Abstract
Modern optogenetics can be tuned to evoke activity that corresponds to naturally occurring local or global activity in timing, magnitude or individual-cell patterning. This outcome has been facilitated not only by the development of core features of optogenetics over the past 10 years (microbial-opsin variants, opsin-targeting strategies and light-targeting devices) but also by the recent integration of optogenetics with complementary technologies, spanning electrophysiology, activity imaging and anatomical methods for structural and molecular analysis. This integrated approach now supports optogenetic identification of the native, necessary and sufficient causal underpinnings of physiology and behaviour on acute or chronic timescales and across cellular, circuit-level or brain-wide spatial scales.
Study of the neural underpinnings of behaviour is rapidly progressing, and many discoveries over the past decade have been enabled by technologies that address a fundamental principle: to determine how neural circuit activity controls behaviour, experimental interventions should be performed with genetic, anatomical and temporal precision. Owing to the development of optogenetics1,2 (in which single genes encoding light-activated ion-conductance regulators or biochemical signalling proteins are introduced into targeted cells; FIG. 1; see Supplementary information S1), researchers can now control activity in defined neuronal populations and projections while examining the consequences on behaviour and physiology. Unlike pharmacological and lesion-based interventions, optogenetics (although readily applicable in bringing cellular specificity to slow or chronic timescales) also opens up causal investigation and specificity for the fast timescales of natural nervous system communication.
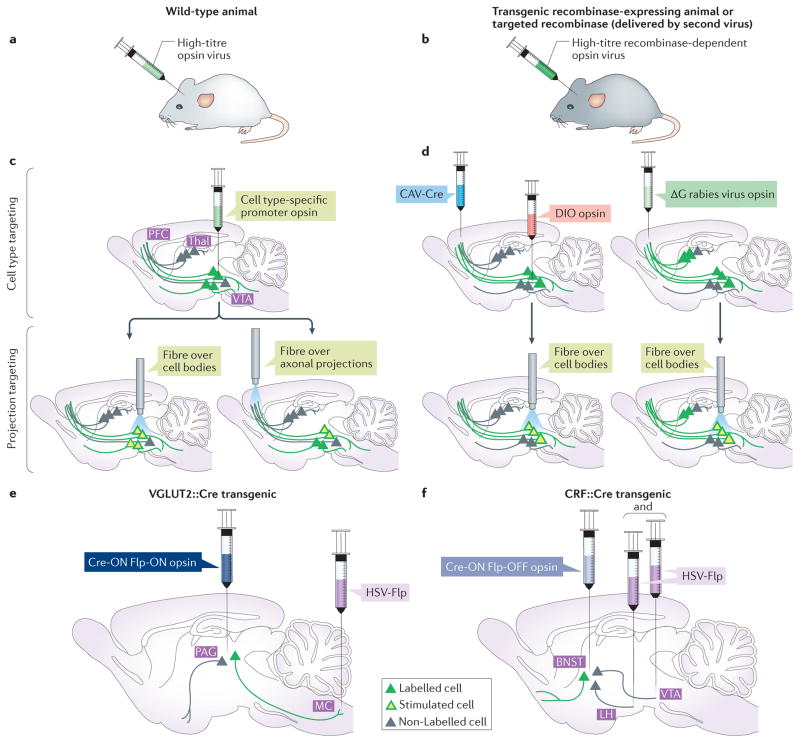
Figure 1. Approaches to opsin targeting with anatomical and cell type specificity.
a,b | The schematics demonstrate the method for expressing opsins in neurons. A DNA vector encoding an opsin is packaged into a high-titre virus (most often adeno-associated virus (AAV)), and this virus is injected into the brain region of interest, inducing opsin expression in target neurons. Cell type specificity of opsin expression can be achieved either by using a cell type-specific promoter virus in a wild-type animal (part a) or by using a recombinase-dependent (for example, Cre-dependent) virus in a transgenic recombinase-driver animal or with a secondary virus containing a targeted recombinase13–15 (part b). c | Following opsin expression in the cell population of interest (green cells; top panel), a light-delivering optical fibre can be placed either over the cell bodies to target all projection neurons (bottom left panel) or in a known downstream region to target a specific projection (bottom right panel). d | A retrograde virus such as canine adenovirus (CAV)-Cre and a Cre-dependent doubly floxed inverted opsin (DIO) virus can be injected into the downstream and upstream region, respectively, to label only a specific projection with opsin. The fibre can then be placed over the cell bodies to manipulate that projection (left panel). Glycoprotein-deleted (ΔG) rabies virus can also be injected into a brain region to retrogradely label all presynaptic inputs. To activate a specific presynaptic input, the fibre can be placed over the input structure studied (right panel). e | The retrograde virus herpes simplex virus (HSV)-Flp can be used in conjunction with combinatorial INTRSECT Cre-dependent and Flp-dependent viruses in transgenic Cre animals (or with Cre viruses) to heighten projection-labelling specificity15. For example, HSV-Flp and a Cre-ON Flp-ON channelrhodopsin (ChR) virus can be used to label vesicular glutamate transporter 2 (VGLUT2)-expressing neurons in a Cre transgenic mouse that project from the periaquaductual grey (PAG) to the magnocellular nucleus (MC) of the medulla. f | This method can also be used to exclude a particular projection target. Corticotropin-releasing factor (CRF)-expressing neurons in the bed nucleus of the stria terminalis (BNST) that do not project to either the lateral hypothalamus (LH) or the ventral tegmental area (VTA) can be labelled in a Cre transgenic mouse using a Cre-ON Flp-OFF ChR virus in conjunction with HSV-Flp. PFC, prefrontal cortex; Thal, thalamus. Part e is adapted with permission from REF. 100, Macmillan Publishers Limited. Part f is adapted with permission from REF. 99, Macmillan Publishers Limited.
As with rigorous experiments in other fields of science in which causal hypotheses can be tested, optogenetic experiments include the triad of disrupting, providing and observing endogenous operation of specific hypothesized causal agents — in this case, activity patterns of neural circuit components during behaviour. Building on the fundamental process of hypothesis testing in other fields of biology — wherein, similarly, for example, a researcher studying a specific protein or gene might inhibit or knock out, activate or deliver, and measure corresponding endogenous activity — the flexibility of optogenetics is unique in further enabling causal tests over a broad range of spatial and temporal timescales to meet the unique challenges of neuroscience. Indeed, to take full advantage of the temporal capabilities of optogenetics, comparably fast behavioural and physiological readouts have been developed; modern optogenetics now includes naturalistic behavioural paradigms with fast quantitative readouts and even closed-loop feedback based on changes in the physiology or actions of the experimental system — enabling an unprecedented level of hypothesis-testing precision2. Such experiments require exchange of light with the nervous system; this can be achieved by implanting a small fibre-optic probe into the brain3–5, which is typically interfaced with a lightweight fibre-optic patch cord coupled to a laser diode or to a light-emitting diode (LED) light source for input and to a fast camera or photomultiplier for readout. This robust hardware is compatible with diverse cognitive and motor tasks such as those measuring social, defensive, aggressive, navigational and indeed virtually all validated freely moving rodent behaviours1; recent developments in fully implantable light devices6,7 may even further facilitate certain increasingly complex behavioural paradigms.
One example (among thousands of other published findings) was the finding that the ventrolateral portion of ventromedial hypothalamus (VMHvl) specifically modulates aggression8. The authors failed to elicit aggression with electric stimulation of the VMHvl, probably because this manipulation activated intermixed subnuclei and axons subserving other (for example, defensive) functions. The authors next optogenetically activated only VMHvl cells and found that this gain-of-function approach elicited a flurry of behavioural attacks on intruder mice, whereas stimulation of nearby surrounding regions did not — instead, it induced freezing, flight or no change. Countless such discoveries (often coupling optogenetic gain-of-function and loss-of-function interventions, measurements of endogenous activity and increasingly rigorous behavioural paradigms) led to results that would not have been achievable through other modes of neural intervention (such as electrical stimulation, pharmacology or lesion).
These earlier methods nevertheless remain powerful and relevant, especially as optogenetic experiments build on this strong foundation that underlies systems and behavioural neuroscience. Interestingly, when expressed nonspecifically in all neurons9–11 to disrupt function, excitatory optogenetic tools are sometimes even used to mimic inhibitory interventions by achieving a ‘lesionlike’ influence with diverse global nonspecific effects9–11. Although this approach leverages neither optogenetic specificity nor optogenetic matching of naturally occurring local and brain-wide patterns, such intentionally nonspecific lesion-type efforts are still potentially interesting, as they could (if properly controlled) capitalize on the unique temporal flexibility of optogenetics over any acute or chronic timescale (from milliseconds to many days or more), to allow intervention-matched comparative testing of dynamical principles across timescales12. Current applications of optogenetics are instead much more commonly designed for ever-increasing specificity of direct targeting and modulation, with careful control experiments confirming that comparable different circuit modulations do not result in the effect of interest (such controls are standard practice in rigorous biology) and with precise matching (or tuning) of native local or global activity patterns.
As neural circuit components are highly diverse, targeting defined cell types or projections on the basis of multiple features was crucial for optogenetics and was enabled by the development of viruses with specially engineered properties, such as axonal transduction and/or retrograde propagation, cell type-specific promoter dependence and/or recombinase dependence (for example, Cre, Flp and Dre recombinases, which can be targeted to specific cell types using recombinase-driver rodent lines or recombinase-expressing viruses). For example, a two-virus strategy developed between 2010 and 2014 (REFS 13–15) entails delivering a targeted recombinase (for example, Cre-expressing) virus together with a strong Cre-dependent double-floxed inverted orientation (DIO) opsin virus to achieve both targeting specificity and high opsin expression levels even in wild-type animals15. Optogenetic control can be further refined by targeting neurons by virtue of their natural activity during behaviour; this is achieved either by recording activity and then manipulating the corresponding neurons with cellular resolution in the same animal or by preferentially labelling active neurons during behaviour via triggered opsin expression (using immediate-early gene (IEG) promoters) and then later reactivating those neurons with optogenetics16–22. Such activity-dependent opsin methods could also be useful for identifying circuit elements that are functionally downstream of specific previously activated populations, by driving or inhibiting those naturalistic populations selectively (all correctly targeted and behaviourally significant activity patterns — to regulate behaviour, just as with naturally occurring activity patterns — must exert long-range effects on other cells and regions across the brain beyond the initial directly targeted population). Cells labelled by virtue of wiring or activity can be later visualized, molecularly characterized and traced across the intact brain using tissue transparency methods, such as those based on hydrogel embedding22–28 (see Supplementary information S1).
In summary, the past decade has witnessed remarkable developments in technologies that can be synergistic with optogenetics. Below we review these advances and highlight key demonstrations of successful integration with optogenetics. These techniques have individually yielded numerous discoveries, but recently some of the most informative lines of investigation have involved combining all of these supporting techniques together with the leverage of optogenetic control.
Natural and engineered opsin diversity
Some of the earliest steps towards genetically targeted optical control of neural activity and behaviour were reported in Drosophila; these elegant experiments were multicomponent in nature, requiring expression of multiple proteins29 or optical uncaging of exogenous chemicals to activate engineered receptors30. By contrast, the single-component optogenetic method that was broadly adopted uses a fundamentally distinct class of molecule: microbial rhodopsin proteins31 that act as unitary light-activated ion pumps or channels (for additional references to this microbial literature — which emerged in 1971 and has included heterologous expression to elicit light-activated ion flow as far back as 1994 — see Supplementary information S1). After the initial demonstration that microbial opsins could be used to control neuronal activity with light1,32, microbial opsin-mediated optical control of neurons was soon reported in various different circuits and species33–38, leading to further development for broad applicability in neuroscience1 (see Supplementary information S1).
Excitatory or inhibitory effects can be elicited by expression of different subclasses of microbial genes encoding opsins; for example, many naturally occurring channelrhodopsins (ChRs) are nonspecific cation channels that depolarize (excite) neurons in response to blue light, whereas halorhodopsin-type Cl− pumps and bacteriorhodopsin-type proton pumps (reviewed in REF. 39) induce hyperpolarization (inhibition) in response to yellow or green light, respectively, by pumping Cl− ions into, or protons out of, the cell. Upon provision of eukaryotic cellular trafficking motifs that were discovered to be useful for safe and effective transport of all three classes of microbial opsins to vertebrate cell membranes13,40–42, now all major classes of these light-activated channel and pump proteins are routinely used in neuroscience for temporally precise circuit manipulation1. As the special suitability of microbial opsins for optogenetics became apparent, ion-selectivity variants were engineered to achieve new functionality — recently exemplified by Cl−-conducting ChRs. These Cl− channels, initially engineered through crystal structure-guided mutation of ChR43,44 and later found as naturally occurring variants45–47, conduct multiple Cl− ions per photon and thus can deliver more efficient and greater light-sensitive inhibition of neural activity48 than Cl− and H+ pumps, which move one ion per photon.
Kinetic opsin variants were also discovered and developed for new domains of application. For example, faster-deactivating ChR variants (including ChETA49, ChIEF50, Chronos51 and others52) have been developed and have been shown to reliably drive spiking up to 200 Hz (REF. 49). These high-speed variants have enabled studies wherein high-frequency modulation is required to mimic naturally occurring dynamics53. By contrast, step-function opsins54 — including the excitatory stabilized step-function opsin (SSFO; a bistable excitatory ChR variant42), the step-waveform inhibitory ChR (SwiChR; a step-function form of Cl−-conducting inhibitory ChRs)43 and others55 — deactivate much more slowly (even over tens of minutes, instead of milliseconds); by inducing prolonged changes with brief light delivery, this class of opsin confers orders-of-magnitude greater light sensitivity to expressing cells and allows orders-of-magnitude reduced duration of light delivery, thus facilitating certain specialized chronic manipulations56,57 (bright light exposure over hours can adversely affect cell health). The properties of these step-function tools also allow subtle modes of modulation. For example, the SwiChR variants43,48 do not strongly hyperpolarize neurons but instead reversibly and stably open a Cl− channel pore, as do native GABA type A receptor (GABAAR) Cl− channels. SwiChR and the second-generation version SwiChR++ by design have much slower kinetics than endogenous GABAAR channels, allowing long-timescale reversible inhibition. SwiChR is thus able to recruit (on long timescales) naturalistic inhibitory membrane properties such as shunting and, similar to native GABAARs, will display physiological sensitivity to natural influences of Cl− balance, resting potential and input resistance43,48. Likewise, when SSFO is expressed, the targeted neurons do not strongly depolarize and do not experience directly light-driven action potentials (which would be useful in many but not all settings); instead, they display altered activity in a manner that is naturally timed or asynchronous in a population42,57 depending on endogenous synaptic input activity.
Although controlling (providing or removing) neural activity with these tools can reveal whether activity in a neural circuit element is necessary or sufficient for behaviour, it is important to understand how the element and its connections across the brain are naturally used and respond to experimental or natural modulation; combining optogenetics with activity readouts such as IEG expression, functional MRI, Ca2+ imaging and electrophysiology is therefore important. Researchers have long integrated optogenetics with global-activity readouts57–60 such as functional MRI, but limitations include slow temporal dynamics, lack of cellular specificity and incompatibility with freely moving behaviour. These limitations can be overcome by integrating optogenetics with genetically encoded Ca2+ indicator (GECI) fluorescence readouts; beyond the kinetic and selectivity opsin variants described above, spectral variants (such as the red-shifted excitatory opsins VChR1 (REF. 61), C1V1 (REF. 42), Chrimson51, ReaChR62 and bReaChES63) are compatible with blue-light-actuated GECIs64–66 to simultaneously manipulate and record from neurons in the same animals. If the spectral properties of GECIs and opsins overlap substantially, the excitation light that is used to image activity causes unwanted cross-stimulation67; red-shifted opsins reduce this effect, enabling all-optical experiments that have taken great steps forward in practicality and application68–72.
Cell type-specific opsin expression
The optogenetic toolbox can be applied with genetic specificity via viral vectors, recombinase-expressing driver animal lines and anatomical targeting strategies. The simplest way to target opsin expression to a specific cell type is to inject a virus expressing the opsin under the control of a cell type-specific promoter (FIG. 1a). Some commonly used promoter fragments that confer cell type preference and are applicable in lentivirus or adeno-associated virus (AAV) vectors are linked to the following markers73: calcium/calmodulin-dependent protein kinase type II subunit-α (CaMKIIα; biased towards excitatory cells in cortical regions3,4), hypocretin74, oxytocin75, D2 dopamine receptor41, glial fibrillary acidic protein (GFAP; for astrocytes), myelin basic protein (MBP; for oligodendrocytes) and somatostatin76. This method has illuminated how hypocretin-producing lateral hypothalamic neurons regulate sleep–wake transitions74 and how nucleus accumbens (NAc) D2 dopamine receptor-expressing cells modulate risk taking41. However, this strategy has not been used to target a wide range of cell types, because the sequences conferring specificity are usually too large to be packaged into viruses.
To address this issue, recombinase-dependent opsin-expressing viral vectors can be injected into transgenic animals or, along with targeted viruses13–15 that drive recombinase expression, in cells of interest. For example, injecting an AAV designed for recombinase-dependent opsin expression from a strong but well-tolerated general promoter (for example, elongation factor 1 alpha (ef1A)), along with a second recombinase-expressing virus13–15 or into a recombinase-driver transgenic mouse (FIG. 1b), will provide strong targeted opsin expression selectively in recombinase-expressing cells in the injected brain region, simultaneously intersecting genetic specificity with regional cell-body targeting (most AAVs do not robustly transduce axons). Hundreds of Cre-recombinase and many Flp-recombinase mouse lines are available, creating ample opportunities. Among the many results enabled by this approach, cortical parvalbumin-expressing interneurons were modulated to test social behaviour regulation42, and septal cholinergic cells were modulated to explore regulation of hippocampal network activity77. ‘Recombinase-off’ viral vectors have also been developed to drive opsin expression in cells that do not express the marker78; for example, such vectors have been used to investigate physiology of frontal-projecting non-cholinergic cells in the globus pallidus externa79.
Projection targeting: light and opsins
Importantly, the strategies described above can be further enhanced to allow targeting of projections between two brain regions, by delivering light to opsin-expressing axon terminals via the fibre-optic interface (‘projection targeting’) (FIG. 1c). Optogenetic projection targeting enables versatile experimental leverage (for example, for selective inhibition of projections between brain regions80–82 or for excitation of cells defined by projecting from one brain region to another1,2,80); however, several considerations must be kept in mind. For example, several weeks are necessary to obtain sufficient opsin expression in long-range axonal projections (although this issue has recently been ameliorated by the discovery that provision of a fused neuritin 3′ untranslated region can substantially accelerate the expression of heterologously expressed proteins in long-range projections22). In addition, if ionic milieu interactions (for example, Cl− exchange, H+ buffering and diverse H+-induced currents) are different in axon terminals compared with somata, opsin effects may be different in the two locations, as would also be the case for native channels such as GABAARs48; these issues suggest that the current best strategy for axonal inhibition is to use Cl− pumps that are both insensitive to ion gradients and designed for robust axonal trafficking13 rather than Cl− channels43 or H+ pumps83.
Additional solutions to these issues can be found in viral targeting of projections, in which opsin expression, rather than light, is targeted selectively to cells that project to a specific downstream population (FIG. 1d). One such strategy uses canine adenovirus (CAV)84–88; CAV driving Cre-recombinase can be injected into a given brain region (X) to transduce (along with local cell bodies) local axon terminals, resulting in expression of Cre in presynaptic cells. Injecting a recombinase-dependent opsin AAV in one of these corresponding presynaptic regions (Y) induces opsin expression exclusively in cells from the upstream region Y that project to the downstream region X. Thus, cells defined by projection can be recruited with light delivery at the soma instead of at the axon terminals. This ‘retrograde’ method can also be performed with engineered AAVs89 or modified strains of herpes simplex virus90,91 (albeit with some toxicity).
These virus-guided (in contrast to light-guided) methods can be stronger and faster in terms of enabling optogenetic control of cells defined by projections, because sufficient accumulation of opsin protein is only required in upstream cell bodies, not in axon terminals. However, true projection-specific control (for example, suppression of activity along a specific tract) cannot be readily achieved without also guiding light via the fibre-optic interface; moreover, most projection-targeting methods involving virus-guided opsin expression require two viruses, each of which will be only partially efficient, potentially resulting in a small number of labelled cells. Alternatively, one can inject a single retrograde vector encoding a recombinase (such as CAV-Cre) into a floxed-STOP (recombinase-dependent) opsin transgenic mouse line, resulting in opsin expression in all presynaptic cells and their fibres (not a single projection defined by origin and target). However, light delivery to a presynaptic population may then affect fibres of passage from multiple areas, potentially confounding the observed behavioural effects. Highly efficient rabies viruses have also been used to retrogradely target opsin expression92,93, but rabies virus infection is toxic for neurons, and thus behavioural experiments must be performed within a few days of infection. The newly developed CVS-N2cΔG rabies virus strain is less toxic, potentially increasing the potential of rabies virus-based opsin targeting94.
Two quantitative points are worthy of consideration regarding the physiology of optogenetic projection inhibition. First, useful information on the natural role of the projection is obtained even when potent but incomplete inhibition of evoked synaptic release is observed; inhibitory-projection targeting is especially important for certain specific experimental questions, as inhibition cannot propagate long distances back to the cell body and down axon collaterals. Second, although behavioural effects are reproducibly observed that are concordant with predicted effects of reducing projection influence in the target brain region (relative to other inputs), the effects of reducing this influence are not expected to be simply reflected in the form of altered mean native spike rates across all cells in the target region. Modern understanding of neural circuit operation in awake animals has progressed far beyond viewing circuit activity as simple summation from excitatory inputs, with local networks waiting passively for an input stimulus. Incoming inputs affect local computations through both excitation and inhibition, and mean spike rate is not expected to be altered in a simply predictable way following inhibition of a single input source among all other inputs to a brain region; instead, this intervention will reduce the influence of the projection in the ongoing computations performed in the local circuit without necessarily altering mean spike rates.
The above viral targeting strategies can be retrograde in nature (travelling from the axonal projection field of a cell to its soma), thus defining cells by virtue of output (creating output-defined elements95). Anterograde genetic targeting of cells defined by input is much less established, and a major limitation is the lack of robust trans-synaptic anterograde viruses. Two trans-synaptic anterograde viruses have been characterized: the H129 strain of the herpes simplex virus, which is polysynaptic96,97, and a vesicular stomatitus virus variant, which can be monosynaptic98 but is not extensively used because of its cytotoxicity. Although widely used monosynaptic anterograde-targeting strategies have not emerged, a method using multiple recombinases to target cell populations intersectionally on the basis of multiple genetic and/or anatomical features and using diverse Boolean logic-based operations has been developed15 (FIG. 1e,f). This method (INTronic Recombinase Sites Enabling Combinatorial Targeting (INTRSECT)) was used to target only dopaminergic cells (but not other cell types) from the ventral tegmental area (VTA) that project to the NAc and only hippocampal interneurons that simultaneously express parvalbumin and somatostatin; later applications of INTRSECT have revealed broad utility99,100 (FIG. 1e,f). Subsequent methods to target neurons defined by both input and output (defining IODES (input/output-defined elements); for example, with TRIO (tracing the relationship between input and output)86–88) have further extended capabilities of genetic and anatomical targeting. Together, these diverse genetic and optical advances have greatly expanded the power of optogenetics itself.
Electrophysiology and optogenetics
Patch-clamp electrophysiology is the unrivalled gold standard for high-speed single-cell monitoring of synaptic input and spiking output, but it is difficult to link the resulting data stream to defined cell types in vivo during behaviour. However, the integration of patch-clamp techniques with projection-targeted optogenetics has circumvented these limitations and allowed researchers to study functional connectivity of long-range projections with cell type specificity. One can ensure that only monosynaptic responses are optically elicited in acute brain slices (taken from the projection-target region after in vivo behavioural experiments and animal sacrifice) by pharmacologically blocking polysynaptic responses with tetrodotoxin while permitting ChR-driven monosynaptic transmitter release enabled by the addition of the stimulatory K+ channel blocker 4-aminopyridine101,102; using this method, it has been shown that (contrary to prior predictions) excitatory projections from the dorsal, but not the ventral, medial prefrontal cortex (mPFC) synapse directly onto fear-suppressing GABAergic intercalated cells in the amygdala82 and that dopaminergic cells in the dorsal raphe release glutamate on extended amygdala cells103. Integration of in vitro patch clamp with optogenetics has produced numerous additional insights into food consumption104, anxiety105, aggression106, reward92 and other behaviours.
Traditional in vivo electrophysiology alone is also difficult to link to specific cell types defined by genetics or connectivity, and therefore combining in vivo electrophysiological recordings with optogenetics has been an important and versatile technological integration, applied to numerous different circuits and behaviours (FIG. 2). For example, optogenetic stimulation of a specific input or cell type during multiunit extracellular recording allows the determination of circuit physiology effects (FIG. 2a,b); extending this approach to behavioural studies allows investigation of whether defined inputs from one region to another also contribute to encoding of a stimulus or behaviour. Using this approach, it was reported that activation of the basolateral amygdala (BLA) excites the NAc to drive reward seeking107, that activation of GABAergic cells from the extended amygdala inhibits lateral hypothalamic neurons leading to increased food consumption81 and that ventral hippocampal input is required by the mPFC to encode goal location108 and aversive (open) versus safe (enclosed) spaces in the elevated-plus maze109. Similar principles can appear across circuits; the bed nucleus of the stria terminalis (BNST) uses BLA input to also encode closed spaces in the same maze105; moreover, optogenetic inhibition of BLA–BNST terminals powerfully reduced BNST representation of closed arms (as noted above, mean spike rate across all cells in the target (recorded) region can be modulated, but this measure is not necessarily expected to be altered by important modulation of a specific input during behaviour). These studies illustrate the power of integrating electrophysiology and optogenetics to create mechanistic explanations that causally link neural activity to functionally significant encoding of behaviour.
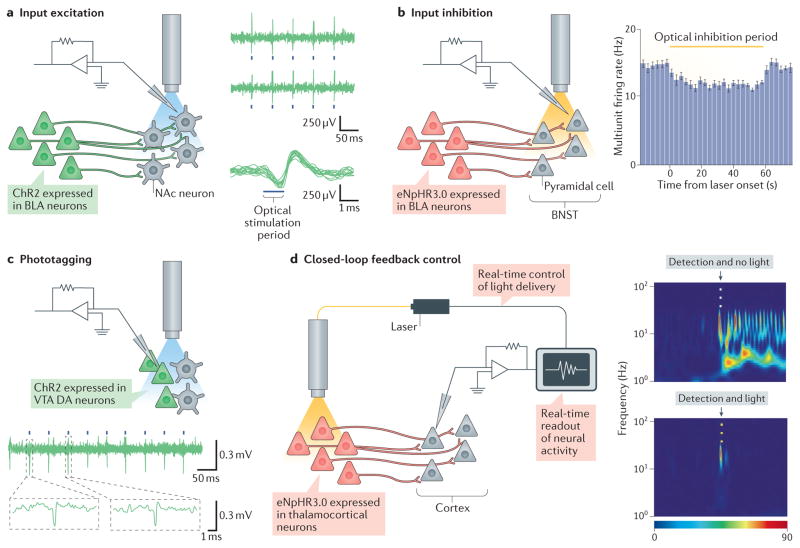
Figure 2. Integrating optogenetic control with in vivo electrophysiology.
a | The left panel shows optogenetic stimulation of axons from the basolateral amygdala (BLA) in which channelrhodopsin 2 (ChR2) has been expressed while simultaneously monitoring downstream activity in the nucleus accumbens (NAc). The right panel shows that optogenetic stimulation of BLA terminals in the NAc results in downstream electrical spiking in local NAc neurons. The top traces are example electrophysiological recordings exhibiting reproducible spiking in response to 1 ms optogenetic stimulation pulses (blue ticks). The bottom trace plots overlay trials of electrical spiking, aligned to the time of optogenetic stimulation (blue bar). b | Optogenetic modulation of bed nucleus of the stria terminalis (BNST) activity through optogenetic inhibition of afferent BLA axons is shown (left panel). Inhibition of BLA terminals in the BNST that express eNpHR3.0 results in net reduction in multiunit firing rate in the local BNST neurons (right panel; the yellow bar indicates the optical stimulation period); in other brain regions, highly efficacious in vivo input modulation may not cause such mean overall rate changes but instead modulates other features of regional computation and behavioural output. c | Phototagging can be used to identify and stimulate genetically specified ventral tegmental area (VTA) dopaminergic (DA) neurons expressing a ChR. ChR-expressing VTA DA cells, in this case, may be identified during in vivo recordings by responsiveness to pulses of blue light (blue ticks in voltage trace) and can be distinguished from non-expressing cells, which do not respond. Two examples of light-triggered spikes are shown in the bottom panel. d | Real-time closed-loop optogenetic inhibition of thalamocortical neurons is triggered when seizure activity is detected in the cortex by electroencephalography (EEG). Incipient seizures are detected in the cortex by EEG (in the figure, indicated by the black arrowhead above spectrogram), which in the absence of optical stimulation result in ongoing seizure activity, shown in pseudocolour (the red end of the spectrum indicates highest activity) by the rapid spiking and intense red spots in the upper panel. When seizure detection triggers yellow-light delivery to thalamocortical neurons, it results in an interruption of the seizure (bottom panel). Part a is reproduced with permission from REF. 107, Macmillan Publishers Limited. Part b is reproduced with permission from REF. 105, Macmillan Publishers Limited. Part c is reproduced with permission from REF. 111, Macmillan Publishers Limited. Part d is adapted with permission from REF. 115, Macmillan Publishers Limited.
A less-commonly applied approach for combining optogenetics with in vivo extracellular physiology is phototagging110–112. This method113 consists of expressing an excitatory opsin in the neuronal population of interest, recording and sorting the spike waveforms of units in that brain region and later attempting to assign individual electrically recorded cells to the genetically targeted population of interest by shortness of latency and low jitter (typically around 0.1 ms) of the corresponding spike waveform apparently evoked by light (FIG. 2c). Although this method will suffer from misidentification errors, in part because spike latency can be variably slow in directly excited cells or fast even in indirectly excited cells, for certain circuits that are well suited to this approach phototagging has been used to study spatial representation, food consumption, aversion and reward responsiveness, and other behaviours91,112. For example, phototagging has been performed with recombinase-driver mouse lines111 to confirm that VTA dopaminergic cells encode reward prediction error and that VTA GABAergic cells are active between cue and reward delivery.
Closed-loop optogenetic interventions
Combining fast readouts (such as electrophysiology) with optogenetics creates the possibility of closed-loop optogenetic interventions, in which optical stimulation is guided by real-time readout of ongoing activity2,114. One study115 recorded neuronal activity in the thalamus and cortex to detect seizure initiation in a cortical lesion-induced rodent model of epilepsy and showed that optogenetically silencing thalamocortical neurons after seizure onset sufficed to terminate ongoing epileptic activity (FIG. 2d); it would have been impossible to selectively silence thalamocortical neurons with an electrode. Similarly, on-demand activation of cerebellar parvalbumin-expressing Purkinje cells was reported to inhibit ongoing medial temporal lobe seizures116. It has also been reported that inhibiting CA1 pyramidal cells can modulate behavioural performance when applied during a specific portion of the task or during a specific phase of ongoing hippocampal theta oscillations: behavioural performance was enhanced either by optogenetic intervention in the encoding segment of the task during theta-wave peaks or by optical inhibition in the retrieval segment during theta-wave troughs117. These results illustrate the diversity and precision of discoveries that can be made at the interface of electrophysiology and optogenetics.
All-optical interrogation of circuits
Electrophysiological recording during behaviour from multiple genetically identified neurons is difficult, but advances in Ca2+ imaging using GECIs64–66 now allow researchers to chronically record correlates of activity across hundreds of genetically defined neurons with either cellular or population-level resolution (see Supplementary information S1). Using cellular-resolution GECI fluorescence imaging, dynamics of local neuronal ensembles have been studied in head-fixed63,69,118,119 or freely moving animals120,121; in general, this technique is limited to a small field of view to maintain cellular resolution and simultaneity, but this limitation is increasingly addressable with advanced wide-field optics. Alternatively, population-level fibre photometry14,122,123 for Ca2+ lacks cellular resolution but is far simpler to implement while delivering genetically specified, behaviourally time-locked signals14,41,72,87,123–125 and is easily scalable to multiple brain regions and axon tracts in the same animal72.
Many studies have used GECI recording and optogenetics, as complementary tools, in separate subjects to study neural circuits deep in the brain of behaving mammals14,41,63. In these studies, naturally occurring neural dynamics during behaviour provide guidance for subsequent precisely timed optogenetic manipulation to mimic or block endogenous activity. For example, fibre photometry was used to identify differences in NAc D2 dopamine receptor-expressing neuron activity during the decision period before making risky or safe choices, and then optogenetic stimulation was applied only during this decision period to achieve single-trial control of risk-seeking behaviour41 (FIG. 3a). Recently, simultaneous use of optogenetics and Ca2+ recording in the same animals has opened entirely new avenues for studying neural circuits, such as exciting or inhibiting axon terminals while imaging neural activity in downstream cell bodies63, or manipulating one defined population of neurons while imaging a separate group of neurons; any of these populations could be defined for either control or imaging, or both, on the basis of genetics, anatomy or activity history during behaviour.
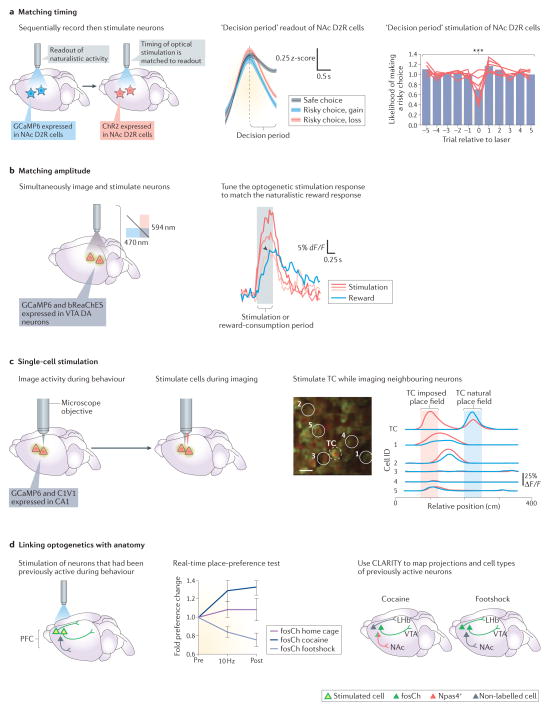
Figure 3. Integrating optogenetic control with optical methods: matching naturally occurring activity patterns and linking to brain-wide projection activity.
a | Matching timing: Ca2+ recording of nucleus accumbens (NAc) D2 dopamine receptor (D2R) neurons expressing the sensor GCaMP6 in one cohort of mice was used to guide optogenetic stimulation timing parameters of channelrhodopsin-expressing NAc D2R neurons in another cohort of mice (left panel). The activity of NAc D2R neurons was recorded during a risky decision-making task (middle panel). The graph shows that the activity during the decision-making period is lower on trials wherein the animal makes a risky versus safe decision, independent of whether the risky decision resulted in a gain or loss. Stimulation of NAc D2R neurons to mimic timing of activity during the task-decision period (using the method shown in panel a) produced an instantaneous, reversible and significant (P < 0.001) reduction in risk seeking (zero timepoint on the x axis); stimulation during other task epochs was much less effective. b | Matching amplitude: this paradigm is designed to simultaneously stimulate and record from ventral tegmental area (VTA) dopaminergic (DA) neurons to match evoked responses to naturally occurring responses (left panel). Right panel: three optogenetically evoked responses (shades of red) could be titrated to closely resemble VTA DA response amplitude to natural reward consumption (blue) in the same animal (in the figure, the black arrowhead indicates similar amplitudes of the evoked response and the natural reward response). c | The left panel shows simultaneous imaging and manipulation of local circuit dynamics in hippocampal CA1 on a cellular level. The example two-photon image shows individual hippocampal neurons expressing GCaMP and C1V1. Cells 1–5 and the target cell (TC) correspond to the traces shown on the right. Optical stimulation of the TC induces network-level changes in the place-cell firing of other neurons, causing some neurons to also fire within the imposed place field of the TC (cells 1 and 2), but not other cells (cells 3–5) (right panel). d | Left panel: integration of optogenetics and activity-dependent immediate-early gene labelling techniques in medial prefrontal cortex (mPFC) populations is shown. Middle panel: compared with the control response in the home cage (fosCh home cage), stimulation of mPFC neurons that had been previously activated by exposure to footshock was sufficient to drive place aversion, whereas stimulation of PFC neurons that had been previously activated by cocaine exposure drove place preference. Right panel: methodologies that enable clearing of brain lipid content after covalent hybridization of other biomolecules to a hydrogel have revealed distinct cell typology and projection patterning between cocaine-activated neurons and footshock-activated neurons. Cocaine-activated neurons (cells expressing the gene neuronal PAS domain-containing protein 4 (Npas4+)) were revealed to have more dense projections to the NAc, whereas footshock-activated neurons project more densely to the lateral habenula (LHb). Part a is adapted with permission from REF. 41, Macmillan Publishers Limited. Part b is adapted with permission from REF. 72, Macmillan Publishers Limited. Part c is adapted with permission from REF. 69, Macmillan Publishers Limited. Part d is adapted with permission from REF. 22, Elsevier.
Perhaps the simplest approach for simultaneously controlling neural activity and monitoring Ca2+ transients in freely behaving mammals is wide-field one-photon activation (through a fibre) of a red-light-excited opsin-expressing population, alongside the use of the same fibre for blue-light-excited GCaMP fluorescence signals to monitor activity. Although this method does not allow control of individual neurons, it is compatible with freely moving animals, maintains genetic specificity, facilitates fast feedback and can be used on multiple brain regions in the same animal, as shown with frame-projected independent fibre photometry (FIP)72. Using FIP, it was possible to record from VTA dopaminergic neurons while stimulating the same neurons with a red-shifted excitatory opsin (bReaChES) for minimal cross-stimulation at the blue GCaMP excitation wavelength. Both perturbing and recording from the same VTA dopaminergic neurons in the same animal enabled titration of optogenetic light power to produce neural activity that matched endogenous VTA dopaminergic responses to reward, thus addressing a key goal of optogenetics: to match optogenetically induced activity to endogenous responses occurring during behaviour (FIG. 3b). Leveraging FIP for accessing multiple regions simultaneously, future developments can now include integration of optogenetics with multisite recording to study natural, causal cell type-specific brain-wide circuit dynamics.
Another method to integrate optogenetics and GECI recording is to use wide-field one-photon optogenetic activation in conjunction with cellular-resolution imaging in the same field; this approach involves a stimulation that is achieved through the same window, cannula or lens that is used to gain optical brain access63. Even more specifically, selective optogenetic stimulation of multiple individual neurons can be carried out while maintaining cellular-resolution Ca2+ imaging of the same and nearby individual neurons67,69,70, enabled by the development of the first red-activated excitatory opsin (C1V1 (REF. 42)) robustly actuated by two-photon illumination126,127. Recently, targeted two-photon stimulation of C1V1 was used in combination with GCaMP two-photon imaging to achieve spectrally separated, all-optical readout and control69,70. In one study using head-fixed mice navigating a virtual reality environment, low-power optogenetic stimulation to a single hippocampal place cell was sufficient to alter firing dynamics and place-field activity of non-stimulated neurons69 (FIG. 3c). Interestingly, these secondary-stimulation effects occurred in neurons that had place fields close to that of the stimulated cell, rather than in neurons that were anatomically in proximity. These results have substantial implications for models determining place-cell dynamics and could not have been obtained without the simultaneity of single-cell optogenetic stimulation and Ca2+ imaging. Newer methods using spatial light modulators70,127 enable recruitment of dozens or more individual neurons at once, which may be important for studying causal effects of multiple individually defined cells on circuit dynamics or behaviour.
Other readout and control modalities
Although combining GECIs and opsins is a powerful approach to circuit analysis, the kinetics of GECIs are too slow to reliably resolve single action potentials in vivo. It may become possible to use genetically encoded voltage indicators (GEVIs) with fast kinetics instead128–130. However, GEVIs result in signal-to-noise ratios that are lower than those resulting from GECIs (membrane-bound, resulting in fewer sensor molecules per cell; and with fast kinetics, demanding high-speed imaging and collection of fewer photons per frame131). Researchers are actively engineering novel variants that may ultimately enable all-optical stimulation and interrogation of large scale networks and microcircuits alike with higher temporal resolution and full spectral separation between sensors and opsins131.
The broad action spectra of microbial opsins may pose fundamental limitations to the number of separable optical channels available for simultaneous control or observation. Adding non-optical axes of control, even if less precise, fast or targetable, could help to enhance increasingly complex forms of optogenetic experimentation. Designer receptors exclusively activated by designer drugs (DREADDs) provide a chemical option, in which G protein-coupled receptors (GPCRs) have been engineered with point mutations to be activated only in the presence of an exogenous ligand (commonly, clozapine-N-oxide). Such ‘chemogenetic’ methods allow activity modulation, although with much reduced spatial and temporal resolution compared with optogenetics132. Non-optical forms of energy delivery have also been explored. In magnetothermal work, magnetic fields cause neuronal depolarization via introduced nanoparticles that generate heat through hysteresis, which in turn activates overexpressed heat-sensitive transient receptor potential cation channels subfamily V member 1 (TRPV1) channels133. These multicomponent systems are currently restricted by limited nanoparticle diffusion in tissue, limited compatibility with free behaviour and potential side effects of TRPV1 expression. Several groups have sought single-component magnetogenetics using non-channel iron-binding proteins, such as ferritin and MagR, to transduce magnetic fields, but possible magnetic or heating mechanisms remain unclear134; moreover, like chemogenetic or ultrasonic135 approaches, the magnetic approaches would be much slower than optogenetics, and also could not be used to control neurons precisely defined by origin and target. Still, efforts like these following the optogenetic principle (genetic introduction of a transducer for external energy) should be pursued to identify new modalities for integrated control.
Activity-guided optogenetic stimulation
As described above, integration of fluorescence-based activity readout with optogenetics now allows experimentally imposed matching of timing and magnitude of activity in cells with particular activity properties during behaviour41,72. Although fluorescence readouts provide high temporal resolution and the option for cellular resolution, these methods do not allow whole-brain single-cell resolution readouts in mammals. Instead, whole-brain single-cell resolution readouts can be achieved by preferentially driving gene expression in previously active neurons via IEG promoter-based transgenic or viral strategies16–22,136,137, and these strategies can be adapted for playing-in activity if the expressed gene is a microbial opsin.
In neurons, IEGs such as FOS or ARC (or engineered promoters such as E-SARE138) are transiently active beginning less than 1 hour after strong bouts of activity or powerful behavioural experiences and therefore are often tracked by investigators using molecular and histochemical approaches (often in combination with optogenetics) to determine which neurons or brain regions were activated by a particular behaviour or intervention (for example, REFS 14,105). Although IEG expression has been demonstrated to correlate with neural activity139, expression can also be seen in non-neuronal cell types. Furthermore, this approach is susceptible to labelling ‘background-activity’ neurons that are active during the broad labelling window of IEGs (on the scale of many hours) but are not involved in the behaviour of interest. However, whereas IEG-driven optogenetics has limited temporal resolution for defining a behavioural experience, the speed of subsequent optogenetic control remains fast, and this approach confers the advantage of simultaneous access to many activity-defined neurons in a large tissue volume, unlike cellular-resolution light delivery. Just as genetically defined and projection-defined optogenetics revealed many previously unknown functions of brain areas and connections in behaviour1, activity-guided optogenetics has the potential for large-volume spatially unbiased discovery of new pathways wherein only a subset of the neurons in a given brain region causally control a behaviour.
In a landmark demonstration of this approach, IEG-defined activity-guided ChR expression was used to preferentially label neurons in the dentate gyrus that were active during the encoding of a fear memory16. The authors designed and used a transgenic mouse strain in which ChR expression from a viral vector was elicited in FOS-expressing dentate neurons activated by fear conditioning. Optogenetic reactivation of these cells was sufficient to induce freezing in a novel, neutral context, showing that contextual memories may be recruited from the dentate gyrus and that stimulation of an ensemble of these neurons is sufficient to reactivate fear behaviour. Importantly, it has been shown that nonspecific stimulation of dentate gyrus granule cells instead disrupts fear-memory encoding and retrieval in mice140, highlighting the value of activity-dependent optogenetics. Related methods have been used not only to reactivate prior fear memory but also to implant a false fear memory in neutral17 or rewarding18 contexts and to improve fear-memory retrieval during chemically induced amnesia20, among other studies19,21,141.
Tissue–hydrogel composites
New techniques allow time-locked labelling of previously activated neurons without use of transgenic mice. Recently, whole-brain optical-access technology via hydrogel embedding142 has been combined with activity-dependent opsin labelling in mice137 to discriminate cell types in the mPFC recruited by behavioural experiences of different valence22. A novel viral vector driving ChR expression under a FOS promoter (linked to a protein-destabilizing sequence for greater temporal precision) enabled temporally precise, activity-dependent labelling of mPFC neurons during either cocaine or footshock exposure and revealed that reactivation of mPFC neurons that had earlier been recruited by footshock induced place aversion, whereas reactivation of neurons recruited earlier by cocaine induced instead place preference. Surprisingly, experimental integration with a hydrogel-based method (in this case, CLARITY24,95) revealed that these two mPFC cell populations not only had different causal effects on behaviour but were in fact different cell types defined by distinct molecular and brain-wide wiring features22 (FIG. 3d). Brain clearing and/or labelling techniques with diverse capabilities continue to be developed that could be partnered with optogenetic techniques (reviewed previously95; see also Supplementary information S1).
Current limitations and future directions
As always, limitations must be considered while designing experiments and interpreting data (reviewed previously1,2,71,95); for example, opsins should be carefully chosen for each application, owing to trade-offs among desirable features (for example, opsins with higher light sensitivity also tend to have slower off-kinetics143). Some limitations specifically affect projection-targeting experiments, such as the time that is required to achieve high levels of opsin expression in long-range axon terminals, or the potential for antidromic spiking that is caused by axon stimulation or nonspecific stimulation of fibres of passage. Integrative activity-tracking solutions can help to circumvent some of these potential caveats. For example, electrophysiology82 and IEG expression measures80 can be used to test whether optogenetic stimulation of axon terminals modulates action potentials in an upstream brain region, and CLARITY-based imaging can be used to demonstrate absence of potentially confounding fibres of passage82. Other controls include verifying that induced effects are abolished by positioning the fibre-optic instead over the potentially confounding fibres of passage themselves82,105 or repeating the experiments while pharmacologically inhibiting different downstream structures to test for blockade of light-induced effects80,144. Another issue that affects projection-targeted experiments is that the most commonly used inhibitory opsins (Cl− or H+ pumps) are less efficient than channels; H+ pumps have an additional issue, as activating these in axons increases spontaneous neurotransmitter release83. Potential solutions include projection targeting by genetics rather than by optics for these cases, so that cell bodies, rather than axons, may be controlled as described above, and future efforts in opsin engineering may continue to advance integrative efforts along these fronts.
Opsin engineering may also be key to solving a remaining challenge for integrating optogenetics with Ca2+ imaging, as all ‘red-shifted’ opsins can still be partially activated by the blue light that is used to image GCaMP2. Although it is possible to circumvent this cross-stimulation issue by titrating down the intensity of GCaMP imaging light69,70,72, this strategy will limit signal-to-noise ratio of Ca2+ transients, and there may also be subthreshold changes in activity that are not detected. Although the use of red Ca2+ indicators with blue-light-activated ChRs could address this issue68,145–147, in general, the red indicators have reduced signal-to-noise ratio and slower kinetics compared with GCaMPs, and some variants exhibit non-Ca2+-related fluorescence changes in response to blue light (photoswitching) 68,147. Future modifications of existing red Ca2+ indicators may allow spectrally independent optical readout and control of neural activity, but currently, issues with photoswitching and photobleaching complicate implementation of this combination in vivo.
The rapidity of targeting-strategy development suggests that the specificity of optogenetic manipulations will continue to grow; emerging opportunities include increasingly diverse AAVs generated by directed evolution89,148 and glycoprotein-deleted rabies viruses generated by pseudotyping94. A second potential future avenue to pursue higher specificity in opsin targeting is to combine different recombinases (for example, Cre, Flp and VCre15) using anatomical and/or genetic criteria to restrict opsin expression to specific cell types and projections. The addition of a third recombinase15 would enable targeting based on intersection of three criteria, and higher targeting specificity can reveal effects not otherwise observable. Methods to express opsins with genetic specificity have led to discoveries about how specific cell types affect behaviour, but future work will need to involve integrated control of multiple cell types, regions and projections. Development of red-shifted opsins and multiregional light-targeting strategies have partially addressed this issue42,63, but spectral overlap among opsins persists, and further discovery and engineering are required for multiple-population control; the development of chimeric ChRs pieced together from known variants149 alongside the ongoing identification of naturally occurring opsins, as well as the structure-guided discovery and utilization of opsin mechanistic principles, may continue to provide new insights and tools.
Beyond precise excitation and inhibition, different kinds of modulation can be achieved. Step-function excitatory and inhibitory tools can alter excitability without directly driving spiking or powerfully hyperpolarizing the membrane, respectively42,43,48, and optogenetic neuromodulation can also be achieved by driving neuromodulatory neurons (for example, dopaminergic14, noradrenergic150, cholinergic77 and peptidergic151 projections). Finally, several strategies have been devised to create neuromodulatory optogenetics in the form of chimaeras of GPCRs and non-microbial rhodopsins, which enable light-activated biochemical signalling152 and have been developed into a generalizable ‘optoXR’ strategy for fast optical control of neuromodulation and behaviour in freely moving animals153 (the X in optoXR stands for the GPCR of interest; these approaches led to the development of tools such as opto-α1AR, opto-β2AR, opto-D1R, and so on, which can be targeted to only those cells naturally expressing the original GPCR using recombinase-dependent AAVs14). For example, the creation and testing of opto-α1AR (which is obtained by replacing the intracellular loops of bovine rhodopsin with those of the mammalian α1 adrenergic receptor) revealed that optical activation of α1 adrenergic receptor pathways in NAc neurons is appetitive by evoked place preference153, whereas optical activation of β2 adrenergic receptor pathways elicits anxiety-related behaviours154. OptoXRs, generated by different groups around the world, also include opto-A2AR (adenosine 2A receptor), opto-μOR (μ-opioid receptor) and opto-mGluR6 (metabotropic glutamate receptor 6; which is obtained by using light-activated domains of the vertebrate melanopsin receptor instead of rhodopsin, resulting in light sensitivity sufficient to partially rescue vision in a mouse model of photoreceptor degeneration155). Rapidly expanding generation and use of optoXRs has opened the door to studying other neurotransmitter systems that are linked to GPCRs, including neuropeptide Y receptors and their ability to regulate pain, obesity and circadian regulation156.
Real-time closed-loop feedback has been performed using optogenetics and electrophysiology115 but has yet to be perfected in all-optical settings with Ca2+ imaging2. By virtue of relative simplicity, fibre photometry provides a ready platform for such real-time feedback control of neural activity, and genetically encoded Ca2+ signals (although slower than electrophysiological signals) enable closed-loop experiments with optogenetics linked to activity in specific cell types or projections. Given the expanded anatomical and genetic toolbox that optogenetics provides, additional possibilities include modulating the activity of one population of neurons on the basis of the Ca2+ readout of a separate population, either in the same or in a different brain region2. The ability to modulate one neural population on the basis of the activity of another population could lead to potential translational insights into disease states such as autism, in which the relative balance of opposing and genetically distinct neural circuit elements is thought to be crucial for maintaining typical behavioural function42. Another avenue would be to use FIP optogenetics to naturalistically stimulate a cell population in one region and to use FIP Ca2+ recording in multiple downstream axon terminal fields to readout activity patterns among different projection neurons. FIP optogenetics could also be used to impose or degrade brain-wide ‘networks’ such as the default mode network or saliency networks or to test causal relevance of brain-wide correlations during behaviour. These and other related approaches may be useful for studying multiple different circuit outputs, as well as for mapping (and testing the causal significance of) brain-wide responses to different behaviours.
Finally, recent developments in brain-clearing techniques22–28 will enable complementary anatomical discoveries to be made22,82,87 (as fibre tracts can now be preserved throughout entire intact brains, and immunostaining22–27 and in situ hybridization28 in this setting can reveal molecular phenotypes of multiple populations of neurons). One pathway to integrate brain clearing with optogenetics and activity imaging would be to first record the activity of a population of neurons during behaviour, selectively manipulate those neurons according to naturally occurring timing and/or magnitude of signals41,72 using cellular-resolution closed-loop optogenetics contingent on activity to evoke changes in behaviour, and then use the tissue–hydrogel method to identify the neurons that were previously imaged and/or optogenetically manipulated and map their brain-wide projection patterns and molecular identities using labelling22 registered back to cellular-resolution activity. Such integrated use of developments in optics, molecular biology, anatomy and behaviour is now feasible, enabling examination of neural circuitry from multiple different perspectives that contribute unique and synergistic information about how cells, connections and circuits modulate local and global network activity in the encoding and causation of behaviour.
Supplementary Material
Acknowledgments
The authors thank the members of the Deisseroth laboratory for helpful discussions; in particular, S.J.Y. and T.J.D. for helpful discussions about fibre photometry analysis. C.K.K. is supported by a National Research Service Award (NRSA) F31 award (NIDA F31DA041795). A.A. is supported by the Walter V. and Idun Berry award, a K99 award (NIMH K99MH106649), and a NARSAD Young Investigator fellowship. K.D. is supported by the National Institutes of Health (NIH), National Science Foundation (NSF), Defense Advanced Research Projects Agency (DARPA) and the Wiegers, Grosfeld, Snyder, Yu, and Woo Foundations.
Glossary
- Fibre-optic patch cord
A flexible and lightweight optical fibre that is used to connect a light source (such as a laser diode or a light-emitting diode (LED)) to a fibre-optic cannula implanted on an animal, allowing light delivery to target cell populations in freely moving animals
- Kinetic opsin variants
Opsin variants that have been engineered to have slower or faster deactivation kinetics, such as the stabilized step-function opsin or ‘ChETA’ (E123T mutation-containing channelrhodopsin) variants, respectively
- Step-function opsins
Opsin proteins with very slow deactivation kinetics, which can thus remain activated for tens of minutes following brief light delivery and can also be switched off in a temporally precise manner with a different wavelength of light
- Red-shifted excitatory opsins
Opsin proteins such as VChR1 and C1V1 that have been discovered and/or engineered to be excited by light of longer wavelengths (that is, red-shifted), in contrast to blue light-activated channelrhodopsins, making them useful for integrating optogenetic excitation with Ca2+ imaging through blue-light-excited GCaMP sensors
- Boolean logic
An algebraic framework in which the basic operations are “OR”, “NOT” and “AND”. These logical operators have been implemented for targeting cell types defined by the presence or absence of multiple features, such as through the use of multiple recombinases and INTRSECT viruses that allow expression of genes encoding opsins in neuronal populations that express Cre NOT Flp recombinase
Footnotes
Author contributions
C.K.K., A.A. and K.D. wrote the paper; C.K.K. and A.A. contributed equally to this work.
Competing interests statement
The authors declare no competing interests.
References
- 1.Deisseroth K. Optogenetics: 10 years of microbial opsins in neuroscience. Nat Neurosci. 2015;18:1213–1225. doi: 10.1038/nn.4091. This recent review covers the history and developments of optogenetics over the past 10 years and addresses potential limitations and standards of practice for application. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grosenick L, Marshel JH, Deisseroth K. Closed-loop and activity-guided optogenetic control. Neuron. 2015;86:106–139. doi: 10.1016/j.neuron.2015.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang F, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravanis AM, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–S156. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- 5.Warden MR, Cardin JA, Deisseroth K. Optical neural interfaces. Annu Rev Biomed Eng. 2014;16:103–129. doi: 10.1146/annurev-bioeng-071813-104733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Montgomery KL, et al. Wirelessly powered, fully internal optogenetics for brain, spinal and peripheral circuits in mice. Nat Methods. 2015;12:969–974. doi: 10.1038/nmeth.3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Park SI, et al. Soft, stretchable, fully implantable miniaturized optoelectronic systems for wireless optogenetics. Nat Biotechnol. 2015;33:1280–1286. doi: 10.1038/nbt.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin D, et al. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–226. doi: 10.1038/nature09736. This study demonstrates the necessity of using cell type-specific optogenetic targeting as opposed to nonspecific electrical stimulation to delineate the hypothalamic neurons that are responsible for controlling aggression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young NP, Deisseroth K. Cognitive neuroscience: in search of lost time. Nature. 2017;542:173–174. doi: 10.1038/nature21497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Licata A, et al. Posterior parietal cortex guides visual decisions in rats. Preprint at. 2016 doi: 10.1523/JNEUROSCI.0105-17.2017. bioRxiv http://dx.doi.org/10.1101/066639. [DOI] [PMC free article] [PubMed]
- 11.Otchy TM, et al. Acute off-target effects of neural circuit manipulations. Nature. 2015;528:358–363. doi: 10.1038/nature16442. [DOI] [PubMed] [Google Scholar]
- 12.Goshen I, et al. Dynamics of retrieval strategies for remote memories. Cell. 2011;147:678–689. doi: 10.1016/j.cell.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 13.Gradinaru V, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunaydin LA, et al. Natural neural projection dynamics underlying social behavior. Cell. 2014;157:1535–1551. doi: 10.1016/j.cell.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenno LE, et al. Targeting cells with single vectors using multiple-feature Boolean logic. Nat Methods. 2014;11:763–772. doi: 10.1038/nmeth.2996. This paper reports a two-virus strategy for delivering a targeted recombinase virus alongside a recombinase-dependent (DIO) opsin-expressing virus; it also reports single viruses implementing Boolean logic on the presence of multiple recombinase types for refined multiple-feature cell type targeting. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–385. doi: 10.1038/nature11028. This study demonstrates optogenetic reactivation of a population of neurons that were labelled by opsin expression during prior experience, using IEG-mediated expression methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramirez S, et al. Creating a false memory in the hippocampus. Science. 2013;341:387–391. doi: 10.1126/science.1239073. [DOI] [PubMed] [Google Scholar]
- 18.Redondo RL, et al. Bidirectional switch of the valence associated with a hippocampal contextual memory engram. Nature. 2014;513:426–430. doi: 10.1038/nature13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Root CM, Denny CA, Hen R, Axel R. The participation of cortical amygdala in innate, odour-driven behaviour. Nature. 2014;515:269–273. doi: 10.1038/nature13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan TJ, Roy DS, Pignatelli M, Arons A, Tonegawa S. Engram cells retain memory under retrograde amnesia. Science. 2015;348:1007–1013. doi: 10.1126/science.aaa5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gore F, et al. Neural representations of unconditioned stimuli in basolateral amygdala mediate innate and learned responses. Cell. 2015;162:134–145. doi: 10.1016/j.cell.2015.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye L, et al. Wiring and molecular features of prefrontal ensembles representing distinct experiences. Cell. 2016;165:1776–1788. doi: 10.1016/j.cell.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiang HL, et al. Manipulating a “cocaine engram” in mice. J Neurosci. 2014;34:14115–14127. doi: 10.1523/JNEUROSCI.3327-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung K, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–337. doi: 10.1038/nature12107. This paper provides the first demonstration of tissue–hydrogel hybrid creation to achieve high-resolution optical access by allowing full delipidation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Renier N, et al. iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell. 2014;159:896–910. doi: 10.1016/j.cell.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 26.Yang B, et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell. 2014;158:945–958. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tomer R, Ye L, Hsueh B, Deisseroth K. Advanced CLARITY for rapid and high-resolution imaging of intact tissues. Nat Protoc. 2014;9:1682–1697. doi: 10.1038/nprot.2014.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sylwestrak EL, Rajasethupathy P, Wright MA, Jaffe A, Deisseroth K. Multiplexed intact-tissue transcriptional analysis at cellular resolution. Cell. 2016;164:792–804. doi: 10.1016/j.cell.2016.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zemelman BV, Lee GA, Ng M, Miesenböck G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 30.Zemelman BV, Nesnas N, Lee GA, Miesenböck G. Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc Natl Acad Sci USA. 2003;100:1352–1357. doi: 10.1073/pnas.242738899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 32.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. This paper offers the first demonstrations of optogenetics using microbial opsins. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- 34.Nagel G, et al. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 35.Li X, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bi A, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae lightgated channels. Neurosci Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Schroll C, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 39.Chow BY, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zalocusky KA, et al. Nucleus accumbens D2R cells signal prior outcomes and control risky decisionmaking. Nature. 2016;531:642–646. doi: 10.1038/nature17400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yizhar O, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477:171–178. doi: 10.1038/nature10360. This study describes the development of C1V1, the first red-light-activated excitatory opsin, which was suitable for integration with blue-light-excited GCaMPs; it also describes the excitatory stabilized step-function opsin SSFO. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berndt A, Lee SY, Ramakrishnan C, Deisseroth K. Structure-guided transformation of channelrhodopsin into a light-activated chloride channel. Science. 2014;344:420–424. doi: 10.1126/science.1252367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wietek J, et al. Conversion of channelrhodopsin into a light-gated chloride channel. Science. 2014;344:409–412. doi: 10.1126/science.1249375. [DOI] [PubMed] [Google Scholar]
- 45.Govorunova EG, Sineshchekov OA, Janz R, Liu X, Spudich JL. Natural light-gated anion channels: a family of microbial rhodopsins for advanced optogenetics. Science. 2015;349:647–650. doi: 10.1126/science.aaa7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wietek J, Broser M, Krause BS, Hegemann P. Identification of a natural green light absorbing chloride conducting channelrhodopsin from Proteomonas sulcata. J Biol Chem. 2016;291:4121–4127. doi: 10.1074/jbc.M115.699637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Govorunova EG, Cunha SR, Sineshchekov OA, Spudich JL. Anion channelrhodopsins for inhibitory cardiac optogenetics. Sci Rep. 2016;6:33530. doi: 10.1038/srep33530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berndt A, et al. Structural foundations of optogenetics: determinants of channelrhodopsin ion selectivity. Proc Natl Acad Sci USA. 2015;113:822–829. doi: 10.1073/pnas.1523341113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gunaydin LA, et al. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 50.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Klapoetke NC, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014;11:338–346. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Berndt A, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci USA. 2011;108:7595–7600. doi: 10.1073/pnas.1017210108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huff ML, Miller RL, Deisseroth K, Moorman DE, LaLumiere RT. Posttraining optogenetic manipulations of basolateral amygdala activity modulate consolidation of inhibitory avoidance memory in rats. Proc Natl Acad Sci USA. 2013;110:3597–3602. doi: 10.1073/pnas.1219593110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 55.Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–278. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- 56.Yamamoto K, et al. Chronic optogenetic activation augments Aβ pathology in a mouse model of Alzheimer disease. Cell Rep. 2015;11:859–865. doi: 10.1016/j.celrep.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 57.Lee JH, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–792. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thanos PK, et al. Mapping brain metabolic connectivity in awake rats with μPET and optogenetic stimulation. J Neurosci. 2013;33:6343–6349. doi: 10.1523/JNEUROSCI.4997-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolodziej A, et al. SPECT-imaging of activitydependent changes in regional cerebral blood flow induced by electrical and optogenetic self-stimulation in mice. Neuroimage. 2014;103:171–180. doi: 10.1016/j.neuroimage.2014.09.023. [DOI] [PubMed] [Google Scholar]
- 60.Ferenczi EA, et al. Prefrontal cortical regulation of brainwide circuit dynamics and reward-related behavior. Science. 2016;351:aac9698. doi: 10.1126/science.aac9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang F, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin JY, Knutsen PM, Muller A, Kleinfeld D, Tsien RY. ReaChR: a red-shifted variant of channelrhodopsin enables deep transcranial optogenetic excitation. Nat Neurosci. 2013;16:1499–1508. [Google Scholar]
- 63.Rajasethupathy P, et al. Projections from neocortex mediate top-down control of memory retrieval. Nature. 2015;526:653–659. doi: 10.1038/nature15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tian L, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–881. doi: 10.1038/nmeth.1398. This study describes the development of GCaMP3-facilitated Ca2+ imaging of activity in awake, behaving mice, allowing for the eventual integration of activity imaging with optogenetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akerboom J, et al. Optimization of a GCaMP calcium indicator for neural activity imaging. J Neurosci. 2012;32:13819–13840. doi: 10.1523/JNEUROSCI.2601-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Szabo V, Ventalon C, De Sars V, Bradley J, Emiliani V. Spatially selective holographic photoactivation and functional fluorescence imaging in freely behaving mice with a fiberscope. Neuron. 2014;84:1157–1169. doi: 10.1016/j.neuron.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Akerboom J, et al. Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Front Mol Neurosci. 2013;6:2. doi: 10.3389/fnmol.2013.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rickgauer JP, Deisseroth K, Tank DW. Simultaneous cellular-resolution optical perturbation and imaging of place cell firing fields. Nat Neurosci. 2014;17:1816–1824. doi: 10.1038/nn.3866. This paper provides the first demonstration of all-optical manipulation at single-cell resolution that provided an activity readout from targeted neurons in an awake, behaving mammal. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Packer AM, Russell LE, Dalgleish HW, Häusser M. Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat Methods. 2015;12:140–146. doi: 10.1038/nmeth.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rajasethupathy P, Ferenczi E, Deisseroth K. Targeting neural circuits. Cell. 2016;165:524–534. doi: 10.1016/j.cell.2016.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim CK, et al. Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain. Nat Methods. 2016;13:325–328. doi: 10.1038/nmeth.3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 74.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, De Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knobloch HS, et al. Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron. 2012;73:553–566. doi: 10.1016/j.neuron.2011.11.030. [DOI] [PubMed] [Google Scholar]
- 76.Mattis J, et al. Frequency-dependent, cell typedivergent signaling in the hippocamposeptal projection. J Neurosci. 2014;34:11769–11780. doi: 10.1523/JNEUROSCI.5188-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vandecasteele M, et al. Optogenetic activation of septal cholinergic neurons suppresses sharp wave ripples and enhances theta oscillations in the hippocampus. Proc Natl Acad Sci USA. 2014;111:13535–13540. doi: 10.1073/pnas.1411233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saunders A, Johnson C, Sabatini B. Novel recombinant adeno-associated viruses for Cre activated and inactivated transgene expression in neurons. Front Neural Circ. 2012;6:47. doi: 10.3389/fncir.2012.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saunders A, et al. A direct GABAergic output from the basal ganglia to frontal cortex. Nature. 2015;521:85–89. doi: 10.1038/nature14179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tye KM, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011;471:358–362. doi: 10.1038/nature09820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jennings JH, Rizzi G, Stamatakis AM, Ung RL, Stuber GD. The inhibitory circuit architecture of the lateral hypothalamus orchestrates feeding. Science. 2013;341:1517–1521. doi: 10.1126/science.1241812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adhikari A, et al. Basomedial amygdala mediates top-down control of anxiety and fear. Nature. 2015;527:179–185. doi: 10.1038/nature15698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mahn M, Prigge M, Ron S, Levy R, Yizhar O. Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat Neurosci. 2016;19:554–556. doi: 10.1038/nn.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soudais C, Laplace-Builhe C, Kissa K, Kremer EJ. Preferential transduction of neurons by canine adenovirus vectors and their efficient retrograde transport in vivo. FASEB J. 2001;15:2283–2285. doi: 10.1096/fj.01-0321fje. [DOI] [PubMed] [Google Scholar]
- 85.Salinas S, et al. CAR-associated vesicular transport of an adenovirus in motor neuron axons. PLoS Pathog. 2009;5:e1000442. doi: 10.1371/journal.ppat.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schwarz LA, et al. Viral-genetic tracing of the input–output organization of a central noradrenaline circuit. Nature. 2015;524:88–92. doi: 10.1038/nature14600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lerner TN, et al. Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell. 2015;162:635–647. doi: 10.1016/j.cell.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Beier KT, et al. Circuit architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell. 2015;162:622–634. doi: 10.1016/j.cell.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tervo DGR, et al. A designer AAV variant permits efficient retrograde access to projection neurons. Neuron. 2016;92:372–382. doi: 10.1016/j.neuron.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stamatakis AM, et al. A unique population of ventral tegmental area neurons inhibits the lateral habenula to promote reward. Neuron. 2013;80:1039–1053. doi: 10.1016/j.neuron.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nieh EH, et al. Decoding neural circuits that control compulsive sucrose seeking. Cell. 2015;160:528–541. doi: 10.1016/j.cell.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lammel S, et al. Input-specific control of reward and aversion in the ventral tegmental area. Nature. 2012;491:212–217. doi: 10.1038/nature11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kiritani T, Wickersham IR, Seung HS, Shepherd GM. Hierarchical connectivity and connection-specific dynamics in the corticospinal–corticostriatal microcircuit in mouse motor cortex. J Neurosci. 2012;32:4992–5001. doi: 10.1523/JNEUROSCI.4759-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reardon TR, et al. Rabies virus CVS-N2c ΔG strain enhances retrograde synaptic transfer and neuronal viability. Neuron. 2016;89:711–724. doi: 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lerner TN, Ye L, Deisseroth K. Communication in neural circuits: tools, opportunities, and challenges. Cell. 2016;164:1136–1150. doi: 10.1016/j.cell.2016.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lo L, Anderson DJ. A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron. 2011;72:938–950. doi: 10.1016/j.neuron.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McGovern A, Davis-Poynter N, Farrell M, Mazzone S. Transneuronal tracing of airways-related sensory circuitry using herpes simplex virus 1, strain H129. Neuroscience. 2012;207:148–166. doi: 10.1016/j.neuroscience.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 98.Beier KT, et al. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci USA. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Marcinkiewcz CA, et al. Serotonin engages an anxiety and fear-promoting circuit in the extended amygdala. Nature. 2016;537:97–101. doi: 10.1038/nature19318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tovote P, et al. Midbrain circuits for defensive behaviour. Nature. 2016;534:206–212. doi: 10.1038/nature17996. [DOI] [PubMed] [Google Scholar]
- 101.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 102.Root DH, et al. Single rodent mesohabenular axons release glutamate and GABA. Nat Neurosci. 2014;17:1543–1551. doi: 10.1038/nn.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matthews GA, et al. Dorsal raphe dopamine neurons represent the experience of social isolation. Cell. 2016;164:617–631. doi: 10.1016/j.cell.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wu Z, et al. GABAergic projections from lateral hypothalamus to paraventricular hypothalamic nucleus promote feeding. J Neurosci. 2015;35:3312–3318. doi: 10.1523/JNEUROSCI.3720-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim SY, et al. Diverging neural pathways assemble a behavioural state from separable features in anxiety. Nature. 2013;496:219–223. doi: 10.1038/nature12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee H, et al. Scalable control of mounting and attack by Esr1+ neurons in the ventromedial hypothalamus. Nature. 2014;509:627–632. doi: 10.1038/nature13169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Stuber GD, et al. Excitatory transmission from the amygdala to nucleus accumbens facilitates reward seeking. Nature. 2011;475:377–380. doi: 10.1038/nature10194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spellman T, et al. Hippocampal–prefrontal input supports spatial encoding in working memory. Nature. 2015;522:309–314. doi: 10.1038/nature14445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Padilla-Coreano N, et al. Direct ventral hippocampal– prefrontal input is required for anxiety-related neural activity and behavior. Neuron. 2016;89:857–866. doi: 10.1016/j.neuron.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lima SQ, Hromádka T, Znamenskiy P, Zador AM. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS ONE. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cohen JY, Haesler S, Vong L, Lowell BB, Uchida N. Neuron-type-specific signals for reward and punishment in the ventral tegmental area. Nature. 2012;482:85–88. doi: 10.1038/nature10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang SJ, et al. Optogenetic dissection of entorhinal-hippocampal functional connectivity. Science. 2013;340:1232627. doi: 10.1126/science.1232627. [DOI] [PubMed] [Google Scholar]
- 113.Cardin JA, et al. Targeted optogenetic stimulation and recording of neurons in vivo using cell-typespecific expression of channelrhodopsin-2. Nat Protoc. 2010;5:247–254. doi: 10.1038/nprot.2009.228. This is a thorough description of phototagging methods to identify specific cell types labelled with opsins in vivo using simultaneous optogenetics and electrophysiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Paz JT, et al. Closed-loop optogenetic control of thalamus as a tool for interrupting seizures after cortical injury. Nat Neurosci. 2013;16:64–70. doi: 10.1038/nn.3269. This paper demonstrates the use of real-time feedback from electrical readouts of cortical activity to trigger optogenetic intervention (in this case, inhibition) to silence both neural activity (seizure-like activity in the thalamus) and behaviour (to block visible signs of seizures) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Krook-Magnuson E, Szabo GG, Armstrong C, Oijala M, Soltesz I. Cerebellar directed optogenetic intervention inhibits spontaneous hippocampal seizures in a mouse model of temporal lobe epilepsy. eNeuro. 2014 doi: 10.1523/ENEURO.0005-14.2014. http://dx.doi.org/10.1523/ENEURO.0005-14.2014. [DOI] [PMC free article] [PubMed]
- 117.Siegle JH, Wilson MA. Enhancement of encoding and retrieval functions through theta phase-specific manipulation of hippocampus. eLife. 2014;3:e03061. doi: 10.7554/eLife.03061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dombeck DA, Harvey CD, Tian L, Looger LL, Tank DW. Functional imaging of hippocampal place cells at cellular resolution during virtual navigation. Nat Neurosci. 2010;13:1433–1440. doi: 10.1038/nn.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Harvey CD, Coen P, Tank DW. Choice-specific sequences in parietal cortex during a virtualnavigation decision task. Nature. 2012;484:62–68. doi: 10.1038/nature10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Flusberg BA, et al. High-speed, miniaturized fluorescence microscopy in freely moving mice. Nat Methods. 2008;5:935–938. doi: 10.1038/nmeth.1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ziv Y, et al. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci. 2013;16:264–266. doi: 10.1038/nn.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schulz K, et al. Simultaneous BOLD fMRI and fiber-optic calcium recording in rat neocortex. Nat Methods. 2012;9:597–602. doi: 10.1038/nmeth.2013. [DOI] [PubMed] [Google Scholar]
- 123.Cui G, et al. Concurrent activation of striatal direct and indirect pathways during action initiation. Nature. 2013;494:238–242. doi: 10.1038/nature11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mandelblat-Cerf Y, et al. Arcuate hypothalamic AgRP and putative POMC neurons show opposite changes in spiking across multiple timescales. eLife. 2015;4:e07122. doi: 10.7554/eLife.07122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Calipari ES, et al. In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc Natl Acad Sci USA. 2016;113:2726–2731. doi: 10.1073/pnas.1521238113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Prakash R, et al. Two-photon optogenetic toolbox for fast inhibition, excitation and bistable modulation. Nat Methods. 2012;9:1171–1179. doi: 10.1038/nmeth.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Packer AM, et al. Two-photon optogenetics of dendritic spines and neural circuits. Nat Methods. 2012;9:1202–1205. doi: 10.1038/nmeth.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.St-Pierre F, et al. High-fidelity optical reporting of neuronal electrical activity with an ultrafast fluorescent voltage sensor. Nat Neurosci. 2014;17:884–889. doi: 10.1038/nn.3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gong Y, Wagner MJ, Li JZ, Schnitzer MJ. Imaging neural spiking in brain tissue using FRETopsin protein voltage sensors. Nat Commun. 2014;5:3674. doi: 10.1038/ncomms4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gong Y, et al. High-speed recording of neural spikes in awake mice and flies with a fluorescent voltage sensor. Science. 2015;350:1361–1366. doi: 10.1126/science.aab0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Vogt N. Voltage sensors: challenging, but with potential. Nat Methods. 2015;12:921–924. doi: 10.1038/nmeth.3591. [DOI] [PubMed] [Google Scholar]
- 132.Lovett-Barron M, et al. Dendritic inhibition in the hippocampus supports fear learning. Science. 2014;343:857–863. doi: 10.1126/science.1247485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen R, Romero G, Christiansen MG, Mohr A, Anikeeva P. Wireless magnetothermal deep brain stimulation. Science. 2015;347:1477–1480. doi: 10.1126/science.1261821. [DOI] [PubMed] [Google Scholar]
- 134.Meister M. Physical limits to magnetogenetics. eLife. 2016;5:e17210. doi: 10.7554/eLife.17210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ibsen S, Tong A, Schutt C, Esener S, Chalasani SH. Sonogenetics is a non-invasive approach to activating neurons in Caenorhabditis elegans. Nat Commun. 2015;6:8264. doi: 10.1038/ncomms9264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Garner AR, et al. Generation of a synthetic memory trace. Science. 2012;335:1513–1516. doi: 10.1126/science.1214985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L. Permanent genetic access to transiently active neurons via TRAP: targeted recombination in active populations. Neuron. 2013;78:773–784. doi: 10.1016/j.neuron.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kawashima T, et al. Functional labeling of neurons and their projections using the synthetic activitydependent promoter E-SARE. Nat Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- 139.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 140.Kheirbek MA, et al. Differential control of learning and anxiety along the dorsoventral axis of the dentate gyrus. Neuron. 2013;77:955–968. doi: 10.1016/j.neuron.2012.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rashid AJ, et al. Competition between engrams influences fear memory formation and recall. Science. 2016;353:383–387. doi: 10.1126/science.aaf0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Deisseroth K. A look inside the brain. Sci Am. 2016;315:30–37. doi: 10.1038/scientificamerican1016-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Mattis J, et al. Principles for applying optogenetic tools derived from direct comparative analysis of microbial opsins. Nat Methods. 2012;9:159–172. doi: 10.1038/nmeth.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Felix-Ortiz AC, et al. BLA to vHPC inputs modulate anxiety-related behaviors. Neuron. 2013;79:658–664. doi: 10.1016/j.neuron.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Inoue M, et al. Rational design of a high-affinity, fast, red calcium indicator R-CaMP2. Nat Methods. 2015;12:64–70. doi: 10.1038/nmeth.3185. [DOI] [PubMed] [Google Scholar]
- 146.Ziegler T, Möglich A. Photoreceptor engineering. Front Mol Biosci. 2015;2:30. doi: 10.3389/fmolb.2015.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Dana H, et al. Sensitive red protein calcium indicators for imaging neural activity. eLife. 2016;5:e12727. doi: 10.7554/eLife.12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deverman BE, et al. Cre-dependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol. 2016;34:204–209. doi: 10.1038/nbt.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang H, et al. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J Biol Chem. 2009;284:5685–5696. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- 150.Carter ME, et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat Neurosci. 2010;13:1526–1533. doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Han S, Soleiman MT, Soden ME, Zweifel LS, Palmiter RD. Elucidating an affective pain circuit that creates a threat memory. Cell. 2015;162:363–374. doi: 10.1016/j.cell.2015.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim JM, et al. Light-driven activation of β2-adrenergic receptor signaling by a chimeric rhodopsin containing the β2-adrenergic receptor cytoplasmic loops. Biochemistry. 2005;44:2284–2292. doi: 10.1021/bi048328i. [DOI] [PubMed] [Google Scholar]
- 153.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 154.Siuda ER, et al. Optodynamic simulation of β-adrenergic receptor signalling. Nat Commun. 2015;6:8480. doi: 10.1038/ncomms9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.van Wyk M, Pielecka-Fortuna J, Löwel S, Kleinlogel S. Restoring the ON switch in blind retinas: opto-mGluR6, a next-generation, cell-tailored optogenetic tool. PLoS Biol. 2015;13:e1002143. doi: 10.1371/journal.pbio.1002143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brothers SP, Wahlestedt C. Therapeutic potential of neuropeptide Y (NPY) receptor ligands. EMBO Mol Med. 2010;2:429–439. doi: 10.1002/emmm.201000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.