential native chemical ligation125,128 and click chemistry,129 as reviewed by others.39,126,127,130 This greatly facilitated structure–functions studies revealing the importance of various features including the nature of the carbohydrate moieties,131 molar mass,125 and an ordered secondary structure132 for activity.
To increase the stability and synthetic accessibility of AFGP mimics, Ben and co-workers prepared series of C-linked antifreeze glycoprotein analogues. The importance of peptide backbone structure,133 C-linker length between backbone and glycoside,134 and glycoside type131 were assayed. A few IRI-active but TH-defective compounds were discovered, which were further investigated for cryopreservation purposes.135,136 C-linked AFGP analogues with either too short, or too long linkers,134 or with glucose, mannose, or talose instead of galactose were (virtually) IRI inactive.131 This led to the proposal of a link between IRI activity and carbohydrate hydration. Incorporation of the monosaccharide galactose, with the highest hydration index and the lowest partial molar compressibility of the four incorporated monosaccharide pyranoses, yielded analogues with IRI activity, while the others (glucose, mannose, talose) did not.131,137 However, this trend did not hold for other carbohydrate-based small molecule IRI inhibitors.135
The most potent IRI active small molecules reported to date are non-ice-binding amphiphilic small molecule carbohydrate derivatives and para-methoxyphenyl-β-d-glycosides. These reduce the mean ice grain size relative to PBS at millimolar concentrations, namely 22 mM for the non-ionic surfactant n-octyl-β-d-galactopyranoside,138 0.5 mM for the hydrogelator N-octyl-d-gluconamide,138 IC50 = 16.3 mM for p-methoxyphenyl β-d-glucopyranoside,68 and IC50 = 14.8 mM for p-bromophenyl β-d-glucopyranoside.68
5.2. Polymeric IBP analogues
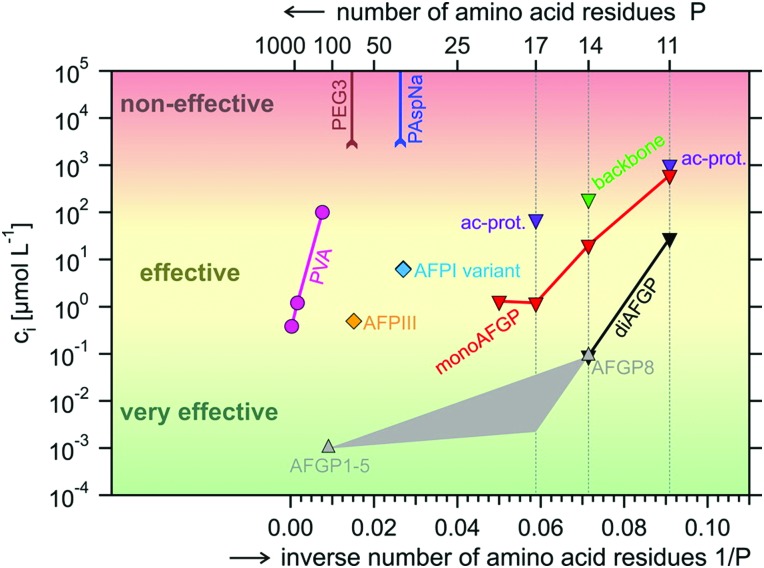
The first synthetic polymer with IRI activity, but without measurable TH activity, was reported in 1995 by Knight and co-workers.90 Since, the TH139 and IRI91,92,140 activity of poly(vinyl alcohol) (PVA) and other polymers has been investigated by several laboratories. In-depth structure–function relations have not yet been established, but it has become clear that chain composition92 (hydroxyl content, co-monomer content) and chain length matter,92,140 while chain architecture appears unimportant.93 Gibson and co-workers showed that PVA becomes IRI active for chain lengths >10 monomers, with higher activities for larger macromolecules,92 as previously observed by Inada et al.140 Similarly, the thermal hysteresis activity of native AFGPs,95 type I AFPs,34 and insect AFPs85 increases with increasing molecular weight. A direct comparison between linear PVA and PVA bottlebrushes (i.e., a graft copolymer with polymeric side-chains tethered to a linear backbone) revealed more or less equal IRI activity,93 while copolymerization with monomers like N-vinyl-pyrrolidone or isopropenyl acetate reduces potency significantly.92 A comprehensive overview of IRI-activity of naturally occurring AF(G)Ps and their mimics (Fig. 6),95 categorizes PVA in the class of effective compounds (10–1 μmol L–1 < Ci < 103 μmol L–1). It is surpassed by the very effective AFGPs (Ci < 10–1 μmol L–1), but more potent than non-effective compounds (Ci > 103 μmol L–1) like poly(ethylene glycol) (PEG), poly(aspartic acid) (PAspNa), and other water-soluble polymers (including glycopolymers). These have weaker or no IRI activity.141
Fig. 6. Overview of the potency of natural and man-made inhibitors of ice recrystallization: natural type III AFPs (AFPIII), natural AFGPs (AFGP1-5, AFGP8), synthetic monosaccharide and disaccharide analogues of AFGPs (monoAFPG, diAFGP), an acetyl-protected monosaccharide peptide (ac-prot), polyvinyl alcohol (PVA), poly(ethylene glycol) (PEG), poly(aspartic acid) (PAspNa). Indicated is the IRI concentration Ci as a function of the inverse number of amino acids (respectively weight-average degree of polymerization) P–1 for (very) effective inhibitors and the maximum concentration tested without signs of activity CLL for non-effective compounds. In grey the anticipated Ci for P–1 intermediate between the values for AFGP1–5 and AFGP8 (no data are available). Reprinted with permission from Budke et al., Cryst. Growth Des., 2014, 14. Copyright (2014) American Chemical Society.
5.3. Other analogues
De novo polypeptides142,143 and peptoids144 with thermal hysteresis and IRI activity have been synthesized, but with lower activity than naturally occurring AFGPs. Laursen and co-workers published two TH active 34-mers with KAAK-motifs.142 Wierzbicki and co-workers reported a 43 residue alanine–lysine-rich peptide with a 32-times lower activity than the shorthorn sculpin (per weight basis).143 Rahaman et al. reported the total chemical synthesis of 4- to 8-mer saccharide subunits of the xylomannan motif of recently discovered non-protein antifreeze glycolipids (AFGLs).145
Davies and co-workers enhanced the activity of TmAFP by extension of its ice-binding site with additional coils.85 An optimum coil length was observed for the nine-coil construct with a maximal TH value of 6.5 °C at 0.7 mg mL–1. Lower activities were measured for the 10- and 11-coil constructs as these presumably have a larger lattice mismatch between their IBS and ice.
The inexpensive metallic salts zirconium(iv) acetate (ZRA) and zirconium(iv) acetate hydroxide (ZRHA) were reported to shape ice and display IRI activity, supposedly through a direct interaction with the ice–water interface.146,147 Similarly, Wang and co-workers reported IRI activity for graphene oxide (GO) due to a direct interaction between GO and ice, which was observed in molecular dynamics simulations.148 Interestingly, Drori and co-workers reported ice shaping into bipyramidal needles and inhibition of ice recrystallization by the synthetic dye Safranine O at millimolar concentrations.149
6. Applications
Control over the nucleation and growth of ice crystals is crucial for the survival of various species,16 atmospheric glaciation processes,18 and decisive for the structural integrity and properties of a broad range of water-based materials.15 This spurred interest in the large-scale production of fully synthetic non-colligative antifreezes for scientific and technological purposes. To this end, current hurdles to (commercial) application of IBPs, such as low yield, high cost, and consumer concerns must be overcome.150 The following sections highlight a number of application areas wherein IBPs and their IRI-active analogues may prove useful.
6.1. Food technology and agriculture
IBPs are attractive candidates for application in food technology and agriculture to e.g. boost cold hardiness,4,5 improve food quality,2,3 and enhance sensorial characteristics.151 In this field of research and technology, AF(G)Ps are commonly referred to as ice-structuring proteins (ISPs), to avoid negative connotations with toxic colligative antifreezes like ethylene glycol.152
Inhibition of ice recrystallization by IBPs used as foodstuff additives may enhance frozen food shelf-life2 and preserve smooth textures in ice cream.151 IBPs may prevent the formation of large ice crystals, which induce morphological and mechanical changes in ice cream and cellular damage in fruits. For example, type III AFP preserved the gel-forming functionality of food muscle proteins,153 AFGP reduced drip loss after thawing in lamb meat,154 and frozen dough treated with a recombinant type I AFP mimic showed better fermentation capacity.155 Inhibition of supercooling by INPs during freeze drying of foods may reduce refrigeration cost and shorten freezing times.156 Summarizing, IBPs that arrest ice recrystallization during production, frozen storage, transport, and thawing of complex food colloids may improve their sensory perception and raise consumer acceptance.2 Notwithstanding, commercial application has so far remained limited to ice cream.157
Several transgenic species of fishes,7 flies158 and crops4 have been generated aiming to promote cold hardiness and frost resistance in e.g. salmon,7Drosophila melanogaster,158 spring wheat,159 and mice.160,161 Thus far however, the achieved freezing protection of a few degrees in transgenic plants is insufficient. Improvements are foreseen if high, targeted expression levels of potent AFPs in model plants (like Arabidopsis) and crops can be realized.5 Also, concerns have been raised about its environmental and sanitary impact.162,163 The US Food and Drug Administration (FDA) recently approved for the first time marketing of a genetically modified animal for food (AquAdvantage Salmon).157
6.2. Biomedical applications
Cryopreservation and hypothermic storage of cells,9 tissues,164 embryos165 and organs164 has long been regarded as one of the most promising application areas of IBPs and their analogues. But, progress is much slower than aspired.9 In part, this is because the mechanisms of freezing damage are not fully understood, let alone how these are affected by cell type, tissue composition, and the presence of cryoprotectants (CPA). Moreover, cryopreservation protocols are widely divergent (e.g., vitrification, slow-freezing) and adapted to a broad spectrum of cell types, which hampers a systematic comparison of results to elucidate the causes underlying success or failure of IBPs to improve preservation strategies.
Cellular damage during cryopreservation is a complex multifactorial process caused by e.g. intracellular ice formation,165,166 mechanical stress,164 freeze-dehydration,164 and ice recrystallization.166,167 These lead to cell death through cell rupture,168 necrosis169 and apoptosis.158,170 Commonly employed cell penetrating CPA, like DMSO and glycerol, are effective but not without disadvantages: cell damage is not fully prevented, while the CPA may be toxic and must be removed after thawing prior to e.g. transplantation.164 This complication inspired widespread research into novel (non-)penetrating CPA like IBPs9,165 and their analogues170 to minimize ice formation,169 to reduce the risk of ice nucleation,171 to prevent ice-induced damage to cell membranes and tissue matrices,172 to limit ice recrystallization,173 and to promote vitrification.171 However, their impact on hypothermic storage and cryopreservation has been discordant.9,170
Biological antifreezes had a negligible effect on the hypothermic storage of spinach thylakoids,168 and an adverse effect on the cryopreservation of bovine oocytes,174 equine embryos,175 and rat cardiac explant.176 For example, AFP type III did not preserve meiotic spindle organization of vitrified in vitro matured bovine oocytes.174 The deleterious effect of AFGPs on cryopreservation of rat cardiomyocytes was attributed to intracellular ice formation and cell lysis due to direct contact between the cells and needle-like ice spicules.177 In fact, type I AFPs may hold promise as chemical adjuvants in cryosurgery.178,179 By contrast, biological antifreezes protected (cell and plasma) membrane integrity and increased cell viability of bovine embryos172 and human hepatoma (HepG2) cells180 during hypothermic storage. Furthermore, IBPs improved the morphological integrity and post-thaw viability of cryopreserved probiotics,181 mouse oocytes,182 sea bream spermatozoa,183 red blood cells,184 mouse ovarian tissue,185 and rat hearts for transplantation.186 The addition of 100 μg mL–1ocean pout type III AFP to semen extender (used to preserve the fertilizing ability of semen) improved the post-thaw motility of spermatozoa chilled at 0–5 °C.187
The observed variations in IBP performance on cryopreservation are due to differences in IBP activity,9 in cell and tissue types,164 in preservation conditions,169 and in freeze–thaw protocols, including CPA concentration as e.g. efficacy and toxicity are dose-dependent.165 As a consequence, consistently successful cryopreservation, especially of whole organs, remains a formidable challenge.164 Future studies should explore a combinatorial approach involving several types of CPAs, exerting both colligative and non-colligative protection, to address the multiplicity of causes of freeze injuries.9
The development of novel IBP analogues for cryopreservation has focused on the (scalable) synthesis of cryopreservants that are non-toxic, TH-inactive, and IRI-active. Various polymers, like poly(vinyl alcohol) PVA and carboxylated ε-poly-l-lysine (PLL), have been prepared and studied as non-penetrating CPAs. Matsumura and co-workers reported higher cryopreservation efficiency and lower cytotoxicity in the cryopreservation of murine L929 cells and rat bone marrow mesenchymal stem cells (RMSCs) for PLL compared to DMSO.188 Cell viability after thawing was found dependent on both PLL dose and the carbonyl mole fraction in the polyampholyte. PVA may assist cryopreservation by vitrification, since small amounts of PVA decreased the glycerol and DMSO concentrations required for vitrification of their solutions.171 Supplementing a 215 mg mL–1 hydroxyethyl starch (HES) solution with 1 mg mL–1 of the biocompatible non-penetrating polymer PVA (9 kDa), increased human red blood cell (HRBC) recovery by 40% upon cryopreservation under rapid cooling.189 Ben and co-workers investigated the potency of several small molecule IRI inhibitors including carbohydrates as cell-penetrating CPA.135,166,173 One of the most potent C-linked AFGP analogues developed by Ben and co-workers was rapidly internalized in human embryonic liver cells. It showed no or little in vitro cytotoxicity and apoptotic death inhibition. Moreover, cell viability increased two-fold relative to cell medium (custodiol histidine–tryptophane–ketoglutarate solution), which was on par with 2.5% DMSO.190,191 Addition of the small molecule IRI inhibitors p-methoxyphenyl (110 mM) and β-d-glucopyranoside (30 mM) increased HRBC post-thaw integrity by 30–50% upon cryopreservation using slow cooling rates at 15% glycerol concentrations.135 In this manner, the glycerol concentration required for effective cryopreservation may be lowered to reduce deglycerolization times, which is important in life-threatening situations warranting fast access for transfusions. Wang and co-workers reported an increase in cryopreserved horse sperm motility from 24.3% to 71.3% upon addition of 0.1 mg mL–1 graphene oxide, while addition of 3.5% glycerol (optimal amount) realized a motility of approx. 55%.148 The hydrogelator N-octyl-d-gluconamide increased the infectivity and thermostability of candidate viral vectors for cancer vaccine development from vaccinia virus (VV) and herpes simplex virus type 1 (HSV-1), and furthermore improves the recovery of cryopreserved vesicular stomatitis virus (VSV).136
Biomedical application of AF(G)Ps in regenerative medicine and in therapeutic strategies against bacterial infections have been proposed recently. Suitable scaffolds for tissue engineering may be manufactured from material precursors supplemented with IBPs.15,192 Heisig et al. reported that a tick AFGP acts as an antivirulence factor preventing the formation of biofilms of several pathogens including Staphylococcus aureus both in vitro and in vivo.193
6.3. Other applications
The interest in IBPs and their analogues for application areas beyond food technology, agriculture, and biomedicine is steadily growing. Rather than providing an extensive overview, specific examples are presented to illustrate future prospects for e.g. materials science,15 petroleum industry,10–12 and climate control.194,195
Ice-templating is an interesting means to tailor the morphology of a broad range of materials including macroporous ceramics,192 polymers, and complex composites15 to produce catalysts, adsorbents, tissue engineering scaffolds, delivery systems, and strong yet light-weight materials, as recently reviewed by Deville.15 For example, porous carrier materials for nutrient and drug delivery, gastronomic applications, and scaffolds for tissue engineering can be manufactured using controlled unidirectional freezing (and subsequent freeze drying).196 As IBPs modulate ice crystal growth rates and habits, these may affect the size (distribution), shape and connectivity of the pores in the matrix.
As IBPs also impact the growth197 and morphology of crystals other than ice,198 they may prove useful as small molecular crystal growth modifiers.199 For example, gas hydrate inhibition by IBPs and their analogues has also become an area of considerable activity.10–12
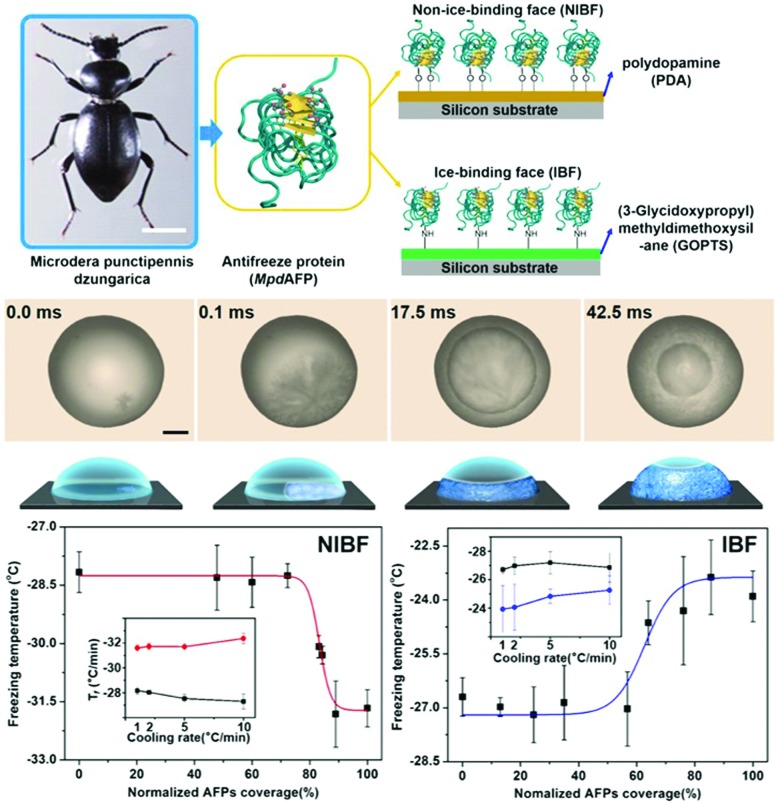
Several studies on anti-icing and de-icing report that AFP surface-tethering may hinder icing and reduce ice adhesion to solid substrates.14,200 But, the opposite effect has been reported for aluminium surfaces functionalized with fish type III AFPs.201 A combined experimental and computational study on three AFPs (an insect AFP from the beetle Microdera punctipennis dzungarica) (MpdAFP), a bacterial AFP from Marinomonas primoryensis (MpAFP), and a type III fish AFP sheds light.13 It demonstrates that the orientation of MpdAFP on the surface is important (Fig. 7). Ice nucleation was promoted when the ice binding site of MpdAFP was exposed to liquid water (IBF). By contrast, ice nucleation was disfavoured in the opposite orientation with the non-ice binding face (NIBF) exposed to the solvent. This is in line with earlier work by Nutt and co-workers who proposed a dual function for the solvation waters of AFPs.110 These promote on the one hand recognition and binding of the ice binding site to ice. On the other hand, engulfment is prevented by perturbing waters in the solvation shell around the remainder of the protein.
Fig. 7. (top) Schematic representation of the tethering of insect AFPs from Microdera punctipennis dzungarica (MpdAFPs) onto polydopamine (PDA) films and (3-glycidoxypropyl) methyldimethoxysilane (GOPTS) coated surfaces exposing respectively the non-ice-binding face (NIBF) and ice-binding face (IBF) of MpdAFPs. (middle) Droplet freezing is recorded by a high-speed camera, which reveals (bottom) MpdAFP coverage dependent suppression and facilitation of ice nucleation on the NIBF and IBF surfaces, respectively. Reproduced (adapted) from Liu et al., Proc. Natl. Acad. Sci. U. S. A., 2016, DOI: 10.1073/pnas.1614379114.
Ice-nucleating agents including INPs receive widespread attention as factors that influence atmospheric glaciation processes and thus impact precipitation, cloud formation, and climate.194,195 INPs have been exploited for artificial snow production; for example, the commercial product Snomax® is based on freeze-dried nonviable cells of Pseudomonas syringae.3 Ice-nucleating proteins have also been utilized as a potent anchoring motif for cell surface display-applications including protein engineering, combinatorial library screening, vaccine development, biocatalysis,202 and bioremediation (see ref. 203 and 204 and refs therein).
7. Concluding remarks
Unveiling how Nature protects against freezing is a grand scientific challenge culminating in the development of powerful synthetic antifreezes for agrifood, materials science, coatings and biomedical applications. Great insights have been obtained in the structure and functioning of IBPs and their analogues, but, many open questions remain. To name but a few: what is the functional role of the various (inactive) isoforms of IBPs? Why are the most active isoforms sometimes least abundant?76,205 What is the link between TH and IRI activity?66,88,206 What is the relation between surface coverage117 and activity? What causes the observed functional differences between IBPs? What is the relation between IBP structure, hydration and interaction with ice? How to design an AFGP analogue with higher IRI activity than its natural counterpart? Furthermore, the commercial potential of IBPs has remained largely unrealized despite great promise. Future experimental and computational studies should address these fundamental and technological challenges in a systematic manner. This will shed light on the relation between IBP (analogue) type, activity, and ice binding characteristics, which is essential for the knowledge-based development of potent bio-inspired mimics that can be manufactured in high yield at low cost for scientific purposes and successful commercialization.
Acknowledgments
This work was financially supported by the European Union (ERC-2014-StG Contract No. 635928), the Dutch Science Foundation (NWO ECHO Grant No. 712.016.002), and the Dutch Ministry of Education, Culture and Science (Gravity Program 024.001.035).
Biography

I. K. Voets
Dr Ilja K. Voets studied Molecular Sciences at Wageningen University in Wageningen, the Netherlands. She performed her PhD research on complex coacervate core micelles assembled from oppositely charged copolymers at the same university, after which she moved to the Adolphe Merkle Institute in Fribourg, Switzerland for a postdoctoral stay. In 2011 she was appointed as Assistant Professor at the Department of Chemical Engineering and Chemistry and the Institute for Complex Molecular Systems of Eindhoven University of Technology (TU/e). Currently she is Associate Professor in the Laboratory of Macromolecular and Organic chemistry and the Laboratory of Physical Chemistry of the TU/e, where she leads an independent research group focusing on self-organized and bioinspired soft matter.
References
- DeVries A. L., Wohlschlag D. E. Science. 1969;163:1073–1075. doi: 10.1126/science.163.3871.1073. [DOI] [PubMed] [Google Scholar]
- Hassas-Roudsari M., Goff H. D. Food Res. Int. 2012;46:425–436. [Google Scholar]
- Cochet N., Widehem P. Appl. Microbiol. Biotechnol. 2000;54:153–161. doi: 10.1007/s002530000377. [DOI] [PubMed] [Google Scholar]
- Gupta R., Deswal R. J. Biosci. 2014;39:931–944. doi: 10.1007/s12038-014-9468-2. [DOI] [PubMed] [Google Scholar]
- Duman J. G., Wisniewski M. J. Environ. Exp. Bot. 2014;106:60–69. [Google Scholar]
- Desjardins M., Le Francois N. R., Fletcher G. L., Blier P. U. Aquaculture. 2007;272:667–674. [Google Scholar]
- Zbikowska H. M. Transgenic Res. 2003;12:379–389. doi: 10.1023/a:1024267416522. [DOI] [PubMed] [Google Scholar]
- Kim H. J. Mar. Drugs. 2017;15:27. doi: 10.3390/md15120372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockbank K. G. M., Campbell L. H., Greene E. D., Brockbank M. C. G., Duman J. G. In Vitro Cell. Dev. Biol.: Anim. 2011;47:210–217. doi: 10.1007/s11626-010-9383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfeldt C. M. Energy Fuels. 2014;28:3666–3672. [Google Scholar]
- Zeng H., Wilson L. D., Walker V. K., Ripmeester J. A. J. Am. Chem. Soc. 2006;128:2844–2850. doi: 10.1021/ja0548182. [DOI] [PubMed] [Google Scholar]
- Tonelli D., Capicciotti C. J., Doshi M., Ben R. N. RSC Adv. 2015;5:21728–21732. [Google Scholar]
- Liu K. Proc. Natl. Acad. Sci. U. S. A. 2016;113:14739–14744. doi: 10.1073/pnas.1614379114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser-Kahn A. P., Trang V., Francis M. B. J. Am. Chem. Soc. 2010;132:13264–13269. doi: 10.1021/ja103038p. [DOI] [PubMed] [Google Scholar]
- Deville S. J. Mater. Res. 2013;28:2202–2219. [Google Scholar]
- Duman J. G. J. Exp. Biol. 2015;218:1846–1855. doi: 10.1242/jeb.116905. [DOI] [PubMed] [Google Scholar]
- Cziko P. A., DeVries A. L., Evans C. W., Cheng C.-H. C. Proc. Natl. Acad. Sci. U. S. A. 2014;111:14583–14588. doi: 10.1073/pnas.1410256111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R. Sci. Adv. 2016;2:e1501630. doi: 10.1126/sciadv.1501630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar Dolev M., Braslavsky I. and Davies P. L., in Annual Review of Biochemistry, Vol 85, Annual Review of Biochemistry, ed. R. D. Kornberg, 2016, pp. 515–542. [DOI] [PubMed] [Google Scholar]
- Oude Vrielink A. S., Aloi A., Olijve L. L. C., Voets I. K. Biointerphases. 2016;11:018906. doi: 10.1116/1.4939462. [DOI] [PubMed] [Google Scholar]
- DeVries A. L., Komatsu S. K., Feeney R. E. J. Biol. Chem. 1970;245:2901–2908. [PubMed] [Google Scholar]
- Yang D. S. C. Biophys. J. 1998;74:2142–2151. doi: 10.1016/S0006-3495(98)77923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu K., Graham L. A., Campbell R. L., Davies P. L. Proc. Natl. Acad. Sci. U. S. A. 2015;112:737–742. doi: 10.1073/pnas.1422272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairley K. J. Biol. Chem. 2002;277:24073–24080. doi: 10.1074/jbc.M200307200. [DOI] [PubMed] [Google Scholar]
- Pertaya N. Biophys. J. 2007;92:3663–3673. doi: 10.1529/biophysj.106.096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haymet A. D. J., Ward L. G., Harding M. M. J. Am. Chem. Soc. 1999;121:941–948. [Google Scholar]
- Lotze S. J. Chem. Phys. 2015;143:201101. doi: 10.1063/1.4936403. [DOI] [PubMed] [Google Scholar]
- Garnham C. P. Biochemistry. 2010;49:9063–9071. doi: 10.1021/bi100516e. [DOI] [PubMed] [Google Scholar]
- Gwak Y. FASEB J. 2014;28:4924–4935. doi: 10.1096/fj.14-256388. [DOI] [PubMed] [Google Scholar]
- Davies P. L. Trends Biochem. Sci. 2014;39:548–555. doi: 10.1016/j.tibs.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Baardsnes J. FEBS Lett. 1999;463:87–91. doi: 10.1016/s0014-5793(99)01588-4. [DOI] [PubMed] [Google Scholar]
- Howard E. I. J. Mol. Recognit. 2011;24:724–732. doi: 10.1002/jmr.1130. [DOI] [PubMed] [Google Scholar]
- Knight C. A., Cheng C. C., DeVries A. L. Biophys. J. 1991;59:409–418. doi: 10.1016/S0006-3495(91)82234-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T., Lin F.-H., Campbell R. L., Allingham J. S., Davies P. L. Science. 2014;343:795–798. doi: 10.1126/science.1247407. [DOI] [PubMed] [Google Scholar]
- Meister K. Proc. Natl. Acad. Sci. U. S. A. 2014;111:17732–17736. doi: 10.1073/pnas.1414188111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham C. P., Campbell R. L., Davies P. L. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7363. doi: 10.1073/pnas.1100429108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotze S., Olijve L. L. C., Voets I. K., Bakker H. J. J. Phys. Chem. B. 2014;118:8962–8971. doi: 10.1021/jp503481e. [DOI] [PubMed] [Google Scholar]
- Gronwald W. Biochemistry. 1996;35:16698–16704. doi: 10.1021/bi961934w. [DOI] [PubMed] [Google Scholar]
- Harding M. M., Anderberg P. I., Haymet A. D. J. Eur. J. Biochem. 2003;270:1381–1392. doi: 10.1046/j.1432-1033.2003.03488.x. [DOI] [PubMed] [Google Scholar]
- Lin Y., Devries A. L., Duman J. G. Biochem. Biophys. Res. Commun. 1972;46:87–92. doi: 10.1016/0006-291x(72)90633-x. [DOI] [PubMed] [Google Scholar]
- Olijve L. L. C. RSC Adv. 2013;3:5903–5908. [Google Scholar]
- Scott G. K., Davies P. L., Shears M. A., Fletcher G. L. Eur. J. Biochem. 1987;168:629–633. doi: 10.1111/j.1432-1033.1987.tb13462.x. [DOI] [PubMed] [Google Scholar]
- Liu Y. PLoS One. 2007;2:e548. doi: 10.1371/journal.pone.0000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronwald W. Biochemistry. 1998;37:4712–4721. doi: 10.1021/bi972788c. [DOI] [PubMed] [Google Scholar]
- Nishimiya Y. J. Mol. Biol. 2008;382:734–746. doi: 10.1016/j.jmb.2008.07.042. [DOI] [PubMed] [Google Scholar]
- Graham L. A., Li J. Y., Davidson W. S., Davies P. L. BMC Evol. Biol. 2012;12:1–13. doi: 10.1186/1471-2148-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamichi M., Nishimiya Y., Miura A., Tsuda S. FEBS J. 2007;274:6469–6476. doi: 10.1111/j.1742-4658.2007.06164.x. [DOI] [PubMed] [Google Scholar]
- Gauthier S. Y. Cryobiology. 2008;57:292–296. doi: 10.1016/j.cryobiol.2008.10.122. [DOI] [PubMed] [Google Scholar]
- Bayer-Giraldi M., Weikusat I., Besir H., Dieckmann G. Cryobiology. 2011;63:210–219. doi: 10.1016/j.cryobiol.2011.08.006. [DOI] [PubMed] [Google Scholar]
- Smallwood M. Biochem. J. 1999;340:385–391. [PMC free article] [PubMed] [Google Scholar]
- Kondo H. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9360–9365. doi: 10.1073/pnas.1121607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentelute B. L. J. Am. Chem. Soc. 2008;130:9695–9701. doi: 10.1021/ja8013538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters K. R., Serianni A. S., Sformo T., Barnes B. M., Duman J. G. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20210–20215. doi: 10.1073/pnas.0909872106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman J. G. Annu. Rev. Physiol. 2001;63:327–357. doi: 10.1146/annurev.physiol.63.1.327. [DOI] [PubMed] [Google Scholar]
- Graether S. P., Jia Z. C. Biophys. J. 2001;80:1169–1173. doi: 10.1016/S0006-3495(01)76093-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olijve L. L. C., Oude Vrielink A. S., Voets I. K. Cryst. Growth Des. 2016;16:4190–4195. [Google Scholar]
- Raymond J. A. Proc. Natl. Acad. Sci. U. S. A. 2011;108:E198–E198. doi: 10.1073/pnas.1106288108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance T. D. R. Biosci. Rep. 2014;34:357–368. doi: 10.1042/BSR20140083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar Dolev M., Bernheim R., Guo S., Davies P. L., Braslavsky I. J. R. Soc., Interface. 2016;13:20160210. doi: 10.1098/rsif.2016.0210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariassen K. E., Kristiansen E. Cryobiology. 2000;41:257–279. doi: 10.1006/cryo.2000.2289. [DOI] [PubMed] [Google Scholar]
- Griffith M., Yaish M. W. F. Trends Plant Sci. 2004;9:399–405. doi: 10.1016/j.tplants.2004.06.007. [DOI] [PubMed] [Google Scholar]
- Duman J. J. Comp. Physiol., B. 2002;172:163–168. doi: 10.1007/s00360-001-0239-7. [DOI] [PubMed] [Google Scholar]
- Drori R., Celik Y., Davies P. L., Braslavsky I. J. R. Soc., Interface. 2014;11:1–10. doi: 10.1098/rsif.2014.0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drori R., Davies P. L., Braslavsky I. RSC Adv. 2015;5:7848–7853. [Google Scholar]
- Gaede-Koehler A., Kreider A., Canfield P., Kleemeier M., Grunwald I. Anal. Chem. 2012;84:10229–10235. doi: 10.1021/ac301946w. [DOI] [PubMed] [Google Scholar]
- Olijve L. L. C. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3740–3745. doi: 10.1073/pnas.1524109113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke C., Heggemann C., Koch M., Sewald N., Koop T. J. Phys. Chem. B. 2009;113:2865–2873. doi: 10.1021/jp805726e. [DOI] [PubMed] [Google Scholar]
- Abraham S. Cryst. Growth Des. 2015;15:5034–5039. [Google Scholar]
- Bar-Dolev M., Celik Y., Wettlaufer J. S., Davies P. L., Braslavsky I. J. R. Soc., Interface. 2012;9:3249–3259. doi: 10.1098/rsif.2012.0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budke C., Koop T. Atmos. Meas. Tech. 2015;8:689–703. [Google Scholar]
- Basu K. Journal of Videotape Experiments. 2014:e51185. doi: 10.3791/51185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight C. A., Devries A. L. Science. 1989;245:505–507. doi: 10.1126/science.245.4917.505. [DOI] [PubMed] [Google Scholar]
- Celik Y. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5423–5428. doi: 10.1073/pnas.0909456107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries A. L. Science. 1971;172:1152–1155. doi: 10.1126/science.172.3988.1152. [DOI] [PubMed] [Google Scholar]
- Scotter A. J. Cryobiology. 2006;53:229–239. doi: 10.1016/j.cryobiol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Marshall C. B., Fletcher G. L., Davies P. L. Nature. 2004;429:153. doi: 10.1038/429153a. [DOI] [PubMed] [Google Scholar]
- Kristiansen E., Zachariassen K. E. Cryobiology. 2005;51:262–280. doi: 10.1016/j.cryobiol.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Haji-Akbari A. Proc. Natl. Acad. Sci. U. S. A. 2016;113:3714–3716. doi: 10.1073/pnas.1602196113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Andorfer C. A., Duman J. G. J. Exp. Biol. 1998;201:2243–2251. doi: 10.1242/jeb.201.15.2243. [DOI] [PubMed] [Google Scholar]
- Kristiansen E., Pedersen S. A., Zachariassen K. E. Cryobiology. 2008;57:122–129. doi: 10.1016/j.cryobiol.2008.07.001. [DOI] [PubMed] [Google Scholar]
- Can O., Holland N. B. Biochemistry. 2013;52:8745–8752. doi: 10.1021/bi401345b. [DOI] [PubMed] [Google Scholar]
- Wu D. W., Duman J. G. J. Comp. Physiol., B. 1991;161:279–283. [Google Scholar]
- Stevens C. A., Drori R., Zalis S., Braslavsky I., Davies P. L. Bioconjugate Chem. 2015;26:1908–1915. doi: 10.1021/acs.bioconjchem.5b00290. [DOI] [PubMed] [Google Scholar]
- Can O., Holland N. B. Bioconjugate Chem. 2011;22:2166–2171. doi: 10.1021/bc2004318. [DOI] [PubMed] [Google Scholar]
- Marshall C. B., Daley M. E., Sykes B. D., Davies P. L. Biochemistry. 2004;43:11637–11646. doi: 10.1021/bi0488909. [DOI] [PubMed] [Google Scholar]
- Ebbinghaus S. J. Am. Chem. Soc. 2010;132:12210–12211. doi: 10.1021/ja1051632. [DOI] [PubMed] [Google Scholar]
- Leiter A. J. Food Eng. 2016;187:53–61. [Google Scholar]
- Gaukel V., Leiter A., Spiess W. E. L. J. Food Eng. 2014;141:44–50. [Google Scholar]
- Knight C. A., Hallett J., Devries A. L. Cryobiology. 1988;25:55–60. doi: 10.1016/0011-2240(88)90020-x. [DOI] [PubMed] [Google Scholar]
- Knight C. A., Wen D. Y., Laursen R. A. Cryobiology. 1995;32:23–34. doi: 10.1006/cryo.1995.1002. [DOI] [PubMed] [Google Scholar]
- Budke C., Koop T. ChemPhysChem. 2006;7:2601–2606. doi: 10.1002/cphc.200600533. [DOI] [PubMed] [Google Scholar]
- Congdon T., Notman R., Gibson M. I. Biomacromolecules. 2013;14:1578–1586. doi: 10.1021/bm400217j. [DOI] [PubMed] [Google Scholar]
- Olijve L. L. C., Hendrix M. M. R. M., Voets I. K. Macromol. Chem. Phys. 2016;217:951–958. [Google Scholar]
- Maruyama M. J. Cryst. Growth. 2005;275:598–605. [Google Scholar]
- Budke C. Cryst. Growth Des. 2014;14:4285–4294. [Google Scholar]
- Wathen B., Kuiper M., Walker V., Jia Z. C. J. Am. Chem. Soc. 2003;125:729–737. doi: 10.1021/ja0267932. [DOI] [PubMed] [Google Scholar]
- Strom C. S., Liu X. Y., Jia Z. C. J. Biol. Chem. 2004;279:32407–32417. doi: 10.1074/jbc.M401712200. [DOI] [PubMed] [Google Scholar]
- Corzana F. J. Am. Chem. Soc. 2007;129:9458–9467. doi: 10.1021/ja072181b. [DOI] [PubMed] [Google Scholar]
- Doxey A. C., Yaish M. W., Griffith M., McConkey B. J. Nat. Biotechnol. 2006;24:852–855. doi: 10.1038/nbt1224. [DOI] [PubMed] [Google Scholar]
- Kobashigawa Y. FEBS Lett. 2005;579:1493–1497. doi: 10.1016/j.febslet.2005.01.056. [DOI] [PubMed] [Google Scholar]
- Wilson P. W., Osterday K. E., Heneghan A. F., Haymet A. D. J. J. Biol. Chem. 2010;285:34741–34745. doi: 10.1074/jbc.M110.171983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vali G. Atmos. Chem. Phys. 2014;14:5271–5294. [Google Scholar]
- Inada T., Koyama T., Goto F., Seto T. J. Phys. Chem. B. 2012;116:5364–5371. doi: 10.1021/jp300535z. [DOI] [PubMed] [Google Scholar]
- Holt C. CryoLetters. 2003;24:323–330. [Google Scholar]
- Zachariassen K. E., Husby J. A. Nature. 1982;298:865–867. [Google Scholar]
- Zachariassen K. E., DeVries A. L., Hunt B., Kristiansen E. Cryobiology. 2002;44:132–141. doi: 10.1016/s0011-2240(02)00014-7. [DOI] [PubMed] [Google Scholar]
- Kubota N. Cryobiology. 2011;63:198–209. doi: 10.1016/j.cryobiol.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Chapsky L., Rubinsky B. FEBS Lett. 1997;412:241–244. doi: 10.1016/s0014-5793(97)00787-4. [DOI] [PubMed] [Google Scholar]
- Smolin N., Daggett V. J. Phys. Chem. B. 2008;112:6193–6202. doi: 10.1021/jp710546e. [DOI] [PubMed] [Google Scholar]
- Nutt D. R., Smith J. C. J. Am. Chem. Soc. 2008;130:13066–13073. doi: 10.1021/ja8034027. [DOI] [PubMed] [Google Scholar]
- Grabowska J., Kuffel A., Zielkiewicz J. J. Chem. Phys. 2016;145:075101. doi: 10.1063/1.4961094. [DOI] [PubMed] [Google Scholar]
- Raymond J. A., DeVries A. L. Proc. Natl. Acad. Sci. U. S. A. 1977;74:2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson P. W. Cryobiology. 1994;31:406–412. [Google Scholar]
- Knight C. A., Wierzbicki A. Cryst. Growth Des. 2001;1:439–446. [Google Scholar]
- Sander L. M., Tkachenko A. V. Phys. Rev. Lett. 2004;93:128102. doi: 10.1103/PhysRevLett.93.128102. [DOI] [PubMed] [Google Scholar]
- Li Q. Z., Luo L. F. Chem. Phys. Lett. 1993;216:453–457. [Google Scholar]
- Grandum S. J. Cryst. Growth. 1999;205:382–390. [Google Scholar]
- Zepeda S., Yokoyama E., Uda Y., Katagiri C., Furukawa Y. Cryst. Growth Des. 2008;8:3666–3672. [Google Scholar]
- Hansen-Goos H., Thomson E. S., Wettlaufer J. S. Planet. Space Sci. 2014;98:169–181. [Google Scholar]
- Meister K. J. Phys. Lett. 2015;6:1162–1167. doi: 10.1021/acs.jpclett.5b00281. [DOI] [PubMed] [Google Scholar]
- Ba Y., Wongskhaluang J., Li J. B. J. Am. Chem. Soc. 2003;125:330–331. doi: 10.1021/ja027557u. [DOI] [PubMed] [Google Scholar]
- Celik Y. Proc. Natl. Acad. Sci. U. S. A. 2013;110:1309–1314. doi: 10.1073/pnas.1213603110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drori R., Davies P. L., Braslavsky I. Langmuir. 2015;31:5805–5811. doi: 10.1021/acs.langmuir.5b00345. [DOI] [PubMed] [Google Scholar]
- Knight C. A., DeVries A. L. Phys. Chem. Chem. Phys. 2009;11:5749–5761. doi: 10.1039/b821256b. [DOI] [PubMed] [Google Scholar]
- Wilkinson B. L. Angew. Chem., Int. Ed. 2012;51:3606–3610. doi: 10.1002/anie.201108682. [DOI] [PubMed] [Google Scholar]
- Urbańczyk M., Góra J., Latajka R., Sewald N. Amino Acids. 2017;49:209–222. doi: 10.1007/s00726-016-2368-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcerzak A. K., Capicciotti C. J., Briard J. G., Ben R. N. RSC Adv. 2014;4:42682–42696. [Google Scholar]
- Pentelute B. L., Gates Z. P., Dashnau J. L., Vanderkooi J. M., Kent S. B. H. J. Am. Chem. Soc. 2008;130:9702–9707. doi: 10.1021/ja801352j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller N., Williams G. M., Brimble M. A. Org. Lett. 2009;11:2409–2412. doi: 10.1021/ol9005536. [DOI] [PubMed] [Google Scholar]
- Peltier R. Chem. Sci. 2010;1:538–551. [Google Scholar]
- Czechura P., Tam R. Y., Dimitrijevic E., Murphy A. V., Ben R. N. J. Am. Chem. Soc. 2008;130:2928–2929. doi: 10.1021/ja7103262. [DOI] [PubMed] [Google Scholar]
- Tachibana Y. Angew. Chem. 2004;116:874–880. [Google Scholar]
- Eniade A., Murphy A. V., Landreau G., Ben R. N. Bioconjugate Chem. 2001;12:817–823. doi: 10.1021/bc0155059. [DOI] [PubMed] [Google Scholar]
- Tam R. Y. J. Am. Chem. Soc. 2009;131:15745–15753. doi: 10.1021/ja904169a. [DOI] [PubMed] [Google Scholar]
- Capicciotti C. J. Sci. Rep. 2015;5:9692. doi: 10.1038/srep09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghobadloo S. M. Sci. Rep. 2014;4:05903. [Google Scholar]
- Tam R. Y., Ferreira S. S., Czechura P., Chaytor J. L., Ben R. N. J. Am. Chem. Soc. 2008;130:17494–17501. doi: 10.1021/ja806284x. [DOI] [PubMed] [Google Scholar]
- Capicciotti C. J. Chem. Sci. 2012;3:1408–1416. [Google Scholar]
- Inada T., Lu S. S. Chem. Phys. Lett. 2004;394:361–365. [Google Scholar]
- Inada T., Lu S. S. Cryst. Growth Des. 2003;3:747–752. [Google Scholar]
- Gibson M. I., Barker C. A., Spain S. G., Albertin L., Cameron N. R. Biomacromolecules. 2009;10:328–333. doi: 10.1021/bm801069x. [DOI] [PubMed] [Google Scholar]
- Zhang W., Laursen R. A. FEBS Lett. 1999;455:372–376. doi: 10.1016/s0014-5793(99)00906-0. [DOI] [PubMed] [Google Scholar]
- Wierzbicki A. Biomacromolecules. 2000;1:268–274. doi: 10.1021/bm000004w. [DOI] [PubMed] [Google Scholar]
- Huang M. L. Proc. Natl. Acad. Sci. U. S. A. 2012;109:19922–19927. doi: 10.1073/pnas.1212826109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crich D., Rahaman M. Y. J. Org. Chem. 2011;76:8611–8620. doi: 10.1021/jo201780e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizrahy O., Bar-Dolev M., Guy S., Braslavsky I. PLoS One. 2013;8:e59540. doi: 10.1371/journal.pone.0059540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deville S. PLoS One. 2011;6:e26474. doi: 10.1371/journal.pone.0026474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng H. Angew. Chem., Int. Ed. 2016;56:997–1001. [Google Scholar]
- Drori R. J. Am. Chem. Soc. 2016;138:13396–13401. doi: 10.1021/jacs.6b08267. [DOI] [PubMed] [Google Scholar]
- James C., Purnell G., James S. J. Food Bioprocess Technol. 2015;8:1616–1634. [Google Scholar]
- Regand A., Goff H. D. J. Dairy Sci. 2006;89:49–57. doi: 10.3168/jds.S0022-0302(06)72068-9. [DOI] [PubMed] [Google Scholar]
- Clarke C. J., Buckley S. L., Lindner N. CryoLetters. 2002;23:89–92. [PubMed] [Google Scholar]
- Boonsupthip W., Lee T. C. J. Food Sci. 2003;68:1804–1809. [Google Scholar]
- Payne S. R., Young O. A. Meat Sci. 1995;41:147–155. doi: 10.1016/0309-1740(94)00073-g. [DOI] [PubMed] [Google Scholar]
- Yeh C. M., Kao B. Y., Peng H. J. J. Agric. Food Chem. 2009;57:6216–6223. doi: 10.1021/jf900924f. [DOI] [PubMed] [Google Scholar]
- Margaritis A., Bassi A. S. Crit. Rev. Biotechnol. 1991;11:277–295. doi: 10.3109/07388559109069185. [DOI] [PubMed] [Google Scholar]
- http://www.fda.gov/AnimalVeterinary/DevelopmentApprovalProcess/GeneticEngineering/GeneticallyEngineeredAnimals/ucm280853.htm .
- Neelakanta G., Hudson A. M., Sultana H., Cooley L., Fikrig E. PLoS One. 2012;7:e33447. doi: 10.1371/journal.pone.0033447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna H. K., Daggard G. E. Plant Cell Rep. 2006;25:1336–1346. doi: 10.1007/s00299-006-0191-9. [DOI] [PubMed] [Google Scholar]
- Heisig M. PLoS One. 2015;10:0116562. doi: 10.1371/journal.pone.0116562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagis H. Mol. Reprod. Dev. 2006;73:1404–1411. doi: 10.1002/mrd.20601. [DOI] [PubMed] [Google Scholar]
- Le Curieux-Belfond O., Vandelac L., Caron J., Seralini G. E. Environ. Sci. Policy. 2009;12:170–189. [Google Scholar]
- Sapkota A. Environ. Int. 2008;34:1215–1226. doi: 10.1016/j.envint.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Bakhach J. Organogenesis. 2009;5:119–126. doi: 10.4161/org.5.3.9583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich A. V. Zygote. 2010;18:145–153. doi: 10.1017/S0967199409990141. [DOI] [PubMed] [Google Scholar]
- Chaytor J. L. Glycobiology. 2012;22:123–133. doi: 10.1093/glycob/cwr115. [DOI] [PubMed] [Google Scholar]
- Huebinger J. Biophys. J. 2016;110:840–849. doi: 10.1016/j.bpj.2015.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hincha D. K., Devries A. L., Schmitt J. M. Biochim. Biophys. Acta. 1993;1146:258–264. doi: 10.1016/0005-2736(93)90364-6. [DOI] [PubMed] [Google Scholar]
- Amir G. Ann. Thorac. Surg. 2004;77:1648–1655. doi: 10.1016/j.athoracsur.2003.04.004. [DOI] [PubMed] [Google Scholar]
- Capicciotti C. J., Doshi M. and Ben R. N., Ice Recrystallization Inhibitors: From Biological Antifreezes to Small Molecules, 2013. [Google Scholar]
- Wowk B. Cryobiology. 2000;40:228–236. doi: 10.1006/cryo.2000.2243. [DOI] [PubMed] [Google Scholar]
- Ideta A. J. Reprod. Dev. 2015;61:1–6. doi: 10.1262/jrd.2014-073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briard J. G. Sci. Rep. 2016;6:23619. doi: 10.1038/srep23619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaves D. F. Cryobiology. 2016;73:324–328. doi: 10.1016/j.cryobiol.2016.10.003. [DOI] [PubMed] [Google Scholar]
- Lagneaux D., Huhtinen M., Koskinen E., Palmer E. Equine Vet. J. 1997;29:85–87. doi: 10.1111/j.2042-3306.1997.tb05108.x. [DOI] [PubMed] [Google Scholar]
- Wang T., Zhu Q., Yang X., Layne Jr J. R., Devries A. L. Cryobiology. 1994;31:185–192. doi: 10.1006/cryo.1994.1022. [DOI] [PubMed] [Google Scholar]
- Mugnano J. A., Wang T., Layne J. R., DeVries A. L., Lee R. E. Am. J. Physiol.: Regul., Integr. Comp. Physiol. 1995;269:R474–R479. doi: 10.1152/ajpregu.1995.269.2.R474. [DOI] [PubMed] [Google Scholar]
- Koushafar H., Pham L., Lee C., Rubinsky B. J. Surg. Oncol. 1997;66:114–121. doi: 10.1002/(sici)1096-9098(199710)66:2<114::aid-jso8>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- Venketesh S., Dayananda C. Crit. Rev. Biotechnol. 2008;28:57–82. doi: 10.1080/07388550801891152. [DOI] [PubMed] [Google Scholar]
- Hirano Y. Cryobiology. 2008;57:46–51. doi: 10.1016/j.cryobiol.2008.05.006. [DOI] [PubMed] [Google Scholar]
- Wu J. H. Eur. Food Res. Technol. 2013;236:637–646. [Google Scholar]
- Jo J. W., Jee B. C., Suh C. S., Kim S. H. PLoS One. 2012;7:e37043. doi: 10.1371/journal.pone.0037043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilli L. PLoS One. 2014;9:e99992. doi: 10.1371/journal.pone.0099992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. G., Koh H. Y., Lee J. H., Kang S. H., Kim H. J. Appl. Biochem. Biotechnol. 2012;167:824–834. doi: 10.1007/s12010-012-9739-z. [DOI] [PubMed] [Google Scholar]
- Lee J. PLoS One. 2015;10:0126252. [Google Scholar]
- Amir G. Cryobiology. 2004;48:273–282. doi: 10.1016/j.cryobiol.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Younis A. I., Rooks B., Khan S., Gould K. G. J. Androl. 1998;19:207–214. [PubMed] [Google Scholar]
- Matsumura K., Hyon S. H. Biomaterials. 2009;30:4842–4849. doi: 10.1016/j.biomaterials.2009.05.025. [DOI] [PubMed] [Google Scholar]
- Deller R. C., Vatish M., Mitchell D. A., Gibson M. I. Nat. Commun. 2014;5:3244. doi: 10.1038/ncomms4244. [DOI] [PubMed] [Google Scholar]
- Leclere M., Kwok B. K., Wu L. K., Allan D. S., Ben R. N. Bioconjugate Chem. 2011;22:1804–1810. doi: 10.1021/bc2001837. [DOI] [PubMed] [Google Scholar]
- Liu S. H. Biomacromolecules. 2007;8:1456–1462. doi: 10.1021/bm061044o. [DOI] [PubMed] [Google Scholar]
- Fukushima M., Tsuda S., Yoshizawa Y. J. Am. Ceram. Soc. 2013;96:1029–1031. [Google Scholar]
- Heisig M. Cell Rep. 2014;9:417–424. doi: 10.1016/j.celrep.2014.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop T., Zobrist B. Phys. Chem. Chem. Phys. 2009;11:10839–10850. doi: 10.1039/b914289d. [DOI] [PubMed] [Google Scholar]
- Pummer B. G. Atmos. Chem. Phys. 2015;15:4077–4091. [Google Scholar]
- Petzold G., Aguilera J. M. Food Biophys. 2009;4:378–396. [Google Scholar]
- Wen X. Proc. Natl. Acad. Sci. U. S. A. 2016;113:6683–6688. doi: 10.1073/pnas.1601519113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wen X., Nikolovski P., Juwita V., Arifin J. F. Chem. Commun. 2012;48:11555–11557. doi: 10.1039/c2cc36264c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. J. Am. Chem. Soc. 2014;136:8973–8981. doi: 10.1021/ja502837t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwak Y. Sci. Rep. 2015;5:1–9. [Google Scholar]
- Charpentier T. V. J., Neville A., Millner P., Hewson R., Morina A. J. Bionic Eng. 2013;10:139–147. [Google Scholar]
- Jung H. C., Lebeault J. M., Pan J. G. Nat. Biotechnol. 1998;16:576–580. doi: 10.1038/nbt0698-576. [DOI] [PubMed] [Google Scholar]
- Chen W., Georgiou G. Biotechnol. Bioeng. 2002;79:496–503. doi: 10.1002/bit.10407. [DOI] [PubMed] [Google Scholar]
- Wernerus H., Stahl S. Biotechnol. Appl. Biochem. 2004;40:209–228. doi: 10.1042/BA20040014. [DOI] [PubMed] [Google Scholar]
- Graham L. A., Davies P. L. Science. 2005;310:461. doi: 10.1126/science.1115145. [DOI] [PubMed] [Google Scholar]
- Yu S. O. Cryobiology. 2010;61:327–334. doi: 10.1016/j.cryobiol.2010.10.158. [DOI] [PubMed] [Google Scholar]