Abstract
Empyema is defined by the presence of bacteria and/or pus in pleural effusions. However, the biology of bacteria within human pleural fluid has not been studied. Streptococcus pneumoniae is the most common cause of pediatric and frequent cause of adult empyema. We investigated whether S. pneumoniae can proliferate within human pleural fluid and if growth is affected by the cellular content of the fluid and/or characteristics of pneumococcal surface proteins. Invasive S. pneumoniae isolates (n = 24) and reference strain recovered from human blood or empyema were inoculated (1.5×106CFU/mL) into sterile human malignant pleural fluid samples (n = 11). All S. pneumoniae (n = 25) strains proliferated rapidly, increasing by a median of 3009 (IQR 1063–9846) from baseline at 24hrs in all pleural effusions tested. Proliferation was greater than in commercial pneumococcal culture media and concentrations were maintained for 48hrs without autolysis. A similar magnitude of proliferation was observed in pleural fluid before and after removal of its cellular content, p = 0.728. S. pneumoniae (D39 strain) wild-type, and derivatives (n = 12), each with mutation(s) in a different gene required for full virulence were inoculated into human pleural fluid (n = 8). S. pneumoniae with pneumococcal surface antigen A (ΔpsaA) mutation failed to grow (2207-fold lower than wild-type), p<0.001, however growth was restored with manganese supplementation. Growth of other common respiratory pathogens (n = 14) across pleural fluid samples (n = 7) was variable and inconsistent, with some strains failing to grow. We establish for the first time that pleural fluid is a potent growth medium for S. pneumoniae and proliferation is dependent on the PsaA surface protein and manganese.
Introduction
Pneumonia affects 450 million patients worldwide each year, [1] with Streptococcus pneumoniae the most common bacterial cause. As many as 20–40% of patients with pneumonia develop a simple parapneumonic pleural effusion [2]. Pleural infection can develop when the pleural fluid is secondarily infected, and affects ~80,000 people in the UK and USA a year with significant morbidity and mortality [3].
Empyema represents the most severe end of the pleural infection spectrum. It is defined by the presence of bacteria, or pus, in the pleural fluid. S. pneumoniae accounts for the vast majority of pediatric empyema and is among the most frequent causative organisms of empyema in adults [4–6]. The incidence of pneumococcal empyema continues to rise globally [7–9].
Compared to most other pathogens that cause pneumonia, S. pneumoniae seems to have a particular affinity for causing pleural infection, but the reasons why are not fully understood. During pleural infection, the pleural fluid is exudative and contains a diverse milieu of biologically active molecules. Despite the clinical importance of empyema, no studies have investigated the biological interactions between bacteria and pleural fluids. It is unknown, for example, if bacteria are merely shed from the pleural tissues into the fluid and if the fluid influences bacterial growth. On one hand, exudative fluids are rich in nutrients that can potentially enhance microbial growth; on the other hand, many proteins and enzymes (eg defensins, and lysozymes etc.) in the fluid can serve to defend against bacterial invasion. These questions are difficult to study in empyema fluid as the presence of bacteria can profoundly alter the pleural fluid composition. In clinical settings, patients are often given systemic antibiotics before the fluid is sampled, taking away the opportunity to study the effects of bacterial growth.
Pleural effusion is not unique to pleural infection but is the common end-product of a variety of other non-infective pleural diseases/inflammation that leads to vascular hyper-permeability and plasma leak into the pleural cavity. In this paper, we studied whether the pleural fluid of patients with other non-infective pleural etiologies could support survival and growth of S. pneumoniae strains. We hypothesized that pleural fluid can serve as a growth medium for common empyema bacteria such as S. pneumoniae. We further investigated the role of common pneumococcal surface proteins, in particular nutrient transporters, on pneumococcal growth and survival within human pleural fluid.
Materials and methods
Pleural fluid samples
Pleural fluid samples were collected from patients attending Sir Charles Gairdner Hospital (SCGH) that required pleural fluid drainage for clinical indications, with approval (SCGH Human Research Ethics Committee 2012–156).
Pleural fluid samples were collected using aseptic techniques from patients with pleural effusions of non-infective etiologies. All samples were subjected to routine bacterial culture for which the fluid was injected into and transported in blood culture bottles and processed in an accredited hospital laboratory (PathWest, Western Australia). Pleural samples confirmed to be culture-negative were used in experiments. Patients who received antibiotics within 72 hours were excluded. Pleural fluid biochemistry (pH, protein, lactate dehydrogenase and glucose) was measured as previously described [10]. Supernatant was obtained from pleural fluid by centrifugation at 1020×g for 10 minutes.
Pleural fluids (n = 53) were collected from 45 patients; effusions obtained from the same individual (n = 8) were collected on separate occasions at least 7 days apart. Most samples were malignant pleural effusions (n = 47, 88.7%) and 10.6% (n = 5) were transudates; (S1 Table). Samples were stored between 2–8°C and used within 96hrs of drainage.
Bacterial strains and inoculums
Clinical isolates of S. pneumoniae (n = 24) from patients with invasive pneumococcal infection were obtained from PathWest (Royal Perth Hospital, WA). Serotypes of the isolates include type 1, 11A and 19F from pleural fluid and 1, 6B, 6C, 8 (n = 3), 10A, 11A, 12F, 19A (n = 8), 21, 22F (n = 2) and 35B from blood cultures. In addition, a S. pneumoniae reference strain of capsular serotype 3 (CIP 104225, ATCC®6303, American Type Culture Collection, Manassas, VA, USA) was used (Table 1).
Table 1. List of Streptococcus pneumoniae serotypes and other bacteria used in this study.
| Source | Capsular Serotype | Bacteria | Source |
|---|---|---|---|
| S. pneumoniae clinical isolates | Reference strains | ||
| Pleural fluid | 1 | ||
| Pleural fluid | 11A | Streptococcus anginosus | ATCC® 10556™ |
| Pleural fluid | 19A | Streptococcus anginosus | ATCC® 33397™ |
| Blood culture | 1 | Streptococcus intermedius | ATCC® 27335™ |
| Blood culture | 6B | Enterococcus faecalis | NCTC® 8213™ |
| Blood culture | 6C | Staphylococcus aureus | ATCC® 9144™ |
| Blood culture | 8 (n = 3) | Escherichia coli | ATCC® 105365™ |
| Blood culture | 10A | Escherichia coli | ATCC® 25922™ |
| Blood culture | 11A | Moraxella catarrhalis | NCTC® 3622™ |
| Blood culture | 12F | Pseudomonas aeruginosa | ATCC® 25668™ |
| Blood culture | 19A (n = 7) | Clinical isolates | |
| Blood culture | 19F | ||
| Blood culture | 21 | Methicillin-resistant Staphylococcus aureus | Clinical isolate |
| Blood culture | 22F (n = 2) | Pseudomonas aeruginosa | Clinical isolate |
| Blood culture | 35B | Pseudomonas aeruginosa | Clinical isolate |
| S. pneumoniae reference strain | Klebsiella oxytoca | Clinical isolate | |
| ATCC® 6303™ | 3 | Klebsiella pneumoniae | Clinical isolate |
ATCC, American Type Culture, Collection, Manassas, VA, US; NCTC, National Collection of Type Cultures, Salisbury UK.
Bacterial reference strains (n = 9) including Streptococcus anginosis ATCC®10556 and ATCC®33397, S. intermedius ATCC®27335, Enterococcus faecalis NCTC®8213 (National collection of type culture, Salisbury UK), Staphylococcus aureus ATCC®9144, Escherichia coli ATCC®105365 and ATCC®25922, Moraxella catarrhalis NCTC®3622, Pseudomonas aeruginosa ATCC®25668 and clinical isolates (n = 5) capable of causing invasive disease including methicillin-resistant S. aureus, P. aeruginosa (n = 2), Klebsiella pneumoniae and Klebsiella oxytoca (PathWest) were used for comparison growth studies with S. pneumoniae (Table 1).
Wild-type D39 S. pneumoniae, a capsule type-2 isolate, and its derivatives (n = 11) with mutations in genes required for full virulence have previously been described [11–20], and those used are listed in Table 2. Mutant strains were selected for using antibiotics (10μg/ml chloramphenicol, 0.2μg/mL erythromycin and/or 500μg/mL kanamycin) where necessary. Bacteria were stored in broth containing 10%(v/v) glycerol at -80°C.
Table 2. Characteristics of D39 S. pneumoniae mutants used across experiments.
| D39 Mutant | Name | Virulence role | Construction | Reference |
|---|---|---|---|---|
| Δply | Pneumolysin |
|
In frame deletion mutant | [11] |
| ΔlytA | Autolysin |
|
Insertion-duplication mutation | [12] |
| ΔpspA | Pneumococcal surface protein A |
|
Insertion-duplication mutation | [13] |
| ΔcbpD | Choline binding protein D |
|
Insertion-duplication mutation | [16] |
| ΔluxS | Biosynthesis of type 2 autoinducer AI-2 |
|
Insertion-duplication mutation | [15] |
| ΔpsaA | Pneumococcal surface antigen A |
|
Insertion-duplication mutation | [14] |
| ΔpitA | Putative iron uptake lipoprotein |
|
Insertion duplication mutation | [17] |
| ΔpiuB/piaA | Iron Uptake ABC transporter |
|
Insertion duplication mutation | [18] |
| ΔadcAI | Zinc uptake ABC transporter |
|
Deletion mutant | [19] |
| ΔadcAII/adcA | Zinc uptake ABC transporter |
|
Deletion mutant | [19] |
| ΔlivH | Branched-chain amino-acid (BCAA) transporter mutant |
|
Deletion mutant | [20] |
To standardize counts between experiments, frozen inocula of S. pneumoniae were prepared as previously described [21]. Inocula for D39 strains and non-pneumococcal bacteria were prepared from 18-hour blood agar to a turbidity of 0.5 MacFarland (PathWest) in sodium chloride 0.85% using a Sensititre Nephelometer (Thermo-Scientific; Waltham, USA).
Pleural fluid inoculation
Pleural fluid samples (in 3mL aliquots) were warmed to 37°C then inoculated with approximately 1.5×106 CFU/mL of bacteria and incubated for 24hrs at 37°C and 5% CO2 unless specified. Baseline inoculum and final concentrations (CFU/mL) were verified by manual counting log serial dilutions from blood agar.
Pleural fluid characteristics and bacteria used in each experiment are presented in Table 3. Pleural fluids before and after centrifugation (n = 11 pairs), to remove cells, were inoculated with S. pneumoniae isolates (n = 25). Six pairs were inoculated within 6 hours from collection and all within 24 hours (range 1 to 18 hours). Growth was compared in parallel to Todd-Hewitt Broth (THB; Thermo-Scientific) containing 17% FCS (Serana Australia, WA), [21], Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen, NY) containing 1000mg/L of glucose and sodium chloride 0.85% (n = 3 each) over 48 hours in six pleural fluid samples at 4, 8, 12, 18, 24, 28 and 48 hours. A further 7 pleural fluids were inoculated with non-pneumococcal bacteria (n = 14) and S. pneumoniae clinical isolate (n = 2) controls, at the same starting inoculum of 1.5×106 CFU/mL, for comparison purposes. The CFU/mL of bacteria failing to grow at 24 hours was subsequently measured at 4 and 8 hours in pleural fluid (n = 3).
Table 3. Pleural fluid characteristics for individual experiments.
| Experiment | S. pneumoniae in pleural fluid | S. pneumoniae in different medium | Other bacterial pathogens | S. pneumoniae D39 mutants* including ΔpsaA | S. pneumoniae D39 nutrient transporters** | S. pneumoniae ΔpsaA + manganese (Mn) |
|---|---|---|---|---|---|---|
| Pleural Fluid Samples, n | 11 | 6 | 7 | 8 | 8 | 16 |
| Pleural Fluid ID † | 1–11 | 12–17 | 47–53 | 18–25 | 26–33 | 31–46 |
| Number of bacterial strains used | 25 S. pneumoniae | 3 S. pneumoniae | 14 non-pneumococci 2 S. pneumoniae |
1 D39 wild-type 6 D39 mutants* |
1 D39 wild-type 5 D39 mutants** |
1 D39 wild 1 D39 ΔpsaA |
| Number of aliquots inoculated | 275 pleural + cells 275 pleural—cells |
18 pleural + cells 27 other media |
112 pleural + cells | 56 pleural + cells | 48 pleural + cells | 32 pleural + cells 32 pleural + cells +Mn |
|
Sex, male (%) Age, mean ±SD |
7 (63.6%) 63 years (SD±)] |
3 (50%) 70 years (SD±14) |
4 (57.1%) 74 years (SD±4) |
3 (37.5%) 70 years (SD±10) |
7 (87.5%) 69 years (SD±8) |
13 (81.2%) 69 years (SD±8) |
| pH, mean (SD) | 7.33 (SD±0.11) | 7.28 (SD±0.11) | 7.37 (SD±0.11) | 7.34 (SD±0.15) | 7.33 (SD±0.12) | 7.32 (SD±0.12) |
| LDH, U/L median (IQR) | 303 (116–4350) | 251 (218–397) | 245 (82.5–392) | 319 (189–752) | 391 (200–574) | 379 (200–574) |
| Protein, g/L median (IQR) | 39 (36–44) | 31 (23–39) | 31 (24–35) | 34 (26–40) | 39 (17–41) | 39 (17–40) |
| Glucose, mmol/L, mean (SD) | 4.09 (SD±1.9) | 4.9 (SD±1.8) | 6.3 (SD±2.3) | 4.9 (SD±2.4) | 8.4 (SD±8.4) | 6.8 (SD±6.0) |
† Individual pleural fluid characteristics are presented in S1 Table.
*Δply, ΔlytA, ΔpspA, ΔpsaA, ΔluxS, ΔcbpD,
** ΔpitA, ΔpiuB/piaA, ΔadcAI, ΔadcAII/adcAI, ΔlivH
D39 wild and mutant pneumococci (n = 12) were tested in pleural effusions (n = 8). Further experiments with ΔpsaA in parallel to wild type in pleural fluids (n = 16) with and without supplementation of 3μM manganese chloride tetrahydrate (Sigma-Aldrich, Sydney, Australia) were performed. The concentration was determined following concentration dependent supplementation experiments ranging from 0.003 to 30μM of manganese.
Statistical analysis
Statistical analyses were performed using SigmaPlot 12.5 (Systat Software, San Jose, CA). Results are presented as mean (SD) or median (IQR) based on normality of data. The Wilcoxon Signed-Rank test and paired t-test (for non-parametric and parametric data respectively) were used to compare baseline to 24 hour pneumococcal concentrations and Student’s t-test to compare growth between pleural fluid and other medium at various time-points over 48 hours. The Mann-Whitney Rank Sum test was used to compare differences between samples containing cells and correspondent supernatant and the Pearson Correlation was used to determine any relationship between spun and unspun samples. Significance was defined as p<0.05.
Results
Optimization and pneumococcal growth conditions
Significant growth was observed when 3mL volumes of pleural fluid were inoculated with 1.5 × 106 CFU/mL S. pneumoniae (n = 3), with a median increase of 6875 (IQR 3494–15744) fold at 24 hours compared to baseline. Volumes ranging from 1 to 10mL in both tubes and multi-well plates demonstrated similar growth, however volumes less than 1mL had inconsistent or poor growth (data not shown). Inoculum concentrations in the range of 1.5 x 103 to 1.5x 106 CFU/mL demonstrated similar bacterial growth at 24 hours; an initial concentration of 106 CFU/mL was used across experiments. Samples were incubated for 24 hours following inoculation as both S. pneumoniae isolates and D39 mutant strains were found to plateau at this time point (data not shown). No difference in CFU/mL of pneumococci was observed in fluids stored for up to 96 hours at 2–8°C following collection.
S. pneumoniae proliferated rapidly in all pleural effusions tested
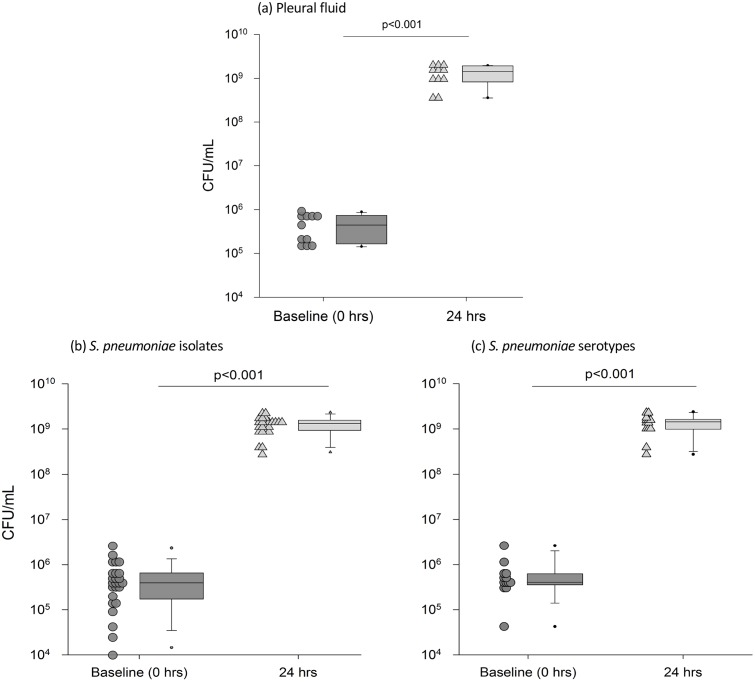
All 25 strains of S. pneumoniae proliferated rapidly in all (n = 11) effusion samples tested (Fig 1) by a median of 3009 fold (IQR 1063–9846) from baseline (from 3.83 × 105 CFU/mL to 1.3 × 109 CFU/mL), p<0.001 after 24 hours. S. pneumoniae growth was consistent with no difference in CFU/mL when data was analyzed as growth per S. pneumoniae isolate, growth in each pleural fluid sample or growth per serotype (p = 0.776).
Fig 1. The median growth of S. pneumoniae (n = 25) in human pleural fluid (n = 11) was consistent across all (a) pleural fluid samples (b) pneumococcal isolates and (c) serotypes.
Each dot-point represents data analyzed as (a) the median growth of all S. pneumoniae in each pleural fluid samples (n = 11), (b) individual S. pneumoniae isolates (n = 25) and the median growth of each isolate across pleural fluid samples and (c) S. pneumoniae grouped according to serotype (n = 13); data from the pleural fluid samples (n = 11) is pooled when more than one serotype is available and includes serotypes 1 (n = 2), 6B, 6C, 8 (n = 3), 10A, 11A (n = 2), 12F, 19A (n = 7), 19F, 21, 22F (n = 2), 35B and 3 reference strain. Proliferation was significant at 24hrs across all pneumococci, serotypes and pleural fluids, p<0.001. The box plot represents the median and IQR of the dot plot data; whiskers represent the 95th percentile. Pleural fluid characteristics are presented in Table 3.
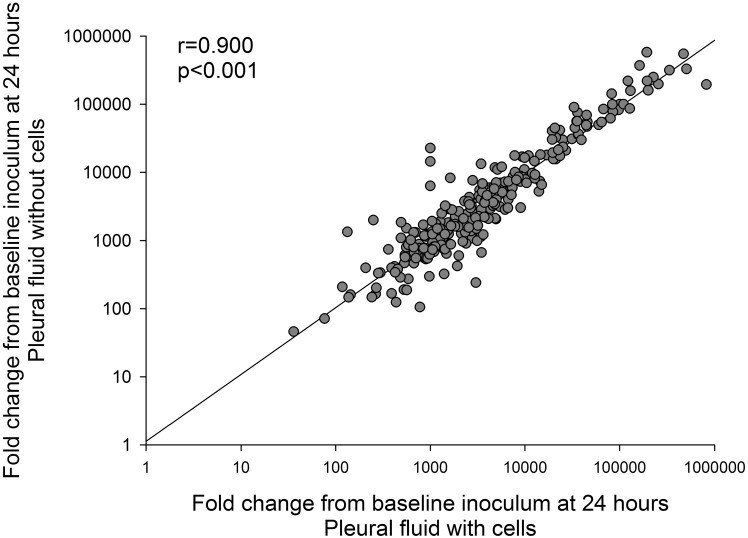
Removal of the cellular content of the pleural fluid by centrifugation did not affect S. pneumoniae growth, with a median fold increase at 24 hours of 2857 (IQR, 1020–9735, p = 0.728 v. growth in uncentrifuged fluid). Growth of S. pneumoniae in pleural fluid with cells correlated closely to growth in pleural fluid without cells present (Fig 2, p<0.001).
Fig 2. Correlation of S. pneumoniae (n = 25) growth in pleural fluid with and without cells (n = 11 pairs) expressed as the fold change from baseline inoculum at 24 hours (n = 275 pairs), p<0.001.
S. pneumoniae reaches high densities in pleural fluid and avoids autolysis
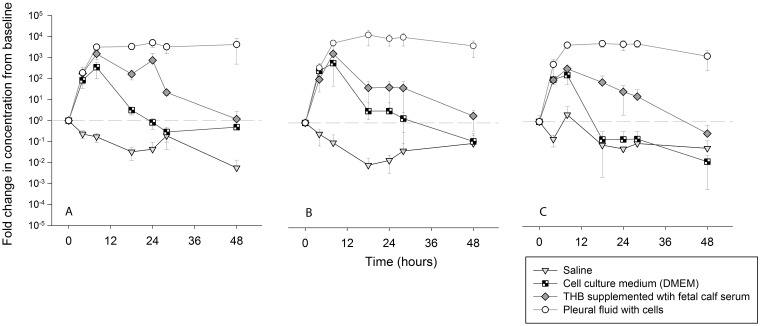
Growth of S. pneumoniae in pleural fluid was compared to growth in 1) THB with fetal calf serum (17%), a supplemented culture medium commonly used to cultivate pneumococci, 2) DMEM, a glucose rich cell culture medium and 3) 0.85% saline as a control. Rapid proliferation of S. pneumoniae was observed in THB with FCS, DMEM and pleural fluid within 8 hours following inoculation (Fig 3). The pneumococci did not proliferate in saline alone. Maximum CFU/mL were obtained at 8-hours post-inoculation in broth medium with higher or similar concentrations observed in pleural fluid at the same time point. From 8 to 18 hours’ post—inoculation, autolysis became evident for bacteria grown in either DMEM or culture medium with S. pneumoniae CFU/mL returning to baseline by 24 to 48 hours. In contrast, concentrations of S. pneumoniae CFU/mL in pleural fluid were maintained at >1000 fold from baseline over 48 hours.
Fig 3. Fold change in CFU/mL from baseline in saline (n = 3), cell-culture medium (DMEM) (n = 3), THB supplemented with fetal calf serum (n = 3) and pleural fluid (n = 6).
Each graph represents a different S. pneumoniae clinical isolate (a) serotype 8 (blood) (b) serotype 19A (blood) and (c) serotype 19A (pleural fluid). Bacterial CFU/mL in each fluid was determined at 4, 8, 12, 18, 24, 28 and 48 hours. Fluid characteristics are listed in Table 3.
S. pneumoniae strains demonstrated consistent growth whereas growth of other bacterial pathogens was variable
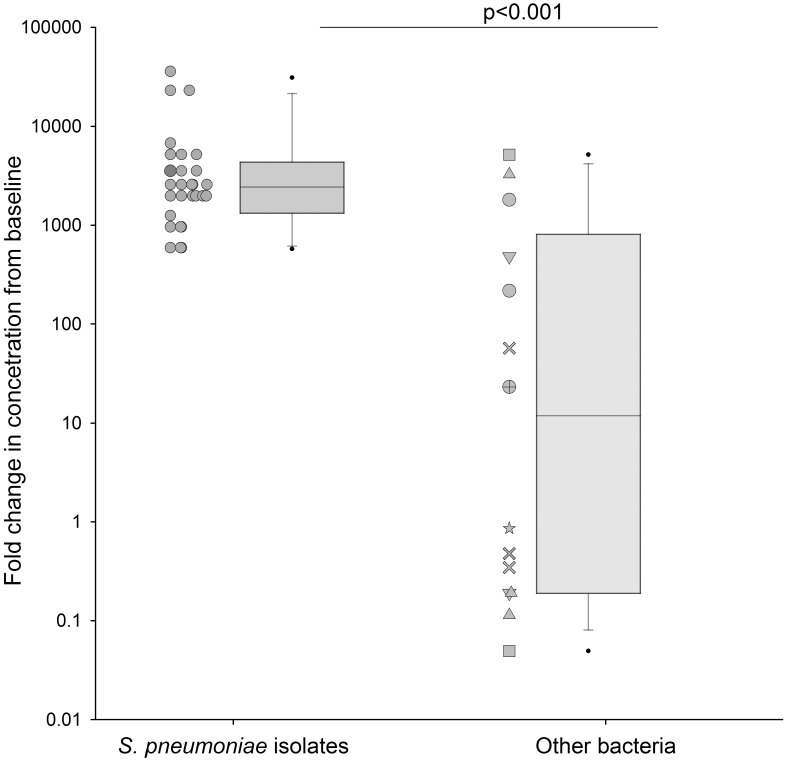
We investigated the growth of other bacterial pathogens (n = 14) across pleural fluid samples (n = 7). Growth measured at 24 hours was variable across other bacterial pathogens (Fig 4) with a median fold change in CFU/mL of 12.0 (IQR 0.2–813.0) compared to 1172 (IQR, 587–1757) for S. pneumoniae controls (n = 2), p<0.001. Furthermore, inconsistent growth between strains of the same bacterial pathogen was evident, including within the viridans streptococci group (n = 3) which are a frequent cause of adult empyema. The bacterial pathogens which failed to grow at 24 hours in pleural fluid did not demonstrate increased growth at earlier time points of 4 or 8 hours (data not shown).
Fig 4. The fold change in CFU/mL of other bacterial pathogens (n = 14) including P. aeruginosa (△), S. aureus (О), S. anginosis group (×), E. coli (□), Klebsiella sp.(▽), E. faecalis (⊕), M. catarrhalis (✯).
Each dot point represents the median fold change of each bacterial strain across pleural fluid (n = 7) samples at 24hrs. Data from other bacterial pathogens is compared to previous results of S. pneumoniae (n = 25) growth in pleural fluid (n = 11), p<0.001. The box plot represents the median and IQR; whiskers represent the 95th percentile.
S. pneumoniae proliferation in pleural fluid is dependent on pneumococcal surface adhesin A (PsaA)
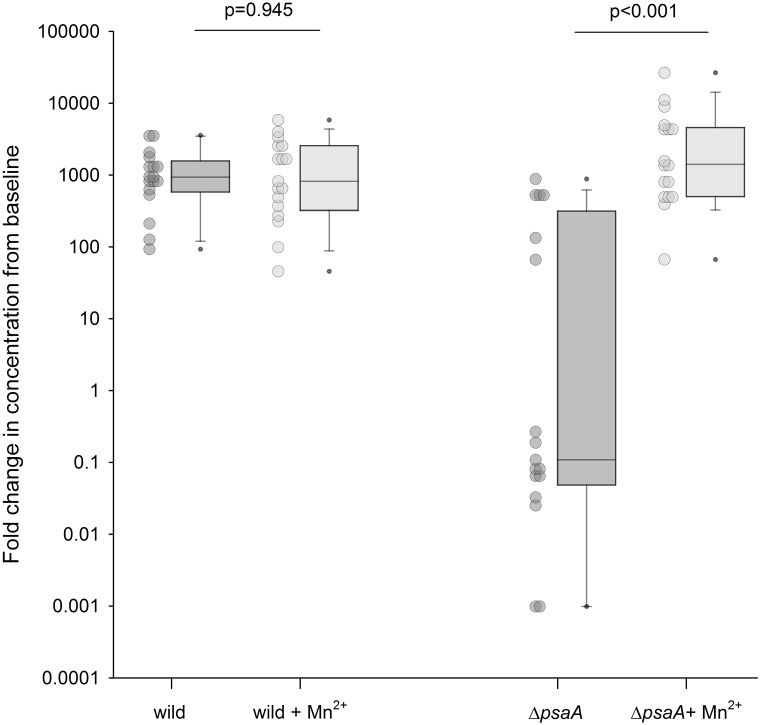
To identify factors required for S. pneumoniae growth in pleural fluid, growth experiments were repeated using the D39 wild-type strain and eleven mutant strains (Δply, ΔlytA, ΔpspA, ΔpsaA, ΔluxS, ΔcbpD, ΔpitA, ΔpiuB/piaA, ΔadcAI, ΔadcAII/adcAI, ΔlivH) in eight pleural fluid samples (Table 2). These mutant strains contain mutations in S. pneumoniae genes required for full virulence either because they encode surface proteins known to be important for host interactions or components of ABC transporters required for nutrient acquisition in vivo [11–17, 19, 20, 22]. All of the mutant strains achieved a similar density of CFU/mL as the wild-type D39 stain with the exception of ΔpsaA. After 24 hours in pleural fluid, the CFU/mL of ΔpsaA had decreased from baseline with a fold change of 0.55 (IQR, 0.08–3.255) whereas the D39 wild strain had increased 1214-fold (503–1746, p<0.001). A decrease in ΔpsaA concentrations from baseline was apparent at 4 hours and further decreased at 8 and 24 hours. Importantly, manganese supplementation of pleural fluid by the addition of 3uM of manganese prior to inoculation restored ΔpsaA proliferation in the pleural fluid, increasing the median fold-change in CFU from 0.11 (0.07–501) to 1546 (509–4775) at 24 hours, p <0.001 (Fig 5). 3μM manganese chloride- tetrahydrate was selected following concentration dependent supplementation experiments ranging from 0.003 to 30μM of manganese. The addition of manganese had no impact on growth of the D39 wild type strain. Iron and zinc supplementation of Δpit and ΔadcAII/I mutants respectively had no impact on growth compared to the D39 wild type control (data not shown).
Fig 5. D39 S. pneumoniae wild type and ΔpsaA growth in pleural fluid (n = 16) at 24 hrs with and without manganese (Mn2+) 3μM supplementation.
ΔpsaA growth was restored with manganese supplementation, p<0.001.
Discussion
S. pneumoniae is a common cause of both pediatric and adult empyema, yet the reasons why this pathogen can cause pleural infections have only been partially investigated. This is the first study to investigate the interactions of bacteria, namely S. pneumoniae, and human pleural fluid ex vivo. We found that pleural fluid provides a rich medium to support the growth of all relevant S. pneumoniae strains tested and that pneumococci appear to be much better adapted to grow in pleural fluid than most other common respiratory pathogens. More importantly, growth of S. pneumoniae in pleural fluid was dependent on PsaA, a manganese transporter making this a potential future therapeutic target. Mutations of other key virulence factors of S. pneumoniae did not affect the growth of pneumococci in human pleural fluid ex vivo.
We have previously shown in our mouse model that S. pneumoniae from infected pneumonia tissues invades the pleural cavity within 4 hours of infection via transcytosis across the mesothelial cells [23]. Little information exists on the fate of bacteria after they invade human pleural fluid. This is the first study to investigate these interactions.
To investigate the capacity of pleural fluid to support bacterial growth we used human pleural effusions, most with malignant etiology. Parapneumonic effusions and malignant pleural effusions are both exudative in nature and have indistinguishable biochemical compositions with elevated protein and lactate dehydrogenase and reduced glucose and pH. The effusions used for this study display the same biochemical features of non-infective simple parapneumonic effusions. Our results show that all 25 isolates of S. pneumoniae consistently proliferated across eleven human exudative pleural fluid samples tested with a median increase of approximately 3000-fold from baseline at 24 hours. In contrast, the median growth of other bacterial pathogens in pleural fluid was approximately 10-fold, and interestingly the growth was variable with some bacterial isolates demonstrating similar levels of growth to S. pneumoniae, whilst other isolates consistently failed to grow.
When tested in parallel, the growth of S. pneumoniae was similar to optimal laboratory media at 8 hours demonstrating that pleural fluid represents an excellent growth medium for S. pneumoniae. Furthermore, pneumococci naturally undergo autolysis after reaching stationary phase during growth which was observed in broth at 18 hours, yet bacterial CFU/mL density was maintained for 48 hours after growth in human pleural fluid. This persistent growth is likely to promote significant neutrophil influx and may be a reason why pneumococci are a frequent cause of empyema. This failure of autolysis would ensure S. pneumoniae strains are capable of maintaining high level infection even in a closed environment such as a pleural effusion. The reasons why autolysis did not occur after growth in pleural fluid are not known. The cellular content of the pleural fluid did not influence S. pneumoniae proliferation, indicating it is the pleural fluid itself that provides the medium for S. pneumoniae growth. The ability to grow in pleural fluid did not vary between individual S. pneumoniae isolates or serotypes investigated.
Multiple cell surface proteins that function as adhesins, in complement resistance, autolytic enzymes and nutrient transporters in S. pneumoniae have been identified as essential for virulence and growth of the bacterium. We investigated eleven of these virulence factors for their role during growth in pleural fluid, using mutants that are known to have reduced growth in blood (eg Δply, ΔlytA, ΔpsaA and ΔlivH) [11, 20, 24–27] or in cation depleted media (ΔpiuB/piaA, ΔadcAI/ΔadcAII) [22, 28, 29]. However, perhaps surprisingly, only one mutant strain demonstrated reduced growth in pleural fluid, the ΔpsaA mutant. PsaA is a lipoprotein required for manganese transport and resistance to oxidative stress [14, 30, 31]. Mutation of PsaA reduces virulence of S. pneumoniae and growth in laboratory medium that has been depleted of manganese. Providing biochemical complementation of the ΔpsaA mutation by supplementing pleural fluid with 3uM manganese was sufficient to restore growth in pleural fluid, confirming that it is the low concentration of manganese that causes failure of growth of the ΔpsaA strain [31]. This suggests that manganese is rate limiting for growth of S. pneumoniae in pleural fluid yet pneumococci are capable of overcoming this using the PsaA transporter. Potential PsaA inhibitors that have recently been identified open opportunities to control S. pneumoniae proliferation in the pleural space [32]. Interestingly, growth of S. pneumoniae with mutations affecting transporters required for iron or zinc uptake was not affected, suggesting that manganese results are specific for growth within pleural fluid.
The study has a number of potential limitations. We were unable to use pleural fluid of infected etiology in the present study for reasons stated in the introduction. However, malignant pleural effusions have a close resemblance in biochemistry to parapneumonic effusions and were therefore used for the majority of our experiments. Although the cellular content of these effusions may differ, we found that the cellular content of the fluid did not influence the growth/proliferation of S. pneumoniae in vitro. Next, due to the nature of the study, experiments were performed in ex vivo pleural fluid and therefore host defenses, including neutrophils that may modify the bacterial proliferation were not accounted for. However, as a proof of principle study our aim was to investigate the direct effect of pleural fluid on S. pneumoniae proliferation. Furthermore, our data showed no difference in proliferation with or without cellular content of the pleural fluid. Finally, we studied 25 invasive strains of S. pneumoniae that had caused empyema or septicemia. Future studies that include other serotypes may be needed, though the very consistent nature of the results indicates that the majority of S. pneumoniae isolates are likely to grow in pleural fluid.
Pleural fluid is a potent growth medium for S. pneumoniae and may be one reason why pneumococci are a common bacterial cause of empyema. The high density and persistent growth of S. pneumoniae in pleural fluid highlights the importance of draining infected effusions. Pneumococcal growth was independent of the cellular content of pleural fluid but dependent on the PsaA surface protein, making PsaA a potential future therapeutic target. The variable growth of other bacterial pathogens in pleural fluid may account for the lower incidence of otherwise common respiratory pathogens in empyema and warrants further exploration in order to understand pathogenesis of pleural infection.
Supporting information
(DOC)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
YCGL is a National Health & Medical Research Council (NHMRC) Career Development Fellow and receives project grant funding from the NHMRC, New South Wales Dust Disease Board (DDB), Sir Charles Gairdner Research Advisory Committee, Institute for Respiratory Health Westcare Alan King Grants and the Cancer Council of Western Australia. JSB is partially funded from the Department of Health’s NIHR Biomedical Research Centre’s funding scheme. JCP is a NHMRC Senior Principal Research Fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Revised global burden of disease 2002 estimates. Geneva; 2004. World Health Organization [Google Scholar]
- 2.Light RW. Parapneumonic effusions and empyema. Proc Am Thorac Soc. 2006;3(1):75–80. doi: 10.1513/pats.200510-113JH [DOI] [PubMed] [Google Scholar]
- 3.Davies HE, Davies RJ, Davies CW. Management of pleural infection in adults: British Thoracic Society Pleural Disease Guideline 2010. Thorax. 2010;65 Suppl 2:ii41–53. [DOI] [PubMed] [Google Scholar]
- 4.Proesmans M, Gijsens B, Van de Wijdeven P, De Caluwe H, Verhaegen J, Lagrou K, et al. Clinical outcome of parapneumonic empyema in children treated according to a standardized medical treatment. Eur J Pediatr. 2014;173(10):1339–45. doi: 10.1007/s00431-014-2319-1 [DOI] [PubMed] [Google Scholar]
- 5.Strachan RE, Cornelius A, Gilbert GL, Gulliver T, Martin A, McDonald T, et al. Bacterial causes of empyema in children, Australia, 2007–2009. Emerg Infect Dis. 2011;17(10):1839–45. doi: 10.3201/eid1710.101825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maskell NA, Batt S, Hedley EL, Davies CW, Gillespie SH, Davies RJ. The bacteriology of pleural infection by genetic and standard methods and its mortality significance. Am J Respir Crit Care Med. 2006;174(7):817–23. doi: 10.1164/rccm.200601-074OC [DOI] [PubMed] [Google Scholar]
- 7.Finley C, Clifton J, Fitzgerald JM, Yee J. Empyema: an increasing concern in Canada. Can Respir J. 2008;15(2):85–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burgos J, Falco V, Pahissa A. The increasing incidence of empyema. Curr Opin Pulm Med. 2013;19(4):350–6. doi: 10.1097/MCP.0b013e3283606ab5 [DOI] [PubMed] [Google Scholar]
- 9.Grijalva CG, Zhu Y, Nuorti JP, Griffin MR. Emergence of parapneumonic empyema in the USA. Thorax. 2011;66(8):663–8. doi: 10.1136/thx.2010.156406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomas R, Cheah HM, Creaney J, Turlach BA, Lee YC. Longitudinal Measurement of Pleural Fluid Biochemistry and Cytokines in Malignant Pleural Effusions. Chest. 2016;149(6):1494–500. doi: 10.1016/j.chest.2016.01.001 [DOI] [PubMed] [Google Scholar]
- 11.Berry AM, Ogunniyi AD, Miller DC, Paton JC. Comparative virulence of Streptococcus pneumoniae strains with insertion-duplication, point, and deletion mutations in the pneumolysin gene. Infect Immun. 1999;67(2):981–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berry AM, Lock RA, Hansman D, Paton JC. Contribution of autolysin to virulence of Streptococcus pneumoniae. Infect Immun. 1989;57(8):2324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Talkington DF, Crimmins DL, Voellinger DC, Yother J, Briles DE. A 43-kilodalton pneumococcal surface protein, PspA: isolation, protective abilities, and structural analysis of the amino-terminal sequence. Infect Immun. 1991;59(4):1285–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berry AM, Paton JC. Sequence heterogeneity of PsaA, a 37-kilodalton putative adhesin essential for virulence of Streptococcus pneumoniae. Infect Immun. 1996;64(12):5255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stroeher UH, Paton AW, Ogunniyi AD, Paton JC. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect Immun. 2003;71(6):3206–12. doi: 10.1128/IAI.71.6.3206-3212.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosink KK, Mann ER, Guglielmo C, Tuomanen EI, Masure HR. Role of novel choline binding proteins in virulence of Streptococcus pneumoniae. Infect Immun. 2000;68(10):5690–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown JS, Gilliland SM, Ruiz-Albert J, Holden DW. Characterization of Pit, a Streptococcus pneumoniae iron uptake ABC transporter. Infect Immun. 2002;70(8):4389–98. doi: 10.1128/IAI.70.8.4389-4398.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown JS, Ogunniyi AD, Woodrow MC, Holden DW, Paton JC. Immunization with components of two iron uptake ABC transporters protects mice against systemic Streptococcus pneumoniae infection. Infect Immun. 2001;69(11):6702–6. doi: 10.1128/IAI.69.11.6702-6706.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bayle L, Chimalapati S, Schoehn G, Brown J, Vernet T, Durmort C. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Mol Microbiol. 2011;82(4):904–16. doi: 10.1111/j.1365-2958.2011.07862.x [DOI] [PubMed] [Google Scholar]
- 20.Basavanna S, Khandavilli S, Yuste J, Cohen JM, Hosie AHF, Webb AJ, et al. Screening of Streptococcus pneumoniae ABC transporter mutants demonstrates that LivJHMGF, a branched-chain amino acid ABC transporter, is necessary for disease pathogenesis. Infect Immun. 2009;77(8):3412–23. doi: 10.1128/IAI.01543-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aaberge IS, Eng J, Lermark G, Lovik M. Virulence of Streptococcus pneumoniae in mice: a standardized method for preparation and frozen storage of the experimental bacterial inoculum. Microb Pathog. 1995;18(2):141–52. [DOI] [PubMed] [Google Scholar]
- 22.Brown JS, Gilliland SM, Holden DW. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol Microbiol. 2001;40(3):572–85. [DOI] [PubMed] [Google Scholar]
- 23.Wilkosz S, Edwards LA, Bielsa S, Hyams C, Taylor A, Davies RJ, et al. Characterization of a new mouse model of empyema and the mechanisms of pleural invasion by Streptococcus pneumoniae. Am J Respir Cell Mol Biol. 2012;46(2):180–7. doi: 10.1165/rcmb.2011-0182OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey RM, Ogunniyi AD, Chen AY, Paton JC. Pneumolysin with low hemolytic activity confers an early growth advantage to Streptococcus pneumoniae in the blood. Infect Immun. 2011;79(10):4122–30. doi: 10.1128/IAI.05418-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canvin JR, Marvin AP, Sivakumaran M, Paton JC, Boulnois GJ, Andrew PW, et al. The role of pneumolysin and autolysin in the pathology of pneumonia and septicemia in mice infected with a type 2 pneumococcus. J Infect Dis. 1995;172(1):119–23. [DOI] [PubMed] [Google Scholar]
- 26.Orihuela CJ, Gao G, Francis KP, Yu J, Tuomanen EI. Tissue-specific contributions of pneumococcal virulence factors to pathogenesis. J Infect Dis. 2004;190(9):1661–9. [DOI] [PubMed] [Google Scholar]
- 27.Marra A, Lawson S, Asundi JS, Brigham D, Hromockyj AE. In vivo characterization of the psa genes from Streptococcus pneumoniae in multiple models of infection. Microbiology. 2002;148(Pt 5):1483–91. doi: 10.1099/00221287-148-5-1483 [DOI] [PubMed] [Google Scholar]
- 28.Brown LR, Gunnell SM, Cassella AN, Keller LE, Scherkenbach LA, Mann B, et al. AdcAII of Streptococcus pneumoniae affects pneumococcal invasiveness. PLoS One. 2016;11(1):e0146785 doi: 10.1371/journal.pone.0146785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plumptre CD, Eijkelkamp BA, Morey JR, Behr F, Counago RM, Ogunniyi AD, et al. AdcA and AdcAII employ distinct zinc acquisition mechanisms and contribute additively to zinc homeostasis in Streptococcus pneumoniae. Mol Microbiol. 2014;91(4):834–51. doi: 10.1111/mmi.12504 [DOI] [PubMed] [Google Scholar]
- 30.Tseng HJ, McEwan AG, Paton JC, Jennings MP. Virulence of Streptococcus pneumoniae: PsaA mutants are hypersensitive to oxidative stress. Infect Immun. 2002;70(3):1635–9. doi: 10.1128/IAI.70.3.1635-1639.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston JW, Myers LE, Ochs MM, Benjamin WH Jr., Briles DE, Hollingshead SK. Lipoprotein PsaA in virulence of Streptococcus pneumoniae: surface accessibility and role in protection from superoxide. Infect Immun. 2004;72(10):5858–67. doi: 10.1128/IAI.72.10.5858-5867.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bajaj M, Mamidyala SK, Zuegg J, Begg SL, Ween MP, Luo Z, et al. Discovery of novel pneumococcal surface antigen A (PsaA) inhibitors using a fragment-based drug design approach. 2015. June 19;10(6):1511–20. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.