Abstract
Neural progenitor cell (NPC) culture within 3D hydrogels is an attractive strategy for expanding a therapeutically-relevant number of stem cells. However, relatively little is known about how 3D material properties such as stiffness and degradability affect the maintenance of NPC stemness in the absence of differentiation factors. Over a physiologically-relevant range of stiffness from ~0.5–50 kPa, stemness maintenance did not correlate with initial hydrogel stiffness. In contrast, hydrogel degradation was both correlated with, and necessary for, maintenance of NPC stemness. This requirement for degradation was independent of cytoskeletal tension generation and presentation of engineered adhesive ligands, instead relying on matrix remodeling to facilitate cadherin-mediated cell-cell contact and promote β-catenin signaling. In two additional hydrogel systems, permitting NPC-mediated matrix remodeling proved to be a generalizable strategy for stemness maintenance in 3D. Our findings have identified matrix remodeling, in the absence of cytoskeletal tension generation, as a previously unknown strategy to maintain stemness in 3D.
Neural progenitor cells (NPCs) are a stem cell population capable of self-renewal and differentiation into the major cell types of the central nervous system1. The potential for NPCs and NPC-derived cells to directly replace damaged tissue and to secrete trophic factors make them attractive therapies for many nervous system disorders1. However, expanding a therapeutically-relevant number of stem cells while retaining their undifferentiated, stem-like phenotype remains a significant challenge2. This is particularly essential for neuronal replacement cell therapies, since once NPCs differentiate down a neuronal lineage, no further cell proliferation can occur. Recently, two separate studies reported decreased regenerative capacity of clinical-grade NPCs relative to research-grade NPCs in pre-clinical rodent models3,4, emphasizing the importance of developing well-defined, scalable systems to expand large numbers of high-quality stem cells for clinical use.
Previous studies on 2D substrates demonstrated that chemical and mechanical matrix cues can maintain stem cells in an undifferentiated state5–8. More recently, 3D materials have been proposed as platforms to expand large numbers of stem cells, as such systems would occupy considerably less space to produce an equivalent number of cells than traditional 2D methods9–12. Thus, the development of 3D materials that support the proliferation and stemness maintenance of NPCs would increase the potential therapeutic relevance of this cell type.
In their native environment, stem cells are maintained in an undifferentiated state by biochemical and biophysical factors collectively termed the stem cell niche, which includes the extracellular matrix (ECM)13–16. Previous studies identified matrix stiffness as a key parameter for maintaining the stemness of several different stem cell populations, including mesenchymal stem cells (MSCs)8,17. However, unlike these other stem cell populations, naïve NPCs are not highly contractile, generating orders of magnitude lower tensional force than MSCs18,19. Thus, we hypothesized that stiffness would not have as profound an effect on NPC stemness in 3D materials as it does for highly contractile stem cell types. This would potentially be different from NPC behavior during differentiation, where it is well-established that lineage specification is biased by matrix stiffness20–24.
For cells embedded in 3D materials, both stiffness and degradability can be critical design variables. Whereas cells cultured in 2D are unrestricted and free to spread, cells embedded within nanoporous 3D hydrogels require matrix remodeling to spread, migrate, and proliferate25,26. In response, proteolytically-degradable hydrogels have been engineered to permit cell proliferation in 3D gels that would not otherwise enable significant cell expansion27,28. As self-renewal is a hallmark of the stem cell phenotype, we further hypothesized that increased 3D matrix degradability may promote NPC proliferation and stemness maintenance.
Here, we investigate the influence of matrix stiffness and degradability on NPC stemness maintenance within 3D protein-engineered hydrogels. We demonstrate that NPC stemness in these materials depends predominantly on degradability over a physiologically relevant range of stiffness (~0.5–50 kPa). In contrast to previous results with differentiating MSCs29, the response of NPCs to matrix degradation is independent of ligand clustering and cytoskeletal tension generation. For NPCs, matrix degradation regulates β-catenin signaling by modulating cadherin-mediated cell-cell contact, emphasizing that matrix degradation can modulate stem cell phenotype through different biochemical mechanisms. To demonstrate the generality of this finding, we employed two additional materials systems: proteolytically-degradable poly(ethylene glycol) hydrogels and physically-remodelable alginate hydrogels. In both systems, stemness varied with matrix remodeling but not stiffness.
NPC stemness is correlated with hydrogel degradability
Modularly designed, elastin-like proteins (ELPs) were utilized to generate a family of hydrogels with a range of stiffness and degradability. These ELPs consist of alternating elastin-like domains and bioactive domains with a cell-adhesive, integrin-binding, arginine-glycine-aspartic acid (RGD) sequence recognized by NPCs30 (Figure 1a). The bioactive domain is also the site within the ELPs that is most susceptible to proteolytic degradation, as elastin-like sequences are relatively insensitive to protease-mediated degradation31. The NPCs used in this study do not secrete significant amounts of active elastase (see Methods), so the majority of NPC-mediated degradation of the ELPs occurs within the bioactive domain. The elastin-like domain provides sites for crosslinking the individual proteins into networks and endows the resulting gels with elastic mechanical properties32.
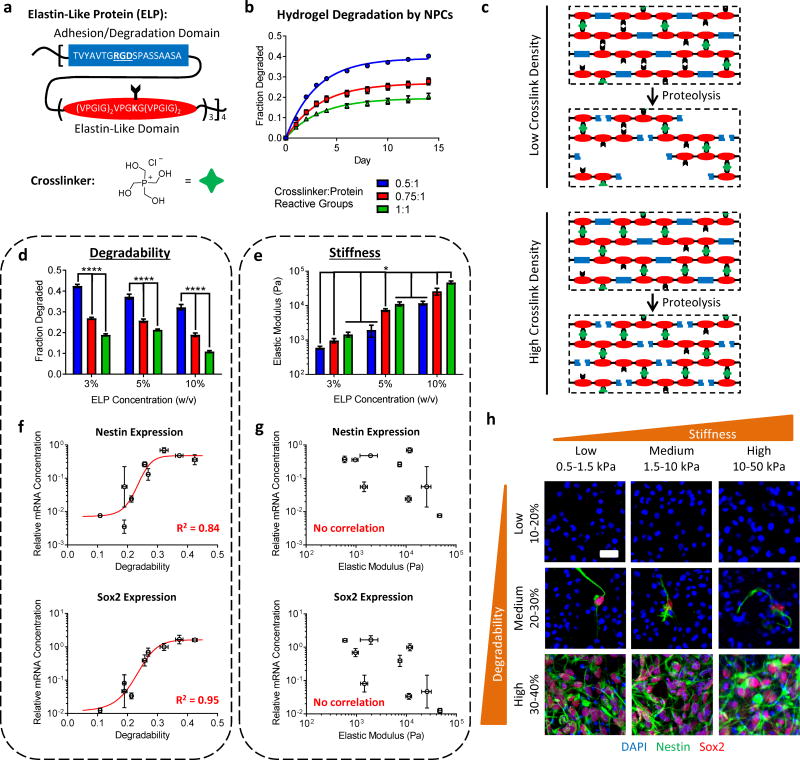
Figure 1. NPC stemness varies as a function of hydrogel degradability.
(a) Modular elastin-like proteins (ELPs) comprised of bioactive domains (containing the integrin-binding RGD motif and protease degradation sites) and of elastin-like structural domains are crosslinked with the amine-reactive crosslinker THPC to produce hydrogels. (b) NPC-mediated degradation of fluorescently-labeled ELP hydrogels is tuned by varying the crosslink density of the network (i.e. the molar ratio of crosslinker to protein reactive sites). A first-order exponential association model was fit to the data to characterize degradation. (c) Simplified schematic depicting the proteolytic degradation of ELP hydrogels by NPC-produced proteases. As there are three crosslinking points in between each bioactive domain, increasing the density of crosslinks in the network decreases the likelihood of any given segment of protein being released from the network, even if all bioactive domains are cleaved. (d) Hydrogel degradability is predominantly tuned by varying the network crosslink density, allowing degradability and initial stiffness to be co-varied. Data are plateau degradation values ± s.e. n = 3 samples per group per time point. ****p< 0.0001, extra sum-of-squares F test. (e) Hydrogels with initial stiffness spanning two orders of magnitude (E~0.5 to 50 kPa) are produced by varying the polymer concentration and network crosslink density. Data are mean ± s.d. n = 3 for 5% and 10% ELP, and 4 for 3% ELP. *p<0.05, one-way ANOVA with Bonferroni post-hoc test. (f) Expression of mRNA for the NPC stemness markers Nestin and Sox2 increases with increasing hydrogel degradability, while (g) Nestin and Sox2 expression is not correlated with initial hydrogel stiffness. Gene expression is presented as geometric mean with 95% confidence intervals. n = 4. (h) Immunocytochemistry confirms that Nestin and Sox2 expression predominantly vary with hydrogel degradability, but not initial hydrogel stiffness. Blue: DAPI (nuclei), Green: Nestin, Red: Sox2. Scale bar: 25 μm.
Because each bioactive domain is separated by three potential crosslinking sites, we hypothesized that the crosslink density could be utilized to tune the maximal degradability of the hydrogel network. To test this, degradation of fluorescently-labeled ELP with encapsulated NPCs was monitored for two weeks. NPCs remained highly viable within the hydrogels in all three crosslink densities (Supplementary Figure 1). As expected, increasing the crosslink density resulted in a decrease in NPC-mediated hydrogel degradation (Figure 1b). Furthermore, the total hydrogel degradation reached distinct plateau values for all three crosslink densities, consistent with the hypothesis that proteolysis occurs primarily in the bioactive domain of the ELPs. At high crosslink densities, a significant fraction of the network will remain intact, even if every proteolytic domain is cleaved (Figure 1c). At low crosslink densities, there is a higher likelihood that portions of the network will become disconnected after proteolytic cleavage, resulting in increased degradation of low crosslink density gels at all polymer concentrations tested (Figure 1d).
Hydrogel stiffness was tuned over two orders of magnitude (E~0.5–50 kPa) by varying both the total polymer content and the crosslink density (Figure 1e). Thus, a family of hydrogels was prepared covering low, medium, and high stiffness ranges, with high, medium, and low degradability gels within each stiffness range (Supplementary Table 1). Changes in NPC stemness within these gels was assessed by measuring mRNA expression for the classical NPC stemness markers Nestin and Sox233,34. Nestin and Sox2 expression increased significantly with increasing hydrogel degradability (p<0.01, Spearman rank correlation, Figure 1f), whereas expression was not correlated with initial hydrogel stiffness (p>0.05, Spearman rank correlation, Figure 1g). Immunostaining confirmed these trends (Figure 1h). Consistent with this observation, at a given polymer concentration, Nestin and Sox2 expression was significantly decreased by increasing crosslink density and thus, decreasing degradability (Supplementary Figure 2a). Furthermore, when comparing gels with similar stiffness but significantly different degradability, Nestin and Sox2 expression varied with degradability but not initial stiffness (Supplementary Figure 2b–d). Taken together, these results indicate that NPC stemness maintenance in these hydrogels correlates with degradation but is insensitive to initial stiffness over a physiologically-relevant range of elastic moduli from ~0.5 to 50 kPa.
NPC stemness maintenance requires matrix degradation
Previous studies on MSCs identified that matrix degradation was required for generation of cytoskeletal tension and subsequent osteogenic differentiation29. Others have demonstrated that clustering of hydrogel-bound, cell-adhesive ligands enhances the osteogenic differentiation of MSCs26,35,36. These results led to the hypothesis that matrix degradation can result in increased ligand clustering, which facilitates cytoskeletal tension generation and osteogenic differentiation37. To determine whether degradation-mediated tension generation was responsible for changes in NPC stemness (Figure 2a), we selected a subset of hydrogels with the stiffness range of brain tissue (E~0.5–1.5 kPa)38,39 (see Supplementary Note 1 and Supplementary Figure 3).
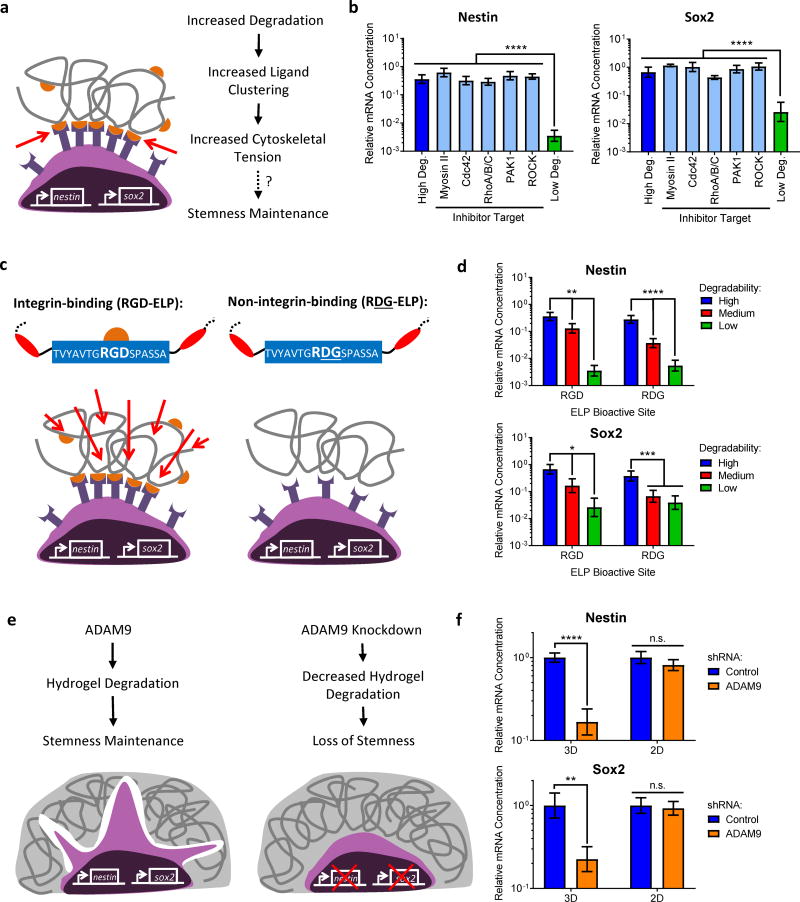
Figure 2. Maintenance of NPC stemness in hydrogels requires matrix degradation but does not depend on cytoskeletal tension or engineered matrix-adhesion ligand interactions.
(a) Schematic depicting a possible role for cytoskeletal tension in directing NPC stemness based on hydrogel degradability. If increased degradability permitted increased ligand clustering, which led to increased cytoskeletal tension and in turn an increase in NPC stemness, we would expect to see a decrease in NPC marker expression in high degradability hydrogels after inhibiting effectors of cytoskeletal tension. (b) However, no decrease in Nestin and Sox2 expression were observed after 7 days of culture in the presence of inhibitors in high degradability hydrogels. (c) Integrin-mediated engagement with the hydrogel matrix is altered by changing the amino acid sequence of the ELP cell-adhesion motif from an integrin-binding RGD sequence to a non-integrin-binding RDG sequence. (d) Expression of mRNA for the NPC markers Nestin and Sox2 in hydrogels with varying degradability, with or without integrin-binding RGD ligands, after 7 days in culture. Nestin and Sox2 expression is increased in hydrogels with increased degradability, irrespective of the presence of integrin-binding ligands. (e) Schematic depicting hypothesis that knocking down ADAM9 will lead to reduced hydrogel degradation, which will in turn lead to decreased NPC stemness. (f) Decreasing hydrogel degradation by shRNA-mediated ADAM9 knockdown decreases expression of Nestin and Sox2 after 3 days in culture in 3D hydrogels. ADAM9 knockdown does not alter expression of the NPC markers Nestin and Sox2 in 2D controls. In b, d, and f, data are presented as geometric means with 95% confidence intervals. n = 4. In b and d, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA with Bonferroni post-hoc test. In f, n.s. = not significant, **p<0.01, ****p<0.0001, two-tailed Student’s t-test.
NPCs in high degradability gels were treated with small molecule inhibitors for effectors of cytoskeletal tension (myosin II, Cdc42, RhoA/B/C, PAK1, and ROCK). Strikingly, inhibition of cytoskeletal tension had no effect on Nestin and Sox2 expression, suggesting that stemness maintenance in high degradability gels was not due to tension generation (Figure 2b). Furthermore, increasing tension in low degradability gels with Rho GTPase activators did not rescue stemness (Supplementary Figure 4). To confirm that ligand clustering was not required for NPC stemness maintenance, cells were encapsulated in ELPs containing a non-integrin-binding, scrambled arginine-aspartic acid-glycine (RDG) sequence (Figure 2c). Nestin and Sox2 trends were similar in the presence or absence of RGD ligands (Figure 2d). Thus, maintenance of NPC stemness in 3D appeared to be insensitive to adhesive ligand concentration and cytoskeletal tension generation. Taken together, these data suggested that NPCs sense changes in matrix degradability by a different mechanism than highly contractile cells such as MSCs.
Next, we sought to confirm that proteolytic degradation of the ELP hydrogels was required for the observed differences in stemness maintenance. We first identified a disintegrin and metalloprotease 9 (ADAM9) as the primary protease involved in NPC-mediated ELP hydrogel remodeling, as described in Supplementary Note 2 (Supplementary Figure 5). As the expression and activity of ADAM9 did not vary with hydrogel degradability or stiffness (Supplementary Figure 6a–d), we hypothesized that knocking down ADAM9 expression in encapsulated NPCs would result in decreased hydrogel degradation, and hence a loss of NPC stemness (Figure 2e).
To test this, NPCs were lentivirally transduced to express a control, non-silencing shRNA or an shRNA targeting ADAM9. As expected, knockdown of ADAM9 resulted in a significant decrease in hydrogel degradation (Supplementary Figure 6e–g). Furthermore, this decrease in hydrogel degradation was correlated with decreased Nestin and Sox2 expression (Figure 2f), supporting the hypothesis that ADAM9-mediated matrix remodeling was responsible for maintenance of NPC stemness in 3D ELP hydrogels. Importantly, ADAM9 knockdown had no impact on NPC stemness in 2D control experiments (Figure 2f), demonstrating that matrix degradation (which is only affected in the 3D experiments) is the primary mechanism by which ADAM9 promotes 3D stemness maintenance. Consistent with these results, protease inhibition also resulted in a significant decrease in degradation (Supplementary Figure 7a,b) and decreased Nestin and Sox2 expression (Supplementary Figure 7c).
NPC self-renewal and differentiation require remodeling
Stem cell maintenance is functionally defined as the capacity for self-renewal and the ability to differentiate into multiple different cell types. Thus, for NPCs, functional outputs of stemness would include proliferation and the ability to differentiate into neural cell types, such as neurons and astrocytes33. Given that NPC stemness markers were correlated with hydrogel degradability, we next tested the hypothesis that matrix remodeling is required for maintenance of NPC stem function. Because hydrogel degradation occurs on the timescale of days (Figure 1b), we anticipated that some time would be required to permit sufficient hydrogel degradation before the NPCs had the capacity to proliferate or the ability to differentiate. Accordingly, NPCs in hydrogels with high degradability were capable of proliferation by 3 days post-encapsulation, whereas NPCs in intermediate degradability hydrogels only exhibited proliferation beginning at day 7 (Figure 3a). NPCs in low degradability hydrogels exhibited no proliferation over 2 weeks. Staining for incorporation of the thymidine analog EdU also indicated enhanced proliferation with increasing degradability (Figure 3b). All newly divided, EdU-positive cells also stained positive for Sox2, indicating symmetric self-renewal and expansion of the stem cell pool (Supplementary Figure 8a,b). Additionally, cells in high degradability gels stained positive for Nestin, but negative for the differentiated neuronal marker microtubule-associated protein 2 (MAP2) and the astrocyte marker glial fibrillary acidic protein (GFAP), further confirming symmetric NPC self-renewal (Supplementary Figure 8c). Blocking hydrogel degradation by ADAM9 knockdown or protease inhibition resulted in decreased proliferation (Supplementary Figure 9). Together, these results indicate that increased matrix remodeling increases NPC self-renewal.
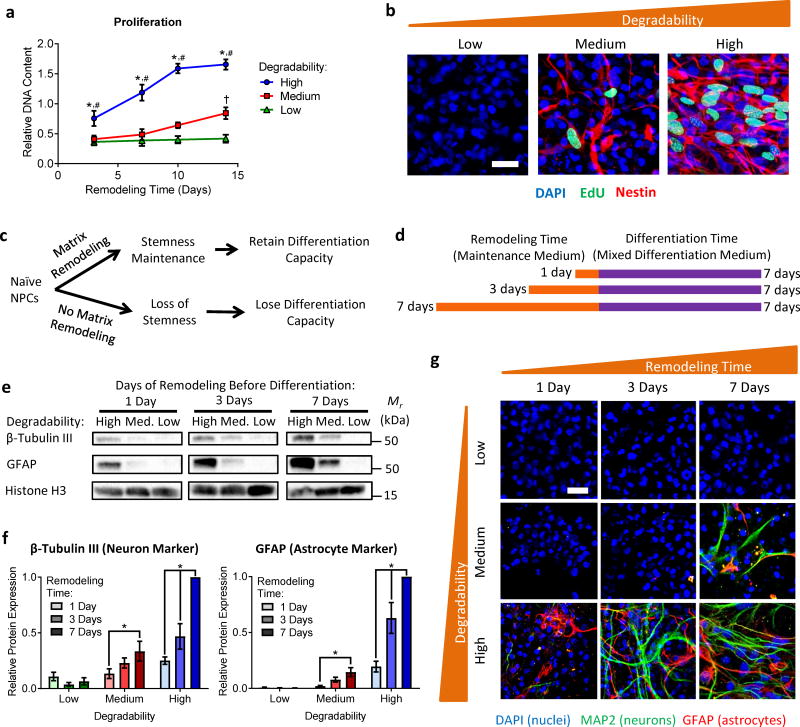
Figure 3. Matrix remodeling is required for NPC proliferation and differentiation capacity.
(a) NPC proliferation increases with increasing hydrogel degradability. Data are mean DNA content relative to low degradability gels at day 3 ± s.e.m. n = 12 for high degradability and 6 for medium and low degradability samples. *p<0.05 for 0.5:1 relative to 0.75:1, #p<0.05 for 0.5:1 relative to 1:1, and †p<0.05 for 0.75:1 relative to 1:1 at a given time point (two-way ANOVA with Bonferroni post-hoc test). (b) Increased DNA synthesis between days 6 and 7 in culture, visualized via EdU incorporation, is observed with increasing hydrogel degradability, confirming enhanced proliferation in gels with increased matrix remodeling. Blue: DAPI (nuclei), Green: EdU, Red: Nestin. Scale bar: 25 μm. (c) Schematic depicting the hypothesis that matrix remodeling is necessary for stemness maintenance and thus required for NPCs to maintain differentiation capacity. (d) To assess the effect of matrix remodeling on NPC differentiation capacity, encapsulated NPCs were cultured in maintenance medium and permitted to remodel the hydrogels for 1, 3, or 7 days prior to an additional 7 days of culture in mixed differentiation medium. (e) Representative Western blots for β-tubulin III (neuron marker) and GFAP (astrocyte marker) expression by NPCs after 1, 3, or 7 days of remodeling followed by 7 days of differentiation. Histone H3 was used as a loading control. (f) Western blot quantification reveals increased differentiation marker expression with both increasing hydrogel degradability and with increased remodeling time. Data are mean ± s.e.m., normalized to Histone H3. n = 4 for β-tubulin III and 5 for GFAP. *p<0.05, two-way ANOVA with Bonferroni post-hoc test. (g) Immunocytochemistry confirms increased capacity for differentiation into neurons and astrocytes with increased remodeling time in high degradability hydrogels. Blue: DAPI (nuclei), Green: MAP2 (neurons), Red: GFAP (astrocytes). Scale bar: 25 μm.
To determine if matrix remodeling also impacted NPC differentiation capacity, NPCs were provided a variable amount of time in which to remodel the matrix prior to induction of differentiation (Figure 3c). NPCs in hydrogels with varying degradability were cultured for 1, 3, or 7 days in maintenance medium prior to culture in mixed differentiation medium for an additional 7 days (Figure 3d). Strikingly, both hydrogel degradability and remodeling time prior to induction of differentiation had significant impacts on NPC differentiation capacity. Extent of differentiation was quantified by Western blot for the neuronal marker β-tubulin III and the astrocytic marker GFAP. NPCs in high degradability gels exhibited the most differentiation regardless of the duration of remodeling time prior to induction (Figure 3e,f). Furthermore, increased remodeling time significantly increased differentiation in both high and intermediate degradability hydrogels. Immunocytochemistry confirmed these results (Figure 3g). Consistent with previous NPC studies using hydrogels spanning this stiffness range, a mix of neurons and astrocytes was observed with no substantial bias towards a particular lineage20,40. Taken together, these results indicate that matrix remodeling is required for maintenance of NPC stemness function (i.e., self-renewal and differentiation capacity) in 3D hydrogels.
Remodeling facilitates cadherin-mediated β-catenin signaling
To investigate the mechanism by which matrix remodeling modulates stemness, we encapsulated GFP-positive NPCs in Cy5-labeled ELP hydrogels to image material remodeling. In low degradability gels, NPCs did not substantially remodel the hydrogels, remaining rounded and isolated (Figure 4a). In contrast, NPCs in high degradability gels reorganized the matrix, permitting significant cell spreading by 3 days post-encapsulation (Figure 4b). Nestin and Sox2 expression tracked closely with matrix remodeling, and no recovery of expression over time was observed in low degradability gels (Figure 4c). However, in high degradability gels, Nestin and Sox2 expression decreased 1 day after encapsulation but recovered by day 3 once remodeling had occurred (Figure 4d). Thus, rounded, isolated cells appear to lose stemness, but regain stemness once the material has been sufficiently remodeled to permit spreading.
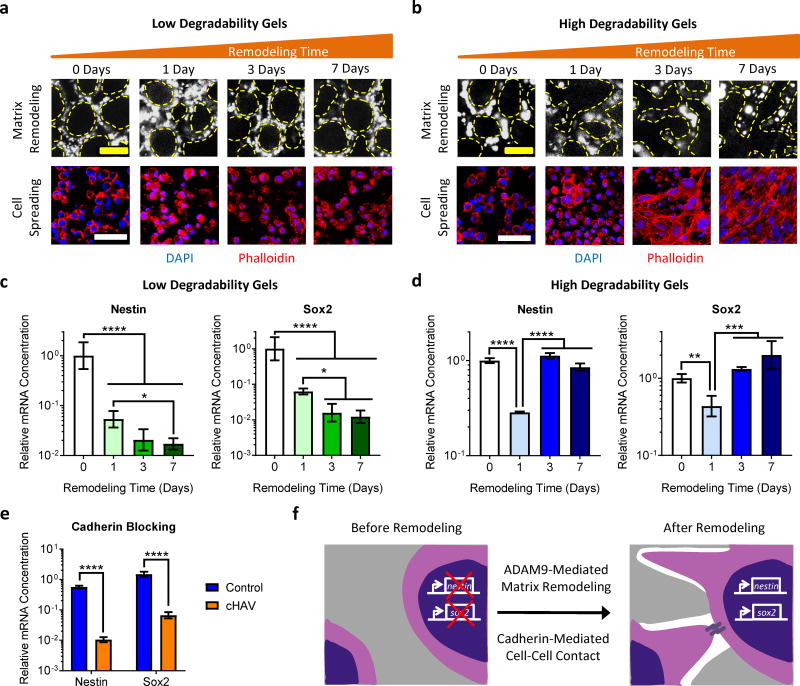
Figure 4. Matrix remodeling regulates NPC stemness by modulating cadherin-mediated cell-cell contact.
(a) Low degradability gels exhibit little remodeling over 7 days in culture, as visualized by confocal microscopy of fluorescently-labeled ELP gels. Without matrix remodeling, the cells are unable to spread and make contacts. (b) In contrast, high degradability ELP gels exhibit significant remodeling over time, resulting in significant cell spreading and cell-cell contact. In a and b, the dashed yellow lines denote the interface of the NPCs with the ELP material. Blue: DAPI, Red: Phalloidin. Scale bars: 10 μm for matrix remodeling, 50 μm for cell spreading. (c) Nestin and Sox2 expression is suppressed at all time points in low degradability gels that do not permit substantial matrix remodeling and cell spreading. (d) Expression of Nestin and Sox2 in high degradability gels is initially suppressed as the cells are confined but rebounds to baseline levels after sufficient matrix remodeling permits cell spreading. (e) Treatment with a cadherin-blocking cyclic-HAV peptide (cHAV) suppresses Nestin and Sox2 expression in high degradability hydrogels after 7 days in culture. (f) Schematic depicting the proposed mechanism through which ADAM9-mediated matrix remodeling regulates NPC stemness by modulating cadherin-mediated cell-cell contact. In c-e, data are presented as geometric means with 95% confidence intervals. n = 3 for cHAV treatment and 4 for all other conditions. In c and d, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, one-way ANOVA with Bonferroni post-hoc test. In e, ****p<0.0001, two-tailed Student’s t-test.
One consequence of enhanced cell spreading is an increase in cell-cell contact. Previous embryogenesis studies demonstrated that cadherin-mediated cell-cell contact can modulate neural progenitor phenotype during development41,42. To test the hypothesis that matrix remodeling enhances stemness by facilitating cadherin-mediated contact, NPCs in high degradability gels were treated with an inhibitor of cadherin binding (cyclic HAV peptide). Cadherin-blocked samples lost Nestin and Sox2 expression similar to cells in matrices that do not permit remodeling (Figure 4e). Because both N- and E-cadherins can play a role in NPC development41,42, their expression was assessed by qRT-PCR and Western blot. The adult NPCs used here predominantly express N-cadherin (Supplementary Figure 10), suggesting that N-cadherin-mediated contact is required for stemness maintenance. An alternative approach to modulating cell-cell contact in 3D hydrogels is to vary the density of encapsulated cells. Within high degradability gels, varying cell density had no effect on stemness maintenance as expected (Supplementary Figure 11a), since cells can extend processes through the remodeled gels to make contact even at low cell density. In contrast, within less degradable gels, increasing cell density resulted in a modest increase in Nestin and Sox2 expression, presumably due to increased probability of random cell-cell contact (Supplementary Figure 11b,c). Together, these results suggest that matrix remodeling facilitates cadherin-mediated cell-cell contact, which in turn promotes stemness maintenance (Figure 4f).
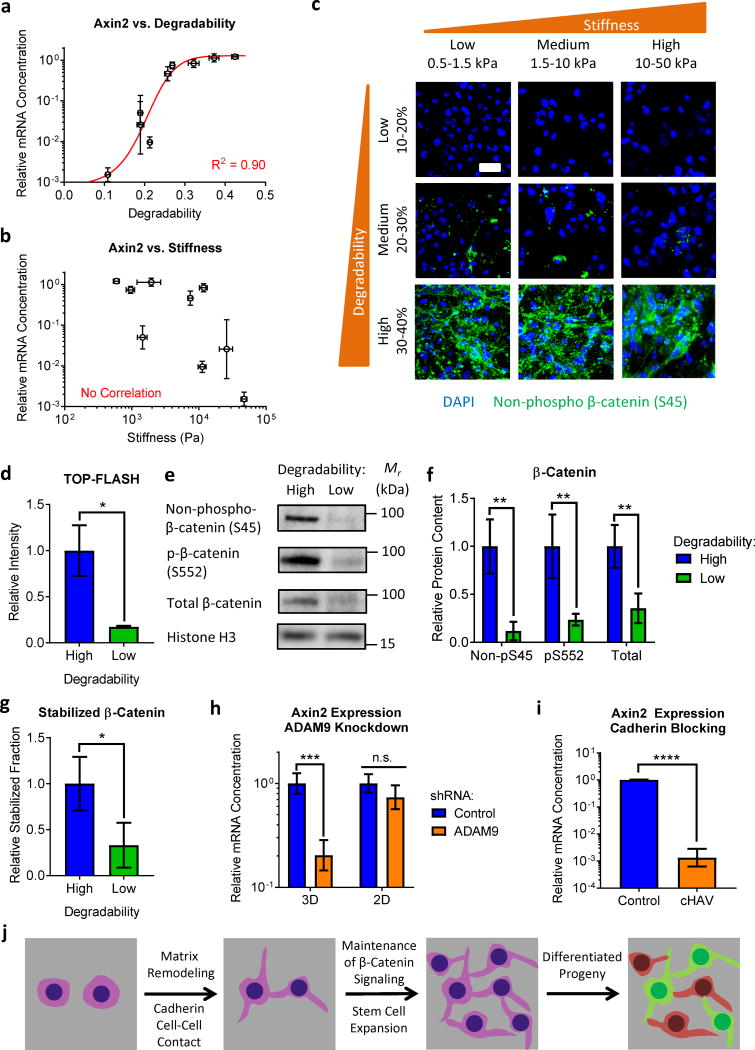
N-cadherin cell-cell contacts can modulate β-catenin signaling41, and β-catenin activation may enhance neural progenitor stemness43. We therefore hypothesized that matrix remodeling was exerting control over NPC stemness by sustaining β-catenin signaling through N-cadherin contacts. Consistent with this, expression of the β-catenin responsive gene Axin2 was strongly correlated with hydrogel degradability (p<0.001, Spearman rank correlation, Figure 5a) but not stiffness (p>0.05, Spearman rank correlation, Figure 5b). At a given stiffness, Axin2 expression was higher in more degradable hydrogels (Supplementary Figure 12). Consistent with these results, immunostaining revealed increased levels of stabilized β-catenin with increased hydrogel degradability across the full range of stiffness (Figure 5c). To confirm β-catenin transcriptional regulation, NPCs were transfected with the TOP-FLASH β-catenin reporter construct. As expected, high degradability gels exhibited significantly more β-catenin-mediated transcription (Figure 5d). Consistent with N-cadherin-mediated regulation of β-catenin signaling, NPCs in high degradability gels exhibited increased levels of stabilized (non-phospho-S45) β-catenin and activated (phospho-S552) β-catenin (Figure 5e–g). Accordingly, blocking cell-cell contact by preventing hydrogel degradation or inhibiting cadherin binding resulted in decreased β-catenin signaling (Figure 5h,i). Therefore, matrix remodeling permits increased cadherin-mediated cell-cell contact, activating β-catenin signaling and maintaining NPC stemness (Figure 5j).
Figure 5. Matrix remodeling modulates β-catenin signaling via cadherin contacts to promote NPC stemness.
(a) Expression of mRNA for the β-catenin responsive gene Axin2 increases with increasing hydrogel degradability, while (b) Axin2 expression is not correlated with initial hydrogel stiffness. (c) Immunocytochemistry confirms that β-catenin activity predominantly varies with hydrogel degradability, but not initial hydrogel stiffness. Blue: DAPI (nuclei), Green: non-phospho β-catenin (S45). Scale bar: 25 μm. (d) NPCs encapsulated in high degradability gels exhibit increased β-catenin-driven transcriptional activity compared to cells in low degradability gels, as measured by the TOP-FLASH reporter assay. (e,f) NPCs in high degradability gels have increased levels of stabilized (non-phospho S45), activated (phospho-S552), and total β-catenin than NPCs in low degradability gels, quantified from Western blot analysis. (g) The fraction of β-catenin in the stabilized (non-phospho S45) state is increased in high degradability gels. (h) Blocking hydrogel degradation by ADAM9 knockdown results in decreased Axin2 expression in 3D gels, but not in 2D control cultures. (i) Inhibiting cadherin-mediated cell-cell contact in high degradability gels with the cHAV peptide results in decreased Axin2 expression. (j) Schematic depicting matrix remodeling regulating β-catenin signaling and stemness maintenance by modulating cadherin-mediated cell-cell contact. In a, b, g, and h, gene expression data are presented as geometric mean with 95% confidence intervals. n = 3 for cHAV and 4 for all other conditions. In d, data are mean ± s.e.m. n = 5. In f and g, data are presented as mean ± s.d. n = 4. In d and f-i, n.s. = not significant, *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001, two-tailed Student’s t-test.
Chemical or physical remodeling can regulate NPC stemness
The ELP hydrogels were deliberately designed to allow stiffness and degradability to be co-varied over a broad range. Biochemical methods then allowed for the effects of stiffness and degradability to be separately interrogated. We next sought to confirm the generality of these results to other biomaterial systems. Specifically, we designed two hydrogel families, one based on covalent crosslinking and one based on physical crosslinking, where stiffness and remodeling could be tuned independently.
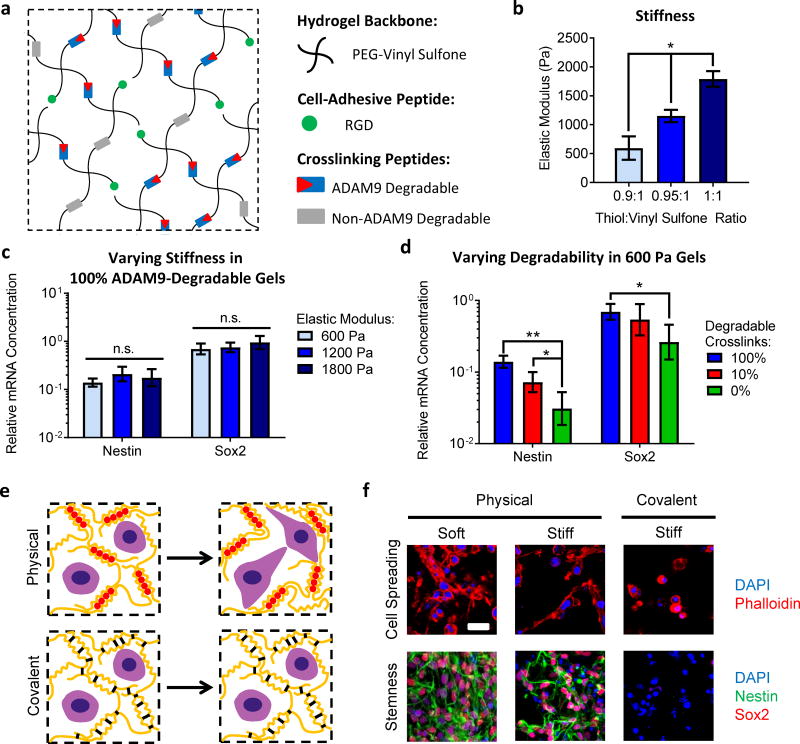
In the first system, ADAM9-degradable, RGD-presenting poly(ethylene glycol) (PEG) hydrogels were prepared with tunable stiffness in the same physiologically-relevant range for NPCs as the ELP hydrogels (E~600–1800 Pa) (Figure 6a,b)38,39. ADAM9-degradability was tuned by varying the fraction of crosslinking peptides that were susceptible to cleavage by ADAM9, and initial stiffness was tuned by varying the overall crosslink density. For NPCs in hydrogels with 100% degradable crosslinks, varying hydrogel stiffness did not alter NPC stemness (Figure 6c). However, even for NPCs in the softest hydrogels, decreasing gel degradability resulted in decreased stemness (Figure 6d). Furthermore, across all stiffness and degradability combinations, Nestin and Sox2 were significantly correlated with degradability but not initial stiffness (Pearson correlation, Supplementary Figure 13). Therefore, in a second material system, NPC stemness was insensitive to initial matrix stiffness over a neural-relevant range but required matrix degradation in 3D. These results demonstrate that control over ADAM9-degradability is a viable strategy for maintaining NPC stemness in covalently-crosslinked hydrogels including both protein-engineered and synthetic polymeric hydrogels.
Figure 6. NPC stemness maintenance varies with remodeling, but not initial stiffness, in both covalently- and physically-crosslinked hydrogels.
(a) Covalently-crosslinked hydrogels with independently tunable ADAM9-degradability and stiffness were prepared by crosslinking 4-arm PEG vinyl sulfone with ADAM9-cleavable and non-ADAM9-cleavable peptides. Hydrogels were also functionalized with RGD peptides to permit integrin-mediated cell-matrix interactions. Hydrogel degradability was tuned by varying the ratio of ADAM9-cleavable to non-cleavable crosslinking peptides. (b) Hydrogel elastic modulus was tuned by varying the total amount of crosslinking peptides (i.e., the molar ratio of thiols on the peptide crosslinkers to vinyl sulfone groups on the PEG macromers). PEG hydrogel stiffness was optimized to cover the same range of physiologically-relevant stiffness tested in the 3% (w/v) ELP hydrogels. (c) Expression of mRNA for the NPC markers Nestin and Sox2 does not vary with hydrogel stiffness in gels that are 100% ADAM9 degradable. (d) However, decreasing degradability even in the softest (E~600 Pa) hydrogels results in decreased Nestin and Sox2 expression. In b, data are presented as mean ± s.d. n = 3. In c and d, data are presented as geometric mean with 95% confidence intervals. n = 4. *p<0.05, **p<0.01, and n.s. = not significant (p>0.05), one-way ANOVA with Bonferroni post-hoc test. (e) Schematic depicting remodeling of physically-crosslinked alginate hydrogels compared to non-remodelable, covalently-crosslinked gels. (f) Physically crosslinked alginate hydrogels facilitate matrix remodeling, cell spreading, and maintenance of Nestin and Sox2 expression at two different stiffness values, while covalent crosslinking blocks matrix remodeling and cell spreading and results in a loss of stemness. Scale bar: 25 μm.
To further demonstrate the generalizability to other biomaterial systems, we next considered whether other modes of matrix remodeling could be used to maintain NPC stemness. We hypothesized that physically-crosslinked gels that undergo physical remodeling of the polymer network in the absence of proteolytic degradation would permit cell spreading and cell-cell contact and hence promote NPC stemness maintenance. Physically-crosslinked alginate gels previously were shown to undergo physical network remodeling to enable 3D cell spreading26. Consistent with results in ELP and PEG materials, NPCs in physically-crosslinked alginate gels maintained Nestin and Sox2 expression irrespective of hydrogel stiffness (Figure 6e–f, Supplementary Figure 14). In contrast, inhibiting physical remodeling by adding covalent crosslinks into the alginate material inhibited cell spreading and decreased NPC stemness (Figure 6e–f, Supplementary Figure 14). Together, these results indicate that matrix remodeling, either through chemical proteolysis or physical network remodeling, facilitates NPC stemness maintenance in 3D hydrogels.
Outlook
These studies have identified matrix remodeling as a requirement for NPC stemness maintenance in 3D hydrogels. Hydrogel remodeling can be achieved either through chemical degradation of covalently-crosslinked networks or physical remodeling of physically-crosslinked networks. Using three different biomaterial platforms, we demonstrated that both remodeling strategies can be used to enable cell spreading, permit cell-cell contact, and initiate downstream β-catenin signaling that maintains NPC stemness. Utilizing matrix remodeling to maintain β-catenin signaling is an attractive strategy to scale up production of NPCs, as these materials permit cell-cell contact even at low initial cell densities without the need to add expensive, exogenous β-catenin agonists. For example, the identification of ADAM9 as a regulator of matrix remodeling may allow for the design of 3D covalently-crosslinked materials for expansion of clinically-relevant numbers of functionally active NPCs. Finally, because remodeling-mediated maintenance of stemness appears to be orthogonal to hydrogel stiffness, mechanosensitive differentiation of NPCs could potentially be employed in tandem with expansion of NPCs in remodelable gels to generate large numbers of differentiated cells within a single material platform.
Methods
Synthesis and characterization of ELP hydrogels
ELPs were expressed in BL21(DE3)pLysS Escherichia coli (Life Technologies), based on a previously published procedure44. Briefly, ELPs were cloned into pET15b plasmids and expressed under control of the T7 promoter. Bacteria containing the plasmids were cultured in Terrific Broth to an OD600 of 0.8, and expression was induced by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 7 hours, the bacteria were harvested by centrifugation, resuspended in TEN buffer (10 mM Tris, 1 mM EDTA, and 100 mM NaCl, pH 8.0), and lysed by repetitive freeze-thaw cycles. Lysates were treated with DNAse I and 1 mM phenylmethanesulfonyl fluoride (PMSF) protease inhibitor. ELPs were purified by inverse temperature cycling and desalted by dialyzing against deionized water. Lyophilization afforded the ELP as a white solid. Protein purity was confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
ELP hydrogels were prepared by dissolving the ELP in phosphate-buffered saline (PBS) to a concentration of 3.75%, 6.25%, or 12.5% (w/v) and then adding the appropriate amount of THPC crosslinker dissolved in PBS to the ELP solution in a 1:4 (THPC solution:ELP solution) volumetric ratio, resulting in hydrogels with a final ELP concentrations of 3%, 5%, or 10% (w/v), respectively. This solution was transferred to a cylindrical silicone mold, and hydrogels were allowed to crosslink at room temperature for 15 minutes, followed by an additional incubation at 37°C for 15 minutes before being submerged in either PBS (for materials characterization) or culture medium (for NPC culture).
For mechanical testing, ELP gels were crosslinked in 8 mm dia. × 2.5 mm thick silicone molds and equilibrated in PBS at 37°C for one hour. The gels were removed from the molds and maintained in PBS at 37°C during the testing. The hydrogel elastic modulus in unconfined compression was determined by generating stress-strain curves to 15% strain using an ARG2 rheometer. Mass swelling ratios were determined by dividing the wet mass of the hydrogels by the dry mass of the hydrogels after lyophilization.
To assess NPC-mediated hydrogel degradation, ELPs were labeled with Cy5 fluorescent dye by reacting Cy5-NHS (Lumiprobe) with ELP in anhydrous DMSO with triethylamine as a basic catalyst. Sufficient Cy5-NHS was added to label at least one primary amine per ELP molecule. Cy5-labeled ELP was purified by dialysis and lyophilized. NPCs were encapsulated and cultured in these gels as described below, and the phenol red-free culture medium was collected and replaced every 1–2 days. After 14 days, the hydrogels were completely degraded by trypsinization, and the fluorescence intensity in the collected medium and trypsinized samples was used to determine the fraction of hydrogel degraded.
Hydrogel microstructure was assessed using label-free coherent anti-Stokes Raman scattering (CARS) microscopy, probing the carbon-hydrogen vibrational mode at 2930 cm−1 to visualize the protein distributions in the ELP samples45. The CARS microscope setup is described in detail elsewhere46. The system is comprised of a Nd:Vanadate laser (Picotrain, HighQ Lasers GmbH, Hohenems, Austria) generating two picosecond pulsed laser beams (532 and 1064 nm, 7 ps, 76 MHz) and an optical parametric oscillator (Levant Emerald OPO, Angewandte Physik & Elektronik GmbH, Berlin, Germany, 690–900 nm). The 532 nm beam pumps the OPO, the output of which was tuned to 811 nm to drive the 2930 cm−1 vibration in the ELP sample. The OPO beam was temporally and spatially overlaid with the 1064 nm beam from the Nd:Vanadate laser, directed into the microscope (Nikon Eclipse TE2000-E, Nikon) through a mirror scanning unit and focused on the sample by an objective (Nikon Plan Fluor 40x/1.30 oil). CARS signals emitted from the focal volume at 656 nm were detected using a single-photon counting photomultiplier tube (PMT; PMC-100-1, Hamamatsu, Japan). Samples were hydrated with PBS and maintained at 37°C throughout imaging. For each sample, a volume of 50 μm × 50 μm × 20 μm was imaged. 3D reconstructions were produced using Volocity software. Fraction polymer-lean space was quantified from individual z-slices using ImageJ software (NIH) to determine the fraction of the imaged area that was not positive for the CARS signal.
Macromolecular diffusivity within the hydrogels was assessed by fluorescence recovery after photobleaching (FRAP), based on a previously published procedure47. Briefly, ELP hydrogels were prepared and then incubated overnight in 4 mg/mL solutions of fluorescein isothiocyanate (FITC) labeled dextrans with molecular weights of 20, 40, 70, 150, 250, and 500 kDa (Sigma). Using a Leica SPE confocal microscope, a 100 μm × 100 μm area of the sample was photobleached (488 nm laser, 100% intensity, 60 seconds). Fluorescent recovery was monitored over the course of 2 minutes with a 1.6 second frame rate. Effective diffusion coefficients were calculated using the open source Matlab code “frap_analysis”48.
Cell culture
Adult murine NPCs from micro-dissected dentate gyrus were kindly provided by Prof. Theo Palmer (Stanford Neurosurgery)49, following a protocol approved by the Stanford Administrative Panel on Laboratory Animal Care. The National Institutes of Health (NIH) guidelines for the care and use of laboratory animals (NIH Publication #85–23 Rev. 1985) were observed. NPCs were expanded in maintenance medium (Neurobasal-A, 2% B27 Supplement, GlutaMAX (Life Technologies), 20 ng/mL FGF-2, and 20 ng/mL EGF (PeproTech)) on polyornithine and laminin-coated tissue culture plastic, following a previously described protocol50. For encapsulation in hydrogels, NPCs were lifted by trypsinization, pelleted, resuspended, and counted. The NPCs were pelleted again and resuspended in a solution of ELP to achieve a cell density of 5×107 cells/mL in the final hydrogels (i.e., after addition of the THPC crosslinker solution). The hydrogels were then crosslinked as described above, cast in 4 mm dia. × 0.5 mm thick silicone molds. Culture medium was replaced every two days. To induce differentiation, NPCs were treated with mixed differentiation medium20, consisting of Neurobasal-A, 2% B27 Supplement, GlutaMAX, 1% fetal bovine serum (FBS), and 1 μM retinoic acid.
Biochemical analysis of stemness markers
For proliferation and cytotoxicity assays, NPC-containing hydrogels were removed from their silicone molds, transferred to lysis buffer (20 mM Tris HCl, 150 mM NaCl, 0.5% Triton X-100, pH 7.4), and disrupted by sonication. For cytotoxicity assays, the culture medium was also collected. DNA content was determined using the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies), following the manufacturer’s instructions. Cytotoxicity was assessed using the CytoTox Glo Cytotoxicity Assay (Promega), following the manufacturer’s instructions. Cytotoxicity was also assessed by Live/Dead staining (Life Technologies), imaged on a Leica SPE confocal microscope.
For mRNA expression analysis, NPC-containing hydrogels were removed from their silicone molds, transferred to TRIzol reagent (Life Technologies), and disrupted by sonication. RNA was isolated by phenol-chloroform extraction using Phase Lock gels (5 Prime). One μg of RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For quantitative polymerase chain reaction (qPCR), 1 μg of the resulting cDNA in 5 μL of nuclease free water was mixed with 10 μL of Fast SYBR Green Master Mix (Applied Biosystems) and run on an Applied Biosystems StepOnePlus Real Time PCR System. Primers are listed in Supplementary Table 2.
For Western blot analysis, NPC-containing hydrogels were removed from their molds, transferred to RIPA buffer (Cell Signaling Technology) supplemented with 1 mM PMSF and protease inhibitor tablets (Roche), and disrupted by sonication. Lysates were clarified by centrifugation. Twenty μg of protein were separated by SDS-PAGE and transferred to PVDF membranes. Membranes were blocked with 5% non-fat milk (NFM) in tris-buffered saline with 2% Tween-20 (TBST) for 1 hour at room temperature and then incubated with primary antibodies diluted in NFM/TBST overnight at 4°C (see Supplementary Table 3 for primary antibodies and dilutions). Membranes were washed with TBST and then incubated with HRP-conjugated secondary antibodies (donkey anti-mouse, donkey anti-rabbit, or donkey anti-chicken, Jackson ImmunoResearch, 1:10,000) for 1 hour at room temperature. The membranes were washed with TBST, developed using SuperSignal West Pico or Femto Chemiluminescent Substrates (Pierce), and imaged using a ChemiDoc MP gel imaging system (Bio-Rad). Densitometry analysis was performed using ImageJ, and protein expression was normalized to expression of Histone H3 as a loading control. Full size Western blots are presented in Supplementary Figure 15.
For immunocytochemistry, NPC-containing hydrogels were fixed with 4% paraformaldehyde in PBS at 37°C for 30 minutes. Samples were permeabilized with PBS plus 0.25% Triton-X (PBST) for 1 hour at room temperature and blocked with 5% bovine serum albumin (BSA) and 5% goat serum (GS) in PBST for 3 hours at room temperature. The samples were then incubated with primary antibodies diluted in PBST with 2.5% BSA and 2.5% GS overnight at 4°C (see Supplementary Table 3 for primary antibodies and dilutions). Samples were thoroughly washed with PBST and then incubated with AlexaFluor-conjugated secondary antibodies (AF488 goat anti-mouse, AF546 goat anti-rabbit (Life Technologies), or AF647 goat anti-chicken (Abcam)) and 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) as a nuclear counterstain overnight at 4°C. Samples were washed thoroughly with PBST and mounted using VECTASHIELD HardSet Antifade Mounting Medium (Vector Laboratories). For the EdU incorporation assay, cell culture medium was supplemented with 10 μM F-ara-EdU51 (Sigma) on day 6, and samples were incubated with 10 μM AF488-azide (Life Technologies) in 100 mM Tris, 1 mM copper(II) chloride, and 100 mM ascorbic acid, pH 8.5 for 1 hour at room temperature prior to blocking52. Samples were imaged using a Leica SPE confocal microscope.
Cytoskeletal tension modulation experiments
NPCs were encapsulated in ELP hydrogels as described above. For tension inhibition studies, NPCs were cultured in maintenance medium treated with inhibitors against myosin II (blebbistatin, 1 μM), Cdc42 (ML141, 10 μM), RhoA/B/C (Rhosin, 30 μM), PAK1 (IPA-3, 10 μM), and ROCK (Y-27632, 10 μM) for 7 days, with inhibitor-treated medium replaced every 2 days. Inhibitors, concentrations, and targets were chosen based on previous studies investigating 2D NPC mechanosensing21. For tension activation studies, cultures in maintenance medium were treated with Rho Activator II and Rho/Rac/Cdc42 Activator I (Cytoskeleton, Inc.) at 1 μg/mL for 7 days, with activator-treated medium replaced every 2 days. Expression of Nestin and Sox2 mRNA was assessed using qRT-PCR as described above.
Identification of ADAM9 as protease responsible for hydrogel degradation
To identify proteases produced by NPCs that may be responsible for NPC-mediated hydrogel degradation, NPCs were encapsulated in ELP hydrogels, cultured in maintenance medium for 2 days, and transferred into antibody array lysis buffer (1% Igepal CA-630, 20 mM Tris, 137 mM NaCl, 10% glycerol, 2 mM EDTA, pH 8.0 supplemented with protease inhibitor tablets (Roche)). Hydrogels were disrupted by sonication and used in the protease antibody array kit (R&D Systems), following the manufacturer’s instructions. Day 2 was chosen as the time point for protease analysis as the greatest amount of hydrogel degradation occurred around this time (Figure 1b). Mean pixel intensity of the antibody array spots was determined using ImageJ.
To identify which of the candidate proteases identified by the antibody array analysis was responsible for hydrogel degradation, protease inhibition studies were performed using NPCs encapsulated in Cy5-labeled ELP hydrogels (prepared as described above). Inhibitors against serine, cysteine, and aspartic proteases and aminopeptidases were purchased as part of the ProteSEEKER kit and used at the manufacturer’s supplied concentrations. Inhibitors of Zn2+ metalloproteases were used at previously reported concentrations (doxycycline (100 μM), GM60001 (25 μM), PD166793 (100 μM), batimastat (100 μM), TAPI-0 (50 μM))47,53. Due to possible off-target effects of doxycycline on mitochondrial activity54, additional inhibitors were included to cover all Zn2+ metalloproteases identified by the antibody array. After 2 days, the fluorescence in the medium was measured to determine relative degradation. As degradation was reduced by treatment with batimastat, which is known to inhibit ADAM9 activity55, but not the broad MMP inhibitor GM6001, which shows poor ADAM9 inhibition56, we concluded that ADAM9 is likely the protease primarily responsible for the NPC-mediated hydrogel degradation.
To measure ADAM9 activity, hydrogels with encapsulated NPCs were transferred into ADAM9 activity assay buffer (25 mM Tris, 2.5 μM ZnCl2, 0.005% Brij-35, pH 9.0) and disrupted by sonication. Lysates were incubated with a fluorogenic ADAM9 peptide substrate (Mca-PLAQAV-Dpa-RSSSR; R&D Systems) at 37°C. Fluorescence was read every 5 minutes for 1 hour, and the time point for determination of relative activity was chosen so that the fluorescence signal had not yet saturated. Relative ADAM9 activity in lysates was normalized to DNA content.
To confirm that NPCs did not produce significant amounts of elastase, elastase activity from both 2D monolayer cultured NPCs and NPCs encapsulated in ELP hydrogels was measured using the SensoLyte Green Elastase Assay Kit (AnaSpec). NPCs were lysed into lysis buffer as above, and samples were also collected from conditioned cell culture medium. The elastase activity assay was run following the manufacturer’s instructions, and fluorescence levels were compared to elastase standards of known activity. Neither cell lysates nor conditioned medium exhibited any detectable elastase activity for NPCs cultured in 2D or 3D.
ADAM9-mediated degradation of ELP
To confirm that ADAM9 can cleave ELP, 18 μg of recombinant mouse ADAM9 (R&D Systems) was added to 50 μL of a 50 mg/ml solution of ELP dissolved in ADAM9 activity assay buffer. The mixture was allowed to incubate a room temperature for 2 days with constant agitation. ELP dissolved in ADAM9 activity assay buffer without addition of rmADAM9 was included as a control. After 2 days, the ELP solutions were separated by SDS-PAGE, transferred to a PVDF membrane, and stained by Coomassie blue to visualize protein bands. The band corresponding to the lowest molecular weight degradation product was cut out and submitted to the Stanford Protein and Nucleic Acid Facility for N-terminal Edman sequencing to identify the ADAM9 cleavage site in ELP.
ADAM9 inhibition and knockdown studies
To inhibit metalloprotease-mediated hydrogel degradation, NPCs encapsulated in high degradability ELP hydrogels were cultured in maintenance medium treated with doxycycline (100 μM) and batimastat (100 μM). Cell culture medium with inhibitors was replaced every 2 days. Cumulative hydrogel degradation after 14 days and relative DNA content and expression of Nestin and Sox2 mRNA after 7 days were measured as described above.
Plasmids for lentiviral transduction of non-silencing and ADAM9-knockdown shRNAs (pGIPZ) were purchased from GE Healthcare (see Supplementary Table 4 for silencing RNA sequences). The pGIPZ plasmids also encode for puromycin resistance and green fluorescent protein (GFP) to allow for selection of transduced cells. To generate lentivirus, pGIPZ plasmids were co-transfected with MISSION Lentiviral Packaging Mix (Sigma) into HEK 293FT cells (Life Technologies) using FuGENE 6 Transfection reagent (Promega), following the manufacturers’ protocols. After 24 hours, the medium on the 293FTs was changed from the standard Dulbecco’s Modified Eagle Medium (DMEM) plus 10% FBS to a serum-free medium consisting of DMEM/F12 medium with PeproGrow-1 Serum Free Cell Culture Supplements (PeproTech), GlutaMAX, and non-essential amino acids for collection of lentivirus. After 24 hours, the lentivirus-conditioned medium was collected and mixed with fresh NPC maintenance medium in a 3:2 volume ratio. This mixture was then applied to NPCs cultured on polyornithine and laminin-coated tissue culture plastic at approximately 70% confluency. After 2 days, the medium was changed to standard NPC maintenance medium with 0.6 μg/mL puromycin. After 3 passages, cells were sorted by FACS to enrich for highly GFP expressing cells, as these cells should also express the highest levels of shRNA, to obtain maximum knockdown efficiency. Knockdown was validated at the mRNA level using qRT-PCR and at the protein level by Western blot. ADAM9-knockdown and non-silencing control NPCs were encapsulated in high degradability ELP gels as described above and cultured in maintenance medium plus 0.6 μg/mL puromycin, with medium replaced every 2 days. Cumulative hydrogel degradation after 14 days and relative DNA content expression of Nestin and Sox2 mRNA after 3 days were measured as described above.
Cadherin blocking studies
For pan-cadherin blocking studies, NPCs were lifted by trypsinization and incubated in maintenance medium with 1 mg/mL cHAV peptide (Exherin, AdooQ Bioscience) at 37°C for 30 minutes with constant agitation. Peptide-treated NPCs were collected by centrifugation and encapsulated in ELP hydrogels as described above. Encapsulated cells were cultured in maintenance medium supplemented with 1 mg/mL cHAV for 7 days, with cHAV-containing medium replaced every 2 days. NPCs treated with DMSO were used as controls.
TOP-FLASH assay
The TOP-FLASH plasmid was kindly provided by Prof. Calvin Kuo (Stanford Medicine). NPCs were seeded on laminin-coated tissue culture plastic and cultured for 2 days in maintenance medium. Lipoplexes for plasmid transfection were prepared by incubating TOP-FLASH plasmid (15 μg per 10 cm dish) with TransIT-LT1 reagent (Mirus Bio) in Opti-MEM at room temperature for 30 minutes in a ratio of 4 μL TransIT-LT1 to 1 μg of DNA. Lipoplexes were added dropwise into cell culture plates with fresh maintenance medium. Cells were cultured for an additional 24 hours in the presence of the lipoplexes before being collected and encapsulated in ELP hydrogels as described above. After 3 days in culture, gels were transferred to Passive Lysis Buffer (Promega) and disrupted by sonication. Luciferase activity was measured using a firefly luciferase assay kit (Promega).
Synthesis and characterization of ADAM9-degradable PEG hydrogels
PEG hydrogels were prepared from 20 kDa, 4-arm PEG vinyl sulfone (JenKem Technology) and thiol-containing, crosslinking peptides, based on a previously published procedure57. ADAM9 cleavable (Ac-GCGPLA↑QAVRSSSRGCG-OH) and scrambled, non-ADAM9-cleavable (Ac-GCGAPSQSVRARSLGCG-OH) crosslinking peptides and RGD cell adhesive peptides (Ac-GCGGGRGDSP-OH) were synthesized by GenScript. The ↑ symbol refers to the predicted ADAM9 cleavage site, based on previous reports55. PEG vinyl sulfone was dissolved to 10% (w/v) in triethanolamine-buffered saline (0.3 M, pH 8.0), and peptides were dissolved in PBS to a concentration of 25 mM. The volume of the PEG solution was adjusted with PBS so that the PEG concentration in the final gels was 2.5% (w/v), and the solution of RGD peptide was mixed with the solution of PEG to afford an RGD concentration in the final gels of 1 mM. Finally, the solution of crosslinking peptides at the appropriate molar ratio of thiol:vinyl sulfone (to control gel stiffness) and of ADAM9-cleavable to non-ADAM9-cleavable peptides (to control gel degradability) was mixed with the RGD+PEG solution, and the gels were crosslinked at 37°C for 30 minutes. Mechanical testing was performed as described above. For cell encapsulation studies, NPCs were collected, counted, and pelleted as described for the ELP hydrogels. Immediately after addition of the crosslinking peptides, the NPC pellet was resuspended in the pre-gel solution to a final concentration of 5×107 cells/mL, and the gels were cast in cylindrical silicone molds as above.
Synthesis and characterization of physically- and chemically-crosslinked alginate gels
Alginate was purchased from FMC Biopolymer (LF20/40) and prepared as described previously26. The peptide sequence GGGGRGDSP was coupled to alginate using carbodiimide chemistry and used at 1 mM final concentration for all gels58. For physically-crosslinked gels, low molecular weight alginate at a final concentration of 2% (w/v) was mixed with 12.2 mM CaSO4 for soft gels and 48.8 mM CaSO4 for stiff gels.
Covalently-crosslinked alginate gels were prepared using biocompatible click reactions as previously described59. Tetrazine-amine (Click Chemistry Tools) and 1-bicyclo[2.2.1]hept-5-en-2-ylmethanamine (norbornene, Matrix Scientific) were separately conjugated to alginate using carbodiimide chemistry. Briefly, 1 g of high molecular weight alginate was dissolved in 100 mL of MES buffer (pH 6.5). In quick succession, 2.17 g N-hydroxysuccinimide (NHS), 2.92 g 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), and either 211.8 mg tetrazine-amine or 139.5 mg norbornene-methylamine were added to the alginate solution and stirred for 24 hours. Hydroxylamine hydrochloride was used to quench the reaction, and the solutions were dialyzed using 10 kDa MWCO tubing for 3 days. The solutions were then sterile filtered and lyophilized. To form covalently crosslinked hydrogels, tetrazine-modified alginate and norbornene-modified alginate were mixed to give a 1:1 ratio of tetrazine to norbornene at a final alginate concentration of 3.5% (w/v). To maintain a constant RGD concentration of 1 mM, RGD-coupled alginate was blended with non-RGD-coupled alginate.
Mechanical testing of the alginate gels was performed using oscillatory shear rheometry. Time sweeps were performed to observe gelation (1% strain, 1 rad/s, 25 mm parallel plate geometry), and stress relaxation tests were performed with 15% constant strain. For cell culture experiments, NPCs were suspended in the alginate solutions prior to gelation and cultured as described above.
Statistical analysis
Comparisons between two experimental groups were made using two-tailed Student’s t-tests. Comparisons among more than two experimental groups with a single varying parameter were performed using one-way analysis of variance (ANOVA) with Bonferroni post-hoc testing. Comparisons among more than two experimental groups with two varying parameters were performed using two-way ANOVA with Bonferroni post-hoc testing. P-values of less than 0.05 were considered statistically significant. Independent biological replicates were used to determine n values. For Western blots, to permit comparisons across multiple blots in replicate experiments, the pixel intensity of all bands was normalized to the high degradability samples. To determine statistical significance for these experiments, one sample Student’s t-tests were performed with the null hypothesis that normalized protein expression was equal to 1 (i.e., the expression in the high degradability hydrogels). For analysis of mRNA expression, because the ΔCT values approximate a normal distribution, statistical analysis was performed prior to transforming to a natural scale. Relative mRNA expression was therefore reported as a geometric mean with asymmetric 95% confidence intervals derived from the non-transformed data60. Only changes in mRNA expression greater than 2-fold were considered significant. For representative immunostaining images, experiments were repeated at least twice. Exact P-values for each data set are reported in Supplementary Table 5. All statistical analyses were performed using GraphPad Prism 6 software. No statistical methods were used to determine sample size.
Supplementary Material
Acknowledgments
The authors thank Prof. Theo Palmer and Dr. Harish Babu (Stanford Neurosurgery) for providing the murine NPCs, Amy Proctor (Stanford Chemical Engineering) for assistance with ELP expression and purification, Karen Dubbin (Stanford Materials Science & Engineering) for assistance with FRAP, and Prof. Calvin Kuo (Stanford Medicine) for providing the TOP-FLASH plasmid. Sorting of the lentivirally transduced NPCs for the ADAM9 knockdown experiments was performed with the assistance of Cathy Crumpton and Bianca Gomez on an instrument in the Stanford Shared FACS Facility obtained using NIH S10 shared instrument grant S10RR025518-01. C.M.M. acknowledges support from an NIH NRSA pre-doctoral fellowship (F31 EB020502) and the Siebel Scholars Program. This work was supported by funding from the National Institutes of Health (S.C.H.: U19 AI116484 and R21 EB018407), National Science Foundation (S.C.H.: DMR 1508006), California Institute for Regenerative Medicine (S.C.H.: RT3-07948), and Trygger Foundation (A.E.).
Footnotes
Author Contributions
Experiments were designed by C.M.M. and S.C.H. and carried out by C.M.M., B.L.L., R.E.D., C.B.D., R.S.S., M.K., K.J.L., and D.N. CARS experiments were performed with D.N. and A.E. Alginate hydrogel experiments were performed with R.S.S. and O.C. The manuscript was written by C.M.M. and S.C.H. The principal investigator is S.C.H.
Competing Financial Interests
The authors declare no competing financial interests.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1.Goldman S. Stem and progenitor cell–based therapy of the human central nervous system. Nat Biotechnol. 2005;23:862–871. doi: 10.1038/nbt1119. [DOI] [PubMed] [Google Scholar]
- 2.Chen KG, Mallon BS, McKay RDG, Robey PG. Human pluripotent stem cell culture: Considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14:13–26. doi: 10.1016/j.stem.2013.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson AJ, Piltti KM, Hooshmand MJ, Nishi RA, Cummings BJ. Preclinical efficacy failure of human CNS-derived stem cells for use in the pathway study of cervical spinal cord injury. Stem Cell Rep. 2017;8:249–263. doi: 10.1016/j.stemcr.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh SE, et al. HuCNS-SC human NSCs fail to differentiate, form ectopic clusters, and provide no cognitive benefits in a transgenic model of Alzheimer’s disease. Stem Cell Rep. 2017;8:235–248. doi: 10.1016/j.stemcr.2016.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Azarin SM, Palecek SP. Matrix revolutions: A trinity of defined substrates for long-term expansion of human ESCs. Cell Stem Cell. 2010;7:7–8. doi: 10.1016/j.stem.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Dzhoyashvili NA, Shen S, Rochev YA. Natural and synthetic materials for self-renewal, long-term maintenance, and differentiation of induced pluripotent stem cells. Adv Healthcare Mater. 2015;4:2342–2359. doi: 10.1002/adhm.201400798. [DOI] [PubMed] [Google Scholar]
- 7.Lutolf MP, Doyonnas R, Havenstrite K, Koleckar K, Blau HM. Perturbation of single hematopoietic stem cell fates in artificial niches. Integr Biol. 2009;1:59–69. doi: 10.1039/b815718a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilbert PM, et al. Substrate elasticity regulates skeletal muscle stem cell self-renewal in culture. Science. 2010;329:1078–1081. doi: 10.1126/science.1191035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDevitt TC. Scalable culture of human pluripotent stem cells in 3D. Proc Natl Acad Sci USA. 2013;110:20852–20853. doi: 10.1073/pnas.1320575111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lei Y, Schaffer DV. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc Natl Acad Sci USA. 2013;110:E5039–E5048. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siti-Ismail N, Bishop AE, Polak JM, Mantalaris A. The benefit of human embryonic stem cell encapsulation for prolonged feeder-free maintenance. Biomaterials. 2008;29:3946–3952. doi: 10.1016/j.biomaterials.2008.04.027. [DOI] [PubMed] [Google Scholar]
- 12.Gerecht S, et al. Hyaluronic acid hydrogel for controlled self-renewal and differentiation of human embryonic stem cells. Proc Natl Acad Sci USA. 2007;104:11298–11303. doi: 10.1073/pnas.0703723104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrison SJ, Spradling AC. Stem cells and niches: Mechanisms that promote stem cell maintenance throughout life. Cell. 2008;132:598–611. doi: 10.1016/j.cell.2008.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keung AJ, Kumar S, Schaffer DV. Presentation counts: Microenvironmental regulation of stem cells by biophysical and material cues. Annu Rev Cell Dev Biol. 2010;26:533–556. doi: 10.1146/annurev-cellbio-100109-104042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Discher DE, Mooney DJ, Zandstra PW. Growth Factors, Matrices, and Forces Combine and Control Stem Cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little L, Healy KE, Schaffer D. Engineering Biomaterials for Synthetic Neural Stem Cell Microenvironments. Chem Rev. 2008;108:1787–1796. doi: 10.1021/cr078228t. [DOI] [PubMed] [Google Scholar]
- 17.Yang C, Tibbitt MW, Basta L, Anseth KS. Mechanical memory and dosing influence stem cell fate. Nat Mater. 2014;13:645–652. doi: 10.1038/nmat3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi P, Shen K, Ghassemi S, Hone J, Kam LC. Dynamic Force Generation by Neural Stem Cells. Cell Mol Bioeng. 2009;2:464–474. doi: 10.1007/s12195-009-0097-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gershlak JR, et al. Mesenchymal Stem Cells Ability to Generate Traction Stress in Response to Substrate Stiffness is Modulated by the Changing Extracellular Matrix Composition of the Heart During Development. Biochem Biophys Res Commun. 2013;439:161–166. doi: 10.1016/j.bbrc.2013.08.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saha K, et al. Substrate Modulus Directs Neural Stem Cell Behavior. Biophys J. 2008;95:4426–4438. doi: 10.1529/biophysj.108.132217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keung AJ, de Juan-Pardo EM, Schaffer DV, Kumar S. Rho GTPases Mediate the Mechanosensitive Lineage Commitment of Neural Stem Cells. Stem Cells. 2011;29:1886–1897. doi: 10.1002/stem.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teixeira AI, et al. The promotion of neuronal maturation on soft substrates. Biomaterials. 2009;30:4567–4572. doi: 10.1016/j.biomaterials.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Leipzig ND, Shoichet MS. The effect of substrate stiffness on adult neural stem cell behavior. Biomaterials. 2009;30:6867–6878. doi: 10.1016/j.biomaterials.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 24.Banerjee A, et al. The influence of hydrogel modulus on the proliferation and differentiation of encapsulated neural stem cells. Biomaterials. 2009;30:4695–4699. doi: 10.1016/j.biomaterials.2009.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baker BM, Chen CS. Deconstructing the third dimension – How 3D culture microenvironments alter cellular cues. J Cell Sci. 2012;125:3015–3024. doi: 10.1242/jcs.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhuri O, et al. Hydrogels with tunable stress relaxation regulate stem cell fate and activity. Nat Mater. 2016;15:326–334. doi: 10.1038/nmat4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel PN, Gobin AS, West JL, Patrick CW. Poly(ethylene glycol) hydrogel system supports preadipocyte viability, adhesion, and proliferation. Tissue Eng. 2005;11:1498–1505. doi: 10.1089/ten.2005.11.1498. [DOI] [PubMed] [Google Scholar]
- 28.Bott K, et al. The effect of matrix characteristics on fibroblast proliferation in 3D gels. Biomaterials. 2010;31:8454–8464. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 29.Khetan S, et al. Degradation-mediated cellular traction directs stem cell fate in covalently crosslinked three-dimensional hydrogels. Nat Mater. 2013;12:458–465. doi: 10.1038/nmat3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saha K, Irwin EF, Kozhukh J, Schaffer DV, Healy KE. Biomimetic interfacial interpenetrating polymer networks control neural stem cell behavior. J Biomed Mater Res Part A. 2007;81A:240–249. doi: 10.1002/jbm.a.30986. [DOI] [PubMed] [Google Scholar]
- 31.Mithieux SM, Weiss AS. Elastin. Adv Protein Chem. 2005;70:437–461. doi: 10.1016/S0065-3233(05)70013-9. [DOI] [PubMed] [Google Scholar]
- 32.Chung C, Lampe KJ, Heilshorn SC. Tetrakis(hydroxymethyl) phosphonium chloride as a covalent cross-linking agent for cell encapsulation within protein-based hydrogels. Biomacromolecules. 2012;13:3912–3916. doi: 10.1021/bm3015279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKay R. Stem cells in the central nervous system. Science. 1997;276:66–71. doi: 10.1126/science.276.5309.66. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed S. The culture of neural stem cells. J Cell Biochem. 2009;106:1–6. doi: 10.1002/jcb.21972. [DOI] [PubMed] [Google Scholar]
- 35.Huebsch N, et al. Harnessing traction-mediated manipulation of the cell/matrix interface to control stem-cell fate. Nat Mater. 2010;9:518–526. doi: 10.1038/nmat2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong X, Yang F. Sliding hydrogels with mobile molecular ligands and crosslinks as 3D stem cell niche. Adv Mater. 2016;28:7257–7263. doi: 10.1002/adma.201601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vincent LG, Engler AJ. Stem cell differentiation: Post-degradation forces kick in. Nat Mater. 2013;12:384–386. doi: 10.1038/nmat3636. [DOI] [PubMed] [Google Scholar]
- 38.Gefen A, Margulies SS. Are in vivo and in situ brain tissues mechanically similar? J Biomech. 2004;37:1339–1352. doi: 10.1016/j.jbiomech.2003.12.032. [DOI] [PubMed] [Google Scholar]
- 39.Taylor Z, Miller K. Reassessment of brain elasticity for analysis of biomechanisms of hydrocephalus. J Biomech. 2004;37:1263–1269. doi: 10.1016/j.jbiomech.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 40.Lim TC, Toh WS, Wang LS, Kurisawa M, Spector M. The effect of injectable gelatin-hydroxyphenylpropionic acid hydrogel matrices on the proliferation, migration, differentiation and oxidative stress resistance of adult neural stem cells. Biomaterials. 2012;33:3446–3455. doi: 10.1016/j.biomaterials.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 41.Zhang J, et al. Cortical neural precursors inhibit their own differentiation via N-cadherin maintenance of β-catenin signaling. Dev Cell. 2010;18:472–479. doi: 10.1016/j.devcel.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karpowicz P, et al. E-cadherin regulates neural stem cell self-renewal. J Neurosci. 2009;29:3885–3896. doi: 10.1523/JNEUROSCI.0037-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 44.Straley KS, Heilshorn SC. Independent tuning of multiple biomaterial properties using protein engineering. Soft Matter. 2009;5:114–124. [Google Scholar]
- 45.Wang H, Cai L, Paul A, Enejder A, Heilshorn SC. Hybrid elastin-like polypeptide–polyethylene glycol (ELP-PEG) hydrogels with improved transparency and independent control of matrix mechanics and cell ligand density. Biomacromolecules. 2014;15:3421–3428. doi: 10.1021/bm500969d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Enejder A, Brackmann C, Svedberg F. Coherent anti-Stokes Raman scattering microscopy of cellular lipid storage. IEEE J Sel Top Quantum Electron. 2010;16:506–515. [Google Scholar]
- 47.Chung C, Pruitt BL, Heilshorn SC. Spontaneous cardiomyocyte differentiation of mouse embryoid bodies regulated by hydrogel crosslink density. Biomaterials Science. 2013;1:1082–1090. doi: 10.1039/C3BM60139K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jönsson P, Jonsson MP, Tegenfeldt JO, Höök F. A Method improving the accuracy of fluorescence recovery after photobleaching analysis. Biophys J. 2008;95:5334–5348. doi: 10.1529/biophysj.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babu H, Cheung G, Kettenmann H, Palmer TD, Kempermann G. Enriched monolayer precursor cell cultures from micro-dissected adult mouse dentate gyrus yield functional granule cell-like neurons. PLoS ONE. 2007;2:e388. doi: 10.1371/journal.pone.0000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Madl CM, Katz LM, Heilshorn SC. Bio-orthogonally crosslinked, engineered protein hydrogels with tunable mechanics and biochemistry for cell encapsulation. Adv Funct Mater. 2016;26:3612–3620. doi: 10.1002/adfm.201505329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neef AB, Luedtke NW. Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. Proc Natl Acad Sci USA. 2011;108:20404–20409. doi: 10.1073/pnas.1101126108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.DiMarco RL, Dewi RE, Bernal G, Kuo C, Heilshorn SC. Protein-engineered scaffolds for in vitro 3D culture of primary adult intestinal organoids. Biomater Sci. 2015;3:1376–1385. doi: 10.1039/c5bm00108k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moullan N, et al. Tetracyclines disturb mitochondrial function across eukaryotic models: A call for caution in biomedical research. Cell Rep. 2015;10:1681–1691. doi: 10.1016/j.celrep.2015.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roghani M, et al. Metalloprotease-disintegrin MDC9: Intracellular maturation and catalytic activity. J Biol Chem. 1999;274:3531–3540. doi: 10.1074/jbc.274.6.3531. [DOI] [PubMed] [Google Scholar]
- 56.Moss ML, Rasmussen FH, Nudelman R, Dempsey PJ, Williams J. Fluorescent substrates useful as high throughput screening tools for ADAM9. Comb Chem High Throughput Screening. 2010;13:358–365. doi: 10.2174/138620710791054259. [DOI] [PubMed] [Google Scholar]
- 57.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Cell-responsive synthetic hydrogels. Adv Mater. 2003;15:888–892. [Google Scholar]
- 58.Rowley JA, Madlambayan G, Mooney DJ. Alginate hydrogels as synthetic extracellular matrix materials. Biomaterials. 1999;20:45–53. doi: 10.1016/s0142-9612(98)00107-0. [DOI] [PubMed] [Google Scholar]
- 59.Desai RM, Koshy ST, Hilderbrand SA, Mooney DJ, Joshi NS. Versatile click alginate hydrogels crosslinked via tetrazine-norbornene chemistry. Biomaterials. 2015;50:30–37. doi: 10.1016/j.biomaterials.2015.01.048. [DOI] [PubMed] [Google Scholar]
- 60.Romano NH, Madl CM, Heilshorn SC. Matrix RGD ligand density and L1CAM-mediated Schwann cell interactions synergistically enhance neurite outgrowth. Acta Biomater. 2015;11:48–57. doi: 10.1016/j.actbio.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.